KPC is currently the most common carbapenemase identified in the United States. More than 40 KPC variants have been described, of which KPC-2 and KPC-3 are the most frequent clinical variants. However, our understanding of the genetic structures and β-lactam resistance profiles of other novel KPC variants remains incomplete. Here, we report a novel blaKPC variant (blaKPC-14) and the complete genome sequence of blaKPC-14-harboring K. pneumoniae strain BK13048, which is susceptible to carbapenems but resistant to ceftazidime-avibactam. To the best of our knowledge, this is one of the earliest KPC-producing K. pneumoniae strains exhibiting resistance to ceftazidime-avibactam.
KEYWORDS: ceftazidime-avibactam, carbapenem, KPC, plasmid, antimicrobial resistance, selection
ABSTRACT
Ceftazidime-avibactam is a potent antibiotic combination against Klebsiella pneumoniae carbapenemase (KPC)-producing Enterobacteriaceae. Here, we describe a unique ceftazidime-avibactam-resistant and carbapenem-susceptible K. pneumoniae strain harboring a novel blaKPC-14 variant. This strain was isolated from a New York City patient in 2003, which predates the introduction of avibactam. Despite resistance to ceftazidime-avibactam, the strain was susceptible to imipenem-relebactam and meropenem-vaborbactam. Comprehensive genomic sequencing revealed that blaKPC-14 is harbored on an ST6 IncN plasmid associated with the early spread of blaKPC.
IMPORTANCE KPC is currently the most common carbapenemase identified in the United States. More than 40 KPC variants have been described, of which KPC-2 and KPC-3 are the most frequent clinical variants. However, our understanding of the genetic structures and β-lactam resistance profiles of other novel KPC variants remains incomplete. Here, we report a novel blaKPC variant (blaKPC-14) and the complete genome sequence of blaKPC-14-harboring K. pneumoniae strain BK13048, which is susceptible to carbapenems but resistant to ceftazidime-avibactam. To the best of our knowledge, this is one of the earliest KPC-producing K. pneumoniae strains exhibiting resistance to ceftazidime-avibactam.
OBSERVATION
The rapid spread of carbapenemases among members of the Enterobacteriaceae family poses a major clinical concern, since it greatly limits therapeutic options. These β-lactamases are capable of hydrolyzing all generations of cephalosporins and carbapenems, the last-resort antibiotics for complicated infections with multidrug-resistant Gram negative bacteria. Among the carbapenemases, Klebsiella pneumoniae carbapenemase (KPC), an Ambler class A serine β-lactamase, is particularly problematic, with major outbreaks in the northeastern United States, followed by its spread throughout the United States and worldwide (1). As a novel combination of a β-lactam and a β-lactamase inhibitor, ceftazidime-avibactam was highly active against KPC-producing bacteria. However, resistance to ceftazidime-avibactam has also been reported in patients who were treated with this combination, primarily due to amino acid substitutions in the KPC β-lactamase (2–4). In this study, we describe a novel KPC variant, KPC-14, isolated from K. pneumoniae strain BK13048, collected in 2003 from a New York City (NYC) patient. Surprisingly, this strain was both susceptible to carbapenems and resistant to ceftazidime-avibactam, a result indicating that this resistant KPC variant existed prior to 2015, when ceftazidime-avibactam was introduced.
Strain BK13048 was identified as a part of retrospective study screening of extended-spectrum cephalosporins and carbapenem-resistant K. pneumoniae from our archived strain collection. A molecular-beacon-based allelic discrimination real-time PCR assay (5) showed that strain BK13048 harbored a blaKPC-6-like variant. PCR and Sanger sequencing of the full-length blaKPC gene revealed a novel blaKPC variant, blaKPC-14. Nucleotide alignment of different blaKPC variants showed that blaKPC-14 differs from blaKPC-2 by a 6-bp deletion (nucleotide positions 721 to 726), resulting in a 2-amino-acid deletion at Ambler positions 242Gly and 243Thr. KPC-28 has the same 242Gly and 243Thr deletion, but an additional His274Tyr substitution distinguishes this variant from KPC-14 (6).
Broth microdilution susceptibility testing showed that BK13048 is resistant to ceftriaxone (MIC, >16 μg/ml), ceftazidime (MIC, >256 μg/ml), piperacillin (MIC, >1,024 μg/ml), aztreonam (MIC, >64 μg/ml), and ceftazidime-avibactam (MICs, >16 and 4 μg/ml) but susceptible to imipenem (MIC, ≤0.25 μg/ml), ertapenem (MIC, ≤0.03 μg/ml), and meropenem (MIC, ≤0.03 μg/ml).
