Abstract
Rationale: Ischemia/reperfusion injury (IRI) is a major cause of acute kidney injury (AKI) that is associated with high morbidity and mortality, and for which specific treatments are lacking. In this study, we investigated the protective effect of human urine-derived stem cells (USCs) and their exosomes against IRI-induced AKI to explore the potential of these cells as a new therapeutic strategy.
Methods: USCs were derived from fresh human urine. Cell surface marker expression was analyzed by flow cytometry to determine the characteristics of the stem cells. Adult male Sprague-Dawley rats were used to generate a lethal renal IRI model. One dose of USCs (2×106 cells/ml) or exosomes (20 µg/1 ml) in the experimental groups or saline (1 ml) in the control group was administered intravenously immediately after blood reperfusion. Blood was drawn every other day for measurement of serum creatinine (sCr) and blood urea nitrogen (BUN) levels. The kidneys were harvested for RNA and protein extraction to examine the levels of apoptosis and tubule injury. In vitro, the hypoxia-reoxygenation (H/R) model in human kidney cortex/proximal tubule cells (HK2) was used to analyze the protective ability of USC-derived exosomes (USC-Exo). Quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR), western blotting, superoxide dismutase activity, and malonaldehyde content analyses were used to evaluate oxidative stress in HK2 cells treated with USC-Exo after H/R. Exosomal microRNA sequencing techniques and bioinformatics analysis were used to search for enriched miRNAs in the exosomes and interacting genes. The interaction between miRNAs and the 3' untranslated region of the target gene was detected using a dual luciferase reporting system. The miRNA mimic and inhibitor were used to regulate the miRNA level in HK2 cells.
Results: Treatment with USCs led to reductions in the levels of sCr, BUN, and renal tubular cell apoptosis; inhibited the infiltration of inflammatory cells; and protected renal function in the rat IRI model. Additionally, USC-derived exosomes protected against IRI-induced renal damage. miR-146a-5p was the most abundant miRNA in exosomes obtained from the conditioned medium (CM) of USCs. miR-146a-5p targeted and degraded the 3'UTR of interleukin-1 receptor-associated kinase 1 (IRAK1) mRNA, subsequently inhibited the activation of nuclear factor (NF)-κB signaling, and protected HK2 cells from H/R injury. USC transplantation also upregulated miR-146a-5p expression, downregulated IRAK1 expression and inhibited nuclear translocation of NF-κB p65 in the kidney of the rat IRI model.
Conclusions: According to our experimental results, USCs could protect against renal IRI via exosomal miR-146a-5p, which could target the 3'UTR of IRAK1 and subsequently inhibit the activation of NF-κB signaling and infiltration of inflammatory cells to protect renal function. As a novel cell source, USCs represent a promising non-invasive approach for the treatment of IRI.
Keywords: urine-derived stem cells, ischemia/reperfusion injury, exosomes, miR-146a-5p, IRAK1
Introduction
Acute kidney injury (AKI) often leads to rapid renal dysfunction and is associated with high mortality and morbility 1. Renal ischemia/reperfusion injury (IRI) is a primary cause of AKI 2. Deprivation and subsequent restoration of blood flow and oxygenation induce IRI, which mainly occurs after infarction, sepsis, and organ transplantation 3. Many physiopathological events are involved in IRI, including reactive oxygen species (ROS) generation, neutrophil infiltration, and inflammatory cytokine production 4, 5. There is a lack of effective treatments for IRI-induced AKI in clinic, and thus, an urgent unmet need for innovative and effective therapies 6, 7.
Stem cell therapy represents a promising new frontier in the treatment of renal diseases 8. Many types of stem cells, including mesenchymal stem cells (MSCs), spermatogonial stem cells, induced pluripotent stem cells (iPSCs) and the cells derived from iPSCs have been found to either attenuate kidney dysfunction or stabilize morphological changes in renal IRI 9, 10. Treatments with MSCs specifically have entered clinical trials 11. Recently, human urine-derived stem cells (hUSCs) were introduced as a new promising candidate in many applications, such as vascular and neural tissue engineering, cartilage regeneration, and bladder/ureter remodeling 12-16. USCs, which are isolated from voided urine, exhibit many characteristics of MSCs and can differentiate along multiple cell lineages to generate endothelial cells, neurocytes, chondrocytes, and myocytes 17. Also, some cell characteristics of USCs will change under the disease state. The patient-derived USCs could be used as a tool to predict the outcome of the kidney disease 18. Additionally, USCs possess the advantages of being easily accessible, consistently producible, and relatively free of ethical concerns in comparison to stem cells from other donor sources 19. Therefore, USCs are emerging as an attractive cell source for a variety of therapies 20.
The therapeutic efficacy of USCs against IRI has been reported; however, the underlying mechanism remains incompletely understood 21. The mechanisms responsible for the beneficial effects of stem cell therapies include both direct pathways (e.g., stem cells may be directly incorporated into injury tissue) and indirect pathways (e.g., stem cells may secrete paracrine factors particularly through production of extracellular vesicles [EVs]) 22. Among the various types of EVs, exosomes are 30- to 150 nm cell membrane-containing vesicles that are secreted by almost all cell types. One study reported that MSC-derived exosomes can mediate repair of kidney injury 23, 24. USCs also secrete exosomes, which were shown to protect against diabetic nephropathy 25, promote of angiogenesis and wound healing in diabetic wound repair 26, and promote the angiogenesis and muscle regeneration in hind-limb ischemia 27. It is well known microRNAs (miRNAs) or small noncoding RNAs in exosomes play important roles in the regulation of target genes 28. We herein hypothesized miRNAs in USC-derived exosomes (USC-Exo) might be vital for the USC-mediated protection against IRI-induced renal injury.
In the present study, we investigated the potential renal protective effect of USCs and their exosomes using in vivo and in vitro models of IRI. We also explored the underlying mechanism of this protective effect by analyzing the miRNAs present in USC-Exo to gain new insights for the application of USCs in relevant therapies.
Materials and Methods
Preparation of USCs
USCs were isolated from two healthy adults. This study was approved by the institutional review board of medical ethics, and written informed consent was obtained from urine donors. A total of 100 ml fresh urine from each healthy adult was collected in one experiment. The urine sample was centrifuged at 400×g for 10 min, and the obtained pellet was washed twice with phosphate-buffered saline (PBS). The cells were resuspended in fresh USC medium and seeded in 48-well plates. The cells were allowed to grow for 4 days, and the culture medium was replaced gently. By day 7, the cells had formed tiny colonies. After culture for another week, USCs were passaged and expanded. USCs of passages 3-5 (P3-P5) were collected for use in the experiments in this study. USC medium consisted of medium A and medium B at a 1:1 ratio. Medium A consisted of high-glucose Dulbecco's Modified Eagle's Medium (DMEM; HyClone) + non-essential amino acid (NEAA) solution (Gibco) + GlutaMAX (Gibco) + 10% fetal bovine serum (FBS; Gibco). Medium B consisted of SingleQuot Kit CC-3191 & CC-4172 renal epithelial cell growth medium (REGM) supplements (Lonza). The expression levels of cell surface markers and specific genes were analyzed by flow cytometric analysis to confirm the characteristics of USCs.
Animal model of IRI
A lethal renal IRI model was established in male adult Sprague-Dawley rats (200-250 g) purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. The rats were anesthetized with intraperitoneal pentobarbital (4 mg/kg). The right kidney was removed, and the left renal pedicle was then clamped for 45 min to induce ischemia. Normal body temperature was maintained using a rectal temperature probe and heat supply. Upon reperfusion of the left kidney, USCs (2×106 cells in 1 ml saline), exosomes (20 μg/ml saline), or saline (1 ml) were injected into the rat via the dorsal vein of the penis. Animals were housed in pathogen-free conditions under a controlled 12-h light-dark cycle and allowed free access to water and chow. Survival rates, serum creatinine (sCr) and blood urea nitrogen (BUN) levels, and renal histopathology were assessed at the designated time points. Renal damage was scored according to the following criteria: 1 = damage area < 25%; 2 = damage area 25-50%; 3 = damage area 50-75%; 4 = damage area >75% 29.
All experiments were conducted in strict accordance with the institutional policies of the First Affiliated Hospital of Sun Yat-sen University. All efforts were made to minimize animal suffering. Animals were housed in pathogen-free conditions with a controlled 12 h light-dark cycle environment and allowed free access to water and chow. This study was approved by the Animal Ethics Committee of the First Affiliated Hospital of Sun Yat-sen University.
In vitro model of hypoxia/reoxygenation (H/R)
HK2 cells (human kidney cortex/proximal tubule cells) were purchased from American Type Culture Collection (ATCC® CRL-2190™). HK2 cells were cultured in DMEM/Nutrient Mixture F12 (DMEM/F12, Gibco) supplemented with 10% FBS, 500 U/ml penicillin, and 500 μg/ml streptomycin. For all culture conditions, the cells were incubated in a humidified incubator containing 5% CO2 at 37 °C. The H/R process was optimized according to the previously described methods 30. H/R injury was induced in normal HK2 human kidney cortex/proximal tubule cells by incubation in glucose- and serum-free medium in a 1% O2 environment for 48 h followed by reoxygenation with normal O2 during culture with USC conditioned medium or medium containing exosomes for 6 h.
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay
Renal sections were stained with the In situ Cell Death Detection Kit (11684817910, Roche) according to the manufacturer's protocol. Briefly, each slide was deparaffinized, rehydrated, and treated with proteinase K (20 mg/L) for 10 min at 37 °C. Endogenous peroxidase was inhibited by treatment with 3% hydrogen peroxide for 5 min, and then the slides were incubated in the TUNEL reaction mixture containing terminal deoxynucleotidyl transferase (TdT) and digoxigenin-11-dUTP for 2 h at 37 °C. Next, the 3,3-diaminobenzidine chromogen was applied, and finally, the number of apoptotic cells was analyzed quantitatively by counting of TUNEL-positive cells in three randomly selected fields of view under a microscope.
Measurement of malondialdehyde (MDA) content and superoxide dismutase (SOD) activity
MDA content and SOD activity were measured in renal tissues and HK2 cells to assess the level of oxidative stress. The intracellular concentration of MDA was detected using a MDA assay kit (TBA Method, Nanjing Jiancheng Bioengineering Institute) and was reported as nmol/mg of extracted protein. SOD activity was detected using the Total Superoxide Dismutase (T-SOD) assay kit (Hydroxylamine method, Nanjing Jiancheng Bioengineering Institute) and was reported as U/mg protein.
Western blot analysis
For western blotting, the soluble proteins were separated on 12% Tris-glycine gels and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore). Protein gel electrophoresis and protein transfer were performed according to standard procedures. Non-specific binding was blocked by addition of 5% non-fat milk for 1 h at room temperature (RT). Then membranes were incubated with specific primary antibodies in a humidified box at 4 °C overnight. After washing with 0.1% Tween-20 in PBS three times, the membranes were incubated with the appropriate horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h at RT. Protein bands were detected using the enhanced chemiluminescence technique (GE AI600, USA). The antibodies employed in the experiment are listed in Table 1.
