Abstract
Protein-lysine methylation is a common posttranslational modification (PTM) throughout the human proteome that plays important roles in diverse biological processes. In humans, there are >100 known and candidate protein lysine methyltransferases (PKMTs), many of which are linked to human diseases. Methyltransferase-like protein 21C (METTL21C) is a PKMT implicated in muscle biology that has been reported to methylate valosin-containing protein/p97 (VCP) and heat shock 70-kDa protein 8 (HSPA8). However, a clear in vitro methyltransferase activity for METTL21C remains yet to be demonstrated, and whether it is an active enzyme that directly methylates substrate(s) in vivo is unclear. Here, we used an unbiased biochemistry-based screening assay coupled to MS, which identified alanine tRNA synthetase 1 (AARS1) as a direct substrate of METTL21C. We found that METTL21C catalyzes methylation of Lys-943 of AARS1 (AARS1-K943me) both in vitro and in vivo. In vitro METTL21C-mediated AARS1 methylation was independent of ATP or tRNA molecules. Unlike for AARS1, and in conflict with previous reports, we did not detect METTL21C methylation of VCP and HSPA8. AARS1-K943 methylation in HEK293T cells depends upon METTL21C levels. Finally, METTL2C was almost exclusively expressed in muscle tissue, and, accordingly, we detected METTL21C-catalyzed methylation of AARS1 in mouse skeletal muscle tissue. These results reveal that AARS1 is a bona fide in vitro substrate of METTL21C and suggest a role for the METTL21C-AARS1 axis in the regulation of protein synthesis in muscle tissue. Moreover, our study describes a straightforward protocol for elucidating the physiological substrates of poorly characterized or uncharacterized PKMTs.
Keywords: methyltransferase-like 21C (METTL21C), aminoacyl-tRNA synthetase, alanine tRNA synthetase, protein methylation, 70-kilodalton heat shock protein (Hsp70), valosin-containing protein (VCP/p97), substrate specificity, subcellular fractionation, posttranslational modification, skeletal muscle, signal transduction, AARS1, lysine methylation, METTL21C
The addition of a methyl moiety to proteins, nucleic acids, and metabolites is one of the most frequent chemical modifications in cells, with hundreds of known and putative methyltransferase enzymes in the human genome (1–5). Proteins can be methylated at several amino acids including arginine, lysine, and histidine residues (3, 6–9). PKMTs utilize S-adenosyl-l-methionine (SAM) as the methyl donor to catalyze the addition of one, two, or three methyl groups to substrate sites (7) and the PKMTs characterized to date contain one of two known catalytic methyltransferase domains: 1) the SET (Su(var)3-9, Enhancer-of-zeste, Trithorax) domain or 2) the 7βS (seven-β strand) domain (3). Protein lysine methylation has been most extensively studied on histone proteins, but a growing list of SET and 7βS containing KMTs methylate nonhistone substrates (6, 10–13). Furthermore, many 7βS-containing enzymes are candidate KMTs, but their potential substrate(s) and biological function(s) are not known.
METTL21C is 7βS-containing domain that is expressed largely in skeletal muscle tissue and belongs to the METTL21 family of proteins, which also includes METTL21A, METTL21B, and METTL21D. All members of the METTL21 family have been reported to methylate nonhistone substrates (and do not methylate histones). METTL21A is reported to methylate several members of the HSP70 family of proteins at a highly conserved lysine (14, 15). Whereas METTL21B methylates the translation elongation factor eEF1A and METTL21D methylates VCP (16, 17). A recent publication also claimed that VCP is methylated by METTL21C at the same site as that catalyzed by METTL21D; however, in this study, the authors could not detect in vitro methylation of recombinant VCP by recombinant METTL21C and in other assays did not directly assay METT21C-catalzyed methylation of VCP (18). A second study reported that METTL21C methylates HSPA8 at the same site as that catalyzed by METTL21A (19). In this study, the authors used an in vitro methylation assay to support their conclusion that could not distinguish a positive signal coming from bona fide methylation of HSPA8 versus autolabeling of METTL21C (see below), making it unclear whether or not METTL21C modifies HSPA8. Overall, the ambiguity around METTL21C activity on VCP and HSPA8 is challenging to evaluate without a clear positive control substrate (18, 19).
Here, to identify bona fide METTL21C substrates and evaluate previously reported substrates, we employed an unbiased biochemical approach coupled to MS (6). Using this strategy, we identify the cytoplasmic aminoacyl-tRNA transferase AARS1 as a METTL21C substrate. We find that METTL21C specifically mono-, di-, and trimethylates AARS1 at lysine 943 in vitro and in cells. Moreover, we observe that AARS1 is methylated in mouse skeletal muscle, the tissue type that shows the highest expression of METTL21C protein. In contrast to AARS1, we find no evidence in vitro or in cells that METTL21C methylates VCP or HSPA8, arguing that more work is required to understand how METTL21C influences biological processes associated with these important proteins and associated pathways. Together, our study identifies a bona fide substrate of METTL21C and highlights the importance of biochemical analyses in assessing candidate activities of PKMTs, a group of enzymes with important roles in human health and disease.
Results
Identification of AARS1 as a candidate substrate of METTL21C
To investigate METTL21C biochemical activity we took advantage of two observations: 1) a crystal structure of METTL21C is available and can be used to generate candidate, structure-guided mutations (PDB code 4MTL) and 2) in in vitro methylation assays using recombinant METTL21C and [3H]SAM as the methyl donor, but no substrate, METTL21C is autolabeled, a phenomenon we have observed for many PKMTs (3, 20, 21) (Fig. 1, A and B). Indeed, this autolabeling may offer an explanation for why indirect assays measuring methylation such as those used in Ref. 18 can complicate conclusions about a putative enzyme's substrates (see “Discussion”). In the structure, tryptophan 92 (Trp-92) and tyrosine 197 (Tyr-197) form contacts with the cofactor SAM (Fig. 1A). Recombinant mutant METTL21C protein harboring substitutions at these two residues (W92F/Y197A) expresses like WT protein but in contrast to the WT protein, shows no autolabeling activity (Fig. 1B). We postulated that METTL21C-W92F/Y197A (METTL21CMut) could be used as a potential negative control in unbiased experiments aimed at identifying a bona fide biochemical activity.
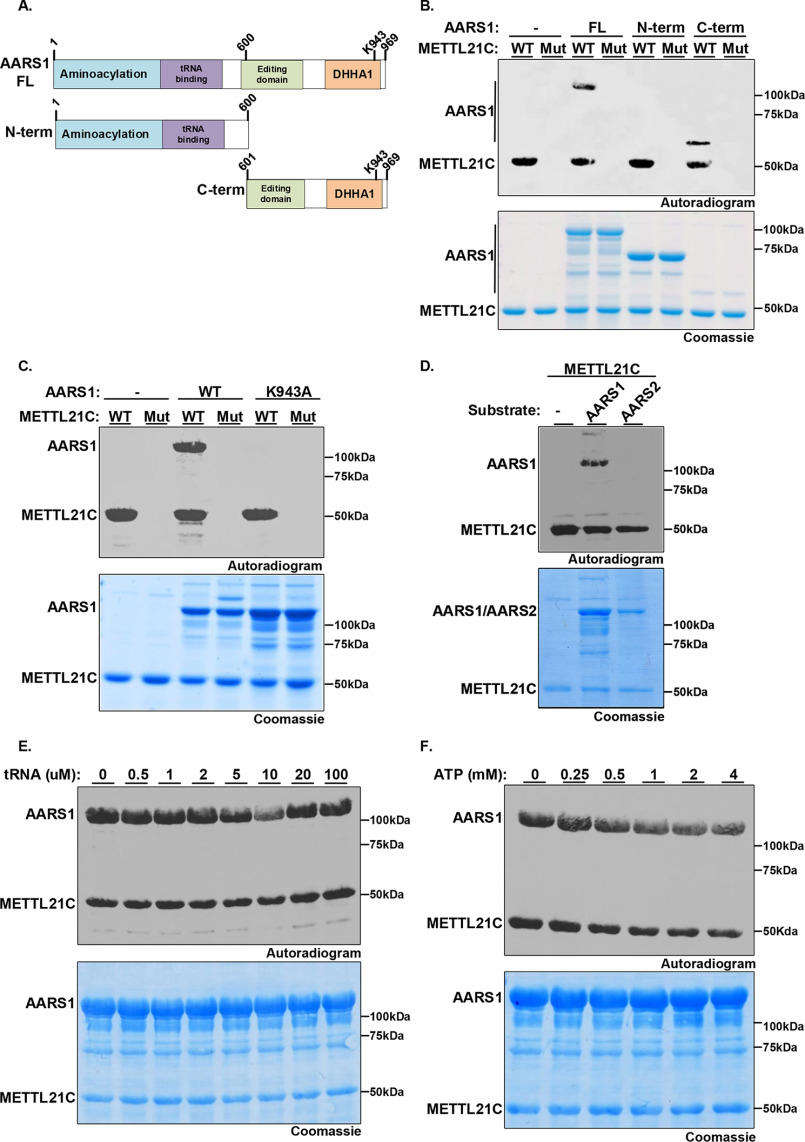
Figure 1.