The MIC results from BK13048 showed an unusual profile: susceptibility to carbapenems but resistance to ceftazidime-avibactam. To investigate this finding, the full-length blaKPC-2, blaKPC-3, and blaKPC-14 genes and the same promoter sequences were cloned into pET28a vectors, followed by electroporation into Escherichia coli DH10B cells (Invitrogen). Susceptibility testing of the blaKPC-14, blaKPC-2, and blaKPC-3 E. coli DH10B constructs showed that the blaKPC-14 construct demonstrated a ceftazidime-avibactam MIC of >16 μg/ml, which is at least 64-fold higher than that of the blaKPC-2 or blaKPC-3 construct (MICs, ≤0.25 μg/ml) (Table 1). Similarly, the MIC of ceftazidime was much higher for the blaKPC-14 plasmid construct (256 μg/ml) than for the cloned blaKPC-2 (4 μg/ml) or blaKPC-3 (16 μg/ml) gene.
TABLE 1.
Susceptibilities of the strains studied to β-lactams
| Strain | Description | Carbapenemase | MICa
(μg/ml) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PIP | TZP | CRO | CAZ | ETP | MEM | IMP | ATM | CAZ-AVI | |||
| BK13048 | Clinical isolate | bla KPC-14 | 1,024 | 8 | 16 | 256 | ≤0.03 | ≤0.03 | 0.25 | 64 | >16 |
| KPC2-pET28 | blaKPC-2-harboring E. coli DH10B | bla KPC-2 | 128 | 32 | 8 | 4 | 1 | 2 | 2 | 16 | 0.25 |
| KPC3-pET28 | blaKPC-3-harboring E. coli DH10B | bla KPC-3 | 128 | 32 | 8 | 16 | 1 | 1 | 2 | 32 | 0.25 |
| KPC14-pET28 | blaKPC-14-harboring E. coli DH10B | bla KPC-14 | 32 | 4 | 8 | 256 | ≤0.03 | ≤0.03 | 0.25 | 32 | >16 |
| E. coli DH10B | 1 | 1 | 0.25 | ≤0.03 | ≤0.03 | ≤0.03 | ≤0.125 | ≤0.03 | |||
PIP, piperacillin; TZP, piperacillin-tazobactam; CRO, ceftriaxone; CAZ, ceftazidime; ETP, ertapenem; MEM, meropenem; IMP, imipenem; ATM, aztreonam; CAZ-AVI, ceftazidime-avibactam.
In contrast, the MICs of the different carbapenems (ertapenem, meropenem, and imipenem) for the blaKPC-14 construct were 8- to 32-fold lower than the MICs for the blaKPC-2 or blaKPC-3 construct (Table 1). The susceptibility testing results presented above were consistent with a previous study by Oueslati et al. testing KPC-14 and KPC-28 using a different plasmid vector (pTOPO) (6). Those results demonstrated that the 242Gly and 243Thr amino acid deletions in KPC-14 decreased carbapenem activity but increased potency against ceftazidime and ceftazidime-avibactam (Table 1), and ceftazidime-avibactam resistance is likely due to increased activity against ceftazidime rather than reduced inhibition against avibactam.
We further characterized and compared the kinetic parameters of KPC-14 and KPC-2. In brief, the sequences without the signal peptide (from blaKPC-14 and blaKPC-2) were obtained by PCR amplification using primers NdeI-KPC-2-F(30–293) (5′-ACGCATATGGCGGAACCATTCGCTAAAC-3′) and Xhol-KPC-2-R-STOPdel (5′-TAACTCGAGCTGCCCGTTGACGCCCAAT-3′), followed by insertion into plasmid pET28a in E. coli DH10B (Invitrogen). The KPC enzymes were then purified, and the steady-state kinetic parameters were determined as described previously (6, 7). The results showed that KPC-14 has a higher catalytic efficiency of ceftazidime and cefepime, but a lower hydrolysis activity of imipenem, than KPC-2 (Table 2). In addition, no meropenem hydrolysis could be detected with purified KPC-14 under current conditions (measurement made over 5 min). The hydrolytic profile of KPC-14 was similar to that in the previous report from Oueslati et al. (6) and was consistent with the MIC observations presented above. Moreover, a previous experiment determining the 50% inhibitory concentrations (IC50) of β-lactamase inhibitors also suggested that the 2-amino-acid 242Gly and 243Thr deletion had no impact on the inhibition properties of avibactam (6).
