The increasing association of the etiological agent of cholera, Vibrio cholerae serogroup O1 and O139, with multiple antibiotic resistance threatens to deprive health practitioners of this effective tool. Drug resistance in cholera results mainly from acquisition of mobile genetic elements. Genomic islands conferring multidrug resistance and mobilizable by IncC conjugative plasmids were reported to circulate in non-O1/non-O139 V. cholerae clinical strains isolated from the 2010 Haitian cholera outbreak. As these genomic islands can be transmitted to pandemic V. cholerae serogroups, their mechanism of transmission needed to be investigated. Our research revealed plasmid- and genomic island-encoded factors required for the resistance island excision, mobilization, and integration, as well as regulation of these functions. The discovery of related genomic islands carrying diverse phage resistance genes but lacking antibiotic resistance-conferring genes in a wide range of marine dwelling bacteria suggests that these elements are ancient and recently acquired drug resistance genes.
KEYWORDS: antibiotic resistance, IncC plasmids, mobilization, relaxase, T4CP, T4SS, Vibrio cholerae, conjugation, genomic islands, horizontal gene transfer, oriT, phage resistance
ABSTRACT
Cholera remains a formidable disease, and reports of multidrug-resistant strains of the causative agent Vibrio cholerae have become common during the last 3 decades. The pervasiveness of resistance determinants has largely been ascribed to mobile genetic elements, including SXT/R391 integrative conjugative elements, IncC plasmids, and genomic islands (GIs). Conjugative transfer of IncC plasmids is activated by the master activator AcaCD whose regulatory network extends to chromosomally integrated GIs. MGIVchHai6 is a multidrug resistance GI integrated at the 3′ end of trmE (mnmE or thdF) in chromosome 1 of non-O1/non-O139 V. cholerae clinical isolates from the 2010 Haitian cholera outbreak. In the presence of an IncC plasmid expressing AcaCD, MGIVchHai6 excises from the chromosome and transfers at high frequency. Herein, the mechanism of mobilization of MGIVchHai6 GIs by IncC plasmids was dissected. Our results show that AcaCD drives expression of GI-borne genes, including xis and mobIM, involved in excision and mobilization. A 49-bp fragment upstream of mobIM was found to serve as the minimal origin of transfer (oriT) of MGIVchHai6. The direction of transfer initiated at oriT was determined using IncC plasmid-driven mobilization of chromosomal markers via MGIVchHai6. In addition, IncC plasmid-encoded factors, including the relaxase TraI, were found to be required for GI transfer. Finally, in silico exploration of Gammaproteobacteria genomes identified 47 novel related and potentially AcaCD-responsive GIs in 13 different genera. Despite sharing conserved features, these GIs integrate at trmE, yicC, or dusA and carry a diverse cargo of genes involved in phage resistance.
IMPORTANCE The increasing association of the etiological agent of cholera, Vibrio cholerae serogroup O1 and O139, with multiple antibiotic resistance threatens to deprive health practitioners of this effective tool. Drug resistance in cholera results mainly from acquisition of mobile genetic elements. Genomic islands conferring multidrug resistance and mobilizable by IncC conjugative plasmids were reported to circulate in non-O1/non-O139 V. cholerae clinical strains isolated from the 2010 Haitian cholera outbreak. As these genomic islands can be transmitted to pandemic V. cholerae serogroups, their mechanism of transmission needed to be investigated. Our research revealed plasmid- and genomic island-encoded factors required for the resistance island excision, mobilization, and integration, as well as regulation of these functions. The discovery of related genomic islands carrying diverse phage resistance genes but lacking antibiotic resistance-conferring genes in a wide range of marine dwelling bacteria suggests that these elements are ancient and recently acquired drug resistance genes.
INTRODUCTION
Cholera is an acute diarrheal disease that leads to severe dehydration and often death in the absence of adequate treatment (1). The seventh cholera pandemic, which began in 1961, is caused by toxigenic strains of the Gammaproteobacteria Vibrio cholerae serogroup O1 biotype El Tor, or more sporadically its O139 variant (1). Since the late 1980s, antibiotic-resistant V. cholerae strains have emerged and spread globally (2). Development of drug resistance in seventh pandemic V. cholerae has been ascribed to mutation but it mostly involves acquisition of mobile genetic elements, including genomic island GI-15, integrative conjugative elements of the SXT/R391 family, and conjugative plasmids of incompatibility group C (IncC) (3–5).
IncC plasmids are large (>120-kbp) broad-host-range conjugative plasmids frequently associated with multidrug resistance in several species of globally distributed pathogenic enterobacteria, and in seventh pandemic V. cholerae strains from Africa, China, and Haiti (4, 6–8). Conjugative transfer of IncC plasmids is controlled by the FlhCD-like heteromeric transcriptional activator AcaCD that they encode (9). AcaCD activates 17 promoters conserved in IncC plasmids, including those driving expression of transfer genes and operons encoding type IV secretion system (T4SS) and conjugative pilus, relaxase TraI, putative type IV coupling protein (T4CP) TraD, and unknown function protein TraJ (9). TraI belongs to the MOBH1 family of relaxases, and together with the product of mobIC, is essential for initiation of conjugative transfer at the origin of transfer (oriT) (10, 11). MobIC is responsible for recognition of the oriT locus of IncC plasmids that is located immediately upstream of mobIC (12). Unlike other transfer genes, mobIC seems to be expressed in an AcaCD-independent manner (9).
Furthermore, AcaCD also triggers excision of at least three types of genomic islands (GIs) shown to be mobilizable in trans by IncC plasmids. The first type, integrated at the 3′ end of the gene of unknown function yicC, is exemplified by the 16.5-kbp MGIVmi1 of Vibrio mimicus (9). The two other types of GIs are inserted into the 3′ end of trmE (also known as mnmE or thdF), a gene encoding the 5-carboxymethylaminomethyluridine-tRNA synthase GTPase subunit (13, 14). One type of GI is illustrated by the 42.4-kbp Salmonella genomic island 1 (SGI1) that confers resistance to ampicillin, chloramphenicol, streptomycin/spectinomycin, sulfamethoxazole, and tetracycline (ACSSuT) in Salmonella enterica (14, 15). The other is illustrated by the 47.4-kbp MGIVchHai6 of V. cholerae HC-36A1 that confers not only the ACSSuT phenotype but also trimethoprim and possibly mercury resistance (Fig. 1A) (13). MGIVchHai6 was found in non-O1/non-O139 V. cholerae strains isolated from patients exhibiting symptoms of cholera at the onset of the 2010 cholera outbreak in Haiti. Despite integrating into the same site as SGI1, MGIVchHai6 encodes a distantly related integrase Int (67% identity) and recombination directionality factor (RDF) Xis (37% identity) (13). MGIVchHai6 also lacks most of the genes and sequences that enable the mobilization of SGI1 by IncC plasmids (13, 16, 17). On the basis of these structural differences, the mechanisms of mobilization of MGIVchHai6 and SGI1 by IncC plasmids are expected to differ considerably. In MGIVchHai6, xis is the last gene of a putative operon-like structure preceded by a putative AcaCD-controlled promoter (Fig. 1A). A second AcaCD binding site is located inside an open reading frame (ORF) that encodes a distant homolog of MobIC (27% identity over two fragments of 109 and 53 amino acid residues) (13). Besides AcaCD, IncC plasmid- and GI-encoded factors involved in MGIVchHai6 mobilization have not been characterized.
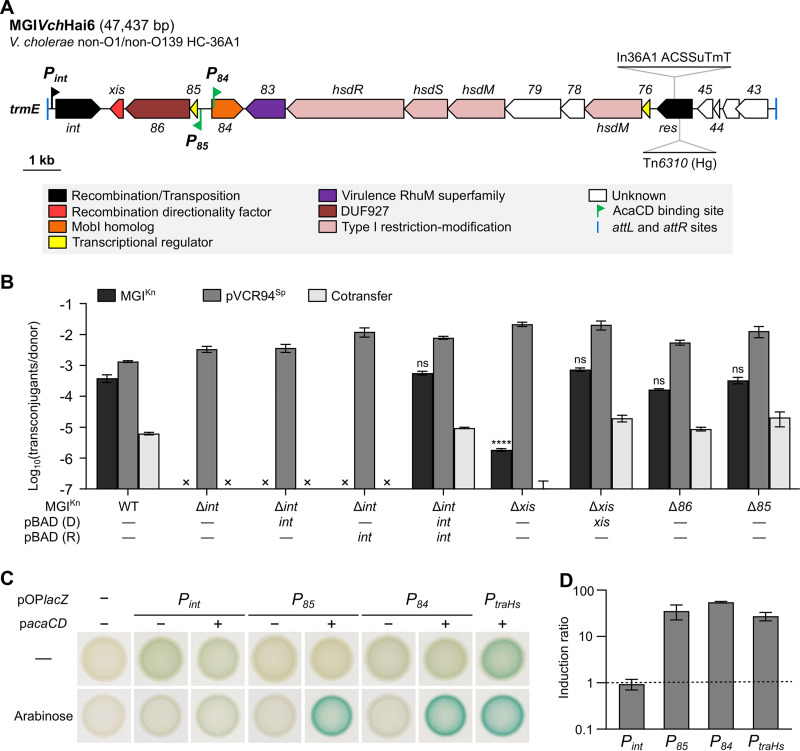
FIG 1.
Role and regulation of int and 85-86-xis in the mobilization of MGIVchHai6. (A) Schematic genetic map of MGIVchHai6 drawn to scale. The left and right junctions (attL and attR) within the host chromosome are indicated by blue ticks at the extremities. ORFs with similar function are color coded as indicated in the figure. Green flags indicate the position and orientation of predicted AcaCD binding sites (13). The black flag indicates the position and orientation of the Pint promoter. The insertion sites of In36A1 integron and Tn6310 transposon are shown. The gene numbers correspond to the last digits of the respective locus tags in GenBank accession no. AXDR01000001. ACSSuTmT, resistance to ampicillin, chloramphenicol, spectinomycin/streptomycin, sulfamethoxazole, trimethoprim, and tetracycline; Hg, mercury resistance. (B) Mobilization assays of MGIKn or its Δint, Δxis, Δ86, or Δ85 mutants were carried out using E. coli GG56 (Nx) bearing pVCR94Sp as the donor strain, and CAG18439 (Tc) as the recipient strain. Complementation assays were performed in the donor (D) or recipient (R) strain by expressing the missing gene from PBAD on pBAD-int or pBAD-xis. An “×” indicates that the transfer frequency was below the detection limit (<10−7). Bars represent the means ± standard errors of the means (error bars) from three independent experiments. Statistical analyses were carried out on the logarithm of the values using a one-way analysis of variance (ANOVA), followed by Dunnett’s multiple-comparison test with the wild-type (WT) MGIKn as the control. Statistical significance is indicated as follows: ****, P < 0.0001; ns, not significant. (C) β-Galactosidase activities of Pint, P85, and P84 transcriptionally fused to lacZ. Colonies were grown on LB agar with or without arabinose to induce acaCD expression from pacaCD. (D) Induction levels of Pint, P85, and P84 in response to AcaCD. β-Galactosidase assays were carried out using the strains of panel C. Ratios between the enzymatic activities in Miller units for the arabinose-induced versus noninduced strains containing pacaCD are shown. The AcaCD-regulated promoter PtraHs of SGI1 served as a positive control, and cells devoid of pOPlacZ served as a negative control. The bars represent the means ± standard errors of the means (error bars) from two independent experiments.
