Abstract
Over the last few decades, advances in our understanding of microbial ecology have allowed us to appreciate the important role of microbial communities in maintaining human health. While much of this research has focused on gut microbes, microbial communities in other body sites and from the environment are increasingly recognized in human disease. Here, we discuss recent advances in our understanding of host-microbiota interactions in the development and manifestation of asthma focusing on three distinct microbial compartments. First, environmental microbes originating from house dust, pets and farm animals have been linked to asthma pathogenesis, which is often connected to their production of bioactive molecules such as lipopolysaccharide. Second, respiratory microbial communities, including newly appreciated populations of microbes in the lung have been associated with allergic airway inflammation. Current evidence suggests that the presence of particular microbes, especially Streptococcus, Haemophilus and Morexella species within the airway may shape local immune responses and alter the severity and manifestations of airway inflammation. Third, the gut microbiota has been implicated in both experimental models and clinical studies in predisposing to asthma. There appears to be a "critical window" of colonization that occurs during early infancy in which gut microbial communities shape immune maturation and confer susceptibility to allergic airway inflammation. The mechanisms by which gut microbial communities influence lung immune responses and physiology, the "gut-lung axis", are still being defined but include the altered differentiation of immune cell populations important in asthma and the local production of metabolites that affect distal sites. Together, these findings suggest an intimate association of microbial communities with host immune development and the development of allergic airway inflammation. Improved understanding of these relationships raises the possibility of microbiota-directed therapies to improve or prevent asthma.
Keywords: Asthma, Allergy, Microbiota, Microbiome, gut-lung axis
Introduction
The recognition that microorganisms could have a profound influence on the health of multicellular life has its roots in the earliest days of microbiology. In the late 1800s, Louis Pasteur, in response to his colleague's investigations into the role of soil microorganisms in plant growth, speculated that complex life was impossible without bacteria [1,2]. While we now know that animals can be reared and grown without microorganisms (i.e. germ-free), Pasteur's prescient interest in these symbiotic relationships mark some of the first theories of symbiosis between multi- and unicellular life.
Much of our current understanding of host-microbial community interactions has come from the study of microbes within the gut, which harbors the largest ecosystem of bacteria in the human body. Facilitated by metagenomic techniques to study microbial communities and next-generation sequencing, we have learned more about the normal development and functions of the gut microbiota. While the configurations of the gut microbiota vary greatly between individuals [3, 4], sampling of healthy people in early infancy to adulthood has revealed many common features of a "normal" gut microbiome. In early infancy, the configuration of the gut microbiota is highly variable and harbors a limited number of bacteria, with a predominance of Bifidobacterium coinciding with breastmilk as the primary dietary component (reviewed in [5]). The complexity of the microbiota gradually increases in early childhood until it reaches an adult-like configuration by approximately three years of age [6]. While the high intrapersonal variability of the gut microbiota has (so far) hampered the development of an unequivocal metric for what constitutes a "healthy" microbiota, we now recognize that a normally functioning gut microbiota performs a number of functions important to human health. These functions include digestion [7], stimulating mucosal immune function [8], exclusion and/or control of pathogens [9] and production of beneficial metabolites [10,11]. Likewise, gut microbial communities that fail to execute these functions have been causally linked to inflammatory bowel disease [12], obesity [13, 14], malnutrition [15] and antibiotic-associated diarrhea [16].
In addition to its importance within the GI tract, metagenomic methods have fueled the recognition that microbial communities influence host physiology outside the gastrointestinal tract. We now know that many other epithelial surfaces, including the airways, genitourinary tract, and epidermis, host distinct consortia of microbes and that these communities shape the function of their respective mucosal surfaces. Further, microbial communities have been demonstrated to influence tissues at distal sites, such as the gut microbiota's influence on multiple sclerosis [17, 18] and atherosclerotic cardiovascular disease [19].
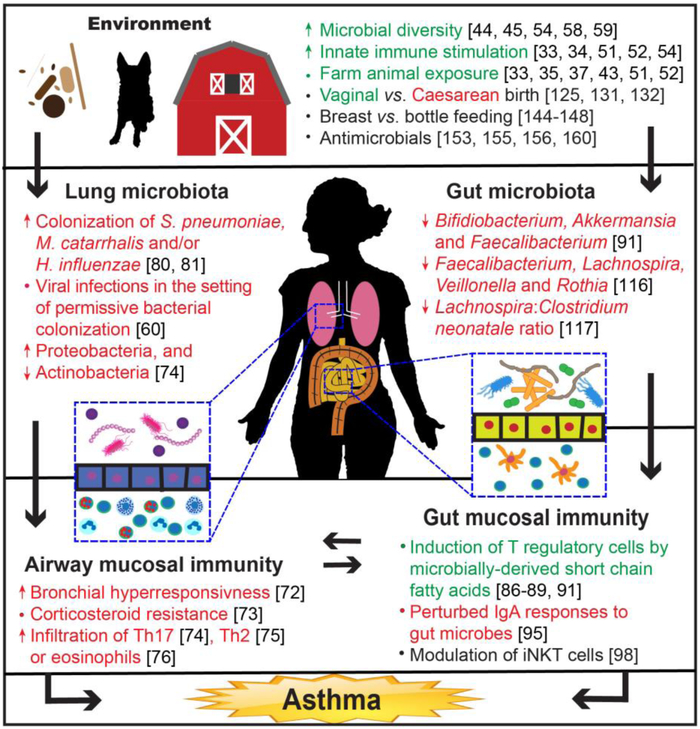
These findings are particularly important for asthma and allergy, where environmental influences are recognized to play an important role in the disease's epidemiology. Beginning with the recognition that infectious exposures can mediate susceptibility to allergy [20], our understanding of how microbes shape susceptibility to atopic diseases has grown substantially [21]. This review will address the role of the microbiota of the environment, lung and gut in the pathogenesis and manifestation of asthma (Figure 1). While the microbiota refers to viral, fungal and bacterial constituents, our discussion will focus primarily on its bacterial components.
Figure 1.
Overview of potential microbial influences on asthma.
The Environmental Microbiome in the Pathogenesis of Asthma
The environment plays a key role in contributing to the risk of asthma and other atopic diseases [22]. This is highlighted in the International Study of Asthma and Allergies in Childhood (ISAAC), which tracked atopic symptoms in over 460,000 children aged 13-14 years in 155 countries. Data from the registry indicate that the prevalence of asthma, allergic rhinoconjunctivitis and atopic eczema symptoms varied up to 60-fold between centers [23]. Children emigrating from countries with a low prevalence of allergic disease to countries with a high prevalence had a lower risk of developing allergy compared to those born in the destination country. However, this protective effect quickly diminished as the children spent more time in the higher-prevalence country, indicating a strong environmental influence on the development of allergy [24].
The influence of environmental microbes on asthma risk may begin before birth. Multiple studies have shown maternal exposure to a farming environment is associated with protection in the offspring from allergic diseases, including asthma [25-29]. Studies comparing the incidence of allergic disease in children raised on farms to those raised in urban environments provide some of the most striking examples of the effects of household and lifestyle on disease susceptibility [30, 31]. Living on a farm is recognized as a protective influence in atopic diseases, including asthma [32-37].
These findings are further supported by animal study data demonstrating a link between microbial exposure in utero and asthma susceptibility [38-40]. For example, intranasal exposure of pregnant mice to a cow shed-derived bacterium, Acinetobacter Iwoffii F78, protected offspring from asthma. This effect was dependent on innate toll-like receptor (TLR) signaling; offspring of mothers without a functional TLR signaling pathway were not protected from asthma [41]. The household environment serves as a major exposure to both microbes and allergens, and heavily influences the risk for asthma and other allergic diseases. Farm animal associated microbes may be particularly protective [38] and are conspicuously scarce in urban environments. Although the majority of the data for the link between microbial burden and allergens exists for house dust, the response to outdoor allergens can also be modulated by associated microbes, as was shown for grass pollen, which contains gram positive bacteria that drive DC maturation and induction of T cell-mediated allergic inflammation [42].
The importance of environmental microbial exposure to asthma risk was compellingly illustrated in a study comparing Amish and Hutterite children. These two populations are genetically similar, but their farming practices diverge markedly. Amish often live on single-family dairy farms, use horses for fieldwork and transportation, and have much higher levels of household endotoxin and increased environmental microbial diversity compared to the Hutterites, who live on large, highly industrialized, communal farms [43]. Exposing mice to dust samples from either Amish or Hutterite households reproduced differences in asthma susceptibility, demonstrating a direct causative link between environmental microbes and asthma. The protective effect of Amish dust was abrogated in mice deficient in the TLR signaling molecules MyD88 or Trif, confirming the importance of microbial activation of innate immune signaling pathways.
As techniques for studying the microbiota have become more sophisticated, we have learned that both quantity and composition of environmental microbes modulate allergic sensitization and asthma risk. In the Urban Environment and Childhood Asthma (URECA) study, increasing allergen exposure was found to correlate with protection in an urban asthma cohort, but only if concurrent microbial exposure was also increased in richness [44, 45]. In contrast, other studies in urban children have found asthma risk to be correlated with allergen exposure, but only if accompanied by concurrent sensitization [46-49]. Surprisingly, despite this known relationship between allergen exposure and asthma risk, pet ownership is reported to be protective for asthma [50]. Based on evidence that increased exposure to endotoxin may mediate the protective effects of farm living [51], it is thought that exposure to environmental microbes (e.g. from pets) may modulate the effect of allergens and be protective in asthma. Consistent with this idea, later studies found that increased levels of endotoxin in mattresses can protect from asthma [52].
