Abstract
Objective
To identify early predictors of disease activity at 18 months in JIA using clinical and biomarker profiling.
Methods
Clinical and biomarker data were collected at JIA diagnosis in a prospective longitudinal inception cohort of 82 children with non-systemic JIA, and their ability to predict an active joint count of 0, a physician global assessment of disease activity of ≤1 cm, and inactive disease by Wallace 2004 criteria 18 months later was assessed. Correlation-based feature selection and ReliefF were used to shortlist predictors and random forest models were trained to predict outcomes.
Results
From the original 112 features, 13 effectively predicted 18-month outcomes. They included age, number of active/effused joints, wrist, ankle and/or knee involvement, ESR, ANA positivity and plasma levels of five inflammatory biomarkers (IL-10, IL-17, IL-12p70, soluble low-density lipoprotein receptor-related protein 1 and vitamin D), at enrolment. The clinical plus biomarker panel predicted active joint count = 0, physician global assessment ≤ 1, and inactive disease after 18 months with 0.79, 0.80 and 0.83 accuracy and 0.84, 0.83, 0.88 area under the curve, respectively. Using clinical features alone resulted in 0.75, 0.72 and 0.80 accuracy, and area under the curve values of 0.81, 0.78 and 0.83, respectively.
Conclusion
A panel of five plasma biomarkers combined with clinical features at the time of diagnosis more accurately predicted short-term disease activity in JIA than clinical characteristics alone. If validated in external cohorts, such a panel may guide more rationally conceived, biologically based, personalized treatment strategies in early JIA.
Keywords: arthritis, childhood arthritis, classification, cytokines, JIA, machine learning, predictors
Rheumatology key messages
Clinical plus biomarker JIA characteristics are more reliable outcome predictors than clinical features only.
Predicting JIA outcomes should be improved by considering clinical and biomarker attributes together.
Validation in another JIA inception cohort is required to confirm our results
Introduction
JIA encompasses a heterogeneous group of diseases that are classified predominantly according to clinical manifestations present within the first 6 months of disease [1]. Only two biomarkers, RF and HLA-B27, are included as criteria in the current JIA classification system. The original intent of the JIA classification system was to define homogeneous groups to facilitate research [1]. A robust classification system might also be expected to predict category-specific disease courses, treatment responses and outcomes and help generate insight into disease aetiology and pathophysiology. However, even within the same JIA category, patients exhibit different disease courses and outcomes [2]. Thus, current JIA category assignment, based on clinical features alone, does not reliably predict which children will experience favourable or unfavourable outcomes [3, 4].
Studies in the era of biologically based pharmacotherapies indicate improving JIA outcomes [5]. Nearly half of children with JIA are estimated to have inactive disease within a year after diagnosis when biologic medications are used sparingly [6]. More liberal use of biologics may be associated with up to 80% of JIA patients having inactive disease in the first year [7, 8]. Treatment intensity, including the early use of biologics, should be informed by knowing the chances disease outcome will be poor if treated less aggressively.
Previous studies have identified clinical predictors of poor prognosis [9–12]. Biomarker profiling, combined with clinical features, might further aid in predicting disease phenotype, severity and course and help refine patient selection for early aggressive treatment [13, 14]. This study’s objective was to identify, in a JIA inception cohort, a combination of clinical attributes and biomarkers that could help predict disease activity 18 months later as reflected by active joint count (AJC), physician global assessment (PGA) and Wallace criteria for inactive disease [15].
Methods
Data collection
Data were from a Canadian prospective, longitudinal inception cohort study [Biologically Based Outcome Predictors in JIA (The BBOP Study)] [16]. Ethics review boards from all sites approved the study (Supplementary Material, section Biomedical Ethics, available at Rheumatology online). Inclusion criteria were a diagnosis of JIA made according to ILAR criteria [1]. As this study evaluated participants at enrolment, assignments of oligoarthritis and polyarthritis categories were presumptive since all patients were enrolled at or <6 months after diagnosis. Participants who were not treated or who had been on non-steroidal anti-inflammatory medication or MTX for ≤6 weeks were eligible for inclusion.
The recruitment strategy for the BBOP Study aimed for a reasonable number of participants in each of the seven JIA categories rather than aspiring to achieve a typical JIA subgroup distribution. To achieve this aim, only participants with polyarthritis or systemic JIA (sJIA), the least prevalent categories, were eligible during the first 6 months of the enrolment period; after 6 months and until the end of the 2-year enrolment period, participants with any JIA category were eligible.
From the entire BBOP cohort, patients were selected for the current study based on availability of complete outcome data at the 18-month follow-up visit. sJIA, a presumed autoinflammatory disorder, is strikingly different from other forms of JIA and may have different pathogeny; consequently, the 11 BBOP participants with sJIA were not included in the current analysis.
BBOP data included 282 clinical characteristics and 48 plasma inflammatory biomarkers. Demographic, clinical and laboratory data were collected at enrolment and were used to predict outcomes 18 months later. Paediatric rheumatologists documented the number of joints with active arthritis and number of effused joints detected on physical examination. Active arthritis was defined as ‘swelling within a joint, or limitation in the range of joint movement with joint pain or tenderness, which persists for at least six weeks, is observed by a physician, and is not due to primarily mechanical disorders or to other identifiable causes’ [1]. Medication use at the time of enrolment and with each subsequent visit was recorded.
Plasma biomarkers
Blood was collected in P100 tubes (BD Biosciences, San Jose, CA, USA) in accord with previously described protocols [17]. Samples were kept at 4°C until shipped on the day of collection by overnight courier at ambient temperature to the central biobank laboratory. On the day of arrival at the destination laboratory, the tubes were centrifuged (1000 g × 15 min) and the plasma stored at −80°C until assayed.
