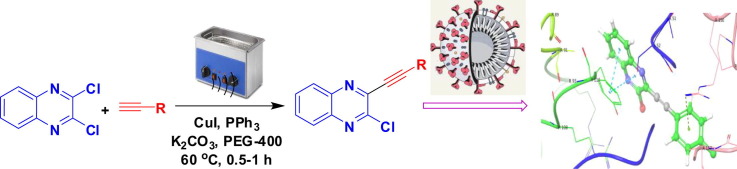
Graphical abstract
We report the in silico studies and ultrasound assisted Cu-catalyzed synthesis of 3-alkynyl substituted 2-chloroquinoxaline derivatives as potential ligands for N-protein of SARS-CoV-2.
Keywords: Chloroquinoxaline, Alkyne, Ultrasound, In silico study, COVID-19
Highlights
-
•
In silico studies were performed using 3-alkynyl 2-chloroquinoxalines.
-
•
These compounds were assessed as ligands for N-protein of SARS-CoV-2.
-
•
Their synthesis involved Cu-catalyzed coupling under ultrasound irradiation.
-
•
In silico studies established a virtual SAR within the series.
-
•
Three compounds appeared as potential agents for further studies.
Abstract
In view of recent global pandemic the 3-alkynyl substituted 2-chloroquinoxaline framework has been explored as a potential template for the design of molecules targeting COVID-19. Initial in silico studies of representative compounds to assess their binding affinities via docking into the N-terminal RNA-binding domain (NTD) of N-protein of SARS-CoV-2 prompted further study of these molecules. Thus building of a small library of molecules based on the said template became essential for this purpose. Accordingly, a convenient and environmentally safer method has been developed for the rapid synthesis of 3-alkynyl substituted 2-chloroquinoxaline derivatives under Cu-catalysis assisted by ultrasound. This simple and straightforward method involved the coupling of 2,3-dichloroquinoxaline with commercially available terminal alkynes in the presence of CuI, PPh3 and K2CO3 in PEG-400. Further in silico studies revealed some remarkable observations and established a virtual SAR (Structure Activity Relationship) within the series. Three compounds appeared as potential agents for further studies.
Being the source of a global pandemic COVID-19 (coronavirus disease 2019) [1], the novel SARS-CoV-2 has already affected the health and economy of several countries severely. Nearly more than 450 thousand people have died throughout the world so far [2] and the number are increasing at a rapid pace. However, there are no promising vaccines and therapeutic drugs available till date to curb the spread of the SARS-CoV-2 worldwide. Thus there is an urgent need to address this global health problem. Multiple studies are in progress, employing diverse approaches to identify effective therapeutics to fight against SARS-CoV-2. For example, in vitro studies have suggested that chloroquine (A, Fig 1 ), an immunomodulant drug traditionally used to treat malaria, might be effective in reducing viral replication in other infections, including the SARS-associated coronavirus (CoV) and MERS-CoV [3], [4]. Its analogue hydroxychloroquine is also being explored as an experimental treatment for COVID-19 [5]. Another drug Favipiravir (T-705, 6-fluoro-3-hydroxypyrazine-2-carboxamide) (B, Fig. 1), an anti-influenza drug which functions to selectively inhibit the RNA- dependent RNA polymerase of influenza virus [6] is being explored for this purpose in Japan. There are several protease inhibitors that are currently in clinical trials for SARS-CoV-2 include Indinavir, Saquinavir, Darunavir, ASC09, Ritonavir and Lopinavir [7].
Fig. 1.

Example of drugs that are being explored against coronavirus.
In 2016 with the goal of finding the potential hit molecules against human coronavirus (CoV-OC43), Chang et al. conducted a molecular docking based virtual screening using nucleocapsid (N)-RNA binding domain as a target protein [8]. The primary function of this protein is to pack the viral RNA within the viral envelope into a ribonucleoprotein (RNP) complex called the capsid, which is a fundamental part of viral self-assembly and replication. Initially, based on docking score eight potential hits were identified, among which a quinoline derivative (C, Fig. 1) was emerged as the most potent compound that was supported by the experimental evidences based on surface plasmon resonance (SPR) analysis. The crystallographic structure of HCoV-OC43 bound with the molecule C was deposited. Very recently, a crystallographic structure of Apo-Nucleocapsid (RNA-binding domain) of SARS-CoV-2/COVID-19 (PDB: 6M3M) has been deposited by Chen et al. [9] Nevertheless, discovery and development of small organic molecule based inhibitor against Nucleocapsid (N) of SARS-CoV-2 is rather uncommon.
