Abstract
Systemic amyloidosis caused by extracellular deposition of insoluble fibrils derived from the pathological aggregation of circulating proteins, such as transthyretin, is a severe and usually fatal condition. Elucidation of the molecular pathogenic mechanism of the disease and discovery of effective therapies still represents a challenging medical issue. The in vitro preparation of amyloid fibrils that exhibit structural and biochemical properties closely similar to those of natural fibrils is central to improving our understanding of the biophysical basis of amyloid formation in vivo and may offer an important tool for drug discovery. Here, we compared the morphology and thermodynamic stability of natural transthyretin fibrils with those of fibrils generated in vitro either using the common acidification procedure or primed by limited selective cleavage by plasmin. The free energies for fibril formation were −12.36, −8.10, and −10.61 kcal mol−1, respectively. The fibrils generated via plasmin cleavage were more stable than those prepared at low pH and were thermodynamically and morphologically similar to natural fibrils extracted from human amyloidotic tissue. Determination of thermodynamic stability is an important tool that is complementary to other methods of structural comparison between ex vivo fibrils and fibrils generated in vitro. Our finding that fibrils created via an in vitro amyloidogenic pathway are structurally similar to ex vivo human amyloid fibrils does not necessarily establish that the fibrillogenic pathway is the same for both, but it narrows the current knowledge gap between in vitro models and in vivo pathophysiology.
Keywords: transthyretin, systemic amyloidosis, amyloid, fibril, protein aggregation, protein stability, thermodynamics, protein misfolding, V122I TTR mutation, mechano-enzymatic mechanism
Understanding the mechanisms of diseases is vital to efficiently tackling unmet medical needs, and this is especially true for diseases of high complexity, such as systemic amyloidosis. The development of in vitro and in vivo models mirroring crucial steps of any pathological process is essential to interpret the natural history of the diseases and offer reliable tools for drug discovery (1).
In the field of amyloid diseases, we have learned how to reshape the conformation of a protein, in a test tube, by simply modifying the buffer's chemical-physical properties, thus driving the transformation of proteins from a globular state to a fibrillar structure (2). A wide range of methods to transform soluble proteins and peptides into amyloid-like fibrils have been established in vitro even for proteins that have no role in amyloid diseases (2).
Indeed, in many cases, fibrillogenesis is carried out in vitro under completely nonphysiological conditions, thus limiting our capacity to speculate on the mechanism of amyloid development in vivo. Development of in vitro fibrillogenesis under biocompatible conditions is particularly important when animal models are unavailable and represents a unique tool for drug discovery. This has been so far the case with transthyretin (TTR) amyloidosis, where there is currently no reliable animal model presenting clinical and pathological features consistent with the human disease and where the identification of potential drug candidates was only based on the inhibition of fibrillogenesis in vitro.
Most of our knowledge of the pathogenesis of TTR amyloidosis derives from the experimental model of in vitro fibrillogenesis at low pH in which protein aggregation is primed by tetramer disassembly (3); this method has been the key tool for identifying drugs, such as tafamidis, currently used to treat the disease (4–7). More recently, based on the observation that truncated forms of TTR are present in natural fibrils (8), we have established a new, more physiological, system of fibrillogenesis in which biomechanical forces combined with specific proteolytic enzymes play a central role (9–11).
Fibrils are essential for work on in vivo imaging tracers and new fibril-clearing drugs, such as β-breakers or specific anti-fibril antibodies (12), but the availability of ex vivo fibrils is very limited. Development of more appropriate methods to generate amyloid-like fibrils, with properties that are closely similar to those of the natural counterpart, is thus of paramount importance. Comparative analysis of structure and function of fibrils derived from different procedures either in vitro or ex vivo has been conducted by methods ranging from light microscopy for tinctorial properties, through EM for low-resolution morphology (13), to cryo-EM for fibrillar ultrastructure analysis (14, 15).
Here we have prepared TTR fibrils generated by the commonly used low-pH procedure and separately by our more physiological mechano-enzymatic approach using plasmin as the protease (11) and compared their structure and thermodynamic stability with natural ex vivo TTR amyloid fibrils. The mechano-enzymatic product was notably more similar to the genuine ex vivo amyloid fibrils and is thus most appropriately suitable for physiologically and therapeutically relevant studies of TTR amyloidosis.
Results and discussion
Amyloid fibril preparation
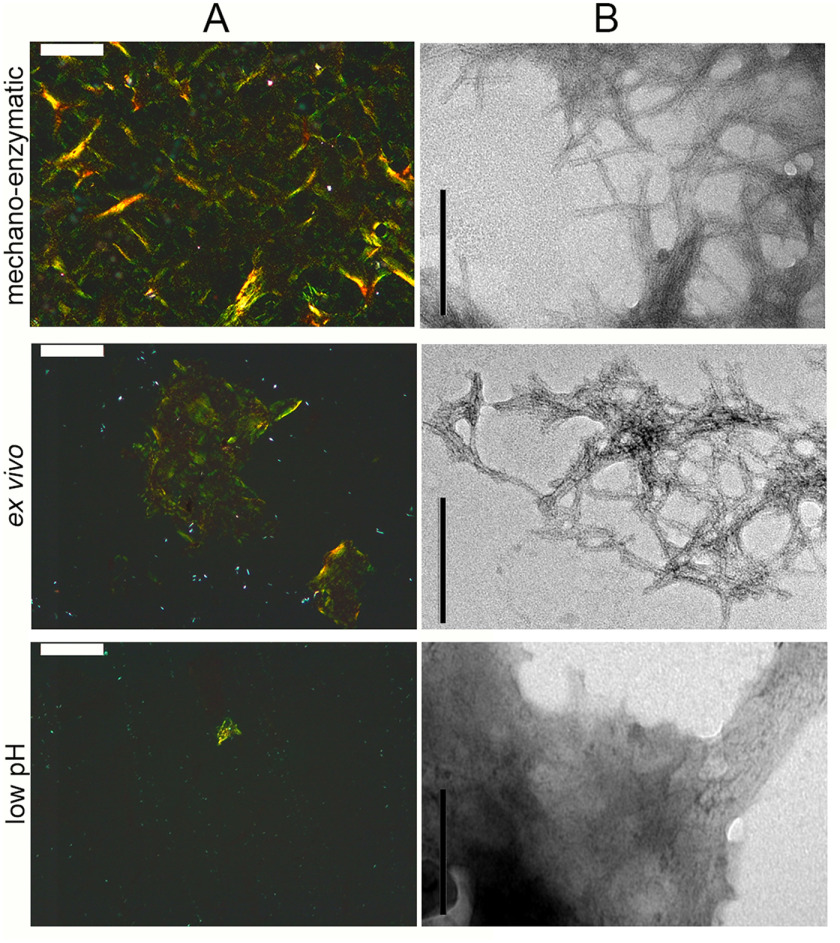
The aggregation of recombinant V122I variant TTR was carried out in vitro using the mechano-enzymatic mechanism we have described recently (11) and also using the widely used low-pH procedure established by Colon and Kelly (3). Both types of synthetic V122I TTR aggregates were characterized in comparison with natural fibrils extracted from the heart of a patient carrying the V122I TTR mutation using the classical water extraction procedure established by Pras et al. (16). The typical pathognomonic amyloid apple-green birefringence under polarized light observed with the Congo red–stained fibrils formed by proteolysis closely resembled that of the ex vivo fibrils, whereas only occasional spots of birefringent material were observed in the low-pH fibrils (Fig. 1A). Transmission EM confirmed the similarity between the mechano-enzymatic and natural fibrils in which an average diameter of ∼7 nm was measured for both types (Fig. 1B and Fig. S1) in contrast to the sparse fibrillar structures buried in a dense background of amorphous material in the aggregates formed at low pH (Fig. 1B and Fig. S1).