The list of biomarkers assayed is shown in Supplementary Table S1, available at Rheumatology online. Cytokine, chemokine, growth factor and metalloproteinase plasma levels were assayed by bead-based immunoassays. Product codes for analytes (Milliplex, Milllipore Sigma, Burlington, MA, USA) were as follows: receptor activator of nuclear factor κB ligand (RANKL; HBN51K1RANKL), regulated on activation, normal T cell expressed and secreted (RANTES; HCYTOMAG-60K-01), osteoprotegerin (OPG; HBN1B-51K-01), tissue inhibitor of metalloproteinases (TIMP)-1/2 (HTIMP1-54K-02), TIMP-3/4 (HTIMP2-54K-01), MMP-3/12/13 (HMMP1-55K-03), MMP-1/2/7/9/10 (HMMP2-55K-05), MMP-8 (HSP2MAG-63K-01), 29-plex cytokine/chemokine panel (HCYTMAG-60K-PX29) and fibroblast growth factor 2 (FGF-2; HCYTOMAG-60K-01). All bead-based analytes were analysed on a Luminex100 LabMAP system (Luminex, Austin, TX, USA; Analytical Facility for Bioactive Molecules, Hospital for Sick Children, Toronto, ON, Canada) according to the manufacturer’s instructions. The soluble low-density lipoprotein 1 (sLRP1) assay is explained in Supplementary Material section ‘Soluble low-density lipoprotein (sLRP1) assay’, available at Rheumatology online.
High mobility group box 1 (HMGB1) and vitamin D assays were performed as previously described [18, 19]. ANA test results were from indirect immunofluorescence assays performed at clinical laboratory facilities at each study site and results dichotomized as positive or negative; ANA patterns and titres were not recorded.
Outcome indicators
The primary outcome was inactive disease at 18 months after enrolment. Three outcome measures were considered to reflect inactive disease: an AJC = 0, a PGA ≤ 1 cm on a 10 cm visual analogue scale, and fulfilment of 2004 Wallace criteria for inactive disease [15]. The three outcomes were evaluated respectively with clinical and biological predictors together and with only clinical predictors.
Feature selection
From 330 BBOP features (variables), 112 were chosen based on disease symptoms and signs and prior knowledge of indicators of JIA pathophysiology. Feature selection (FS), which can be applied to a large dataset to select the optimal features for class (outcome) prediction, is valuable for analysing high-dimensional data (i.e. datasets in which the number of variables exceeds the number of subjects) [20]. By eliminating redundant and irrelevant features, FS techniques improve prediction accuracy. FS aims to identify the most informative features. In this study, we used two filter-based FS approaches: correlation-based feature selection (CFS) and ReliefF [21]. CFS and ReliefF are computationally efficient multivariate methods that consider relationships among features. CFS is based on the rationale ‘a good feature subset is one that contains features highly correlated with the class, yet uncorrelated with each other’ [22]. It assesses both redundancy of features by applying correlation algorithms and the predictive ability of a subset of features [23]. ReliefF chooses the features that are distinct among different classes. The basic idea of ReliefF is to select subjects randomly, compute their nearest neighbours (nearest subjects), and identify features that discriminate the subject from neighbours of different classes (in this study classes are active JIA disease and inactive JIA disease). Specifically, ReliefF randomly draws a subject (A) and then identifies its two nearest neighbours: one from the same class (nearest hit, H) and the other from the different class (nearest miss, M). It then calculates differences between features from subjects A and H and between A and M. A desirable scenario is when the subjects A and M have different values of a particular feature so that the feature is able to discriminate two subjects with different class values, and subjects A and H have similar values of the individual feature.
To estimate a weight for each feature (f), the algorithm uses the following probability equation:
The operation is iterative and gives more weight to the features that discriminate the subject from the neighbours of a different class [24, 25]. High-ranked features identified by both CFS and ReliefF were selected for further analysis.
Predicting outcome in JIA based on clinical and biological features
To determine how well a constellation of selected features predicts JIA outcome, the random forest classification algorithm was applied [26]. A prediction model was trained using 90% of the data (training set) randomly, and then the model was tested on the remaining 10% of the data (test set). This procedure is iterative and is termed 10-fold cross-validation. The ultimate goal of the random forest classification algorithm was to maximize the predictive accuracy of the trained model on the new data (test set) [27].
The random forest algorithm generates many decision trees (each of which predicts the class value by learning simple decision rules inferred from the selected features) from randomly selected subsets of the subjects. At each subset, decision trees are constructed using the best split among randomly selected features [26]. This additional layer of randomness makes random forest more robust than other ensemble classification methods. There are two assumptions: first, most of the trees correctly predict the class for most of the subjects, and second, the trees make mistakes at different places. According to these assumptions the algorithm conducts voting for each of the classes and collectively ranks the importance of variables in predicting the correct class [26, 28].
Using Waikato Environment for Knowledge Analysis (Weka) machine learning software, randomness was injected into the training procedure by randomly selecting log2(number-of-features + 1) subjects from the dataset prior to training each decision tree.
The mean decrease in accuracy of a variable was determined during the cross-validation. A single variable was excluded from the test set, then the accuracy rate of the model was calculated. The variable was considered more important if its removal caused a relatively large decrease in the model performance indicators.
Data analysis
Statistical analyses were performed using SPSS Statistics Professional v23 (IBM Corp., Armonk, NY, USA), and R v3.2.2 (R Foundation for Statistical Computing, Vienna, Austria). Weka was used for FS and prediction [29]. The outlier-labelling rule was applied to identify and remove extreme values [30]. Protein concentrations and continuous variables were log and Z-score transformed.
