Abstract
Magnetotactic bacteria (MTB) are prokaryotes that possess genes for the synthesis of membrane-bounded crystals of magnetite or greigite, called magnetosomes. Despite over half a century of studying MTB, only about 60 genomes have been sequenced. Most belong to Proteobacteria, with a minority affiliated with the Nitrospirae, Omnitrophica, Planctomycetes, and Latescibacteria. Due to the scanty information available regarding MTB phylogenetic diversity, little is known about their ecology, evolution and about the magnetosome biomineralization process. This study presents a large-scale search of magnetosome biomineralization genes and reveals 38 new MTB genomes. Several of these genomes were detected in the phyla Elusimicrobia, Candidatus Hydrogenedentes, and Nitrospinae, where magnetotactic representatives have not previously been reported. Analysis of the obtained putative magnetosome biomineralization genes revealed a monophyletic origin capable of putative greigite magnetosome synthesis. The ecological distributions of the reconstructed MTB genomes were also analyzed and several patterns were identified. These data suggest that open databases are an excellent source for obtaining new information of interest.
Subject terms: Bacterial genetics, Microbial ecology, Evolutionary genetics, Data mining
Introduction
The amount of data obtained from genome and metagenome sequencing has been sharply increasing for the last several years1. These data are kept in open databases, such as the widely used NCBI2 and IMG3 databases. In the case of IMG, the number of entries for metagenomic data greatly exceeds that for genomic ones3. In most cases, scientists use only a part of the sequencing information uploaded to the databases, leaving large quantities of information essentially unanalyzed. This gives the possibility that the obtained data may contribute to other studies and shorten the time and efforts of other scientists. In the present study, data stored in open genomic and metagenomic databases were used to search for magnetosome biomineralization genes related to magnetotactic bacteria (MTB).
The MTB are a group of organisms characterized by the ability to synthesize magnetosomes, which are crystals of magnetite (Fe3O4) or greigite (Fe3S4) enveloped by a lipid membrane4. These crystals can be applied in medicine as contrast agents for MRI5 and for treating tumors using magnetic hyperthermia6, and they are also of great interest in geology7–9 and astrobiology10. The synthesis of magnetosomes is controlled by the magnetosome gene cluster (MGC), previously called the magnetosome island or MAI. The MGC comprises genes that control magnetosome biosynthesis and that determine magnetosome morphology and chemical composition. The MGCs are unique and are associated only with MTB. The genes essential to the biomineralization process are called mam (magnetosome membrane) genes. Nine of them (mamA, -В, -M, -K, -P, -Q, -E, -O, and -I), are present in all MGCs11,12. In addition to the mam genes, genes specific to certain groups may also occur; for instance, mad genes are found in MTB from the Deltaproteobacteria and Nitrospirae, while man genes are present only in the Nitrospirae13.
At present, only about 60 MTB genomes are known, and most are affiliated with the phyla Proteobacteria, Nitrospirae, and Ca. Omnitrophica. Recently, MTB genomes associated with Latescibacteria14 and Planctomycetes12 have been found in open databases, implying that these databases could contain substantial amounts of new information about MTB.
To date, due to the lack of sufficient amounts of genomic data, little is known about the origin and evolution of MGCs15. Thus, additional investigations are needed to determine the mono- or polyphyletic origin of the MGCs, their evolutionary history, and whether the original MGCs were responsible for magnetite or greigite biomineralization.
This article describes the first large-scale search of magnetosome biomineralization genes in open genomic and metagenomic databases. Bioinformatics analysis of the search results allowed new MTB genomes to be obtained. Taxonomic assignments for the studied genomes provided the first evidence of their affiliation to new for MTB taxonomic ranks, including three new phyla. These results significantly expanded the knowledge of MTB diversity. The analysis of the ecological distribution of the reconstructed MTB genomes helped to identify several new patterns. Further comparative analysis of MGCs and marker genes of studied genomes allowed new data to be obtained concerning the origin and evolution of magnetosome biomineralization genes.
Results
The search for magnetosome biomineralization genes in open databases
The search for MTB genomes in open databases was guided by detecting MGCs unique to magnetotactic bacteria. Unfortunately, MGC sequences are not annotated as magnetosomal in open databases. This necessitated the use of previously known sequences of MGCs as search targets. The search was further complicated by the low identity values between the sequences of the same MGC gene in different MTB taxonomic groups. To cover the maximum number of new MTB representatives, MGC protein sequences were drawn from all known taxonomic groups where MTB were found previously. For this purpose, a database was created of known MGC protein sequences12–43 (Supplementary Table S1). The database included 67 MGCs from Proteobacteria, Nitrospirae, Ca. Omnitrophica, Latescibacteria, and Planctomycetes. The sequences of nine Mam proteins present in all MGCs were used to conduct BLASTp with genomic data from the NCBI and IMG databases. This resulted in the detection of four new genomes containing magnetosome biomineralization genes (Table 1, Supplementary Table S2).
Table 1.
Characteristics of genomes with MGCs obtained from the NCBI and IMG database genomic data.
| Organism | Phylum/Class | Accession in NCBI/IMG | Size (bp) | Scaffolds (no.) | GC (%) | N50 (bp) | CheckM completeness (%) | CheckM contamination (%) |
|---|---|---|---|---|---|---|---|---|
| Magnetovibrio sp. ARS851,83 | Alphaproteobacteria | GCA_002686765.1 | 2019305 | 197 | 59.64 | 10605 | 62.87 | 1.00 |
| Elusimicrobia bacterium NORP12264,84 | Elusimicrobia | GCA_002401485.1 | 2913226 | 191 | 54.93 | 19622 | 74.06 | 1.82 |
| Unclassified Nitrospina Bin 2545,114 | Nitrospinae | 2651870060 | 4158979 | 431 | 37.69 | 11956 | 92.31 | 4.27 |
| Planctomycetes bacterium SCGC_JGI090-P21115 | Planctomycetes | 2264265205 | 1230646 | 242 | 49.20 | 12722 | 38.87 | 2.19 |
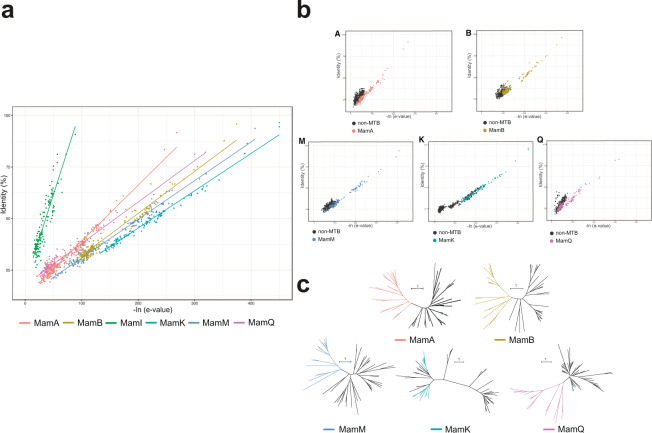
The use of all nine Mam proteins in metagenomic databases is complicated by the fact that much more data is kept in metagenomic than in genomic ones. To hasten the search process, one Mam protein out of nine common ones that met the required parameters was chosen for further BLAST analysis. The first chosen parameter was the identity between sequences from different taxonomic groups in each protein. The low values of these identities allowed exclusion of MamE, MamO, and MamP proteins from the analysis. The remaining MamA, -B, -M, -K, -I, and -Q proteins were assessed for sequences with the highest -ln of e-values, in addition to high identities (Fig. 1a). MamI was the least consistent with these requirements and was not used in further analyses. By contrast, MamK was the most consistent.
Fig. 1.
The choice of Mam protein for further searching for MGCs in open databases. (a) Correlations between –ln of e-values (x axis) and identities (y axis) among MamA, -B, -M, -K, -I, and -Q proteins sequences. (b) Correlations between identities and –ln of e-values among Mam protein sequences with their homologs. (c) Phylogenetic trees based on investigated sequences. Trees were reconstructed by the maximum-likelihood method with LG + F + I + G4 substitution model. Bootstrap values were calculated based on 1000 resamplings. Bar represents one substitution per 100 amino acid positions.
Each Mam protein has its homologs in non-MTB that are not involved in the magnetosomes biomineralization process. These homologs should be avoided when searching for MGCs. For this, Mam protein was chosen whose identities and -ln of e-values were significantly varied from these parameters in homologs (Fig. 1b). MamK showed the best result in this case, and its minimum identity and –ln e-value between sequences were 30 and 135, respectively. However, part of homologs had identities and –ln of e-values similar to the values found between Mam protein sequences. These homologs were confirmed not to be Mam sequences by verifying their phylogenetic separation (Fig. 1c). The sequences of each Mam protein formed monophyletic clades, while MamK formed two clades. Despite this, no homologs were observed inside the MamK clades. Based on all the investigated parameter results, the MamK protein sequences were chosen for the MGC gene search in the open databases.
The MamK protein sequences were used for BLAST for 10587 metagenomes from water, terrestrial, engineered, and host-associated ecosystems. The analysis revealed 2798 sequences potentially affiliated with the MamK protein (Supplementary Fig. S1a). Their scaffolds were checked for the presence of other Mam protein sequences. After that, 227 MamK sequences referring to 135 metagenomes were obtained (Supplementary Tables S3 and S4). These and previously known MamK sequences were used to construct a phylogenetic tree (Supplementary Fig. S1b), which revealed that the identified MamK sequences were not closely related to previously known sequences. This assumes that they could refer to taxonomic groups in which MTB were not found before.
