Abstract
SIX2 (SIX homeobox 2)–positive nephron progenitor cells (NPCs) give rise to all epithelial cell types of the nephron, the filtering unit of the kidney. NPCs have a limited lifespan and are depleted near the time of birth. Epigenetic factors are implicated in the maintenance of organ-restricted progenitors such as NPCs, but the chromatin-based mechanisms are incompletely understood. Here, using a combination of gene targeting, chromatin profiling, and single-cell RNA analysis, we examined the role of the murine histone 3 Lys-27 (H3K27) methyltransferases EZH1 (enhancer of zeste 1) and EZH2 in NPC maintenance. We found that EZH2 expression correlates with NPC growth potential and that EZH2 is the dominant H3K27 methyltransferase in NPCs and epithelial descendants. Surprisingly, NPCs lacking H3K27 trimethylation maintained their progenitor state but cycled slowly, leading to a smaller NPC pool and formation of fewer nephrons. Unlike Ezh2 loss of function, dual inactivation of Ezh1 and Ezh2 triggered overexpression of the transcriptional repressor Hes-related family BHLH transcription factor with YRPW motif 1 (Hey1), down-regulation of Six2, and unscheduled activation of Wnt4-driven differentiation, resulting in early termination of nephrogenesis and severe renal dysgenesis. Double-mutant NPCs also overexpressed the SIX family member Six1. However, in this context, SIX1 failed to maintain NPC stemness. At the chromatin level, EZH1 and EZH2 restricted accessibility to AP-1–binding motifs, and their absence promoted a regulatory landscape akin to differentiated and nonlineage cells. We conclude that EZH2 is required for NPC renewal potential and that tempering of the differentiation program requires cooperation of both EZH1 and EZH2.
Keywords: nephron progenitors, polycomb, histone methylation, development, kidney, epigenetics, chromatin, enhancer of zeste (Ezh), H3K27 methyltransferase, SIX homeobox 2 (SIX2), nephrology
The developing mammalian kidney is endowed with thousands of mesenchymal multipotent nephron progenitor cells (NPCs), which express Six2 (mouse) or SIX1 and SIX2 (human) (1, 2). NPCs undergo mesenchyme-to-epithelium transition to form pretubular aggregates and renal vesicles, the earliest epithelial precursors of nephrons, the filtering units of the kidney. NPCs generate thousands of nephrons before they are consumed around the time of birth (1, 3). Balancing rates of cell renewal and differentiation is essential to maintain NPC's normal lifespan and to prevent congenital disorders such as renal hypoplasia or uncontrolled growth (e.g. Wilms tumor) (4, 5). NPC stemness is governed by a core set of transcription factors, e.g. SIX1/SIX2, OSR1, WT1, SALL1, EYA1, and PAX2, and secreted growth factors, e.g. FGF9, FGF20, and BMP7. A principal mechanism by which Six2 maintains NPC stemness is via sequence-specific binding and repression of Wnt4, the major driver of the nephron differentiation program (6–8).
In addition to transcription factors, several lines of evidence implicate chromatin-based mechanisms in the control of NPC maintenance. Gene targeting in mice has demonstrated that inactivation of members of the HDAC–NURD complex or DNA methyltransferases impair the balance between NPC renewal and differentiation, leading to low nephron endowment later in life (9–13). Chromatin profiling of metanephric mesenchyme cell lines has shown that Wnt-induced differentiation is accompanied by loss of repressive H3K27me3 from bivalent (K4me3+/K27me3+) domains in lineage-specific developmental genes that are poised for activation such as Pax8, Wnt4, and Fgf8 (14). Conversely, Wilms tumor cells, which are differentiation-arrested metanephric mesenchyme cells, retain large domains of repressive H3K27me3 on developmental enhancers (15). Profiling the accessible genome utilizing ATAC-seq/ChIP-seq in native mouse NPCs demonstrated dynamic age-related changes in chromatin accessibility and enhancer activity and suggested that the age-related decline in NPC stemness may reflect differential accessibility to developmental regulators (16).
Studies in which the polycomb repressive complex 2 (PRC2) member EED was genetically ablated in NPCs revealed that EED deficiency induced loss of H3K27me3 from NPCs and descendant renal epithelial cells and caused depletion of Six2+ progenitors accompanied by up-regulation of the differentiation gene Lhx1 (11). NPCEed−/− mice developed a nephron deficit as a result of the imbalance between NPC renewal and differentiation. However, EED does not possess methyltransferase activity and acts by reading H3K27me2/3 and recruiting the catalytic components of PRC2: Ezh1 and Ezh2 (Enhancer of zeste1 and 2). Accordingly, the present study was designed to delineate the specific component of PRC2 responsible for H3K27me3 deposition in NPCs and their individual contributions to NPC stemness and maintenance. Our studies reveal that although Ezh2 is the dominant (if not only) H3K27 trimethyltransferase in nephron progenitors and epithelial progeny, its role is limited to cell renewal. Although Ezh1 is dispensable, it cooperates with Ezh2 in restraining the differentiation program, maintenance of lineage identity, and control of chromatin accessibility.
Results
Ezh2 is the dominant H3K27 methyltransferase in Six2+NPCs
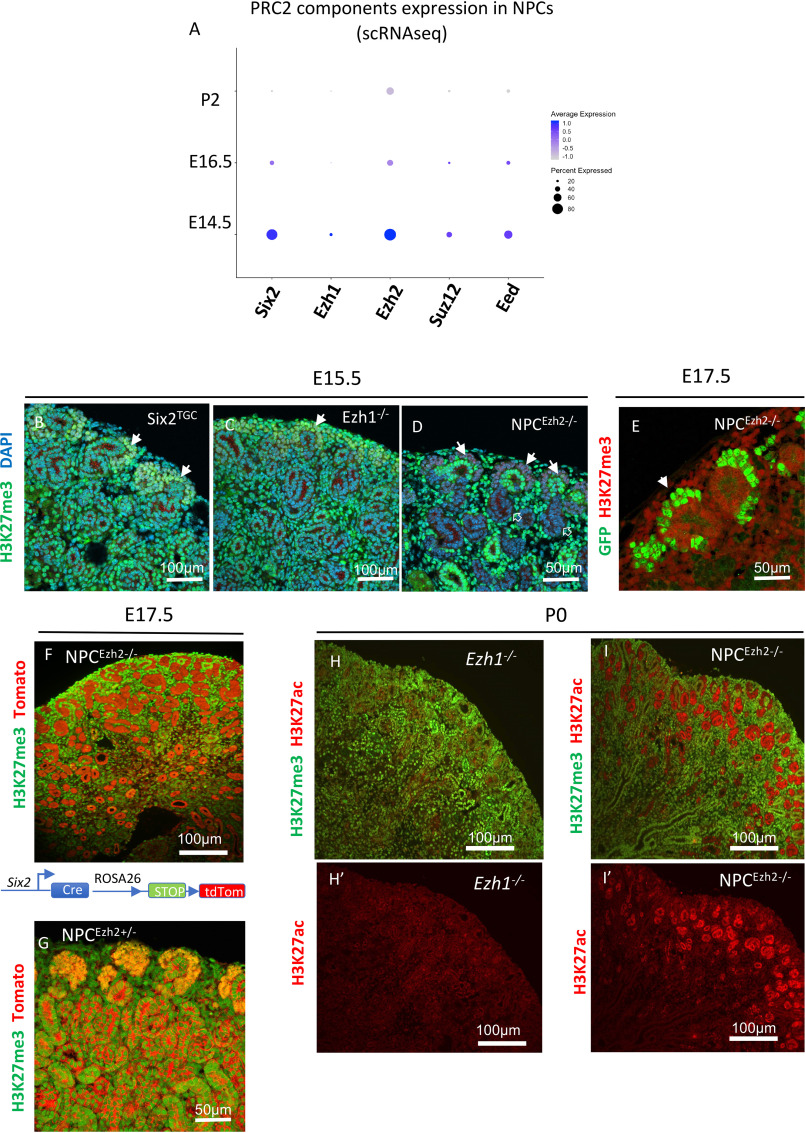
The life span of Six2+NPC in mice extends from embryonic day 10.5 (E10.5) to postnatal day 4 (P4) (17, 18). We compared the developmental expression of PRC2 components in embryonic (E14.5 and E16.5) and neonatal (P2) Six2GFP+NPCs and found that the temporal expression of Ezh1, Ezh2, Suz12, and Eed is similar to that of Six2: high in younger NPCs and declining with time in more committed NPCs (Fig. 1A).
Figure 1.
Ezh2 is the dominant H3K27 methyltransferase in nephron progenitors and descendant renal epithelial cells. A, dot plot depicting developmental expression of PRC2 components in NPCs from representative stages. Single-cell RNA values from E14.5 NPCs were obtained from GSE130606. ScRNA data from E16.5 and P2 are from the present study (GSE144384). B and C, at E15.5 Six2TGC and Ezh1−/− kidneys show abundant expression of H3K27me3 in the nephrogenic cortex. Solid arrows point to cap mesenchyme. D, in E15.5 NPCEzh2−/− kidneys (Six2GFPCre;Ezh2fl/fl), H3K27me3 is absent in the cap mesenchyme (solid arrows) and derived nascent nephrons (open arrows) but present in other structures such as surrounding stroma and ureteric bud branches and tips. E, H3K27me3 is absent in the cap mesenchyme of E17.5 GFP+NPCEzh2−/− (filled arrow). F and G, permanent genetic labeling shows absence of H3K27me3 in tdTomato-labeled tubules derived from NPCEzh2−/− but not NPCEzh2+/− kidneys. H, H′, I, and Í, H3K27Ac replaces H3K27me3 in NPCs and tubular derivatives of NPCEzh2−/− but not Ezh1−/− kidneys. DAPI, 4′,6-diamidino-2-phenylindole.
Both Ezh1 and Ezh2 harbor a catalytic Set domain that methylates H3K27 moieties. However, their individual roles vary by age, tissue, and cell type (19). To determine the functional redundancy between Ezh1 and Ezh2 in NPCs, we crossed BAC-transgenic Six2GFP-Cre(TGC) and R26R:Ezh2fl/fl:Ezh1−/−(germline) mice to generate NPCEzh2−/− and compound Ezh1+/− NPCEzh2−/− and Ezh1−/− NPCEzh2−/− mice and to allow permanent labeling of NPCEzh2−/− progenitors.
In developing Six2TGC kidneys (control E15.5), nuclear H3K27me3 is enriched in NPCs and to a lesser extent in the rest of the nephrogenic zone (Fig. 1B). Unexpectedly, H3K27me3 expression was unaffected in Ezh1−/− kidneys (Fig. 1C). In comparison, NPCEzh2−/− kidneys showed a complete lack of H3K27me3 expression in NPCs and descendant nascent tubular epithelia, whereas H3K27me3 staining remained intact in the surrounding stroma and ureteric bud tips/collecting ducts (Fig. 1D). Co-staining of H3K27me3 and GFP (a surrogate marker of Six2Cre expression) clearly shows a complete absence of H3K27me3 in NPCEzh2−/− (Fig. 1E). In addition, co-staining of H3K27me3 with the R26R tdTomato reporter confirmed a lack of H3K27me3 in NPCEzh2−/− and descendant renal tubules (Fig. 1F), as compared with NPCEzh2+/− (Fig. 1G). Because H3K27ac and H3K27me3 are two mutually exclusive marks, we also demonstrate that inactivation of Ezh2, but not Ezh1, results in a net gain of H3K27ac (Fig. 1, H, I, H′, and I′). Of note, enhanced H3K27Ac was more pronounced in nascent epithelia than mesenchymal progenitors of NPCEzh2−/− kidneys (Fig. 1, H′ and I′). This may be related to higher abundance of histone deacetylases 1 and 2 in NPCs than nascent nephrons (10). Thus, Ezh2 is the dominant (if not sole) H3K27 methyltransferase in NPCs and nascent nephrons during nephrogenesis.