TABLE 2.
Steady-state kinetic parameters of purified KPC-2 and KPC-14 β-lactamases
| β-Lactam | KPC-2 |
KPC-14a
|
||||
|---|---|---|---|---|---|---|
| kcat (s−1) | Km (μM) | kcat/Km (μM−1·s−1) | kcat (s−1) | Km (μM) | kcat/Km (μM−1·s−1) | |
| Meropenem | 8.078 | 16.263 | 0.497 | NDb | ND | ND |
| Imipenem | 28.797 | 98.350 | 0.293 | 19.490 | 548.805 | 0.036 |
| Ceftazidime | 3.274 | 590.717 | 0.006 | 24.600 | 73.860 | 0.333 |
| Aztreonam | 12.601 | 2398.451 | 0.005 | 2.875 | 192.335 | 0.015 |
| Cefepime | 4.748 | 310.480 | 0.015 | 7.588 | 70.406 | 0.108 |
| Piperacillin | 7.709 | 793.526 | 0.010 | 1.084 | 45.767 | 0.024 |
| Cefazolin | 65.877 | 110.746 | 0.595 | 27.126 | 287.930 | 0.094 |
KPC-14 differs from KPC-2 by a 2-amino-acid 242Gly and 243Thr deletion.
ND, not detectable due to a low initial rate of hydrolysis.
Additional testing of BK13048 and the blaKPC-14 plasmid construct against other novel β-lactam–β-lactamase combinations, i.e., imipenem-relebactam and meropenem-vaborbactam (by disk diffusion assay), showed that they were susceptible to imipenem-relebactam (inhibition zone diameter, >28 mm for both strains) and meropenem-vaborbactam (inhibition zone diameter, >30 mm).
To better understand the genetic structure associated with the blaKPC-14 gene, comprehensive whole-genome sequencing was performed using a combination of the Oxford Nanopore MinION and Illumina HiSeq platforms, followed by hybrid assembly using Unicycler (8). The complete sequencing of BK13048 showed that it contains a 5,213,293-bp chromosome with an average G+C content of 57.6% and harbors 5,311 predicted open reading frames. In addition, it contains seven plasmids ranging from 5 kbp to 82 kbp. In silico multilocus sequencing typing (MLST) revealed that BK13048 belongs to sequence type (ST) 16 (9), which has been reported to cause nosocomial infections worldwide and is associated with blaNDM-1-encoded carbapenemase and the presence of the extended-spectrum beta-lactamase (ESBL) gene blaCTX-M-15 (10). Analysis of acquired antimicrobial resistance (11) identified 14 antimicrobial resistance genes encoding resistance to β-lactams, aminoglycosides, fluoroquinolones, fosfomycin, sulfonamide, and trimethoprim (Table 3). In addition, in silico plasmid replicon typing (12) indicated that the seven plasmids belong to IncA/C, F, M, N, R, ColRNAI, and a novel incompatibility group (Table 3).
TABLE 3.
Key features of the genome and plasmids harbored by BK13048
| Characteristic | Chromosome | pBK13048_1 | pBK13048_2 | pBK13048_3 | pBK13048_KPC | pBK13048_5 | pBK13048_6 | pBK13048_7 |
|---|---|---|---|---|---|---|---|---|
| Size (bp) | 5,213,293 | 82,240 | 61,331 | 51,887 | 50,635 | 44,850 | 28,729 | 5,251 |
| % G+C | 57.6 | 51.9 | 50.8 | 52.8 | 53 | 53.6 | 53.5 | 49.2 |
| β-Lactamase(s) | bla SHV-1 | blaOXA-9, blaTEM-1A | bla KPC-14 | |||||
| Other resistance genes | fosA, oqxB, oqxA | aadA1, strA, strB, aac(6′)-Ib, sul2 | dfrA14 | |||||
| Plasmid incompatibility (Inc) | A/C | M | New | N | R | F | ColRNAI |
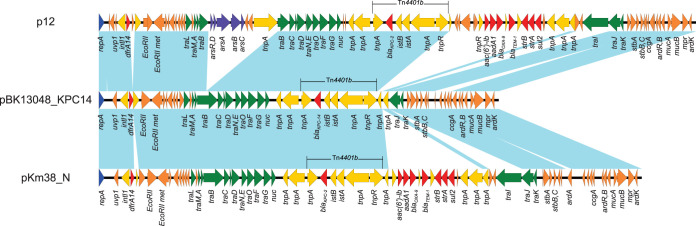
The blaKPC-14 gene is located on an IncN plasmid, pBK13048_KPC14 (Table 1). pBK13048_KPC14 is 50,635 bp long with an average G+C content of 53% and carries blaKPC-14 on the Tn4401b transposon (Fig. 1). Full plasmid sequence BLAST against NCBI GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi) showed that pBK13048_KPC14 is highly similar to plasmid pKm38_N from Klebsiella oxytoca, which was isolated in 1997 in New York City (13), with 100% query coverage and overall 99.98% sequence identity (Fig. 1). In addition, pBK13048_KPC14 showed 94% query coverage and overall 99.97% sequence identity to one of the first sequenced blaKPC-harboring IncN plasmids, plasmid 