Figure 1.
Microscopic analysis of in vitro and ex vivo TTR fibrils. A, amyloid was identified by light microscopy of Congo red–stained specimens viewed under crossed polarized light (scale bar, 100 μm). B, negatively stained transmission EM (scale bar, 200 nm) of TTR fibrils prepared in vitro with the mechano-enzymatic mechanism, at low pH or extracted from human amyloidotic tissue. Additional images are shown in Fig. S1. Only the mechano-enzymatically generated fibrils showed similar morphology to natural fibrils with an estimated diameter of 6.9 ± 0.80 and 7.0 ± 0.64 nm, respectively, as described under “Experimental procedures.”
Identification of TTR species in the mechano-enzymatic and ex vivo fibrils
MS analysis was carried out to identify the main species in the mechano-enzymatic and ex vivo fibrils, in contrast to the material treated at low pH, which is homogenously composed of full-length recombinant V122I TTR only. The pellet of plasmin-digested TTR fibrils was resuspended in acetonitrile and TFA as described under “Experimental procedures” and directly analyzed by MALDI-TOF MS in linear mode (Fig. S2). The three main species identified were the 49–127 and 81–127 peptides together with the full-length V122I TTR protomer (Table 1 and Fig. S2).
Table 1.
Composition of mechano-enzymatic and ex vivo fibrils
Shown are the identified TTR protein/peptides with residue numbers. All the species identified in the natural fibrils contained both WT and V122I variant TTR, as demonstrated after further digestion of each band with trypsin or AspN proteases (data not shown).
a Main components of the fibrils generated in vitro using the mechano-enzymatic mechanism of recombinant V122I TTR fibrillogenesis with plasmin were identified by linear mode MALDI MS as described under “Experimental procedures” (Fig. S2).
bMain components of the ex vivo fibrils extracted from a V122I ATTR patient were identified after separation in SDS-PAGE (Fig. S3A) and MALDI-TOF spectra of the fragments eluted from the electrophoretic bands (Fig. S3B). In this sample, the residue 81–127 peptide could not be detected in the material derived from the electrophoretic band, consistent with the apparent size of the fragment.
Natural fibrils comprise a complex mixture of ubiquitous nonfibrillar components, including proteoglycans, glycosaminoglycans, and lipids, which affect desorption and/or ionization of the sample (17) so that, not unexpectedly, direct linear mode MALDI-TOF MS analysis of the solubilized material was not possible. The solubilized pellet was therefore first separated by SDS-PAGE (Fig. S3A), and the main protein bands at the apparent size of the full-length TTR monomer and its fragments were excised, eluted, and analyzed by MALDI-TOF MS in linear mode (Table 1 and Fig. S3B).
Our results confirm the presence of both full-length and 49–127 TTR in both the mechano-enzymatic and ex vivo fibrils, but a detailed quantification of full-length TTR and its fragments will require an alternative MS approach, such as parallel reaction monitoring. The 81–127 peptide fragment was detected in the plasmin-digested material, and it was not observed in the eluted electrophoretic band with its apparent size from the ex vivo fibrils (Fig. S3). Another fragment corresponding to the residue 46–127 TTR fragment was identified in the natural fibrils as described previously (18).
Stability of V122I TTR precursor and fibrillar structures
We used the strong chaotropic agent guanidine thiocyanate (Gdn-SCN) to determine the stability of amyloid fibrils obtained by the different procedures.