Metagenome binning, phylogenomic inferences, and MGC reconstruction
The phylogenetic position of genomes to which the MamK sequences belonged was assessed by conducting metagenome binning, and it yielded 14688 metagenome-assembled genomes (MAGs) (Supplementary Table S3). Two metagenomes were also determined to be single-cell amplified genomes (SAGs), so no binning procedures were required for them. Of all the MAGs obtained in this study, only 140 contained previously detected MamK sequences. For those of the 140 whose completeness was >45% decontamination was conducted. This left 32 MAGs with completeness >45% and contamination <10% that contained MGCs (Table 2, Supplementary Table S6). The phylogenomic affiliations of the obtained MAGs, SAGs, and genomes were then determined, the MGСs genes were reconstructed, and the ecological distributions were studied.
Table 2.
Characteristics of reconstructed MAGs with MGCs obtained from the IMG metagenomic data.
| Organism | Phylum/Class | Metagenome accession in NCBI/IMG | Size (bp) | Scaffolds (no.) | GC (%) | N50 (bp) | CheckM completeness (%) | CheckM contamination (%) |
|---|---|---|---|---|---|---|---|---|
| Ca. Hydrogenedentes bacterium MAG_17963_hgd_11185 | Ca. Hydrogenedentes | 3300017963 | 3018788 | 288 | 60.18 | 11662 | 71.11 | 1.46 |
| Ca. Hydrogenedentes bacterium MAG_17971_hgd_13044 | Ca. Hydrogenedentes | 3300017971 | 2683901 | 240 | 60.43 | 12541 | 60.01 | 1.16 |
| Deltaproteobacteria bacterium MAG_00134_naph_00686,119 | Deltaproteobacteria | 3300000134 | 1498667 | 692 | 49.54 | 2676 | 60.69 | 3.87 |
| Deltaproteobacteria bacterium MAG_00241_naph_01087,119 | Deltaproteobacteria | 3300000241 | 1547003 | 324 | 49.45 | 6761 | 55.59 | 2.41 |
| Deltaproteobacteria bacterium MAG_00792_naph_01688,119 | Deltaproteobacteria | 3300000792 | 3032840 | 409 | 49.74 | 11269 | 89.28 | 5.86 |
| Deltaproteobacteria bacterium MAG_09788_naph_3789 | Deltaproteobacteria | 3300009788 | 899797 | 137 | 47.24 | 7579 | 49.08 | 0.97 |
| Deltaproteobacteria bacterium MAG_15370_dsfb_8190,120 | Deltaproteobacteria | 3300015370 | 3868622 | 334 | 48.42 | 14397 | 89.68 | 5.59 |
| Deltaproteobacteria bacterium MAG_17929_sntb_2691 | Deltaproteobacteria | 3300017929 | 2777907 | 276 | 53.10 | 17193 | 62.13 | 5.10 |
| Deltaproteobacteria bacterium MAG_17996_sntb_2092 | Deltaproteobacteria | 3300017996 | 1691080 | 454 | 53.11 | 4033 | 50.53 | 2.33 |
| Deltaproteobacteria bacterium MAG_22204_dsfv_00193 | Deltaproteobacteria | 3300022204 | 2675335 | 75 | 52.74 | 60141 | 89.52 | 0.36 |
| Deltaproteobacteria bacterium MAG_22309_dsfv_02248 | Deltaproteobacteria | 3300022309 | 2902378 | 66 | 55.15 | 78905 | 91.60 | 1.79 |
| Gammaproteobacteria bacterium MAG_00150_gam_01094 | Gammaproteobacteria | 3300000150 | 2847655 | 486 | 49.07 | 8986 | 98.17 | 3.96 |
| Gammaproteobacteria bacterium MAG_00160_gam_00995 | Gammaproteobacteria | 3300000160 | 2903803 | 318 | 49.10 | 15339 | 99.39 | 4.88 |
| Gammaproteobacteria bacterium MAG_00172_gam_01896 | Gammaproteobacteria | 3300000172 | 2866084 | 274 | 48.97 | 18904 | 96.95 | 3.05 |
| Gammaproteobacteria bacterium MAG_00188_gam_00697 | Gammaproteobacteria | 3300000188 | 2672010 | 567 | 48.83 | 6818 | 95.12 | 4.19 |
| Gammaproteobacteria bacterium MAG_00212_gam_198 | Gammaproteobacteria | 3300000212 | 2103212 | 955 | 48.40 | 2901 | 78.43 | 5.08 |
| Gammaproteobacteria bacterium MAG_00215_gam_02099 | Gammaproteobacteria | 3300000215 | 2931288 | 507 | 49.02 | 8845 | 95.73 | 5.34 |
| Magnetococcales bacterium MAG_21055_mgc_1100 | Ca. Etaproteobacteria | 3300021055 | 3585593 | 930 | 52.41 | 5203 | 84.82 | 3.65 |
| Nitrospinae bacterium MAG_09705_ntspn_70101 | Nitrospinae | 3300009705 | 2024644 | 120 | 42.63 | 30902 | 67.25 | 2.56 |
| Nitrospirae bacterium MAG_10313_ntr_31102 | Nitrospirae | 3300010313 | 1933163 | 344 | 35.33 | 7568 | 90.20 | 3.64 |
| Pelobacteraceae bacterium MAG_21601_9_030103 | Deltaproteobacteria | 3300021601 | 2536371 | 232 | 54.11 | 20074 | 78.15 | 8.39 |
| Pelobacteraceae bacterium MAG_13126_9_058104 | Deltaproteobacteria | 3300013126 | 3576562 | 72 | 52.01 | 83631 | 91.61 | 1.29 |
| Pelobacteraceae bacterium MAG_21600_9_004105 | Deltaproteobacteria | 3300021600 | 3430740 | 60 | 51.50 | 87025 | 90.32 | 0.65 |
| Planctomycetes bacterium MAG_11118_pl_115106 | Planctomycetes | 3300011118 | 3767441 | 157 | 48.98 | 33372 | 89.44 | 1.24 |
| Planctomycetes bacterium MAG_17991_pl_60107 | Planctomycetes | 3300017991 | 1289005 | 144 | 49.53 | 10179 | 64.20 | 0.00 |
| Planctomycetes bacterium MAG_18080_pl_157108 | Planctomycetes | 3300018080 | 3144921 | 139 | 48.44 | 34208 | 90.91 | 3.41 |
| Rhodospirillaceae bacterium MAG_01419_mvb_30 | Alphaproteobacteria | 3300001419 | 2811682 | 477 | 55.72 | 7268 | 94.58 | 4.10 |
| Rhodospirillaceae bacterium MAG_04806_tlms_2109 | Alphaproteobacteria | 3300004806 | 2085124 | 309 | 57.51 | 8435 | 87.64 | 2.12 |
| Rhodospirillaceae bacterium MAG_05422_2-02_14110 | Alphaproteobacteria | 3300005422 | 2281835 | 255 | 61.09 | 11800 | 85.45 | 0.50 |
| Rhodospirillaceae bacterium MAG_05596_2-02_51111 | Alphaproteobacteria | 3300005596 | 1831947 | 329 | 61.19 | 6777 | 76.91 | 0.25 |
| Rhodospirillaceae bacterium MAG_06104_tlms_034112 | Alphaproteobacteria | 3300006104 | 3186839 | 353 | 64.25 | 13005 | 89.59 | 2.53 |
| Rhodospirillaceae bacterium MAG_22225_2-02_112113 | Alphaproteobacteria | 3300022225 | 2547095 | 147 | 61.01 | 26510 | 91.17 | 5.22 |
| Ca. Omnitrophica bacterium SCGC AG-290-C17 (SAG)116 | Ca. Omnitrophica | 3300015153 | 1712617 | 171 | 48.60 | 13921 | 62.84 | 0.00 |
| Uncultured microorganism SbSrfc.SA12.01.D19 (SAG)117 | Deltaproteobacteria | 3300022116 | 2501480 | 175 | 52.60 | 25257 | 49.13 | 0.00 |
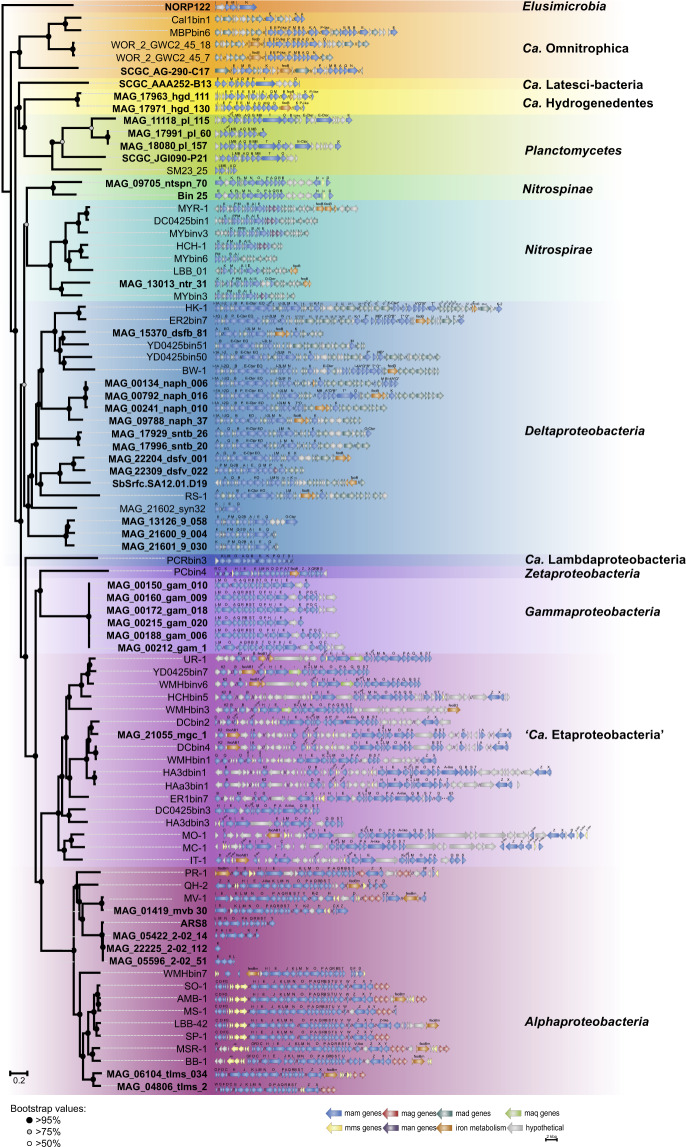
The identification of the phylogenomic position of the studied genomes revealed, for the first time, their affiliation to the phyla Elusimicrobia, Ca. Hydrogenedentes, and Nitrospinae (Supplementary Fig. S2, Supplementary Tables S2 and S5). One genome was affiliated with the phylum Elusimicrobia and referred to order UBA1565 in the Elusimicrobia class. After MGC reconstruction, the mamI, -B, -M, and -N genes were revealed in the investigated genome (Fig. 2). Two MAGs from Ca. Hydrogenedentes belonged to the same species (98.70% average nucleotide identity), but they were obtained independently from different metagenomes. These MAGs referred to the GCA-2746185 family in the order Hydrogenedentiales. The 16S rRNA gene from the Ca. Hydrogenedentes bacterium MAG_17971_hgd_13044 had 90% similarity with the closest non-MTB Ca. Hydrogenedentes bacterium YC-ZSS-LKJ63. All these data confirmed that the obtained binning results were regular and did not represent a computational error. Only mam genes were found in the MGCs of the studied genomes.