12, isolated from NYC in 2005 (Fig. 1) (14). In agreement with the structure of other IncN plasmids (13, 15), pBK13048_KPC14 contains a 2-kb acquired region integrated downstream of uvp1 and harbors dfrA14, encoding trimethoprim resistance. In addition, pBK13048_KPC14 contains a second acquired region downstream of the nuc gene and carries blaKPC-14 (Fig. 1). This highlights the important role played by IncN plasmids in the spread of blaKPC during the early years of the carbapenem resistance epidemic. In silico IncN plasmid MLST showed that pKm38_N (isolated in 1997), pBK13048_KPC14 (2003), and p12 (2005) all belong to ST6 (repN-traJ-korA, allele profile 2-4-2), which is different from the sequence type harboring the blaKPC-28-containing IncN plasmid pWI2-KPC28 (ST15, allele profile 7-6-3) from E. coli. Even though both KPC-14 and KPC-28 have the same 242Gly and 243Thr amino acid deletions, their genomic history suggests that pBK13048_KPC14 and pWI2-KPC28, as well as the blaKPC-14 and blaKPC-28 genes, likely evolved independently on different IncN plasmid backgrounds.
FIG 1.
Structures of plasmids p12 (GenBank accession no. FJ223605), pBK13048_KPC14 (accession no. CP045022), and pKm38_N (accession no. KY128483). Colored arrows indicate open reading frames, with blue, orange, green, red, and purple arrows representing replication genes, plasmid backbone genes, mobile elements, plasmid transfer genes, and antimicrobial and heavy metal resistance genes, respectively. Blue shading indicates regions of shared homology among different elements.
A recent study from Italy described the emergence of two ceftazidime-avibactam-resistant subpopulations of K. pneumoniae ST1685 (unrelated to the ST16 of BK14038), carrying KPC-14 and KPC-31 (Asp179Tyr substitution within the KPC Ω-loop), in a patient following prolonged ceftazidime-avibactam treatment (16). Our study also suggested that the ceftazidime-avibactam-resistant KPC variants, e.g., KPC-14, could exist even without ceftazidime-avibactam exposure. These KPC variants, with reduced carbapenem hydrolytic capacities, raise a challenge for phenotypic and genotypic carbapenemase detection tests, since some of these assays may classify KPC-14 strains as carbapenemase producers (6, 16). Consequently, molecular testing followed by a phenotypic carbapenemase activity assay has been proposed to detect and differentiate KPC variants associated with carbapenem susceptibility and ceftazidime-avibactam resistance (16).
Taken together, we identified, completely sequenced, and characterized a novel blaKPC variant from K. pneumoniae BK13048, designated blaKPC-14, that revealed an unexpected resistance to ceftazidime-avibactam. Comprehensive sequence analysis and assembly using both the Illumina and Oxford Nanopore platforms revealed the genetic changes in blaKPC-14 and its plasmid structure. In contrast to other ceftazidime-avibactam-resistant blaKPC variants, the blaKPC-14 gene was not under ceftazidime-avibactam selection pressure, as evidenced by the fact that the isolation of BK13048 predated the U.S. introduction of this novel β-lactam and β-lactamase inhibitor combination in 2015.
Accession number(s).
The complete nucleotide sequence of strain BK13048 has been deposited in GenBank as accession no. CP045015 to CP045022.
ACKNOWLEDGMENTS
This study was supported by a grant from the National Institutes of Health (grant R01AI090155 to B.N.K.). This work was also supported by grants R01AI100560, R01AI063517, and R01AI072219 (to R.A.B.) from the National Institutes of Health and by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, the Veterans Affairs Merit Review Program, and the Geriatric Research Education and Clinical Center VISN 10 (to R.A.B.).
