Abstract
Background
Vaccines to prevent coronavirus disease 2019 (Covid-19) are urgently needed. The effect of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccines on viral replication in both upper and lower airways is important to evaluate in nonhuman primates.
Methods
Nonhuman primates received 10 or 100 μg of mRNA-1273, a vaccine encoding the prefusion-stabilized spike protein of SARS-CoV-2, or no vaccine. Antibody and T-cell responses were assessed before upper- and lower-airway challenge with SARS-CoV-2. Active viral replication and viral genomes in bronchoalveolar-lavage (BAL) fluid and nasal swab specimens were assessed by polymerase chain reaction, and histopathological analysis and viral quantification were performed on lung-tissue specimens.
Results
The mRNA-1273 vaccine candidate induced antibody levels exceeding those in human convalescent-phase serum, with live-virus reciprocal 50% inhibitory dilution (ID50) geometric mean titers of 501 in the 10-μg dose group and 3481 in the 100-μg dose group. Vaccination induced type 1 helper T-cell (Th1)–biased CD4 T-cell responses and low or undetectable Th2 or CD8 T-cell responses. Viral replication was not detectable in BAL fluid by day 2 after challenge in seven of eight animals in both vaccinated groups. No viral replication was detectable in the nose of any of the eight animals in the 100-μg dose group by day 2 after challenge, and limited inflammation or detectable viral genome or antigen was noted in lungs of animals in either vaccine group.
Conclusions
Vaccination of nonhuman primates with mRNA-1273 induced robust SARS-CoV-2 neutralizing activity, rapid protection in the upper and lower airways, and no pathologic changes in the lung. (Funded by the National Institutes of Health and others.)
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (Covid-19), is responsible for the 2020 global pandemic.1,2 Developing a vaccine that is safe, effective, and rapidly deployable is an urgent global health priority. The majority of vaccine candidates have focused on inducing antibody responses against the trimeric SARS-CoV-2 spike (S) protein, a class I fusion protein that facilitates binding to the angiotensin-converting–enzyme 2 (ACE2) receptor and triggers virus–cell-membrane fusion.3 A variety of vaccine approaches4 and formulations for targeting the SARS-CoV-2 S protein are being pursued, including nucleic acid vaccines (RNA and DNA),5-8 human and simian replication-defective adenoviral vaccines,9,10 whole-inactivated SARS-CoV-2,11,12 and subunit protein vaccines.13
In assessing the immunogenicity and protection of vaccines in preclinical animal models, nonhuman primates provide several advantages for clinical translation. They are outbred, have greater similarity to humans than rodents in innate immune responses and B-cell and T-cell repertoires, and allow for the use of clinically relevant vaccine doses. Recent studies have shown that SARS-CoV-2 targets similar replication sites and recapitulates some aspects of Covid-19–like disease in nonhuman primates.7,14 After SARS-CoV-2 infection, nonhuman primates have transient viral replication in the upper and lower airways and mild inflammation in the lung that resolves within 14 days.7 Thus, nonhuman primates are a useful animal model for assessing vaccine-mediated protection against early viral replication.7,14
The use of messenger RNA (mRNA) is a promising approach for Covid-19 vaccination, since it combines rapid manufacturing and expeditious modification of the encoded immunogen, both of which accelerate vaccine development.14 RNA vaccines encoding viral antigens have been shown to be safe and immunogenic in several clinical trials,5,15 including in a recent phase 1 clinical trial of mRNA-1273, a SARS-CoV-2 vaccine candidate.16 Data from a mouse model showing that a low dose of mRNA-1273 induced a robust neutralizing antibody response and high-level protection against SARS-CoV-26 raised the possibility that vaccination with mRNA-1273 could prevent or limit both upper- and lower-airway infection in nonhuman primates.
Methods
Vaccine mRNA and Lipid Nanoparticle Production
We synthesized a sequence-optimized mRNA encoding prefusion-stabilized SARS-CoV-2 S-2P protein in vitro. The mRNA was purified by oligo-dT affinity purification and encapsulated in a lipid nanoparticle through a modified ethanol-drop nanoprecipitation process, as described previously.17
Rhesus Macaque Model
Experiments in animals were performed in compliance with National Institutes of Health (NIH) regulations and with approval from the Animal Care and Use Committee of the Vaccine Research Center and from Bioqual (Rockville, MD). Challenge studies were conducted at Bioqual. The authors vouch for the accuracy and completeness of the data in this report.
Female and male Indian-origin rhesus macaques (12 of each sex; age range, 3 to 6 years) were sorted according to sex, age, and weight (see Supplementary Appendix 2, available with the full text of this article at NEJM.org) and then stratified into groups of three. Within each stratum, one of the three animals was assigned to each study group arbitrarily. Animals were vaccinated intramuscularly at week 0 and at week 4 with either 10 or 100 μg of mRNA-1273 in 1 ml of 1× phosphate-buffered saline (PBS) into the right hind leg. Unvaccinated control animals were administered an equal volume of 1× PBS. At week 8 (4 weeks after the second vaccination), all animals were challenged with a total dose of 7.6×105 plaque-forming units (PFU). The stock of 1.9×105 PFU per milliliter SARS-CoV-2 (USA-WA1/2020 strain) was administered in a volume of 3 ml by the intratracheal route and in a volume of 1 ml by the intranasal route (0.5 ml per nostril). Pre- and postchallenge specimen collection is detailed in Figure S1 in Supplementary Appendix 1 (note that all supplementary figures and tables cited in the main article text are located in Supplementary Appendix 1).