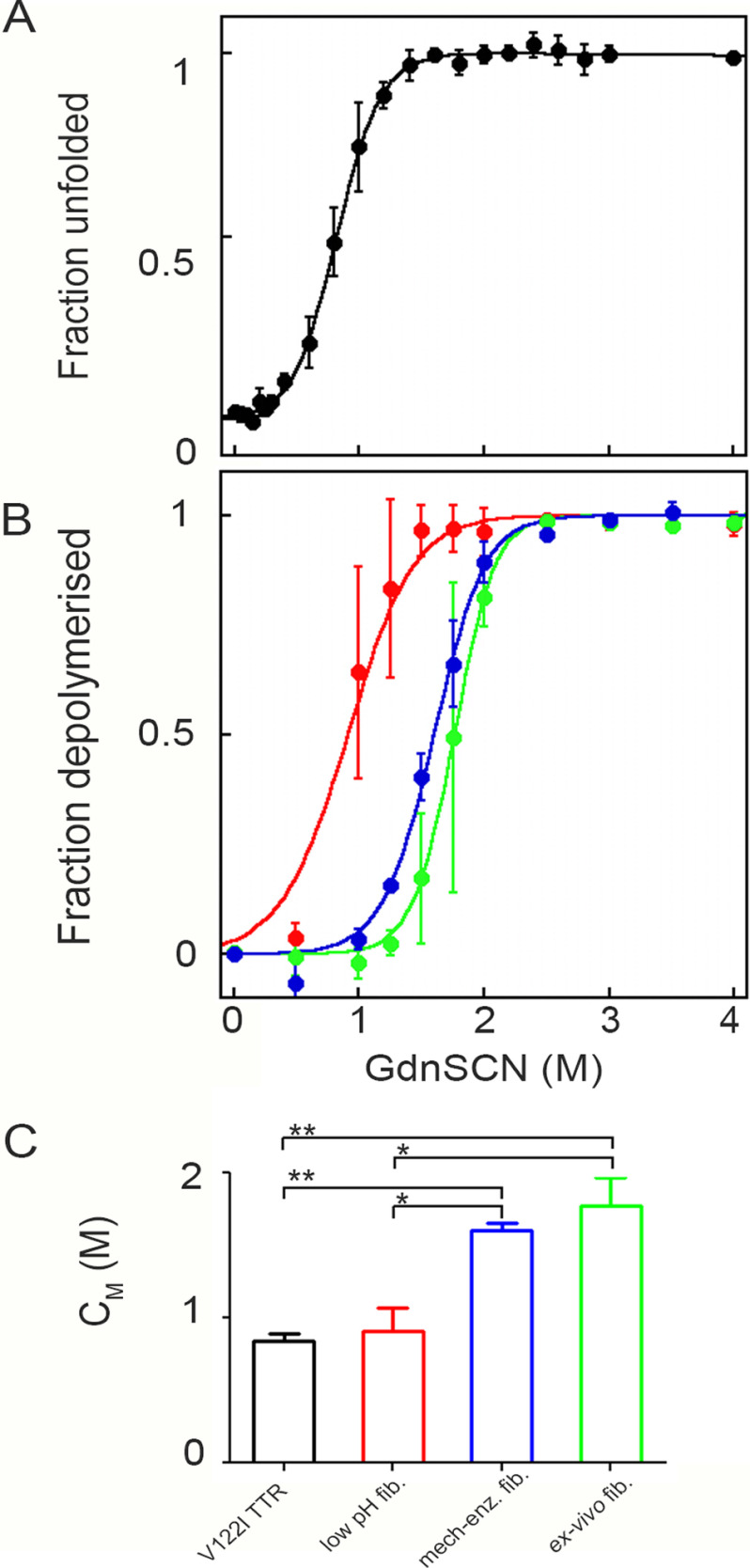
Exposure of native TTR to increasing concentrations of Gdn-SCN was monitored by change in the tryptophan fluorescence emission. The ratio between the intrinsic fluorescence intensities of the unfolded monomer at 355 nm and the folded tetramer at 335 nm (9, 19) as a function of the denaturant concentration was analyzed by the method of Santoro and Bolen (9, 19, 20), to determine the main thermodynamic parameters. Experimental data are shown as the fraction of unfolded protein (Fig. 2A) in which the sharp transition between 0.6 and 1.0 m denaturant concentration can be observed. Values of 2.95 ± 0.38 kcal mol−1 and 0.84 ± 0.06 m were obtained for the free energy of unfolding in the absence of denaturant (ΔGH2O) and the midpoint denaturant concentration (CM), respectively (Table 2).
Figure 2.
Thermodynamic stability of V122I TTR precursor and fibrils. A, denaturation profile for V122I TTR is derived from the change in fluorescence (F355/F335) following 295 nm excitation calculated according to Equation 2 (20) under “Experimental procedures.” B, denaturation of in vitro V122I TTR fibrils formed at low pH (red), mechano-enzymatic mechanism (blue), or ex vivo ATTR V122I TTR fibrils (green) is based on a ThT fluorescence assay of the residual pellet after denaturation and centrifugation and analyzed with Equation 4 described under “Experimental procedures.” Curves are shown as mean ± S.D. (error bars) of three independent experiments. C, values of midpoint Gdn-SCN concentration are shown as mean ± S.D. of three experiments. Two-way analysis of variance for multiple comparison gave p < 0.01 (**) for V122I TTR precursor versus mechano-enzymatic as well as ex vivo fibrils and p < 0.05 (*) for low pH fibrils versus both mechano-enzymatic and ex vivo fibrils.
Table 2.
Thermodynamic parameters for Gdn-SCN–induced unfolding of V122I TTR precursor and fibrils
All measurements are reported as mean ± S.D. of three independent experiments.
| Precursor | ΔGH2Oa | CMa |
|---|---|---|
| V122I TTR | 2.95 ± 0.38 | 0.84 ± 0.06 |
| V122I TTR fibrils | ΔG0 elb | CMb |
| Low pH | −8.10 ± 0.56 | 0.91 ± 0.16 |
| Mechano-enzymatic | −10.61 ± 0.53 | 1.60 ± 0.05 |
| Ex vivo | −12.36 ± 1.13 | 1.77 ± 0.20 |
aValues of CM (m), midpoint concentration of Gdn-SCN and ΔGH2O (kcal mol−1), free energy of unfolding in the absence of denaturant for equilibrium denaturation of globular V122I TTR variant were determined using a two-state model described previously (20).
bValues of CM (m) and ΔGel0 (kcal mol−1), free energy of elongation in the absence of denaturant, were calculated following the linear polymerization mode as described under “Experimental procedures.”
