Abstract
Viperin plays an important and multifaceted role in the innate immune response to viral infection. Viperin is also notable as one of very few radical SAM–dependent enzymes present in higher animals; however, the enzyme appears broadly conserved across all kingdoms of life, which suggests that it represents an ancient defense mechanism against viral infections. Although viperin was discovered some 20 years ago, only recently was the enzyme's structure determined and its catalytic activity elucidated. The enzyme converts CTP to 3′-deoxy-3′,4′-didehydro-CTP, which functions as novel chain-terminating antiviral nucleotide when misincorporated by viral RNA-dependent RNA polymerases. Moreover, in higher animals, viperin interacts with numerous other host and viral proteins, and it is apparent that this complex network of interactions constitutes another important aspect of the protein's antiviral activity. An emerging theme is that viperin appears to facilitate ubiquitin-dependent proteasomal degradation of some of the proteins it interacts with. Viperin-targeted protein degradation contributes to the antiviral response either by down-regulating various metabolic pathways important for viral replication or by directly targeting viral proteins for degradation. Here, we review recent advances in our understanding of the structure and catalytic activity of viperin, together with studies investigating the interactions between viperin and its target proteins. These studies have provided detailed insights into the biochemical processes underpinning this unusual enzyme's wide-ranging antiviral activity. We also highlight recent intriguing reports that implicate a broader role for viperin in regulating nonpathological cellular processes, including thermogenesis and protein secretion.
Keywords: radical SAM enzyme; virus; innate immune signaling; thermogenesis; lipid metabolism; protein secretion; protein ubiquitination; viperin; RNA-dependent RNA polymerase (RdRp); 3′,4′-didehydro-3′-deoxy-CTP; innate immunity; viral replication; protein degradation; enzyme mechanism; ubiquitylation (ubiquitination); antiviral agent
Viperin (virus-inhibitory protein, endoplasmic reticulum–associated, interferon-inducible) is an interferon-stimulated gene (ISG) product that restricts the infectivity of a wide range of viruses (1–4). The enzyme was first identified in 1997 as one of several genes up-regulated in response to infection by human cytomegalovirus (HCMV) and was therefore initially referred to as cig5 (cytomegalovirus-inducible gene 5) (5). Early work also identified the gene as up-regulated in viral infections of fish (6). In 2001, the gene was isolated from HCMV-infected fibroblasts (7). When stably expressed, viperin was shown to restrict HCMV replication and abolish the expression of glycoproteins from the viral capsid. Later it became apparent that viperin is identical to the radical SAM domain–containing protein 2 (RSAD2) located on chromosome 2 of the human genome (7). Subsequently, this highly species-conserved protein has been identified in the genomes of various mammals, fish, and reptiles.
Viperin is expressed at low basal levels in most cell types but is strongly induced by numerous viruses (6, 8–10). Table 1 summarizes the different viruses that viperin has been found to restrict and its mode of action, where known. Among the viruses documented to induce viperin are DNA viruses such as cytomegalovirus (CMV) (7, 48, 49) and retroviruses such as HIV-1 (45). Viperin also restricts many positive-strand RNA viruses such as flaviviruses, including hepatitis C virus (HCV) (9, 12, 13), West Nile virus (15), Zika virus (18, 21, 22), and dengue fever virus (15, 16), and alphaviruses that cause Sindbis (25, 26) and Chikungunya fevers (23, 24, 26). Last, viperin exhibits antiviral properties against negative-strand RNA viruses, including orthomyxoviruses such as influenza A virus (11), rhabdoviruses including rabies virus (1, 39), and paramyxoviruses such as Sendai virus (1, 27).
Table 1.
Viruses reported to be restricted by viperin
| Virus | Virus family | Mechanism of restriction | References |
|---|---|---|---|
| ssRNA viruses | |||
| Influenza A | Orthomyxoviridae | Inhibits viral budding from cell membrane | 8, 11 |
| Hepatitis C | Flaviviridae | Inhibits formation of replication complex by promoting degradation of viral protein NS5A | 12–14 |
| Dengue (type II) | Flaviviridae | Inhibits viral genome replication interacting with viral protein NS3 | 3, 15–17 |
| Tick-borne encephalitis | Flaviviridae | Promotes degradation of viral protein NS3 through proteasome-mediated pathway | 18–20 |
| Zika | Flaviviridae | Promotes degradation of viral protein NS3 through proteasome-mediated pathway | 3, 17, 18, 21, 22 |
| West Nile | Flaviviridae | Regulates viral replication; exact molecular mechanism not known | 3, 15, 17 |
| Chikungunya | Togaviridae | Reduces expression of viral RNA helicase protein NSP2; exact mechanism not known | 23, 24 |
| Sindbis | Togaviridae | Regulates viral replication; exact mechanism not known | 25, 26 |
| Sendai | Paramyxoviridae | Controls viral replication; exact mechanism not known | 10, 27 |
| Vesicular stomatitis virus | Rhabdoviridae | Inhibits viral replication in IFN-independent pathway | 28–30 |
| Classical swine fever | Flaviviridae | Inhibits CSFV replication by interacting with viral nonstructural protein NS5A and E2 | 31, 32 |
| Viral hemorrhagic septicemia | Rhabdoviridae | Inhibits RNA transcription and replication | 6, 33 |
| Measles | Paramyxoviridae | Inhibits viral release | 34 |
| Newcastle disease | Paramyxoviridae | Regulates viral replication, potentially by interacting with viral matrix proteins, involved in viral assembly and budding | 29, 31, 35 |
| Enterovirus A71 | Picornaviridae | Inhibits viral replication by interacting with viral protein 2C, important for membrane binding of virus. | 36 |
| Lymphocytic choriomeningitis | Arenaviridae | Exact mechanism not known | 37 |
| Junin mammarena | Arenaviridae | Impairs viral budding by perturbing lipid droplet morphogenesis and mis localization of viral glycoproteins | 38 |
| Rabies | Rhabdoviridae | Impairs viral budding by disrupting cholesterol/sphingomyelin synthesis and up-regulating TLR4 | 28, 39 |
| Porcine reproductive and respiratory syndrome | Arteriviridae | Impairs early steps of viral entry and genome replication and translation by interacting with viral protein N | 40 |
| Respiratory syncytial | Pneumoviridae | Impairs viral transmission by inhibiting virus filament formation | 41 |
| Bunyamwera | Peri Bunyaviridae | Inhibits viral replication; exact mechanism not known | 42 |
| Rhino | Picornaviridae | May inhibit viral replication; exact mechanism not known | 43 |
| Yellow fever | Flaviviridae | May inhibit viral replication; exact mechanism not known | 44 |
| Retroviruses | |||
| Human immunodeficiency | Retroviridae | Inhibits viral egress by perturbing lipid raft formation | 45, 46 |
| Equine infectious anemia | Retroviridae | Reduces viral budding by interacting with viral envelope proteins | 47 |
| dsDNA viruses | |||
| Human cytomegalovirus | Herpesviridae | Co-opted by the virus to inhibit β-oxidation of fatty acids by mitochondrial trifunctional protein | 6, 7, 48–50 |
| Herpes simplex 1 | Herpesviridae | Inhibits viral replication by interacting with viral glycoprotein D, found in the virion envelope | 51, 52 |
| Singaporean grouper iridovirus | Iridoviridae | Restricts viral replication by up-regulating interferon and inflammatory response; exact mechanism not known | 53 |
Viperin expression is also induced by a wide range of extracellular macromolecules that trigger the innate immune response by engaging various cell-surface receptors; these include type I, II, and III interferons, dsDNA and RNA, and lipopolysaccharides (8). In particular, the expression of mammalian viperin is induced through direct activation of Toll-like receptor (TLR) or RIG-I receptors by viruses (10, 54–56). The interferon-stimulating gene factor 3 (ISGF3) shows strong regulation of viperin expression in the interferon-dependent pathway, whereas in the interferon-independent pathway, the expression of viperin is controlled by interferon-regulatory factors (IRF1 and IRF3) (10).
Viperin is increasingly regarded as playing a central role in the cellular antiviral response and, more broadly, in innate immune signaling; evidence is also emerging that it plays a role in regulating metabolic processes under nonpathological conditions. Although the antiviral properties of the enzyme have been recognized for some time, it is only recently that the biochemistry underpinning viperin's biological function has begun to be elucidated. Viperin was recently shown to synthesize the antiviral ribonucleotide 3′-deoxy-3′,4′-didehydrocytidine triphosphate (ddhCTP), which inhibits the replication of some RNA viruses by acting as a chain terminator when misincorporated by viral RNA-dependent RNA polymerases (Fig. 1) (57). Viperin also interacts with numerous cellular and viral proteins within the cell. These interactions are important in immune signaling, in down-regulating metabolic pathways that are important for viral replication, and in ubiquitin-dependent degradation of viral proteins (Fig. 1). Here we review recent progress in our understanding of viperin from the biochemist's perspective; we refer the reader to other recent reviews that discuss viperin more in the context of virology and immunology (2, 3, 58).
Figure 1.
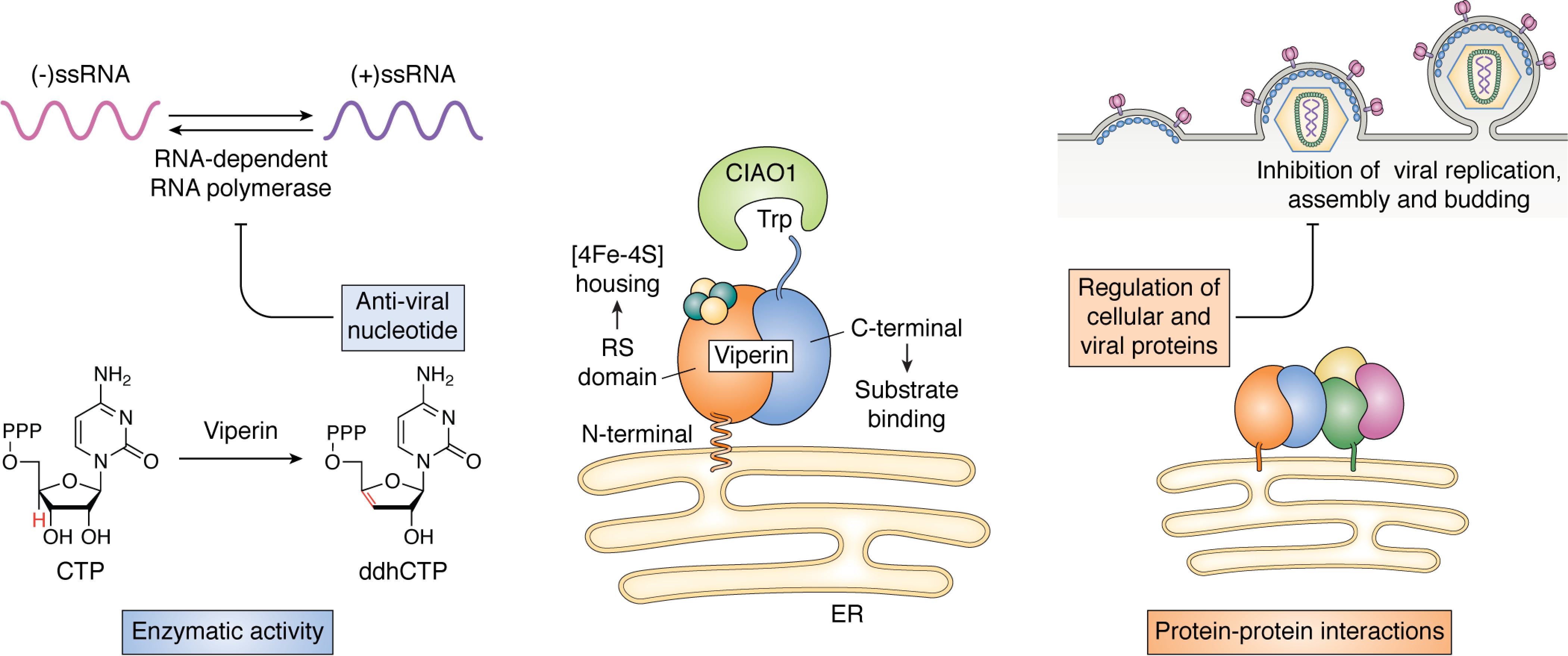
The two faces of viperin's antiviral activity. Center, viperin associates with the cytosolic face of the ER membrane through an N-terminal amphipathic helix. The radical SAM domain (orange) houses the active-site Fe-S cluster, whereas the C-terminal (blue) domain is important for substrate binding. The C-terminal tryptophan residue is an important recognition factor for the Fe-S cluster–installing protein, CIAO1. Left, viperin catalyzes the dehydration of CTP to ddhCTP through a radical mechanism; for some viral RNA-dependent RNA polymerases, ddhCTP may inhibit genome replication by acting as a chain terminator. Right, many of viperin's antiviral effects arise from its interactions with a wide variety of host and viral proteins; these variously inhibit viral replication, assembly, and budding by regulating metabolic and signaling pathways.
