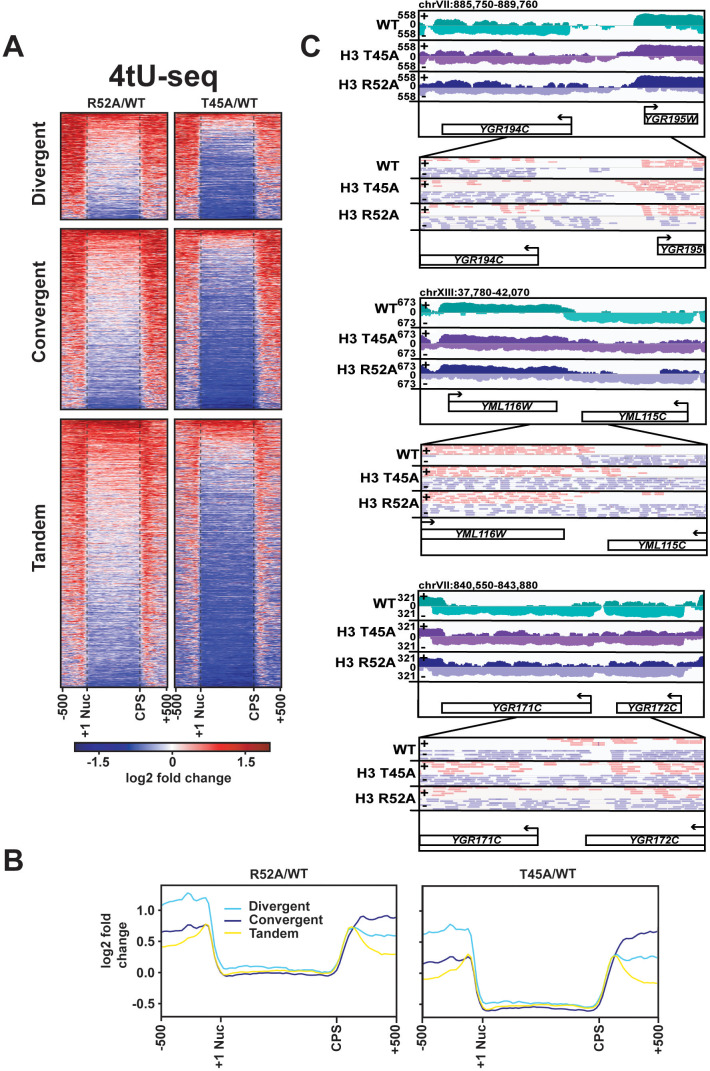
Figure 4. DNA entry-exit site mutants display aberrant transcription 5’ and 3’ to protein-coding genes.
(A) Heatmaps showing log2-fold change in spike-in normalized 4tU-seq read counts in H3 R52A or H3 T45A mutants relative to WT at divergent, convergent, and tandem genes. Heatmap rows represent 1186 divergent, 1981 convergent, and 2976 tandem protein-coding genes showing 500 bp upstream of the +1 nucleosome (Brogaard et al., 2012) and 500 bp downstream of the CPS (Ozsolak et al., 2010). (B) Metagene plots show averaged intensity over regions displayed in the difference heatmaps shown in panel A. (C) Browser tracks visualized in IGV (Thorvaldsdóttir et al., 2013) depicting aberrant transcription 5’ and 3’ to divergent (top), convergent (middle), and tandem (bottom) gene pairs in the indicated strains. Data represent log-scaled, spike-in normalized 4tU-seq read density over the plus (+) and minus (-) strands. Regions were chosen to be void of neighboring ncRNA loci. IGV-visualized BAM-file snapshots depict presence or absence of strand-specific read-pairs between genes. Arrows above gene names indicate directionality of transcription. All heatmaps and metagene plots were generated using deepTools2 (Ramírez et al., 2014; Ramírez et al., 2016) using 25 bp bins.
