Abstract
The emergence and spread of cryptococcosis caused by the Cryptococcus gattii species complex has become a major public concern worldwide. C. deuterogattii (VGIIa) outbreaks in the Pacific Northwest region demonstrate the expansion of this fungal infection to temperate climate regions. However, infections due to the C. gattii species complex in China have rarely been reported. In this study, we studied eleven clinical strains of the C. gattii species complex isolated from Guangxi, southern China. The genetic identity and variability of these isolates were analyzed via multi-locus sequence typing (MLST), and the phylogenetic relationships among these isolates and global isolates were evaluated. The mating type, physiological features and antifungal susceptibilities of these isolates were also characterized. Among the eleven isolates, six belonged to C. deuterogattii, while five belonged to C. gattii sensu stricto. The C. deuterogattii strains from Guangxi, southern China were genetically variable and clustered with different clinical isolates from Brazil. All strains were MATα, and three C. deuterogattii isolates (GX0104, GX0105 and GX0147) were able to undergo sexual reproduction. Moreover, most strains had capsule and were capable of melanin production when compared to the outbreak strain from Canada. Most isolates were susceptible to antifungal drugs; yet one of eleven immunocompetent patients died of cryptococcal meningitis caused by C. deuterogattii (GX0147). Our study indicated that the highly pathogenic C. deuterogattii may be emerging in southern China, and effective nationwide surveillance of C. gattii species complex infection is necessary.
Author summary
Cryptococcosis is a fatal systemic fungal disease caused by Cryptococcus neoformans/gattii species complexes. As a former member of the C. neoformans, C. gattii had been easily neglected before being elevated to species level. Human C. gattii species complex infection was previously confined to the tropical and subtropical regions worldwide. However, in 1999, an outbreak of C. gattii species complex occurred on Vancouver Island in Canada then expanded to the Pacific Northwest in the USA, causing over 200 infections. The highly virulent, highly pathogenic and more resistant to antifungal drugs of this species have become a therapeutic problem. To initiate a better understanding of the infection characteristics and pathogenicity of C. gattii species complex in Guangxi, southern China, the current study aimed to characterize the C. gattii species complex isolates genetically and phenotypically. The ISHAM consensus MLST scheme was utilized to investigate the genetic structure of C. gattii species complex and to correlate their geographic origin, clinical source, virulence factors and antifungal susceptibility. The authors expect that this work can support surveillance and encourage more research and public health initiatives to prevent and control the cryptococcosis cause by C. gattii species complex.
Introduction
Cryptococcus is a life-threatening fungal pathogen of humans and animals[1]. The infection process of Cryptococcus is usually via the inhalation of airborne spores (or yeast cells) into the respiratory tract and their subsequent dissemination to the central nervous system, causing pulmonary cryptococcosis and cryptococcal meningoencephalitis[1, 2]. During the past two decades, considerable genetic heterogeneity has been demonstrated to occur in the C. neoformans/gattii species complexes by a plethora of molecular methods. Various molecular biological techniques have been used to study the epidemiology and population structure of the Cryptococcus gattii/neoformans species complexes, including random amplification of polymorphic DNA (RAPD) analysis, PCR fingerprinting, amplified fragment length polymorphism (AFLP) analysis, multilocus microsatellite typing (MLMT) analysis and multi-locus sequence typing (MLST) analysis[3]. Recently, next-generation sequencing (NGS) technology has been utilized to investigate the epidemiology of C. gattii isolates[4]. However, the identification of the species is commonly based on the MLST protocol standardized by the International Society for Human and Animal Mycology (ISHAM)[5]. The MLST scheme has become an important tool for the characterization of the population genetic structure of the Cryptococcus species. Since the taxonomy of the polyphyletic genus Cryptococcus has been thoroughly revised, two varieties of C. neoformans have been recognized as species: C. neoformans (formerly C. neoformans variety grubii) and C. deneoformans (formerly C. neoformans variety neoformans)[6–8]. However, the molecular types of C. gattii species complex have been elevated to the species level as C. gattii sensu stricto (AFLP4/VGI), C. deuterogattii (AFLP6/VGII), C. bacillisporus (AFLP5/VGIII), C. tetragattii (AFLP7/VGIV) and C. decagattii (AFLP10/VGIV)[7]. The C. deuterogattii subtype (AFLP6A/VGIIa, AFLP6B/VGIIb and AFLP6C/VGIIc) caused an outbreak in the Pacific Northwest (PNW) region of Canada and the United States[9, 10].
Cryptococcosis is a global and invasive systematic mycosis caused by C. gattii/neoformans species complexes, leading to morbidity and mortality in both immunocompetent and immunocompromised individuals, such as those with acquired immune deficiency syndrome (AIDS) or those undergoing organ transplantation[11]. Infections due to C. neoformans species complex occur worldwide while cryptococcosis caused by C. gattii species complex was traditionally considered an endemic disease, and associated with tropical and subtropical climates[11]. However, the outbreaks of C. deuterogattii in humans and a wide range of mammals in Vancouver Island and the PNW of the USA demonstrates that the fungus has adapted to environments beyond the endemic (sub)tropical regions[9, 10]. The source of infection is usually traced back to Eucalyptus and other species of trees, such as Ficus spp. and Terminalia spp. (almond) trees[12, 13].
In Asia, the first case of C. gattii species complex infection was reported by Wanqing Liao in 1980 by the strain formerly named as S8012 and now identified as C. gattii s.s.[14]. C. gattii s.s. is the most frequently encountered species worldwide[3], and only a few sporadic cases of C. deuterogattii infection have been reported in Asian countries such as India, Thailand, and Malaysia[15]. In 2007, Okamoto et al. reported the first case of cryptococcosis caused by a highly virulent C. deuterogattii subgenotype (AFLP6A/VGIIa) in Japan[16]. However, to our knowledge, reports of C. gattii species complex infection in China are limited. A study in 2008 showed that nine of 115 (7.8%) clinical Cryptococcus isolates were members of the C. gattii species complex, consisting of eight strains of C. gattii s.s. and one strain of C. deuterogattii[17]. Several sporadic C. deuterogattii infections have also been reported in mainland China, mainly in southern China[18, 19]. However, the genetic identity and variability of these isolates and the phylogenetic relationships among these clinical isolates and global isolates have yet to be thoroughly investigated.
In this study, we described eleven cases of cryptococcosis caused by C. gattii species complex infections between 2014 and 2018 in Guangxi, southern China. We determined their genotypes by MLST and studied their phylogenetic relationships with the global strains. We also characterized the mating type, physiological characteristics, virulence factors and antifungal susceptibility of these clinical isolates.
Materials and methods
Ethics statement
This study was approved by the Medical Ethics Committee of First Affiliated Hospital of Guangxi Medical University. All participants were adults. The clinical data in this study was obtained with the written consent of the patient or the patient’s family and data collected concerning them was anonymized.
