Experience-induced differences in behavior, brain plasticity, and epigenetics persist even after stimulus withdrawal.
Abstract
Individuals differ in their response to environmental stimuli, but the stability of individualized behaviors and their associated changes in brain plasticity are poorly understood. We developed a novel model of enriched environment to longitudinally monitor 40 inbred mice exploring 35 connected cages over periods of 3 to 6 months. We show that behavioral individuality that emerged during the first 3 months of environmental enrichment persisted when mice were withdrawn from the enriched environment for 3 additional months. Behavioral trajectories were associated with stable interindividual differences in adult hippocampal neurogenesis and persistent epigenetic effects on neuronal plasticity genes in the hippocampus. Using genome-wide DNA methylation sequencing, we show that one-third of the DNA methylation changes were maintained after withdrawal from the enriched environment. Our results suggest that, even under conditions that control genetic background and shared environment, early-life experiences result in lasting individualized changes in behavior, brain plasticity, and epigenetics.
INTRODUCTION
The ability of animals to change their behavior in response to experiences is crucial for their adaptation and fitness and, thus, has evolutionary and ecological consequences (1, 2). Experience-dependent behavioral changes can differ between individuals within one population (3). This behavioral variability is widespread among species, including humans (4), and has been attributed to genetic differences, gene-environment interactions, and developmental stochasticity (5–7). A characteristic feature of individual behaviors (or personalities) is their intraindividual stability over time and in different contexts (8). However, environmental aspects that trigger persistent individualized behaviors and their underlying neurobiological and molecular basis are still poorly understood.
We have previously shown that environmental enrichment (ENR) promotes the development of behavioral variability between inbred C57BL/6JRj mice (9, 10). In those experiments, interindividual differences among inbred mice emerged when they were housed in the same ENR cage, i.e., even in the absence of nominal genetic and environmental variation. The complexity of ENR allows the individual experience of nonshared environmental components, which amplifies and reinforces subtle initial differences between animals over time (9). However, since our previous studies were restricted to an ENR housing period of 3 months, the within-animal stability of the behavioral changes over longer periods of time and their plasticity toward environmental change remained unknown. Do ENR-induced interindividual differences persist within animals for long term? Would prolonged ENR lead to even further phenotypic divergence or does the individualizing effect plateau?
Activity-dependent brain plasticity is a neurobiological basis for stable behavioral changes that develop in response to environmental stimulation (11). ENR leads to structural changes in the brain that correlate with increased synaptic plasticity and improved cognitive performance (11–13). A prime example of ENR-induced structural brain plasticity is the greater number of new neurons that are generated in the hippocampus of ENR-housed mice (14). Our previous study has shown that, among a number of measures related to structural brain changes, adult hippocampal neurogenesis was the only phenotype which, in ENR animals, showed enhanced variance in addition to increases in group means that have been reported in essentially all previous studies (10). Furthermore, we have found that a substantial part of the variance in adult hippocampal neurogenesis (22%) was explained by individualized behavioral trajectories that emerged during time in ENR (9), linking individual behavior with brain plasticity. Whether variability in responses to early-life stimulation would influence levels of adult hippocampal neurogenesis in later life was, however, unexplored. We hypothesized that ENR would lead to persistent brain individualization through long-term individuality in adult hippocampal neurogenesis, which could potentially serve as a substrate for interindividual differences in brain function during aging.
To analyze the long-term stability of ENR-induced interindividual differences in behavior and adult hippocampal neurogenesis, we designed a novel cage system that allows the automated longitudinal monitoring of laboratory mice in a large enriched environment over longer time periods. The established system offers greatly improved temporospatial resolution of animal activity compared to the enclosure that we had used to establish the emergence of behavioral variability in our previous study (9). Using this system, we here show that behavioral and structural brain individualization are maintained with prolonged stimulation in ENR and even after withdrawal of the stimulus for 3 months. Thereby, our study demonstrated how subtle differences in early-life experiences determine individual life trajectories even in genetically identical animals.
RESULTS
Longitudinal monitoring of mice in a stimulus-rich environment
To investigate the effects of long-term ENR on behavioral variability, groups of 40 female C57BL/6JRj mice were housed in ENR or standard housing (STD) for a period of 6 months starting at an age of 5 weeks (Fig. 1A). To analyze the maintenance of individual behaviors after stimulus withdrawal, a third equally sized animal group was housed in ENR for 3 months (experimental phase 1) and afterward returned to STD cages where the animals stayed for another 3 months (experimental phase 2; ENR-STD mice).
Fig. 1. Experimental setup, enriched environment, and longitudinal monitoring of body weight.

(A) At an age of 5 weeks, 120 female C57BL/6JRj mice were split into three equally sized groups: 40 mice lived in ENR for 6 months, while a second group stayed in standard housing cages (STD) for 6 months, and the third group (ENR-STD) lived in ENR for the first 3 months (phase 1) and in STD for the last 3 months of the experiment (phase 2). Behavioral testing was performed with all animals in the last weeks of both phases. For quantification of adult hippocampal neurogenesis, all mice were injected with the thymidine analogs 5-iododeoxyuridine (IdU) and 5-chlorodeoxyuridine (CldU) 4 weeks before the ends of phase 1 and phase 2, respectively. (B) Image of the ENR cage system used for automated behavioral tracking of mice by RFID technology (photo credit: Susan Schilling, DZNE Dresden). (C) Schematic representation of the ENR cage depicting cage and tunnel access for the ENR group (light blue; top) and ENR-STD group (violet; bottom). RFID antenna were located around every tunnel and are highlighted in red. (D) ENR-induced reduction in body weight rebound to STD levels after returning ENR-STD mice to STD cages. Data points indicate means and SEM. Arrow marks return of ENR-STD to STD cages. Depicted P values from Brown-Forsythe test refer to housing effects. Full information on statistical tests is presented in Table 1 and data file S1.
To longitudinally monitor the behavior of individual mice, we used a novel ENR cage system that enabled automated behavioral tracking of animals in a seminaturalistic environment (Fig. 1B). The ENR cage consisted of 70 interconnected cages arranged on seven levels. Cages were connected by tunnels equipped with antenna for radio frequency identification (RFID)–based detection of animal movements between adjacent cages or levels. During experimental phase 1, ENR and ENR-STD mice stayed in separated subcompartments of the ENR system, each consisting of 35 cages that covered a total area of 1.37 m2 (Fig. 1C). ENR mice remained in the same subcompartment during experimental phase 2.
To monitor the general health status of mice, we measured body weight of all mice throughout the experiment and found an expected age-related increase in all animal groups (Fig. 1D). At the end of phase 1, the body weight of ENR and ENR-STD mice was reduced compared to STD mice (Fig. 1D and Table 1), which is consistent with the previously reported decrease of body weight by ENR (10, 14). However, within 4 weeks after returning ENR-STD mice to STD, their body weights increased to the levels observed in STD mice, while the long-term ENR group maintained lower body weights throughout phase 2 (Fig. 1D). This result showed that ENR-induced changes in body weight as a gross effect of the stimulation did not persist for long-term after environmental change.
