Abstract
Mammalian cell transfection is a powerful technique commonly used in molecular biology to express exogenous DNA or RNA in cells and study gene and protein function. Although several transfection strategies have been developed, there is a wide variation with regards to transfection efficiency, cell toxicity and reproducibility. Thus, a sensitive and robust method that can optimize transfection efficiency based not only on expression of the target protein of interest but also on the uptake of the nucleic acids, can be an important tool in molecular biology. Herein, we present a simple, rapid and robust flow cytometric method that can be used as a tool to optimize transfection efficiency while overcoming limitations of prior established methods that quantify transfection efficiency.
Keywords: Transfection, Flow cytometry, Nucleic acids, Protein expression, DNA labeling
Background
Transfection is one of the most commonly used techniques in molecular biology (Stoll and Calos, 2002; Kim and Eberwine, 2010). Transfection is the process of introducing nucleic acid (DNA that carries a gene of interest or mRNA) into target cells that then eventually express the desired nucleic acid or protein. There are several biological, chemical, and physical methods for introducing nucleic acids into cells (Stoll and Calos, 2002; Kim and Eberwine, 2010; Zhou et al., 2016 ). However, all these methods are variable and don’t assess the cell transfection efficiency, cell toxicity and the level of gene expression within the same experiment. To truly optimize cellular transfection, a sensitive and robust detection assay is required to quantify and optimize the efficiency of different transfection methods to deliver the target gene into the cytosol and facilitate protein expression, while reducing cell toxicity.
Herein, we demonstrate the development of a flow-cytometric assay to determine transfection efficiency by labeling a reporter plasmid with Label IT® TrackerTM ( Homann et al., 2017 ) (Figure 1). This method does not depend on co-transfection of two different plasmids and simultaneously quantifies cell death, uptake of the labeled plasmid during transient transfection, and expression of the target protein. We demonstrate that this method can be used as a tool to i) optimize transfection efficiency in 2 independent standard cell lines, ii) quantify cellular toxicity of different transfection methods, iii) determine uptake of DNA into difficult to transfect cells via electroporation without the need to use co-transfection of GFP plasmid that can further reduce the efficiency of transfection. This flow cytometric method can be directly applied to optimize several transfection methods including gene therapy strategies (e.g., CRISPR/Cas system).
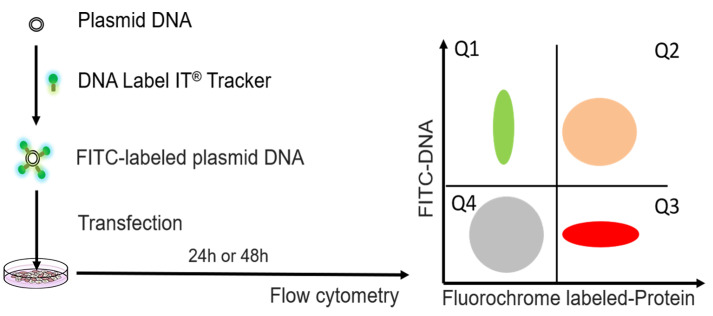
Figure 1. Experimental design for determination of transfection efficiency by flow cytometric method.
The plasmid DNA was labeled with FITC by DNA label IT@ tracker. After transfection, cells were detected by flow cytometry. The FITC fluorochrome is used to detect intracellular levels of the transfected plasmid that has been labeled with FITC (Label IT tracker, green). The second fluorochrome is used to quantify expression of the target protein (by directly measuring fluorescence of the expressed protein if the target protein is fluorescent or by using a fluorescent-labeled antibody against the target protein, red). Either Q1+Q2 (DNA signal) or Q2+Q3 (protein signal) should be used as readouts of transfection efficiency.