For each outcome, receiver operating characteristic curves were plotted, where the y-axis represents true positive rate (sensitivity/recall) and the x-axis represents false-positive rate or 1 − specificity. Each outcome was evaluated based on the area under its receiver operating characteristic curve (AUC) where a value of 1 represents perfect discrimination and 0.5 represents performance at chance level. AUC is a threshold-independent measure of overall classification accuracy. Accuracy, precision, recall and F-measure (the harmonic average of the precision and recall) were calculated for each model as additional measures of model performance.
Results
Demographic and clinical characteristics at enrolment are shown in Table 1, categorized by participation in analyses for each outcome. A total of 82 patients were included: 77 were entered in analyses for AJC = 0, 82 in analyses for PGA ≤ 1 cm and 48 in analyses for inactive disease by Wallace criteria. In the cohort 73% were female. The median age at enrolment was 11 (interquartile range = 6–14) years. Most of the participants were enrolled into the study on the date of JIA diagnosis. Twenty participants were enrolled ≥30 days after diagnosis. The median time between diagnosis and enrolment was 0 (interquartile range = 0–26) days.
Table 1.
Patient characteristics at enrolment
| JIA category | Outcome | M/F | Age, median (IQR), years | AJC, mean (s.d.) | PGA, mean (s.d.) | ESR, mean (s.d.), mm/hr |
|---|---|---|---|---|---|---|
| Oligoarthritis | 1 (n = 17) | 5/12 | 10 (4–14) | 2 (2) | 3.1 (2.0) | 24 (20) |
| 2 (n = 21) | 8/13 | 8 (4–13) | 2 (2) | 3.0 (2.0) | 25 (23) | |
| 3 (n = 9) | 3/6 | 14 (5–15) | 3 (4) | 3.0 (2.0) | 32 (21) | |
| Polyarthritis (RF-negative) | 1 (n = 32) | 5/27 | 11 (4–14) | 9 (8) | 4.4 (2.0) | 28 (24) |
| 2 (n = 33) | 5/28 | 11 (5–14) | 10 (8) | 4.5 (2.0) | 31 (26) | |
| 3 (n = 20) | 3/17 | 10 (6–14) | 8 (8) | 4.0 (2.0) | 28 (26) | |
| Polyarthritis (RF-positive) | 1 (n = 10) | 2/8 | 13 (7–15) | 18 (11) | 4.8 (2.0) | 38 (30) |
| 2 (n = 10) | 1/9 | 12 (7–15) | 17 (11) | 4.3 (2.0) | 41 (30) | |
| 3 (n = 6) | 1/5 | 12 (4–14) | 10 (10) | 5.0 (3.0) | 18 (12) | |
| Psoriatic arthritis | 1 (n = 6) | 2/4 | 9 (5–14) | 11 (7) | 5.3 (2.0) | 27 (22) |
| 2 (n = 6) | 2/4 | 9 (5–14) | 8 (7) | 4.5 (2.0) | 27 (22) | |
| 3 (n = 4) | 1/3 | 9 (3–13) | 8 (9) | 6.0 (1.0) | 14 (7) | |
| Enthesitis-related arthritis | 1 (n = 7) | 4/3 | 13 (9–15) | 6 (5) | 3.2 (3.0) | 33 (28) |
| 2 (n = 7) | 4/3 | 13 (9–15) | 6 (5) | 3.2 (3.0) | 43 (37) | |
| 3 (n = 6) | 3/3 | 12 (9–14) | 7 (3) | 4.0 (2.0) | 36 (18) | |
| Undifferentiated arthritis | 1 (n = 5) | 1/4 | 14 (10–15) | 3 (2) | 3.7 (2.0) | 20 (16) |
| 2 (n = 5) | 1/4 | 14 (10–15) | 3 (2) | 3.7 (2.0) | 20 (16) | |
| 3 (n = 3) | 0/3 | 15 (14–15) | 12 (14) | 5.0 (1.0) | 20 (23) |
Outcome numbers: 1: active joint count of 0; 2: physician global assessment of ≤1 cm; 3: inactive disease by Wallace criteria [15]. AJC: active joint count; F: female; IQR: interquartile range; M: male; PGA: physician global assessment.
Feature selection
From an initial set of 112 features considered at enrolment (Supplementary Table S2, available at Rheumatology online), 81 uninformative and redundant features were removed using CFS and ReliefF. The remaining 31 features were then tested in prediction models. After dropping features that added little predictive ability in the model, 13 predictors remained. The final sets of clinical and biomarker features for predicting each outcome are shown in Table 2. In addition to clinical disease manifestations, five inflammation-related plasma biomarkers were identified as important predictors of inactive disease 18 months later (Table 2). These included IL-10, IL-12p70, IL-17, sLRP1 and vitamin D. The number of active and effused joints, age at enrolment, ANA and IL-12p70 were common predictors of all three outcomes. Wrist and ankle joint involvement, ESR and IL-17, in addition to the above variables, were common predictors of AJC and PGA outcomes. The selected features were used as input for the random forest algorithm. The same analysis was applied to clinical predictors alone (after exclusion of plasma biomarkers); the resulting clinical-only predictors are shown in Table 3.
Table 2.
Clinical and biological predictors
| Outcome 1 (AJC = 0) | Outcome 2 (PGA ≤ 1 cm) | Outcome 3 (inactive disease by Wallace criteria) |
|---|---|---|
| Number of active joints | Number of active joints | Number of active joints |
| Number of effused joints | Number of effused joints | Number of effused joints |
| Wrist | Knee | Age |
| Ankle | Ankle | ANA |
| Age | Age | IL-10 |
| ANA | ANA | IL-12p70 |
| ESR | ESR | Vitamin D |
| IL-10 | IL-12p70 | sLRP1 |
| IL-12p70 | IL-17 | |
| IL-17 |
Features at enrolment predictive of arthritis outcomes at 18 months after enrolment. See Table 1 for definitions of outcomes. AJC: active joint count; PGA: physician global assessment; sLRP1: soluble low-density lipoprotein receptor-related protein 1.