Table 1. Summary statistics for individual group comparisons.
| Phenotype | Statistical test |
Adjusted P value (STD vs. ENR) |
Adjusted P value (STD vs. ENR-STD) |
Adjusted P value (ENR vs. ENR-STD) |
| Body weight phase 1 | Longitudinal model | 0.012 | 0.00016 | 0.27 |
| Body weight phase 2 | Longitudinal model | 0.045 | 0.50 | 0.50 |
| Body weight phase 2 | Brown-Forsythe | 0.07 | 0.00019 | 0.19 |
| Neurogenesis phase 1 (IdU) | Wilcoxon | 6.2 × 10−10 | 1.9 × 10−08 | 0.32 |
| Neurogenesis phase 1 (IdU) | Brown-Forsythe | 0.00096 | 0.00018 | 1.0 |
| Neurogenesis phase 2 (CldU) | Wilcoxon | 1.4 × 10−05 | 0.27 | 6.8 × 10−08 |
| Neurogenesis phase 2 (CldU) | Brown-Forsythe | 0.16 | 0.64 | 0.14 |
| Locomotion phase 1 | Wilcoxon | 1.4 × 10−13 | 4.6 × 10−15 | 0.46 |
| Locomotion phase 2 | Wilcoxon | 9.0 × 10−09 | 0.46 | 6.7 × 10−13 |
| Object exploration phase 1 | Wilcoxon | 0.0011 | 0.0014 | 1 |
| Object exploration phase 1 | Brown-Forsythe | 0.00090 | 0.17 | 0.12 |
| Object exploration phase 2 | Wilcoxon | 8.6 × 10−06 | 3.0 × 10−05 | 1 |
| Object exploration phase 2 | Brown-Forsythe | 0.00054 | 0.00011 | 0.91 |
ENR mice develop stable individualized trajectories in exploratory behavior
We have previously shown that behavioral individualization in ENR can be detected using roaming entropy (RE) as a measure of territorial coverage and spatial exploration of the environment by individual mice (9). Using the longitudinal mouse activity data obtained from the RFID recordings in the ENR cage, we calculated REs for the 40 mice of the ENR group and analyzed the development and stability of individual behavior during the 6 months of ENR housing.
Mean REs decreased with time in ENR (Fig. 2A), presumably as a result of the habituation of mice to the ENR cage. Despite this decline in exploratory activity, total variances of nightly REs increased during ENR housing (Fig. 2B), which was accompanied by the divergence of the behavioral trajectories of the individual mice with time (Fig. 2C). To assess the consistency of individual behavior, we estimated the variance fraction that is explained by interindividual behavioral differences and calculated the repeatability of REs within time blocks of 21 calendar days. Repeatability is a measure used in behavioral ecology to detect stable differences between animals in a population that are high relative to intraindividual fluctuations (15, 16). We found that, during the first 6 weeks in ENR (time blocks T1 and T2), the behavior of mice was characterized by repeatability and interindividual variance not significantly different from zero (Fig. 2, D and E, and table S1). After this initial phase, repeatability of behavior and interindividual variance continuously increased until the fourth month of ENR housing (time block T5), suggesting a progressive behavioral individualization of mice in ENR. Accordingly, the model assuming heterogeneous interindividual variance of REs between time blocks was better supported than the model with homogeneous variance [∆ deviance information criteria (DIC) = 105.1; table S2]. Thereafter, repeatability did not further increase, resulting in a stabilization of behavioral trajectories, which was supported by the high interindividual correlations of REs between time blocks in later periods of ENR housing (Fig. 2F and table S3).
Fig. 2. Mice in ENR develop consistent interindividual differences in behavior.

(A) Mean nightly RE of all ENR mice decreased with time. Data prior night 50 was incomplete as a result of technical problems. (B) SD of nightly REs increased with time. R2 and P values in (A) and (B) are derived from linear regression. (C) Individual mouse trajectories of REs aggregated over time blocks of 21 calendar days diverged with time. Numbers of nights per time block: T1, 6; T2, 3; T3, 11; T4, 17; T5, 15; T6, 17; T7, 16; and T8, 14. (D) Repeatability of REs between time blocks increased until T5 (fourth month of ENR housing) and remained stable thereafter. Posterior densities depict probabilities of the variance values obtained from generalized linear mixed model. (E) Variance due to interindividual behavioral differences increased until T6. Shown are modes of the posterior density with 95% confidence intervals (CIs). (F) Interindividual posterior correlations of RE were significant between later time blocks. Color code highlights the coefficient of Pearson correlation. (n = 39). See tables S1 to S3 for statistical details.
The results of the longitudinal activity monitoring reproduce our previously observed development of behavioral individuality in ENR (9) in a novel ENR cage system and additionally demonstrate that behavioral trajectories get progressively individualized until the fourth month in ENR and stabilize thereafter.
Stability of interindividual differences in adult neurogenesis
ENR enhances variance in adult hippocampal neurogenesis compared to STD mice, which we have previously related to the development of stable behavioral trajectories (9, 10). To analyze whether ENR has lasting consequences on individual rates of adult neurogenesis, we injected mice 4 weeks before the end of each phase with the thymidine analog 5-iododeoxyuridine (IdU; phase 1) and 5-chlorodeoxyuridine (CldU; phase 2), which can be separately detected by immunohistochemistry (Fig. 1A). ENR and ENR-STD mice showed a greater than twofold increase in the numbers of IdU-positive cells in the dentate gyrus and a significantly higher variance of cell numbers compared to STD mice (Fig. 3, A and B, table 1, and fig. S1A). Together with the previously observed increase in mean and variance of adult neurogenesis after 3 months of ENR (10), these results indicated that individual differences in numbers of neurons generated in the hippocampus during early-life ENR are stable with time and maintained even after withdrawal of ENR.
Fig. 3. Long-term maintenance of interindividual differences in adult-born neurons in the hippocampus.
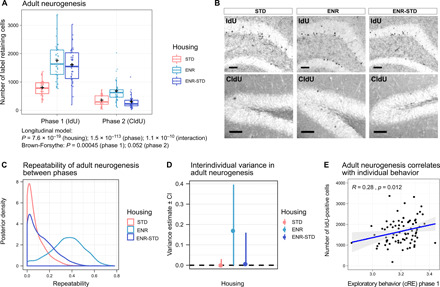
Adult neurogenesis during phase 1 and phase 2 was analyzed by injection of IdU (4 weeks before the end of phase 1) and CldU (4 weeks before the end of phase 2) and quantified at the end of the experiment. (A) Increased mean and variance in numbers of new hippocampal neurons produced during phase 1 (IdU) were observed in ENR and ENR-STD mice. ENR but not ENR-STD mice showed increased levels of adult neurogenesis compared to STD mice in phase 2 (CldU). [n = 36 (STD), 39 (ENR), and 40(ENR-STD)]. Box and whisker plots: center line, median; plus sign, mean; upper and lower hinges, first and third quartiles; whiskers, highest and lowest values within 1.5 times the interquartile range outside hinges; dots, individual data points. (B) Representative images of immunohistochemical stainings for detection of IdU- and CldU-positive cells. Scale bars, 50 μm. (C) Adult hippocampal neurogenesis is repeatable between phases only in ENR mice. (D) Variance fraction that is explained by interindividual differences shows posterior distribution shifted toward positive values in ENR compared to STD and ENR-STD mice. Modes of posterior densities with 95% credible intervals are shown. (E) Phenotypic correlation of adult neurogenesis to cumulative RE (cRE) in phase 1. See table S1 and data file S1 for statistical details. See also fig. S1.
When we compared the numbers of new hippocampal cells generated in both phases, we found an expected age-related decrease in adult hippocampal neurogenesis, as well as an interaction between phase and housing condition (Fig. 3A). Accordingly, ENR-STD mice exhibited a lower number of cells generated during experimental phase 2 (CldU-positive) compared to ENR mice but no difference compared to STD mice (Fig. 3A and Table 1), indicating that ENR-stimulated neurogenic activity is not maintained after withdrawal of environmental stimulation. In contrast, ENR mice showed a significant twofold increase in CldU-positive cell numbers in the dentate gyrus compared to STD mice. In addition, numbers of new hippocampal cells were repeatable and significantly correlated between phases only in ENR mice (Fig. 3C and fig. S1, B to D), which suggested that ENR mice developed consistent interindividual differences in adult neurogenesis. Although the lower 95% confidence intervals for interindividual variance in ENR mice abutted zero (Fig. 3D), the model, which estimated separate interindividual variance in housing groups, was better supported than a model with homogeneous variance (∆ DIC = 13.6; table S2). Note that the survival periods of IdU- and CldU-labeled cells were different (4 months and 1 month, respectively). Our results indicate that, despite the discontinuation of the environmental stimulation in the ENR-STD group, the new neurons generated in response to the initial ENR phase were maintained, while the de novo production of new neurons was declined to baseline levels.