Fig. 2.
Comparison of the MGC regions in the MAGs and SAGs (in bold) obtained in this study versus previously known MTB genomes. Full names for MTB strains can be found in Supplementary Table S1.
In the Nitrospinae phylum, two MAGs were affiliated with different genera of the order Nitrospinales. Their MGCs revealed the presence of mam and mms (magnetic particle-membrane specific) genes. Samples for the metagenomes of the obtained MAGs were collected from the Gulf of Mexico45 and Arctic Ocean waters. Non-MTB representatives of this phylum were also detected only in marine habitats46,47, indicating that bacteria from the Nitrospinae could prefer to inhabit marine environments.
The 14 reconstructed MAGs belonged to different families of Deltaproteobacteria. Of the 14, three MAGs were affiliated with the UBA8499 genus in the Pelobacteraceae family. In their MGCs, apart from the mam and mad genes, which are typical for Deltaproteobacteria, the man genes were detected for the first time. Previously, the man genes were associated only with MTB from the Nitrospirae. Another two MAGs were affiliated with the Syntrophobacteraceae family, where MTB were discovered previously41. This is further evidence that binning was conducted correctly and that MTB representatives are indeed present in this family.
Three genomes also belonged to the Desulfobulbales order. Of these, the Deltaproteobacteria bacterium MAG_22309_dsfv_02248 contained man3 gene in addition to the mam and mad genes, thereby confirming the routine presence of man genes in Deltaproteobacteria. A further four MAGs were related to the NaphS2 family in the Desulfatiglanales order. Analysis of their MGCs revealed genes responsible for putative greigite magnetosome synthesis. Metagenomic samples of the studied genomes were obtained from marine sediments, as well as all other known non-MTB genomes of this family49,50.
In Alphaproteobacteria, three MAGs and one genome were related to a 2-02-FULL-58-16 family in the Rhodospirillales order. Metagenomic samples of the studied genomes were isolated from marine ecosystems. The other non-MTB genomes of this family were also detected only in marine ecosystems51. For the first time, two MAGs containing MGCs were also detected in Telmatospirillum genus. Their metagenomic samples were collected from a freshwater bog. Telmatospirillum siberiense, the only known representative of this genus, was also isolated from freshwater peat soil52. Thus, this group possibly tends to inhabit freshwater ecosystems. Reconstruction of the MGCs revealed mam and mms genes in the studied MAGs. One MAG was referred to the Ca. Etaproteobacteria class. Genomes from this class previously were found in both saline and freshwater habitats15,31,53. The obtained MAG clustered with genomes isolated from freshwater environments. The MGC of the recovered MAG revealed a standard gene set inherent to MTB from this class. A further six MAGs were affiliated with the Gammaproteobacteria. All of these were sampled from one source and had 100% identity between their genes. Only the mam genes were detected in their MGCs.
The Nitrospirae phylum was affiliated with one MAG. A metagenomic sample of this phylum was obtained from a hot spring. Previously, other MTB and non-MTB from this phylum were also detected in hot springs54,55. Three of the recovered MAGs belonged to the SG8-4 order in the Phycisphaerae class of Planctomycetes. Apart from the reconstructed MAGs, one SAG was also obtained from the UBA1845 order in Phycisphaerae class. The completeness of this SAG was very low (39%), but it was also taken into analyses due to the large number of mam genes detected in the MGC. Another detected SAG was affiliated with Сa. Omnitrophica and was referred to the GWA2-52-8 family in the Omnitrophales order. The MGC of this genome had a set of genes that were specific to all magnetotactic representatives from this phylum.
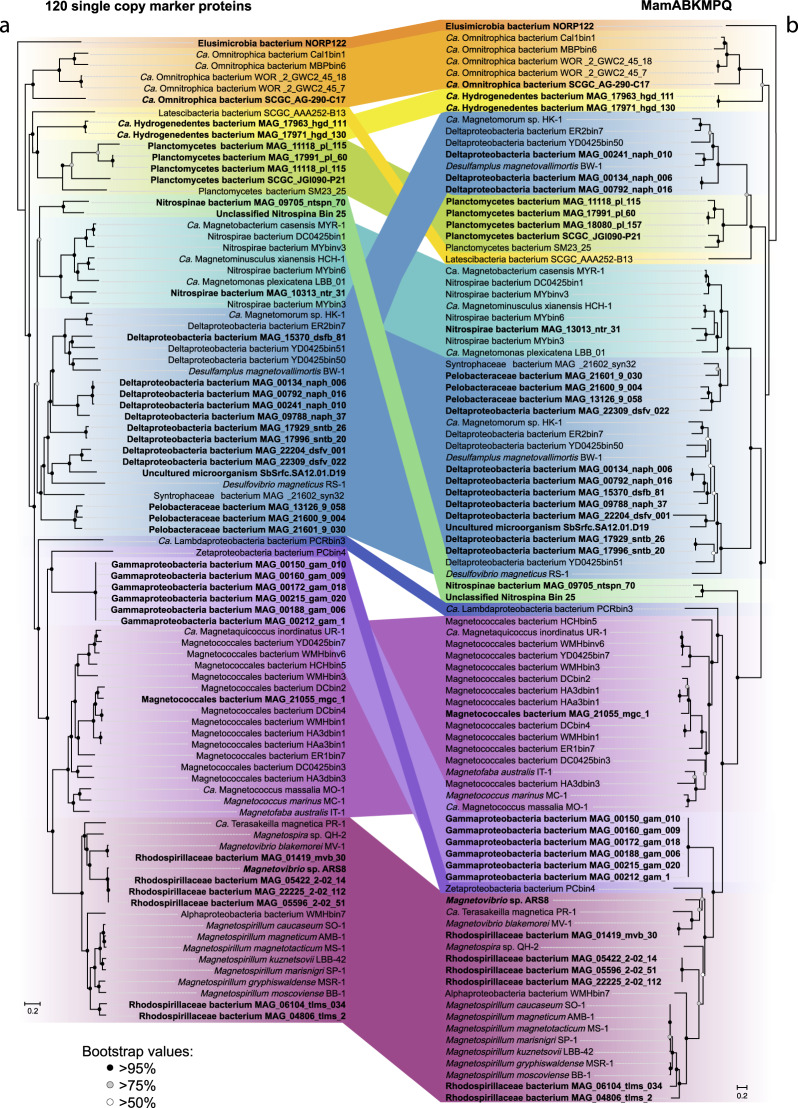
Reconstruction of the evolutionary pathways for MGCs
The identification of putative genes involved in magnetosome biomineralization allowed investigation of MGC evolutionary pathways. These were analyzed by constructing a phylogenetic tree of concatenated MamABKMPQ sequences (“Mam tree”, Fig. 3b) and comparing this tree with one based on 120 single-copy marker gene proteins (“core genome tree”, Fig. 3a). Comparative analysis of the MTB position on the trees revealed some incongruences. For instance, the Deltaproteobacteria group from “core genome tree” was divided into three subgroups on the “Mam tree.” The first subgroup comprised representatives capable of putative greigite magnetosome synthesis, while the other two subgroups included representatives with MGCs for magnetite magnetosome biomineralization. One of the magnetite subgroups included representatives of the Pelobacteraceae, Syntrophia, and Desulfurivibrionaceae families, which clustered with the Nitrospirae. According to the “Mam tree” topology, the man genes could be assumed to have originated in the Deltaproteobacteria and were inherited by the Nitrospirae through horizontal gene transfer. The compared trees also indicated vertical inheritance in the Alpha- and Ca. Etaproteobacteria groups, although the occurrence of horizontal transfer events was previously established in these groups27,31. These types of transfers have been confirmed to have occurred recently, which is why they cannot be detected through the tree topology analysis.