Equal amounts of amyloid fibrils, either prepared in vitro or extracted from the heart of a patient with V122I TTR amyloidosis, were incubated in solutions containing increasing concentrations of Gdn-SCN at neutral pH and room temperature for 96 h until samples reached an apparent equilibrium. The unfolding of fibrils was monitored by thioflavin T (ThT) emission fluorescence of pellets separated from their supernatants in which there was no turbidity detectable by spectrophotometric absorbance in the 350–400-nm range. As the disassembly of fibrils parallels the loss of the specific ThT/fibril complex (21), fractional loss of ThT signal at increasing concentrations was converted into the fraction of TTR species dissociated from amyloid fibrils. Data were fitted with the linear polymerization model using Equation 4 as described under “Experimental procedures” (21, 22) (Fig. 2B) to yield the main thermodynamic parameters, including the midpoint denaturant concentration and the change in the Gibbs free energy of elongation in the absence of denaturant, ΔG0el. Fibrils prepared at low pH were less stable than both the mechano-enzymatic and natural fibrils, with a midpoint denaturant concentration of 0.91 ± 0.16 m, which is close to the CM measured for the unfolding of the precursor V122I TTR and significantly lower than the midpoint Gdn-SCN concentration measured for the mechano-enzymatic (1.60 ± 0.05 m) and natural fibrils (1.77 ± 0.20 m) (Fig. 2C and Table 2). A value of ΔG0el = −8.10 ± 0.56 kcal mol−1 was obtained for the less stable acid-mediated fibrils compared with both the mechano-enzymatic in vitro (−10.61 ± 0.53 kcal mol−1) fibrils and the natural ex vivo (−12.36 ± 1.13 kcal mol−1) fibrils, both of which contain cleaved TTR fragments (Table 2).
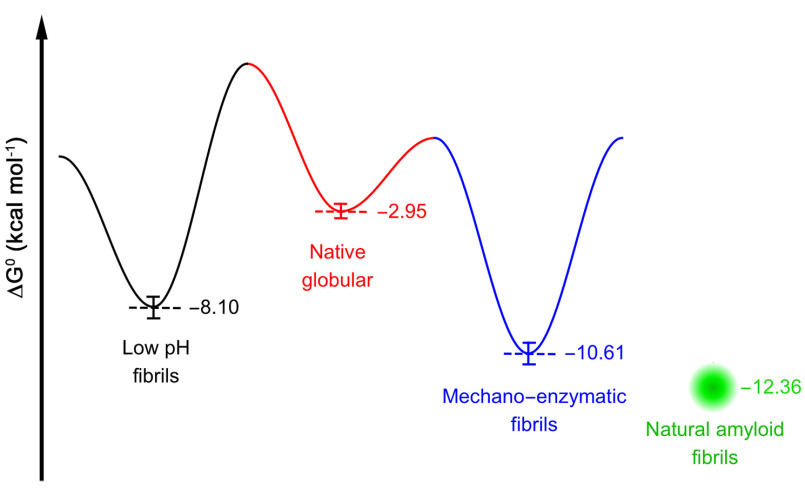
We are aware that the linear polymerization model (21–23) represents a simplification based on the assumption that all equilibria in a solution of polymers have the same equilibrium constant. However, having kept the total protein concentration constant across the different types of sample, the equilibrium unfolding curves can be fitted with this model to compare the free energy of elongation in the absence of denaturant, ΔG0el, and the midpoint denaturant concentration, CM, obtained for each type of fibrillar aggregate. Based on the same model, we have previously studied equilibrium denaturation curves to compare the stability of fibrils formed by WT and H50Q α-synuclein (with ΔG0el values of −7.36 ± 0.02 and −8.46 ± 0.2 kcal mol−1, respectively) (24) as well as characterized the thermodynamics of WT and D76N β2-microglobulin fibrils (25) grown in different experimental conditions (yielding ΔG0el values of −9.3 ± 0.36 and −12.8 ± 0.35 kcal mol−1, respectively). As shown previously for synuclein and β2-microglobulin, all of the TTR fibrillar species are more stable than the corresponding globular precursor, confirming that the aggregation pathway moves toward the most stable structures, consistent with the landscape of the energetic diagram of different conformations of a polypeptide proposed previously by Hartl and Hayer-Hartl (26). We have adapted this scheme of the funnel-shaped free energy landscape explored by proteins in their native and fibrillar state by showing the energy levels determined for amyloid fibrils prepared at low pH or by the mechano-enzymatic mechanism or extracted from human amyloidotic tissue, all having a minimum much lower than that of the globular precursor (Fig. 3).
Figure 3.
Energy landscape scheme of native TTR folding and fibrillation. The scheme reports ΔGH2O of (un)folding of tetrameric V122I TTR (red) and the ΔG of elongation in the absence of denaturant of the fibrillar species produced by low pH (black) and mechano-enzymatic (blue) methods, respectively. The presence of mechanical forces and traces of protease are likely to reduce the energetic barriers due to the cleavage of the Lys48–Thr49 peptide bond. The ΔG of elongation of ex vivo fibrils is indicated as a green dot, and the surrounding shade indicates the uncertainty.
As thermodynamic stability of fibrils is determined by intermolecular bonds, this information may be complementary to the structural observations on fibrils obtained by solid-state NMR (27) and cryo-EM, which have recently been applied to a number of natural pathologic filaments (28) and fibrils (14, 15, 29). Ultrastructural polymorphism of amyloid fibrils has been observed among fibrils derived from the different monoclonal immunoglobulin light chains and different mutations; different sources of natural fibrils may lead to significant structural differences. A good example is offered by the structures of amyloid fibrils of two λ light chains, a λVI by Swuec et al. (30) and a λI by Radamaker et al. (15), in which the contribution of the N-terminal part to the fibril core is different as well as the geometry of protofilament assembly. The heterogeneity observed in light chains is particularly relevant, because apparently fibrils are made from precursors that present similar generic folds in their globular state, being both λ light chains, although of different subclasses (I or VI). We can therefore assume that specific sequences, truncations, and co-factors may influence the final structure of fibrils. The heterogeneity in structure may correlate with the thermodynamic stability of fibrils and subsequently with their kinetics of growth and resistance to solubilization. Recently, Fändrich's team has released the first cryo-EM structure of natural fibrils extracted from the heart of a patient with V30M variant TTR amyloidosis (29). In this case, apparently, the fibrils were composed of a mixture of full-length monomers and C-terminal truncated fragments plus the N-terminal cleaved polypeptide. It may be possible to compare the cryo-EM structures of natural fibrils generated by other TTR variants and compare them with the structures of fibrils prepared in vitro through different methods.
In conclusion, comparative thermodynamic analysis of fibrils made in vitro through different methods with natural fibrils extracted from different organs, and formed by different TTR variants, is a useful and important probe that will complement forthcoming structural information from cryo-EM and solid-state NMR. In addition, the thermodynamic similarity of mechano-enzymatic TTR amyloid fibrils to natural ex vivo TTR amyloid fibrils provides further strong support for use of the mechano-enzymatic method in the design and development of novel drugs targeting TTR amyloidosis.