To relate adult hippocampal neurogenesis to the emerged behavioral trajectories, we correlated exploration in ENR (cumulative RE) with adult neurogenesis (number of IdU-positive cells) for all mice that were housed in ENR during phase 1 (ENR and ENR-STD group). In agreement with our previous observations (9), adult neurogenesis showed a positive correlation with individual levels of exploratory behavior (Fig. 3E).
Together, these results showed that (i) individual behavioral trajectories and correlated levels of adult hippocampal neurogenesis developed in ENR during phase 1, (ii) individuality in behavior and adult neurogenesis remained stable with continued ENR in phase 2, and (iii) structural individuality of the hippocampus through adult neurogenesis in phase 1 was maintained after withdrawal of ENR in phase 2.
Selective maintenance of ENR-induced individuality in object exploration after environmental change
We next asked whether the stability of the emerged individual behaviors with time was dependent on continuous stimulation in ENR or whether behavioral patterns were also maintained after returning mice to STD cages. We have recently shown that ENR-induced behavioral variability can be detected using open-field and object exploration tests in a cross-sectional experimental design (10). Therefore, to investigate the long-lasting effects of ENR on behavioral variability, all mice were analyzed in these tests at the end of both experimental phases (Fig. 1A).
ENR changed activity patterns in the open field in phase 1, which were preserved with prolonged time in ENR but not maintained after ENR-STD mice returned to STD cages in phase 2 (fig. S2). For instance, locomotor activity in the open-field arena was reduced in ENR mice compared to STD mice at the end of phases 1 and 2 (Fig. 4A and Table 1). In contrast, while locomotion of ENR-STD mice was reduced compared to STD mice in phase 1, it was similar to STD mice and increased compared to ENR mice in phase 2. Direct comparison of locomotion between phases 1 and 2 further showed that individual locomotor activity was stable within animals in STD and ENR mice, but not in ENR-STD mice (Fig. 4, B and C, and fig. S3, A to C).
Fig. 4. Stability of ENR-induced individuality in object exploration with time and maintenance after withdrawal from ENR.
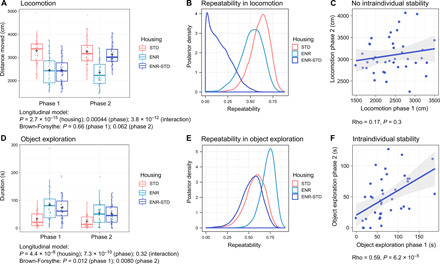
(A to C) Locomotion in the open-field test is depicted as s an example of a behavior that is not maintained after withdrawal from ENR. (A) At the end of phase 1, ENR and ENR-STD mice traveled shorter distances in the open-field test compared to STD mice. At the end of phase 2, ENR mice maintained reduced levels of locomotion, while ENR-STD mice traveled similar distances as STD mice. (B) High repeatability of locomotion between phase 1 and phase 2 is observed in ENR and STD mice but not in ENR-STD mice. (C) Lack of correlation of individual locomotion between phases 1 and 2 in ENR-STD mice suggests behavioral change after withdrawal from ENR. (D) ENR increased duration and variance of object exploration in trial 1 of the object exploration test. Increases in mean and variance of initial object exploration induced by ENR can still be observed after withdrawal from ENR. (E) High repeatability of initial object exploration between phases 1 and 2 in ENR and ENR-STD mice indicates maintenance of individual levels of object exploration after withdrawal from ENR. (F) Significant correlation of initial object exploration between phases in ENR-STD mice confirms intraindividual stability of behavior. [n = 38 (STD), 39 (ENR), and 40 (ENR-STD)]. Box and whisker plots in (A) and (D) as described in Fig. 3. See table S1 and data file 1 for statistical details. See also fig. S2 and S3.
In contrast to the animals’ activity in the open-field test, we found that the ENR-induced variability in object exploration was not only stable over time in ENR but also maintained after withdrawal from ENR. In the object exploration tests of phases 1 and 2, ENR mice spent more time around objects and showed a significantly higher variance of the duration of object exploration compared to STD mice (Fig. 4D, Table 1, and figs. S2, F and G, and S3D). In both phases, ENR-STD mice showed greater means and variances of object exploration compared to STD mice and no differences compared to ENR mice, suggesting that ENR-induced changes in object exploration are maintained after returning mice to STD cages. Individual levels of initial object exploration were highly repeatable and significantly correlated between the two experimental phases not only in STD and ENR mice but also in the ENR-STD group (Fig. 4, E and F, and fig. S3, E and F). Together, these results confirm the previously observed variability in object exploration after 3 months of ENR (10) and additionally demonstrate that the variance-enhancing effect of ENR in object exploration is preserved with prolonged periods of ENR housing. Moreover, these results show that ENR-induced increases in object exploration are stably maintained within animals even after withdrawal of animals from the enriched environment.
To analyze the influence of ENR on relationships between behaviors and brain plasticity, we correlated individual levels of object exploration with adult hippocampal neurogenesis and spatial exploration (RE) in the ENR cage. In both phases, ENR mice tended to show positive correlations of object exploration with RE and adult neurogenesis, which were, however, not statistically significant at the conventional levels of P = 0.05 (fig. S4). The lack of a strong association might suggest that consequences of ENR on object exploration involve processes that are largely independent of adult hippocampal neurogenesis and spatial exploration.
In summary, we showed that ENR-induced individualization in behavior and adult hippocampal neurogenesis in phase 1 resulted in lasting behavioral and structural brain individuality that was not only preserved with continuous ENR housing in phase 2 but also maintained for 3 months after withdrawal from ENR.
Maintenance of ENR-induced DNA methylation changes in the dentate gyrus after environmental change
Experience-dependent epigenetic changes, such as DNA methylation, have been linked to stable behavioral differences (17). Dynamic DNA methylation changes in neurons contribute to synaptic plasticity and memory formation (18, 19). To identify molecular mechanisms underlying the maintenance of individual behavior after withdrawal of ENR, we performed genome-wide DNA methylation profiling on microdissected dentate gyrus tissue by reduced representation bisulfite sequencing (RRBS) (20).
We detected significant methylation differences between ENR and STD mice at 12,167 CpGs (2.67% of all CpGs) and 1927 CpHs (0.087% of CpHs; fig. S5, A and B, and data file S2). Genes containing ENR-induced differentially methylated cytosines were enriched in pathways related to axon guidance and neuronal plasticity (fig. S5, C and D). To relate ENR-induced DNA methylation changes to gene expression, we integrated them with previously published RNA sequencing results from the dentate gyrus of mice housed in ENR for 2 months (21). Genes up-regulated by ENR were enriched among genes containing ENR-induced hypomethylated cytosines located within gene bodies (fig. S5, E and F), suggesting an association between ENR-induced DNA methylation and transcriptional changes. Comparing ENR-STD with STD mice, we identified 10,216 differentially methylated CpGs (2.24% of CpGs) and 1315 differentially methylated CpHs (0.053% of CpHs; fig. S5, G and H, and data file S2), indicating that early-life ENR led to stable DNA methylation changes in the dentate gyrus that persisted after withdrawal of the stimulus. Genes with differentially methylated cytosines after ENR-STD showed a similar enrichment in neuronal plasticity pathways and a similar relationship to gene expression as long-term ENR mice (fig. S5, I to L).