A mechanistic understanding of how environmental microbial products, like lipopolysaccharide (LPS) alter susceptibility to asthma continues to emerge. For example, polymorphisms in the LPS co-receptor CD14 mediate differing effects of environmental interventions on the risk of atopy and asthma. The Dutch Prevention and Incidence of Asthma and Mite Allergy (PIAMA) study initially found no effect of early use of mite-allergen impermeable mattress covers in preventing atopy or asthma. However, subsequent analysis revealed that children with an alternate allele in CD14 had higher risk of atopic dust mite sensitization and asthma with use of impermeable covers [53]. Variations in the structure of endotoxin itself have also been shown to mediate beneficial versus detrimental effects of LPS exposure [54].
Role of Respiratory Microbes in Asthma
The upper airway, especially the oropharynx and sinuses, has long been recognized to harbor communities of microbes that influence human health, but more recently, molecular techniques for microbial discovery have revolutionized our understanding of microbial communities that reside within the lower airway. The airway consists of the tissues of the nares, sinuses, naso- and oropharynx, trachea, and bronchioles, which have distinct embryologic and structural properties that create unique microenvironments for microbial colonization. Temperature, pH, mucin quality, ciliary function, oxygen tension, and local mucosal immune responses shape the microbial ecology of each of these sites. As a result, microbial communities within the airways demonstrate significant biogeographical variation. Distinct communities inhabit the nasal cavity, oropharynx and bronchial mucosa reflecting adaptation of microbial consortia to different anatomic sites within the airway [55]. Further microbial community specialization to regions within the lung are also likely: lung explants and post-mortem analyses of patients with cystic fibrosis and chronic obstructive pulmonary disease have demonstrated that unique communities can be discerned between different segments of the lung [56, 57]. The effect of these biogeographical variations in microbial community composition within the airway is not fully understood but have practical and function implications that are important to understanding their role in asthma.
Acquisition of upper airway microbes commences at birth, where early seeding of the respiratory tract (e.g. naso- and oropharynx) occurs during delivery. The types of microbes that initiate early colonization depend on the mode of delivery, with children born vaginally harboring a different consortium of microbes than babies born by Caesarian section [58]. After birth, respiratory microbial community assembly continues to occur over the first two years of life, with stable nasopharyngeal bacterial community structures associated with Dolosigranulum and Moraxella species [59]. Additionally, daycare exposure, viral infections and antibiotic treatment can modulate the composition of the nasopharyngeal microbiota [60].
In contrast to the upper airway microbiota, the origins and functions of the lower airway microbiota have been much more challenging to study. Obtaining samples from the lower airway requires either invasive bronchoscopy or induced sputum, which requires additional analysis and processing to account for contamination by the upper airway microbiota. These efforts have consistently shown that the lower airway harbors a population of bacteria that is detectable above what is found in controls performed to rule out environmental contamination [61]. The constituents of the lung microbiota are thought be delivered from the oropharynx through micro-aspiration events and/or mucosal dispersion from contiguous tissues [62]. Microbial density within the lungs is low, with estimates of the densities of microbes within the lower airway being approximately 103-104 less abundant in the lung compared to the upper airway [63]. These findings have led to the notion that the bacterial community ecology of the lungs in healthy individuals is determined by the equilibrium established by of immigration of microbes from the upper airway and their elimination, rather than microbial persistence and proliferation in the lower respiratory tract [64].
Recognition that maladaptive configurations of the gut microbiota increases risk for certain types of gastrointestinal diseases has led to considerable interest in determining if the airway microbiota contributes to asthma pathogenesis. Studying the airway microbiota, however, presents a number of challenges. First, it is unclear what anatomic location(s) within the respiratory tract are most relevant to the pathogenesis asthma. While many of the defining pathologic features of asthma occur within the distal bronchial tree, the density of bacteria in these locations in the lung is very low compared to the upper airway [63]. Similarly, although the upper airway is not usually thought to play a central role in asthma, the higher bacterial density and the potential for mucosal immune responses to be shared along the entirety of the airways make the upper airway another viable location to examine for host-microbe interactions critical to asthma. Second, both the complex anatomy of the respiratory tract and the relatively low biomass of microbes in the airway make investigating the respiratory microbiota technically challenging and difficult to generalize across studies. The anatomy of the airways has led to numerous different methods for sampling the respiratory tract (e.g. bronchoalveolar lavage vs. endobronchial brushings), which are known to lead to different microbiota profiles, even when samples are obtained from the same patients [65]. Furthermore, the low biomass of bacteria within healthy airways leads to higher levels of background or contaminating data and prevents the application of many genomic techniques (e.g. metagenomic sequencing for microbial RNA-seq) that could aid in identifying and characterizing the airway microbiota. Third, while defined animal models to study the single airway microbes in asthma have been explored [66-70], causal relationships between human airway microbial communities and allergic airway inflammation are net yet firmly established.
Despite these challenges, data from multiple human clinical studies demonstrate an association between alterations in the lung microbiota and various asthma phenotypes [65, 71-77]. For example, markers of allergic inflammation (from the three-gene mean panel including CLCA1, SERPINB2 and PSTN) are associated with differences in the composition of the airway microbiota [76]. Likewise, eosinophilic inflammation, Th17 gene expression and changes in steroid responsiveness (as measured by FKBP5) also correlate with an altered lung microbiota community [74]. Neutrophilic inflammation in asthmatics may also be linked to components of the airway microbiota. Neutrophilic asthma is regarded as a specific endotype of asthma that has a distinct immunopathogenesis from allergic or eosinophilic asthma and is generally more difficult to treat. Neutrophil abundance in the sputum of asthmatics has been linked to levels particular taxa, including Moraxella, which has been reported in multiple studies [78, 79]. Treatment with inhaled corticosteroids, a mainstay of therapy in asthma, is associated with airway community alterations [65]. Moreover, Haemophilus colonization is associated with a diminished response to inhaled corticosteroids, [76], which may act to blunt the effect of steroids on airway macrophages [73].
Changes in the airway microbiota may precede the development of asthma in early childhood [60, 80, 81]. Early colonization with Streptococcus pneumoniae, M. catarrhalis and/or H. influenzae at one month predicts later wheezing, hospitalization and ultimately, the diagnosis of asthma at five years of age [80]. Enrichment of these same taxa within the upper airway microbiota also appears to occur during acute wheezing episodes in children and is a risk factor for wheezing episodes, independent from viral infections [81]. Bacterial colonization also modifies the risk and severity of viral infections--airway microbiota dominated by Moraxella predisposes to lower respiratory tract infections and increases the risk of fever when in the presence of respiratory syncytial virus (RSV) [60]. Febrile respiratory illnesses, in turn, increase the risk of wheezing before the age of five and suggest that both airway respiratory microbes and viruses contribute to the risk of later wheeze.
The Gut Microbiota and Asthma
While the potential for airway microbes to modulate asthma is readily appreciated due to the close proximity of respiratory microbial communities and the sites of allergic inflammation, gut microbes, despite their anatomical separation, are now appreciated to play important roles in asthma. There is an extensive body of literature documenting the roles of the gut microbes in health and perturbations in the gut microbiota are known to have indirect physiologic consequences at remote anatomic sites, including the lung [82]. This crosstalk between the gut and the lungs has been termed the gut-lung axis, which emphasizes the interconnection between gut and lung function [83, 84]. Although a comprehensive understanding of the mechanisms underlying this axis has not yet been achieved, the gut microbiota is thought to play an important role in altering lung function and several contributing pathways have been identified (Fig 1).
Treg cells
T regulatory cells (Tregs) play a crucial role in immune homeostasis, particularly in allergy. In mice, blockade of peripheral Treg induction results in Th2-type inflammation at mucosal sites in model systems [85]. The size and functions of the murine Treg pool are, in turn, regulated by short chain fatty acids (SCFAs, primarily acetate, propionate and butyrate) produced in the gut by anaerobic bacterial fermentation of indigestible dietary fiber [11, 86, 87]. Supplementing the diet of mice with SCFAs reverses allergic lung inflammation [88]. Dietary SCFAs may also protect from asthma through mechanisms separate from Treg induction. Diets high in fiber boost levels of circulating SCFAs and increase infiltration of dendritic cells in the lungs of mice. These dendritic cells have greater phagocytic activity, but decreased ability to promote Th2 inflammation and allergic airway inflammation [89]. In contrast, mice fed a low fiber diet have less exposure to SCFAs and increased allergic airway inflammation. In humans, Treg responses directly mediate tolerance to allergens [90]. The gut microbiota and their metabolites have been implicated in allergy, in part, due to their ability to modulate the differentiation of Treg cells [91]. However, while there is firm data in mice demonstrating microbe-mediated induction of Tregs in the gut and also an established role of Tregs in modulating allergen tolerance, a direct link between the two remains a compelling, yet unproven, concept.
Tregs also modulate production of the major mucosal antibody, IgA, through the production of TGfβ. In healthy mice, Tregs direct IgA secretion at mucosal surfaces to exclude microbial ligands, which lowers systemic inflammation due to decreased overall CD4+ T cell activation [92]. In human disease, IgA responses to the microbiota can be disordered, reflecting perturbations in the microbial ecology characteristic of dysbiosis, and transfer of these microbiota to healthy mice can recapitulate elements of the original human phenotype [93, 94]. Similarly, alterations in IgA targeting patterns, evident by lower levels of IgA-bound bacteria, as well as differences in individual taxa targeted, are present in children at risk for developing asthma [95].
iNKT cells
Induced NKT (iNKT) cells fill an important niche, bridging both innate and adaptive immune functions. They are capable of massive cytokine release that, depending on the context, can be either Th1 or Th2 predominant, and iNKT dysfunction is implicated in multiple inflammatory disorders [96, 97]. In animals lacking a gut microbiota (germ-free mice), iNKT cells accumulate in the intestines and lungs and confer increased susceptibility to inflammation at these sites compared animals harboring a normal intestinal microbiota [98]. Reintroduction of the intestinal microbiota into germ-free mice can restrict this inflammatory predilection, but only if this occurs in the neonatal period, implying a critical window in the development of the mucosal immune system. While the role of iNKT cells in the pathogenesis of asthma remains somewhat unclear in humans [99, 100], animal studies illustrate the potential of gut microbiota to direct differentiation and function of immune populations in the lungs.