Table 1.
Antibodies used in this study
| Protein name | Manufacturer | Catalog No. |
|---|---|---|
| Rabbit monoclonal anti-WT1 antibody | Abcam | 89901 |
| Rabbit polyclonal anti-Nephrin antibody | Abcam | 72908 |
| Mouse Anti-Human CD29 | BD | 555443 |
| Mouse Anti-Human CD31 | BD | 560984 |
| Mouse Anti-Human CD34 | BD | 560940 |
| Mouse Anti-Human CD44 | BD | 550989 |
| Mouse Anti-Human CD45 | BD | 340943 |
| Mouse Anti-Human CD73 | BD | 550257 |
| Mouse Anti-Human CD90 | BD | 561971 |
| Mouse Anti-Human CD146 | BD | 550315 |
| Mouse Anti-Human HLA-DR antibody [MEM-12] | Abcam | 28323 |
| Anti-cleaved Caspase-3 antibody | Abcam | 49822 |
| Bcl-2 (D55G8) Rabbit mAb (human specific) | Cell Signaling | 4223 |
| Bcl-2 Polyclonal Antibody (for rat) | Invitrogen | PA5-27094 |
| Bax Rabbit Antibody | Cell Signaling | 2772 |
| ExoAb Antibody Kit (CD9, CD63, CD81, HSP70 antibodies, rabbit anti-human) | SBI | #EXOAB-KIT-1 |
| Anti-Human Nuclear Antigen antibody [235-1] | Abcam | 191181 |
| IRAK1 Polyclonal Antibody | Invitrogen | PA5-19855 |
| NF-κB p65 (D14E12) XP® Rabbit mAb | Cell Signaling | 8242S |
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis
qRT-PCR was performed using the 2*Super SYBR Green qPCR Master Mix (ES Science) according to the manufacturer's instructions. The reaction volumes contained 8 μl diluted cDNA solution, 10 μl Mix, and 2 µl each of forward and reverse primer. qRT-PCR was performed on the CFX96 touch (BIO-RAD) with the following cycling scheme: 10 min at 95 °C followed by 40 cycles of 15 sec at 95 °C and 60 sec at 60 °C. Ct-values were calculated with automatically set thresholds and baselines, and those higher than 38 were excluded from the analysis. The primers used for qRT-PCR are listed in Table 2.
Table 2.
Primers used for RT-qPCR analysis
| Gene name | Forward sequence (5′-3′) | Reverse sequence (5′-3′) |
|---|---|---|
| Human gene | ||
| h-ACTIN | CACCCAGCACAATGAAGATCAAGAT | CACCCAGCACAATGAAGATCAAGAT |
| h-OCT4 | GAGTGAGAGGCAACCTGGAGAAT | ACCGAGGAGTACAGTGCAGTGAA |
| h-NANOG | GAGAAGAGTGTCGCAAAAAAGGA | TGAGGTTCAGGATGTTGGAGAGT |
| h-IRAK1 | CGGTGCCAGGACCAAGTATCT | CCTCTCGTACACCTGGGTCATAG |
| hsa-miR-146a-5p | ACACTCCAGCTGGGTGAGAACTGAATTCCA | TGGTGTCGTGGAGTCG |
| hsa-miR-16-5p | ACACTCCAGCTGGGTAGCAGCACGTAAATA | TGGTGTCGTGGAGTCG |
| hsa-miR-21-5p | ACACTCCAGCTGGGTAGCTTATCAGACTGA | TGGTGTCGTGGAGTCG |
| hsa-miR-30a-5p | ACACTCCAGCTGGGTGTAAACATCCTCGAC | TGGTGTCGTGGAGTCG |
| hsa-let-7b-5p | ACACTCCAGCTGGGTGAGGTAGTAGGTTGT | TGGTGTCGTGGAGTCG |
| hsa-miR-92a-3p | ACACTCCAGCTGGGTATTGCACTTGTCCCG | TGGTGTCGTGGAGTCG |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
| Rat gene | ||
| R-Gapdh | TTCCTACCCCCAATGTATCCG | CATGAGGTCCACCACCCTGTT |
| R-Irak1 | GACTTTGGTCTGGCTCGTTTCA | TCACTCCCACCTCTTCAGCCT |
| rno-miR-146a-5p | ACACTCCAGCTGGGTGAGAACTGAATTCCA | TGGTGTCGTGGAGTCG |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
Exosome isolation from CM or serum
During cell passage, digestion was stopped with exosome-depleted FBS (System Biosciences, SBI). Briefly, when the cells reached 80% confluency, they were digested with 0.25% EDTA-trypsin, centrifuged at 200×g for 5 min, and washed with medium supplemented with exosome-free FBS (SBI). After washing, 5×105 cells were seeded in 10 cm plates. After 3 days in culture, the supernatant of the culture medium, USC conditioned medium (CM), was collected for exosome separation by the co-precipitation method (ExoQuick-TC, SBI). Before isolation, the culture medium was centrifuged at 3,000×g for 15 min to remove dead cells and cell debris. Next, the supernatant was collected, mixed well with a 1/5 volume of the exosome isolation solution, and kept at 4 °C overnight. Next, the mixture was centrifuged at 1,500×g at 4 °C for 30 min, and the supernatant was discarded. The pellet of exosomes was resuspended in saline and stored at -80 °C. The protein concentration of exosomes was determined by the BCA protein assay kit (UBIO, UW0202A). The morphology of exosomes was examined by transmission electron microscopy (TEM), and the mean diameter of the exosomes was measured by particle size analysis.
Serum exosomes were extracted with according to the instructions of the serum exosome extraction kit (EXOQ5A-1, SBI). Briefly, 500 μl serum was centrifuged at 3,000×g for 15 min to remove cells and cell debris. The supernatant was collected, mixed well with 126 μl exosome precipitation solution, and kept at 4 °C for 30 min. Next, the mixture was centrifuged at 1,500×g at 4 °C for 30 min, and the supernatant was discarded. The exosome precipitate was lysed using a specific exosome RNA purification kit (EZBioscience, EZB-exo-RN1), and RNA extraction was carried out according to the kit instructions. Because the amount of RNA obtained from the serum exosomes was very small, we reverse transcribed all the extracted RNA and diluted it 2 times for qRT-PCR analysis.
In vitro exosome tracing
The PKH67 Green Fluorescent Cell Linker Mini Kit (Sigma-Aldrich, Mini67-1KT) was used to stain the membrane of isolated exosomes, and DAPI (4',6-diamidino-2-phenylindole) was used to stain HK2 cell nuclei. All dyeing steps were carried out according to the protocols recommended by the manufacturers. The stained exosomes in Diluent C were extracted again using the exosome extraction kit (as mentioned above). Fluorescently labeled exosomes were added to the supernatant of HK2 cells. After incubation for 30 min, 1 h, or 3 h, the supernatant was discarded and the cells were washed three times with PBS. The fluorescence signals among the adherent cells were detected by fluorescence microscopy to determine whether fusion had occurred between the added exosomes and plated cells.
miRNA isolation and high-throughput sequencing
To perform miRNA-sequencing analysis, exosomal total RNA was extracted using the miRNeasy Mini Kit (Qiagen). The quality and purity of the extracted RNA were determined using the Agilent 2100 Bioanalyzer (Agilent Technologies). The miRNA sequencing library was obtained using the NEXTflex®Small RNA-Seq Kit v3 (Bioo Scientific Corp, NOVA-5132-05), and we used a sample input of 20 ng total RNA. Finally, we profiled the expression of miRNAs in the library using Hiseq2500 (Illumina, Inc.).
Cell transfection and dual-luciferase reporting system
HEK293T cells were plated at 1×105 cells/well in a 24-well plate, and transfection was performed once the cells achieved 60-70% confluency. Thirty minutes before transfection, the complete medium was replaced with serum-free medium. Lipofectamine 2000 (Invitrogen Life Technologies, 780373) was used to transfect the cells. The cells were lysed 48 h after transfection, and miRNA expression was detected by chemiluminescence using the Dual-Luciferase Reporter Assay System (Promega). With firefly luciferase as the internal reference, the fluorescence ratio was calculated by dividing the relative light units (RLU) for Renilla luciferase by the RLU for firefly luciferase. The inhibitory effect of the miRNA on target gene expression was compared according to the obtained ratio.
Fluorescence in situ hybridization (FISH)
For FISH analysis, tissue samples were harvested, washed, and immediately placed in the fixative solution for 12 h. After gradient alcohol dehydration and paraffin embedding, tissue slices were cut and then baked in a 62 °C oven for 2 h. Subsequently, the tissue sections were immersed in dimethylbenzene xylene I for 15 min, dimethylbenzene xylene II for 15 min, anhydrous ethanol I for 5 min, anhydrous ethanol II for 5 min, dry naturally and soak in diethylpyrocarbonate (DEPC)-treated water. Then the slices were boiled in the repair solution for 15 min and cooled naturally. Protease K (20 µg/ml) was added for protein digestion at 37 °C for 30 min. After washing with pure water, the slices were washed three times with PBS for 5 min. Pre-hybridization solution was added and incubated at 37 °C for 1 h. After removal of the pre-hybridization solution, the hybridization solution containing an 8 ng/µl miR-146a-5p probe was dropped onto the slides for hybridization at 37 °C overnight. Washing with 2*SSC in 37 °C for 10 min, 1*SSC in 37 °C for 10 min, 0.5*SSC in 37 °C for 10 min after hybridization. After the solution was removed, the sealing solution (bovine serum albumin, BSA) was added for incubation at RT for 30 min. After removal of the sealing solution, anti-DIG-488 solution was added for incubation at 37 °C for 50 min. Next the slices were washed with PBS four times, 5 min each, and DAPI solution was added for incubation in darkness for 8 min. After final rinsing, anti-fluorescence quenching sealant was added to seal the slices. The slices were observed under a fluorescence microscope (Olympus), and images were collected.
Immunofluorescence staining
For immunofluorescence analysis, HK2 cells were seeded on coverslips in 24-well plates. After H/R injury, the cells were washed with PBS twice and fixed in 4% paraformaldehyde at RT for 30 min. After washing with PBS for 5 min to quench the aldehyde group, the cells were permeabilized with PBS containing 0.2% Triton X-100 for 10 min at RT. After washing twice with PBS, the cells were blocked with 10% normal goat serum in PBS for 60 min at RT with gentle shaking. The cells were then incubated with primary NF-κB p65 antibody (CST, 8242S, 1:50 dilution) at 4 °C overnight with gentle shaking. The next day the cells were washed with PBS containing 0.05% Tween 20 and 1% bovine serum albumin (BSA) three times with shaking before incubation with secondary antibody (Alexa Fluor 488-conjugated goat anti-rabbit IgG, ABclonal, AS053, 1:100 dilution) at RT for 1 h in darkness with gentle shaking. The cells were again washed with PBS containing 0.05%Tween 20 and 1% BSA three times with shaking and then incubated in 0.2 mg/ml DAPI solution for 2 min for nuclear staining, followed by a final rinse with PBS. Immunofluorescence staining of NF-κB p65 within the cells was observed under a fluorescence microscope (Olympus).