Ezh2 is required for NPC proliferation
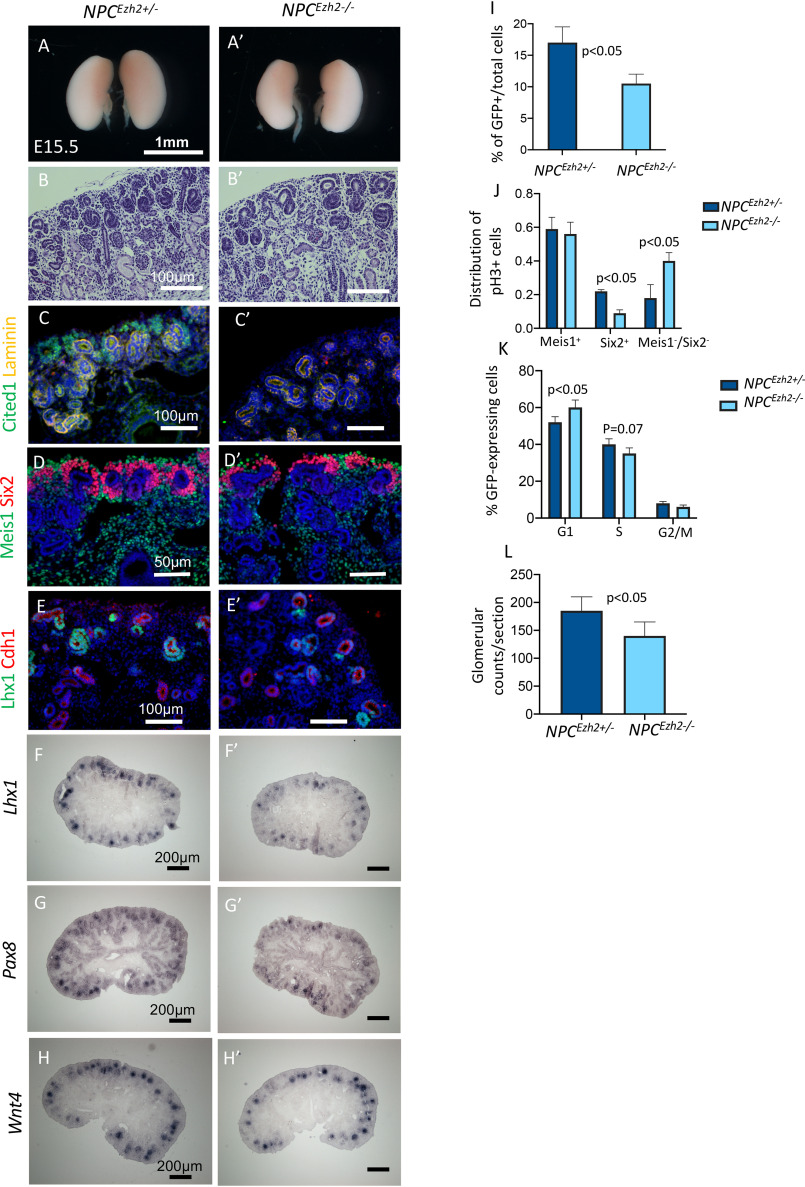
Histologic analysis and immunostaining for markers of NPCs, induced mesenchyme and proximal tubules in germline Ezh1−/− mice revealed normal nephrogenesis as compared with controls (Fig. S1). At E15.5, developing NPCEzh2−/− kidneys, on the other hand, had a thinner nephrogenic zone and exhibited a loss of Cited1 (a marker of the self-renewing NPCs) and a thinner Six2+ cap than NPCEzh2+/− kidneys (Fig. 2, A–D′). Moreover, NPCEzh2−/− kidneys had fewer nascent nephrons expressing Lhx1, Pax8, and Wn4 (Fig. 2, E–H′). FACS analysis of GFP+ cells revealed 30% reduction in the percentage of GFP+/total cells (Fig. 2I) and proliferating GFP+ cells in the Six2+ compartment of NPCEzh2−/− as compared with NPCEzh2+/− kidneys (Fig. 2J). Cell cycle analysis also showed that GFP+NPCEzh2−/− have longer G1–S and shorter S and G2–M phases (Fig. 2K). There was no significant difference in the number of cells undergoing apoptosis (cleaved caspase 3+) between the two groups (not shown). At 30 days of postnatal life, NPCEzh2−/− kidneys had ∼30% lower glomerular counts/section than NPCEzh2+/− kidneys (Fig. 2L). Thus, whereas inactivation of Ezh1 has no discernable effects on nephrogenesis, inactivation of Ezh2 impairs NPC proliferation and reduces the final nephron number. Although expression of Cited1 (a marker of self-renewing NPCs) was markedly down-regulated in NPCEzh2−/− kidneys, there was only a modest reduction (20–30%) in GFP+ cells and glomeruli. We do not have an explanation at present for the dissociation between Cited1 expression and the number of nephrons in NPCEzh2−/−. However, it is important to note that although Cited1 is an important marker of the uncommitted NPCs, its absence has no functional implications, and it does not mean that NPCs are not present. Indeed Six2-expressing cells are not completely missing, which may account for the number of nephrons still present in the Ezh2 mutants. It should be pointed out that NPCEzh2−/− mice survive into adulthood and are fertile.
Figure 2.
NPC-specific inactivation of Ezh2 impairs proliferation potential and nephron formation. E15.5 NPCEzh2± (Six2TGC;Ezh2fl/+) are compared with NPCEzh2−/− (Six2TGC;Ezh2fl/fl) kidneys. A, Á, B, and B′, gross view and hematoxylin-stained sections. C and C′, co-staining for Cited1 (a marker of self-renewing NPCs) and Laminin (marks basement membrane of nascent nephrons and tubules). D and D′, co-staining of the stroma marker Meis1 and NPC marker Six2. E and É, co-staining of the epithelial cell marker E-cadherin and nephron differentiation marker Lhx1. F–H′, section in situ hybridization for the differentiation genes Lhx1, Pax8, and Wnt4. I, FACS isolated Six2GFP+ cells were counted and divided by the total number of kidney cells. J, phospho-H3+ cells were counted and factored for total 4′,6-diamidino-2-phenylindole (nuclei) in co-stained sections identifying stroma (Meis1), NPC (Six2), and tubular compartments (negative for Six2 and Meis1). K, cell cycle analysis of Six2GFP+ cells. L, glomerular counts/tissue section at P30. n = 3 animals/group.
Ezh1 and Ezh2 cooperate to restrain the NPC differentiation program
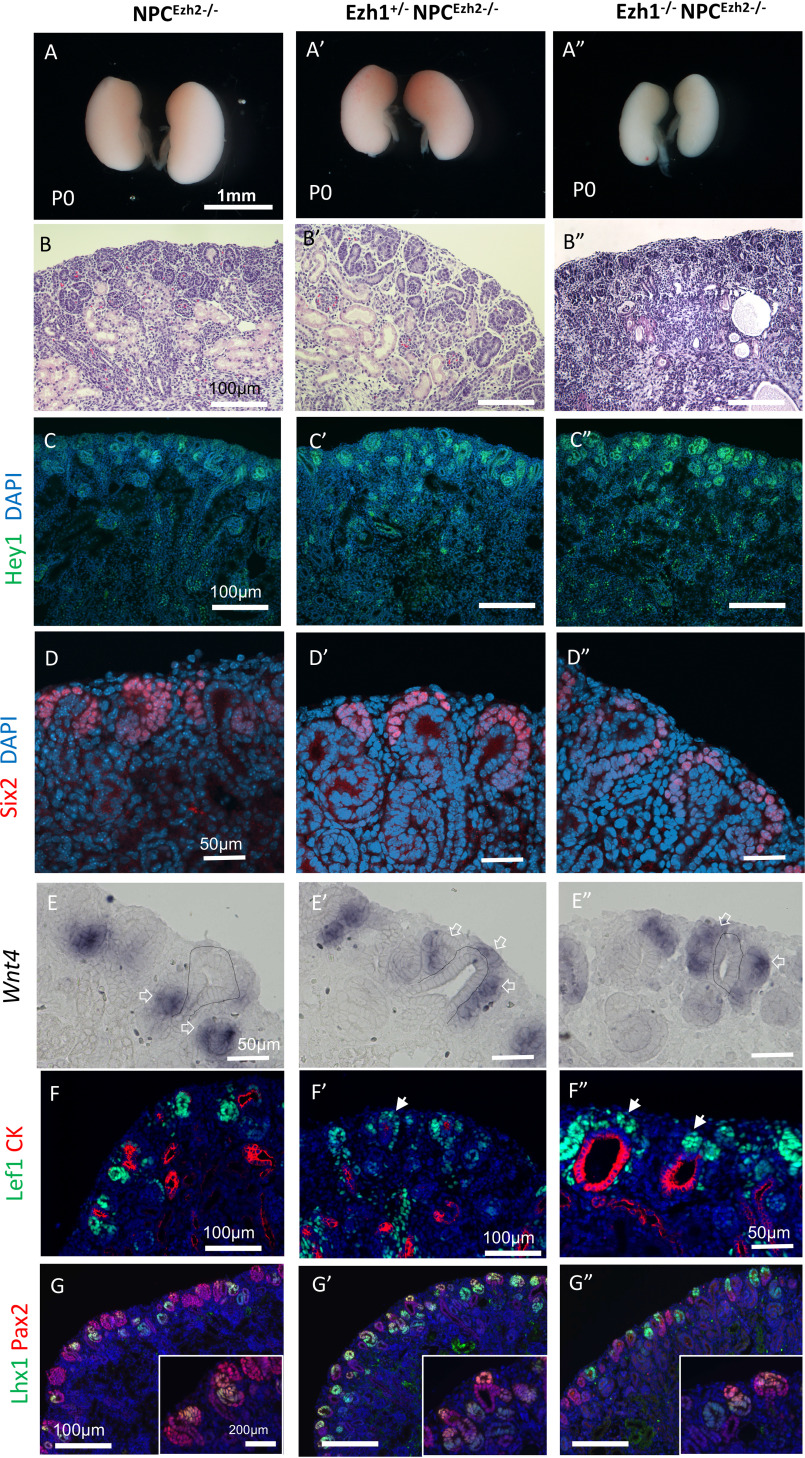
Previous studies have demonstrated that Ezh1 can mediate chromatin compaction in a H3K27me-independent manner (20). This function may be masked by Ezh2. To uncover the redundancy between Ezh1 and Ezh2 in nephrogenesis, we compared NPCEzh2−/− kidneys with those of Ezh1+/− NPCEzh2−/− and Ezh1−/− NPCEzh2−/− kidneys. Deletion of all four Ezh1/2 alleles caused early postnatal demise associated with severe kidney dysgenesis, poorly developed nephrogenic zone, and scattered cysts (Fig. 3, A–B″). These structural changes are accompanied by up-regulation of the Notch transcriptional repressor, Hey1 (Fig. 3, C–C″), and depletion of Six2+NPCs (Fig. 3, D–D″′).
Figure 3.
Ezh1 and Ezh2 are essential for tempering the differentiation program in NPCs. Phenotypic comparison of P0 kidneys from NPCEzh2−/− (Six2TGC;Ezh2fl/fl), compound heterozygous Ezh1+/−NPCEzh2−/− (Ezh1+/−;Six2TGC;Ezh2fl/fl), and homozygous Ezh1−/−NPCEzh2−/− (Ezh1−/−;Six2TGC;Ezh2fl/fl) mice. A, gross view. B, hematoxylin and eosin section staining showing gene dosage–dependent disorganization of the outer nephrogenic cortex and cystic dysplasia. C–D´´, section IF of Hey1 (C–C´´) and Six2 (D–D´´). E–E´´, section ISH for Wnt4: Wnt4 is expressed in renal vesicles (open arrows) beneath the UB branch (dotted line) as seen in NPCEzh2−/− kidneys but is ectopically expressed in NPCs dorsal to UB tip in double mutant kidneys. F–G´´, section IF of Lef1 (F–F´´) and Lhx1 (G–G´´) denoting premature burst of NPC differentiation in double-mutant kidneys. Solid white arrows in F´ and F´´ point to ectopic Lef1-expressing NPCs. DAPI, 4′,6-diamidino-2-phenylindole.