Table 3.
Clinical features predictive of outcomes
| Outcome 1 (AJC = 0) | Outcome 2 (PGA ≤ 1 cm) | Outcome 3 (inactive disease by Wallace criteria) |
|---|---|---|
| Number of active joints | Number of active joints | Number of active joints |
| Number of effused joints | Number of effused joints | Number of effused joints |
| TMJ | Wrist | TMJ |
| Wrist | Knee | Wrist |
| Knee | Ankle | Knee |
| Ankle | Age | Ankle |
| Age | ANA | Age |
| ANA | ESR | ANA |
| ESR | Platelet | ESR |
| Sex |
Clinical features at enrolment predictive of arthritis outcomes at 18 months when applying the models on clinical-only features only. See Table 1 for definitions of outcomes. AJC: active joint count; PGA: physician global assessment; TMJ: temporomandibular joint.
Prediction models
Eighteen months after enrolment, 62%, 71% and 83% of patients had inactive disease defined by the three outcomes, AJC = 0, PGA ≤ 1 and Wallace 2004 criteria, respectively. For each outcome, the random forest model was trained. The performances of the predictive model are reported in Table 4 and the directions of outcomes are shown in Table 5. The corresponding receiver operating characteristic curves are shown in Fig. 1. Combined clinical/biomarker models predicted AJC = 0, PGA ≤ 1 cm, and inactive disease by Wallace 2004 criteria with 0.79, 0.80 and 0.83 accuracy, respectively. When outcome was defined as inactive disease by Wallace 2004 criteria, the model achieves relatively higher performance measures (as reflected by AUC, sensitivity, specificity, precision and F-measure) than the other outcomes (Table 4). The model for predicting an AJC of 0 had the lowest predictive performance. The performance of prediction models that included only clinical variables was lower in most instances (Table 4).
Table 4.
Performance measures of predictors of each outcome direction of the prediction
| Performance measure | Predictor | Outcome |
||
|---|---|---|---|---|
| AJC = 0 (n = 77) | PGA < 1 (n = 82) | Inactive disease by Wallace criteria (n = 48) | ||
| AUC | Clinical–biological | 0.84 | 0.83 | 0.88 |
| Clinical | 0.81 | 0.78 | 0.83 | |
| F-measure | Clinical–biological | 0.75 | 0.80 | 0.82 |
| Clinical | 0.73 | 0.72 | 0.79 | |
| Accuracy (CI) | Clinical–biological | 0.79 (0.712, 0.868) | 0.80 (0.724, 0.876) | 0.83 (0.741, 0.919) |
| Clinical | 0.75 (0.667, 0.833) | 0.72 (0.635, 0.805) | 0.80 (0.705, 0.895) | |
| Precision (CI) | Clinical–biological | 0.75 (0637, 0.863) | 0.80 (0.697, 0.903) | 0.82 (0.678, 0.962) |
| Clinical | 0.73 (0.614, 0.846) | 0.72 (0.601, 0.839) | 0.79 (0.639, 0.941) | |
| Specificity (CI) | Clinical–biological | 0.75 (0.622, 0.860) | 0.80 (0.683, 0.917) | 0.83 (0.716, 0.944) |
| Clinical | 0.72 (0.594, 0.846) | 0.71 (0.594, 0.826) | 0.82 (0.702, 0.938) | |
| Sensitivity/recall (CI) | Clinical–biological | 0.75 (0.640, 0.860) | 0.80 (0.700, 0.900) | 0.82 (0.672, 0.968) |
| Clinical | 0.73 (0.613, 0.847) | 0.72 (0.592, 0.848) | 0.79 (0.636, 0.944) | |
Performance measures of models using combined clinical and biological predictors and clinical predictors only for the outcomes AJC = 0, PGA < 1, and inactive disease. AJC: active joint count; AUC: area under the curve; PGA: physician global assessment.
Table 5.
Direction of outcome for each predictor performance measure
| Predictor at enrolment | 18-month outcome |
|---|---|
| Higher number of active joints | Worse |
| Higher number of effused joints | Worse |
| TMJ involvement | Worse |
| Wrist involvement | Worse |
| Knee involvement | Better |
| Ankle involvement | Better |
| Older age | Worse |
| Male | Better |
| Female | Worse |
| ANA positivity | Better |
| High ESR | Worse |
| Higher levels of IL-10 | Better |
| Higher levels of IL-12p70 | Worse |
| Higher levels of IL-17 | Worse |
| Higher levels of vitamin D | Better |
| Higher levels of sLRP1 | Better |
sLRP1: soluble low-density lipoprotein receptor-related protein 1; TMJ: temporomandibular joint.
Fig. 1.
