Summary
Local translation is a conserved mechanism conferring cells the ability to quickly respond to local stimuli. In the brain, it has been recently reported in astrocytes, whose fine processes contact blood vessels and synapses. Yet the specificity and regulation of astrocyte local translation remain unknown. We study hippocampal perisynaptic astrocytic processes (PAPs) and show that they contain the machinery for translation. Using a refined immunoprecipitation technique, we characterize the entire pool of ribosome-bound mRNAs in PAPs and compare it with the one expressed in the whole astrocyte. We find that a specific pool of mRNAs is highly polarized at the synaptic interface. These transcripts encode an unexpected molecular repertoire, composed of proteins involved in iron homeostasis, translation, cell cycle, and cytoskeleton. Remarkably, we observe alterations in global RNA distribution and ribosome-bound status of some PAP-enriched transcripts after fear conditioning, indicating the role of astrocytic local translation in memory and learning.
Keywords: astrocyte, perisynaptic astrocyte processes, local translation, neuroglial interactions
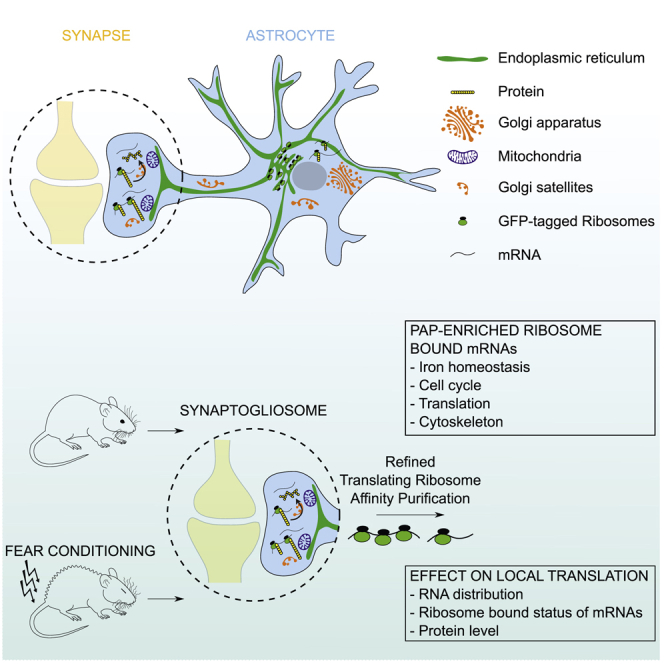
Graphical Abstract
Highlights
-
•
Local translation occurs in perisynaptic astrocytic processes (PAPs)
-
•
The repertoire of ribosome-bound mRNAs enriched in hippocampal PAPs is specific
-
•
RNA distribution and local translation change in PAPs after fear conditioning
Local translation is an evolutionary conserved mechanism that allows quick protein delivery in polarized cellular compartments. Here, Mazaré et al. characterize the entire repertoire of ribosome-bound mRNAs in astroglial perisynaptic processes of the dorsal hippocampus and its changes after contextual fear conditioning.
Introduction
Astrocytes constitute the most abundant population of glial cells in the mammalian brain. They are morphologically complex cells, with many ramifications extending toward both blood vessels and neurons (Verkhratsky and Toescu, 2006). Specific domains called endfeet contact the blood vessels and thus enable astrocytes to modulate important vascular functions, such as blood-brain barrier integrity, immunity (Alvarez et al., 2013) and cerebral blood flow (Iadecola, 2017). The perisynaptic astrocytic processes (PAPs) interact with both synapses and dendrites and regulate synaptic transmission (Ghézali et al., 2016). Furthermore, PAPs can sense changes in the composition of the perisynaptic extracellular space produced by active neurotransmission (Dallérac et al., 2018). The PAPs prevent prolonged neuronal activation and excitotoxicity by clearing ions and neurotransmitters released by the synapse. Astroglial processes are indeed equipped with transporters and channels, such as glutamate transporters and inward rectifying potassium channels, which tightly control the levels of perisynaptic glutamate (Yang et al., 2009) and potassium (Sibille et al., 2014), respectively. PAPs also release neuroactive factors such as ATP and D-serine through various pathways, including connexin hemichannels (Chever et al., 2014) and vesicles (Sultan et al., 2015). Astrocytes also influence synaptic function by dynamically modulating their synaptic coverage (Bernardinelli et al., 2014; Pannasch et al., 2011). Last, these cells eliminate weak synapses and actively refine neuronal circuits during development and orchestrate synaptogenesis in the mature brain (Allen and Eroglu, 2017; Chung et al., 2015; Stogsdill and Eroglu, 2017). Understanding how astrocytes control this wide variety of synaptic functions is crucial because aberrant communication between neurons and astrocytes is known to contribute to several brain diseases. Characterization of the underlying molecular mechanisms may not only reveal the brain’s fundamental workings but also facilitate the development of new therapeutic tools.
Specialized cell polarity is a hallmark of astrocytes, but the underlying mechanisms are still unknown. The evolutionarily conserved cellular strategies involved in functional polarization notably include compartmentalization of mRNAs in distal regions of the cytoplasm and local translation for spatiotemporal targeting of protein delivery (Besse and Ephrussi, 2008). In the brain, both processes have been described in oligodendrocytes (Wake et al., 2011) and have been extensively studied in neurons, the cytoplasmic processes of which may be more than 1,000 times longer than the cell body (Van Driesche and Martin, 2018; Holt et al., 2019). It has been shown that local translation provides synapses with a rapid access to new proteins and has a crucial role in synaptic function and plasticity (Glock et al., 2017; Sutton and Schuman, 2006). Importantly, the impairment of local translation in neurons has been implicated in several neuropathological diseases, such as fragile-X syndrome (Kao et al., 2010), amyotrophic lateral sclerosis (ALS) (Alami et al., 2014; Fallini et al., 2012), and spinal muscular atrophy (Jablonka et al., 2001; Zhang et al., 2003).
We recently showed that local translation occurs in perivascular astrocyte processes (PvAPs) and characterized the locally translated molecular repertoire (Boulay et al., 2017). Local translation was also highlighted in radial glia during brain development (Pilaz et al., 2016). A third study focused on astrocyte-enriched or specific mRNAs translated in PAPs of the adult cortex (Sakers et al., 2017). It has been suggested that impaired local translation in astrocytes is involved in ALS (Barton et al., 2019).
In the present study, we characterized local translation in PAPs in the dorsal hippocampus, a region of the brain involved in memory and learning. We first checked for the presence in PAPs of mRNAs, ribosomes, local translation, the endoplasmic reticulum (ER)-Golgi intermediate compartment (ERGIC), and elements of the Golgi apparatus that might be crucial for local translation and protein maturation. We then used our recently refined translating ribosome affinity purification (TRAP) protocol (Boulay et al., 2019) to characterize for the first time the global ribosome-bound mRNA repertoire in PAPs and, notably, the mRNAs present at higher levels in the processes than in the astrocyte as a whole. Last, we probed the physiological relevance of local translation in PAPs by characterizing changes in levels of memory-related, PAP-enriched, ribosome-bound mRNAs after contextual memory acquisition.
Results
Hippocampal PAPs Contain Protein Synthesis and Maturation Organelles
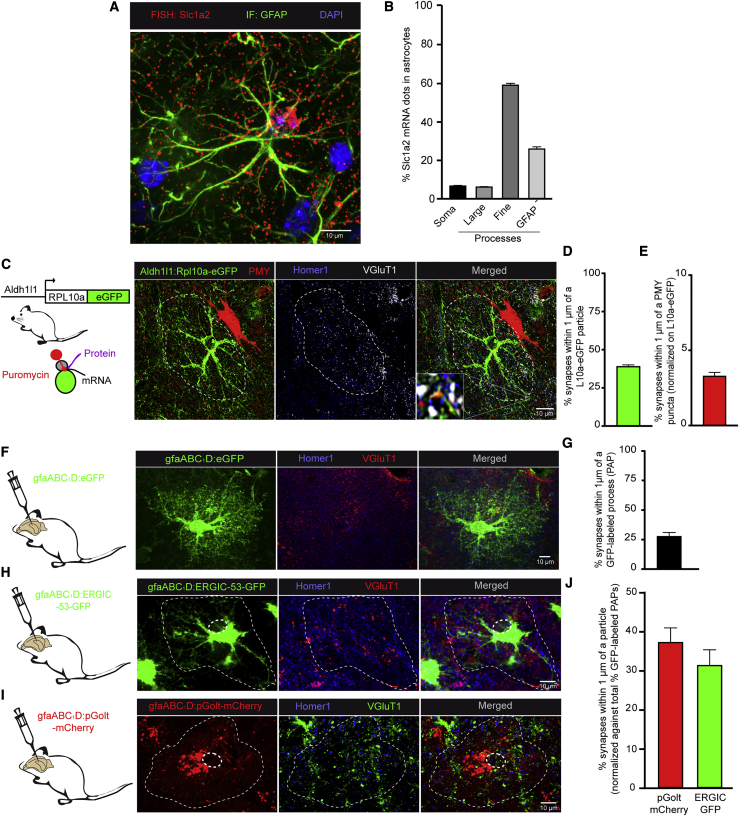
To test for the existence of local translation in hippocampal PAPs, we first characterized the mRNA distribution in astrocytes. We performed a fluorescence in situ hybridization (FISH) experiment on adult mouse CA1 hippocampal sections in order to detect Slc1a2 mRNA, a transcript coding for the astrocyte-specific glutamate transporter GLT1. The samples were co-immunostained for the astrocyte-specific intermediate filament glial fibrillary acidic protein (GFAP) (Oudart et al., 2020). As shown in Figure 1A, Slc1a2 mRNA FISH dots appeared to be distributed throughout the astrocyte. We used our recently developed AstroDot ImageJ plugin to characterize the mRNA distribution by counting the number of FISH dots localized on GFAP-immunolabeled intermediate filament in the somata and the large and fine processes (Oudart et al., 2020). As Slc1a2 is specifically expressed by astrocytes in the CA1 region, we approximated that mRNA FISH dots localized outside GFAP-immunolabeled processes could be attributed to PAPs, where GFAP is known to be less present (Bushong et al., 2002). The estimated mean ± standard error (SEM) proportion of Slc1a2 mRNAs was 7.2% ± 0.4% in the astrocyte somata, 6.7% ± 1.3% in GFAP-positive large processes, 59.6% ± 1.0% in GFAP-positive fine processes, and 26.5% ± 1.1% in GFAP-negative areas (Figure 1B). Overall, these results indicate that Slc1a2 mRNAs are preferentially distributed in distal areas of astrocytes that probably include PAPs. We next sought to quantify the number of synapses contacted by PAPs containing ribosomes on hippocampal sections from Aldh1l1:L10a-eGFP transgenic mice, which express the eGFP-tagged ribosomal protein Rpl10a specifically in astrocytes (Heiman et al., 2008; Figures 1C and 1D). To visualize synapses, the presynaptic VGluT1 protein and the postsynaptic Homer1 protein were co-immunolabeled. Astrocyte ribosomes were present in somata and distal processes. Among VGluT1/Homer1-positive synapses, 39.7% ± 1.2% were contacted by PAPs containing eGFP-labeled ribosomes (Figure 1D). In parallel, we detected local translation events by ribopuromycylation of nascent chains in hippocampal CA1 PAPs (Bastide et al., 2018; Figure 1C). Puromycin (PMY) is an aminoglycoside antibiotic that mimics charged tRNATyr and is incorporated into the ribosome A site. It induces premature translation termination by ribosome-catalyzed covalent incorporation into the nascent carboxy-terminal chain (Pestka and Brot, 1971). Freshly prepared hippocampal sections from Aldh1l1:L10a-eGFP mice were incubated in the presence or absence (negative control, not shown) of PMY. The PMY signal was detected by immunofluorescence (Figure 1C), and co-localization with eGFP-labeled astrocytic ribosomes was assessed. We then quantified the percentage of synapses within 1 μm of a PMY/L10a-eGFP signal. Results were normalized to the total number of VGluT1/Homer1-positive synapses contacted by L10a-eGFP-positive PAPs (Figure 1D). VGlut1/Homer1-positive synapses within 1 μm of a PMY/L10a-eGFP signal represented 3.3% ± 0.3% of the synapses contacted by RPL10a-eGFP-labeled astrocytic ribosomes (Figure 1E), thus indicating the presence of local translation in PAPs.
Figure 1.
The Perisynaptic Processes of Hippocampal Astrocytes Synthesize Proteins and Contain Protein Maturation Organelles
(A) Representative confocal microscopy image of the FISH detection of Slc1a2 mRNAs (red dots) in a dorsal hippocampal CA1 astrocyte immunolabeled for GFAP (green). Nuclei were stained with DAPI (blue).
(B) AstroDot analysis of the distribution of Slc1a2 mRNA in somata, large (>0.3 μm) and fine (<0.3 μm) GFAP-immunolabeled processes, and GFAP-negative processes of CA1 astrocytes (n = 55 cells in three mice).
(C) Detection of CA1 astrocyte ribosomes and local translation events in PAPs by puromycylation (+PMY, with puromycin) in Aldh1l1:L10a-eGFP transgenic mice. Astrocyte ribosomes were immunolabeled for GFP (green) and PMY (red). Synapses were co-immunolabeled for VGluT1 (pre-synapse) (gray) and Homer1 (post-synapse) (blue). The white square is a magnified view (10×) of an astrocyte translation event near synapses. The astrocyte domain is indicated by a dotted line.
(D) Quantification of the percentage of Homer1/VGluT1-positive synapses within 1 μm of a L10A-eGFP signal. n = 128 cells in three mice.
(E) Quantification of the percentage of VGluT1/Homer1-positive synapses within 1 μm of a L10A-eGFP/PMY-positive signal. n = 128 cells in three mice. PMY labeling is intense in neurons (red cell on the right of the astrocyte) compared with astrocytes.
(F) gfaABC1D-eGFP-AAV-infected hippocampal astrocytes immunolabeled with Homer1 (blue) and VGluT1 (red).
