Abstract
This contribution describes the current state of experimental model development and use as a strategy for gaining insight into the form and function of certain types of host-microbe associations. Development of quality models for the study of symbiotic systems will be critical not only to facilitate an understanding of mechanisms underlying symbiosis, but also for providing insights into how drug development can promote healthy animal-microbe interactions as well as the treatment of pathogenic infections. Because of the growing awareness over the last decade of the importance of symbiosis in biology, a number of model systems has emerged to examine how these partnerships are maintained within and across generations of the host. The focus here will be upon host-bacterial symbiotic systems that, as in humans, (i) are acquired from the environment each generation, or horizontally transmitted, and (ii) are defined by interactions at the interface of their cellular boundaries, i.e., extracellular symbiotic associations. As with the use of models in other fields of biology where complexity is daunting (e.g., developmental biology or brain circuitry), each model has its strengths and weaknesses, i.e., no one model system will provide easy access to all the questions defining what is conserved in cell-cell interactions in symbiosis and what creates diversity within such partnerships. Rather, as discussed here, the more models explored, the richer our understanding of these associations is likely to be.
Keywords: horizontally transmitted symbioses, extracellular microbial partners
Graphical Abstract
1. Introduction
With new sequencing technology, biologists over the past two decades are getting the first ‘close look’ at the microbial world, now known to be more diverse and more critical to biosphere structure and function than ever imagined. Animals and plants are often highly complex assemblages, with the host organisms accommodating a vast array of associating viruses, fungi, bacteria, and archaea. A principal question in this research arena is: how do the symbiotic systems function to generate and sustain a healthy condition in the partnership? Biomedical science, of course, aims to reveal the mechanistic underpinnings of human health and disease. The new findings about the importance of the microbiome change our understanding of our true nature, as well as our understanding of our position in the biosphere [1].
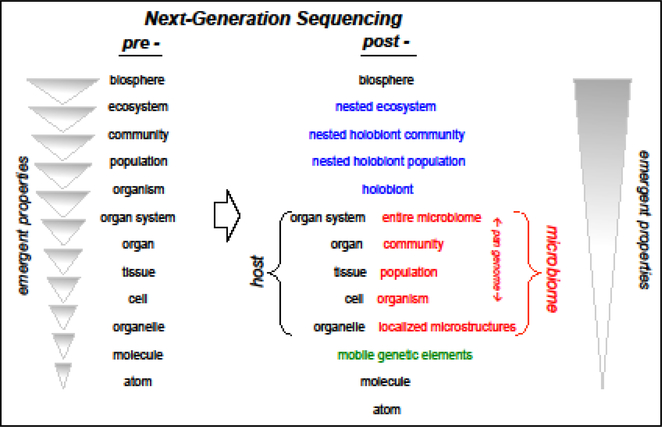
This anthropocentric view is embedded in major changes in the conceptual framework of the field of biology as a whole; i.e., very basic theories of biology will be recast as a result of the new horizons presented by microbiome research. For example, it is no longer accurate to generate constructs of the hierarchy of life that ignore the complexity of the holobiont [2], i.e., the nested and integrated partnerships that animal or plant hosts have with the microbial world, including mobile genetic elements (e.g., viruses; Fig. 1). The holobiont comprises complementary host and microbe levels of hierarchical complexity, with the characteristic of emergent properties, i.e., features of a given level that cannot be predicted by a thorough knowledge of the lower levels of the hierarchy. This intricacy of relationships within and between animal and plant holobionts demands development of new conceptual and technical frameworks with which to gain an understanding of the structure and function of these systems, specifically: (i) how does the microbiota affect the host and (ii) how does the resulting holobiont interface with the higher levels of the hierarchy (Fig. 1)? The inclusion of the microbes into this conceptual framework may make an understanding of emergent properties more accessible and integrated. It should be noted here that the term ‘holobiont’, unlike the controversial term ‘hologenome’, refers only to the composite of the host and its microbial partners and does not hold meaning as to whether selection pressure occurs on the individual constituents of the holobiont or, as with the concept of the hologenome, on the holobiont as a whole [3].
Fig. 1. The larger arena in which model systems of symbiosis are developing.
A growing recognition of the complexity of symbiotic systems drives a conceptual change in our view of the hierarchy of life, one that reflects the nested nature of biological systems. Left: Before next-generation sequencing (‘pre-’), the increasing complexity of the hierarchical organization of biological systems was presumed to be one dimensional, with ‘emergent properties’ arising with each successive level of organization. The lack of continuity between levels (discrete triangles) represents the tendency for the field to be in sub-disciplinary silos focused on one or a few levels. Right: New data (‘post-’) have revealed a more complex hierarchy that incorporates the inherent nesting of the macro- and microbial worlds; blue, synthetic levels that represent the nesting of each successive level of the hierarchy; red, microbial elements that join with corresponding host levels to create the holobiont; green, a proposed additional level. While more complex, this view of the hierarchy could render predictions between levels more approachable, and lead to development of a more integrated view of the biosphere, represented by the single image of emergent properties.