In this report, we established a model of mobilization of MGIVchHai6 by helper IncC plasmids and compared this model to SGI1 mobilization. Deletion mutants of the helper plasmid and MGI were used in mating assays to characterize the contribution of each element in MGIVchHai6 mobilization. The presence of AcaCD binding sites upstream of xis and inside the putative gene encoding a MobIC homolog suggests the presence of AcaCD-responsive promoters that were verified using lacZ reporter fusions. By analogy with IncC plasmids, we hypothesized that the MobIC homolog encoded by MGIVchHai6 recognizes a cognate oriT locus located upstream of its gene. The ability of IncC plasmids to mobilize chromosomal DNA through MGIVchHai6 was also tested and provided the directionality of transfer initiated at oriT. Finally, phylogenetic analyses based on the mobilization factor revealed the existence of a large class of potential IncC-mobilized GIs that are integrated at three different chromosomal sites in mostly marine dwelling species of Gammaproteobacteria. Our results show that MGIVchHai6-like GIs share a mechanism of mobilization by helper IncC plasmids that differs from the one used by SGI1-like GIs.
RESULTS
int and xis are essential for excision and mobilization of MGIVchHai6.
MGIVchHai6 carries a large cargo of antibiotic and mercury resistance genes. To make it more amenable for this study, we constructed a kanamycin (Kn)-resistant mutant of MGIVchHai6 that lacks In36A1, Tn6310, and res (Fig. 1A). The resulting 19.5-kb mutant, named hereafter MGIKn, was mobilized by the helper IncC plasmid pVCR94Sp at the same frequency as MGIVchHai6 ([5.2 ± 0.3] × 10−4 versus [5.7 ± 1.2] × 10−4, P = 0.622, Student’s t test).
To establish whether int is required for mobilization of MGIVchHai6, we carried out mobilization assays using pVCR94Sp and a Δint mutant of MGIKn. Deletion of int abolished MGIKn mobilization (Fig. 1B). Complementation assays done by expressing int from the arabinose-inducible promoter PBAD on pBAD-int restored mobilization to the wild-type level only when pBAD-int was present in both the donor and recipient strains. Therefore, int is likely required for excision of MGIKn in donor cells and for its integration into the chromosome of recipient cells.
The predicted (RDF) gene xis encodes a 114-amino-acid (aa)-residue protein containing a predicted helix-turn-helix (HTH_17, Pfam accession no. PF12728) domain (position 39 to 89). To assess the role of xis, we mobilized the Δxis mutant of MGIKn using pVCR94Sp. Transfer of this mutant was reduced ∼200-fold compared to the wild type (Fig. 1B). Complementation using pBAD-xis in donor cells was sufficient to restore the mobilization of MGIKn Δxis to the wild-type level, thereby confirming that Xis is required only in donor cells, likely to facilitate Int-mediated excision of the GI.
xis is located downstream of two open reading frames (ORFs), 85 and 86 (the gene numbers correspond to the last digits of the respective locus tags in GenBank accession no. AXDR01000001), of unknown function. The predicted translation products of 85 is a 65-aa-residue protein that, like xis, contains an HTH_17 domain (position 11 to 62). 86 encodes a predicted 558-aa-residue protein containing a domain of unknown function (DUF927, Pfam accession no. PF06048) in its N-terminal half. Mobilization assays using Δ85 and Δ86 mutants of MGIKn and pVCR94Sp revealed that neither 85 nor 86 is involved in the excision, mobilization, or integration of MGIKn under laboratory conditions, as the frequency of mobilization remained unaffected by deletions (Fig. 1B).
To validate the involvement of int and xis in the excision step, we carried out PCR excision assays to detect the attP site on the plasmid-like form of excised MGIKn, using E. coli GG56 (nalidixic acid) bearing MGIKn or MGIKn Δint. Spontaneous excision was undetectable, as shown by the absence of attP PCR product (see Fig. S1 in the supplemental material). Overexpression of int from Pint did not trigger excision, whereas overexpression of xis resulted in excision of MGIKn, but not of MGIKn Δint. Thus, excision of MGIVchHai6 is induced by Xis and requires Int, in line with their proposed roles of RDF and integrase, respectively.
Ethidium bromide-stained 1.5% agarose gel of PCR products for detection of MGIKn excision. Excision assays were carried out in E. coli GG56 bearing MGIKn or its Δint mutant and, where indicated, arabinose-inducible vector pBAD-int or pBAD-xis. Download FIG S1, PDF file, 0.08 MB (87KB, pdf) .
Copyright © 2020 Rivard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
AcaCD-dependent activation of mobility genes of MGIVchHai6.
AcaCD binding sites were previously detected upstream of 85 and inside 84 (13). To test whether AcaCD activates promoter sequences upstream of int and 85 or inside 84, Pint, P85, and P84 were introduced upstream of a single-copy, chromosomal promoterless lacZ gene cassette that is transcriptionally isolated by the terminator sequences rgnB and tl3 (9). The AcaCD-responsive promoter sequence PtraHs of SGI1 (17) was used as a positive control. The β-galactosidase activity of each promoter was monitored in the presence and absence of ectopically expressed acaCD.
Pint yielded a weak yet constitutive β-galactosidase activity regardless of the presence of AcaCD (Fig. 1C), confirming that the promoter that drives expression of the integrase gene is not controlled by AcaCD. In contrast, P85, which likely drives expression of the RDF gene xis, did not appear to produce detectable β-galactosidase activity in the absence of AcaCD (Fig. 1C). When acaCD was expressed, P85 activity increased 35-fold (Fig. 1D). Finally, P84 exhibited a weak, constitutive expression similar to Pint and weaker than PtraHs in the absence of AcaCD (Fig. 1C). Like PtraHs, P84 was strongly induced (55-fold increase) upon expression of acaCD (Fig. 1D).
These results confirm that the two promoter sequences P85 and P84 containing the predicted AcaCD binding sites are activated by AcaCD. In contrast, Pint drives low-level constitutive expression of the integrase gene.
MobIM is required for MGIVchHai6 mobilization.
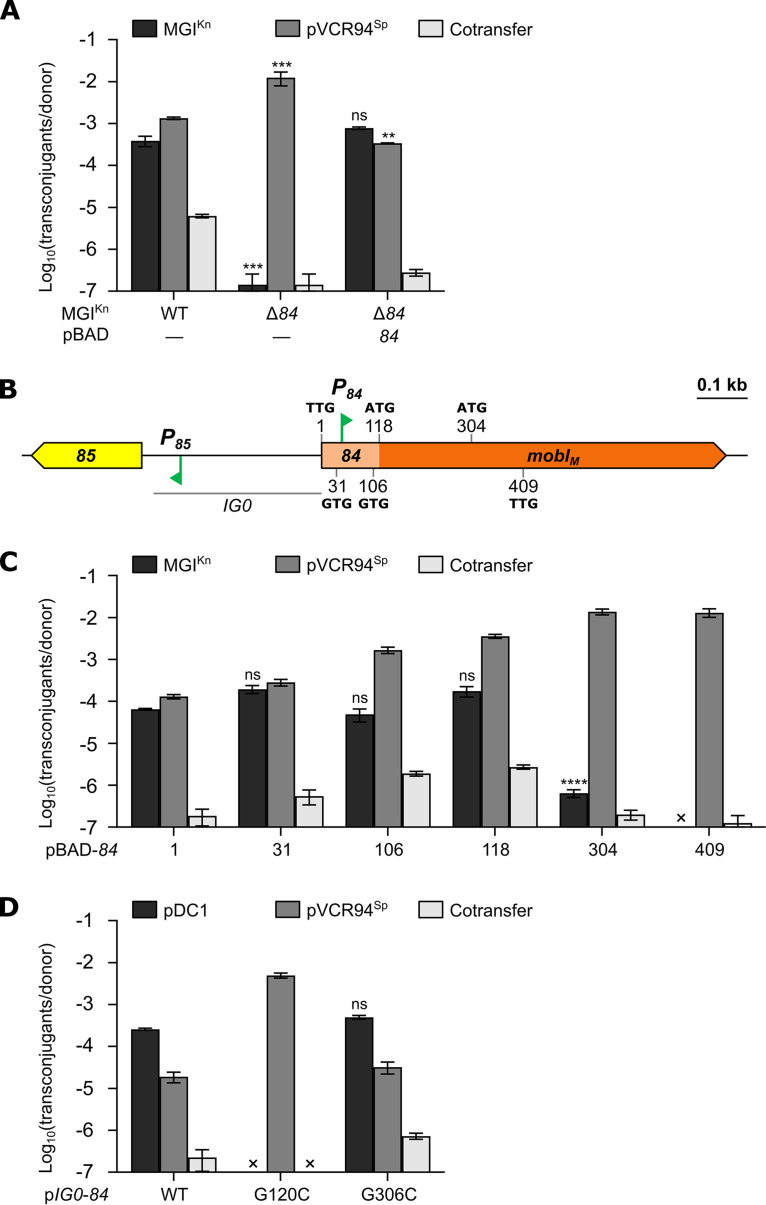
The 795-bp open reading frame 84 (locus VCHC36A1_0084 of V. cholerae HC-36A1) of MGIVchHai6 encodes a distant homolog of MobIC (27% identity). Since MobIC is a key factor for IncC plasmid transfer, we wondered whether 84 could play an important role in mobilization of MGIVchHai6. To test the hypothesis, we constructed MGIKn Δ84 and carried out mobilization assays with pVCR94Sp. MGIKn transfer was strongly impaired by this deletion and reduced ∼2,700-fold (Fig. 2A). In this context, we observed a slight (12-fold), yet statistically significant, increase of pVCR94Sp transfer. Complementation of the Δ84 mutation from pBAD-84 restored MGIKn mobilization to the wild-type level, while reducing transfer of pVCR94Sp ∼40-fold, suggesting that 84 is essential for MGIVchHai6 mobilization. However, the position of the AcaCD-responsive P84 promoter inside 84 suggested that the protein effective for mobilization is smaller than predicted. To test this hypothesis, we constructed complementation plasmids containing fragments of 84 starting at the following alternative start codons: +31, GTG; +106, GTG; +118, ATG; +304, ATG; and +409, TTG (Fig. 2B). All plasmids but pBAD-84-304 and pBAD-84-409 restored mobilization of MGIKn Δ84 by pVCR94Sp to the wild-type level (Fig. 2C). Therefore, the ORF located downstream of P84 and starting at ATG118 in 84 is the likely gene that allows MGIVchHai6 mobilization. This gene, hereafter referred to as mobIM, produces a putative 225-aa-residue protein.
FIG 2.
Role of 84 in MGIVchHai6 mobilization. (A) Mobilization of MGIKn or its Δ84 mutant by pVCR94Sp. (B) Schematic representation of the 85-84 region of MGIVchHai6. Genes are color coded as indicated in Fig. 1A. The position and sequence of alternative start codons within 84 are indicated. (C) Complementation assays of the Δ84 mutation by alternative ORFs within 84. When indicated, donor strains contained the complementation plasmid pBAD-84 or one of its derivatives (Table 1). (D) Confirmation of ATG118 as the genuine start codon of mobIM. Conjugation assays were performed using E. coli GG56 (Nx) containing the specified elements as donor strains and either CAG18439 (Tc) (A and C) or VB112 (Rf) (D) as the recipient strain. The bars represent the means ± standard errors of the means from three independent experiments. Statistical analyses were carried out on the logarithm of the values using a one-way ANOVA followed by Dunnett’s multiple-comparison test with the WT MGIKn (A), pBAD-84 (C), or pIG0-84 (D) as the control. Statistical significance is indicated as follows: ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; ns, not significant.
Localization and characterization of the origin of transfer (oriT).
In IncC plasmids, oriT is located directly upstream of mobIC (10, 12). By analogy, we predict that oriT of MGIVchHai6 is located at the corresponding position, i.e., upstream of mobIM. To confirm this hypothesis, we first cloned the intergenic region located between 85 and 84 (IG0) as well as 84 into the low-copy-number nonmobilizable plasmid pDC1. The resulting plasmid, pIG0-84, was mobilized by pVCR94Sp at a frequency comparable to that of MGIKn (Fig. 2D), thereby confirming the cloned fragment sufficient to support mobilization by the IncC plasmid even in the absence of MGIVchHai6. Site-directed mutagenesis G120C (ATG to ATC) and G306C (ATG to ATC) in 84 confirmed that ATG118 is the start codon of mobIM since pIG0-84 carrying the mutation G120C was not mobilizable, whereas mutation G306C had no effect on transfer (Fig. 2D).