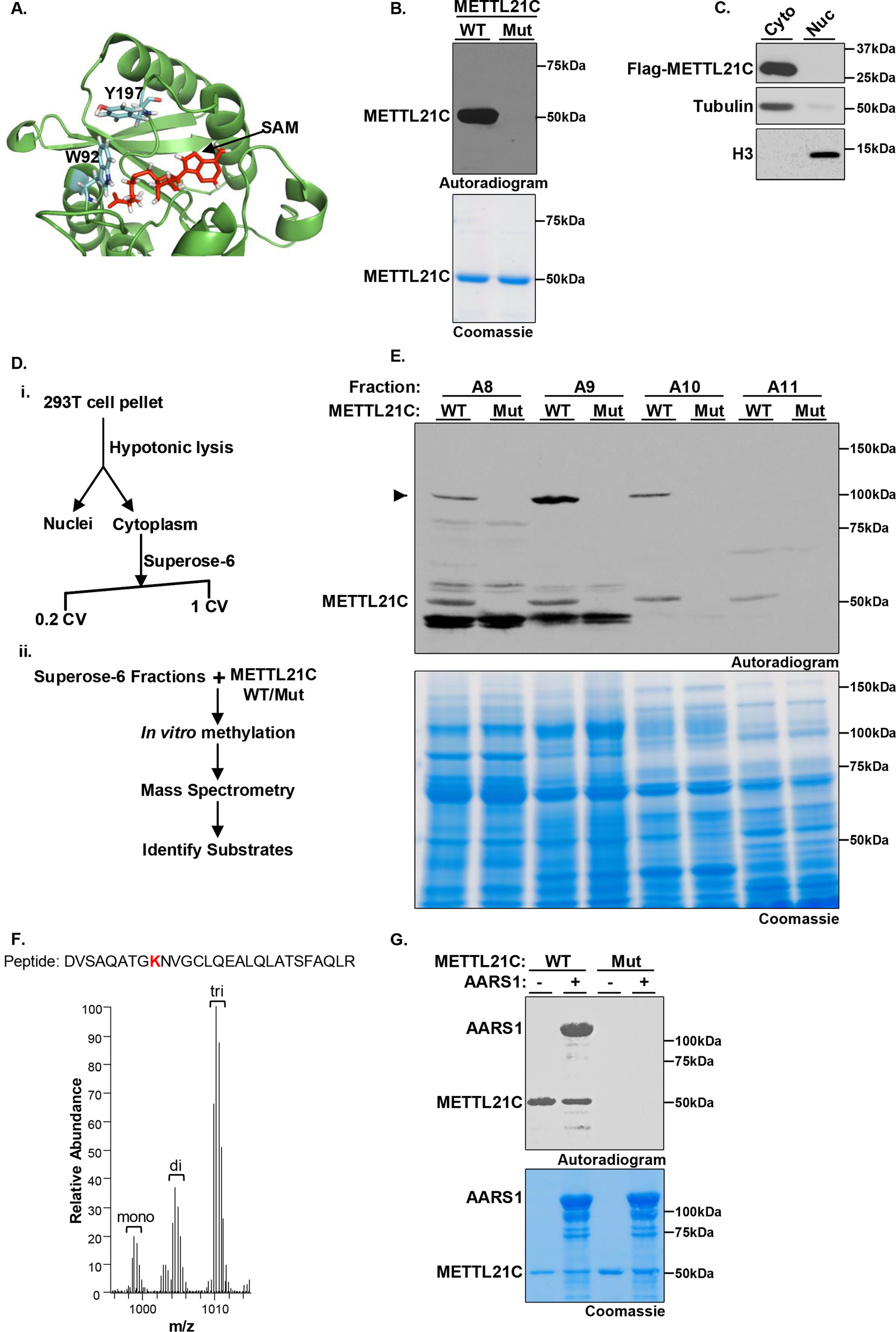
Identification of AARS1 as a bona fide in vitro substrate of METTL21C. A, structural model of METTL21C catalytic domain. The co-factor by product SAH bound to the MTase domain is shown in red. Two of the SAH-interacting residues Tyr-179 and Trp-92 are shown in stick representation. B, identification of point mutations that abrogate METTL21C enzymatic activity. In vitro methylation assay with recombinant GST-WT or the double catalytic mutant W92A/Y197A (Mut) METTL21C as indicated and [3H]SAM as substrate. Top panel, autoradiography; bottom panel, Coomassie Blue staining of proteins in the reaction. C, ectopically expressed METTL21C is detected in the cytoplasm but not the nucleus of 293T cells. Lysates of Flag- METTL21C–transfected cells were biochemically fractionation to separate cytoplasmic and nuclear proteins. Western blots of the indicated fractions and using the indicated antibodies is shown. Tubulin and H3 are shown to ascertain integrity of the fractionation. D, schematic of strategy to identify the methylated substrates of METTL21C; (i) the cytoplasmic fraction of 293T cell lysates was separated by size exclusion chromatography; (ii) SEC fractions were used as substrate in an in vitro methylation assay with WT or Mut METTL21C and then methylation detected by radiolabeling or MS. E, in vitro methylation reactions as described in B but using substrates of the indicated fractions as described in D. Top panel: autoradiography of fractions A8-A11; a candidate substrate indicated by an arrow is detected in fractions A8, A9, and A10 (most enriched in A9) when WT but not Mut METTL21C is used. Bottom panel: Coomassie Blue staining of proteins in the reaction. F, an averaged MS1 full scan of the AARS1 peptides modified by deuterated SAM. The peptide sequence is shown on top of the spectrum, with the methylated lysine (Lys-943) highlighted in bold red. The monoisotopic m/z with a charge of 3 ions is shown: monomethyl 998.5233; dimethyl 1004.2014; and trimethyl 1009.8796. G, recombinant AARS1 is methylated by METTL21C in vitro. Methylation assays are as shown in B with the indicated enzymes and recombinant AARS1. Top panel, autoradiogram. Bottom panel, Coomassie stain of proteins in the reactions.
In cellular localization studies, we observed that Flag-METTL21C (stably expressed) is principally detected in the cytoplasm (Fig. 1C). We were not able to determine the cellular distribution of endogenous METTL21C as it is not detectable in a number of standard human cell lines that we have examined. Based on these results, we focused on identifying cytoplasmic proteins as candidate METTL21C substrates using a two-step fractionation protocol coupled to MS (see schematic in Fig. 1D). In vitro methylation assays were performed with WT or mutant METTL21C and [3H]SAM on fractions from total cytoplasmic extract size separated by gel filtration chromatography to screen for a METTL21C-dependent activity; defined as a radiolabeled band specifically detected in reactions with WT but not mutant METTL21C (6). As shown in Fig. 1E, a band detected at 100 kDa that meets these criteria was clearly observed in three fractions (A8-10), with no other specific band detected in these extracts. To identify the candidate protein and residue(s) modified by METTL21C, in vitro methylation assays were performed on three fractions (fractions 8-10) and the products were analyzed by liquid chromatography-tandem MS (LC–MS/MS); deuterated SAM was used as the methyl donor to distinguish recombinant METTL21C-catalyzed methylation from other sources, including chemical methylation (21, 22). MS analysis identified a peptide from the 100-kDa protein alanine-tRNA synthetase 1 (AARS1) in which deuterated methyl (me1-3) was incorporated at lysine 943 (Lys-943), whereas no other deuterated peptides were identified in the fractions (Fig. 1F, Fig. S1 and Tables S1–S4). We note that a corresponding unmodified peptide containing Lys-943 of AARS1 is cleaved by trypsin after Lys-943 and thus elutes earlier than di- and trimethylated peptides as well as the miscleaved monomethylated peptide. Finally, recombinant AARS1 is directly methylated by METT21C (Fig. 1G). Based on these data, we postulated that AARS1-K943 is a candidate substrate site of METTL21C.
