Supplemental Digital Content is available in the text.
Keywords: biomarkers, coronavirus disease 2019, host response, inflammation, intensive care unit
Objectives:
Coronavirus disease 2019 patients admitted to the ICU have high mortality. The host response to coronavirus disease 2019 has only been partially elucidated, and prognostic biomarkers have not been identified. We performed targeted proteomics on critically ill coronavirus disease 2019 patients to better understand their pathophysiologic mediators and to identify potential outcome markers.
Design:
Blood was collected at predetermined ICU days for proximity extension assays to determine the plasma concentrations of 1,161 proteins.
Setting:
Tertiary care ICU and academic laboratory.
Subjects:
All patients admitted to the ICU suspected of being infected with severe acute respiratory syndrome coronavirus 2, using standardized hospital screening methodologies, had blood samples collected until either testing was confirmed negative on ICU day 3 (coronavirus disease 2019 negative) or until ICU day 10 if the patient positive (coronavirus disease 2019 positive).
Interventions:
None.
Measurements and Main Results:
Age- and sex-matched healthy control subjects and ICU patients who were either coronavirus disease 2019 positive or coronavirus disease 2019 negative were enrolled. Cohorts were well-balanced with the exception that coronavirus disease 2019 positive patients suffered bilateral pneumonia more frequently than coronavirus disease 2019 negative patients. Mortality rate for coronavirus disease 2019 positive ICU patients was 40%. Feature selection identified the top performing proteins for identifying coronavirus disease 2019 positive ICU patients from both healthy control subjects and coronavirus disease 2019 negative ICU patients (classification accuracies 100%). The coronavirus disease 2019 proteome was dominated by interleukins and chemokines, as well as several membrane receptors linked to lymphocyte-associated microparticles and/or cell debris. Mortality was predicted for coronavirus disease 2019 positive patients based on plasma proteome profiling on both ICU day 1 (accuracy 92%) and ICU day 3 (accuracy 83%). Promising prognostic proteins were then narrowed down to six, each of which provided excellent classification performance for mortality when measured on ICU day 1 CMRF-35-like molecule, interleukin receptor-12 subunit B1, cluster of differentiation 83 [CD83], family with sequence similarity 3, insulin-like growth factor 1 receptor and opticin; area-under-the-curve =1.0; p = 0.007).
Conclusions:
Targeted proteomics with feature classification easily distinguished both healthy control subjects and coronavirus disease 2019 tested negative ICU patients from coronavirus disease 2019 tested positive ICU patients. Multiple proteins were identified that accurately predicted coronavirus disease 2019 tested positive patient mortality.
Coronavirus disease 2019 (COVID19) is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Confirmed cases of COVID19 worldwide continue to grow (1), with a reported case-fatality rate of approximately 3.4% (2). COVID19 primarily affects lungs and in the most severe cases results in acute respiratory distress syndrome associated with or without multiple organ dysfunction (3–6). Once COVID19 patients are admitted to the ICU, the mortality rate is reported to be 31% with a median of 9 days to ICU death (7). There are no specific therapies for COVID19, and patients are provided only supportive care. Identification of pathophysiologic mediators, as well as prognostic biomarkers and/or therapeutic targets, is essential for improving COVID19 patient outcomes.
A “cytokine storm” may determine the severity of COVID19 (8), which is the excessive or uncontrolled release of cytokines in response to a pathologic event, such as a viral infection (9). Using a multiplex immunoassay platform, we have previously determined that COVID19 is associated with a unique inflammatory profile dominated by tumor necrosis factor (TNF), granzyme B, heat shock protein 70 (HSP70), interleukin (IL) 18, interferon-gamma inducible protein 10, and elastase 2 (10). Potential therapies may, therefore, target TNF and/or serine proteases with antibodies or small molecule inhibitors. In addition, thrombosis profiling in COVID19 patients identified microvascular injury, including endothelial activation and glycocalyx degradation, as a contributor to mortality (11). These latter findings suggest platelet adhesion to the injured pulmonary microvasculature could be an important COVID19 pathologic mechanism. Protection and/or restoration of the endothelial glycocalyx, as well as platelet adhesion inhibitors, may improve microvascular functioning during COVID19. Although the more common inflammatory and thrombotic proteins contributing to COVID19 pathophysiology have been identified (10, 11), a plethora of protein mediators have yet to be investigated and may hold critically important information to improve COVID19 outcomes.
The overall aim of this study was to assess a targeted proteome from critically ill COVID19 patients over the first 10 days of their ICU stay to identify potential candidates as prognostic biomarkers and/or therapeutic targets. Our specific objectives were (1) to determine protein alterations between coronavirus disease 2019 positive (COVID19+) ICU patients and healthy control subjects (2) to determine proteins that most accurately differentiated COVID19+ from coronavirus disease 2019 negative (COVID19–) ICU patients; and (3) to determine which proteins predict outcome shortly after ICU admission and how these proteins change over time in COVID19+ ICU patients.