Domain structure and function
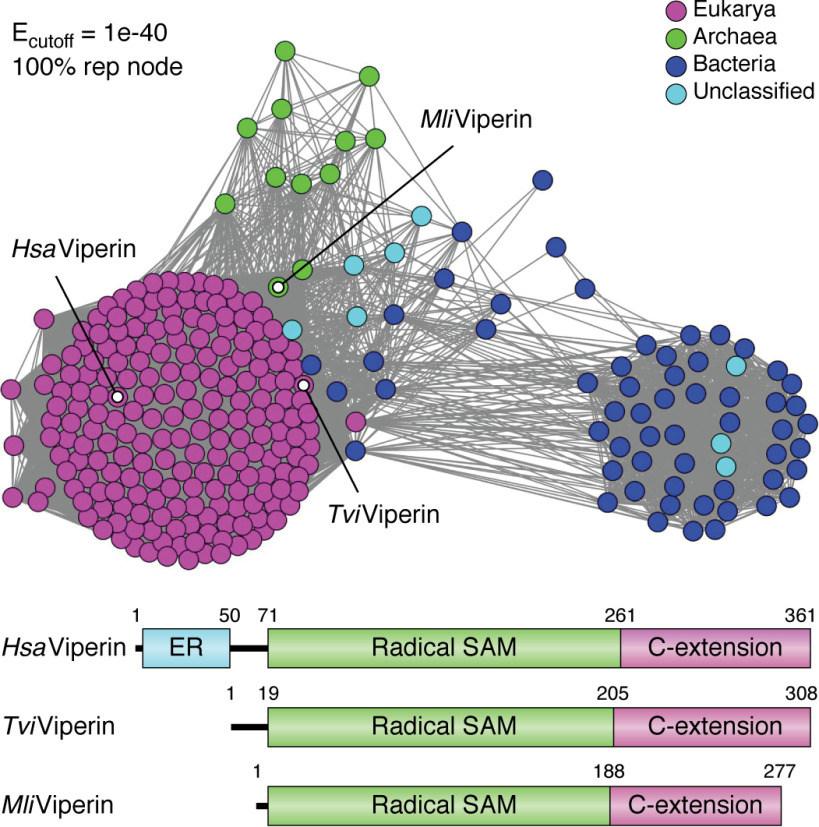
Sequence analysis of viperin shows it to be a 361-residue protein (human enzyme), with Mr ∼ 42,000. The enzyme appears to be broadly conserved across all animal phyla, and recent bio-informatic analyses have identified viperin-like sequences across all six kingdoms of life, including fungi, bacteria, and archaea (59, 60) (Fig. 2). The protein sequence contains three distinct regions (often loosely referred to as domains in the literature). The N-terminal extension, which comprises ∼50 residues, shows considerable variability between species in both length and sequence and is absent in most viperin-like sequences from microbes. This sequence is predicted to adopt an amphipathic α-helical structure and aids in localizing viperin to the cytosolic face of the endoplasmic reticulum (ER) and lipid droplets (61, 62); these are the sites at which viruses, such as flaviviruses, are assembled in the cell (3). Accordingly, truncating the N-terminal extension of viperin results in its delocalization from the ER membrane, which severely compromises viperin's ability to restrict the replication of some viruses (12, 19).
Figure 2.
Viperin-like sequences are found across all domains of life. Shown is a sequence similarity network of viperin, showing that in addition to eukaryotes (magenta nodes), viperin-like sequences also cluster in archaea (green nodes) and bacteria (blue nodes). The highlight nodes represent the sequences from Homo sapiens (HsaViperin), the fungus Trichoderma virens (TviViperin), and the archaeon Methanofollis liminatans (MliViperin), which have been the subject of biochemical characterization. Notably, the microbial enzymes lack the ER-localizing domain. Reproduced from Ref. 60 with permission. This research was originally published in the Journal of Biological Chemistry. Chakravarti, A., Selvadurai, K., Shahoei, R., Lee, H., Fatma, S., Tajkhorshid, E., and Huang, R. H. Reconstitution and substrate specificity for isopentenyl pyrophosphate of the antiviral radical SAM enzyme viperin. Journal of Biological Chemistry. 2018;293:14122–14133. © the American Society for Biochemistry and Molecular Biology.
The central SAM-binding domain and C-terminal extension of viperin are, in contrast, highly conserved across species. The central domain contains four sequence motifs associated with the radical SAM superfamily, including the canonical tricysteine motif (CXXXCXXC), responsible for binding the catalytic [4Fe-4S] cluster. As discussed below, the central domain adopts the “partial β-barrel” (βα)6 fold common to most radical SAM enzymes, whereas the C-terminal extension is important for binding CTP.
Last, the extreme C-terminal residues of viperin have been implicated as important for mediating installation of the [4Fe-4S] cluster by the iron-sulfur cluster–installing protein CIAO1 (19, 63). Studies with human viperin, transiently transfected in HEK 293T cells, showed that mutating the C-terminal tryptophan residue prevented CIAO1 from binding to viperin. Subsequently, in vivo labeling studies employing 55Fe demonstrated that the mutated viperin no longer contained an iron-sulfur cluster after it was immunoprecipitated from cell lysates.
Elucidation of the substrate for viperin
Radical SAM enzymes catalyze a remarkably diverse range of chemical transformations (64–69). These include key steps in the biosynthesis of various enzyme cofactors and the biosynthesis of a wide range of natural products. They also catalyze various post-translational and post-transcriptional modifications of proteins and tRNAs and are involved in the fermentation of various carbon sources by bacteria. Initially, however, there was skepticism that viperin might be a radical SAM enzyme.
Several factors contributed to this skepticism. Although more than 600,000 sequences currently fall in the radical SAM enzyme superfamily (70–73), at the time of viperin's discovery, far fewer radical SAM enzymes were known, and those were confined to bacteria. All radical SAM enzymes are oxygen-sensitive, making it seem unlikely that they would be active in aerobic organisms. Also, the various enzyme cofactors synthesized by radical SAM enzymes are obtained by animals in their diets as vitamins, obviating the need for the complex chemical transformations catalyzed by radical SAM enzymes. Furthermore, the literature contained conflicting information regarding whether the radical SAM domain of viperin was or was not required for the protein's antiviral activity, which seemed to depend upon the virus in question. For example, mutation of the conserved cysteine residues that ligate the catalytic [4Fe-4S] cluster was found to abolish viperin's antiviral activity against tick-borne encephalopathy virus (19) but had no effect on the enzyme's ability to restrict the infectivity of influenza A virus (11).
It is now established that viperin genuinely is a radical SAM enzyme—one of only eight annotated in the human genome; the others are involved in molybdopterin biosynthesis, fatty acid biosynthesis, and tRNA modifications (65). The first evidence supporting this view came from the demonstration that viperin catalyzed the reductive cleavage of SAM to form 5′-deoxyadenosine (5′-dA), which is a hallmark of radical SAM enzymes, albeit in a slow uncoupled reaction (74). However, none of the studies demonstrating viperin's antiviral properties provided any clear insight into the substrate(s) for the enzyme. The identification of the substrate for viperin therefore relied on educated guesswork.
Two key observations aided in identifying the substrate. They were (i) that the gene encoding viperin is found adjacent to the nucleoside monophosphate–phosphorylating enzyme cytidylate monophosphate kinase 2 (CMPK2) and (ii) viperin's similarity to the molybdopterin biosynthetic enzyme MoaA (59, 75). This enzyme catalyzes the cyclization of GTP to 3′,8-cyclo-GTP as the first step of molybdopterin biosynthesis (76). It is one of the few other radical SAM enzymes found in animals and is most similar to viperin in both sequence and structure, which are compared in more detail below. In particular, both viperin and MoaA possess the C-terminal extension to the radical SAM domain involved in binding their respective nucleoside triphosphate substrates, which is not generally conserved with other radical SAM enzymes.
After surveying a large number of viperin constructs, a stably folded N-terminally truncated version of rat viperin was identified that formed 5′-dA in vitro when CTP was used as a co-substrate (57). Subsequent analysis of the reaction products revealed that CTP was converted to ddhCTP, by a dehydration reaction that introduces a double bond between the 3′ and 4′ carbons of CTP (see Fig. 5B). Further confirmation that ddhCTP is produced by viperin was obtained by showing that mammalian cell lines that overexpress viperin produce high levels of ddhCTP (57). We note here that viperin also catalyzes the dehydration of UTP to form ddhUTP but binds this substrate much more weakly, with a Km ∼40-fold higher than that of CTP (75).
Figure 5.
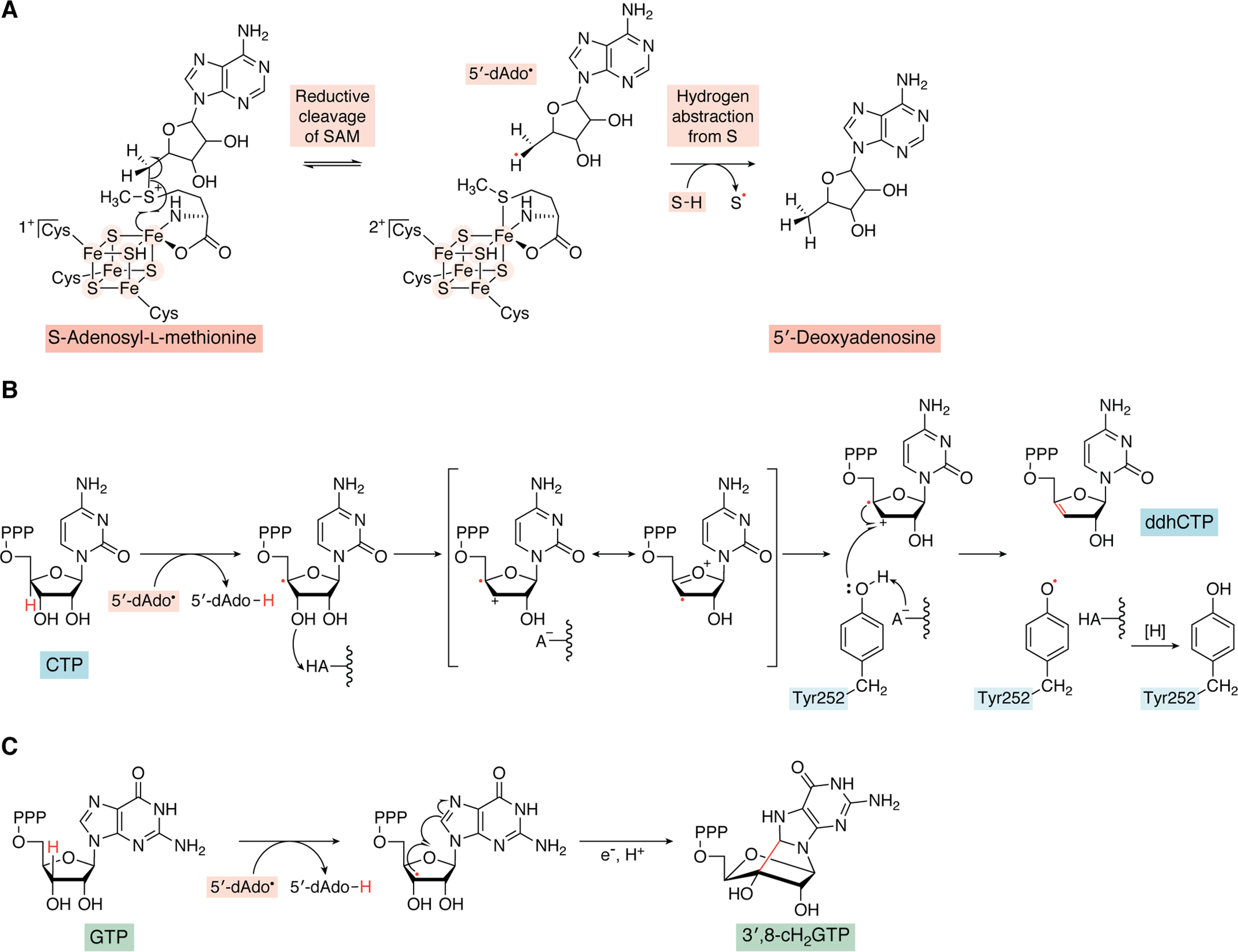
Proposed mechanism for the radical-mediated dehydration of CTP catalyzed by viperin. A, 5′-deoxyadenosyl radical is generated by reductive cleavage of SAM followed by hydrogen abstraction from the substrate (S), as occurs with other radical SAM enzymes. B, a mechanistic proposal for the dehydration of CTP in which the 5'-deoxyadenosyl radical first abstracts the 4′-hydrogen of CTP; this is followed by the loss of the 3′-hydroxyl group, likely assisted by a protein side chain (HA-) acting as a general acid, leading to a resonance-stabilized radical-cation intermediate. Tyr-252 is proposed to act as the intermediate source of the electron needed to complete the reaction and yield ddhCTP product. C, reaction mechanism of the formation of 3′,8-cH2GTP from GTP by MoaA in which the 5′-deoxyadenosyl radical abstracts the 3′-hydrogen of GTP, followed by the radical-mediated formation of a five-membered ring between the carbons at the 3′- and 8-positions. In the last step, an electron from the auxiliary [4Fe-4S] cluster reduces the radical, generating the product.
ddhCTP appears to exert its antiviral effects by acting as a chain-terminating inhibitor of genome replication by RNA viruses (Fig. 1). When ddhCTP is misincorporated by the viral RNA-dependent RNA polymerase, RNA polymerization cannot proceed as the growing chain now lacks a 3′-hydroxyl group. In vitro studies established that ddhCTP is efficiently misincorporated by the RNA-dependent RNA polymerases from flaviviruses such as the hepatitis C virus, dengue virus, West Nile virus, and Zika virus (57). Furthermore, in cell lines infected with Zika virus, the addition of ddhC to the culture medium was found to effectively suppress the viral titer. From these experiments, it appears that ddhCTP may be effective against flaviviruses. However, ddhCTP was not effective against RNA-dependent RNA polymerases from viruses from the picornavirus family, such as poliovirus and human rhinovirus (57), even though viperin is reported to be induced in response to these viruses (2). This latter observation suggests that ddhCTP production alone only partially explains viperin's antiviral properties.