Selection of C. gattii species complex from clinical isolates
To study the epidemiological characteristics of cryptococcosis caused by C. gattii species complex in Guangxi, southern China, all the clinical strains of Cryptococcus spp. stored in the Fungal Diseases Survey Center of Guangxi, the First Affiliated Hospital of Guangxi Medical University Guangxi, southern China, have been evaluated. In total, one hundred and twenty Cryptococcus strains were isolated from patients with clinically confirmed cryptococcosis between 2014 and 2018. All but three were isolated from the patients hospitalized in First Affiliated Hospital of Guangxi Medical University, which is the largest tertiary care hospital in the Guangxi Autonomous Region in southern China and has 2750 beds. Two strains were from The Fourth People's Hospital of Nanning and one strain was from the People's Hospital of Guangxi Zhuang Autonomous Region. Among the 120 strains, most of them were from incident patients, except one (GX0049) was a relapse patient. Six strains isolated from the bronchoalveolar lavage fluid (BALF), three from lung tissue and two from skin tissue; the other strains isolated from CSF. The data of the patients was collected using a standardized form that was based entirely on the medical reports of each patient, including demographic information (age and gender), domiciles, birth and development details, medical history, clinical manifestations, laboratory data, imaging changes, diagnoses, treatments, and prognoses.
All strains were subcultured on Sabouraud Dextrose Agar (SDA) plates and incubated at 25°C, and then transferred onto L-Canavanine-glycine-bromothymol blue (CGB) medium for three to seven days to differentiate C. neoformans from C. gattii species complex as previously described [20]. In the CGB medium, eleven isolates had a positive reaction and turned the medium blue, while the rest of isolates failed to produce a color change, which were further identified as C. neoformans var. grubii by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). Among the eleven strains of C. gattii species complex, ten strains were isolated from CSF, and one strain was isolated from lung tissue.
Reference Cryptococcus strains R265 (AFLP6A/VGIIa) and H99 (AFLP1/VNI) were acquired from Shanghai Key Laboratory of Molecular Medical Mycology, Changzheng Hospital, Second Military Medical University, Shanghai, China. MATa strains (AFLP2/VNIV, AFLP1/VNI and AFLP6/VGII) were obtained from State Key Laboratory of Mycology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China.
MLST and phylogenetic analysis
Isolates were cultured on SDA for 72 h prior to DNA extraction. Genomic DNA was extracted with the NuClean Plant Genomic DNA Kit (CWBIO, Beijing, China). Based on the ISHAM consensus MLST scheme, seven loci including capsule polysaccharide (CAP59), glycerol 3-phosphate dehydrogenase, (GPD1), laccase (LAC1), the intergenic spacer (IGS1) region, phospholipase B1 (PLB1), superoxide dismutase (SOD1), and orotidine monophosphate pyrophosphorylase (URA5) genes were amplified and sequenced[5]. The alleles were analyzed, and the STs were determined based on the MLST database (http://mlst.mycologylab.org). The sequences of the seven MLST loci have been deposited in the MLST database and GenBank.
Phylogenetic analysis of the C. gattii species complex was performed based on the alignment of seven concatenated nucleotide sequences (CAP59, GPD1, LAC1, IGS1, PLB1, SOD1 and URA5)[21]. The genetic relationships among these Chinese clinical strains and strains in different countries[21–23] were investigated by MEGA7[24]; a phylogenetic tree was generated using the maximum likelihood method with a bootstrap analysis using 1000 replicates[24–26]. Principal component analysis (PCA) was performed with the Adegenet 2.1.1 package for software R (version 3.4.4) to explore the genetic relationships and geographic patterns of the strains[27].
Mating type determination and physiological analysis
Mating types were determined by using specific primers targeting the MFα and MFa pheromone genes as previously described[28]. Mating experiments were performed by pairing eleven clinical strains and two reference strains (MATα; AFLP6A/VGIIa R265 and AFLP1/VNI H99) with three strains of the opposite mating type (MATa; AFLP2/VNIV, AFLP1/VNI and AFLP6/VGII) on V8 medium at 25°C in darkness for two weeks. The experiment was carried out twice, and the formation of hyphae and sexual structures was observed and investigated under a microscope.
Since certain strains were able to mate in vitro, we also studied and compared their virulence factors to the standard reference strain (R265). Melanin production, and capsule formation were evaluated using slightly modified protocols that were published previously[29–31]. Visual analysis of melanin production was performed on caffeic acid agar; strains were grown on agar plates incubated at 30°C and 37°C for 72 h and observed the appearance of brown yeast colonies. Capsule formation was induced with RPMI-1640 medium for 72 h at 37°C and 5% CO2. The capsule size of at least 100 cells was quantified by light microscopy using encapsulated to naked yeast size (cell wall to cell wall diameter) ratios.
Antifungal susceptibility testing
Antifungal agents fluconazole (FLC), fluorocytosine (5FC), amphotericin B (AMB), itraconazole (ITC), voriconazole (VOR), posaconazole (POS) and isavuconazole (ISA) were used in the susceptibility tests. The broth microdilution method was performed following the CLSI M27-A3 guidelines[32] to assess the antifungal susceptibility of all clinical isolates in vitro. The concentration ranges were 0.125–64 μg/mL for FLC, 0.008–4 μg/mL for 5FC, 0.016–8 μg/mL for AMB and 0.002–1 μg/mL for ITC, VOR, POS and ISA. The MIC values were recorded after incubation at 37°C for 72 h. The MIC of AMB was defined as the lowest concentration of drug showing no yeast growth, while the MICs for other antifungal agents were defined as low drug concentrations that caused a prominent reduction in growth (≥50%) compared with the drug-free growth control. Since there was no established breakpoints standard of antifungal drugs for Cryptococcus, therefore, epidemiological cutoff values (ECVs) has been offered to determine whether a strain is wild type (in vitro susceptible) or non-wild type (in vitro resistant). Based on the previous recommendation[33–35], the ECVs of C. gattii s.s. for FLC were 8 μg/mL; 4 μg/mL for 5FC; 0.5 μg/mL for AMB, ITC, VOR and POS; and 0.25 μg/mL for ISA. The ECVs of C. deuterogattii were 32 μg/mL for FLC; 16 μg/mL for 5FC; 1 μg/mL for AMB; 0.5 μg/mL for ITC and POS; and 0.25 μg/mL for VOR and ISA[33–35]. Candida parapsilosis ATCC22019 was used for quality control strain.
Statistical analysis
Statistical analysis was performed using SPSS 17.0 (SPSS Inc., Chicago, IL, USA). The p values of the relative capsule size data were calculated using one-way ANOVA statistical analysis. p < 0.05 was considered statistically significant.
Accession numbers
MLST nucleotide sequences for the eleven clinical isolates determined in this study have been deposited in the GenBank nucleotide sequence database under accession numbers
Results
Clinical characteristics of C. gattii species complex cases
The clinical manifestations and medical histories of the 11 patients are summarized in Table 1. A total of ten cryptococcal meningitis (CM) patients and one pulmonary cryptococcosis (PC) patients were enrolled. The population included eight males and three females, with a mean age of 38.0±7.8 years (range 29–54 years). Among ten CM patients, the common clinical manifestation was headache, fever, vomiting and nausea. All of the ten CM patients were positive for CSF ink staining and nine of them were positive for CSF cryptococcal antigen latex agglutination test (titer 1:16–1:1024). Chest pain was the most obvious symptom in the patient with PC (GX0717). Chest CT showed a nodular shadow (0.6 cm in diameter) in the right lower lobe and multiple patchy shadow of the lungs; pathologic examination of the lung tissue biopsy demonstrated the cryptococcal yeast forms. Only one patient (GX0080) was previously diagnosed as systemic lupus erythematosus (SLE) and received corticosteroid treatment. Whereas, no significant immune abnormalities was detected in other patients.