Th17 cells
The Th17 axis is important for maintaining barrier function and clearing pathogens at mucosal surfaces, while on the other hand its dysregulation has been implicated in a range of inflammatory disorders [101]. In asthma, Th17 activation reciprocally regulates Th2 inflammation in mice and humans [102] and is associated with a separate endotype of asthma characterized by neutrophilic inflammation and decreased responsiveness to steroids. Hence, it is a potential therapeutic target in severe asthma [103, 104], although the only clinical trial completed to date targeting the Th17 axis showed no treatment effect [105].
The gut microbiota plays a key role in the early development and conditioning of the Th17 axis. Much of the data available linking the influence of the microbiota on Th17 responses comes from animal studies. In mice, a single species of Firmicute, Segmented Filamentous Bacteria, is sufficient to induce Th17 development in the gut [106], which in turn confers protection from subsequent mucosal infections. Th17 inflammatory responses are an example of the gut-lung axs where immune responses originating in the gut lead to lung pathology [107], while respiratory infections inducing a Th17 response can result in intestinal injury [108]. These studies illustrate the potential for perturbations in the gut microbiota to influence inflammation at distant sites, such as the lungs, and highlight their potential to influence asthma.
The mechanistic link between gut microbiota and regulation of the Th17 axis in humans is less clear than in mice. To date, no corresponding human gut microbiota species have been directly linked to Th17 induction, although certain species from the human gut microbiota have been shown to induce Th17 responses when transferred to mice [109] . As our understanding of the role the microbiota plays in Th17 development improves, it is possible that new therapies may be developed targeting the microbiota to influence Th17 respones to prevent and treat asthma.
Early establishment of the gut microbiome
Many studies have described how the establishment and development of the human gut microbiota early in life can be affected by variables such as diet, mode of delivery, and geography [6, 110-112]. The first several years of life are marked by considerable changes in composition and diversity of the gut microbial community, and for most individuals the microbiota will have developed a mature, adult-like configuration by three years of age [6, 113, 114].
Altered development of the microbiota, evidenced by low gut microbiota diversity in infancy, has been correlated with development of atopy and asthma later in life [115]. Recently, the Canadian Healthy Infant Longitudinal Development (CHILD) Study [116] demonstrated that infants who would eventually develop atopy and wheeze had lower levels of several bacterial genera, including Faecalibacterium, Lachnospira, Veillonella, and Rothia (together, shortened to FLVR), relative to healthy controls. These differences corresponded to a shift in the metabolic capacity of the microbiome, with decreased levels of the SCFA acetate in the feces of infants at three months of age. Importantly, the changes observed in infants at three months normalized by the age of one year, and the authors inferred from this observation that the first 100 days of life represent a “critical window” for the establishment of a healthy microbiota that would avert asthma risk later in life. Expanding on these initial observations, the authors colonized germ-free mice with feces from one of the children with atopy and wheeze, with or without supplementation from cultures of FLVR. Mice that did not receive FLVR had lower levels of the SCFA butyrate and increased airway inflammation after allergen sensitization and challenge. However, mice receiving FLVR-supplementation were protected from allergen induced airway inflammation. A follow up study of the same CHILD cohort found that the ratio of Lachnospira to Clostridium neonatale (L/C) in the gut at three months of age predicted asthma diagnosis at four years. The odds ratio was as high as 15 for those with the lowest L/C ratio, suggesting that this measurement may be useful as a biomarker to assess asthma risk [117].
Other studies have also found alterations in the fecal microbiota that precede the development of asthma. In a U.S. pediatric birth cohort, the configuration of the gut microbiota before the age of one year correlated with risk of multisensitized atopy at age two and doctor-diagnosed asthma at age four [91]. Lower relative abundance of several bacterial genera, including Bifidiobacterium, Akkermansia and Faecalibacterium, as well as higher levels of fungi (Candida and Rhodotorula) were associated with the highest risk for later developing allergic disease. Ex vivo incubation of sterile fecal water from the highest risk patients with adult T cells led to an increased proportion of IL-4 secreting T cells and proportionally decreased numbers of Tregs, demonstrating a direct link between altered microbiota composition and enhanced allergic immune responses.
Shifts in the fecal microbiota are also present in at-risk asthmatic children from developing countries, who have different environmental exposures including reduced exposure to antibiotics. Consistent with what has been observed in developed countries, atopic wheeze at the age of five years was associated with alterations in the gut bacterial as well as fungal populations and decreased fecal SCFAs at three months of age relative to controls [118]. However, the associated bacterial taxa were different from those identified in previous studies, highlighting the potential for a number of different microbes to occupy a similar functional niche.
Birth Mode
Differences in the gut microbiota based on birth mode are associated with development of asthma and allergy. Initial establishment of the gastrointestinal microbiota has traditionally been thought to occur during or very shortly after birth. In children born via vaginal delivery, the first microbial contact occurs during the descent through the vaginal tract, along with incidental exposure to the maternal fecal microbiota. This results in an overall increase in microbial diversity in the infant gut microbiota that is enriched, among others, for Lactobacillus, Prevotella, and Bacteroides, in children born vaginally compared to those born by caesarean section [58, 112, 113, 119, 120]. In contrast, colonization patterns in caesarean-delivered neonates most closely resemble the skin microbiota, and may be no more specific to the birth mother’s skin microbes than to those of other caesarean-delivered neonates [58]. Other studies have also shown alterations in the gut microbiota between neonates delivered vaginally and those born via caesarean section [121-126], including a systematic analysis of literature on the subject through 2015 [127].
Modern trends of increasing rates of caesarean section both in the U.S. [128] and worldwide [129] may be causally linked to rising rates of autoimmune and allergic conditions [130]. Indeed, multiple studies have found an association between caesarean section and allergic disorders, including asthma. Meta-analyses of these studies have found about a 20% increased risk of asthma associated with delivery by caesarean section [131, 132]. It is still unclear, however, if the microbiota mediates the differences in asthma risk attributable to birth mode.
Breastfeeding
Breastfeeding plays an important role in shaping the early gut microbiota [112, 113, 133]. In addition to the marked difference between breast-fed and bottle-fed infants, there is a rapid shift in the microbiome of breast-fed infants to a more “adult-like” composition after weaning, suggesting a dominant effect of breast milk components on microbiota composition [112]. Breast milk can also guide the development of the infant gut microbiota by serving as a source of nutrients for microorganisms [134], delivering immune-active compounds, including maternally secreted IgA, to the infant gut [135-137] and transmitting microbes present in the breast milk itself to the nascent microbial community [138-141]. While decreased rates of breastfeeding [142, 143] show a similar temporal association with increased prevalence of autoimmune and allergic disorders, studies investigating this link have arrived at conflicting results [144-148]. Similar to caesarean section, there are tantalizing associations between breastfeeding, the microbiota and asthma, but establishing a causal role between these three variables will require additional investigation.
Antimicrobials
Antibiotics profoundly affect the microbiota [149-151] and consequently exert broad influences on the physiology of the host [152]. There is intense interest in the effect of antibiotic use in infancy or early childhood and subsequent asthma risk [153-156]. Currently, no universal agreement exists regarding the role of antibiotics in the risk of asthma in humans, but the potential negative effects of antibiotics on the microbiome and physiology have been demonstrated in mice. Antibiotic exposure during a “critical window” of gut microbial development can result in lasting consequences for the host [157-159]. This critical window is known to be important for allergic airway inflammation: antibiotic treatment of neonatal mice increases susceptibility to asthma, while treatment of adult mice does not[160]. This effect also extends to the perinatal period, where treatment with antibiotics can increase susceptibility to allergic asthma [161, 162]. Although there is little evidence in humans linking intrapartum antibiotics to specific disease states, there is evidence that they can disrupt the establishment of a normal neonatal microbiome [113, 163, 164]. In future studies it will also be important to differentiate the direct effects of antibiotics on immune cell populations from those exerted via alteration of the microbiota.
MICROBIOTA DIRECTED THERAPEUTICS
The increasing evidence of a relationship between alterations in the microbiota and asthma support the idea that the microbiota could be harnessed to treat allergic airway inflammation. Administration of probiotics appears to be beneficial in treating certain conditions, such as acute infectious diarrhea [165], but their role in treating asthma is uncertain. Early trials examining the prenatal administration of Lactobacillus GG showed a decrease in early atopic disease in high-risk children [166]. However, in a follow up study four years later, those same subjects had persistent protection from eczema, but no significant difference in subsequent development of asthma [167]. A systematic review in 2007 found insufficient evidence to support pre- or postnatal probiotic use to prevent allergic disease [168]. Despite additional studies in the interim, more recent meta-analyses continue to find insufficient evidence to support probiotic use in prevention of asthma or wheeze [169-171]. Interestingly, these studies observe that while the probiotics examined to date have not prevented asthma, they have consistently reduced atopic sensitization and eczema. Correspondingly, the World Allergy Organization recommends probiotics for the primary prevention of eczema in pregnancy and during breastfeeding when there is high risk of allergic disease (based on a positive family history) and in high risk infants [172], although other societies find insufficient evidence to do so (see West, et al for a more in-depth review [173]). This suggests that the premise of protection from asthma using probiotics is not necessarily wrong; rather, our current knowledge of the microbiota is insufficient to inform effective treatments. Indeed, a recent study found a protective effect of probiotics against allergic diseases, including asthma, in adolescents, but not in younger children. This seems to contradict other studies proposing an early critical window for influencing asthma risk through the microbiota, and serves to highlight that many complexities regarding the role of probiotics in preventing or treating asthma still need to be resolved. [174]. Other approaches, including prebiotics (compounds that function as nutrient sources for specific subsets of the microbiota) and synbiotics, (co-administration of a probiotic paired with a specifically selected prebiotic) hold promise for the prevention and treatment of asthma [175, 176] but currently lack sufficient evidence to support their recommendation [177, 178]. As we gain a better understanding of the mechanisms underlying the development of the microbiota and its interaction with the host, new, more effective microbiota directed interventions might be developed.