Fig. 3.
Maximum-likelihood phylogenomic trees of MTB genomes. Trees were inferred from a comparison of 120 concatenated single-copy marker proteins of MTB genomes (a) and concatenated magnetosome associated protein sequences (MamABKMPQ) (b). Both trees were reconstructed with evolutionary model LG + F + I + G4. Branch supports were obtained with 1000 ultrafast bootstraps. The scale bar represents amino acid substitutions per site.
A further investigation examined whether MGC originated once or more than once. This was done by adding the Mam protein sequences recovered in this study to previously known Mam protein sequences and their non-MTB homologs and then constructing phylogenetic trees (Supplementary Fig. S3). Analysis of the constructed trees confirmed the previous results15 showing that all Mam protein sequences, except for MamK, formed monophyletic clades and that these clades did not contain any homolog sequences. This indicates that the MGCs for magnetite and greigite synthesis are likely to have a common origin.
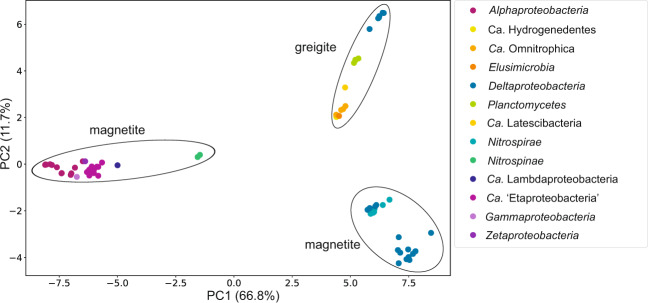
The magnetosome chemical composition in genomes of every phylum where MTB were found for the first time were predicted by counting the phylogenetic distances of the concatenated sequences of six essential Mam proteins (MamA, -B, -K, -M, -P, and -Q) and conducting a principal component analysis (Fig. 4). All values clustered to three groups. First was the group that comprised Planctomycetes, and Latescibacteria, which are known to have genes for putative greigite magnetosome synthesis12,14,56. The NaphS2 family of Deltaproteobateria, Ca. Hydrogenedentes, Сa. Omnitrophica, and Elusimicrobia also clustered with this group. The other two groups comprised representatives with magnetite magnetosome synthesis genes. The first magnetite group included Nitrospinae and all classes of Proteobacteria where MTB were known. The exception was the remaining studied classes of Deltaproteobateria, which clustered with the second magnetite group, together with Nitrospirae.
Fig. 4.
The prediction of magnetosome chemical composition for phyla in which MTB genomes were found for the first time. Predictions were made using principal component analysis for a maximum-likelihood distance matrix of concatenated Mam protein sequences.
Discussion
This study represents the first large-scale search of magnetosome biomineralization genes in open databases. Bioinformatic analysis of the gathered data almost doubled the number of MTB genomes from the 60 previously known; 4 genomes, 2 SAGs, and 32 MAGs were obtained as a result of this research. Besides, analysis of the database of collected MGC protein sequences revealed MamK as the most appropriate protein for MGC searching in open databases. This finding will allow the use of these putative protein sequences as markers for MTB detection in environmental samples.
This study also provides the first description of magnetosome biomineralization genes in the genomes of Elusimicrobia, Nitrospinae, and Ca. Hydrogenedentes. Non-MTB representatives of Elusimicrobia phylum were previously found as free-living57 and ecto- and endosymbionts58,59 of multicellular eukaryotes. MTB living symbiotically with eukaryotes have been detected previously60,61. Further investigations are needed to solve the enigma of whether MTB from Elusimicrobia free-living or symbiotic organisms are.
To date, little is known about Ca. Hydrogenedentes, except for its genome presence62–64. More is known about Nitrospinae, where one axenic culture was previously described65. However, these reports do not give an extensive understanding of the capabilities of this phylum’s representatives. Thus, the detection of MGCs in genomes that belong to these phyla significantly supplements the knowledge of MTB diversity and evolution, while also providing new information about these phyla.
This work also gives much new information about groups where MTB were previously recognized. For instance, the relatively few genomes were affiliated with Alpha- and Ca. Etaproteobacteria, while the current belief is that representatives of these classes dominate among MTB in all natural environments12. In addition, within the Alphaproteobacteria class, the presence of MGCs was discovered for the first time in genomes belonging to the Telmatospirillum genus. This may indicate a common origin for magnetosome biomineralization genes among the Magnetospirillum, Magnetospira, and Magnetovibrio genera.
Furthermore, for the first time the presence of man genes was revealed in MGCs of the Deltaproteobacteria. Previously, these genes were found only in Nitrospinae. Whether horizontal gene transfer events occurred between representatives of these phylogenetic groups or their MGCs shared a common origin is not known. Further studies are required to determine which possibility is correct.
The genomes with magnetosome biomineralization genes obtained in this study allowed the investigation of the origin and evolution of the MGCs. A comparison of the “core genome” and “Mam” trees revealed clustering of the Deltaproteobacteria greigite subgroup sequences with the Planctomycetes, Latescibacteria, Ca. Hydrogenedentes, Сa. Omnitrophica, and Elusimicrobia phyla. Of these, Latescibacteria14 and Planctomycetes12 were already known to have MGCs for putative greigite synthesis. Note that Ca. Omnitrophica was also associated with the greigite subgroup, although it is believed that they biomineralize magnetite magnetosomes43. Such assumptions are based on Сa. Omnitrophus magneticus SKK-01 however, this genome is highly contaminated (Supplementary Table S1). Thus, further investigations are needed to study Ca. Omnitrophica magnetosome chemical composition.
In addition to all mentioned findings, the latest version of the bacterial tree of life66, based on GTDB R04-RS89 reference data (Supplementary Fig. S4) helped to reveal the most ancient phylum in which MTB representatives were known. It was indicated that the Elusimicrobia phylum is the most closely related to the last universal common ancestor (LUCA). If the MTB of this phylum are assumed capable of greigite magnetosome synthesis, then greigite MGCs could have appeared much earlier than commonly believed, and the first MTB could have greigite, not magnetite, MGCs. The other phyla with MTB representatives in the vicinity of LUCA are Ca. Omnitrophica and Proteobacteria, although Nitrospirae MTB was previously thought to be the most ancient40.
Considering the existing data regarding the presence of horizontal transfer events among MTB and analyzing the discrepancies in “core genome” and “Mam” trees, the proposal could be made that horizontal gene transfers occur much more often than previously thought and are of great importance in MGC evolution.
The genomes obtained in this work require further confirmation by morphological identification. Once confirmed, these data will allow a more thorough study of the contribution of vertical and horizontal gene transfer events with respect to MGC inheritance. The data obtained in the present work will allow the study of the environmental and metabolic preferences of newly discovered MTB genomes, which may become the key to isolating them in axenic cultures. Moreover, a detailed MGC analysis could help to find as yet unidentified genes that are involved in magnetosome synthesis and to reveal much about the biomineralization process.
Generally, in this work, it was shown that MamK is the most appropriate protein for MGCs detecting in open databases. The search results allowed to receive 38 new genomes containing MGCs, that were affiliated to both taxonomic groups where MTB were found before and three new phyla. Thus, received MTB genomes permitted to unravel the MTB diversity and can be used in further MTB studies or in receiving new information about these phyla. Also, a comparison of MTB position on “mam tree” and “core genome tree” helped to reveal signs of putative horizontal gene transfers. This led to assumptions that such MGC transfers could occur with higher frequency and probably play a much more important role in MGC evolution than it was previously thought. Moreover, a proposal was made that the origin of MGC probably is more ancient than it was suggested earlier and possibly was capable of greigite magnetosomes biomineralization rather than magnetite.
Thus, all received data allowed the expansion of knowledge about MTB diversity, ecology, and evolution and has opened up new opportunities for further searches for and investigations of magnetotactic bacteria.