Statistical analysis
Data are presented as mean ± standard error of the mean (SEM). First, we determined whether the data followed a normal distribution. When data for two independent conditions followed a normal distribution, the parameter test was used. When variance was homogeneous, the unpaired Student's t test was used. For the comparison of data among multiple groups, one-way analysis of variance (ANOVA) was applied. All of the statistical tests were performed using GraphPad 7.0 (GraphPad Software), and the level of significance was set at P < 0.05 for all comparisons.
Results
hUSCs were isolated from human urine samples
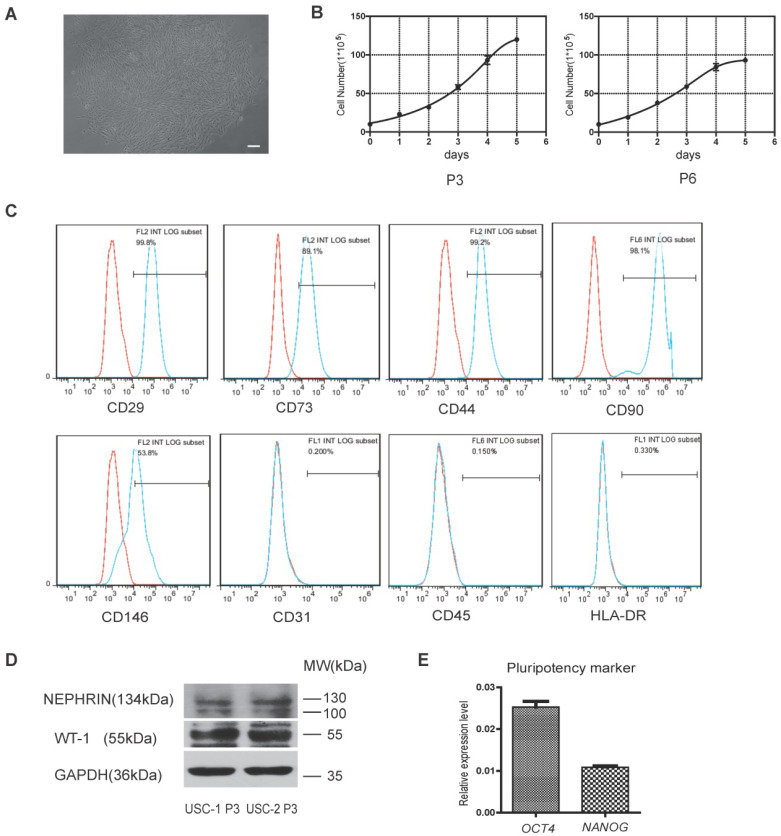
Cells isolated from human urine samples were cultured for 3-5 days, and tiny colonies of slender spindle-like cells were observed (Figure 1A). The cells proliferated rapidly in vitro, with cells of passages 3 and 6 (P3 and P6) exhibiting rapid proliferation capacity (Figure 1B). Flow cytometric analysis showed that USCs expressed common surface markers of MSCs, including CD29, CD73, CD44, CD90, and CD146, but not endothelial or hematopoietic cell surface markers, including CD31 and CD45. Low expression of human leukocyte antigen (HLA)-DR suggested the low immunogenicity of USCs (Figure 1C). Western blot analysis showed USCs expressed specific renal markers including nephrin and Wilms' tumor-1 (WT1; Figure 1D). qRT-PCR analysis showed that USCs expressed pluripotency markers, including OCT4 and NANOG (Figure 1E).
Figure 1.
Characterization of human urine-derived stem cells (USCs). (A) Spindle-shaped morphology of USCs (scale bar = 50 µm). (B) Growth curves for USCs of different passages (P3 and P6). (C) After 3 passages, the majority of isolated USCs expressed MSC markers CD29, CD73, CD44, CD90, and CD146 but not CD31, C45, and HLA-DR. (D) Protein expression of renal markers Nephrin and WT-1 in USCs from two healthy people. (E) Relative expression levels of pluripotency markers OCT4 and NANOG in USCs.
USCs reduced renal tubular injury and cell apoptosis and inhibited local inflammation in the renal IRI model
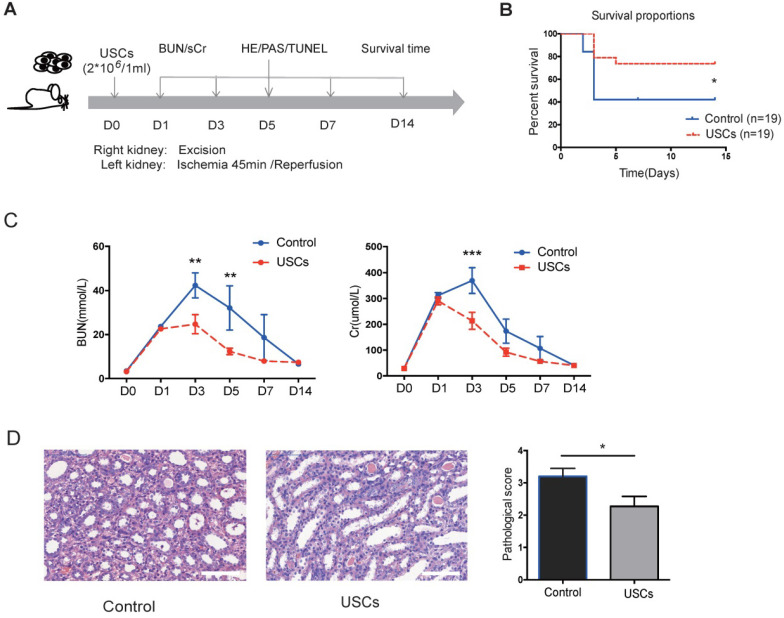
Compared to the control treatment, USC treatment significantly increased rat survival (P < 0.05), decreased sCr and BUN levels at day 3 post-injection (P < 0.05), and reduced the pathological score for renal injury at day 7 post-injection (P < 0.05; Figure 2B-D). These results indicated that intravenous transfusion of USCs ameliorated renal injury and protected renal function in the rat IRI model.
Figure 2.
The protective effects of USCs on renal function after renal IRI. (A) Diagram of the ischemia-reperfusion injury (IRI) animal model. (B) Survival curves for the control and USC-treated groups (Control group, n=19; USC-treated group, n=19; P= 0.0328). (C) Time-dependent changes in blood urea nitrogen (BUN) and serum creatinine (sCr) in control (n=25) and USC-treated groups (USCs, n=26) at days 1, 3, 5, 7 and 14 after IRI (BUN: P= 0.0028 on day 3, P= 0.0066 on day 5; sCr: P<0.0001 on day 3). Data represent the mean ± SEM, *P<0.05, **P<0.01, ***P<0.001. (D) Histopathological scores for control (n=10) and USC-treated groups (USCs, n=11) on day 7 (P= 0.0311). Scale bars =100 µm. Data represent the mean ± SEM, *P<0.05, **P<0.01.
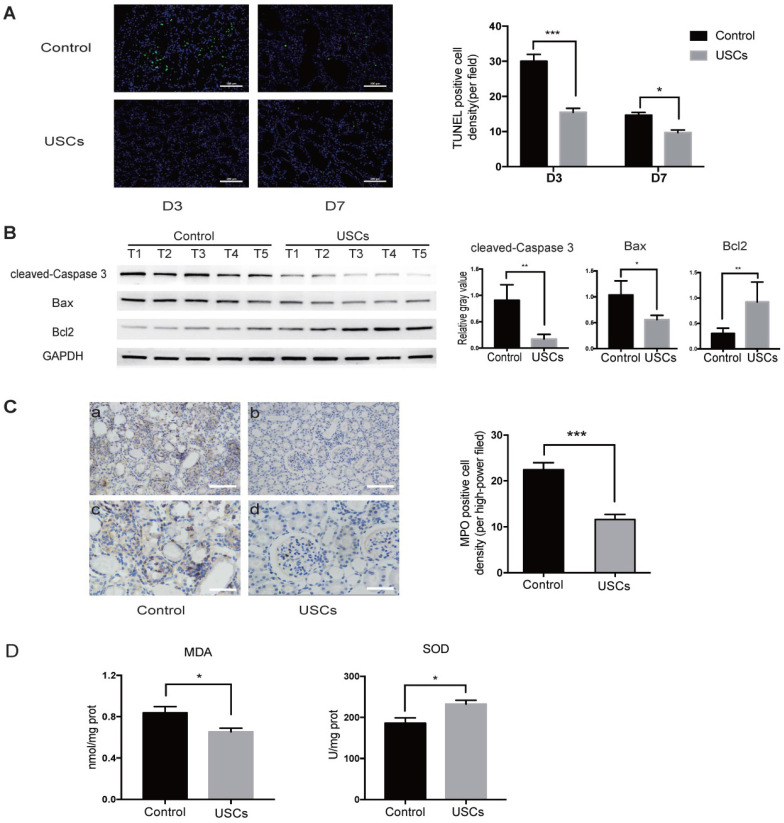
Compared to the control treatment, USC treatment also significantly decreased apoptosis in the IRI kidney at days 3 and 7 as indicated by TUNEL staining (Figure 3A). Western blot analysis showed that expression of pro-apoptotic cleaved-caspase-3 and Bax was decreased, whereas anti-apoptotic Bcl2 expression was increased in the kidney of USC-treated rats at day 3 (Figure 3B). MPO staining showed significantly fewer infiltrating neutrophils in the IRI kidney of hUSC-treated rats at day 3 (P < 0.001; Figure 3C). Additionally, the MDA content was significantly reduced (P < 0.05) and SOD activity was significantly increased (P < 0.05) at day 3 in the kidney of the USC-treated rats (Figure 3D). Taken together, these results suggest that USC treatment reduced cell apoptosis, neutrophil infiltration, and oxidative stress in the IRI kidney.
Figure 3.