CONCLUSIONS
A promising literature from observational clinical studies and basic science research now exists to support a role for the microbiota in shaping the pathogenesis of asthma. These studies point toward the microbiota as a key modulator of immune, metabolic and cellular functions that responds to inflammatory signals associated with asthma and likely mediates asthma susceptibility, severity and phenotype (summarized in Fig 1). These studies are transformative for both their scientific impact on our understanding of asthma and the potential clinical advances they suggest.
Nevertheless, while many common themes are beginning to emerge, further efforts are clearly needed to establish the mechanisms by which microbial exposure and colonization mediate their effects on asthma. First, coordination within the clinical and scientific community to standardize and share sequencing data will catalyze efforts to identify and confirm important findings across study types and locations. Second, moving away from strictly metagenomic characterization of the microbiota to culture-based efforts at identification, isolation and characterization of asthma-modifying bacterial species will allow the development of new approaches to study and manipulate the microbiota in asthma. Third, further developing clinical and pre-clinical capacity to demonstrate causal relationships between the microbiota and asthma will be essential to realize the potential of new microbiota-targeting therapies and diagnostics.
New modalities to diagnose, phenotype and predict response to therapies in asthma have been demonstrated in principle as an outgrowth of our increased understanding of the role of commensal microbes on asthma. Perhaps even more exciting is the potential of our new view of microbial ecology to lead to new therapeutic approaches to prevent and treat asthma. Probiotics have been investigated for their ability to prevent allergy [166, 167], but as we further dissect the microbe-host interaction it is likely that we will identify "next-generation" probiotics [179] derived from commensals with known, specific effects on host physiology. Similarly, pharmacologic manipulation of the microbiota [72] to reshape its structure or to target specific host pathways impacted by the microbiota [180] in asthma also remain promising therapeutic approaches.
Acknowledgements
The authors would like to thank Anne Rosen for her helpful input during the writing of this review.
Footnotes
Disclosure of potential conflicts of interest Aaron Ver Heul, MD,PhD has received research support from the National Institutes of Health (5T32DK077653-27). Joseph Planer, MD/PhD is a resident in internal medicine at Massachusetts General Hospital. He has no other conflicts of interest to declare.
Andrew L. Kau, MD/PhD has received research support from the National Institutes of Health (K08AI113184) and the AAAAI Foundation. Dr. Kau also reports equity interest in Gilead Sciences, Inc.
Research involving Human Participants and/or Animals This article does not contain any studies with human participants performed by any of the authors
Informed Consent This is a review article. Informed consent is not required.
Conflicts of Interest
ALK has received research support from the National Institutes of Health, Institute for Allergy, Immunology and Infectious Diseases and the AAAAI Foundation.
BIBLIOGRAPHY
- 1.Duclaux É (1885) Presentée par M. Pasteur, Physiologie végétale. Sur la germination dans un sol riche en matières organiques, mais exempt de microbes. . Comptes rendus de Académie des sciences 100:66–68 [Google Scholar]
- 2.Pasteur L (1885) Observations relatives à la Note précédente de. M. Duclaux. Comptes rendus de Académie des sciences 100:68 [Google Scholar]
- 3.Falony G, Joossens M, Vieira-Silva S, et al. (2016) Population-level analysis of gut microbiome variation. Science 352:560–564. 10.1126/science.aad3503 [DOI] [PubMed] [Google Scholar]
- 4.Zhernakova A, Kurilshikov A, Bonder MJ, et al. (2016) Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352:565–569. 10.1126/science.aad3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gritz eC, and Bhandari V (2015) The human neonatal gut microbiome: a brief review. Front Pediatr 3:17 10.3389/fped.2015.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yatsunenko T, Rey FE, Manary MJ, et al. (2012) Human gut microbiome viewed across age and geography. Nature 486:222–227. 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marchesi JR, Adams DH, Fava F, et al. (2016) The gut microbiota and host health: a new clinical frontier. Gut 65:330–339. 10.1136/gutjnl-2015-309990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Honda K, and Littman DR (2016) The microbiota in adaptive immune homeostasis and disease. Nature 535:75–84. 10.1038/nature18848 [DOI] [PubMed] [Google Scholar]
- 9.Lawley TD, and Walker AW (2013) Intestinal colonization resistance. Immunology 138:1–11. 10.1111/j.1365-2567.2012.03616.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maslowski KM, Vieira AT, Ng A, et al. (2009) Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461:1282–1286. 10.1038/nature08530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furusawa Y, Obata Y, Fukuda S, et al. (2013) Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504:446–450. 10.1038/nature12721 [DOI] [PubMed] [Google Scholar]
- 12.Hall AB, Tolonen AC, and Xavier RJ (2017) Human genetic variation and the gut microbiome in disease. Nat Rev Genet 18:690–699. 10.1038/nrg.2017.63 [DOI] [PubMed] [Google Scholar]
- 13.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, and Gordon JI (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1031. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- 14.Ridaura VK, Faith JJ, Rey FE, et al. (2013) Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341:1241214 10.1126/science.1241214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith MI, Yatsunenko T, Manary MJ, et al. (2013) Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 339:548–554. 10.1126/science.1229000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mullish BH, and Williams HR (2018) Clostridium difficile infection and antibiotic-associated diarrhoea. Clin Med (Lond) 18:237–241. 10.7861/clinmedicine.18-3-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berer K, Gerdes LA, Cekanaviciute E, et al. (2017) Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc Natl Acad Sci U S A 114:10719–10724. https://doi.Org/10.1073/pnas.1711233114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cekanaviciute E, Yoo BB, Runia TF, et al. (2017) Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc Natl Acad Sci U SA 114:10713–10718. https://doi.Org/10.1073/pnas.1711235114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, and Hazen SL (2013) Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med 368:1575–1584. 10.1056/NEJMoa1109400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strachan DP (1989) Hay fever, hygiene, and household size. BMJ 299:1259–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rook GAW (2010) 99th Dahlem Conference on Infection, Inflammation and Chronic Inflammatory Disorders: Darwinian medicine and the ‘hygiene’ or ‘old friends’ hypothesis Clinical & Experimental Immunology 160:70–79. https://doi.Org/10.1111/j.1365-2249.2010.04133.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dick S, Friend A, Dynes K, AlKandari F, Doust E, Cowie H, Ayres JG, and Turner SW (2014) A systematic review of associations between environmental exposures and development of asthma in children aged up to 9 years. BMJ open 4:e006554 10.1136/bmjopen-2014-006554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asthma TISo, and Committee AiCIS (1998) Worldwide variation in prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and atopic eczema: ISAAC. Lancet (London, England) 351:1225–1232 [PubMed] [Google Scholar]
- 24.Garcia-Marcos L, Robertson CF, Ross Anderson H, Ellwood P, Williams HC, Wong GW, and Group tIPTS (2014) Does migration affect asthma, rhinoconjunctivitis and eczema prevalence? Global findings from the international study of asthma and allergies in childhood. Int J Epidemiol 43:1846–1854. 10.1093/ije/dyu145 [DOI] [PubMed] [Google Scholar]
- 25.Roduit C, Wohlgensinger J, Frei R, et al. (2011) Prenatal animal contact and gene expression of innate immunity receptors at birth are associated with atopic dermatitis. J Allergy Clin Immunol 127:179–185– 185.e171. 10.1016/j.jaci.2010.10.010 [DOI] [PubMed] [Google Scholar]
- 26.Ege MJ, Bieli C, Frei R, et al. (2006) Prenatal farm exposure is related to the expression of receptors of the innate immunity and to atopic sensitization in school-age children. Journal of Allergy and Clinical Immunology 117:817–823. https://doi.Org/10.1016/j.jaci.2005.12.1307 [DOI] [PubMed] [Google Scholar]
- 27.Loss G, Bitter S, Wohlgensinger J, et al. (2012) Prenatal and early-life exposures alter expression of innate immunity genes: the PASTURE cohort study. J Allergy Clin Immunol 130:523–530.e529. https://doi.Org/10.1016/j.jaci.2012.05.049 [DOI] [PubMed] [Google Scholar]