METHODS
This study was approved by the Western University, Human Research Ethics Board (HREB). Given the unprecedented pandemic situation and the restricted hospital access for substitute decision makers, waived consent was approved for a short, defined period of time. In keeping with the Society for Critical Care Medicine statement on “Waiver of Informed Consent in Emergency Situations” (12), the following criteria were considered relevant for HREB approval of waived consent: the subjects were admitted to the ICU with a life-threatening condition; the subjects had impaired decisional capacity; the research staff encountered significant obstacles and delays when attempting to contact the absent substitute decision-makers; the study risk was minimal; the research knowledge gained on this new, lethal disease offered an eventual chance of benefit; and community consultation had been implemented.
Study Participants and Clinical Data
We enrolled consecutive patients who were admitted to our level-3 academic ICUs at London Health Sciences Centre (London, Ontario) and were suspected of having COVID19 based on standard hospital screening procedures (13). Blood sampling began on ICU admission for up to 3 days in COVID19– patients or up to 7 days in COVID19+ patients (1 additional blood draw on day 10). COVID19 status was confirmed as part of standard hospital testing by detection of two SARS-CoV-2 viral genes using polymerase chain reaction (14). Patient baseline characteristics were recorded at admission and included age, sex, comorbidities, medications, hematologic labs, creatinine, Pao2 to Fio2 ratio, and chest radiograph findings. We calculated Multiple Organ Dysfunction Score (MODS) (15) and Sequential Organ Failure Assessment score (16) for both COVID19+ and COVID19– patient groups to enable objective comparison of their illness severity. Both patient groups were characterized as having confirmed or suspected sepsis diagnosis using Sepsis 3.0 criteria (16). We also recorded clinical interventions received during the observation period including use of antibiotics, antiviral agents, systemic corticosteroids, vasoactive medications, venous thromboembolism prophylaxis, antiplatelet or anticoagulation treatment, renal replacement therapy, high flow oxygen therapy, and mechanical ventilation (invasive and noninvasive). Final participant groups were constructed by age- and sex-matching COVID19+ patients with COVID19– patients and healthy control subjects without disease, acute illness, or prescription medications that were previously banked in the Translational Research Centre, London, ON (Directed by Dr. D.D. Fraser; https://translationalresearchcentre.com/) (17, 18).
Blood Draws
Standard operating procedures were used to ensure all samples were treated rapidly and equally. Blood was obtained from critically ill ICU patients via indwelling catheters daily in the morning and placed immediately on ice. If a venipuncture was required, research blood draws were coordinated with a clinically indicated blood draw. In keeping with accepted research phlebotomy protocols for adult patients, blood draws did not exceed maximal volumes (19). Once transferred to a negative pressure hood, blood was centrifuged and plasma isolated, aliquoted at 250 µL and frozen at –80°C. All samples remained frozen until use and freeze/thaw cycles were avoided.
Proximity Extension Assay
A total of 1,161 plasma proteins were measured using an immunoassay based on proximity extension assay (PEA) technology (Olink Proteomics, Sweden) (20, 21). A 0.25 mL aliquot of trisodium citrate anticoagulated plasma was obtained from each subject and was transported frozen on dry ice to the Clinical Research Laboratory and Biobank (Hamilton, ON). The data generated were expressed as relative quantification on the log2 scale of normalized protein expression (NPX) values. Individual samples were screened based on quality controls for immunoassay and detection, as well as degree of hemolysis. NPX values were rank-based normal transformed for further analyses. Following proteomic quality control, all 30 participants were deemed suitable for analysis.
Population Statistics
Medians (interquertile ranges [IQRs]) and frequency (%) were used to report ICU patient baseline characteristics for continuous and categorical variables, respectively; continuous variables were compared using Mann-Whitney U tests (or Kruskal-Wallis tests, as appropriate), and categorical variables were compared using Fisher exact chi-square, with p values less than 0.05 considered statistically significant. Receiver operating characteristic (ROC) curves were conducted to determine sensitivity and specificity of individual proteins for predicting outcome (alive or dead). Area-under-the-curve (AUC) was calculated as an aggregate measure of protein performance across all possible classification thresholds. All analyses were conducted using SPSS version 26 (IBM Corp., Armonk, NY).
Machine Learning
COVID19 analyte data were visualized with a nonlinear dimensionality reduction on the full data matrix using the t-distributed stochastic nearest neighbor embedding (t-SNE) algorithm (10). t-SNE assumes that the “optimal” representation of the data lies on a manifold with complex geometry, but low dimension, embedded in the full dimensional space of the raw data (22). For feature selection, the raw data for each subject were ingested and normalized within each feature, across subjects. More specifically, the data for each marker were scaled to have unit norm. A random forest classifier was trained on the variables to predict COVID status. A random forest is a set of decision trees, and consequently, we were able to interrogate this collection of trees to identify the features that have the highest predictive value (namely, those features that frequently appear near the top of the decision tree). The feature matrix for day 1 COVID19+ ICU patients was classified for patient outcome using a three-fold cross validation with a Random Forest of 100 trees and maximum depth of 6 to reduce overfitting (23).