In recent years, a strong emphasis has been placed on research focused toward understanding the dynamics of the human microbiome. The complexity of interactions among the human-holobiont networks presents perhaps the most intricate landscape that biologists have encountered [4]. To consider an analogy, the human brain’s neocortex has an estimated 1014 synapses [5], about the same as the number of microbes in the human gut. Determining how the brain functions has been a daunting challenge, even though, unlike the microbiome, the cells of the brain are all derived from the human genetic background. In the more diverse symbiotic systems such as the vertebrate gut microbiome, these invisible, often uncultured partners, complement the host’s biology with an equivalent number of cells, and orders of magnitude more genes and the metabolic pathways/metabolites they encode [4]. As such, the numbers and types of potential linkages between and among the cells, populations, and communities of the human holobiont dwarf those of neural networks of the brain.
Although humans and other vertebrates harbor these highly complex microbiomes, greater than 97% of all animal species are outside of the vertebrate subphylum. In contrast to vertebrates, with the exception of a few species, such as termites and their relatives the cockroaches [6], most invertebrate species have comparatively simple microbiomes [7], with only one or a few microbial partners in the symbiosis (Fig. 2). Further, because microbes have been associated with animals during and through their diversification over the last several hundred million years [1], it is likely that conserved features driving the form and function of symbioses will be common across the animal kingdom. Thus, a way forward toward understanding these conserved, critical features is to couple observations and descriptions of complex microbiomes with experimental approaches performed in simpler model systems (see, e.g., [8]). Here, we will consider the value of models for the study of symbiosis, including highlighting some of the advances they have made possible.
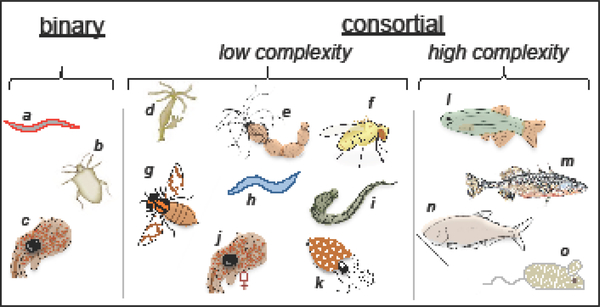
Fig. 2. Some experimental models of extracellular, environmentally acquired, animal symbioses.
Living with a single animal host species -Left: the binary symbioses, one principal bacterial phylotype in a specific organ [hosts – a, nematode Steindernema carpocapsae; b, stink bug, various species; c, bobtail squid, Euprymna scolopes. Middle: low complexity consortial symbioses, with only a few to dozens of phylotypes [hosts – d, Hydra spp.; e, starlet sea anemone, Nematostella vectensis; f, fruit fly, Drosophila melanogaster; g, honey bee, Apis mellifera; h, nematode, Caenorhaditis elegans; i, leech, Hirudo verbana; j, female bobtail squid, Euprymna scolopes; k, cuttlefish, Sepia officianalis.] Right: high complexity consortial symbioses, with hundreds to thousands of phylotypes [hosts –l, zebrafish, Danio rerio; m, three- spine stickleback, Gasterosteus aculeatus; n, blind cavefish, Astyanax mexicanus; o, mouse, Mus musculus.
2. Types of models for studies of symbiosis
“...A model’s just an imitation of the real thing.” –Mae West
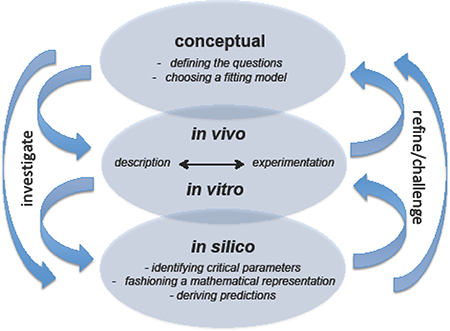
Of the many different types of models that might be considered, a useful set that can be applied readily to symbiotic systems was developed in an essay by Emily Griffiths https://sites.qooqle.com/a/ncsu.edu/emilv-qriffiths/whatisamodel.pdf In this contribution, Griffiths distinguishes four types of models: (i) conceptual – an outline or diagram of a particular subject; (ii) in vivo – whole, live organisms; (iii) in vitro – derivatives of the in vivo model, such as cell cultures or tissue extracts; and, (iv) in silico – a mathematical treatment of the data that generates predictions (Graphical Abstract). Producing a conceptual model might be considered the first step, wherein the research questions to be asked are defined and an experimental model that will address the question is chosen. In vivo and in vitro models offer the opportunity to describe and manipulate the system experimentally, and collect data under different conditions. The results of these activities are essential to providing the numerical information that drives the production of predictive mathematical models.
Such models are often applied sequentially to define biological systems and to achieve specific outcomes. Notably, as mentioned above, simpler systems have the potential to shed light on the ‘rules’ governing more complex systems, e.g., wiring of the brain, developmental processes from fertilized egg to adult, and, relevant here, the dynamics of the mammalian-microbiome axis. In addition, comparative analyses of the diversity of natural ‘experiments’ in symbiosis through evolutionary time allow biologists to define those features that are conserved and fundamental to animal-microbe interaction. For example, because epithelial surfaces are common throughout the eumetazoans, conserved mechanisms underlying the chronic colonization of epithelia by bacteria can be studied across the animal kingdom. In contrast, an identification of examples of convergence in form and function also provides powerful insights. For example, in much the same way that the study of bird, bat, and insect wings, while not homologous, shed light on biomechanical requirements for flight, differences among symbiotic systems can provide information about how similar outcomes are achieved by different means, e.g., various modes of host-symbiont nutritional exchange. Whatever the focus of a given research program, the application of these types of models in an iterative fashion has provided great insights into a wide array of different animal symbiosis (Fig. 2; Table 1).