METTL21C directly methylates AARS1 at lysine 943 in vitro
AARS1 plays a critical role in translating the genetic code via the high fidelity linkage of alanine to its cognate tRNA (tRNAAla) (23). To map the site(s) of METTL21C-catalyzed methylation of AARS1, in vitro methylation assays were performed using recombinant WT or catalytic mutant METLL21C and full-length (FL) AARS1 or two AARS1 derivatives spanning either the N terminus 600 residues (N-term, amino acids 1-600) or the C terminus (C-term, amino acids 601-969) as substrates (Fig. 2, A and B). WT METTL21C, but not the catalytic mutant, methylates full-length and C-term AARS1, but not N-term AARS1, indicating that the C-terminal region, which contains Lys-943, is a sufficient substrate for METTL21C (Fig. 2B). Next, substitution of lysine 943 to alanine (K943A) on full-length AARS1 abrogated METTL21C methylation of AARS1 (though METTL21C retained autolabeling, see “Discussion”) (Fig. 2C). Similar results were observed by LC–MS/MS analysis of the in vitro methylation assay using the full-length recombinant proteins and deuterated-SAM; we note that trimethylation was not observed in these reactions (Fig. S2, Table S5). Based on these data we conclude that AARS1 is in vitro methylated at Lys-943 by METTL21C.
Figure 2.
METTL21C specifically methylates AARS1 at Lys-943. A, schematic diagram of AARS1 protein including functional domains and the two derivatives used to map site of methylation: N-term, the N-terminal region (1-600 amino acids) containing the aminoacylation domain and the tRNA-binding domain; C-term, the C-terminal region (601-969 amino acids) containing the editing domain and oligomerization DHHA1 domain. AARS1 full-length is named AARS1-FL. B, the C-terminal region of AARS1 is necessary and sufficient to serve as substrate for METTL21C. In vitro methylation assays as shown in Fig. 1B with WT or Mut METTL21C as indicated on AARS1 FL, N-term, or C-term as indicated. Top panel, autoradiogram. Bottom panel, Coomassie stain of proteins in the reactions. C, METTL21C specifically methylates AARS1 at Lys-943. In vitro methylation assays as in B with the indicated enzyme and substrate. D, METTL21C methylates AARS1 but not AARS2. In vitro methylation assays as in B with METTL21C on substrates AARS1 or the mitochondrial proteins AARS2. E and F, tRNA and ATP do not impact on METTL21C methylation activity on AARS1. In vitro methylation assays as in B using METTL21C and AARS1 as substrate in the presence of tRNA (E) or ATP (F) at the indicated concentrations.
In addition to AARS1, cells possess a second alanine tRNA-synthetase named AARS2 that is specific for mitochondrial synthetized proteins. Consistent with the fact that METTL21C is not located in the mitochondria, METTL21C does not methylate AARS2 in vitro, suggesting specificity for AARS1 (Fig. 2D). A critical step in AARS1-catalyzed formation of activated alanine tRNA, is alanine activation by ATP, which facilitates transfer to the tRNA molecule (24, 25). We therefore next asked whether tRNA or ATP impacts METTL21C methylation of AARS1. As shown in Fig. 2, E and F, the intensity of METT21C-catalyzed AARS1 methylation is not altered by the presence of tRNA or ATP molecules. Taken together, these results demonstrate that AARS1, but not AARS2, is a substrate that is directly methylated by METTL21C, and this methylation reaction occurred independently of ATP and total tRNA in vitro.
METTL21C does not methylate VCP or HSPA8 in vitro
Key residues that constitute the catalytic pocket are conserved within the METTL21 family (METTL21A-D; see Fig. S3A for alignment). We therefore asked whether the other METTL21 proteins (A, B, and D) share METTL21C's activity and can methylate AARS1. In in vitro methylation assays, only METTL21C, but not METTL21A, METTL21B, or METTL21D methylates AARS1 (Fig. 3A). In this context, METTL21C was claimed to methylate the METTL21A substrate HSPA8 and the METTL21D substrate VCP, although in both cases, direct evidence of METTL21C specifically catalyzing the addition of methyl to substrate was not reported (14, 15, 17–19). Leveraging AARS1 as a positive control, we compared the activity of METTL21C on VCP and HSPA8 relative to its activity AARS1, as well as relative to the positive control enzymes METTL21A and METTL21D methylation of HSPA8 and VCP, respectively (Fig. 3, B and C). Notably, in contrast to the positive control enzymes, METTL21C, which is active on AARS1, does not methylate HSPA8 (Fig. 3B) or VCP (Fig. 3C). These data suggest that METTL21C does not directly regulate VCP or HSPA8 and therefore the reported physiologic functions of METTL21C in regulating these proteins may need reevaluation (see “Discussion”).
Figure 3.
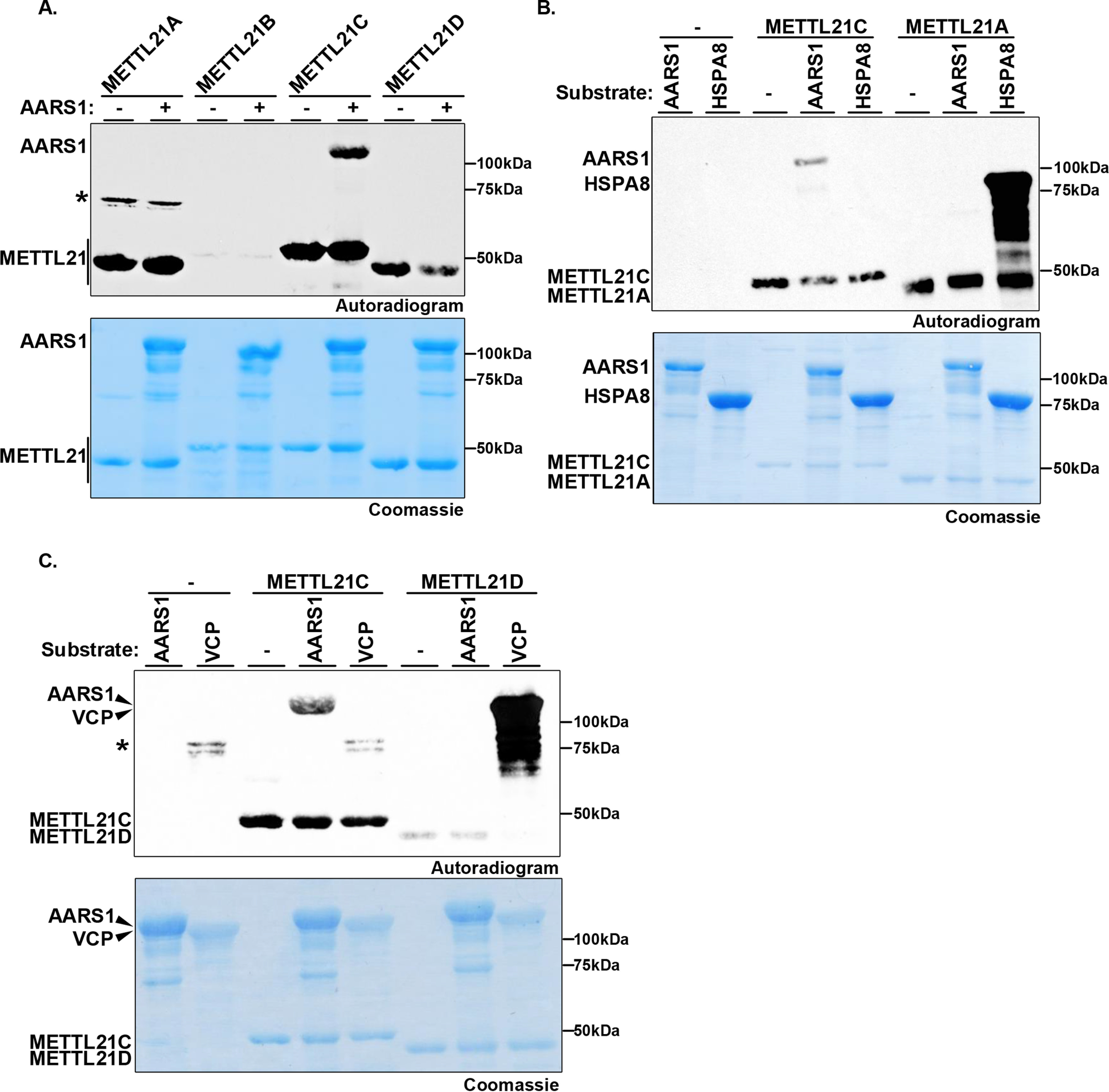
In vitro methylation of AARS1, but not VCP or HSPA8 by METTL21C. A, METTL21C is the only member of the METTL21 family that methylates AARS1. In vitro methylation assays as in Fig. 2B of AARS1 with the indicated enzymes. Autolabeling by the METTL21 enzymes is indicated. Asterisk indicates dimerized GST-METTL21A that is autolabeled. B, METTL21C does not methylate HSPA8 in vitro. In vitro methylation assays as in A with METTL21C or METTL21A as indicated on substrates AARS1 or HSPA8. C, METTL21C does not methylate VCP in vitro. In vitro methylation assays as in A with METTL21C or METTL21D as indicated on substrates AARS1 or VCP. Asterisk shows a bacterial protein that co-purifies with VCP in Escherichia coli that is labeled. We have not been able to ID the band, but it is detected irrespective of the absence or presence of enzyme in the reaction.