(G) Quantification of the percentage of VGluT1/Homer1-positive synapses within 1 μm of eGFP staining, representing the total number of synapses contacted by PAPs detected by this technique. n = 20 cells in three mice.
(H) Visualization of ERGIC in CA1 astrocytes infected with a gfaABC1D-ERGIC-GFP-expressing AAV (green). Synapses were immunolabeled for VGluT1 (red) and Homer1 (blue).
(I) Visualization of Golgi particles in CA1 astrocytes infected with a gfaABC1D-pGolt-Cherry-expressing AAV (red). Synapses were immunolabeled for VGluT1 (green) and Homer1 (blue). In (H) and (I), the astrocyte domain and its nucleus are indicated by a dotted line.
(J) Quantification of the percentage of synapses within 1 μm of astrocytic ERGIC or pGolt particles in (H) and (I). n = 30 cells from three mice for ERGIC and n = 38 cells from three mice for pGolt. The values were normalized against the total number of VGluT1/Homer1-positive synapses within 1 μm of a cytosolic eGFP-labeled PAP in mice infected with a gfaABC1D-eGFP expressing AAV in control experiments (G).
Scale bars: 10 µm; error bars: mean ± SEM.
PAPs are extremely thin structures (<50 nm) (Reichenbach et al., 2010), the subcellular organization of which has not been substantially addressed. Nevertheless, were a membrane protein such as GLT1 to be translated inside a PAP, it would have to pass through the ER and Golgi to be properly folded and functional at the plasma membrane. Here, we addressed the presence of these organelles in PAPs. We used an adeno-associated virus (AAV) bearing the gfaABC1D synthetic promoter (derived from Gfap; Lee et al., 2008) to drive the expression of the Golgi tracker pGolt (Mikhaylova et al., 2016) and ERGIC-53 (an integral membrane protein localized in the ERGIC) (Ben-Tekaya et al., 2005). These proteins were tagged with mCherry and GFP, respectively. The AAVs were injected separately into the CA1 region of the dorsal hippocampus of adult mice (Figures 1H and 1I). VGluT1 and Homer1 proteins were co-immunolabeled to visualize the synapses. As a control, adult mice were injected with an AAV driving the expression of eGFP alone in astrocytes (gfaABC1D-eGFP) (Figure 1F), and we determined that 28.6% ± 3.2% of VGlut1/Homer1-positive synapses were contacted by eGFP-labeled PAPs within an astrocyte’s territory (Figure 1G). Co-immunofluorescent detection of GFAP with gfaABC1D-eGFP demonstrated the astrocytic specificity of infection and transgene expression (Figure S1). We then quantified the percentage of VGluT1/Homer1-positive synapses within 1 μm of ERGIC-53-GFP and pGolt-mCherry-positive PAPs over the total number of VGluT1/Homer1-positive synapses within the astrocyte’s territory. ERGIC-53-GFP was detected homogeneously throughout astrocytes, suggesting the presence of a continuous ERGIC network, and 32.0% ± 4.0% of VGluT1/Homer1-positive synapses within the territory of an astrocyte were contacted by PAPs containing ERGIC particles (Figures 1H and 1J). In contrast, pGolt-mCherry fluorescence was discontinuous, and 38.0% ± 4.0% of VGluT1/Homer1-positive synapses within the territory of an astrocyte were contacted by PAPs containing pGolt particles (Figures 1I and 1J). Thus, it appeared that some PAPs contain ERGIC and elements of the Golgi apparatus. Taken as a whole, these results indicate that local translation and possibly post-translational modifications occur in hippocampal PAPs.
Identification of the PAPome, the Pool of Ribosome-Bound mRNAs in PAPs of the Dorsal Hippocampus
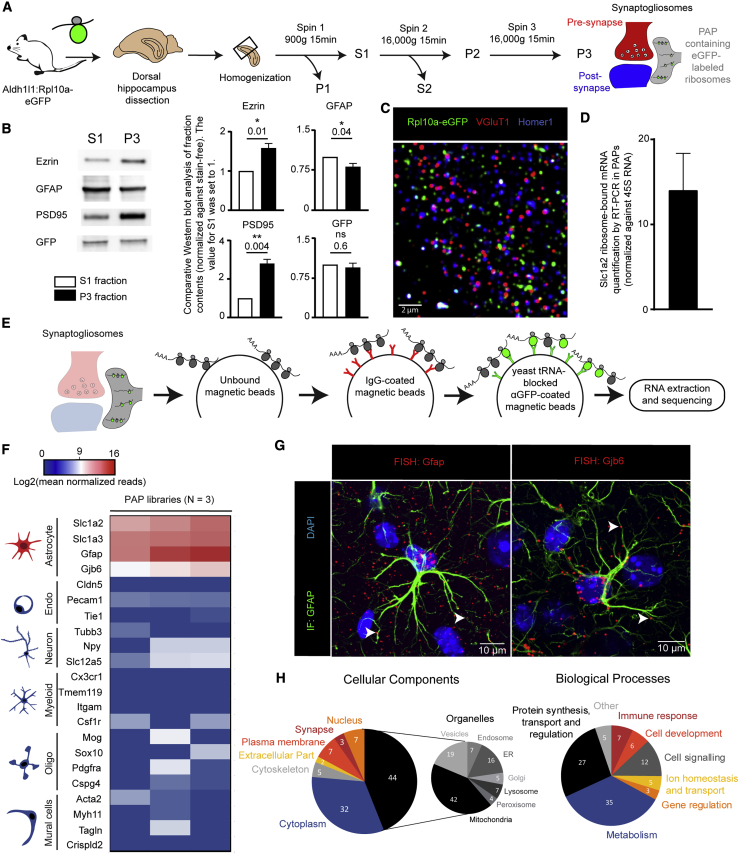
To further study local translation in PAPs, we next characterized the pool of ribosome-bound mRNAs inside these processes. It has been shown that synaptogliosome preparations, consisting of apposed pre- and postsynaptic membranes, contain PAPs that remain attached to synaptic neuronal membranes (Carney et al., 2014). Here, we purified synaptogliosomes from the dorsal hippocampus of Aldh1l1:L10a-eGFP mice and characterized the various fractions of our preparation by western blots (Figures 2A and 2B). Compared with the first supernatant obtained from hippocampal homogenate (S1), the synaptogliosome fraction (P3) contained low levels of GFAP and high levels of the postsynaptic protein PSD95 and the cytosolic PAP protein Ezrin (Derouiche and Frotscher, 2001; Lavialle et al., 2011). Rpl10a-eGFP was also detected in this fraction (Figure 2B). These results indicated that the P3 fraction comprised ribosome-containing PAPs. We also checked for the presence of astrocyte ribosomes in the P3 fraction via the immunofluorescent detection of eGFP, VGluT1, and Homer1 (Figure 2C). As we previously observed Slc1a2 mRNA in fine astrocyte processes (Figure 1A), we used TRAP and qRT-PCR (see below) to test for the presence of Slc1a2 ribosome-bound mRNAs in PAPs in the P3 fraction (Figure 2D). Taken as a whole, these results indicate that PAPs co-purify with hippocampal synapses and contain ribosome-bound mRNAs.
Figure 2.
Identification of the PAPome, the Pool of Ribosome-Bound mRNAs in PAPs from the Dorsal Hippocampus
(A) Flowchart for the preparation of synaptogliosomes from the dorsal hippocampus of Aldh1l1:L10a-eGFP transgenic mice. S, supernatant; P, pellet.
(B) Western blot comparison of S1 and P3 fractions. ∗p ≤ 0.05, ∗∗p ≤ 0.01, and ns (not significant) in a two-tailed paired t test. The data are presented as mean ± SEM (n = 4).
(C) A confocal immunofluorescence microscopy image of synaptogliosomes from an Aldh1l1:L10a-eGFP mouse. The ribosomes in PAPs were immunolabeled with eGFP (green). Pre- and postsynaptic areas were immunolabeled for VGluT1 (red) and Homer1 (blue), respectively. Scale bar: 2 µm.
(D) RT-PCR detection of Slc1a2 ribosome-bound mRNAs in synaptogliosomes. The levels were normalized against 45S RNA (n = 4). The data are presented as mean ± SEM.
(E) Flowchart for TRAP and analysis of PAP ribosome-bound mRNAs extracted from dorsal hippocampal synaptogliosomes. The raw sequencing data are given in Table S1.
(F) Purity heatmap of the RNA-seq data for a selection of mRNAs specific for each type of brain cell. The centered value represents mRNAs with log2(500 normalized reads), which corresponds to our threshold for the presence of an mRNA. Each column represents an independent cDNA library (n = 3 libraries).
(G) Confocal microscopy images (FISH detection) of Gfap and Gjb6 mRNAs (red dots) in dorsal hippocampal astrocytes immunolabeled for GFAP (green). Nuclei were stained with DAPI. White arrowheads indicate FISH dots in fine astrocytic processes. Scale bars: 10 µm.
(H) Gene Ontology analysis of the PAPome.
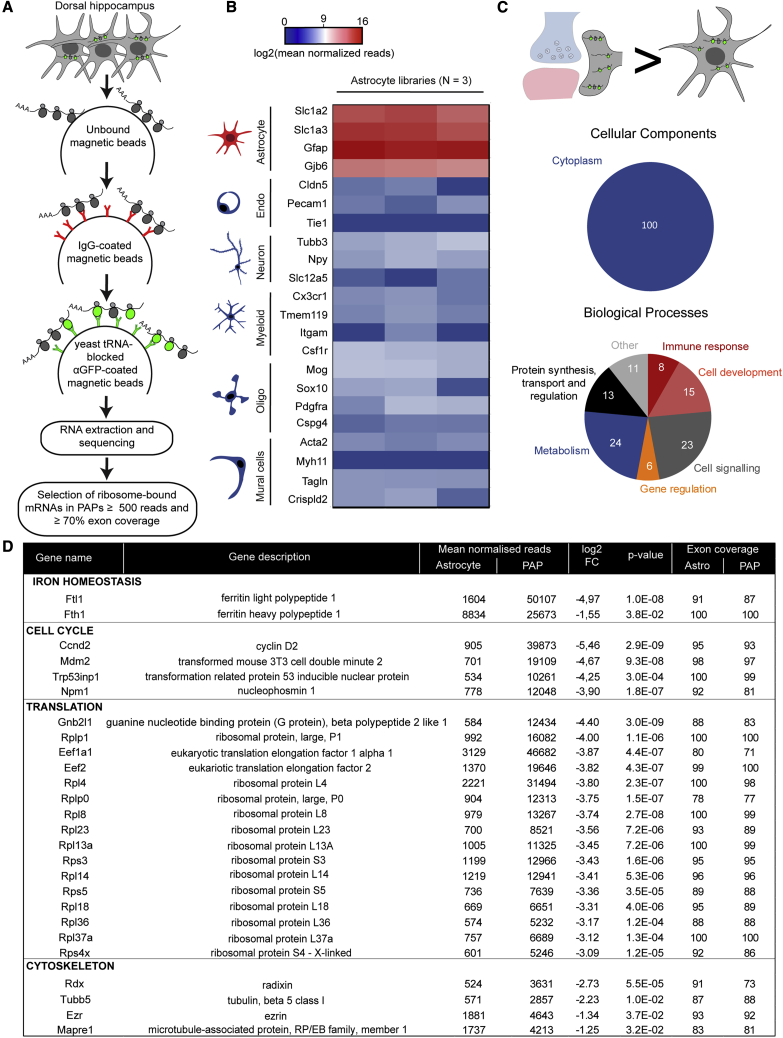
We next aimed to purify and characterize ribosome-bound mRNAs present in PAPs by TRAP (Heiman et al., 2008). To preclude the copurification of mRNAs from brain cells other than astrocytes (which would result from non-specific affinity of mRNAs to the TRAP column), we added pre-cleaning and blocking steps to the TRAP protocol (Boulay et al., 2019; Figure 2E). Indeed, the previous attempt to characterize PAP ribosome-bound mRNAs used indirect subtractive approaches to remove contaminating neuronal mRNAs, which suggested that only astrocyte-enriched or astrocyte-specific transcripts were considered (Sakers et al., 2017). We then immunoprecipitated GFP-tagged ribosomes from PAPs sampled from the dorsal hippocampus of adult Aldh1l1:L10a-eGFP mice. After reverse transcription, cDNA libraries were sequenced, and the reads were aligned against the Mus musculus genome, in order to identify the corresponding genes. We first checked that our libraries were enriched in astrocyte-specific genes and depleted in genes specific to neurons, myeloid cells, oligodendrocytes, mural cells (vascular smooth muscle cells and pericytes), and endothelial cells (Clarke et al., 2018; Figure 2F). To further assess the purity of our libraries with regard to neuronal transcripts, we extended our analysis of neuron-specific mRNAs (Zhang et al., 2014; Figure S2). In particular, we investigated mRNAs such as Grin1, Slc12a5, Cacna1a, and Bsn, which have previously been detected in the synapses or in the neuropil of the hippocampus (Hafner et al., 2019; Cajigas et al., 2012). All showed a very small number of reads and exon coverage in our PAP-TRAP libraries (Figure S2A). Next, we compared their levels using qPCR in the PAP-TRAP and the PAP unbound fraction (Figures S2B–S2D). Considering the different nature of these extracts, two different markers, Gfap and Gapdh, were used for normalization. In both cases, compared with the unbound fraction, all neuronal mRNAs were barely present in the PAP-TRAP fraction, thus confirming the absence of neuronal contamination in our TRAP experiments (Figures S2B–S2D). The raw RNA sequencing list was then refined by removing transcripts with less than 70% exon coverage and fewer than 500 normalized reads; this selected the most highly represented, complete mRNAs corresponding to potential translational events (for an exhaustive list, see Table S1). A total of 844 mRNAs were detected in PAPs. This set included most of the known astrocyte-specific and astrocyte-enriched mRNAs (e.g., Slc1a2, Slc1a3, Mlc1, Hepacam, Gfap, Ezr, Gjb6, Gja1, Gpr37l1) (Zhang et al., 2014; Table S1). These results were validated by FISH experiments evidencing the distal distribution of some of these mRNAs (Gfap and Gjb6) (Figure 2G). Interestingly, a number of astrocyte-specific mRNAs (Sox9, Aldh1l1, Slc25a18, and Ptprz1) were not found, suggesting that mRNAs are selectively distributed in PAPs (Table S1). A Gene Ontology (GO) analysis indicated that most of the dorsal hippocampal PAP mRNAs encode cytosolic (32%) and intracellular organelle (44%) proteins. Large proportions are involved in protein synthesis (27%), metabolism (35%), and cell signaling (12%) (Figure 2H). Consistently, some of the most abundant mRNAs in PAPs coded for ribosomal proteins (such as Rpl4 and Rplp1) or proteins involved in mRNA stability and translation (such as the elongation factors eEF1A1 and eEF2 and the Poly[A] binding protein PABPC1) (Table S1). This indicates that the molecular machinery for translation itself might be translated in PAPs. Last, mRNAs encoding the two ferritin isoforms Ftl1 and Fth1 were also highly represented, suggesting that local translation has a role in local iron storage and reduction (Dringen et al., 2007).