Structural similarity between different fibrils obviously does not necessarily imply that the respective in vitro and in vivo processes of fibrillogenesis are identical. Nevertheless, our present findings are fully consistent with all of the other available evidence that plasmin likely plays a crucial role in vivo. Unequivocal in vivo confirmation will eventually have to come from a robust new animal model.
Let us say that when we generate models of diseases, mimicking the in vivo pathological process, we should never forget the timeless recommendation by René Descartes: “Whenever people notice some similarity between two things they are in the habit of ascribing to the one what they find true of the other, even when the two are not in that respect similar.”
Experimental procedures
Amyloid fibril preparation in vitro
Two sets of TTR fibrils were prepared in vitro by the low-pH procedure (3) or the mechano-enzymatic method (11).
Acidic-mediated protein aggregation was initiated by the addition of stock solution of recombinant tetrameric V122I TTR (0.4 mg/ml in PBS, pH 7) to an equivalent volume of 200 mm sodium acetate, 0.1% (w/v) NaN3, pH 4.4, and incubation at 37 °C was carried out in glass vials for 7 days.
Proteolysis-mediated fibrillogenesis of V122I TTR was carried out in glass vials (air/interface of 1.5 cm2) stirred at 1,500 rpm (IKA magnetic stirrer) and 37 °C for 96 h using 1 mg/ml TTR in PBS, pH 7.4, in the presence of 20 ng/µl plasmin (Sigma–Aldrich, P1867).
Fibrillar aggregates separated after a 20 min centrifugation at 20,800 × g were quantified at the end of each procedure by the bicinchoninic acid (BCA) assay.
Extraction of natural fibrils ex vivo
Amyloid fibrils were isolated from the heart of a patient with cardiac amyloidosis associated with the V122I TTR variant. The study was carried out in accordance with the Declaration of Helsinki and the written consent of the patient. Fibrils were isolated from the cardiac tissue (∼100 mg) by water extraction in the presence of 1.5 mm phenylmethylsulfonyl fluoride after repeated homogenization in the presence of 10 mm Tris-HCl containing 140 mm NaCl, 10 mm EDTA, 0.1% (w/v) NaN3, 1.5 mm phenylmethylsulfonyl fluoride, pH 8.0, and a 30 min centrifugation at 60,000 × g. The yield in fibrils was monitored by microscopic analysis of the extracted material stained with Congo red. Quantification of total protein was performed by a BCA assay (Pierce).
Microscopic analysis
The pathognomonic amyloid apple-green birefringence in both in vitro and ex vivo fibrils was evaluated under high-intensity cross-polarized light after alkaline alcoholic Congo red staining (31).
Samples were also examined by negative staining transmission EM. Briefly, a drop of each sample was allowed to dry onto a Formvar/carbon-coated copper grid 3 min before blotting with filter paper to remove excess solvent and staining with 2% (w/v) uranyl acetate for 3 min. After further washing, blotting, and drying in air, transmission electron microscope (Joel JEM-1400 Flash) images were obtained at 120 kV. Estimation of diameter of both mechano-enzymatic and ex vivo fibrils was performed with ImageJ software. Only fibrils that appeared to be single filaments were selected for measurements, which were taken directly in pixels and then converted into nanometers using the 200 nm bar.
Identification of TTR species in the mechano-enzymatic and ex vivo fibrils
MALDI-TOF MS analysis was carried out to identify TTR species in both the mechano-enzymatic and ex vivo fibril samples. A 10 μl mixture containing mechano-enzymatic fibrils was centrifuged at 20,800 × g for 20 min, and the resulting pellet was resuspended in 10 μl of α-cyano-4-hydroxycinnamic acid (5 mg/ml in acetonitrile, 0.2% TFA (70:30, v/v)), and finally 1 µl was left to dry onto the target plate.
Ex vivo fibrils were subjected to SDS 15% PAGE under reducing conditions. After staining with colloidal Coomassie Blue, the bands were excised and passively eluted from the gel. Each band was crushed in 30 µl of methanol/isopropyl alcohol/water (30:30:40, v/v/v) (32) and then vortexed and stirred overnight at room temperature. Finally, samples were sonicated and centrifuged at 20,800 × g for 10 min.
The resulting supernatants were mixed with α-cyano-4-hydroxycinnamic acid (5 mg/ml in acetonitrile, 0.2% TFA (70:30, v/v)), and 1 µl was left to dry onto the target plate.
All spectra were recorded in linear mode by MALDI-TOF/TOF 5800 system (AB SCIEX, Framingham, MA, USA) and analyzed using TOF/TOF Series Explorer acquisition software. Calibration standards were recombinant V122I TTR or WT β2-microglobulin.
Preparation of recombinant V122I TTR
Site-directed mutagenesis of peTM11 plasmid encoding hexahistidine-tagged WT TTR was carried out using the QuikChange kit (Stratagene) and the primer CC ACC ACG GCT GTC ATC ACC AAT CCC AAG G containing the underlined codon for isoleucine at position 122. Transformed BL21 star (DE3) cells (Thermo Fisher Scientific) were then plated onto Luria broth (LB) medium containing agar, 30 µg/ml kanamycin for overnight incubation at 37 °C. A single colony was isolated and cultured overnight at 37 °C in 5 ml of LB medium containing 30 μg/ml kanamycin under shaking conditions (LB/kan). This preparation was inoculated into 1 liter of LB/kan for an initial growth at 37 °C. When the culture reached A600 = 0.5, the temperature was reduced to 30 °C. Protein expression was induced at A600 = 0.6 by adding isopropylthiogalactoside (1 mm final concentration) for overnight incubation. The cells were harvested by centrifugation; suspended in lysis buffer containing 20 mm Tris-HCl, pH 8, 250 mm NaCl, 3 mm imidazole; and finally sonicated at 4 °C. The supernatant was clarified after 30 min of centrifugation at 18,000 × g and loaded onto a HisTrap FF crude nickel affinity chromatography column (GE Healthcare) equilibrated in lysis buffer. After extensive washing with 20 mm Tris-HCl, 10 mm imidazole, containing stepwise increasing concentrations of NaCl (250 mm, 500 mm, and 1 m, respectively), the column was eluted with 20 mm Tris-HCl, 250 mm NaCl, 250 mm imidazole, pH 8.0. His-tagged tobacco etch virus protease (Sigma–Aldrich) was added at 1% (w/w) during dialysis to selectively cleave the hexahistidine tag, which was then removed by affinity chromatography, together with the enzyme. Fractions containing TTR were pooled and subjected to size-exclusion chromatography using a Superdex 75 Hi Load 26/60 column (GE Healthcare) equilibrated and eluted with 25 mm Tris-HCl, 100 mm NaCl, pH 8.0. Fractions containing TTR were dialyzed against water at 4 °C for at least 3 days and then lyophilized. Purity and molecular weight were determined by SDS-PAGE analysis and MS, respectively.