Methylation of AARS1 by METTL21C in cells
Enzyme-substrate interactions in a physiologic context are frequently transient but still strong enough to detect. In this context, endogenous AARS1 is present in Flag-METTL21C, but not control IgG, co-immunoprecipitates (co-IPs) from 293T cells (Fig. 4A). To ascertain whether METTL21C could methylate AARS1 in cells, GFP-AARS1 was co-expressed with or without Flag-METTL21C in 293T cells and the methylation status of Lys-943 was determined by LC–MS/MS. In the absence of METTL21C expression, GFP-AARS1 is not methylated on Lys-943 (Fig. 4B). In cells expressing Flag-METTL21C, the majority of GFP-AARS1 is methylated at Lys-943, with trimethylation being the most abundant species (Fig. 4B, Figs. S4 and S5E, and Table S6). In addition to GFP-AARS1, we also found that endogenous AARS1 is mono-, di-, and trimethylated upon expression of Flag-METTL21C (Fig. 4C, Fig. S5, A–D and F, Table S7).
Figure 4.
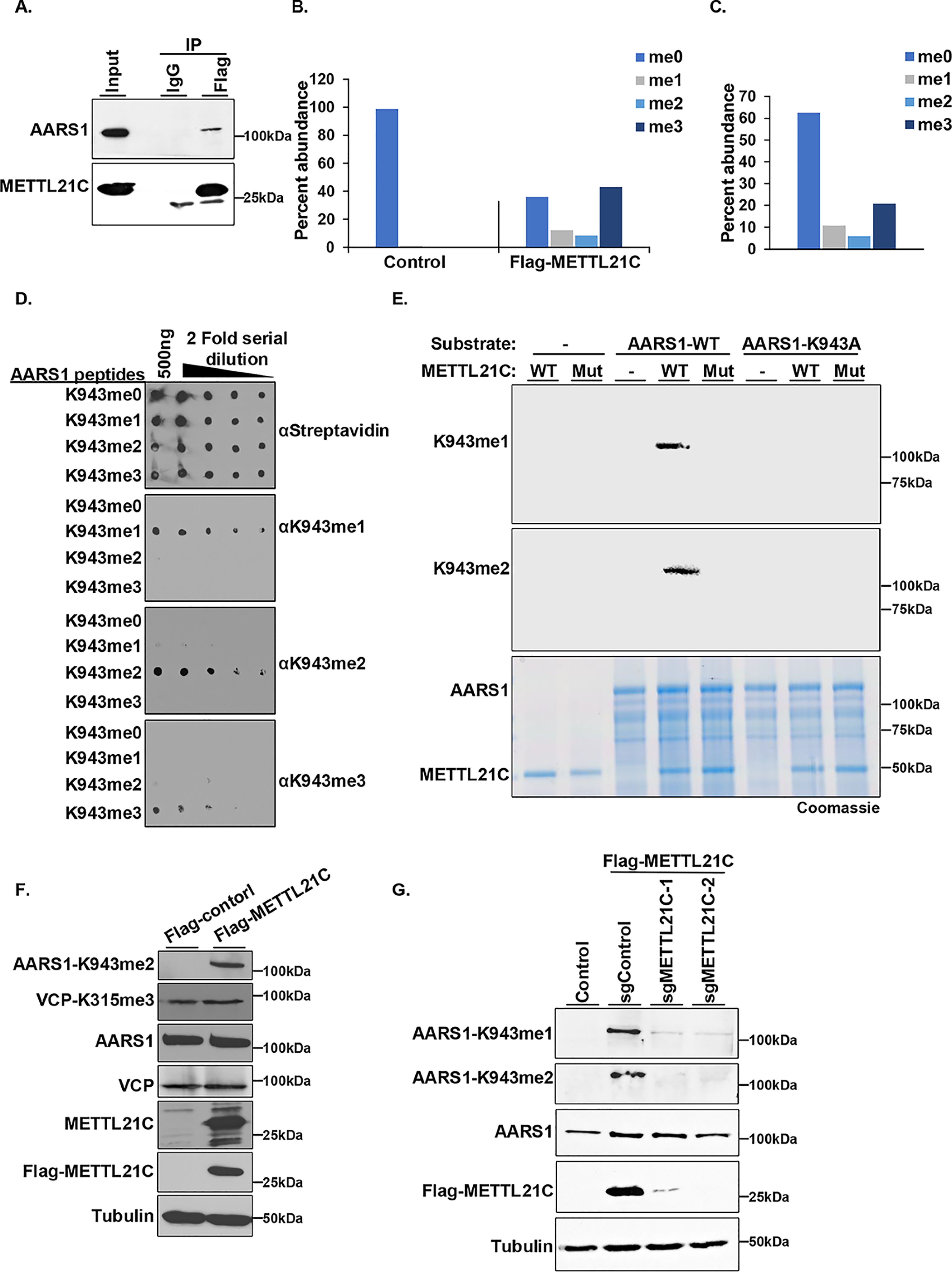
METTL21C methylates AARS1 in cells. A, METTL21C interacts with AARS1 in cells. Western blotting of Flag-METTL21C IPs with the indicated antibodies. IgG used as a negative control. B and C, AARS1 is methylated by METTL21C in cells. MS quantification of (B) GFP-AARS1 or (C) AARS1 methylation at Lys-943 in 293T cells with or without Flag-METTL21C. LC–MS/MS analysis of purified AARS1 in-gel digested with Glu-C. D, generation of selective AARS1-K943me1/me2/me3 antibodies. Dot blot analysis of ΑARS-K943me1/me2/me3 antibodies reactivity with the indicated serial diluted biotinylated peptides. Blot probed with horseradish peroxidase-conjugated streptavidin (strep-HRP) is shown for equal loading of peptides. E, anti-AARS1-me1/me2 antibodies recognize in vitro METTL21C-methylated recombinant AARS1. In vitro methylation assay as above but with nonradiolabeled SAM. Top panel, Western blotting of reactions with anti-AARS1-K943me1. Middle panel, Western blotting of reaction with anti-AARS1-K943me2. Bottom panel, Coomassie stain of proteins in the reaction. F, AARS1 methylates endogenous AARS1, but not VCP, in cells. Western blots with the indicated antibodies in 293T cells with or without Flag-METTL21C expression. VCPK315me3 is the site of methylation reported to be modified in cells by METTL21C (18) but we do not detect any change upon METTL21C expression. Tubulin is shown as a loading control. G, METTL21C is required for AARS1 methylation in cells. Western blotting analysis with the indicated antibodies of whole cell lysates of Flag-METTL21C stable 293T cells expressing the indicated single guide RNA (control or two independent METTL21C guides). Tubulin is shown as a loading control.