After characterizing the pool of ribosome-bound mRNAs present in dorsal hippocampal PAPs, we decided to refer to it as the “PAPome,” by analogy to our recent description of the “endfeetome,” the ribosome-bound transcriptome found in perivascular astrocyte endfeet (Boulay et al., 2017).
Ribosome-Bound mRNAs Are Differentially Distributed in Hippocampal PAPs
After having characterized the PAPome, we determined which of its mRNA components were enriched in the PAP relative to the astrocyte as a whole. After homogenizing dorsal hippocampus tissue from Aldh1l1:L10a-eGFP mice, we used TRAP to purify all the ribosome-bound mRNAs in astrocytes (Figure 3A). As described above, three independent cDNA libraries were synthesized and sequenced. Astrocyte-specific mRNAs were highly enriched, whereas other brain cell-specific RNAs were depleted (Clarke et al., 2018; Figure 3B). In particular, these libraries were devoid of neuronal contamination, as previously observed in the PAP (Figure S2A). Application of the criteria used in the previous experiment (≥500 reads and ≥70% exon coverage) to the raw list gave a refined list of 2,545 mRNAs, including those coding for most of the known astrocyte-specific or astrocyte-enriched proteins, such as Sox9, Aldh1l1, Slc25a18, and Ptprz1, which were not present in the PAPome (Table S1). The distribution of ribosome-bound mRNAs within the astrocyte was then analyzed by comparing the PAPome with the whole-astrocyte repertoire (Table S1). We found that 557 mRNAs were significantly enriched in whole astrocytes (p ≤ 0.05, log2 fold change ≥ 1), and 82 were enriched in PAPs (p ≤ 0.05, log2 fold change ≤ −1). The astrocyte-specific transcripts Gfap, Slc1a2, Slc1a3, Mlc1, Hepacam, Gjb6, Gja1, and Gpr37ll1 were all equally detected in PAPs and whole astrocytes, indicating that they are probably homogeneously translated in hippocampal astrocytes. The pool of PAP-enriched mRNAs had an unexpected profile; a GO analysis indicated that most of them encoded cytosolic proteins involved in metabolism (24%), cell signaling (23%), development (15%), or protein synthesis (13%) (Figure 3C). Twenty-seven of the transcripts coded for ribosomal proteins, such as Rplp1 and Rpl4 (Figure 3D). We also noted the presence of the two ferritin subunit-encoding mRNAs Fth1 and Ftl1, as well as mRNAs related to cell cycle mechanisms (e.g., Ccnd2, Mdm2) (Figure 3D). The PAPs were also rich in mRNAs coding for cytoskeletal proteins, including not only microtubule-related proteins (Tubb5 and Mapre1) but also the Ezrin and Radixin proteins involved in linking the cell membrane to the actin cytoskeleton (Figure 3D). The qPCR results for a selection of the most abundant PAP-enriched mRNAs validated these findings (Table S2). Overall, our data show that the ribosome-bound mRNA distribution has a highly specific molecular polarity in PAPs compared with the rest of the astrocyte. Moreover, PAP-enriched ribosome-bound mRNAs code for proteins involved in specific functions related to protein synthesis, cell cycle regulation, iron homeostasis, and cytoskeleton dynamics.
Figure 3.
Identification of Ribosome-Bound mRNAs Enriched in PAPs from the Dorsal Hippocampus
(A) Flowchart for the TRAP and analysis of ribosome-bound mRNAs in astrocytes from the dorsal hippocampus. Sequencing raw data are in Table S1.
(B) Purity heatmap of the RNA-seq data for a selection of mRNAs specific for each type of brain cell. The centered value represents mRNAs with log2(500 normalized reads), which corresponds to our threshold for the presence of an mRNA. Each column represents an independent cDNA library (n = 3 libraries).
(C) Gene Ontology analysis of mRNAs enriched in PAPs, relative to whole astrocytes (p ≤ 0.05, log2 fold change [FC] ≤ −1).
(D) Raw sequencing data for a selection of the most PAP-enriched mRNAs sorted by log2 fold change (p ≤ 0.05, log2 fold change ≤ −1).
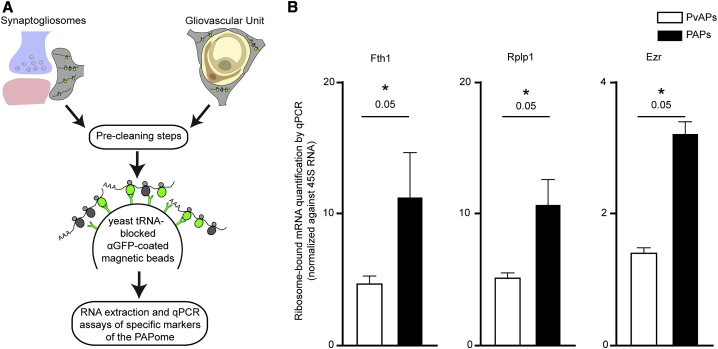
Differences in Ribosome-Bound mRNA Distribution between PAPs and PvAPs
We have previously reported that local translation occurs in PvAPs (Boulay et al., 2017). Although PvAPs contact synapses and therefore also qualify as PAPs (Boulay et al., 2017), most PAPs are far from blood vessels and thus are likely to be dedicated to specific neuroglial interactions. We hypothesized that if local translation sustains astrocyte functional polarity, it would differ between PvAPs and PAPs. To address this question, we used qPCR assays to compare the expression of ribosome-bound mRNAs in hippocampal PAPs and PvAPs. Importantly, we took our PAP-enriched ribosome-bound mRNA list as a reference, as the equivalent repertoire in dorsal hippocampus PvAPs is unknown. Ribosome-bound mRNAs were extracted from adult Aldh1l1:Rpl10a-eGFP whole hippocampus, using TRAP. For PAPs, we isolated synaptogliosomes (as described above). For PvAPs, we purified gliovascular units (Boulay et al., 2019; Figure 4A). Interestingly, Ezr, Fth1, and Rplp1 mRNAs were more enriched in PAPs than in PvAPs, while all other mRNAs were equally detected in PAPs and PvAPs (Figure 4B; Table S3). These results suggest that local translation differs in PAPs and PvAPs and may thus sustain distinct molecular identities at each of these astrocytic interfaces.
Figure 4.
Comparison of Enriched Ribosome-Bound mRNA Levels in PAPs versus PvAPs
(A) Flowchart for the comparison of a selection of enriched mRNAs in PAPs versus PvAPs. Ribosome-bound mRNAs were isolated using TRAP from Aldh1l1:L10a-eGFP whole hippocampal synaptogliosomes or purified gliovascular units. The raw qPCR data are given in Table S3.
(B) qPCR results for Fth1, Rplp1, and Ezr, demonstrating their enrichment in PAPs versus PvAPs. ∗p ≤ 0.05, one-tailed Mann-Whitney test. The data are expressed as mean ± SEM (n = 3).
The PAPome Changes after Fear Conditioning
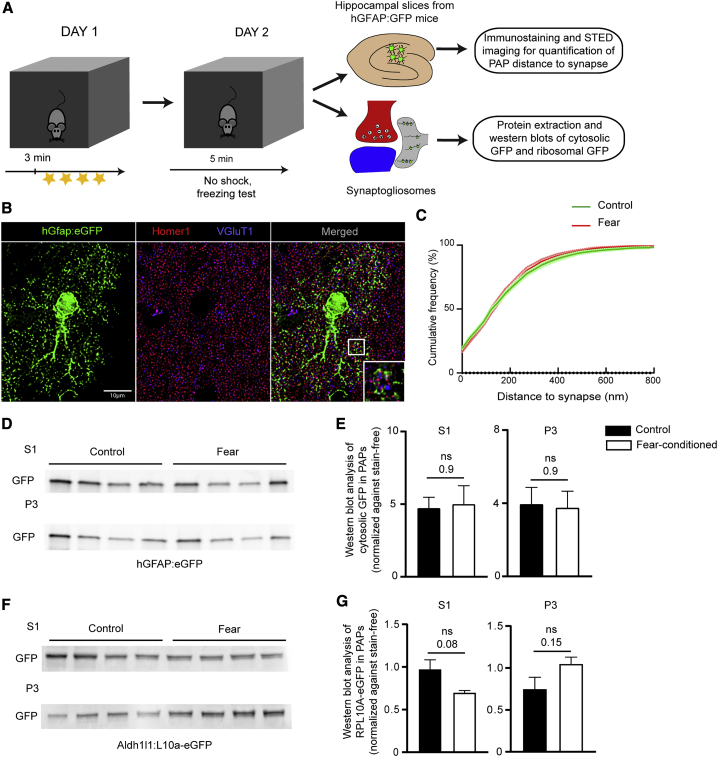
Recent studies have shown that astrocytes directly regulate memory formation through their contribution to local synaptic transmission and plasticity via exchange of regulatory signals with synapses (Adamsky et al., 2018; Martin-Fernandez et al., 2017; Santello et al., 2019). Although mRNA distribution and local translation in neurons have already been explored in the context of synaptic plasticity (Huber et al., 2000; Kang and Schuman, 1996) and memory formation (Alberini, 2008; Jones et al., 2018; Miller et al., 2002), these mechanisms have not yet been evaluated in astrocytes. Strikingly, several of the ribosome-bound mRNAs found to be enriched in PAPs, including Flt1, Fth1 (Dringen et al., 2007; Zheng et al., 2018), Ccnd2 (Ben Abdallah et al., 2013; Gilani et al., 2014; Jedynak et al., 2012), Mdm2 (Li et al., 2016; Liu et al., 2017; Tsai et al., 2012, 2017; Zhu et al., 2015), Gnb2l1 (Sklan et al., 2006; Yaka et al., 2002; Zhu et al., 2016), and Eef1a1 (Blumenstock et al., 2019; Giustetto et al., 2003; Tsokas et al., 2005), have been linked to learning, memory, or cognitive decline. We therefore investigated whether the local translation of these mRNAs in PAPs might be regulated by learning and memory processes. Hence, we exposed mice to a simple fear conditioning task (Figure 5A): on day 1, they were placed in a specific context (a new cage with white noise [5,000 Hz, 60 dB] and an acetic acid [1%] smell). They were left to explore for 3 min before being exposed to four consecutive foot shocks. The mice thus learned to associate the cage with the unpleasant stimulus. On day 2, the mice were placed in the same cage without receiving any foot shocks. Re-exposure of the animal to the same aversive context allowed us to check the efficacy of the conditioning induction, as estimated by the duration of freezing (>70 s). Mice showing no or short freezing time (<70 s) were excluded. Before comparing mRNAs in normal versus conditioned mice, we first checked that the distance between the PAP and the synapse had not been affected by the conditioning protocol (Figures 5B and 5C). Indeed, it has been reported that the consolidation of fear conditioning induces morphological rearrangements in astrocytes from the lateral amygdala, notably with the emergence of synapses lacking PAPs (Ostroff et al., 2014). Here, using super-resolution stimulated emission depletion (STED) microscopy, we first measured individual PAP-synapse distances (ranging from 0 to 800 nm) in the CA1 region of the dorsal hippocampus from hGfap:eGFP transgenic mice (in which eGFP fills the astrocyte cytoplasm) under control and fear conditions (Nolte et al., 2001). STED imaging and co-immunostaining of eGFP with the pre- and postsynaptic markers VGluT1 and Homer1 generated data on the cumulative frequency of the distances between eGFP and VGluT1/Homer1 co-labeled synapses (Figures 5B and 5C). There was no significant difference between control and fear-conditioned mice, indicating that the PAP-to-synapse distance was unchanged 24 h after exposure to contextual fear conditioning. Next, we performed western blot analysis on the S1 (whole dorsal hippocampus) and P3 (synaptogliosome) fractions (Figure 2A) from hGfap:eGFP mice. We did not observe any difference in eGFP content in either fraction, which suggests that the same amount of PAPs was purified from our synaptogliosomes (Figures 5D and 5E). Last, we analyzed Aldh1l1:L10a-eGFP synaptogliosomes and again did not observe any intercondition differences in the ribosomal content of PAPs (Figures 5F and 5G). Overall, these results indicate that PAPs from the dorsal hippocampus did not retract, and retained their cytosolic and ribosomal content 24 h after fear conditioning.
Figure 5.
Characterization of PAPs upon Fear Conditioning
(A) Flowchart for the fear-conditioning protocol, followed by analyses of the PAP-to-synapse distance and PAP proteins in dorsal hippocampus synaptogliosomes.