Six2 maintains NPC stemness partly via transcriptional repression of Wnt4 (6,8). Therefore, we examined the effects of Ezh1/2 inactivation on Wnt4 gene expression by in situ hybridization. P0 NPCEzh2−/− kidneys maintained the WT pattern of Wnt4 in pretubular aggregates and renal vesicles located beneath the ureteric bud branch (Fig. 3E). By contrast, P0 Ezh1+/− NPCEzh2−/− and Ezh1−/− NPCEzh2−/− kidneys exhibited ectopic expression of Wnt4 in NPCs located dorsal to the ureteric bud branch tip (Fig. 3, E and E″). Similarly, the differentiation markers, Lef1 and Lhx1, are ectopically expressed in NPCs (Fig. 3, F–G″). Importantly, we determined that ectopic Wnt4 expression in Ezh1−/− NPCEzh2−/− cells is not due to ectopic expression of its upstream activator, Wnt9b, which remains confined to the ureteric bud stalk as in control Six2TGC kidneys (Fig. S2). At E15.5 and E17.5, compared with NPCEzh2−/−, inactivation of all four Ezh1/2 alleles caused noticeable reductions in Six2+NPCs, nascent nephrons and cell proliferation (Figs. S3 and S4). Of note, ectopic expression of Lef1 in NPCs was apparent at E17.5 (Fig. S4, D–D″). Thus, the concerted action of Ezh1 and Ezh2 is required to restrain unscheduled activation of the differentiation program toward the end of nephrogenesis.
To determine whether Ezh1/2 inactivation promotes cell autonomous NPC differentiation, we cultured Six2GFP+ NPCs from control Six2TGC and Ezh1−/−NPCEzh2−/− cells in F12/Dulbecco's modified Eagle's medium–based KO medium that lacks renewal factors such as Fgf9 and thus favors differentiation. At 24 h of culture, staining for Six2 and E-cadherin revealed that Ezh1−/−NPCEzh2−/− cells underwent a much more robust mesenchyme-to-epithelium transition than controls (Fig. S5).
Analysis of NPC renewal–differentiation imbalance by single-cell profiling
To gain a deeper insight into how Ezh1/2 inactivation affects NPC gene expression, Six2GFP+ NPCs from E16.5 Six2TGC (control), NPCEzh2−/−, Ezh1+/−NPCEzh2−/−, and Ezh1−/− NPCEzh2−/− kidneys were profiled using 10× Chromium scRNA-seq (10× Genomics). Sorted cells (ranging in number from 6574 to 11,382 cells/group) were initially analyzed together using Seurat's version 3.0 data set integration approach (21). Unbiased clustering using t-distributed stochastic neighborhood embedding (t-SNE) revealed eight cell clusters (Fig. 4A and Fig. S6). Our NPC clustering data were in close agreement to those recently reported by Combes et al. (22) in Six2GFP+ from E14.5 Six2GCE mouse kidney. Cluster NPC0 had high expression of uncommitted progenitor genes such as Cited1; NPC1, 2, and 5 had high levels of cell cycle genes; NPC3 had high expression of early committed cells, whereas NPC6 had cells committed to differentiation (e.g. Wnt4, Tmem100); NPC4 cluster featured cells rich in Spry2 and Notch2. Spry2 is a negative regulator of FGF signaling that is important in NPC maintenance (23). Notch signaling is also known to regulate NPC maintenance (24). A Spry2/Notch2-enriched cluster was previously described by Combes et al. (22) in E14.5 mouse NPCs and is thought to represent a transitional state between nephron progenitor and early nascent nephrons. Lastly, the NPC7 cluster represents nephron progenitor-stromal cluster. A previous scRNA-seq study has shown that individual NPCs exhibit stochastic expression of stroma markers (25). The identity of these clusters can be further seen in the heat maps in Fig. 4 (B–D), which depict representative markers of each cluster. Fig. 4E depicts feature plots, generated using the feature plot function of Seurat 3.0, of the relative enrichment of differentiation markers (Ccnd1, Jag1, and Lhx1) (cluster 6) as compared with Six2.
Figure 4.
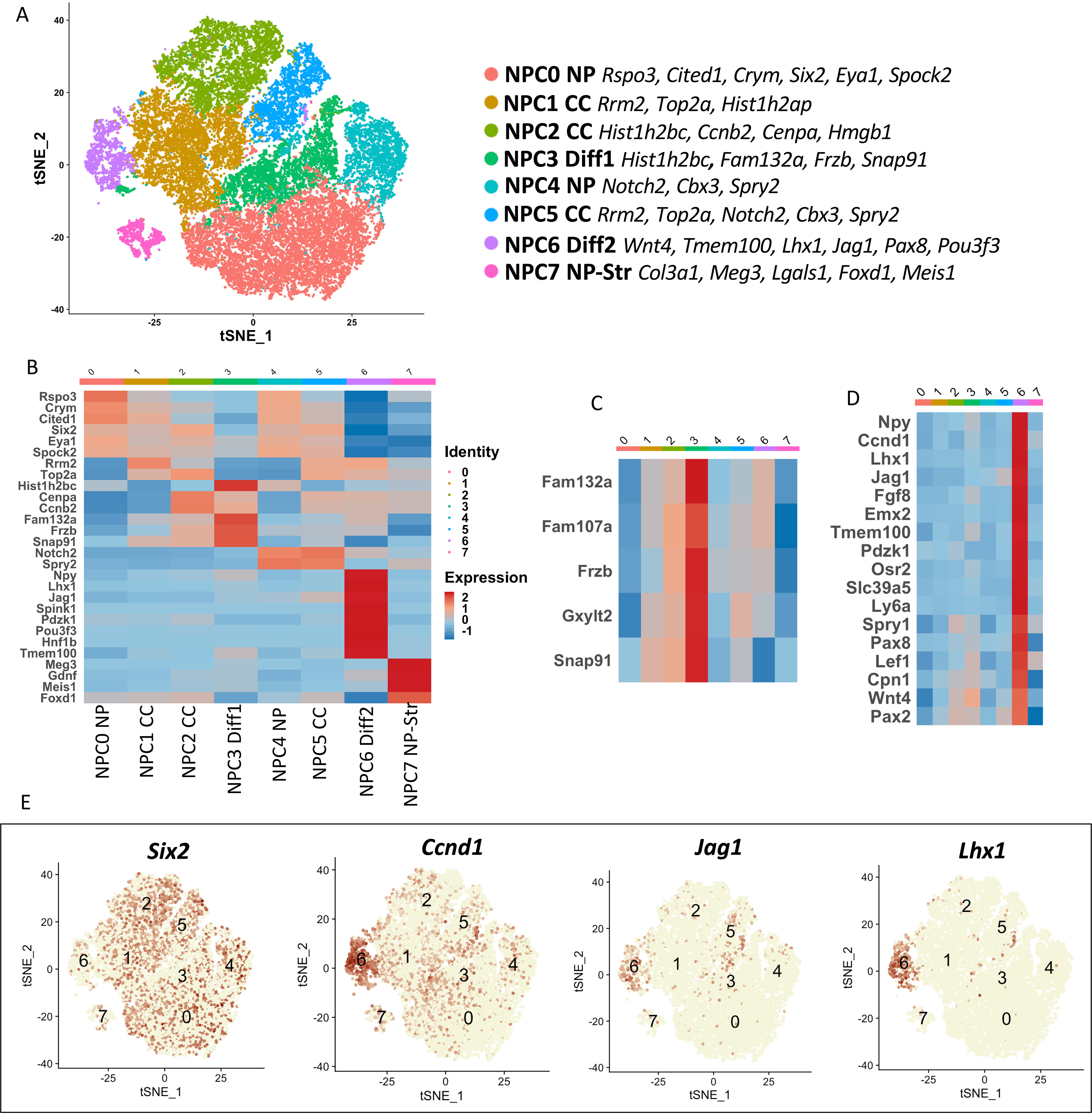
Single-cell RNA profiling of NPCs. Six2GFP+ NPCs from E16.5 Six2TGC (control), NPCEzh2−/−, Ezh1+/−NPCEzh2−/−, and Ezh1−/−NPCEzh2−/− kidneys were profiled using 10× chromium scRNA-seq. Sorted NPCs (ranging in number from 6574 to 11,382 cells/group) were analyzed together using Seurat's version 3.0 data set integration approach (21). A, t-SNE plot of integrated data. Clusters are referred to as NPC clusters 0–7 (NPC0–NPC7). Top expressed markers are listed next to each cluster ID. NPC0 cluster contains “uncommitted” progenitor genes such as Cited1. Clusters NPC1, NPC2, and NPC5 are enriched with cell cycle (CC) genes. NPC3 cluster includes genes expressed in committed progenitors, whereas NPC6 cluster features of differentiating markers. Cluster NPC3 expressed Spry2, a negative regulator of FGF signaling that is important NPC maintenance (23). Notch signaling is also known to regulate NPC maintenance (24). A Spry2/Notch2-enriched cluster was described by Combes et al. (22) and is thought to represent a transitional state between nephron progenitor and early nascent nephrons. B, heat map of representative genes of the various NPC clusters shown in A. C and D, heat maps highlighting NPC3 and NPC6 clusters. E, feature plots showing the landscape of cells enriched in the progenitor transcription factor Six2 and pro-differentiation genes Ccnd1, Jag1, and Lhx1.
A heat map and volcano plots of representative markers of progenitor and differentiating cells in our scRNA-seq analysis are depicted in Fig. 5 (A and B) and show that deletion of one or two Ezh1 alleles on Ezh2-null background decreases progenitor gene expression and increases differentiation gene expression, indicating that Ezh1/2 inactivation reprograms the timing of NPC differentiation. A previous study in which the Polycomb member Eed was deleted in NPCs found persistent expression of Six2 mRNA in mutant nascent epithelial cells (11). Similarly, our scRNA analysis revealed a higher number of Six2-expressing cells in the NPCEzh2−/− group as compared with other genotypes (Fig. 5B). The biological significance of this finding is unknown at present because we do not observe a corresponding persistence of Six2 protein in mutant NPCs (Fig. 3, D–D″). The imbalance of progenitor and differentiation gene expression in Ezh1/2 double-mutant NPCs is further illustrated in the dot plots shown in Fig. 5 (C–E). In addition, consistent with the findings in Fig. 3 (C–C″), we confirmed up-regulation of Hey1 gene expression in Ezh1−/−NPCEzh2−/− cells (Fig. 5F), which may contribute to Six2 repression (26). Other interesting changes include activation of the immediate early genes and members of the AP-1 transcription factor family (c-fos, c-Jun, and JunB) and down-regulation of JunD in Ezh1−/−NPCEzh2−/− as compared with control NPCs (Fig. 5G), which could stimulate cell renewal or differentiation, depending on the composition of the AP-1 complex. Notably, the observed induction of AP-1 component expression in double-mutant NPCs occurs independently of upstream growth factor expression (Bmp7, Fgf9, and Fgf20) (Fig. 5A) previously shown to mediate AP-1 activation in NPCs (27).
Figure 5.