TABLE 1.
Examples of model symbioses for the study of horizontally transmitted symbioses.
| Type of symbiosis | Host-symbiont(s) | Culturable1 | Molecular genetics | Genome sequenced | Selected references | |||
|---|---|---|---|---|---|---|---|---|
| H | S | H | S | H | S | |||
| Binary (host/symbiont) | Steinernema carpocapsae–Xenorhabdus nematophilus | + | + | -2 | + | + | + | [9,10] |
| Stinkbugs – Riptortus pedestris/Burkholderia spp. & Plautia stali/Pantoea spp. | + | + | + | + | -3 | + | [11–13] | |
| Bobtail squid - Euprymna scolopes/Vibrio fischeri | + | + | -3 | + | + | + | [14,15] | |
| Consortial - low complexity | Hydra spp. | + | +/−4 | + | +/− | + | +/− | [16,17] |
| Starlet sea anemone - Nematostella vectensis | + | +/− | + | - | + | - | [18,19] | |
| Fruit fly - Drosophila melanogaster | + | +/− | + | + | + | +/− | [20,21] | |
| Honey bee - Apis mellifera | + | + | + | + | + | + | [22–24] | |
| Caenorhabditis elegans | + | +/− | + | +/− | + | - | [25,26] | |
| Leech - Hirudo spp. | -2 | + | +/− | + | + | + | [27,28] | |
| Female bobtail squid - Euprymna scolopes | + | +/− | -3 | - | + | +/− | [29,30] | |
| Cuttlefish - Sepia officianalis | - | - | - | - | -3 | - | [31] | |
| Consortial - high complexity | Zebrafish - Danio rerio | + | +/− | + | +/− | + | +/− | [32,33] |
| Three-spine stickleback - Gasterosteus aculeatus | + | +/− | + | - | + | +/− | [34] | |
| Blind cavefish – Astyanax mexicanus | + | +/− | + | - | + | - | [35,36] | |
| Mouse - Mus musculus | + | +/− | + | +/− | + | +/− | [37,38] | |
The partner can be grown in a non-symbiotic state.
- = Not yet available.
Currently in progress or in press.
+/− = Present in some, but not all, of the symbionts.
3. Conceptual models of symbiosis all questions have been converted from bullet points
3.1.-. Defining the questions
One approach to developing conceptual models of horizontally transmitted symbiotic systems is to consider how establishment and maintenance of the association is managed through ontogeny of the host (for a comprehensive review of transmission of microbial symbionts between generations, see [39]). In addition, questions beyond the processes through the ontogeny of an individual are also presented and can be deeply characterized using a variety of models.
Recruitment of the symbionts from the environment with fidelity each generation
As these types of symbiosis are acquired anew each generation from the environment (i.e., the symbionts do not participate in the process of embryogenesis), the following questions for initial encounter of the would-be partners are presented: i) What features of the host – genetic to morphological - are ‘hard wired’ into host embryogenesis to ensure that post-embryogenic interactions with environmental reservoirs of the symbiont(s) will promote successful colonization? ii) What behaviors of the colonized host serve to supply symbionts to their environmental reservoirs, rendering them available to the next generation of hosts? iii) Does the host employ biomechanical mechanisms to recruit its symbionts? And, iv) What features promote specificity of interaction during initial colonization?
Because interaction with an environmental reservoir is critical to success in horizontally transmitted symbiotic systems, evolution has selected for divergent strategies in aquatic and terrestrial animal hosts. Aquatic microbial reservoirs are supported in the environment, where they exist as free-living members of the nanoplankton. As such, aquatic animals and their symbionts have highly refined recognition mechanisms that promote specificity within the high background of non-specific cells. In contrast, terrestrial animals typically facilitate horizontal transmission by the involvement of conspecifics in the transfer of their specific symbionts between generations (e.g., direct interactions with the mother, siblings, or nest mates). Thus, models suitable for the study of the onset of symbiosis in aquatic and terrestrial animals will reflect these principal differences, and some key features of symbiont recruitment will not be well conserved across this major habitat divide.
Processes following recruitment
Once the symbiosis is established, often along the apical surfaces of host epithelia, many of the critical research questions are similar between aquatic and terrestrial systems, and a differentiation between the models of these two habitat types is not longer as meaningful. In both types of systems, the following questions arise: i) How do the symbionts influence the developmental trajectory of the host? ii) Does establishment of a localized symbiosis have broader effects on host development; if so, what are those effects and how is the symbiont’s influence on development mediated? iii) In consortial symbioses, is there a developmental succession of the symbiont community? iv) What are the host-symbiont dynamics that underlie persistence of the symbiosis and maintenance of a healthy state? v) In any given consortial symbioses, what is the relative role of the autochthonous (resident, coevolved) to allochthonous (‘tourist’) microbiota to the overall function of the association? vi) How are other organ systems of the host integrated into the function of the organ system in which a given microbiota is located? And, vii) What is the principal function of the symbiosis (e.g., nutritional, defense, communication), and how do the details of the host’s biology reflect the promotion of that function through ontogeny?