RESULTS
We investigated 10 COVID19+ ICU patients (median years old = 61.0; IQR = 54.8–67.0), 10 age- and sex-matched COVID19- ICU patients (median years old = 58.0; IQR = 52.5–63.0) and 10 age- and sex-matched healthy control subjects (median years old = 57.5; IQR = 52.8–62.8; p = 0.686). Baseline demographic characteristics, comorbidities, laboratory values, and chest radiograph findings are reported in Table 1. The COVID19– ICU patients had significantly higher unilateral pneumonia, whereas COVD19+ ICU patients were more likely to have bilateral pneumonia. Sepsis was “confirmed” by infectious pathogen identification in only 20% of COVID19– ICU patients, whereas sepsis was “suspected” in the remaining 80%. A mortality rate of 40% was determined for COVID19+ ICU patients.
Table 1.
Subject Demographics and Clinical Data
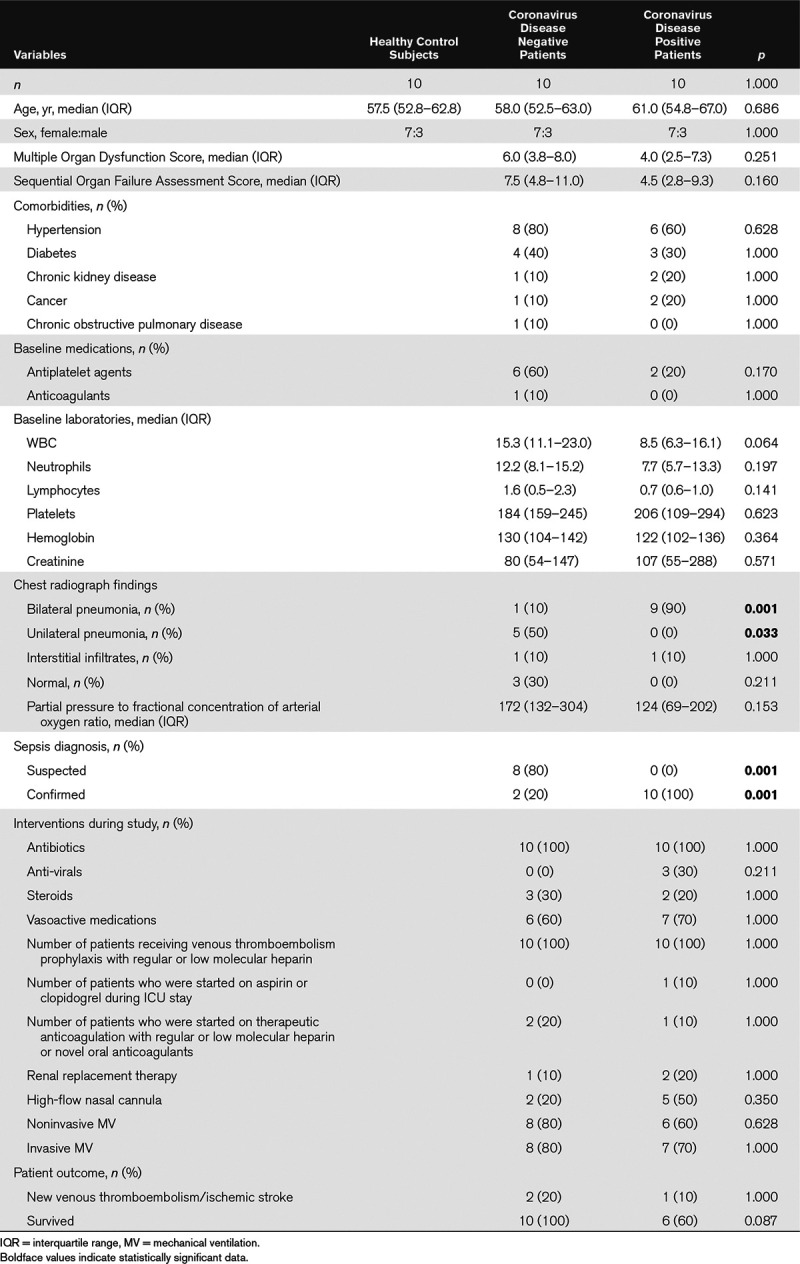
We measured 1,161 plasma proteins using PEA immunoassays. Figure 1A shows a t-SNE plot illustrating that the COVID19+ ICU patient proteome was distinct and easily separable from age- and sex-matched healthy control subjects. Feature classification identified the top 20 proteins underlying these differences between cohorts and are shown in Figure 1B with their associated importance. Classification accuracy was 100%. The biological functions of these leading 20 proteins are described in Supplemental Table 1 (Supplemental Digital Content 1, http://links.lww.com/CCX/A287.
Figure 1.
Targeted proteomics accurately differentiates coronavirus disease 2019 positive (COVID19+) ICU patients from healthy control subjects. A, Subjects plotted in 2D following dimensionality reduction of their respective proteomes by stochastic neighbor embedding. Yellow dots represent healthy control subjects, whereas purple dots represent age- and sex-matched COVID19+ ICU patients (ICU day 1 plasma). The dimensionality reduction shows that based on the plasma proteome, the two cohorts are distinct and easily separable (100% classification accuracy). The axes are dimension less. B, Feature classification demonstrating the top 20 plasma proteins that classify coronavirus disease 2019 status with their % association.