Somewhat controversially, it has recently been suggested that ddhCTP does not exert its antiviral effects by acting as a chain terminator of viral replication (77). This proposal was based on the high IC50 values for ddhCTP competing with CTP measured for several viral RNA polymerases, which are several orders of magnitude higher than most synthetic chain-terminating nucleotide analogs (57, 77). Instead, it was proposed that by synthesizing ddhCTP, viperin depletes the cellular pool of CTP and UTP nucleotides available to the viral polymerases and that ddhCTP may furthermore interfere with mitochondrial metabolism. However, even low levels of misincorporation (less than 1 in 1000) would effectively terminate synthesis of a viral genome that contains several thousand cytidine nucleotides. Furthermore, co-localization of viperin and the viral RNA polymerase at the ER membrane would be expected to increase the local concentration of ddhCTP. Other studies from the same laboratory provided preliminary evidence that ddhCTP may act as an inhibitor of several NAD+-dependent dehydrogenases, including glyceraldehyde-3-phosphate dehydrogenase, lactate dehydrogenase, and malate dehydrogenase (78). But whether the inhibitory effects of ddhCTP have physiological relevance and how such broad-brush inhibition of primary metabolism might restrict viral infection are unclear. Further studies on the effects of ddhCTP on viral genome replication in vivo will be valuable to better establish its mode of action.
Other reactions of viperin and viperin-like enzymes
The viperin-like enzymes from fungi and archaea lack the N-terminal ER-localizing domain and have proved easier to overexpress in Escherichia coli than the mammalian enzymes. For these enzymes, other reactions were identified prior to the discovery of viperin's ddhCTP-synthesizing activity, which are summarized in Fig. 3. Following the observation that viperin may interfere with the synthesis of viral glycoproteins, a study using an enzyme from a thermophilic fungus (Thielavia terrestris) found that viperin catalyzed the addition of uridine diphosphate glucose (UDP-glucose) to the adenosine moiety of SAM through a radical mechanism (79). More recently, it has been shown that this enzyme, like mammalian viperin, also catalyzes the dehydration of CTP and UTP. It also appears to catalyze nonproductive radical formation at the 5′-position of ATP, which is trapped by reaction with the radical anion generated from the sodium dithionite used to reduce the enzyme (80). In a similar vein, the observation that viperin may inhibit farnesyl pyrophosphate synthase (FPPS) led to another study that demonstrated that fungal (Trichoderma virens) and archaeal (Methanofollis liminatans) paralogs of viperin catalyze the addition of the 5′-deoxyadenosyl radical to the double bond of isopentenyl pyrophosphate (60) (Fig. 3).
Figure 3.
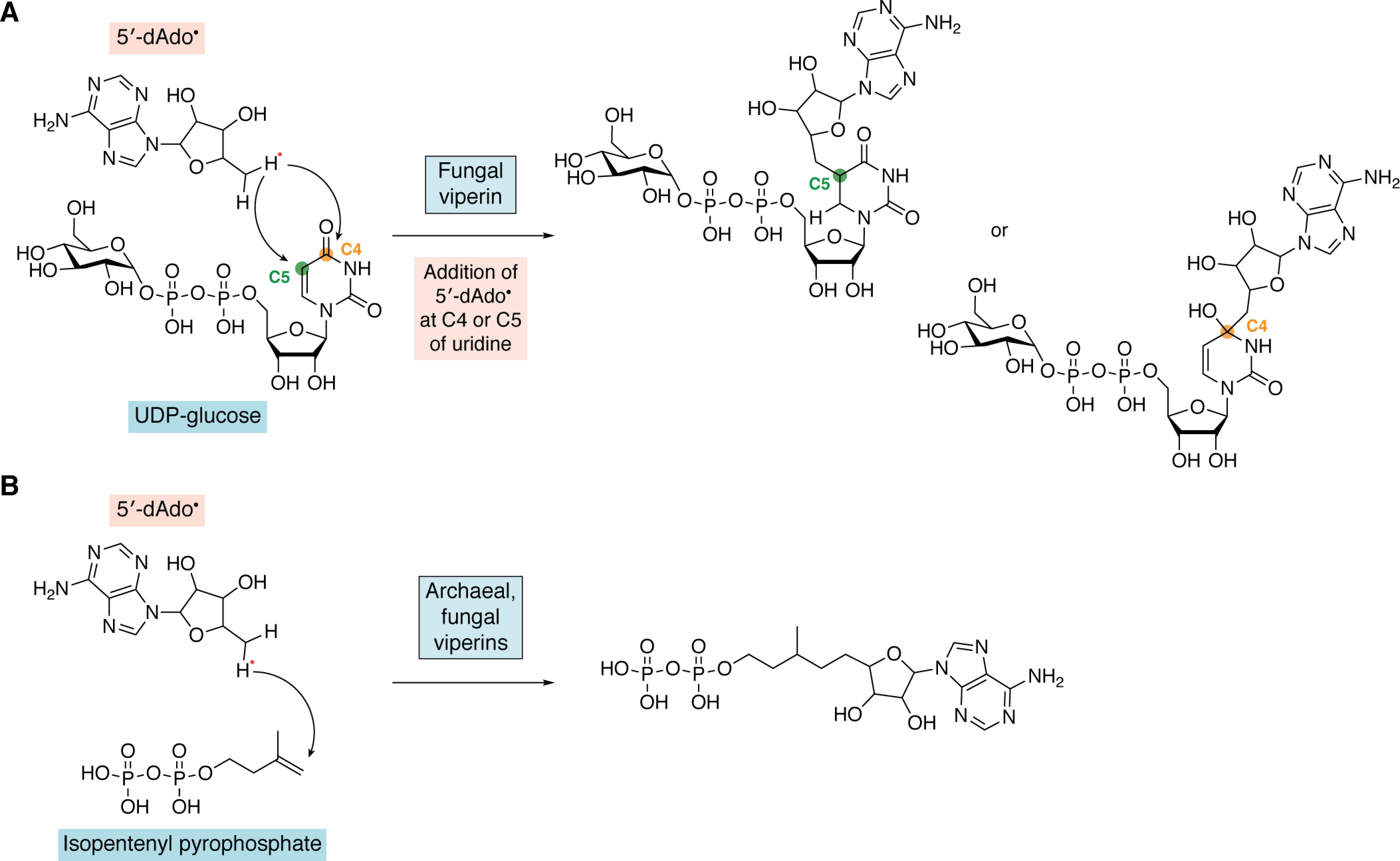
Radical addition reactions catalyzed by fungal and archaeal viperins. A, addition reaction of 5′-deoxyadenosyl radical to UDP-glucose at either C4 or C5 of uridine by a fungal viperin. B, addition reaction of 5′-deoxyadenosyl radical to isopentenyl pyrophosphate to form adenylated isopentenyl pyrophosphate catalyzed by archaeal and fungal viperins.
Neither UDP-glucose nor isopentenyl pyrophosphate were found to be substrates for the mammalian enzymes (57), and the physiological significance, if any, of these reactions remains unclear as nothing is known about the antiviral response in these organisms. Absent a clear biological rationale, it seems plausible that these reactions may result from adventitious binding of these substrates by the enzyme due to their negatively charged phosphate groups. Furthermore, this type of radical addition reaction is a departure from canonical radical SAM reactions that proceed through abstraction of a hydrogen atom from the substrate.
Last, it has been reported that viperin catalyzes the oxidation of protein methionine residues in DNA and RNA helicases (81). In particular, viperin was found to bind to Kaposi's sarcoma–associated herpesvirus helicase, the human DNA helicase, MCM7, and the human RNA helicase, RIG-I. However, the claim that viperin catalyzes methionine oxidation relies on indirect evidence (i.e. co-expression of viperin with these helicases resulted in oxidation of methionine at several positions in the helicases (81)). No mechanism was proposed for how such an oxidation would occur. Given what is known about the structure and mechanism of viperin, it would seem rather unlikely that protein methionine residues are a genuine substrate for the enzyme and more likely that this is an indirect effect of viperin overexpression. Confusingly, this protein modification was found to stabilize the helicases against degradation; therefore, if genuinely an effect of viperin, methionine oxidation seems unlikely to be an antiviral response.
Structure and mechanism of viperin
The crystal structure of mouse viperin lacking the first 43 residues of the ER-localizing sequence has been solved to 2.0 Å resolution with SAH bound in place of SAM (59). Subsequently, the structure with CTP also bound was solved to 1.45 Å resolution. (75) The structures are shown in Fig. 4. The N-terminal residues (residues 45–73) appear to be intrinsically disordered and are not resolved in any of the structures. Residues 75–284 of the radical SAM domain adopt the canonical “partial β-barrel” (βα)6 fold that characterizes other radical SAM enzymes (82). The catalytic [4Fe-4S] cluster is ligated by three cysteinyl side chains of the CXXXCXXC motif and projects into the lumen of the barrel, suspended by a loop between the first β-strand and α-helix of the barrel domain. The amino and carboxylate groups of SAH chelate the fourth (vacant) iron atom of the [4Fe-4S] cluster, and SAH binds in a conformation very similar to that seen in the structures of other radical SAM enzymes. The geometry of the interaction is arranged to facilitate reductive cleavage of SAM by electron transfer from the cluster to the sulfonium ion of SAM, which (based on the structure with SAH bound) is 3.6 Å away from the fourth, unique iron atom (75).
Figure 4.
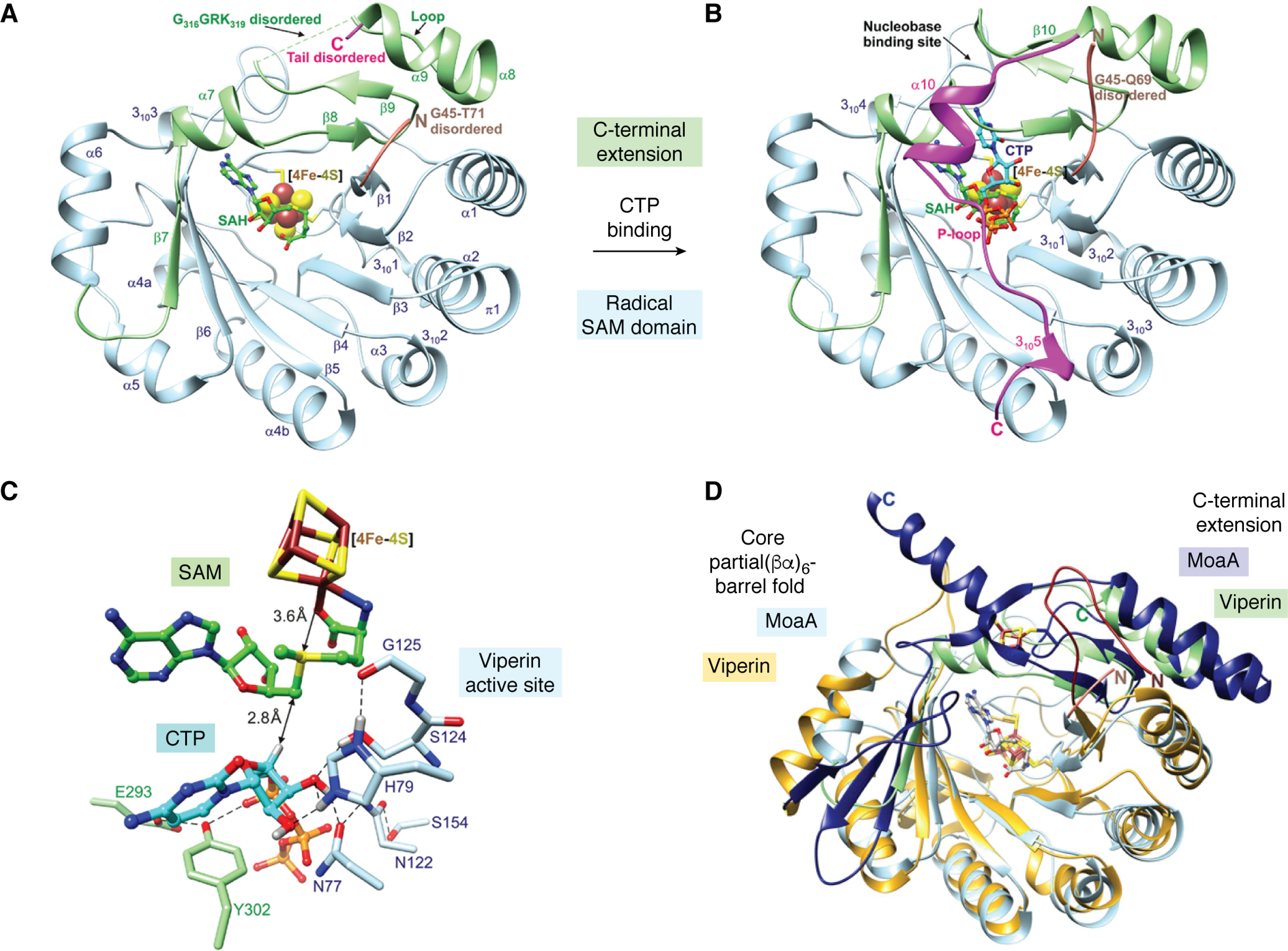
Structure of viperin. A, the radical SAM domain, in blue, adopts the “partial β-barrel” (βα)6 fold similar to radical SAM enzymes, but in the absence of CTP, the C-terminal extension remains disordered. B, with CTP bound, the C-terminal extension, in magenta, folds over the radical SAM domain to complete the binding site. C, detail of the active site with SAM modeled in place of SAH; the positioning of the substrates is ideally set up to facilitate reductive cleavage of SAM and abstraction of the 4′-hydrogen from CTP. D, overlay of the structures of MoaA (Protein Data Bank code 2FB3) and viperin (5VSL). The core partial (βα)6 fold is colored gold for viperin and pale blue for MoaA; the C-terminal extension is colored green for viperin and dark blue for MoaA; viperin contains an additional short N-terminal extension that is colored pink. The catalytic [4Fe-4S] clusters, auxiliary [4Fe-4S] cluster of MoaA, and bound SAH are shown as sticks. A–C are reproduced from Ref. 75, and D is reproduced from Ref. 59 with permission. This research was originally published in Biochemistry. Fenwick, M. K., Su, D., Dong, M., Lin, H., and Ealick, S. E. Structural basis of the substrate selectivity of viperin. Biochemistry. 2020;59:652–662. © American Chemical Society; Proceedings of the National Academy of Sciences of the United States of America. Fenwick, M. K., Li, Y., Cresswell, P., Modis, Y., and Ealick, S. E. Structural studies of viperin, an antiviral radical SAM enzyme. Proceedings of the National Academy of Sciences of the United States of America. 2017; 14, 6806–6811. © United States National Academy of Sciences.