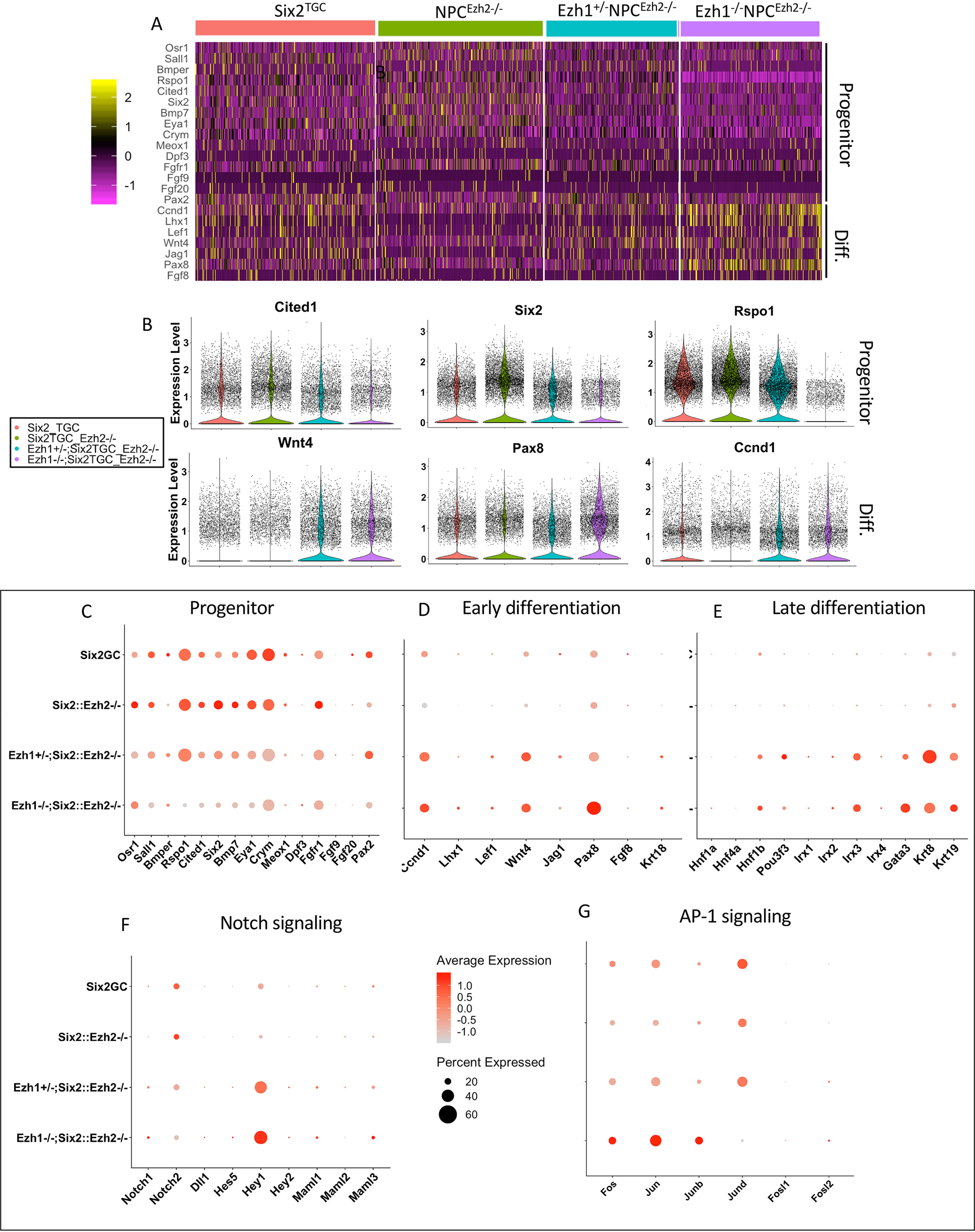
scRNA profiling reveals imbalance in progenitor and differentiation gene expression. A and B, heat map and volcano plot representations of progenitor and differentiation gene expression. Progenitor gene expression is maintained in E16.5 NPCEzh2−/− but is down-regulated in Ezh1−/−NPCEzh2−/− NPCs, which also express higher levels of differentiation markers than control Six2TGC NPCs. C–G, scRNA dot plots representing expression of progenitor, early and late differentiation, Notch and AP-1 transcription factor family genes. The intensity of the red color represents normalized expression, and the size of the dot represents percentage of cells expressing individual genes.
To further determine the effects of Ezh1/2 inactivation on timing of NPC differentiation, we used Monocle 2.0 to construct pseudo-time trajectories in control and mutant NPCs after selecting for cells enriched in the differentiation gene, Lhx1. In control Six2TGC NPCs, Lhx1-enriched cells are clustered toward the end of trajectory (Fig. 6A). In comparison, Ezh1−/− NPCEzh2−/− NPCs exhibit two major clusters of Lhx1-expressing cells at both ends of the trajectory (Fig. 6B).
Figure 6.
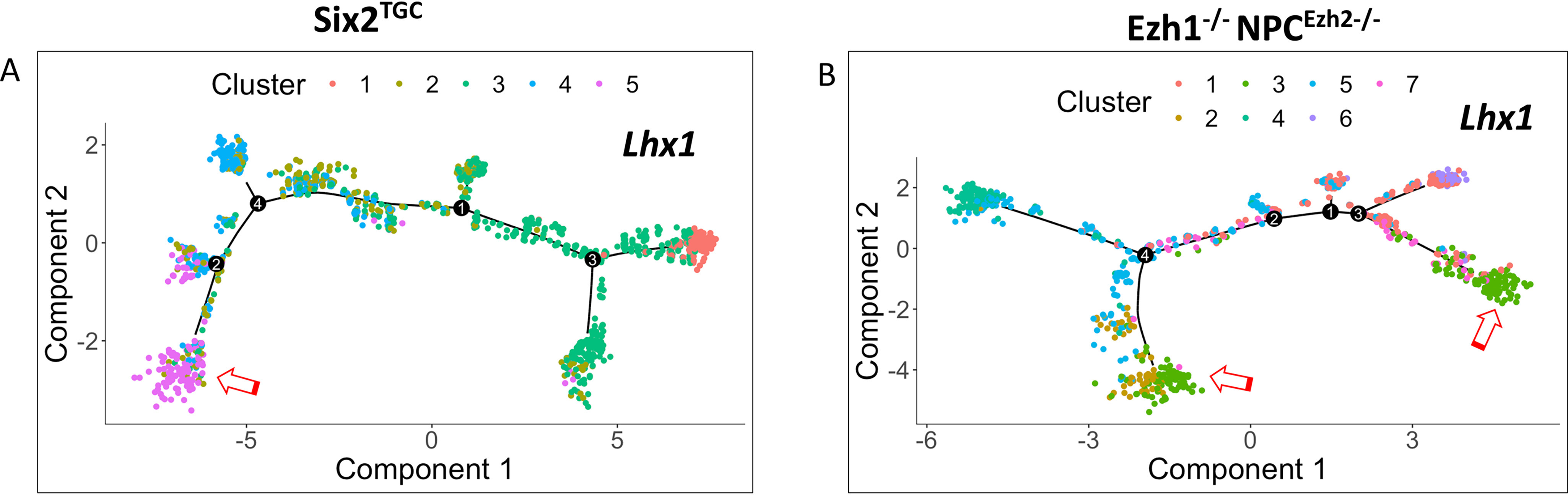
Single-cell trajectory analysis. A and B, monocle 2.0-based pseudo-time trajectory analysis of Lhx1-enriched NPCs. Open red arrows denote the locations of Lhx1high cells along the trajectory. In control, Six2TGC NPCs, Lhx1high cells (purple) are largely confined to subcluster 5 at the end of trajectory; in Ezh1−/−NPCEzh2−/− NPCs, there are early and late subclusters of Lhxhigh cells (green, subcluster #3) along the pseudo-time trajectory. FDR, false discovery rate.
Ezh1 and Ezh2 co-regulate chromatin accessibility in NPCs
We next examined whether reprograming of Ezh1/2-mutant NPCs is accompanied by reorganization of ATAC-accessible chromatin. We identified ∼73,000–98,000 peaks representing accessible “open” chromatin regions in the four groups of E16.5 GFP+ NPCs (Fig. 7A). We then focused on comparing Six2TGC and Ezh1−/−NPCEzh2−/− cells, which show the most significant differences by principal component analysis (Fig. 7B). We next compared the accessible chromatin regions of Six2TGC and Ezh1−/−NPCEzh2−/− cells using DiffBind R (RRID:SCR_012918). The affinity analysis is a quantitative approach to assess for differential chromatin access at consensus peaks. This method takes read densities computed over consensus peak regions and provides a statistical estimate of the difference in read concentration between the two conditions. Differential chromatin accessibility depicted in Fig. 7C is expressed as a log fold change of at least 2-fold and a p value of < 0.05 and reveals the relative gain of open chromatin regions in Six2TGC as compared with the gain in Ezh1−/−NPCEzh2−/−. Direct comparison of ATAC-seq tracks revealed increased chromatin accessibility at promoter and en-hancer regions of Wnt4, Pax8, and Lhx1 in Ezh1−/−NPCEzh2−/− cells (Fig. 7D), concordant with increased gene expression (Fig. 5D). We also noted a loss of chromatin accessibility at a putative regulatory region upstream of Notch2 but a gain in open chromatin in Hey1 in Ezh1−/−NPCEzh2−/− cells (Fig. 7E), in support of gene expression changes (Fig. 5G). These findings suggest that Ezh1 and Ezh2 restrain chromatin accessibility in NPCs to prevent unscheduled activation of the differentiation program.
Figure 7.
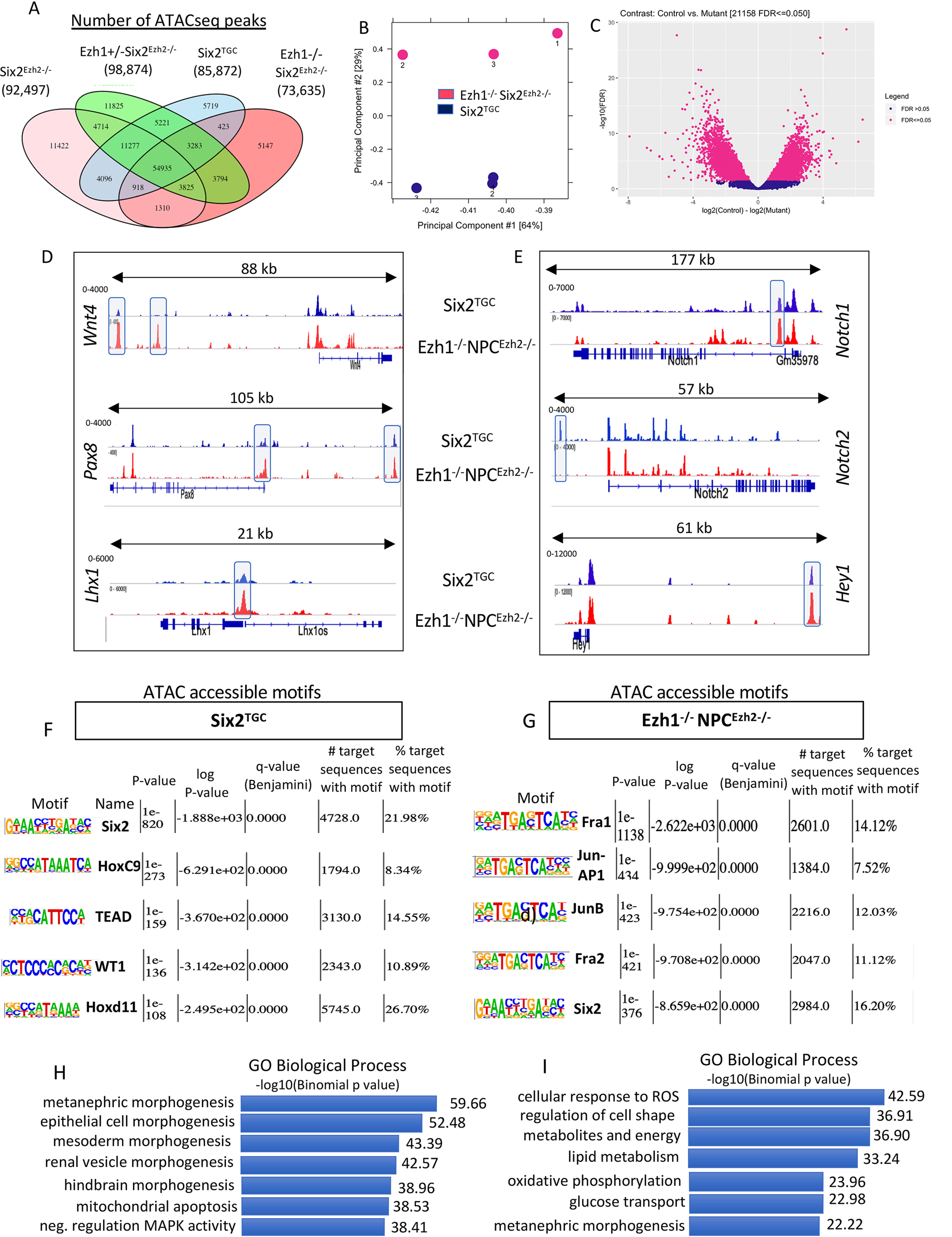
Ezh1 and Ezh2 regulate chromatin accessibility in NPCs. Ezh1 and Ezh2 regulate chromatin accessibility in NPCs. A, number of ATAC-seq accessible chromatin regions in the four NPC genotypes. B, principal component analysis of control and mutant samples. C, differential motif accessibility in control and mutant NPCs using DiffBind R. D and E, ATACseq tracks showing open chromatin regions of representative genes comparing Six2TGC and double mutant Ezh1−/−NPCEzh2−/− groups. F and G, HOMER-based ranking of top accessible motifs in control and mutant NPCs. The p values represent the statistical significance within each group. H and I, GREAT identification of Gene Ontology biological terms associated with ATAC-accessible chromatin.