Equilibrium unfolding of native TTR
Samples containing tetrameric V122I TTR (0.1 mg/ml) were incubated at increasing concentrations of Gdn-SCN in 50 mm sodium phosphate, 1 mm EDTA, 1 mm DTT, pH 7.0, at 25 °C for 24 h. Tryptophan fluorescence spectra were recorded between 310 and 410 nm with excitation at 295 nm using a 1 cm light path cell in a PerkinElmer LS55 spectrofluorimeter. All spectra were blank-subtracted. Because maximum emission of native TTR is between 337 and 338 nm, whereas the unfolded protein shows maximum emission at 355–358 nm, we used the fluorescence ratio between 355 (unfolded) and 335 nm (folded) to generate denaturation curves as a function of Gdn-SCN (19) for further analysis.
Depolymerization of TTR fibrils
V122I TTR fibrils (1.5 mg/ml) were resuspended in sodium phosphate 50 mm, EDTA 1 mm, DTT 1 mm buffer, pH 7, at increasing concentrations of Gdn-SCN. Samples were mixed by vortexing before incubation at room temperature for 96 h to allow the samples to reach equilibrium. To separate nonaggregated fraction from aggregated protein, samples were centrifuged for 20 min at 20,800 × g.
A ThT assay was used to determine the persistent fibrillar material in the insoluble fraction after the incubation with Gdn-SCN (Mfib). Briefly, the pellets were resuspended in 100 µl of PBS containing 10 μm ThT, and fluorescence emission was monitored at 485 nm, following excitation at 445 nm in a CLARIOstar microplate reader (BMG Labtech). Values of ThT fluorescence at each denaturant concentration were then normalized to the value of the corresponding fibrillar sample in the absence of denaturant to determine the fibrillar content (Mfib/MT). The fraction of depolymerization was then derived considering the following,
| (Eq. 1) |
where MT is the total amount of protein and M is the protein content in the soluble fraction at each denaturant concentration.
Thermodynamic stability parameters of tetrameric TTR
Gdn-SCN denaturation curves were analyzed according to a two-state unfolding model as described previously for the transition from native folded tetramer to unfolded monomer for the V122I TTR precursor. Experimental data were fit according the Santoro–Bolen equation (20).
| (Eq. 2) |
where the yobs is the fluorescence ratio at each denaturant concentration; y0N and y0U are the signals of the native and denaturated states, respectively, in the absence of denaturant; and [D] is the Gdn-SCN concentration. The linear dependence of pre- and post-transition with denaturant concentration is defined by mN and mU, respectively. The model provides quantitative measurements of the difference in free energy between the folded and unfolded state in the absence of denaturant (ΔGH2O), the midpoint denaturant concentration (CM), and the dependence of ΔG on Gdn-SCN (m).
Experimental data were reported as apparent unfolded fraction using the formula,
| (Eq. 3) |
where y is the experimental value observed at a given denaturant concentration, and yN and yU are the values of the native and unfolded protein, respectively, extrapolated from the pre- and post-transition baselines defined by Equation 2 (20). All data were fitted using Kaleidagraph 4.0 (Synergy Software, Reading, PA, USA).
Thermodynamic stability parameters of TTR amyloid fibrils
The Gdn-SCN equilibrium unfolding curves of TTR fibrils were analyzed using a linear polymerization model (21, 22, 33),
in which [M] and [Fi] represent the concentration of monomers and fibrillar aggregates of size i, respectively, where it is assumed that the equilibrium constants K are equal for all of the interactions between monomers and other species in solutions.
Based on this model, the fraction of species released during denaturation can be expressed as function of the total concentration [M]/[MT] (fraction depolymerized) as follows.
| (Eq. 4) |
The equilibrium constant K can also be expressed as follows,
| (Eq. 5) |
in which ΔGel is the free energy of elongation, R is the gas constant, and T is the absolute temperature. In the presence of chemical denaturants (i.e. Gdn-SCN), ΔGel is linearly dependent on the concentration of denaturant, [D], according to the following,
| (Eq. 6) |
where m is a cooperativity coefficient and ΔGel0 is the free energy of elongation in the absence of denaturant (21, 22). By introducing these terms in Equation 4, we can describe the fibril depolymerization curve as a function of the denaturant concentration. Experimental data of the equilibrium unfolding of in vitro and natural TTR fibrils were fitted to Equation 4 to obtain the main thermodynamic parameters using KaleidaGraph 4.0 (Synergy Software). Values of midpoint denaturant concentration, CM, were also calculated. All measurements are reported as mean ± S.D. of three independent experiments, and two-way analysis of variance was performed using GraphPad Prism 5 for pairwise multiple comparison.
Data availability
All data are contained within the article and associated supporting information. MS data sets and additional information are available upon request from Sofia Giorgetti(s.giorgetti@unipv.it).
Supplementary Material
Acknowledgments
We thank the Parco Tecnico Scientifico (PTS) of the University of Pavia and Polymerix S.r.l. (Pavia, Italy) for providing access to mass spectrometry facilities. We also thank Nicola Botcher (National Amyloidosis Centre, University College London and Royal Free Hospital, London, UK) for technical support.
This article contains supporting information.
Author contributions—S. R., P. P. M., G. V., D. C., P. N., L. M., R. P., V. M., G. F., G. W. T., J. D. G., A. C., M. B. P., and S. G. investigation; S. R., G. V., D. C., P. N., L. M., V. M., and G. F. methodology; S. R., P. P. M., G. W. T., and M. B. P. writing-review and editing; P. P. M. data curation; G. V. and S. G. formal analysis; S. G. and V. B. supervision; V. B. conceptualization; V. B. writing-original draft.