To evaluate whether DNA methylation patterns of ENR-STD mice resemble those in ENR animals, we first correlated the methylation differences of ENR and ENR-STD compared to STD for all individual cytosines. Significant positive correlations were detected for CpG (R = 0.55) and CpH (R = 0.55) contexts (Fig. 5A), suggesting that ENR and ENR-STD showed similar methylation changes at individual cytosines on a genome-wide scale. Next, we overlapped significantly differentially methylated cytosines by ENR with cytosines differentially methylated by ENR-STD (Fig. 5B). In total, 27.30% of ENR-induced differentially methylated CpGs and 31.40% of CpHs also showed similar methylation changes after ENR-STD. Plotting the absolute DNA methylation percentages for those overlapping cytosines showed that the magnitude of the methylation changes was similar in ENR and ENR-STD mice (Fig. 5C). While STD mice showed accumulations of CpGs and CpHs with intermediate methylation percentages, ENR led to long-lasting hypermethylation of those CpGs and a hypomethylation of the CpHs with intermediate methylation percentages (Fig. 5C). These results suggested that ENR led to DNA methylation changes that are maintained for at least 3 months after withdrawal from ENR.
Fig. 5. ENR-induced long-lasting DNA methylation changes at ephrin signaling-related genes in the dentate gyrus.

(A) Differences in CpG and CpH methylation between ENR and STD mice correlate with methylation differences between ENR-STD and STD mice at individual cytosines (all cytosines sequenced). P values from Pearson correlation (n = 5 per group). (B) Overlap of 27.30% of differentially methylated CpGs and 31.40% of differentially methylated CpHs between ENR-induced and ENR-STD–induced methylation changes. (C) ENR-STD mice show absolute DNA methylation percentages of the overlapping 3321 CpGs and 605 CpHs similar to ENR mice but different to STD mice. Median methylation percentages for individual samples are highlighted. (D) Overlap of genes with differentially methylated CpGs or CpHs in ENR compared to STD and ENR-STD compared to STD mice. (E) The 3214 genes with long-lasting ENR-induced DNA methylation changes are enriched in biological processes (Gene Ontology) and pathways (Reactome) related to neuronal plasticity, including ephrin signaling, neuron migration, and synaptic plasticity pathways. (F) STRING interaction network of genes with enrichment in ephrin receptor signaling pathways. (G) ENR-induced DNA methylation changes at CpGs located in proximity to exon 1 of the Efnb1 gene are maintained in ENR-STD mice. Green bar depicts a CpG island. Bars represent mean and SEM for each CpG. See also fig. S5 and data file S2.
Context-specific differences were detected in the directionality of the persistent methylation changes. While in the CpG context, 22.30 and 31.72% of ENR-induced differentially methylated cytosines also changed in ENR-STD mice, in the CpH context, 59.07% of hypomethylated CpHs but only 3.64% of hypermethylated CpHs were maintained (Fig. 5B). This indicates that, at CpHs, maintenance of ENR effects is restricted to hypomethlyation and that ENR-induced CpH hypermethylation is dependent on continuous stimulation in ENR and does not persist after withdrawal of ENR.
Gene annotation showed that 36.14% of ENR-induced differentially methylated genes persisted in ENR-STD mice (Fig. 5D). Gene Ontology and Reactome pathway analyses suggested that genes with maintained methylation changes were involved in processes related to ephrin signaling, neuron migration, and synaptic signaling (Fig. 5E). Twenty-eight genes with known role in ephrin signaling showed persistent ENR-induced DNA methylation changes (Fig. 5F). One example of these was a differentially methylated region in proximity to the transcription start site of the ephrin B1 ligand Efnb1 (Fig. 5G). Ephrin signaling has multiple roles in neuronal plasticity such as axon guidance, control of dendritic growth, synapse formation, synaptic plasticity (22), neuronal differentiation, and adult hippocampal neurogenesis (23). Furthermore, ephrin signaling controls behaviors related to anxiety, stress, and depression (24, 25). Hence, the ENR-induced regulation of the ephrin signaling pathway in the hippocampus is a potential molecular link between persistent individual behavior and brain plasticity after withdrawal of ENR.
DISCUSSION
A characteristic feature of behavioral individuality is the stability of behavioral patterns over time (1, 3). Phenotypic variability can be an indication of persistent interindividual differences or within-individual fluctuations, and these two components can only be distinguished in longitudinal studies. Here, we showed that variability in behavior and adult hippocampal neurogenesis induced by ENR is stable within animals over prolonged periods of ENR housing. Furthermore, structural individualization of the hippocampus through long-term integrated adult-born neurons was maintained even after withdrawal of ENR, together with sustained individualized behavior in object exploration. Persistent behavioral differences after ENR were accompanied by changes in hippocampal DNA methylation patterns, which were maintained after stimulus withdrawal. Our data reveal the long-term maintenance of behavioral individuality in ENR and its relation to structural plasticity and the epigenetic state of the hippocampus.
ENR increased interindividual differences in spatial exploration (RE) and variance in object exploration and adult hippocampal neurogenesis, which confirmed the findings from our previous studies (9, 10). The fact that we detected ENR-induced interindividual differences in all three studies, despite using structurally distinct enriched environment cages, highlights the robustness of the development of individuality in ENR. While both previous studies used wide span cages that allowed exploration and interaction in an open space, the current study used a novel compartmentalized ENR system in which mice can navigate through a network of separated cages. Moreover, in the present study, the two groups, ENR and ENR-STD, were similarly housed in ENR during experimental phase 1 and tested in open-field and object exploration tests afterward, which served as an internal replication of the initial ENR effect. Both groups showed increased variance in object exploration and adult neurogenesis compared to STD in phase 1, and no significant behavioral differences were observed between them. The robustness and reproducibility of the individualization in behavior and hippocampal structure in ENR underpin the suitability of enriched environments as a tool to study mechanisms of individuality development independent of genetic variation.
Behavioral individualization, as measured by increasing interindividual variance and repeatability in RE, progressed until the fourth month of ENR housing and plateaued thereafter. We have previously proposed that individuality in ENR develops by the amplification of initial differences through a self-reinforced positive feedback loop between behavior and adult hippocampal neurogenesis (9, 10), which was supported by the positive phenotypic correlation between neurogenesis and exploratory activity. Other studies also suggested self-reinforcement as drivers of animal variability (26). The here detected stability of interindividual differences in adult hippocampal neurogenesis and the significant repeatability of this phenotype only in ENR animals further support this model. The observed age-related decrease in adult hippocampal neurogenesis and the reduction of exploratory activity due to the habituation to the ENR cage might have reduced the strength of the positive feedback loop with time and resulted in the detected plateauing of behavioral individualization. Despite this plateauing effect, behavioral trajectories, once developed, were stable over time in ENR, which was highlighted by the high interindividual correlations of RE between time blocks and by the high repeatability ENR mice showed in object exploration between phases. Future studies should address whether these behavioral trajectories remain stable throughout the animals’ life and result in individualized brain health during aging.
Not all tracking data from the first experimental period could be used because the recordings from a number of nights were incomplete due to technical difficulties or because the cage area available to the animals had been inadvertently larger than intended. As a result, the complete data were available from only six nights in the first week for the T1 block and from three nights in T2 block, which could result in less accurate estimates of parameters. The technical issues were resolved from night 50 onward. Nevertheless, the regression patterns of RE did not change, when data prior night 50 was removed from the analysis [mean RE: R2 (coefficient of determination) = 0.44, P = 2 × 10−12; SD RE: R2 = 0.13, P = 0.0004]. Furthermore, the support for the model with heterogeneous interindividual variance compared to the model with homogenous interindividual variance was not affected by exclusion of the potentially noisy data from T1 and T2 time blocks (∆ DIC = 54.73). T2 also showed positive interindividual correlations with later time blocks, suggesting a progressive development of rank-order stability of mice within ENR. Therefore, the main conclusions regarding patterns of RE were not influenced by the data loss.