Beyond the ontogeny of an individual
Finally, all phases of a symbiosis are impacted by common elements that are captured by several overarching questions, including: i) How have the characteristic features of a symbiosis evolved; i.e., how and when did they arise and how have they changed? ii) Does the specificity of the association occur at the level of phylotype or guild? [Here guild is used in the classical ecological sense, i.e., to refer to species that exploit the same resource, overlapping in niche requirements.] iii) How are these features reflected in the genomes of the host and symbiont(s)? And, iv) How do strain differences among symbiont species influence the form and function of the association; do those differences lead to distinct biogeographic distributions?
3.2.–. Selecting symbiosis models for the study of horizontally acquired, extracellular associations
The above set of questions is not meant to be exhaustive, but rather to give the reader an idea of the broad scope of the field. In approaching these facets of symbiosis, the ideal model will present features that foster determination of the mechanisms underlying a certain phenomenon, remembering that no one model will be suitable for addressing all questions. The frontier field of microbiome studies has required, and will continue to require, the development of new models, so that we go beyond ‘looking for the keys under the lamp post’. Thus, although Drosophila melanogaster (fruit fly), Caenorhabditis elegans (nematode worm), and Danio rerio (zebrafish) have proven to be good systems for the study of symbiosis, we should not restrict ourselves to already existing, well developed biological models such as these, i.e., we must develop new, powerful models for the study of questions specific to symbiosis. In addition, to obtain a realistic picture of the dynamics of a symbiosis, both partners should be considered in any study. For example, an accurate understanding of the biology of the gut consortium cannot be gained by considering the consortium’s activities as analogous by all measures to a free-living bacterial community (e.g., in seawater or soil); unlike such a community, the gut consortium occurs within the context of the host, which is a highly dynamic entity that reacts quickly to the microbes with specific co-evolved responses. A full understanding of this rich environment will require unprecedented collaboration between micro- and macrobiologists, as well as with colleagues from across the other STEM disciplines.
4. Some available model systems
This review highlights 15 model systems that have been, or are being, established for the study of horizontally transmitted symbioses (Fig. 2; Table 1), the vast majority of which have been developed over the last dozen years. The array of symbioses presented here can be divided into three groups: (i) binary, (ii) low complexity consortia, and (iii) high complexity consortia (associated references in Table 1).
4.1. Binary associations
The value of the naturally occurring binary symbioses is that they allow unparalleled resolution of the ‘conversation’ between host and symbiont, and are often the first platform on which newly developed tools are applied. The symbiosis between the nematode, Steindernema carpocapsae and its bacterial symbiont, Xenorhabdus nematophilus, proves a unique window into a tripartite interaction, one that involves both mutualism and pathogenesis. The ‘infective juveniles’ (IJs) of the nematode carry the symbiont in a receptacle that is an outpocketing of the gut. The symbiotic IJs infect and kill a variety of arthropods by invading the host arthropod’s body cavity. The symbiont cells are released from the nematode, grow and produce toxins that kill the arthropod. The nematode then grows and reproduces within the insect carcass, producing IJs that are released to begin this cycle again.
Another set of binary associations occurs in stinkbugs, where the symbionts promote growth and, on occasion, confer host resistance to insecticides. As such, they provide the opportunities to understand the precise roles of the symbiont in host nutrition, as well as mechanisms by which host animals can modify and detoxify environmental agents. These symbioses are particularly valuable models as genetic approaches have been developed in both partners.
The squid-vibrio symbiosis has been studied for over 30 years, providing insights into host-microbe interactions, from molecular mechanisms underlying formation and persistence of the association with each generation to the ecology and evolution of these systems. The short time period, hours to days, over which the system proceeds from recruitment to a fully established symbiosis, coupled with the ability to view each stage of this process by confocal microscopy, has allowed for detailed analysis of the associated interactions between partners. In addition, the physical and biomechanical landscape of colonization spans over about 100 μm of at least five biomechanical and biochemical environments that serve to mediate the exquisite specificity of this association. Among other findings, the squid-vibrio system was the first to report a role for symbiont MAMPs in the morphogenesis of the tissues with which they associate, the function of the symbionts in driving host circadian rhythms, and the requirement for only a single genetic change in the symbiont to alter host range.
The systems mentioned here represent a sampling of those available and under development. In addition, it should be noted here that certain consortial symbioses, such as those in gnotobiotic vertebrates, can be manipulated to create a binary association. This strategy has proven to be a valuable companion to the study of naturally occurring binary associations and a window into host responses to a specific partner of a consortium. However, conclusions drawn about the natural relationship using such artificial constructs must be made with caution.
4.2. The relatively low complexity consortia of invertebrates
When considering the whole body microbiota, the low complexity consortial symbioses are restricted to invertebrates. These associations provide a window into how animals maintain populations and communities of microbes along the apical surfaces of epithelia. In particular, the extreme low-diversity systems (i.e., with fewer than a dozen core phylotypes, e.g., the fruit fly, honey bee, leech, and cuttlefish) offer the opportunity to explore the contributions of individual phylotypes to the function of a consortium; as such, they provide valuable models for the study of the more complex consortial symbioses.