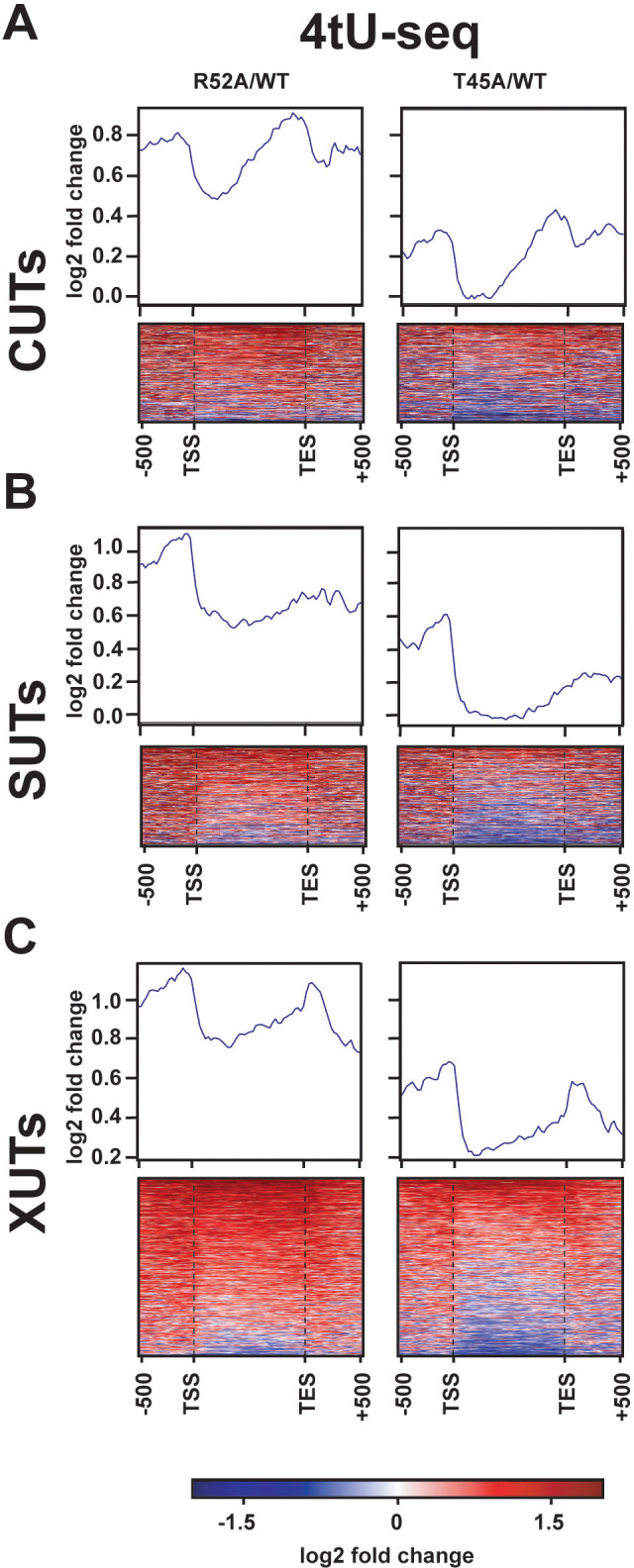
Figure 4—figure supplement 1. Some genes in DNA entry-exit site mutants show little to no change in 5’ and 3’ expression.
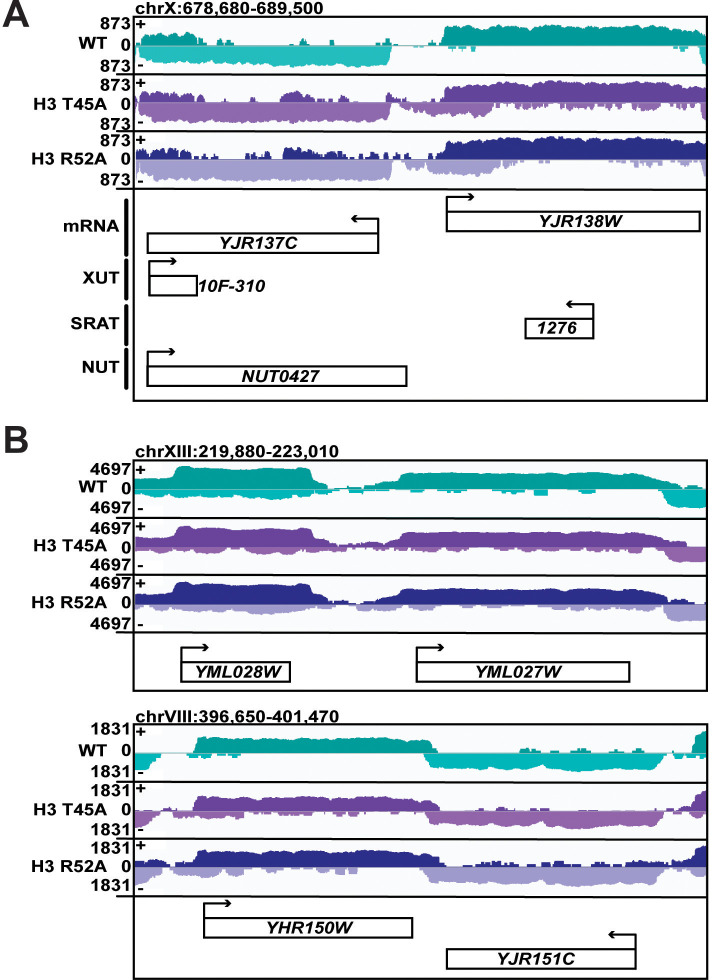
Figure 4—figure supplement 2. Transcriptional changes at ncRNA loci in DNA entry-exit site mutants.