Materials and methods
The search for magnetosome biomineralization genes in open databases
The search for magnetosome biomineralization genes was conducted by collecting a database of MGC protein sequences based on currently known MTB genomes (Supplementary Table S1). The search was provided using BLASTp analysis, with identity >30% and e-value >1e−05. Searches of the IMG and NCBI genomic databases used sequences of nine essential Mam proteins from different taxonomic groups as targets. The IMG metagenomic database was searched by BLASTp using MamK sequences. The sequences obtained from BLAST analysis were further checked to separate MGC proteins from their homologs. For this, each Mam protein sequence was checked for joint clustering on the phylogenetic trees. The presence of other Mam proteins in the same scaffold provided additional support for choosing those scaffolds for further analysis. The search was conducted in April 2018.
Genome reconstruction and analyses
Metagenome assembled genome (MAG) reconstruction was conducted using the Busybee web67, Maxbin268, and MyCC69 with standard parameters. The DAS Tool70 was used for choosing consensus assemblies for the obtained MAGs. Completeness and contamination values of genomes were obtained using lineage-specific marker genes and default parameters in CheckM v. 1.0.1271. RefineM v. 0.0.2450 was used to remove contamination based on taxonomic assignments. This process, called ‘decontamination’, involves the classification of obtained genes and scaffolds in each MAG relative to the gene base with a known taxonomic classification. After that, scaffolds with incongruent taxonomic classifications are removed from the MAGs. The quality metrics were assessed using the QUAST72 tool. The average nucleotide identity (ANI) was calculated using fastANI73. The MGCs were determined using local BLAST and comparison with reference sequences of magnetotactic bacteria.
Phylogenetic analyses
Taxonomic assignments for the studied genomes 16S rRNA genes were obtained using the GTDB 16S r89 dataset in IDTAXA74. The GTDB-Tk v.0.1.375 ‘classify_wf’ command was used to find 120 single-copy bacterial marker protein sequences, to construct their multiple alignments and to get the taxonomic assignment using the GTDB r86 database76. Amino acid sequence sets of the MamA, -B, -M, -K, -P, and -Q proteins were independently aligned using MAFFT77, curated with Gblocks v. 0.91b78 with an option that allows gap positions within the final blocks, and then concatenated. These Mam protein sequences were also used to build trees with their homologs. Maximum-likelihood trees were inferred with IQ-TREE79 using evolutionary models selected by ModelFinder80. Branch supports were obtained with 1000 ultrafast bootstraps81. Trees were visualized with iTOL v482. The genomes of Ca. Omnitrophus magneticus SKK-01, Ca. Magnetoglobus multicellularis str. Araruama, Ca. Magnetobacterium bavaricum TM-1, and Ca. Magnetoovum chiemensis CS-04 were not subjected to phylogenetic analyses because they had failed the quality check (Supplementary Table S1). Taxonomic classification of the obtained genomes on phylum rank was performed using NCBI taxonomy; other ranks were named using GTDB.
Supplementary information
Acknowledgements
We thank David Walsh (contributor of metagenomic data with accession 3300009705), Frank Stewart (3300022225,3300005596,3300005422), Nikos Kyrpides (3300001419), Katherine McMahon (3300004806, 3300006104), Ramunas Stepanauskas (3300010313), Hebe Dionisi (3300000134, 3300000241, 3300000792), Emiley Eloe-Fadrosh (3300009788, 3300011118), Josh Neufeld (3300021600, 3300021601), Steven Hallam (3300000150, 3300000160, 3300000172, 3300000188, 3300000212, 3300000215), Kelly Wrighton (3300021055), David Valentine (3300017971, 3300017963, 3300018080), Sean Crowe (3300013126), Erik Lilleskov (3300017929, 3300017996), Mark Dopson (3300015370, sequences were generated by the JGI community sequencing program project 502935), Christopher Francis (3300022309, 3300022204) for permission to use metagenomic data in this study. The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science Facility, was supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. This study was performed using scientific equipment at the Core Research Facility ‘Bioengineering’ (Research Center of Biotechnology RAS). This study was funded by the Russian Foundation for Basic Research as research project no. 18-34-01005 and by the Ministry of Science and Higher Education of the Russian Federation.
Author contributions
M.U. and L.A. created MGC protein sequences database. M.U. conducted MGCs search, analyzed obtained data and wrote the manuscript. L.A. reconstructed MGCs of obtained genomes. M.K. conducted metagenomes binning. D.G. had the initial idea for the analysis. V.K. and D.G. discussed and interpreted the results and revised the manuscript. All authors read and approved the final manuscript.
Data availability
The genomes and metagenomes used during the current study are publicly available in NCBI (https://www.ncbi.nlm.nih.gov/)44,48,83–113 and IMG (https://img.jgi.doe.gov/cgi-bin/m/main.cgi)114–117 databases. Scaffolds of obtained MAGs could be found in Supplementary Table S6, hosted at figshare118. All data generated and analyzed in this study are also available in figshare118 and in supplementary information accompany this paper. Assembly of Rhodospirillaceae bacterium MAG_01419_mvb_30 could be found in RAST (https://rast.nmpdr.org/) using ‘guest’ as login and as password.
Code availability
The following tools were used for the presented analysis and described in the main text:
Busybee web, Maxbin2, MyCC, and DAS Tool with standard parameters were used for the reconstruction of metagenome-assembled genomes (MAGs).
1. Busybee web https://ccb-microbe.cs.uni-saarland.de/busybee
2. Maxbin2 https://sourceforge.net/projects/maxbin2/
3. MyCC https://sourceforge.net/projects/sb2nhri/files/MyCC/
4. DAS Tool https://github.com/cmks/DAS_Tool
5. CheckM was used to estimate obtained genomes completeness and contamination https://github.com/Ecogenomics/CheckM
6. RefineM was used to remove contamination https://github.com/dparks1134/RefineM
7. QUAST helped to access quality metrics http://cab.spbu.ru/software/quast/
8. fastANI was used to calculate ANI https://github.com/ParBLiSS/FastANI
9. IDTAXA helped to obtain taxonomic assignments for the studied genomes 16S rRNA genes http://www2.decipher.codes/Classification.html
10. GTDB-Tk was used to find 120 single-copy bacterial marker protein sequences, to construct their multiple alignments and to get the taxonomic assignment using the GTDB r86 database https://github.com/Ecogenomics/GTDBTk
11. MAFFT was used for aligning amino acid sequence sets of the MamA, -B, -M, -K, -P, and -Q proteins https://mafft.cbrc.jp/alignment/server/
12. Gblocks helped to curate sequences aligned in MAFFT http://molevol.cmima.csic.es/castresana/Gblocks_server.html
13. Phylogenetic trees were inferred with IQ-TREE http://www.iqtree.org/
14. Obtained trees were visualized with iTOL https://itol.embl.de/
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41597-020-00593-0.