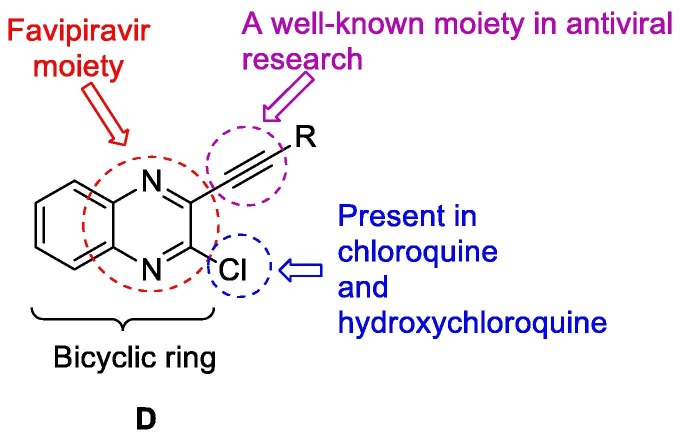
In our effort for the identification of new and potential agents for COVID-19 we planned to evaluate the library of compounds based on N-heterocycles. Accordingly, we focused on compounds based on the template D (Fig 2 ) at the initial stage. We reasoned that the template D containing the chloro group and pyrazine ring of existing experimental agents A and B (Fig. 1) and the alkyne moiety studied in antiviral research earlier [10], [11] would be worthy to explore. Notably, we preferred the bicyclic ring over the monocyclic one because of the fact that the earlier molecule C targeted towards the human coronavirus nucleocapsid protein was also a bicyclic compound. Nevertheless, to substantiate our qualitative reasoning in favor of choosing the template D we performed the related docking studies in silico. Firstly, we tried to compare the N-protein of SARS-CoV-2 with HCoV-OC43 both sequentially and structurally to find out the common regions between them. While a 52% sequence identity was observed in the conserve region based sequence alignment (See Fig. S-6A in ESI) our major interest was to see secondary structure (which reveals structural pattern) common in them. Hence we conducted the secondary structure based sequence alignment (See Fig. S-6B in ESI) using PRALINE web-tool [12] and performed the structural comparison in PyMol [13] (Fig. 3 ). The RMSD was found to be only 0.9 Å whereas the active site residues were appeared to be conserved. While these data clearly established the binding site of SARS-CoV-2 nucleocapsid protein however for further confirmation the computational binding site prediction was conducted by using fconv program [14] where the same binding cavity was found with the volume of 279.31 Å3.
Fig. 2.
The 3-alkynyl substituted 2-chloroquinoxaline as a potential template to target COVID-19.
Fig. 3.
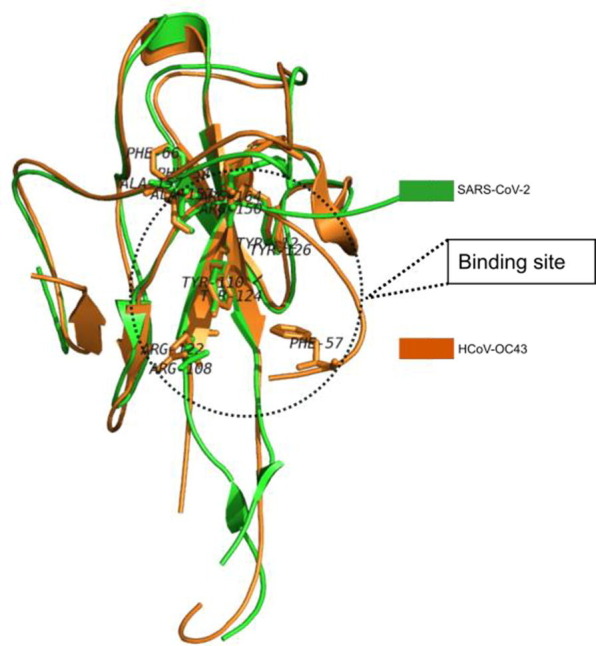
Structural alignment between RNA-binding domain of nucleocapsid protein of SARS-CoV-2 and HCoV-OC43. Active site residues are shown in stick representation.
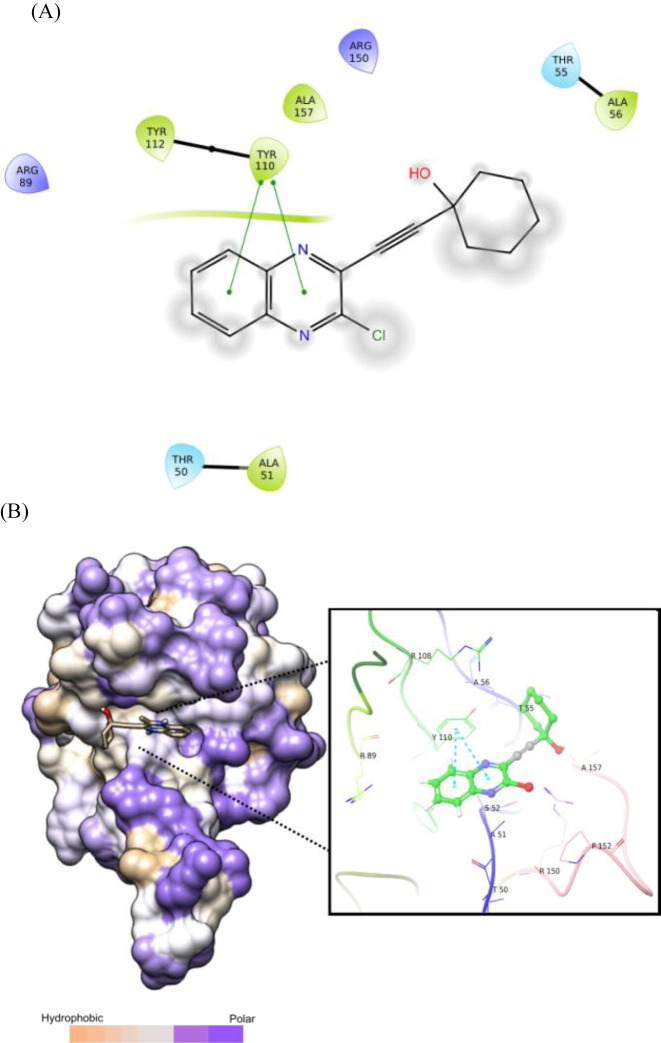
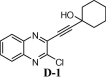
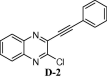
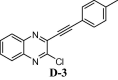
To assess the binding affinity of some representative molecules, e.g. D-1, D-2, D-3 and D-4 (related to the template D) against N protein of SARS-CoV-2, we performed molecular docking study at the nucleotide binding (active) site (PDB: 6M3M). The protein (PDB: 6M3M) as well as all molecules were prepared (e.g. energy optimization, charge calculation and addition of hydrogen etc.) using AutoDock tool [15] and all molecules were docked using reliable open-source tool AutoDock Vina [16]. The binding energy of best pose of each molecule is presented in Table 1 . Notably, for further understanding of the polarity and coulombic electrostatic potential of the binding site, the hydrophobicity and electrostatic surface representation was generated using UCSF Chimera [17] (see the following figures). While a balance between hydrophilic and hydrophobic residues was evident in case of N-protein and the binding site was somewhat hydrophobic in nature however it’s positive charge was essential for nucleotide binding.