Table 1. Epidemiological and clinical characteristics of eleven patients with Cryptococcus gattii species complex infections in Guangxi, southern China*.
| Case no. | Age,y/ gender | Occupation | Clinical syndrome | Physical signs | Specimen cultured | Latex agglutination test | Clinical diagnosis | Treatment | Outcome |
|---|---|---|---|---|---|---|---|---|---|
| GX0049 | 37/M | Worker | Convulsion, altered consciousness, limb weakness, fever | Meningeal irritation and pathological reflex positive | CSF | 1:64 | CM | AMB(1275mg)+FLC(13.2g)+5FC(270g) | Survived |
| GX0079 | 40/M | Truck driver | Headache, vomiting, blurred vision | Neck stiffness | CSF | 1:256 | CM | Unknown | Survived |
| GX0080 | 31/F | Farmer | Fever, headache, nausea, vomiting, chills | Normal | CSF | 1:16 | CM | AMB(2035mg)+FLC(36.4g) | Survived |
| GX0158 | 54/F | Farmer | Headache, nausea, vomiting, fever | Normal | CSF | 1:128 | CM | AMB(2580mg)+FLC(22g)+5FC(299.5g) | Survived |
| GX1622 | 29/M | Farmer | Headache, vomiting, fever, blurred vision | Normal | CSF | 1:80 | CM | AMB(740mg)+FLC(8g) | Survived |
| GX0104 | 30/M | Farmer | Headache, nausea, vomiting, fever | Normal | CSF | 1:1024 | CM | AMB(1800mg)+FLC(39.6g)+5FC(52g) | Survived |
| GX0105 | 29/F | Unknown | Headache, fever, dizzy, chills | Neck stiffness, Kernig’s sign positive | CSF | 1:64 | CM | AMB(1625mg)+FLC(26.4g)+5FC(248g) | Survived |
| GX0147 | 44/M | Farmer | Headache, fever, chills | Neck stiffness | CSF | 1:1024 | CM | AMB(189mg)+FLC(5g)+5FC(30g) | Died |
| GX0476 | 42/M | Unknown | Headache, fever, cough | Normal | CSF | None | CM | Unknown | Survived |
| GX0903 | 36/M | Aquiculture | Headache, dizzy, nausea, vomiting, fever | Neck stiffness | CSF | 1:32 | CM | AMB(2200mg)+FLC(20.8g)+5FC(278g) | Survived |
| GX0717 | 47/M | Civil servant | Chest pain, cough, expectoration | Normal | Lung tissue | None | PC | AMB(140mg)+FLC(2.4g) | Survived |
*Cerebrospinal fluid, CSF; CM, Cryptococcal meningitis; PC, Pulmonary cryptococcosis
According to Infectious Diseases Society of America (IDSA) guidelines and China's expert consensus on the diagnosis and treatment of cryptococcal meningitis[36, 37], most of the patients have been significantly relieved after the cryptococcosis induction period treatment with combination therapy of two (AMB and FLC) or three (AMB, FLC and 5FC) antifungal drugs. Median cumulative dose and duration were 1625 mg (range 140–2580 mg) and 47 days (range 5–63 days) for AMB, 259 g (range 30–299.5 g) and 45.5 days (range 5–63 days) for 5FC, 20.8 g (range 2.4–39.6 g) and 50 days (range 6–66 days) for FLC. Even after the infection was treated and discharged from hospital, most patients continued the antifungal treatment with 0.2 g-0.4 g/day of FLC. Despite antifungal treatment, one patient died (GX0147) after giving up treatment because of no significant improvement. No patient had travel history outside China.
Identification of sequence type
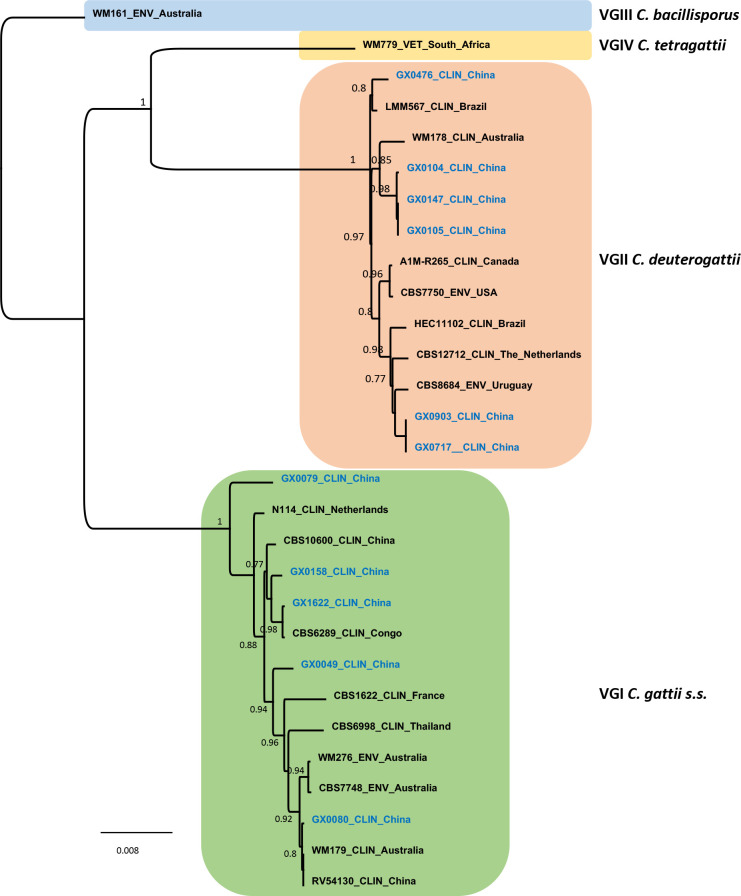
MLST analysis of the eleven isolates indicated that five isolates were C. gattii s.s., and the other six isolates belonged to C. deuterogattii (Figs 1 and 2, Table 2). Five and three different sequence types (STs) were detected in C. gattii s.s. and C. deuterogattii, respectively; essentially all C. gattii s.s. isolates were genetically different and represented by a single isolate when MLST sequences were analyzed (Fig 1); sample GX0079 was quite divergent from the selected AFLP4/VGI strains. In contrast, C. deuterogattii isolates were divided into three major groups, with ST169 and ST129 represented by more than one isolate (Table 2).
Fig 1. Phylogenetic analysis of eleven C. gattii species complex.
Phylogenetic relationships inferred from a maximum likelihood analysis of CAP59, GPD1, LAC1, IGS1, PLB1, SOD1 and URA5 sequences of eleven C. gattii species complex from Guangxi, southern China (in blue) and 18 reference strains, covering the four major molecular types in C. gattii species complex. The branches with bootstrap support higher than 70% are shown.