- 28.Ege MJ, Herzum I, Büchele G, et al. (2008) Prenatal exposure to a farm environment modifies atopic sensitization at birth. J Allergy Clin Immunol 122:407–412– 412.e401-404. 10.1016/j.jaci.2008.06.011 [DOI] [PubMed] [Google Scholar]
- 29.Douwes J, Cheng S, Travier N, et al. (2008) Farm exposure in utero may protect against asthma, hay fever and eczema. The European respiratory journal 32:603–611. 10.1183/09031936.00033707 [DOI] [PubMed] [Google Scholar]
- 30.Milligan KL, Matsui E, and Sharma H (2016) Asthma in Urban Children: Epidemiology, Environmental Risk Factors, and the Public Health Domain. Curr Allergy Asthma Rep 16:33 10.1007/s11882-016-0609-6 [DOI] [PubMed] [Google Scholar]
- 31.Jie Y, Isa ZM, Jie X, Ju ZL, and Ismail NH (2013) Urban vs. rural factors that affect adult asthma. Rev Environ Contam Toxicol 226:33–63. 10.1007/978-1-4614-6898-1_2 [DOI] [PubMed] [Google Scholar]
- 32.von Mutius E, Schmid S, and Group PS (2006) The PASTURE project: EU support for the improvement of knowledge about risk factors and preventive factors for atopy in Europe. Allergy 61:407–413. 10.1111/j.1398-9995.2006.01009.x [DOI] [PubMed] [Google Scholar]
- 33.Alfvén T, Braun-Fahrländer C, Brunekreef B, et al. (2006) Allergic diseases and atopic sensitization in children related to farming and anthroposophic lifestyle--the PARSIFAL study. Allergy 61:414–421. 10.1111/j.1398-9995.2005.00939.x [DOI] [PubMed] [Google Scholar]
- 34.Ege MJ, Mayer M, Normand A-C, et al. (2011) Exposure to environmental microorganisms and childhood asthma. New England Journal of Medicine 364:701–709. 10.1056/NEJMoa1007302 [DOI] [PubMed] [Google Scholar]
- 35.Brunekreef B, Von Mutius E, Wong GK, Odhiambo JA, and Clayton TO (2012) Early life exposure to farm animals and symptoms of asthma, rhinoconjunctivitis and eczema: an ISAAC Phase Three Study. Int J Epidemiol 41:753–761. 10.1093/ije/dyr216 [DOI] [PubMed] [Google Scholar]
- 36.Gehring U, Strikwold M, Schram-Bijkerk D, et al. (2008) Asthma and allergic symptoms in relation to house dust endotoxin: Phase Two of the International Study on Asthma and Allergies in Childhood (ISAAC II). Clinical & Experimental Allergy 38:1911–1920. 10.1111/j.1365-2222.2008.03087.x [DOI] [PubMed] [Google Scholar]
- 37.Illi S, Depner M, Genuneit J, et al. (2012) Protection from childhood asthma and allergy in Alpine farm environments-the GABRIEL Advanced Studies. J Allergy Clin Immunol 129:1470–1477.e1476. 10.1016/j.jaci.2012.03.013 [DOI] [PubMed] [Google Scholar]
- 38.Debarry J, Garn H, Hanuszkiewicz A, et al. (2007) Acinetobacter lwoffii and Lactococcus lactis strains isolated from farm cowsheds possess strong allergy-protective properties. Journal of Allergy and Clinical Immunology 119:1514–1521. 10.1016/j.jaci.2007.03.023 [DOI] [PubMed] [Google Scholar]
- 39.Peters M, Kauth M, Scherner O, Gehlhar K, Steffen I, Wentker P, Von Mutius E, Holst O, and Bufe A (2010) Arabinogalactan isolated from cowshed dust extract protects mice from allergic airway inflammation and sensitization. J Allergy Clin Immunol 126:648–656.e641-644. 10.1016/j.jaci.2010.05.011 [DOI] [PubMed] [Google Scholar]
- 40.Vogel K, Blümer N, Korthals M, et al. (2008) Animal shed Bacillus licheniformis spores possess allergy-protective as well as inflammatory properties. J Allergy Clin Immunol 122:307–312– 312.e301-308. 10.1016/j.jaci.2008.05.016 [DOI] [PubMed] [Google Scholar]
- 41.Conrad ML, Ferstl R, Teich R, et al. (2009) Maternal TLR signaling is required for prenatal asthma protection by the nonpathogenic microbe Acinetobacter lwoffii F78. J Exp Med 206:2869–2877. 10.1084/jem.20090845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heydenreich B, Bellinghausen I, König B, Becker W-M, Grabbe S, Petersen A, and Saloga J (2012) Gram-positive bacteria on grass pollen exhibit adjuvant activity inducing inflammatory T cell responses. Clin Exp Allergy 42:76–84. 10.1111/j.1365-2222.2011.03888.x [DOI] [PubMed] [Google Scholar]
- 43.Stein MM, Hrusch CL, Gozdz J, et al. (2016) Innate Immunity and Asthma Risk in Amish and Hutterite Farm Children. New England Journal of Medicine 375:411–421. 10.1056/NEJMoa1508749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lynch SV, Wood RA, Boushey H, et al. (2014) Effects of early-life exposure to allergens and bacteria on recurrent wheeze and atopy in urban children. J Allergy Clin Immunol 134:593–601.e512. 10.1016/j.jaci.2014.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Connor GT, Lynch SV, Bloomberg GR, et al. (2017) Early-life home environment and risk of asthma among inner-city children. J Allergy Clin Immunol. 10.1016/j.jaci.2017.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Call RS, Smith TF, Morris E, Chapman MD, and Platts-Mills TA (1992) Risk factors for asthma in inner city children. J Pediatr 121:862–866. 10.1016/S0022-3476(05)80329-4 [DOI] [PubMed] [Google Scholar]
- 47.Gelber LE, Seltzer LH, Bouzoukis JK, Pollart SM, Chapman MD, and Platts-Mills TA (1993) Sensitization and exposure to indoor allergens as risk factors for asthma among patients presenting to hospital. The American review of respiratory disease 147:573–578. https://doi.Org/10.1164/ajrccm/147.3.573 [DOI] [PubMed] [Google Scholar]
- 48.Rosenstreich DL, Eggleston P, Kattan M, et al. (1997) The role of cockroach allergy and exposure to cockroach allergen in causing morbidity among inner-city children with asthma. New England Journal of Medicine 336:1356–1363. 10.1056/NEJM199705083361904 [DOI] [PubMed] [Google Scholar]
- 49.Lau S, llli S, Sommerfeld C, Niggemann B, Bergmann R, von Mutius E, and Wahn U (2000) Early exposure to house-dust mite and cat allergens and development of childhood asthma: a cohort study. Multicentre Allergy Study Group. Lancet (London, England) 356:1392–1397 [DOI] [PubMed] [Google Scholar]
- 50.Ownby DR, Johnson CC, and Peterson EL (2002) Exposure to Dogs and Cats in the First Year of Life and Risk of Allergic Sensitization at 6 to 7 Years of Age. JAMA : the journal of the American Medical Association 288:963–972. https://doi.Org/10.1001/jama.288.8.963 [DOI] [PubMed] [Google Scholar]
- 51.von Mutius E, Braun-Fahrländer C, Schierl R, Riedler J, Ehlermann S, Maisch S, Waser M, and Nowak D (2000) Exposure to endotoxin or other bacterial components might protect against the development of atopy. Clinical & Experimental Allergy 30:1230–1234 [DOI] [PubMed] [Google Scholar]
- 52.Braun-Fahrländer C, Riedler J, Herz U, et al. (2002) Environmental exposure to endotoxin and its relation to asthma in school-age children. New England Journal of Medicine 347:869–877. 10.1056/NEJMoa020057 [DOI] [PubMed] [Google Scholar]
- 53.Kerkhof M, Daley D, Postma DS, et al. (2012) Opposite effects of allergy prevention depending on CD14 rs2569190 genotype in 3 intervention studies. J Allergy Clin Immunol 129:256–259. https://doi.Org/10.1016/j.jaci.2011.08.040 [DOI] [PubMed] [Google Scholar]
- 54.Vatanen T, Kostic AD, d’Hennezel E, et al. (2016) Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell 165:842–853. https://doi.Org/10.1016/j.cell.2016.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Durack J, Huang YJ, Nariya S, et al. (2018) Bacterial biogeography of adult airways in atopic asthma. Microbiome 6:104 10.1186/s40168-018-0487-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Erb-Downward JR, Thompson DL, Han MK, et al. (2011) Analysis of the lung microbiome in the "healthy" smoker and in COPD. PLoS One 6:e16384 10.1371/journal.pone.0016384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willner D, Haynes MR, Furlan M, Schmieder R, Lim YW, Rainey PB, Rohwer F, and Conrad D (2012) Spatial distribution of microbial communities in the cystic fibrosis lung. Isme J 6:471–474. 10.1038/ismej.2011.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, and Knight R (2010) Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A 107:11971–11975. 10.1073/pnas.1002601107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Biesbroek G, Tsivtsivadze E, Sanders EA, Montijn R, Veenhoven RH, Keijser BJ, and Bogaert D (2014) Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med 190:1283–1292. 10.1164/rccm.201407-1240OC [DOI] [PubMed] [Google Scholar]
- 60.Teo SM, Mok D, Pham K, et al. (2015) The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 17:704–715. 10.1016/j.chom.2015.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dickson RP, Erb-Downward JR, Martinez FJ, and Huffnagle GB (2016) The Microbiome and the Respiratory Tract. Annu Rev Physiol 78:481–504. 10.1146/annurev-physiol-021115-105238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dickson RP, Erb-Downward JR, and Huffnagle GB (2015) Homeostasis and its disruption in the lung microbiome. Am J Physiol Lung Cell Mol Physiol 309:L1047–1055. 10.1152/ajplung.00279.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, Bushman FD, and Collman RG (2011) Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med 184:957–963. 10.1164/rccm.201104-0655OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Beck JM, Huffnagle GB, and Curtis JL (2015) Spatial Variation in the Healthy Human Lung Microbiome and the Adapted Island Model of Lung Biogeography. Ann Am Thorac Soc 12:821–830. 10.1513/AnnalsATS.201501-029OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Denner DR, Sangwan N, Becker JB, et al. (2016) Corticosteroid therapy and airflow obstruction influence the bronchial microbiome, which is distinct from that of bronchoalveolar lavage in asthmatic airways. J Allergy Clin Immunol 137:1398–1405 e1393. 10.1016/j.jaci.2015.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nembrini C, Sichelstiel A, Kisielow J, Kurrer M, Kopf M, and Marsland BJ (2011) Bacterial-induced protection against allergic inflammation through a multicomponent immunoregulatory mechanism. Thorax 66:755–763. 10.1136/thx.2010.152512 [DOI] [PubMed] [Google Scholar]