The C-terminal extension forms a subdomain that is important for binding CTP. This subdomain is flexible and folds over the barrel, mimicking a closed (βα)8 fold. In the absence of CTP, the final 26 residues are disordered, but on binding CTP, all but the last 2 residues become structured to form an additional α-helical region, followed by a P-loop that binds the γ-phosphate (Pγ) of CTP and, last, a region of 310 helix (Fig. 4B). When bound, the 4′-hydrogen of CTP is only 2.8 Å away from the 5′-carbon of SAM and ideally positioned for its abstraction by the adenosyl radical (Fig. 4C). Interestingly, although the residues comprising the P-loop are highly conserved across all viperin and viperin-like sequences, those that make contact with the cytidine base are much less so (75). This suggests that other viperin-like enzymes might have selectivity toward nucleobases other than cytidine and possibly even modify deoxyribonucleotides. The ability of mammalian viperins to use UTP as a substrate, albeit a poor one, in addition to CTP provides further support for this idea.
Overall, the three-dimensional structure of viperin most closely resembles that of MoaA (76, 83), as shown in the overlay of the two proteins in Fig. 4D. In particular, the residues involved in binding the triphosphate groups of the substrates are highly conserved between the two enzymes, and their structures overlay closely in this region. One important difference is that viperin lacks the auxiliary C-terminal [4Fe-4S] cluster found in the C-terminal domain of MoaA (Fig. 4D). Many radical SAM enzymes contain such auxiliary clusters, and they often function to accept or donate an additional electron that is required in the reaction (84). As shown in Fig. 5, the reactions catalyzed by viperin and MoaA (83) share some mechanistic similarities, including the requirement for a final one-electron transfer to reduce the product radical, so it is perhaps surprising that viperin lacks an auxiliary cluster that could perform this reduction.
The mechanism by which CTP is converted to ddhCTP has not been investigated in great detail, but a plausible mechanism is shown in Fig. 5. Isotope labeling has established that the 4′-hydrogen of CTP is transferred to 5′-dA during the reaction (57). The C4′-CTP radical thus generated has the effect of making the 3′-hydroxyl of the ribose ring a much better leaving group by stabilizing the resulting positive charge at the C3′ position (Fig. 5, A and B). A similar catalytic strategy is employed in the radical-mediated dehydration and deamination reactions catalyzed by ribonucleotide reductases and adenosylcobalamin-dependent enzyme, such as diol dehydratase and ethanolamine ammonia lyase (66, 85, 86). However, in these enzymes, radicals are employed catalytically, with the radical-generating cofactor reconstituted after each turnover. In contrast, SAM is consumed stoichiometrically in the viperin-catalyzed reaction, and therefore an additional electron is needed to reduce the ddhCTP radical in the last step of the reaction.
In this regard, the viperin reaction is most similar to the deamination of TDP-4-amino-4,6-dideoxy-d-glucose catalyzed by the radical SAM enzyme TDP-4,6-dideoxyhexose 3,4-enoyl reductase (DesII) (87). However, in contrast to MoaA, neither viperin nor DesII contains an auxiliary [4Fe-4S] cluster that could supply this electron. This implies that the additional electron required by the reaction must be supplied by another route. The pathway by which the final one-electron reduction step occurs remains unclear, but interestingly recent EPR studies suggest that a conserved active-site tyrosine residue may serve as the immediate electron donor in the viperin reaction (Fig. 5). The evidence for this proposal comes from EPR studies on the viperin-like enzyme from T. terrestris, which when reacted with UTP generated an EPR spectrum characteristic of a protein tyrosyl radical. Mutation of the active-site Tyr residue to Phe abolished both the EPR signal and enzyme activity, suggesting that this residue is responsible for the EPR signal and essential for the reaction (80).
Interactions with other proteins
Although the synthesis of the antiviral ribonucleotide ddhCTP explains how viperin may inhibit the replication of some viruses, it is also clear that viperin interacts with numerous other cellular and viral proteins and that these interactions are vitally important to the broad-spectrum antiviral activity associated with viperin's expression. Table 2 summarizes the various different proteins that viperin interacts with and the effects of these interactions, where known. Broadly speaking, the targets of viperin may be divided into three groups: cellular proteins involved in innate immune signaling; proteins involved in cellular metabolic pathways that are exploited during the viral life cycle; and structural and nonstructural viral proteins. However, it is not always clear whether the effects attributed to viperin expression arise from a direct interaction between viperin and the other protein under investigation or are an indirect consequence of viperin expression. This ambiguity often arises from the differing nature of the experimental techniques employed, which in some cases leads to apparently contradictory findings. Here we discuss some of the better characterized interactions of viperin with other proteins, some of which are summarized in Fig. 6, that provide insights into how the enzyme is integrated into broader cellular antiviral response.
Table 2.
Proteins reported to interact with viperin
| Protein | Role of protein | Effect of interaction on viperin | Effect of interaction on the target protein | References |
|---|---|---|---|---|
| Cellular proteins | ||||
| IRAK 1 | Protein kinase involved in immune signaling | Co-activator of ddhCTP synthesis | Promotes Lys-63–linked ubiquitination of IRAK1 | 51, 55, 88 |
| TRAF6 | Lys-63–linked E3 ubiquitin ligase, involved in immune signaling | Co-activator of ddhCTP synthesis | Activates TRAF6 toward ubiquitination of IRAK1 | 51, 55, 88 |
| HADHB | β-Subunit of mitochondrial trifunctional protein, | Activator of ddhCTP synthesis | Inhibits thiolase activity of HADHB; promotes degradation of HADHB | 49, 50, 89 |
| FPPS | catalyzes synthesis of farnesyl diphosphate, a precursor for sterols | None observed | Promotes intracellular degradation of FPPS | 11, 90 |
| GBF1 | Guanine nucleotide exchange factor involved in protein trafficking in the early secretory pathway | Not known | Inhibits the cellular function of GBF1, leading to the selective release of TBEV particles | 91 |
| MAVS | Downstream adaptor of the cytosolic RIG-I and MDA-5 receptors; induces the IFN-β expression | Not known | Suppresses MAVS-dependent IFN-β production. May act indirectly | 10, 27, 92 |
| Helicase MCM2 | DNA helicase | Not known | Promotes methionine oxidation of MCM2, enhancing protein stability | 81 |
| UBE4A | E3 ubiquitin ligase mediating Lys-48–linked polyubiquitination of proteins | Promotes polyubiquitination and degradation through proteasome-mediated pathway | Not known | 30 |
| Viral proteins | ||||
| NS5A | Important for formation of viral replication complex | HCV NS5A deactivates viperin to catalyze the conversion reaction of CTP to ddhCTP | Promotes degradation of NS5A through proteasome-mediated pathway | 12–14, 17, 61, 93 |
| NS3 | Acts as protease/helicase in viral replication | Not known | Promotes proteasome-mediated degradation of NS3 in some viruses | 16–18, 93 |
| pp28 and pp65 (HCMV) | Tegument proteins involved in immune evasion or viral assembly and egress | Not known | Not known | 7, 94 |
| vMIA | Suppresses apoptosis by blocking permeabilization of the mitochondrial outer membrane | Relocalizes viperin from ER to mitochondria | Not known | 49, 95 |
| Glycoprotein B/D | Binds to membrane receptor on host cells facilitating viral entry | Disrupts interaction between viperin and TRAF6 in Herpesvirus infection | Direct modification of the proteins was not observed | 6, 7, 51 |
| C or N capsid proteins | Protects viral nucleic acids from host cell enzymes | Not known | May impair viral replication by interacting with capsid proteins | 16, 36, 40, 91 |
Figure 6.
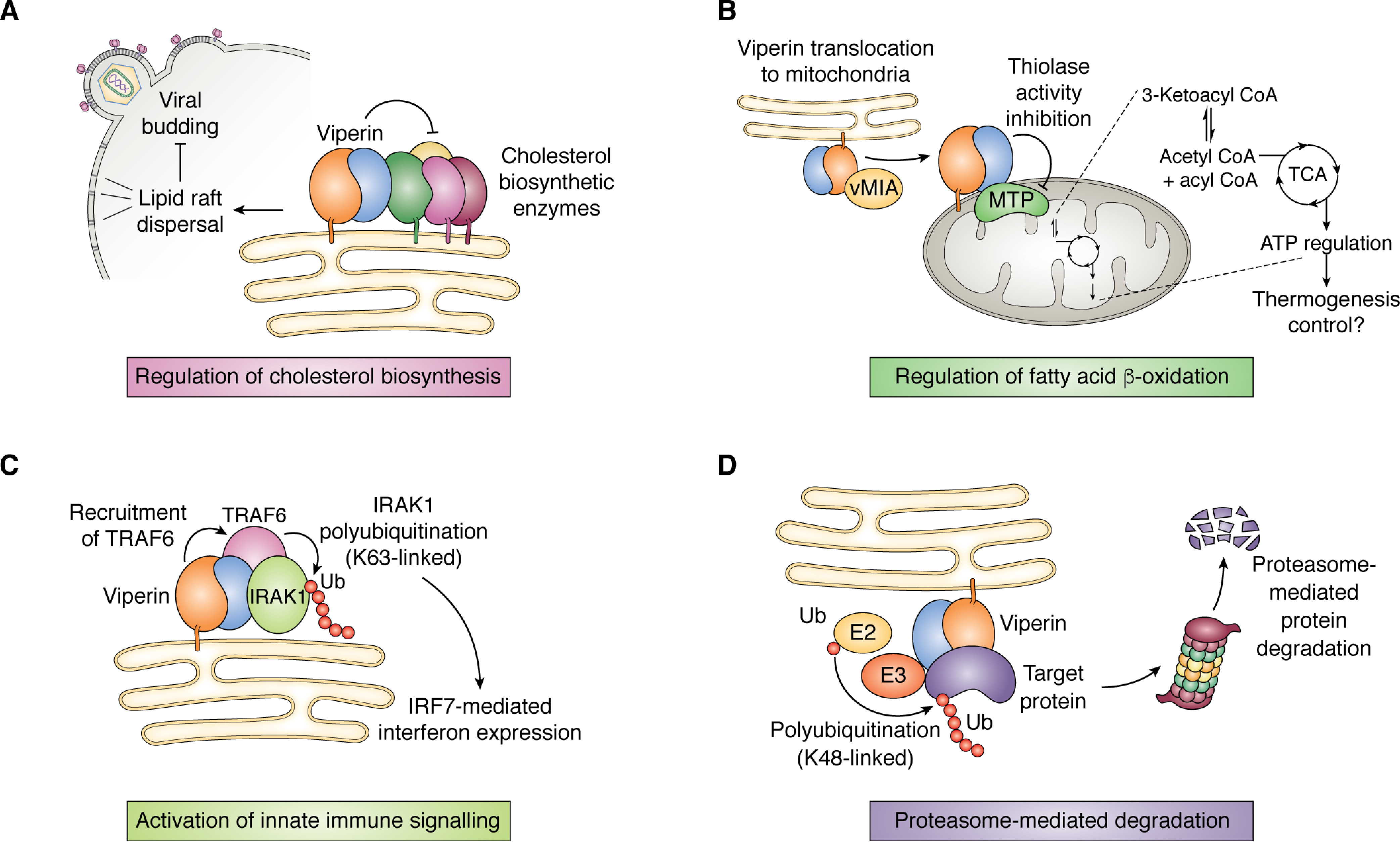
Overview of viperin's interaction with other proteins. A, viperin perturbs the formation of cholesterol-rich lipid rafts, which stalls budding of enveloped viruses such as influenza A. This is thought to occur through the down-regulation of cholesterol biosynthetic enzymes, including FPPS. B, translocation of viperin to the mitochondrion results in down-regulation of the thiolase activity of HADHB, the β-subunit of the MTP complex involved in fatty acid β-oxidation, which impacts ATP production. C, viperin interacts with the protein kinase IRAK1 and E3 ubiquitin ligase TRAF6 to facilitate Lys-63–linked polyubiquitination of IRAK1 by TRAF6, thereby activating the IRF7-mediated interferon expression. D, viperin interacts with various structural and nonstructural viral proteins and promotes their degradation through the ubiquitin-dependent proteasomal pathway.
