The crystal structure of YeeE shows a sophisticated hourglass shape for thiosulfate uptake.
Abstract
We have demonstrated that a bacterial membrane protein, YeeE, mediates thiosulfate uptake. Thiosulfate is used for cysteine synthesis in bacteria as an inorganic sulfur source in the global biological sulfur cycle. The crystal structure of YeeE at 2.5-Å resolution reveals an unprecedented hourglass-like architecture with thiosulfate in the positively charged outer concave side. YeeE is composed of loops and 13 helices including 9 transmembrane α helices, most of which show an intramolecular pseudo 222 symmetry. Four characteristic loops are buried toward the center of YeeE and form its central region surrounded by the nine helices. Additional electron density maps and successive molecular dynamics simulations imply that thiosulfate can remain temporally at several positions in the proposed pathway. We propose a plausible mechanism of thiosulfate uptake via three important conserved cysteine residues of the loops along the pathway.
INTRODUCTION
Sulfur is an essential element in living organisms. The synthesis of L-cysteine (Cys) via the assimilation of inorganic sulfur sources in bacteria and plants plays a pivotal role in the biological sulfur cycle on Earth. In the global market, more than 5000 metric tons/year of Cys are used for functional materials in the development of such products as foods, cosmetics, and pharmaceuticals, among others (1). Most Cys is industrially produced via extraction from the keratin hydrolysis product and asymmetric hydrolysis of a precursor, DL-2-amino-thiazoline-4-carboxylic acid, by a bacterial enzyme. Although certain methods use microorganisms for Cys production, their costs are not currently industrially acceptable (2). However, if the secretion efficiency of Cys by microorganisms can be markedly improved, this procedure will be superior to existing methods, such as in the production of glutamic acid by Corynebacterium glutamicum. Understanding Cys synthesis in microorganisms is therefore biologically important and will provide valuable insights into enhancing its production efficiency by microorganisms (3–5).
The Escherichia coli CysUWA (also called CysTWA) complex (the gene product of cysU, cysW, and cysA), an adenosine triphosphate–binding cassette transporter, takes up sulfate and thiosulfate ions from the environment as a sulfur source in combination with periplasmic Sbp and CysP, respectively (Fig. 1A) (2, 6). These transported ions are converted to Cys in a stepwise manner by several enzymes present in the cytoplasm (2, 7). The energy efficiency of Cys synthesis from thiosulfate is better than that from sulfate because of the lower number of synthesis steps in the cytoplasm (2, 4). Therefore, thiosulfate is considered a better sulfur source than sulfate in E. coli (7–9), and in the yeast Saccharomyces cerevisiae (10). Here, by performing growth tests of an E. coli strain lacking an unknown functional gene, yeeE, we have demonstrated that bacterial YeeE imports thiosulfate as a sulfur source. In addition, we have elucidated the crystal structure of YeeE at 2.5-Å resolution, which provides insight into the mechanics of thiosulfate uptake and molecular mechanism in YeeE.
Fig. 1. Thiosulfate uptake depending on YeeE in E. coli.
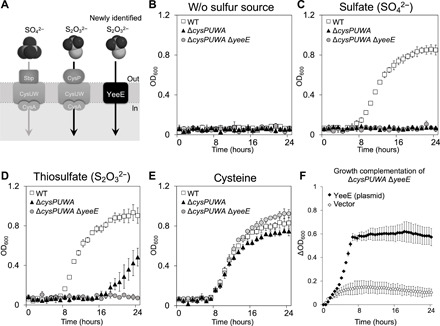
(A) Uptake of sulfate and thiosulfate in the inner membrane. (B to E) Survivability of E. coli WT (MG1655) and its derivatives (ΔcysPUWA and ΔcysPUWA ΔyeeE) in nonsulfur medium (B) or single sulfur source minimal media containing 100 μM sulfate (C), 500 μM thiosulfate (D), or 100 μM cysteine (E). The OD600 (optical density at 600 nm) was monitored every hour. (F) Growth complementation of ΔcysPUWA ΔyeeE (DE3) in the single sulfur source minimal media containing thiosulfate by expression of E. coli YeeE. OD600 was monitored every 30 min. Error bars indicate the SD (n = 3). The growth patterns shown in (B) to (F) indicate that YeeE transports thiosulfate.
RESULTS
Thiosulfate uptake mediated by E. coli YeeE
We monitored the growth of the E. coli ΔcysPUWA strain under different sulfur sources [wild type (WT) versus ΔcysPUWA; Fig. 1, B to E]. We found that the ΔcysPUWA strain grew in the presence of thiosulfate as a sulfur source but not in the presence of sulfate, suggesting that E. coli has another pathway for thiosulfate uptake, which differs from the CysUWA/CysP pathway. According to the EcoliWiki database (11), the bacterial YeeE, belonging to the YeeE/YedE family (12), is a putative transporter for sulfur-containing ions and has three conserved Cys residues, which may be involved in ion transport. Here, we deleted yeeE from the E. coli ΔcysPUWA strain and found that the resulting ΔcysPUWA ΔyeeE strain did not grow, even in the presence of thiosulfate (Fig. 1D). Using a plasmid to express E. coli YeeE restored this growth defect (Fig. 1F). Notably, all Δ strains shown in Fig. 1 grew in the presence of cysteine as a sulfur source (Fig. 1E), suggesting that YeeE transports thiosulfate for use in Cys synthesis. Although structural analysis of YeeE is required to uncover the mechanism of this selective transport, no structural information of the YeeE/YedE family is available in Protein Data Bank (PDB) as confirmed by the XtalPred server (13). The small size of the YeeE protein [molecular weight (MW), ~40,000] makes determination of its structure at high resolution difficult using cryo–electron microscopy. Here, we present the structure of YeeE by x-ray crystallography.