Quantification of SARS-CoV-2 RNA and Subgenomic RNA
Polymerase chain reaction (PCR) was used to quantify viral RNA, and subgenomic RNA was used to quantify replicating RNA in bronchoalveolar-lavage (BAL) fluid and nasal swab specimens, as described previously.18
Histopathology
Nonhuman primate SARS-CoV-2–infected lung tissue specimens were fixed, processed, embedded in paraffin, sectioned, and stained with hematoxylin and eosin. Immunohistochemical staining with the use of rabbit polyclonal SARS-CoV-2 (GeneTex) was performed with formalin-fixed, paraffin-embedded lung-tissue sections. Chromogenic in situ hybridization (CISH) was performed with RNAscope technology, as described previously.19 The RNAscope 2.5 LS Probe V-nCoV2019-S (Advanced Cell Diagnostics) was used as the target probe to detect positive-sense SARS-CoV-2.
Human Convalescent-Phase Serum
A panel of 42 human convalescent-phase serum specimens were obtained from persons between 18 and 84 years of age who had mild, moderate, or severe Covid-19 under institutional review board–approved specimen-collection protocols at the NIH Clinical Center (Bethesda, MD) (ClinicalTrials.gov number, NCT00067054), Aaron Diamond AIDS Research Center, Columbia University (New York) (NCT04342195), and the University of Washington (Seattle) (Hospitalized or Ambulatory Adults with Respiratory Viral Infection [HAARVI] study and STUDY00000959). Written informed consent was provided by all participants. Participants had a history of laboratory-confirmed SARS-CoV-2 infection roughly 1 to 2 months before they provided specimens.
Serum Antibody Measurements
Total SARS-CoV-2 S-2P–specific or N-specific IgG in serum was quantified by enzyme-linked immunosorbent assay (ELISA); the methods used were similar to previously published methods.6 ACE2 binding inhibition was completed, as described previously,16 on 1:40 diluted serum samples with the use of Mesoscale Discovery 384-well, 4-Spot Custom Serology SECTOR plates precoated with SARS-CoV-2 receptor-binding domain. Binding was detected with SULFO-TAG–labeled ACE2. Both reagents were supplied by the manufacturer (Meso Scale Diagnostics) free of charge.
Pseudoviruses were produced by cotransfection of plasmids encoding SARS-CoV-2 S (Wuhan-1, GenBank accession number, MN908947.3), luciferase reporter, lentivirus backbone, and human transmembrane protease, serine 2 (TMPRSS2), into HEK293T/17 cells (ATCC), and neutralization was assessed as described previously.6,16 Live-virus neutralization was assessed with a full-length SARS-CoV-2 virus based on the Seattle isolate, which was designed to express luciferase and was recovered by means of reverse genetics, as described previously.20-22
Intracellular Cytokine Staining
Cryopreserved peripheral-blood mononuclear cells were thawed, rested overnight, and stimulated with SARS-CoV-2 S protein (S1 and S2, homologous to the vaccine insert) and nucleoprotein (N) peptide pools (JPT Peptide Technologies) and costimulatory antibodies anti-CD28 and anti-CD49d (clones CD28.2 and 9F10, BD Biosciences). Negative controls received an equal concentration of dimethyl sulfoxide (without peptides) and costimulatory antibodies. Cytokine staining was performed as described previously,23 with amendments to the monoclonal antibodies used as detailed in Supplementary Appendix 1.
Statistical Analysis
For the prespecified primary analysis of viral load in the BAL fluid and nasal swab specimens, the peak over days 2 through 7 for each animal was compared with Mann–Whitney tests and a hierarchical testing procedure in which the 100-μg dose group was compared with the control group first, and only if that comparison was significant at a P value of less than 0.05 was the 10-μg dose group then compared with both the control group and the 100-μg dose group. For all other measures, groups were compared by Kruskal–Wallis test, followed by pairwise Mann–Whitney tests with Holm’s adjustment on the set of pairwise tests if the Kruskal–Wallis test indicated significance. Correlations were estimated and tested with the use of Spearman’s nonparametric method. Positivity with respect to intracellular cytokine responses was determined with the MIMOSA (Mixture Models for Single-Cell Assays) algorithm.24 Primary data for all graphs and tables are provided in Supplementary Appendix 2.
Results
Antibody Responses after mRNA-1273 Vaccination
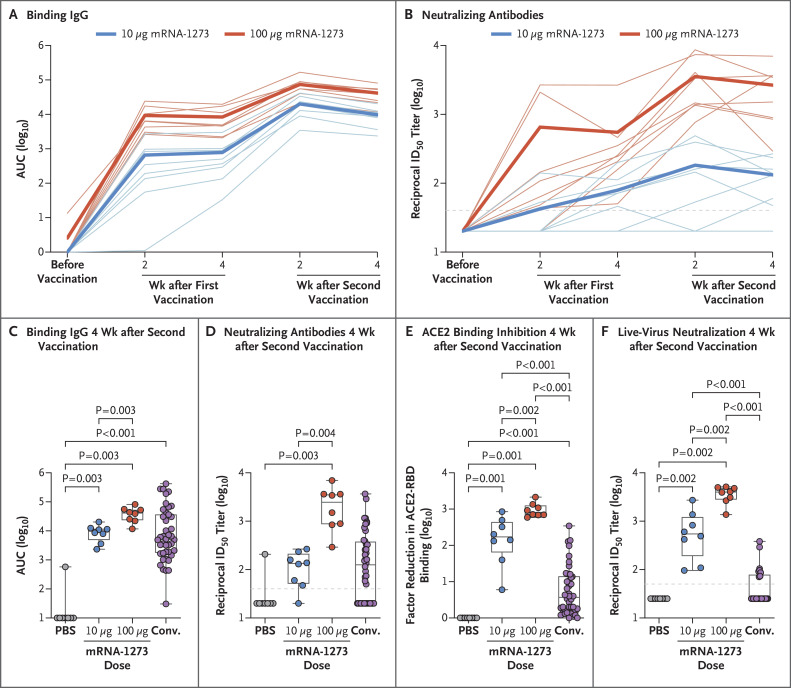
First, we evaluated temporal SARS-CoV-2 S-2P–specific antibody responses after vaccination. IgG binding to the conformationally defined prefusion S-2P protein25,26 was increased over baseline in a dose-dependent manner after two vaccinations, reaching an area under the curve of 8241 and 36,186 by 4 weeks after the second vaccination with 10 and 100 μg of mRNA-1273, respectively (Figure 1A). Similarly, there was a dose-dependent increase in neutralizing activity measured with a pseudotyped lentivirus reporter. Animals vaccinated with 10 μg of mRNA-1273 had a reciprocal 50% inhibitory dilution (ID50) geometric mean titer (GMT) of 63 at 4 weeks after the first vaccination, which increased to 103 by 4 weeks after the second vaccination. Neutralizing activity at 4 weeks after the first vaccination in animals that received 100 μg (GMT, 305) was 5 times that seen at the lower dose and rose to a GMT of 1862 after the second vaccination (Figure 1B). The S-specific IgG binding (Figure 1C) and neutralizing GMT (Figure 1D) elicited by vaccination with 100 μg of mRNA-1273 at 4 weeks after the second vaccination were 5 times and 15 times as high, respectively, as in convalescent-phase serum specimens from a panel of 42 humans representing a full range of disease severity (see Supplementary Appendix 2).