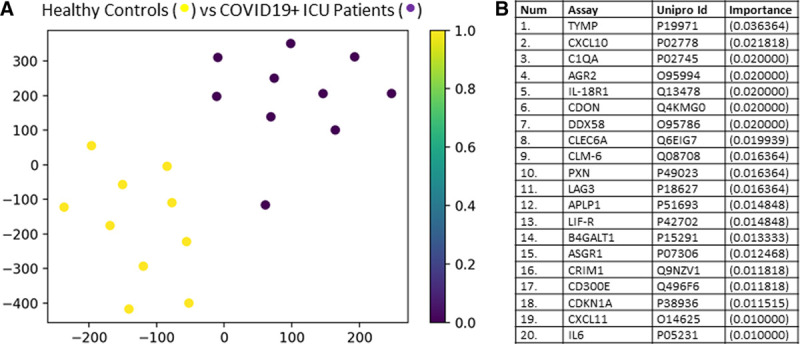
Figure 2A shows a t-SNE plot illustrating that the COVID19+ ICU patient proteome was distinct and easily separable from age- and sex-matched COVID19– ICU patients. Feature classification identified the top 20 proteins underlying these differences between cohorts and are shown in Figure 2B with their associated importance. Classification accuracy was 100%. The biological functions of these leading 20 proteins are described in Supplemental Table 2 (Supplemental Digital Content 2, http://links.lww.com/CCX/A288.
Figure 2.
Targeted proteomics accurately differentiates coronavirus disease 2019 positive (COVID19+) ICU patients from coronavirus disease 2019 negative (COVID19–) ICU patients. A, Subjects plotted in 2D following dimensionality reduction of their respective proteomes by stochastic neighbor embedding. Yellow dots represent COVID19+ ICU patients, whereas purple dots represent age- and sex-matched COVID19– ICU patients (ICU day 1 plasma for both populations). The dimensionality reduction shows that based on the plasma proteome, the two cohorts are distinct and easily separable (100% classification accuracy). The axes are dimension less. B, Feature classification demonstrating the top 20 plasma proteins that classify coronavirus disease 2019 status with their % association.
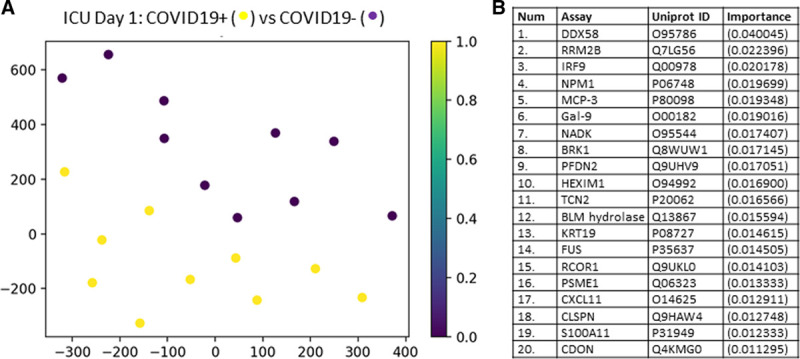
We then determined the ability of the plasma proteome to predict mortality in COVID19+ patients on either ICU days 1 or 3. Figure 3A shows a t-SNE plot demonstrating that the proteome between COVID19+ patients on ICU day 1 that either survived or died were distinct and easily separable. The leading 21 proteins underlying these outcome differences are shown, and their biological functions are described in Supplemental Table 3 (Supplemental Digital Content 3, http://links.lww.com/CCX/A289). Figure 3B shows a t-SNE plot demonstrating that the proteome between COVID19+ patients on ICU day 3 that who survived or died were distinct and separable. The leading 21 proteins underlying these outcome differences are shown, and their biological functions are described in Supplemental Table 4 (Supplemental Digital Content 4, http://links.lww.com/CCX/A290. The classification accuracy to predict outcome with the entire 1,161 proteins in COVID19+ patients on ICU days 1 and 3 was 92% and 83%, respectively.
Figure 3.
Targeted proteomics accurately differentiates coronavirus disease 2019 positive (COVID19+) patients that lived or died on ICU days 1 and 3. A, COVID19+ ICU patients plotted in 2D following dimensionality reduction of their respective outcomes (alive or dead) by stochastic neighbor embedding. Green dots represent COVID19+ ICU patients who survived, whereas red dots represent COVID19+ ICU patients who died (ICU day 1 plasma for both populations). The dimensionality reduction shows that based on the plasma proteome, the two cohorts are distinct and easily separable (92% classification accuracy). The axes are dimension less. B, COVID19+ ICU patients plotted in 2D following dimensionality reduction of their respective outcomes (alive or dead) by stochastic neighbor embedding (ICU day 3 plasma for both populations). Green dots represent COVID19+ ICU patients who survived, whereas red dots represent COVID19+ ICU patients who died. The dimensionality reduction shows that based on the plasma proteome, the two cohorts are reasonably distinct and separable (83% classification accuracy). The axes are dimension less. C, Feature classification demonstrating the top 21 plasma proteins obtained on ICU day 1 that classify outcome for COVID19+ ICU patients with their % association. D, Feature classification demonstrating the top 21 plasma proteins obtained on ICU day 3 that classify outcome for COVID19+ ICU patients with their % association.