REFERENCES
- 1.Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. 2014. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol 22:686–696. doi: 10.1016/j.tim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shields RK, Chen L, Cheng S, Chavda KD, Press EG, Snyder A, Pandey R, Doi Y, Kreiswirth BN, Nguyen MH, Clancy CJ. 2017. Emergence of ceftazidime-avibactam resistance due to plasmid-borne blaKPC-3 mutations during treatment of carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob Agents Chemother 61:e02097-16. doi: 10.1128/AAC.02097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giddins MJ, Macesic N, Annavajhala MK, Stump S, Khan S, McConville TH, Mehta M, Gomez-Simmonds A, Uhlemann AC. 2018. Successive emergence of ceftazidime-avibactam resistance through distinct genomic adaptations in blaKPC-2-harboring Klebsiella pneumoniae sequence type 307 isolates. Antimicrob Agents Chemother 62:e02101-17. doi: 10.1128/AAC.02101-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shields RK, Potoski BA, Haidar G, Hao B, Doi Y, Chen L, Press EG, Kreiswirth BN, Clancy CJ, Nguyen MH. 2016. Clinical outcomes, drug toxicity, and emergence of ceftazidime-avibactam resistance among patients treated for carbapenem-resistant Enterobacteriaceae infections. Clin Infect Dis 63:1615–1618. doi: 10.1093/cid/ciw636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen L, Mediavilla JR, Endimiani A, Rosenthal ME, Zhao Y, Bonomo RA, Kreiswirth BN. 2011. Multiplex real-time PCR assay for detection and classification of Klebsiella pneumoniae carbapenemase gene (blaKPC) variants. J Clin Microbiol 49:579–585. doi: 10.1128/JCM.01588-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oueslati S, Iorga BI, Tlili L, Exilie C, Zavala A, Dortet L, Jousset AB, Bernabeu S, Bonnin RA, Naas T. 2019. Unravelling ceftazidime/avibactam resistance of KPC-28, a KPC-2 variant lacking carbapenemase activity. J Antimicrob Chemother 74:2239–2246. doi: 10.1093/jac/dkz209. [DOI] [PubMed] [Google Scholar]
- 7.Barnes MD, Winkler ML, Taracila MA, Page MG, Desarbre E, Kreiswirth BN, Shields RK, Nguyen MH, Clancy C, Spellberg B, Papp-Wallace KM, Bonomo RA. 2017. Klebsiella pneumoniae carbapenemase-2 (KPC-2), substitutions at Ambler position Asp179, and resistance to ceftazidime-avibactam: unique antibiotic-resistant phenotypes emerge from β-lactamase protein engineering. mBio 8:e00528-17. doi: 10.1128/mBio.00528-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Espinal P, Nucleo E, Caltagirone M, Mattioni Marchetti V, Fernandes MR, Biscaro V, Rigoli R, Carattoli A, Migliavacca R, Villa L. 2019. Genomics of Klebsiella pneumoniae ST16 producing NDM-1, CTX-M-15, and OXA-232. Clin Microbiol Infect 25:385.e1–385.e5. doi: 10.1016/j.cmi.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eilertson B, Chen L, Chavda KD, Kreiswirth BN. 2017. Genomic characterization of two KPC-producing Klebsiella isolates collected in 1997 in New York City. Antimicrob Agents Chemother 61:e02458-16. doi: 10.1128/AAC.02458-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gootz TD, Lescoe MK, Dib-Hajj F, Dougherty BA, He W, Della-Latta P, Huard RC. 2009. Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York City hospital. Antimicrob Agents Chemother 53:1998–2004. doi: 10.1128/AAC.01355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L, Hu H, Chavda KD, Zhao S, Liu R, Liang H, Zhang W, Wang X, Jacobs MR, Bonomo RA, Kreiswirth BN. 2014. Complete sequence of a KPC-producing IncN multidrug-resistant plasmid from an epidemic Escherichia coli sequence type 131 strain in China. Antimicrob Agents Chemother 58:2422–2425. doi: 10.1128/AAC.02587-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bianco G, Boattini M, Iannaccone M, Cavallo R, Costa C. 2020. Bloodstream infection by two subpopulations of Klebsiella pneumoniae ST1685 carrying KPC-33 or KPC-14 following ceftazidime/avibactam treatment: considerations regarding acquired heteroresistance and choice of carbapenemase detection assay. J Antimicrob Chemother doi: 10.1093/jac/dkaa283. [DOI] [PubMed] [Google Scholar]