Next, we raised state-specific antibodies against AARS1-K943me1, -me2, and -me3 epitopes, which showed selective recognition of the cognate Lys-943 mono-, di-, and trimethylated peptides and no cross-reactivity with unmodified AARS1 peptide (Fig. 4D). Furthermore, aAARS1-K943me1 and aAARS1-K943me2 in Western blotting assays recognized in vitro methylated AARS1 (Fig. 4E). The aAARS1-K943me3 antibody did not work in this application, likely because there is little trimethylation generated in the in vitro reactions as described above. Finally, we generated 293T stable cell lines expressing Flag-METTL21C and detected METTL21C-dependent methylation of AARS1-K943me2 (Fig. 4F). Notably, in these cell lines, in which there is a clear increase in AARS1 methylation, no change was observed for methylation at VCP-K315, the purported site catalyzed by METTL21C (Fig. 4F) (18). The methylation of AARS1 is almost certainly due to METTL21C, as depletion of Flag-METTL21C using two independent CRISPR guide RNAs resulted in depletion of AARS1-K943me (Fig. 4G). Based on these results we conclude that METTL21C methylates AARS1-K943 in a cellular context.
Evidence that AARS1 is a physiological substrate of METTL21C in muscle tissue
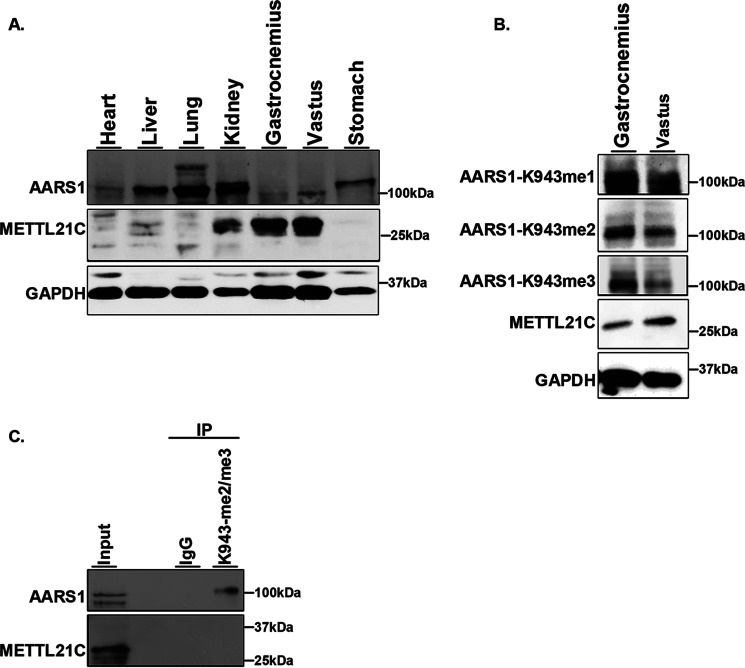
The mRNA levels of METTL21C are reported to be significantly higher in muscle compared with other tissues in mice (18, 26–28). Analysis of endogenous METTL21C and AARS1 levels in different mouse tissue types confirmed that METTL21C protein levels are elevated in gastrocnemius and vastus skeletal muscle compared with other tissue (Fig. 5A). Interestingly, AARS1 protein expression does not follow this pattern, with AARS1 protein levels lower in muscle tissue compared with other tissue types (Fig. 5A). We detect AARS1-943 methylation (me1-3) signal on endogenous AARS1 from skeletal muscle (Fig. 5B). Indeed, in IPs of skeletal muscle tissue lysate, which express METTL21C, the aAARS1-K943me2/3 antibodies pulldown endogenous AARS1, but the control IP does not (Fig. 5C). We do not detect AARS1-K943 methylation in other tissues and cell lines that do not express METTL21C. Together, these results suggest that AARS1 is methylated at Lys-943 in mouse skeletal muscle tissue, a tissue type in which METTL21C is expressed.
Figure 5.
AARS1 is methylated mouse muscle tissue. A, analysis of METTL21C and AARS1 protein levels in mouse tissues. Western blots of the indicated tissue with the indicated antibodies is shown. GAPDH is shown as a loading control. B and C, AARS1 is methylated in skeletal mouse tissue. B, Western blots of the indicated skeletal muscle tissues and the indicated antibodies. GAPDH is shown as a loading control. C, AARS1 is detected in IPs using the anti-AARS1-K943me2 and -me3 antibodies. Western blotting of the indicated IPs with the indicated antibodies is shown. IgG is used as negative control.
Discussion
There is a growing appreciation that methylation of proteins beyond histones by PKMTs represents a major mechanism for regulating key cellular pathways (3). The 7βS subfamily of METTL enzymes family are important members of the expanding list of nonhistone PKMTs (3, 13). For instance, methylation of the essential translational elongation factor eEF1A by five different METTL enzymes likely represent a signaling nexus with functions that could emulate the biological complexity associated with histone H3 methylation (13, 29–31). For example, METTL13, which dimethylates eEF1A at Lys-55, increases protein synthesis to strongly promote oncogenic Ras-driven pancreatic and lung cancers (11, 32). Indeed, developing inhibitors of METTL13 and almost certainly other METTL enzymes should prove to be a promising therapeutic strategy in oncology and other disease applications. Another eEF1A-modifying enzyme is METTL21B, one of the four METTL21 family members (16). Notably, despite the homology between the METTL21 enzymes (see Fig. S3), their substrate specificities differ. METTL21A methylates HSP70 proteins, METTL21B methylates eEF1a, and METTL21D methylates VCP (13, 15–17). Although previous studies reported that METTL21C methylates HSPA8 and VCP (18, 19), our findings here do not support these proteins being direct targets and argue for other substrates such as AARS1 being more physiologically relevant. Regardless, future work investigating potential regulation of HSPA8 and particularly VCP by METT21C may want to consider our negative results and independently test the basic findings before embarking on more in-depth studies.
Here, we used an unbiased biochemical strategy to identify AARS1 as a bona fide in vitro substrate of METTL21C. Our data comparing METTL21C activity on AARS1 side-by-side with HSPA8 and VCP (and including METTL21A and METT21D as positive controls) failed to reproduce the published work on METTL21C (18, 19). The absence of a clear positive control in the previous studies emphasizes the importance of showing a purported activity with recombinant proteins and the methylation on the specific substrate lysine ideally detected by LC–MS/MS with deuterated SAM to eliminate the risk of chemical methylation during sample preparation (33). We have not been able to obtain muscle tissue from METTL21C knockout mice (18). Nonetheless, we speculate that as AARS1 levels are low in muscle tissue, the methylation by METTL21C may positively regulate its activity and contribute to METTL21C physiologic functions in skeletal muscles. We speculate that METTL21C, via AARS1, may promote protein synthesis to help meet increased demand during times of intense muscle usage. At the molecular level, we can speculate that Lys-943 trimethylation regulates AARS1 function either in cis (e.g. modulates AARS1 homodimerization) or in trans by either blocking binding of an AARS1-interacting partner or via its recognition by a protein with a K941me3-binding reader domain. Finally, a biochemical strategy for elucidating substrates of candidate PKMTs has previously proven to be a successful one (for example, see Refs. 6 and 34–36) and can also be used to identify other potential METTL21C substrates in muscle or other tissue that may link METTL21C to new functions. In summary, our study has identified AARS1 as a potential physiologic substrate of METTL21C and suggests that future work is needed to reconcile the discrepancy in this enzyme's catalytic functions and the relationship to muscle biology.