Role of viperin in immune signaling
Interactions with IRAK1 and TRAF6
Viperin is increasingly recognized to play an important role in innate immune signaling (2, 20). It has been shown to enhance the activation of key signaling molecules in both the ssRNA-sensing Toll-like receptor 7 (TLR7) and CpG DNA-sensing TLR9 pathways that lead to type I interferon production (55, 96). Various lines of evidence point to viperin stimulating Lys-63–linked polyubiquitination of interleukin-1 receptor–associated kinase (IRAK1) (97–99) by the E3 ubiquitin ligase, tumor necrosis factor receptor (TNFR)-associated factor 6 (TRAF6), which are downstream signaling components of the TLR7 and TLR9 pathways (55, 98). Lys-63–linked polyubiquitination of IRAK1 activates this kinase to phosphorylate interferon-regulatory factor 7 (IRF7), causing IRF7 to migrate to the nucleus, where it activates transcription of type I interferons (Fig. 6C) (99, 100).
The interactions between viperin, IRAK1, and TRAF6 have been investigated by immunoprecipitation experiments, which demonstrated that viperin, IRAK1, and TRAF6 form a complex. Viperin did not bind TRAF6 in the absence of IRAK1, suggesting that IRAK1 mediates the interaction between viperin and TRAF6 (88). Using transfected proteins in HEK 293T cells, viperin was found to be necessary to stimulate the polyubiquitination of IRAK1 by TRAF6, when the three proteins form a complex. In contrast, an inactive mutant, viperinΔ3C, that lacks the [4Fe-4S] cluster, failed to stimulate polyubiquitination of IRAK1 by TRAF6. It appeared that removing the [4Fe-4S] cluster destabilized viperin, resulting in a significantly shorter cellular t½; however, its t½ was restored when cells were cultured in the presence of sinefungin, a tight-binding inhibitor of many SAM-dependent enzymes. Sinefungin presumably stabilizes the structure of the enzyme (88). Significantly, sinefungin also rescued the ability of viperinΔ3C to stimulate polyubiquitination of IRAK1, demonstrating that the radical SAM activity of viperin is not required to activate TRAF6 to ubiquitinate IRAK1 (88). Rather, sinefungin acts by stabilizing viperin in an active conformation.
Interactions with STING and TBK1
Recently it was shown that viperin interacts with the signaling adaptor proteins, STING (stimulator of interferon genes) and the threonine-serine protein kinase, TBK1 (101). STING plays a central role in sensing cytosolic dsDNA and as an adaptor protein involved in type I interferon-signaling pathways (102, 103). Its downstream target is TBK1. STING binding to TBK1 activates its kinase activity by stimulating Lys-63–linked polyubiquitination of TBK1 on lysine residues 30 and 401 through the action of an E3 ubiquitin ligase, yet unidentified. Once activated, TBK1 phosphorylates transcription factor IRF3, which is translocated to the nucleus and promotes production of IFN-I. Viperin binding to STING was found to enhance the activation of TBK1 by increasing the polyubiquitination of TBK1 (101). Although STING is not itself a ubiquitin ligase, viperin's interactions with STING and TBK1, which up-regulate polyubiquitination, clearly parallel those observed with IRAK1 and TRAF6. In this case, the [4Fe-4S] cluster of viperin appears to be important for its activating effect because the viperinΔ3C mutant failed to stimulate polyubiquitination of TBK1. By analogy with the activation of IRAK1/TRAF6, the [4Fe-4S] cluster most likely plays a structural role, maintaining viperin in a conformation that can bind STING and TBK1, rather than reflecting the need for catalytically active enzyme.
Activation of viperin by immune signaling components
As discussed in the examples above, viperin modulates the biological activity of many of its binding partners. It would seem likely, therefore, that these proteins would also affect the activity of viperin, and indeed this seems to be the case.
In common with many radical SAM enzymes, viperin turns over very slowly; for example, for the recombinantly expressed and purified truncated rat enzyme, kcat is ∼0.2 min−1 (57). The full-length human enzyme has a turnover number of only 0.04 min−1 when overexpressed and assayed in HEK 293T cell lysates. However, in lysates prepared from cells co-expressing IRAK1 and TRAF6, the specific activity of viperin increases ∼10-fold to 0.36 min−1. This activation is primarily due to viperin binding IRAK1 but is further enhanced by TRAF6 (88). A similar result is observed when viperin is co-expressed with STING and TBK1. In this case, STING and TBK1 increase the specific activity of viperin by ∼10 fold, with STING providing the majority of the activation (101).
These observations suggest a regulatory mechanism that links the synthesis of the antiviral nucleotide, ddhCTP, by viperin to innate immune signaling pathways mediated by proteins, such as IRAK1, TRAF6, STING, and TBK1. Through this mechanism, the up-regulation of various proteins necessary for the cellular antiviral response would be coordinated with the production of ddhCTP. In both cases, viperin stimulates polyubiquitination of a protein kinase to potentiate the signaling pathway—as discussed further below, it seems plausible that activation of E3 ubiquitin ligase may underpin other aspects of viperin's antiviral activity.
Interaction of viperin with metabolic enzymes
Farnesyl-pyrophosphate synthase
FPPS was one of the first enzymes identified as a possible target of viperin (before the catalytic activity of viperin had been elucidated) as a result of yeast two-hybrid screening to detect potential interaction partners (11). Farnesyl pyrophosphate is a key intermediate in the pathway leading to isoprenoids and cholesterol (104, 105). Many enveloped viruses exploit cholesterol-rich lipid rafts in the membrane to bud from the host cell (2, 62, 106). Cell-based studies implicated FPPS inhibition by viperin as contributing to viperin's antiviral activity against influenza A, as viperin expression reduced the activity of FPPS and resulted in viral particles becoming trapped in the cell membrane (Fig. 6A) (11). It is thought that by inhibiting FPPS, and thus cholesterol production, viperin retards the virus from budding from the cell.
Subsequent studies, employing recombinantly expressed enzymes purified from E. coli, failed to find any direct interaction between viperin and FPPS as judged by co-immunoprecipitation assays (90). Further experiments, employing FPPS and viperin transiently co-expressed in HEK 293T cells, demonstrated that the enzymatic activity of viperin and FPPS is unchanged by co-expression. Instead, it was found that co-expression of viperin significantly reduces cellular FPPS levels, most likely by accelerating the rate at which FPPS is degraded (90). Unexpectedly, inactive viperin mutants, in which the conserved cysteinyl residues chelating the [4Fe-4S] cluster were mutated to alanine, still effectively suppressed FPPS expression. Although the molecular mechanism by which viperin accelerates the degradation of FPPS was not established, these studies were among the first to demonstrate that viperin's dual biological functions as radical SAM enzyme and agonist of protein degradation may be independent of each other.
Mitochondrial trifunctional protein
Another metabolic target identified for viperin is the mitochondrial trifunctional protein (MTP), which is involved in fatty acid β-oxidation (Fig. 6B) (7). Interestingly, this interaction was identified in studies of cytomegalovirus infection and arises through the virus-induced relocalization of viperin to the mitochondrion (49). Translocation into the mitochondrion occurs through the interaction of viperin with the virally encoded protein, vMIA (mitochondrion-localized inhibitor of apoptosis), although the details of this process are not understood. It is thought that by inhibiting β-oxidation, and hence ATP production, the virus weakens the cytoskeleton to facilitate its escape from the cell (49).
Studies using purified enzymes in vitro showed that viperin directly binds to the β-subunit of MTP (HADHB) and inhibits the thiolase activity, which is performed by this subunit of the multienzyme complex (50). This interaction was further investigated in vivo by directly targeting viperin to the mitochondrion by appending a mitochondrial leader sequence to the enzyme. Localizing viperin to the mitochondria decreased the thiolase activity of cell extracts, consistent with the in vitro experiments, and consequently caused cellular ATP levels to fall significantly (50). These effects may partly be attributed to viperin binding HADHB and inhibiting its activity, but targeting viperin to the mitochondrion also lowered HADHB protein levels, most likely by increasing the rate at which HADHB was retro-translocated out of mitochondria and degraded by the proteasome. These functions appear to depend on the enzyme possessing a [4Fe-4S] cluster, as a viperin mutant lacking the [4Fe-4S] cluster still bound HADHB but failed to stimulate HADHB degradation, decrease thiolase activity, or depress cellular ATP levels (50).
A recent report showed that viperin is intrinsically expressed at low levels in adipocytes and intriguingly pointed to viperin decreasing fatty acid β-oxidation in rodents as part of the nonpathological regulation of thermogenesis (89). Viperin knockout mice were found to generate more heat and to have reduced body fat and improved high-fat diet–induced glucose tolerance. These observations are consistent with viperin acting to regulate β-oxidation through its interaction with HADHB, as discussed above. These observations may also explain the rather surprising manner in which CMV co-opts viperin to facilitate the virus's escape from the cell. We speculate that CMV may have evolved to exploit to its advantage existing mechanisms for targeting viperin to the mitochondrion and regulating mitochondrial metabolism. However, it is not clear whether viperin is translocated to mitochondria in nonpathogenic conditions.
Surprisingly, although viperin inhibits HADHB, HADHB was found to activate viperin toward the synthesis of ddhCTP severalfold (50). It is unclear why viperin should be activated to synthesize ddhCTP in mitochondria because this is not a site of viral replication. However, given that viperin appears to be involved in nonpathogenic regulation of mitochondrial metabolism, an intriguing possibility is that ddhCTP may play an unrecognized role in modulating mitochondrial transcription. ddhCTP does not seem to be incorporated by nuclear RNA polymerases, but the mitochondrial enzyme most closely resembles the bacteriophage T7 RNA polymerase (107) and so might be susceptible to ddhCTP.
Interaction of viperin with viral proteins
Viperin has been shown to interact with various viral proteins, both structural and nonstructural, from a number of viruses (Fig. 6D). Its interactions with flaviviruses, which are responsible for diseases such as yellow fever, dengue fever, West Nile fever, Zika fever, and tick-borne and Japanese encephalitis, have been most extensively documented and are the subject of a recent review (3). Notably, even within the flavivirus family, viperin's interaction with viral proteins appears idiosyncratic. For example, viperin binds nonstructural protein 3 (NS3), the protease that cleaves the viral polyprotein into mature components, from many flaviviruses. For tick-borne encephalitis virus (TBEV) and Zika virus (18), viperin causes NS3 to be degraded by the proteasomal degradation pathway (26); in contrast, NS3 proteins from Japanese encephalitis and yellow fever viruses were not degraded, and interestingly, these latter viruses are reported not to be inhibited by viperin. In another example, the dengue virus coat protein was shown to interact with viperin, but the TBEV coat protein did not (16, 91). Complicating matters, some of viperin's interactions with viral proteins may occur indirectly (e.g. co-expression of the TBEV NS3 protein with viperin results in other TBEV proteins, including prM, E, NS2A, and NS2B, being degraded, although these proteins are not degraded by viperin alone). This may be explained by the fact that all five viral proteins associate as part of the viral replication complex (91).
The flavivirus protein nonstructural protein 5A, NS5A, plays a crucial but incompletely understood role in viral replication; it binds both viral RNA and the viral RNA polymerase, NS5B, among other proteins (9, 12). The NS5A protein from hepatitis C has been investigated and found to form a complex with the 33-kDa vesicle-associated protein, VAP33, and viperin (13). All three proteins are membrane-associated and reside at the ER membrane or on lipid droplets, which are the sites for replication of flaviviruses such as HCV (61). The association of these proteins with the ER membrane or lipid droplets appears to be important for their interaction: when expressed without their membrane-localizing domains, NS5A, VAP33, and viperin failed to form a complex (14).