Determination of YeeE crystal structure
We attempted purification and crystallization of E. coli YeeE and YeeEs from 12 bacteria highly homologous to E. coli YeeE and successfully crystallized the Spirochaeta thermophila YeeE (StYeeE) in the presence of thiosulfate, but not in its absence. The amino acid residues of StYeeE are approximately 30% identical to those of E. coli YeeE (fig. S1). The expression of StYeeE also restored the growth defect of E. coli ΔcysPUWA ΔyeeE strain in the presence of thiosulfate, but not sulfate (fig. S2, A and C). In addition, we confirmed the interaction between purified StYeeE and thiosulfate ion by isothermal titration calorimetry experiments (fig. S2B). These data support that StYeeE conducts thiosulfate in the same way as E. coli YeeE. The x-ray diffraction dataset of YeeE was collected from YeeE crystals at 2.5-Å resolution. Because the structural model of YeeE/YedE has not been reported, the phase was calculated by the single isomorphous replacement with anomalous scattering (SIRAS) method using merged diffraction data from 326 crystals of SeMet-substituted YeeE, which is a rare case of Se-SIRAS phase determination using merged diffraction data from numerous crystals for an unsolved protein. The native structure (Fig. 2 and Table 1) showed alternative conformations derived from the oxidized and reduced forms between C22 and C91 in the cytoplasmic side (fig. S3). Approximately half of the purified YeeE showed disulfide formation (fig. S3A). Disulfide bonds are unfavorable in the cytoplasm of E. coli cells, which is a reductive environment. To determine the reduced conformation of YeeE, we determined the structure of the C91A mutant, without disulfide bonds, providing an accurate reduced conformation around C22 and C91 in the reduced form (fig. S3B). On the basis of the mutant structure, we determined a more appropriate structure of the native YeeE in a reduced state and discuss this structure here (WT-B; fig. S3B). In particular, a structural similarity search using the Dali server (14) suggested that the crystal structure of YeeE showed a previously unknown fold.
Fig. 2. Crystal structure of YeeE.
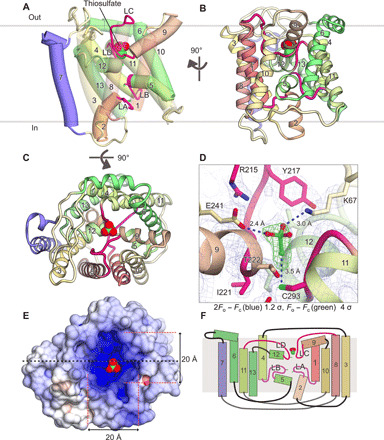
(A and B) Overall structure of StYeeE in cartoon (A) and ribbon (B) representations from the membrane side. The transparency of H1, H3, H8, and H10 in (A) is 30% to clearly display the inside. Numbers of α helices are indicated, and YeeE characteristic loops (LA–LD) are highlighted in red and magenta. Thiosulfate ions are shown as a space-filling model. (C) YeeE structure in ribbon representation from the outside. (D) Close-up view of thiosulfate-binding site. 2Fo − Fc map and thiosulfate-omit (Fo − Fc) map are shown with 1.2 σ and 4.0 σ, respectively. (E) Surface model of YeeE viewed from the outside, colored to indicate electrostatic potential ranging from blue (+10 kT/e) to red (−10 kT/e). (F) Schematic topology model of YeeE. YeeE has four short α helices (H2, H5, H9, and H12), the LA–LD loop, and nine transmembrane α helices (H1, H3, H4, H6, H7, H8, H10, H11, and H13).
Table 1. Data collection and refinement.
Statistics for the highest-resolution shell are shown in parentheses.
| PDB ID | YeeE (WT) | YeeE (C91A) | SeMet YeeE |
| 6LEO | 6LEP | ||
| Data collection | |||
| X-ray source | SPring-8 BL32XU |
SPring-8 BL32XU |
SPring-8 BL32XU |
| Wavelength (Å) | 1.44 | 1.00 | 0.979 |
| Space group | C2221 | C2221 | C2221 |
| a, b, and c (Å) | 73.51, 95.32, and 101.4 |
73.91, 94.61, and 101.5 |
73.60, 95.68, and 101.5 |
| Resolution range (Å) |
43.1–2.52 (2.61–2.52) |
42.9–2.60 (2.69–2.60) |
43.3–2.80 (2.90–2.80) |
| No. of merged datasets |
515 × (10°, Δφ = 0.100) |
93 × (10°, Δφ = 0.100) |
326 × (7°, Δφ = 0.100) |
| Total reflections | 2,037,612 (200,477) |
301,559 (15,399) |
673,420 (68,965) |
| Unique reflections |
12,366 (1224) | 10,299 (898) | 9,125 (901) |
| Multiplicity | 164.8 (163.8) | 29.3 (17.1) | 73.8 (76.5) |
| Completeness (%) |
99.85 (99.59) | 89.62 (76.73) | 99.84 (99.67) |
| I/σ(I) | 20.49 (2.31) | 10.15 (1.28) | 19.40 (5.31) |
| R-meas | 0.6336 (8.719) | 0.3629 (3.059) | 0.3533 (1.72) |
| R-pim | 0.04322 (0.6173) |
0.05311 (0.6174) |
0.03531 (0.1729) |
| CC1/2 | 0.994 (0.522) | 0.981 (0.213) | 0.997 (0.859) |
| Refinement | |||
| No. of reflections | 12,353 (1219) | 10,104 (841) | |
| Rwork/Rfree | 0.202/0.252 | 0.235/0.286 | |
| (0.259/0.34) | (0.336/0.366) | ||
| No. of atoms | 2,784 | 2,523 | |
| Protein | 2,531 | 2,396 | |
| Ligand | 5 | 5 | |
| Monoolein | 225 | 100 | |
| Solvent | 23 | 22 | |
| RMS derivations | |||
| Bond length (Å) | 0.002 | 0.002 | |
| Bond angles (°) | 0.49 | 0.56 | |
| Ramachandran | |||
| Favored | 97.55 | 97.44 | |
| Allowed | 1.53 | 1.92 | |
| Outliers | 0.92 | 0.64 | |
| Average B factor | 36.55 | 43.75 | |
| Protein | 35.52 | 43.24 | |
| Ligand | 42.48 | 93.02 | |
| Monoolein | 48.43 | 53.48 | |
| Solvent | 32.17 | 43.18 |
Overall architecture of YeeE
YeeE is composed of loops and 13 helices (H1–H13) including 9 transmembrane α helices, most of which are membrane embedded, with the N terminus on the outside and the C terminus on the inside (Fig. 2, fig. S3, and movie S1). H1–H6 and H8–H13 are linked by H7 and the C-terminal region of H6, which reverses the topology of H1–H6 and H8–H13 (Figs. 2 and 3A). Each of the H1–H3, H4–H6, H8–H10, and H11–H13 elements shows a similar architecture that has two transmembrane helices linked by a loop and short helix structure (Fig. 3C), although the sequence identity is not high (Fig. 3D). The four elements are assembled in an intramolecular pseudo 222 symmetry manner (Fig. 3, A and B). In other words, there are the orthogonal pseudo dyad axes in YeeE molecule as a, b, and c in Fig. 3B, referred to as the intramolecular 222 fold. The loops ahead of H2, H5, H9, and H12 are designated as loops A to D (LA–LD), which form the characteristic architecture of YeeE. The loops are buried toward the center of YeeE and form its central region surrounded by other transmembrane helices (Fig. 2). Conserved amino acid residues including Cys in YeeEs are at the LA–LD loops and their peripheral region (Fig. 4A), which may be critical to the function of YeeE.