(B) Deconvoluted confocal microscopy image of a single plane containing an astrocyte and synapses in the dorsal hippocampus of a control hGfap-eGFP mouse. The astrocyte is immunolabeled for eGFP. Pre- and post- synapses are immunolabeled for VGluT1 (blue) and Homer1 (red), respectively. The magnified area shows the STED image for VGluT1 and Homer1 merged with deconvoluted confocal image for eGFP. Scale bar: 10 µm
(C) Cumulative frequency of eGFP as a function of the distance to the synapse (from 0 to 800 nm). Solid lines represent the calculated means, and dotted lines represent the SEM for astrocytes from control mice (green) and fear-conditioned (red) mice. n ≥ 270 synapses per cell, three cells per mouse, and three mice per condition. Two-way ANOVA, interaction value, p = 0.5 (not significant).
(D) Western blot analysis of S1 (whole dorsal hippocampus extracts) and P3 (synaptogliosomes) (Figure 2) from hGfap:eGFP control mice or fear-conditioned mice.
(E) Analysis of the results in (D), indicating that the amount of eGFP in astrocytes and PAPs was stable upon fear conditioning. Signals were normalized against stain-free membranes. The data are presented as mean ± SEM (n = 4 per condition); ns, not significant in an unpaired two-tailed t test.
(F) Western blot analysis of S1 (whole dorsal hippocampus extracts) and P3 (synaptogliosomes) from Aldh1l1:L10a-eGFP control mice and fear-conditioned mice.
(G) Analysis of the results in (F), indicating that the amount of eGFP-tagged ribosomes was stable in astrocytes and PAPs. The signals were normalized against stain-free membrane. The data are presented as mean ± SEM (n = 4 per condition); ns, not significant in an unpaired two-tailed t test.
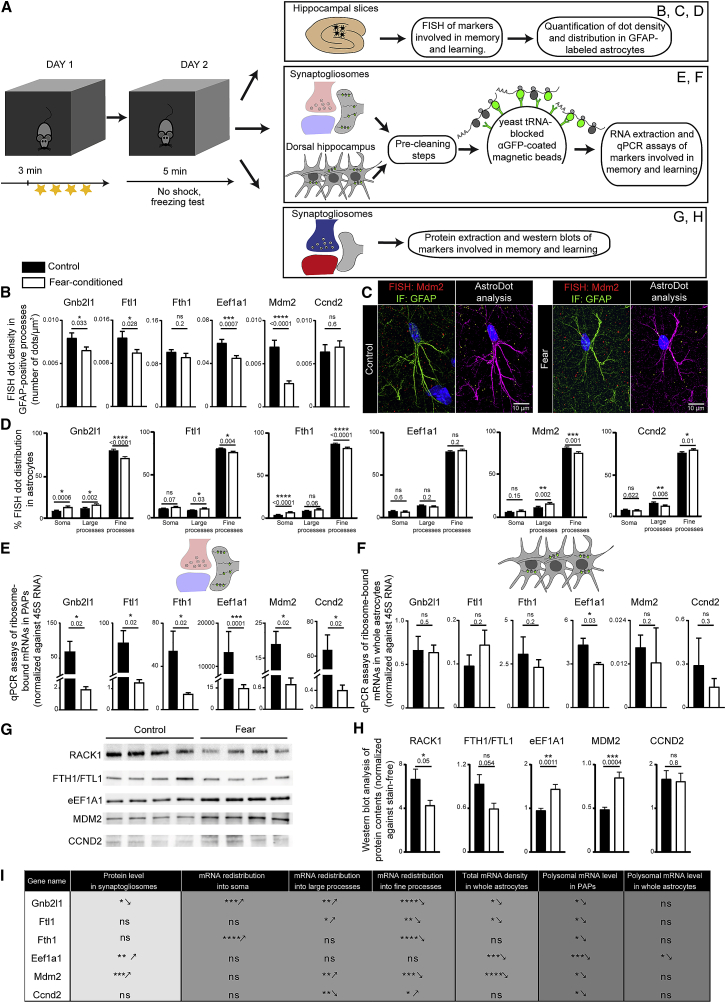
We next assessed the impact of learning on local translation of our selected markers in astrocytes (Figure 6A). First, we looked at whether the density and distribution of these mRNAs were modified by fear learning (Figures 6B and 6C; Table S4). To this end, we used our AstroDot technique (Oudart et al., 2020). In contrast to our previous analysis (Figures 1A and 1B), FISH dots present in GFAP-negative processes were not included in this study. Indeed, analyzed mRNAs are ubiquitous, and we therefore could not differentiate mRNAs in GFAP-negative processes from mRNAs in other brain cells. The FISH procedure includes a RNA-unmasking-protease digestion step, which does not conserve small fluorescent proteins, hence precluding us from using astrocyte reporter mouse strains or viral strategies. Therefore, we restricted our analysis to the distribution of fluorescently labeled mRNAs in astrocytes in GFAP-immunolabeled intermediate filaments in control and conditioned mice (Oudart et al., 2020). With the exception of Fth1 and Ccnd2, the densities of astrocyte mRNAs were lower in the fear-conditioned group (Figures 6B and 6C; Table S4). The distribution differed for almost all the mRNAs, with higher levels in large processes and lower levels in fine processes; the only exceptions were Eef1a1 (no difference in distribution) and Ccnd2 (lower levels in large processes and higher levels in fine processes) (Figure 6D; Table S4). These results suggest that fear conditioning induces changes in the density and/or distribution of these mRNAs in astrocytes.
Figure 6.
Local Translation Changes in PAPs after Fear Conditioning
(A) Flowchart for the fear-conditioning protocol, followed by analyses of mRNAs and proteins in astrocytes and PAPs from the dorsal hippocampus.
(B) AstroDot analysis of the density of PAP-enriched mRNAs encoding RACK1 (Gnb2l1), FTL1, FTH1, eEF1A1 MDM2, and CCND2 in GFAP-immunolabeled astrocytic processes in the dorsal hippocampus of control mice and fear-conditioned mice. ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, and ns (not significant) in an unpaired two-tailed t test; n ≥ 35 cells from three mice per condition. The data are presented as mean ± SEM, and the raw data are given in Table S4.
(C) FISH detection of Mdm2 mRNAs in dorsal hippocampal astrocytes immunolabeled for GFAP (green) in a control mouse and a fear-conditioned mouse. Left panels: confocal microscopy image of the FISH detection of Mdm2 mRNAs (red dots). GFAP-immunolabeled astrocyte processes are in green. Nuclei are stained with DAPI (blue). Right panels: AstroDot analysis of the left-panel image for each condition. The green dots were located in the soma or in GFAP-immunolabeled large processes; the yellow dots were located in GFAP-immunolabeled fine processes. GFAP immunostaining is shown in magenta. Scale bar: 10 µm
(D) AstroDot analysis of the distribution of PAP-enriched mRNAs encoding RACK1 (Gnb2l1), FTL1, FTH1, eEF1A1 MDM2, and CCND2 in somata, large (>0.3 μm) and fine (<0.3 μm) GFAP-immunolabeled astrocyte processes from the dorsal hippocampus of control and fear-conditioned mice. ∗p ≤ 0.05, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001, and ns (not significant) in a two-tailed unpaired t test; n ≥ 35 cells from three mice per condition. Data are presented as mean ± SEM, and the raw data are given in Table S4.
(E and F) qPCR quantification of ribosome-bound mRNAs in dorsal hippocampal PAPs (E) and whole astrocytes (F) from control mice and fear-conditioned mice. Signals are normalized against 45S RNA. ∗p ≤ 0.05, ∗∗∗p ≤ 0.0001, and ns (not significant) in a one-tailed Mann-Whitney test. The data are presented as mean ± SEM (n = 3–5 per condition), and the raw data are given in Table S5.
(G) Western blots of RACK1, FTH1/FTL1, CCND2, MDM2, and eEF1A1 in dorsal hippocampal synaptogliosomes prepared from control mice and fear-conditioned mice.
(H) Analysis of the western blots in (G); signals were normalized against stain-free membranes. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, and ns (not significant) in an unpaired two-tailed paired t test. The data are presented as mean ± SEM (n = 4–5 per condition).
(I) A summary of the results.
We next determined whether the translation of these mRNAs in PAPs was modified upon fear conditioning. We used TRAP to extract ribosome-bound mRNAs from dorsal hippocampus astrocytes and synaptogliosomes sampled from control and fear-conditioned Aldh1l1:L10a-eGFP mice and then used qPCR to compare our panel of markers in PAPs and whole astrocytes (Figures 6E and 6F; Table S5). In whole astrocytes, levels of all markers were similar, with the exception of Eef1a1, which appeared to be slightly downregulated (Figure 6F; Table S5). In marked contrast, the PAP levels of all markers were very low in the fear-conditioned group (Figure 6E; Table S5). These results indicated that fear conditioning has a major impact on the ribosome-bound status of these mRNAs and thus possibly modifies their translation specifically in PAPs. We finally analyzed the overall amount of each protein (using western blots) in fear-conditioned and control synaptogliosomes extracted from the dorsal hippocampus (Figures 6G and 6H). In the fear-conditioned groups compared with controls, levels of RACK1 (encoded by Gnb2l1) and ferritins (although not significantly) were lower, while levels of MDM2 and eEF1A1 were higher and levels of CCND2 remained unchanged. These results suggested that the lower translation of RACK1 and the ferritins in PAPs could be responsible for the lower level of these proteins at the neuroglial interface. In contrast, the higher or unchanged levels of eEF1A1, MDM2, and CCND2 suggested that opposite processes might take place in the synaptic compartment.
In summary, levels of PAP-enriched transcripts may be regulated in different ways (i.e., mRNA transcription, mRNA redistribution, and local changes in ribosome binding) (Figure 6I). Taken as a whole, these results suggest that learning and memory processes could affect local translation in PAPs from the dorsal hippocampus.
Discussion
The role of mRNA distribution and local translation in establishing cell functional polarity is widely established (Besse and Ephrussi, 2008). In the central nervous system, these mechanisms contribute to neuronal development and synaptic transmission (Holt et al., 2019), and their potential involvement in the control of distal synaptic and vascular functions in astrocytes (a highly ramified type of glial cell) has recently been suggested (Boulay et al., 2017; Sakers et al., 2017). In the present study, we focused on the perisynaptic astrocytic interface in the dorsal hippocampus, a region of the brain involved in memory and learning. We identified the pool of ribosome-bound mRNAs in PAPs and showed that it is locally modified after a behavioral task related to learning and memory.
To study local translation in PAPs, we used TRAP to purify ribosome-bound mRNAs in PAPs from synaptogliosomes. A modification of the TRAP protocol developed in our laboratory (the inclusion of cytosolic extract pre-cleaning and column-blocking steps) enabled us to eliminate non-specific mRNA binding events (Boulay et al., 2019). As a result, we were not restrained to the analysis of astrocyte-enriched or astrocyte-specific mRNAs only. Instead, we purified for the first time the entire range of astrocytic ribosome-bound mRNAs, without using their cell specificity or enrichment as a selection criteria (Sakers et al., 2017). We were thus also able to directly compare for the first time whole astrocytes and PAPs to identify PAP-enriched ribosome-bound mRNAs.
The nature of enriched ribosome-bound mRNAs in dorsal hippocampus PAPs suggests a role for local translation in specific biological processes: (1) The structural plasticity of PAPs: Radixin and Ezrin are actin-binding proteins that have already been involved in PAP morphology and motility (Carney et al., 2014; Derouiche and Frotscher, 2001; Lavialle et al., 2011). The cytoskeleton-associated protein category contained β-tubulin (encoded by Tubb5 mRNA, and which was recently described as being essential for neuronal differentiation and dendritic spine formation in vivo; Ngo et al., 2014) and EB1 (encoded by Mapre1, and which binds to the plus end of microtubules and regulates their dynamics; Nehlig et al., 2017). In vitro, microtubules have been implicated in the regulation of intermediate filament polarization in astrocytes (Leduc and Etienne-Manneville, 2017). Although microtubules have not been observed in PAPs in vivo, microtubule dynamics in these structures might also regulate their polarity. (2) Iron homeostasis: Ferritin sequesters iron ions and regulates their toxicity through its ferroxidase activity. The local synthesis of Ferritin in PAPs might therefore allow astrocytes to locally buffer iron and protect the synaptic environment from oxidative stress (Codazzi et al., 2015; Dringen et al., 2007). (3) Translation: a large proportion of PAP-enriched mRNAs are thought to be involved in protein synthesis; these mRNAs code for ribosomal proteins, elongation factors, MDM2 (a ubiquitin E3 ligase that binds to ribosomal proteins [Liu et al., 2014] and acts as a translational repressor [Liu et al., 2017]), and NPM1 (a nucleolar phosphoprotein that regulates ribosome biogenesis [Pfister and D’Mello, 2015]). Similar observations were made in neuronal processes (Deglincerti and Jaffrey, 2012; Giustetto et al., 2003; Moccia et al., 2003; Si et al., 2003). Moreover, it was recently reported that newly translated ribosomal proteins in axons of primary cultured neurons incorporate into axonal ribosomes and sustain local translation (Shigeoka et al., 2019). These convergent results strongly suggest that in astrocytes, as in neurons, local translation is sustained by the local synthesis of translation machinery, with either the de novo assembly of translational complexes or the replacement of damaged proteins. It is also possible that these proteins display non-canonical functions in PAPs, as has been described for eEF1A1 (Abbas et al., 2015; Migliaccio et al., 2016).
In the present study, we did not look for alternative mRNA isoforms. In fact, our Illumina sequencing was based on the reconstruction of 75 bp sequences, which is not well suited to isoform analysis. Nevertheless, by comparing the results in PAPs versus whole astrocytes, we noted that the C-terminal exons of Gfap α and δ (Kamphuis et al., 2012) were differentially distributed: Gfap α was more abundant in PAPs, and GFAP δ was more abundant in the cell soma (Table S1). These findings corroborate our recent observation (based on FISH) that GFAP δ mRNA is more somatic than GFAP α mRNA (Oudart et al., 2020) and suggest that alternative splicing may contribute to the PAPs’ molecular identity.