Whether viperin binding to NS5A is a mechanism by which viperin inhibits viral replication or, conversely, NS5A inhibits viperin is still not entirely clear. On the one hand, co-expression of viperin with VAP33 and NS5A in HEK 293T cells was found to promote the degradation of NS5A through the proteasomal protein degradation pathway; this activity was not dependent on viperin possessing its [4Fe-4S] cluster. As NS5A is essential for viral replication, its degradation would inhibit this process. On the other hand, when co-expressed with VAP33 and NS5A, viperin's ddhCTP-synthesizing activity was found to be lower (14). This may be viewed as an adaptation by the virus to partially neutralize viperin's antiviral effects, but further studies are needed to establish the significance of this observation in relation to viperin's ability to restrict HCV infection in cells.
Regulation of protein secretion
Some studies point to viperin playing a role in regulating the translocation of proteins into the ER and hence their subsequent export from the cell. This is the route that some viruses, such as flaviviruses, exploit to exit the cell. An early study found that overexpression of viperin inhibits the secretion of soluble proteins (62), and the N-terminal amphipathic helix of viperin appeared to be important for this to occur. It was speculated that its amphipathic helix may induce changes in the morphology of the ER that inhibited protein secretion. Another study identified an interaction between viperin and the cellular protein Golgi brefeldin A–resistant guanine nucleotide exchange factor 1 (GBF1) (91). This protein is involved in vesicle trafficking in the secretory pathway and is known to be very important in the life cycle of many different viruses (3). It was hypothesized that by sequestering GBF1, viperin may impede viral budding into the ER and, in turn, the exit of virus particles from the cell. Another study implicated viperin in regulating secretion of soluble proteins in differentiating chondrocytes (108), which are the cells responsible for the production and maintenance of cartilage. In particular, it was found to regulate the secretion of the cytokine CXC motif chemokine ligand 10 (CXCL10). This appears to be another example of viperin acting as a regulatory protein under nonpathological conditions. The mechanism by which viperin may regulate protein secretion is currently unexplored.
Outlook
Although viperin had been known for some 20 years to be induced in response to viral infection and by interferon (as its name implies), it is only recently that its enzymatic activity has been elucidated. The conservation of this radical SAM enzyme across all kingdoms of life indicates that the synthesis of ddhCTP (and ddhUTP by fungal enzymes) is an ancient antiviral mechanism. It is notable that this arm of the antiviral response has been preserved even in higher animals, in which few other radical SAM enzymes have been retained. Based on their conserved sequences, it seems likely that the viperin-like enzymes found in unicellular organisms also synthesize ddhCTP and/or ddhUTP, although at this point, it cannot be ruled out that they may also catalyze other reactions. The mechanism by which viperin catalyzes the dehydration of CTP to ddhCTP has yet to be investigated in great detail, but the principle of generating a substrate radical to activate an adjacent hydroxyl group is well-established in the mechanism of ribonucleotide reductase.
The synthesis of ddhCTP provides a simple and elegant explanation for viperin's antiviral activity, but it is increasingly clear that in animals viperin is integrated into the much broader cellular antiviral response through a network of protein-protein interactions. The significance of these various interactions is only just beginning to be understood. For example, it is well-established that viperin is regulated at the transcriptional level, but only recently has it become apparent that viperin's activity is also modulated by its interactions with other proteins, as illustrated by the ∼10-fold activation of ddhCTP-synthesizing activity when viperin is complexed with IRAK1 and TRAF6 (88). It is also evident that many of viperin's functional interactions with other proteins do not depend on its radical SAM-based catalytic activity. As one example, discussed above, an inactive viperin mutant can be rescued to efficiently stimulate TRAF6-catalyzed polyubiquitination of IRAK1 by adding the tight-binding SAM analog, sinefungin.
Among the diverse antiviral properties reported for viperin, one emerging theme is that viperin promotes the degradation of proteins, both cellular and viral, to impede the replication of viruses. Various lines of indirect evidence point to viperin stimulating ubiquitin-dependent protein degradation of its targets, which would imply that viperin may engage with a subset of the E3 ubiquitin ligases responsible for tagging proteins with Lys-48–linked polyubiquitin chains to mark them for degradation. This hypothesis chimes with the observation that viperin stimulates Lys-63–linked polyubiquitination as part of innate immune signaling pathways. Here there is still much to learn, including how viperin recognizes its target proteins and which of the many ubiquitin ligases it interacts with.
An emerging function of viperin is that it may play a role in regulating cellular metabolism under nonpathological conditions, as viperin is constitutively expressed in some tissues at low levels. So far, several reports have linked viperin to the regulation of protein secretion through the endoplasmic reticulum. Preliminary evidence also exists for viperin regulating thermogenesis through its interaction with HADHB in mitochondria. As the interactome of viperin becomes better established, we believe that other protein interactions will be identified that may serve a regulatory function outside of the antiviral response.
So far, the only documented function of ddhCTP is as a chain-terminating inhibitor of certain viral RNA-dependent RNA polymerases. However, as viperin is now implicated in regulating an increasing number of cellular processes (e.g. protein secretion and thermogenesis), which are unrelated to the innate immune response, it seems reasonable to ask whether there may be additional functions for ddhCTP. One recently suggested role is that ddhCTP, synthesized when viperin is translocated to the mitochondria, may alter the transcription of mitochondrial genes by acting as a chain terminator of mitochondrial mRNA (50, 77).
The elucidation of viperin's catalytic activity—synthesizing the antiviral nucleotide ddhCTP—provided a major advance in our understanding of this ancient radical SAM enzyme that otherwise seemed out of place in modern, aerobic animals. However, it is also clear that, like many enzymes in higher animals, viperin serves important regulatory functions beyond its role as a catalyst. A major challenge for the field is to rationalize how one protein can participate in so many apparently unrelated cellular processes. Although viperin is known to interact with numerous proteins, how viperin and its interacting partners recognize each other remains completely unknown. The crystal structure of the enzyme provides no clues for how other proteins might bind to it, and no structures have yet been determined for viperin complexed with any of its interaction partners. This represents a significant bottleneck in our understanding of viperin's modes of action. Going forward, detailed structural information for the complexes formed by viperin with other proteins will be a prerequisite for understanding how viperin recognizes its target proteins and regulates their biological activities.
Acknowledgments
We thank Prof. Jennifer Bridwell-Rabb, Prof. Karla Helbig, Dr. Timothy Grunkemeyer, and Ayesha Patel for critical reading of the manuscript.
Funding and additional information—Work on viperin in the authors' laboratory is supported by National Institutes of Health Grant GM 093088 (to E. N. G. M.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- ISG
- interferon-stimulated gene
- CMV
- cytomegalovirus
- HCMV
- human CMV
- HCV
- hepatitis C virus
- TLR
- Toll-like receptor
- ddhCTP
- 3′-deoxy-3′,4′-didehydrocytidine triphosphate
- ddhUTP
- 3′-deoxy-3′,4′-didehydrouridine triphosphate
- FPPS
- farnesyl pyrophosphate synthase
- TBEV
- tick-borne encephalitis virus
- ER
- endoplasmic reticulum
- IFN
- interferon.
References
- 1. Seo J.-Y., Yaneva R., and Cresswell P. (2011) Viperin: a multifunctional, interferon-inducible protein that regulates virus replication. Cell Host Microbe 10, 534–539 10.1016/j.chom.2011.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Helbig K. J., and Beard M. R. (2014) The role of viperin in the innate antiviral response. J. Mol. Biol. 426, 1210–1219 10.1016/j.jmb.2013.10.019 [DOI] [PubMed] [Google Scholar]
- 3. Lindqvist R., and Överby A. K. (2018) The role of viperin in antiflavivirus Responses. DNA Cell Biol. 37, 725–730 10.1089/dna.2018.4328 [DOI] [PubMed] [Google Scholar]
- 4. Mattijssen S., and Pruijn G. J. (2012) Viperin, a key player in the antiviral response. Microbes Infect. 14, 419–426 10.1016/j.micinf.2011.11.015 [DOI] [PubMed] [Google Scholar]
- 5. Zhu H., Cong J.-P., and Shenk T. (1997) Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc. Natl. Acad. Sci. U. S. A. 94, 13985–13990 10.1073/pnas.94.25.13985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boudinot P., Massin P., Blanco M., Riffault S., and Benmansour A. (1999) vig-1, a new fish gene induced by the rhabdovirus glycoprotein, has a virus-induced homologue in humans and shares conserved motifs with the MoaA family. J. Virol. 73, 1846–1852 10.1128/JVI.73.3.1846-1852.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chin K. C., and Cresswell P. (2001) Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 98, 15125–15130 10.1073/pnas.011593298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fitzgerald K. A. (2011) The interferon inducible gene: viperin. J. Interferon Cytokine Res. 31, 131–135 10.1089/jir.2010.0127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Helbig K. J., Lau D. T. Y., Semendric L., Harley H. A. J., and Beard M. R. (2005) Analysis of ISG expression in chronic hepatitis C identifies viperin as a potential antiviral effector. Hepatology 42, 702–710 10.1002/hep.20844 [DOI] [PubMed] [Google Scholar]
- 10. Severa M., Coccia E. M., and Fitzgerald K. A. (2006) Toll-like receptor-dependent and -independent viperin gene expression and counter-regulation by PRDI-binding factor-1/BLIMP1. J. Biol. Chem. 281, 26188–26195 10.1074/jbc.M604516200 [DOI] [PubMed] [Google Scholar]
- 11. Wang X., Hinson E. R., and Cresswell P. (2007) The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2, 96–105 10.1016/j.chom.2007.06.009 [DOI] [PubMed] [Google Scholar]
- 12. Helbig K. J., Eyre N. S., Yip E., Narayana S., Li K., Fiches G., McCartney E. M., Jangra R. K., Lemon S. M., and Beard M. R. (2011) The antiviral protein viperin inhibits hepatitis C virus replication via interaction with nonstructural protein 5A. Hepatology 54, 1506–1517 10.1002/hep.24542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang S., Wu X., Pan T., Song W., Wang Y., Zhang F., and Yuan Z. (2012) Viperin inhibits hepatitis C virus replication by interfering with binding of NS5A to host protein hVAP-33. J. Gen. Virol. 93, 83–92 10.1099/vir.0.033860-0 [DOI] [PubMed] [Google Scholar]
- 14. Ghosh S., Patel A. M., Grunkemeyer T. J., Dumbrepatil A. B., Zegalia K., Kennedy R. T., and Marsh E. N. G. (2020) Interactions between viperin, vesicle-associated membrane protein A, and Hepatitis C virus protein NS5A modulate viperin activity and NS5A degradation. Biochemistry 59, 780–789 10.1021/acs.biochem.9b01090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jiang D., Weidner J. M., Qing M., Pan X.-B., Guo H., Xu C., Zhang X., Birk A., Chang J., Shi P.-Y., Block T. M., and Guo J.-T. (2010) Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infections. J. Virol. 84, 8332–8341 10.1128/JVI.02199-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Helbig K. J., Carr J. M., Calvert J. K., Wati S., Clarke J. N., Eyre N. S., Narayana S. K., Fiches G. N., McCartney E. M., and Beard M. R. (2013) Viperin is induced following dengue virus type-2 (DENV-2) infection and has anti-viral actions requiring the C-terminal end of viperin. PLoS Negl. Trop. Dis. 7, e2178 10.1371/journal.pntd.0002178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cedillo-Barrón L., García-Cordero J., Shrivastava G., Carrillo-Halfon S., León-Juárez M., Bustos Arriaga J., León Valenzuela P., and Gutiérrez Castañeda B. (2018) The role of flaviviral proteins in the induction of innate immunity. Subcell. Biochem. 88, 407–442 10.1007/978-981-10-8456-0_17 [DOI] [PubMed] [Google Scholar]