USCs reduced the expression of apoptosis-related proteins, inflammatory cell infiltration, and oxidative stress level in the kidney after IRI. (A) TUNEL staining analysis of the apoptosis among renal tubular epithelial cells in the control (n=5) and USC-treated group (USCs, n=5) on days 3 and 7 after IRI. Scale bars = 100 µm, P<0.001 on day 3, P= 0.0139 on day 7. (B) Western blot analysis of cleaved-caspase-3 and Bcl2/Bax expression in the control group (n=5) and USC-treated group (n=5) on day 3 after IRI. GAPDH was used as a loading control. (C) MPO staining analysis of neutrophil infiltration in the kidney tissue on day 3. n=5 in each group. Scale bars = 200 µm in a,b; 100 µm in c,d; P<0.001. (D) SOD and MDA analysis of oxidative stress in kidney tissue on day 3 after IRI. n=4 in each group. P= 0.0378 for MDA, P= 0.0262 for SOD. Data represent the mean ± SEM. *P<0.05, **P<0.01, ***P<0.001.
Most injected hUSCs were trapped in lung tissue and not found in damaged kidney tissue
To test whether transfused USCs integrate into the injured tissue, we labeled USCs with PKH26 before injection into IRI rats through the dorsal vein of the penis and harvested rat organs, including the lungs, liver, spleen, and damaged kidneys, 24 h later. Fluorescence imaging showed that most of the injected hUSCs were trapped in lung tissue and did not reach the damaged kidney tissue (Figure S1A).
We also performed human nuclear antigen (hNA) staining of IRI kidney slices to distinguish human cells from rat tissues at day 7 post-injection. The results demonstrated that, while USCs were found in the kidney after infusion into the renal artery after IRI (positive control); few USCs had integrated into the tubular epithelial lining of the kidney after injection via the dorsal vein of the penis in rats (Figure S1B). This result supported the idea that direct contact between injected USCs and injured renal cells might not be required the protective action of the injected USCs.
USC-Exo protected against IRI in vivo and inhibited oxidative stress in HK-2 cells after H/R injury in vitro
We collected CM from USCs to test whether USC-secreted soluble factors affect renal tubular cell injury in vitro. H/R injury in normal human kidney cortex/proximal tubule cells HK2 was induced by incubation in glucose- and serum-free medium in 1% O2 for 48 h followed by reoxygenation with normal O2 in either USC CM or control medium for 6 h (Figure S2A). The SOD activity and MDA content were analyzed to determine the level of oxidative stress in the injured cells. The results showed that USC CM reduced oxidative stress by increasing SOD activity and decreasing MDA content (Figure S2B).
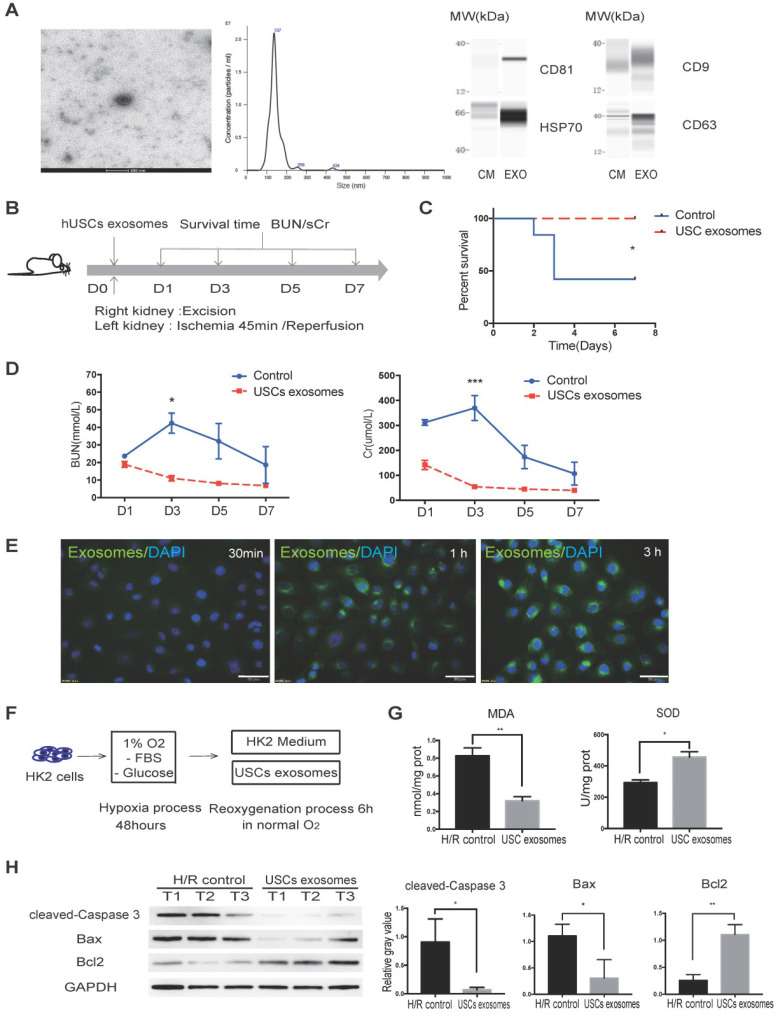
Previous studies reported that exosomal contents secreted from MSCs are protective against IR injury 31, 32. We, therefore, aimed to investigate whether USCs protect HK2 cells from H/R-induced injury via secreted exosomal factors. Following the latest guidelines for exosome isolation and characterization 33, we isolated USC-secreted EVs and examined them by TEM. We found that the USC-EVs showed a sphere-like morphology with diameters of 100-150 nm. We further carried out particle size analysis on the isolated exosomes and measured a mean diameter of 144.9 ± 41.6 nm. The exosomes were found to express exosomal markers, including CD81, CD9, CD63, and HSP70, by western blot analysis (Figure 4A). These results demonstrated the exosomal identify of the EVs isolated from USCs.
Figure 4.
USC-Exo protect renal function in vivo and HK2 cells after H/R injury in vitro. (A) Characterization of USC-Exo. TEM images of exosomes isolated from USC CM (scale bar = 200 nm). Size analysis of USC-Exo (mean diameter = 144.9 ± 41.6 nm). Western blot analysis of exosomal markers CD81, CD9 CD63, and HSP70. CM served as the control. (B) Diagram of IRI animal model with exosome treatment. (C) Survival curve analysis for the two groups (control group, n=19; USC-Exo-treated group, n=6; P=0.0201). (D) Time-dependent changes in BUN and sCr in the control group (n=25) and USC-Exo-treated group (USC-Exo, n=6) on days 1, 3, 5, and 7 after IRI (BUN: P= 0.0228 on day 3; sCr: P= 0.0589 on day 1, P<0.001 on day 3). (E) After incubation of HK2 cells with USC-Exo (PKH67, green fluorescence) for 30 min, 1 h, or 3 h, and washing with PBS, the fusion of exosomes and cells was observed fluorescence microscopy (scale bars = 50 µm). (F) Schematic diagram of H/R-induced injury in HK2 cells with or without USC-Exo treatment during the reoxygenation process. (G) Oxidative stress levels in HK2 cells after H/R in the two groups were determined by measuring MDA and SOD (P=0.0073 for MDA, P=0.0132 for SOD). Each experiment was repeated three times. (H) Protein expression of cleaved-Caspase-3 and BCL2/BAX in the indicated groups. After USC-Exo treatment, the expression levels of cleaved-Caspase-3 and BAX in HK2 cells were decreased significantly. Each experiment was repeated three times. GAPDH was used as loading control. Data represent the mean ± SEM. *P<0.05, **P<0.01, ***P<0.001.
To further evaluate the protective effect of USC-Exo against renal IR injury, we again used the lethal rat renal IRI model and injected USC-Exo into the IRI rat at the time of reperfusion (Figure 4B). We found that the levels of BUN and sCr in the middle-dose group (20 μg/rat) were similar to those in the high-dose group (40 μg/rat) and lower than those in the low-dose group (10 μg/rat; data not shown). Therefore, in the subsequent experiments, we used the middle dose (20 μg/rat) of USC-Exo.
We found that the rats in the USC-Exo treated group did not die during the observation period (Figure 4C). We continuously monitored the changes in BUN and sCr levels and found that these levels in the USC-Exo treated group were significantly lower than those in the control group (Figure 4D). These results demonstrated that treatment with USC-Exo could also ameliorate renal damage, similarly to USCs, in the rat IRI model.
We further examined the protective effect of USC-Exo in vitro. To demonstrate the fusion of exosomes with the target cells, we labeled USC-Exo with green fluorescent dye (PKH67) and incubated DAPI-stained HK2 cells with the labeled exosomes for 30 min, 1 h, or 3 h. After these different periods, we washed the cells with PBS and detected the fluorescence signals among the plated cells. We found that the intracellular fluorescence signal gradually increased with the extension of the incubation time. After incubation for 3 h, the fluorescence signal of USC-Exo could be detected in most cells (Figure 4E). After exposure of the cells to hypoxia for 48 h, we added 100 μL USC-Exo solution (20 μg/mL) into the HK2 medium during the re-oxygenation process for 6 h (Figure 4F). The MDA content was significantly lower (P < 0.05) and SOD activity significantly greater (P < 0.05) in the USC-Exo-treated group compared with the control group (Figure 4G). Western blot analysis showed that treatment with USC-Exo reduced H/R-induced expression of cleaved caspase-3 and BAX, but increased BCL2 expression (Figure 4H). These results demonstrate the protective effect of USC-Exo against H/R injury in HK2 cells.
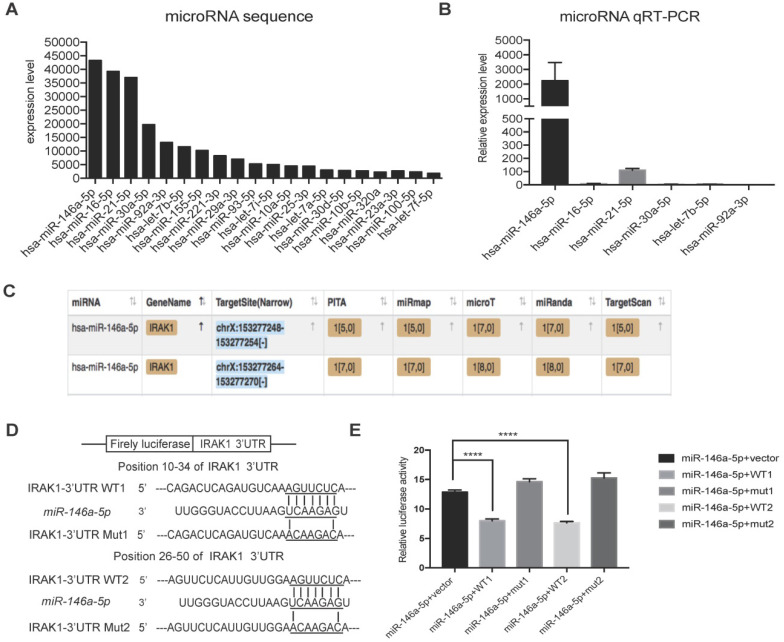
miR-146a-5p in USC-Exo inhibited IRAK1 expression
To further investigate the mechanism responsible for the protective effect of USC-Exo, we hypothesized that miRNAs might play important roles based on reports that exosomal miRNAs are involved in multiple physiological and pathological processes 34. RNA sequencing analysis revealed that 20 annotated miRNAs were enriched in the USC-Exo (Figure 5A). We next confirmed the expression levels of the top six enriched miRNAs by qRT-PCR and identified miR-146a-5p as the most highly expressed miRNA in USC-Exo (Figure 5B). To further determine the functions of miR-146a-5p, we searched the miRNA target prediction analysis database Starbase v2.0 (http://starbase.sysu.edu.cn) and found 451 genes as potential targets of miR-146a-5p. Interleukin-1 receptor-associated kinase 1 (IRAK1) was ranked highest among the predicted targets of miR-146a-5p, with two predicted target sites for miR-146a-5p in the 3' untranslated region (UTR) of the transcript of IRAK1 (Figure 5C). In order to verify the direct binding of miR-146a-5p to the 3'UTR region of IRAK1 gene, we then cloned the wild-type and mutant 3'UTR of IRAK1 downstream of a firefly luciferase cassette in a luciferase reporter vector. Co-transfection of miR-146a-5p mimic with the wild-type reporter plasmids in HEK293T cells significantly reduced the luciferase activity, which was significantly reversed by co-transfection with two mutant reporter plasmids (Figure 5D, E). These results indicated that miR-146a-5p from USC-Exo may bind the IRAK1 mRNA 3'UTR region and thereby inhibit IRAK1 expression via post-translational repression.