Since we were able to use pBAD-84-118 to complement and mobilize the Δ84 mutant of MGIKn that lacks the 117-bp segment containing the AcaCD-responsive promoter P84, the possibility that oriT could be located in this sequence or in mobIM was ruled out. Therefore, we cloned IG0 and the region extending upstream of mobIM (IG10) into pDC1 and carried out mobilization assays using pVCR94Sp to provide the conjugative machinery. Donor cells also carried pT84 to provide MobIM. The empty vector and the same vector bearing IG0 in the absence of pT84 were both unable to transfer (Fig. 3A). In contrast, when pT84 was present in donor cells, the vector bearing IG10 or IG0 transferred at frequencies that were comparable to that of MGIKn Δ84 complemented with pT84. This result indicated that oriT is located within IG0.
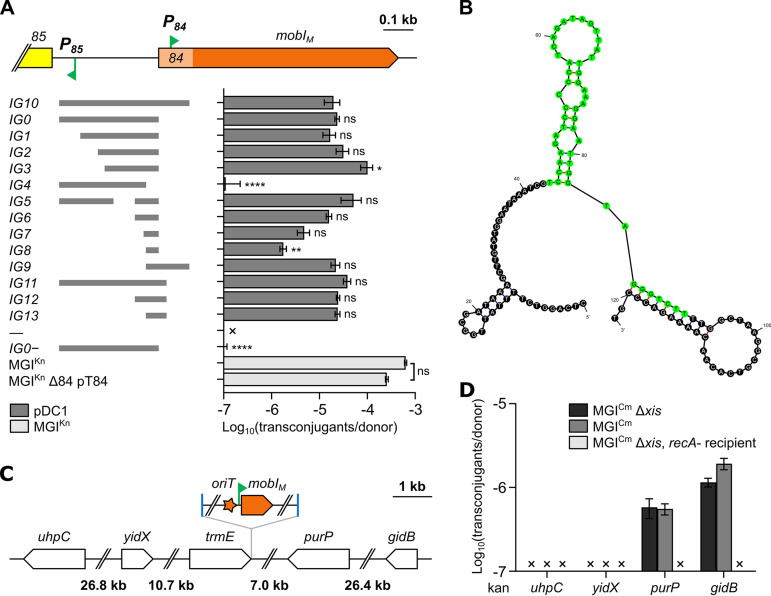
FIG 3.
Localization of the oriT locus of MGIVchHai6. (A) On the left, fragments of the 85-mobI region that were cloned into the nonmobilizable vector pDC1 are represented by gray bars. The resulting transfer frequencies for the corresponding fragments are presented on the right side. Mobilization assays of pDC1 derivatives were performed using E. coli GG56 (Nx) bearing pVCR94Sp and pT84 as the donor and VB112 (Rf) as the recipient. “—” indicates an empty pDC1. “IG0−” indicates that the transfer of pIG0 was assessed in the absence of pT84. Statistical analyses were carried out on the logarithm of the values using a one-way ANOVA followed by Dunnett’s multiple-comparison test with pIG10 or MGIKn as the control. Statistical significance is indicated as follows: ****, P < 0.0001; **, P < 0.01; *, P < 0.05; ns, not significant. (B) Predicted folding of IG12, with IG8 highlighted in green. Folding of the upper strand (panel A) was predicted using the Mfold web server (74). (C) Schematic map of the chromosomal region surrounding trmE in E. coli K-12. The position and orientation of MGICm are indicated. (D) MGICm-mediated mobilization of chromosomal markers from E. coli JW3642, JW3692, JW3718, or JW5858 (Kn) bearing pVCR94Sp and MGICm or its Δxis mutant to CAG18439 or its ΔrecA mutant (Tc). In panels A and D, the bars represent the means ± standard errors of the means from three independent experiments. “×” indicates that the transfer frequency was below the detection limit (<10−7).
Insert reduction was then performed to find the minimal sequence of IG0 allowing mobilization by pVCR94Sp. The smallest insert capable of acting as a suitable oriT, IG8, was 49 bp long and located immediately upstream of 84 (Fig. 3A). Although functional, IG8 provided only 1/10th of the mobilization activity of IG0 or larger inserts such as IG6 or IG13 that provided additional upstream or downstream sequences. The addition of either 43 bp upstream (IG6) or 30 bp downstream (IG13) led to transfer frequencies equivalent to that obtained with IG10, and addition of both fragments (IG12) did not enhance transfer further. Predicted folding of IG12 revealed three potential stem-loop structures within IG8 and on either side, highlighting the presence of repeated sequences potentially involved in relaxosome assembly (Fig. 3B).
Directionality of transfer initiated at oriT.
To determine the direction of conjugative transfer initiated at oriT of MGIVchHai6, chromosomal markers located upstream and downstream of trmE, the integration site of MGIVchHai6, were tested for mobilization. Accordingly, MGICm or the excision-defective mutant MGICm Δxis were introduced together with pVCR94Sp into Escherichia coli BW25113 derivatives carrying a kanamycin resistance (Knr) marker integrated at uhpC, yidX, purP, or gidB (Keio knockout collection) (18). These genes are located between 7 and 39.5 kb on either side of trmE (Fig. 3C). Mobilization assays failed to produce any transconjugants when the Knr marker was inserted upstream of trmE. In contrast, transfer was easily detectable for Knr insertions at purP and gidB that are located downstream of trmE, regardless of the ability of MGICm to excise from the chromosome (Fig. 3D). This result demonstrates that transfer of MGIVchHai6 initiated at oriT progresses downstream of mobIM and that the last genes translocated into the recipient cells are xis-86-85.
Involvement of IncC DNA processing genes in MGIVchHai6 mobilization.
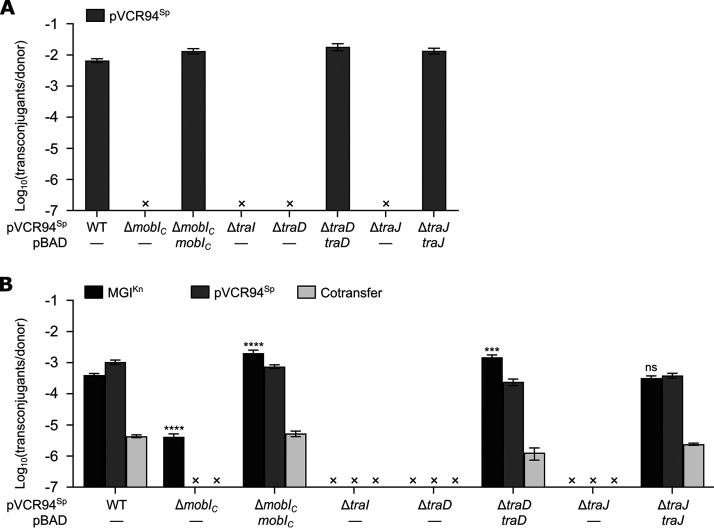
To test whether DNA processing genes of IncC plasmids are involved in the mobilization of MGIVchHai6, we constructed nonpolar deletion mutants of mobIC, traI, traD, and traJ in pVCR94Sp and carried out conjugation assays. Each individual deletion abolished transfer of pVCR94Sp. Except for traI, all deletions could be complemented by ectopic expression of the corresponding gene (Fig. 4A). Complementation of traI could not be tested as attempts to clone an intact copy of this gene failed, suggesting that expression under PBAD, even in the absence of arabinose, is toxic.
FIG 4.
Mobilization of MGIVchHai6 relies on the T4CP and relaxase encoded by IncC plasmids. (A) Impact of mobIC, traI, traD, and traJ deletions on pVCR94Sp transfer. (B) Mobilization of MGIKn by the mobIC, traI, traD, and traJ deletion mutants of pVCR94Sp. Conjugation assays were performed with E. coli GG56 (Nx) containing the specified elements as donor strains and CAG18439 (Tc) as the recipient strain. The bars represent the means ± standard errors of the means from three independent experiments. “×” indicates that the transfer frequency was below the detection limit (<10−7). Statistical analyses were carried out on the logarithm of the values using a one-way ANOVA followed by Dunnett’s multiple-comparison test with WT MGIKn as the control. Statistical significance is indicated as follows: ****, P < 0.0001; ***, P < 0.001; ns, not significant.
As observed for pVCR94Sp, deletion of traI, traD, and traJ abolished MGIKn mobilization. Complementation of each mutation restored mobilization to the wild-type level (Fig. 4B). Therefore, these genes appear to be essential for processing and transfer of excised MGIKn DNA. Interestingly, although MGIVchHai6 encodes its own MobIM factor, deletion of mobIC resulted in ∼100-fold reduction of MGIKn mobilization (Fig. 4B), whereas complementation with pBAD-mobIC restored mobilization to the wild-type level.
Likewise, we confirmed that MGIVchHai6 relies exclusively on the T4SS encoded by IncC plasmids as deletion of traHC, traGC, or traNC abolished mobilization of MGIKn (Fig. S2). Moreover, mobilization of MGIKn by pVCR94Sp into a recipient strain carrying pVCR94Cm was abolished (Fig. S2), thus confirming that MGIVchHai6 does not evade IncC entry exclusion.
Mobilization of MGIKn by pVCR94Sp T4SS mutants. “pVCR94Cm (R)” indicates that the element was present in the recipient strain. Donor and recipient strains are the same as in Fig. 4. Statistical analyses were carried out on the logarithm of the values using a one-way ANOVA followed by Dunnett’s multiple comparison test with WT MGIKn as the control. Statistical significance is indicated as follows: **, P < 0.01; ns, not significant. Download FIG S2, PDF file, 0.05 MB (47.9KB, pdf) .
Copyright © 2020 Rivard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MGIVchHai6 is the prototype of a large and diverse subfamily of GIs mobilizable by IncC plasmids.
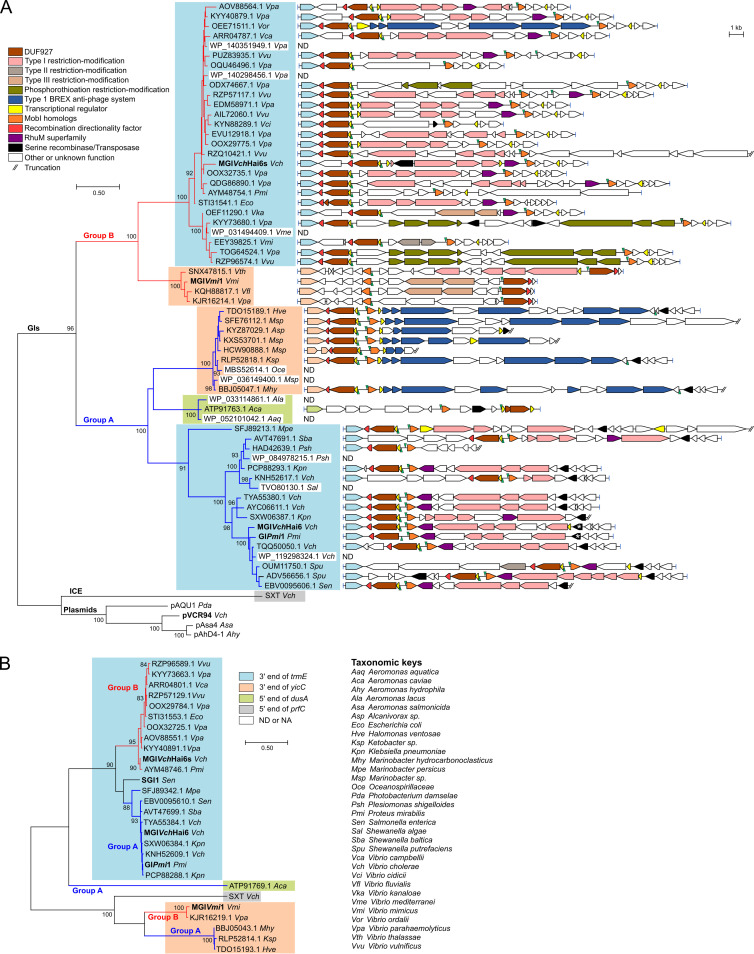
Given the importance of MobI-like factors, we searched the GenBank database for MobI homologs and extracted associated sequences to assess diversity of the GIs related to MGIVchHai6 (see Table S1 in the supplemental material). Phylogenetic analyses of MobI proteins revealed that MobI homologs encoded by GIs are distinct from those encoded by conjugative plasmids and cluster in two major groups (Fig. 5A). Group A contains GIs integrated at the 3′ end of trmE or yicC, and at the 5′ end of dusA. With the exception of the GI inserted at dusA, all GIs encoding group A MobI proteins exhibited the same structure: int-xis-86-85-ig-mobI, where ig likely contains the oriT, and xis-86-85 and mobI are divergent and preceded by AcaCD-like binding sites. In a few GIs, variable DNA is inserted between the convergent int and xis genes. MGIVchHai6 and GIPmi1 belong to group A. GIPmi1 of Proteus mirabilis resembles MGIVchHai6, and it contains a large multidrug resistance cluster inserted at the same position but lacks the mercury resistance transposon Tn6310 (19). Most trmE-specific group A GIs encode predicted type I restriction-modification (R/M) systems. In contrast, the yicC-specific group A GIs encode predicted type 1 BREX antiphage systems.