The fruit fly, of course, offers a vast array of tools for the study of the mechanisms of host-symbiont interaction. That said, the diversity and constancy of the fruit fly microbiome continues to be controversial. A recent study, however, confirmed that D. melanogaster has a relatively simple composition of gut-associated microbes, which have been shown to strongly impact its development and viability under different nutritional conditions. Bacteria also affect the fly life span, gut homeostasis, interaction with pathogens, and behavior. Further, these are species-specific associations that can be fostered through microbial ‘farming’; D. melanogaster can continuously disseminate the symbiont into the environment where these bacteria are beneficial for larval development.
The honey bee gut community is dominated by five core phylotypes (or species) that are found in every healthy honey bee worker worldwide and that are not found in other habitats. These are transmitted through direct social contact among individuals and form a stable, spatially organized community within the hindgut. Based on phylogenetic analyses and comparisons with related social bee species (bumble bees and stingless bees), these lineages have coevolved with hosts and with each other, having colonized honey bee ancestors at least 80 million years ago. Because newly emerged bees lack a gut microbiota, they can be colonized with defined communities, enabling a variety of experiments. Experimental results have shown that the microbiota affects insulin signaling, appetite, weight gain, and protection against opportunistic pathogens. Disruption of an intact microbiota, with antibiotics or glyphosate, results in higher host mortality under field conditions, and increase susceptibility to pathogens.
Two models presented here are systems dominated by only two symbiotic phylotypes - the leech and the cuttlefish. In the leech these two phylotypes are co-occurring in the gut, offering another quality model for the comparison of the dynamics of symbioses in animal digestive tracts. Studies of the leech digestive-tract symbiosis revealed that similar molecular mechanisms are critical for host colonization in beneficial and pathogenic associations. Further, although the leech has the consistent diet of blood, the nutritional dynamics of the gut microbiome are complex, with nutrients being provided by ingested food, by the host producing mucin, and microbes producing nutritive metabolites that are available to other members of the microbiota. The populations of the leech gut can also be easily manipulated. For example, the presence of very low levels of antibiotics in the leech digestive tract can alter the gut composition and allow strains to colonize that are normally outcompeted by the native symbiont. In contrast with the leech, the two symbiont phylotypes of the cuttlefish, which have just recently been defined, do not co-occur; one is associated with the esophageal epithelium and one with the gills. No other portions of the body, including the gut, have stable associations with microbes. As such, the cuttlefish promises to shed light on the processes that restrict microbial phylotypes to specific regions of an animal’s body.
While comparatively low in complexity when compared with the vertebrate microbiota, invertebrates also offer consortia with dozens of phylotypes. Mentioned here is the gut symbiosis of C. elegans. This host has been grown under laboratory conditions for decades, where it has been used as a genetic model system. Under these conditions, the host animal does not harbor a gut symbiosis. However, recent studies with field-collected nematodes have demonstrated a rich microbiota that profoundly influences the biology of the host.
The other three invertebrate systems with a richer microbiota that are mentioned, i.e., the surfaces of the hydra and the starlet sea anemone, and the tubules of the accessory nidamental gland of the female bobtail squid, are defensive symbioses. They protect the host from fouling by the production of antimicrobials, often directed at environmental fungi.
The freshwater polyp hydra and the starlet anemone are excellent models for studying host-microbe interactions and how metaorganisms function in vivo. In both species, a stable multi-species bacterial community densely colonizes the epithelial surface. In hydra, the presence and structure the microbiota is critical for tissue homeostasis and health of the polyps. Remarkably, each hydra species supports long-term associations with a different set of bacteria, suggesting that the host imposes specific selection pressure onto its microbiota. The findings reveal that epithelia and components of the innate immune system play an active role in selecting the resident microbiota via a complex genetic network. Further, the work on hydra has been instrumental in changing our view of the immune system; components of the hydra innate immune system with its host-specific antimicrobial peptides appear to have evolved in early branching metazoans to control the resident beneficial microbes rather dissuading invasive pathogens. Recent findings with this model also show that microbes affect the animal’s behavior by directly interfering with neuronal receptors. These observations provide new insight into the original role of the nervous system, and suggest that it emerged to orchestrate multiple functions including host-microbiome interactions. The more recently developed starlet anemone model offers the opportunity to study the effects of the geographic range on symbiosis. This host species spans four distinct coastlines types, with large temperature differences, along the Atlantic coast of North America. A wide variety of tools have been developed for the study of both of these cnidarians.
The accessory nidamental gland (ANG) symbiosis of female Euprymna scolopes is an emerging model for understanding the role of beneficial bacteria in egg defense in aquatic environments. The ANG is colonized by multiple bacterial phylotypes that are segregated in dense epithelium-lined tubules. The symbionts are deposited into eggs where they are hypothesized to protect embryos from fouling and pathogenic environmental microbes. Many ANG bacteria can be cultured in the lab, and genomic and chemical analyses have revealed the potential for secondary metabolite production. The expansion of E. scolopes as a model host for studying both binary and consortia! associations also presents unique opportunities. For example, a recent analysis of the host genome revealed that the light organ and ANG likely evolved by different mechanisms; the light organ, with a light modulating function, shares expression of many genes with the eye, whereas the ANG expresses a significantly greater number of orphan genes. Comparative approaches to studying these two systems may also reveal shared and unique mechanisms by which environmentally transmitted symbioses are established and maintained in the same host.