Figure 5.
miRNA sequencing of USC-Exo contents and potential targets of miR-146a-5p.(A) The top 20 most-enriched miRNAs in USCs-Exo. (B) qRT-PCR analysis of the expression levels of the top 6 most-enriched miRNAs in USCs-Exo. (C) StarBase analysis of the target genes of miR-146a-5p. (D) Sequence alignments of miR-146a-5p and its two candidate target sites in the 3'UTR of IRAK1. (E) Luciferase reporter assay of miR-146a-5p mimic-treated HEK293T cells, which overexpressed either IRAK1-wildtype 3'UTR (WT1 and WT2) or IRAK1-mutant 3'UTR (mut1 and mut2). Data represent the mean ± SEM. *P<0.05, **P<0.01, ***P<0.001.
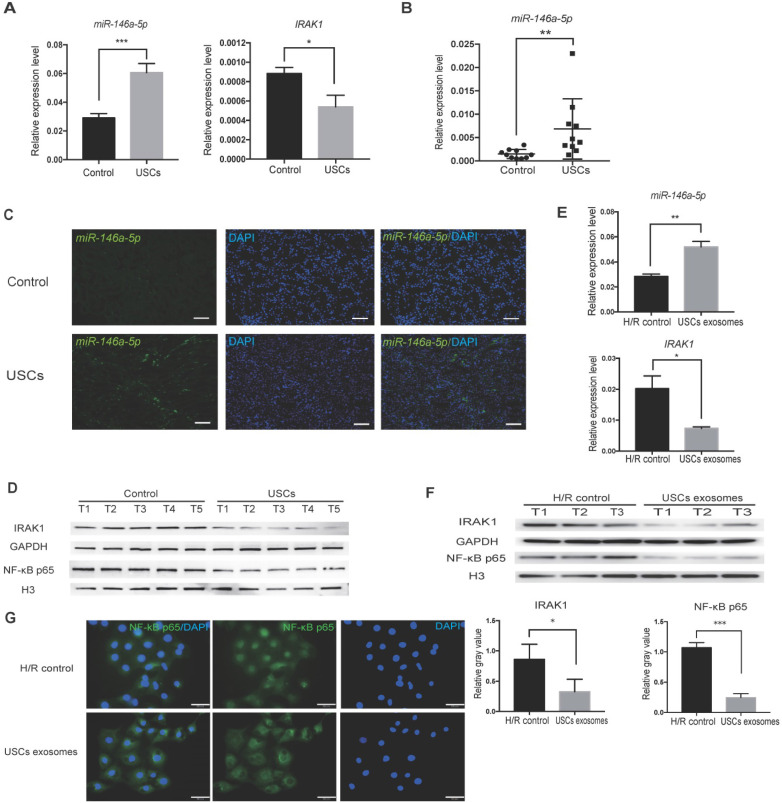
USCs enhanced miR-146a-5p expression and inhibited IRAK1 expression and nuclear translocation of the NF-κB p65 subunit in vivo and in vitro
To further confirm the hypothesis that miR-146a-5p is critical to the protective effect of USCs in IRI, we first identified the expression level of miR-146a-5p in the kidney tissue of USC-treated IRI rats. qRT-PCR analysis revealed upregulated miR-146a-5p expression in the kidney of the USC-treated IRI group (Figure 6A). To further determine whether USC-secreted miR-146a-5p reaches the damaged kidney tissue via exosomal delivery, we collected serum samples from the rats on day 3 after USC treatment, extracted the exosomes, and detected the expression level of miR-146a-5p. The results showed greater expression of exosomal miR-146a-5p in the serum of the USC-treated group compared with the control group (Figure 6B). FISH images further demonstrated the increased expression of miR-146a-5p in damaged kidney tissue (Figure 6C). The mRNA (Figure 6A) and protein (Figure 6D) expression levels of IRAK1 were further examined and shown to be downregulated in the kidney of USC-treated IRI rats.
Figure 6.
USCs or USC-Exo upregulate miR-146a-5p expression, which targets the IRAK1 and NF-κB signaling in vivo and in vitro. Rat IRI was induced before treatment with or without USCs (annotated as USCs and control, respectively). (A) qRT-PCR analysis of the relative expression levels of miR-146a-5p (P=0.0004) and IRAK1 (P=0.0227). (B) qRT-PCR analysis of the relative expression levels of miR-146a-5p in rat serum exosomes on day 3 (n=10 in each group, P=0.0012). (C) FISH images of miR-146a-5p expression in kidney sections (scale bars = 100 µm). (D) Western blot analysis of the protein levels of IRAK1 and nuclear NF-κB p65. n=5 in each group. GAPDH and H3 were used as loading controls, respectively. H/R injury was induced in HK2 cells in the absence or presence of USC-Exo (annotated as HK2 medium and HK2 medium+exosomes, respectively). (E) qRT-PCR analysis of the relative expression levels of miR-146a-5p (P=0.0078) and IRAK1 (P=0.0358). (F) Western blot assay of the protein levels of IRAK1 and nuclear NF-κB p65. n=3 in each group. GAPDH and H3 were used as loading controls, respectively. (G) Immunofluorescence analysis showed that NF-κB p65 in HK2 cells was transferred from the cytoplasm to the nucleus after H/R treatment, and this nuclear translocation of NF-κB p65 could be inhibited by USC-Exo treatment (scale bars = 50 µm). Each experiment was repeated three times. Data represent the mean ± SEM. *P<0.05, **P<0.01.
A corresponding in vitro experiment was conducted using H/R-induced injury in HK2 cells. Treatment with USC-Exo significantly upregulated miR-146a-5p expression and downregulated IRAK1 expression in these cells (Figure 6E). In agreement with the in vivo observations, western blotting showed that IRAK1 protein expression as well as nuclear translocation of the NF-κB p65 subunit was reduced by treatment with USC-Exo (Figure 6F). We further performed staining of NF-κB p65 in the USC-Exo treated cells and found that in the control HK2 cells after H/R, NF-κB p65 was localized in the nucleus. In contrast, in the USC-Exo treated HK2 cells after H/R, some NF-κB p65 remained in the cytoplasm, and its nuclear localization level was significantly reduced (Figure 6G). These results suggested that the protective effect of USCs against renal injury may occur via exosomal miR-146a-5p-mediated downregulation of NF-κB signaling.
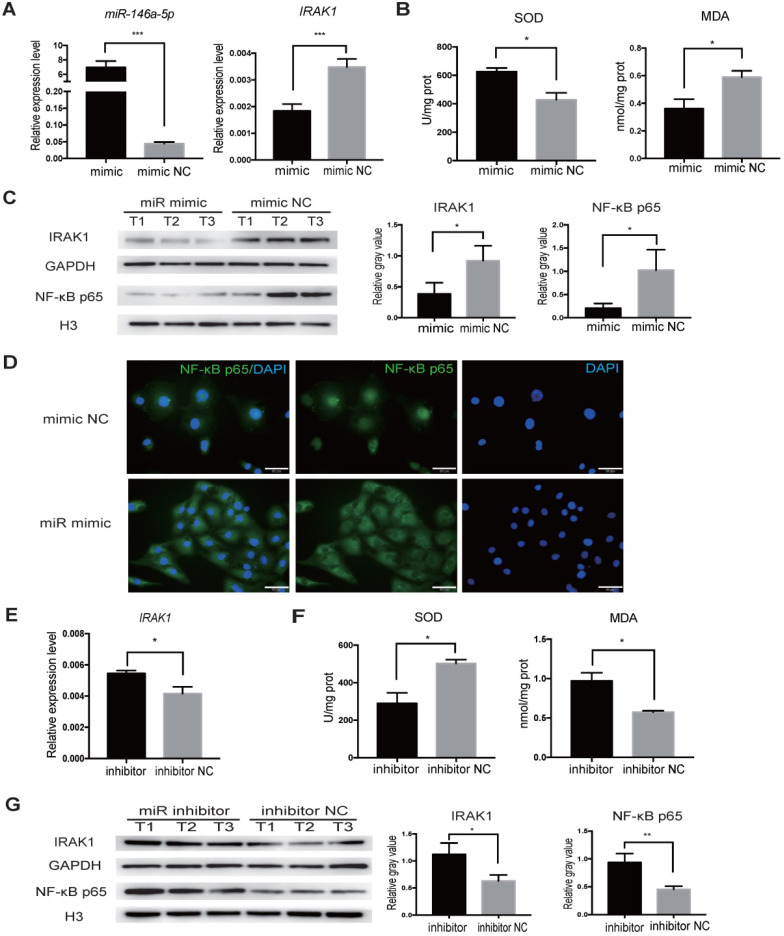
miR-146a-5p inhibited IRAK1 expression and nuclear translocation of NF-κB p65 in response to H/R in HK2 cells
We next aimed to delineate the regulatory relationship between miR-146a-5p and IRAK1 using the H/R-induced injury model in vitro. We transfected HK2 cells with miR-146a-5p mimic, and increased miR-146a-5p expression was confirmed by qRT-PCR (Figure 7A). The transfected cells were subjected to H/R injury at the same time, and qRT-PCR analysis showed that miR-146a-5p mimic resulted in a significant decrease in IRAK1 mRNA expression (Figure 7A). Furthermore, transfection with the miR-146a-5p mimic could reduce oxidative stress, increase SOD activity, and decrease MDA content (Figure 7B). Western blot analysis showed that transfection with the miR-146a-5p mimic resulted in a significant decrease in IRAK1 protein expression and NF-κB p65 subunit nuclear translocation (Figure 7C). The results of immunofluorescence staining also showed that the miR-146a-5p mimic could reduce the nuclear localization of NF-κB p65 (Figure 7D). We also treated HK2 cells with a miRNA inhibitor and found that the expression level of IRAK1 in the inhibitor-treated group was higher than that in control group (Figure 7E). miR-146a-5p inhibitor could aggravate the oxidative stress, decrease SOD activity, and increase MDA content (Figure 7F). Western blot analysis also showed that miR-146a-5p inhibitor could support IRAK1 protein expression and NF-κB p65 subunit nuclear translocation (Figure 7G).