FIG 5.
Diversity of genomic islands encoding MobI homologs. (A and B) Maximum likelihood phylogenetic analysis of MobI (A) and Int (B) homologs. Trees with the highest likelihoods (−7,967.92 and −5,688.94 for MobI and Int, respectively) are shown. Bootstrap supports are indicated as percentages at the branching points only when >80%. Branch lengths represent the number of substitutions per site over 201 and 311 amino acid positions for MobI and Int, respectively. Only one representative per cluster of similar proteins (>95% identity threshold) is shown in each tree. In panel A, the schematic structure of the genomic island encoding the corresponding MobI protein is shown next to each node. ORFs with similar function are color coded as indicated in the panel. Each node and the integrase gene of the corresponding node are color coded based on the integration site of the genomic island (refer to panel B for color key [ND, not determined; NA, not available]). Vertical green arrowheads indicate position and orientation of AcaCD binding sites. attL and attR attachment sites flanking each GI are represented by blue bars. The asterisks in MGIVchHai6 and GIPmi1 indicate the insertion site of the complex resistance integrons. Additional details on GIs and host strains are provided in Table S1 in the supplemental material.
Features of GIs presented in Fig. 5 and accession numbers. Download Table S1, XLSX file, 0.03 MB (33KB, xlsx) .
Copyright © 2020 Rivard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
GIs coding for group B MobI proteins are integrated either at the 3′ end of trmE or 3′ end of yicC. In group B GIs, the xis-86-85 cluster is separated from mobI by a large region of variable DNA often encoding predicted type I or phosphorothioation R/M systems. Furthermore, while trmE-specific group B GIs conserved the overall structure of group A GIs, yicC-specific group B GIs lack 85 and have undergone an inversion of the xis-86-to-mobI region relative to the int gene. These GIs encode predicted type I and type II R/M systems. Surprisingly, analysis undertaken in this study led to discovery of MGIVchHai6s, a group B GI inserted at trmE in a tandem fashion directly downstream of MGIVchHai6 in V. cholerae HC-36A1. Unlike MGIVchHai6, MGIVchHai6s does not carry antibiotic or heavy metal resistance genes. The group B GI of P. mirabilis JN40 encoding AYM48754.1 is also integrated in a tandem fashion downstream of SGI1-PmJN40, an SGI1-like element that confers multidrug resistance (20). Thus, tandem integration of different GIs in trmE is not a rare occurrence.
Phylogenetic analysis of integrases encoded by respective GIs revealed similar clustering into groups A and B within trmE- and yicC-specific integrase clusters (Fig. 5B). Although the integrase encoded by SGI1 also mediates integration at trmE, it could not be ascribed to either group with confidence (<80 bootstraps), which is consistent with the considerable structural and genetic differences between SGI1-like and MGIVchHai6-like GIs.
DISCUSSION
Bacterial conjugation typically results from concerted action of the cytoplasmic relaxosome responsible for DNA processing initiated at oriT, and the type IV secretion system (T4SS) that translocates the processed DNA across cell membranes into the recipient cell (21, 22). The type IV coupling protein (T4CP), an inner membrane-anchored protein, acts as a relaxosome docking station at the T4SS. In most cases, the relaxosome comprises a relaxase working together with auxiliary proteins that either help or are required for the relaxase activity. Most of the auxiliary factors hitherto described alter DNA topology by either locally bending DNA at the oriT or unwinding DNA through a helicase activity (23–29). Several oriT-binding factors, such as TraJ of RP4, TrwA of R388, and Int of Tn916, take part in specific recruitment of the relaxase at this locus (30–34). Other known auxiliary factors are involved in recognition of the T4CP or in relaxosome stabilization by protein-protein interactions (34–36).
Together with SXT/R391 elements, IncC and IncA plasmids share a set of transfer genes encoding a T4SS of the MPFF family, a coupling protein (TraD), a relaxase of the MOBH1 family (TraI), and two essential factors thought to be part of the relaxosome (TraJ [DUF4400, PF14348] and MobI) (8, 10, 11, 37). We have also shown here that TraJ is essential to transfer of IncC plasmids and MGIVchHai6 (Fig. 4A and B). While results obtained with SXT and the IncHI1 plasmid R27 suggest a role as a relaxosome component (37–39), the exact function of TraJ remains unknown. MobI is required for conjugative transfer of IncC plasmids and SXT/R391 integrative and conjugative elements (ICEs) as well as GIs that mimic the oriT locus of the latter but lack a mobI gene (10, 40, 41). In contrast, deletion of mobI in the helper element does not abolish transfer of pCloDF13 and SGI1, which possess unique oriT loci and cognate mobilization proteins MobBC for pCloDF13 and MpsAB for SGI1 (12, 16, 40, 42). MGIVchHai6 lacks the mobilization genes mpsAB and the oriT locus that are essential for SGI1 mobilization (13, 16). Instead, we showed that MGIVchHai6 encodes MobIM, a distant homolog of the IncC plasmid-encoded MobIC that provides some independence from mobIC (Fig. 1A, 2A, and 4B). Deletion of mobIC in the IncC plasmid R16a was shown to enhance (45-fold increase) mobilization of SGI1-C, suggesting competition between SGI1-C and its helper plasmid for the conjugative machinery (12). In contrast, we found that the absence of mobIC impaired transfer of MGIVchHai6 (∼100-fold decrease) (Fig. 4B), suggesting that MobIC enhances initiation of transfer mediated by TraI and MobIM at oriT of MGIVchHai6. However, deletion of mobIM, while abolishing MGIVchHai6 mobilization, also enhanced transfer of the helper plasmid (12-fold increase) (Fig. 2A), consistent with competition between the two elements.
We identified the oriT locus of MGIVchHai6 within the intergenic region upstream of mobIM (IG0) and found that a 49-bp region (IG8) was sufficient to promote mobilization of a nonmobilizable plasmid. While IG0 shares low nucleotide identity (45%) with the oriT of pVCR94 (10, 12), the 49-bp IG8 shares 63% identity with this oriT, with the last 32 bp of IG8 sharing 84% with the corresponding region in pVCR94 oriT. Nevertheless, this level of conservation is notably limited compared to the similarity reported between oriT of SXT and the GIs it mobilizes (>63% over ∼300 bp) (41). This lack of conservation likely accounts for the requirement for MobIM to achieve optimal transfer of MGIVchHai6. Together with the absence of a MobI homolog in SXT-mobilizable GIs, the specificity of MobIM and MobIC for their respective elements further supports the proposed role for MobI as an auxiliary relaxosome component involved in oriT recognition (40, 43).
SGI1 encodes three functional T4SS subunits, TraHS, TraGS, and TraNS, that displace the homologous subunits encoded by IncC plasmids despite strong amino acid sequence divergences (64, 37, and 78% identity, respectively) (17). This alteration of the mating channel is crucial to enhance SGI1 mobilization (13, 17). Incorporation of TraGS into the IncC mating pore allows SGI1 to circumvent entry exclusion exerted by an IncC plasmid in the recipient cells, allowing SGI1 to spread freely, even in a population of cells carrying IncC plasmids (17, 44). In contrast, MGIVchHai6 lacks genes encoding T4SS subunits. Predictably, MGIVchHai6 was shown here to conform to IncC entry exclusion and thus is unable to transfer to a strain already containing an IncC plasmid (see Fig. S2 in the supplemental material).
In SGI1, AcaCD binding sites have been identified upstream of xis, S004-rep, traNS, traHGS, and S018 and corresponding promoters shown to respond to AcaCD activation (9, 17, 45–47). In MGIVchHai6, putative AcaCD binding sites were predicted at the 5′ end of 84 and upstream of an operon-like gene cluster containing xis, which encodes a putative protein sharing only 37% identity with the RDF Xis of SGI1 (13). In this study, Xis is indeed an RDF acting together with Int to catalyze excision under the control of AcaCD (Fig. 1B and C and Fig. S1). Regulation of xis and mobI by AcaCD is consistent with the previously proposed model in which these GIs remain quiescent in the chromosome in the absence of an IncC plasmid (43).
MGIVchHai6-like elements have been detected in silico in environmental and clinical O1 and non-O1/non-O139 V. cholerae isolates from the Indian subcontinent and South America, and in Shewanella sp. from North America, although those seem to be devoid of antibiotic resistance genes (13). IDH‐03944, a cotrimoxazole resistance-conferring GI related to MGIVchHai6, was recently reported in a 2011 isolate of V. cholerae O44 recovered from diarrheal patients in Kolkata, India (48). Comparative genomics revealed a much more diverse set of GIs related to MGIVchHai6 in the genome of marine dwelling species and integrated at the 3′ end of trmE and yicC or at the 5′ end of dusA (Fig. 5). The gene cargo of these GIs is predominantly associated with DNA modification (methylation, phosphorothioation) and restriction systems, as well as antiphage systems such as BREX (Fig. 5A). Only MGIVchHai6 and GIPmi1, together with IDH‐03944 (48), contained integrons carrying antibiotic resistance genes, suggesting that these elements are an ancient, large reservoir of antiphage systems, recently hijacked as vectors for drug resistance genes. Most GIs of the MGIVchHai6 and MGIVchHai6s clades share a gene encoding a predicted RhuM-like virulence factor, usually immediately upstream or downstream of mobIM (Fig. 5). A mutant of rhuM located in S. enterica SPI-3 pathogenicity island is deficient for epithelial cell invasion, neutrophil transmigration, and killing of its Caenorhabditis elegans host (49). While these results suggest that MGIVchHai6-like elements may be involved in virulence modulation, the molecular function of RhuM is not known and its potency in the Vibrionaceae has not been assessed. One striking feature of MGIVchHai6-like elements is the syntenic conservation of an operon-like region typically including a predicted AcaCD binding site followed by genes encoding a putative transcriptional regulator containing a helix-turn-helix (HTH) domain, a DUF927 domain-containing protein, and the xis gene (Fig. 5A). While the DUF927 gene is ubiquitous, GIs of the MGIVmi1 clade lack the upstream transcriptional regulator gene and the AcaCD binding site is located directly upstream of the DUF927 gene. Such high conservation is even more surprising since 85 and 86 are dispensable for mobilization of MGIVchHai6 by its helper plasmid (Fig. 1B). Why such factors should be under the control of the IncC transfer activator AcaCD is puzzling. Interestingly, a similar region exists in GIs mobilized by ICEs of the SXT/R391 family (Fig. S3). In prototypical MGIVflInd1, a binding site of the transfer activator SetCD lies upstream of rdfM and cds8, which encode a predicted transcriptional regulator and a DUF927 domain-encoding protein. While the role of cds8 remains elusive, RdfM shares weak amino acid identity (25%) with the product of 85 and acts as an RDF, facilitating excision of MGIVflInd1 upon setCD expression (50). In addition to the aforementioned factors, MGIVflInd1 and several MGIVchHai6-like GIs share an integration site at the 3′ end of yicC (Fig. 5B), an observation that prompted us to hypothesize that Xis and 85/RdfM act as alternative RDFs, each allowing excision from a specific integration site. To test this, a ΔrdfM mutant of MGIVflInd1 was complemented by overexpressing 85 in a strain also containing ICEVflInd1 to provide setCD. However, 85 failed to restore excision and an attP or attB junction was not detected (data not shown), suggesting that 85 has a different function or is too divergent to promote excision of MGIVflInd1.