References
- 1.Mukherjee S, et al. Genomes OnLine database (GOLD) v.7: Updates and new features. Nucleic Acids Res. 2019;47:D649–D659. doi: 10.1093/nar/gky977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agarwala R, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018;46:D8–D13. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen IMA, et al. IMG/M v.5.0: An integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 2019;47:D666–D677. doi: 10.1093/nar/gky901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blakemore RP. Magnetotactic Bacteria. Science. 1975;190:377–379. doi: 10.1126/science.170679. [DOI] [PubMed] [Google Scholar]
- 5.Benoit MR, et al. Visualizing implanted tumors in mice with magnetic resonance imaging using magnetotactic bacteria. Clin Cancer Res. 2009;15:5170–5177. doi: 10.1158/1078-0432.CCR-08-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alphandéry E, Chebbi I, Guyot F, Durand-Dubief M. Use of bacterial magnetosomes in the magnetic hyperthermia treatment of tumours: A review. Int. J. Hyperth. 2013;29:801–809. doi: 10.3109/02656736.2013.821527. [DOI] [PubMed] [Google Scholar]
- 7.Chang S, Kirschvink JL. Magnetofossils, the magnetization of sediments, and the evolution of magnetite biomineralization. Annu. Rev. Earth Planet. Sci. 1989;17:169–95. [Google Scholar]
- 8.Kodama KP, Moeller RE, Bazylinski DA, Kopp RE, Chen AP. The mineral magnetic record of magnetofossils in recent lake sediments of Lake Ely, PA. Glob. Planet. Change. 2013;110:350–363. [Google Scholar]
- 9.Kopp RE, Kirschvink JL. The identification and biogeochemical interpretation of fossil magnetotactic bacteria. Earth-Science Rev. 2008;86:42–61. [Google Scholar]
- 10.Mckay CP, Friedmann EI, Frankel RB, Bazylinski DA. Magnetotactic bacteria on Earth and on Mars. Astrobiology. 2003;3:263–271. doi: 10.1089/153110703769016361. [DOI] [PubMed] [Google Scholar]
- 11.Uebe R, Schüler D. Magnetosome biogenesis in magnetotactic bacteria. Nature Reviews Microbiology. 2016;14:621–637. doi: 10.1038/nrmicro.2016.99. [DOI] [PubMed] [Google Scholar]
- 12.Lin W, Pan Y, Bazylinsky DA. Diversity and ecology of and biomineralization by magnetotactic bacteria. Environ. Microbiol. Rep. 2017;9:345–356. doi: 10.1111/1758-2229.12550. [DOI] [PubMed] [Google Scholar]
- 13.Lin W, et al. Genomic insights into the uncultured genus ‘Candidatus Magnetobacterium’ in the phylum Nitrospirae. ISME J. 2014;8:2463–2477. doi: 10.1038/ismej.2014.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin W, Pan Y. A putative greigite-type magnetosome gene cluster from the candidate phylum Latescibacteria. Environ. Microbiol. Rep. 2015;7:237–242. doi: 10.1111/1758-2229.12234. [DOI] [PubMed] [Google Scholar]
- 15.Lin W, et al. Genomic expansion of magnetotactic bacteria reveals an early common origin of magnetotaxis with lineage-specific evolution. ISME J. 2018. 2018;12:1508–1519. doi: 10.1038/s41396-018-0098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji B, et al. Comparative genomic analysis provides insights into the evolution and niche adaptation of marine Magnetospira sp. QH-2 strain. Environ. Microbiol. 2014;16:525–544. doi: 10.1111/1462-2920.12180. [DOI] [PubMed] [Google Scholar]
- 17.Koziaeva VV, et al. Magnetospirillum kuznetsovii sp. nov., a novel magnetotactic bacterium isolated from a lake in the Moscow region. Int. J. Syst. Evol. Microbiol. 2019;69:1953–1959. doi: 10.1099/ijsem.0.003408. [DOI] [PubMed] [Google Scholar]
- 18.Matsunaga T, et al. Complete genome sequence of the facultative anaerobic magnetotactic bacterium Magnetospirillum sp. strain AMB-1. DNA Res. 2005;12:157–166. doi: 10.1093/dnares/dsi002. [DOI] [PubMed] [Google Scholar]
- 19.Smalley MD, Marinov GK, Bertani LE, DeSalvo G. Genome sequence of Magnetospirillum magnetotacticum strain MS-1. Genome Announc. 2015;3:e00233–15. doi: 10.1128/genomeA.00233-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koziaeva VV, et al. Draft Genome sequences of two magnetotactic bacteria, Magnetospirillum moscoviense BB-1 and Magnetospirillum marisnigri SP-1. Genome Announc. 2016;4:e00814–16. doi: 10.1128/genomeA.00814-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ke L, Liu P, Liu S, Gao M. Complete genome sequence of Magnetospirillum sp. ME-1, a novel magnetotactic bacterium isolated from East Lake, Wuhan, China. Genome Announc. 2017;5:e00485–17. doi: 10.1128/genomeA.00485-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, et al. Complete genome sequence of Magnetospirillum sp. Strain XM-1, isolated from the Xi’an City Moat. China. Genome Announc. 2016;4:e01171–16. doi: 10.1128/genomeA.01171-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grouzdev DS, et al. Draft genome sequence of Magnetospirillum sp. Strain SO-1, a freshwater magnetotactic bacterium isolated from the Ol’khovka River, Russia. Genome Announc. 2014;2:e00235–14. doi: 10.1128/genomeA.00235-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ullrich S, Kube M, Schübbe S, Reinhardt R, Schüler D. A hypervariable 130-kilobase genomic region of Magnetospirillum gryphiswaldense comprises a magnetosome island which undergoes frequent rearrangements during stationary growth. J. Bacteriol. 2005;187:7176–7184. doi: 10.1128/JB.187.21.7176-7184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trubitsyn D, et al. Draft genome sequence of Magnetovibrio blakemorei strain MV-1, a marine vibrioid magnetotactic bacterium. Genome Announc. 2016;4:e01330–16. doi: 10.1128/genomeA.01330-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jogler C, et al. Comparative analysis of magnetosome gene clusters in magnetotactic bacteria provides further evidence for horizontal gene transfer. Environ. Microbiol. 2009;11:1267–1277. doi: 10.1111/j.1462-2920.2009.01854.x. [DOI] [PubMed] [Google Scholar]
- 27.Monteil CL, et al. Genomic study of a novel magnetotactic Alphaproteobacteria uncovers the multiple ancestry of magnetotaxis. Environ. Microbiol. 2018;20:4415–4430. doi: 10.1111/1462-2920.14364. [DOI] [PubMed] [Google Scholar]
- 28.Schübbe S, et al. Complete genome sequence of the chemolithoautotrophic marine magnetotactic coccus strain MC-1. Appl. Environ. Microbiol. 2009;75:4835–4852. doi: 10.1128/AEM.02874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji B, et al. The chimeric nature of the genomes of marine magnetotactic coccoid-ovoid bacteria defines a novel group of Proteobacteria. Environ. Microbiol. 2017;19:1103–1119. doi: 10.1111/1462-2920.13637. [DOI] [PubMed] [Google Scholar]
- 30.Morillo V, et al. Isolation, cultivation and genomic analysis of magnetosome biomineralization genes of a new genus of South-seeking magnetotactic cocci within the Alphaproteobacteria. Front. Microbiol. 2014;5:72. doi: 10.3389/fmicb.2014.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koziaeva V, et al. Genome-based metabolic reconstruction of a novel uncultivated freshwater magnetotactic coccus “Ca. Magnetaquicoccus inordinatus” UR-1, and proposal of a candidate family “Ca. Magnetaquicoccaceae”. Front. Microbiol. 2019;10:2290. doi: 10.3389/fmicb.2019.02290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abreu F, et al. Deciphering unusual uncultured magnetotactic multicellular prokaryotes through genomics. ISME J. 2014;8:1055–1068. doi: 10.1038/ismej.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kolinko S, Richter M, Glöckner FO, Brachmann A, Schüler D. Single-cell genomics reveals potential for magnetite and greigite biomineralization in an uncultivated multicellular magnetotactic prokaryote. Environ. Microbiol. Rep. 2014;6:524–531. doi: 10.1111/1758-2229.12198. [DOI] [PubMed] [Google Scholar]
- 34.Lefèvre CT, et al. Comparative genomic analysis of magnetotactic bacteria from the Deltaproteobacteria provides new insights into magnetite and greigite magnetosome genes required for magnetotaxis. Environ. Microbiol. 2013;15:2712–2735. doi: 10.1111/1462-2920.12128. [DOI] [PubMed] [Google Scholar]
- 35.Nakazawa H, et al. Whole genome sequence of Desulfovibrio magneticus strain RS-1 revealed common gene clusters in magnetotactic bacteria. Genome Res. 2009;19:1801–1808. doi: 10.1101/gr.088906.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lefèvre CT, et al. Novel magnetite-producing magnetotactic bacteria belonging to the Gammaproteobacteria. ISME J. 2012;6:440–450. doi: 10.1038/ismej.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baker, B. J., Lazar, C. S., Teske, A. P. & Dick, G. J. Genomic resolution of linkages in carbon, nitrogen, and sulfur cycling among widespread estuary sediment bacteria. Microbiome3 (2015). [DOI] [PMC free article] [PubMed]
- 38.Jogler C, et al. Cultivation-independent characterization of ‘Candidatus Magnetobacterium bavaricum’ via ultrastructural, geochemical, ecological and metagenomic methods. Environ. Microbiol. 2010;12:2466–2478. doi: 10.1111/j.1462-2920.2010.02220.x. [DOI] [PubMed] [Google Scholar]
- 39.Kolinko S, Richter M, Glöckner FO, Brachmann A, Schüler D. Single-cell genomics of uncultivated deep-branching magnetotactic bacteria reveals a conserved set of magnetosome genes. Environ. Microbiol. 2016;18:21–37. doi: 10.1111/1462-2920.12907. [DOI] [PubMed] [Google Scholar]
- 40.Lin W, et al. Origin of microbial biomineralization and magnetotaxis during the Archean. Proc. Natl. Acad. Sci. 2017;114:2171–2176. doi: 10.1073/pnas.1614654114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koziaeva, V. V. et al. Biodiversity of magnetotactic bacteria in the freshwater lake Beloe Bordukovskoe, Russia. Microbiology89, 348–358, 10.1134/S002626172003008X (2020).