- 67.McCann JR, Mason SN, Auten RL, St Geme JW 3rd, and Seed PC (2016) Early-Life Intranasal Colonization with Nontypeable Haemophilus influenzae Exacerbates Juvenile Airway Disease in Mice. Infect Immun 84:2022–2030. 10.1128/IAI.01539-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Preston JA, Essilfie AT, Horvat JC, Wade MA, Beagley KW, Gibson PG, Foster PS, and Hansbro PM (2007) Inhibition of allergic airways disease by immunomodulatory therapy with whole killed Streptococcus pneumoniae. Vaccine 25:8154–8162. 10.1016/j.vaccine.2007.09.034 [DOI] [PubMed] [Google Scholar]
- 69.Preston JA, Thorburn AN, Starkey MR, et al. (2011) Streptococcus pneumoniae infection suppresses allergic airways disease by inducing regulatory T-cells. Eur Respir J 37:53–64. 10.1183/09031936.00049510 [DOI] [PubMed] [Google Scholar]
- 70.Thorburn AN, Foster PS, Gibson PG, and Hansbro PM (2012) Components of Streptococcus pneumoniae suppress allergic airways disease and NKT cells by inducing regulatory T cells. J Immunol 188:4611–4620. 10.4049/jimmunol.1101299 [DOI] [PubMed] [Google Scholar]
- 71.Hilty M, Burke C, Pedro H, et al. (2010) Disordered microbial communities in asthmatic airways. PLoS One 5:e8578 10.1371/journal.pone.0008578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang YJ, Nelson CE, Brodie EL, et al. (2011) Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. Journal of Allergy and Clinical Immunology 127:372–381.e373. 10.1016/j.jaci.2010.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goleva E, Jackson LP, Harris JK, et al. (2013) The effects of airway microbiome on corticosteroid responsiveness in asthma. Am J Respir Crit Care Med 188:1193–1201. 10.1164/rccm.201304-0775OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang YJ, Nariya S, Harris JM, Lynch SV, Choy DF, Arron JR, and Boushey H (2015) The airway microbiome in patients with severe asthma: Associations with disease features and severity. J Allergy Clin Immunol 136:874–884. 10.1016/j.jaci.2015.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Q, Cox M, Liang Z, et al. (2016) Airway Microbiota in Severe Asthma and Relationship to Asthma Severity and Phenotypes. PLoS One 11:e0152724 10.1371/journal.pone.0152724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Durack J, Lynch SV, Nariya S, et al. (2017) Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. Journal of Allergy and Clinical Immunology 140:63–75. 10.1016/j.jaci.2016.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sverrild A, Kiilerich P, Brejnrod A, Pedersen R, Porsbjerg C, Bergqvist A, Erjefalt JS, Kristiansen K, and Backer V (2017) Eosinophilic airway inflammation in asthmatic patients is associated with an altered airway microbiome. J Allergy Clin Immunol 140:407–417 e411. 10.1016/j.jaci.2016.10.046 [DOI] [PubMed] [Google Scholar]
- 78.Green BJ, Wiriyachaiporn S, Grainge C, Rogers GB, Kehagia V, Lau L, Carroll MP, Bruce KD, and Howarth PH (2014) Potentially pathogenic airway bacteria and neutrophilic inflammation in treatment resistant severe asthma. PLoS One 9:e100645 10.1371/journal.pone.0100645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Taylor SL, Leong LEX, Choo JM, et al. (2018) Inflammatory phenotypes in patients with severe asthma are associated with distinct airway microbiology. J Allergy Clin Immunol 141:94–103 e115. 10.1016/j.jaci.2017.03.044 [DOI] [PubMed] [Google Scholar]
- 80.Bisgaard H, Hermansen MN, Buchvald F, et al. (2007) Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med 357:1487–1495. 10.1056/NEJMoa052632 [DOI] [PubMed] [Google Scholar]
- 81.Bisgaard H, Hermansen MN, Bonnelykke K, Stokholm J, Baty F, Skytt NL, Aniscenko J, Kebadze T, and Johnston SL (2010) Association of bacteria and viruses with wheezy episodes in young children: prospective birth cohort study. BMJ 341:c4978 10.1136/bmj.c4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lynch SV, and Pedersen O (2016) The Human Intestinal Microbiome in Health and Disease. N Engl J Med 375:2369–2379. 10.1056/NEJMra1600266 [DOI] [PubMed] [Google Scholar]
- 83.Budden KF, Gellatly SL, Wood DL, Cooper MA, Morrison M, Hugenholtz P, and Hansbro PM (2017) Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol 15:55–63. 10.1038/nrmicro.2016.142 [DOI] [PubMed] [Google Scholar]
- 84.Marsland BJ, Trompette A, and Gollwitzer ES (2015) The Gut-Lung Axis in Respiratory Disease. Ann Am Thorac Soc 12 Suppl 2:S150–156. 10.1513/AnnalsATS.201503-133AW [DOI] [PubMed] [Google Scholar]
- 85.Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y, Umetsu DT, and Rudensky AY (2012) Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature 482:395–399. 10.1038/nature10772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, and Garrett WS (2013) The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis Science (New York, N.Y.) 341:569–573. 10.1126/science.1241165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Arpaia N, Campbell C, Fan X, et al. (2013) Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504:451–455. 10.1038/nature12726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cait A, Hughes MR, Antignano F, et al. (2017) Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol 32:164 10.1038/mi.2017.75 [DOI] [PubMed] [Google Scholar]
- 89.Trompette A, Gollwitzer ES, Yadava K, et al. (2014) Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med 20:159–166. 10.1038/nm.3444 [DOI] [PubMed] [Google Scholar]
- 90.Bacher P, Heinrich F, Stervbo U, et al. (2016) Regulatory T Cell Specificity Directs Tolerance versus Allergy against Aeroantigens in Humans. Cell 167:1067–1078 e1016. 10.1016/j.cell.2016.09.050 [DOI] [PubMed] [Google Scholar]
- 91.Fujimura KE, Sitarik AR, Havstad S, et al. (2016) Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med 22:1187–1191. 10.1038/nm.4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cong Y, Feng T, Fujihashi K, Schoeb TR, and Elson CO (2009) A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci U S A 106:19256–19261. 10.1073/pnas.0812681106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Palm NW, de Zoete MR, Cullen TW, et al. (2014) Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158:1000–1010. 10.1016/j.cell.2014.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kau AL, Planer JD, Liu J, et al. (2015) Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci Transl Med 7:276ra224–276ra224. 10.1126/scitranslmed.aaa4877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dzidic M, Abrahamsson TR, Artacho A, Björkstén B, Collado MC, Mira A, and Jenmalm MC (2017) Aberrant IgA responses to the gut microbiota during infancy precede asthma and allergy development. Journal of Allergy and Clinical Immunology 139:1017–1025.e1014. 10.1016/j.jaci.2016.06.047 [DOI] [PubMed] [Google Scholar]
- 96.Bendelac A, Savage PB, and Teyton L (2007) The biology of NKT cells. Annu Rev Immunol 25:297–336. 10.1146/annurev.immunol.25.022106.141711 [DOI] [PubMed] [Google Scholar]
- 97.Kumar A, Suryadevara N, Hill TM, Bezbradica JS, Van Kaer L, and Joyce S (2017) Natural Killer T Cells: An Ecological Evolutionary Developmental Biology Perspective. Frontiers in immunology 8:1858 10.3389/fimmu.2017.01858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Olszak T, An D, Zeissig S, et al. (2012) Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336:489–493. 10.1126/science.1219328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Akbari O, Faul JL, Hoyte EG, Berry GJ, Wahlström J, Kronenberg M, Dekruyff RH, and Umetsu DT (2006) CD4+ invariant T-cell-receptor+ natural killer T cells in bronchial asthma. New England Journal of Medicine 354:1117–1129. 10.1056/NEJMoa053614 [DOI] [PubMed] [Google Scholar]
- 100.Vijayanand P, Seumois G, Pickard C, Powell RM, Angco G, Sammut D, Gadola SD, Friedmann PS, and Djukanovic R (2007) Invariant natural killer T cells in asthma and chronic obstructive pulmonary disease. New England Journal of Medicine 356:1410–1422. 10.1056/NEJMoa064691 [DOI] [PubMed] [Google Scholar]
- 101.Weaver CT, Elson CO, Fouser LA, and Kolls JK (2013) The Th17 pathway and inflammatory diseases of the intestines, lungs, and skin. Annu Rev Pathol 8:477–512. 10.1146/annurev-pathol-011110-130318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Choy DF, Hart KM, Borthwick LA, et al. (2015) TH2 and TH17 inflammatory pathways are reciprocally regulated in asthma. Sci Transl Med 7:301ra129–301ra129. 10.1126/scitranslmed.aab3142 [DOI] [PubMed] [Google Scholar]