Our results show that most activity measures, such as locomotion and frequency of center crossings in the open-field test and body weight, are not maintained after withdrawal from ENR. Object exploration stood out as the only activity measure where the ENR-induced changes were maintained within mice for at least 3 months after they were withdrawn from the enriched environment. During ENR housing, the weekly change of object location and type (toys, houses, and tunnels) in the cage provides repeated cognitive and novelty stimulation and, thus, represents a core aspect of ENR. Previous studies by others have shown stable changes in hippocampal synaptic plasticity (13), cerebral cortex weight, and cortical acetylcholinesterase activity (27) after withdrawal from ENR, which could contribute to the observed maintenance of individual behavior. We here demonstrated that, although the proneurogenic effect of ENR was depended on continuous stimulation, early-life ENR led to persistent individualization of the hippocampal network through adult neurogenesis. Because adult hippocampal neurogenesis has a known role in promoting cognitive flexibility and influencing affective behaviors (28, 29), the lasting behavioral changes could be mediated (at least in part) by the long-term integration of adult-born neurons generated during early-life ENR. Future experiments should investigate the causal role of adult hippocampal neurogenesis and other brain plasticity measures in the development and maintenance of individual behavioral trajectories.
In the present study, we analyzed the intraindividual stability of those behaviors and brain plasticity measures for which we had detected ENR-induced increases in variance in our previous multivariate study (10). Our behavioral analysis was deliberately focused on the animals’ exploratory activity; however, we do not exclude lasting effects of ENR on other behavioral traits. Previous studies have shown that ENR also reduces anxiety levels (30) and promotes cognitive abilities, including hippocampus-dependent spatial navigation (12) and context discrimination (31). The causal relationship between ENR-induced individuality in exploratory behavior and cognitive performance or affective behaviors should be unraveled in the future, in specifically designed experiments. To assess brain plasticity, we here focused on adult hippocampal neurogenesis for two reasons. First, our previous research revealed a robust increase in variability of this brain plasticity measure. Second, adult neurogenesis, to a certain extent, can be measured at separate time points within one animal. The individuality-promoting effects of ENR on other forms of activity-dependent plasticity, including dendritic branching and synaptic plasticity, and on brain connectivity are currently unknown and should be addressed in future research.
A number of studies suggested epigenetic factors as potential molecular drivers of individuality development (7, 26). DNA methylation profiling in humans identified tissue-independent, interindividual DNA methylation differences, many of which were attributed to environmental variation, including nonshared environmental influences, rather than genetic differences (32, 33). Whether ENR increases interindividual differences in DNA methylation patterns in the dentate gyrus and whether those correlate with exploratory behavior and brain plasticity would be an exciting research area for future studies. Here, we have profiled DNA methylation changes that might be associated with the persistence of individuality in exploratory behavior after environmental change. We found that almost one-third of the ENR-induced DNA methylation changes in the hippocampal dentate gyrus were maintained for at least 3 months after returning mice to STD cages. Although DNA methylation patterns of the dentate gyrus are influenced by numbers and maturation stages of newborn neurons, the proportion of newborn neurons among all granule cells is very low (2 to 3%) and below the applied threshold for detection of differentially methylated cytosines (25%). Therefore, the here identified persistent ENR-induced differentially methylated genes reflect changes in mature hippocampal neurons rather than changes in the cellular composition of the dentate gyrus due to adult hippocampal neurogenesis.
Future investigations into the functional role of the detected ENR-induced DNA methylation changes could provide vital links to gene expression, additional layers of gene regulation, brain plasticity, and emergent behavioral patterns. Other molecular players, synergistically with ENR-induced DNA methylation changes or in isolation, could potentiate individualized behavior. For instance, preexisting interindividual DNA methylation differences induced by early-life experiences such as maternal care (34), ENR-induced epigenetic changes other than DNA methylation (35), or environmentally regulated somatic mosaicism in the hippocampus (36) are potential drivers of behavioral variability. Furthermore, differential microRNA (miRNA) levels in the hippocampus have been linked to lasting behavioral changes after ENR (13) and could play a role in the observed maintenance of ENR-induced individuality. Because DNA methylation controls many molecular processes, including expression of miRNAs (37), somatic mosaicism (38), and other epigenetic modifications in the brain (39), the here-identified DNA methylation changes likely interact with other molecular players to facilitate the development and maintenance of individual behavior.
The present study showed that female mice develop persistent individualized behavior when housed in ENR from an age of 5 weeks onward. Whether the emergence of individuality induced by exposure to ENR is dependent on the animal’s age is not yet known. Since the phase of early postnatal development is characterized by high levels of brain plasticity and sensitivity to environmental influences, starting ENR before the age of 5 weeks would likely have similar, if not stronger, effects on brain and behavioral individualization. This would be consistent with previous studies showing that preweaning ENR modulates behavior of mice in adulthood (40). Although ENR similarly influences behavior and adult neurogenesis in middle-aged and aged animals (41), if ENR were started in late adulthood, then the age-related reduction in brain plasticity that underlies experience-dependent behavioral effects might lead to a more subtle and less stable individualization. Whether the development of stable individual behavior in ENR is restricted to a sensitive period early in life will be tested in future experiments. Furthermore, as all of our published previous reports on ENR, this study was done using female mice, both to achieve consistency and to avoid confounding effects of territorial and dominance behavior of male mice. The question of potential sex differences, however, is important and interesting. It remains to be tested in studies with even larger cage size, a start time earlier in life and cohorts of closely related males.
Our study suggests that a longitudinal version of the enriched environment paradigm can also be applied to study the neurobiological mechanisms underlying fundamental concepts such as “brain reserves” and “brain maintenance.” While brain reserve refers to the accumulation of neural resources during early life that attenuate later functional decline during aging, brain maintenance describes the preservation of brain integrity over time (42, 43). We show that early exposure to enriched sensory, cognitive, and social stimulation induces long-lasting changes in behavior, brain plasticity, and the hippocampal epigenome and thus highlight the potential of environmental experiences to determine life trajectories. At the same time, we identify plastic responses to ENR that depend on continuous stimulation. Our paradigm thus offers an opportunity to dissect neurobiological mechanisms underlying brain resilience by opening the related animal research to the inclusion of longitudinal trajectories.
MATERIALS AND METHODS
Animal husbandry
Female C57BL/6JRj mice were purchased at an age of 4 weeks from Janvier Labs and housed in groups of five animals in standard polycarbonate cages (Type III, Tecniplast). In the first week upon arrival, all animals were subcutaneously injected into their neck with a glass-coated microtransponder (SID 102/A/2; Euro I.D.) under brief isoflurane anesthesia. At an age of 5 weeks, mice were randomly assigned to three experimental groups using the online tool “Research Randomizer” (www.randomizer.org). Two animal groups of 40 mice each were housed in a cage system custom-built to our specifications (PhenoSys GmbH, now marketed as “PhenoSys ColonyRack Individuality 3.0”), which was divided into two equally sized enriched environments (Fig. 1, B and C). Each enriched environment covered an area of 1.37 m2 and consisted of 35 polycarbonate cages (1264C Type II, Tecniplast) that were connected via transparent tunnels and distributed on four levels. Food and water were provided on every level of the cage system. For cognitive stimulation, the cages were equipped with plastic toys, tunnels, and hideouts, which were replaced and rearranged once per week. Additional 40 mice stayed in standard polycarbonate cages (36.5 cm by 20.7 cm by 14 cm; Type III, Tecniplast) in groups of five animals per cage. Dirty cages in the enriched environment and the standard housing cages were cleaned once per week.
Mice were maintained on a 12-hour light/12-hour dark cycle with 55 ± 10% humidity at the animal facility of the Center for Regenerative Therapies Dresden. Control and enriched animals received the same fortified chow (no. V1534; Sniff) with 9% of energy from fat, 24% from protein, and 67% from carbohydrates. Food and water were provided freely. All mice received intraperitoneal injections of IdU (57.5 mg/kg) 9 weeks after start of the experiment and of CldU (42.5 mg/kg) 4 weeks before perfusion. IdU and CldU were dissolved in 0.9% sodium chloride and injected three times at 6-hour time intervals. All experiments were conducted in accordance with the applicable European and national regulations (Tierschutzgesetz) and were approved by the local authority (Landesdirektion Sachsen; file number 25-5131/354/63).