We next used the Hypergeometric Optimization of Motif EnRichment (HOMER) tool to identify putative DNA-binding motifs in open chromatin regions of E16.5 Six2TGC and Ezh1−/− NPCEzh2−/− cells. Six2TGC NPCs showed enrichment for the binding motifs of core transcription factors Six2, Hoxc9, Hoxd11, WT1, and TEAD (Fig. 7F). In comparison, open chromatin regions in Ezh1−/−NPCEzh2−/− cells displayed preferential access to binding motifs of the AP-1 transcription factor family (Fig. 7G). This finding is quite interesting because RNA profiling has shown enhanced expression of several members of the AP-1 family (Fig. 5G), raising the possibility that Ezh1 and Ezh2 link growth factor signaling to the AP-1–responsive epigenome.
We next used the Genomic Regions Enrichment of Annotations Tool (GREAT) to compare the cis-regulatory elements within the accessible chromatin regions. Gene Ontology biological terms predicted processes related to nephrogenesis, apoptosis, and mitogen-activated protein kinase activity in Six2TGC NPCs (Fig. 7H). In comparison, the accessible regulatory regions in Ezh1−/−NPCEzh2−/− cells predicted more differentiated functions such as regulation of cell shape and metabolic/energy functions (Fig. 7I).
Ezh1 and Ezh2 maintain NPC lineage fidelity
Ezh1 and Ezh2 maintain embryonic lineage fidelity via repression of non–cell type–specific genes (28). We asked whether this function is also important in organ-restricted progenitors such as NPCs. scRNA analysis revealed that nonlineage genes, many of which are PRC2-regulated genes such as Hoxd13, Esx, Six1, Foxa1, Tbx15, and Nkx genes, are induced in Ezh1−/−NPCEzh2−/− but not in NPCEzh2−/− cells (Fig. 8, A and B). Derepression of nonlineage genes was confirmed by an independent analysis of NPC RNA using microarray hybridization (n = 3 biological replicates/genotype) (Table 1). The transcription factor Six1 and the cell cycle inhibitor Cdkn2a/p16 are known Ezh2-targets in other developing organs such as hair follicles, heart, and retina (29–32), so we examined them more closely. As expected, these normally silent genes are occupied by large domains of H3K27me3 in NPCs (Fig. 8C, top panels), and Ezh1/2 inactivation elicits enhanced chromatin accessibility at these loci (Fig. 8C, bottom panels). Section immunostaining revealed that Ezh2 inactivation alone had a negligible effect on Six1 and p16 expression in NPCs (Fig. 8, D and G). In contrast, combined inactivation of Ezh1 and Ezh2 resulted in a striking gene dosage–dependent up-regulation of p16 and Six1 expression (Fig. 8, E, F, H, and I). Thus, Six1 and Cdkn2a are bona fide targets for PRC2-mediated repression in NPCs.
Figure 8.
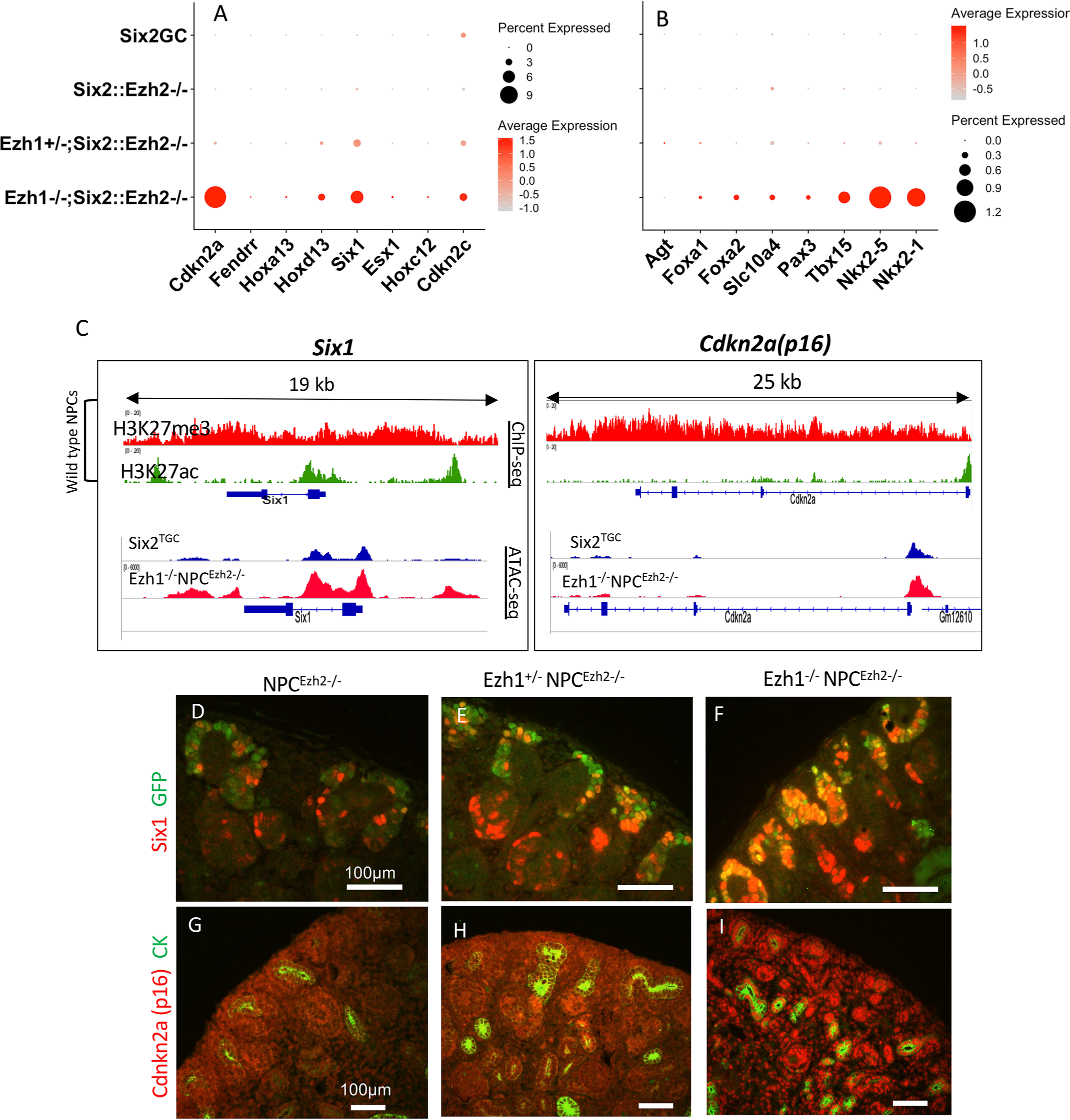
Nonlineage genes are targets for Ezh1/2-mediated repression in NPCs. A and B, scRNA-seq dot plot depicting expression of nonlineage genes in control and double-mutant Ezh1−/−NPCEzh2−/− cells. C, top panels, P0 WT NPCs - ChIP-seq tracks of Six1 and Cdkn2a/p16: both genes are occupied by large domains of repressive H3K27me3. Bottom panels, ATAC-seq showing increased chromatin accessibility in Six1 and Cdkn2a genes in Ezh1−/−NPCEzh2−/− as compared with control NPCs. D–I, section IF: gene dosage–dependent derepression of Six1 and p16 expression.
Table 1.
Top differentially expressed genes in NPCEzh2−/− and Ezh1−/−NPCEzh2−/− by microarray analysis as compared with control Six2TGC NPCs (n = 3 independent samples/genotype; p < 0.001). CNS, central nervous system; TF, transcription factor; SAM, sterile alpha motif
| Molecules | Fold change | Comments | |
|---|---|---|---|
| NPCEzh2−/− | Lin28b | +37.0 | Repressor of Let-7 microRNAs; overexpressed in tumors |
| Slc25a37 | +10.5 | Mitochondrial solute carrier and iron importer; erythropoiesis | |
| Bcl11a | +9.5 | Transcriptional co-repressor; hematopoiesis and CNS development | |
| Agt | +9.5 | Angiotensinogen; made in liver and mature proximal tubule; tubular differentiation | |
| Six1 | +8.4 | Homeobox TF; cardiac, ear, limb, and kidney development | |
| Lypd1 | +8.2 | Modulator of nicotinic receptors; CNS function | |
| Nfia | +7.8 | TF; CNS and urinary tract development | |
| Zc3hav11 | +4.6 | Zinc finger protein; innate immune response | |
| Vstm2l | +4.0 | Modulator of Wnt signaling | |
| Nr1h5 | −25.5 | Pseudogene | |
| Ezh2 | −24.6 | Enhancer of Zeste2 | |
| Hsd3b1 | −8.7 | Steroid metabolism | |
| Kyat3 | −4.1 | Kynurenine aminotransferase1; metabolism of cysteine conjugates | |
| Sac3 | −3.6 | CNS development | |
| Rprd2 | −3.4 | Viral immune response | |
| Tas2r3 | −3.3 | G protein–coupled receptor expressed in taste buds | |
| Dcx | −2.9 | Doublecortin; CNS development | |
| Ezh1−/− NPCEzh2−/− | Cdkn2a/p16 | +220 | Cell cycle inhibitor; senescence |
| Hoxd13 | +59.9 | Homeobox TF; limb patterning | |
| Six1 | +42.7 | Homeobox TF; cardiac, ear, limb, and kidney development | |
| Foxa2 | +22.8 | Forkhead Box TF; endoderm differentiation | |
| Hoxa13 | +20.2 | Homeobox TF; hand, foot, and genital patterning | |
| FoxG1 | +19.3 | Forkhead Box TF; CNS development | |
| Bcl11a | +18.5 | Transcriptional co-repressor; hematopoiesis and CNS development. | |
| Foxa1 | +17.9 | Pioneer factor; tissue-specific gene expression during development | |
| Ezh1 | −36.6 | Enhancer of Zeste 1 | |
| Ezh2 | −23.0 | Enhancer of Zeste 2 | |
| Nr1h5 | −21.0 | Pseudogene | |
| ARL16 | −13.7 | ADP-ribosylation factor; immune signaling | |
| Hsd3b1 | −7.3 | Steroid metabolism | |
| Rsad2 | −3.7 | Radical SAM domain-2; immune signaling | |
| OCM | −3.0 | oncodevelopmental protein found in early embryonic cells in the placenta and also in tumors | |
| Dcx | −2.9 | Doublecortin; CNS development |
We next asked whether ectopic Six1 expression contributed to impaired maintenance of Ezh1/2-mutant NPCs. To address this question, we generated crosses of Ezh1−/−;NPCEzh2−/−;Six1+/−(germline) mice to reduce Six1 gene dosage. As shown earlier, Ezh1−/−NPCEzh2−/−;Six1+/+ kidneys have a thin nephrogenic zone, lack mature nephrons, overexpress Six1 in NPCs, and show fewer GFP+NPCs (Fig. S7). Although Six1 gene dosage reduction in Ezh1−/−Six2Ezh2−/−;Six1+/− mice reduced the overall abundance of Six1, it failed to rescue NPCs or improve nephrogenesis (n = 4 mice/group) (Fig. S7).