- 42.Wrighton KC, et al. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science (80−). 2012;337:1661–1665. doi: 10.1126/science.1224041. [DOI] [PubMed] [Google Scholar]
- 43.Kolinko S, et al. Single-cell analysis reveals a novel uncultivated magnetotactic bacterium within the candidate division OP3. Environ. Microbiol. 2012;14:1709–1721. doi: 10.1111/j.1462-2920.2011.02609.x. [DOI] [PubMed] [Google Scholar]
- 44.2020. BioSample of Candidatus Hydrogenedentes bacterium MAG_17971_hgd_130. NCBI BioSample. SAMN14911668
- 45.Thrash CJ, et al. Metagenomic assembly and prokaryotic metagenome-assembled genome sequences from the northern Gulf of Mexico “Dead Zone”. Microbiol. Resour. Announc. 2018;7:4–6. doi: 10.1128/MRA.01033-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watson SW, Waterbury JB. Characteristics of two marine nitrite oxidizing bacteria, Nitrospina gracilis nov. gen. nov. sp. and Nitrococcus mobilis nov. gen. nov. sp. Arch. Microbiol. 1971;77:203–230. [Google Scholar]
- 47.Tian RM, et al. The deep-sea glass sponge Lophophysema eversa harbours potential symbionts responsible for the nutrient conversions of carbon, nitrogen and sulfur. Environ. Microbiol. 2016;18:2481–2494. doi: 10.1111/1462-2920.13161. [DOI] [PubMed] [Google Scholar]
- 48.2020. BioSample of Deltaproteobacteria bacterium MAG_22309_dsfv_022. NCBI BioSample. SAMN14911677
- 49.Didonato RJ, et al. Genome sequence of the deltaproteobacterial strain NaphS2 and analysis of differential gene expression during anaerobic growth on naphthalene. PLos One. 2010;5:e14072. doi: 10.1371/journal.pone.0014072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parks DH, et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2017;2:1533–1542. doi: 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- 51.Tully BJ, Graham ED, Heidelberg JF. The reconstruction of 2,631 draft metagenome-assembled genomes from the global oceans. Sci. Data. 2018;5:1–8. doi: 10.1038/sdata.2017.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sizova MV, Panikov NS, Spiridonova EM, Slobodova NV, Tourova TP. Novel facultative anaerobic acidotolerant Telmatospirillum siberiense gen. nov. sp. nov. isolated from mesotrophic fen. Syst. Appl. Microbiol. 2007;30:213–220. doi: 10.1016/j.syapm.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 53.Bazylinski DA, et al. Magnetococcus marinus gen. nov., sp. nov., a marine, magnetotactic bacterium that represents a novel lineage (Magnetococcaceae fam. nov., Magnetococcales ord. nov.) at the base of the Alphaproteobacteria. Int. J. Syst. Evol. Microbiol. 2013;63:801–808. doi: 10.1099/ijs.0.038927-0. [DOI] [PubMed] [Google Scholar]
- 54.Lebedeva EV, et al. Isolation and characterization of a moderately thermophilic nitrite-oxidizing bacterium from a geothermal spring. FEMS Microbiol. Ecol. 2011;75:195–204. doi: 10.1111/j.1574-6941.2010.01006.x. [DOI] [PubMed] [Google Scholar]
- 55.Lefèvre CT, et al. Moderately thermophilic magnetotactic bacteria from hot springs in Nevada. Appl. Environ. Microbiol. 2010;76:3740–3743. doi: 10.1128/AEM.03018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lefèvre CT, et al. Comparative genomic analysis of magnetotactic bacteria from the Deltaproteobacteria provides new insights into magnetite and greigite magnetosome genes required for magnetotaxis. Syst. Appl. Microbiol. 2017;40:280–289. doi: 10.1111/1462-2920.12128. [DOI] [PubMed] [Google Scholar]
- 57.Mikaelyan A, et al. High-resolution phylogenetic analysis of Endomicrobia reveals multiple acquisitions of endosymbiotic lineages by termite gut flagellates. Environ. Microbiol. Rep. 2017;9:477–483. doi: 10.1111/1758-2229.12565. [DOI] [PubMed] [Google Scholar]
- 58.Izawa K, et al. Discovery of ectosymbiotic Endomicrobium lineages associated with protists in the gut of stolotermitid termites. Environ. Microbiol. Rep. 2017;9:411–418. doi: 10.1111/1758-2229.12549. [DOI] [PubMed] [Google Scholar]
- 59.Ohkuma M, et al. The candidate phylum ‘Termite Group 1’ of bacteria: Phylogenetic diversity, distribution, and endosymbiont members of various gut flagellated protists. FEMS Microbiol. Ecol. 2007;60:467–476. doi: 10.1111/j.1574-6941.2007.00311.x. [DOI] [PubMed] [Google Scholar]
- 60.Dufour SC, et al. Magnetosome-containing bacteria living as symbionts of bivalves. ISME J. 2014;8:2453–2462. doi: 10.1038/ismej.2014.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Monteil CL, et al. Ectosymbiotic bacteria at the origin of magnetoreception in a marine protist. Nat. Microbiol. 2019;4:1088–1095. doi: 10.1038/s41564-019-0432-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rinke C, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 63.Probst AJ, et al. Genomic resolution of a cold subsurface aquifer community provides metabolic insights for novel microbes adapted to high CO2 concentrations. Environ. Microbiol. 2016;19:459–474. doi: 10.1111/1462-2920.13362. [DOI] [PubMed] [Google Scholar]
- 64.Tully BJ, Wheat CG, Glazer BT, Huber JA. A dynamic microbial community with high functional redundancy inhabits the cold, oxic subseafloor aquifer. ISME J. 2018;12:1–16. doi: 10.1038/ismej.2017.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lücker S, Nowka B, Rattei T, Spieck E, Daims H. The genome of Nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer. Front. Microbiol. 2013;4:27. doi: 10.3389/fmicb.2013.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mendler K, et al. Annotree: Visualization and exploration of a functionally annotated microbial tree of life. Nucleic Acids Res. 2019;47:4442–4448. doi: 10.1093/nar/gkz246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Laczny CC, et al. BusyBee Web: Metagenomic data analysis by bootstrapped supervised binning and annotation. Nucleic Acids Res. 2017;45:W171–W179. doi: 10.1093/nar/gkx348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu YW, Simmons BA, Singer SW. MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics. 2016;32:605–607. doi: 10.1093/bioinformatics/btv638. [DOI] [PubMed] [Google Scholar]
- 69.Lin HH, Liao YC. Accurate binning of metagenomic contigs via automated clustering sequences using information of genomic signatures and marker genes. Sci. Rep. 2016;6:12–19. doi: 10.1038/srep24175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sieber CMK, et al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 2018;3:836–843. doi: 10.1038/s41564-018-0171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Murali A, Bhargava A, Wright ES. IDTAXA: A novel approach for accurate taxonomic classification of microbiome sequences. Microbiome. 2018;6:140. doi: 10.1186/s40168-018-0521-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chaumeil, P., Mussig, A. J., Parks, D. H. & Hugenholtz, P. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 1–3, 10.1093/bioinformatics/btz848 (2019). [DOI] [PMC free article] [PubMed]
- 76.Parks DH, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018;36:996. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 77.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 79.Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kalyaanamoorthy S, Minh BQ, Wong TKF, Haeseler AV, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.2013. ASM268676v1 assembly for Magnetovibrio sp. NCBI Assembly. GCA_002686765.1
- 84.2017. ASM240148v1assembly for Elusimicrobia bacterium NORP122. NCBI Assembly. GCA_002401485.1
- 85.2020. BioSample of Candidatus Hydrogenedentes bacterium MAG_17963_hgd_111. NCBI BioSample. SAMN14911667
- 86.2020. BioSample of Deltaproteobacteria bacterium MAG_00134_naph_006. NCBI BioSample. SAMN14911648
- 87.2020. BioSample of Deltaproteobacteria bacterium MAG_00241_naph_010. NCBI BioSample. SAMN14911655
- 88.2020. BioSample of Deltaproteobacteria bacterium MAG_00792_naph_016. NCBI BioSample. SAMN14911656
- 89.2020. BioSample of Deltaproteobacteria bacterium MAG_09788_naph_37. NCBI BioSample. SAMN14911662
- 90.2020. BioSample of Deltaproteobacteria bacterium MAG_15370_dsfb_81. NCBI BioSample. SAMN14911665
- 91.2020. BioSample of Deltaproteobacteria bacterium MAG_17929_sntb_26. NCBI BioSample. SAMN14911666
- 92.2020. BioSample of Deltaproteobacteria bacterium MAG_17996_sntb_20. NCBI BioSample. SAMN14911670
- 93.2020. BioSample of Deltaproteobacteria bacterium MAG_22204_dsfv_001. NCBI BioSample. SAMN14911675
- 94.2020. BioSample of Gammaproteobacteria bacterium MAG_00150_gam_010. NCBI BioSample. SAMN14911649
- 95.2020. BioSample of Gammaproteobacteria bacterium MAG_00160_gam_009. NCBI BioSample. SAMN14911650
- 96.2020. BioSample of Gammaproteobacteria bacterium MAG_00172_gam_018. NCBI BioSample. SAMN14911651