Figure 1. Antibody Responses after mRNA-1273 Vaccination in Rhesus Macaques.
Animals were administered phosphate-buffered saline (PBS) as a control or 10 μg or 100 μg of mRNA-1273. Serum specimens were assessed for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) S-specific IgG by enzyme-linked immunosorbent assay (ELISA) (Panel A) and SARS-CoV-2 pseudovirus neutralization (Panel B) at all time points after the first and second vaccinations. Data in Panel A are the area under the curve (AUC) and indicate the amount of IgG binding to S-2P over time, and data in Panel B are the reciprocal 50% inhibitory dilution (ID50). Faint lines in Panels A and B represent individual animals, and bold lines represent the geometric mean titer for each group. S-specific IgG (Panel C), pseudovirus neutralization (Panel D), inhibition of angiotensin-converting enzyme 2 (ACE2) binding to the receptor-binding domain (RBD) (Panel E), and live-virus neutralization by NanoLuc reporter assay (Promega) (Panel F) were assessed at 4 weeks after the second vaccination, immediately before challenge. Results were compared with the antibody responses in a panel of human convalescent-phase serum specimens (Conv.) (42 specimens in Panels C, D, and E and 26 specimens in Panel F). In Panel E, the amount of signal emitted in wells containing no specimen was used as the maximal binding response against which each factor reduction was measured. In the box-and-whisker plots, the horizontal line indicates the median, the top and bottom of the box the interquartile range, and the whiskers the range. Symbols represent individual animals and overlap with one another for equal values where constrained. Dashed lines indicate the assay limit of detection.
We extended those analyses to assess antibody responses to specific domains of S and used two additional orthogonal in vitro approaches to measure functional viral inhibition. First, since a critical mechanism for productive infection in vivo is the interaction of the S receptor-binding domain with viral receptor ACE2, we explored antibodies against this target of vulnerability. We determined whether serum from mRNA-1273–vaccinated animals could bind the receptor-binding domain in ELISA or inhibit binding to ACE2. Indeed, mRNA-1273–vaccinated nonhuman primates produced more potent receptor-binding domain–specific serum antibodies than were seen in convalescent-phase human serum specimens (Fig. S2A). Moreover, serum from animals in the 100-μg dose group had inhibition of ACE2 binding to the receptor-binding domain that was 938 times as high as that in serum from animals in the control group and 348 times as high as that in human convalescent-phase serum (Figure 1E).
Binding to the N-terminal domain of S1 was then assessed, because a potential major benefit of targeting domains other than the receptor-binding domain is to establish a polyclonal antibody response that recognizes multiple functional S domains to achieve inhibition of viral attachment,27-31 additive neutralizing activity,32 and postattachment fusion inhibition.33 Targeting multiple epitopes may also mitigate the possibility of immune escape by antigenic drift.27 Here, mRNA-1273 elicited more S1 N-terminal domain–specific antibody responses than human convalescent-phase serum (Fig. S2B). Lastly, neutralizing activity was measured with a live SARS-CoV-2 reporter virus. Animals vaccinated with 10 or 100 μg of mRNA-1273 had reciprocal ID50 GMTs of 501 and 3481, respectively, values that are 12 times and 84 times as high, respectively, as in human convalescent-phase serum (Figure 1F). These data show that mRNA-1273 induced robust S-specific antibody responses, targeting both the receptor-binding domain and the N-terminal subdomains of S1 with potent neutralizing capacity.