Materials and Reagents
4 mm cuvettes (Gene Pulser cuvettes, Biorad, Hercules, CA)
-
Cell lines of interest
293T cell line (ATCC, catalog number: A-498)
Jurkat E6-1 (obtained through the NIH AIDS Reagent Program, Division of AIDS, NIH)
Jurkat Clone E6-1 (from Dr. Arthur Weiss)
pNL4-3, received from AIDS reagent program (#114, NIH)
pUltraHot encoding mCherry (8,314 bp) was a kind gift of Jeff F. Miller, California Nano Systems Institute
One ShotTM Stbl3TM Chemically Competent E. coli (Thermo Scientific, catalog number: C737303)
Stellar bacteria (Clontech, catalog number: 636763)
Dulbecco's modified Eagle medium (DMEM) (Thermo Scientific, catalog number: 11965-092)
Roswell Park Memorial Institute (RPMI) 1640 medium (Thermo Scientific, catalog number: 61870127)
Fetal bovine serum (FBS) (Omega Scientific, catalog number: FB-02)
OptiMEM (Thermo Scientific, catalog number: 31985-070)
Penicillin and streptomycin (Thermo Scientific, catalog number: 15-140-122)
L-glutamine (Invitrogen, catalog number: 125030-081)
Phosphate-buffered saline (PBS) (Omega Scientific, catalog number: FB-02)
4% paraformaldehyde (PFA) (Thermo Scientific, catalog number: J19943-K2)
Tween 20 (Sigma, catalog number: P9416)
PureLink HiPure Midiprep Kit (Thermo Scientific, catalog number: K210004)
DNA Label IT® TrackerTM [Fluorescein isothiocyanate (FITC)] (Mirus, catalog number: MIR7025)
TransIT-X2 (Mirus, catalog number: MIR6004)
Jet Prime (Polyplus, catalog number: 114-07)
Lipofectamine 2000 (Thermo Scientific, catalog number: 11668019)
Fugene HD (Promega, catalog number: E2311)
Quantum Molecules of Equivalent Soluble Fluorochrome (MESF) (Bangs Laboratories, catalog number: 647-B)
Ghost Violet 450 Live/dead dye (Tonbo Biosciences, catalog number: 13-0863-T100)
Anti-human immunodeficiency virus (HIV)-1 p24 monoclonal antibody (71-31) (NIH AIDS reagent program #530)
Human IgG1 (Biolegend, catalog number: 409302)
Mix-N-Stain CF647 dye (Biotium, catalog number: 92238)
10% FBS complete culturing medium for 293T (see Recipes)
10% FBS complete culturing medium for Jurkat cells (see Recipes)
0.02% Tween/PBS (see Recipes)
Equipment
37 °C, 5% CO2 humidified incubator (Thermoforma, model: 3110)
Centrifuge (Thermo scientific, model: ST8)
BioRad Gene Pulser Xcell electroporation system
LSRII Fortessa flow cytometer (BD Biosciences, San Jose, CA, USA)
Spectrophotometer (Thermo Fisher, NanoDropTM 2000)
Software
Acquisition Software: BDFACSDiVaTM Software (BD Biosciences)
Analysis Software: FlowJo version 10.6
Procedure
-
Plasmid labeling
Extract pNL4-3 and pUltraHot encoding mCherry for transfection from Stbl3TM (Thermo Fisher, Waltham, MA) or Stellar (Clontech, Mountain View, CA) bacteria using the PureLink HiPure Midiprep Kit (Thermo Fisher, Waltham, MA). Determine concentration and purity (260/280 ratio) of nucleic acid using spectrophotometry.
Label Plasmid DNA with DNA Label IT® Tracker (Mirus, Madison, WI) the day before transfection using 0.5 μl FITC/1 μg DNA according to the manufacturer’s protocol. Remove unreacted Label IT® Tracker reagent using ethanol precipitation. According to the manufacturer, 1 μl of labeling dye per 1 μg plasmid DNA will yield labeling efficiencies of approximately one Label molecule every 40 base pairs (on average) of double-stranded DNA. Determine the purity and concentration of labeled DNA using spectrophotometry.
-
Antibody conjugation
Conjugate Anti-HIV-1 p24 monoclonal antibody (71-31) with Mix-n-Stain CF647 dye, according to the manufacturer’s protocol.
Conjugate Human IgG1 control with Mix-n-Stain CF647 dye according to the manufacturer’s protocol.
-
Transfection of 293T cells with FITC labeled DNA by different transfection reagents
Plate 3 x 105 293T cells of < 20 passages into 12-well plates the day before transfection. At this density, cells are usually attached to the bottom of the well at a confluency of 70%-80% after 24 h.
Twenty-four hours after seeding, transfect cells with FITC-labeled and unlabeled pNL4-3 or pUltraHot encoding mCherry. The transfections can be performed in 12-well plates using 1 μg plasmid DNA and 3 μl TransIT ×2 in serum-free media, according to the manufacturer’s protocol.
-
Culture cells in complete media after adding transfection complexes for 6 h. Then change medium to 10% FBS complete culturing medium.
Note: For experiments comparing different transfection reagents, the cells can be transfected with 1 μg DNA per well with varying amounts of transfection reagents to facilitate the recommended ratio of transfection reagent to DNA for each reagent: 3 μl TransIT ×2, 4 μl Lipofectamine 2000, 2 μl Jet Prime and 3 μl Fugene HD.
-
Harvest cells at 24 h post-transfection and wash one time in PBS.