Fig. 3. Intramolecular pseudo 222 fold in YeeE.
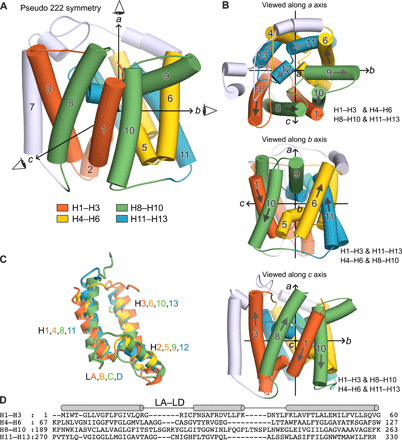
(A) Structure of YeeE with three orthogonal pseudo dyad axes a, b, and c. The pseudo symmetrical elements H1–H3, H4–H6, H8–H10, and H11–H13 are colored in orange, green, yellow, and blue, respectively. (B) Each intramolecular pseudo twofold symmetry. YeeE structure viewed along one of the pseudo twofold axes (top, a axis; middle, b axis; bottom, c axis). Each pair of pseudo twofold symmetry elements are shown, e.g., H1–H3 and H4–H6 (top). (C) Superimposition of the H1–H3, H4–H6, H8–H10, and H11–H13. (D) Structure-based sequence alignment of H1–H3, H4–H6, H8–H10, and H11–H13, generated from (C).
Fig. 4. Conserved residues and crystallographic B factor of YeeE.
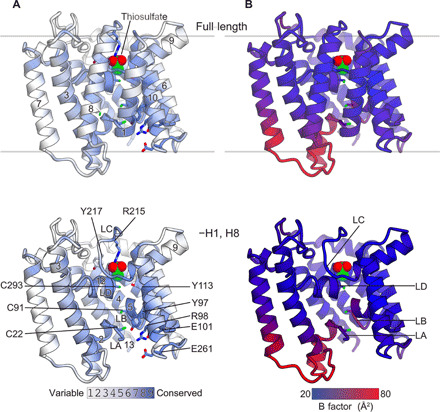
(A) Mapping of conserved residues onto the crystal structure of YeeE. The amino acids are colored according to their ConSurf conservation grades, as shown in fig. S1. (B) Crystallographic B factor of YeeE. The crystal structure of YeeE is colored ranging from 20 to 80 Å2. The top and bottom figures show the full-length structures and H1- and H8-omitted structures, respectively.
Characteristic indentations
Both the outer and inner surfaces of YeeE are indented toward the center, forming an hourglass-shaped architecture (Figs. 2E and 5C). The periplasmic indentation is positively charged and holds a molecule, depicted by gourd-like electron density map in the omit map, adjacent to C293 (Fig. 2D). This electron density was assigned to the substrate thiosulfate, which was added for crystallization of YeeE. The three side chains of nonconserved K67, E241, and conserved C293 surround the thiosulfate, and the thiosulfate-occluded site (position I) is partly covered by the conserved R215 from the periplasmic side (Fig. 2D). Given the negative charge of thiosulfate and the distance (2.4 to 3.5 Å) between the thiosulfate and the oxygen, nitrogen, or sulfur atoms of E241, K67, or C293, respectively, the interactions are presumably hydrogen bonds. Although PROPKA3.1 (15) assumed that the pKa (where Ka is the acid dissociation constant) of E241 is ~4.0, the side chain should take part in the hydrogen bond and be protonated. The thickness of the thinnest region comprising the LA–LD is approximately 15 Å because of the indentations (Fig. 5C). In this region, three highly conserved Cys residues, C22, C91, and C293 in the LA, LB, and LD, respectively, are linearly located at intervals of ~7 Å. A growth complement assay using the ΔcysPUWA ΔyeeE (DE3) strain indicated that these cysteines are essential for E. coli YeeE activity (Fig. 5, A and B, and fig. S4). In addition, unassignable electron densities along this region in the 2Fo − Fc map, which are larger than water molecules, are located near the Cys residues (Fig. 5C). These positions are termed as positions II and III. Although the densities at position III of the WT are ambiguous due to their alternative conformations, those of the C91A mutant showed an ellipse density. In addition, the unassignable densities may be related to the cysteines, as the density at position II of the C91A mutation is missing compared with that of the WT (Fig. 5C and fig. S3C). The density at positions II and III imply that some YeeE molecules in the crystals form other conformations such as transport intermediates including the substrate.
Fig. 5. Mutational analysis and working model of YeeE.