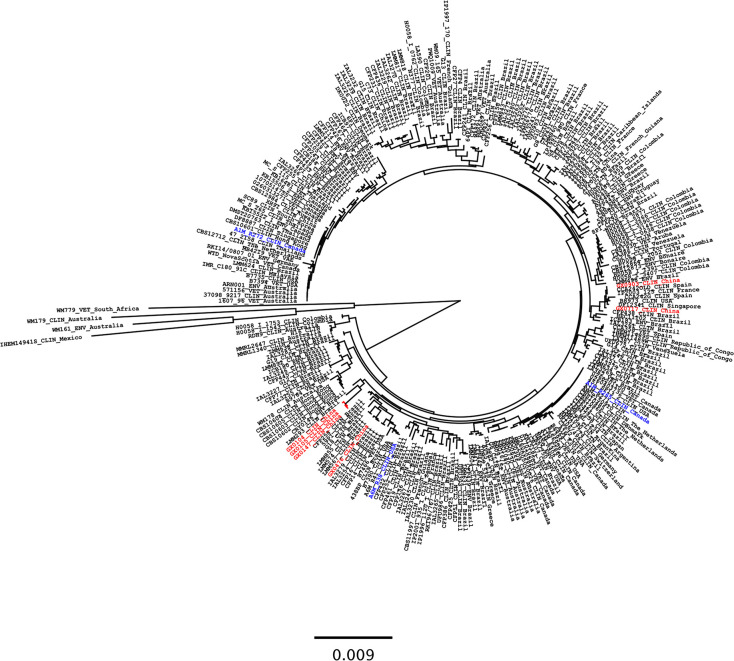
Fig 2. Phylogenetic analysis of C. deuterogattii isolates.
The figure showing the relationships between the Chinese (strains of current investigation in red) and global C. deuterogattii strains inferred from the maximum likelihood analysis based on the combined ISHAM consensus MLST loci using MEGA7. Standard reference strains C. gattii s.s. (WM179), C. bacillisporus (WM161), C. tetragattii (WM779) and C. decagattii (IHEM14941) were used as outgroups. Epidemiologically significant isolates from the Pacific Northwest outbreaks were in blue. The branches with bootstrap support of more than 70% are indicated in bold.
Table 2. The STs and mating types of clinical Cryptococcus gattii species complex isolates in Guangxi, southern China*.
| Strains | Allele type | ST | Molecular type | Mating type | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CAP59 | GPD1 | IGS1 | LAC1 | PLB1 | SOD1 | URA5 | ||||
| GX0049 | 36 | 11 | 13 | 5 | 13 | 36 | 14 | 106 | VGI | α |
| GX0079 | 49 | 11 | 59 | 13 | 13 | 71 | 24 | 227 | VGI | α |
| GX0080 | 16 | 5 | 3 | 5 | 5 | 32 | 12 | 51 | VGI | α |
| GX0158 | 36 | 11 | 83 | 13 | 13 | 47 | 15 | 222 | VGI | α |
| GX1622 | 53 | 11 | 13 | 13 | 13 | 68 | 15 | 232 | VGI | α |
| GX0104 | 40 | 35 | 57 | 4 | 1 | 59 | 2 | 129 | VGII | α |
| GX0105 | 8 | 6 | 25 | 4 | 1 | 16 | 6 | 129 | VGII | α |
| GX0147 | 40 | 35 | 57 | 4 | 1 | 59 | 2 | 129 | VGII | α |
| GX0476 | 2 | 35 | 57 | 4 | 1 | 104 | 2 | 309 | VGII | α |
| GX0903 | 2 | 25 | 26 | 21 | 9 | 8 | 7 | 169 | VGII | α |
| GX0717 | 2 | 25 | 26 | 21 | 9 | 8 | 7 | 169 | VGII | α |
*ST, sequence type
Genetic analysis of the Chinese and global C. deuterogattii isolates indicated that four Chinese isolates (GX0104, GX0105, GX0147 and GX0476) clustered closely with the C. deuterogattii AFLP6/VGII strains from Brazil (LMM293 and ILA3279), while the clinical isolates GX0903 and GX0717 clustered in a group containing C. deuterogattii AFLP6/VGII strains from Spain (CCA242OLD), Singapore (DF12341), the USA (B8973) and France (IP2003/125) (Fig 2). These isolates did not cluster with the highly virulent C. deuterogattii strains (AFLP6A/VGIIa R265 and AFLP6C/VGIIc A6M-R38) reported in the PNW outbreaks in Canada and the USA, but isolate GX0476 clustered in a group forming a sister relationship with the group harboring AFLP6C/VGIIc (A6M-R38) from Oregon, the USA.
Comparison of Chinese and global isolates
PCA (based on ISHAM-MLST) was also used to assess the genetic relationship between Chinese and global isolates (S1 Fig). Approximately 41.8% of the genetic variation can be explained in C. gattii s.s. (PC1 30.0%, PC2 11.8%) (S1A Fig), and 35% of the variation was explained in C. deuterogattii (PC1 17.9%, PC2 17.1%) (S1B Fig). The PCA did not group C. gattii species complex strains according to their origin, which was consistent with the results of the phylogenetic analysis (S1 Fig). Among the C. deuterogattii isolates, the four C. deuterogattii isolates appeared to be originated from South America.
Physiological characterization
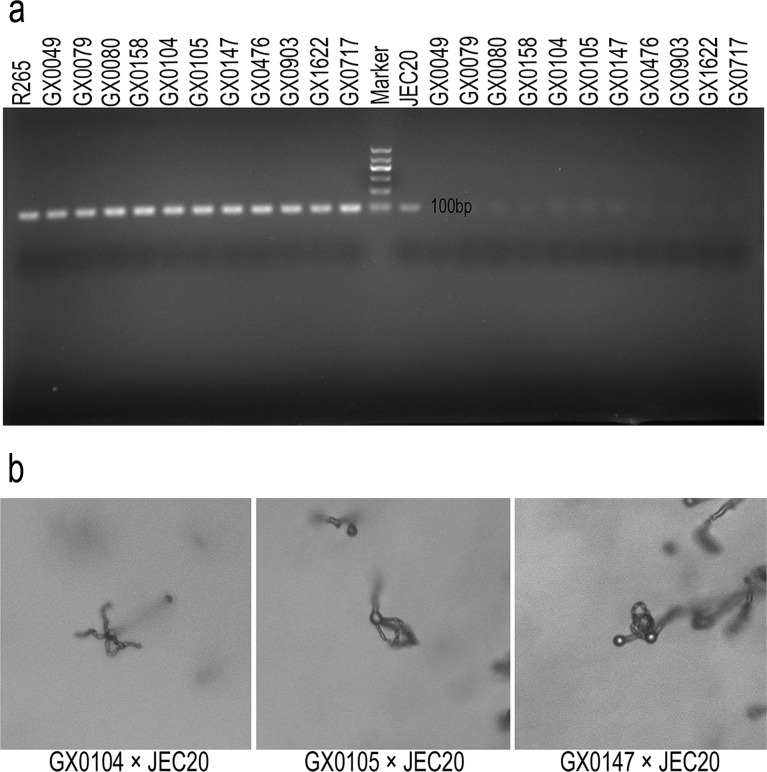
All of the clinical isolates analyzed were MATα (Fig 3A). Based on the crossing experiment with JEC20 (AFLP2/VNIV), three C. deuterogattii strains (GX0104, GX0105 and GX0147) were able to undergo sexual reproduction in vitro, and basidia and basidiospores were formed during sexual reproduction (Fig 3B). Two C. deuterogattii isolates (GX0105 and GX0147) exhibited a weak mating response, with only a few filaments, basidia and basidiospores developed.