Materials and methods
Cell culture
HEK293T (ATCC) cells were grown in Dulbecco's modified Eagle's medium (Life Technologies) supplemented with 10% fetal calf serum 2 mm l-glutamine and penicillin–streptomycin (Life Technologies). Transfections were performed with either TransIT-293 or TransIT-LT1 (Mirus) in HEK293T cells. Flag-METTL21C cDNA cloned into pLenti CMV Hygro DEST (W117-1) (Addgene). Stable cell lines were generated using lentiviral transductions. HEK293T cells were co-transfected with the lentiviral plasmid, pCMV-Δ8.2, and pCMV-VSVg in a ratio of 9:8:1 by mass. After 48 h of transfection, target cells were transduced with 0.45-μm of filtered viral supernatants supplemented with 8 μg/ml of Polybrene. 293T-Flag-METTL21C stable cell lines were treated with 200 µg/ml of hygromycin for 10 days for selection.
Immunoblot analysis
Protein lysates were prepared using RIPA lysis buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 0.5% deoxycholate, 0.1% SDS, 1% (v/v) Nonidet P-40) with 1 mm phenylmethylsulfonyl fluoride and protease inhibitor mixture. Then, lysate was sonicated and centrifuged at 14,000 rpm for 20 min at 4 °C, the supernatant was recovered. The protein concentration was determined using the DC protein assay (Bio-Rad), and 25–100 μg were resolved on an SDS-PAGE and transferred to a polyvinylidene difluoride membrane. Dot blot analysis was performed by directly loading 1 μl of the indicated amounts of biotinylated peptides onto a polyvinylidene difluoride membrane (0.2 mm). Immunoblots were performed using the appropriate antibodies: anti-AARS1 (Novus), anti-FLAG (M2, Sigma), anti-AARS1-K943me1/me2/me3 (ABclonal), anti-tubulin (Millipore), anti-PAN-actin (Cytoskeleton), anti-VCP (Abcam), anti-VCP-K315me (Millipore), and anti-METTL21C antibody was generated by immunizing recombinant full-length METTL21C (Genemed Synthesis). All secondary antibodies were used at a 1:10,000 dilution. Protein bands were visualized using Amersham Biosciences ECL or Amersham Biosciences ECL Prime Western blotting Detection Reagent.
Site-directed mutagenesis
The PCR mixture in a volume of 50 μl was 3.5 μl of ddH2O, 5 μl of 10× Pfu Turbo Reaction mix, 2 μl of 2.5 mm dNTP, 2.5 µl of DMSO, 2 μl of Primer mix (10 μm each) (final 0.4 μm each), 2 μl of Primer mix (10 μm each) (final 0.4 μm each), 2 μl of template DNA (10-200 ng), and 1 μl of Pfu turbo DNA polymerase. The PCR reaction was: step 1: heating 95 °C for 2 min; step 2: heating 95 °C for 20 s; step 3: annealing 55 °C for 30 s; step 4: elongation 68 °C for 14 min; step 5: repeat steps 2-4 for 19 cycles; step 6: elongation 68 °C for 10 min; step 7: 4 °C forever; step 8: end. After PCR analysis, samples were incubated with 1 µl of DpnI for 4 h at 37 °C. Then, 10 μl of a DpnI-digested mixture was used for transformation in 100 µl of DH5a.
Immunoprecipitation
For immunoprecipitation, protein lysates were prepared by Nonidet P-40 lysis buffer (50 mm Hepes, pH 7.4, 100 mm NaCl, 0.5% Nonidet P-40, 10 mm EDTA, 20 mm β-glycerophosphate, 0.1 mg/ml of phenylmethylsulfonyl fluoride, 1.2 mm NaVO4, 5 mm NaF, 1 mm DTT, protease inhibitor mixture, and 25 units/ml of Benzonase (Novagen)) for 30 min on ice and centrifuged at 14,000 rpm for 25 min at 4 °C, and the supernatant was recovered. The indicated antibodies were incubated with protein G magnetic beads at 4 °C for 3 h, then another 1 h at room temperature. Antibody-protein G-conjugated were washed 3 times with lysis buffer then incubated with equal amounts of whole cell extracts overnight at 4 °C. The beads were washed with cell lysis buffer three times at 4 °C, boiled in Laemmli buffer, and then run in SDS-PAGE gel for MS or Western blot analysis as described.
Expression and purification of recombinant proteins
GST fusion proteins were expressed in BL21 Escherichia coli by overnight culture at 20 °C in LB medium (10 g/liter of tryptone, 5 g/liter of yeast extract, and 10 g/liter of NaCl) supplemented with 0.1 mm IPTG (isopropyl 1-thio-β-d-galactopyranoside, Sigma), purified using GSH Sepharose 4B (GE Healthcare), and eluted in 10 mm reduced GSH (Sigma). Protein concentrations were measured using Pierce Coomassie Plus Assay.
Cellular fractionation
Cytoplasmic cellular extracts were prepared using established protocols (37). In brief, cells were disrupted by hypotonic lysis (25 mm KCl, 1.5 mm MgCl2, 10 mm Hepes–KOH, pH 7.9, and protease inhibitors) and Dounce homogenization. Size-exclusion chromatography (Superose 6) was performed in hypotonic lysis buffer. Fractions were concentrated with centrifugal filter units (Amicon, 3,000 Da molecular mass cut-off) before being used as substrate in in vitro reactions with radiolabeled SAM or deuterated SAM. Chromatography for substrate enrichment was performed with an AKTA FPLC (GE Healthcare).
In vitro methylation assay
In vitro methylation assays were performed similar to as described by Mazur et al. (38), by combining 3-5 µg of recombinant proteins and equal amounts of recombinant enzymes in a methyltransferase buffer (50 mm Tris, pH 8.0, 20 mm KCl, 5 mm MgCl2, and 10% glycerol) supplemented with 1 mm SAM (New England Biolabs) or deuterated Ado- Met (C/D/N Isotopes) or 2 mCi of tritiated AdoMet (American Radiolabeled Chemicals). The reaction mixtures were incubated overnight at 37 °C. Reactions were resolved by SDS-PAGE, followed by autoradiography, Coomassie stain, or MS analysis.