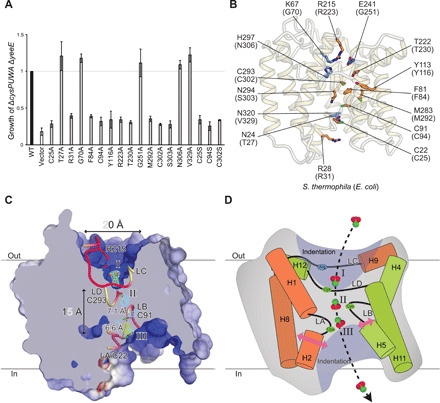
(A) Complementation of ΔcysPUWA ΔyeeE (DE3) growth by the indicated YeeE mutants as in Fig. 1F. Growth (ΔOD600) at 24 hours was normalized to that of WT YeeE-expressing cells. Error bars indicate the SD (n = 3). (B) Mapping of the mutational analysis. The side chains of the mutated positions in (A) are shown as a stick model. The orange and blue residues are essential and nonessential, respectively. (C) Cross-sectional model of YeeE along the dashed line in Fig. 2E. The LA–LD loops are shown as a tube model. Thiosulfate, conserved R215, and three cysteines in the loops are shown as a stick model. The thiosulfate-omit (Fo − Fc) map of WT and C91A mutant with 4.0 σ are shown in blue and green mesh, respectively. Position I is the thiosulfate-binding site. Positions II and III are predicted binding sites. (D) Schematic working model of YeeE. The LA–LD loops and their vicinity helices are shown. The conserved arginine in the LC and three conserved cysteines in the LA, LB, and LD are depicted. Thiosulfate ions may be recognized at position I and passed through the membrane along the arrow.
Thiosulfate-binding site
On the basis of the YeeE crystal structure with a thiosulfate ion at position I, the result of molecular dynamics (MD) simulation (MD position I) showed that the thiosulfate ion stably occupied position I (fig. S5A). If the intermediate points of the thiosulfate-conducting pathway are positions II and III, then thiosulfate should remain temporarily at positions II and III. Here, we performed 100-ns MD simulations of the YeeE models, including a thiosulfate ion manually placed at positions II or III according to the electron density map (MD position II or MD position III) (fig. S5, B and C). In each case, the exposed sulfur atom of thiosulfate was placed toward the periplasmic side in the initial model, but the direction of the thiosulfate was inverted and stably existed during the MD simulation. Because there is an uncertainty in the initial models for MD position II and MD position III due to the manual arrangement of thiosulfate, and the time of the MD simulations was only 100 ns, it is inappropriate to discuss the detailed interactions of thiosulfate of the MD structures. However, the MD simulations at least showed that the thiosulfate ion, which is not forming a disulfide bond with the Cys residue, can stably exist even if it is incorporated at position II or III without large conformational changes of YeeE. In comparing the crystal structure with the final models of MD position II and MD position III (fig. S5D), the overall root mean square deviations (RMSDs) for Cα atoms are less than 1.5 Å. In detail, the RMSDs of the central region (LA–LD and H2, H5, H9, and H12) were higher than those of the surrounding transmembrane helices (H1, H3, H4, H6, H7, H8, H10, H11, and H13), indicating that the surrounding transmembrane helices do not need large conformational changes while holding thiosulfate, although the central region can fluctuate more. In addition, the crystallographic B factors of loops between H2–H3 and H7–H8 at the cytoplasmic side showed higher values than those of the periplasmic loops (Fig. 4B). Notably, the LA exhibited higher B factors than the LB and may undergo conformational transitions such as dislocation of the LA when YeeE transports thiosulfate.
Important residues around the center
A series of E. coli YeeE mutational analyses illustrated the importance of amino acids near these positions. (Fig. 5, A and B, and fig. S4). In this assay, despite the accumulation variations of E. coli YeeE mutants (fig. S4B), low-accumulated N306A showed the growth complementation activity of E. coli ΔcysPUWA ΔyeeE, while high-accumulated C94A did not (Fig. 5A and fig. S4). Because the activity did not depend on the difference in accumulation, this experiment proved to be an applicable method to qualitatively show E. coli YeeE activity. The three conserved cysteines, C25, C94, and C302, in E. coli, corresponding to C22, C91, and C293 in S. thermophila, respectively, which are thought to be located along the thiosulfate-conducting pathway (Fig. 5C), were not able to be functionally replaced by Ser and were essential. Presumably, these cysteines may interact with thiosulfate during transportation via S─H─S hydrogen bonds. We also found the R233 in E. coli, the sole positive charge in the periplasmic indentation, to be important. Although in the crystal structure of StYeeE the thiosulfate ion is hydrogen bonded to K67 and E241 (Fig. 2D), the corresponding residues G70 and G251 in E. coli were not important.
In addition, we established an assay system to address the importance of amino acid residues of StYeeE (fig. S2). The mutants C91A and C293A did not complement the growth defect of E. coli ΔcysPUWA ΔyeeE, but C22A complemented growth. The residue C22, located on the cytoplasmic side of the pathway, may be involved in releasing thiosulfate into the cytoplasm, although it is not crucial for the StYeeE function. The StYeeE mutants K67A and E241A showed less influence on YeeE activity, which is consistent with the results from E. coli YeeE mutants. Unlike in the case of E. coli mutant R223A, the mutation of R215A in StYeeE rescued the growth defect, possibly due to two positively charged residues in the periplasmic indentation of StYeeE. Meanwhile, the two mutations K67A and R215A in StYeeE did not completely rescue the growth defect, implying the importance of positive charges. Thus, at the initial phase of thiosulfate uptake, providing the adequate size and positive charge with the conserved Cys at the periplasmic side may be important for the activity of YeeE. Collectively, the conserved cysteines and their surrounding residues play a pivotal role in YeeE functions, presumably implying that the conserved region is involved in the thiosulfate pathway.
DISCUSSION
We propose that thiosulfate is transported into cells through this diminished region while being guided by conserved cysteines (Fig. 5D). In the crystal structure of WT (fig. S3A), the disulfide bond between C22 and C91 at the cytoplasmic side was formed, but the cytoplasm is in a reduced environment. Therefore, in our model, the cysteine residues are in reduced states. Initially, a thiosulfate ion can be captured at the center of the positively charged indentation (position I) on the periplasmic side by some residues, including conserved C293 and R215, via hydrogen bonds (Fig. 2D). Next, the substrate may be transferred to position II and then may successively be relayed to position III. During this process, the sulfur atom of thiosulfate may temporarily interact with each thiol group of the conserved C293, C91, and C22 by S─H─S hydrogen bond as shown in the crystal structure (C293-thiosulfate; Fig. 2D), and the MD simulation results indicate that the orientation of thiosulfate may be variable during transportation. Eventually, thiosulfate will be released into the cytoplasm. While we introduced the plausible model in which YeeE transports thiosulfate, our current data cannot exclude the possibility that YeeE forms a channel for anions including thiosulfate ion.