Fig 3.
(A) Simultaneous amplification of all clinical strains and two reference strains using primers targeting the MATα and MATa genes and (B) mating reactions of the isolates showing typical basidia and basidiospores. (A) Reference strains: R265 (MATα, AFLP6A/VGIIa) and JEC20 (MATa, AFLP2/VNIV). Clinical isolates were all MATα. (B) Sexual reproduction cultures including hyphae, basidia and basidiospores, incubated on V8 agar (pH = 5) at 25°C for 2 weeks in the dark.
The tested clinical isolates produced melanin at both 30°C and 37°C, while most strains had greater melanin production at 30°C than at 37°C (S2A Fig). Two C. deuterogattii strains (GX0104 and GX0903) appeared to have greater melanin production than the reference strain (AFLP6A/VGIIa R265) at 37°C. Moreover, these clinical strains produced capsule of different size in vitro (S2B Fig); however, our data revealed no significant difference (p > 0.05) in the capsule size between GX0104 and R265 (S2C Fig).
Antifungal drug susceptibility
As the MICs shown in Table 3, none of the strains demonstrated resistance to the antifungal drugs. The susceptibility ranges of the five C. gattii s.s. isolates were 1–4 μg/mL for FLC, 0.125–0.5 μg/mL for 5FC, 0.25–0.5 μg/mL for AMB, 0.125–0.25 μg/mL for ITC, 0.0156–0.125 μg/mL for VOR, 0.0156–0.25 μg/mL for POS, and 0.0078–0.125 μg/mL for ISA. For the six C. deuterogattii isolates, the susceptibility ranges were 1–16 μg/mL for FLC, 0.0625–1 μg/mL for 5FC, 0.25–1 μg/mL for AMB, 0.0625–0.25 μg/mL for ITC, 0.0156–0.125 μg/mL for VOR, 0.0156–0.25 μg/mL for POS, and 0.0078–0.125 μg/mL for ISA. According to the clinical efficacy, combination of two or three drugs in AMB, FLC, and 5FC were effective for treatment. There were no significant differences between the C. gattii s.s. and C. deuterogattii isolates against these seven antifungal drugs (p > 0.05).
Table 3. Minimal inhibitory concentrations (MICs) of all the Cryptococcus gattii species complex isolates in Guangxi, southern China*.
| Strains | Minimal inhibitory concentration (μg/mL) | ||||||
|---|---|---|---|---|---|---|---|
| FLC | 5FC | AMB | ITC | VOR | POS | ISA | |
| GX0049 | 1 | 0.5 | 0.25 | 0.125 | 0.0313 | 0.0625 | 0.0313 |
| GX0079 | 2 | 0.125 | 0.5 | 0.25 | 0.0625 | 0.0625 | 0.0313 |
| GX0080 | 1 | 0.25 | 0.5 | 0.25 | 0.0156 | 0.0156 | 0.0078 |
| GX0158 | 2 | 0.125 | 0.25 | 0.125 | 0.0313 | 0.0625 | 0.0313 |
| GX1622 | 4 | 0.5 | 0.5 | 0.25 | 0.125 | 0.25 | 0.125 |
| GX0104 | 1 | 0.0625 | 1 | 0.0625 | 0.0156 | 0.0156 | 0.0078 |
| GX0105 | 4 | 0.5 | 0.5 | 0.25 | 0.0625 | 0.0625 | 0.0625 |
| GX0147 | 4 | 1 | 0.5 | 0.25 | 0.0625 | 0.0625 | 0.0625 |
| GX0476 | 16 | 0.5 | 0.5 | 0.125 | 0.0625 | 0.25 | 0.125 |
| GX0903 | 2 | 0.25 | 0.25 | 0.25 | 0.0625 | 0.125 | 0.125 |
| GX0717 | 4 | 0.5 | 0.5 | 0.25 | 0.125 | 0.125 | 0.125 |
| MIC range | 1–16 | 0.125–1 | 0.25–1 | 0.0625–0.25 | 0.0156–0.125 | 0.0156–0.25 | 0.0078–0.125 |
| MIC50 | 2 | 0.25 | 0.5 | 0.25 | 0.0625 | 0.0625 | 0.0625 |
| MIC90 | 4 | 0.5 | 0.5 | 0.25 | 0.125 | 0.25 | 0.125 |
| Geometric mean | 2.5733 | 0.2836 | 0.4695 | 0.1824 | 0.0486 | 0.0709 | 0.0456 |
* FLC, fluconazole; 5FC, fluorocytosine; AMB, amphotericin B; ITC, itraconazole; VOR, voriconazole; POS, posaconazole; ISA, isavuconazole; MIC, minimal inhibitory concentration
Discussion
Our current study showed that C. gattii species complex infection in Guangxi, southern China were caused by both C. gattii s.s. (AFLP4/VGI) and C. deuterogattii (AFLP6/VGII), with different MLST genotypes and virulence factor characteristics. Similar pattern has also been reported in other countries such as Canada, Brazil, Australia and the USA[29–31]. Although C. deuterogattii in this study was genetically indistinguishable from the outbreak genotypes AFLP6A/VGIIa (R265) and AFLP6C/VGIIc (A6M-R38), they were related to other global C. deuterogattii isolates isolated from Brazil (LMM293 and ILA3279), Singapore (DF12341) and Spain (CCA242OLD). C. deuterogattii strains are genetically diverse, and there was great genotypic variability among C. deuterogattii subtypes (AFLP6A/VGIIa and AFLP6C/VGIIc), particularly those in Brazil[38]. Despite the small sample size in this study, most Chinese clinical strains were genetically diverse, with eight STs reported. The variability could be due to special ecological interactions, adaptations and evolutionary mechanisms. Cryptococcus species are able to adapt to different ecological niches and temperatures and they can adapt within the host and environment through microevolution. The Cryptococcus genome is also dynamic in its plasticity[39].
In the present study, we found that all isolates harbored the MATα gene at the mating type locus, and three of six C. deuterogattii strains (GX0104, GX0105 and GX0147) were able to undergo sexual reproduction, thus recombination could be one of the causes for the genetic diversity among the Chinese C. deuterogattii strains. Moreover, offspring produced by sexual reproduction/recombination may be able to adapt to and colonize new environment[40, 41]. In the mating assays of eleven clinical strains with the opposite mating type strains (AFLP6/VGII, AFLP2/VNIV and AFLP1/VNI), only the cross of AFLP6/VGII × AFLP2/VNIV exhibited mating response. Hybridization between the two cryptococcal species complexes seems to be a much rarer event in nature compared to hybridization between C. neoformans species complex lineages[42]. Even though the mating between the parental lineages can be induced in the laboratory, their progenies were not commonly reported in the environmental and clinical samples[43]. Recombination may also occur between the AFLP4/VGI and AFLP6/VGII isolates in China because previous research has highlighted possible gene transfer (introgression) between different C. gattii s.l. clades, either bisexually or unisexually, thereby contributing to the production of virulence subtypes[41, 44]. Sex can contribute to de novo diversity[45], which may be the reason for the diversity in the MLST analysis of clinical C. deuterogattii isolates. In addition, the capacity for mating could generate additional infectious propagules, leading to increased exposure and, ultimately, an enhanced infection rate[31].