Figure 7.
miR-146a-5p mimic reduces oxidative stress and inhibits IRAK1 and NF-κB signaling in H/R-induced injury of HK2 cells. HK2 cells were transfected with miR-146a-5p mimic or its inhibitor before the H/R process. (A) qRT-PCR analysis of the relative expression levels of miR-146a-5p and IRAK1 in the mimic and mimic negative control (mimic NC) groups (P<0.001). (B) Analysis of SOD activity and MDA content in the mimic and mimic NC groups after H/R (P=0.0216 for SOD, P=0.0497 for MDA). (C) Western blot analysis of the protein levels of IRAK1 and nuclear translocation of NF-κB p65 in the mimic and mimic NC groups. n=3 in each group. GAPDH and H3 were used as loading controls, respectively. (D) Immunofluorescence analysis of the nuclear translocation of NF-κB p65 in the mimic and mimic NC groups after H/R treatment (scale bars=50 µm). (E) qRT-PCR analysis of the relative expression levels of IRAK1 in the inhibitor and inhibitor negative control (NC) groups (P=0.015). (F) Analysis of SOD activity and MDA content in the inhibitor and inhibitor NC groups after H/R (P=0.0228 for SOD, P=0.0192 for MDA). (G) Western blot analysis of the protein levels of IRAK1 and nuclear translocation of NF-κB p65 in the inhibitor and inhibitor NC groups. n=3 in each group. GAPDH and H3 were used as loading controls, respectively. Each experiment was repeated three times. Data represent the mean ± SEM. *P<0.05, **P<0.01, ***P<0.001.
Together these results indicated the indispensable role of miR-146a-5p in regulating IRAK1 expression and NF-κB p65 subunit nuclear translocation in H/R-induced injury.
Knock down of IRAK1 inhibited the nuclear translocation of NF-κB p65 in response to H/R in HK2 cells
To further test whether IRAK1 could regulate the nuclear translocation of NF-κB p65, we knocked down IRAK1 expression by siRNA. Among three target siRNAs (target sequence 1: GGACATCCTTCTGGGTACA, target sequence 2: GAGCCACCGCAGATTATCA, target sequence 3: GCCCGAAGAAAGTGATGAA), we found that si-2 and si-3 had the higher inhibition efficiencies (Figure S3A), and we choose to use si-2 and -3 in combination (Figure S3B). Western blot analysis showed that the nuclear level of NF-κB p65 was reduced upon knock down of IRAK1 (Figure S3C). These results indicate that knock down of IRAK1 inhibited the nuclear translocation of NF-κB p65 in HK2 cells after H/R injury.
Discussion
In this study, we demonstrated the renal protective effect of USCs in vitro and in vivo. In a rat renal IRI model, USC therapy significantly reduced mortality, decreased sCr and BUN levels, and attenuated renal tubular injury. We further showed that USCs possessed anti-inflammatory, anti-oxidative, and anti-apoptotic effects, which are known to contribute to the protective effects of other types of stem cells against renal IRI 35. Most importantly, we revealed a novel mechanism of USC function: USC-Exo delivery of miR-146a-5p to downregulate IRAK1 expression and subsequently inhibit NF-κB signaling.
The potential therapeutic efficacy of MSCs in kidney diseases has been widely reported 11, 36. Due to the invasive collection process and limited proliferation abilities of MSCs, we aimed to explore the utility of USCs in renal IR injury repair. USCs expressed stem cell markers such as SSEA4, Sox2, and Oct3/4 at the early stage of isolation. USCs stably expressed MSC-specific surface molecules (CD29, CD44, CD73, CD90, etc.) and did not express hematopoietic stem cell markers (CD133, CD34, CD45, etc.) or endothelial cell markers (CD31, etc.), suggesting that USCs are not hematopoietic stem cells or endothelial precursor cells, but a special type of MSCs. A previous study found that most USCs expressed cytokeratin 7 (CK7), suggesting that these USCs may be uroplakin basal cells. However, the same group also found that USCs expressed renal markers (CD224, CD13, NR3C2, Pax2, and Pax8) and renal epithelial cell-specific markers (CD146 and podocin), but did not express markers of renal tubular epithelial cells, ureteral epithelial cells, or urethral epithelial cell markers. This suggested that USCs may be derived from the junction of visceral and parietal epithelial cells of the renal glomerulus, rather than urethral epithelial basal cells 13, 37. However, the real source of USCs remains to be explored. In our experiments, the surface molecular expression patterns of the USCs were consistent with those described previously. The major advantages of USCs include the non-invasive collection process and non-costly isolation/expansion protocol. In agreement, Tian et al. 21 and we reported that USCs exert anti-inflammatory and anti-apoptotic effects to facilitate the repair of ischemic acute kidney injury in rats. Tian et al. detected USCs in tubular epithelium, because they injected USCs directly into the upper, middle and lower cortex of the kidney. We, however, did not observe the incorporation of USCs into the renal tissue after intravenous administration. This difference may be related to the different injection routes employed by the studies. A recent study also concluded that hUSCs have a nephron-protective effect on renal function via anti-inflammatory, anti-oxidative stress, and anti-fibrotic activities in a rat model of chronic kidney disease, which was induced by IRI and gentamicin administration 38. Another recent study also described the therapeutic effects of USCs in a rat model of cisplatin-induced AKI 39. Both of these studies together with our study support the effectiveness of USCs in the treatment of renal-related diseases.
Thus, our present study also aimed to explore the indirect mechanism by which USCs protect against IRI, given that we did not found evidence of direct-targeted migration of USCs into the damaged tissue. The effects of MSCs are primarily mediated via complex paracrine actions and the transfer of EVs/exosomes packaging various molecules, such as bioactive lipids, proteins, and RNAs 40. MSC-derived EVs could restore renal structure and function via their immunomodulatory, anti-apoptosis, and proliferation stimulation abilities in AKI models 41-44. We, therefore, investigated the role of EVs/exosomes in USC therapy for renal IRI. USC-Exo were isolated from USC CM and identified as exosomes based on size and the expression of exosome-specific markers. Our results showed that USC-Exo could preserve renal function after IRI, improve the survival rate of IRI rats, and reduce the levels of sCr and BUN in vivo. Consistently, USC-Exo also protected HK2 cells from H/R injury and reduced oxidative stress in vitro.
One component of exosomes that is found at extremely high levels in miRNA. Exosomal miRNAs have been implicated in many processes such as cartilage repair, renal fibrosis inhibition, and IRI in myocardium cells 45-47. Thus, we performed miRNA sequencing of the contents of USC-Exo and identified miR-146a-5p as the most enriched miRNA. The role of miR-146a-5p has been reported in renal diseases, and treatment with miR-146a-5p was shown to inhibit renal fibrosis in vivo 48. miR-146a-5p expression in the kidneys as well as its urinary excretion are specifically associated with the development of interstitial lesions and correlated with inflammatory cell infiltration 49. miR-146a(-/-) mice exhibit more extensive tubular injury, inflammatory infiltrates, and fibrosis than wild-type mice 50. miR-146a-5p expression is decreased in the glomeruli of patients with type 2 diabetes, which correlates with increased albuminuria and glomerular damage 51. One study also found that the expression levels of nine miRNAs (miR-21, miR-20a, miR-146a, miR-199a-3p, miR-214, miR-192, miR-187, miR-805, and miR-194) were altered in kidney tissues of C57BL/6 mice exposed to unilateral thermal warm ischemia compared with their levels in the control group. Among these miRNAs, miR-146a showed significantly increased expression at 14 days after IRI 52. In our experiment, the time point chosen for miR-146a detection was before day 7 after IRI. Therefore, the expression level of miR-146a in the IRI control group was still at a low level, while the expression level of miR-146a in the USC-treated group was significantly increased. These results suggest a protective role of miR-146a-5p against renal injury in both mice and human.
We further investigated the downstream mechanism of USC-Exo derived miR-146a-5p and showed that it could downregulate the expression of IRAK1. As a serine-threonine kinase, IRAK1 mediates the Toll-like receptor (TLR) and interleukin-1 (IL-1) signaling pathways and therefore plays a critical role in innate immunity 53. miR-146a-mediated downregulation of IRAK1 is involved in a wide spectrum of diseases, including autoimmune disorders, cancer, diabetes, or neuropathic pain 54-57, as well as in cerebral, hepatic, or intestinal IRI 58-60. Considering research showing that downregulation or inhibition of IRAK1 can attenuate high glucose- or lipopolysaccharide (LPS)-induced renal injury (podocyte apoptosis) 61, 62, in combination with our current results, a critical role for IRAK1 in renal IRI is strongly supported.
We also showed that miR-146a-5p is implicated in USC-Exo suppression of IRAK1 expression and NF-κB signaling. miR-146a-5p was also reported to be an NF-κB dependent gene that can directly downregulate the production of pro-inflammatory cytokines, in control of TLR and cytokine signaling through a negative feedback regulation loop involving down-regulation of tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6) and IRAK1 protein levels 63, 64. Previous studies have demonstrated that miR-146a-deficient mice develop severe gouty arthritis via TRAF6, IRAK1 and NALP3 (NACHT, LRR and PYD domain-containing protein 3)-induced inflammasome dysregulation 65. Inhibition of chemokine signaling prevents the development of inflammation and fibrosis after IRI in miR-146a(-/-) mice 50. Our experiments also showed that miR-146a-5p regulates downstream IRAK1 and NF-κB signaling. Taken together, our results indicate that the role of miR-146a-5p in limiting inflammation in renal IRI may occur though IRAK1/NF-κB signaling.
Some limitations of our study should be considered. The most recognized gold standard for separating exosomes is "differential ultracentrifugation", in which cells, debris, and large-diameter vesicles are removed step-by-step by differential centrifugation, and exosomes are precipitated and enriched by ultracentrifugation. In this study, a more convenient and easy-to-operate commercial kit was utilized for exosome extraction, which may have affected the purity of the isolated exosomes. Also, if USCs can be treated by gene editing, key factors of the exosome secretion pathway can be knocked out or inhibited, thereby blocking exosome secretion. USCs that lose the ability to secrete exosomes can be injected into animal models, and investigations of the changes in the renal protection ability of such cells can further prove the important role of exosomes in the renal protective effects of USCs. Finally, due to the limitations of our experimental conditions, it was impossible to trace and detect the exosomes released by USCs in vivo. If USC-secreted exosomes could be specifically labeled and directionally tracked in vivo, the enrichment of exosomes in the kidney instead of whole cells could be detected. This design could more directly prove that USCs offer renal protection through exosomes.