To establish whether the astrocyte’s functional polarity is mediated by local translation, we compared the levels of PAP-enriched ribosome-bound mRNAs in PAPs and in PvAPs. Interestingly, some mRNAs appeared to be enriched in PAPs compared with PvAPs, indicating that the distribution and local translation of mRNAs differ at these two interfaces. The enrichment of Ezrin mRNAs in PAPs further illustrates the involvement of this specific cytoskeleton-associated protein to control PAPs morphology and motility (Carney et al., 2014; Derouiche and Frotscher, 2001; Lavialle et al., 2011). The enrichment of Rplp1 mRNA suggests that PvAPs and PAPs have different repertoires of ribosomal proteins and thus may follow different translational rules. The enrichment of Fth1 but not Ftl1 ribosome-bound mRNAs in PAPs suggests that iron reduction rather than storage prevails in PAPs (Dringen et al., 2007). Another striking difference between PAPs and PvAPs probably relates to their ultrastructure. Although both processes display a continuous ER network, PvAPs (which are much larger than PAPs) can contain a full Golgi apparatus; in contrast, only pGolt-containing Golgi particles were observed in PAPs. Moreover, most of the mRNAs in the previously identified PvAP-enriched translatome code for secreted proteins and membrane proteins, whereas those in the PAP code mainly for cytoplasmic proteins. These results indicate that post-translational protein modifications might be quite different in the two compartments and might again confer different functions to locally translated proteins.
Some of the mRNAs that are most highly enriched in PAPs code for proteins involved in learning and memory, namely, FTH1, FTL1, eEF1A1, CCND2, MDM2, and RACK1. We therefore investigated the influence of fear conditioning on the translation of the corresponding mRNAs. Importantly, we performed our molecular characterization 24 h post-fear conditioning (immediately after the retrieval session) in order to selectively choose “good learners” with high freezing scores (>70 s). As a consequence, we could not distinguish whether the observed molecular effects resulted from the learning process or memory retrieval. We found that the level of these ribosome-bound mRNAs dropped specifically in PAPs 24 h after fear conditioning. Although the level of ribosome-bound mRNAs does not necessarily reflect the level of translation, the observed drop was very drastic, suggesting a potential effect on local translation. Interestingly, not all studied mRNAs behaved the same way. Some showed a transcriptional effect and, in some cases, a redistribution of mRNAs within astrocytes, suggesting a role for mRNA transport in astrocyte processes. Overall, mechanisms underlying these changes seem to vary greatly from one mRNA to another, suggesting the necessity to develop specific approaches toward individual mRNAs. Finally, the lower level of ribosome-bound mRNAs in PAPs after fear conditioning was not always accompanied by a lower level of protein in synaptogliosomes, which indicates the possible presence of opposing mechanisms in synapses. This was particularly striking for Eef1a1, which displayed higher protein levels and lower levels of both total mRNA and ribosome-bound mRNA in the synaptogliosomes after fear conditioning. Remarkably, Eef1a1 mRNA has been detected in axons, where it might be locally translated to maintain synaptic plasticity (Giustetto et al., 2003) and long-term potentiation (Tsokas et al., 2005). Its decrease at the synaptic level has been related to α-synucleinopathy (Blumenstock et al., 2019). Since long-term potentiation and fear conditioning are intimately linked (Rogan et al., 1997), opposite changes might occur in neurons and astrocytes (i.e., an increase in Eef1a1 levels on the neuronal side and a decrease on the astrocytic side).
Overall, our findings provide unexpected insights into the PAP’s molecular identity, particularly the preferential translation of mRNAs related to iron homeostasis, cytoskeletal dynamics, and the translation machinery in these compartments. They suggest that astrocyte polarity might be sustained by local translation. Our findings also constitute the first evidence of a correlation between fear conditioning and translational changes in PAPs, changes that might be linked to the regulation of synaptic and circuit functions underlying complex behaviors.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-CCND2 (B-6) | Santa Cruz | Cat#sc376676; RRID:AB_11151389 |
| Rabbit monoclonal anti-EEF1A1 | Abcam | Cat#ab157455 |
| Mouse monoclonal anti-Ezrin (3C12) | Abcam | Cat#ab4069; RRID:AB_304261 |
| Rabbit monoclonal anti-FTH1 (D1D4) | Cell Signaling | Cat#4393S; RRID:AB_11217441 |
| Rabbit polyclonal anti-GFAP | Sigma | Cat#G9269; RRID:AB_477035 |
| Chicken polyclonal anti-GFP | Aves | Cat#1020; RRID:AB_10000240 |
| Rabbit polyclonal anti-Homer1 | Synaptic Systems | Cat#160003; RRID:AB_887730 |
| Mouse monoclonal anti-MDM2 (2A10) | Abcam | Cat#ab16895; RRID:AB_2143534 |
| Mouse monoclonal anti-PSD95 | BD Biosciences | Cat#610495; RRID:AB_397861 |
| Mouse monoclonal anti-RACK1 (B-3) | Clinisciences | Cat#17754; RRID:AB_2247471 |
| Mouse monoclonal anti-VGluT1 | Synaptic Systems | Cat#135511; RRID:AB_887879 |
| Mouse monoclonal anti-PMY-647 (12D10) | Sigma | Cat#MABE343-AF647; RRID:AB_2736876 |
| FISH Probes | ||
| Ftl1 | Bio-Techne | NM_010240.2 |
| Gfap | Bio-Techne | NM_001131020.1 |
| Gjb6 | Bio-Techne | NM_001010937.2 |
| Slc1a2 | Bio-Techne | NM_001077514.3 |
| Gnb2l1 | Bio-Techne | NM_008143.3 |
| Mdm2 | Bio-Techne | NM_010786.4 |
| Eef1a1 | Bio-Techne | NM_010106.2 |
| Ccnd2 | Bio-Techne | NM_009829.3 |
| Fth1 | Bio-Techne | NM_010239.2 |
| qPCR probes | ||
| Ccnd2 | Thermofisher | Mm00438070_m1 |
| Ezr | Thermofisher | Mm00447761_m1 |
| Mdm2 | Thermofisher | Mm00487656_m1 |
| Rdx | Thermofisher | Mm00501337_m1 |
| Rps3 | Thermofisher | Mm00656272_m1 |
| Rpl8 | Thermofisher | Mm00657299_g1 |
| Hsp90ab1 | Thermofisher | Mm00833431_g1 |
| Rps14 | Thermofisher | Mm00849906_g1 |
| Fth1 | Thermofisher | Mm00850707_g1 |
| Slc1a2 | Thermofisher | Mm01275814_m1 |
| Gnb2l1 | Thermofisher | Mm01291968_g1 |
| Eef1a1 | Thermofisher | Mm01973893_g1 |
| Npm1 | Thermofisher | Mm02391781_g1 |
| Rplp1 | Thermofisher | Mm02601846_g1 |
| Ftl1 | Thermofisher | Mm03030144_g1 |
| Eef2 | Thermofisher | Mm05700170_g1 |
| Rpl4 | Thermofisher | Mm05781370_g1 |
| Cacna1a | Thermofisher | Mm00432190_m1 |
| Slc12a5 | Thermofisher | Mm00803929_m1 |
| Grin1 Bsn |
Thermofisher Thermofisher |
Mm00433790_m1 Mm00464452_m1 |
| RNA45S (18S) | Thermofisher | Mm04277571_s1 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J wildtype | Janvier labs | SC-C57J-M |
| Mouse: Tg(Aldh1l1-eGFP/Rpl10a) JD130Htz | Nathaniel Heintz’s laboratory (Rockefeller University, New York City, NY) | MGI:5496674www.bactrap.org |
| Mouse: hGfap:eGFP | Max Delbrück Center for Molecular Medicine (Berlin-Buch, Germany) | (Nolte et al., 2001) |
| Software and Algorithms | ||
| ImageJ | https://imagej.nih.gov/ij/ | https://imagej.nih.gov/ij/ |
| AstroDot plugin for ImageJ | Our laboratory | Oudart et al., 2020 |
| Virus Strains | ||
| Serotype 9 AAV GfaABC1D:eGFP | This paper | N/A |
| Serotype 9 AAV GfaABC1D:ERGIC-GFP | This paper | N/A |
| Serotype 9 AAV GfaABC1D:pGolt-mCherry | This paper | N/A |
| Deposited Data | ||
| Raw data and analysis | This paper | GEO: GSE143531 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Puromycin dihydrochloride | Sigma | Cat#P7255 |
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Martine Cohen-Salmon (martine.cohen-salmon@college-de-france.fr).
Material availability
Requests for plasmids generated in the study should be directed to and will be fulfilled by the Lead Contact.
Data and Code Availability
RNA sequencing data are recorded in the Gene Expression Omnibus (GEO) website. The accession number for the data reported in this paper is GEO: GSE143531
Experimental Model and Subject Details
Tg(Aldh1l1-eGFP/Rpl10a) JD130Htz (MGI: 5496674) (Aldh1l1:L10a-eGFP) mice were obtained from Nathaniel Heintz’s laboratory (Rockefeller University, New York City, NY) and kept under pathogen-free conditions (Heiman et al., 2014). The genotyping protocol is described on the bacTRAP project’s web site (www.bactrap.org). hGfap-eGFP mice were obtained from the Max Delbrück Center for Molecular Medicine (Berlin-Buch, Germany) (Nolte et al., 2001). C57BL6 mice were purchased from Janvier Labs (France) and kept in pathogen-free conditions. All animal experiments were carried out in compliance with the European Directive 2010/63/EU on the protection of animals used for scientific purposes and the guidelines issued by the French National Animal Care and Use Committee (reference: 2013/118). The project was also approved by the French Ministry for Research and Higher Education’s institutional review board (reference 2018051809274047).
The TRAP and fear conditioning experiments were performed on male mice only, as hippocampal astrocytes display sex-difference (Conejo et al., 2003). All other mouse experiments included both sexes.
All experiments were performed on 2-month-old mice. Slc1a2 FISH experiments were performed on 3 C57BL6 mice. Puromycilation experiments were performed on 3 Aldh1l1:L10a-eGFP mice. Immunofluorescent detection and quantification of synapses were performed on 3 Aldh1l1:L10a-eGFP mice. Viral injections and immunofluorescent detection of ERGIC, eGFP and pGolt were performed on 9 C57BL6 mice (3 mice per condition). TRAP experiments and RNA sequencing were performed on 6 Aldh1l1:L10a-eGFP mice (2 mice per library, 3 libraries) for PAPs and on 3 Aldh1l1:L10a-eGFP mice (1 mouse per library, 3 libraries) for the whole astrocytes. qPCR validation of the TRAP was performed on 6 Aldh1l1:L10a-eGFP mice (2 mice per sample) for PAPs, and 3 Aldh1l1:L10a-eGFP mice (1 mice per sample) for whole astrocytes. Western blot analysis of the synaptogliosome was performed on 4 Aldh1l1:L10a-eGFP mice. Western blot analysis in the fear conditioning study was performed on 4 hGfap:eGFP and 4 Aldh1l1:L10a-eGFP mice per condition. FISH experiments in the fear conditioning study was performed on 3 C57BL6 mice per condition. qPCR quantification in the fear conditioning study was done on 3 to 5 Aldh1l1:L10a-eGFP mice per condition.
Method Details
High-resolution fluorescent in situ hybridization and GFAP co-immunofluorescent detection and analysis
Fluorescent in situ hybridization (FISH) was performed on floating brain sections obtained from mice perfused with 4% PFA/PBS, according to the v2 Multiplex RNAscope technique (Advanced Cell Diagnostics, Inc., Newark, CA, USA). After the FISH procedure, GFAP was immunofluorescently detected. Astrocyte-specific FISH dots were recognized on the base of their localization on the GFAP immunolabeling, using the AstroDot ImageJ plug-in. This method has been described in detail elsewhere (Oudart et al., 2020). The Bacillus subtilis dihydrodipicolinate reductase (dapB) gene was used as a negative control. The probes are listed in the Key Resources Table. Independent experiments were performed on three animals and two brain slices per animal.
Gene ontology (GO) analyses
We used the text-mining Pathway Studio ResNet database (Ariadne Genomics, Rockville, MD) and the gene set enrichment analysis (GSEA) tool (Subramanian et al., 2005) in Pathway Studio (version 12.1.0.9) (Nikitin et al., 2003) to identify signaling pathways and biological processes that were overrepresented in our list of differentially expressed genes. To determine pathways for PAPs and whole astrocytes, we used the GO tool (categories: Biological Processes and Cellular Components; p value threshold: 0.01; percentage overlap: > 15%). To compare the lists from PAPs versus whole astrocytes, we selected the Kolmogorov-Smirnov test in the GSEA tool (p value threshold: 0.05).
Preparation of synaptogliosomes
Synaptogliosomes were prepared from the dorsal hippocampi of 2-month-old mice. All steps were performed at 4°C. Dorsal hippocampi were dissected and homogenized with a tight glass homogenizer (20 strokes) in buffer solution (0.32 M sucrose and 10 mM HEPES in diethyl-dicarbonate-treated water, with 0.5 mM DTT, Protease Inhibitors (Complete-EDTA free) 1 mini tablet/10 mL, RNasin 1 μL/mL, CHX 100 μg/mL added extratemporaneously). The homogenate was then centrifuged at 900 g for 15 min. The pellet P1 was discarded, and the supernatant S1 was centrifuged at 16,000 g for 15 min. The supernatant S2 was discarded, and the pellet P2 (containing synaptogliosomes) was diluted in 600 μl of buffer solution and centrifuged again at 16,000 g for 15 min. The final pellet P3 contained the synaptogliosomes.
Aldh1l1:L10a-eGFP TRAP, and RNA sequencing and analysis
The TRAP experiments were performed on male mice only since hippocampal astrocytes display sex-difference (Conejo et al., 2003). Two dorsal hippocampi were used for whole astrocyte ribosome-bound mRNA extraction (n = 3 libraries). For PAP ribosome-bound mRNAs, synaptogliosomes were isolated from dorsal hippocampi pooled from two mice (n = 3 libraries). Ribosome-bound mRNAs were extracted using our recently published TRAP protocol (Boulay et al., 2019). In order to avoid non-specific binding of the mRNAs to the beads or to IgG, and in contrast to the initial TRAP protocol (Heiman et al., 2014), the cytosolic extracts were pre-cleaned more extensively prior to immunoprecipitation. Lysates were first placed on an antibody-free pre-cleaning column and then on a column with bound non-specific mouse IgGs. Non-specific sites on the immunoprecipitation column were blocked with yeast tRNA and 2% BSA (Boulay et al., 2019).