Clinical data from this study showed that almost all patients had no obvious immunodeficiency, except one patient with SLE, which was consistent with the fact that the C. gattii species complex causes infection in immunocompetent hosts[11]. Most strains in Guangxi, southern China had the capabilities to produce melanin and capsule, as well as to undergo sexual reproduction. These are major virulence factors known to contribute to Cryptococcus pathogenesis and have been well characterized in C. gattii species complex. However, very few past studies have investigated these virulence factors in Chinese strains. Our data showed that most Chinese strains, similar to the reference outbreak strain R265 from Canada, have the abilities to be virulent. However, animal model studies need to be performed in order to establish their virulence levels to human hosts and whether their virulence would be comparable to the highly virulent outbreak (R265) and less virulent strain (R272) from the PNW[29].
Most patients significantly relieved after combination therapy while one patient (GX0147) passed away after giving up treatment because of no significant improvement. However, antifungal susceptibility testing indicated that strains in this study are not resistant to antifungal drugs because their MIC values were within the susceptible range[33–35]. Therefore, although none of the strains demonstrated resistance to these drugs, other factors may complicate the therapeutic outcomes of these patients.
Cryptococcus can be dispersed through the movement of trees and wood products, air currents, water currents, and biotic sources, such as birds, animals and insects[13]. Unlike C. neoformans species complex, which is found commonly in soils contaminated with wild and pet bird droppings, especially pigeon droppings, C. gattii species complex prefer to grow within and underneath moist bark, tree hollows, tree trunks and in soil debris near specific trees that are often found in nature (including Eucalyptus, Azadirachta, Castanopsis, Prunus dulcis, Pinus canariensis and Pseudotsuga menziesii)[46]. The evidence for C. gattii species complex dispersal by wind and air currents is limited, but fungal isolations from air samples have been obtained around positive trees in Canada and India[13]. “Guangxi Autonomous Region” belongs to southern China and lays on the southeastern corner of the Yunnan-Guizhou Plateau, situated from 20º54′N to 26º24′N and from 104º26′E to 112º04′E. This region borders Vietnam to the southwest and is surrounded by Guangdong, Guizhou, Yunnan, and Hunan Provinces in China. The region has a terraced topography sloping from the northwest to the southeast, with hilly land constituting 85% of its total area and plains constituting 15%. The region has a subtropical humid monsoon climate, with average daily temperatures of 16°C to 23°C. The rainy season lasts from April until September, with an annual rainfall of 1500 mm to 2000 mm[47]. The warm, subtropical monsoon climate in Guangxi is also favorable for the growth and reproduction of the C. gattii species complex. In this study, most patients were farmers in rural areas and had a history of contact with above plants and soil, especially Eucalyptus, but without travel record to the epidemic areas such as the PNW region, or the endemic area of C. gattii species complex such as South America and Australia, indicating that their cryptococcosis infection was spread through contact with the local environment. Frequent and regular contact with the natural environment may explain how these patients acquired the infections[48]. The genetic variabilities of Chinese C. gattii species complex members also suggested the possibility of multiple independent origins. Combined with the phylogenetic analysis of this study, some Chinese strains were related to those from Brazil. Therefore, this may be traced to the wood/seedlings imported from elsewhere, such as South America and Australia[49]. However, additional isolation and investigation of the C. gattii species complex from the environment such as trees, soil and wild animals is necessary to establish the environment as the source of infection in Guangxi Province, China.
Cryptococcosis infections caused by the C. gattii species complex are relatively rare in comparison to those caused by the C. neoformans species complex (11.4% versus 88.6%)[15]; however, recent studies have indicated that they may be mis- or underdiagnosed globally[50]. Similarly, the number of C. gattii species complex infections in China may be underestimated. Compared with the data from other provinces in China[14, 51], the rate of C. gattii species complex infection in Guangxi was approximately 9% (11/120), and most infection were caused by C. deuterogattii; however, additional epidemiological and surveillance studies are required.
There are several limitations of this study. First, a small number of patients were included in the study. Second, due to the retrospective nature of study, there was no long-term follow-up assessment of these patients, which have led to some missing data. Third, the level of virulence among these strains still needs further study on animal models. Fourth, environmental isolates should be sampled to assess the relationship of environmental niche for C. gattii species complex in this area. Thus, performing a prospective study with a larger study population are expected to elucidate the population structure and mechanisms of C. gattii species complex. Presently, we are in collaboration with other hospitals to carry out regional surveillance of Cryptococcus infections in China.
In summary, the C. gattii species complex should receive substantial attention in China due to its genetic variability, ability to infect immunocompetent hosts and propensity to undergo sexual reproduction and cause outbreaks, even the number of infections was low. Given that all patients in this investigation may have acquired the infection from nature, the environmental distribution, genetic variability and virulence level of the C. gattii species complex should not be underestimated. At the same time, we strive to improve the differential diagnosis of the C. gattii species complex in the early stages of infection and to use targeted treatment programs to reduce the risk of infection.
Supporting information
(a) and C. deuterogattii (b) between Guangxi, southern China, and global isolates illustrated by principal component analysis (PCA).
(PDF)
(a)Visual analysis of melanin production after fungal growth on caffeic acid agar at 30°C and 37°C for three days. (b) Polysaccharide capsule surrounding the cells of C. deuterogattii isolates under microscopy. (c) Capsule production test in RPMI-1640 with 5% CO2 at 37°C representing the average capsule-capsule:cell wall-cell wall ratio of the six clinical isolates and the reference strain (AFLP6A/VGIIa R265). (X400, p < 0.001) (error bars ± SE = 2 SE).
(PDF)
Acknowledgments
We acknowledge all microbiologists at the First Affiliated Hospital of Guangxi Medical University for technical assistance and the isolation of Cryptococcus cultures. We also thank Ferry Hagen (Westerdijk Fungal Biodiversity Institute, the Netherlands) for discussion and sharing the sequence alignment of global C. deuterogattii (AFLP6/VGII) isolates.
Data Availability
MLST nucleotide sequences for the eleven clinical isolates determined in this study have been deposited in the GenBank nucleotide sequence database under accession numbers MK344035-MK344111.