Comparison of MGIVchHai6 and the conserved core of MGIVflInd1. The maps are drawn to scale. The left and right junctions (attL and attR) within the host chromosome are indicated by blue ticks at the extremities. ORFs with similar function are color coded as indicated in the legend. Green arrows indicate the position and orientation of AcaCD and SetCD binding sites. Homologous regions are bracketed and linked by a gray line with the corresponding percentage of nucleotide identity. MGIVflInd1 can be accessed through GenBank accession number KC117176. Download FIG S3, PDF file, 0.07 MB (67.6KB, pdf) .
Copyright © 2020 Rivard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
SXT/R391 ICEs have been shown to promote conjugative transfer of chromosomal DNA located 3′ of prfC, their integration site, and remotely 5′ of yicC, the integration site of the GIs they mobilize (41, 51). Thus, these elements are able to mobilize large stretches (≥1Mbp) of chromosomal DNA locally and remotely in an Hfr-like manner, suggesting that SXT/R391 ICEs play an evolutionary role that extends beyond their own dissemination. Our results show that IncC plasmids can mobilize chromosomal DNA located downstream of trmE by mobilization of MGIs without their prior excision from the chromosome (Fig. 3). Given the presence of MGIVchHai6-like elements integrated at different chromosomal sites and across a wide range of Vibrionaceae and other species of Gammaproteobacteria, IncC plasmids and their subordinate GIs can be concluded to comprise a potent driving force in the gene flow circulating in many bacterial pathogens. In fact, this is superbly exemplified by the recently reported presence of an MGIVchHai6-like GI in V. cholerae Santiago-089, a non-O1/non-O139 clinical isolate harboring many virulence genes scattered throughout chromosome 1 (52). Not only is the GI itself poised to be mobilized by an incoming IncC plasmid—along with the GI-borne antibiotic and mercury resistance genes—but it is also plausible that it may, in fact, usher transfer of downstream elements GIVch-T3SS and VPI-2, thus simultaneously contributing to dissemination of virulence determinants.
Cholera continues to cause epidemics that include millions of cases worldwide (53). The geographical range of V. cholerae is expected to expand dramatically as climate change renders the marine environment increasingly hospitable to this pathogen (54, 55). While the ability to promote epidemic outbreaks was traditionally regarded as an appanage of O1 and O139 strains (56), it is becoming increasingly clear that the actual picture is far more nuanced (52, 57–60). In various species, cumulative acquisition of antibiotic resistance and/or virulence determinants through exchange of genomic islands has time and again allowed emergence of virulent strains, some of which lack canonical virulence hallmarks (61–63). IncC plasmids circulating in non-O1/non-O139 V. cholerae populations may prove to comprise the perfect trigger for emergence of unforeseen pandemics.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains and plasmids used in this study are described in Table 1. The strains were routinely grown in lysogeny broth (LB) (EMD) at 37°C in an orbital shaker and stored at −80°C in LB broth with 15% (vol/vol) glycerol. The following antibiotics and concentrations were employed: ampicillin (Ap), 50 μg/ml; chloramphenicol (Cm), 20 μg/ml; kanamycin (Kn), 10 μg/ml for single-copy integrants of pOPlacZ-derived constructs, 50 μg/ml otherwise; nalidixic acid (Nx), 40 μg/ml; rifampin (Rf), 50 μg/ml: spectinomycin (Sp), 50 μg/ml; streptomycin (Sm), 200 μg/ml; tetracycline (Tc), 12 μg/ml. Conjugation assays were performed as previously described (17). However, donors and recipients were selected according to their sole chromosomal markers. When required, mating experiments were performed using LB plates with 0.02% arabinose to induce expression of pBAD30-derived complementation vectors. The frequencies of transconjugant formation were computed as ratios of transconjugant per donor CFU from three independent mating experiments.
TABLE 1.
Strains and plasmids used in this study
| Strain, plasmid, or element |
Relevant genotype or phenotypea | Reference |
|---|---|---|
| Vibrio cholerae | ||
| HC-36A1 | Clinical, non-O1/non-O139, Haiti, 2010 (Ap Cm Kn Sp Sm Su Tc Tm) | 57 |
| Escherichia coli | ||
| BW25113 | F− Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− rph-1 Δ(rhaD-rhaB)568 hsdR514 | 75 |
| GG56 | Nxr derivative of BW25113 (Nx) | 76 |
| VB112 | Rfr derivative of MG1655 (Rf) | 40 |
| CAG18439 | MG1655 lacZU118 lacI42::Tn10 (Tc) | 77 |
| VB47 | CAG18439 ΔrecA ΔgalK | 78 |
| JW3642 | BW25113 ΔuhpC772::aph (Kn) | 18 |
| JW3692 | BW25113 ΔpurP745::aph (Kn) | 18 |
| JW3718 | BW25113 ΔgidB769::aph (Kn) | 18 |
| JW5858 | BW25113 ΔyidX732::aph (Kn) | 18 |
| Plasmids | ||
| pVCR94 | IncC conjugative plasmid, V. cholerae O1 El Tor (Su Tm Cm Ap Tc Sm) | 10 |
| pVCR94Sp | Spr derivative of pVCR94 (pVCR94ΔX2) (Su Sp) | 9 |
| pVCR94Kn | Knr derivative of pVCR94 (pVCR94ΔX3) (Su Kn) | 9 |
| pVCR94Cm | Cmr derivative of pVCR94 (pVCR94ΔX4) (Su Cm) | 17 |
| pVCR94ΔmobIC | mobIC deletion mutant of pVCR94Sp (Su Sp) | This study |
| pVCR94ΔtraI | traI deletion mutant of pVCR94Sp (Su Sp) | This study |
| pVCR94ΔtraD | traD deletion mutant of pVCR94Sp (Su Sp) | This study |
| pVCR94ΔtraJ | traJ deletion mutant of pVCR94Sp (Su Sp) | This study |
| pVCR94ΔtraN | traN deletion mutant of pVCR94Sp (Su Sp) | 9 |
| pVCR94ΔtraG | traG deletion mutant of pVCR94Sp (Su Sp) | 9 |
| pVCR94ΔtraH | traH deletion mutant of pVCR94Sp (Su Sp) | 9 |
| pBAD30 | orip15A bla araC PBAD (Ap) | 79 |
| pacaCD | pBAD30::acaDC (Ap) | 9 |
| pBAD-mobIC | pBAD30::mobIC (Ap) | This study |
| pBAD-traD | pBAD30::traD (Ap) | This study |
| pBAD-traJ | pBAD30::traJ (Ap) | This study |
| pBAD-traN | pBAD30::traN (Ap) | 17 |
| pBAD-traG | pBAD30::traG (Ap) | 17 |
| pBAD-traH | pBAD30::traH (Ap) | 17 |
| pBAD-int | pBAD30::int (Ap) | This study |
| pBAD-xis | pBAD30::xis (Ap) | This study |
| pT84 | pBAD-TOPO::84 (Ap) | This study |
| pBAD-84 | pBAD30::84 (Ap) | This study |
| pBAD-84-31 | pBAD30::84 starting at position +31 of 84 (Ap) | This study |
| pBAD-84-106 | pBAD30::84 starting at position +106 of 84 (Ap) | This study |
| pBAD-84-118 | pBAD30::84 starting at position +118 of 84 (Ap) | This study |
| pBAD-84-304 | pBAD30::84 starting at position +304 of 84 (Ap) | This study |
| pBAD-84-409 | pBAD30::84 starting at position +409 of 84 (Ap) | This study |
| pOPlacZ | oriR6Kγ attPλ aph lacZ (Kn) | 9 |
| pPromint | pOPlacZ containing −163 to −12 of MGIVchHai6 Pint (Kn) | This study |
| pProm85 | pOPlacZ containing −389 to −11 of MGIVchHai6 P85 (Kn) | This study |
| pPrommobI | pOPlacZ containing −345 to −11 of MGIVchHai6 P84 (Kn) | This study |
| pPromtraHS | pOPlacZ containing −243 to −14 of SGI1 PtraHs (Kn) | 17 |
| pDC1 | pACYC184 Δcat (ΔMscI-PvuII) (Tc) | 40 |
| pIG0-84 | pDC1::IG0-84 (Tc) | This study |
| pIG0-84G120C | pDC1::IG0-84 G120C (Tc) | This study |
| pIG0-84G306C | pDC1::IG0-84 G306C (Tc) | This study |
| pIG0 | pDC1::IG0 (Tc) | This study |
| pIG1 | pDC1::IG1 (Tc) | This study |
| pIG2 | pDC1::IG2 (Tc) | This study |
| pIG3 | pDC1::IG3 (Tc) | This study |
| pIG4 | pDC1::IG4 (Tc) | This study |
| pIG5 | pDC1::IG5 (Tc) | This study |
| pIG6 | pDC1::IG6 (Tc) | This study |
| pIG7 | pDC1::IG7 (Tc) | This study |
| pIG8 | pDC1::IG8 (Tc) | This study |
| pIG9 | pDC1::IG9 (Tc) | This study |
| pIG10 | pDC1::IG10 (Tc) | This study |
| pIG11 | pDC1::IG11 (Tc) | This study |
| pIG12 | pDC1::IG12 (Tc) | This study |
| pIG13 | pDC1::IG13 (Tc) | This study |
| pINT-ts | oriR101 cI857 λpR-intλ (ts, Ap) | 80 |
| pSIM6 | λRed recombination thermo-inducible encoding plasmid (ts, Ap) | 81 |
| pMS1 | λRed recombination thermo-inducible encoding plasmid (ts, Gm) | 10 |
| pKD3 | cat (Cm) template for one-step chromosomal gene inactivation | 75 |
| pKD4 | aph (Kn) template for one-step chromosomal gene inactivation | 75 |
| pCP20 | Flp recombinase thermo-inducible encoding plasmid (ts, Ap Cm) | 82 |
| pCP20-Gm | Gmr derivative of pCP20 (ts, Gm Cm) | 83 |
| Genomic islands | ||
| MGIVchHai6 | Drug resistance island, V. cholerae HC-36A1 (Ap Cm Sm Su Tm Tc) | 13 |
| MGIKn | Knr derivative of MGIVchHai6 lacking In36A1 and Tn6310 (Kn) | This study |
| MGICm | Cmr derivative of MGIVchHai6 lacking In36A1 and Tn6310 (Cm) | This study |
| MGIKnΔint | int deletion mutant of MGIKn (Kn) | This study |
| MGIKnΔxis | xis deletion mutant of MGIKn (Kn) | This study |
| MGICmΔxis | xis deletion mutant of MGICm (Cm) | This study |
| MGIKnΔ86 | 86 deletion mutant of MGIKn (Kn) | This study |
| MGIKnΔ85 | 85 deletion mutant of MGIKn (Kn) | This study |
| MGIKnΔ84 | 84 deletion mutant of MGIKn (Kn) | This study |
Ap, ampicillin; Cm, chloramphenicol; Gm, gentamicin, Kn, kanamycin; Nx, nalidixic acid; Rf, rifampin; Sp, spectinomycin; Sm, streptomycin; Su, sulfamethoxazole; Tc, tetracycline; Tm, trimethoprim; ts, thermosensitive.