To optimize outcome prediction in COVID19+ patients, we then narrowed the number of proteins from 1,161 using ROC analyses. The top six proteins for predicting ICU survival/death using only day 1 plasma samples are shown in Figure 4, also shown is their associated time course over 10 ICU days. There were no deaths during the 10 ICU days for either cohort; however, one COVID19+ ICU survivor was discharged on day 7 with no further plasma measurements. With all six proteins, the COVID19+ patients who died had elevated levels relative to those COVID19+ patients who survived to ICU discharge. All six proteins provided excellent classification performance with AUCs equals to 1.0 (p = 0.007) for outcome when measured in plasma on ICU day 1 with the following cutoffs: CMRF-35-like molecule [CLM-1, 7.8], interleukin receptor-12 subunit B1 [IL12RB1, 3.3], cluster of differentiation 83 [CD83, 3.3], family with sequence similarity 3 [FAM3B, 4.7], insulin-like growth factor 1 receptor [IGF1R, 3.8] and opticin [OPTC], 3.6.
Figure 4.
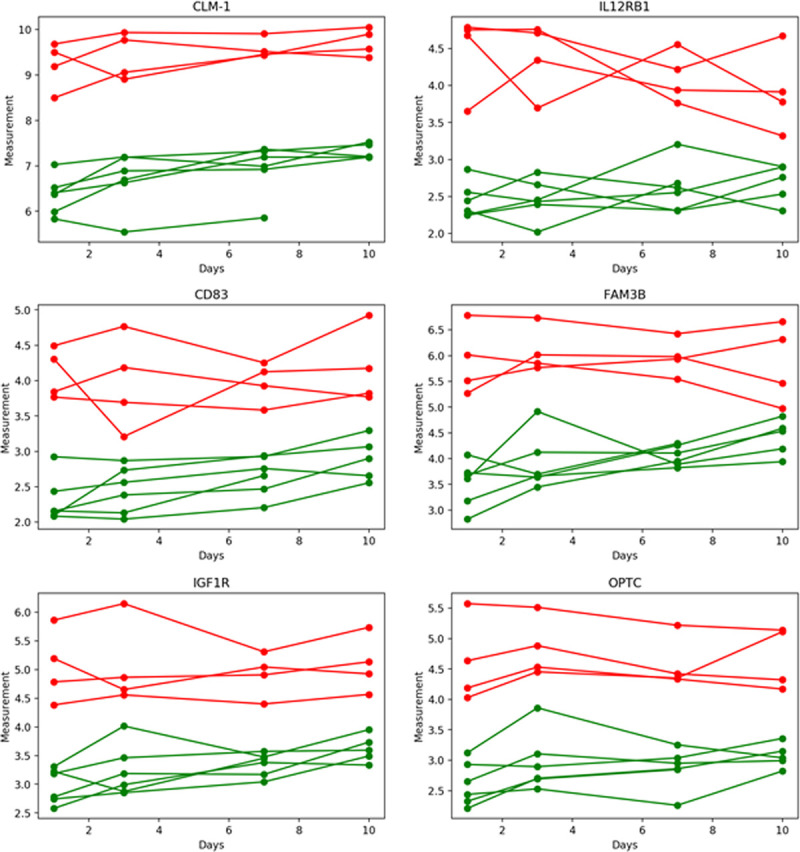
Time course for the top six plasma proteins that predicted coronavirus disease 2019 (COVID19) outcome. Green lines represent six coronavirus disease 2019 positive (COVID19+) ICU patients who survived (one patient was discharged by ICU day 7), whereas red lines represent four COVID19+ ICU patients who died. Receiver operating characteristic analyses for all six proteins measured on ICU day 1 had area-under-the-curves of 1.0 (p = 0.007), indicating excellent classification performance. Their respective cutoff values were: CMRF-35-like molecule (CLM-1, 7.8), interleukin receptor-12 subunit B1 (IL12RB1, 3.3), cluster of differentiation 83 (CD83, 3.3), family with sequence similarity 3 (FAM3B, 4.7), insulin-like growth factor 1 receptor (IGF1R, 3.8) and opticin (OPTC, 3.6).
DISCUSSION
In this study, we measured 1,161 proteins in plasma obtained from ICU patients, both COVID19+ and COVID19-, as well as age- and sex-matched healthy control subjects. Given the number of analytes measured, we analyzed the data with state-of-the-art machine learning. Our data indicate the presence of a unique COVID19 plasma proteome with six proteins predicting ICU mortality with 100% accuracy. Despite the exploratory nature of our study, the data generated suggest that these six proteins could be considered for further investigation as potential disease severity and/or outcome biomarkers, and they may be useful for patient stratification in clinical interventional trials.