Funding Statement
This study was supported by grants from the National Natural Science Foundation of China (Nos. 81571971 and 81271804) and the Natural Science Foundation of Guangxi Province of China (2017GXNSFAA198004, AB18221017 and 2018GXNSFAA294090). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Bielska E, May RC. What makes Cryptococcus gattii a pathogen? FEMS yeast research. 2016;16(1):fov106 10.1093/femsyr/fov106 . [DOI] [PubMed] [Google Scholar]
- 2.Kwon-Chung KJ, Fraser JA, Doering TL, Wang Z, Janbon G, Idnurm A, et al. Cryptococcus neoformans and Cryptococcus gattii, the Etiologic Agents of Cryptococcosis. Cold Spring Harb Perspect Med. 2014;4(7):a019760 10.1101/cshperspect.a019760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen SC, Meyer W, Sorrell TC. Cryptococcus gattii Infections. Clinical microbiology reviews. 2014;27(4):980–1024. 10.1128/CMR.00126-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, Frazzitta AE, Litvintseva AP, Fang C, Mitchell TG, Springer DJ, et al. Next generation multilocus sequence typing (NGMLST) and the analytical software program MLSTEZ enable efficient, cost-effective, high-throughput, multilocus sequencing typing. Fungal genetics and biology: FG & B. 2015;75:64–71. 10.1016/j.fgb.2015.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer W, Aanensen DM, Boekhout T, Cogliati M, Diaz MR, Esposto MC, et al. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Medical mycology. 2009;47(6):561–70. 10.1080/13693780902953886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu XZ, Wang QM, Theelen B, Groenewald M, Bai FY, Boekhout T. Phylogeny of tremellomycetous yeasts and related dimorphic and filamentous basidiomycetes reconstructed from multiple gene sequence analyses. Studies in mycology. 2015;81:1–26. 10.1016/j.simyco.2015.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hagen F, Khayhan K, Theelen B, Kolecka A, Polacheck I, Sionov E, et al. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal genetics and biology: FG & B. 2015;78:16–48. 10.1016/j.fgb.2015.02.009 . [DOI] [PubMed] [Google Scholar]
- 8.Liu XZ, Wang QM, Goker M, Groenewald M, Kachalkin AV, Lumbsch HT, et al. Towards an integrated phylogenetic classification of the Tremellomycetes. Studies in mycology. 2015;81:85–147. 10.1016/j.simyco.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, et al. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proceedings of the National Academy of Sciences of the United States of America. 2004;101(49):17258–63. 10.1073/pnas.0402981101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datta K, Bartlett KH, Baer R, Byrnes E, Galanis E, Heitman J, et al. Spread of Cryptococcus gattii into Pacific Northwest region of the United States. Emerging infectious diseases. 2009;15(8):1185–91. 10.3201/eid1508.081384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herkert PF, Hagen F, Pinheiro RL, Muro MD, Meis JF, Queiroz-Telles F. Ecoepidemiology of Cryptococcus gattii in Developing Countries. Journal of fungi. 2017;3(4). 10.3390/jof3040062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagen F, Chowdhary A, Prakash A, Yntema JB, Meis JF. Molecular characterization of Cryptococcus gattii genotype AFLP6/VGII isolated from woody debris of divi-divi (Caesalpinia coriaria), Bonaire, Dutch Caribbean. Revista iberoamericana de micologia. 2014;31(3):193–6. 10.1016/j.riam.2013.10.007 . [DOI] [PubMed] [Google Scholar]
- 13.Springer DJ, Chaturvedi V. Projecting Global Occurrence of Cryptococcus gattii. Emerging infectious diseases. 2010;16(1):14–20. 10.3201/eid1601.090369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen M, Pan WH, Boekhout T. Cryptococcus gattii infections in China: extent of the problem? Chinese Medical Journal. 2013;126(2):203–5. 10.3760/cma.j.issn.0366-6999.20122407 [DOI] [PubMed] [Google Scholar]
- 15.Cogliati M. Global Molecular Epidemiology of Cryptococcus neoformans and Cryptococcus gattii: An Atlas of the Molecular Types. Scientifica. 2013;2013:675213 10.1155/2013/675213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okamoto K, Hatakeyama S, Itoyama S, Nukui Y, Yoshino Y, Kitazawa T, et al. Cryptococcus gattii Genotype VGIIa Infection in Man, Japan, 2007. Emerging infectious diseases. 2010;16(7):1155–7. 10.3201/eid1607.100106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng X, Yao Z, Ren D, Liao W, Wu J. Genotype and mating type analysis of Cryptococcus neoformans and Cryptococcus gattii isolates from China that mainly originated from non-HIV-infected patients. FEMS yeast research. 2008;8(6):930–8. 10.1111/j.1567-1364.2008.00422.x . [DOI] [PubMed] [Google Scholar]
- 18.Wu SY, Lei Y, Kang M, Xiao YL, Chen ZX. Molecular characterisation of clinical Cryptococcus neoformans and Cryptococcus gattii isolates from Sichuan province, China. Mycoses. 2015;58(5):280–7. 10.1111/myc.12312 . [DOI] [PubMed] [Google Scholar]
- 19.Dou HT, Xu YC, Wang HZ, Li TS. Molecular epidemiology of Cryptococcus neoformans and Cryptococcus gattii in China between 2007 and 2013 using multilocus sequence typing and the DiversiLab system. European journal of clinical microbiology & infectious diseases: official publication of the European Society of Clinical Microbiology. 2015;34(4):753–62. 10.1007/s10096-014-2289-2 . [DOI] [PubMed] [Google Scholar]
- 20.Klein KR, Hall L, Deml SM, Rysavy JM, Wohlfiel SL, Wengenack NL. Identification of Cryptococcus gattii by Use of L-Canavanine Glycine Bromothymol Blue Medium and DNA Sequencing. Journal of clinical microbiology. 2009;47(11):3669–72. 10.1128/JCM.01072-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herkert PF, Hagen F, de Oliveira Salvador GL, Gomes RR, Ferreira MS, Vicente VA, et al. Molecular characterisation and antifungal susceptibility of clinical Cryptococcus deuterogattii (AFLP6/VGII) isolates from Southern Brazil. European journal of clinical microbiology & infectious diseases: official publication of the European Society of Clinical Microbiology. 2016;35(11):1803–10. 10.1007/s10096-016-2731-8 . [DOI] [PubMed] [Google Scholar]
- 22.Kinne J, Joseph M, Wernery U, Nogradi N, Hagen F. Disseminated Cryptococcus deuterogattii (AFLP6/VGII) infection in an Arabian horse from Dubai, United Arab Emirates. Revista iberoamericana de micologia. 2017;34(4):229–32. 10.1016/j.riam.2017.02.007 . [DOI] [PubMed] [Google Scholar]
- 23.Souto AC, Bonfietti LX, Ferreira-Paim K, Trilles L, Martins M, Ribeiro-Alves M, et al. Population Genetic Analysis Reveals a High Genetic Diversity in the Brazilian Cryptococcus gattii VGII Population and Shifts the Global Origin from the Amazon Rainforest to the Semi-arid Desert in the Northeast of Brazil. PLoS Negl Trop Dis. 2016;10(8):e0004885 10.1371/journal.pntd.0004885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Molecular biology and evolution. 2016;33(7):1870–4. 10.1093/molbev/msw054 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price MN, Dehal PS, Arkin AP. FastTree 2 –Approximately Maximum-Likelihood Trees for Large Alignments. PLoS One. 2010;5(3):e9490 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rocha DFS, Cruz KS, Santos C, Menescal LSF, Neto J, Pinheiro SB, et al. MLST reveals a clonal population structure for Cryptococcus neoformans molecular type VNI isolates from clinical sources in Amazonas, Northern-Brazil. PLoS One. 2018;13(6):e0197841 10.1371/journal.pone.0197841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24(11):1403–5. 10.1093/bioinformatics/btn129 . [DOI] [PubMed] [Google Scholar]