Molecular biology methods.
Plasmid DNA was extracted using the EZ-10 Spin Column Plasmid DNA Minipreps kit (Bio Basic) following the manufacturer’s instructions. Enzymes used in this study were purchased from New England Biolabs. PCR assays were performed with the primers listed in Table 2. PCR conditions were as follows: (i) 3 min at 94°C; (ii) 30 cycles, with 1 cycle consisting of 30 s at 94°C, 30 s at the appropriate annealing temperature, and 1 min/kb at 68°C; and (iii) 5 min at 68°C. When required, the resulting products were purified using the EZ-10 Spin Column PCR Products purification kit (Bio Basic) following the manufacturer’s instructions. E. coli strains were transformed by electroporation as described previously (64) in a Bio-Rad Gene Pulser Xcell device set at 25 μF, 200 V, and 1.8 kV using 1-mm gap electroporation cuvettes.
TABLE 2.
DNA sequences of the primers used in this study
| Primer name | Nucleotide sequence (5′ to 3′)a | Reference |
|---|---|---|
| 94delmobI.for | ATGTGGATAGGAATTGGATAGGAATTGGGAGGGTATTGAGGTGTAGGCTGGAGCTGCTTC | This study |
| 94delmobI.rev | TCCCCAGTTTCGCCAATTCAGTGGCCGCTACAGATGCTGTCATATGAATATCCTCCTTA | This study |
| 94deltraI.for | TGTAGCGACGAGAAATACGATAGGAATATGGTCAACCTACGTGTAGGCTGGAGCTGCTTC | This study |
| 94deltraI.rev | CACGGCATCTCGTAGGCGAGCGGGTCATAACTCATTGTCATATGAATATCCTCCTTA | This study |
| 94deltraD.for | CAGGGAAAATTGTATTTAGAGGTAAATGAAACATGAGTGTAGGCTGGAGCTGCTTC | This study |
| 94deltraD.rev | TCTCTATACTTCTTGTCACACATAGCTCTGCTCTTACATATGAATATCCTCCTTA | This study |
| 94deltraJ.for | GCCGTGGTTGCTGGTCGCATGGCTGCTGGTTATTGAGTGTAGGCTGGAGCTGCTTC | This study |
| 94deltraJ.rev | ATTTATAACGGAGTAGCTCATCCTCCCTCCCTATACCATATGAATATCCTCCTTA | This study |
| Hai6delWE.for | TGCCGTAATCAAGGAAATGATACGGCAGAAAGTCGTGGAACGGAATAGGAACTTCAAGAT | This study |
| Hai6delWE.rev | TGATTACGACTACCGAAACCATATTGGGTTTAATGACTGAAGGAACTTCAGAGCGCTT | This study |
| Hai6delint.for | CTGGCGCACATATGGCGCACAAGATGAGAAAAACCTGTGTAGGCTGGAGCTGCTTC | This study |
| Hai6delint.rev | TTGTAAAGATGTTGTCGTTTGATCGTAATAATGCTCTTACATATGAATATCCTCCTTA | This study |
| Hai6delxis.for | TCCCACTTTAAGTTGGCAGTGTAGGAGAGTATCAACGTGTAGGCTGGAGCTGCTTC | This study |
| Hai6delxis.rev | CCATTCATATTTATTTTCAAAAGGTTAAATCTCAACTTACATATGAATATCCTCCTTA | This study |
| Hai6del86.for | GCTGCATGGTTGAGTGATTTATAAAAGGGGGAAGTGGTGTAGGCTGGAGCTGCTTC | This study |
| Hai6del86.rev | TTAAAGTGGGATAAACGGGAAACTGGGAACACTAAGTTACATATGAATATCCTCCTTA | This study |
| Hai6del85.for | TTACTCCTGCAATCTAGATAATAAACAGGAGCCATCGTGTAGGCTGGAGCTGCTTC | This study |
| Hai6del85.rev | CTGCAACTTTTTTTGCATCACTTCCCCCTTTTATAACATATGAATATCCTCCTTA | This study |
| Hai6delmobI.for | ATCAGATAGTTATTGGAAAGGAATTGGTAGGGTCTTGTGTAGGCTGGAGCTGCTTC | This study |
| Hai6delmobI.rev | CTCTAATGAGTTGAGGGCTGTGTTGTAAGATCATAATTACATATGAATATCCTCCTTA | This study |
| 94mobIEcoRI.for | NNNNNN GAATTC AAGGAGGAATAATAAATGAGTCTACCAACAGAGCGATGGCT | This study |
| 94mobIEcoRI.rev | NNNNNN GAATTC TCACACCTCGTCGCTATGTGTCTT | This study |
| 94traD61EcoRI.for | NNNN GAATTC AAGGAGGAATAATAAATGACAATGAGTTATGAC | This study |
| 94traD61EcoRI.rev | NNNN GAATTC TTACTGGGTCAATATGGGCAGAC | This study |
| 94traJ63EcoRI.for | NNNN GAATTC AAGGAGGAATAATAAATGAAGAAGCCGTGGTTGCTG | This study |
| 94traJ63EcoRI.rev | NNNN GAATTC CTATACCCGTTTCTGGAGGTTG | This study |
| Hai6intEcoRI.for | NNNN GAATTC AAGGAGGAATAATAAATGAAAATCAGCATACATAAAC | This study |
| Hai6intEcoRI.rev | NNNN GAATTC TTAGTCTGATGACTCATCGAAG | This study |
| Hai6xisEcoRI.for | NNNN GAATTC AAGGAGGAATAATAAATGACTGAAAAAGAGATGCTTC | This study |
| Hai6xisEcoRI.rev | NNNN GAATTC CTAATATCTCCGGCCATAC | This study |
| Hai6mobIXbaI.for | NNNNNN TCTAGA AAGGAGGAATAATAAATGGCTAAGGCGTCACAACAAAA | This study |
| Hai6mobIXbaI.rev | NNNNNN TCTAGA TTATAAAGGATTATCCATCTTAAAATCGTCGAT | This study |
| Hai6mobINcoI.for | CCATGG CTAAGGCGTCACAAC | This study |
| Hai6mobI.rev | TTATAAAGGATTATCCATCTTAAAATCGTCGAT | This study |
| Hai6promintPstI.f | NNNNNN CTGCAG AATAGTAAAACACATCAAACC | This study |
| Hai6promintPstI.r | NNNNNN CTGCAG ATCTTGTGCGCCATATGTGCG | This study |
| Hai6prom85PstI.f | NNNNNN CTGCAG AATAACTATCTGATGGGGGAT | This study |
| Hai6prom85PstI.r | NNNNNN CTGCAG GTTTATTATCTAGATTGCAGG | This study |
| Hai6prom84PstI.f | NNNNNN CTGCAG AATGAGTTGCTCGACATTAC | This study |
| Hai6prom84PstI.r | NNNNNN CTGCAG ACTGTATGTTGAATACAGAG | This study |
| 43_attPF | GCAATTAATGATAAAGACGGGTA | 13 |
| Ec104D.rev | AACCATTTTGAGGTCACACA | 15 |
| Hai6_attPR | TCAAATCACTTCCCACCAAG | This study |
| Start_mobI_F1 | GTGCCCCAAAAGGGCACGAAG | This study |
| Start_mobI_F2 | GTGGATTATGTCATGACCAA | This study |
| Start_mobI_F3 | ATGACCAAAAATTATTCATTTTTAG | This study |
| Start_mobI_F4 | ATGCTTTTTTCATTATCAGTTTCAG | This study |
| Start_mobI_F5 | TTGTTCTGGCAGCACAACCGA | This study |
| Start_mobI_Rv2 | TTATTATTCCTCCTTTCTAGAGGATC | This study |
| Hai6oriT84XbaI.for | NNNNNN TCTAGA TTGCAGGAGTAATCATCTCGAAAAA | This study |
| mobI120_GC.F | ATTATGTCATCACCAAAAATTATTCATTTTTAG | This study |
| mobI120_GC.R | CCACTGTATGTTGAATACAG | This study |
| mobI306_GC.F | ACGATGCTATCCTTTTTTCATTATC | This study |
| mobI306_GC.R | CTTGATTAACGTTTGTGC | This study |
| Hai6oriTXbaI.rev | NNNNNN TCTAGA AAGACCCTACCAATTCCTTTC | This study |
| minoriT1.F | CACGCTGATATTTAATGATATTTATTC | This study |
| minoriT1-4a.R | TCTAGAAATATTTTATCTGATTAATAAGATG | This study |
| minoriT2.F | AGCCACCAATGAGTTGCT | This study |
| minoriT3.F | CCGTATCAATTTATGCGC | This study |
| minoriT4b.F | TCTAGATTTCAGTGCAATTTATC | This study |
| minoriT4b.R | ACGATTTATTCCATACAAGC | This study |
| minoriT5.F | CTCAGGTCTTTTATTGGC | This study |
| minoriT5.R | CTCTGGTTATTCACTGGC | This study |
| minoriT7.F | AATAAATCGTCCAAGATCC | This study |
| minoriT8.F | TCCAAGATCCCCCATCAG | This study |
| minoriT8XbaI.F | NNNN TCTAGA TCCAAGATCCCCCATCAG | This study |
| minoriT9XbaI.R | NNNN TCTAGA GACATAATCCACTGTATGTTG | This study |
| minoriT11.R | ACGGGTCTTTTGTTGTGAC | This study |
| Hai6delWEcm.for | TGCCGTAATCAAGGAAATGATACGGCAGAAAGTCGTGGAATACCTGTGACGGAAGATCAC | This study |
| Hai6delWEcm.rev | TGATTACGACTACCGAAACCATATTGGGTTTAATGACTGATAGGAACTTCATTTAAATGG | This study |
Restriction sites are underlined.
Plasmid and strain construction.
The plasmids and primers used in this study are listed in Tables 1 and 2, respectively. Detailed description of plasmid and strain construction is provided in Text S1 in the supplemental material.
Plasmid and strain construction. Download Text S1, DOCX file, 0.02 MB (21.7KB, docx) .
Copyright © 2020 Rivard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Detection of MGIVchHai6 excision.
Excision of MGIVchHai6 was detected by PCR on genomic DNA of the appropriate strains using primers listed in Table 2. The attR and attP sites were respectively amplified using primer pairs 43_attPF/Ec104D.rev and 43_attPF/Hai6_attPR.
β-Galactosidase assays.
Qualitative assays were performed by depositing 10-μl aliquots of overnight cultures with appropriate antibiotics on solid agar supplemented with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) with or without 0.02% arabinose. The plates were observed after an overnight incubation at 37°C.
Quantitative assays were performed with 2-nitrophenyl-β-d-galactopyranoside (ONPG) according to a protocol adapted from Miller (65). After an overnight incubation at 37°C with appropriate antibiotics, cultures were diluted 1:100 in 50 ml LB broth supplemented with 50 μg/ml ampicillin and grown until an optical density at 600 nm (OD600) of 0.2 was reached. Two series of 1/10 dilutions were then prepared in total volumes of 5 ml LB broth supplemented with 50 μg/ml ampicillin with or without 0.2% arabinose and incubated for 2 h at 37°C.
Phylogenetic analyses.