Funding and additional information—This work was supported by investment from the University College London Technology Fund and United Kingdom Medical Research Council Grant MR/R016984/1 (to V. B.), Rosetrees Trust/Royal Free Charity PhD Programme Grant M427 (to G. V.), Italian Ministry of Health Ricerca Finalizzata RF 2013 02355259 (to V. B.), the Italian Ministry of Research and University Dipartimenti di Eccellenza 2018–2022 grant to the Molecular Medicine Department (University of Pavia), and the Istituto Nazionale di Biostrutture e Biosistemi. Core support for the Wolfson Drug Discovery Unit is provided by the United Kingdom National Institute for Health Research Biomedical Research Centre and Unit Funding Scheme via the UCLH/UCL Biomedical Research Centre and by the UCL Amyloidosis Research Fund.
Conflict of interest—The authors declared that they have no conflict of interest with the contents of this article.
- TTR
- transthyretin
- ThT
- thioflavin T
- Gdn-SCN
- guanidine thiocyanate
- ΔGH2O
- free energy of unfolding in the absence of denaturant
- CM
- midpoint denaturant concentration
- ΔG0el
- free energy of elongation in the absence of denaturant
- LB
- Luria broth
- kan
- kanamycin
- BCA
- bicinchoninic acid.
References
- 1. Barker R. (2016) Bioscience—Lost in Translation? How Precision Medicine Closes the Innovation Gap, Oxford University Press, Oxford [Google Scholar]
- 2. Chiti F., and Dobson C. M. (2017) Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu. Rev. Biochem. 86, 27–68 10.1146/annurev-biochem-061516-045115 [DOI] [PubMed] [Google Scholar]
- 3. Colon W., and Kelly J. W. (1992) Partial denaturation of transthyretin is sufficient for amyloid fibril formation in vitro. Biochemistry 31, 8654–8660 10.1021/bi00151a036 [DOI] [PubMed] [Google Scholar]
- 4. Berk J. L., Suhr O. B., Obici L., Sekijima Y., Zeldenrust S. R., Yamashita T., Heneghan M. A., Gorevic P. D., Litchy W. J., Wiesman J. F., Nordh E., Corato M., Lozza A., Cortese A., Robinson-Papp J., et al. (2013) Repurposing diflunisal for familial amyloid polyneuropathy: a randomized clinical trial. JAMA 310, 2658–2667 10.1001/jama.2013.283815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bulawa C. E., Connelly S., Devit M., Wang L., Weigel C., Fleming J. A., Packman J., Powers E. T., Wiseman R. L., Foss T. R., Wilson I. A., Kelly J. W., and Labaudinière R. (2012) Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade. Proc. Natl. Acad. Sci. U. S. A. 109, 9629–9634 10.1073/pnas.1121005109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Penchala S. C., Connelly S., Wang Y., Park M. S., Zhao L., Baranczak A., Rappley I., Vogel H., Liedtke M., Witteles R. M., Powers E. T., Reixach N., Chan W. K., Wilson I. A., Kelly J. W., et al. (2013) AG10 inhibits amyloidogenesis and cellular toxicity of the familial amyloid cardiomyopathy-associated V122I transthyretin. Proc. Natl. Acad. Sci. U. S. A. 110, 9992–9997 10.1073/pnas.1300761110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sant'Anna R., Gallego P., Robinson L. Z., Pereira-Henriques A., Ferreira N., Pinheiro F., Esperante S., Pallares I., Huertas O., Rosario Almeida M., Reixach N., Insa R., Velazquez-Campoy A., Reverter D., Reig N., et al. (2016) Repositioning tolcapone as a potent inhibitor of transthyretin amyloidogenesis and associated cellular toxicity. Nat. Commun. 7, 10787 10.1038/ncomms10787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ihse E., Rapezzi C., Merlini G., Benson M. D., Ando Y., Suhr O. B., Ikeda S., Lavatelli F., Obici L., Quarta C. C., Leone O., Jono H., Ueda M., Lorenzini M., Liepnieks J., et al. (2013) Amyloid fibrils containing fragmented ATTR may be the standard fibril composition in ATTR amyloidosis. Amyloid 20, 142–150 10.3109/13506129.2013.797890 [DOI] [PubMed] [Google Scholar]
- 9. Mangione P. P., Porcari R., Gillmore J. D., Pucci P., Monti M., Porcari M., Giorgetti S., Marchese L., Raimondi S., Serpell L. C., Chen W., Relini A., Marcoux J., Clatworthy I. R., Taylor G. W., et al. (2014) Proteolytic cleavage of Ser52Pro variant transthyretin triggers its amyloid fibrillogenesis. Proc. Natl. Acad. Sci. U. S. A. 111, 1539–1544 10.1073/pnas.1317488111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marcoux J., Mangione P. P., Porcari R., Degiacomi M. T., Verona G., Taylor G. W., Giorgetti S., Raimondi S., Sanglier-Cianférani S., Benesch J. L., Cecconi C., Naqvi M. M., Gillmore J. D., Hawkins P. N., Stoppini M., et al. (2015) A novel mechano-enzymatic cleavage mechanism underlies transthyretin amyloidogenesis. EMBO Mol. Med. 7, 1337–1349 10.15252/emmm.201505357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mangione P. P., Verona G., Corazza A., Marcoux J., Canetti D., Giorgetti S., Raimondi S., Stoppini M., Esposito M., Relini A., Canale C., Valli M., Marchese L., Faravelli G., Obici L., et al. (2018) Plasminogen activation triggers transthyretin amyloidogenesis in vitro. J. Biol. Chem. 293, 14192–14199 10.1074/jbc.RA118.003990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Giorgetti S., Greco C., Tortora P., and Aprile F. A. (2018) Targeting amyloid aggregation: an overview of strategies and mechanisms. Int. J. Mol. Sci. 19, 2677 10.3390/ijms19092677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Merlini G., and Bellotti V. (2003) Molecular mechanisms of amyloidosis. N. Engl. J. Med. 349, 583–596 10.1056/NEJMra023144 [DOI] [PubMed] [Google Scholar]