Conclusions
In summary, we demonstrated that USCs protect against renal injury in a lethal rat IRI model via the interplay of anti-oxidative, anti-inflammatory, and anti-apoptotic cytoprotective effects. Furthermore, using an H/R-induced oxidative stress cell model in vitro, we identified the key player, miR-146a-5p in the USC-Exo, which mediates the cytoprotective effects by downregulating IRAK1/NF-κB signaling. These findings provide a theoretical foundation for the possibility of using the non-invasive and cost-effective USCs or USC-Exo for the treatment of AKI.
Supplementary Material
Supplementary figures.
Acknowledgments
We greatly acknowledge Dr. Xianchang Li (Houston Methodist Hospital, Weill Cornell Medical College of Cornell University) for the expert linguistic assistance and technical assistance during the preparation of this manuscript. This project was supported by the National Natural Science Foundation of China (81870511, 81670680, 81700655, 81300623, 81770749), Science and Technology Planning Project of Guangdong Province, China (2014B020212006, 2015B020226002, 2017A020215012), Guangdong Natural Science Foundation (2018A030313016, 2015A030313135), Guangdong Basic and Applied Basic Research Foundation (2020A1515010884), Key Scientific and Technological Program of Guangzhou City (201803040011), Guangdong Provincial Key Laboratory on Organ Donation and Transplant Immunology (2013A061401007, 2017B030314018), and Guangdong Provincial International Cooperation Base of Science and Technology (Organ Transplantation, 2015B050501002).
Author Contributions
X.L., J.L., and X.S., conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; W.L., Y.G., experiment instruction and technique support, final approval of manuscript; Z.B. and J.L., animal model and collection and assembly of data, final approval of manuscript; J.W., Q.S., H.H., and Y.W., collection and/or assembly of data, final approval of manuscript; L.L., and C.W., conception and design, financial support, data interpretation, manuscript writing, final approval of manuscript.
Abbreviations
- AKI
acute kidney injury
- BUN
blood urea nitrogen
- DMEM
dulbecco's modified eagle's medium
- HK2
human kidney cortex/proximal tubule cells
- H/R
hypoxia-reoxygenation
- USCs
urine-derived stem cells
- USC-Exo
exosomes derived from human urine-derived stem cells
- IRAK1
interleukin-1 receptor-associated kinase 1
- IRI
ischemia reperfusion injury
- miRNAs
microRNAs
- NEAA
non-essential amino acid
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- qRT-PCR
quantitative reverse-transcriptase polymerase chain reaction
- sCr
serum creatinine
- SOD
superoxide dismutase activity
- MDA
malonaldehyde
- TUNEL
terminal-deoxynucleoitidyl transferase-mediated nick end labeling
References
- 1.Lameire NH, Bagga A, Cruz D, De Maeseneer J, Endre Z, Kellum JA. et al. Acute kidney injury: an increasing global concern. Lancet. 2013;382:170–9. doi: 10.1016/S0140-6736(13)60647-9. [DOI] [PubMed] [Google Scholar]
- 2.Sharfuddin AA, Molitoris BA. Pathophysiology of ischemic acute kidney injury. Nat Rev Nephrol. 2011;7:189–200. doi: 10.1038/nrneph.2011.16. [DOI] [PubMed] [Google Scholar]
- 3.Salvadori M, Rosso G, Bertoni E. Update on ischemia-reperfusion injury in kidney transplantation: Pathogenesis and treatment. World J Transplant. 2015;5:52–67. doi: 10.5500/wjt.v5.i2.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malek M, Nematbakhsh M. Renal ischemia/reperfusion injury; from pathophysiology to treatment. J Renal Inj Prev. 2015;4:20–7. doi: 10.12861/jrip.2015.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rovcanin B, Medic B, Kocic G, Cebovic T, Ristic M, Prostran M. Molecular Dissection of Renal Ischemia-Reperfusion: Oxidative Stress and Cellular Events. Curr Med Chem. 2016;23:1965–80. doi: 10.2174/0929867323666160112122858. [DOI] [PubMed] [Google Scholar]
- 6.Kellum JA, Prowle JR. Paradigms of acute kidney injury in the intensive care setting. Nat Rev Nephrol. 2018;14:217–30. doi: 10.1038/nrneph.2017.184. [DOI] [PubMed] [Google Scholar]
- 7.Guo C, Dong G, Liang X, Dong Z. Epigenetic regulation in AKI and kidney repair: mechanisms and therapeutic implications. Nat Rev Nephrol. 2019;15:220–39. doi: 10.1038/s41581-018-0103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rota C, Morigi M, Imberti B. Stem Cell Therapies in Kidney Diseases: Progress and Challenges. Int J Mol Sci. 2019. 20. [DOI] [PMC free article] [PubMed]
- 9.Patschan D, Buschmann I, Ritter O, Kribben A. Cell-Based Therapies in Acute Kidney Injury (AKI) Kidney Blood Press Res. 2018;43:673–81. doi: 10.1159/000489624. [DOI] [PubMed] [Google Scholar]
- 10.Shen WC, Chou YH, Huang HP, Sheen JF, Hung SC, Chen HF. Induced pluripotent stem cell-derived endothelial progenitor cells attenuate ischemic acute kidney injury and cardiac dysfunction. Stem Cell Res Ther. 2018;9:344. doi: 10.1186/s13287-018-1092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yun CW, Lee SH. Potential and Therapeutic Efficacy of Cell-based Therapy Using Mesenchymal Stem Cells for Acute/chronic Kidney Disease. Int J Mol Sci. 2019. 20. [DOI] [PMC free article] [PubMed]
- 12.Liu G, Wu R, Yang B, Deng C, Lu X, Walker SJ. et al. Human Urine-Derived Stem Cell Differentiation to Endothelial Cells with Barrier Function and Nitric Oxide Production. STEM CELL TRANSL MED. 2018;7:686–98. doi: 10.1002/sctm.18-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bharadwaj S, Liu GH, Shi YG, Wu RP, Yang B, He TC. et al. Multipotential Differentiation of Human Urine-Derived Stem Cells: Potential for Therapeutic Applications in Urology. Stem Cells. 2013;31:1840–56. doi: 10.1002/stem.1424. [DOI] [PubMed] [Google Scholar]
- 14.Qin DN, Long T, Deng JH, Zhang YY. Urine-derived stem cells for potential use in bladder repair. Stem Cell Res Ther. 2014. 5. [DOI] [PMC free article] [PubMed]
- 15.Chen L, Li L, Xing F, Peng J, Peng K, Wang Y. et al. Human Urine-Derived Stem Cells: Potential for Cell-Based Therapy of Cartilage Defects. Stem Cells Int. 2018;2018:4686259. doi: 10.1155/2018/4686259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan JJ, Niu X, Gong FX, Hu B, Guo SC, Lou YL. et al. Biological characteristics of human-urine-derived stem cells: potential for cell-based therapy in neurology. Tissue Eng Part A. 2014;20:1794–806. doi: 10.1089/ten.tea.2013.0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji X, Wang M, Chen F, Zhou J. Urine-Derived Stem Cells: The Present and the Future. Stem Cells Int. 2017;2017:4378947. doi: 10.1155/2017/4378947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiong G, Tang W, Zhang D, He D, Wei G, Atala A. et al. Impaired Regeneration Potential in Urinary Stem Cells Diagnosed from the Patients with Diabetic Nephropathy. Theranostics. 2019;9:4221–32. doi: 10.7150/thno.34050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu C, Chen L, Huang YZ, Huang Y, Parolini O, Zhong Q. et al. Comparison of the Proliferation and Differentiation Potential of Human Urine-, Placenta Decidua Basalis-, and Bone Marrow-Derived Stem Cells. Stem Cells Int. 2018;2018:7131532. doi: 10.1155/2018/7131532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pavathuparambil Abdul Manaph N, Al-Hawwas M, Bobrovskaya L, Coates PT, Zhou XF. Urine-derived cells for human cell therapy. Stem Cell Res Ther. 2018;9:189. doi: 10.1186/s13287-018-0932-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian SF, Jiang ZZ, Liu YM, Niu X, Hu B, Guo SC. et al. Human urine-derived stem cells contribute to the repair of ischemic acute kidney injury in rats. Mol Med Rep. 2017;16:5541–8. doi: 10.3892/mmr.2017.7240. [DOI] [PubMed] [Google Scholar]
- 22.Han C, Sun X, Liu L, Jiang H, Shen Y, Xu X. et al. Exosomes and Their Therapeutic Potentials of Stem Cells. Stem Cells Int. 2016;2016:7653489. doi: 10.1155/2016/7653489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aghajani Nargesi A, Lerman LO, Eirin A. Mesenchymal stem cell-derived extracellular vesicles for kidney repair: current status and looming challenges. Stem Cell Res Ther. 2017;8:273. doi: 10.1186/s13287-017-0727-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruno S, Grange C, Deregibus MC, Calogero RA, Saviozzi S, Collino F. et al. Mesenchymal stem cell-derived microvesicles protect against acute tubular injury. J Am Soc Nephrol. 2009;20:1053–67. doi: 10.1681/ASN.2008070798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang ZZ, Liu YM, Niu X, Yin JY, Hu B, Guo SC. et al. Exosomes secreted by human urine-derived stem cells could prevent kidney complications from type I diabetes in rats. Stem Cell Res Ther. 2016;7:24. doi: 10.1186/s13287-016-0287-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen CY, Rao SS, Ren L, Hu XK, Tan YJ, Hu Y. et al. Exosomal DMBT1 from human urine-derived stem cells facilitates diabetic wound repair by promoting angiogenesis. Theranostics. 2018;8:1607–23. doi: 10.7150/thno.22958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu Q, Li Q, Niu X, Zhang G, Ling X, Zhang J. et al. Extracellular Vesicles Secreted by Human Urine-Derived Stem Cells Promote Ischemia Repair in a Mouse Model of Hind-Limb Ischemia. Cell Physiol Biochem. 2018;47:1181–92. doi: 10.1159/000490214. [DOI] [PubMed] [Google Scholar]
- 28.Stoorvogel W. Functional transfer of microRNA by exosomes. Blood. 2012;119:646–8. doi: 10.1182/blood-2011-11-389478. [DOI] [PubMed] [Google Scholar]