The primary sequence of homologs of MobI proteins encoded by MGIVchHai6 and MGIVmi1 were obtained using the NCBI blastp algorithm (66) against the nr/nt database restricted to Gammaproteobacteria (taxid: 1236). Primary sequences sharing less than 45% identity and under 85% minimum coverage were filtered out of subsequent analyses. Distant MobI homologs from SXT (GenBank accession no. EET25017.1), pVCR94 (GenBank WP_001447712.1), pAhD4-1 (GenBank ALZ82609.1), pAsa4 (GenBank ABO92354.1), and pAQU1 (GenBank WP_014386842.1) were introduced manually in the data set as an outgroup. MobI homologs were first clustered with CD-HIT (67) to the best cluster that met the 0.95 identity cutoff prior to alignment. The predicted primary sequences of Int homologs were recovered from a representative sample of GIs encoding MobI homologs. Int primary sequences of SXT and SGI1 (GenBank accession no. AF261825.2) were introduced manually. Phylogenetic analyses were computed using amino acid alignments generated by MUSCLE (68). Prior to phylogenetic analysis, poorly aligned regions were discarded using trimAl v1.3 software with the automated heuristic approach (69). Evolutionary analyses were performed within MEGA X (v 10.0.5) (70) using the maximum likelihood method (PhyML) (71) and either the JTT (MobI) or LG (Int) matrix-based models (72, 73). The initial tree(s) for the heuristic search was obtained automatically by applying neighbor-joining and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model and then selecting the topology with superior log likelihood value.
ACKNOWLEDGMENTS
We are grateful to Maude Chabot for technical assistance and members of the Burrus laboratory for insightful discussions.
This work was supported by Discovery Grant (2016-04365) from the Natural Sciences and Engineering Council of Canada (NSERC) and project grant (PJT-153071) from the Canadian Institutes of Health Research (CIHR) to V.B., and FRQS MSc scholarship and Alexander Graham Bell Canada Graduate Scholarship from the NSERC to N.R.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Harris JB, LaRocque RC, Qadri F, Ryan ET, Calderwood SB. 2012. Cholera. Lancet 379:2466–2476. doi: 10.1016/S0140-6736(12)60436-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Das B, Verma J, Kumar P, Ghosh A, Ramamurthy T. 2020. Antibiotic resistance in Vibrio cholerae: understanding the ecology of resistance genes and mechanisms. Vaccine 38(Suppl 1):A83–A92. doi: 10.1016/j.vaccine.2019.06.031. [DOI] [PubMed] [Google Scholar]
- 3.Verma J, Bag S, Saha B, Kumar P, Ghosh TS, Dayal M, Senapati T, Mehra S, Dey P, Desigamani A, Kumar D, Rana P, Kumar B, Maiti TK, Sharma NC, Bhadra RK, Mutreja A, Nair GB, Ramamurthy T, Das B. 2019. Genomic plasticity associated with antimicrobial resistance in Vibrio cholerae. Proc Natl Acad Sci U S A 116:6226–6231. doi: 10.1073/pnas.1900141116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weill F-X, Domman D, Njamkepo E, Tarr C, Rauzier J, Fawal N, Keddy KH, Salje H, Moore S, Mukhopadhyay AK, Bercion R, Luquero FJ, Ngandjio A, Dosso M, Monakhova E, Garin B, Bouchier C, Pazzani C, Mutreja A, Grunow R, Sidikou F, Bonte L, Breurec S, Damian M, Njanpop-Lafourcade B-M, Sapriel G, Page A-L, Hamze M, Henkens M, Chowdhury G, Mengel M, Koeck J-L, Fournier J-M, Dougan G, Grimont PAD, Parkhill J, Holt KE, Piarroux R, Ramamurthy T, Quilici M-L, Thomson NR. 2017. Genomic history of the seventh pandemic of cholera in Africa. Science 358:785–789. doi: 10.1126/science.aad5901. [DOI] [PubMed] [Google Scholar]
- 5.Spagnoletti M, Ceccarelli D, Rieux A, Fondi M, Taviani E, Fani R, Colombo MM, Colwell RR, Balloux F. 2014. Acquisition and evolution of SXT-R391 integrative conjugative elements in the seventh-pandemic Vibrio cholerae lineage. mBio 5:e01356-14. [CrossRef] doi: 10.1128/mBio.01356-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Folster JP, Katz L, McCullough A, Parsons MB, Knipe K, Sammons SA, Boncy J, Tarr CL, Whichard JM. 2014. Multidrug-resistant IncA/C plasmid in Vibrio cholerae from Haiti. Emerg Infect Dis 20:1951–1953. doi: 10.3201/eid2011.140889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang R, Yu D, Zhu L, Li J, Yue J, Kan B. 2015. IncA/C plasmids harboured in serious multidrug-resistant Vibrio cholerae serogroup O139 strains in China. Int J Antimicrob Agents 45:249–254. doi: 10.1016/j.ijantimicag.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 8.Harmer CJ, Hall RM. 2015. The A to Z of A/C plasmids. Plasmid 80:63–82. doi: 10.1016/j.plasmid.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Carraro N, Matteau D, Luo P, Rodrigue S, Burrus V. 2014. The master activator of IncA/C conjugative plasmids stimulates genomic islands and multidrug resistance dissemination. PLoS Genet 10:e1004714. doi: 10.1371/journal.pgen.1004714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carraro N, Sauvé M, Matteau D, Lauzon G, Rodrigue S, Burrus V. 2014. Development of pVCR94ΔX from Vibrio cholerae, a prototype for studying multidrug resistant IncA/C conjugative plasmids. Front Microbiol 5:44. doi: 10.3389/fmicb.2014.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcillán-Barcia MP, Francia MV, de la Cruz F. 2009. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev 33:657–687. doi: 10.1111/j.1574-6976.2009.00168.x. [DOI] [PubMed] [Google Scholar]
- 12.Hegyi A, Szabó M, Olasz F, Kiss J. 2017. Identification of oriT and a recombination hot spot in the IncA/C plasmid backbone. Sci Rep 7:10595. doi: 10.1038/s41598-017-11097-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carraro N, Rivard N, Ceccarelli D, Colwell RR, Burrus V. 2016. IncA/C conjugative plasmids mobilize a new family of multidrug resistance islands in clinical Vibrio cholerae non-O1/non-O139 isolates from Haiti. mBio 7:e00509-16. doi: 10.1128/mBio.00509-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyd D, Peters GA, Cloeckaert A, Boumedine KS, Chaslus-Dancla E, Imberechts H, Mulvey MR. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J Bacteriol 183:5725–5732. doi: 10.1128/JB.183.19.5725-5732.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doublet B, Boyd D, Mulvey MR, Cloeckaert A. 2005. The Salmonella genomic island 1 is an integrative mobilizable element. Mol Microbiol 55:1911–1924. doi: 10.1111/j.1365-2958.2005.04520.x. [DOI] [PubMed] [Google Scholar]
- 16.Kiss J, Szabó M, Hegyi A, Douard G, Praud K, Nagy I, Olasz F, Cloeckaert A, Doublet B. 2019. Identification and characterization of oriT and two mobilization genes required for conjugative transfer of Salmonella genomic island 1. Front Microbiol 10:457. doi: 10.3389/fmicb.2019.00457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carraro N, Durand R, Rivard N, Anquetil C, Barrette C, Humbert M, Burrus V. 2017. Salmonella genomic island 1 (SGI1) reshapes the mating apparatus of IncC conjugative plasmids to promote self-propagation. PLoS Genet 13:e1006705. doi: 10.1371/journal.pgen.1006705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siebor E, de Curraize C, Neuwirth C. 2018. Genomic context of resistance genes within a French clinical MDR Proteus mirabilis: identification of the novel genomic resistance island GIPmi1. J Antimicrob Chemother 73:1808–1811. doi: 10.1093/jac/dky126. [DOI] [PubMed] [Google Scholar]
- 20.Bie L, Fang M, Li Z, Wang M, Xu H. 2018. Identification and characterization of new resistance-conferring SGI1s (Salmonella genomic island 1) in Proteus mirabilis. Front Microbiol 9:3172. doi: 10.3389/fmicb.2018.03172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Llosa M, Gomis-Rüth FX, Coll M, de la Cruz Fd F. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol 45:1–8. doi: 10.1046/j.1365-2958.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 22.Ilangovan A, Connery S, Waksman G. 2015. Structural biology of the Gram-negative bacterial conjugation systems. Trends Microbiol 23:301–310. doi: 10.1016/j.tim.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 23.Luo Y, Gao Q, Deonier RC. 1994. Mutational and physical analysis of F plasmid TraY protein binding to oriT. Mol Microbiol 11:459–469. doi: 10.1111/j.1365-2958.1994.tb00327.x. [DOI] [PubMed] [Google Scholar]
- 24.Tsai MM, Fu YH, Deonier RC. 1990. Intrinsic bends and integration host factor binding at F plasmid oriT. J Bacteriol 172:4603–4609. doi: 10.1128/jb.172.8.4603-4609.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ziegelin G, Pansegrau W, Lurz R, Lanka E. 1992. TraK protein of conjugative plasmid RP4 forms a specialized nucleoprotein complex with the transfer origin. J Biol Chem 267:17279–17286. [PubMed] [Google Scholar]
- 26.Zhang S, Meyer R. 1997. The relaxosome protein MobC promotes conjugal plasmid mobilization by extending DNA strand separation to the nick site at the origin of transfer. Mol Microbiol 25:509–516. doi: 10.1046/j.1365-2958.1997.4861849.x. [DOI] [PubMed] [Google Scholar]
- 27.Lee CA, Babic A, Grossman AD. 2010. Autonomous plasmid-like replication of a conjugative transposon. Mol Microbiol 75:268–279. doi: 10.1111/j.1365-2958.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas J, Lee CA, Grossman AD. 2013. A conserved helicase processivity factor is needed for conjugation and replication of an integrative and conjugative element. PLoS Genet 9:e1003198. doi: 10.1371/journal.pgen.1003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fekete RA, Frost LS. 2002. Characterizing the DNA contacts and cooperative binding of F plasmid TraM to its cognate sites at oriT. J Biol Chem 277:16705–16711. doi: 10.1074/jbc.M111682200. [DOI] [PubMed] [Google Scholar]
- 30.Nelson WC, Howard MT, Sherman JA, Matson SW. 1995. The traY gene product and integration host factor stimulate Escherichia coli DNA helicase I-catalyzed nicking at the F plasmid oriT. J Biol Chem 270:28374–28380. doi: 10.1074/jbc.270.47.28374. [DOI] [PubMed] [Google Scholar]
- 31.Ziegelin G, Fürste JP, Lanka E. 1989. TraJ protein of plasmid RP4 binds to a 19-base pair invert sequence repetition within the transfer origin. J Biol Chem 264:11989–11994. [PubMed] [Google Scholar]
- 32.Moncalián G, Grandoso G, Llosa M, de la Cruz F. 1997. oriT-processing and regulatory roles of TrwA protein in plasmid R388 conjugation. J Mol Biol 270:188–200. doi: 10.1006/jmbi.1997.1082. [DOI] [PubMed] [Google Scholar]
- 33.Rocco JM, Churchward G. 2006. The integrase of the conjugative transposon Tn916 directs strand- and sequence-specific cleavage of the origin of conjugal transfer, oriT, by the endonuclease Orf20. J Bacteriol 188:2207–2213. doi: 10.1128/JB.188.6.2207-2213.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pansegrau W, Balzer D, Kruft V, Lurz R, Lanka E. 1990. In vitro assembly of relaxosomes at the transfer origin of plasmid RP4. Proc Natl Acad Sci U S A 87:6555–6559. doi: 10.1073/pnas.87.17.6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parker C, Meyer RJ. 2007. The R1162 relaxase/primase contains two, type IV transport signals that require the small plasmid protein MobB. Mol Microbiol 66:252–261. doi: 10.1111/j.1365-2958.2007.05925.x. [DOI] [PubMed] [Google Scholar]