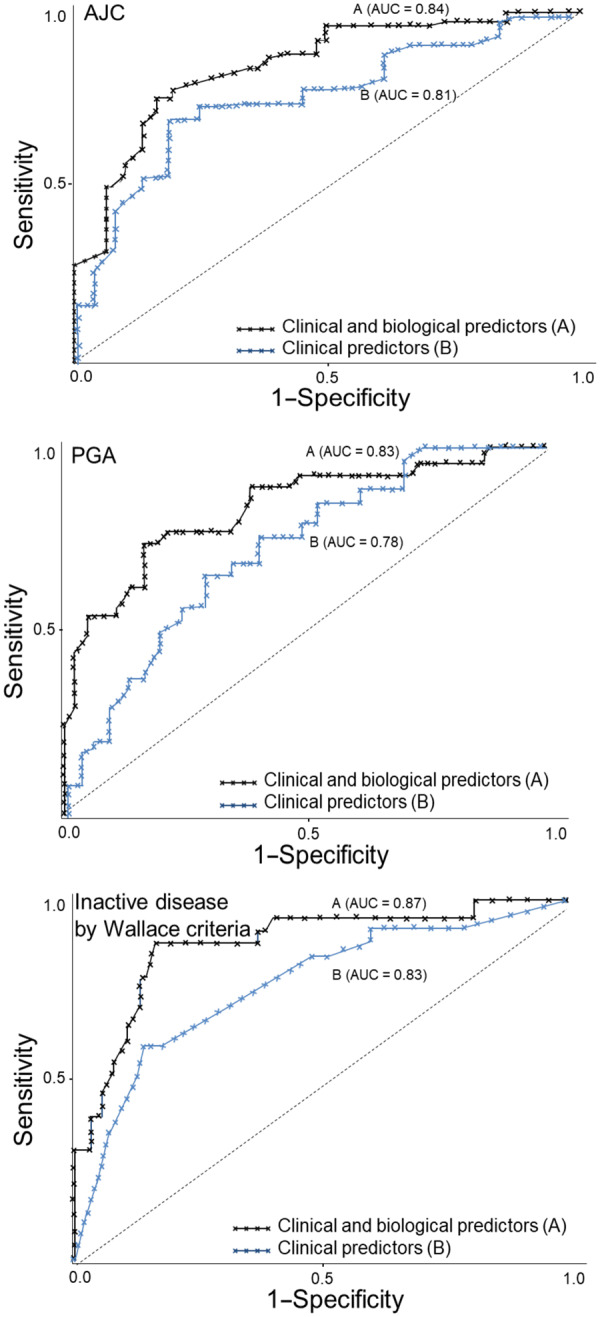
Receiver operating characteristic curve of predictors for three outcomes
Clinical and biological predictors (A) and clinical predictors (B) for AJC = 0 (upper), PGA < 1(middle), and inactive disease by Wallace criteria (lower). The diagonal line denotes the expected performance of a tool that uses random guessing. AJC: active joint count; PGA: physician global assessment.
Effect of treatment
Treatments used over the study period are shown in Table 6. Thirty-five participants had received medication within 6 weeks prior to enrolment including 27 who had received a non-steroidal anti-inflammatory medication, eight who received MTX and one who received oral prednisone. Considering PGA ≤ 1 cm as the 18-month outcome measure, there was no difference in AUC, accuracy, precision or sensitivity comparing the group that was treatment naïve and the group that had received prior medications (Table 7). There were insufficient data for meaningful comparison of treatment naïve and treated groups for AJC and Wallace criteria outcomes.
Table 6.
Treatments received during the study
| Visit | NSAIDs, n (%) | DMARDs, n (%) | Intra-articular corticosteroid injections, n (%) | Biologic medications, n (%) |
|---|---|---|---|---|
| Enrolment | 71 (86.6) | 31 (37.8) | 9 (11.0) | 0 (0.0) |
| 6-Month visit | 63 (76.8) | 48 (58.5) | 8 (9.8) | 6 (8.5) |
| 12-Month visit | 49 (51.3) | 43 (52.4) | 2 (2.4) | 11 (17.7) |
| 18-Month visit | 36 (47.4) | 52 (63.4) | 5 (6.1) | 12 (15.4) |
Numbers are the number of patients (%) who were on the medication at that visit.
Table 7.
Comparison of performance measures between treatment-naïve participants and those who received a medication prior to enrolment
| Performance measures | Predictor | Outcome: PGA < 1 cm |
|
|---|---|---|---|
| Treatment naïve (n = 46) | Treatment prior to enrolment (NSAIDs, MTX, prednisone) (n = 36) | ||
| AUC | Clinical–biological | 0.80 | 0.80 |
| F-measure | Clinical–biological | 0.74 | 0.75 |
| Accuracy | Clinical–biological | 0.81 | 0.81 |
| Precision | Clinical–biological | 0.75 | 0.75 |
| Specificity | Clinical–biological | 0.76 | 0.73 |
| Sensitivity/recall | Clinical–biological | 0.75 | 0.75 |
AUC: area under the curve; PGA: physician global assessment.
Discussion
Using a composite panel of clinical and biomarker variables in non-sJIA patients shortly after diagnosis, we found improved prediction of inactive disease 18 months later compared with conventional clinical variables alone.
The panel was developed from a set of clinical and biomarker variables by applying FS and random forest techniques. Random forest is a robust machine learning classification algorithm that can investigate prediction power of variables in a compound (quantitative and categorical) high-dimensional dataset and can handle missing data. It is among the most accurate methods of classification and provides both a measure of the relative importance of features and a prediction algorithm.
Previous studies have focused on the utility of clinical or laboratory characteristics separately [31]. From reported clinical and laboratory JIA outcome predictors [4, 11, 32–34] the current study confirmed AJC, effused joint count, wrist involvement, age at diagnosis and ESR as important predictors and added ankle and knee joint involvement, and ANA as clinical predictors in the composite panel. Consistent with the report of Guzman et al. [35] we found that JIA categories were not predictors of short-term JIA outcome.