- 103.Newcomb DC, and Peebles RS (2013) Th17-mediated inflammation in asthma. Curr Opin Immunol 25:755–760. https://doi.Org/10.1016/j.coi.2013.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nembrini C, Marsland BJ, and Kopf M (2009) IL-17-producing T cells in lung immunity and inflammation. J Allergy Clin Immunol 123:986–994-quiz 995-986. 10.1016/j.jaci.2009.03.033 [DOI] [PubMed] [Google Scholar]
- 105.Busse WW, Holgate S, Kerwin E, Chon Y, Feng J, Lin J, and Lin S-L (2013) Randomized, double-blind, placebo-controlled study of brodalumab, a human anti-IL-17 receptor monoclonal antibody, in moderate to severe asthma. Am J Respir Crit Care Med 188:1294–1302. 10.1164/rccm.201212-2318OC [DOI] [PubMed] [Google Scholar]
- 106.Ivanov II, Atarashi K, Manel N, et al. (2009) Induction of Intestinal Th17 Cells by Segmented Filamentous Bacteria. Cell 139:485–498. 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bradley CP, Teng F, Felix KM, et al. (2017) Segmented Filamentous Bacteria Provoke Lung Autoimmunity by Inducing Gut-Lung Axis Th17 Cells Expressing Dual TCRs. Cell host & microbe 22:697–704.e694. 10.1016/j.chom.2017.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang J, Li F, Wei H, Lian Z-X, Sun R, and Tian Z (2014) Respiratory influenza virus infection induces intestinal immune injury via microbiota-mediated Th17 cell-dependent inflammation. J Exp Med 211:2397–2410. 10.1084/jem.20140625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tan TG, Sefik E, Geva-Zatorsky N, et al. (2016) Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc Natl Acad Sci U S A 113:E8141–E8150. 10.1073/pnas.1617460113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, and Knight R (2009) Bacterial community variation in human body habitats across space and time Science (New York, N.Y.) 326:1694–1697. 10.1126/science.1177486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Faith JJ, Guruge JL, Charbonneau M, et al. (2013) The long-term stability of the human gut microbiota Science (New York, N.Y.) 341:1237439–1237439. 10.1126/science.1237439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bäckhed F, Roswall J, Peng Y, et al. (2015) Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell host & microbe 17:690–703. 10.1016/j.chom.2015.04.004 [DOI] [PubMed] [Google Scholar]
- 113.Bokulich NA, Chung J, Battaglia T, et al. (2016) Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med 8:343ra382–343ra382. 10.1126/scitranslmed.aad7121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, and Ley RE (2011) Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 108 Suppl 1:4578–4585. 10.1073/pnas.1000081107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, and Jenmalm MC (2014) Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy 44:842–850. 10.1111/cea.12253 [DOI] [PubMed] [Google Scholar]
- 116.Arrieta M-C, Stiemsma LT, Dimitriu PA, et al. (2015) Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med 7:307ra152–307ra152. 10.1126/scitranslmed.aab2271 [DOI] [PubMed] [Google Scholar]
- 117.Stiemsma LT, Arrieta M-C, Dimitriu PA, et al. (2016) Shifts in Lachnospira and Clostridium sp. in the 3-month stool microbiome are associated with preschool age asthma Clinical science (London, England: : 1979) 130:2199–2207. 10.1042/CS20160349 [DOI] [PubMed] [Google Scholar]
- 118.Arrieta M-C, Arévalo A, Stiemsma L, et al. (2017) Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J Allergy Clin Immunol. 10.1016/j.jaci.2017.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA, and Stobberingh EE (2006) Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118:511–521. 10.1542/peds.2005-2824 [DOI] [PubMed] [Google Scholar]
- 120.Hyman RW, Fukushima M, Diamond L, Kumm J, Giudice LC, and Davis RW (2005) Microbes on the human vaginal epithelium. Proceedings of the National Academy of Sciences 102:7952–7957. 10.1073/pnas.0503236102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gronlund MM, Lehtonen OP, Eerola E, and Kero P (1999) Fecal microflora in healthy infants born by different methods of delivery: permanent changes in intestinal flora after cesarean delivery. J Pediatr Gastroenterol Nutr 28:19–25 [DOI] [PubMed] [Google Scholar]
- 122.Salminen S, Gibson GR, McCartney AL, and Isolauri E (2004) Influence of mode of delivery on gut microbiota composition in seven year old children. Gut 53:1388–1389. 10.1136/gut.2004.041640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hällström M, Eerola E, Vuento R, Janas M, and Tammela O (2004) Effects of mode of delivery and necrotising enterocolitis on the intestinal microflora in preterm infants. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology 23:463–470. 10.1007/s10096-004-1146-0 [DOI] [PubMed] [Google Scholar]
- 124.Adlerberth I, Lindberg E, Aberg N, Hesselmar B, Saalman R, Strannegård I-L, and Wold AE (2006) Reduced enterobacterial and increased staphylococcal colonization of the infantile bowel: an effect of hygienic lifestyle? Pediatr Res 59:96–101. 10.1203/01.pdr.0000191137.12774.b2 [DOI] [PubMed] [Google Scholar]
- 125.Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, Björkstén B, Engstrand L, and Andersson AF (2014) Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut 63:559–566. 10.1136/gutjnl-2012-303249 [DOI] [PubMed] [Google Scholar]
- 126.Dogra S, Sakwinska O, Soh S-E, et al. (2015) Dynamics of infant gut microbiota are influenced by delivery mode and gestational duration and are associated with subsequent adiposity. mBio 6:e02419–02414. 10.1128/mBio.02419-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rutayisire E, Huang K, Liu Y, and Tao F (2016) The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants' life: a systematic review. BMC Gastroenterology 16:86 10.1186/s12876-016-0498-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Martin JA, Hamilton BE, Osterman MJK, Driscoll AK, and Mathews TJ (2017) Births: Final Data for 2015. National vital statistics reports : from the Centers for Disease Control and Prevention, National Center for Health Statistics, National Vital Statistics System 66:1. [PubMed] [Google Scholar]
- 129.Betran AP, Torloni MR, Zhang J, et al. (2015) What is the optimal rate of caesarean section at population level? A systematic review of ecologic studies. Reproductive health 12:57 10.1186/s12978-015-0043-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sevelsted A, Stokholm J, Bønnelykke K, and Bisgaard H (2015) Cesarean section and chronic immune disorders. Pediatrics 135:e92–98. 10.1542/peds.2014-0596 [DOI] [PubMed] [Google Scholar]
- 131.Thavagnanam S, Fleming J, Bromley A, Shields MD, and Cardwell CR (2008) A meta-analysis of the association between Caesarean section and childhood asthma. Clinical & Experimental Allergy 38:629–633. 10.1111/j.1365-2222.2007.02780.x [DOI] [PubMed] [Google Scholar]
- 132.Bager P, Wohlfahrt J, and Westergaard T (2008) Caesarean delivery and risk of atopy and allergic disease: meta-analyses. Clin Exp Allergy 38:634–642. 10.1111/j.1365-2222.2008.02939.x [DOI] [PubMed] [Google Scholar]
- 133.Planer JD, Peng Y, Kau AL, Blanton LV, Ndao IM, Tarr PI, Warner BB, and Gordon JI (2016) Development of the gut microbiota and mucosal IgA responses in twins and gnotobiotic mice. Nature 534:263–266. 10.1038/nature17940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Martens EC, Chiang HC, and Gordon JI (2008) Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell host & microbe 4:447–457. 10.1016/j.chom.2008.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rogier EW, Frantz AL, Bruno MEC, Wedlund L, Cohen DA, Stromberg AJ, and Kaetzel CS (2014) Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc Natl Acad Sci U S A 111:3074–3079. 10.1073/pnas.1315792111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Newburg DS (2005) Innate immunity and human milk. J Nutr 135:1308–1312 [DOI] [PubMed] [Google Scholar]
- 137.Isaacs CE (2005) Human milk inactivates pathogens individually, additively, and synergistically. J Nutr 135:1286–1288 [DOI] [PubMed] [Google Scholar]
- 138.Jost T, Lacroix C, Braegger CP, Rochat F, and Chassard C (2014) Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ Microbiol 16:2891–2904. 10.1111/1462-2920.12238 [DOI] [PubMed] [Google Scholar]
- 139.Martín V, Maldonado-Barragán A, Moles L, Rodriguez-Baños M, Campo RD, Fernández L, Rodríguez JM, and Jiménez E (2012) Sharing of bacterial strains between breast milk and infant feces. J Hum Lact 28:36–44. 10.1177/0890334411424729 [DOI] [PubMed] [Google Scholar]
- 140.Gueimonde M, Laitinen K, Salminen S, and Isolauri E (2007) Breast milk: a source of bifidobacteria for infant gut development and maturation? Neonatology 92:64–66. 10.1159/000100088 [DOI] [PubMed] [Google Scholar]
- 141.Fernández L, Langa S, Martin V, Maldonado A, Jiménez E, Martín R, and Rodríguez JM (2013) The human milk microbiota: Origin and potential roles in health and disease. Pharmacological Research 69:1–10. 10.1016/j.phrs.2012.09.001 [DOI] [PubMed] [Google Scholar]
- 142.Wolf JH (2003) Low breastfeeding rates and public health in the United States. American journal of public health 93:2000–2010. 10.2105/AJPH.93.12.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.(1989) Protecting, promoting and supporting breast-feeding: The special role of maternity services. A Joint WHO/UNiCeF Statement. World Health Organization 1–36 [Google Scholar]