- 36.Disqué-Kochem C, Dreiseikelmann B. 1997. The cytoplasmic DNA-binding protein TraM binds to the inner membrane protein TraD in vitro. J Bacteriol 179:6133–6137. doi: 10.1128/jb.179.19.6133-6137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beaber JW, Hochhut B, Waldor MK. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J Bacteriol 184:4259–4269. doi: 10.1128/jb.184.15.4259-4269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wozniak RAF, Fouts DE, Spagnoletti M, Colombo MM, Ceccarelli D, Garriss G, Déry C, Burrus V, Waldor MK. 2009. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet 5:e1000786. doi: 10.1371/journal.pgen.1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawley TD, Gilmour MW, Gunton JE, Standeven LJ, Taylor DE. 2002. Functional and mutational analysis of conjugative transfer region 1 (Tra1) from the IncHI1 plasmid R27. J Bacteriol 184:2173–2180. doi: 10.1128/jb.184.8.2173-2180.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ceccarelli D, Daccord A, René M, Burrus V. 2008. Identification of the origin of transfer (oriT) and a new gene required for mobilization of the SXT/R391 family of integrating conjugative elements. J Bacteriol 190:5328–5338. doi: 10.1128/JB.00150-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daccord A, Ceccarelli D, Burrus V. 2010. Integrating conjugative elements of the SXT/R391 family trigger the excision and drive the mobilization of a new class of Vibrio genomic islands. Mol Microbiol 78:576–588. doi: 10.1111/j.1365-2958.2010.07364.x. [DOI] [PubMed] [Google Scholar]
- 42.Cabezón E, Sastre JI, de la Cruz F. 1997. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol Gen Genet 254:400–406. doi: 10.1007/s004380050432. [DOI] [PubMed] [Google Scholar]
- 43.Carraro N, Rivard N, Burrus V, Ceccarelli D. 2017. Mobilizable genomic islands, different strategies for the dissemination of multidrug resistance and other adaptive traits. Mob Genet Elements 7:1–6. doi: 10.1080/2159256X.2017.1304193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Humbert M, Huguet KT, Coulombe F, Burrus V. 2019. Entry exclusion of conjugative plasmids of the IncA, IncC, and related untyped incompatibility groups. J Bacteriol 201:e00731-18. doi: 10.1128/JB.00731-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiss J, Papp PP, Szabó M, Farkas T, Murányi G, Szakállas E, Olasz F. 2015. The master regulator of IncA/C plasmids is recognized by the Salmonella genomic island SGI1 as a signal for excision and conjugal transfer. Nucleic Acids Res 43:8735–8745. doi: 10.1093/nar/gkv758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murányi G, Szabó M, Olasz F, Kiss J. 2016. Determination and analysis of the putative AcaCD-responsive promoters of Salmonella genomic island 1. PLoS One 11:e0164561. doi: 10.1371/journal.pone.0164561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huguet KT, Rivard N, Garneau D, Palanee J, Burrus V. 2020. Replication of the Salmonella genomic island 1 (SGI1) triggered by helper IncC conjugative plasmids promotes incompatibility and plasmid loss. PLoS Genet 16:e1008965. doi: 10.1371/journal.pgen.1008965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morita D, Takahashi E, Morita M, Ohnishi M, Mizuno T, Miyoshi S-I, Dutta D, Ramamurthy T, Chowdhury G, Mukhopadhyay AK, Okamoto K. 2020. Genomic characterization of antibiotic resistance-encoding genes in clinical isolates of Vibrio cholerae non-O1/non-O139 strains from Kolkata, India: generation of novel types of genomic islands containing plural antibiotic resistance genes. Microbiol Immunol 64:435–444. doi: 10.1111/1348-0421.12790. [DOI] [PubMed] [Google Scholar]
- 49.Tenor JL, McCormick BA, Ausubel FM, Aballay A. 2004. Caenorhabditis elegans-based screen identifies Salmonella virulence factors required for conserved host-pathogen interactions. Curr Biol 14:1018–1024. doi: 10.1016/j.cub.2004.05.050. [DOI] [PubMed] [Google Scholar]
- 50.Daccord A, Mursell M, Poulin-Laprade D, Burrus V. 2012. Dynamics of the SetCD-regulated integration and excision of genomic islands mobilized by integrating conjugative elements of the SXT/R391 family. J Bacteriol 194:5794–5802. doi: 10.1128/JB.01093-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hochhut B, Marrero J, Waldor MK. 2000. Mobilization of plasmids and chromosomal DNA mediated by the SXT element, a constin found in Vibrio cholerae O139. J Bacteriol 182:2043–2047. doi: 10.1128/jb.182.7.2043-2047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arteaga M, Velasco J, Rodriguez S, Vidal M, Arellano C, Silva F, Carreño LJ, Vidal R, Montero DA. 2020. Genomic characterization of the non-O1/non-O139 Vibrio cholerae strain that caused a gastroenteritis outbreak in Santiago, Chile, 2018. Microb Genom 6:e000340. doi: 10.1099/mgen.0.000340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ali M, Nelson AR, Lopez AL, Sack DA. 2015. Updated global burden of cholera in endemic countries. PLoS Negl Trop Dis 9:e0003832. doi: 10.1371/journal.pntd.0003832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Escobar LE, Ryan SJ, Stewart-Ibarra AM, Finkelstein JL, King CA, Qiao H, Polhemus ME. 2015. A global map of suitability for coastal Vibrio cholerae under current and future climate conditions. Acta Trop 149:202–211. doi: 10.1016/j.actatropica.2015.05.028. [DOI] [PubMed] [Google Scholar]
- 55.Vezzulli L, Grande C, Reid PC, Hélaouët P, Edwards M, Höfle MG, Brettar I, Colwell RR, Pruzzo C. 2016. Climate influence on Vibrio and associated human diseases during the past half-century in the coastal North Atlantic. Proc Natl Acad Sci U S A 113:E5062–E5071. doi: 10.1073/pnas.1609157113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaper JB, Morris JG, Levine MM. 1995. Cholera. Clin Microbiol Rev 8:48–86. doi: 10.1128/CMR.8.1.48-86.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hasan NA, Choi SY, Eppinger M, Clark PW, Chen A, Alam M, Haley BJ, Taviani E, Hine E, Su Q, Tallon LJ, Prosper JB, Furth K, Hoq MM, Li H, Fraser-Liggett CM, Cravioto A, Huq A, Ravel J, Cebula TA, Colwell RR. 2012. Genomic diversity of 2010 Haitian cholera outbreak strains. Proc Natl Acad Sci U S A 109:E2010–E2017. doi: 10.1073/pnas.1207359109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bik EM, Gouw RD, Mooi FR. 1996. DNA fingerprinting of Vibrio cholerae strains with a novel insertion sequence element: a tool to identify epidemic strains. J Clin Microbiol 34:1453–1461. doi: 10.1128/JCM.34.6.1453-1461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li M, Shimada T, Morris JG, Sulakvelidze A, Sozhamannan S. 2002. Evidence for the emergence of non-O1 and non-O139 Vibrio cholerae strains with pathogenic potential by exchange of O-antigen biosynthesis regions. Infect Immun 70:2441–2453. doi: 10.1128/iai.70.5.2441-2453.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dalsgaard A, Serichantalergs O, Forslund A, Lin W, Mekalanos J, Mintz E, Shimada T, Wells JG. 2001. Clinical and environmental isolates of Vibrio cholerae serogroup O141 carry the CTX phage and the genes encoding the toxin-coregulated pili. J Clin Microbiol 39:4086–4092. doi: 10.1128/JCM.39.11.4086-4092.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Montero DA, Canto FD, Velasco J, Colello R, Padola NL, Salazar JC, Martin CS, Oñate A, Blanco J, Rasko DA, Contreras C, Puente JL, Scheutz F, Franz E, Vidal RM. 2019. Cumulative acquisition of pathogenicity islands has shaped virulence potential and contributed to the emergence of LEE-negative Shiga toxin-producing Escherichia coli strains. Emerg Microbes Infect 8:486–502. doi: 10.1080/22221751.2019.1595985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peacock SJ, Moore CE, Justice A, Kantzanou M, Story L, Mackie K, O’Neill G, Day NPJ. 2002. Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect Immun 70:4987–4996. doi: 10.1128/iai.70.9.4987-4996.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deshpande LM, Fritsche TR, Moet GJ, Biedenbach DJ, Jones RN. 2007. Antimicrobial resistance and molecular epidemiology of vancomycin-resistant enterococci from North America and Europe: a report from the SENTRY antimicrobial surveillance program. Diagn Microbiol Infect Dis 58:163–170. doi: 10.1016/j.diagmicrobio.2006.12.022. [DOI] [PubMed] [Google Scholar]
- 64.Dower WJ, Miller JF, Ragsdale CW. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res 16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 66.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 67.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 68.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 72.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 73.Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol Biol Evol 25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 74.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grenier F, Matteau D, Baby V, Rodrigue S. 2014. Complete genome sequence of Escherichia coli BW25113. Genome Announc 2:e01038-14. doi: 10.1128/genomeA.01038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Singer M, Baker TA, Schnitzler G, Deischel SM, Goel M, Dove W, Jaacks KJ, Grossman AD, Erickson JW, Gross CA. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev 53:1–24. doi: 10.1128/MMBR.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garriss G, Waldor MK, Burrus V. 2009. Mobile antibiotic resistance encoding elements promote their own diversity. PLoS Genet 5:e1000775. doi: 10.1371/journal.pgen.1000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Haldimann A, Wanner BL. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol 183:6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Datta S, Costantino N, Court DL. 2006. A set of recombineering plasmids for gram-negative bacteria. Gene 379:109–115. doi: 10.1016/j.gene.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 82.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 83.Doublet B, Douard G, Targant H, Meunier D, Madec J-Y, Cloeckaert A. 2008. Antibiotic marker modifications of lambda Red and FLP helper plasmids, pKD46 and pCP20, for inactivation of chromosomal genes using PCR products in multidrug-resistant strains. J Microbiol Methods 75:359–361. doi: 10.1016/j.mimet.2008.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ethidium bromide-stained 1.5% agarose gel of PCR products for detection of MGIKn excision. Excision assays were carried out in E. coli GG56 bearing MGIKn or its Δint mutant and, where indicated, arabinose-inducible vector pBAD-int or pBAD-xis. Download FIG S1, PDF file, 0.08 MB (87KB, pdf) .
Copyright © 2020 Rivard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mobilization of MGIKn by pVCR94Sp T4SS mutants. “pVCR94Cm (R)” indicates that the element was present in the recipient strain. Donor and recipient strains are the same as in Fig. 4. Statistical analyses were carried out on the logarithm of the values using a one-way ANOVA followed by Dunnett’s multiple comparison test with WT MGIKn as the control. Statistical significance is indicated as follows: **, P < 0.01; ns, not significant. Download FIG S2, PDF file, 0.05 MB (47.9KB, pdf) .
Copyright © 2020 Rivard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Features of GIs presented in Fig. 5 and accession numbers. Download Table S1, XLSX file, 0.03 MB (33KB, xlsx) .
Copyright © 2020 Rivard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparison of MGIVchHai6 and the conserved core of MGIVflInd1. The maps are drawn to scale. The left and right junctions (attL and attR) within the host chromosome are indicated by blue ticks at the extremities. ORFs with similar function are color coded as indicated in the legend. Green arrows indicate the position and orientation of AcaCD and SetCD binding sites. Homologous regions are bracketed and linked by a gray line with the corresponding percentage of nucleotide identity. MGIVflInd1 can be accessed through GenBank accession number KC117176. Download FIG S3, PDF file, 0.07 MB (67.6KB, pdf) .
Copyright © 2020 Rivard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Plasmid and strain construction. Download Text S1, DOCX file, 0.02 MB (21.7KB, docx) .
Copyright © 2020 Rivard et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.