YeeE appears to be a sophisticated structure evolved for thiosulfate uptake that can allow the thiosulfate transport by minimal structural changes of the LA–LD loop at the constrict site, at the center, while maintaining membrane permeability. Notably, in this proposed model, YeeE does not require large conformational changes in the surrounding transmembrane helices (as demonstrated by the MD simulation), compared with those of other transporter proteins, such as conformational transitions between inward- and outward-facing forms (16–18) and “rocking bundle” motions of LeuT (19). If the S─H─S hydrogen bond is critical for the thiosulfate transport activity, then YeeE would be structurally unable to transport sulfates in the same manner because of the unexposed sulfur atom of sulfate (Fig. 1A). It remains unclear whether YeeE requires other energies such as a concentration gradient. It is possible that YeeE spontaneously allows the uptake of thiosulfate by inherent interactions between the sulfurs of the conserved cysteines and that of thiosulfate via hydrogen bond. Moreover, because thiosulfate is constantly used as a sulfur source in the cytoplasm, the intracellular thiosulfate concentration is generally low. Therefore, when thiosulfate is abundant in the environment, a concentration gradation of thiosulfate would form, which may drive thiosulfate uptake. Thus, YeeE may act as a thiosulfate-conducting channel through the positions I to III.
In conclusion, the crystal structure of YeeE shows unprecedented folding properties, and our revealing of this structure improves understanding of the molecular mechanisms of transporters and channels while providing a foundation for constructing an efficient cysteine synthesis system using microorganisms.
MATERIALS AND METHODS
E. coli strains
Gene deletion in E. coli WT MG1655 was performed using the λ-Red recombination system developed by Datsenko and Wanner (20). The plasmid pKD46, which carries an arabinose-inducible λ-Red gene, was used to allow for Red recombination, and the plasmid pMW-λattL-KmR-λattR (21) was used as templates to provide polymerase chain reaction (PCR)–generated gene disruption cassettes for ::kan. The PCRs were performed using the following primers: 5′-ATG TTT TCA ATG ATA TTA AGC GGG CTA ATT TGT GGT GCT CTG CTG GGA TTT GAA GCC TGC TTT TTT ATA CTA AGT TGG CA-3′ and 5′-TTA ATT TGC CGC AGC AGT TGC CAG TCG CGC CTT ACG CTG CGG TCG AAC ATC GCT CAA GTT AGT ATA AAA AAG CTG AAC GA-3′, and 5′-TTA TAA ATA TGA TGG CTA TTA GAA AGT CAT TAA ATT TAT AAG GGT GCG CAT GAA GCC TGC TTT TTT ATA CTA AGT TGG CA-3′ and 5′-AGC CCG GAT CGC GGG AAG CGC CTC CGG GCG TTT AAC ATT CAC TCA ACC TAC GCT CAA GTT AGT ATA AAA AAG CTG AAC GA-3′ for deletion of yeeE and cysPUWA genes, respectively. To excise markers from the cassettes flanked by attL/R, we used the phage λ site-specific Int/Xis system (21), wherein the plasmid pMW-int-xis encoding the Int/Xis recombinase was used. The integration of λ (DE3) into the E. coli chromosome was performed using λDE3 Lysogenization Kit (Novagen).
Sulfur source requirement of E. coli strains
Isolated E. coli strains (MG1655, MG1655 ΔcysPUWA::kan, and MG1655 ΔcysPUWA ΔyeeE::kan) were cultured at 37°C for 16 hours in LB Broth Lennox (NACALAI TESQUE) medium containing 0 or 25 μg/ml kanamycin for WT and Δ strains, respectively, and then diluted 100-fold with LB medium and cultured at 37°C for 8 hours. The cultures were washed with S-free medium [42 mM Na2HPO4, 22 mM KH2PO4, 8.6 mM NaCl, 19 mM NH4Cl, 1 mM MgCl2, 0.2% (w/v) glucose, 0.01% (w/v) thiamine hydrochloride, and 0.1 mM CaCl2] twice and then suspended in S-free medium with the same volume as the preculture. The cultures were diluted 100-fold with S-free medium or S-free medium supplemented with 100 μM cysteine, 100 μM Na2SO4, or 500 μM Na2S2O3 as a sulfur source and kept shaking at 37°C for 24 hours. OD600 (optical density at 600 nm) was manually measured every hour using Biowave CO8000 Cell Density Meter (WPA).
Growth complementation tests of E. coli YeeE mutants
E. coli MG1655 ΔcysPUWA ΔyeeE::kan (DE3) cells harboring a pET16b (Novagen)–based plasmid expressing N-terminal MGH10SSGENLYFQGSH-tagged E. coli YeeE or mutant YeeE were used. A DNA fragment including yeeE amplified from E. coli genomic DNA (JCM 20135, RIKEN BRC) using primers (5′-AAA TTT ATA TTT TCA AGG ATC CCA TAT GTT TTC AAT GAT ATT AAG-3′ and 5′-GGC TTT GTT AGC AGC CCT CGA GTC AGG CTT TTT GAA CGG-3′) was inserted between the restriction sites Bam HI and Xho I of pET16b. The mutations in yeeE were introduced by site-directed mutagenesis. Isolated E. coli cells were cultured at 37°C for 16 hours in LB medium containing kanamycin (25 μg/ml) and ampicillin (50 μg/ml) and then diluted 100-fold with LB medium and cultured at 37°C for 8 hours. The cultures were washed twice with S-free medium. Then, the bacterial suspension was adjusted to OD600 = 0.5 in S-free medium supplemented with 500 μM Na2S2O3 or 100 μΜ Na2SO4 and kept shaking at 37°C for 24 hours. OD600 was automatically monitored every half hour using OD-Monitor ODBox-C (TAITEC). The accumulation of YeeE in isolated membrane fraction was confirmed by anti–His-tag immunoblotting using monoclonal antibody (anti-His-tag mAb D291, MBL).