Our COVID19+ ICU patients were similar to those reported in earlier cohorts (3–6) with respect to demographic, comorbidities, and clinical presentation. In contrast to COVID19– ICU patients, our COVID19+ ICU patients had a higher occurrence rate of bilateral pneumonia. Previous work by our study group in these same patients have determined a unique inflammatory and thrombotic profile. Characterized by elevated TNF and serine proteases (10) and in association with endothelial activation and glycocalyx degradation (11), COVID19 represents a severe illness with a high mortality rate. Indeed, in our cohort of COVID19 patients, ICU death was 40%.
Our study has identified a unique proteome in COVID19+ ICU patients that is hypothesis-generating for future biomarker studies of disease severity and/or outcome. Not only have we provided rank order listing of proteins important for COVID19 status, we also identified six proteins that accurately determined COVID19 outcome. The former represents diverse proteins that are dominated by ILs, CXCLs/chemokines, membrane receptors linked to lymphocyte-associated microparticles and/or cell debris, pattern-recognition receptors, cytoplasmic/cytoskeletal proteins, and/or nuclear proteins/transcription factors. The six novel protein outcome predictors that were elevated on ICU day 1 in COVID19+ patients are reviewed individually below.
CLM-1, a type-1 transmembrane glycoprotein with an extracellular immunoglobulin G domain (24), accurately predicted COVID19 ICU outcome. CLM-1 is expressed predominantly in myeloid cells where it can impair IL-6 production in bone marrow–derived mast cells (25) and promotes phagocytosis of dead cells by binding phosphatidylserine (26, 27), which serves as a common apoptotic cell surface recognition cue. The removal of apoptotic cells by CLM-1 expressing macrophages may prevent the generation of secondary necrosis and the release of potentially toxic or immunogenic components from necrotic cells, reducing the likelihood of an inflammatory reaction (28).
IL12RB1, one of two subunits within the IL-12 receptor, is expressed on natural killer cells and activated T-cells. Essential for resistance to intracellular pathogens, IL12RB mediates the proinflammatory response to IL-12 that is released by antigen presenting cells (29). Individual variability in IL12RB1 function is introduced at the epigenetic, genomic polymorphism, and messenger RNA splicing levels, thereby conferring disease susceptibility and variable outcomes (30).
CD83, a member of the immunoglobulin superfamily, is expressed on a variety of activated immune cells (31). Providing selective immunosuppression when membrane bound on antigen presenting cells, soluble CD83 inhibits proliferation and function of T-cells. Viral infection leads to the degradation of dendritic cell CD83, a mechanism described as a viral immune escape mechanism (32).
FAM3B expression is induced by glucose and proinflammatory cytokines in the islets of Langerhans of the endocrine pancreas (33). Under physiologic conditions, FAM3B facilitates insulin secretion (33); however, it is also a secreted cytokine-like protein that can induce cellular apoptosis (34). Increased FAM3B secretion is associated with pancreatic β cell dysfunction, hyperglycemia, and insulin resistance, suggesting its role in the regulation of glucose and lipid metabolism (35).
IGF1R, a transmembrane tyrosine kinase receptor that is activated by insulin-like growth factor 1 and 2, is expressed on lymphocytes and macrophages where it can cause proliferation, cytokine production, and priming/activation of target cells (36). In particular, phosphorylation of the IGF1R exaggerates inflammation, and its overexpression increases cytokine levels during influenza infection (37). Conversely, IGF1R deficiency attenuates the acute inflammatory response in a mouse model of acute lung injury (38).
OPTC is highly expressed in the eye nonpigmented ciliary epithelium that secretes it into the vitreous cavity where it associates with vitreous collagen and adjacent basement membranes (39). As a small leucine-rich protein, opticin binds collagen fibrils and regulates extracellular matrix adhesiveness to suppress capillary morphogenesis and inhibit endothelial invasion (40). OPTC is also expressed in lymphocytes (41) and articular joints where it may be degraded by matrix metalloproteinases 1, 2, 3, 7, 8, and 9, and a disintegrin and metalloproteinase with thrombospondin motifs-4 and -5 (42), but its role in infection and inflammation is unknown.
The plasma concentrations of the six protein outcome markers described above remained remarkably consistent over the first 10 days of ICU care and despite the eventual bifurcation of outcome to survival or death. We have reported a similar trend in the plasma concentrations of TNF, HSP70, and IL-18 (10). These temporal observations on plasma biomarker concentrations may suggest that outcome is determined early with COVID19 disease onset or perhaps changes in biomarker concentrations precede death, which was beyond our sampling time frame of 10 ICU days.
Despite the novelty of the biomarkers discovered, our study has several limitations. First, we only studied critically ill patients, and we cannot determine the proteome changes contributing to ICU admissions. Second, we acknowledge that our COVD19 study population was limited; however, we still identified strong associations between individual proteins and outcomes, and we fulfilled an urgent need for exploratory data to focus future hypothesis-driven studies on larger cohorts. Third, we report only mortality as our primary clinical outcome. Future studies with larger sample sizes can explore whether reported changes in inflammatory analytes correlate with additional clinical outcomes such as functional status in survivors. Finally, our analyses employed a cross-validation methodology in which the classifier was trained multiple times, each time on a different subset of the data, with the remainder of the data withheld for use only in testing. The reported accuracy is the mean accuracy of all such trials; this is a standard, accepted technique in the machine learning literature but should be validated on a larger testing set that is used only once. Overfitting was minimized by using a very small number of trees, and the very limited depth is protective against over-fitting (23).