Note: For time course experiments, the cells can be transfected at the same time and harvested at 0, 6, 12, 24, 36, 48 and 72 h.
Fix cells in 4% PFA for 15 min at room temperature.
Permeabilize cells in 0.2% Tween/PBS for 15 min.
Stain cells for HIV-1 p24 protein with 1 μg anti-p24 antibody conjugated to CF647 in 100 μl staining medium containing PBS/2% BSA for 30 min at 4 °C.
Wash 293T cells once with PBS.
Fix cells in 4% PFA for 15 min.
-
Electroporation of Jurkat cells with FITC labeled pUltraHot
Wash Jurkat E6-1 cells 2 times in OptiMEM before resuspending cells in OptiMEM at 1 x 106 cells/100 μl.
Transfer 1 x 106 cells into 4 mm cuvettes and add 4 μg of labeled or unlabeled pUltraHot plasmid to the suspension.
Electroporate Jurkat E6-1 cells with a BioRad Gene Pulser Xcell electroporation system using exponential protocol with 250 voltage, 350 μF capacitance, 1,000 Resistance at room temperature.
Transfer cells immediately into pre-warmed media and incubate cells for 24 h before staining for viability and Flow cytometric analysis.
Collect Jurkat cells transfected with pUltraHot. Stain Jurkat cells with 1 μl Ghost Violet 450 Live/dead dye in 1 ml PBS incubate 30 min at room temperature.
Wash Jurkat cells with 1% FBS in PBS.
Fix Jurkat cells in 4% PFA for 15 min.
-
Flow cytometry analysis
Acquire samples on an LSRII Fortessa flow cytometer (BD Biosciences, San Jose, CA, USA) with BDFACSDiVaTM Software (BD Biosciences). The flow cytometer is equipped with 405, 488, 561 and 635 nm lasers, and emission filters for Pacific blue (LP: −, BP:450/50), Alexa fluor-488 (LP: 505, BP: 530/30), PE (LP: −, BP: 582/15), mCherry (LP: 600 BP: 610/20), PerCP-Cy5.5 (LP: 685, BP: 695/40), APC (LP: −, BP: 670/14). FITC fluorescence intensity (y-axis) is plotted on a log scale against the fluorescence intensity (x-axis) of fluorochrome that is used to quantify protein expression (e.g., mCherry) (Figure 2).
The mean fluorescence intensity (MFIs) of FITC and Alexa Fluor® 647 can be further standardized using Quantum MESF (Molecules of Equivalent Soluble Fluorochrome) kits for Alexa Fluor® 488 and Alexa Fluor® 647 as previously described ( Kapoor et al., 2009 ).
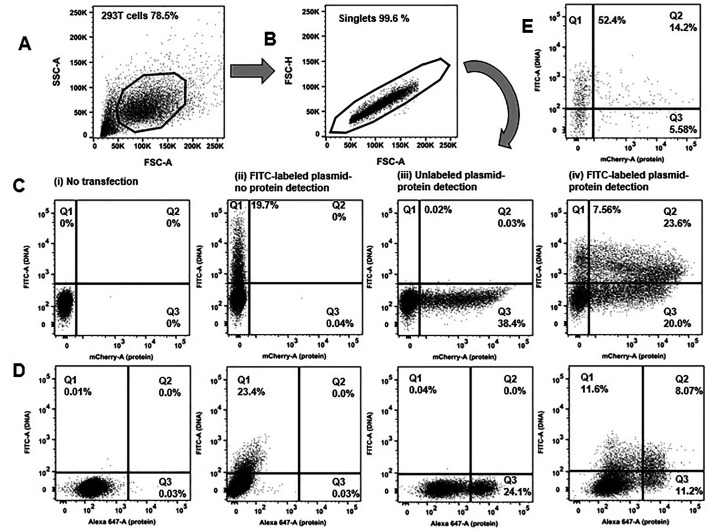
Figure 2. Flow cytometric determination of transfection efficiency based on two independent readouts (DNA plasmid uptake and protein expression).