Discussion
Although coordinated deployment of lineage-specific transcription factors establishes tissue- and cell type–specific expression patterns, the maintenance of expression patterns through generations of cell divisions is largely accomplished by epigenetic regulators that set heritable chromatin states even in the absence of the initial transcription factors. PRC2-mediated repression ensures strict temporal control of progenitor cell differentiation in response to environmental cues while maintaining their identity and stemness. Aberrant PRC2 function has been implicated in a variety of disease states, ranging from congenital syndromes to cancer including the pediatric Wilms tumor (28, 33).
A recent study demonstrated that Six2-Cre mediated deletion of Eed, a PRC2 component required for reading H3K27me3 and recruitment of PRC2 complexes, eliminates H3K27me3 in NPCs and disrupts the balance between NPC proliferation and differentiation (11). Eed is a component of both Ezh1–PRC2 and Ezh2–PRC2 complexes; therefore the relative contributions of Ezh1 and Ezh2 to H3K27 methylation and NPC fate could not be discerned with certainty. In other tissue cell types such as hepatocytes, germ cells, and epidermal cells (30, 34, 35), it was found that both enzymes catalyze the deposition of H3K27me3. Thus, the present study demonstrates a unique dominant role of Ezh2 as the dominant H3K27 methyltransferase in the Six2-derived nephron lineage.
Ezh2 is essential for mouse development (36) and is a major developmental regulator of progenitor stemness in various developing organs such as lung, bone, epidermis, retina, neurons, liver, and pancreas (28, 37, 38). In these tissues, Ezh2 is indispensable for progenitor cell maintenance by sustaining expression of stemness factors and tempering the developmental differentiation program. By comparison, the present study demonstrates that in the developing kidney, Ezh2 inactivation predominantly affects progenitor renewal potential, whereas tempering the differentiation program requires the concerted actions of Ezh1 and Ezh2.
The finding that deletion of Ezh2 in NPCs and progeny results in a relatively mild phenotype (25% reduction in nephron number) was somewhat surprising. We do not believe this is due to residual H3K27me3 because of the striking reduction if not loss of H3K27me3 in NPCs and descendants in the Ezh2 mutants as seen in Fig. 1 Our strategy deleted the histone methyltransferase domain of Ezh2, so one speculation is that the rest of the Ezh2 protein may have methyltransferase-independent functions in NPCs. This is purely speculative, and we do not have any data to support this hypothesis. Additionally, our preliminary observations using Hoxb7-cre to inactivate Ezh2 in the ureteric bud lineage showed that loss of H3K27me3 alone resulted in a severe phenotype,3 suggesting that Ezh2 functions are lineage-dependent.
Six2-mediated repression of Wnt4 is a principal mechanism for maintenance of NPC stemness. Indeed, genetic ablation of Six2 results in ectopic Wnt4 expression and premature epithelial differentiation (1, 7). In the present study, NPC-specific Ezh2 inactivation (and loss of H3K27me3) failed to significantly alter Wnt4 expression. In contrast, dual inactivation of Ezh1 and Ezh2 led to down-regulation of Six2 and ectopic Wnt4 expression provoking unscheduled mesenchyme-to-epithelium differentiation. In vitro assays in cultured NPCs also support the idea that loss of Ezh1/2-mediated repression facilitates NPC differentiation. The mechanisms of Six2 down-regulation in Ezh1/2 double-mutant NPCs are likely multifactorial, including reduced expression of activators (e.g. Pax2, Wt1, Sall1) but also up-regulation of transcriptional repressors such as Hey1. Mice in which Notch2 is overexpressed under the Six2 regulatory elements exhibit Six2 repression and aberrant up-regulation of Hey1, a transcriptional repressor and a central transducer of Notch signaling (26). Interestingly, Zhang et al. (11) found up-regulation of Hey1 in Eed-mutant NPCs. In sum, we surmise that loss of PRC2-mediated repression up-regulates Hey1, which in turn represses Six2, leading to unscheduled Wnt4 activation and premature epithelial differentiation.
A previous study carried out in epidermis stem cells has shown that Ezh2 promotes the proliferative potential of progenitors by repressing the Ink4 locus (encoding Cdkn2a/p16) (29). In the present study, Ezh2 inactivation in NPCs did not trigger Cdkn2a expression, whereas combined inactivation of Ezh1 and Ezh2 did, which may have contributed further to loss of growth potential of NPCs. Given the important role of Cdkn2a/p16 in cell cycle arrest and senescence, further studies are needed to determine whether Cdkn2a gene dosage reduction can rescue the growth impairment in Ezh1/2 mutant progenitors.
ChIP-seq analysis in epidermis stem cells revealed that Ezh2 prevents the recruitment of the AP-1 transcriptional activator complex to structural genes necessary for epidermal differentiation and that stimulation or inhibition of AP-1 activity can modulate the proliferative and differentiation responses in Ezh2-deficient epidermis (29). In the present study, we found that Ezh1/2 inactivation induces c-Fos, c-Jun, and Junb in NPCs. Moreover, using ATAC-seq to uncover accessible regulatory genomic regions, we found that Ezh1/2 cooperate to restrict chromatin accessibility to AP-1–binding motifs. In a previous study, we reported increased chromatin accessibility to AP-1 in older differentiation-prone NPCs (16). AP-1 complexes are implicated in balancing NPC maintenance in response to niche growth factors: Fgf9 stimulates c-fos, whereas Bmp7 stimulates c-jun expression (27). However, the composition of AP-1 complex determines the cell fate: in this study both c-jun and junb were induced in Ezh1/2 mutant NPCs. It is conceivable that c-jun–junb complexes promote differentiation and/or cell cycle arrest, thereby antagonizing the growth promoting effects of c-fos:c-jun. Indeed, Junb–AP-1 complexes are known to induce cell cycle arrest and senescence through induction of Cdkn2a/p16 (39). In the future, it will be important to determine the composition of the AP-1 complex induced by Ezh1/2 deficiency in NPCs and to test whether Junb/AP-1 links growth factor signaling to the epigenome to mediate or facilitate the effects of PRC2 on NPC maintenance.
Considering that Ezh1-mediated H3K27 methylation is negligible in NPCs, an important question that ensues is how does Ezh1 accomplish its epigenetic regulation of NPC fate? Margueron et al. (20) shed some lights on this issue: these investigators found that Ezh1 inactivation in stem cells had minimal effects on global H3K27me2/3 levels, whereas Ezh2 inactivation markedly affected global H3K27me2/3 levels. They further demonstrated using nucleosomal templates that Ezh1 is capable of compacting nucleosomes in the absence of methyl group donors and independently of Ezh1 methyltransferase activity. Thus, we reason that Ezh1 and Ezh2 likely act via distinct epigenetic mechanisms to maintain NPC stemness. Future studies investigating the dynamic genomic occupancy of Ezh1 and Ezh2 during NPC maturation will be required to test this hypothesis.
O'Brien et al. (40) demonstrated that unlike mouse NPCs, which express Six2, human NPCs express both SIX1 and SIX2; O'Brien et al. proposed that this may be important to accommodate generation of hundreds of thousands of nephrons over a more extended period of nephrogenesis in humans as opposed to mice. Intriguingly, we found that Six1 up-regulation in Ezh1/2-mutant NPCs is ineffective in sustaining Six2 expression. We considered that the high Six1 levels may act as dominant-negative to repress Six2; however, tempering Six1 expression failed to restore Six2 expression. It is also conceivable that Six1 lacks the necessary modifications and/or partners to bind/activate the Six2 enhancer in mouse NPCs. Regardless of the mechanism, our findings provide conclusive evidence that PRC2 mediates repression of Six1 in mouse NPCs.
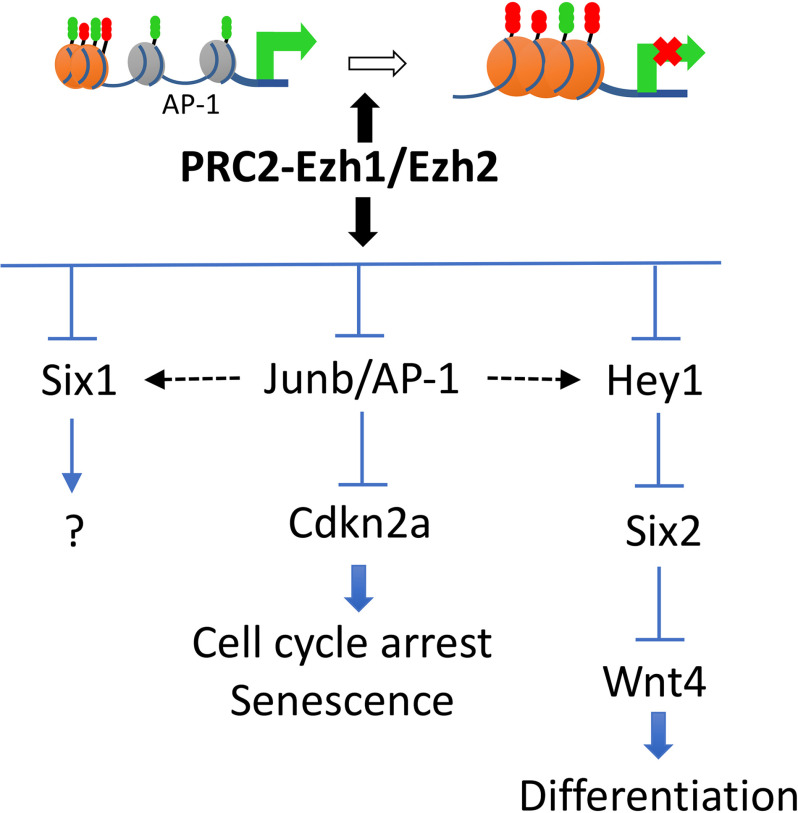
In this study, we used single-cell RNA-Seq to better understand how interference with repressive epigenetic mechanisms mediated by PRC2 in nephron progenitors can lead to morphologic and developmental defects. Assessment of chromatin accessibility added a new dimension to the overall analysis by showing how PRC2 inactivation affects the regulatory landscape governing the NPC fate. The combination of single-cell RNA analysis with ATAC-seq therefore offers a powerful tool to assess the developmental basis of congenital defects in organogenesis. Our results support a hypothesis in the epigenetic regulation of NPC maintenance by PRC2 through co-regulation of growth and differentiation signals (Fig. 9).
Figure 9.
Schematic depicting a working model for the role of PRC2 in nephron progenitor maintenance. PRC2-Ezh1/Ezh2 complexes restrict chromatin access to AP-1 motifs. This action prevents unscheduled/unwanted activation of genes that disrupt the balance between NPC stemness and differentiation. Note: the composition of the AP-1 complex is based on gene expression results and thus should be considered hypothetical at this point.