Ribosome-bound mRNAs were purified using the RNeasy Lipid tissue kit (QIAGEN). Ten nanograms of total RNA were amplified and converted into cDNA using a SMART-Seq v4 Ultra Low Input RNA kit (Clontech). Next, an average of 150 pg of amplified cDNA was used per library, with a Nextera XT DNA kit (Illumina). Libraries were multiplexed on two high-output flow cells and sequenced (75 bp reads) on a NextSeq 500 device (Illumina). The mean ± standard deviation number of reads per sample meeting the Illumina quality criterion was 23 ± 6 million.
The data were analyzed (read filtering, mapping, alignment filtering, read quantification, normalization and differential analysis) using the Eoulsan pipeline (Jourdren et al., 2012). Before mapping, adapters were removed (using TrimGalore (Dobin et al., 2013) version 0.4.1), poly N read tails were trimmed, reads ≤ 40 bases were removed, and reads with a mean quality score ≤ 30 were discarded. The reads were then aligned against the Mus musculus genome (Ensembl version 84) using STAR (version 2.5.2b) (Li et al., 2009). Alignments from reads matching more than once on the reference genome were removed using a Java version of samtools (Anders et al., 2015). The annotated Mus musculus gene transfer format file (Ensembl version 84) was used to compute gene expression. The regions in which alignments and referenced exons overlapped were counted using HTSeq-count (version 0.5.3) (Anders et al., 2015).
The RNA-seq gene expression data and raw fastq files are available on the GEO repository (https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE143531. The sample counts were normalized using DESeq2 (version 1.8.1) (Love et al., 2014). Statistical processing including a hierarchical clustering and differential analyses were also performed using DESeq2 (version 1.8.1). p values were corrected for multiple testing using the Benjamini-Hochberg (BH) adjustment. The presence of an mRNA in whole astrocytes or in PAPs was defined by a read count ≥ 500 and ≥ 70% exon coverage. Enrichment in PAPs was defined as a log2 fold-change ≤ −1 and a p value ≤ 0.05, whereas depletion was defined as a log2 fold-change ≥ 1 and a p value ≤ 0.05.
Quantitative RT-PCR
RNA was extracted using the Rneasy Lipid tissue mini kit (QIAGEN, Hilden, Germany). cDNA was then generated using the Superscript III Reverse Transcriptase kit and pre-amplified with a mixture of the tested TaqMan® probes (Key Resources Table) using the SsoAdvanced PreAmp Supermix (Biorad). Differential levels of cDNA expression were measured using the droplet digital PCR (ddPCR) system (Biorad) and TaqMan® copy number assay probes. Briefly, cDNA and 6-carboxyfluorescein (FAM) probes and primers were distributed into approximately 10,000 to 20,000 droplets. The nucleic acids were then PCR-amplified in a thermal cycler and read (as the number of positive and negative droplets) with a QX200 Droplet Digital PCR System. The ratio for each tested gene was normalized against the total number of positive droplets against 45S RNA.
Western blots
Synaptogliosome pellets were sonicated three times for 10 s at 20 Hz (Vibra cell VCX130) and boiled in Laemmli loading buffer. Protein content was measured using the Pierce 660 nm protein assay reagent (Thermo Scientific, Waltham, MA, USA). Equal amounts of proteins were separated by denaturing electrophoresis in Mini-Protean TGX stain-free gels (Biorad) and then electrotransferred to nitrocellulose membranes using the Trans-blot Turbo Transfer System (Biorad). Membranes were hybridized as described previously (Ezan et al., 2012). The antibodies used in this study are listed in the Key Resources Table. Horseradish peroxidase activity was visualized by enhanced chemiluminescence in a Western Lightning Plus system (Perkin Elmer, Waltham, MA, USA). Chemiluminescent imaging was performed on a LAS4000 system (Fujifilm, Minato-ku, Tokyo, Japan). At least four independent samples were analyzed in each experiment. The level of chemiluminescence for each antibody was normalized against stain-free signal on membranes.
Viral vectors and stereotaxic injection
Two independent transgenes (composed of ERGIC-53 and pGolt cDNAs fused with GFP or mCherry respectively) were inserted under the control of the gfaABC1D synthetic promoter (Lee et al., 2008) (to drive their expression in astrocytes) in an AAV shuttle plasmid containing the inverted terminal repeats of AAV2. Pseudotyped serotype 9 AAV particles were produced by transient co-transfection of HEK293T cells, as described previously (Fol et al., 2016). Viral titles were determined by quantitative PCR amplification of the inverted terminal repeats on DNase-resistant particles, and expressed in vector genomes (vg) per ml.
Two- to five-month-old mice were anesthetized with a mixture of ketamine (95 mg/kg; Merial) and xylazine (10 mg/kg; Bayer) in 0.9% NaCl and placed on a stereotaxic frame with constant body temperature monitoring. Adeno-associated viruses were diluted in PBS with 0.01% Pluronic F-68 at a concentration of 9x1012 vg/ml and 1 μl of virus was injected unilaterally into the right hippocampus at a rate of 0.1 μl/min, using a 29-gauge blunt-tip needle linked to a 2 μl Hamilton syringe (Phymep). The stereotaxic coordinates relative to the bregma were: anteroposterior, −2 mm; mediolateral, +1.5 mm; and dorsoventral, −1.5 mm. The needle was left in place for 5 min and was then slowly removed. The skin was glued back in place, and the animals’ recovery was checked regularly for the next 24 h. After 3 weeks, the mice were sacrificed and processed for immunostaining (see below).
Immunohistolabeling, puromycylation, confocal imaging and analysis
Brain sections were fixed in PBS/PFA 4% for 15 min, rinsed in PBS, incubated for 1 h at room temperature in blocking solution (2% goat serum, 0.2% Triton X-100 in PBS). Next, the sections were incubated with primary antibodies diluted in the blocking solution for 12 h at 4°C, rinsed for 5 min in PBS three times, incubated with secondary antibodies diluted in blocking solution for 2h at room temperature, rinsed for 5 min in PBS three times, and mounted in Fluoromount (Southern Biotech, Birmingham, AL). Brain sections were imaged on X1 and W1 spinning-disk confocal microscopes (Yokogawa). Images were acquired with a 63X oil immersion objective (Zeiss). To determine the number of synapses contacted by PAPs containing ERGIC or pGolt particles, we developed an ImageJ plugin. We first defined a synapse as a VGluT1 particle sharing at least one common voxel with a Homer1 particle. The center of a synapse was calculated as the half distance between VGluT1 and Homer1 particle centers. Regions of interest (ROI) were manually defined around individual astrocytes and the total number of synapses within each ROI was determined. Synapses presenting an ERGIC or pGolt particle within a range between 0 and 1 μm were assessed and the percentage of synapses contacted by PAPs containing ERGIC or pGolt was calculated over the total number of synapses within each astrocyte’s territory.
Synaptogliosomes were immobilized on a glass slide coated with Cell-Tak (Corning) and immunostained as described previously. In order to measure the distance from the PAP to the synapse, we performed super-resolution stimulated emission depletion (STED) microscopy on a home-built, time-gated system. The latter was based on a commercial point scanning microscope (RESOLFT, Abberior Instruments) comprising a microscope frame (IX83, Olympus), galvanometer mirrors (Quad scanner, Abberior Instruments), and a detection unit consisting of two avalanche photodiodes (SPCM-AQRH, Excelitas Technologies). Images were acquired with a 100X/1.4 NA oil immersion objective lens (UPLSAPO 100X, Olympus). Confocal images were then deconvolved using the Huygens software and combined with the STED images in one file for analysis. For each astrocyte, 4 regions of interest of about 600 μm2 were imaged. Homer1 and VGluT1 stainings were acquired in both confocal and STED resolution. The analysis was performed using an in-house developed plugin on ImageJ to analyze the distance from an identified synapse to the closest astrocyte process. In brief, maxima intensities were identified in STED images and compared to the deconvolved confocal images to avoid false positive punctae. A synapse was defined as two punctae from Homer1 and VGluT1 found within 300 μm of each other. Distances were calculated from the center of the synapse to the closest eGFP-labeled astrocyte process.
For puromycylation, transverse slices of 150μm from the hippocampi of P14-P17 Aldh1l1:L10a-eGFP mice were prepared using a vibrating blade microtome (Leica) in cold (4°C) Artificial cerebrospinal fluid (aCSF) containing (in mM): 119 NaCl, 2.5 KCl, 2.5 CaCl2, 1.3 MgSO4, 1 NaH2PO4, 26.2 NaHCO3 and 11 glucose saturated with 95% O2 and 5% CO2. Slices were left to rest in storage chamber for 1 h at room temperature, then incubated at 32°C for 1 h. Half of the slices were placed in a chamber with 91 μM puromycin (Sigma) at 32°C for 10 min. The other half was left untouched and used as negative control. Slices were then washed out for 2 min in fresh aCSF, fixed in PBS/PFA 4% overnight at 4°C, rinsed for 3min in PBS three times and permeabilized and blocked in 4% goat serum, 0.5% Triton X-100 in PBS overnight at 4°C. Next, slices were incubated overnight at 4°C with primary antibodies in the blocking solution without Triton, rinsed 3 times in PBS for 20 min, and incubated with secondary antibodies for 3h at room temperature. Slices were rinsed again with PBS and mounted in Fluoromount (Southern Biotech, Birmingham, AL) on glass coverslips. The number of synapses within 1 μm of L10A-eGFP signal or a puromycin puncta was determined as previously described for ERGIC and pGolt, except colocalization was first assessed between puromycin and L10A-eGFP to isolate puromycin signal originating from astrocytes only. The number of synapses within 1 μm of L10A-eGFP particles used as normalization for puromycin calculations. The image presented in Figure 1C was acquired using Zeiss LSM 980 Confocal – Airyscan 2, with a 63X oil immersion objective (Zeiss). Antibodies and applications are listed in the Key Resources Table.
Fear conditioning
Behavioral tests were performed on two-month-old male Aldh1l1:L10a-eGFP, hGFAP-GFP and wild-type mice. On day 1, the fear-conditioning group was placed in a conditioning chamber (characterized by white noise (5000 Hz, 60 dB) and a smell of acetic acid (1%)) and left to explore for 3 min. The mice were exposed to four consecutive 1 s scrambled foot shocks (current: 0.6 mA; inter-shock interval: ∼60 s) and then returned to their home cage. On day 2, the mice were put back in the conditioning chamber to assess contextual conditioning. The time spent freezing during a 5 min session was recorded automatically using a camera Sony effio-e 700TVL and infrared tracking device. It was quantified using the PolyFear Software. The mice were sacrificed shortly after the freezing test. Home-caged animals were used as controls.
Quantification and Statistical Analysis
Except for the RNaseq data analyses, all statistical tests were performed in GraphPad Prism (Prism 8 for windows 64bit, Version 8.0.2 (263)). Statistical details for the RNaseq and Gene ontology studies (Figures 2F, 2H, and 3) are all given in the Methods details section. Raw sequencing data are presented in Table S1. Three independent libraries were sequenced for PAPs and whole astrocytes. Each PAP library was prepared from the dorsal hippocampi from 2 mice. Each whole astrocyte library was prepared from the dorsal hippocampi from one mouse.
Western blot characterization of the synaptogliosome (Figure 2B) is presented in the Figure 2 legend as the mean ± SEM. Results were analyzed using a two-tailed, paired t test. Four independent samples prepared from 4 mice were analyzed. Western blot characterizations of the synaptogliosome upon fear conditioning (Figures 5D–5G, 6G, and 6H) are presented in the Figure 5 and 6 legends as the mean ± SEM. Results were analyzed using an unpaired, two-tailed t test. Four or five independent samples prepared from 4 or 5 mice were analyzed. AstroDot analysis of the mRNA density in PAPs after fear conditioning (Figures 6B–6D) is presented in the Figure 6 legend as the mean ± SEM. Raw data are given in Table S4. Results were analyzed using a two-tailed, unpaired t test. n ≥ 35 cells from 3 mice per condition were analyzed. qPCR experiments (Figures 4B, 6E, and 6F; Figures S2C and S2D; Tables S2, S3, and S5) are all presented as the mean ± SEM. Results were analyzed using a one-tailed Mann-Whitney test. For each experiments, 3 to 5 independent samples prepared from 3 to 5 mice were analyzed. Raw data and analysis details are presented in the Figure 4 and 6 and S2 legends and Tables S2, S3, and S5. A one-tailed Mann-Whitney test was applied for all qPCR experiments. STED analysis for PAP-synapse distance (Figure 5C) is presented as the mean ± SEM n ≥ 270 synapses per cell, 3 cells per mouse, and 3 mice per condition were analyzed. Details of the analysis are presented in the Figure 5 legend. Results were analyzed using a Two-way ANOVA.
Acknowledgments
This work was funded by Fondation pour la Recherche Médicale to M.C.-S. (AJE20171039094) and N.M. (FDT201904008077), the ED3C doctoral school to N.M., M.O., and J.M., the European Research Council (Consolidator grant 683154) and European Union’s Horizon 2020 research and innovation program (Marie Skłodowska-Curie Innovative Training Networks, grant 722053, EU-GliaPhD) to N.R., the FP7-PEOPLE Marie Curie Intra-European Fellowship for career development (grant 622289) to G.C., and “Journées de Neurologie de Langue Française” to R.T. The creation of the Center for Interdisciplinary Research in Biology (CIRB) was funded by the Fondation Bettencourt Schueller. We thank Carole Escartin and Océane Guillemaud for helpful discussions and all members of the Orion imaging facility for their high-quality technical support. The École Normale Supérieure genomic core facility was supported by the France Génomique national infrastructure, funded as part of the “Investissements d’Avenir” program managed by Agence Nationale de la Recherche (contract ANR-10-INBS-09).