The plasma biomarker panel we identified is pertinent to JIA. IL-17-expressing T cells are abundant in JIA joints and correlate with the number of involved joints [36]. Increased IL-17 levels in synovial fluid of patients with enthesitis-related arthritis correlate with disease activity [37]. IL-12p70 promotes the induction and activation of both Th1-cells and Th17-cells, key mediators in the pathophysiology of JIA [38].
sLRP1 is an integral regulator of inflammation and immune responses [39]. As a receptor for a multitude of potentially pathogenic ligands, LRP1 is involved in antigen presentation to T cells [40]. sLRP1 is biologically active, mediating cell signalling and promoting expression of regulatory cytokines. Elevated levels of sLRP1 have been found in adults with rheumatoid arthritis suggesting that sLRP1 could be a circulating biomarker reflective of inflammation status and could function as an endogenous inhibitor of inflammation by scavenging pathogenic peptides and by down-regulating certain pro-inflammatory cytokines (TNF-α, IL-6, MCP-1 as examples) and nuclear factor-κB pathway activation [41, 42]. Our results, which report sLRP1 levels in JIA for the first time, show that elevated sLRP1 levels at diagnosis predict a more favourable outcome.
Consistent with observations in this study, inadequate vitamin D levels in patients with JIA have been associated with increased disease activity [43].
Thus, we identified a set of biomarkers as predictors of disease outcomes that may be implicated in JIA pathogenesis or have been associated with outcome.
Van Dijkhuizen et al. [14] could not reliably predict inactive disease in a JIA cohort using clinical, cytokine and microbiome inputs. However, when certain JIA categories were considered separately (oligoarticular, polyarthritis RF negative and those with ANA positivity), prediction of inactive disease was moderately robust. The discrepancy between the results reported by Van Dijkhuizen et al. and our results might be due to different FS methods. In the current study, we did not investigate predictors for individual JIA categories as the number in each group was insufficient for meaningful analysis.
Rypdal et al. [44] evaluated long-term predictive ability of clinical characteristics in a Nordic JIA cohort study. They assessed four outcomes 8 years after diagnosis: non-achievement of remission off medications, functional disability by two measures, and joint damage. They reported AUCs across the four outcomes between 0.73 and 0.78. In our study, AUCs were higher using only clinical predictors in the model but a combination of clinical and biomarker variables resulted in even better model performance. However, future larger-scale studies are required to determine whether the improved performance is clinically meaningful enough to influence changes in treatment approaches and to justify any added cost.
In contrast to observations by Al-Matar et al. [9], we observed ankle involvement to be associated with a more favourable outcome. However, Al-Matar et al. reported observations only in oligoarticular JIA. The under-sampling of oligoarthritis and the short-term follow-up in our study might explain the discrepant observations.
In our analyses, JIA ILAR category was not retained as a predictor in any of the models. This observation could suggest that JIA categories might not align precisely with category-specific pathobiological processes that mediate outcomes. However, analyses of larger cohorts will be required to confidently ascertain whether JIA category is or is not an outcome predictor.
The results of this study, if confirmed in an independent JIA validation cohort, will inform the development of a clinically useful tool for early prediction of JIA outcomes and thereby aid in treatment selection. We found that readily accessible clinical measures alone had reasonable performance statistics. Adding biomarkers improved accuracy and should add a more personalized approach to assessing individual patients. However, reliable biomarker assays are not easily accessible in current clinical settings. Until such time as evidenced-based biomarker panels become integrated into usual clinical care, a two-step approach to prognostication and treatment selection could be applied. Under such a model, clinical features could be considered first in all patients and then, if indicated, targeted biomarker assessments undertaken with reference to the respective clinical contexts.
One limitation of this study is insufficient numbers of patients to stratify into treatment groups or JIA categories. Thus, effects of different medication groups on outcomes were not assessed and differences among JIA categories were not examined in depth. Another limitation is that outcome measured at a single time ignores the occurrence of flares.
Fluctuations in biomarkers can be influenced by diurnal variations, physical activity, sleep, stress and food intake [45], variables that were not controlled for in this study. Although blood sample processing protocols were standardized within this study based on our earlier quality control experiments [17] minimal variations in time to plasma separation and transport time and temperatures could potentially influence cytokine levels. Accumulating new information about the influences on biomarker expression and stability should help improve the development of reliable, clinically accessible biomarker assessment protocols [46]. Recent studies suggest joint ultrasound features are predictive of inactive disease [47]; however, we did not study imaging findings as potential predictors. Our study did not include genetic markers (such as HLA and single nucleotide polymorphisms) or gene expression and metabolomic profiling. Considering these additional biological markers could further enhance and refine panels of outcome predictors. Further, this study did not include an external validation cohort; the generalizability of the results requires validation in an independent JIA cohort.
In this study, we used three clinical indicators of disease activity; a broader array of outcome measures and longer duration of follow-up should further strengthen the reliability of clinical-biomarker predictive panels.
Conclusion
Data mining and supervised machine learning algorithms are enabling us to overcome limitations of conventional statistical models especially when large datasets are available in small study populations. We used methods that can evaluate the predictive ability of a relatively small panel of clinical measures and inflammation-related biomarkers simultaneously. We have shown that combined clinical and biological measures of JIA disease activity at or shortly after diagnosis may predict clinically important 18-month outcomes. Further validation studies are required for confirmation of our results.
Supplementary Material
Acknowledgements
We thank Dr Marianna Newkirk, McGill University, Montreal, Canada, for conducting sLRP1 assays. The participation of patients and their families is gratefully acknowledged.
Funding: This research was supported by funding from the Canadian Institutes of Health Research (Institute of Musculoskeletal Health and Arthritis and Institute of Infection and Immunity), the Arthritis Society, the Canadian Arthritis Network, the University of Saskatchewan, the Manitoba Institute of Child Health, McGill University (Division of Pediatric Rheumatology), Memorial University, The University of British Columbia (Division of Pediatric Rheumatology), and the Clinical Research Centre of the Centre Hospitalier Universitaire de Sherbrooke.