4.3. The high complexity microbiomes of vertebrates
The data to date suggest that a shared, derived character of all vertebrates is the carriage of complex microbial consortia of hundreds to thousands of species. Because the interactions between vertebrates and their microbes have unique features that are not present in the invertebrates, most notably an adaptive immune system, development of vertebrate models has been essential to an understanding of symbiosis within this clade. Mice, of course, allow the study of vertebrate characters specific to mammals, such as birth mode and lactation. Experimental studies of mice have provided valuable insights into every aspect of mammalian symbiosis and, as such, have significantly broadened our understanding of the human microbiome.
Fishes comprise 40% of the vertebrate species. This tremendous diversity offers a rich palette for the study of host-microbe interactions among the vertebrates. As an experimental model with well developed host genetics, zebrafish has been established as a powerful mode system for studying vertebrate microbiome assembly, dynamics, and function. The optical transparency of zebrafish larvae allows visualization of bacterial colonization dynamics and biogeography in living animals. These studies have demonstrated how small scale microbial interactions, such as aggregation, can have profound impacts on how bacterial populations experience flow forces in the gut, determining their organ-level biogeography. The zebrafish’s fecundity and ex-utero development enables the derivation of large numbers of germ-free animals that can be used for well-powered gnotobiotic studies to identify bacterial strains and products necessary for aspects of the animals’ development. These studies have uncovered novel bacterial secreted proteins that impact aspects of host development, such as pancreatic beta cell mass and intestinal inflammatory tone.
Recently, other fish models have been under development for the study of symbiosis. The blind cavefish is being exploited as a model for the effects of the microbiome on host behavior, particular the aberrant behaviors associated with autism. The three-spined stickleback is used by biologists to understand molecular genetics of evolutionary change in wild populations. With many tools available in this host animal, it promises to be a powerful model for the study of evolution of microbial consortia in the vertebrates.
4.4. Conserved trends of symbiotic systems revealed by the study of models
What are some of the general messages that we have learned from these models thus far? One major change has been in our view of the relationship of the microbial world to animals and plants. The presence of a normal, persistent microbiota is now well recognized. However, we have a long legacy of thinking of microbes as either compromising health (i.e., since the development of the germ theory by Robert Koch in the mid-19th century, biologists have focused on microbes as pathogens), or as generally being of little importance to health (i.e., ‘commensal’). Relevant here is that most drug discovery related to host-microbe interactions has focused upon pathogenesis. Work with symbiosis models, as well as descriptions of non-model animal symbioses, have shown us that the most common relationships of animals and plants is with coevolved non-pathogenic microbes, and that pathogens are interlopers into a pre-existing conversation that the host has with its microbial partners. Notably, most pathogens are congeners of members of the host’s normal microbiota and often, as in Neisseria spp. [40], pathogens appear to be evolutionarily derived from the normal microbiota. Further, members of the normal microbiota can become opportunistic pathogens when the relationship goes out of balance. With the recognition of this complex conceptual landscape, some microbiologists are abandoning the concept of pathogen, commensal, and mutualist, as for any given species the effect on host fitness is context dependent [41]. Instead, a focus on the outcome more closely reflects the nature of host-microbe interactions, wherein a state is pathogenic to the host and beneficial for the microbe; or ‘commensal’, where one partner is not affected by the presence of the other; or mutually beneficially to both partners. This shift in the conceptual framework around host-microbe interactions will be invaluable in our strategies for the development of drugs that will promote the healthy state and discourage pathogenesis.
Support for these views has come from discoveries in both invertebrate and vertebrate model systems. For example, many features of microbes that have traditionally been called ‘virulence determinants’ (e.g., microbe-associated molecular patterns, such as lipopolysaccharide and peptidoglycan derivatives, type III and type VI secretion systems, and quorum signaling), and host responses termed animal and plant ‘defenses’ (e.g., ‘antimicrobial peptides’, the mucociliary surfaces, immunoglobulins, and other features of the immune system) have evolved principally to shape communities of the normal host microbiota. Basically, it appears that much of the molecular ‘language’ of host-microbe interactions is the same in pathogenic and beneficial associations; it is merely used differently, either with a different tuning or within a different tissue context.
This change in our general view of microbial associations with animals and plants has also led to a dramatic change in our view of the immune system [42]. At one time, the prevailing concept of the immune system was as a non-self recognition system evolved to control pathogenic infection. We now know that a principal role of the immune system is for maintaining persistent and healthy relationships with the normal microbiota. Those animals that have relatively simple microbiota, i.e., most invertebrates, rely on the innate immune system, whereas the highly complex consortia of vertebrates coevolved with the adaptive immune system. Studies of vertebrate model systems have shown us that the normal microbiota is essential for the proper development of the immune system and for its normal function.