- 28.S C, B R, J F, CM M, BL W, V. C. Direct PCR of Cryptococcus neoformans MATɑ and MATa Pheromones To Determine Mating Type, Ploidy, and Variety: a Tool for Epidemiological and Molecular Pathogenesis Studies. Journal of clinical microbiology. 2000;38(5):2007–9. 10.1128/JCM.38.5.2007-2009.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ngamskulrungroj P, Price J, Sorrell T, Perfect JR, Meyer W. Cryptococcus gattii Virulence Composite: Candidate Genes Revealed by Microarray Analysis of High and Less Virulent Vancouver Island Outbreak Strains. PLoS One. 2011;6(1):e16076 10.1371/journal.pone.0016076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barcellos VA, Martins LMS, Fontes ACL, Reuwsaat JCV, Squizani ED, de Sousa Araujo GR, et al. Genotypic and Phenotypic Diversity of Cryptococcus gattii VGII Clinical Isolates and Its Impact on Virulence. Frontiers in microbiology. 2018;9:132 10.3389/fmicb.2018.00132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ngamskulrungroj P, Serena C, Gilgado F, Malik R, Meyer W. Global VGIIa isolates are of comparable virulence to the major fatal Cryptococcus gattii Vancouver Island outbreak genotype. Clinical Microbiology and Infection. 2011;17(2):251–8. 10.1111/j.1469-0691.2010.03222.x [DOI] [PubMed] [Google Scholar]
- 32.Hong N, Chen M, Xu N, Al-Hatmi AMS, Zhang C, Pan WH, et al. Genotypic diversity and antifungal susceptibility of Cryptococcus neoformans isolates from paediatric patients in China. Mycoses. 2019;62(2):171–80. 10.1111/myc.12863 . [DOI] [PubMed] [Google Scholar]
- 33.Espinel-Ingroff A, Chowdhary A, Cuenca-Estrella M, Fothergill A, Fuller J, Hagen F, et al. Cryptococcus neoformans-Cryptococcus gattii Species Complex: an International Study of Wild-Type Susceptibility Endpoint Distributions and Epidemiological Cutoff Values for Amphotericin B and Flucytosine. Antimicrobial agents and chemotherapy. 2012;56(6):3107–13. 10.1128/AAC.06252-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Espinel-Ingroff A, Aller AI, Canton E, Castanon-Olivares LR, Chowdhary A, Cordoba S, et al. Cryptococcus neoformans-Cryptococcus gattii Species Complex: an International Study of Wild-Type Susceptibility Endpoint Distributions and Epidemiological Cutoff Values for Fluconazole, Itraconazole, Posaconazole, and Voriconazole. Antimicrobial agents and chemotherapy. 2012;56(11):5898–906. 10.1128/AAC.01115-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Espinel-Ingroff A, Chowdhary A, Gonzalez GM, Guinea J, Hagen F, Meis JF, et al. Multicenter Study of Isavuconazole MIC Distributions and Epidemiological Cutoff Values for the Cryptococcus neoformans-Cryptococcus gattii Species Complex Using the CLSI M27-A3 Broth Microdilution Method. Antimicrobial agents and chemotherapy. 2015;59(1):666–8. 10.1128/AAC.04055-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perfect JR, Dismukes WE, Dromer F, Goldman DL, Graybill JR, Hamill RJ, et al. Clinical Practice Guidelines for the Management of Cryptococcal Disease: 2010 Update by the Infectious Diseases Society of America. Clinical Infectious Diseases. 2010;50(3):291–322. 10.1086/649858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu ZY, Wang GQ, Zhu LP, Lv XJ, Zhang QQ, Yu YS, et al. Expert consensus on the diagnosis and treatment of cryptococcal meningitis. Chinese Journal of Internal Medicine. 2018;57(5):317–23. 10.3760/cma.j.issn.0578-1426.2018.05.003 [DOI] [PubMed] [Google Scholar]
- 38.Lockhart SR, Iqbal N, Harris JR, Grossman NT, DeBess E, Wohrle R, et al. Cryptococcus gattii in the United States: Genotypic Diversity of Human and Veterinary Isolates. PLoS One. 2013;8(9):e74737 10.1371/journal.pone.0074737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y, Farrer RA, Giamberardino C, Sakthikumar S, Jones A, Yang T, et al. Microevolution of Serial Clinical Isolates of Cryptococcus neoformans var. grubii and C. gattii. mBio. 2017;8(2):e00166–17. 10.1128/mBio.00166-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maruyama FH, de Paula DAJ, Menezes IG, Favalessa OC, Hahn RC, de Almeida A, et al. Genetic Diversity of the Cryptococcus gattii Species Complex in Mato Grosso State, Brazil. Mycopathologia. 2019;184(1):45–51. 10.1007/s11046-018-0313-2 . [DOI] [PubMed] [Google Scholar]
- 41.Billmyre RB, Croll D, Li W, Mieczkowski P, Carter DA, Cuomo CA, et al. Highly Recombinant VGII Cryptococcus gattii Population Develops Clonal Outbreak Clusters through both Sexual Macroevolution and Asexual Microevolution. mBio. 2014;5(4):e01494–14. 10.1128/mBio.01494-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samarasinghe H, Xu J. Hybrids and hybridization in the Cryptococcus neoformans and Cryptococcus gattii species complexes. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2018;66:245–55. 10.1016/j.meegid.2018.10.011 . [DOI] [PubMed] [Google Scholar]
- 43.Samarasinghe H, You M, Jenkinson TS, Xu J, James TY. Hybridization Facilitates Adaptive Evolution in Two Major Fungal Pathogens. Genes. 2020;11(1). 10.3390/genes11010101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Engelthaler DM, Hicks ND, Gillece JD, Roe CC, Schupp JM, Driebe EM, et al. Cryptococcus gattii in North American Pacific Northwest: Whole-Population Genome Analysis Provides Insights into Species Evolution and Dispersal. mBio. 2014;5(4):e01464–14. 10.1128/mBio.01464-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu C, Sun S, Billmyre RB, Roach KC, Heitman J. Unisexual versus bisexual mating in Cryptococcus neoformans: Consequences and biological impacts. Fungal genetics and biology: FG & B. 2015;78:65–75. 10.1016/j.fgb.2014.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diaz JH. The Disease Ecology, Epidemiology, Clinical Manifestations, and Management of Emerging Cryptococcus gattii Complex Infections. Wilderness & environmental medicine. 2020;31(1):101–9. 10.1016/j.wem.2019.10.004 . [DOI] [PubMed] [Google Scholar]
- 47.Cao C, Liang L, Wang W, Luo H, Huang S, Liu D, et al. Common reservoirs for Penicillium marneffei infection in humans and rodents, China. Emerging infectious diseases. 2011;17(2):209–14. 10.3201/eid1702.100718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.May RC, Stone NR, Wiesner DL, Bicanic T, Nielsen K. Cryptococcus: from environmental saprophyte to global pathogen. Nature reviews Microbiology. 2016;14(2):106–17. 10.1038/nrmicro.2015.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang GQ, Zhao QG. The history, status quo, ecological problems and countermeasures of Eucalyptus plantations in Guangxi. Acta Ecologica Sinica. 2014;34(18):5142–52. [Google Scholar]
- 50.Tintelnot K, Hagen F, Han CO, Seibold M, Rickerts V, Boekhout T. Pitfalls in Serological Diagnosis of Cryptococcus gattii Infections. Medical mycology. 2015;53(8):874–9. 10.1093/mmy/myv061 . [DOI] [PubMed] [Google Scholar]
- 51.Fang W, Fa Z, Liao W. Epidemiology of Cryptococcus and cryptococcosis in China. Fungal genetics and biology: FG & B. 2015;78(2015):7–15. 10.1016/j.fgb.2014.10.017 . [DOI] [PubMed] [Google Scholar]