Disclosure statement: The authors have declared no conflicts of interest.
Supplementary data
Supplementary data are available at Rheumatology online.
References
- 1. Petty RE, Southwood TR, Baum J. et al. Revision of the proposed classification criteria for juvenile idiopathic arthritis: Durban, 1997. J Rheumatol 1998;10:1991–4. [PubMed] [Google Scholar]
- 2. Guzman J, Henrey A, Loughin T. et al. Predicting which children with juvenile idiopathic arthritis will have a severe disease course. Results from the ReACCh-out Cohort. J Rheumatol 2017;44:230–40. [DOI] [PubMed] [Google Scholar]
- 3. Minden K, Kiessling U, Listing J. et al. Prognosis of patients with juvenile chronic arthritis and juvenile spondyloarthropathy. J Rheumatol 2000;27:2256–63. [PubMed] [Google Scholar]
- 4. Oen K, Malleson PN, Cabral DA. et al. Disease course and outcome of juvenile rheumatoid arthritis in a multicenter cohort. J Rheumatol 2002;29:1989–99. [PubMed] [Google Scholar]
- 5. Berard RA, Laxer RM.. Early aggressive therapy for patients with juvenile idiopathic arthritis: are we there yet? J Rheumatol 2014;41:2343–6. [DOI] [PubMed] [Google Scholar]
- 6. Guzman J, Oen K, Tucker LB. et al. The outcomes of juvenile idiopathic arthritis in children managed with contemporary treatments: results from the ReACCh-Out Cohort. Ann Rheum Dis 2015;74:1854–60. [DOI] [PubMed] [Google Scholar]
- 7. Sengler C, Klotsche J, Niewerth M. et al. The majority of newly diagnosed patients with juvenile idiopathic arthritis reach an inactive disease state within the first year of specialised care: data from a German inception cohort. RMD Open 2015;1:e000074.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ringold S, Seidel KD, Koepsell TD, Wallace CA.. Inactive disease in polyarticular juvenile idiopathic arthritis: current patterns and associations. Rheumatology 2009;48:972–7. [DOI] [PubMed] [Google Scholar]
- 9. Al-Matar MJ, Petty RE, Tucker LB. et al. The early pattern of joint involvement predicts disease progression in children with oligoarticular (pauciarticular) juvenile rheumatoid arthritis. Arthritis Rheumatol 2002;46:2708–15. [DOI] [PubMed] [Google Scholar]
- 10. Oen K. Long-term outcomes and predictors of outcomes for patients with juvenile idiopathic arthritis. Best Pract Res Clin Rheumatol 2002;16:347–60. [PubMed] [Google Scholar]
- 11. Oen K, Malleson PN, Cabral DA. et al. Early predictors of longterm outcome in patients with juvenile rheumatoid arthritis: subset-specific correlations. J Rheumatol 2003;30:585–93. [PubMed] [Google Scholar]
- 12. Spiegel LR, Schneider R, Lang BA. et al. Early predictors of poor functional outcome in systemic-onset juvenile rheumatoid arthritis: a multicenter cohort study. Arthritis Rheumatol 2000;43:2402.. [DOI] [PubMed] [Google Scholar]
- 13. Duurland CL, Wedderburn LR.. Current developments in the use of biomarkers for juvenile idiopathic arthritis. Curr Rheumatol Rep 2014;16:406.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Van Dijkhuizen EHP, Aidonopoulos O, ter Haar NM. et al. Prediction of inactive disease in juvenile idiopathic arthritis: a multicentre observational cohort study. Rheumatology 2018;57:1752–60. [DOI] [PubMed] [Google Scholar]
- 15. Wallace CA, Ruperto N, Giannini E; Childhood Arthritis and Rheumatology Research Alliance; Pediatric Rheumatology International Trials Organization; Pediatric Rheumatology Collaborative Study Group. Preliminary criteria for clinical remission for select categories of juvenile idiopathic arthritis. J Rheumatol 2004;31:2290–4. [PubMed] [Google Scholar]
- 16. Eng SW, Duong TT, Rosenberg AM, Morris Q, Yeung RS.. The biologic basis of clinical heterogeneity in juvenile idiopathic arthritis. Arthritis Rheumatol 2014;66:3463–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Matheson LA, Duong TT, Rosenberg AM, Yeung RS.. Assessment of sample collection and storage methods for multicenter immunologic research in children. J Immunol Methods 2008;339:82–9. [DOI] [PubMed] [Google Scholar]
- 18. Burlingame RW, Rubin RL, Rosenberg AM.. Autoantibodies to chromatin components in juvenile rheumatoid arthritis. Arthritis Rheumatol 1993;36:836–41. [DOI] [PubMed] [Google Scholar]
- 19. McNally JD, Matheson LA, Sankaran K, Rosenberg AM.. Capillary blood sampling as an alternative to venipuncture in the assessment of serum 25 hydroxyvitamin D levels. J Steroid Biochem Mol Biol 2008;112:164–8. [DOI] [PubMed] [Google Scholar]
- 20. Hira ZM, Gillies DF.. A review of feature selection and feature extraction methods applied on microarray data. Adv Bioinformatics 2015;2015:198363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kira K, Rendell LA.. The feature selection problem: traditional methods and a new algorithm. In: Tenth National Conference on Artificial Intelligence. MIT Press, Cambridge, 1992:129–34. [Google Scholar]
- 22. Hall MA. Correlation-based feature selection of discrete and numeric class machine learning In: Proceedings of the Seventeenth International Conference on Machine Learning (ICML-2000), Stanford University, Stanford, CA, USA. San Francisco: Morgan Kaufmann Publishers Inc, 2000: 359–66. [Google Scholar]