Author Contributions
Conceptualization, M.C.-S. and N.R.; Methodology, M.C.-S., N.M., and N.R.; Investigation, N.M., M.O., J.M., G.C., R.T., P.M., D.M., A.-P.B., A.-C.B., C.B., L.J., and S.L.C.; Writing – Original Draft, M.C.-S.; Funding Acquisition, M.C.-S. and N.R.; Supervision, M.C.-S.
Declaration of Interests
The authors declare no conflict of interest.
Published: August 25, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108076.
Supplemental Information
This table contains 7 pages: 1. RNA-seq raw data of ribosome-bound mRNAs extracted from whole astrocytes and PAPs; 2. PAPome: list of ribosome-bound transcripts in PAPs with ≥ 500 reads and exon coverage ≥ 70%; 3. Whole astrocytes: list of ribosome-bound transcripts in whole astrocytes with ≥ 500 reads and exon coverage ≥ 70%; 4. PAP-enriched: list of ribosome-bound transcripts enriched in PAPs: BH-adjusted p value ≤ 0.05; log2 fold-change ≤ −1; 5. Astrocyte-enriched: list of ribosome-bound transcripts enriched in whole astrocytes: BH-adjusted p value ≤ 0.05; log2 fold-change ≥ 1; 6. GFAP exon study: comparison of whole astrocytes versus PAPs for each GFAP exon. Id: identity number in Ensembl.org.
References
- Abbas W., Kumar A., Herbein G. The eEF1A proteins: at the crossroads of oncogenesis, apoptosis, and viral infections. Front. Oncol. 2015;5:75. doi: 10.3389/fonc.2015.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamsky A., Kol A., Kreisel T., Doron A., Ozeri-Engelhard N., Melcer T., Refaeli R., Horn H., Regev L., Groysman M. Astrocytic activation generates de novo neuronal potentiation and memory enhancement. Cell. 2018;174:59–71.e14. doi: 10.1016/j.cell.2018.05.002. [DOI] [PubMed] [Google Scholar]
- Alami N.H., Smith R.B., Carrasco M.A., Williams L.A., Winborn C.S., Han S.S.W., Kiskinis E., Winborn B., Freibaum B.D., Kanagaraj A. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron. 2014;81:536–543. doi: 10.1016/j.neuron.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberini C.M. The role of protein synthesis during the labile phases of memory: revisiting the skepticism. Neurobiol. Learn. Mem. 2008;89:234–246. doi: 10.1016/j.nlm.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen N.J., Eroglu C. Cell biology of astrocyte-synapse interactions. Neuron. 2017;96:697–708. doi: 10.1016/j.neuron.2017.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez J.I., Katayama T., Prat A. Glial influence on the blood brain barrier. Glia. 2013;61:1939–1958. doi: 10.1002/glia.22575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton S.K., Gregory J.M., Chandran S., Turner B.J. Could an impairment in local translation of mRNAs in glia be contributing to pathogenesis in ALS? Front. Mol. Neurosci. 2019;12:124. doi: 10.3389/fnmol.2019.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastide A., Yewdell J.W., David A. The ribopuromycylation method (RPM): an immunofluorescence technique to map translation sites at the sub-cellular level. Bio. Protoc. 2018;8:e2669. doi: 10.21769/BioProtoc.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Abdallah N.M., Filipkowski R.K., Pruschy M., Jaholkowski P., Winkler J., Kaczmarek L., Lipp H.P. Impaired long-term memory retention: common denominator for acutely or genetically reduced hippocampal neurogenesis in adult mice. Behav. Brain Res. 2013;252:275–286. doi: 10.1016/j.bbr.2013.05.034. [DOI] [PubMed] [Google Scholar]
- Ben-Tekaya H., Miura K., Pepperkok R., Hauri H.P. Live imaging of bidirectional traffic from the ERGIC. J. Cell Sci. 2005;118:357–367. doi: 10.1242/jcs.01615. [DOI] [PubMed] [Google Scholar]
- Bernardinelli Y., Muller D., Nikonenko I. Astrocyte-synapse structural plasticity. Neural Plast. 2014;2014:232105. doi: 10.1155/2014/232105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse F., Ephrussi A. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat. Rev. Mol. Cell Biol. 2008;9:971–980. doi: 10.1038/nrm2548. [DOI] [PubMed] [Google Scholar]
- Blumenstock S., Angelo M.F., Peters F., Dorostkar M.M., Ruf V.C., Luckner M., Crux S., Slapakova L., Arzberger T., Claverol S. Early defects in translation elongation factor 1α levels at excitatory synapses in α-synucleinopathy. Acta Neuropathol. 2019;138:971–986. doi: 10.1007/s00401-019-02063-3. [DOI] [PubMed] [Google Scholar]
- Boulay A.C., Saubaméa B., Adam N., Chasseigneaux S., Mazaré N., Gilbert A., Bahin M., Bastianelli L., Blugeon C., Perrin S. Translation in astrocyte distal processes sets molecular heterogeneity at the gliovascular interface. Cell Discov. 2017;3:17005. doi: 10.1038/celldisc.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulay A.C., Mazaré N., Saubaméa B., Cohen-Salmon M. Preparing the astrocyte perivascular endfeet transcriptome to investigate astrocyte molecular regulations at the brain-vascular interface. Methods Mol. Biol. 2019;1938:105–116. doi: 10.1007/978-1-4939-9068-9_8. [DOI] [PubMed] [Google Scholar]
- Bushong E.A., Martone M.E., Jones Y.Z., Ellisman M.H. Protoplasmic astrocytes in CA1 stratum radiatum occupy separate anatomical domains. J. Neurosci. 2002;22:183–192. doi: 10.1523/JNEUROSCI.22-01-00183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajigas I.J., Tushev G., Will T.J., tom Dieck S., Fuerst N., Schuman E.M. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74:453–466. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney K.E., Milanese M., van Nierop P., Li K.W., Oliet S.H., Smit A.B., Bonanno G., Verheijen M.H. Proteomic analysis of gliosomes from mouse brain: identification and investigation of glial membrane proteins. J. Proteome Res. 2014;13:5918–5927. doi: 10.1021/pr500829z. [DOI] [PubMed] [Google Scholar]
- Chever O., Lee C.Y., Rouach N. Astroglial connexin43 hemichannels tune basal excitatory synaptic transmission. J. Neurosci. 2014;34:11228–11232. doi: 10.1523/JNEUROSCI.0015-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W.S., Allen N.J., Eroglu C. Astrocytes control synapse formation, function, and elimination. Cold Spring Harb. Perspect. Biol. 2015;7:a020370. doi: 10.1101/cshperspect.a020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L.E., Liddelow S.A., Chakraborty C., Münch A.E., Heiman M., Barres B.A. Normal aging induces A1-like astrocyte reactivity. Proc. Natl. Acad. Sci. U S A. 2018;115:E1896–E1905. doi: 10.1073/pnas.1800165115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codazzi F., Pelizzoni I., Zacchetti D., Grohovaz F. Iron entry in neurons and astrocytes: a link with synaptic activity. Front. Mol. Neurosci. 2015;8:18. doi: 10.3389/fnmol.2015.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conejo N.M., González-Pardo H., Pedraza C., Navarro F.F., Vallejo G., Arias J.L. Evidence for sexual difference in astrocytes of adult rat hippocampus. Neurosci. Lett. 2003;339:119–122. doi: 10.1016/s0304-3940(02)01484-2. [DOI] [PubMed] [Google Scholar]
- Dallérac G., Zapata J., Rouach N. Versatile control of synaptic circuits by astrocytes: where, when and how? Nat. Rev. Neurosci. 2018;19:729–743. doi: 10.1038/s41583-018-0080-6. [DOI] [PubMed] [Google Scholar]
- Deglincerti A., Jaffrey S.R. Insights into the roles of local translation from the axonal transcriptome. Open Biol. 2012;2:120079. doi: 10.1098/rsob.120079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derouiche A., Frotscher M. Peripheral astrocyte processes: monitoring by selective immunostaining for the actin-binding ERM proteins. Glia. 2001;36:330–341. doi: 10.1002/glia.1120. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dringen R., Bishop G.M., Koeppe M., Dang T.N., Robinson S.R. The pivotal role of astrocytes in the metabolism of iron in the brain. Neurochem. Res. 2007;32:1884–1890. doi: 10.1007/s11064-007-9375-0. [DOI] [PubMed] [Google Scholar]
- Ezan P., André P., Cisternino S., Saubaméa B., Boulay A.C., Doutremer S., Thomas M.A., Quenech’du N., Giaume C., Cohen-Salmon M. Deletion of astroglial connexins weakens the blood-brain barrier. J. Cereb. Blood Flow Metab. 2012;32:1457–1467. doi: 10.1038/jcbfm.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallini C., Bassell G.J., Rossoll W. The ALS disease protein TDP-43 is actively transported in motor neuron axons and regulates axon outgrowth. Hum. Mol. Genet. 2012;21:3703–3718. doi: 10.1093/hmg/dds205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fol R., Braudeau J., Ludewig S., Abel T., Weyer S.W., Roederer J.P., Brod F., Audrain M., Bemelmans A.P., Buchholz C.J. Viral gene transfer of APPsα rescues synaptic failure in an Alzheimer’s disease mouse model. Acta Neuropathol. 2016;131:247–266. doi: 10.1007/s00401-015-1498-9. [DOI] [PubMed] [Google Scholar]
- Ghézali G., Dallerac G., Rouach N. Perisynaptic astroglial processes: dynamic processors of neuronal information. Brain Struct. Funct. 2016;221:2427–2442. doi: 10.1007/s00429-015-1070-3. [DOI] [PubMed] [Google Scholar]
- Gilani A.I., Chohan M.O., Inan M., Schobel S.A., Chaudhury N.H., Paskewitz S., Chuhma N., Glickstein S., Merker R.J., Xu Q. Interneuron precursor transplants in adult hippocampus reverse psychosis-relevant features in a mouse model of hippocampal disinhibition. Proc. Natl. Acad. Sci. U S A. 2014;111:7450–7455. doi: 10.1073/pnas.1316488111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giustetto M., Hegde A.N., Si K., Casadio A., Inokuchi K., Pei W., Kandel E.R., Schwartz J.H. Axonal transport of eukaryotic translation elongation factor 1alpha mRNA couples transcription in the nucleus to long-term facilitation at the synapse. Proc. Natl. Acad. Sci. U S A. 2003;100:13680–13685. doi: 10.1073/pnas.1835674100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glock C., Heumüller M., Schuman E.M. mRNA transport & local translation in neurons. Curr. Opin. Neurobiol. 2017;45:169–177. doi: 10.1016/j.conb.2017.05.005. [DOI] [PubMed] [Google Scholar]
- Hafner A.S., Donlin-Asp P.G., Leitch B., Herzog E., Schuman E.M. Local protein synthesis is a ubiquitous feature of neuronal pre- and postsynaptic compartments. Science. 2019;364:364. doi: 10.1126/science.aau3644. [DOI] [PubMed] [Google Scholar]
- Heiman M., Schaefer A., Gong S., Peterson J.D., Day M., Ramsey K.E., Suárez-Fariñas M., Schwarz C., Stephan D.A., Surmeier D.J. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M., Kulicke R., Fenster R.J., Greengard P., Heintz N. Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP) Nat. Protoc. 2014;9:1282–1291. doi: 10.1038/nprot.2014.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C.E., Martin K.C., Schuman E.M. Local translation in neurons: visualization and function. Nat. Struct. Mol. Biol. 2019;26:557–566. doi: 10.1038/s41594-019-0263-5. [DOI] [PubMed] [Google Scholar]
- Huber K.M., Kayser M.S., Bear M.F. Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science. 2000;288:1254–1257. doi: 10.1126/science.288.5469.1254. [DOI] [PubMed] [Google Scholar]
- Iadecola C. The neurovascular unit coming of age: a journey through neurovascular coupling in health and disease. Neuron. 2017;96:17–42. doi: 10.1016/j.neuron.2017.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonka S., Bandilla M., Wiese S., Bühler D., Wirth B., Sendtner M., Fischer U. Co-regulation of survival of motor neuron (SMN) protein and its interactor SIP1 during development and in spinal muscular atrophy. Hum. Mol. Genet. 2001;10:497–505. doi: 10.1093/hmg/10.5.497. [DOI] [PubMed] [Google Scholar]
- Jedynak P., Jaholkowski P., Wozniak G., Sandi C., Kaczmarek L., Filipkowski R.K. Lack of cyclin D2 impairing adult brain neurogenesis alters hippocampal-dependent behavioral tasks without reducing learning ability. Behav. Brain Res. 2012;227:159–166. doi: 10.1016/j.bbr.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Jones K.J., Templet S., Zemoura K., Kuzniewska B., Pena F.X., Hwang H., Lei D.J., Haensgen H., Nguyen S., Saenz C. Rapid, experience-dependent translation of neurogranin enables memory encoding. Proc. Natl. Acad. Sci. U S A. 2018;115:E5805–E5814. doi: 10.1073/pnas.1716750115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdren L., Bernard M., Dillies M.A., Le Crom S. Eoulsan: a cloud computing-based framework facilitating high throughput sequencing analyses. Bioinformatics. 2012;28:1542–1543. doi: 10.1093/bioinformatics/bts165. [DOI] [PubMed] [Google Scholar]