T-Cell Responses after mRNA-1273 Vaccination
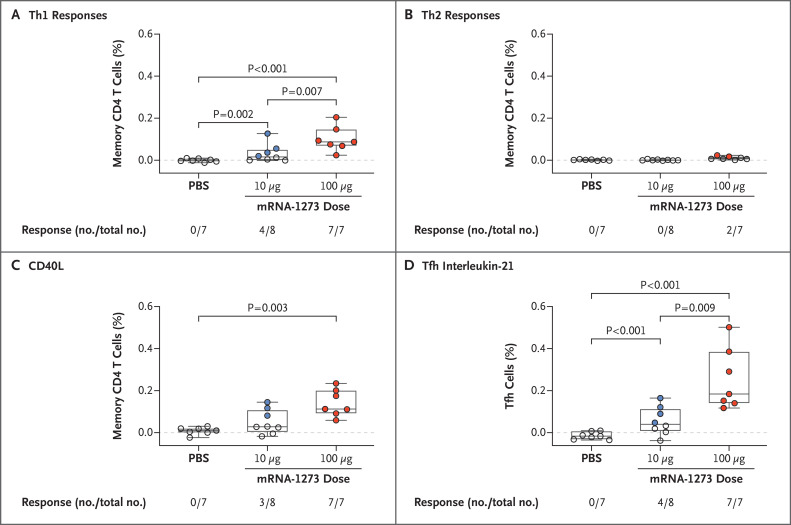
SARS-CoV-2–specific T-cell immunity may have a role in pathogenesis or protection against SARS-CoV-2 and can influence the humoral immune response.34-36 Activated CD4 T cells are critical for B-cell activation and antibody production and can be segregated into functional subsets on the basis of their production of specific cytokines. The induction of CD4 type 2 helper T-cell (Th2) (interleukin-4, -5, or -13) responses has been associated with vaccine-associated enhanced respiratory disease (VAERD), as seen in some patients with respiratory virus infections, such as respiratory syncytial virus infection and measles,19,20 as well as in animal models of Middle East respiratory syndrome coronavirus (MERS-CoV).21 VAERD is generally not observed when a CD4 Th1 (interferon-γ, interleukin-2, tumor necrosis factor α) response occurs in the absence of a Th2 response.35,37-39 Thus, we used 19-color multiparameter flow cytometry to assess the functional heterogeneity of S-specific CD4 and CD8 T-cell cytokine responses after mRNA-1273 vaccination.
A dose-dependent increase in Th1 responses was noted 4 weeks after the second vaccination; four of eight animals in the 10-μg dose group and seven of seven animals in the 100-μg dose group (one animal in this group could not be evaluated for technical reasons) had Th1 responses. Th1 response levels were higher in the 100-μg dose group than in the control group or in the 10-μg dose group. Even at the 10-μg dose, Th1 responses were higher than in the control group (Figure 2A). In contrast, Th2 responses were low to undetectable in both vaccine dose groups (Figure 2B). CD8 T-cell responses were also low to undetectable after mRNA-1273 vaccination.
Figure 2. T-Cell Responses after mRNA-1273 Vaccination in Rhesus Macaques.
Intracellular staining was performed on peripheral blood mononuclear cells at 8 weeks, immediately before challenge, to assess T-cell responses to the SARS-CoV-2 S1 peptide pool. Panel A shows type 1 helper T-cell (Th1) responses (interferon-γ, interleukin-2, or tumor necrosis factor α), Panel B Th2 responses (interleukin-4 or 13), Panel C CD40L up-regulation, and Panel D interleukin-21 from peripheral follicular helper T (Tfh) cells (central memory CXCR5+PD-1+ICOS+ CD4 T cells). Positivity with respect to intracellular cytokine responses was determined with the MIMOSA algorithm; numbers of animals positive and total numbers of animals are shown as fractions below each group. In the box-and-whisker plots, the horizontal line indicates the median, the top and bottom of the box the interquartile range, and the whiskers the range. Open symbols represent animals with a probable nonresponse, and solid symbols represent animals with a probable response. Dashed lines are used to highlight 0.0%.
We extended the analysis of CD4 T-cell responses, given their importance in regulating antibody responses. CD40L is a cell-surface marker expressed after CD4 T-cell activation that mediates B-cell activation for efficient isotype switching; three of eight animals in the 10-μg dose group and seven of seven animals in the 100-μg dose group had S-specific CD40L+ CD4 T-cell responses, and 100 μg of mRNA-1273 induced greater responses than the PBS control (Figure 2C). Since nucleoside-modified RNA vaccines encoding various viral antigens have been shown to induce robust antibody responses in nonhuman primates in association with increased CD4 T follicular helper (Tfh) cells,40 we measured interleukin-21, the canonical cytokine produced by Tfh cells. Tfh cells are critical for the formation of germinal centers and generation of long-term B-cell memory. Four of eight animals in the 10-μg dose group and seven of seven animals in the 100-μg dose group had interleukin-21 responses; the responses differed between the 100-μg dose group and the control and 10-μg dose groups, as well as between the 10-μg dose group and the control group (Figure 2D). These data show that mRNA-1273 induced Th1 and interleukin-21–producing Tfh-cell responses. We did not find evidence of Th2 or CD8 T-cell responses in this study.
Protective Efficacy against Upper- and Lower-Airway SARS-CoV-2 Infection
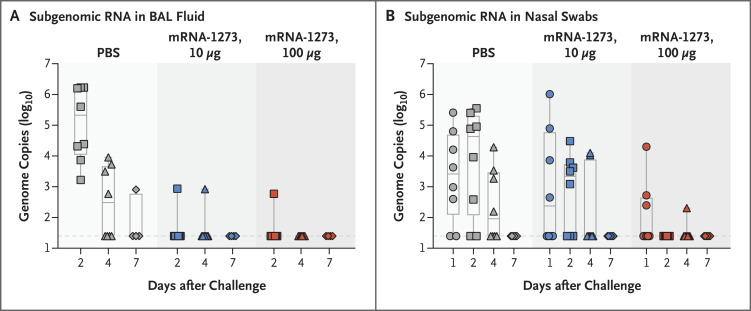
To evaluate the protective efficacy of mRNA-1273, all animals were challenged by combined intratracheal and intranasal routes with a total dose of 7.6×105 PFU, approximately equivalent to 106 50% tissue-culture infectious doses (TCID50), administered 4 weeks after the second vaccination. This challenge route and dose were based on a model development study in which we challenged nonhuman primates that had no previous exposure to the virus with different doses of SARS-CoV-2 administered by the intratracheal–intranasal or endobronchial route (see Supplementary Appendix 2). The objective was to deliver virus to both the upper and lower airways in order to detect levels of virus that were similar to what has been detected in nasal secretions of humans after SARS-CoV-2 infection.41 The predefined primary end points of the study were the difference in the viral load in BAL fluid between the vaccine groups and the control group. The analysis of the efficacy end point involved PCR and subgenomic RNA PCR, similar to other studies of SARS-CoV2 vaccines in nonhuman primates,10,14 to quantify input virus and replicating virus, respectively.