Studies with models of symbiosis have also demonstrated a vast array of strongly conserved mechanisms of host-bacterial interactions across the animal kingdom. In light of the fact that all animal phyla arose in the microbe-rich environment of the oceans, it is not surprising that this biotic force has played a continuing role in shaping animal biology across evolutionary time. For example: the microbiota participates in the shaping of circadian rhythms, from bobtail squid [43] to mammals [44]; cancer driven by pathological dynamics of the microbiome has been described from hydra [45] to mammals [46]; and, the same cell-surface molecules of bacteria drive aspects of development in invertebrates [47] and vertebrates [48].
5. Conclusions and Horizons
As in other areas of biology where models have provided the critical, Nobel-prize-winning insights into a field [49], models have proven invaluable in the study of symbiotic systems. The above-mentioned contributions of experimental model to the field of symbiosis represent a selected few. Thus far, biologists have only begun to take advantage of the experiments that evolution has done with host-microbe interactions. The discipline is in its infancy, but these early studies of the holobiont have already shown that the recognition of symbiosis as central to biology will change the foundations of the field.
6. Acknowledgments
We thank E Ruby for helpful discussions and comments on the manuscript, and my colleagues in the field for text on their model systems that was included in this manuscript. This work was funded by NIH grants R37AI050661 to MM-N and E Ruby, and OD011024 and GM135254 to E Ruby and MM-N.
Footnotes
[Abbreviations: none]
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McFall-Ngai M et al. (2013) Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U. S. A 110, 3229–3236, doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bordenstein SR and Theis KR (2015) Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol. 13, e1002226, doi: 10.1371/journal.pbio.1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morris JJ (2018) What is the hologenome concept of evolution? F1000Res. doi: 10.12688/f1000research.14385.1. eCollection 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lozupone CA, et al. (2012) Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230, doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pakkenberg B et al. (2003) Aging and the human neocortex. Exp. Gerontol 38, 95–99. [DOI] [PubMed] [Google Scholar]
- 6.Berlanga M (2015) Functional symbiosis and communication in microbial ecosystems. The case of the wood-eating termites and cockroaches. Int Microbiol 18, 159–169. doi: 10.2436/20.1501.01.246. [DOI] [PubMed] [Google Scholar]
- 7.McFall-Ngai M (2007) Adaptive immunity: care for the community. Nature 445, 153, doi: 10.1038/445153a. [DOI] [PubMed] [Google Scholar]
- 8.Bosch TCG et al. (2019) Evolutionary “experiments” in symbiosis: the study of model animals provides insights into the mechanisms underlying the diversity of host-microbe interactions. Bioessays, e1800256, doi: 10.1002/bies.201800256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodrich-Blair H (2007) They’ve got a ticket to ride: Xenorhabdus nematophila-Steinernema carpocapsae symbiosis. Curr. Opin. Microbiol 10, 225–230, doi: 10.1016/j.mib.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 10.Richards GR and Goodrich-Blair H (2009) Masters of conquest and pillage: Xenorhabdus nematophila global regulators control transitions from virulence to nutrient acquisition. Cellul. Microbiol 11, 1025–1033, doi: 10.1111/j.1462-5822.2009.01322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hosokawa T et al. (2016) Obligate bacterial mutualists evolving from environmental bacteria in natural insect populations. Nat. Microbiol 1, 15011, doi: 10.1038/nmicrobiol.2015.11. [DOI] [PubMed] [Google Scholar]
- 12.Takeshita K and Kikuchi Y (2017) Riptortus pedestris and Burkholderia symbiont: an ideal model system for insect-microbe symbiotic associations. Res. Microbiol 168, 175–187, doi: 10.1016/J.RESMIC.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 13.Ohbayashi T et al. (2019) Comparative cytology, physiology and transcriptomics of Burkholderia insecticola in symbiosis with the bean bug Riptortus pedestris and in culture. ISME J. 13, 1469–1483, doi: 10.1038/s41396-019-0361-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mandel MJ and Dunn AK (2016) Impact and influence of the natural Vibrio-squid symbiosis in understanding bacterial-animal interactions. Front. Microbiol 7, 1982, doi: 10.3389/fmicb.2016.01982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McFall-Ngai MJ (2014) The importance of microbes in animal development: lessons from the squid-vibrio symbiosis. Annu. Rev. Microbiol 68, 177–194, doi: 10.1146/annurev-micro-091313-103654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deines P et al. (2017) Competing forces maintain the Hydra metaorganism. Immunol. Rev 279, 123–136, doi: 10.1111/imr.12564. [DOI] [PubMed] [Google Scholar]
- 17.Schroder K and Bosch TCG (2016) The origin of mcosal immunity: lessons from the holobiont Hydra. mBio 7, e1184–1116, doi: 10.1128/mBio.01184-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Domin H et al. (2018) Predicted bacterial interactions affect in vivo microbial colonization dynamics in Nematostella. Front. Microbiol 9, 728, doi: 10.3389/fmicb.2018.00728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraune S et al. (2016) Using Nematostella vectensis to study the interactions between genome, epigenome, and bacteria in a changing environment. Front. Mar. Sci 3, 148, doi: 10.3389/fmars.2016.00148. [DOI] [Google Scholar]