Analysis of RFID data
Antenna contacts were recorded using the software PhenoSoft Control (PhenoSys GmbH), which saved antenna identifier and mouse identifier together with the time stamp of the antenna contact into a database. During the course of the experiment, over 24.13 million (phase 1) and 8.52 mln (phase 2) events were registered. Data between the 40th and 49th day of ENR were removed from the analysis due to technical problems resulting in incomplete recordings. Between the 9th and 35th day of ENR, animals repeatedly forced the locks introduced in the tunnels between two compartments of the cage system and thus ENR and ENR-STD group mixed. Because the number and locations at which animal can be recorded influences its distribution and entropy value, the data from this period could not be compared with the rest of the data and were therefore excluded from further analysis. These technical problems, which occurred because the present experiment was the first large-scale experiment with our custom-built cage system, were resolved after night 49.
Data reduction and calculation of RE were performed as previously described (9). Because mice are nocturnal animals, only the events recorded during the dark phase were retained. Each night was divided into 8640 segments of 5-s length, and for each mouse and each time segment, the last registered antenna was recorded into a time series. Frequencies of antenna in this time series were converted to probabilities pi,j,t of a mouse i being at an antenna j at a night t. Shannon entropy of the roaming distribution was calculated as , where k is the number of antenna. Dividing the entropy by log(k) scales the RE to the range between zero and one. Data from nights following events that could disturb patterns of exploration, such as cleaning of the cage or behavioral testing, were excluded. The reduced dataset comprised 99 nights, which were partitioned into eight time blocks, each spanning 21 calendar days. The number of nights per each time block in the final dataset was as follows: T1, 6; T2, 3; T3, 11; T4, 17; T5, 15; T6, 17; T7, 16; and T8, 14. Cumulative RE at the end of phase 1 was calculated by cumulative addition of mean RE from the first four time blocks.
Open-field and object exploration tests
The open-field and object exploration tests were performed similarly as previously described (10). Briefly, mice were placed into a square arena (60 cm by 60 cm), and their exploration was recorded using a camera (Logitech) and EthoVision software (Noldus). In total, four trials were performed on two consecutive days with a trial length of 5 min each (fig. S3A). In the first trial (open-field test), mice were put in the empty arena, which also served as habituation for the object exploration tests. In the following two trials, mice were presented with two identical objects. In the fourth trial, one object was replaced with a “new” object. To avoid preference for object properties or placement, the use of object A and object B as “old” or new object and the position of the new object were randomized. Object A was a composite of a red and yellow plastic tube, each with a diameter of 55 mm and a height of 98 mm. Object B was a gray cuboid of 13 cm by 10 cm by 6 cm with a blue surface and holes on one side. Data analysis of open-field and object exploration tests was performed as previously described (10).
Tissue preparation and immunohistochemistry
Tissue fixation and immunohistochemistry for the analysis of adult neurogenesis were performed as previously described (10). Briefly, mice were anesthetized with ketamine (WDT) (100 mg/kg) and xylazin (10 mg/kg) (Serumwerk Bernburg AG) and transcardially perfused with 0.9% sodium chloride. Brains were removed from the skull, and one hemisphere was fixed in 4% paraformaldehyde prepared in phosphate buffer (pH 7.4) overnight at 4°C. Brains were incubated in 30% sucrose in phosphate buffer for 2 days and cut into 40-μm coronal sections using a dry ice–cooled copper block on a sliding microtome (Leica, SM2000R). Sections were stored at 4°C in cryoprotectant solution [25% ethylene glycol and 25% glycerol in 0.1 M phosphate buffer (pH 7.4)].
For detection of IdU- and CldU-positive cells, the peroxidase method was applied. Briefly, free-floating sections were incubated in 0.6% hydrogen peroxide for 30 min to inhibit endogenous peroxidase activity. For antigen retrieval, sections were incubated in prewarmed 2.5 M hydrochloric acid for 30 min at 37°C, followed by extensive washes. Unspecific binding sites were blocked in tris-buffered saline (TBS) supplemented with 10% donkey serum (Jackson ImmunoResearch Labs) and 0.2% Triton X-100 (Carl Roth) for 1 hour at room temperature. Primary antibodies were applied overnight at 4°C as follows: monoclonal mouse anti–5-bromo-2′-deoxyuridine (BrdU) (1:500; BD Biosciences) for IdU detection and monoclonal rat anti-BrdU (1:500; Serotec) for CldU detection. Biotinylated secondary antibodies (Jackson ImmunoResearch Labs) were incubated for 2 hours at room temperature. Antibodies were diluted in TBS supplemented with 3% donkey serum and 0.2% Triton X-100. Detection was performed using the VECTASTAIN Elite ABC Reagent (9 μg/ml of each component: Vector Laboratories, LINARIS) with diaminobenzidine (0.075 mg/ml; Sigma-Aldrich) and 0.04% nickel chloride as a chromogen. All washing steps were performed in TBS. Stained sections were mounted onto glass slides, cleared with Neo-Clear (Millipore), and cover-slipped using Neo-Mount (Millipore). IdU- and CldU-positive cells were counted on every sixth section along the entire rostro-caudal axis of the dentate gyrus using a bright-field microscope (Leica DM 750).
Statistical analysis
Experiments were carried out with the experimenter blind to the experimental group. Statistical analyses were performed using the statistical software R (R Core Team, 2014). To compare means of longitudinal data, we used a rank-based nonparametric test applying the nparLD function from the nparLD package, which reports analysis of variance (ANOVA)–type statistics for time, group, and time:group interaction, followed by pairwise comparison with Wilcoxon’s test. To compare variances between groups, Brown-Forsythe test was performed using the leveneTest function from the car package. Ninety-five percent confidence intervals for variance ratios between groups were derived from a product of ratio of sample variances and reciprocals of quantiles of F distribution with n1 – 1, n2 – 1 degrees of freedom . All tests were two-tailed, and differences were considered to be statistically significant at P < 0.05. Multiple testing correction was performed using the Holm method. Data were visualized using the ggplot2 package. In the box-whisker plots, center line and plus sign mark the median and mean, respectively. Upper and lower hinges indicate first and third quartiles, respectively. The upper whisker extends from the hinge to the largest value no more than 1.5 times the interquartile range (IQR; a distance between the first and third quartiles); the lower whisker extends from the hinge to the smallest value at most 1.5 times IQR. Full results of statistical tests are available in data file S1.
Mixed linear models and repeatability estimation
Repeatability (R) is the fraction of total variance, which can be attributed to the differences between individuals , where Vres is a residual, i.e., within individual variance (16, 44). To decompose the phenotypic variance into interindividual and within-individual components, we used generalized linear mixed models in a Bayesian framework implemented in the MCMCglmm package v2.29 in R v3.4.4.
Behavioral phenotypes from open-field and object exploration tests were square root transformed to correct the skew, and the normal distribution of the data was confirmed using Shapiro-Wilk test. RE and neurogenesis were mean centered and scaled to unity variance. The full model for RE in the ENR animals included the time block as a fixed effect and an interaction between time block and an individual identifier as a random effect, which was specified using the function us. This specification of the random term allowed to estimate interindividual variance for each time block, as well as covariance between the time blocks. The residual variance was estimated separately in each time block using the function idh. The models for behavioral parameters and neurogenesis included an interaction between housing and the phase as fixed effects, and an interaction between housing and an individual identifier as a random effect, as well as heterogeneous residual variance specified with the function idh. To assess whether interindividual variance differed between time blocks (RE) or housing conditions (behavior and neurogenesis), we omitted respective terms in the model and compared the DIC between the full and simpler models, assuming that a DIC score reduced by at least two units indicates better fit to the data.