Table 1.
Docking of molecules into N-terminal RNA-binding domain (NTD) of N-protein of SARS-CoV-2.a
| Molecules | AutoDock Vina score (Kcal/mol) |
|---|---|
 |
−6.4 |
| D-1 | |
 |
−6.2 |
| D-2 | |
 |
−6.5 |
| D-3 | |
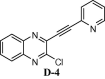 |
−6.1 |
| D-4 | |
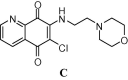 |
−5.6 |
| C |
Docking of each individual molecule was performed for 5 times and maximum difference in score was found to be ±0.2.
Nevertheless, the comparable binding energies of these molecules with the known inhibitor C suggested that compounds based on framework D might interact with the nucleocapsid (N) of SARS-CoV-2 too. Indeed, the quinoline ring of molecule D-1 showed effective pi-pi interactions with residue TYR110 (Fig. 4 A). Additionally, it participated in hydrophobic / van der Waals contacts with residues ALA157, ARG150, THR55, ALA56, ARG89 etc (Fig 4B, see also Fig. S-1 in ESI).
Fig. 4.
(A) 2D interaction diagram between N-terminal RNA-binding domain (NTD) of N-protein of SARS-CoV-2 and compound D-1 (where pi-pi interaction shown in green line), prepared in Maestro visualizer (Schrödinger, LLC). (B) Hydrophobic surface representation along with 3D interaction diagram (where pi-pi interaction shown in cyan dashed line).
However, the most effective interactions were observed in case of molecule D-3 as evident from its binding energy (Table 1). Its ability to form pi-cation interaction with ARG150, pi-pi interactions with TYR110 and comparatively more hydrophobic interactions with residues ARG108, TYR112, PRO152, SER52, ALA51, and ARG89 etc (Fig 5 , see also Fig S-2 in ESI) could be the reason for this observation. The molecule D-2 participated in a pi-cation interaction with ARG150 and a pi-pi stacking with TYR110. In addition, it formed hydrophobic contacts with other residues like ARG108, TYR112, PRO152, SER52, ALA51 etc (see the Supplementary data). Similar interactions were observed in case of molecule D-4. Notably, the reference compound C showed relatively low docking score perhaps due to the lack of proper aromaticity thereby related pi-interactions. Nevertheless, the molecule was involved in hydrophobic contacts mainly with residues such as ALA157, ARG150, THR55, ALA56, ARG89, TYR110 etc (see the Supplementary data).
Fig. 5.
(A) 2D interaction diagram between compound D-3 and N-protein of SARS-CoV-2. (B) Electrostatic surface representation followed by 3D interaction diagram.
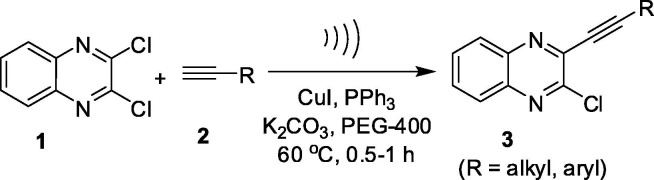
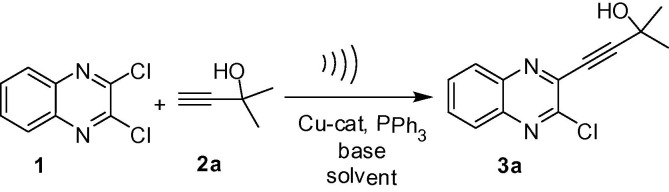
The alkynyl substituted quinoxaline or related derivatives have been prepared via Sonogashira type coupling under Pd(0)-Cu catalysis in the presence of Et3N or Et2NH [18], [19], [20], [21], [22]. In our effort the synthesis of 3-alkynyl substituted 2-chloroquinoxaline has been carried out via a selective mono alkynylation of 2,3-dichloroquinoxaline (1) earlier [23], [24]. The reactions were carried out using 10%Pd/C-CuI-PPh3 as a catalyst system, Et3N as a base and EtOH as a solvent and the duration was 2–4 h. However, all these methodologies involved the use of bi-metallic salts as catalysts and environmentally harmful alkylamine as a base. Moreover, the organic solvents used in some of these cases are not environmentally friendly. All these concerns prompted us to explore a more convenient and environmentally safer method for the synthesis of compounds based on D (Fig. 2) including D-1, D-2, D-3 and D-4 (Table 1). Thus the 3-alkynyl substituted 2-chloroquinoxaline derivatives (3) were synthesized via a Cu-catalyzed coupling of 2,3-dichloroquinoxaline (1) with commercially available terminal alkynes (2) under ultrasound irradiation (Scheme 1 ). The methodology involved the use of PEG-400 as a solvent under mild reaction conditions and does not require the use of any co-catalyst. We now present the details of this study.