Growth complementation tests of StYeeE mutants
E. coli MG1655 ΔcysPUWA ΔyeeE::kan (DE3) was transformed by a pCGFP-BC–based plasmid (22) (pAZ101), which expresses full-length StYeeE linked to the C-terminal polyhistidine tag. The mutations in yeeE were introduced by site-directed mutagenesis. Precultures were prepared as described above. The bacterial suspension was adjusted to OD600 = 0.5 in S-free medium–HCl (pH 5.8) supplemented with 500 μM Na2S2O3 or 100 μΜ Na2SO4 and kept shaking at 37°C for 12 hours. Other methods were the same as those used for E. coli YeeE mutational analysis.
Purification
E. coli C41 (DE3) (Lucigen) harboring a modified pCGFP-BC (22) vector, pKK550, encoding S. thermophila YeeE(1–328)–GSSGENLYFQFTS–H8 was cultivated in LB Broth (Lennox, Nacalai) supplemented with ampicillin (50 μg/ml) at 37°C. When OD600 nm reached 0.8, the protein expression was induced by 1 mM isopropyl β-D-thiogalactopyranoside (IPTG), and then the culture was continued at 37°C for 15 hours. The cells were harvested at 7500g for 15 min; suspended in 10 mM tris-HCl (pH 8.0), 1 mM EDTA-Na (pH 8.0), and 0.1 mM phenylmethylsulfonyl fluoride (PMSF); and disrupted using an M-110EH Microfluidizer (Microfluidics) three times at 15,000 psi. The resulting lysate was centrifuged at 22,500g for 30 min. The supernatant was ultracentrifuged at 140,000g for 60 min. Membrane fraction as the pellet was resuspended in 20 mM tris-HCl (pH 8.0), 300 mM NaCl, 0.1 mM PMSF, 1 mM β–mercaptoethanol (ME), 20 mM imidazole (pH 8.0), and 1% n-dodecyl β-maltoside (DDM), and rotated for 45 min at 4°C. After ultracentrifugation at 140,000g for 30 min, the supernatant was mixed with Ni-NTA agarose resin (Qiagen) equilibrated with buffer A [20 mM tris-HCl (pH 8.0), 300 mM NaCl, and 0.1% DDM] containing 20 mM imidazole-HCl (pH 8.0) for 45 min. The resin was washed with buffer A containing 20 mM imidazole (pH 8.0), and then proteins were eluted with buffer A containing 40, 60, 100, and 300 mM imidazole (pH 8.0) in a stepwise manner. Mixture of the eluted YeeE and TEV(S219V) protease (23) with a molar ratio of 10:1 was dialyzed against 20 mM tris-HCl (pH 8.0), 300 mM NaCl, 1 mM β-ME, 0.05% DDM, and 20 mM imidazole-HCl (pH 8.0) at 4°C for 16 hours to remove the His tag at the C terminus of YeeE. The solution was mixed with Ni-NTA resin equilibrated with buffer A containing 20 mM imidazole (pH 8.0) and incubated at 4°C for 45 min. The flow-through solution containing YeeE was concentrated using an Amicon Ultra 30K filter (Millipore) and applied to a Superdex 200 Increase 10/300 GL (GE Healthcare) equilibrated with buffer [20 mM tris-HCl (pH 8.0), 300 mM NaCl, 0.1% DDM, 1 mM β-ME, and 0.1 mM PMSF]. The peak fractions were concentrated to approximately 20 mg/ml for crystallization. For the expression of SeMet-labeled YeeE, the E. coli cells were grown in a salt medium (24) instead of the LB medium. The purification of SeMet-labeled YeeE was performed in the same manner as that of native YeeE.
Crystallization
Crystals of YeeE appeared in lipidic cubic phase (LCP). A solution of 300 mM sodium thiosulfate was added to the purified protein solution (about 20 mg/mL YeeE) to the final concentration of 75 mM. The solution was mixed with monoolein (Nu-Chek Prep) at a 2:3 ratio (w/w) using the twin-syringe mixing method (25). Each 50 nl of the mixture was spotted on crystallization plates and covered by 3 μl of a reservoir solution using a crystallization robot (Gryphon LCP, Art Robbins Instruments). Crystals of native YeeE and SeMet-labeled YeeE were grown up to about 80 μm (long axis) and 30 μm (short axis) at 20°C in a reservoir solution containing 24 to 26% Pentaerythritol-propoxylate (5/4 PO/OH), 100 mM 2-morpholinoethanesulfonic acid (MES)–NaOH (pH 6.5 to 7.0), and 100 to 150 mM NaCl for 5 days. The crystals were harvested using Crystal Mounts and Loops (MiTegen), directly flash cooled in liquid nitrogen, and stored in liquid nitrogen until the x-ray diffraction experiments.
Data collection and processing
The x-ray diffraction experiments were performed at 100 K at beamline BL32XU of SPring-8. The complete datasets were obtained by merging multiple small-wedge datasets. For the native dataset, diffraction images of 10° rotation range with a pitch of 0.1° oscillation range were collected from each of the 515 crystals. For the dataset of the C91A mutant, diffraction images of 10° rotation range with a pitch of 0.1° oscillation range were collected from each of the 93 crystals. For the SeMet dataset, diffraction images of 7° rotation range with a pitch of 0.1° oscillation range were collected from each of the 326 crystals. All the diffraction images were collected automatically by the Zoo system (26) and then processed using KAMO (27) with XDS (28).
Experimental phasing and structural refinement
The SIRAS method (29) using the SeMet derivative was applied to solve the crystallographic phase problem. Initially, seven Se sites were identified using SHELXC and SHELXD (30). Heavy-atom parameter refinement and SIRAS phasing were carried out using the program AutoSol (31). The resulting initial model of SeMet YeeE was refined using COOT (32) and PHENIX (33) to Rfree = ~0.28. The phase of the native dataset was calculated by molecular replacement using PHASER (34) with the SeMet model. The structural model of native YeeE(1–328) was stepwise refined using COOT and PHENIX to Rwork/Rfree = 0.202/0.252 with space group C2221 at 2.52-Å resolution. The structure model of the YeeE C91A mutant was initially calculated by molecular replacement using PHASER with native YeeE and refined to Rwork/Rfree = 0.235/0.286 at 2.60-Å resolution. Ramachandran plots were constructed using MolProbity (35), and molecular graphics were generated using CueMol2 (www.cuemol.org/en/).