- 23. Wang Y, Tetko IV, Hall MA. et al. Gene selection from microarray data for cancer classification—a machine learning approach. Comput Biol Chem 2005;29:37–46. [DOI] [PubMed] [Google Scholar]
- 24. Wang Y, Makedon F.. Application of ReliefF feature filtering algorithm to selecting informative genes for cancer classification using microarray data. In: Proceedings of the 2004 IEEE Computational Systems Bioinformatics Conference (CBS 2004), Stanford, CA, USA. Piscataway: IEEE, 2004: 497–8. [Google Scholar]
- 25. Robnik-Šikonja M, Kononenko I.. Theoretical and empirical analysis of ReliefF and RReliefF. Mach Learn 2003;53:23–69. [Google Scholar]
- 26. Breiman L. Random forests. Mach Learn 2001;45:5–32. [Google Scholar]
- 27. Dietterich T. Overfitting and undercomputing in machine learning. ACM Comput Surv 1995;27:326–7. [Google Scholar]
- 28. Díaz-Uriarte R, De Andres SA.. Gene selection and classification of microarray data using random forest. BMC Bioinformatics 2006;7:3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Witten IH, Frank E, Hall MA, Pal CJ.. Data mining: practical machine learning tools and techniques. Burlington, MA: Morgan Kaufmann, 2016: 403–584. [Google Scholar]
- 30. Hoaglin DC, Iglewicz B, Tukey JW.. Performance of some resistant rules for outlier labeling. J Am Stat Assoc 1986;81:991–9. [Google Scholar]
- 31. Van Dijkhuizen EP, Wulffraat NM.. Early predictors of prognosis in juvenile idiopathic arthritis: a systematic literature review. Ann Rheum Dis 2015;74:1996–2005. [DOI] [PubMed] [Google Scholar]
- 32. Oen K, Tucker L, Huber AM. et al. Predictors of early inactive disease in a juvenile idiopathic arthritis cohort: results of a Canadian multicenter, prospective inception cohort study. Arthritis Care Res 2009;61:1077–86. [DOI] [PubMed] [Google Scholar]
- 33. Flatø B, Lien G, Smerdel A. et al. Prognostic factors in juvenile rheumatoid arthritis: a case-control study revealing early predictors and outcome after 14.9 years. J Rheumatol 2003;30:386–93. [PubMed] [Google Scholar]
- 34. Flatø B, Hoffmann-Vold AM, Reiff A. et al. Long-term outcome and prognostic factors in enthesitis-related arthritis: a case–control study. Arthritis Rheumatol 2006;54:3573–82. [DOI] [PubMed] [Google Scholar]
- 35. Guzman J, Henrey A, Loughin T. et al. Predicting which children with juvenile idiopathic arthritis will not attain early remission with conventional treatment: results from the ReACCh-Out cohort. J Rheumatol 2019;46:628. [DOI] [PubMed] [Google Scholar]
- 36. Nistala K, Moncrieffe H, Newton KR. et al. Interleukin‐17 producing T cells are enriched in the joints of children with arthritis, but have a reciprocal relationship to regulatory T cell numbers. Arthritis Rheumatol 2008;58:875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Agarwal S, Misra R, Aggarwal A.. Interleukin-17 levels are increased in juvenile idiopathic arthritis synovial fluid and induce synovial fibroblasts to produce proinflammatory cytokines and matrix metalloproteinases. J Rheumatol 2008;35:515–9. [PubMed] [Google Scholar]
- 38. Müller-Berghaus J, Kern K, Paschen A. et al. Deficient IL-12p70 secretion by dendritic cells based on IL-12b promoter genotype. Genes Immun 2004;5:431–4. [DOI] [PubMed] [Google Scholar]
- 39. Gonias SL, Campana WM.. LDL receptor-related protein-1: a regulator of inflammation in atherosclerosis, cancer, and injury to the nervous system. Am J Pathol 2014;184:18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Binder RJ, Srivastava PK.. Essential role of CD91 in re-presentation of gp96-chaperoned peptides. Proc Natl Acad Sci USA 2004;101:6128–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gorovoy M, Gaultier A, Campana WM, Firestein GS, Gonias SL.. Inflammatory mediators promote production of shed LRP1/CD91, which regulates cell signaling and cytokine expression by macrophages. J Leukoc Biol 2010;88:769–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gaultier A, Arandjelovic S, Niessen S. et al. Regulation of tumor necrosis factor receptor-1 and the IKK-NF-kappaB pathway by LDL receptor-related protein explains the antiinflammatory activity of this receptor. Blood 2008;111:5316–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Finch SL, Rosenberg AM, Vatanparast H.. Vitamin D and juvenile idiopathic arthritis. Pediatr Rheumatol Online J 2018;16:34.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rypdal V, Arnstad ED, Aalto K. et al. Predicting unfavorable long-term outcome in juvenile idiopathic arthritis: results from the Nordic cohort study. Arthritis Res Ther 2018;20:91.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kong S, Stabler T, Criscione L. et al. Diurnal variation of serum and urine biomarkers in patients with radiographic knee osteoarthritis. Arthritis Rheumatol 2006;54:2496–504. [DOI] [PubMed] [Google Scholar]
- 46. Yeung RS, Albani S, Feldman BM. et al. Enhancing translational research in paediatric rheumatology through standardization. Nat Rev Rheumatol 2016;12:684–90. [DOI] [PubMed] [Google Scholar]
- 47. Gremese E, Fedele AL, Alivernini S, Ferraccioli G.. Ultrasound assessment as predictor of disease relapse in children and adults with arthritis in clinical stable remission: new findings but still unmet needs. Ann Rheum Dis 2018;77:1391–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