- 18. Panayiotou C., Lindqvist R., Kurhade C., Vonderstein K., Pasto J., Edlund K., Upadhyay A. S., and Överby A. K. (2018) Viperin restricts Zika virus and tick-borne encephalitis virus replication by targeting NS3 for proteasomal degradation. J. Virol. 92, e02054–17 10.1128/jvi.02054-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Upadhyay A. S., Vonderstein K., Pichlmair A., Stehling O., Bennett K. L., Dobler G., Guo J.-T., Superti-Furga G., Lill R., Överby A. K., and Weber F. (2014) Viperin is an iron-sulfur protein that inhibits genome synthesis of tick-borne encephalitis virus via radical SAM domain activity. Cell. Microbiol. 16, 834–848 10.1111/cmi.12241 [DOI] [PubMed] [Google Scholar]
- 20. Lindqvist R., Upadhyay A., and Överby A. K. (2018) Tick-borne flaviviruses and the type I interferon response. Viruses 10, 340 10.3390/v10070340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Van der Hoek K. H., Eyre N. S., Shue B., Khantisitthiporn O., Glab-Ampi K., Carr J. M., Gartner M. J., Jolly L. A., Thomas P. Q., Adikusuma F., Jankovic-Karasoulos T., Roberts C. T., Helbig K. J., and Beard M. R. (2017) Viperin is an important host restriction factor in control of Zika virus infection. Sci. Rep. 7, 4475 10.1038/s41598-017-04138-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vanwalscappel B., Tada T., and Landau N. R. (2018) Toll-like receptor agonist R848 blocks Zika virus replication by inducing the antiviral protein viperin. Virology 522, 199–208 10.1016/j.virol.2018.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carissimo G., Teo T.-H., Chan Y.-H., Lee C. Y.-P., Lee B., Torres-Ruesta A., Tan J. J., Chua T.-K., Fong S.-W., Lum F.-M., and Ng L. F. (2019) Viperin controls chikungunya virus-specific pathogenic T cell IFNγ Th1 stimulation in mice. Life Sci. Alliance 2, e201900298 10.26508/lsa.201900298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Teng T.-S., Foo S.-S., Simamarta D., Lum F.-M., Teo T.-H., Lulla A., Yeo N. K. W., Koh E. G. L., Chow A., Leo Y.-S., Merits A., Chin K.-C., and Ng L. F. P. (2012) Viperin restricts chikungunya virus replication and pathology. J. Clin. Invest. 122, 4447–4460 10.1172/JCI63120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang Y., Burke C. W., Ryman K. D., and Klimstra W. B. (2007) Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J. Virol. 81, 11246–11255 10.1128/JVI.01282-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chan Y.-L., Chang T.-H., Liao C.-L., and Lin Y.-L. (2008) The cellular antiviral protein viperin is attenuated by proteasome-mediated protein degradation in Japanese encephalitis virus-infected cells. J. Virol. 82, 10455–10464 10.1128/JVI.00438-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Horner S. M., Wilkins C., Badil S., Iskarpatyoti J., and Gale M. Jr. (2015) Proteomic analysis of mitochondrial-associated ER membranes (MAM) during RNA virus infection reveals dynamic changes in protein and organelle trafficking. PLoS ONE 10, e0117963 10.1371/journal.pone.0117963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Boudinot P., Riffault S., Salhi S., Carrat C., Sedlik C., Mahmoudi N., Charley B., and Benmansour A. (2000) Vesicular stomatitis virus and pseudorabies virus induce a vig1/cig5 homologue in mouse dendritic cells via different pathways. J. Gen. Virol. 81, 2675–2682 10.1099/0022-1317-81-11-2675 [DOI] [PubMed] [Google Scholar]
- 29. Stirnweiss A., Ksienzyk A., Klages K., Rand U., Grashoff M., Hauser H., and Kröger A. (2010) IFN regulatory factor-1 bypasses IFN-mediated antiviral effects through viperin gene induction. J. Immunol. 184, 5179–5185 10.4049/jimmunol.0902264 [DOI] [PubMed] [Google Scholar]
- 30. Yuan Y., Miao Y., Qian L., Zhang Y., Liu C., Liu J., Zuo Y., Feng Q., Guo T., Zhang L., Chen X., Jin L., Huang F., Zhang H., Zhang W., et al. (2020) Targeting UBE4A revives viperin protein in epithelium to enhance host antiviral defense. Mol. Cell 77, 734–747.e7 10.1016/j.molcel.2019.11.003 [DOI] [PubMed] [Google Scholar]
- 31. Li W., Mao L., Cao Y., Zhou B., Yang L., Han L., Hao F., Lin T., Zhang W., and Jiang J. (2017) Porcine viperin protein inhibits the replication of classical swine fever virus (CSFV) in vitro. Virol. J. 14, 202 10.1186/s12985-017-0868-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu C., Feng L., Chen P., Li A., Guo S., Jiao X., Zhang C., Zhao Y., Jin X., Zhong K., Guo Y., Zhu H., Han L., Yang G., Li H., et al. (2020) Viperin inhibits classical swine fever virus replication by interacting with viral nonstructural 5A protein. J. Med. Virol. 92, 149–160 10.1002/jmv.25595 [DOI] [PubMed] [Google Scholar]
- 33. Shanaka K. A. S. N., Tharuka M. D. N., Priyathilaka T. T., and Lee J. (2019) Molecular characterization and expression analysis of rockfish (Sebastes schlegelii) viperin, and its ability to enervate RNA virus transcription and replication in vitro. Fish Shellfish Immunol. 92, 655–666 10.1016/j.fsi.2019.06.015 [DOI] [PubMed] [Google Scholar]
- 34. Kurokawa C., Iankov I. D., and Galanis E. (2019) A key anti-viral protein, RSAD2/VIPERIN, restricts the release of measles virus from infected cells. Virus Res. 263, 145–150 10.1016/j.virusres.2019.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shah M., Bharadwaj M. S. K., Gupta A., Kumar R., and Kumar S. (2019) Chicken viperin inhibits Newcastle disease virus infection in vitro: a possible interaction with the viral matrix protein. Cytokine 120, 28–40 10.1016/j.cyto.2019.04.007 [DOI] [PubMed] [Google Scholar]
- 36. Wei C., Zheng C., Sun J., Luo D., Tang Y., Zhang Y., Ke X., Liu Y., Zheng Z., and Wang H. (2018) Viperin inhibits enterovirus A71 replication by interacting with viral 2C protein. Viruses 11, 13 10.3390/v11010013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hinson E. R., Joshi N. S., Chen J. H., Rahner C., Jung Y. W., Wang X., Kaech S. M., and Cresswell P. (2010) Viperin is highly induced in neutrophils and macrophages during acute and chronic lymphocytic choriomeningitis virus infection. J. Immunol. 184, 5723–5731 10.4049/jimmunol.0903752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Peña Cárcamo J. R., Morell M. L., Vázquez C. A., Vatansever S., Upadhyay A. S., Överby A. K., Cordo S. M., and García C. C. (2018) The interplay between viperin antiviral activity, lipid droplets and Junín mammarenavirus multiplication. Virology 514, 216–229 10.1016/j.virol.2017.10.012 [DOI] [PubMed] [Google Scholar]
- 39. Tang H.-B., Lu Z.-L., Wei X.-K., Zhong T.-Z., Zhong Y.-Z., Ouyang L.-X., Luo Y., Xing X.-W., Liao F., Peng K.-K., Deng C.-Q., Minamoto N., and Luo T. R. (2016) Viperin inhibits rabies virus replication via reduced cholesterol and sphingomyelin and is regulated upstream by TLR4. Sci. Rep. 6, 30529–30529 10.1038/srep30529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fang J., Wang H., Bai J., Zhang Q., Li Y., Liu F., and Jiang P. (2016) Monkey viperin restricts porcine reproductive and respiratory syndrome virus replication. PLoS ONE 11, e0156513 10.1371/journal.pone.0156513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jumat M. R., Huong T. N., Ravi L. I., Stanford R., Tan B. H., and Sugrue R. J. (2015) Viperin protein expression inhibits the late stage of respiratory syncytial virus morphogenesis. Antiviral Res. 114, 11–20 10.1016/j.antiviral.2014.11.007 [DOI] [PubMed] [Google Scholar]
- 42. Carlton-Smith C., and Elliott R. M. (2012) Viperin, MTAP44, and protein kinase R contribute to the interferon-induced inhibition of Bunyamwera Orthobunyavirus replication. J. Virol. 86, 11548–11557 10.1128/JVI.01773-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Proud D., Turner R. B., Winther B., Wiehler S., Tiesman J. P., Reichling T. D., Juhlin K. D., Fulmer A. W., Ho B. Y., Walanski A. A., Poore C. L., Mizoguchi H., Jump L., Moore M. L., Zukowski C. K., et al. (2008) Gene expression profiles during in vivo human Rhinovirus infection. Am. J. Respir. Crit. Care Med. 178, 962–968 10.1164/rccm.200805-670OC [DOI] [PubMed] [Google Scholar]
- 44. Khaiboullina S. F., Rizvanov A. A., Holbrook M. R., and St. Jeor S. (2005) Yellow fever virus strains Asibi and 17D-204 infect human umbilical cord endothelial cells and induce novel changes in gene expression. Virology 342, 167–176 10.1016/j.virol.2005.07.035 [DOI] [PubMed] [Google Scholar]
- 45. Nasr N., Maddocks S., Turville S. G., Harman A. N., Woolger N., Helbig K. J., Wilkinson J., Bye C. R., Wright T. K., Rambukwelle D., Donaghy H., Beard M. R., and Cunningham A. L. (2012) HIV-1 infection of human macrophages directly induces viperin which inhibits viral production. Blood 120, 778–788 10.1182/blood-2012-01-407395 [DOI] [PubMed] [Google Scholar]
- 46. Lim E. S., Wu L. I., Malik H. S., and Emerman M. (2012) The function and evolution of the restriction factor Viperin in primates was not driven by lentiviruses. Retrovirology 9, 55–55 10.1186/1742-4690-9-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tang Y.-D., Na L., Zhu C.-H., Shen N., Yang F., Fu X.-Q., Wang Y.-H., Fu L.-H., Wang J.-Y., Lin Y.-Z., Wang X.-F., Wang X., Zhou J.-H., and Li C.-Y. (2014) Equine viperin restricts equine infectious anemia virus replication by inhibiting the production and/or release of viral Gag, Env, and receptor via distortion of the endoplasmic reticulum. J. Virol. 88, 12296–12310 10.1128/JVI.01379-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Seo J.-Y., and Cresswell P. (2013) Viperin regulates cellular lipid metabolism during human cytomegalovirus infection. PLoS Pathog. 9, e1003497 10.1371/journal.ppat.1003497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Seo J.-Y., Yaneva R., Hinson E. R., and Cresswell P. (2011) Human cytomegalovirus directly induces the antiviral protein viperin to enhance infectivity. Science 332, 1093–1097 10.1126/science.1202007 [DOI] [PubMed] [Google Scholar]
- 50. Dumbrepatil A. B., Zegalia K. A., Sajja K., Kennedy R. T., and Marsh E. N. G. (2020) Targeting viperin to the mitochondrion inhibits the thiolase activity of the trifunctional enzyme complex. J. Biol. Chem. 295, 2839–2849 10.1074/jbc.RA119.011526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li M., Liao Z., Xu Z., Zou X., Wang Y., Peng H., Li Y., Ou X., Deng Y., Guo Y., Gan W., Peng T., Chen D., and Cai M. (2019) The interaction mechanism between herpes simplex virus 1 glycoprotein D and host antiviral protein viperin. Front. Immunol. 10, 2810 10.3389/fimmu.2019.02810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shen G., Wang K., Wang S., Cai M., Li M-L., and Zheng C. (2014) Herpes simplex virus 1 counteracts viperin via its virion host shutoff protein UL41. J. Virol. 88, 12163–12166 10.1128/JVI.01380-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang Y., Lv S., Zheng J., Huang X., Huang Y., and Qin Q. (2019) Grouper viperin acts as a crucial antiviral molecule against iridovirus. Fish Shellfish Immunol. 86, 1026–1034 10.1016/j.fsi.2018.12.038 [DOI] [PubMed] [Google Scholar]
- 54. Rivieccio M. A., Suh H. S., Zhao Y., Zhao M. L., Chin K. C., Lee S. C., and Brosnan C. F. (2006) TLR3 ligation activates an antiviral response in human fetal astrocytes: a role for viperin/cig5. J. Immunol. 177, 4735–4741 10.4049/jimmunol.177.7.4735 [DOI] [PubMed] [Google Scholar]
- 55. Saitoh T., Satoh T., Yamamoto N., Uematsu S., Takeuchi O., Kawai T., and Akira S. (2011) Antiviral protein viperin promotes Toll-like receptor 7- and Toll-like receptor 9-mediated type I interferon production in plasmacytoid dendritic cells. Immunity 34, 352–363 10.1016/j.immuni.2011.03.010 [DOI] [PubMed] [Google Scholar]
- 56. Wang B., Fang Y., Wu Y., Koga K., Osuga Y., Lv S., Chen D., Zhu Y., Wang J., and Huang H. (2015) Viperin is induced following toll-like receptor 3 (TLR3) ligation and has a virus-responsive function in human trophoblast cells. Placenta 36, 667–673 10.1016/j.placenta.2015.03.002 [DOI] [PubMed] [Google Scholar]