Scheme 1.
Ultrasound assisted synthesis of 3-alkynyl substituted 2-chloroquinoxalines (3) under Cu-catalysis.
Over the years the ultrasound-assisted reactions have emerged as one of the popular approaches in organic synthesis which is evident from a wide range of applications of these methodologies both in academia and industrial organizations [25]. The features and advantages of these reactions include their (i) involvement as green approaches in organic synthesis [26], (ii) reduction of waste generation as well as energy requirements [27] and (iii) efficiency and effectiveness for the synthesis of desired products via employing shorter reaction time and milder conditions at the same time increasing the product yields [28], [29]. The PEG-400 on the other hand being a high boiling, non-hazardous and polar solvent has found applications in various reactions. Indeed, due to its easy recovery from the reaction mixture and recyclability PEG-400 is considered as an environmentally friendly solvent [30]. As part of our ongoing effort in the use of ultrasound as an alternative source of energy and PEG-400 as a greener solvent in various organic reactions we became interested in exploring these reaction conditions in our current endeavor. Notably, while the CuI/PPh3-catalyzed Sonogashira coupling of iodoarene with terminal alkynes (i) in PEG-water under microwave heating or reflux in oil bath [31] (at 120 °C) or (ii) in water [32] (at 120 °C) or (iii) under biphasic conditions [33] (water/organic substrates) have been reported earlier, a similar coupling of chloroheteroarene with terminal alkynes under ultrasound is not known.
In order to find the optimized reaction conditions for the coupling of 2,3-dichloroquinoxaline (1) with the alkyne i.e. 2-methylbut-3-yn-2-ol (2a) was examined under a range of reaction conditions and the results are summarized in Table 2 . The reaction proceeded well when carried out using 10 mol% CuI as a catalyst, 30 mol% PPh3 as a ligand and K2CO3 as a base in PEG-400 under ultrasound using a laboratory ultrasonic bath SONOREX SUPER RK 510H model producing irradiation of 35 kHz (entry 1, Table 2). However, the desired coupled product 3a was obtained in 54% yield after 4 h. The increase of CuI loading from 10 mol% to 15 mol% improved the product yield significantly and the reaction was completed within 1 h (entry 2, Table 2). Encouraged by this observation we continued our study for possibility of further improvement in product yield. Thus the quantity of CuI used was increased further from 15 mol% to 20 mol% but no significant increase in yield of 3a was observed (entry 3, Table 2). The use of other solvent e.g. EtOH or n-BuOH (entry 4 and 5, Table 2) in place of PEG-400 or other base e.g. Et3N (entry 6, Table 2) in place of K2CO3 did not improve the product yield. The use of pure water as a solvent was not successful as the partial hydrolysis of 1 was observed under the conditions employed. We also examined the use of other Cu-catalysts e.g. CuBr or CuCl in the present coupling reaction but these were found to be less effective (entry 7 and 8, Table 2). Notably, the C—C coupling did not proceed in the absence of a catalyst (entry 9, Table 2) indicating key role played by the Cu-salt in the current alkynylation method. Moreover, though the reaction proceeded in the absence of ligand PPh3 the product yield was not particularly high (entry 10, Table 2). The reaction was also found to be less efficient in terms of product yield when carried out in the absence of ultrasound (entry 11, Table 2). While the reaction temperature was maintained at 50 °C during all reactions as mentioned above the decrease and increase of temperature was examined. The reaction did not proceed at lower temperature e.g. at 40 °C and no improvement in product yield was observed at higher temperature e.g. at 70 °C though the reaction progressed well at this temperature. Overall, the condition of entry 2 of Table 2 (i.e. the combination of CuI, PPh3 and K2CO3 in PEG-400 at 50 °C under ultrasound) appeared to be optimum and was used for the preparation of analogues of 3a.
Table 2.
| Entry | Cu-cat (mol%) | Base | Solvent | Time (h) | Yieldb |
|---|---|---|---|---|---|
| 1. | CuI (10) | K2CO3 | PEG-400 | 4 | 54 |
| 2. | CuI (15) | K2CO3 | PEG-400 | 1 | 73 |
| 3. | CuI (20) | K2CO3 | PEG-400 | 1 | 75 |
| 4. | CuI (15) | K2CO3 | EtOH | 1 | 66 |
| 5. | CuI (15) | K2CO3 | n-BuOH | 1 | 62 |
| 6. | CuI (15) | Et3N | PEG-400 | 4 | 61 |
| 7. | CuBr (15) | K2CO3 | PEG-400 | 4 | 49 |
| 8. | CuCl (15) | K2CO3 | PEG-400 | 4 | 37 |
| 9. | No catalyst | K2CO3 | PEG-400 | 4 | No reaction |
| 10. | CuI (15) | K2CO3 | PEG-400 | 1 | 60c |
| 11. | CuI (15) | K2CO3 | PEG-400 | 4 | 43d |
All reactions were carried out using the chloro compound 1 (1 equiv.), alkyne 2a (1 equiv.), a Cu-catalyst, PPh3 (30 mol%) and base (2 equiv.) in a solvent (5.0 mL) at 50 °C under ultrasound irradiation.