Representative transfections are shown. 293T cells underwent chemical transfection using the TransITX2 transfection reagent as described in Procedure. The same amount (1 μg) of DNA was used for two independent plasmids: a small (B. pUltraHot expressing mCherry, 8.3 kb) and a large (C. pNL4-3 expressing p24, 14.0 kb) DNA plasmid. Gating strategy is shown: A) forward and side scatters B) discrimination of doublets C, D) two independent readouts of transfection efficiency. FITC fluorescence corresponds to the uptake of FITC-labeled plasmid DNA (y-axis). A fluorochrome that has no spectral overlap with FITC is used to quantify protein expression. Either a fluorescent protein can be used (e.g., mCherry; shown in C) or a protein labeled with a fluorescent-labeled antibody (e.g., intracellular expression of HIV-1 p24 protein was detected by a CF647-labeled anti-p24 antibody; shown in D). Co-expression of DNA taken up by cells and target protein were analyzed 24 h after transfection. The numbers in the quadrants indicate the percentages of viable cells that took up the FITC labeled DNA plasmid versus the expressed protein that was detected. The following dot plots are shown for each chemical transfection in 293T cells: i) untransfected cells (negative control), ii) cells transfected with FITC-labeled DNA plasmid harvested before protein expression occurred (3 h post transfection), iii) cells transfected with unlabeled plasmid harvested 24 h after transfection (when protein expression can be quantified) iv) cells transfected with FITC-labeled DNA plasmid and harvested 24 h after transfection (when protein expression can be quantified). In this plot Q3 quadrant demonstrates many cells that express protein but do not show any fluorescence associated with uptake of the plasmid DNA. Either Q1+Q2 (DNA signal) or Q2+Q3 (protein signal) should be used as readouts of transfection efficiency. E. Transfection efficiency was quantified in human lymphocytes (Jurkat E6 cells) harvested 24 h after electroporation with FITC-labeled DNA mCherry plasmid without the need to use co-transfection of 2 different plasmids and GFP reporter.
Data analysis
Results of the peak fluorescence measurements are blank-corrected with signal from non-transfected cells and normalized to 1 x 104 cells with the corresponding cell density value from the same well. Values are presented as max fluorescence (median fluorescence intensity; MFI) or relative fluorescence units (rfu) per 10 cells. Cell viability at 24 and 48 h is expressed as % of viable cells (based on the death dye) of total number of cells and can be compared to the viability from the non-transfected control cells. This method takes into consideration cell toxicity as a direct result of the transfection and the nucleic acid per se and uses two independent readouts of transfection efficiency: a) the amount of plasmid nucleic acid that cells have taken up during transfection b) the amount of the encoded expressed protein. In summary, we provide a relatively simple, rapid and robust flow cytometric method that can be used as a tool to optimize transfection efficiency.
Recipes
-
10% FBS complete culturing medium for 293T
DMEM supplemented with:
10% heat-inactivated FBS
100 unit/ml Penicillin and streptomycin
2 mM L-glutamine
-
10% FBS complete culturing medium for Jurkat cells
RPMI1640 supplemented with:
10% heat-inactivated FBS
100 unit/ml Penicillin and Streptomycin
2 mM L-glutamine
-
0.02% Tween/PBS
4 μl of Tween 20 in 20 ml PBS
Acknowledgments
We would like to acknowledge the Center for AIDS Research Virology Core Lab and the Gene and Cellular Therapy Core that are supported by the National Institutes of Health [UCLA Center for AIDS Research (CFAR) NIH/NIAID 5P30 AI028697] and by the UCLA AIDS Institute and the UCLA Council of Bioscience Resources. This research was supported by NIH grants NIH/NCATS grant # UL1TR000124, NIH K08AI08272 and by grant from the UCLA AIDS Institute/UCLA Center for AIDS Research (CFAR) (NIH/NIAID AI028697). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or any of the funders. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing Interests
The authors have declared that no competing interests exist.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Stoll S. M. and Calos M. P.(2002). Extrachromosomal plasmid vectors for gene therapy. Curr Opin Mol Ther 4(4): 299-305. [PubMed] [Google Scholar]
- 2. Kim T. K. and Eberwine J. H.(2010). Mammalian cell transfection: the present and the future. Anal Bioanal Chem 397(8): 3173-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou Z. L., Sun X. X., Ma J., Man C. H., Wong A. S., Leung A. Y. and Ngan A. H.(2016). Mechanical oscillations enhance gene delivery into suspended cells. Sci Rep 6: 22824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Homann S., Hofmann C., Gorin A. M., Nguyen H. C. X., Huynh D., Hamid P., Maithel N., Yacoubian V., Mu W., Kossyvakis A., Sen Roy S., Yang O. O. and Kelesidis T.(2017). A novel rapid and reproducible flow cytometric method for optimization of transfection efficiency in cells. PLoS One 12(9): e0182941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kapoor V., Hakim F. T., Rehman N., Gress R. E. and Telford W. G.(2009). Quantum dots thermal stability improves simultaneous phenotype-specific telomere length measurement by FISH-flow cytometry. J Immunol Methods 344(1): 6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]