In summary, we report a unique proteome in COVID19+ ICU patients, with identification of six novel proteins that appear to be accurate outcome biomarkers for future studies. Given the high morbidity and mortality associated with COVID19 critical illness, our exploratory data may be invaluable for guiding resource mobilization and/or goals of care discussion, but only after validation in larger COVID19+ cohorts. Furthermore, patient stratification is critically important for future COVID19 interventional trials.
ACKNOWLEDGMENTS
We thank Ms. Shana Lamers for performing PEA (Hamilton, ON), Mr. Cole Zimmerman for providing Olink support, and the entire Lawson COVID 2019 Study Team for their assistance (Dr. Robert Arntfield, Dr. Ian Ball, Mr, Gordon Barkwell, Ms. Tracey Bentall, Dr. Karen Bosma, Ms. Saoirse Cameron, Ms. Eileen Campbell, Mr. David Carter, Dr. Carolina Gillio-Meina, Dr. Robert Hegele, Ms. Natalya Odoardi, Dr. Ram Singh, Dr. Kelly Summers, and Ms. Sue Tereschyn). We are grateful for the enthusiastic support of the frontline Critical Care Nursing Staff at London Health Sciences Centre.
Supplementary Material
Footnotes
We acknowledge funding from Western University (Research), the Departments of Medicine and Pediatrics at Western University, the Lawson Health Research Institute (https://www.lawsonresearch.ca/), the London Health Sciences Foundation (https://lhsf.ca/), and the AMOSO Innovation Fund.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (http://journals.lww.com/ccejournal).
The authors disclosed a patent pending (COVID-19 Outcome Prediction; #63041670).
Contributor Information
Collaborators: Robert Arntfield, Ian Ball, Gordon Barkwell, Tracey Bentall, Karen Bosma, Saoirse Cameron, Eileen Campbell, David Carter, Carolina Gillio-Meina, Robert Hegele, Natalya Odoardi, Ram Singh, Kelly Summers, and Sue Tereschyn
REFERENCES
- 1.John Hopkins University & Medicine: COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). Available at: https://coronavirus.jhu.edu/map.html. Accessed August 18, 2020.
- 2.World Health Organization: WHO Director-General’s opening remarks at the media briefing on COVID-19 - 3 March 2020. Available at: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---3-march-2020. Accessed August 18, 2020
- 3.Bhatraju PK, Ghassemieh BJ, Nichols M, et al. Covid-19 in critically ill patients in the Seattle region - case series. N Engl J Med. 2020; 382:2012–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet. 2020; 395:1054–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu C, Chen X, Cai Y, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020; 180:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grasselli G, Zangrillo A, Zanella A, et al. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy Region, Italy. JAMA. 2020; 323:1574–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Auld SC, Caridi-Scheible M, Blum JM, et al. ICU and ventilator mortality among critically ill adults with coronavirus disease 2019. Crit Care Med. 2020. May 26. [online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tisoncik JR, Korth MJ, Simmons CP, et al. Into the eye of the cytokine storm. Microbiol Mol Biol Rev. 2012; 76:16–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehta P, McAuley DF, Brown M, et al. ; HLH Across Speciality Collaboration, UK. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet. 2020; 395:1033–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser DD, Cepinskas G, Slessarev M, et al. Inflammation profiling of critically ill coronavirus disease 2019 patients. Crit Care Explor. 2020; 2:e0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraser DD, Patterson EK, Slessarev M, et al. Endothelial injury and glycocalyx degradation in critically ill Covid19 patients: Implications for microvascular platelet aggregation. Crit Care Expl. 2020; 2:e0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bauer PR, Tate JA. Society of Critical Care Medicine: Waiver of Informed Consent in Emergency Situations. Available at: https://www.sccm.org/Communications/Critical-Connections/Archives/2018/Waiver-of-Informed-Consent-in-Emergency-Situations. Accessed August 18, 2020
- 13.Centers for Disease Control and Prevention: Overview of Testing for SARS-CoV-2. Available at: https://www.cdc.gov/coronavirus/2019-nCoV/hcp/clinical-criteria.html. Accessed August 18, 2020.