Isolated yields.
The reaction was performed in absence of PPh3
The reaction was performed in the absence of ultrasound.
A range of terminal alkynes (2) were employed to couple with the chloro compound (1) under the optimized conditions. The alkyne may contain a primary, secondary or tertiary hydroxyl group or an aliphatic chain such as n-butyl, n-pentyl, n-hexyl etc or an aryl or heteroaryl group. The Cu-catalyzed C—C bond forming reaction proceeded smoothly in all these cases affording the desired coupled product in good to acceptable yield (Table 3 ). It is worthy to mention that in none of the cases the product yield was high due to the partial dimerization of the terminal alkyne used. It is well known that dimerization of terminal alkynes to the corresponding diyne is often a side reaction under the Sonogashira coupling conditions and Cu-salts play a key role in such oxidative homocoupling (Glaser coupling) of terminal alkynes [34], [35], [36]. Additionally, the formation of bis-alkynylated product in some cases (particularly in case of 3b, 3c, 3g etc) albeit in trace quantity perhaps affected the yield of desired monoalkynylated product. Generally, to avoid the formation of unwanted bis-alkynylated product the use of reactant alkyne (2) was restricted to 1 equivalent. However, a slow evaporation of the corresponding terminal alkyne i.e. 3,3-dimethylbut-1-yne (due to the low boiling point i.e. 37–38 °C) was observed under the reaction conditions employed in case of 3b and hence the use of higher quantity of alkyne (1.5 equiv.) was necessary in this case. Nevertheless, all the compounds were characterized by using common spectral (1H and 13C NMR and Mass) data (See ESI).
Table 3.
Synthesis of 3-alkynyl substituted 2-chloroquinoxaline derivatives (3)a,b (Scheme 1).
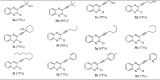 |
aAll reactions were carried out using the chloro compound 1 (1 equiv.), alkyne 2 (1 equiv.), CuI (15 mol%), PPh3 (30 mol%) and K2CO3 (2 equiv.) in PEG-400 (5.0 mL) at 50 °C under ultrasound irradiation.
bFigure in the bracket represents isolated yield.
c1.5 equivalent of alkyne (3,3-dimethylbut-1-yne) was used in this case.
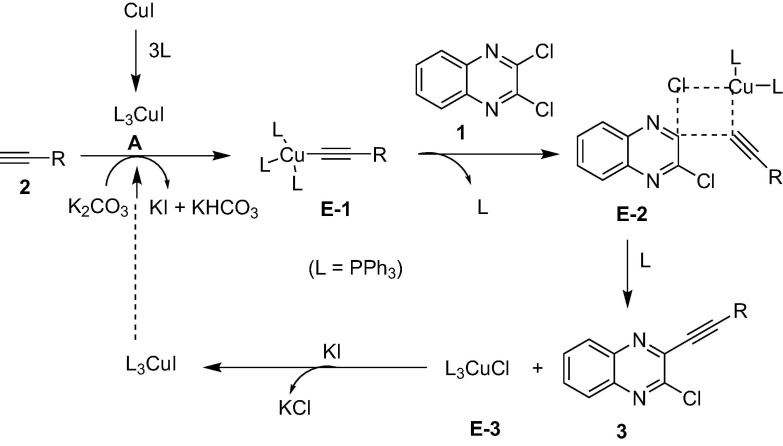
Based on the results of Table 2 and the earlier reports [31], [32], [33] a proposed reaction mechanism for the Cu-catalyzed coupling of 1 with 2 under ultrasound irradiation is presented in Scheme 2 . Initially, a Cu(I) complex (A) was formed via the interaction of CuI with the ligand PPh3 under ultrasound irradiation. Indeed, the complex A was the actual catalytic species that catalyzed the present C—C bond forming reaction. On interaction with the terminal alkyne (2) in the presence of K2CO3 the complex A afforded the acetylide intermediate E-1 with the generation of KI. Subsequently, a copper cluster linked with both alkyne as well as heteroarene moiety (E-2) was formed via the interaction of E-1 with the chloro compound (1). The Cu-complex E-2 then furnished the desired mono-alkynylated product 3 along with the regeneration of the catalyst A (via E-3 in the presence of KI) thereby completing the catalytic cycle. The results of Table 2 suggested that the present Cu-catalyzed coupling reaction was accelerated greatly by the ultrasound irradiation. It is known that the compression of the liquid and then rarefaction (expansion) caused by ultrasound results in a sudden pressure drop that forms small, oscillating bubbles of gaseous substances. Subsequently, with each cycle of the applied ultrasonic energy these bubbles continue to expand till they reach to an unstable size. At this stage these cavitation bubbles can collide and/or collapse violently which can cause the increase of local temperature within the reaction medium. As a result of this the crossing of energy barrier is facilitated [37] that allows the faster conversion of reactants to intermediates and subsequently to product(s) within short reaction time. The acceleration of various steps of Scheme 2 by ultrasound in this way explains the rapid formation of product 3 from the chloro compound (1) and alkyne (2).