Two days after challenge, only one of eight animals in each of the vaccine dose groups had detectable subgenomic RNA in BAL fluid, as compared with eight of eight animals in the control group (Figure 3A). By day 2, none of the eight animals in the 100-μg dose group had detectable subgenomic RNA detected in nasal swab specimens, as compared with five of eight animals in the 10-μg dose group and six of eight animals in the control group (Figure 3B). On day 4, two of eight animals in the 10-μg dose group and one of eight animals in the 100-μg dose group had low levels of subgenomic RNA detected in the nose. In accordance with the prespecified statistical analysis, the peak levels of subgenomic RNA over days 2 through 7 were significantly lower in both the 100-μg dose group and the 10-μg dose group than in the control group, in both BAL fluid specimens (P<0.001 for both comparisons) and nasal swab specimens (P=0.009 and P=0.03, respectively). Furthermore, total RNA levels were significantly lower in BAL fluid in both the 100-μg dose group and the 10-μg dose group than in the control group (P<0.001 for both comparisons) (Fig. S3). With regard to nasal secretions, animals vaccinated with 100 μg of mRNA-1273 also had lower total RNA levels than animals in the control group (P=0.03) or in the 10-μg dose group (P=0.003).
Figure 3. Efficacy of mRNA-1273 against Upper and Lower Respiratory Viral Replication.
Bronchoalveolar-lavage (BAL) fluid (Panel A) and nasal swab (Panel B) specimens were obtained on days 1, 2, 4, and 7 after challenge, where applicable, and viral replication was assessed by analysis of SARS-CoV-2 subgenomic RNA. In the box-and-whisker plots, the horizontal line indicates the median, the top and bottom of the box the interquartile range, and the whiskers the range. Symbols represent individual animals and overlap with one another for equal values where constrained. Dashed lines indicate the assay limit of detection.
To extend these analyses and provide additional evidence for limited viral infection in the lung, a panel of innate cytokines and chemokines were assessed in BAL fluid at days 2 and 4 after challenge. Inflammatory cytokine induction was limited in both dose groups, which suggests that there was rapid control of virus sufficient to limit innate immune activation (Fig. S4). These data show rapid control of viral replication within 2 days in both the upper and lower airways.
To assess potential immune correlates of protection, mRNA-1273–induced serum neutralization activity at the time of challenge was analyzed in the context of protection as defined by viral PCR and subgenomic RNA in BAL fluid and nasal secretions. Overall, the antibody measurements, including measurements of in vitro viral neutralization, were negatively correlated with nasal secretion viral PCR results at day 2 after challenge (Fig. S5). To explore potential immune mechanisms mediating rapid control of viral replication in the lung, we measured postchallenge antibody levels in BAL fluid; a dose-dependent increase in S-specific IgG was noted in BAL fluid from vaccine recipients as compared with animals in the control group. S-specific IgA responses in BAL fluid were lower than IgG responses but were also increased in the 100-μg dose group (Fig. S6). Postchallenge humoral S- and N-specific IgG increased in control animals within 2 weeks after challenge, whereas antibody levels in mRNA-1273–vaccinated animals remained stable; thus, no anamnestic response was found after challenge (Fig. S7).
Pathology and Viral Load in Lung Tissue after Challenge
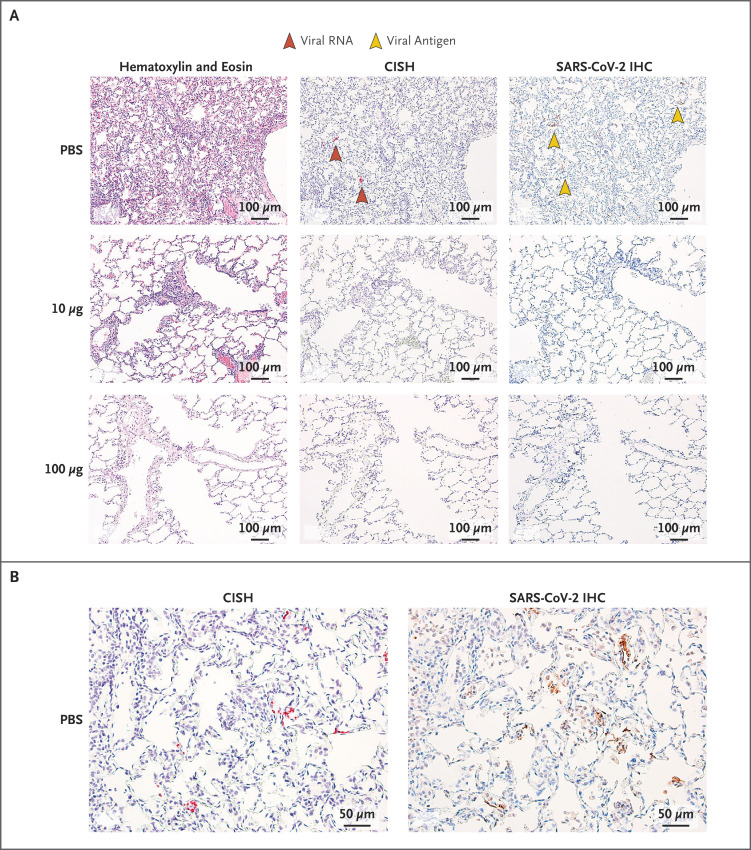
Consistent with previous reports,14,42,43 SARS-CoV-2 infection in the control animals caused moderate-to-severe inflammation that often involved the small airways and the adjacent alveolar interstitia. Alveolar air spaces occasionally contained inflammatory cell infiltrates, alveolar capillary septa were moderately thickened, and moderate and diffuse type II pneumocyte hyperplasia was seen. Multiple pneumocytes in the lung sections from the control group were positive for both SARS-CoV-2 viral RNA and antigen, as assessed by CISH and immunohistochemical analysis (Figure 4; results of these analyses for days 7 through 15 are summarized in Table S1). At day 7, in animals vaccinated with 10 μg of mRNA-1273, inflammation was mild, and no viral RNA was detected (Figure 4A). However, at day 8, one animal in the 10-μg dose group had a single pneumocyte that was positive for viral antigen (Fig. S8). No substantial inflammation was observed in the lungs of nonhuman primates vaccinated with 100 μg of mRNA-1273, and neither viral RNA nor antigen was detected at day 7 or 8 after challenge (Figure 4A). In addition to the lung sections from these earlier time points, lung sections from animals that were killed at day 14 or 15 after challenge had no evidence of substantial inflammation, and neither viral RNA nor viral antigen was detected in any of the groups, including the control group. Vaccine-associated immunopathologic changes were not observed in any of the sections examined.