- Kamphuis W., Mamber C., Moeton M., Kooijman L., Sluijs J.A., Jansen A.H., Verveer M., de Groot L.R., Smith V.D., Rangarajan S. GFAP isoforms in adult mouse brain with a focus on neurogenic astrocytes and reactive astrogliosis in mouse models of Alzheimer disease. PLoS ONE. 2012;7:e42823. doi: 10.1371/journal.pone.0042823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Schuman E.M. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- Kao D.I., Aldridge G.M., Weiler I.J., Greenough W.T. Altered mRNA transport, docking, and protein translation in neurons lacking fragile X mental retardation protein. Proc. Natl. Acad. Sci. U S A. 2010;107:15601–15606. doi: 10.1073/pnas.1010564107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavialle M., Aumann G., Anlauf E., Pröls F., Arpin M., Derouiche A. Structural plasticity of perisynaptic astrocyte processes involves ezrin and metabotropic glutamate receptors. Proc. Natl. Acad. Sci. U S A. 2011;108:12915–12919. doi: 10.1073/pnas.1100957108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc C., Etienne-Manneville S. Regulation of microtubule-associated motors drives intermediate filament network polarization. J. Cell Biol. 2017;216:1689–1703. doi: 10.1083/jcb.201607045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Messing A., Su M., Brenner M. GFAP promoter elements required for region-specific and astrocyte-specific expression. Glia. 2008;56:481–493. doi: 10.1002/glia.20622. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Stockton M.E., Bhuiyan I., Eisinger B.E., Gao Y., Miller J.L., Bhattacharyya A., Zhao X. MDM2 inhibition rescues neurogenic and cognitive deficits in a mouse model of fragile X syndrome. Sci. Transl. Med. 2016;8:336ra61. doi: 10.1126/scitranslmed.aad9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., He Y., Jin A., Tikunov A.P., Zhou L., Tollini L.A., Leslie P., Kim T.H., Li L.O., Coleman R.A. Ribosomal protein-Mdm2-p53 pathway coordinates nutrient stress with lipid metabolism by regulating MCD and promoting fatty acid oxidation. Proc. Natl. Acad. Sci. U S A. 2014;111:E2414–E2422. doi: 10.1073/pnas.1315605111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.C., Seimetz J., Lee K.Y., Kalsotra A., Chung H.J., Lu H., Tsai N.P. Mdm2 mediates FMRP- and Gp1 mGluR-dependent protein translation and neural network activity. Hum. Mol. Genet. 2017;26:3895–3908. doi: 10.1093/hmg/ddx276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Fernandez M., Jamison S., Robin L.M., Zhao Z., Martin E.D., Aguilar J., Benneyworth M.A., Marsicano G., Araque A. Synapse-specific astrocyte gating of amygdala-related behavior. Nat. Neurosci. 2017;20:1540–1548. doi: 10.1038/nn.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliaccio N., Martucci N.M., Ruggiero I., Sanges C., Ohkubo S., Lamberti A., Agostinelli E., Arcari P. Ser/Thr kinases and polyamines in the regulation of non-canonical functions of elongation factor 1A. Amino Acids. 2016;48:2339–2352. doi: 10.1007/s00726-016-2311-3. [DOI] [PubMed] [Google Scholar]
- Mikhaylova M., Bera S., Kobler O., Frischknecht R., Kreutz M.R. A dendritic Golgi satellite between ERGIC and retromer. Cell Rep. 2016;14:189–199. doi: 10.1016/j.celrep.2015.12.024. [DOI] [PubMed] [Google Scholar]
- Miller S., Yasuda M., Coats J.K., Jones Y., Martone M.E., Mayford M. Disruption of dendritic translation of CaMKIIalpha impairs stabilization of synaptic plasticity and memory consolidation. Neuron. 2002;36:507–519. doi: 10.1016/s0896-6273(02)00978-9. [DOI] [PubMed] [Google Scholar]
- Moccia R., Chen D., Lyles V., Kapuya E., e Y., Kalachikov S., Spahn C.M., Frank J., Kandel E.R., Barad M., Martin K.C. An unbiased cDNA library prepared from isolated Aplysia sensory neuron processes is enriched for cytoskeletal and translational mRNAs. J. Neurosci. 2003;23:9409–9417. doi: 10.1523/JNEUROSCI.23-28-09409.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehlig A., Molina A., Rodrigues-Ferreira S., Honoré S., Nahmias C. Regulation of end-binding protein EB1 in the control of microtubule dynamics. Cell. Mol. Life Sci. 2017;74:2381–2393. doi: 10.1007/s00018-017-2476-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo L., Haas M., Qu Z., Li S.S., Zenker J., Teng K.S., Gunnersen J.M., Breuss M., Habgood M., Keays D.A., Heng J.I. TUBB5 and its disease-associated mutations influence the terminal differentiation and dendritic spine densities of cerebral cortical neurons. Hum. Mol. Genet. 2014;23:5147–5158. doi: 10.1093/hmg/ddu238. [DOI] [PubMed] [Google Scholar]
- Nikitin A., Egorov S., Daraselia N., Mazo I. Pathway studio—the analysis and navigation of molecular networks. Bioinformatics. 2003;19:2155–2157. doi: 10.1093/bioinformatics/btg290. [DOI] [PubMed] [Google Scholar]
- Nolte C., Matyash M., Pivneva T., Schipke C.G., Ohlemeyer C., Hanisch U.K., Kirchhoff F., Kettenmann H. GFAP promoter-controlled EGFP-expressing transgenic mice: a tool to visualize astrocytes and astrogliosis in living brain tissue. Glia. 2001;33:72–86. [PubMed] [Google Scholar]
- Ostroff L.E., Manzur M.K., Cain C.K., Ledoux J.E. Synapses lacking astrocyte appear in the amygdala during consolidation of Pavlovian threat conditioning. J. Comp. Neurol. 2014;522:2152–2163. doi: 10.1002/cne.23523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudart M., Tortuyaux R., Mailly P., Mazaré N., Boulay A.C., Cohen-Salmon M. AstroDot: a new method for studying the spatial distribution of mRNA in astrocytes. J. Cell Sci. 2020;133:cs239756. doi: 10.1242/jcs.239756. [DOI] [PubMed] [Google Scholar]
- Pannasch U., Vargová L., Reingruber J., Ezan P., Holcman D., Giaume C., Syková E., Rouach N. Astroglial networks scale synaptic activity and plasticity. Proc. Natl. Acad. Sci. U S A. 2011;108:8467–8472. doi: 10.1073/pnas.1016650108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S., Brot N. Studies on the formation of transfer ribonucleic acid-ribosome complexes. IV. Effect of antibiotics on steps of bacterial protein synthesis: some new ribosomal inhibitors of translocation. J. Biol. Chem. 1971;246:7715–7722. [PubMed] [Google Scholar]
- Pfister J.A., D’Mello S.R. Insights into the regulation of neuronal viability by nucleophosmin/B23. Exp. Biol. Med. (Maywood) 2015;240:774–786. doi: 10.1177/1535370215579168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilaz L.J., Lennox A.L., Rouanet J.P., Silver D.L. Dynamic mRNA transport and local translation in radial glial progenitors of the developing brain. Curr. Biol. 2016;26:3383–3392. doi: 10.1016/j.cub.2016.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach A., Derouiche A., Kirchhoff F. Morphology and dynamics of perisynaptic glia. Brain Res. Brain Res. Rev. 2010;63:11–25. doi: 10.1016/j.brainresrev.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Rogan M.T., Stäubli U.V., LeDoux J.E. Fear conditioning induces associative long-term potentiation in the amygdala. Nature. 1997;390:604–607. doi: 10.1038/37601. [DOI] [PubMed] [Google Scholar]
- Sakers K., Lake A.M., Khazanchi R., Ouwenga R., Vasek M.J., Dani A., Dougherty J.D. Astrocytes locally translate transcripts in their peripheral processes. Proc. Natl. Acad. Sci. U S A. 2017;114:E3830–E3838. doi: 10.1073/pnas.1617782114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santello M., Toni N., Volterra A. Astrocyte function from information processing to cognition and cognitive impairment. Nat. Neurosci. 2019;22:154–166. doi: 10.1038/s41593-018-0325-8. [DOI] [PubMed] [Google Scholar]
- Shigeoka T., Koppers M., Wong H.H., Lin J.Q., Cagnetta R., Dwivedy A., de Freitas Nascimento J., van Tartwijk F.W., Strohl F., Cioni J.M. On-site ribosome remodeling by locally synthesized ribosomal proteins in axons. Cell Rep. 2019;29:3605–3619.e10. doi: 10.1016/j.celrep.2019.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K., Giustetto M., Etkin A., Hsu R., Janisiewicz A.M., Miniaci M.C., Kim J.H., Zhu H., Kandel E.R. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell. 2003;115:893–904. doi: 10.1016/s0092-8674(03)01021-3. [DOI] [PubMed] [Google Scholar]
- Sibille J., Pannasch U., Rouach N. Astroglial potassium clearance contributes to short-term plasticity of synaptically evoked currents at the tripartite synapse. J. Physiol. 2014;592:87–102. doi: 10.1113/jphysiol.2013.261735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklan E.H., Podoly E., Soreq H. RACK1 has the nerve to act: structure meets function in the nervous system. Prog. Neurobiol. 2006;78:117–134. doi: 10.1016/j.pneurobio.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Stogsdill J.A., Eroglu C. The interplay between neurons and glia in synapse development and plasticity. Curr. Opin. Neurobiol. 2017;42:1–8. doi: 10.1016/j.conb.2016.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan S., Li L., Moss J., Petrelli F., Cassé F., Gebara E., Lopatar J., Pfrieger F.W., Bezzi P., Bischofberger J., Toni N. Synaptic integration of adult-born hippocampal neurons is locally controlled by astrocytes. Neuron. 2015;88:957–972. doi: 10.1016/j.neuron.2015.10.037. [DOI] [PubMed] [Google Scholar]
- Sutton M.A., Schuman E.M. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Tsai N.P., Wilkerson J.R., Guo W., Maksimova M.A., DeMartino G.N., Cowan C.W., Huber K.M. Multiple autism-linked genes mediate synapse elimination via proteasomal degradation of a synaptic scaffold PSD-95. Cell. 2012;151:1581–1594. doi: 10.1016/j.cell.2012.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai N.P., Wilkerson J.R., Guo W., Huber K.M. FMRP-dependent Mdm2 dephosphorylation is required for MEF2-induced synapse elimination. Hum. Mol. Genet. 2017;26:293–304. doi: 10.1093/hmg/ddw386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsokas P., Grace E.A., Chan P., Ma T., Sealfon S.C., Iyengar R., Landau E.M., Blitzer R.D. Local protein synthesis mediates a rapid increase in dendritic elongation factor 1A after induction of late long-term potentiation. J. Neurosci. 2005;25:5833–5843. doi: 10.1523/JNEUROSCI.0599-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Driesche S.J., Martin K.C. New frontiers in RNA transport and local translation in neurons. Dev. Neurobiol. 2018;78:331–339. doi: 10.1002/dneu.22574. [DOI] [PubMed] [Google Scholar]
- Verkhratsky A., Toescu E.C. Neuronal-glial networks as substrate for CNS integration. J. Cell. Mol. Med. 2006;10:826–836. doi: 10.1111/j.1582-4934.2006.tb00527.x. [DOI] [PubMed] [Google Scholar]
- Wake H., Lee P.R., Fields R.D. Control of local protein synthesis and initial events in myelination by action potentials. Science. 2011;333:1647–1651. doi: 10.1126/science.1206998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaka R., Thornton C., Vagts A.J., Phamluong K., Bonci A., Ron D. NMDA receptor function is regulated by the inhibitory scaffolding protein, RACK1. Proc. Natl. Acad. Sci. U S A. 2002;99:5710–5715. doi: 10.1073/pnas.062046299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Gozen O., Watkins A., Lorenzini I., Lepore A., Gao Y., Vidensky S., Brennan J., Poulsen D., Won Park J. Presynaptic regulation of astroglial excitatory neurotransmitter transporter GLT1. Neuron. 2009;61:880–894. doi: 10.1016/j.neuron.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.L., Pan F., Hong D., Shenoy S.M., Singer R.H., Bassell G.J. Active transport of the survival motor neuron protein and the role of exon-7 in cytoplasmic localization. J. Neurosci. 2003;23:6627–6637. doi: 10.1523/JNEUROSCI.23-16-06627.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen K., Sloan S.A., Bennett M.L., Scholze A.R., O’Keeffe S., Phatnani H.P., Guarnieri P., Caneda C., Ruderisch N. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Jiang R., Chen M., Maimaitiming Z., Wang J., Anderson G.J., Vulpe C.D., Dunaief J.L., Chen H. Multi-copper ferroxidase-deficient mice have increased brain iron concentrations and learning and memory deficits. J. Nutr. 2018;148:643–649. doi: 10.1093/jn/nxy012. [DOI] [PubMed] [Google Scholar]
- Zhu D., Li C., Swanson A.M., Villalba R.M., Guo J., Zhang Z., Matheny S., Murakami T., Stephenson J.R., Daniel S. BAI1 regulates spatial learning and synaptic plasticity in the hippocampus. J. Clin. Invest. 2015;125:1497–1508. doi: 10.1172/JCI74603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Chen X., Song Y., Zhang Y., Zhou L., Wan L. Deficit of RACK1 contributes to the spatial memory impairment via upregulating BECLIN1 to induce autophagy. Life Sci. 2016;151:115–121. doi: 10.1016/j.lfs.2016.02.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This table contains 7 pages: 1. RNA-seq raw data of ribosome-bound mRNAs extracted from whole astrocytes and PAPs; 2. PAPome: list of ribosome-bound transcripts in PAPs with ≥ 500 reads and exon coverage ≥ 70%; 3. Whole astrocytes: list of ribosome-bound transcripts in whole astrocytes with ≥ 500 reads and exon coverage ≥ 70%; 4. PAP-enriched: list of ribosome-bound transcripts enriched in PAPs: BH-adjusted p value ≤ 0.05; log2 fold-change ≤ −1; 5. Astrocyte-enriched: list of ribosome-bound transcripts enriched in whole astrocytes: BH-adjusted p value ≤ 0.05; log2 fold-change ≥ 1; 6. GFAP exon study: comparison of whole astrocytes versus PAPs for each GFAP exon. Id: identity number in Ensembl.org.
Data Availability Statement
RNA sequencing data are recorded in the Gene Expression Omnibus (GEO) website. The accession number for the data reported in this paper is GEO: GSE143531