Models were fitted by Markov chain Monte Carlo estimation with Gibbs sampling using the MCMCglmm package. We used weakly informative default proper priors for fixed effects and assumed a Gaussian error distribution. For the residual variance, we used an inverse Wishart distribution prior with a low belief parameter nu = 0.002. To estimate interindividual variances and covariances, we used parameter-expanded proper priors (45). We confirmed that results were not affected by changes in the prior specification. For each model, we fitted five chains using at least 600,000 iterations, 100,000 burn-in, and a thinning interval of 100. Chain mixing and convergence were first inspected visually. To confirm model convergence, we ensured that the Gelman-Rubin potential scale reduction factor statistic, calculated with the gelman.diag function from the R package coda, was close to 1. The R code with the prior and model specifications is available in data file S3. The estimates of repeatability and interindividual correlations and their 95% credible intervals were derived from posterior (co)variance distributions using a mode of the posterior density with the function posterior.mode and the parameter adjust set to 1, and the function HPDinterval from the MCMCglmm package, respectively. Because variance must have a positive value, significant repeatability is indicated by confidence intervals not abutting zero, while confidence intervals for significant correlations are not including zero. Full results are reported in tables S1 to S3.
Reduced representation bisulfite sequencing
Animals used for DNA methylation analysis were selected randomly. Genomic DNA was isolated from microdissected dentate gyrus tissue using the QIAamp DNA Micro Kit (Qiagen) following the manufacturer’s manuals. Libraries for RRBS were prepared from 100 ng DNA per sample using the Premium RRBS Kit (Diagenode). Final libraries were purified twice using Agencourt AMPure XP beads (1× bead volume; Beckman Coulter). The quality and concentration of RRBS libraries were determined using the High Sensitivity NGS Fragment Analysis Kit (Advanced Analytical) and a fragment analyzer with capillary size of 33 cm (Advanced Analytical). Sequencing was performed using a NextSeq 5000 platform in a 75–base pair single-end mode with a minimum sequencing depth of 15 million reads per sample.
Bioinformatic data analysis of DNA methylation data
FASTQ reads were trimmed using Trim Galore 0.4.4 and the function Cutadapt 1.8.1 in RRBS mode and mapped against mm10 using Bismark 0.19.0. Detection of differentially methylated cytosines was performed using methylKit v1.5.2 (46). Briefly, methylation levels were extracted from sorted Binary Alignment Map files using the function processBismarkAln. Data were filtered for cytosines with a minimum coverage of 10 reads and a maximum coverage of 99.9% percentile in at least three samples per group using the functions filterByCoverage and unite. Differentially methylated cytosines were identified using the methDiff function applying the Chi-squared test with multiple testing correction using the sliding linear model (SLIM) method, a significance threshold of q < 0.001 and a threshold for absolute cytosine methylation differences higher than 25%. Overlaps of cytosines were performed using the function subsetByOverlaps of the R package Genomic Ranges 3.7. Venn diagrams were generated using the function draw.pairwise.venn of the VennDiagram package. Violin plots were created using the R package ggplot2.
Differentially methylated cytosines were annotated to the gene with the nearest transcription start site using data tables downloaded from Ensembl BioMart (downloaded as of 28 May 2019). Functional gene enrichment analysis was performed using the R packages TopGO and ReactomePA. Gene Ontology and pathway enrichment analyses were performed with differentially methylated genes as query lists and all genes covered by RRBS as background lists.
For comparison of DNA methylation with gene expression, we used genes that significantly changed expression (adjusted P < 0.05) in either ventral or dorsal dentate gyrus after ENR housing as reported in (21). Genomic coordinates for promoters, gene bodies, and enhancers were retrieved from the University of California Santa Cruz Genome Browser. Enrichment analysis was performed using hypergeometric testing applying the phyper/dhyper function in R. P values were adjusted using false discovery rate correction. Background lists were normalized for genes detected in DNA methylation analysis and RNA sequencing analysis.
Supplementary Material
Acknowledgments
We thank all members of the Kempermann laboratory for assistance in the logistics of the experiment, N. Rund for help with CldU immunohistochemistry, and the DRESDEN-concept Genome Center for excellent sequencing service. Funding: This study was financed from basic institutional funds (Helmholtz Association and TU Dresden) and partly supported by the EnergI project funded by Bundesministerium für Bildung und Forschung (BMBF). S.Z. was supported by a fellowship from the International Max Planck Research School on the Life Course, Berlin. J.B.L. was supported by Coordination for the Improvement of Higher Education Personnel (CAPES), Brazil. Author contributions: Conceptualization: S.Z., A.N.G., V.S.A., and G.K.; Methodology: S.Z., A.N.G., Y.W., S.S., R.W.O., and G.K. Animal experiment: S.S., S.Z., J.B.L., and S.G. Formal analysis and visualization: S.Z., A.N.G., and S.S. Data curation: S.Z. and A.N.G. Writing (original draft): S.Z. and G.K. Writing (review and editing): S.Z., A.N.G., R.W.O., V.S.A., J.B.L., and G.K. Supervision: S.Z. and G.K. Competing interests: The authors declare that they have no competing interests. Data and materials availability: Sequencing data have been deposited at Gene Expression Omnibus (accession number GSE139051). Code for bioinformatic analysis is available upon request. Data from behavioral analysis and adult neurogenesis have been deposited at Dryad (https://datadryad.org/stash/share/ZZthabWxmPjNe0TFRVoST3HP9pHrZCrQl7cOaqPXiak). All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/35/eabb1478/DC1
REFERENCES AND NOTES
- 1.Dall S. R. X., Bell A. M., Bolnick D. I., Ratnieks F. L. W., An evolutionary ecology of individual differences. Ecol. Lett. 15, 1189–1198 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolf M., Weissing F. J., Animal personalities: Consequences for ecology and evolution. Trends Ecol. Evol. 27, 452–461 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Dingemanse N. J., Kazem A. J. N., Réale D., Wright J., Behavioural reaction norms: Animal personality meets individual plasticity. Trends Ecol. Evol. 25, 81–89 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Pennisi E., The power of personality. Science 352, 644–647 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Beets I., Zhang G., Fenk L. A., Chen C., Nelson G. M., Félix M.-A., de Bono M., Natural variation in a dendritic scaffold protein remodels experience-dependent plasticity by altering neuropeptide expression. Neuron 105, 106–121.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bach J.-F., The biological individual – The respective contributions of genetics, environment and chance. C. R. Biol. 332, 1065–1068 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Honegger K., de Bivort B., Stochasticity, individuality and behavior. Curr. Biol. 28, R8–R12 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Trillmich F., Müller T., Müller C., Understanding the evolution of personality requires the study of mechanisms behind the development and life history of personality traits. Biol. Lett. 14, 20170740 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freund J., Brandmaier A. M., Lewejohann L., Kirste I., Kritzler M., Krüger A., Sachser N., Lindenberger U., Kempermann G., Emergence of individuality in genetically identical mice. Science 340, 756–759 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Körholz J. C., Zocher S., Grzyb A. N., Morisse B., Poetzsch A., Ehret F., Schmied C., Kempermann G., Selective increases in inter-individual variability in response to environmental enrichment in female mice. eLife 7, e35690 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kempermann G., Environmental enrichment, new neurons and the neurobiology of individuality. Nat. Rev. Neurosci. 20, 235–245 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Garthe A., Roeder I., Kempermann G., Mice in an enriched environment learn more flexibly because of adult hippocampal neurogenesis. Hippocampus 26, 261–271 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benito E., Kerimoglu C., Ramachandran B., Pena-centeno T., Jain G., Stilling R. M., Islam M. R., Capece V., Zhou Q., Edbauer D., Dean C., Fischer A., RNA-dependent intergenerational inheritance of enhanced synaptic plasticity after environmental enrichment. Cell Rep. 23, 546–554 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kempermann G., Kuhn H. G., Gage F. H., More hippocampal neurons in adult mice living in an enriched environment. Nature 386, 493–495 (1997). [DOI] [PubMed] [Google Scholar]