- 14.Centers for Disease Control and Prevention: CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. Available at: https://www.fda.gov/media/134922/download. Accessed August 18, 2020
- 15.Priestap F, Kao R, Martin CM. External validation of a prognostic model for intensive care unit mortality: A retrospective study using the Ontario Critical Care Information System. Can J Anaesth. 2020; 67:981–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus definitions for sepsis and septic shock (sepsis-3). JAMA. 2016; 315:801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brisson AR, Matsui D, Rieder MJ, et al. Translational research in pediatrics: Tissue sampling and biobanking. Pediatrics. 2012; 129:153–162 [DOI] [PubMed] [Google Scholar]
- 18.Gillio-Meina C, Cepinskas G, Cecchini EL, et al. Translational research in pediatrics II: Blood collection, processing, shipping, and storage. Pediatrics. 2013; 131:754–766 [DOI] [PubMed] [Google Scholar]
- 19.NIH Human Research Protection Program. POLICY: Guidelines for Limits of Blood Drawn for Research Purposes in the Clinical Center, M95-9 (rev). National Institutes of Health (NIH) Clinical Center, 2009
- 20.Assarsson E, Lundberg M, Holmquist G, et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 2014; 9:e95192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lundberg M, Eriksson A, Tran B, et al. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res. 2011; 39:e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008; 9:2579–2605 [Google Scholar]
- 23.Tang C, Garreau D, von Luxburg U. When do random forests fail 2018Proceedings of the 32nd International Conferenceon Neural Information Processing Systems 20182987–2997 [Google Scholar]
- 24.Chung DH, Humphrey MB, Nakamura MC, et al. CMRF-35-like molecule-1, a novel mouse myeloid receptor, can inhibit osteoclast formation. J Immunol. 2003; 171:6541–6548 [DOI] [PubMed] [Google Scholar]
- 25.Izawa K, Kitaura J, Yamanishi Y, et al. An activating and inhibitory signal from an inhibitory receptor LMIR3/CLM-1: LMIR3 augments lipopolysaccharide response through association with FcRgamma in mast cells. J Immunol. 2009; 183:925–936 [DOI] [PubMed] [Google Scholar]
- 26.Choi SC, Simhadri VR, Tian L, et al. Cutting edge: Mouse CD300f (CMRF-35-like molecule-1) recognizes outer membrane-exposed phosphatidylserine and can promote phagocytosis. J Immunol. 2011; 187:3483–3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian L, Choi SC, Murakami Y, et al. p85α recruitment by the CD300f phosphatidylserine receptor mediates apoptotic cell clearance required for autoimmunity suppression. Nat Commun. 2014; 5:3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee HN, Tian L, Bouladoux N, et al. Dendritic cells expressing immunoreceptor CD300f are critical for controlling chronic gut inflammation. J Clin Invest. 2017; 127:1905–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ford NR, Miller HE, Reeme AE, et al. Inflammatory signals direct expression of human IL12RB1 into multiple distinct isoforms. J Immunol. 2012; 189:4684–4694 [DOI] [PubMed] [Google Scholar]
- 30.Robinson RT. IL12Rβ1: The cytokine receptor that we used to know. Cytokine. 2015; 71:348–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z, Ju X, Silveira PA, et al. CD83: Activation marker for antigen presenting cells and its therapeutic potential. Front Immunol. 2019; 10:1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heilingloh CS, Mühl-Zürbes P, Steinkasserer A, et al. Herpes simplex virus type 1 ICP0 induces CD83 degradation in mature dendritic cells independent of its E3 ubiquitin ligase function. J Gen Virol. 2014; 95:1366–1375 [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Yang W, Wang J, et al. FAM3 gene family: A promising therapeutical target for NAFLD and type 2 diabetes. Metabolism. 2018; 81:71–82 [DOI] [PubMed] [Google Scholar]
- 34.Cao X, Gao Z, Robert CE, et al. Pancreatic-derived factor (FAM3B), a novel islet cytokine, induces apoptosis of insulin-secreting beta-cells. Diabetes. 2003; 52:2296–2303 [DOI] [PubMed] [Google Scholar]
- 35.Chi Y, Meng Y, Wang J, et al. FAM3B (PANDER) functions as a co-activator of FOXO1 to promote gluconeogenesis in hepatocytes. J Cell Mol Med. 2019; 23:1746–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomson AW, Lotze MT. 2003FourthEdition, Amsterdam, The Netherlands; Boston, MA: Academic Press [Google Scholar]
- 37.Li G, Zhou L, Zhang C, et al. Insulin-like growth factor 1 regulates acute inflammatory lung injury mediated by influenza virus infection. Front Microbiol. 2019; 10:2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piñeiro-Hermida S, López IP, Alfaro-Arnedo E, et al. IGF1R deficiency attenuates acute inflammatory response in a bleomycin-induced lung injury mouse model. Sci Rep. 2017; 7:4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramesh S, Bonshek RE, Bishop PN. Immunolocalisation of opticin in the human eye. Br J Ophthalmol. 2004; 88:697–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Goff MM, Sutton MJ, Slevin M, et al. Opticin exerts its anti-angiogenic activity by regulating extracellular matrix adhesiveness. J Biol Chem. 2012; 287:28027–28036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palani S, Elima K, Ekholm E, et al. Monocyte stabilin-1 suppresses the activation of Th1 lymphocytes. J Immunol. 2016; 196:115–123 [DOI] [PubMed] [Google Scholar]
- 42.Tío L, Martel-Pelletier J, Pelletier JP, et al. Characterization of opticin digestion by proteases involved in osteoarthritis development. Joint Bone Spine. 2014; 81:137–141 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