Experimental procedures
Mice
All mouse studies were performed and approved in accordance with the guidance of the Institutional Animal Care and Use Committee at Tulane University School of Medicine. Ezh2 floxed allele mice (41) and germline Ezh1 KO mice were obtained from Dr. Alexander Tarakhovsky (Rockefeller University, NY). R26R-tdTomato mice were purchased from the Jackson Laboratory (catalog no. 007909), and BAC transgenic Six2-TGC mice (1) were obtained from Andrew McMahon (formerly Harvard University). Germline Six1Lacz(knock-in) mice were obtained from Drs. Sean Li and Ruirong Tan (Harvard University).
Antibodies and reagents
The antibodies and reagents used in the study were as follows: anti-cytokeratin (1:200, catalog no. C2562, Sigma–Aldrich), anti-Six2 (1:200, catalog no. 11562, Proteintech), anti-Pax2 (1:200, catalog no. 616000, Invitrogen), anti-cleaved caspase 3 (1:100, catalog no. 9661s, Cell Signaling Technology), anti-Jag1 (H-114; 1:100, catalog no. sc-8303, Santa Cruz Biotechnology), Lhx1 (1:100, catalog no. 4F2-C, Developmental Studies Hybridoma Bank), anti-Lef1 (1:100, catalog no. 2230s, Cell Signaling Technology), anti-GFP (1:100, catalog no. ab13970, Abcam), anti–E-cadherin (1:100, catalog no. 610181, BD Biosciences), anti-WT1 (1:100, catalog no. ab15247, Abcam), anti-laminin (1:100, catalog no. L9393, Sigma–Aldrich), anti-H3K27me2/3 (1:200, catalog no. 39535, Active Motif), and anti-H3K27ac (1:200, catalog no. ab4729, Abcam).
Histology and immunohistochemistry
The kidneys were fixed in 10% buffered formalin, embedded in paraffin, and sectioned at 4 μm. Histological analyses were performed by standard hematoxylin and eosin staining. Section IF was performed as previously described (42). Antigen retrieval was accomplished by placing slides in 10 mm boiling sodium citrate, pH 6.0, for 20 min. In negative controls, the primary antibody was omitted or replaced by nonimmune serum. For IF, the secondary antibodies were Alexa Fluor 488–conjugated anti-rabbit and Alexa Fluor 594–conjugated anti-rabbit (1:2000, Invitrogen) and anti-mouse FITC (1:200, Sigma–Aldrich). In addition, FITC-conjugated Lotus tetragonolobus lectin agglutinin (1:100, Vector Laboratories) was used to label the apical brush border of proximal tubules. The nuclei were counterstained by 4′,6-diamidino-2-phenylindole (1:500, D1306, Invitrogen). The immunofluorescent images were captured using a 3D or deconvolution microscope (Leica DMRXA2). Numbers of glomeruli were quantified at P30 by counting total glomeruli in a single histological section from the widest diameter of the kidney that includes the medulla. At least three kidneys from littermate individual mice were analyzed.
Section in situ hybridization (ISH)
ISH was performed using digoxigenin-labeled antisense probes on kidney tissue fixed with 4% paraformaldehyde (PFA) as previously described (10). The kidney tissues were collected in diethyl pyrocarbonate–treated PBS, fixed in 4% PFA in diethyl pyrocarbonate–treated PBS overnight at 4 °C, dehydrated in a series of alcohol, cleared in xylene, and embedded in paraffin wax. Sections were cut to 10-μm thickness. After rehydration in 0.1% Tween in PBS, the samples were digested with proteinase K and then refixed in 4% PFA, followed by 0.2% glutaraldehyde, followed by three washes in PBS. After a 3-h incubation in hybridization solution, the explants were hybridized with the digoxigenin-labeled antisense probes (∼1 μg of probe/vial) overnight at 65 °C. The next day, the samples were sequentially washed with hybridization solution, 2× saline sodium citrate, pH 4.5, 2× saline sodium citrate, pH 7.0, 0.1% CHAPS, maleic acid buffer, and PBS at room temperature. The slides were incubated with preblocked antibody (1:10,000, anti-Dig alkaline phosphatase, Roche Applied Science) at 4 °C overnight. The following day, after sequential washes of 0.1% BSA in PBS, PBS, and AP-1 buffer at room temperature, the samples were stained by BM Purple (Roche Applied Science) at 4 °C. When the desired level of staining was reached, the reaction was stopped by two washes of stop solution for 15 min each. The experimental and control samples were put in the same reaction vessel to allow for proper comparison. All the experiments, including ISH and immunostaining, were repeated at least three times.
Cell culture
Metanephric mesenchyme isolated from E13.5 Six2TGC and Ezh1−/−NPCEzh2−/− mice were digested with collagenase/pancreatin at 37 °C for 5 min and cultured on Transwell membrane in knockout Dulbecco's modified Eagle's medium/F-12 medium (Thermo Fisher catalog no. 12660012). The cells were stained with anti–E-cadherin (1:200) and anti-Six2 (1:200) antibodies.
Assay for transposase-accessible chromatin-sequencing (ATAC-seq)
We followed the procedures we previously described (16). For sample library preparation, we followed the Omni-ATAC method outlined in Refs. 43 and 44). Briefly 50,000 nuclei from FACS-sorted cells were processed for Tn5 transposase-mediated tagmentation and adaptor incorporation at sites of accessible chromatin. This reaction was carried out using the Nextera DNA library prep kit (catalog no. FC-121-1030, Illumina) at 37°C for 30 min. Following tagmentation the DNA fragments were purified using the Zymo DNA clean and concentrator kit (catalog no. D4014, ZYMO Research). Library amplification was performed using the Ad1 and any of Ad2.1 through Ad2.12 barcoded primers (44). The quality of the purified DNA library was assessed on 6% Tris-Borate-EDTA (TBE) gels as well as on a Bioanalyzer (2100 Expert software, Agilent Technologies) using high-sensitivity DNA chips (catalog no. 5067-4626, Agilent Technologies Inc.). The appropriate concentration of sample was determined using the Qubit fluorometer (Molecular Probes). 4 nm samples were pooled and run on a NextSeq 500/550 high-output kit (catalog no. 20024907, Illumina, Inc. San Diego, CA) and the NextSeq 500 Illumina Sequencer to obtain paired end reads of 75 bp. Three to four independent biological replicates were sequenced per sample. The paired-end reads for each sample run across four lanes of the flow cell (catalog no. 20022408, Illumina) were concatenated to obtain one forward and one reverse fastq.gz files each. The quality of the reads was assessed using FASTQC (v0.11.7). The paired end reads were aligned to the mouse reference genome mm10 using Bowtie 2. The properly aligned reads were filtered for mitochondrial reads (Sam tools) and cleared of duplicates (Picard-tools, version 1.77). Only paired reads with high mapping quality (>30) were included in the downstream analysis. The narrow peaks were called using MACS2 using the following parameters (effective genome size = 1.87e + 09; no model p = 0.001, no lambda; bandwidth = 300, d = 200; p values cutoff = 1.00e-03). Normalized bigwig files were generated using bedtools. Annotation and Known, as well as de novo Motif, discovery was achieved with HOMER. Gene ontology analysis was performed using GREAT analysis 3.0.
Single-cell RNA-seq
The single-cell suspensions of FACS-sorted NPCs from the four genotypes (ranging from 6574 to 11,382 cells, total of 44,339 cells) were processed into barcoded scRNA-seq libraries using the Chromium single-cell 3′ library kit (10× Genomics) according to the manufacturer's protocol. Indexed libraries were sequenced on an Illumina HiSeq4000 and mapped to the mouse genome (build mm10) using CellRanger (10× Genomics, version 2.1.1), resulting in a postnormalization average 12,728 reads/cell and a median of 1,669 genes/cell and a saturation rate of 89.8% on average. We then used Cell Seurat suite version 3.1 and Monocle 3.0 for downstream analysis. Filtering was performed to remove multiplets and broken cells, and uninteresting sources of variation were regressed out. Variable genes were determined by iterative selection based on the dispersion versus average expression of the gene. For clustering, principal-component analysis was performed for dimension reduction. The top 10 principal components were selected by using a permutation-based test implemented in Seurat and passed to t-SNE for visualization of clusters. Genes expressed by <50 cells, cells that had either <200 or >5000 genes, or >5% of UMIs derived from the mitochondrial genome were removed.
Microarray analysis
Fluorescently labeled cRNA was generated from 0.5 μg of total RNA in each reaction using a fluorescent direct label kit (Agilent) and 1.0 mm cyanine 3′- or 5′-labeled dCTP (PerkinElmer). Hybridization was performed using an oligonucleotide microarray hybridization and in situ hybridization plus kit (Agilent). The labeled cRNA was hybridized to Agilent 44K whole mouse genome oligonucleotide microarray (containing ∼41,000 probes) as previously described (45). The arrays were scanned using a dual-laser DNA microarray scanner (Agilent). The data were then extracted from images using feature extraction software 6.1 (Agilent). MultiExperiment Viewer version 4.9 software was used to generate lists of genes differentially expressed between control and mutant kidneys, using p < 0.001 and a minimum 2.3-fold change in gene expression. Genes were classified according to their function using IPA software.
Statistics
All data are presented as means ± S.E. Unpaired two-tailed Student's t test was used for data presented in Fig. 3 to determine statistical significance. p values of <0.05 were considered to be statistically significant.
Data availability
Microarray data, scRNA-seq, and ATAC-seq data are available at the Gene Expression Omnibus repository under accession numbers GSE110925 and GSE144384. All other data are found within the article.
Supplementary Material
Acknowledgments
We thank Alexander Tarakhovsky for Ezh1 KO and Ezh2 floxed alleles mice, Andrew McMahon for Six2-TGC mice, and Sean Li and Ruirong Tan for the Six1Lacz/+ mice. We also thank Erik Flemington and Melody Baddoo from the Tulane Cancer Center and Alanna Wanek from the Tulane Translational Center of Immunity and Inflammation for assistance with pipeline analysis of ATACseq and scRNA-seq, Yuwen Li for assistance in analysis of microarray data, and the Tulane Renal and Hypertension Center of Excellence for use of the core facilities. We also thank the Tulane Center for Translational Research in Immunity and Inflammation Core for assistance with ATACseq and scRNAseq.
This article contains supporting information.
Author contributions—H. L., S. H., and E. K. data curation; H. L. and S. H. validation; H. L., S. H., E. K., C.-H. C., and Z. S. investigation; H. L., S. H., E. K., and C.-H. C. methodology; H. L., S. H. writing-original draft; S. H. and S. S. E.-D. resources; S. H. software; S. H. and S. S. E.-D. formal analysis; S. H. visualization; Z. S. and S. S. E.-D. supervision; Z. S. and S. S. E.-D. project administration; S. S. E.-D. conceptualization; S. S. E.-D. funding acquisition; S. S. E.-D. writing-review and editing.
Funding and additional information—This work was supported by National Institutes of Health Grants 1RO1 DK114500 and 1P50 DK096373 and American Heart Association Grant 17SDG33660072. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
H. Liu and S. S. El-Dahr, unpublished observations.
- NPC
- nephron progenitor cell
- PRC
- polycomb repressive complex
- En
- embryonic day n
- Pn
- postnatal day n
- t-SNE
- t-distributed stochastic neighborhood embedding
- HOMER
- Hypergeometric Optimization of Motif Enrichment
- GREAT
- Genomic Regions Enrichment of Annotations Tool
- IF
- immunofluorescent
- ISH
- in situ hybridization
- PFA
- paraformaldehyde
- ATAC-seq
- assay for transposase-accessible chromatin sequencing
- RNA-seq
- RNA sequencing.