- 15.Bell A. M., Hankison S. J., Laskowski K. L., The repeatability of behaviour: A meta-analysis. Anim. Behav. 77, 771–783 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dingemanse N. J., Dochtermann N. A., Quantifying individual variation in behaviour: Mixed-effect modelling approaches. J. Anim. Ecol. 82, 39–54 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Kundakovic M., Champagne F. A., Early-life experience, epigenetics, and the developing brain. Neuropsychopharmacology 40, 141–153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaas G. A., Zhong C., Eason D. E., Ross D. L., Vachhani R. V., Ming G.-l., King J. R., Song H., Sweatt J. D., TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron 79, 1086–1093 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng J., Zhou Y., Campbell S. L., Le T., Li E., Sweatt J. D., Silva A. J., Fan G., Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 13, 423–430 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyle P., Clement K., Gu H., Smith Z. D., Ziller M., Fostel J. L., Holmes L., Meldrim J., Kelley F., Gnirke A., Meissner A., Gel-free multiplexed reduced representation bisulfite sequencing for large-scale DNA methylation profiling. Genome Biol. 13, R92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang T.-Y., Keown C. L., Wen X., Li J., Vousden D. A., Anacker C., Bhattacharyya U., Ryan R., Diorio J., O’toole N., Lerch J. P., Mukamel E. A., Meaney M. J., Environmental enrichment increases transcriptional and epigenetic differentiation between mouse dorsal and ventral dentate gyrus. Nat. Commun. 9, 298 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grunwald I. C., Korte M., Wolfer D., Wilkinson G. A., Unsicker K., Lipp H.-P., Bonhoeffer T., Klein R., Kinase-independent requirement of EphB2 receptors in hippocampal synaptic plasticity. Neuron 32, 1027–1040 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Ashton R. S., Conway A., Pangarkar C., Bergen J., Il Lim K., Shah P., Bissell M., Schaffer D. V., Astrocytes regulate adult hippocampal neurogenesis through ephrin-B signaling. Nat. Neurosci. 15, 1399–1406 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheleg M., Yu Q., Go C., Wagner G. C., Kusnecov A. W., Zhou R., Decreased maternal behavior and anxiety in ephrin-A5−/− mice. Genes Brain Behav. 16, 271–284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang R.-X., Han Y., Chen C., Xu L.-Z., Li J.-L., Chen N., Sun C.-Y., Chen W.-H., Zhu W.-L., Shi J., Lu L., EphB2 in the medial prefrontal cortex regulates vulnerability to stress. Neuropsychopharmacology 41, 2541–2556 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vogt G., Facilitation of environmental adaptation and evolution by epigenetic phenotype variation: Insights from clonal, invasive, polyploid, and domesticated animals. Environ. Epigenetics. 3, dvx002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bennett E. L., Rosenzweig M. R., Diamond M. C., Morimoto H., Hebert M., Effects of successive environments on brain measures. Physiol. Behav. 12, 621–631 (1974). [DOI] [PubMed] [Google Scholar]
- 28.Garthe A., Huang Z., Kaczmarek L., Filipkowski R. K., Kempermann G., Not all water mazes are created equal: Cyclin D2 knockout mice with constitutively suppressed adult hippocampal neurogenesis do show specific spatial learning deficits. Genes Brain Behav. 13, 357–364 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.David D. J., Samuels B. A., Rainer Q., Wang J.-W., Marsteller D., Mendez I., Drew M., Craig D. A., Guiard B. P., Guilloux J.-P., Artymyshyn R. P., Gardier A. M., Gerald C., Antonijevic I. A., Leonardo E. D., Hen R., Neurogenesis-dependent and -independent effects of fluoxetine in an animal model of anxiety/depression. Neuron 62, 479–493 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benaroya-Milshtein N., Hollander N., Apter A., Kukulansky T., Raz N., Wilf A., Yaniv I., Pick C. G., Environmental enrichment in mice decreases anxiety, attenuates stress responses and enhances natural killer cell activity. Eur. J. Neurosci. 20, 1341–1347 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Clemenson G. D., Lee S. W., Deng W., Barrera V. R., Iwamoto K. S., Fanselow M. S., Gage F. H., Enrichment rescues contextual discrimination deficit associated with immediate shock. Hippocampus 25, 385–392 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Busche S., Shao X., Caron M., Kwan T., Allum F., Cheung W. A., Ge B., Westfall S., Simon M.-M.; The Multiple Tissue Human Expression Resource, Barrett A., Bell J. T., McCarthy M. I., Deloukas P., Blanchette M., Bourque G., Spector T. D., Lathrop M., Pastinen T., Grundberg E., Population whole-genome bisulfite sequencing across two tissues highlights the environment as the principal source of human methylome variation. Genome Biol. 16, 290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gunasekara C. J., Scott C. A., Laritsky E., Baker M. S., MacKay H., Duryea J. D., Kessler N. J., Hellenthal G., Wood A. C., Hodges K. R., Gandhi M., Hair A. B., Silver M. J., Moore S. E., Prentice A. M., Li Y., Chen R., Coarfa C., Waterland R. A., A genomic atlas of systemic interindividual epigenetic variation in humans. Genome Biol. 20, 105 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weaver I. C. G., Cervoni N., Champagne F. A., D’Alessio A. C., Sharma S., Seckl J. R., Dymov S., Szyf M., Meaney M. J., Epigenetic programming by maternal behavior. Nat. Neurosci. 7, 847–854 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Fischer A., Sananbenesi F., Wang X., Dobbin M., Tsai L.-H., Recovery of learning and memory is associated with chromatin remodelling. Nature 447, 178–182 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Muotri A. R., Zhao C., Marchetto M. C. N., Gage F. H., Environmental influence on L1 retrotransposons in the adult hippocampus. Hippocampus 19, 1002–1007 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi C., Kim T., Chang K. T., Min K.-T., DSCR 1-mediated TET 1 splicing regulates miR-124 expression to control adult hippocampal neurogenesis. EMBO J. 38, e101293 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muotri A. R., Marchetto M. C. N., Coufal N. G., Oefner R., Yeo G., Nakashima K., Gage F. H., L1 retrotransposition in neurons is modulated by MeCP2. Nature 468, 443–446 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell R. R., Wood M. A., How the epigenome integrates information and reshapes the synapse. Nat. Rev. Neurosci. 20, 133–147 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baldini S., Restani L., Baroncelli L., Coltelli M., Franco R., Cenni M. C., Maffei L., Berardi N., Enriched early life experiences reduce adult anxiety-like behavior in rats: A role for insulin-like growth factor 1. J. Neurosci. 33, 11715–11723 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kempermann G., Gast D., Gage F. H., Neuroplasticity in old age: Sustained fivefold induction of hippocampal neurogenesis by long-term environmental enrichment. Ann. Neurol. 52, 135–143 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Cabeza R., Albert M., Belleville S., Craik F. I. M., Duarte A., Grady C. L., Lindenberger U., Nyberg L., Park D. C., Reuter-Lorenz P. A., Rugg M. D., Steffener J., Rajah M. N., Maintenance, reserve and compensation: The cognitive neuroscience of healthy ageing. Nat. Rev. Neurosci. 19, 701–710 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stern Y., Arenaza-Urquijo E. M., Bartr D., Belleville S., Cantilon M., Chetelat G., Ewers M., Franzmeier N., Kempermann G., Kremen W. S., Okonkwo O., Scarmeas N., Soldan A., Udeh-momoh C., Valenzuela M., Vemuri P., Vuoksimaa E., Whitepaper: Defining and investigating cognitive reserve, brain reserve, and brain maintenance. Alzheimers Dement. S1552-5260, 33491–33495 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakagawa S., Schielzeth H., Repeatability for Gaussian and non-Gaussian data: A practical guide for biologists. Biol. Rev. 85, 935–956 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Gelman A., Prior distributions for variance parameters in hierarchical models (comment on article by Browne and Draper). Bayesian Anal. 1, 515–534 (2006). [Google Scholar]
- 46.Akalin A., Kormaksson M., Li S., Garrett-Bakelman F. E., Figueroa M. E., Melnick A., Mason C. E., methylKit: A comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13, R87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/35/eabb1478/DC1