MS
Recombinant or immunoprecipitated proteins were separated by SDS-PAGE and stained using InstantBlue Protein Stain (Expedeon). Bands were cut and destained in 50% acetonitrile, 50% ammonium bicarbonate (NH4HCO3, 50 mm) for 10 min twice. Gel pieces were incubated in 50 mm NH4HCO3 containing 10 mm DTT at 60 °C for 30 min, followed by treatment with 25 mm iodoacetamide in 50 mm NH4HCO3 at room temperature for 45 min, results in the covalent addition of a carbamidomethyl group to cystines (57.07 Da). In-gel digestion was performed using 100 ng/ml of Glu-C in 50 mm phosphate buffer (pH 7.8) at 37 °C or 10 ng/ml of trypsin in 50 mm NH4HCO3 overnight at 37 °C (Promega). Two consecutive peptide extractions were processed with 5% formic acid, 49% water, and 50% acetonitrile. The resulting peptides were dried by SpeedVac and desalted using C18 StageTips (Thermo Fisher Scientific). Desalted peptides were resuspended in 0.1% formic acid and analyzed by online capillary nanoLC-MS/MS. Samples were separated on an in-house made 20-cm reversed phase column (100 μm inner diameter, packed with ReproSil-Pur C18-AQ 3.0 μm resin (Dr. Maisch GmbH)) equipped with a laser-pulled nanoelectrospray emitter tip. Peptides were eluted at a flow rate of 400 nl/min using a two-step linear gradient including 2–25% buffer B in 70 min and 25–40% B in 20 min (buffer A: 0.2% formic acid and 5% DMSO in water; buffer B: 0.2% formic acid and 5% DMSO in acetonitrile) in a Dionex Ultimate 3000 LC-system (Thermo Scientific). Peptides were then analyzed using a LTQ Orbitrap Elite mass spectrometer (Thermo Scientific). Data acquisition was executed in data dependent mode with full MS scans acquired in the Orbitrap mass analyzer with a resolution of 60,000 and m/z scan range of 340-1600. The top 20 most abundant ions with intensity threshold above 500 counts and charge states 2 and above were selected for fragmentation using collision-induced dissociation with an isolation window of 2 m/z, normalized collision energy of 35%, activation Q of 0.25, and activation time of 5 ms. The collision-induced dissociation fragments were analyzed in the ion trap with rapid scan rate. Dynamic exclusion was enabled with repeat count of 1 and exclusion duration of 30 s. The AGC target was set to 1,000,000 and 5,000 for full FTMS scans and ITMSn scans, respectively. The maximum injection time was set to 250 ms and 100 ms for full FTMS scans and ITMSn scans, respectively. Data were searched using MaxQuant version 1.5.5.1 platform, against a database combining Uniprot human (downloaded 2016-08-24) containing 21,014 proteins and common contaminants or against AARS1 protein sequence, a full table of MaxQuant parameters is included (Table S8). The precursor mass range was set to 350-10,000 Da, the mass error tolerance was set to FTMS of 20 ppm, and deisotoping tolerance of 7 ppm, and the fragment mass error tolerance; ITMS MS/MS match tolerance of 0.5 Da, and deisotoping tolerance of 0.15 Da. Enzyme specificity was set to trypsin or Glu-C with Max miss cleavage of 2. Carbamidomethylation of cysteines (57.021) was set as fixed modification, whereas oxidation of methionines and acetylation of protein N terminus (+42.011): methyl (K deuterated +3), dimethyl (K deuterated +3), and trimethyl (K deuterated +3) were set as variable modifications. MaxQuant was used to filter peptides and proteins to estimation of false discovery rate (FDR); PSM FDR 0.01, protein FDR 0.01. Abundance quantification was based on intensities. Selected ion chromatograms for peptides spanning were extracted using Xcalibur Qual Browser (Thermo).
Viral transduction
For CRISPR-Cas9 knockouts, virus particles were produced by co-transfection of 293T cells with the pLenti-U6-sgRNA-SFFV-Cas9-2A-Puro construct (abmgood) expressing the indicated sgRNAs, pCMV-VSV-G, and pCMV-dR8.2 dvpr in a ratio of 9:8:1 by mass. 48 h after transfection, target cells were transduced with 0.45-mm filtered viral supernatant and 4 µg/ml of Polybrene. Cells were selected 24 h after media replacement with 2 µg/ml of puromycin.
Data availability
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (39), partner repository with the data set identifier PXD019886. All other data are described in the manuscript.
Supplementary Material
This article contains supporting information.
Author contributions—M. Z., S. M. C., and O. G. conceptualization; M. Z. data curation; M. Z., L. Z., T-M. L., and O. G. formal analysis; M. Z. and O. G. funding acquisition; M. Z., T-M. L., and S. M. C. investigation; M. Z., L. Z., and T-M. L. methodology; M. Z. writing-original draft; J. E. E. and O. G. resources; O. G. supervision; O. G. project administration; O. G. writing-review and editing.
Funding and additional information–This work was supported in part by National Institutes of Health Grant R01 CA236118 (to O. G.) and IDT's Foundation for Advance Technology and Community Giving (ATCG Foundation). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—O. G. is a co-founder of EpiCypher, Inc. and Athelas Therapeutics, Inc.
- PKMTs
- protein-lysine methyltransferases
- 7βS
- seven-β strand
- co-IPs
- co-immunoprecipitates
- C-term
- C terminus
- FL
- full-length
- HSPA8
- heat shock protein family A (Hsp70) member 8
- K943me1
- monomethylated lysine 943
- K943me2
- dimethylated lysine 943
- K943me3
- trimethylated lysine 943
- METTL21C
- methyltransferase-like protein 21C
- METTL21CMut
- METTL21C-W92F/Y197A
- Mut
- mutant
- N-term
- N terminus
- PTM
- post-translational modification
- SET
- Su(var)3-9, Enhancer-of-zeste, Trithorax
- VCP
- valosin-containing protein/p97
- FDR
- false discovery rate
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- HSP
- heat shock protein.
References
- 1. Michalak E. M., Burr M. L., Bannister A. J., and Dawson M. A. (2019) The roles of DNA, RNA and histone methylation in ageing and cancer. Nat. Rev. Mol. Cell Biol. 20, 573–589 10.1038/s41580-019-0143-1 [DOI] [PubMed] [Google Scholar]
- 2. Du J., Johnson L. M., Jacobsen S. E., and Patel D. J. (2015) DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 16, 519–532 10.1038/nrm4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carlson S. M., and Gozani O. (2016) Nonhistone lysine methylation in the regulation of cancer pathways. Cold Spring Harb. Perspect. Med. 6, a026435 10.1101/cshperspect.a026435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xiang Y., Laurent B., Hsu C. H., Nachtergaele S., Lu Z., Sheng W., Xu C., Chen H., Ouyang J., Wang S., Ling D., Hsu P. H., Zou L., Jambhekar A., He C., et al. (2017) RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature 543, 573–576 10.1038/nature21671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Husmann D., and Gozani O. (2019) Histone lysine methyltransferases in biology and disease. Nat. Struct. Mol. Biol. 26, 880–889 10.1038/s41594-019-0298-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wilkinson A. W., Diep J., Dai S., Liu S., Ooi Y. S., Song D., Li T. M., Horton J. R., Zhang X., Liu C., Trivedi D. V., Ruppel K. M., Vilches-Moure J. G., Casey K. M., Mak J., et al. (2019) SETD3 is an actin histidine methyltransferase that prevents primary dystocia. Nature 565, 372–376 10.1038/s41586-018-0821-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mozzetta C., Boyarchuk E., Pontis J., and Ait-Si-Ali S. (2015) Sound of silence: the properties and functions of repressive Lys methyltransferases. Nat. Rev. Mol. Cell Biol. 16, 499–513 10.1038/nrm4029 [DOI] [PubMed] [Google Scholar]
- 8. Biggar K. K., and Li S. S. (2015) Non-histone protein methylation as a regulator of cellular signalling and function. Nat. Rev. Mol. Cell Biol. 16, 5–17 10.1038/nrm3915 [DOI] [PubMed] [Google Scholar]
- 9. Moore K. E., and Gozani O. (2014) An unexpected journey: lysine methylation across the proteome. Biochim. Biophys. Acta 1839, 1395–1403 10.1016/j.bbagrm.2014.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clarke S. G. (2013) Protein methylation at the surface and buried deep: thinking outside the histone box. Trends Biochem. Sci. 38, 243–252 10.1016/j.tibs.2013.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu S., Hausmann S., Carlson S. M., Fuentes M. E., Francis J. W., Pillai R., Lofgren S. M., Hulea L., Tandoc K., Lu J., Li A., Nguyen N. D., Caporicci M., Kim M. P., Maitra A., et al. (2019) METTL13 methylation of eEF1A increases translational output to promote tumorigenesis. Cell 176, 491–504.e421 10.1016/j.cell.2018.11.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Serre N. B. C., Alban C., Bourguignon J., and Ravanel S. (2018) An outlook on lysine methylation of non-histone proteins in plants. J. Exp. Bot. 69, 4569–4581 10.1093/jxb/ery231 [DOI] [PubMed] [Google Scholar]