- 97.2020. BioSample of Gammaproteobacteria bacterium MAG_00188_gam_006. NCBI BioSample. SAMN14911652
- 98.2020. BioSample of Gammaproteobacteria bacterium MAG_00212_gam_1. NCBI BioSample. SAMN14911653
- 99.2020. BioSample of Gammaproteobacteria bacterium MAG_00215_gam_020. NCBI BioSample. SAMN14911654
- 100.2020. BioSample of Magnetococcales bacterium MAG_21055_mgc_1. NCBI BioSample. SAMN14911672
- 101.2020. BioSample of Nitrospinae bacterium MAG_09705_ntspn_70. NCBI BioSample. SAMN14911661
- 102.2020. BioSample of Nitrospirae bacterium MAG_10313_ntr_31. NCBI BioSample. SAMN14911663
- 103.2020. BioSample of Desulfuromonadales bacterium MAG_21601_9_030. NCBI BioSample. SAMN14911674
- 104.2020. BioSample of Desulfuromonadales bacterium MAG_13126_9_058. NCBI BioSample. SAMN14911678
- 105.2020. BioSample of Desulfuromonadales bacterium MAG_21600_9_004. NCBI BioSample. SAMN14911673
- 106.2020. BioSample of Planctomycetes bacterium MAG_11118_pl_115. NCBI BioSample. SAMN14911664
- 107.2020. BioSample of Planctomycetes bacterium MAG_17991_pl_60. NCBI BioSample. SAMN14911669
- 108.2020. BioSample of Planctomycetes bacterium MAG_18080_pl_157. NCBI BioSample. SAMN14911671
- 109.2020. BioSample of Rhodospirillaceae bacterium MAG_04806_tlms_2. NCBI BioSample. SAMN14911657
- 110.2020. BioSample of Rhodospirillaceae bacterium MAG_05422_2-02_14. NCBI BioSample. SAMN14911658
- 111.2020. BioSample of Rhodospirillaceae bacterium MAG_05596_2-02_51. NCBI BioSample. SAMN14911659
- 112.2020. BioSample of Rhodospirillaceae bacterium MAG_06104_tlms_034. NCBI BioSample. SAMN14911660
- 113.2020. BioSample of Rhodospirillaceae bacterium MAG_22225_2-02_112. NCBI BioSample. SAMN14911676
- 114.2016. Assembly for unclassified Nitrospina Bin 25. IMG. 2651870060
- 115.2015. Assembly for Planctomycetes bacterium SCGC JGI090-P21. IMG Assembly. 2264265205
- 116.2017. Assembly for Omnitrophica bacterium SCGC_AG-290-C17. IMG Assembly. 3300015153
- 117.2017. Assembly for uncultured microorganism SbSrfc.SA12.01.D19. IMG Assembly. 3300022116
- 118.Uzun, M., Alekseeva, L., Krutkina, M., Koziaeva, V. & Grouzdev, D. Analysis: unravelling the diversity of magnetotactic bacteria through analysis of open genomic databases. fighsare10.6084/m9.figshare.c.4883706 (2020). [DOI] [PMC free article] [PubMed]
- 119.Espínola F, et al. Metagenomic Analysis of Subtidal Sediments from Polar and Subpolar Coastal Environments Highlights the Relevance of Anaerobic Hydrocarbon Degradation Processes. Microb. Ecol. 2018;75:123–139. doi: 10.1007/s00248-017-1028-5. [DOI] [PubMed] [Google Scholar]
- 120.Wu X, et al. Microbial metagenomes from three aquifers in the Fennoscandian shield terrestrial deep biosphere reveal metabolic partitioning among populations. ISME J. 2016;10:1192–1203. doi: 10.1038/ismej.2015.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- 2020. BioSample of Candidatus Hydrogenedentes bacterium MAG_17971_hgd_130. NCBI BioSample. SAMN14911668
- 2020. BioSample of Deltaproteobacteria bacterium MAG_22309_dsfv_022. NCBI BioSample. SAMN14911677
- 2013. ASM268676v1 assembly for Magnetovibrio sp. NCBI Assembly. GCA_002686765.1
- 2017. ASM240148v1assembly for Elusimicrobia bacterium NORP122. NCBI Assembly. GCA_002401485.1
- 2020. BioSample of Candidatus Hydrogenedentes bacterium MAG_17963_hgd_111. NCBI BioSample. SAMN14911667
- 2020. BioSample of Deltaproteobacteria bacterium MAG_00134_naph_006. NCBI BioSample. SAMN14911648
- 2020. BioSample of Deltaproteobacteria bacterium MAG_00241_naph_010. NCBI BioSample. SAMN14911655
- 2020. BioSample of Deltaproteobacteria bacterium MAG_00792_naph_016. NCBI BioSample. SAMN14911656
- 2020. BioSample of Deltaproteobacteria bacterium MAG_09788_naph_37. NCBI BioSample. SAMN14911662
- 2020. BioSample of Deltaproteobacteria bacterium MAG_15370_dsfb_81. NCBI BioSample. SAMN14911665
- 2020. BioSample of Deltaproteobacteria bacterium MAG_17929_sntb_26. NCBI BioSample. SAMN14911666
- 2020. BioSample of Deltaproteobacteria bacterium MAG_17996_sntb_20. NCBI BioSample. SAMN14911670
- 2020. BioSample of Deltaproteobacteria bacterium MAG_22204_dsfv_001. NCBI BioSample. SAMN14911675
- 2020. BioSample of Gammaproteobacteria bacterium MAG_00150_gam_010. NCBI BioSample. SAMN14911649
- 2020. BioSample of Gammaproteobacteria bacterium MAG_00160_gam_009. NCBI BioSample. SAMN14911650
- 2020. BioSample of Gammaproteobacteria bacterium MAG_00172_gam_018. NCBI BioSample. SAMN14911651
- 2020. BioSample of Gammaproteobacteria bacterium MAG_00188_gam_006. NCBI BioSample. SAMN14911652
- 2020. BioSample of Gammaproteobacteria bacterium MAG_00212_gam_1. NCBI BioSample. SAMN14911653
- 2020. BioSample of Gammaproteobacteria bacterium MAG_00215_gam_020. NCBI BioSample. SAMN14911654
- 2020. BioSample of Magnetococcales bacterium MAG_21055_mgc_1. NCBI BioSample. SAMN14911672
- 2020. BioSample of Nitrospinae bacterium MAG_09705_ntspn_70. NCBI BioSample. SAMN14911661
- 2020. BioSample of Nitrospirae bacterium MAG_10313_ntr_31. NCBI BioSample. SAMN14911663
- 2020. BioSample of Desulfuromonadales bacterium MAG_21601_9_030. NCBI BioSample. SAMN14911674
- 2020. BioSample of Desulfuromonadales bacterium MAG_13126_9_058. NCBI BioSample. SAMN14911678
- 2020. BioSample of Desulfuromonadales bacterium MAG_21600_9_004. NCBI BioSample. SAMN14911673
- 2020. BioSample of Planctomycetes bacterium MAG_11118_pl_115. NCBI BioSample. SAMN14911664
- 2020. BioSample of Planctomycetes bacterium MAG_17991_pl_60. NCBI BioSample. SAMN14911669
- 2020. BioSample of Planctomycetes bacterium MAG_18080_pl_157. NCBI BioSample. SAMN14911671
- 2020. BioSample of Rhodospirillaceae bacterium MAG_04806_tlms_2. NCBI BioSample. SAMN14911657
- 2020. BioSample of Rhodospirillaceae bacterium MAG_05422_2-02_14. NCBI BioSample. SAMN14911658
- 2020. BioSample of Rhodospirillaceae bacterium MAG_05596_2-02_51. NCBI BioSample. SAMN14911659
- 2020. BioSample of Rhodospirillaceae bacterium MAG_06104_tlms_034. NCBI BioSample. SAMN14911660
- 2020. BioSample of Rhodospirillaceae bacterium MAG_22225_2-02_112. NCBI BioSample. SAMN14911676
- 2016. Assembly for unclassified Nitrospina Bin 25. IMG. 2651870060
- 2015. Assembly for Planctomycetes bacterium SCGC JGI090-P21. IMG Assembly. 2264265205
- 2017. Assembly for Omnitrophica bacterium SCGC_AG-290-C17. IMG Assembly. 3300015153
- 2017. Assembly for uncultured microorganism SbSrfc.SA12.01.D19. IMG Assembly. 3300022116
Supplementary Materials
Data Availability Statement
The genomes and metagenomes used during the current study are publicly available in NCBI (https://www.ncbi.nlm.nih.gov/)44,48,83–113 and IMG (https://img.jgi.doe.gov/cgi-bin/m/main.cgi)114–117 databases. Scaffolds of obtained MAGs could be found in Supplementary Table S6, hosted at figshare118. All data generated and analyzed in this study are also available in figshare118 and in supplementary information accompany this paper. Assembly of Rhodospirillaceae bacterium MAG_01419_mvb_30 could be found in RAST (https://rast.nmpdr.org/) using ‘guest’ as login and as password.
The following tools were used for the presented analysis and described in the main text:
Busybee web, Maxbin2, MyCC, and DAS Tool with standard parameters were used for the reconstruction of metagenome-assembled genomes (MAGs).
1. Busybee web https://ccb-microbe.cs.uni-saarland.de/busybee
2. Maxbin2 https://sourceforge.net/projects/maxbin2/
3. MyCC https://sourceforge.net/projects/sb2nhri/files/MyCC/
4. DAS Tool https://github.com/cmks/DAS_Tool
5. CheckM was used to estimate obtained genomes completeness and contamination https://github.com/Ecogenomics/CheckM
6. RefineM was used to remove contamination https://github.com/dparks1134/RefineM
7. QUAST helped to access quality metrics http://cab.spbu.ru/software/quast/
8. fastANI was used to calculate ANI https://github.com/ParBLiSS/FastANI
9. IDTAXA helped to obtain taxonomic assignments for the studied genomes 16S rRNA genes http://www2.decipher.codes/Classification.html
10. GTDB-Tk was used to find 120 single-copy bacterial marker protein sequences, to construct their multiple alignments and to get the taxonomic assignment using the GTDB r86 database https://github.com/Ecogenomics/GTDBTk
11. MAFFT was used for aligning amino acid sequence sets of the MamA, -B, -M, -K, -P, and -Q proteins https://mafft.cbrc.jp/alignment/server/
12. Gblocks helped to curate sequences aligned in MAFFT http://molevol.cmima.csic.es/castresana/Gblocks_server.html
13. Phylogenetic trees were inferred with IQ-TREE http://www.iqtree.org/
14. Obtained trees were visualized with iTOL https://itol.embl.de/