Isothermal titration calorimetry
Purified StYeeE protein (~20 mg/ml) was dialyzed against isothermal titration calorimetry (ITC) buffer [20 mM tris-HCl (pH 7.0), 300 mM NaCl, 0.1% DDM, and 1 mM tris(2-carboxyethyl)phosphine] overnight at 4°C. Dialyzed YeeE sample was collected and centrifuged at 13,000 rpm for 10 min at 4°C. The concentration of YeeE protein was adjusted to 56.2 μM with the ITC buffer that had been used for dialysis (used ITC buffer). Sodium thiosulfate was dissolved in the used ITC buffer to prepare 500 μM sodium thiosulfate solution. The ITC experiment was carried out using MicroCal iTC200 (GE Healthcare) at 20°C. To subtract the heat of the dilution, 500 μM sodium thiosulfate solution was titrated against the used ITC buffer. Data were analyzed using Origin 7 software.
MD simulations
Simulations were carried out using GROMACS version 2019.3 simulation suite (36). The start model for MD position I was the crystal structure of StYeeE (WT-B in fig. S2B; PDB ID: 6LEO). To prepare the start models for MD position II and MD position III, we removed the thiosulfate from the model WT-B and manually placed thiosulfate on the electron density map at positions II and III, respectively. The Charmm36 force field (37) was applied. During the model building for MD position I by the above process, we changed the deprotonated state of E241 to its protonated state because the length between E241 and thiosulfate in Fig. 2D represents a hydrogen bond. The YeeE model was embedded into a POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphorylcholine) bilayer generated by the CHARMM-GUI Membrane Builder (38, 39) and solvated in an 80 × 80 × 80 Å3 box of the TIP3P (40) water molecules. The water molecules were replaced with sodium and chloride ions to neutralize the simulation system. The resulting concentration of NaCl was estimated at approximately 40 mM. The thiosulfate topology file was prepared using ATB (https://atb.uq.edu.au) (41). The energy minimization with 50,000 steps before NVT (constant number of particles, volume, and temperature) equilibration was performed for two successive target Fmax of no greater than 1000 and then 200 kJ mol−1 nm−1. Then, the simulations were performed with equilibrium for 100 ps in the NVT ensemble and 1 ns in the NPT (constant number of particles, pressure, and temperature) ensemble at 300 K. The MD simulations were carried out for 100 ns (0.002 ps × 50,000,000 steps) for StYeeE with thiosulfate and lipid bilayer in water. The interactions were analyzed by jsPISA (42), and RMSD values were calculated using CueMol (www.cuemol.org/en/).
Supplementary Material
Acknowledgments
We thank K. Abe for secretarial assistance, K. Kobayashi for technical support, I. Ohtsu (University of Tsukuba) for useful suggestions, S. Kubo (Kyoto University) for helping with the MD simulation, and the beamline scientists at BL32XU of SPring-8 (Hyogo, Japan) for helping with data collection. The synchrotron radiation experiments were performed at BL32XU of SPring-8 with the approval of the Japan Synchrotron Radiation Research Institute (JASRI) (proposal nos. 2017A2557, 2017B2557, 2018A2542, 2018B2542, and 2019A2518). E. coli DNA (JCM 20135) was provided by RIKEN BRC, which is participating in the National BioResource Project of the MEXT, Japan. Funding: This work was supported by the JSPS/MEXT KAKENHI (grant nos. JP17H05669 and JP19K06526 to Y.T.; the JSPS/MEXT KAKENHI (grant nos. JP26119007, JP18H02405, JP18KK0197, and JP19K22395) and private research foundations (Mitsubishi Foundation, Noguchi Institute, Naito Foundation, the Uehara Memorial Foundation, Takeda Science Foundation, and G-7 Scholarship Foundation) to T.T.; and the JSPS KAKENHI (grant no. JP19K23726) to M.I. Author contributions: Conceptualization: H.T., G.N., and T.T. Methodology and investigation: Y.T., K.Y., A.T., M.I., T.M., S.U., Y.S., T.H., and G.N. Writing, original draft: Y.T., H.T., and T.T. Writing, review and editing: M.I. and T.T. Supervision: T.T. Competing interests: The authors declare that they have no competing interests. Data and materials availability: Coordinates and structure factors have been deposited in the PDB under accession nos. 6LEO and 6LEP. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/35/eaba7637/DC1
REFERENCES AND NOTES
- 1.Ikeda M., Amino acid production processes. Adv. Biochem. Eng. Biotechnol. 79, 1–35 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Takagi H., Ohtsu I., l-Cysteine metabolism and fermentation in microorganisms. Adv. Biochem. Eng. Biotechnol. 159, 129–151 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Wada M., Takagi H., Metabolic pathways and biotechnological production of l-cysteine. Appl. Microbiol. Biotechnol. 73, 48–54 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Takumi K., Ziyatdinov M. K., Samsonov V., Nonaka G., Fermentative production of cysteine by Pantoea ananatis. Appl. Environ. Microbiol. 83, e0250-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.N. Kredich, Biosynthesis of cysteine, in Escherichia Coli and Salmonella Typhimurium: Cellular and Molecular Biology, F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznicoff, M. Riley, M. Schaechter, J. E. Umbarger, Eds. (ASM, Washington DC, ed. 2, 1996), pp. 514–527.