- 57. Gizzi A. S., Grove T. L., Arnold J. J., Jose J., Jangra R. K., Garforth S. J., Du Q., Cahill S. M., Dulyaninova N. G., Love J. D., Chandran K., Bresnick A. R., Cameron C. E., and Almo S. C. (2018) A naturally occurring antiviral ribonucleotide encoded by the human genome. Nature 558, 610–614 10.1038/s41586-018-0238-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Crosse K. M., Monson E. A., Beard M. R., and Helbig K. J. (2018) Interferon-stimulated genes as enhancers of antiviral innate immune signaling. J. Innate Immun. 10, 85–93 10.1159/000484258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fenwick M. K., Li Y., Cresswell P., Modis Y., and Ealick S. E. (2017) Structural studies of viperin, an antiviral radical SAM enzyme. Proc. Natl. Acad. Sci. U. S. A. 114, 6806–6811 10.1073/pnas.1705402114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chakravarti A., Selvadurai K., Shahoei R., Lee H., Fatma S., Tajkhorshid E., and Huang R. H. (2018) Reconstitution and substrate specificity for isopentenyl pyrophosphate of the antiviral radical SAM enzyme viperin. J. Biol. Chem. 293, 14122–14133 10.1074/jbc.RA118.003998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hinson E. R., and Cresswell P. (2009) The antiviral protein, viperin, localizes to lipid droplets via its N-terminal amphipathic alpha-helix. Proc. Natl. Acad. Sci. U. S. A. 106, 20452–20457 10.1073/pnas.0911679106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hinson E. R., and Cresswell P. (2009) The N-terminal amphipathic α-helix of viperin mediates localization to the cytosolic face of the endoplasmic reticulum and inhibits protein secretion. J. Biol. Chem. 284, 4705–4712 10.1074/jbc.M807261200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Upadhyay A. S., Stehling O., Panayiotou C., Rösser R., Lill R., and Överby A. K. (2017) Cellular requirements for iron-sulfur cluster insertion into the antiviral radical SAM protein viperin. J. Biol. Chem. 292, 13879–13889 10.1074/jbc.M117.780122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Holliday G. L., Akiva E., Meng E. C., Brown S. D., Calhoun S., Pieper U., Sali A., Booker S. J., and Babbitt P. C. (2018) Atlas of the radical SAM superfamily: divergent evolution of function using a “Plug and Play” domain. Methods Enzymol. 606, 1–71 10.1016/bs.mie.2018.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Landgraf B. J., McCarthy E. L., and Booker S. J. (2016) Radical S-adenosylmethionine enzymes in human health and disease. Annu. Rev. Biochem. 85, 485–514 10.1146/annurev-biochem-060713-035504 [DOI] [PubMed] [Google Scholar]
- 66. Frey P. A. (2014) Travels with carbon-centered radicals. 5′-deoxyadenosine and 5′-deoxyadenosine-5′-yl in radical enzymology. Acc. Chem. Res. 47, 540–549 10.1021/ar400194k [DOI] [PubMed] [Google Scholar]
- 67. Frey P. A., Hegeman A. D., and Ruzicka F. J. (2008) The radical SAM superfamily. Crit. Rev. Biochem. Mol. Biol. 43, 63–88 10.1080/10409230701829169 [DOI] [PubMed] [Google Scholar]
- 68. Zhang Q., and Liu W. (2011) Complex biotransformations catalyzed by radical S-adenosylmethionine enzymes. J. Biol. Chem. 286, 30245–30252 10.1074/jbc.R111.272690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wang J., Woldring R. P., Román-Meléndez G. D., McClain A. M., Alzua B. R., and Marsh E. N. G. (2014) Recent advances in radical SAM enzymology: new structures and mechanisms. ACS Chem. Biol. 9, 1929–1938 10.1021/cb5004674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Broderick J. B., Duffus B. R., Duschene K. S., and Shepard E. M. (2014) Radical S-adenosylmethionine enzymes. Chem. Rev. 114, 4229–4317 10.1021/cr4004709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Marsh E. N. G., Patterson D. P., and Li L. (2010) Adenosyl radical: reagent and catalyst in enzyme reactions. Chembiochem 11, 604–621 10.1002/cbic.200900777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sofia H. J., Chen G., Hetzler B. G., Reyes-Spindola J. F., and Miller N. E. (2001) Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 29, 1097–1106 10.1093/nar/29.5.1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yokoyama K., and Lilla E. A. (2018) C-C bond forming radical SAM enzymes involved in the construction of carbon skeletons of cofactors and natural products. Nat. Prod. Rep. 35, 660–694 10.1039/c8np00006a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Duschene K. S., and Broderick J. B. (2010) The antiviral protein viperin is a radical SAM enzyme. FEBS Lett. 584, 1263–1267 10.1016/j.febslet.2010.02.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Fenwick M. K., Su D., Dong M., Lin H., and Ealick S. E. (2020) Structural basis of the substrate selectivity of viperin. Biochemistry 59, 652–662 10.1021/acs.biochem.9b00741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hänzelmann P., and Schindelin H. (2006) Binding of 5'-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism. Proc. Natl. Acad. Sci. U. S. A. 103, 6829–6834 10.1073/pnas.0510711103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ebrahimi K. H., Howie D., Rowbotham J. S., McCullagh J., Armstrong F. A., and James W. S. (2020) Viperin, through its radical-SAM activity, depletes cellular nucleotide pools and interferes with mitochondrial metabolism to inhibit viral replication. FEBS Lett. 594, 1624–1630 10.1002/1873-3468.13761 [DOI] [PubMed] [Google Scholar]
- 78. Ebrahimi K. H., Vowles J., Browne C., McCullagh J., and James W. S. (2020) ddhCTP produced by the radical-SAM activity of RSAD2 (viperin) inhibits the NAD+-dependent activity of enzymes to modulate metabolism. FEBS Lett. 594, 1631–1644 10.1002/1873-3468.13778 [DOI] [PubMed] [Google Scholar]
- 79. Honarmand Ebrahimi K., Carr S. B., McCullagh J., Wickens J., Rees N. H., Cantley J., and Armstrong F. A. (2017) The radical-SAM enzyme Viperin catalyzes reductive addition of a 5'-deoxyadenosyl radical to UDP-glucose in vitro. FEBS Lett. 591, 2394–2405 10.1002/1873-3468.12769 [DOI] [PubMed] [Google Scholar]
- 80. Ebrahimi K. H., Rowbotham J. S., McCullagh J., and James W. S. (2020) Mechanism of diol dehydration by a promiscuous radical-SAM enzyme homologue of the antiviral enzyme viperin (RSAD2). Chembiochem 21, 1605–1612 10.1002/cbic.201900776 [DOI] [PubMed] [Google Scholar]
- 81. Bai L., Dong J. Z., Liu Z. Q., Rao Y. L., Feng P. H., and Lan K. (2019) Viperin catalyzes methionine oxidation to promote protein expression and function of helicases. Sci. Adv. 5, eaax1031 10.1126/sciadv.aax1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Vey J. L., and Drennan C. L. (2011) Structural insights into radical generation by the radical SAM superfamily. Chem. Rev. 111, 2487–2506 10.1021/cr9002616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hover B. M., Loksztejn A., Ribeiro A. A., and Yokoyama K. (2013) Identification of a cyclic nucleotide as a cryptic intermediate in molybdenum cofactor biosynthesis. J. Am. Chem. Soc. 135, 7019–7032 10.1021/ja401781t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lanz N. D., and Booker S. J. (2015) Auxiliary iron-sulfur cofactors in radical SAM enzymes. Biochim. Biophys. Acta 1853, 1316–1334 10.1016/j.bbamcr.2015.01.002 [DOI] [PubMed] [Google Scholar]
- 85. Banerjee R. (2003) Radical carbon skeleton rearrangements: catalysis by coenzyme B12-dependent mutases. Chem. Rev. 103, 2083–2094 10.1021/cr0204395 [DOI] [PubMed] [Google Scholar]
- 86. Marsh E. N. G., and Meléndez G. D. R. (2012) Adenosylcobalamin enzymes: theory and experiment begin to converge. Biochim. Biophys. Acta 1824, 1154–1164 10.1016/j.bbapap.2012.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ruszczycky M. W., Ogasawara Y., and Liu H-W. (2012) Radical SAM enzymes in the biosynthesis of sugar-containing natural products. Biochim. Biophys. Acta 1824, 1231–1244 10.1016/j.bbapap.2011.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Dumbrepatil A. B., Ghosh S., Zegalia K. A., Malec P. A., Hoff J. D., Kennedy R. T., and Marsh E. N. G. (2019) Viperin interacts with the kinase IRAK1 and the E3 ubiquitin ligase TRAF6, coupling innate immune signaling to antiviral ribonucleotide synthesis. J. Biol. Chem. 294, 6888–6898 10.1074/jbc.RA119.007719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Eom J., Kim J. J., Yoon S. G., Jeong H., Son S., Lee J. B., Yoo J., Seo H. J., Cho Y., Kim K. S., Choi K. M., Kim I. Y., Lee H. Y., Nam K. T., Cresswell P., et al. (2019) Intrinsic expression of viperin regulates thermogenesis in adipose tissues. Proc. Natl. Acad. Sci. U. S. A. 116, 17419–17428 10.1073/pnas.1904480116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Makins C., Ghosh S., Román-Meléndez G. D., Malec P. A., Kennedy R. T., and Marsh E. N. G. (2016) Does viperin function as a radical S-adenosyl-l-methionine-dependent enzyme in regulating farnesylpyrophosphate synthase expression and activity? J. Biol. Chem. 291, 26806–26815 10.1074/jbc.M116.751040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Vonderstein K., Nilsson E., Hubel P., Nygård Skalman L., Upadhyay A., Pasto J., Pichlmair A., Lundmark R., and Överby A. K. (2017) Viperin targets flavivirus virulence by inducing assembly of noninfectious capsid particles. J. Virol. 92, e01751–17 10.1128/JVI.01751-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Hee J. S., and Cresswell P. (2017) Viperin interaction with mitochondrial antiviral signaling protein (MAVS) limits viperin-mediated inhibition of the interferon response in macrophages. PLoS ONE 12, e0172236 10.1371/journal.pone.0172236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Chen S., Wu Z., Wang M., and Cheng A. (2017) Innate immune evasion mediated by Flaviviridae non-structural proteins. Viruses 9, 291 10.3390/v9100291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Tomtishen J. P. (2012) Human cytomegalovirus tegument proteins. Virol J. 9, 22–22 10.1186/1743-422X-9-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Goldmacher V. S. (2002) vMIA, a viral inhibitor of apoptosis targeting mitochondria. Biochimie 84, 177–185 10.1016/S0300-9084(02)01367-6 [DOI] [PubMed] [Google Scholar]
- 96. Jiang X., and Chen Z. J. (2011) Viperin links lipid bodies to immune defense. Immunity 34, 285–287 10.1016/j.immuni.2011.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wang L., Qiao Q., Ferrao R., Shen C., Hatcher J. M., Buhrlage S. J., Gray N. S., and Wu H. (2017) Crystal structure of human IRAK1. Proc. Natl. Acad. Sci. U. S. A. 114, 13507–13512 10.1073/pnas.1714386114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Conze D. B., Wu C. J., Thomas J. A., Landstrom A., and Ashwell J. D. (2008) Lys63-linked polyubiquitination of IRAK-1 is required for interleukin-1 receptor- and toll-like receptor-mediated NF-κB activation. Mol. Cell Biol. 28, 3538–3547 10.1128/MCB.02098-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Honda K., Yanai H., Mizutani T., Negishi H., Shimada N., Suzuki N., Ohba Y., Takaoka A., Yeh W. C., and Taniguchi T. (2004) Role of a transductional-transcriptional processor complex involving MyD88 and IRF-7 in Toll-like receptor signaling. Proc. Natl. Acad. Sci. U. S. A. 101, 15416–15421 10.1073/pnas.0406933101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Honda K., Takaoka A., and Taniguchi T. (2006) Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 25, 349–360 10.1016/j.immuni.2006.08.009 [DOI] [PubMed] [Google Scholar]
- 101. Crosse K. M., Monson E. A., Dumbrepatil A. B., Smith M., Tseng Y.-Y., Van der Hoek K. H., Revill P. A., Tscharke D. C., Marsh E. N. G., Beard M. R., and Helbig K. J. (2019) Viperin binds STING and enhances the type-I interferon response following dsDNA detection. bioRxiv 10.1101/493098 [DOI] [PubMed]
- 102. Barber G. N. (2015) STING: infection, inflammation and cancer. Nat. Rev. Immunol. 15, 760–770 10.1038/nri3921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ng K. W., Marshall E. A., Bell J. C., and Lam W. L. (2018) cGAS-STING and cancer: dichotomous roles in tumor immunity and development. Trends Immunol. 39, 44–54 10.1016/j.it.2017.07.013 [DOI] [PubMed] [Google Scholar]
- 104. Kellogg B. A., and Poulter C. D. (1997) Chain elongation in the isoprenoid biosynthetic pathway. Curr. Opin. Chem. Biol. 1, 570–578 10.1016/S1367-5931(97)80054-3 [DOI] [PubMed] [Google Scholar]
- 105. Ogura K., and Koyama T. (1998) Enzymatic aspects of isoprenoid chain elongation. Chem. Rev. 98, 1263–1276 10.1021/cr9600464 [DOI] [PubMed] [Google Scholar]
- 106. Miyanari Y., Atsuzawa K., Usuda N., Watashi K., Hishiki T., Zayas M., Bartenschlager R., Wakita T., Hijikata M., and Shimotohno K. (2007) The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9, 1089–1097 10.1038/ncb1631 [DOI] [PubMed] [Google Scholar]
- 107. Schwinghammer K., Cheung A. C. M., Morozov Y. I., Agaronyan K., Temiakov D., and Cramer P. (2013) Structure of human mitochondrial RNA polymerase elongation complex. Nat. Struct. Mol. Biol. 20, 1298–1303 10.1038/nsmb.2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Steinbusch M. M. F., Caron M. M. J., Surtel D. A. M., van den Akker G. G. H., van Dijk P. J., Friedrich F., Zabel B., van Rhijn L. W., Peffers M. J., and Welting T. J. M. (2019) The antiviral protein viperin regulates chondrogenic differentiation via CXCL10 protein secretion. J. Biol. Chem. 294, 5121–5136 10.1074/jbc.RA119.007356 [DOI] [PMC free article] [PubMed] [Google Scholar]