- 20.Broderick NA and Lemaitre B (2012) Gut-associated microbes of Drosophila melanogaster. Gut Microbes 3, 307–321, doi: 10.4161/gmic.19896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pais IS et al. (2018) Drosophila melanogaster establishes a species-specific mutualistic interaction with stable gut-colonizing bacteria. PLoS Biology 16, e2005710, doi: 10.1371/journal.pbio.2005710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonilla-Rosso G and Engel P (2018) Functional roles and metabolic niches in the honey bee gut microbiota. Curr. Opin. Microbiol 43, 69–76, doi: 10.1016/j.mib.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Romero S et al. (2019) The honey bee gut microbiota: strategies for study and characterization. Insect Mol. Biol 28, 455–472, doi: 10.1111/imb.12567. [DOI] [PubMed] [Google Scholar]
- 24.Zheng H et al. (2018) Honey bees as models for gut microbiota research. Lab Animal 47, 317–325, doi:https:// 10.1038/s41684-018-0173-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang F et al. (2017) Caenorhabditis elegans as a model for microbiome research. Front. Microbiol 8, 485, doi: 10.3389/fmicb.2017.00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zimmermann J et al. (2019) The functional repertoire encoded within the native microbiome of the model nematode Caenorhabditis elegans. ISME J. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marden JN et al. (2016) Host matters: medicinal leech digestive-tract symbionts and their pathogenic potential. Front. Microbiol 7, 1569, doi: 10.3389/fmicb.2016.01569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson MC and Graf J (2012) Bacterial symbioses of the medicinal leech Hirudo verbana. Gut Microbes 3, 322, doi: 10.4161/gmic.20227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collins AJ et al. (2012) Diversity and partitioning of bacterial populations within the accessory nidamental gland of the squid Euprymna scolopes. Appl. Environ. Microbiol 78, 4200–4208, doi: 10.1128/AEM.07437-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kerwin AH and Nyholm SV (2018) Reproductive system symbiotic bacteria are conserved between two distinct populations of Euprymna scolopes from Oahu, Hawaii. mSphere 3, doi: 10.1128/msphere.00531-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lutz HL et al. (2019) A simple microbiome in the European common cuttlefish, Sepia officinalis. mSystems 4, e00177–00119, doi:0.1128/mSystems.00177-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burns AR and Guillemin K (2017) The scales of the zebrafish: host-microbiota interactions from proteins to populations. Curr. Opin. Microbiol 38, 137–141, doi: 10.1016/j.mib.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melancon E et al. (2017) Best practices for germ-free derivation and gnotobiotic zebrafish husbandry. Meth. Cell. Biol 138, 61–100, doi: 10.1016/bs.mcb.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith CCR et al. (2015) Dietary input of microbes and host genetic variation shape among-population differences in stickleback gut microbiota. ISME J. 9, 2515–2526, doi: 10.1038/ismej.2015.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeffery WR (2009) Evolution and development in the cavefish Astyanax. Curr. Topics Devel. Biol 86, 191–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keene AC et al. (2016) Biology and evolution of the Mexican cavefish. 412, doi:doi.org/ 10.1016/C2014-0-01426-8. [DOI] [Google Scholar]
- 37.Hugenholtz F and de Vos WM (2018) Mouse models for human intestinal microbiota research: a critical evaluation. Cellul. Mol. Life Sci 75, 149–160, doi: 10.1007/s00018-017-2693-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen TLA et al. (2015) How informative is the mouse for human gut microbiota research? Dis. Models Mech 8, 1–16, doi: 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bright M and Bulgheresi S (2010) A complex journey: transmission of microbial symbionts. Nat. Rev. Microbiol 8, 218–230, doi: 10.1038/nrmicro2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marri PR et al. (2010) Genome sequencing reveals widespread virulence gene exchange among human Neisseria species. PLoS One 5, e11835, doi: 10.1371/journal.pone.0011835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Casadevall A and Pirofski LA (1999) Host-pathogen interactions: redefining the basic concepts of virulence and pathogenicity. Infect. Immun 67, 3703–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Belkaid Y and Hand TW (2014) Role of the microbiota in immunity and inflammation. Cell 157, 121–141, doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwartzman JA and Ruby EG (2016) A conserved chemical dialog of mutualism: lessons from squid and vibrio. Microbes Infect. 18, 1–10, doi: 10.1016/j.micinf.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nobs SP et al. (2019) Microbiome diurnal rhythmicity and its impact on host physiology and disease risk. EMBO Rep. 20, doi: 10.15252/embr.201847129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Domazet-Loso T et al. (2014) Naturally occurring tumours in the basal metazoan Hydra. Nat. Commun 5, 4222, doi: 10.1038/ncomms5222. [DOI] [PubMed] [Google Scholar]
- 46.Elinav E et al. (2019) The cancer microbiome. Nat. Rev. Cancer 19, 371–376, doi: 10.1038/s41568-019-0155-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koropatnick TA et al. (2004) Microbial factor-mediated development in a host-bacterial mutualism. Science 306, 1186–1188, doi: 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- 48.Bouskra D et al. (2008) Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 456, 507–510, doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- 49.Ruby EG (2008) Symbiotic conversations are revealed under genetic interrogation. Nat. Rev. Microbiol 6, 752–762, doi: 10.1038/nrmicro1958. [DOI] [PMC free article] [PubMed] [Google Scholar]