Scheme 2.
Proposed reaction mechanism for the Cu-catalyzed coupling of 1 with 2 under ultrasound irradiation.
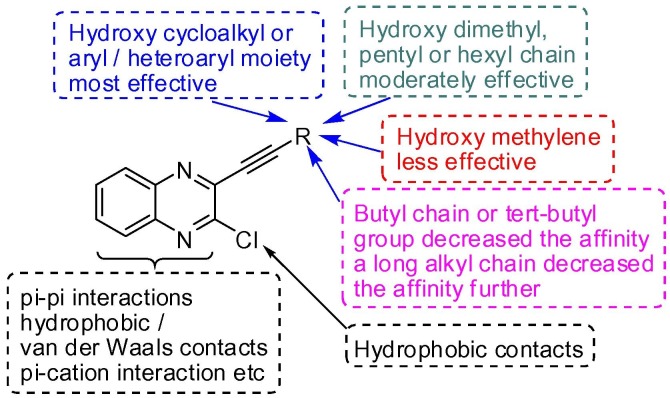
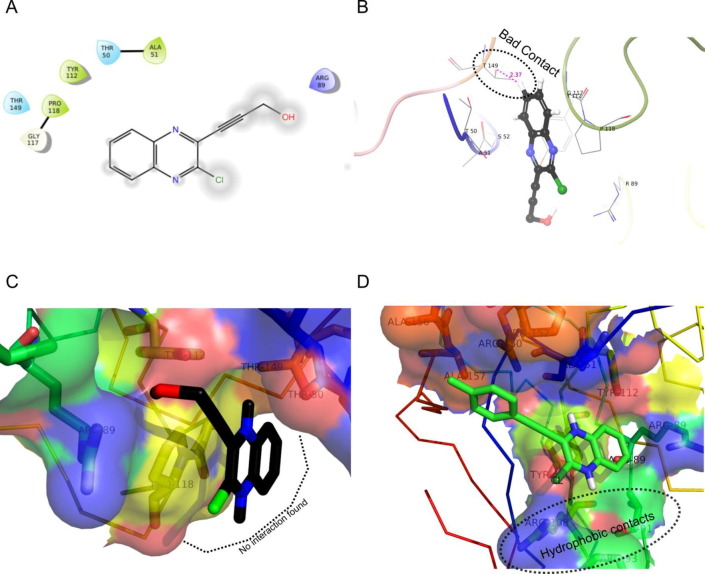
Having synthesized a small library of molecules (3) based on D (Fig. 2) we re-focused on assessing their binding affinities via docking into the N-terminal RNA-binding domain (NTD) of N-protein of SARS-CoV-2 (PDB: 6M3M). The purpose of this study was to gain some insights regarding the virtual SAR (Structure-Activity-Relationship) within this series of compounds. The results are presented in Table 4 including that of Table 1 for head-to-head comparison and discussion. It was evident that most of the compounds showed good to acceptable or moderate binding affinities (>5.0 Kcal/mol) except the compound 3c. The nature and size of the substituent (other than the chloroquinoxalin moiety) attached to the alkyne moiety appeared to have played a key role in binding with the protein in silico (Fig 6 ). For example, the in silico study of compound 3c (Fig 7 A-B) suggested flipping of the molecule to adopt a different orientation (aided by the high affinity of its hydroxyl group towards ARG89) that resulted in losing of several hydrophobic contacts (Fig 7C) in compared to that of 3k (Fig 7D). Notably, these hydrophobic contacts contributed significantly towards overall binding affinity of the most effective compound 3k. Secondly, a steric clash (termed as bad contact, see Fig 7B) observed between a non-polar H atom of 3c and the residue THR149 at a distance of 2.37 Å. Nevertheless, generally a bulky hydroxy cycloalkyl group (3e) or aryl/heteroaryl moiety (3j-l) was found to be beneficial for binding because of favorable hydrophobic and/or pi-interactions. Among rest of the molecules those possessing the hydroxy dimethyl group (3b) and pentyl or hexyl chain (3g and 3h) were found to be moderately effective. The affinity was decreased further in case of butyl chain (e.g. 3f) and tert-butyl group (e.g. 3a). The relatively less number of pi-interactions in all these cases (only one in each case) could be the reason (see Fig. S-3A and B in ESI). Notably, a too long alkyl chain (e.g. 3i) was found to be somewhat less effective due to a bad steric clash observed between a non-polar H atom of compound 3i and TYR 112 at a distance of 2.46 Å (see Fig. S-3C and D in ESI). While the role of quinoxalin ring has been discussed earlier however the role of its chloro group was not highlighted. Indeed, the chloro group was prominently involved in hydrophobic contacts mainly with residues that were in close proximity e.g. TYR110, ARG108, ARG93 and ALA56 etc (see Fig. S-4 in ESI). Nevertheless, the compound 3e, 3j and 3k appeared as potential agents for further studies.
Table 4.
Docking of molecules into N-terminal RNA-binding domain (NTD) of N-protein of SARS-CoV-2.a
Fig. 6.
Summary of in silico binding affinities of compound 3.
Fig. 7.
(A) The 2D and (B) 3D interaction diagram of compound 3c with N-protein of SARS-CoV-2. (C) The surface representation of docked 3c into the N-protein of SARS-CoV-2 where no interaction found at the bottom site. (D) The surface representation of docked 3k into the N-protein of SARS-CoV-2 showing hydrophobic contacts at the bottom site.