- 6.Aguilar-Barajas E., Díaz-Pérez C., Ramírez-Díaz M. I., Riveros-Rosas H., Cervantes C., Bacterial transport of sulfate, molybdate, and related oxyanions. Biometals 24, 687–707 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Nakatani T., Ohtsu I., Nonaka G., Wiriyathanawudhiwong N., Morigasaki S., Takagi H., Enhancement of thioredoxin/glutaredoxin-mediated L-cysteine synthesis from S-sulfocysteine increases L-cysteine production in Escherichia coli. Microb. Cell Fact. 11, 62 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawano Y., Onishi F., Shiroyama M., Miura M., Tanaka N., Oshiro S., Nonaka G., Nakanishi T., Ohtsu I., Improved fermentative l-cysteine overproduction by enhancing a newly identified thiosulfate assimilation pathway in Escherichia coli. Appl. Microbiol. Biotechnol. 101, 6879–6889 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Jez J. M., Dey S., The cysteine regulatory complex from plants and microbes: What was old is new again. Curr. Opin. Struct. Biol. 23, 302–310 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Funahashi E., Saiki K., Honda K., Sugiura Y., Kawano Y., Ohtsu I., Watanabe D., Wakabayashi Y., Abe T., Nakanishi T., Suematsu M., Takagi H., Finding of thiosulfate pathway for synthesis of organic sulfur compounds in Saccharomyces cerevisiae and improvement of ethanol production. J. Biosci. Bioeng. 120, 666–669 (2015). [DOI] [PubMed] [Google Scholar]
- 11.McIntosh B. K., Renfro D. P., Knapp G. S., Lairikyengbam C. R., Liles N. M., Niu L., Supak A. M., Venkatraman A., Zweifel A. E., Siegele D. A., Hu J. C., EcoliWiki: A wiki-based community resource for Escherichia coli. Nucleic Acids Res. 40, D1270–D1277 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gristwood T., McNeil M. B., Clulow J. S., Salmond G. P., Fineran P. C., PigS and PigP regulate prodigiosin biosynthesis in Serratia via differential control of divergent operons, which include predicted transporters of sulfur-containing molecules. J. Bacteriol. 193, 1076–1085 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slabinski L., Jaroszewski L., Rychlewski L., Wilson I. A., Lesley S. A., Godzik A., XtalPred: A web server for prediction of protein crystallizability. Bioinformatics 23, 3403–3405 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Holm L., Laakso L. M., Dali server update. Nucleic Acids Res. 44, W351–W355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H., Robertson A. D., Jensen J. H., Very fast empirical prediction and rationalization of protein pKa values. Proteins 61, 704–721 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Quistgaard E. M., Löw C., Guettou F., Nordlund P., Understanding transport by the major facilitator superfamily (MFS): Structures pave the way. Nat. Rev. Mol. Cell Biol. 17, 123–132 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Yan N., Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem. Sci. 38, 151–159 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Zakrzewska S., Mehdipour A. R., Malviya V. N., Nonaka T., Koepke J., Muenke C., Hausner W., Hummer G., Safarian S., Michel H., Inward-facing conformation of a multidrug resistance MATE family transporter. Proc. Natl. Acad. Sci. U.S.A. 116, 12275–12284 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joseph D., Pidathala S., Mallela A. K., Penmatsa A., Structure and gating dynamics of Na+/Cl− coupled neurotransmitter transporters. Front. Mol. Biosci. 6, 80 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Datsenko K. A., Wanner B. L., One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.G. Nonaka, K. Takumi, L-cysteine producing bacterium and a method for producing L-cysteine. U. S. Patent 8383372 B2 (2013).
- 22.Kawate T., Gouaux E., Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Kapust R. B., Tözsér J., Fox J. D., Anderson D. E., Cherry S., Copeland T. D., Waugh D. S., Tobacco etch virus protease: Mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 14, 993–1000 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Tsukazaki T., Mori H., Fukai S., Numata T., Perederina A., Adachi H., Matsumura H., Takano K., Murakami S., Inoue T., Mori Y., Sasaki T., Vassylyev D. G., Nureki O., Ito K., Purification, crystallization and preliminary X-ray diffraction of SecDF, a translocon-associated membrane protein, from Thermus thermophilus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 376–380 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caffrey M., Cherezov V., Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 4, 706–731 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirata K., Yamashita K., Ueno G., Kawano Y., Hasegawa K., Kumasaka T., Yamamoto M., ZOO: An automatic data-collection system for high-throughput structure analysis in protein microcrystallography. Acta Crystallogr. D Struct. Biol. 75, 138–150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamashita K., Hirata K., Yamamoto M., KAMO: Towards automated data processing for microcrystals. Acta Crystallogr. D Struct. Biol. 74, 441–449 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blow D. M., Rossmann M. G., The single isomorphous replacement method. Acta Crystallogr. 14, 1195–1202 (1961). [Google Scholar]
- 30.Schneider T. R., Sheldrick G. M., Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Terwilliger T. C., Adams P. D., Read R. J., McCoy A. J., Moriarty N. W., Grosse-Kunstleve R. W., Afonine P. V., Zwart P. H., Hung L.-W., Decision-making in structure solution using Bayesian estimates of map quality: The PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 65, 582–601 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., Adams P. D., Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J., Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams C. J., Headd J. J., Moriarty N. W., Prisant M. G., Videau L. L., Deis L. N., Verma V., Keedy D. A., Hintze B. J., Chen V. B., Jain S., Lewis S. M., Arendall W. B. III, Snoeyink J., Adams P. D., Lovell S. C., Richardson J. S., Richardson D. C., MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., Berendsen H. J. C., GROMACS: Fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Klauda J. B., Venable R. M., Freites J. A., O’Connor J. W., Tobias D. J., Mondragon-Ramirez C., Vorobyov I., MacKerell A. D. Jr., Pastor R. W., Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jo S., Kim T., Iyer V. G., Im W., CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Wu E. L., Cheng X., Jo S., Rui H., Song K. C., Dávila-Contreras E. M., Qi Y., Lee J., Monje-Galvan V., Venable R. M., Klauda J. B., Im W., CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 35, 1997–2004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.H. J. C. Berendsen, J. P. M. Postma, W. F. Van Gunsteren, J. Hermans, Interaction models for water in relation to protein hydration, in Intermolecular Forces, B. Pullman, Ed. (Springer, Dordrecht, 1981), pp. 331–342.
- 41.Malde A. K., Zuo L., Breeze M., Stroet M., Poger D., Nair P. C., Oostenbrink C., Mark A. E., An automated force field topology builder (ATB) and repository: Version 1.0. J. Chem. Theory Comput. 7, 4026–4037 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Krissinel E., Stock-based detection of protein oligomeric states in jsPISA. Nucleic Acids Res. 43, W314–W319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/35/eaba7637/DC1