- 14. Fitzpatrick A. W., and Saibil H. R. (2019) Cryo-EM of amyloid fibrils and cellular aggregates. Curr. Opin. Struct. Biol. 58, 34–42 10.1016/j.sbi.2019.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Radamaker L., Lin Y. H., Annamalai K., Huhn S., Hegenbart U., Schonland S. O., Fritz G., Schmidt M., and Fandrich M. (2019) Cryo-EM structure of a light chain-derived amyloid fibril from a patient with systemic AL amyloidosis. Nat. Commun. 10, 1103 10.1038/s41467-019-09032-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pras M., Schubert M., Zucker-Franklin D., Rimon A., and Franklin E. C. (1968) The characterization of soluble amyloid prepared in water. J. Clin. Invest. 47, 924–933 10.1172/JCI105784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang M. Z., and Fitzgerald M. C. (2001) A solid sample preparation method that reduces signal suppression effects in the MALDI analysis of peptides. Anal. Chem. 73, 625–631 10.1021/ac0009090 [DOI] [PubMed] [Google Scholar]
- 18. Bergström J., Gustavsson A., Hellman U., Sletten K., Murphy C. L., Weiss D. T., Solomon A., Olofsson B. O., and Westermark P. (2005) Amyloid deposits in transthyretin-derived amyloidosis: cleaved transthyretin is associated with distinct amyloid morphology. J. Pathol. 206, 224–232 10.1002/path.1759 [DOI] [PubMed] [Google Scholar]
- 19. Hammarström P., Schneider F., and Kelly J. W. (2001) Trans-suppression of misfolding in an amyloid disease. Science 293, 2459–2462 10.1126/science.1062245 [DOI] [PubMed] [Google Scholar]
- 20. Santoro M. M., and Bolen D. W. (1988) Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl α-chymotrypsin using different denaturants. Biochemistry 27, 8063–8068 10.1021/bi00421a014 [DOI] [PubMed] [Google Scholar]
- 21. Narimoto T., Sakurai K., Okamoto A., Chatani E., Hoshino M., Hasegawa K., Naiki H., and Goto Y. (2004) Conformational stability of amyloid fibrils of β2-microglobulin probed by guanidine-hydrochloride-induced unfolding. FEBS Lett. 576, 313–319 10.1016/j.febslet.2004.09.024 [DOI] [PubMed] [Google Scholar]
- 22. Baldwin A. J., Knowles T. P., Tartaglia G. G., Fitzpatrick A. W., Devlin G. L., Shammas S. L., Waudby C. A., Mossuto M. F., Meehan S., Gras S. L., Christodoulou J., Anthony-Cahill S. J., Barker P. D., Vendruscolo M., and Dobson C. M. (2011) Metastability of native proteins and the phenomenon of amyloid formation. J. Am. Chem. Soc. 133, 14160–14163 10.1021/ja2017703 [DOI] [PubMed] [Google Scholar]
- 23. Vettore N., and Buell A. K. (2019) Thermodynamics of amyloid fibril formation from chemical depolymerization. Phys. Chem. Chem. Phys. 21, 26184–26194 10.1039/c9cp04524d [DOI] [PubMed] [Google Scholar]
- 24. Porcari R., Proukakis C., Waudby C. A., Bolognesi B., Mangione P. P., Paton J. F., Mullin S., Cabrita L. D., Penco A., Relini A., Verona G., Vendruscolo M., Stoppini M., Tartaglia G. G., Camilloni C., et al. (2015) The H50Q mutation induces a tenfold decrease in the solubility of α-synuclein. J. Biol. Chem. 290, 2395–2404 10.1074/jbc.M114.610527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Natalello A., Mangione P. P., Giorgetti S., Porcari R., Marchese L., Zorzoli I., Relini A., Ami D., Faravelli G., Valli M., Stoppini M., Doglia S. M., Bellotti V., and Raimondi S. (2016) Co-fibrillogenesis of wild-type and D76N β2-microglobulin: the crucial role of fibrillar seeds. J. Biol. Chem. 291, 9678–9689 10.1074/jbc.M116.720573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hartl F. U., and Hayer-Hartl M. (2009) Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 16, 574–581 10.1038/nsmb.1591 [DOI] [PubMed] [Google Scholar]
- 27. Jaroniec C. P. (2019) Two decades of progress in structural and dynamic studies of amyloids by solid-state NMR. J. Magn. Reson. 306, 42–47 10.1016/j.jmr.2019.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fitzpatrick A. W. P., Falcon B., He S., Murzin A. G., Murshudov G., Garringer H. J., Crowther R. A., Ghetti B., Goedert M., and Scheres S. H. W. (2017) Cryo-EM structures of tau filaments from Alzheimer's disease. Nature 547, 185–190 10.1038/nature23002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schmidt M., Wiese S., Adak V., Engler J., Agarwal S., Fritz G., Westermark P., Zacharias M., and Fandrich M. (2019) Cryo-EM structure of a transthyretin-derived amyloid fibril from a patient with hereditary ATTR amyloidosis. Nat. Commun. 10, 5008 10.1038/s41467-019-13038-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Swuec P., Lavatelli F., Tasaki M., Paissoni C., Rognoni P., Maritan M., Brambilla F., Milani P., Mauri P., Camilloni C., Palladini G., Merlini G., Ricagno S., and Bolognesi M. (2019) Cryo-EM structure of cardiac amyloid fibrils from an immunoglobulin light chain AL amyloidosis patient. Nat. Commun. 10, 1269 10.1038/s41467-019-09133-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Puchtler H., Waldrop F. S., and Meloan S. N. (1985) A review of light, polarization and fluorescence microscopic methods for amyloid. Appl. Pathol. 3, 5–17 [PubMed] [Google Scholar]
- 32. Cohen S. L., and Chait B. T. (1997) Mass spectrometry of whole proteins eluted from sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels. Anal. Biochem. 247, 257–267 10.1006/abio.1997.2072 [DOI] [PubMed] [Google Scholar]
- 33. Oosawa F., and Kasai M. (1962) A theory of linear and helical aggregations of macromolecules. J. Mol. Biol. 4, 10–21 10.1016/S0022-2836(62)80112-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article and associated supporting information. MS data sets and additional information are available upon request from Sofia Giorgetti(s.giorgetti@unipv.it).