In conclusion, the 3-alkynyl substituted 2-chloroquinoxaline framework has been explored as a potential template for the design of molecules targeting COVID-19. Initially few representative molecules were evaluated in silico via assessing their binding affinities against the N-terminal RNA-binding domain (NTD) of N-protein of SARS-CoV-2. The encouraging outcome of this study prompted further evaluation of these molecules. Thus building of a small library of molecules based on the said template was undertaken. Accordingly, a convenient and environmentally safer method has been developed based on Cu-catalyzed C—C bond forming reaction under ultrasound irradiation. The methodology allowed a rapid synthesis of 3-alkynyl substituted 2-chloroquinoxaline via the coupling of 2,3-dichloroquinoxaline with commercially available terminal alkynes in the presence of CuI, PPh3 and K2CO3 in PEG-400. This simple and straightforward method does not involve the use of bi-metallic salts as catalysts and afforded a range of target compounds smoothly. All these compounds were assessed in silico to establish a virtual SAR (Structure Activity Relationship) within the series. Future study will focus more on validation of identified compounds in vitro and in vivo. To sum up, the current report not only described an ultrasound assisted rapid and greener approach towards 3-alkynyl substituted 2-chloroquinoxaline derivatives under Cu-catalysis but also revealed a potential template for the design and identification of new agents for fighting against COVID-19.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors thank the management of Dr. Reddy’s Institute of Life Sciences, Hyderabad, India, for continuous support and encouragement.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tetlet.2020.152336.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronavirus (COVID-19); World Health Organization; https://who.sprinklr.com/, accessed on May 7, 2020.
- 3.Savarino A., Boelaert J.R., Cassone A., Majori G., Cauda R. Effects of chloroquine on viral infections: an old drug against today’s diseases? Lancet Infect. Dis. 2003;3:722–727. doi: 10.1016/S1473-3099(03)00806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colson P., Rolain J.-M., Raoult D. Chloroquine for the 2019 novel coronavirus SARS-CoV-2. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cortegiani A., Ingoglia G., Ippolito M., Giarratano A., Einav S. A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J. Critical Care. 2020 doi: 10.1016/j.jcrc.2020.03.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furuta Y., Gowen B.B., Takahashi K., Shiraki K., Smee D.F., Barnard D.L. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antivir. Res. 2013;1002:446–454. doi: 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrison C. Coronavirus puts drug repurposing on the fast track. Nature Biotechnol. 2020;38:379–381. doi: 10.1038/d41587-020-00003-1. [DOI] [PubMed] [Google Scholar]
- 8.Chang C.K., Jeyachandran S., Hu N.J., Liu C.L., Lin S.Y., Wang Y.S., Chang Y.-M., Hou M.H. Structure-based virtual screening and experimental validation of the discovery of inhibitors targeted towards the human coronavirus nucleocapsid protein. Mol. BioSystems. 2016;12:59–66. doi: 10.1039/c5mb00582e. [DOI] [PubMed] [Google Scholar]
- 9.S. Chen, S. Kang. 6M3M: Structural insights of SARS-CoV-2 nucleocapsid protein RNA binding domain reveal potential unique drug targeting sites. RSCB PDB; doi: 10.2210/pdb6m3m/pdb. [DOI] [PMC free article] [PubMed]
- 10.De Clercq E., Cools M., Balzarini J., Snoeck R., Andrei G., Hosoya M., Shigeta S., Ueda T., Minakawa N., Matsuda A. Antiviral activities of 5-ethynyl-1-beta-D-ribofuranosylimidazole-4-carboxamide and related compounds. Antimicrob. Agents Chemother. 1991;35:679–684. doi: 10.1128/aac.35.4.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shealy Y.F., O'Dell C.A., Arnett G., Shannon W.M. Synthesis and antiviral activity of the carbocyclic analogs of 5-ethyl-2'-deoxyuridine and of 5-ethynyl-2'-deoxyuridine. J. Med. Chem. 1986;29:79–84. doi: 10.1021/jm00151a013. [DOI] [PubMed] [Google Scholar]
- 12.V.A. Simossis, J. Heringa. PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information, Nucleic Acids Res. 2005, 33, W289–W294 (Issue suppl_2); https://doi.org/10.1093/nar/gki390. [DOI] [PMC free article] [PubMed]
- 13.DeLano W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsletter Protein Crystal. 2002;40:82–92. [Google Scholar]
- 14.Neudert G., Klebe G. fconv: format conversion, manipulation and feature computation of molecular data. Bioinformatics. 2011;27:1021–1022. doi: 10.1093/bioinformatics/btr055. [DOI] [PubMed] [Google Scholar]
- 15.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity. J. Comput. Chem. 2009;16:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 18.M. Armengol, J.A. Joule. Synthesis of thieno[2,3-b]quinoxalines and pyrrolo[1,2-a]quinoxalines from 2-haloquinoxalines. J. Chem. Soc., Perkin Trans. 1, 2001, 978-984; doi: 10.1039/B101458G.