Figure 4. Lung Histopathological Analysis and Viral Detection 7 Days after Challenge in mRNA-1273–Vaccinated Rhesus Macaques.
Seven days after challenge, lungs were harvested from two animals per group for histopathological analysis and assessment of evidence of viral infection; representative images taken at different degrees of magnification are shown for localization of virus by chromogenic in situ hybridization (CISH) and SARS-CoV-2 immunohistochemical analysis (IHC) in serial tissue sections. The images in Panel B are shown at twice the magnification of the images in Panel A.
Discussion
Previous vaccine studies in nonhuman primates with recombinant chimpanzee-derived adenovirus vector vaccine from Oxford (ChAdOX) or DNA vaccine showed protection against lower-airway viral replication and against pathologic changes in the lung after challenge with approximately 106 TCID50 (for the ChAdOX vaccine)10 or 104 PFU (for the DNA vaccine)7 of SARS-CoV-2. However, these studies provided no evidence of a reduction of viral replication in nasal tissue, raising questions as to whether these vaccines could affect virus transmission. In contrast, our study showed early prevention of viral replication in the upper and lower airways after a high-dose challenge (approximately 8×105 PFU) with SARS-CoV-2. The ability to limit viral replication in both the lower and the upper airways has important implications for vaccine-induced prevention of both SARS-CoV-2 disease and transmission.
This study shows that mRNA-1273 induced robust S-specific antibody and neutralizing activity, which we confirmed with several orthogonal serologic assays. The antibody-binding activity and neutralizing activity were substantially higher than previously reported in nonhuman primates vaccinated with whole-inactivated,11,12 DNA,7 or adenovirus vector vaccines,9,10 all of which were shown to provide protection of the lower airways after a range of different SARS-CoV-2 challenge doses. The mRNA-1273 vaccine candidate induced higher ACE2 binding inhibition (348 times as high), more potent receptor-binding domain and N-terminal domain antibody responses, and higher neutralizing activity (12 to 84 times as high) than was measured in a panel of convalescent-phase serum specimens obtained from patients with Covid-19 of various levels of clinical severity. On the basis of recent data in humans that show a reduction in antibodies over time,44,45 vaccine-induced immunity that exceeds the antibody response to primary infection may be needed for durable protection. Studies are now under way to determine the durability of immunity and protection over 1 year after vaccination.
We hypothesize that the potent neutralizing activity induced by mRNA-1273 is based on two major factors. First, structure-based vaccine design was used to stabilize the S protein encoded by the mRNA vaccine.25 Stabilizing the prefusion conformation of class I fusion proteins has successfully improved immunogenicity of these important vaccine targets for respiratory syncytial virus,46,47 parainfluenza virus,48 Nipah virus,49 MERS-CoV,6,25 and human immunodeficiency virus.50,51 This improvement is based on preserving neutralization-sensitive epitopes at the apex of prefusion structures52 and improved protein expression from transduced cells.6,25 Furthermore, anchoring the S-2P protein immunogen in the membrane also contributes to maintenance of the native conformation and antigenicity that improves immunogenicity as compared with secreted protein.6
The second major influence on the immune response is the formulation, purification, and delivery of the RNA. Accordingly, nucleoside modifications and the process of purifying RNA can limit the innate immune stimulation triggered by RNA, thereby facilitating translation and prolonging protein production in vivo.53 Moreover, the modified RNAs have been shown to increase the frequency of CD4 Tfh cells in tissues, which has promoted antibody responses to other viral antigens in nonhuman primates.40 In our study, it was notable that mRNA-1273 induced S-specific CD4 T cells that produce interleukin-21, the canonical cytokine that defines Tfh cell responses, which suggests this as an additional mechanism for generating potent antibody responses.
Both vaccine groups had high-level protection and limited variation in detectable viral replication, as estimated by analysis of subgenomic RNA, and therefore we were not able to define specific immune correlates with this measurement as a protective end point; however, neutralizing potency was negatively correlated with viral loads in the nose, as detected by PCR. On the basis of the rapid reduction in viral replication within 24 to 48 hours after challenge and the detection of antibodies in BAL fluid, we postulate that antibodies are the primary mechanism of protection for this vaccine. Passive-transfer studies involving serum from vaccinated animals will be needed to assess whether antibodies are necessary and sufficient to mediate protection. Furthermore, studies encompassing lower, subprotective doses will be necessary to define a protective threshold and to evaluate antibody specificities or functions that correlate with protection.