References
- 1. Kobayashi A., Valerius M. T., Mugford J. W., Carroll T. J., Self M., Oliver G., and McMahon A. P. (2008) Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell 3, 169–181 10.1016/j.stem.2008.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lindström N. O., McMahon J. A., Guo J., Tran T., Guo Q., Rutledge E., Parvez R. K., Saribekyan G., Schuler R. E., Liao C., Kim A. D., Abdelhalim A., Ruffins S. W., Thornton M. E., Basking L., et al. (2018) Conserved and divergent features of human and mouse kidney organogenesis. J. Am. Soc. Nephrol. 29, 785–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Little M. H. (2015) The life cycle of the nephron progenitor. Dev. Cell 35, 5–6 10.1016/j.devcel.2015.09.023 [DOI] [PubMed] [Google Scholar]
- 4. Zohdi V., Sutherland M. R., Lim K., Gubhaju L., Zimanyi M. A., and Black M. J. (2012) Low birth weight due to intrauterine growth restriction and/or preterm birth: effects on nephron number and long-term renal health. Int. J. Nephrol. 2012, 136942 10.1155/2012/136942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Luyckx V. A., and Brenner B. M. (2019) Clinical consequences of developmental programming of low nephron number. Anat. Rec. (Hoboken) 10.1002/ar.24270 [DOI] [PubMed] [Google Scholar]
- 6. Park J. S., Ma W., O'Brien L. L., Chung E., Guo J. J., Cheng J. G., Valerius M. T., McMahon J. A., Wong W. H., and McMahon A. P. (2012) Six2 and Wnt regulate self-renewal and commitment of nephron progenitors through shared gene regulatory networks. Dev. Cell 23, 637–651 10.1016/j.devcel.2012.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Self M., Lagutin O. V., Bowling B., Hendrix J., Cai Y., Dressler G. R., and Oliver G. (2006) Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. EMBO J. 25, 5214–5228 10.1038/sj.emboj.7601381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu J., Liu H., Park J. S., Lan Y., and Jiang R. (2014) Osr1 acts downstream of and interacts synergistically with Six2 to maintain nephron progenitor cells during kidney organogenesis. Development 141, 1442–1452 10.1242/dev.103283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Denner D. R., and Rauchman M. (2013) Mi-2/NuRD is required in renal progenitor cells during embryonic kidney development. Dev. Biol. 375, 105–116 10.1016/j.ydbio.2012.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu H., Chen S., Yao X., Li Y., Chen C. H., Liu J., Saifudeen Z., and El-Dahr S. S. (2018) Histone deacetylases 1 and 2 regulate the transcriptional programs of nephron progenitors and renal vesicles. Development 145, dev153619 10.1242/dev.153619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang L., Ettou S., Khalid M., Taglienti M., Jain D., Jung Y. L., Seager C., Liu Y., Ng K. H., Park P. J., and Kreidberg J. A. (2018) EED, a member of the polycomb group, is required for nephron differentiation and the maintenance of nephron progenitor cells. Development 145, dev157149 10.1242/dev.157149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li S. Y., Park J., Guan Y., Chung K., Shrestha R., Palmer M. B., and Susztak K. (2019) DNMT1 in Six2 progenitor cells is essential for transposable element silencing and kidney development. J. Am. Soc. Nephrol. 30, 594–609 10.1681/ASN.2018070687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wanner N., Vornweg J., Combes A., Wilson S., Plappert J., Rafflenbeul G., Puelles V. G., Rahman R. U., Liwinski T., Lindner S., Grahammer F., Kretz O., Wlodek M. E., Romano T., Moritz K. M., et al. (2019) DNA methyltransferase 1 controls nephron progenitor cell renewal and differentiation. J. Am. Soc. Nephrol. 30, 63–78 10.1681/ASN.2018070736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McLaughlin N., Yao X., Li Y., Saifudeen Z., and El-Dahr S. S. (2013) Histone signature of metanephric mesenchyme cell lines. Epigenetics 8, 970–978 10.4161/epi.25753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aiden A. P., Rivera M. N., Rheinbay E., Ku M., Coffman E. J., Truong T. T., Vargas S. O., Lander E. S., Haber D. A., and Bernstein B. E. (2010) Wilms tumor chromatin profiles highlight stem cell properties and a renal developmental network. Cell Stem Cell 6, 591–602 10.1016/j.stem.2010.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hilliard S., Song R., Liu H., Chen C. H., Li Y., Baddoo M., Flemington E., Wanek A., Kolls J., Saifudeen Z., and El-Dahr S. S. (2019) Defining the dynamic chromatin landscape of mouse nephron progenitors. Biol. Open 8, bio042754 10.1242/bio.042754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brunskill E. W., Lai H. L., Jamison D. C., Potter S. S., and Patterson L. T. (2011) Microarrays and RNA-Seq identify molecular mechanisms driving the end of nephron production. BMC Dev. Biol. 11, 15 10.1186/1471-213X-11-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Short K. M., Combes A. N., Lefevre J., Ju A. L., Georgas K. M., Lamberton T., Cairncross O., Rumballe B. A., McMahon A. P., Hamilton N. A., Smyth I. M., and Little M. H. (2014) Global quantification of tissue dynamics in the developing mouse kidney. Dev. Cell 29, 188–202 10.1016/j.devcel.2014.02.017 [DOI] [PubMed] [Google Scholar]
- 19. Kouznetsova V. L., Tchekanov A., Li X., Yan X., and Tsigelny I. F. (2019) Polycomb repressive 2 complex: molecular mechanisms of function. Protein Sci. 28, 1387–1399 10.1002/pro.3647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Margueron R., Li G., Sarma K., Blais A., Zavadil J., Woodcock C. L., Dynlacht B. D., and Reinberg D. (2008) Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell 32, 503–518 10.1016/j.molcel.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Butler A., Hoffman P., Smibert P., Papalexi E., and Satija R. (2018) Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Combes A. N., Phipson B., Lawlor K. T., Dorison A., Patrick R., Zappia L., Harvey R. P., Oshlack A., and Little M. H. (2019) Single cell analysis of the developing mouse kidney provides deeper insight into marker gene expression and ligand-receptor crosstalk. Development 146, dev178673 10.1242/dev.178673 [DOI] [PubMed] [Google Scholar]
- 23. Barak H., Huh S. H., Chen S., Jeanpierre C., Martinovic J., Parisot M., Bole-Feysot C., Nitschké P., Salomon R., Antignac C., Ornitz D. M., and Kopan R. (2012) FGF9 and FGF20 maintain the stemness of nephron progenitors in mice and man. Dev. Cell 22, 1191–1207 10.1016/j.devcel.2012.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chung E., Deacon P., and Park J. S. (2017) Notch is required for the formation of all nephron segments and primes nephron progenitors for differentiation. Development 144, 4530–4539 10.1242/dev.156661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brunskill E. W., Park J. S., Chung E., Chen F., Magella B., and Potter S. S. (2014) Single cell dissection of early kidney development: multilineage priming. Development 141, 3093–3101 10.1242/dev.110601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fujimura S., Jiang Q., Kobayashi C., and Nishinakamura R. (2010) Notch2 activation in the embryonic kidney depletes nephron progenitors. J. Am. Soc. Nephrol. 21, 803–810 10.1681/ASN.2009040353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Muthukrishnan S. D., Yang X., Friesel R., and Oxburgh L. (2015) Concurrent BMP7 and FGF9 signalling governs AP-1 function to promote self-renewal of nephron progenitor cells. Nat. Commun. 6, 10027 10.1038/ncomms10027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Deevy O., and Bracken A. P. (2019) PRC2 functions in development and congenital disorders. Development 146, dev181354 10.1242/dev.181354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ezhkova E., Pasolli H. A., Parker J. S., Stokes N., Su I. H., Hannon G., Tarakhovsky A., and Fuchs E. (2009) Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 136, 1122–1135 10.1016/j.cell.2008.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ezhkova E., Lien W. H., Stokes N., Pasolli H. A., Silva J. M., and Fuchs E. (2011) EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 25, 485–498 10.1101/gad.2019811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Delgado-Olguín P., Huang Y., Li X., Christodoulou D., Seidman C. E., Seidman J. G., Tarakhovsky A., and Bruneau B. G. (2012) Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nat. Genet. 44, 343–347 10.1038/ng.1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yan N., Cheng L., Cho K., Malik M. T., Xiao L., Guo C., Yu H., Zhu R., Rao R. C., and Chen D. F. (2016) Postnatal onset of retinal degeneration by loss of embryonic Ezh2 repression of Six1. Sci. Rep. 6, 33887 10.1038/srep33887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim K. H., and Roberts C. W. (2016) Targeting EZH2 in cancer. Nat. Med. 22, 128–134 10.1038/nm.4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mu W., Starmer J., Shibata Y., Yee D., and Magnuson T. (2017) EZH1 in germ cells safeguards the function of PRC2 during spermatogenesis. Dev. Biol. 424, 198–207 10.1016/j.ydbio.2017.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bae W. K., Kang K., Yu J. H., Yoo K. H., Factor V. M., Kaji K., Matter M., Thorgeirsson S., and Hennighausen L. (2015) The methyltransferases enhancer of zeste homolog (EZH) 1 and EZH2 control hepatocyte homeostasis and regeneration. FASEB J. 29, 1653–1662 10.1096/fj.14-261537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. O'Carroll D., Erhardt S., Pagani M., Barton S. C., Surani M. A., and Jenuwein T. (2001) The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell Biol. 21, 4330–4336 10.1128/MCB.21.13.4330-4336.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kavaliauskaitė D., Stakišaitis D., Martinkutė J., Šlekienė L., Kazlauskas A., Balnytė I., Lesauskaitė V., and Valančiütė A. (2017) The effect of sodium valproate on the glioblastoma U87 cell line tumor development on the chicken embryo chorioallantoic membrane and on EZH2 and p53 expression. Biomed. Res. Int. 2017, 6326053 10.1155/2017/6326053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dupret B., Völkel P., Vennin C., Toillon R. A., Le Bourhis X., and Angrand P. O. (2017) The histone lysine methyltransferase Ezh2 is required for maintenance of the intestine integrity and for caudal fin regeneration in zebrafish. Biochim. Biophys. Acta 1860, 1079–1093 10.1016/j.bbagrm.2017.08.011 [DOI] [PubMed] [Google Scholar]
- 39. Piechaczyk M., and Farràs R. (2008) Regulation and function of JunB in cell proliferation. Biochem. Soc. Trans. 36, 864–867 10.1042/BST0360864 [DOI] [PubMed] [Google Scholar]
- 40. O'Brien L. L., Guo Q., Lee Y., Tran T., Benazet J. D., Whitney P. H., Valouev A., and McMahon A. P. (2016) Differential regulation of mouse and human nephron progenitors by the Six family of transcriptional regulators. Development 143, 595–608 10.1242/dev.127175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Su I. H., Basavaraj A., Krutchinsky A. N., Hobert O., Ullrich A., Chait B. T., and Tarakhovsky A. (2003) Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat. Immunol. 4, 124–131 10.1038/ni876 [DOI] [PubMed] [Google Scholar]
- 42. Hilliard S. A., Yao X., and El-Dahr S. S. (2014) Mdm2 is required for maintenance of the nephrogenic niche. Dev. Biol. 387, 1–14 10.1016/j.ydbio.2014.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Corces M. R., Trevino A. E., Hamilton E. G., Greenside P. G., Sinnott-Armstrong N. A., Vesuna S., Satpathy A. T., Rubin A. J., Montine K. S., Wu B., Kathiria A., Cho S. W., Mumbach M. R., Carter A. C., Kasowski M., et al. (2017) An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 10.1038/nmeth.4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Buenrostro J. D., Wu B., Chang H. Y., and Greenleaf W. J. (2015) ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schanstra J. P., Bachvarova M., Neau E., Bascands J. L., Bachvarov D. (2007) Gene expression profiling in the remnant kidney model of wild type and kinin B1 and B2 receptor knockout mice. Kidney. Int. 72, 442–454 10.1038/sj.ki.5002172 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data, scRNA-seq, and ATAC-seq data are available at the Gene Expression Omnibus repository under accession numbers GSE110925 and GSE144384. All other data are found within the article.