- 19.Armengol M., Joule J.A. Synthesis of thieno[2,3-b]quinoxalines from 2-haloquinoxalines. J. Chem. Soc., Perkin Trans. 1. 2001:154–158. [Google Scholar]
- 20.Ames D.E., Broch M.I. Alkynyl- and dialkynyl-quinoxalines. Synthesis of condensed quinoxalines. J. Chem. Soc. Perkins Trans. 1. 1980:1384–1389. doi: 10.1039/P19800001384. [DOI] [Google Scholar]
- 21.Arcadi A., Cacchi S., Fabrizi G., Parisi L.M. 2,3-Disubstituted pyrrolo[2,3-b]quinoxalines via aminopalladation-reductive elimination. Tetrahedron Lett. 2004;45:2431–2434. [Google Scholar]
- 22.Nxumalo W., Dinsmore A. Preparation of 6-ethynylpteridine derivatives by sonogashira coupling. Heterocycles. 2013;87:79. doi: 10.3987/com-12-12610. [DOI] [Google Scholar]
- 23.Nakhi A., Rahman M.S., Seerapu G.P.K., Banote R.K., Kumar K.L., Kulkarni P., Haldar D., Pal M. Transition metal free hydrolysis/cyclization strategy in a single pot: synthesis of fused furo N-heterocycles of pharmacological interest. Org. Biomol. Chem. 2013;11:4930–4934. doi: 10.1039/c3ob41069b. [DOI] [PubMed] [Google Scholar]
- 24.Nakhi A., Rahman M.S., Kishore R., Meda C.L.T., Deora G.S., Parsa K.V.L., Pal M. Pyrrolo[2,3-b]quinoxalines as inhibitors of firefly luciferase: Their Cu-mediated synthesis and evaluation as false positives in a reporter gene assay. Bioorg. Med. Chem. Lett. 2012;22:6433–6441. doi: 10.1016/j.bmcl.2012.08.056. [DOI] [PubMed] [Google Scholar]
- 25.Mason T.J. Ultrasound in synthetic organic chemistry. Chem. Soc. Rev. 1997;26:443–451. [Google Scholar]
- 26.R. Cella, H.A. Stefani. Ultrasonic Reactions, in Green Techniques for Organic Synthesis and Medicinal Chemistry (eds W. Zhang and B. W. Cue), John Wiley & Sons, Ltd, Chichester, UK. doi: 10.1002/9780470711828.ch13, 2012.
- 27.L. Pizzuti, M.S.F. Franco, A.F.C. Flores, F.H. Quina, C.M.P. Pereira. Recent Advances in the Ultrasound-Assisted Synthesis of Azoles, Green Chem. - Environ. Benign Approaches. Kidwai, M.; Mishra, N. K. IntechOpen 2012; doi:10.5772/35171.
- 28.Puri S., Kaur B., Parmar A., Kumar H. Applications of ultrasound in organic synthesis - a green approach. Curr. Org. Chem. 2013;17:1790–1828. [Google Scholar]
- 29.Reddy D.N.K., Chandrasekhar K.B., Siva Ganesh Y.S., Reddy G.R., Kumar J.P., Kapavarapu R.K., Pal M. FeF3-catalyzed MCR in PEG-400: ultrasound assisted synthesis of N-substituted 2-aminopyridines. RSC Adv. 2016;6:67212–67217. doi: 10.1039/c6ra14228a. [DOI] [Google Scholar]
- 30.Chen J., Spear S.K., Huddleston J.G., Rogers R.D. Polyethylene glycol and solutions of polyethylene glycol as green reaction media. Green Chem. 2005;7:64–82. [Google Scholar]
- 31.Chen G., Xie J., Weng J., Zhu X., Zheng Z., Cai J., Wan Y. CuI/PPh3/PEG–water: an efficient catalytic system for cross-coupling reaction of aryl iodides and alkynes. Synth. Commun. 2011;41:3123–3133. doi: 10.1080/00397911.2010.517363. [DOI] [Google Scholar]
- 32.Guan J.T., Yu G.-A., Chen L., Weng T.Q., Yuan J.J., Liu S.H. CuI/PPh3-catalyzed Sonogashira coupling reaction of aryl iodides with terminal alkynes in water in the absence of palladium. Appl. Organometal. Chem. 2009;23:75–77. doi: 10.1002/aoc.1474. [DOI] [Google Scholar]
- 33.Liu Y., Blanchard V., Danoun G., Zhang Z., Tlili A., Zhang W., Monnier F., Lee A.V.D., Mao J., Taillefer M. Copper-catalyzed Sonogashira reaction in water. Chem. Select. 2017;2:11599–11602. doi: 10.1002/slct.201702854. [DOI] [Google Scholar]
- 34.Krause N., Thorand S. J. Org. Chem. 1998;63:8551–8553. [Google Scholar]
- 35.Rosenblum S.B., Huynh T., Afonso A., Davis Jr H.R. Synthesis of 3-arylpropenyl, 3-arylpropynyl and 3-arylpropyl 2-azetidinones as cholesterol absorption inhibitors: application of the palladium-catalyzed arylation of alkenes and alkynes. Tetrahedron. 2000;56:5735–5742. [Google Scholar]
- 36.Brimble M.A., Pavia G.S., Stevenson R.J. A facile synthesis of aryldihydropyrans using a Sonogashira–selenoetherification strategy. Tetrahedron Lett. 2002;43:1735–1738. [Google Scholar]
- 37.Suslick K.S., Hammerton D.A., Cline R.E. Sonochemical hot spot. J. Am. Chem. Soc. 1986;108:5641–5642. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.