A major potential concern in SARS-CoV-2 vaccine development is VAERD, which is associated with induction of nonneutralizing antibodies that can lead to immune complex formation, complement activation, Th2-biased responses, and immunopathologic complications. Mitigating approaches that may be used to avoid vaccine-enhanced disease syndromes include eliciting potently neutralizing antibodies with functional activity commensurate with binding, as well as avoiding Th2-biased CD4 T-cell responses.54 Here, we show that mRNA-1273 induces high levels of neutralizing activity and Th1 responses with low-to-undetectable Th2 responses and no pathologic changes in the lung in either of the mRNA-1273 vaccine groups 1 week after challenge.
A final issue addressed here is whether a nonhuman primate vaccine and infection model can inform clinical vaccine development. Although the inoculum of SARS-CoV-2 that is required for efficient human transmission is not known, the amount of virus detected by PCR in the upper airways of humans immediately after infection is approximately 106 RNA copies per nasal swab.18 Consistent with that finding, after challenge with a dose of 7.6×105 PFU in this study, unvaccinated nonhuman primates had approximately 106 RNA copies per milliliter detected in their noses at day 1 after challenge. The results reported here provide data on mRNA-1273 immunogenicity and protection of the upper and lower airways in nonhuman primates that complement the immunogenicity and safety data established by a phase 1 clinical study involving humans.
Acknowledgments
We thank Karin Bok, Kevin Carlton, and additional members of all included laboratories for discussions and advice pertaining to experiments included in this report; Judy Stein and Monique Young for technology transfer and administrative support, respectively; members of the VRC Translational Research Program, including Diana Scorpio, Hana Bao, Elizabeth McCarthy, Jay Noor, Alida Taylor, and Ruth Woodward, for technical and administrative assistance with experiments in animals; Huihui Mu and Michael Farzan for providing angiotensin-converting enzyme 2 (ACE2)–overexpressing 293 cells; David Ho and Helen Chu for providing human convalescent-phase serum specimens; and Jon Fintzi and Keith Lumbard for implementing the MIMOSA algorithm for data on intracellular cytokine staining.
Supplementary Appendix 1
Supplementary Appendix 2
Disclosure Forms
This article was published on July 28, 2020, and updated on August 7, 2020, at NEJM.org.
Footnotes
Supported by the Intramural Research Program of the Vaccine Research Center (VRC), National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH); and the Office of the Assistant Secretary for Preparedness and Response, Biomedical Advanced Research and Development Authority, Department of Health and Human Services (contract 75A50120C00034). Dr. Corbett is the recipient of a research fellowship that was partially funded by the Undergraduate Scholarship Program, Office of Intramural Training and Education, Office of the Director, NIH. Dr. Martinez was funded by grants from the NIAID (T32-AI007151 and F32 AI152296) and a Burroughs Wellcome Fund Postdoctoral Enrichment Program Award.
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Cucinotta D, Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed 2020;91:157-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Callaway E, Cyranoski D, Mallapaty S, Stoye E, Tollefson J. The coronavirus pandemic in five powerful charts. Nature 2020;579:482-483. [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181(2):271.e8-280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amanat F, Krammer F. SARS-CoV-2 vaccines: status report. Immunity 2020;52:583-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mulligan MJ, Lyke KE, Kitchin N, et al. Phase 1/2 study to describe the safety and immunogenicity of a COVID-19 RNA vaccine candidate (BNT162b1) in adults 18 to 55 years of age: interim report. July 1, 2020. (https://www.medrxiv.org/content/10.1101/2020.06.30.20142570v1). preprint.
- 6.Corbett KS, Edwards D, Leist SR, et al. SARS-CoV-2 mRNA vaccine development enabled by prototype pathogen preparedness. June 11, 2020. (https://www.biorxiv.org/content/10.1101/2020.06.11.145920v1). preprint. [DOI] [PMC free article] [PubMed]
- 7.Yu J, Tostanoski LH, Peter L, et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 2020. May 20 (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith TRF, Patel A, Ramos S, et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat Commun 2020;11:2601-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu F-C, Li Y-H, Guan X-H, et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet 2020;395:1845-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Doremalen N, Lambe T, Spencer A, et al. ChAdOx1 nCoV-19 vaccination prevents SARS-CoV-2 pneumonia in rhesus macaques. May 13, 2020. (https://www.biorxiv.org/content/10.1101/2020.05.13.093195v1). preprint. [DOI] [PubMed]
- 11.Gao Q, Bao L, Mao H, et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science 2020;369:77-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Zhang Y, Huang B, et al. Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell. June 6, 2020. (https://www.sciencedirect.com/science/article/pii/S0092867420306954). [DOI] [PMC free article] [PubMed]
- 13.Chen W-H, Strych U, Hotez PJ, Bottazzi ME. The SARS-CoV-2 vaccine pipeline: an overview. Curr Trop Med Rep 2020. March 3 (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munster VJ, Feldmann F, Williamson BN, et al. Respiratory disease and virus shedding in rhesus macaques inoculated with SARS-CoV-2. March 21, 2020. (https://www.biorxiv.org/content/10.1101/2020.03.21.001628v1). preprint. [DOI] [PMC free article] [PubMed]
- 15.Bahl K, Senn JJ, Yuzhakov O, et al. Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Mol Ther 2017;25:1316-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson LA, Anderson EJ, Rouphael NG, et al. An mRNA vaccine against SARS-CoV-2 — preliminary report. N Engl J Med. DOI: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hassett KJ, Benenato KE, Jacquinet E, et al. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol Ther Nucleic Acids 2019;15:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020;581:465-469. [DOI] [PubMed] [Google Scholar]