- 29.Li L, Wang R, Jia Y, Rong R, Xu M, Zhu T. Exosomes Derived From Mesenchymal Stem Cells Ameliorate Renal Ischemic-Reperfusion Injury Through Inhibiting Inflammation and Cell Apoptosis. Front Med (Lausanne) 2019;6:269. doi: 10.3389/fmed.2019.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H, Wang L, Weng X, Chen H, Du Y, Diao C. et al. Inhibition of Brd4 alleviates renal ischemia/reperfusion injury-induced apoptosis and endoplasmic reticulum stress by blocking FoxO4-mediated oxidative stress. Redox Biol. 2019;24:101195. doi: 10.1016/j.redox.2019.101195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai RC, Arslan F, Lee MM, Sze NS, Choo A, Chen TS. et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010;4:214–22. doi: 10.1016/j.scr.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 32.Yuan X, Li D, Chen X, Han C, Xu L, Huang T. et al. Extracellular vesicles from human-induced pluripotent stem cell-derived mesenchymal stromal cells (hiPSC-MSCs) protect against renal ischemia/reperfusion injury via delivering specificity protein (SP1) and transcriptional activating of sphingosine kinase 1 and inhibiting necroptosis. Cell Death Dis. 2017;8:3200. doi: 10.1038/s41419-017-0041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R. et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng L, Zhang K, Wu S, Cui M, Xu T. Focus on Mesenchymal Stem Cell-Derived Exosomes: Opportunities and Challenges in Cell-Free Therapy. Stem Cells Int. 2017;2017:6305295. doi: 10.1155/2017/6305295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee KH, Tseng WC, Yang CY, Tarng DC. The Anti-Inflammatory, Anti-Oxidative, and Anti-Apoptotic Benefits of Stem Cells in Acute Ischemic Kidney Injury. Int J Mol Sci. 2019. 20. [DOI] [PMC free article] [PubMed]
- 36.Bochon B, Kozubska M, Surygala G, Witkowska A, Kuzniewicz R, Grzeszczak W, Mesenchymal Stem Cells-Potential Applications in Kidney Diseases. Int J Mol Sci. 2019. 20. [DOI] [PMC free article] [PubMed]
- 37.Zhang YY, McNeill E, Tian H, Soker S, Andersson KE, Yoo JJ. et al. Urine Derived Cells are a Potential Source for Urological Tissue Reconstruction. J Urology. 2008;180:2226–33. doi: 10.1016/j.juro.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 38.Zhang C, George SK, Wu R, Thakker PU, Abolbashari M, Kim TH. et al. Reno-protection of Urine-derived Stem Cells in A Chronic Kidney Disease Rat Model Induced by Renal Ischemia and Nephrotoxicity. Int J Biol Sci. 2020;16:435–46. doi: 10.7150/ijbs.37550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun B, Luo X, Yang C, Liu P, Yang Y, Dong X. et al. Therapeutic Effects of Human Urine-Derived Stem Cells in a Rat Model of Cisplatin-Induced Acute Kidney Injury In vivo and In vitro. Stem Cells Int. 2019;2019:8035076. doi: 10.1155/2019/8035076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spees JL, Lee RH, Gregory CA. Mechanisms of mesenchymal stem/stromal cell function. Stem Cell Res Ther. 2016;7:125. doi: 10.1186/s13287-016-0363-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bruno S, Grange C, Collino F, Deregibus MC, Cantaluppi V, Biancone L, Microvesicles Derived from Mesenchymal Stem Cells Enhance Survival in a Lethal Model of Acute Kidney Injury. Plos One. 2012. 7. [DOI] [PMC free article] [PubMed]
- 42.He J, Wang Y, Lu XY, Zhu B, Pei XH, Wu JQ. et al. Micro-vesicles derived from bone marrow stem cells protect the kidney both in vivo and in vitro by microRNA-dependent repairing. Nephrology. 2015;20:591–600. doi: 10.1111/nep.12490. [DOI] [PubMed] [Google Scholar]
- 43.Bai M, Zhang L, Fu B, Bai J, Zhang Y, Cai G. et al. IL-17A improves the efficacy of mesenchymal stem cells in ischemic-reperfusion renal injury by increasing Treg percentages by the COX-2/PGE2 pathway. Kidney Int. 2018;93:814–25. doi: 10.1016/j.kint.2017.08.030. [DOI] [PubMed] [Google Scholar]
- 44.Zhang G, Zou X, Huang Y, Wang F, Miao S, Liu G. et al. Mesenchymal Stromal Cell-Derived Extracellular Vesicles Protect Against Acute Kidney Injury Through Anti-Oxidation by Enhancing Nrf2/ARE Activation in Rats. Kidney Blood Press Res. 2016;41:119–28. doi: 10.1159/000443413. [DOI] [PubMed] [Google Scholar]
- 45.Zhang S, Chuah SJ, Lai RC, Hui JHP, Lim SK, Toh WS. MSC exosomes mediate cartilage repair by enhancing proliferation, attenuating apoptosis and modulating immune reactivity. Biomaterials. 2018;156:16–27. doi: 10.1016/j.biomaterials.2017.11.028. [DOI] [PubMed] [Google Scholar]
- 46.Wang B, Yao K, Huuskes BM, Shen HH, Zhuang JL, Godson C. et al. Mesenchymal Stem Cells Deliver Exogenous MicroRNA-let7c via Exosomes to Attenuate Renal Fibrosis. Mol Ther. 2016;24:1290–301. doi: 10.1038/mt.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li YX, Shao LB, Zhang Y, Zhang ZW, Wang JJ, Lan BB, MSC Exosome Mediated Myocardial Repair is Inhibited by Knockdown of Igf-1 Signaling Pathway Through CRISPR/Cas9 Genome Editing. Circulation. 2016. 134.
- 48.Morishita Y, Imai T, Yoshizawa H, Watanabe M, Ishibashi K, Muto S. et al. Delivery of microRNA-146a with polyethylenimine nanoparticles inhibits renal fibrosis in vivo. Int J Nanomed. 2015;10:3475–88. doi: 10.2147/IJN.S82587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ichii O, Otsuka S, Sasaki N, Namiki Y, Hashimoto Y, Kon Y. Altered expression of microRNA miR-146a correlates with the development of chronic renal inflammation. Kidney Int. 2012;81:280–92. doi: 10.1038/ki.2011.345. [DOI] [PubMed] [Google Scholar]
- 50.Amrouche L, Desbuissons G, Rabant M, Sauvaget V, Nguyen C, Benon A. et al. MicroRNA-146a in Human and Experimental Ischemic AKI: CXCL8-Dependent Mechanism of Action. J Am Soc Nephrol. 2017;28:479–93. doi: 10.1681/ASN.2016010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee HW, Khan SQ, Khaliqdina S, Altintas MM, Grahammer F, Zhao JL. et al. Absence of miR-146a in Podocytes Increases Risk of Diabetic Glomerulopathy via Up-regulation of ErbB4 and Notch-1. J BIOL CHEM. 2017;292:732–47. doi: 10.1074/jbc.M116.753822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Godwin JG, Ge X, Stephan K, Jurisch A, Tullius SG, Iacomini J. Identification of a microRNA signature of renal ischemia reperfusion injury. Proc Natl Acad Sci U S A. 2010;107:14339–44. doi: 10.1073/pnas.0912701107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gottipati S, Rao NL, Fung-Leung WP. IRAK1: a critical signaling mediator of innate immunity. Cell Signal. 2008;20:269–76. doi: 10.1016/j.cellsig.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 54.Long JP, Dong LF, Chen FF, Fan YF. miR-146a-5p targets interleukin-1 receptor-associated kinase 1 to inhibit the growth, migration, and invasion of breast cancer cells. Oncol Lett. 2019;17:1573–80. doi: 10.3892/ol.2018.9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lo WY, Peng CT, Wang HJ. MicroRNA-146a-5p Mediates High Glucose-Induced Endothelial Inflammation via Targeting Interleukin-1 Receptor-Associated Kinase 1 Expression. Front Physiol. 2017;8:551. doi: 10.3389/fphys.2017.00551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chatzikyriakidou A, Voulgari PV, Georgiou I, Drosos AA. The role of microRNA-146a (miR-146a) and its target IL-1R-associated kinase (IRAK1) in psoriatic arthritis susceptibility. Scand J Immunol. 2010;71:382–5. doi: 10.1111/j.1365-3083.2010.02381.x. [DOI] [PubMed] [Google Scholar]
- 57.Wang Z, Liu F, Wei M, Qiu Y, Ma C, Shen L. et al. Chronic constriction injury-induced microRNA-146a-5p alleviates neuropathic pain through suppression of IRAK1/TRAF6 signaling pathway. J Neuroinflammation. 2018;15:179. doi: 10.1186/s12974-018-1215-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chu B, Zhou Y, Zhai H, Li L, Sun L, Li Y. The role of microRNA-146a in regulating the expression of IRAK1 in cerebral ischemia-reperfusion injury. Can J Physiol Pharmacol. 2018;96:611–7. doi: 10.1139/cjpp-2017-0586. [DOI] [PubMed] [Google Scholar]
- 59.Jiang W, Kong L, Ni Q, Lu Y, Ding W, Liu G. et al. miR-146a ameliorates liver ischemia/reperfusion injury by suppressing IRAK1 and TRAF6. Plos One. 2014;9:e101530. doi: 10.1371/journal.pone.0101530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chassin C, Hempel C, Stockinger S, Dupont A, Kubler JF, Wedemeyer J. et al. MicroRNA-146a-mediated downregulation of IRAK1 protects mouse and human small intestine against ischemia/reperfusion injury. EMBO Mol Med. 2012;4:1308–19. doi: 10.1002/emmm.201201298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y, Chen X, Yuan L, Zhang Y, Wu J, Guo N. et al. Down-regulation of IRAK1 attenuates podocyte apoptosis in diabetic nephropathy through PI3K/Akt signaling pathway. Biochem Biophys Res Commun. 2018;506:529–35. doi: 10.1016/j.bbrc.2018.09.175. [DOI] [PubMed] [Google Scholar]
- 62.Holterman CE, Boisvert NC, Thibodeau JF, Kamto E, Novakovic M, Abd-Elrahman KS. et al. Podocyte NADPH Oxidase 5 Promotes Renal Inflammation Regulated by the Toll-Like Receptor Pathway. Antioxid Redox Signal. 2019;30:1817–30. doi: 10.1089/ars.2017.7402. [DOI] [PubMed] [Google Scholar]
- 63.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He X, Jing Z, Cheng G. MicroRNAs: new regulators of Toll-like receptor signalling pathways. Biomed Res Int. 2014;2014:945169. doi: 10.1155/2014/945169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Q, Qing Y, Yin C, Li Z, Liu X, Mi Q, Mice with miR-146a deficiency develop severe gouty arthritis via dysregulation of TRAF 6, IRAK 1 and NALP3 inflammasome. ARTHRITIS RES THER. 2018. 20. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures.