Abstract
During unfavorable conditions (e.g. tumor hypoxia or viral infection), canonical, cap-dependent mRNA translation is suppressed in human cells. Nonetheless, a subset of physiologically important mRNAs (e.g. hypoxia-inducible factor 1α [HIF-1α], fibroblast growth factor 9 [FGF-9], and p53) is still translated by an unknown, cap-independent mechanism. Additionally, expression levels of eukaryotic translation initiation factor 4GI (eIF4GI) and of its homolog, death-associated protein 5 (DAP5), are elevated. By examining the 5′ UTRs of HIF-1α, FGF-9, and p53 mRNAs and using fluorescence anisotropy binding studies, luciferase reporter-based in vitro translation assays, and mutational analyses, we demonstrate here that eIF4GI and DAP5 specifically bind to the 5′ UTRs of these cap-independently translated mRNAs. Surprisingly, we found that the eIF4E-binding domain of eIF4GI increases not only the binding affinity but also the selectivity among these mRNAs. We further demonstrate that the affinities of eIF4GI and DAP5 binding to these 5′ UTRs correlate with the efficiency with which these factors drive cap-independent translation of these mRNAs. Integrating the results of our binding and translation assays, we conclude that eIF4GI or DAP5 is critical for recruitment of a specific subset of mRNAs to the ribosome, providing mechanistic insight into their cap-independent translation.
Keywords: 5′ cap-independent translation enhancer (CITE), internal ribosome entry site (IRES), eukaryotic translation initiation factor 4 GI (eIF4GI), death-associated protein 5 (DAP5), fluorescence anisotropy, protein synthesis, gene expression, cell stress, hypoxia-inducible factor 1α (HIF-1α), eukaryotic translation initiation factor 4G (eIF4G), protein-nucleic acid interaction, gene regulation, mRNA, hypoxia-inducible factor (HIF), cap-independent translation
Translation of mRNAs into proteins is the most energy-consuming process in the cell (1, 2) and plays a major role in the regulation of gene expression. In eukaryotes, initiation of cellular mRNA translation generally occurs via a cap-dependent pathway in which eukaryotic initiation factor 4E (eIF4E) binds to the N7-methylguanosine-triphosphate (m7GpppN, where N is any nucleotide) cap at the 5′ end of the mRNA to be translated (3, 4). Cap-bound eIF4E subsequently recruits eIF4G, which, together with eIF4A, recruits a ribosomal 43S preinitiation complex (PIC) composed of the 40S ribosomal subunit, a methionylated initiator tRNA (Met-tRNAiMet), and additional eIFs to the m7GpppN cap. Subsequent scanning of the resulting 48S PIC to find the AUG start codon on the mRNA and joining of the 60S ribosomal subunit to the 48S PIC results in the formation of an elongation-competent 80S IC that can go on to translate the mRNA.
In addition to undergoing cap-dependent initiation, many cellular mRNAs can also initiate translation via cap-independent pathways in response to changes in cellular conditions (5). The ability of these mRNAs to switch from cap-dependent to cap-independent modes of translation initiation plays an important role in maintaining normal cellular physiology (6) as well as in the cellular response to diseases such as cancer, diabetes, and, possibly, neurological disorders (7–12). A subset of cellular mRNAs, for example, has been shown to successfully bypass a global suppression of translation initiation that is caused by stress conditions such as tumor hypoxia, viral infection, and nutrient deprivation (3, 5, 13) and that is driven by the sequestration of eIF4E by hypophosphorylated 4E-binding proteins (4E-BPs). Whereas translation initiation of these mRNAs under these conditions is often referred to as cap independent, it may be more accurately described as eIF4E independent. Nonetheless, we will use the term cap-independent here to refer to translation initiation that does not involve eIF4E-based recruitment of other eIFs to the m7GpppN cap. Many of the stress conditions that result in the suppression of cap-dependent initiation also result in increased expression levels of eIF4GI (7, 8) and/or death-associated protein 5 (DAP5) (also called p97, NAT1, or eIF4G2) (14, 15), suggesting that these two proteins are involved in a cap-independent initiation mechanism(s).
The subset of mRNAs that is translated cap independently under stress conditions as described above are predicted to contain highly stable structures in their 5′ untranslated regions (UTRs) that may act as internal ribosome entry sites (IRES) or cap-independent translation enhancers (CITEs) (5, 16). IRES-like mechanisms involve direct recruitment of the ribosome to structured IRES that are close to the AUG start codon (5, 17), whereas CITE-like mechanisms involve direct recruitment of eIFs to structured CITEs near the 5′ end of the mRNA, where the eIFs are then thought to initiate cap-independent translation via 48S PIC scanning to the AUG start codon (18, 19). Although chemical and enzymatic probing of cellular mRNAs thought to contain IRESs/CITEs have revealed stem loops, pseudoknots, and other structures (20, 21), no common sequence or structural motifs have been identified to allow prediction of cellular IRES/CITEs from mRNA sequence data. Regardless of whether their structured 5′ UTRs act as IRESs or CITEs, the ability of these mRNAs to bypass the 4E-BP-mediated global suppression of cap-dependent initiation has been linked to enhanced tumor development and cancer progression (2).
The overexpression of eIF4GI and DAP5 during stress conditions in which global cap-dependent translation initiation is suppressed implicates these two factors in cap-independent translation. DAP5 is a member of the eIF4G family that is homologous to the C-terminal two-thirds of eIF4GI (Fig. 1A) and that interacts with other known eIFs in manners that are both similar to and distinct from that of eIF4GI. Specifically, DAP5 and eIF4GI share 39% sequence identity in the central core region that comprises the eIF4A-, eIF3-, and RNA-binding domains (22) (Fig. 1B). Consistent with its similarity to the domain structure of eIF4GI, DAP5 interacts with eIF4A and the eIF3 component of the 43S PIC (22). Notably, however, DAP5 lacks the N-terminal eIF4E- and polyadenine binding protein (PABP)-binding domains that are present in eIF4GI and, consequently, does not interact with eIF4E or PABP (Fig. 1A). In addition, the β subunit of eIF2 (eIF2β) that interacts with the ternary complex formed by eIF2, GTP, and Met-tRNAi (23) binds to the C-terminal domain of DAP5 but not to the corresponding domain in eIF4GI. Collectively, these differences between DAP5 and eIF4GI suggest differences in translation initiation mechanisms involving these two eIFs (14). For example, rather than interacting with eIF4E to recruit factors to the m7GpppN cap in cap-dependent initiation of mRNAs, DAP5 has instead been shown to mediate the cap-independent initiation of a subset of mRNAs, including those encoding Bcl2, Apaf-1, p53, and DAP5 itself (24, 25). With the exception of p53 mRNA, which contains a structural element that functions as an IRES and has been shown to bind DAP5 using electrophoretic mobility shift assay studies (24), there have not yet been any studies aimed at investigating whether mRNAs translated cap independently directly recruit eIF4GI and/or DAP5 to the 5′ UTRs and how such recruitments might drive the switch from cap-dependent to cap-independent initiation in response to cellular stress.
Figure 1.
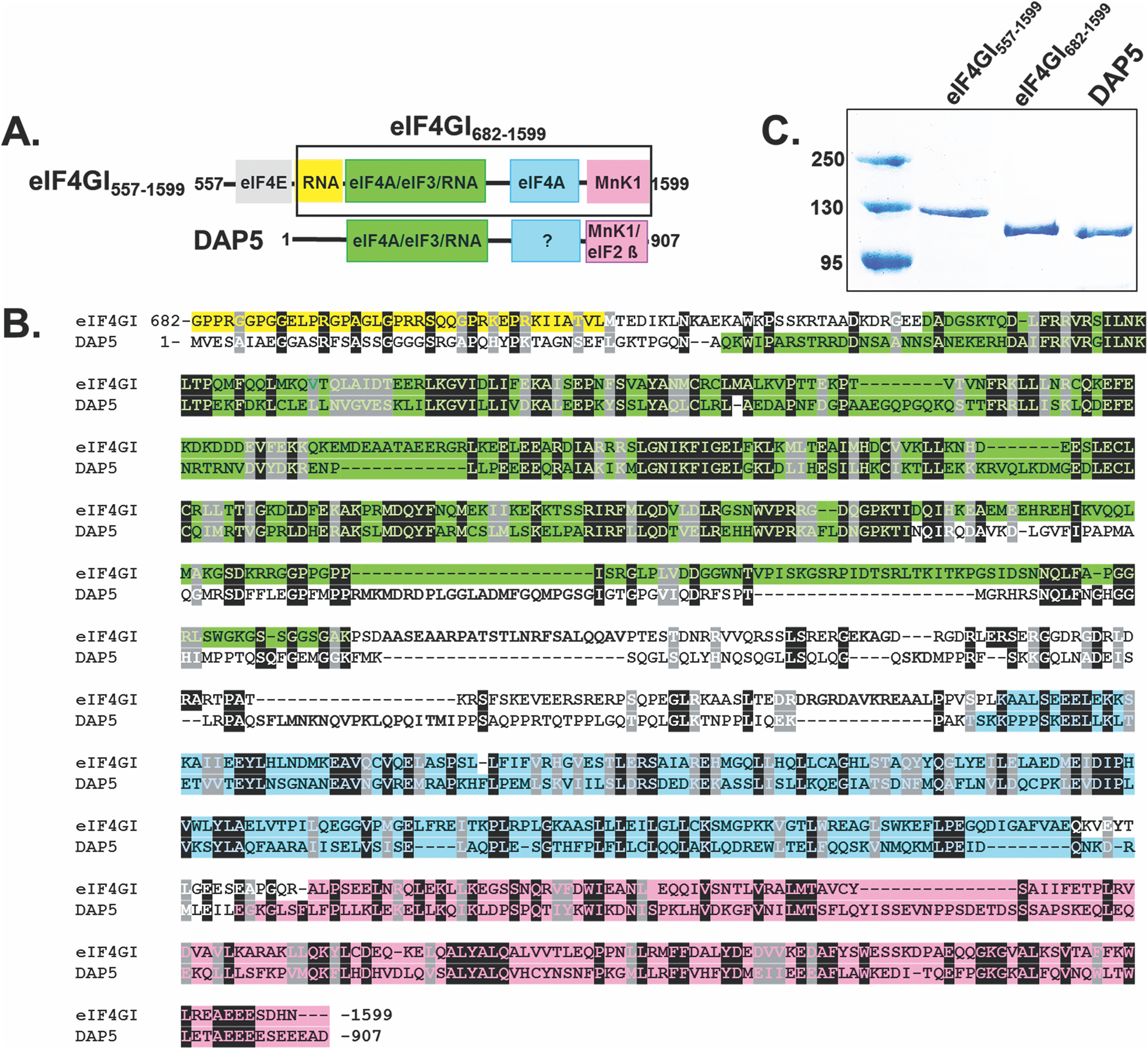
Domain structure and sequence alignment between eIF4GI and DAP5. A, cartoons showing the domain architecture of eIF4GI557-1599 and full-length DAP5. eIF4GI contains a second eIF4A binding region in the MA3 domain (blue box in the eIF4GI cartoon), but the corresponding domain in DAP5 (blue box with question mark) does not bind to eIF4A and has unknown function (22). The shorter construct of eIF4GI, eIF4GI682-1599, is highlighted in the black box. B, sequence alignment encompassing similar domains in eIF4GI682-1599 and DAP5. The sequence alignment was performed using T-coffee (61). Residues are color-coded according to their domain organization shown in panel A. C, a 10% SDS-PAGE gel showing purity of eIF4GI557-1599, eIF4GI682-1599, and DAP5 used for this study.
To address these gaps in our understanding, we used a fluorescence anisotropy-based equilibrium binding assay to measure the affinities with which two variants of human eIF4GI, one that lacks the N-terminal, eIF4E-binding domain (eIF4GI682-1599) and one that contains the eIF4E binding domain (eIF4GI557-1599), as well as full-length human DAP5, bind to RNA oligonucleotides corresponding to the 5′ UTRs of a representative set of mRNAs. These mRNAs bypass (2) the 4E-BP-mediated global suppression of cap-dependent initiation and encode HIF-1α, FGF-9, and p53. These mRNAs were chosen because HIF-1α and FGF-9 have been shown to be translationally upregulated in hypoxic conditions where cap-dependent translation is suppressed and where eIF4GI and 4EBPI levels are overexpressed (2, 26). p53 mRNAs were chosen because these mRNAs are known to interact with DAP5 as part of a cap-independent translation mechanism (24). Complementing these binding assays, a luciferase-based gene expression reporter assay was used to characterize whether and to what extent binding of eIF4GI682-1599, eIF4GI557-1599, and/or DAP5 promotes the translation of luciferase-encoding mRNAs containing these same 5′ UTRs (UTR-Luc mRNAs) in a rabbit reticulocyte lysate-based cap-dependent in vitro translation system. We further used this assay with UTR-Luc mRNA constructs that either lack or have a highly stable, engineered hairpin (stem-loop) near the 5′ cap of the 5′ UTR that blocks initiation and scanning from the 5′ end. These experiments have further allowed us to investigate whether these mRNAs use an IRES-like or CITE-like mechanism of translation initiation. The results of our experiments demonstrate that eIF4GI682-1599, eIF4GI557-1599, and DAP5 exhibit differential binding affinities to the 5′ UTRs of the mRNAs encoding HIF-1α, FGF-9, and p53; that the eIF4E-binding domain of eIF4GI confers additional binding affinity and specificity; and that binding affinity positively correlates with the abilities of these eIFs to drive translation of these same UTR-Luc mRNAs. Moreover, the inhibition, or lack thereof, of translation initiation by the engineered hairpin suggests that some of these mRNAs use an IRES-like mechanism of translation initiation that does not require an exposed 5′ end, whereas others use a CITE-like mechanism (27). Based on our observations, we propose that unlike cap-dependent translation, where binding of eIF4E to the m7G-cap is the initial event in protein synthesis, an important initial event in cap-independent initiation of eukaryotic cellular mRNAs with structured 5′ UTRs is the binding of eIF4GI or DAP5. These 5′ UTR structured mRNAs selectively recruit eIF4GI or DAP5 to drive expression of the proteins they encode, similar to the cap-independent initiation of CITE- or IRES-containing viral mRNAs (28–30).
Results
eIF4GI and DAP5 bind specifically and with differential affinities to the 5′ UTRs of a subset of cellular mRNAs
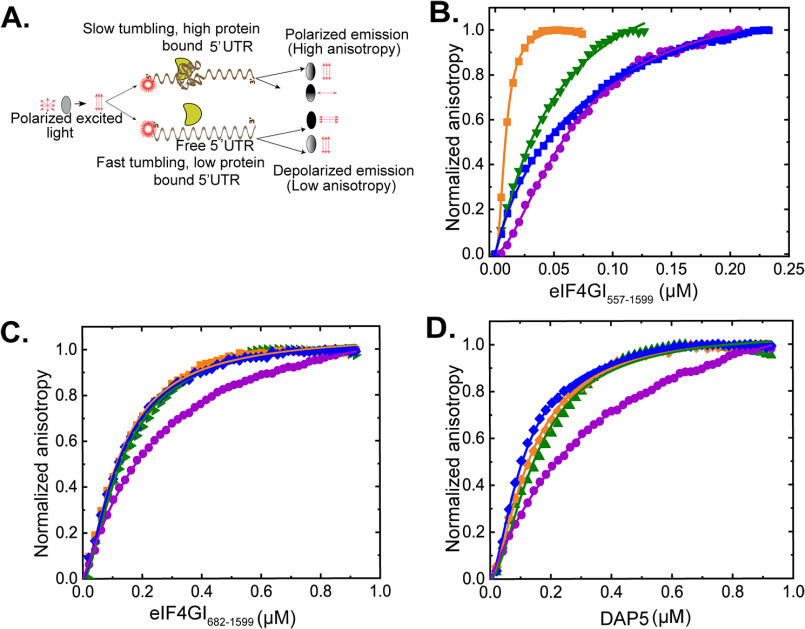
To characterize the binding of eIF4GI and DAP5 to the 5′ UTRs of the mRNAs encoding HIF-1α, FGF-9, and p53, we used a fluorescence anisotropy-based equilibrium binding assay developed in our laboratories (Fig. 2A). This assay produces binding curves from fluorescence anisotropy changes that arise as titrated proteins bind to RNA oligonucleotides that are covalently 5′-end labeled with fluorescein and that lack a m7GpppN cap. Four RNA oligonucleotides comprising the 5′ UTRs of mRNAs encoding HIF-1α, FGF-9, and the two 5′ UTRs of p53 were assayed. The two p53 5′ UTRs represent the two 5′ UTRs involved in translation of the two distinct isoforms of p53. Each p53 5′ UTR contains elements that select for translation initiation at either an upstream start codon (p53A) or a downstream start codon (p53B), which produce the full-length p53 (FL-p53) or an N-terminal-truncated isoform (ΔN-p53), respectively (31). When eIF4GI682-1599, eIF4GI557-1599, or DAP5 binds to one of the fluorescein-labeled 5′ UTRs, there is an increase in molecular weight of the 5′ UTR that increases the rotational restriction as the system becomes more rigid, resulting in a decrease in the rotational speed of the fluorescein-labeled 5′ UTR. The slower rotation is observed as increased fluorescence anisotropy of the fluorescein reporter. To quantify the affinity of eIF4GI557-1599, eIF4GI557-1599, and DAP5 binding to the four 5′ UTRs, we recorded the change in the fluorescence anisotropy of each fluorescein-labeled 5′ UTR as a function of increasing concentrations of eIF4GI682-1599, eIF4GI557-1599, and DAP5 and fitted the resulting data points with a single-site, equilibrium-binding isotherm (Fig. 2B, C, and D). The results of these experiments (Table 1) demonstrate that eIF4GI682-1599, eIF4GI557-1599, and DAP5 bind to the four 5′ UTRs with equilibrium dissociation constants (Kds) ranging from 12–290 nm (Table 1).
Figure 2.
Equilibrium binding titrations of 5′ UTRs with eIF4GI557-1599, eIF4GI682-1599, and DAP5. A, cartoon showing the fluorescence anisotropy-based equilibrium binding assay. Normalized anisotropy changes for the interaction of fluorescein-labeled FGF-9 (γ), HIF-1α (▾), p53A (●), and p53B (♦) uncapped 5′ UTRs, with eIF4GI557-1599 (B), eIF4GI682-1599 (C), and DAP5 (D). Briefly, 10 nm (B) or 100 nm (C and D) fluorescein-labeled uncapped RNA oligonucleotides were titrated with increasing concentrations of eIF4GI557-1599, eIF4GI682-1599, or DAP5 in the titration buffer at 25 °C, and the anisotropy at each titration point was measured using excitation and emission wavelengths of 495 nm and 520 nm, respectively. Data points correspond to the average from three independent anisotropy measurements, and the curves represent the nonlinear fits that were used to obtain the averages and standard deviations for the corresponding Kd values presented in Table 1.
Table 1.
Parameters describing the equilibrium binding of eIF4GI constructs and DAP5 to the 5′ UTRsa
| 5′ UTR | eIF4GI557-1599 |
eIF4GI682-1599 |
DAP5 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Kd ± S.D. (nm) | Amp. (rmax–rmin) | χ2 | Kd ± S.D. (nm) | Amp. (rmax–rmin) | χ2 | Kd ± S.D. (nm) | Amp. (rmax–rmin) | χ2 | |
| HIF1α | 50 ± 6.0 | 0.045 | 0.996 | 139 ± 23.0 | 0.034 | 0.994 | 154 ± 18.0 | 0.061 | 0.988 |
| FGF-9 | 12 ± 2.0 | 0.079 | 0.995 | 120 ± 7.0 | 0.027 | 0.996 | 136 ± 10.0 | 0.039 | 0.998 |
| p53A | 86 ± 4.0 | 0.021 | 0.998 | 248 ± 16.0 | 0.036 | 0.999 | 290 ± 14.0 | 0.044 | 0.999 |
| p53B | 68 ± 19.0 | 0.025 | 0.998 | 126 ± 4.0 | 0.017 | 0.998 | 112 ± 6.0 | 0.048 | 0.998 |
a Kd is the equilibrium dissociation constant, rmax–rmin is the amplitude (Amp.) that indicates change in anisotropy, and χ2 represents the goodness of fit.
Comparative analyses of our results demonstrate that eIF4GI682-1599, eIF4GI557-1599, and DAP5 exhibit differential binding affinities among the 5′ UTRs. Specifically, eIF4GI682-1599 binds to the p53A 5′ UTR with a Kd that is ∼2-fold higher (i.e. binding that is ∼2-fold weaker) than that with which it binds to the other 5′ UTRs (Table 1). DAP5 exhibited an even greater difference in binding affinity among the 5′ UTRs, with a Kd for the p53B 5′ UTR that is more than 2.5-fold lower than the Kd for the p53A 5′ UTR. It is important to note, however, that the trend of the differences in binding affinities between the two translation factors (eIF4GI682-1599 and DAP5) for the 5′ UTRs is similar. However, dramatic differences in binding among the 5′ UTRs were observed for eIF4GI557-1599. Inclusion of the eIF4E-binding domain in eIF4GI557-1599 relative to eIF4GI682-1599 increased the binding affinity of eIF4GI557-1599 from 1.8-fold (p53B 5′ UTR) to 10-fold (FGF-9 5′ UTR) compared with those measured for eIF4GI682-1599. Further, the differences in binding affinity among the 5′ UTRs were as much as 7-fold different compared with an ∼2-fold difference among the 5′ UTRs for eIF4GI682-1599. Taken together, these results suggest much of the specificity of eIF4GI binding to the 5′ UTRs is conferred by the eIF4E-binding domain (i.e. residues 557–682) of eIF4GI.
The binding of eIF4GI and DAP5 is specific to the 5′ UTRs of the selected mRNAs
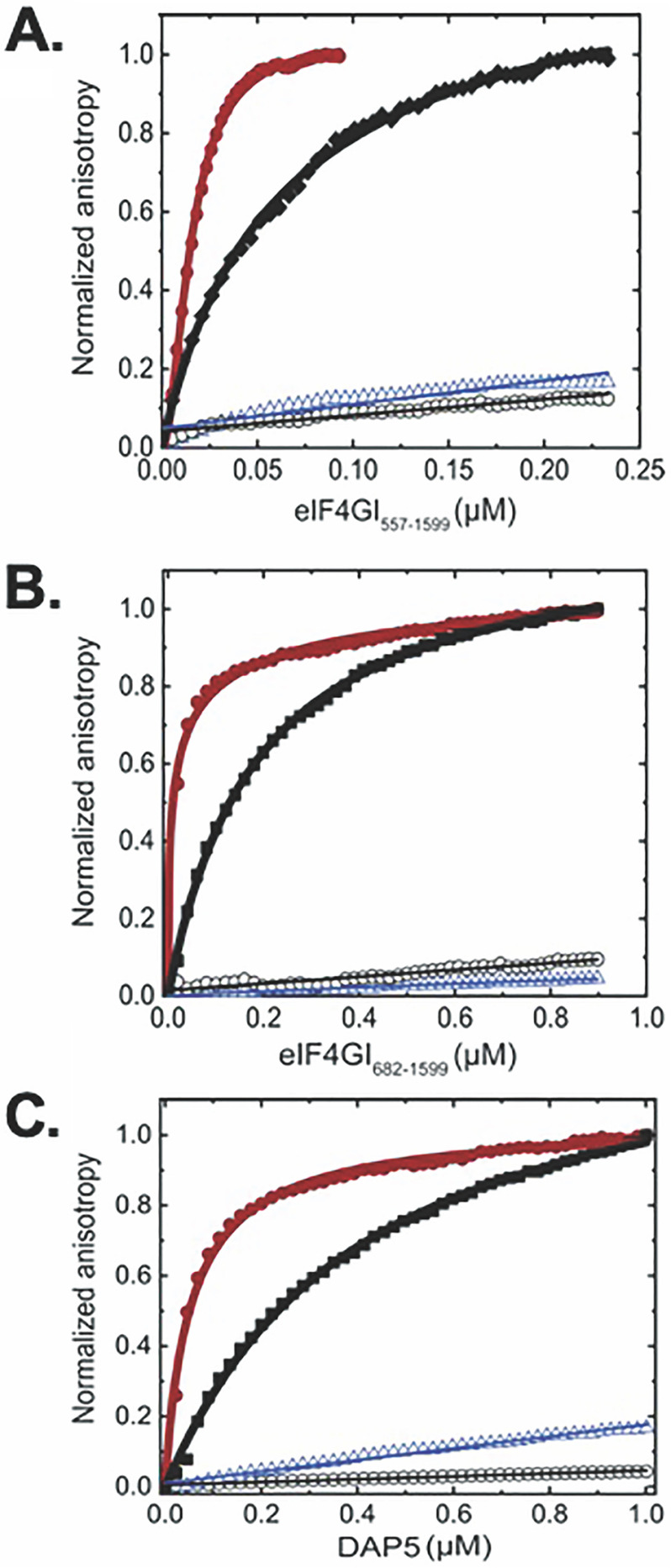
To further investigate whether binding of eIF4GI and DAP5 to RNA depends on structural features within the RNA and to contextualize and validate the results of the binding studies described above, we assessed the binding of eIF4GI682-1599, eIF4GI557-1599, and DAP5 to a presumably unstructured, 101-nucleotide poly(UC) RNA oligonucleotide and an oligonucleotide encompassing the 5′ UTR of the mRNA encoding β-actin, which has been reported to utilize a cap-dependent mechanism for translation initiation (32). The results of these experiments demonstrate that eIF4GI682-1599, eIF4GI557-1599, and DAP5 do not exhibit appreciable binding to the polyUC oligonucleotide (Fig. 3A, B, and C and Table S1) or to the β-actin 5′ UTR, findings consistent with the hypothesis that structural features within the 5′ UTRs of our subset of RNAs act as specific recognition elements and binding sites for eIF4GI and DAP5.
Figure 3.
Equilibrium binding titrations of fluorescein-labeled ferritin IRE (●), EMCV J/K IRES (γ), β-actin UTR (○), and polyUC (∇) binding to eIF4GI557-1599 (A), eIF4GI682-1599 (B), and DAP5 (C). Briefly, 10 nm (A) or 100 nm (B and C) fluorescein-labeled uncapped RNA oligonucleotides were titrated with increasing concentrations of eIF4GI557-1599, eIF4GI682-1599, or DAP5 in the titration buffer at 25 °C, and the anisotropy at each titration point was measured using excitation and emission wavelengths of 495 nm and 520 nm, respectively. Averages from 3 independent experiments were performed, and the curves represent the nonlinear fits that were used to obtain the averages and standard deviations for the corresponding Kd values given in Table S1.
Previously, we have shown that eIF4F (a complex composed of eIF4GI, eIF4A, and eIF4E) binds to the 30-nucleotide iron-responsive element (IRE) stem-loop within the 5′ UTR of the mRNA encoding ferritin with a Kd of 9 nm, a binding interaction that stimulates ferritin mRNA translation in response to elevated cellular concentrations of iron (33). Using the fluorescence anisotropy-based equilibrium binding assay, here we show that eIF4GI682-1599, eIF4GI557-1599, and DAP5 bind to the IRE with Kds of 18 nm, 15 nm, and 35 nm, respectively (Fig. 3 and Table S1). Similarly, we performed experiments in which we quantified the affinity of eIF4GI682-1599, eIF4GI557-1599, and DAP5 for the J/K domain of the IRES in the 5′ UTR of the positive-strand genomic RNA, encoding the encephalomyocarditis virus (EMCV) polyprotein (Fig. 3 and Table S1). These experiments show that eIF4GI682-1599, eIF4GI557-1599, and DAP5 bind to the EMCV J/K IRES with Kds of 175 nm, 63 nm, and 519 nm, respectively. The Kd for binding of eIF4GI682-1599 to EMCV J/K IRES RNA is similar to what has been previously reported for the binding of human eIF4GI (643–1076) to the EMCV J/K IRES using electrophoretic mobility shift assay (170 nm [34]) as part of the mechanism through which eIF4GI drives expression of the EMCV polyprotein in human cells (35). The same report showed that a construct containing an N-terminal portion of DAP5 (62–330) did not bind to the EMCV J/K IRES, consistent with our much higher Kd (519 nm) for full-length DAP5 than for eIF4GI construct binding.
The 5′ UTRs of a subset of cellular mRNAs that bind eIF4GI and DAP5 can drive cap-independent translation
Having demonstrated that eIF4GI and DAP5 bind specifically and with relatively high affinity to the 5′ UTRs of HIF-1α, FGF-9, p53A, and p53B, we examined the cap-independent translation of a set of reporters containing these 5′ UTRs. To quantify the cap-independent activities of our selected mRNAs, the 5′ UTRs of the HIF-1α, FGF-9, p53A, and p53B mRNAs and, as a control, the 5′ UTR of β-actin mRNA were cloned upstream of a luciferase reporter gene (Fig. 4A). The resulting capped, polyadenylated UTR-Luc mRNAs were translated using a nuclease-treated, rabbit reticulocyte (RRL)-based, cap-dependent, in vitro translation system containing a natural abundance of the eIFs. To assess cap-independent luciferase expression, a nonfunctional cap analog (ApppG) was used to cap the UTR-Luc mRNAs (here referred to as the ApppG-capped transcripts). The cap-independent expression levels of the ApppG-capped transcripts were compared with the expression observed when these transcripts were capped with a functional m7GpppA cap. We observed that the activities of the ApppG-capped transcripts ranged from ∼70% for FGF-9 UTR-Luc mRNA to 8% for p53B UTR-Luc mRNA compared with their corresponding m7GpppA-capped transcripts (Fig. S1). Expression of the ApppG-capped β-act-UTR-Luc mRNA was reduced to less than 2% of the corresponding m7GpppA-capped construct (Fig. S1A). These results further support the notion that the 5′ UTR of a subset of cellular mRNAs can drive cap-independent translation, albeit at a relatively lower efficiency than cap-dependent translation. During stressful conditions in which cap-dependent translation is inhibited and the expression of eIF4GI and DAP5 are elevated, expression from these 5′ UTRs can help cells mitigate the effects of the stressor (2).
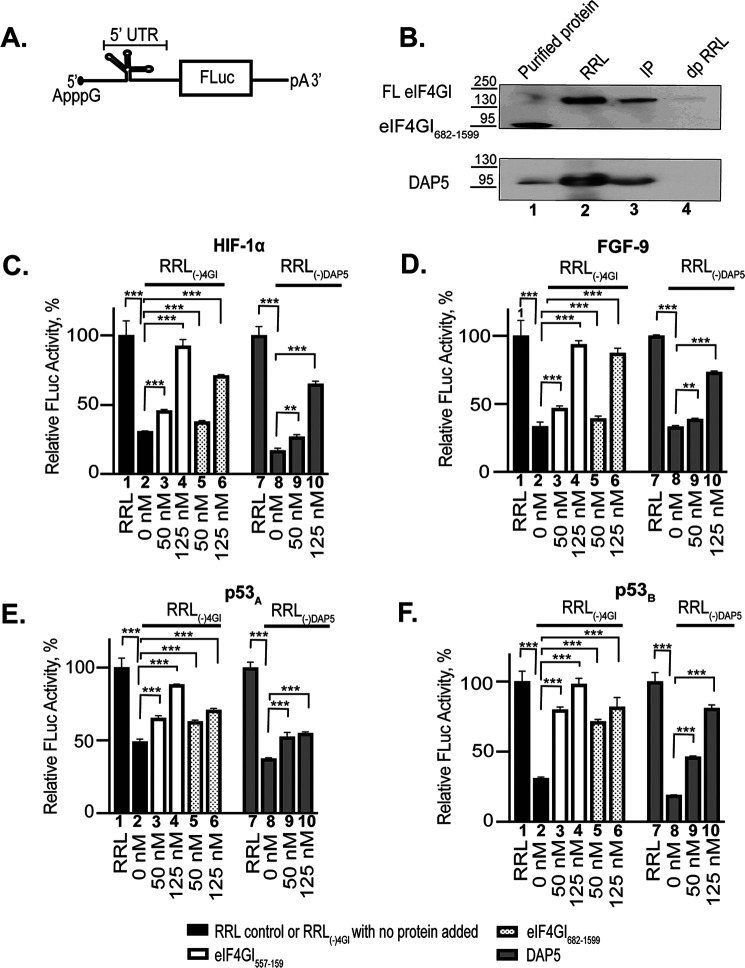
Figure 4.
Effect of eIF4GI557-1599, eIF4GI682-1599, and DAP5 on the expression of ApppG-capped-UTR-Luc mRNAs. A, cartoon showing the design of ApppG-capped-UTR-Luc mRNA reporter constructs containing a structural element upstream of the firefly luciferase gene. B, Western blotting of eIF4GI- and DAP5-depleted RRL probed using specific eIF4GI and DAP5 antibodies. Purified recombinant eIF4GI682-1599 or DAP5 protein used for the study were included as positive controls (lane 1). The endogenous eIF4GI detected in RRL is full length (FL eIF4GI). Lane 2 shows nondepleted RRL; lane 3, immunoprecipitate pulled down by antibodies for either eIF4GI (top) or DAP5 (lower). dpRRL is the depleted RRL [RRL(-)4GI or RRL(-)DAP5]. Lysates were run on 4–12% SDS gradient gels and transferred to a nitrocellulose membrane. The effect of increasing concentrations of eIF4GI557-1599, eIF4GI682-1599, and DAP5 on the translation yields of ApppG-capped transcripts of HIF-1α (C), FGF-9 (D), p53A (E), and p53B (F) ApppG-capped UTR-Luc mRNAs. Relative luciferase activity was measured in RRL (bar 1 or bar 7) or RRL(-)4GI or RRL(-)DAP5 (bar 2 or bar 8) in the presence of increasing concentrations of either eIF4GI557-1599 (bar 3–4), eIF4GI682-1599 (bar 5–6), and DAP5 (bar 9–10). Relative luciferase activity was normalized to the respective controls (ApppG-capped UTR-Luc mRNA) for each reporter performed in nondepleted RRL. Bar heights and error bars correspond to the average and standard deviations, respectively, from three independent luciferase activity measurements. Data were analyzed by two-tailed unpaired Student's t test: **, p = 0.002; ***, p < 0.001.
The eIF4E–eIF4GI interaction allows recruitment of the eIF4F complex to the m7G cap of mRNAs (36). 4EGI-1 is a small molecule that binds to the same site on eIF4E that interacts with eIF4GI and inhibits the interaction of eIF4GI and eIF4E (37, 38). To further support the idea that translation initiation of these mRNAs was independent of eIF4E, we determined the extent to which translation of the m7GpppA- and ApppG-capped transcripts was affected by 4EGI-1. Cap-dependent initiation was significantly suppressed through the addition of 4EGI-1 to m7GpppA-capped β-act-UTR-Luc mRNA but had no effect on the ApppG-capped transcripts of our selected 5′ UTRs, with the exception of the ApppG-capped HIF-1α transcript, which exhibited an ∼44% reduction in translation (Fig. S2). These results show that, to a large extent, translation of the ApppG-capped transcripts proceeds via an eIF4E-independent mechanism. The observation that the ApppG-capped HIF-1α transcript expression is partially reduced suggests that eIF4E does not function exclusively during cap-dependent translation initiation but also plays an important role in the cap-independent translation initiation of some mRNAs. For example, eIF4E has been shown to stimulate the helicase activity of eIF4A in the case of some highly structured 5′ UTRs (39), such as HIF-1α, and/or to induce conformational changes in eIF4G that enhance its binding to mRNAs (40).
eIF4GI and DAP5 stimulate and restore cap-independent in vitro translation of the selected 5′ UTR-Luc mRNAs
Having established that eIF4GI and DAP5 bind specifically and with relatively high affinity to the HIF-1α, FGF-9, p53A, and p53B 5′ UTRs and that the corresponding UTR-Luc mRNAs are translated through a cap-independent pathway, we sought to establish the extent to which this cap-independent translation initiation of these mRNAs depended on eIF4GI and DAP5. To accomplish this, we used our luciferase-based reporter assay and first determined the effects on translation of depletion of either eIF4GI or DAP5. We measured the luciferase activity produced by each ApppG-capped transcript in an RRL that had been depleted of either endogenous eIF4GI [RRL(-)4GI] or DAP5 [RRL(-)DAP5] using antibodies directed against these proteins. Western blots confirmed the successful depletion of eIF4GI or DAP5 (Fig. 4B) from the RRLs. As a control to test for the indirect depletion of other eIFs, eIF4E, eIF2β, and eIF4A levels were tested in RRL(-)4GI or RRL(-)DAP5 and were found to be similar to, or only slightly reduced from, those seen in RRL (Fig. S3). Using either RRL(-)4GI or RRL(-)DAP5, we measured the luciferase activity produced by each translation reaction of the ApppG-capped transcript and compared it to RRL translation assays of the same transcript without depletion of endogenous eIF4GI or DAP5. To confirm the eIF4GI or DAP5 dependence on the translation initiation of these UTR-Luc mRNAs, we added increasing concentrations of exogenous eIF4GI557-1599, eIF4GI682-1599, or DAP5 to the corresponding depleted RRL. Translation of UTR-Luc mRNAs was significantly reduced in the depleted RRLs compared with those of the RRL control (Fig. 4, C–F). Specifically, the translation output of HIF-1α, FGF-9, p53A, and p53B ApppG-capped UTR-Luc mRNAs were reduced by 70%, 67%, 51%, and 70%, respectively, in RRL(-)4GI (Fig. 4, C–F). Addition of eIF4GI557-1599 or eIF4GI682-1599 significantly rescued and stimulated the cap-independent translation initiation of all four UTR-Luc mRNAs (Fig. 4, C–F). For RRL(-)4GI, eIF4GI557-1599 restored translation of the ApppG-capped transcripts to ∼85–100% of the levels observed in RRL, whereas eIF4GI682-1599 was slightly less effective, restoring levels to ∼70–90% of the RRL levels for the same transcripts (Fig. 4, C–F). In the case of RRL(-)DAP5, the translation outputs of HIF-1α, FGF-9, p53A, and p53B ApppG-capped UTR-Luc mRNAs were reduced by 83%, 67%, 63%, and 81%, respectively. Addition of DAP5 was also able to rescue cap-independent translation initiation of all four UTR-Luc mRNAs, although with somewhat less efficiency than the eIF4GI constructs. DAP5 restored translation to ∼45–65% of the levels for the same transcripts in RRL (Fig. 4, C–F). These data follow the same overall trend as our fluorescence anisotropy-based binding assay data, where eIF4GI557-1599 showed the highest binding affinity and DAP5 the lowest affinity to the 5′ UTRs (Table 1).
Further demonstrating the cap dependence of translation, the ApppG-capped β-act-UTR-Luc mRNA showed less than 2% of the translation output of that of the corresponding m7GpppA-capped transcript (Fig. S2). In agreement with our binding results, eIF4GI682-1599, eIF4GI557-1599, and DAP5 did not show any significant stimulation of the cap-independent translation of this ApppG-capped β-act-UTR-Luc mRNA (data not shown). Significantly, although depletion of eIF4GI and DAP5 from the lysate decreased the translation of m7GpppA-capped β-act-UTR-Luc mRNA to ∼18 and ∼67%, respectively, only the eIF4GI557-1599 construct containing the eIF4E binding domain rescued its translation (Fig. S4), results that are consistent with the previous and widespread use of this mRNA as a control for cap-dependent initiation and translation (32). As expected, neither eIF4GI682-1599 nor DAP5, which lack the eIF4E binding domain, rescued the translation of the control m7GpppA-capped β-act-UTR-Luc mRNA (Fig. S4), as demonstrated by the absence of any significant change in luciferase activity, indicating the specificity of the stimulation.
An exposed 5′ end is important for the cap-independent translation activities of HIF-1α and p53A UTR-Luc mRNAs but not FGF-9 and p53B UTR-Luc mRNAs
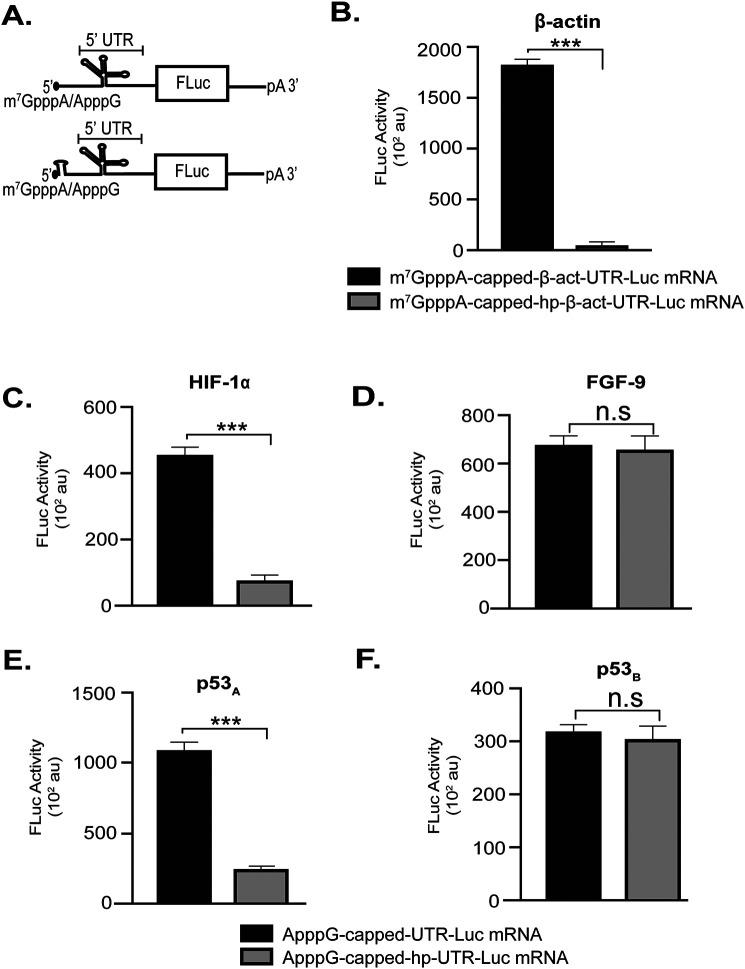
To gain a better mechanistic understanding of the cap-independent translation initiation of our selected mRNAs, we introduced a stable hairpin at the 5′ end of our mRNAs (Fig. 5A). This same hairpin at this same position previously was used to block scanning from the exposed 5′ end of an mRNA while having no effect on the internal translation initiation mediated by an IRES (25, 27, 41). We found that the hairpin structure significantly repressed translation of m7GpppA-capped β-act-UTR-Luc mRNA (Fig. 5B), which served as a positive control for inhibition of 5′-end scanning-dependent translation initiation. Comparative analyses of our selected UTR-Luc mRNAs showed that the hairpin did not affect the translation initiation activities of FGF-9 UTR-Luc mRNA (Fig. 5D) and p53B UTR-Luc mRNA (Fig. 5F), suggesting that internal initiation occurred on these UTR-Luc mRNAs. In contrast, the translation efficiencies of HIF-1α Luc mRNA (Fig. 5C) and p53A UTR-Luc mRNA (Fig. 5E) were significantly reduced by the hairpin, suggesting that translation of these UTR-Luc mRNAs required an exposed 5′ end for translation initiation and that these 5′ UTRs contain structural elements that act as CITEs rather than IRESs.
Figure 5.
Effect of a stable 5′ hairpin (hp) inserted at the 5′ terminal of UTR-Luc mRNAs. A, cartoon representation of the reporter construct used to test the effects of 5′ UTR accessibility of HIF-1α, FGF-9, p53A, and p53B UTR-Luc mRNAs. B, inhibition of 5′-end-dependent translation of m7GpppA-capped β-act-UTR-Luc mRNA by a 5′ hairpin. C–F, comparison of the translation output of ApppG-capped-UTR-Luc mRNAs versus ApppG-capped-hp-UTR-Luc mRNA. Data were analyzed by two-tailed unpaired Student's t test: n.s, p = 0.12; ***, p < 0.001.
Discussion
In this study, using a fluorescence anisotropy-based equilibrium binding assay and a luciferase-based gene expression reporter assay, we have demonstrated that eIF4GI and DAP5 directly bind to and stimulate cap-independent translation initiation of a subset of cellular mRNAs. The truncated eIF4GI proteins, eIF4GI557-1599 and eIF4GI682-1599, as well as DAP5 bind to the 5′ UTRs of this subset of mRNAs with Kds in the range of 12–290 nm (Fig. 2 and 3 and Table 1) and stimulate cap-independent translation of the corresponding UTR-Luc mRNAs by 2- to 5-fold over lysates depleted of either eIF4GI or DAP5 (Fig. 4, C–F). These Kds are comparable with those observed for the binding of eIF4GI constructs to the EMCV J/K IRES (Fig. 3 and Table S1), a well-characterized system in which it has been previously shown that binding of truncated eIF4GI643-1076 to a well-defined structural feature within the 5′ UTR of EMCV mRNA stimulates the cap-independent translation of this mRNA (34). In line with cellular data (2, 15), our observations strongly suggest that both eIF4GI and DAP5 recognize and bind to specific structural features within the 5′ UTRs of our selected mRNAs. Moreover, the observation that eIF4GI682-1599 and DAP5 bound to our selected 5′ UTRs with similar binding affinities (Fig. 2 and Table 1) suggests that eIF4GI and DAP5 recognize similar elements within the various 5′ UTRs. Furthermore, the intriguing differences between the Kds for the larger construct of eIF4GI557-1599 and DAP5 (Table 1) suggest that, to bring about translational outcomes, cellular mRNAs with structured 5′ UTRs may preferably recruit eIF4GI, depending on its cellular availability, which may change according to the type of stress conditions, cell type, or stability of the proteins (2, 42, 43).
Several reports have demonstrated that a subset of cellular mRNAs possess IRES-like elements in addition to the cap structure, and that these mRNAs act to recruit ribosomal PICs and, presumably, key eIFs internally to the 5′ UTR of these mRNAs to initiate cap-independent translation (2, 5, 44). However, no common structural motifs were identified among the cellular IRES elements, and, compared with viral IRES, cellular IRES elements appear to be much more diverse and less stable in terms of Gibbs free energy of folding (45). In contrast to these reports, it has recently been proposed that other cellular mRNAs recruit ribosomal PICs in a cap-independent manner to the 5′ end using a CITE that recruits eIFs, and that the ribosomal PIC then scans from the 5′ end rather than using an IRES to initiate translation internally (19). Given the fact that some 5′ UTRs may possess either IRESs and/or CITEs, we were prompted to test the hypothesis that the exposed 5′ end of our selected 5′ UTRs is required for translation. Our data showed that the presence of a highly stable, scanning-inhibiting hairpin (41, 46) at the 5′ end of the FGF-9 and p53B ApppG-capped hp-UTR-Luc mRNAs did not affect the absolute translation levels of these mRNAs (Fig. 5), demonstrating that they were cap-independently translated, presumably using IRES-like mechanisms. In contrast with this, the hairpin at the 5′ end of the HIF-1α and p53A ApppG-capped hp-UTR-Luc mRNAs repressed the cap-independent translation initiation activities of these UTR-Luc mRNAs (Fig. 5), suggesting that accessibility of the 5′ end of these cellular mRNAs is necessary and, correspondingly, a CITE-like mechanism is important for the initiation of these mRNAs. Because the subset of cellular mRNAs we have investigated here employ different systems of cap-independent translation, we speculate that this phenomenon provides additional regulation for the expression of these mRNAs.
Our findings are reminiscent of the manner in which viruses use highly structured IRESs to directly recruit eIFs, the 40S subunit, and/or the 43S PIC to drive the cap-independent translation of viral mRNA (5, 47, 48). Although the mechanisms through which viral mRNAs use IRESs to drive cap-independent initiation have been extensively characterized using genetic, biochemical, and structural approaches (16, 49, 50), the mechanisms through which eukaryotic cellular mRNAs drive cap-independent initiation remain largely unknown. Nonetheless, an increasing body of evidence suggests eukaryotic cellular mRNAs can employ noncanonical initiation mechanisms that are distinct from those employed by IRES-containing viral mRNAs. For example, Cate and coworkers have recently shown that the d subunit of eIF3 (eIF3d) targets mRNAs encoding proteins involved in cell proliferation and serves as a transcript-specific, cap-binding protein (51). Using the mRNA encoding the c-Jun transcription factor as an example, these authors showed that, under conditions in which an RNA structure in the 5′ UTR blocks eIF4E from binding to the 5′ cap, eIF3d binds directly to the 5′ cap and serves as an alternative cap-binding factor (52). Even more recently, it has been shown that DAP5 is a direct binding partner of eIF3d, and it has been proposed that DAP5 uses eIF3 to direct the eIF4E-independent, cap-dependent translation of a subset of cellular mRNAs when cellular stress conditions lead to the inactivation of eIF4E (53). In addition to these mechanisms in which RNAs utilize IRES elements, eIF3d, or other eIFs that function as alternative cap-binding proteins, the methylation of adenosine residues in the 3′ and 5′ UTRs of eukaryotic cellular mRNAs have been shown to stimulate translation by an unknown mechanism (54, 55). The cellular IRES- and CITE-based mechanisms we have investigated here are distinct from these other types of mechanisms that make use of an alternative cap-binding protein or posttranscriptional modification of the mRNA to be translated. Here, eIF4GI or DAP5 completely bypasses any cap-dependent processes and is instead directly recruited to IRESs or CITEs within the mRNAs. Indeed, because the binding studies we present here employ purified eIF4GI557-1599, eIF4GI682-1599, and DAP5 and uncapped RNA oligonucleotides, our results show these translation factors can bind directly to 5′ UTRs of select mRNAs in a completely cap-independent manner and without the need of alternative cap-binding proteins or mRNA methylation. Similarly, because the gene expression assays we present here also employed purified mRNAs with nonfunctional cap analogs and are performed in RRL, RRL(-)4GI, and RRL(-)DAP5, we can be confident that the stimulation of translation we observe is because of the direct interaction of eIF4GI or DAP5 with the IRES or CITE elements in the 5′ UTRs of our selected mRNAs rather than to the indirect effects of alternative cap-binding proteins or methylation of the mRNA.
Based on our observations, we propose that an important initial event in cap-independent initiation of eukaryotic cellular IRES- or CITE-containing mRNAs is binding of eIF4GI or DAP5, similar to the cap-independent initiation of CITE- or IRES-containing viral mRNAs (28–30). Specifically, we propose that eIF4GI or DAP5, perhaps with the aid of additional factors, bind to elements within the 5′ UTRs and subsequently recruit additional eIFs, the 40S subunit, and/or the 43S PIC to these mRNAs. In analogy to the cap-independent initiation of several CITE-containing viral mRNAs (28, 30, 56), we propose that the resulting 48S PIC scans to the AUG start codon. Our data suggest that this is the case for the HIF-1α and p53A mRNAs. Alternatively, these factors may be bound to elements near the AUG, acting as IRES-like structures, as appears to be the case for FGF-9 and p53B (Fig. 6). It is not surprising that multiple mechanisms and eIF requirements may be present, with different and possibly dynamic RNA structures in the 5′ UTR having different requirements. This is certainly the case for viral cap-independent translation (48).
Figure 6.
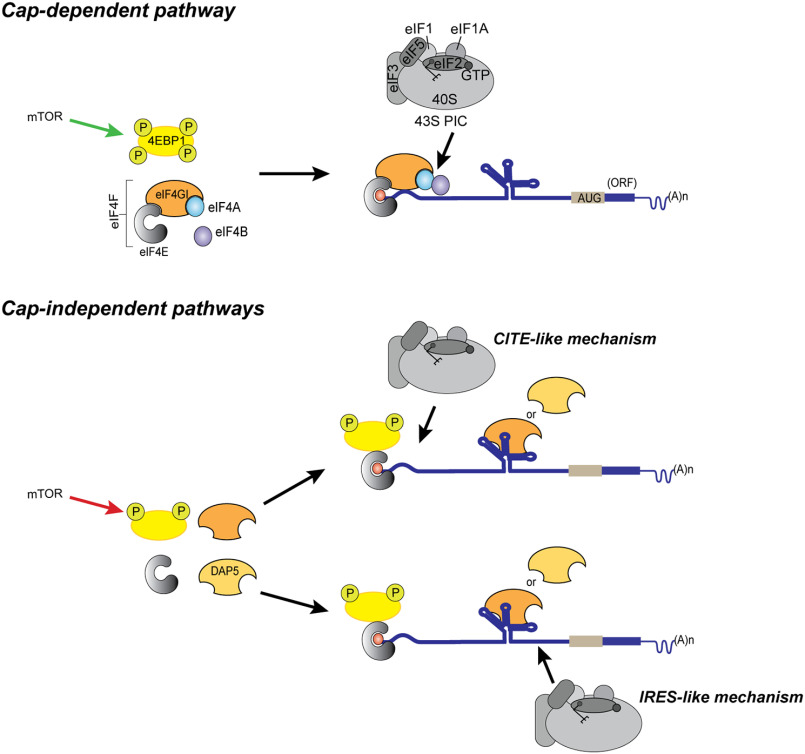
Cartoon summarizing models for canonical (cap-dependent) and proposed cap/eIF4E-independent translation initiation of the mRNAs containing structured 5′ UTRs. The top panel depicts normal physiological conditions where activated mTOR (mTOR) hyperphosphorylates 4EBP1, making it inactive. eIF4E binds to the m7G-capped mRNA to recruit other eIFs and the 43S PIC to initiate canonical cap-dependent translation. In the lower panel, inactive mTOR leads to hypophosphorylation of 4EBP1 which it blocks eIF4E's interaction with eIF4GI. The 4EBP1-eIF4E complex may interact with the m7G-capped mRNA but is unable to interact with eIF4GI. Our model shows eIF4GI or DAP5 binding the 5′ UTR and recruiting the 43S PIC by either a CITE-like mechanism or an IRES-like mechanism.
The transition from cap-dependent to cap-independent translation of cellular mRNAs under stress conditions has important physiological consequences, particularly in disease states, such as diabetes, cancer, and possibly neurological disorders (7–12). Under stress conditions, the levels of 4EBP1, eIF4GI, and DAP5 are elevated (2, 7, 8, 14, 15). Here, we have identified key interactions of eIF4GI and DAP5 through which cellular mRNAs containing either IRESs or CITEs may bypass the 4EBP1-mediated depletion of eIF4E and undergo cap-independent initiation and expression. Identification of specific interactions that facilitate the transition from cap-dependent to cap-independent translation promises to facilitate a better understanding of noncanonical translation mechanisms and to inform the potential development of novel therapies that modulate this transition to regulate the cellular response to stress conditions.
Experimental procedures
Preparation of RNAs for fluorescence anisotropy-based equilibrium binding studies
DNA templates corresponding to the 5′ UTR of HIF-1α (294 nucleotides [nt]; GenBank accession no. AH006957.2) (57), the 5′ UTR of FGF-9 (177 nt; GenBank accession no. AY682094.1) (26), the 5′ UTR for one isoform of p53 (p53A) (136 nt; GenBank accession no. JN900492.1), the 5′ UTR for a second isoform of p53 (p53B) (117 nt; GenBank accession no. MG595994.1) (31, 50), the ferritin IRE (58), the EMCV J/K IRES (nt 680–786) (34, 49), the 101-nt polyUC, and the 5′ UTR of β-actin (84 nt; GenBank accession no. AK301372.1) (32) (Table S2) were purchased from Integrated DNA Technology (IDT), and the corresponding RNAs were synthesized via in vitro transcription using the HiScribeTM T7 quick high-yield RNA synthesis kit (New England Biolabs, Inc.) by following the manufacturer's protocol. RNAs from transcription reactions were purified using the RNA Clean and Concentrator kit from Zymo Research by following the manufacturer's protocol. Purified RNA transcripts were labeled with fluorescein at their 5′ termini using the 5′ EndTag DNA/RNA labeling kit from Vector Laboratories by following the manufacturer's protocol. RNA concentrations were determined using a nano-drop UV-visible spectrometer, and integrity was verified by 1.5% agarose gel electrophoresis.
Preparation of eIF4GI557-1599, eIF4GI682-1599, and DAP5
The plasmids for expression of eIF4GI557-1599, and eIF4GI682-1599 were a generous gift from Dr. Christopher Fraser (University of California at Davis). Full-length eIF4GI purified from cells as part of the eIF4F complex is easily degraded during purification and is contaminated with various amounts of eIF4E and/or eIF4A. Therefore, we used the stable, functional constructs described here (59–61). The codon-optimized eIF4GI682-1599 construct (34) includes the minimal sequence for IRES-mediated cap-independent translation initiation (31) and is similar in domain structure to full-length DAP5. This construct has an N-terminal, 6× histidine tag followed by a Flag tag in the pET28c vector. We introduced a tobacco etch virus (TEV) protease cleavage site following the Flag tag using the Q5® site-directed mutagenesis kit from New England Biolabs, Inc., by following the manufacturer's protocol. The eIF4GI557-1599 construct was in a fastback vector. It was PCR amplified using a forward primer containing an NcoI site and a 6× histidine tag followed by a TEV protease site and a reverse primer containing an XhoI site. The amplified PCR product was subcloned in pET28c at the NcoI-XhoI site and used to express and purify the eIF4GI557-1599 protein. The plasmid encoding full-length human DAP5 with an N-terminal 6× histidine tag was purchased from GenScript (Piscataway, NJ). All of the proteins were recombinantly expressed in Escherichia coli BL21-CodonPlus (DE3)-RIL cells (Agilent) and were purified using a combination of Ni2+-nitrilotriacetic acid (Ni-NTA) affinity and heparin affinity columns, as previously described (25, 34). Briefly, the proteins were first purified from bacterial cell lysates using His-Trap HP (Ni-NTA) columns (GE Healthcare Life Sciences) per the manufacturer's instructions. The purified 6× histidine-tagged proteins were dialyzed overnight against storage buffer (20 mm HEPES-KOH, pH 7.6, 200 mm KCl, 10 mm β-mercaptoethanol, 10% glycerol) in the presence of TEV protease to cleave off the tags. The untagged proteins were further purified and concentrated using 1 ml HiTrapTM heparin HP columns (GE Healthcare Life Sciences). The eluted proteins were analyzed on 10% SDS-PAGE gels, and pure fractions (>95% purity) were pooled and dialyzed overnight against storage buffer. The concentrations of the purified and concentrated proteins were quantified using Coomassie protein assay reagent (Thermo Scientific) and were aliquoted and stored at −80 °C.
Fluorescence anisotropy-based equilibrium binding assays
Fluorescein-labeled RNAs were diluted to 100 nm using folding buffer (20 mm HEPES-KOH, pH 7.5, and 100 mm KCl), heated to 90 °C for 2 min, and slowly cooled over 1 h to room temperature. MgCl2 was then added to the solution to a final concentration of 1 mm, and the solution was gently mixed and incubated on ice for about 1 h. Fluorescence anisotropy measurements for assessing the binding of eIF4GI constructs or DAP5 to the fluorescein-labeled RNAs were performed using the equilibrium titration module of an SF-300X stopped-flow fluorimeter (KinTek Corporation, Austin, TX). Fluorescein-labeled RNAs were excited at 495 nm, and emission was detected using a 515-nm high-pass filter (Semrock, Rochester, NY). Equilibrium binding titrations began with a 200-μl sample of either 10 nm (Fig. 2B and 3A) or 100 nm (Fig. 2, C–D, and Fig. 3, B–C) fluorescein-labeled RNA in the titration buffer (20 mm HEPES-KOH, pH 7.6, 100 mm KCl, and 1 mm MgCl2), and 20–50 data points were collected for each anisotropy measurement by automated continuous injection of 20 μl of 2.5 μm eIF4GI557-1599, 10 μm eIF4GI682-1599, or 10 μm DAP5 over a period of 30 min at a temperature of 25 °C. Note that the first reading is taken in the absence of protein. Using the Origin 2018b software package, the data were fitted to a nonlinear, single-site equilibrium binding equation of the form:
where robs is the observed anisotropy value, rmin is the minimum anisotropy value in the absence of eIF4GI or DAP5, rmax is the final saturated anisotropy value, [eIF4GI or DAP5] is the concentration of eIF4GI557-1599, eIF4GI682-1599, or DAP5, and Kd is the equilibrium dissociation constant. The chi-squared values (χ2) that represented the statistical goodness of fit were always close to 1 and are reported in Table 1. Fitting data to a two-site model did not improve the fit, as judged by χ2 values. The equilibrium binding titration of each 5′ UTR was performed three times and fit independently for Kd. The fitted Kds were then averaged and the standard deviations were calculated (Table 1).
Preparation of UTR-Luc reporter mRNAs for luciferase-based gene expression reporter assays
The UTR-Luc mRNA constructs for the luciferase gene expression reporter assays were generated from the BlucB plasmid (62), which contains a firefly luciferase gene flanked by 5′- and 3′ UTR sequences of the barley yellow dwarf virus genomic RNA. To generate each UTR-Luc mRNA reporter construct, a sequence containing the T7 promoter followed by the target 5′ UTR was cloned into the BlucB plasmid vector upstream of the firefly luciferase coding region, after removing the barley yellow dwarf virus 5′ UTR as follows. Briefly, all UTR-Luc reporter constructs were PCR amplified with the forward primers containing a NotI site and reverse primers containing a BssHII site, as shown in Table 2, and then digested with NotI and BssHII. All the restriction enzymes were purchased from New England Biolabs (NEB), and restriction digestions were performed according to the manufacturer's instructions. The PCR-amplified sequences were ligated into a NotI- and BssHII-digested BlucB plasmid using DNA ligase (NEB) and transformed into E. coli DH5-α competent cells. Five colonies were selected, grown overnight in Luria-Bertani (LB) growth medium supplemented with 100 μg/ml ampicillin, and used to isolate plasmid DNA using the QIAprep® Spin miniprep kit from Qiagen by following the manufacturer's protocol.
Table 2.
Primers used
| Reporter | Forward primer | Reverse primer |
|---|---|---|
| Used for the cloning of the UTRs in reporter constructs | ||
| HIF1α | 5′-CTAGGCGGCCGCTAATACGAC-3′ | 5′-CTAGGCGCGCGGTGAATCGGTC-3′ |
| FGF9 | 5′-CTAGGCGGCCGCTAATACGAC-3′ | 5′-CTAGGCGCGCCAGAGGACTCGGC-3′ |
| p53A | 5′-CTAGGCGGCCGCTAATACGAC-3′ | 5′-CTAGGCGCGCGGCAGTGAC-3′ |
| p53B | 5′-CTAGGCGGCCGCTAATACGAC-3′ | 5′-CTAGGCGCGCTGCTTGGGAC-3′ |
| β-actin | 5′-CTAGGCGGCCGCTAATACGAC-3′ | 5′-CTAGGCGCGCGGTGAGCTGG-3′ |
| Used for insertion of stable hairpin at 5′UTR end of UTR-Luc constructs | ||
| HIF1α | 5′-AGGCCGTGCACAGTGCTGCCTCG-3′ | 5′-AGGCCAAGCCTATAGTGAGTCGTATTAG-3′ |
| FGF9 | 5′-AGGCCGAAACAGCAGATTACTTTTATTTATG-3′ | 5′-AGGCCTATAGTGAGTCGTATTAGC-3′ |
| p53A | 5′-AGGCCGGTCTAGAGCCACCGTCC-3′ | 5′-AGGCCCTATAGTGAGTCGTATTAGCGG-3′ |
| p53B | 5′-AGGCCATGGAGGAGCCGCAGTCA-3′ | 5′-AGGCCCCCTATAGTGAGTCGTATTAGCG-3′ |
| β-actin | 5′-AGGCCACCGCCGAGACCGCGTCC-3′ | 5′-AGGCCCCTATAGTGAGTCGTATTAGCGCGCG-3′ |
To assess whether the availability of a 5′ end is required for the translation of these reporter mRNAs, a highly stable hairpin (41) was inserted at the 5′ end of each UTR-Luc reporter (hp-UTR-Luc mRNA) using site-directed mutagenesis with the primers in Table 1. All clones were confirmed by sequencing (Genewiz). To generate a linearized plasmid DNA template for in vitro transcription, plasmid DNAs were linearized using KpnI so as to remove the 3′ UTR from the UTR-Luc mRNA reporter construct. The resulting linearized DNA was purified using the GeneJET gel extraction and DNA cleanup micro kit from GeneJET per the manufacturer's instructions. DNA templates were in vitro transcribed using a T7 RiboMax large-scale RNA production kit (Promega) by following the manufacturer's protocol. ApppG (NEB) or Ribo m7GpppA cap analog (Promega) was added to the transcription mix in an ApppG or Ribo m7GpppA:GTP ratio of 10:1 to get mRNA transcripts with nonfunctional and functional caps, respectively. Capped RNAs were poly(A) tailed using the poly(A) tailing kit (Invitrogen) by following the manufacturer's protocol. The resulting capped and polyadenylated mRNAs were then purified using an RNA Clean and Concentrator kit (Zymo) by following the manufacturer's protocol. RNA concentrations were determined using a nano-drop UV-visible spectrometer, and integrity was verified by 1.5% agarose gel electrophoresis.
Western blots and depletion of eIF4GI and DAP5 from the rabbit reticulocyte lysate
Monoclonal mouse primary antibodies for DAP5 (catalog number sc-137011), eIF4GI (catalog number sc-373892), eIF4E (catalog number sc-9976), eIF2β (catalog number sc-9978), and eIF4AI/II (catalog number sc-377315) (Santa Cruz Antibodies) were used for Western blotting and/or immunoprecipitation experiments. For the Western blotting experiments, the specific antibodies were diluted to 1:1000 in PBST (PBS-Tween) buffer containing 1% BSA. The blots were incubated overnight in the primary antibodies at 4 °C with constant shaking. The membranes were washed three times with PBST and then incubated with horseradish peroxidase-goat anti-mouse secondary antibody (1:5000 dilution, Invitrogen, catalog number 31430) for 1 h at room temperature. After three subsequent washes in PBST, the membrane was developed using enhanced chemiluminescent substrate (SuperSignalTM West Femto, ThermoScientific, catalog number 34094). For the depletion of eIF4GI and DAP5 from the RRL, 20 μl of either eIF4GI or DAP5 capture antibodies were incubated with PureProteomeTM protein A/G mix magnetic beads (EMD Millipore Corporation) at room temperature with continuous mixing for 1 h. The bead-antibody complexes were washed for 10 s with 500 μl PBS. The bead-antibody complexes were captured using a magnetic stand (Promega), and the suspensions were removed. The washing step was repeated 2 more times with PBS and then with the wash buffer (20 mm HEPES-KOH, pH 7.5, 50 mm KCl, 75 mm KOAc, 1 mm MgCl2). The nuclease-treated RRLs (Promega) were incubated with these preformed bead-antibody complexes at 4 °C with continuous shaking for 1 h. The magnet was reengaged to capture the bead-antibody-protein complex, and the resulting depleted RRL [RRL(-)4GI or RRL(-)DAP5] was collected and used for translation. To determine the extent of depletion, Western blotting assays were performed after resolving the samples on a 4–15% Tris-HCl gradient gel (Bio-Rad Laboratories).
Luciferase-based gene expression reporter assays
Gene expression was achieved by translating the UTR-Luc mRNAs in vitro, using the nuclease-treated RRL in vitro translation system from Promega. The RRL was made more cap dependent by addition of 75 mm KCl (63). Each 25-μl reaction mixture contained 70%, v/v, RRL, RRL(-)4GI, or RRL(-)DAP5 (Promega) supplemented with 0.5 mm MgCl2, 0.02 mm amino acid mixture, 10 units/μl RiboLock RNase inhibitor (ThermoScientific), and various concentrations of purified eIF4GI constructs or DAP5, as indicated in the figure legends. 4EGI-1 chemical (10 mm stock in DMSO) (Selleck Chemicals, catalog number S7369) was added to the translation mix when indicated at a concentration of 0.2 mm. Briefly, 1 μg of UTR-Luc mRNA was added to the RRL, RRL(-)4GI, or RRL(-)DAP5 in vitro translation mixture that had been preincubated at 30 °C for 10 min following the addition of the specified concentration of eIF4GI constructs or DAP5. The resulting in vitro translation reaction then was incubated at 30 °C for 1 h and stopped by the addition of 60 μm puromycin. Firefly luciferase activities were then assayed using a GloMax 96 microplate illuminometer (Promega). To achieve this, 3 μl of translation reaction mixture was added to 30 μl Bright-Glo luciferase assay reagent (Promega), and the resulting luminescence was measured in the illuminometer over a spectral wavelength of 350–650 nm and an integration time of 10 s at room temperature. After subtracting the background, measured using an in vitro translation reaction to which no UTR-Luc mRNA had been added, the luminescence data were analyzed and plotted using the Prism 8 software package. At least 3 different batches of RRLs were used. The translation data for each UTR-Luc mRNA were reported as an average from three independent experiments. Each independent experiment was done in triplicate, and the means ± S.D. were calculated using GraphPad Prism 8. Statistical significance between the mean values was analyzed using two-tailed unpaired Student's t test (GraphPad Prism 8 software). The statistical significance was set at a p value of <0.05, and the p values were calculated. The calculated p values for the analyses are indicated above the bar graphs (Fig. 4 and 5 and Figs. S1, S2, and S4, n.s. [nonsignificant], p = 0.12; *, p = 0.033; **, p = 0.002; ***, p < 0.001).
Data availability
All data are presented in the manuscript.
Supplementary Material
Acknowledgments
We thank Paul Powell for helpful discussions. We thank Prof. Christopher S. Fraser (University of California, Davis) for the recombinant eIF4GI clones. We thank Maya Ramachandran for carrying out some of the preliminary binding studies.
This article contains supporting information.
Author contributions—S. A. H., U. B., R. L. G., S. M., and D. J. G. conceptualization; S. A. H. data curation; S. A. H., U. B., and D. J. G. formal analysis; S. A. H. and U. B. investigation; S. A. H., U. B., and S. M. methodology; S. A. H. and S. M. writing-original draft; U. B., R. L. G., and D. J. G. writing-review and editing; R. L. G. and D. J. G. funding acquisition; D. J. G. supervision; D. J. G. project administration.
Funding and additional information—The research reported in this publication was supported by the Susan G. Komen Foundation for the Cure Postdoctoral Fellowship PDF12231199 (to S. M.), the National Institute of General Medical Science of the National Institutes of Health under award numbers GM R01 084288 (to R. L. G.) and GM R01 128239 (to D. J. G.), and the National Center for Advancing Translational Sciences 1UL1TR002384-01 Seed Project (to D. J. G.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- IRES
- internal ribosome entry site
- CITE
- 5′ cap-independent translation enhancer
- PIC
- preinitiation complex
- EMCV
- encephalomyocarditis virus
- RRL
- rabbit reticulocyte
- TEV
- tobacco etch virus.
References
- 1. Sonenberg N., and Hinnebusch A. G. (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Braunstein S., Karpisheva K., Pola C., Goldberg J., Hochman T., Yee H., Cangiarella J., Arju R., Formenti S. C., and Schneider R. J. (2007) A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol. Cell 28, 501–512 10.1016/j.molcel.2007.10.019 [DOI] [PubMed] [Google Scholar]
- 3. Hinnebusch A. G., and Lorsch J. R. (2012) The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb. Perspect. Biol. 4, a011544 10.1101/cshperspect.a011544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jackson R. J., Hellen C. U., and Pestova T. V. (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev Mol. Cell Biol. 11, 113–127 10.1038/nrm2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Komar A. A., and Hatzoglou M. (2011) Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle 10, 229–240 10.4161/cc.10.2.14472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hinnebusch A. G., Ivanov I. P., and Sonenberg N. (2016) Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 352, 1413–1416 10.1126/science.aad9868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dennis M. D., Shenberger J. S., Stanley B. A., Kimball S. R., and Jefferson L. S. (2013) Hyperglycemia mediates a shift from cap-dependent to cap-independent translation via a 4E-BP1-dependent mechanism. Diabetes 62, 2204–2214 10.2337/db12-1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Silvera D., Arju R., Darvishian F., Levine P. H., Zolfaghari L., Goldberg J., Hochman T., Formenti S. C., and Schneider R. J. (2009) Essential role for eIF4GI overexpression in the pathogenesis of inflammatory breast cancer. Nat. Cell Biol. 11, 903–908 10.1038/ncb1900 [DOI] [PubMed] [Google Scholar]
- 9. Dhungel N., Eleuteri S., Li L.-B., Kramer N. J., Chartron J. W., Spencer B., Kosberg K., Fields J. A., Stafa K., Adame A., Lashuel H., Frydman J., Shen K., Masliah E., Gitler A. D., et al. (2015) Parkinson's disease genes VPS35 and EIF4G1 interact genetically and converge on alpha-synuclein. Neuron 85, 76–87 10.1016/j.neuron.2014.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dyer J., and Sossin W. S. (2013) Characterization of the role of eIF4G in stimulating cap- and IRES-dependent translation in aplysia neurons. PLoS ONE 8, e74085 10.1371/journal.pone.0074085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Olszewska D. A., McCarthy A., and Lynch T. (2016) Commentary: Parkinson's Disease genes VPS35 and EIF4G1 interact genetically and converge on alpha-synuclein. Front. Neurosci. 10, 162 10.3389/fnins.2016.00162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schrufer T. L., Antonetti D. A., Sonenberg N., Kimball S. R., Gardner T. W., and Jefferson L. S. (2010) Ablation of 4E-BP1/2 prevents hyperglycemia-mediated induction of VEGF expression in the rodent retina and in Muller cells in culture. Diabetes 59, 2107–2116 10.2337/db10-0148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Graber T. E., and Holcik M. (2007) Cap-independent regulation of gene expression in apoptosis. Mol. Biosyst. 3, 825–834 10.1039/b708867a [DOI] [PubMed] [Google Scholar]
- 14. Lee S. H., and McCormick F. (2006) p97/DAP5 is a ribosome-associated factor that facilitates protein synthesis and cell proliferation by modulating the synthesis of cell cycle proteins. EMBO J. 25, 4008–4019 10.1038/sj.emboj.7601268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Alard A., Musa F., and Schneider R. (2015) Evidence that the translational initiation factor DAP5 plays a critical role in breast cancer metastasis. Cancer Res. 75, 2262 10.1158/1538-7445.AM2015-2262 [DOI] [Google Scholar]
- 16. Plank T. D., and Kieft J. S. (2012) The structures of nonprotein-coding RNAs that drive internal ribosome entry site function. Wiley Interdiscip. Rev. RNA 3, 195–212 10.1002/wrna.1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johnson A. G., Grosely R., Petrov A. N., and Puglisi J. D. (2017) Dynamics of IRES-mediated translation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160177 10.1098/rstb.2016.0177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lacerda R., Menezes J., and Romao L. (2017) More than just scanning: the importance of cap-independent mRNA translation initiation for cellular stress response and cancer. Cell Mol. Life Sci. 74, 1659–1680 10.1007/s00018-016-2428-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shatsky I. N., Terenin I. M., Smirnova V. V., and Andreev D. E. (2018) Cap-independent translation: what's in a name? Trends Biochem. Sci. 43, 882–895 10.1016/j.tibs.2018.04.011 [DOI] [PubMed] [Google Scholar]
- 20. Grover R., Sharathchandra A., Ponnuswamy A., Khan D., and Das S. (2011) Effect of mutations on the p53 IRES RNA structure: implications for de-regulation of the synthesis of p53 isoforms. RNA Biol. 8, 132–142 10.4161/rna.8.1.14260 [DOI] [PubMed] [Google Scholar]
- 21. Leppek K., Das R., and Barna M. (2018) Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 19, 158–174 10.1038/nrm.2017.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Virgili G., Frank F., Feoktistova K., Sawicki M., Sonenberg N., Fraser C. S., and Nagar B. (2013) Structural analysis of the DAP5 MIF4G domain and its interaction with eIF4A. Structure 21, 517–527 10.1016/j.str.2013.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thakur A., Marler L., and Hinnebusch A. G. (2019) A network of eIF2beta interactions with eIF1 and Met-tRNAi promotes accurate start codon selection by the translation preinitiation complex. Nucleic Acids Res. 47, 2574–2593 10.1093/nar/gky1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Weingarten-Gabbay S., Khan D., Liberman N., Yoffe Y., Bialik S., Das S., Oren M., and Kimchi A. (2014) The translation initiation factor DAP5 promotes IRES-driven translation of p53 mRNA. Oncogene 33, 611–618 10.1038/onc.2012.626 [DOI] [PubMed] [Google Scholar]
- 25. Liberman N., Gandin V., Svitkin Y. V., David M., Virgili G., Jaramillo M., Holcik M., Nagar B., Kimchi A., and Sonenberg N. (2015) DAP5 associates with eIF2beta and eIF4AI to promote internal ribosome entry site driven translation. Nucleic Acids Res. 43, 3764–3775 10.1093/nar/gkv205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen T. M., Shih Y. H., Tseng J. T., Lai M. C., Wu C. H., Li Y. H., Tsai S. J., and Sun H. S. (2014) Overexpression of FGF9 in colon cancer cells is mediated by hypoxia-induced translational activation. Nucleic Acids Res. 42, 2932–2944 10.1093/nar/gkt1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Terenin I. M., Andreev D. E., Dmitriev S. E., and Shatsky I. N. (2013) A novel mechanism of eukaryotic translation initiation that is neither m7G-cap-, nor IRES-dependent. Nucleic Acids Res. 41, 1807–1816 10.1093/nar/gks1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee K. M., Chen C. J., and Shih S. R. (2017) Regulation mechanisms of viral IRES-driven translation. Trends Microbiol. 25, 546–561 10.1016/j.tim.2017.01.010 [DOI] [PubMed] [Google Scholar]
- 29. Banerjee B., and Goss D. J. (2014) Eukaryotic initiation factor (eIF) 4F binding to barley yellow dwarf virus (BYDV) 3'-untranslated region correlates with translation efficiency. J. Biol. Chem. 289, 4286–4294 10.1074/jbc.M113.530329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sharma S. D., Kraft J. J., Miller W. A., and Goss D. J. (2015) Recruitment of the 40S ribosome subunit to the 3'-untranslated region (UTR) of a viral mRNA, via the eIF4 complex, facilitates cap-independent translation. J. Biol. Chem. 290, 11268–11281 10.1074/jbc.M115.645002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sharathchandra A., Katoch A., and Das S. (2014) IRES mediated translational regulation of p53 isoforms. Wiley Interdiscip. Rev. RNA 5, 131–139 10.1002/wrna.1202 [DOI] [PubMed] [Google Scholar]
- 32. Dmitriev S. E., Terenin I. M., Dunaevsky Y. E., Merrick W. C., and Shatsky I. N. (2003) Assembly of 48S translation initiation complexes from purified components with mRNAs that have some base pairing within their 5′ untranslated regions. Mol. Cell. Biol. 23, 8925–8933 10.1128/MCB.23.24.8925-8933.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ma J., Haldar S., Khan M. A., Sharma S. D., Merrick W. C., Theil E. C., and Goss D. J. (2012) Fe2+ binds iron responsive element-RNA, selectively changing protein-binding affinities and regulating mRNA repression and activation. Proc. Natl. Acad. Sci. U S A 109, 8417–8422 10.1073/pnas.1120045109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lomakin I. B., Hellen C. U., and Pestova T. V. (2000) Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 20, 6019–6029 10.1128/mcb.20.16.6019-6029.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pestova T. V., Shatsky I. N., and Hellen C. U. (1996) Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 16, 6870–6878 10.1128/mcb.16.12.6870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gruner S., Peter D., Weber R., Wohlbold L., Chung M. Y., Weichenrieder O., Valkov E., Igreja C., and Izaurralde E. (2016) The structures of eIF4E-eIF4G complexes reveal an extended interface to regulate translation initiation. Mol. Cell 64, 467–479 10.1016/j.molcel.2016.09.020 [DOI] [PubMed] [Google Scholar]
- 37. Moerke N. J., Aktas H., Chen H., Cantel S., Reibarkh M. Y., Fahmy A., Gross J. D., Degterev A., Yuan J., Chorev M., Halperin J. A., and Wagner G. (2007) Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell 128, 257–267 10.1016/j.cell.2006.11.046 [DOI] [PubMed] [Google Scholar]
- 38. Sekiyama N., Arthanari H., Papadopoulos E., Rodriguez-Mias R. A., Wagner G., and Leger-Abraham M. (2015) Molecular mechanism of the dual activity of 4EGI-1: dissociating eIF4G from eIF4E but stabilizing the binding of unphosphorylated 4E-BP1. Proc. Natl. Acad. Sci. U S A 112, E4036–E4045 10.1073/pnas.1512118112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Feoktistova K., Tuvshintogs E., Do A., and Fraser C. S. (2013) Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc. Natl. Acad. Sci. U S A 110, 13339–13344 10.1073/pnas.1303781110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao P., Liu Q., Miller W. A., and Goss D. J. (2017) Eukaryotic translation initiation factor 4G (eIF4G) coordinates interactions with eIF4A, eIF4B, and eIF4E in binding and translation of the barley yellow dwarf virus 3' cap-independent translation element (BTE). J. Biol. Chem. 292, 5921–5931 10.1074/jbc.M116.764902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pelletier J., and Sonenberg N. (1985) Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell 40, 515–526 10.1016/0092-8674(85)90200-4 [DOI] [PubMed] [Google Scholar]
- 42. Alard A., Marboeuf C., Fabre B., Jean C., Martineau Y., Lopez F., Vende P., Poncet D., Schneider R. J., Bousquet C., and Pyronnet S. (2019) Differential regulation of the three eukaryotic mRNA translation initiation factor (eIF) 4Gs by the proteasome. Front. Genet. 10, 254 10.3389/fgene.2019.00254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nevins T. A., Harder Z. M., Korneluk R. G., and Holcik M. (2003) Distinct regulation of internal ribosome entry site-mediated translation following cellular stress is mediated by apoptotic fragments of eIF4G translation initiation factor family members eIF4GI and p97/DAP5/NAT1. J. Biol. Chem. 278, 3572–3579 10.1074/jbc.M206781200 [DOI] [PubMed] [Google Scholar]
- 44. Svitkin Y. V., Herdy B., Costa-Mattioli M., Gingras A. C., Raught B., and Sonenberg N. (2005) Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol. Cell. Biol. 25, 10556–10565 10.1128/MCB.25.23.10556-10565.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xia X., and Holcik M. (2009) Strong eukaryotic IRESs have weak secondary structure. PLoS ONE 4, e4136 10.1371/journal.pone.0004136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Babendure J. R., Babendure J. L., Ding J. H., and Tsien R. Y. (2006) Control of mammalian translation by mRNA structure near caps. RNA 12, 851–861 10.1261/rna.2309906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Terenin I. M., Smirnova V. V., Andreev D. E., Dmitriev S. E., and Shatsky I. N. (2017) A researcher's guide to the galaxy of IRESs. Cell Mol. Life Sci. 74, 1431–1455 10.1007/s00018-016-2409-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kieft J. S. (2008) Viral IRES RNA structures and ribosome interactions. Trends Biochem. Sci. 33, 274–283 10.1016/j.tibs.2008.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pestova T. V., Hellen C. U., and Shatsky I. N. (1996) Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16, 6859–6869 10.1128/mcb.16.12.6859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ray P. S., Grover R., and Das S. (2006) Two internal ribosome entry sites mediate the translation of p53 isoforms. EMBO Rep. 7, 404–410 10.1038/sj.embor.7400623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lee A. S., Kranzusch P. J., and Cate J. H. (2015) eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature 522, 111–114 10.1038/nature14267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lee A. S., Kranzusch P. J., Doudna J. A., and Cate J. H. (2016) eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 536, 96–99 10.1038/nature18954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. de la Parra C., Ernlund A., Alard A., Ruggles K., Ueberheide B., and Schneider R. J. (2018) A widespread alternate form of cap-dependent mRNA translation initiation. Nat. Commun. 9, 3068 10.1038/s41467-018-05539-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Meyer K. D., Patil D. P., Zhou J., Zinoviev A., Skabkin M. A., Elemento O., Pestova T. V., Qian S. B., and Jaffrey S. R. (2015) 5′ UTR m(6)A promotes cap-independent translation. Cell 163, 999–1010 10.1016/j.cell.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhou J., Wan J., Gao X., Zhang X., Jaffrey S. R., and Qian S. B. (2015) Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594 10.1038/nature15377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Guo L., Allen E. M., and Miller W. A. (2001) Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol. Cell 7, 1103–1109 10.1016/S1097-2765(01)00252-0 [DOI] [PubMed] [Google Scholar]
- 57. Yasuda M., Hatanaka T., Shirato H., and Nishioka T. (2014) Cell type-specific reciprocal regulation of HIF1A gene expression is dependent on 5′- and 3'-UTRs. Biochem. Biophys. Res. Commun. 447, 638–643 10.1016/j.bbrc.2014.04.058 [DOI] [PubMed] [Google Scholar]
- 58. Khan M. A., Ma J., Walden W. E., Merrick W. C., Theil E. C., and Goss D. J. (2014) Rapid kinetics of iron responsive element (IRE) RNA/iron regulatory protein 1 and IRE-RNA/eIF4F complexes respond differently to metal ions. Nucleic Acids Res. 42, 6567–6577 10.1093/nar/gku248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. De Gregorio E., Preiss T., and Hentze M. W. (1999) Translation driven by an eIF4G core domain in vivo. EMBO J. 18, 4865–4874 10.1093/emboj/18.17.4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Özeş A. R., Feoktistova K., Avanzino B. C., and Fraser C. S. (2011) Duplex unwinding and ATPase activities of the DEAD-box helicase eIF4A are coupled by eIF4G and eIF4B. J. Mol. Biol. 412, 674–687 10.1016/j.jmb.2011.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Morino S., Imataka H., Svitkin Y. V., Pestova T. V., and Sonenberg N. (2000) Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol. Cell. Biol. 20, 468–477 10.1128/mcb.20.2.468-477.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Treder K., Kneller E. L., Allen E. M., Wang Z., Browning K. S., and Miller W. A. (2008) The 3' cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation. RNA 14, 134–147 10.1261/rna.777308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chu L. Y., and Rhoads R. E. (1978) Translational recognition of the 5′-terminal 7-methylguanosine of globin messenger RNA as a function of ionic strength. Biochemistry 17, 2450–2455 10.1021/bi00605a032 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are presented in the manuscript.