- 13. Falnes P. Ø., Jakobsson M. E., Davydova E., Ho A., and Małecki J. (2016) Protein lysine methylation by seven-β-strand methyltransferases. Biochem. J. 473, 1995–2009 10.1042/BCJ20160117 [DOI] [PubMed] [Google Scholar]
- 14. Jakobsson M. E., Moen A., Bousset L., Egge-Jacobsen W., Kernstock S., Melki R., and Falnes P. O. (2013) Identification and characterization of a novel human methyltransferase modulating Hsp70 protein function through lysine methylation. J. Biol. Chem. 288, 27752–27763 10.1074/jbc.M113.483248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sieber J., Wieder N., Ostrosky-Frid M., Dvela-Levitt M., Aygun O., Udeshi N. D., Carr S. A., and Greka A. (2017) Lysine trimethylation regulates 78-kDa glucose-regulated protein proteostasis during endoplasmic reticulum stress. J. Biol. Chem. 292, 18878–18885 10.1074/jbc.M117.797084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Malecki J., Aileni V. K., Ho A. Y. Y., Schwarz J., Moen A., Sorensen V., Nilges B. S., Jakobsson M. E., Leidel S. A., and Falnes P. O. (2017) The novel lysine specific methyltransferase METTL21B affects mRNA translation through inducible and dynamic methylation of Lys-165 in human eukaryotic elongation factor 1α (eEF1A). Nucleic Acids Res. 45, 4370–4389 10.1093/nar/gkx002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kernstock S., Davydova E., Jakobsson M., Moen A., Pettersen S., Mælandsmo G. M., Egge-Jacobsen W., and Falnes P. Ø. (2012) Lysine methylation of VCP by a member of a novel human protein methyltransferase family. Nat. Commun. 3, 1038 10.1038/ncomms2041 [DOI] [PubMed] [Google Scholar]
- 18. Wiederstein J. L., Nolte H., Gunther S., Piller T., Baraldo M., Kostin S., Bloch W., Schindler N., Sandri M., Blaauw B., Braun T., Holper S., and Kruger M. (2018) Skeletal muscle-specific methyltransferase METTL21C trimethylates p97 and regulates autophagy-associated protein breakdown. Cell Rep 23, 1342–1356 10.1016/j.celrep.2018.03.136 [DOI] [PubMed] [Google Scholar]
- 19. Wang C., Arrington J., Ratliff A. C., Chen J., Horton H. E., Nie Y., Yue F., Hrycyna C. A., Tao W. A., and Kuang S. (2019) Methyltransferase-like 21c methylates and stabilizes the heat shock protein HSPA8 in type I myofibers in mice. J. Biol. Chem. 294, 13718–13728 10.1074/jbc.RA119.008430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Levy D., Kuo A. J., Chang Y., Schaefer U., Kitson C., Cheung P., Espejo A., Zee B. M., Liu C. L., Tangsombatvisit S., Tennen R. I., Kuo A. Y., Tanjing S., Cheung R., Chua K. F., et al. (2011) Lysine methylation of the NF-κB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NF-κB signaling. Nat. Immunol. 12, 29–36 10.1038/ni.1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carlson S. M., Moore K. E., Sankaran S. M., Reynoird N., Elias J. E., and Gozani O. (2015) A proteomic strategy identifies lysine methylation of splicing factor snRNP70 by the SETMAR enzyme. J. Biol. Chem. 290, 12040–12047 10.1074/jbc.M115.641530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garcia B. A., Mollah S., Ueberheide B. M., Busby S. A., Muratore T. L., Shabanowitz J., and Hunt D. F. (2007) Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat. Protoc. 2, 933–938 10.1038/nprot.2007.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peacock J. R., Walvoord R. R., Chang A. Y., Kozlowski M. C., Gamper H., and Hou Y. M. (2014) Amino acid-dependent stability of the acyl linkage in aminoacyl-tRNA. RNA 20, 758–764 10.1261/rna.044123.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ibba M., and Soll D. (2000) Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69, 617–650 10.1146/annurev.biochem.69.1.617 [DOI] [PubMed] [Google Scholar]
- 25. Swairjo M. A., Otero F. J., Yang X. L., Lovato M. A., Skene R. J., McRee D. E., Ribas de Pouplana L., and Schimmel P. (2004) Alanyl-tRNA synthetase crystal structure and design for acceptor-stem recognition. Mol. Cell 13, 829–841 10.1016/S1097-2765(04)00126-1 [DOI] [PubMed] [Google Scholar]
- 26. Yang G., Lu H., Wang L., Zhao J., Zeng W., and Zhang T. (2019) Genome-wide identification and transcriptional expression of the METTL21C gene family in chicken. Genes (Basel) 10, 628 10.3390/genes10080628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jerez-Timaure N., Gallo C., Ramirez-Reveco A., Greif G., Strobel P., Pedro A. V. F., and Morera F. J. (2019) Early differential gene expression in beef longissimus thoracis muscles from carcasses with normal (<5.8) and high (>5.9) ultimate pH. Meat Sci. 153, 117–125 10.1016/j.meatsci.2019.03.013 [DOI] [PubMed] [Google Scholar]
- 28. Huang J., Hsu Y. H., Mo C., Abreu E., Kiel D. P., Bonewald L. F., Brotto M., and Karasik D. (2014) METTL21C is a potential pleiotropic gene for osteoporosis and sarcopenia acting through the modulation of the NF-κB signaling pathway. J. Bone Miner. Res. 29, 1531–1540 10.1002/jbmr.2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cornett E. M., Ferry L., Defossez P. A., and Rothbart S. B. (2019) Lysine methylation regulators moonlighting outside the epigenome. Mol. Cell 75, 1092–1101 10.1016/j.molcel.2019.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hamey J. J., and Wilkins M. R. (2018) Methylation of elongation factor 1A: where, who, and why? Trends Biochem. Sci. 43, 211–223 10.1016/j.tibs.2018.01.004 [DOI] [PubMed] [Google Scholar]
- 31. Jakobsson M. E., Małecki J., and Falnes P. Ø. (2018) Regulation of eukaryotic elongation factor 1α (eEF1A) by dynamic lysine methylation. RNA Biol. 15, 314–319 10.1080/15476286.2018.1440875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jakobsson M. E., Małecki J. M., Halabelian L., Nilges B. S., Pinto R., Kudithipudi S., Munk S., Davydova E., Zuhairi F. R., Arrowsmith C. H., Jeltsch A., Leidel S. A., Olsen J. V., and Falnes P. Ø. (2018) The dual methyltransferase METTL13 targets N terminus and Lys55 of eEF1A and modulates codon-specific translation rates. Nat. Commun. 9, 3411 10.1038/s41467-018-05646-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zee B. M., Levin R. S., Xu B., LeRoy G., Wingreen N. S., and Garcia B. A. (2010) In vivo residue-specific histone methylation dynamics. J. Biol. Chem. 285, 3341–3350 10.1074/jbc.M109.063784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Małecki J. M., Willemen H. L. D. M., Pinto R., Ho A. Y. Y., Moen A., Eijkelkamp N., and Falnes P. Ø. (2019) Human FAM173A is a mitochondrial lysine-specific methyltransferase that targets adenine nucleotide translocase and affects mitochondrial respiration. J. Biol. Chem. 294, 11654–11664 10.1074/jbc.RA119.009045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jakobsson M. E., Małecki J., Nilges B. S., Moen A., Leidel S. A., and Falnes P. O. (2017) Methylation of human eukaryotic elongation factor α (eEF1A) by a member of a novel protein lysine methyltransferase family modulates mRNA translation. Nucleic Acids Res. 45, 8239–8254 10.1093/nar/gkx432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Małecki J., Jakobsson M. E., Ho A. Y. Y., Moen A., Rustan A. C., and Falnes P. Ø. (2017) Uncovering human METTL12 as a mitochondrial methyltransferase that modulates citrate synthase activity through metabolite-sensitive lysine methylation. J. Biol. Chem. 292, 17950–17962 10.1074/jbc.M117.808451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Baymaz H. I., Spruijt C. G., and Vermeulen M. (2014) Identifying nuclear protein-protein interactions using GFP affinity purification and SILAC-based quantitative mass spectrometry. Methods Mol. Biol. 1188, 207–226 10.1007/978-1-4939-1142-4_15 [DOI] [PubMed] [Google Scholar]
- 38. Mazur P. K., Reynoird N., Khatri P., Jansen P. W. T. C., Wilkinson A. W., Liu S., Barbash O., Van Aller G. S., Huddleston M., Dhanak D., Tummino P. J., Kruger R. G., Garcia B. A., Butte A. J., Vermeulen M., et al. (2014) SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature 510, 283–287 10.1038/nature13320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Pérez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., et al. (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (39), partner repository with the data set identifier PXD019886. All other data are described in the manuscript.