- 144.Dogaru CM, Nyffenegger D, Pescatore AM, Spycher BD, and Kuehni CE (2014) Breastfeeding and childhood asthma: systematic review and meta-analysis. American journal of epidemiology 179:1153–1167. 10.1093/aje/kwu072 [DOI] [PubMed] [Google Scholar]
- 145.Lodge CJ, Tan DJ, Lau MXZ, Dai X, Tham R, Lowe AJ, Bowatte G, Allen KJ, and Dharmage SC (2015) Breastfeeding and asthma and allergies: a systematic review and meta-analysis Acta paediatrica (Oslo, Norway: : 1992) 104:38–53. 10.1111/apa.13132 [DOI] [PubMed] [Google Scholar]
- 146.Ahmadizar F, Vijverberg SJH, Arets HGM, de Boer A, Garssen J, Kraneveld AD, and Maitland-van der Zee AH (2017) Breastfeeding is associated with a decreased risk of childhood asthma exacerbations later in life. Pediatric allergy and immunology : official publication of the European Society of Pediatric Allergy and Immunology 28:649–654. 10.1111/pai.12760 [DOI] [PubMed] [Google Scholar]
- 147.Brew BK, Allen CW, Toelle BG, and Marks GB (2011) Systematic review and meta-analysis investigating breast feeding and childhood wheezing illness. Paediatric and perinatal epidemiology 25:507–518. 10.1111/j.1365-3016.2011.01233.x [DOI] [PubMed] [Google Scholar]
- 148.Sharma ND (2017) Breastfeeding and the risk of childhood asthma: A two-stage instrumental variable analysis to address endogeneity. Pediatric allergy and immunology : official publication of the European Society of Pediatric Allergy and Immunology 28:564–572. 10.1111/pai.12750 [DOI] [PubMed] [Google Scholar]
- 149.Maurice CF, Haiser HJ, and Turnbaugh PJ (2013) Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152:39–50. 10.1016/j.cell.2012.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ferrer M, Méndez-García C, Rojo D, Barbas C, and Moya A (2017) Antibiotic use and microbiome function. Biochemical Pharmacology 134:114–126. 10.1016/j.bcp.2016.09.007 [DOI] [PubMed] [Google Scholar]
- 151.Dethlefsen L, and Relman DA (2011) Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A 108 Suppl 1:4554–4561. 10.1073/pnas.1000087107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Langdon A, Crook N, and Dantas G (2016) The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome medicine 8:39 10.1186/s13073-016-0294-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Murk W, Risnes KR, and Bracken MB (2011) Prenatal or early-life exposure to antibiotics and risk of childhood asthma: a systematic review. Pediatrics 127:1125–1138. 10.1542/peds [DOI] [PubMed] [Google Scholar]
- 154.Kew KM, Undela K, Kotortsi I, and Ferrara G (2015) Macrolides for chronic asthma. Cochrane Database Syst RevCD002997. 10.1002/14651858.CD002997.pub4 [DOI] [PubMed] [Google Scholar]
- 155.Penders J, Kummeling I, and Thijs C (2011) Infant antibiotic use and wheeze and asthma risk: a systematic review and meta-analysis. Eur Respir J 38:295–302. 10.1183/09031936.00105010 [DOI] [PubMed] [Google Scholar]
- 156.Zhao D, Su H, Cheng J, Wang X, Xie M, Li K, Wen L, and Yang H (2015) Prenatal antibiotic use and risk of childhood wheeze/asthma: A meta-analysis. Pediatric allergy and immunology : official publication of the European Society of Pediatric Allergy and Immunology 26:756–764. 10.1111/pai.12436 [DOI] [PubMed] [Google Scholar]
- 157.Cox LM, Yamanishi S, Sohn J, et al. (2014) Altering the Intestinal Microbiota during a Critical Developmental Window Has Lasting Metabolic Consequences. Cell 158:705–721. 10.1016/j.cell.2014.05.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cho I, Yamanishi S, Cox L, et al. (2012) Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 488:621–626. 10.1038/nature11400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Nobel YR, Cox LM, Kirigin FF, et al. (2015) Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat Commun 6:7486 10.1038/ncomms8486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Russell SL, Gold MJ, Hartmann M, et al. (2012) Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO reports 13:440–447. 10.1038/embor.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Russell SL, Gold MJ, Willing BP, Thorson L, McNagny KM, and Finlay BB (2013) Perinatal antibiotic treatment affects murine microbiota, immune responses and allergic asthma. Gut Microbes 4:158–164. 10.4161/gmic.23567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Miyoshi J, Bobe AM, Miyoshi S, Huang Y, Hubert N, Delmont TO, Eren AM, Leone V, and Chang EB (2017) Peripartum Antibiotics Promote Gut Dysbiosis, Loss of Immune Tolerance, and Inflammatory Bowel Disease in Genetically Prone Offspring. Cell reports 20:491–504. 10.1016/j.celrep.2017.06.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gomez-Arango LF, Barrett HL, McIntyre HD, Callaway LK, Morrison M, and Dekker Nitert M (2017) Antibiotic treatment at delivery shapes the initial oral microbiome in neonates. Sci Rep 7:43481 10.1038/srep43481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nogacka A, Salazar N, Suárez M, et al. (2017) Impact of intrapartum antimicrobial prophylaxis upon the intestinal microbiota and the prevalence of antibiotic resistance genes in vaginally delivered full-term neonates. 1–10. 10.1186/s40168-017-0313-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Allen SJ, Martinez EG, Gregorio GV, and Dans LF (2010) Probiotics for treating acute infectious diarrhoea. Cochrane Database Syst RevCD003048. 10.1002/14651858.CD003048.pub3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kalliomäki M, Salminen S, Arvilommi H, Kero P, Koskinen P, and Isolauri E (2001) Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet (London, England) 357:1076–1079. 10.1016/S0140-6736(00)04259-8 [DOI] [PubMed] [Google Scholar]
- 167.Kalliomäki M, Salminen S, Poussa T, Arvilommi H, and Isolauri E (2003) Probiotics and prevention of atopic disease: 4-year follow-up of a randomised placebo-controlled trial. Lancet (London, England) 361:1869–1871. 10.1016/S0140-6736(03)13490-3 [DOI] [PubMed] [Google Scholar]
- 168.Osborn DA, and Sinn JK (2007) Probiotics in infants for prevention of allergic disease and food hypersensitivity. Cochrane Database Syst RevCD006475. 10.1002/14651858.CD006475.pub2 [DOI] [PubMed] [Google Scholar]
- 169.Elazab N, Mendy A, Gasana J, Vieira ER, Quizon A, and Forno E (2013) Probiotic administration in early life, atopy, and asthma: a meta-analysis of clinical trials. Pediatrics 132:e666–676. 10.1542/peds.2013-0246 [DOI] [PubMed] [Google Scholar]
- 170.Cabana MD (2014) No consistent evidence to date that prenatal or postnatal probiotic supplementation prevents childhood asthma and wheeze. Evid Based Med 19:144 10.1136/eb-2014-101721 [DOI] [PubMed] [Google Scholar]
- 171.Davies G, Jordan S, Brooks CJ, et al. (2018) Long term extension of a randomised controlled trial of probiotics using electronic health records. Sci Rep 8:7668 10.1038/s41598-018-25954-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Fiocchi A, Pawankar R, Cuello-Garcia C, et al. (2015) World Allergy Organization-McMaster University Guidelines for Allergic Disease Prevention (GLAD-P): Probiotics. World Allergy Organ J 8:4 10.1186/s40413-015-0055-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.West CE, Dzidic M, Prescott SL, and Jenmalm MC (2017) Bugging allergy; role of pre-, pro- and synbiotics in allergy prevention. Allergol Int 66:529–538. 10.1016/j.alit.2017.08.001 [DOI] [PubMed] [Google Scholar]
- 174.Krzych-Falta E, Furmanczyk K, Tomaszewska A, Olejniczak D, Samolinski B, and Samolinska-Zawisza U (2018) Probiotics: Myths or facts about their role in allergy prevention. Adv Clin Exp Med 27:119–124. 10.17219/acem/65476 [DOI] [PubMed] [Google Scholar]
- 175.van de Pol MA, Lutter R, Smids BS, Weersink EJ, and van der Zee JS (2011) Synbiotics reduce allergen-induced T-helper 2 response and improve peak expiratory flow in allergic asthmatics. Allergy 66:39–47. https://doi.Org/10.1111/j.1398-9995.2010.02454.x [DOI] [PubMed] [Google Scholar]
- 176.Williams NC, Johnson MA, Shaw DE, Spendlove I, Vulevic J, Sharpe GR, and Hunter KA (2016) A prebiotic galactooligosaccharide mixture reduces severity of hyperpnoea-induced bronchoconstriction and markers of airway inflammation. Br J Nutr 116:798–804. 10.1017/S0007114516002762 [DOI] [PubMed] [Google Scholar]
- 177.Osborn DA, and Sinn JK (2013) Prebiotics in infants for prevention of allergy. Cochrane Database Syst RevCD006474. 10.1002/14651858.CD006474.pub3 [DOI] [PubMed] [Google Scholar]
- 178.Cuello-Garcia C, Fiocchi A, Pawankar R, et al. (2017) Prebiotics for the prevention of allergies: A systematic review and meta-analysis of randomized controlled trials. Clin Exp Allergy 47:1468–1477. 10.1111/cea.13042 [DOI] [PubMed] [Google Scholar]
- 179.Plovier H, Everard A, Druart C, et al. (2017) A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med 23:107–113. 10.1038/nm.4236 [DOI] [PubMed] [Google Scholar]
- 180.Casale TB, Kessler J, and Romero FA (2006) Safety of the intranasal toll-like receptor 4 agonist CRX-675 in allergic rhinitis. Ann Allergy Asthma Immunol 97:454–456. 10.1016/S1081-1206(10)60934-9 [DOI] [PubMed] [Google Scholar]