- 19.Wang F, Flanagan J, Su N, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 2012;14:22-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou YJ, Okuda K, Edwards CE, et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 2020;182(2):429-446.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scobey T, Yount BL, Sims AC, et al. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc Natl Acad Sci U S A 2013;110:16157-16162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yount B, Curtis KM, Fritz EA, et al. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc Natl Acad Sci USA 2003;100:12995-13000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donaldson MM, Kao S-F, Foulds KE. OMIP-052: an 18-color panel for measuring Th1, Th2, Th17, and Tfh responses in rhesus macaques. Cytometry A 2019;95:261-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finak G, McDavid A, Chattopadhyay P, et al. Mixture models for single-cell assays with applications to vaccine studies. Biostatistics 2014;15:87-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pallesen J, Wang N, Corbett KS, et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci U S A 2017;114:E7348-E7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020;367:1260-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Shi W, Chappell JD, et al. Importance of neutralizing monoclonal antibodies targeting multiple antigenic sites on the Middle East respiratory syndrome coronavirus spike glycoprotein to avoid neutralization escape. J Virol 2018;92(10):e02002-17-e02002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang N, Rosen O, Wang L, et al. Structural definition of a neutralization-sensitive epitope on the MERS-CoV S1-NTD. Cell Rep 2019;28(13):3395.e6-3405.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Lu S, Jia H, et al. A novel neutralizing monoclonal antibody targeting the N-terminal domain of the MERS-CoV spike protein. Emerg Microbes Infect 2017;6(5):e37-e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogers TF, Zhao F, Huang D, et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 2020. June 15 (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi R, Shan C, Duan X, et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 2020. May 26 (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 32.Wang W, Wang H, Deng Y, et al. Characterization of anti-MERS-CoV antibodies against various recombinant structural antigens of MERS-CoV in an imported case in China. Emerg Microbes Infect 2016;5(11):e113-e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Widjaja I, Wang C, van Haperen R, et al. Towards a solution to MERS: protective human monoclonal antibodies targeting different domains and functions of the MERS-coronavirus spike glycoprotein. Emerg Microbes Infect 2019;8:516-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neidleman J, Luo X, Frouard J, et al. SARS-CoV-2-specific T cells exhibit unique features characterized by robust helper function, lack of terminal differentiation, and high proliferative potential. June 8, 2020. (https://www.biorxiv.org/content/10.1101/2020.06.08.138826v1). preprint. [DOI] [PMC free article] [PubMed]
- 35.Weiskopf D, Schmitz KS, Raadsen MP, et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci Immunol 2020;5:eabd2071-eabd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meckiff BJ, Ramírez-Suástegui C, Fajardo V, et al. Single-cell transcriptomic analysis of SARS-CoV-2 reactive CD4 + T cells. June 13, 2020. (https://www.biorxiv.org/content/10.1101/2020.06.12.148916v1). preprint. [DOI] [PMC free article] [PubMed]
- 37.Grifoni A, Weiskopf D, Ramirez SI, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 2020;181(7):1489.e15-1501.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng Y, Mentzer AJ, Liu G, et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent COVID-19 patients. June 8, 2020. (https://www.biorxiv.org/content/10.1101/2020.06.05.134551v1). preprint. [DOI] [PMC free article] [PubMed]
- 39.Sekine T, Perez-Potti A, Rivera-Ballesteros O, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. June 29, 2020. (https://www.biorxiv.org/content/10.1101/2020.06.29.174888v1). preprint. [DOI] [PMC free article] [PubMed]
- 40.Pardi N, Hogan MJ, Naradikian MS, et al. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J Exp Med 2018;215:1571-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zou L, Ruan F, Huang M, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med 2020;382:1177-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chandrashekar A, Liu J, Martinot AJ, et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science 2020. May 20 (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rockx B, Kuiken T, Herfst S, et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science 2020;368:1012-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edridge AWD, Kaczorowska JM, Hoste ACR, et al. Coronavirus protective immunity is short-lasting. June 16, 2020. (https://www.medrxiv.org/content/10.1101/2020.05.11.20086439v2). preprint. [DOI] [PubMed]
- 45.Long Q-X, Tang X-J, Shi Q-L, et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med 2020. June 18 (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 46.Crank MC, Ruckwardt TJ, Chen M, et al. A proof of concept for structure-based vaccine design targeting RSV in humans. Science 2019;365:505-509. [DOI] [PubMed] [Google Scholar]
- 47.Graham BS, Gilman MSA, McLellan JS. Structure-based vaccine antigen design. Annu Rev Med 2019;70:91-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stewart-Jones GBE, Chuang G-Y, Xu K, et al. Structure-based design of a quadrivalent fusion glycoprotein vaccine for human parainfluenza virus types 1-4. Proc Natl Acad Sci U S A 2018;115:12265-12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loomis RJ, Stewart-Jones GBE, Tsybovsky Y, et al. Structure-based design of Nipah virus vaccines: a generalizable approach to paramyxovirus immunogen development. Front Immunol 2020;11:842-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanders RW, Vesanen M, Schuelke N, et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol 2002;76:8875-8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chuang GY, Geng H, Pancera M, et al. Structure-based design of a soluble prefusion-closed HIV-1 Env trimer with reduced CD4 affinity and improved immunogenicity. J Virol 2017;91(10):e02268-16-e02268-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Killikelly AM, Kanekiyo M, Graham BS. Pre-fusion F is absent on the surface of formalin-inactivated respiratory syncytial virus. Sci Rep 2016;6:34108-34108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karikó K, Muramatsu H, Welsh FA, et al. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther 2008;16:1833-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Graham BS. Rapid COVID-19 vaccine development. Science 2020;368:945-946. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.