Abstract
Introduction:
The glucocorticoid receptor (GR) is one of the most widely studied ligand-dependent nuclear receptors. The combination of transcriptional regulatory factors required for the expression of individual genes targeted by GR varies across cell types; however, the mechanisms underlying this cell type–specific regulation of gene expression are not yet clear.
Methods:
Here, we investigated genes regulated by GR in two different cell lines, A549 and ARPE-19, and examined how gene expression varied according to the effect of pioneer factors using RNA-seq and RT-qPCR.
Results:
Our RNA-seq results identified 19 and 63 genes regulated by GR that are ARPE-19-specific and A549-specific, respectively, suggesting that GR induces the expression of different sets of genes in a cell type–specific manner. RT-qPCR confirmed that the epithelial sodium channel (ENACα) gene is an ARPE-19 cell-specific GR target gene, whereas the FK506 binding protein 5 (FKBP5) gene was A549 cell-specific. There was a significant decrease in ENACα expression in FOXA1-deficient ARPE-19 cells, suggesting that FOXA1 might function as a pioneer factor enabling the selective expression of ENACα in ARPE-19 cells but not in A549 cells.
Conclusion:
These findings indicate that ENACα expression in ARPE-19 cells is regulated by FOXA1 and provide insights into the molecular mechanisms of cell type–specific expression of GR-regulated genes.
Keywords: ENACα, FOXA1, glucocorticoid receptor, pioneer factor
Introduction
The glucocorticoid receptor (GR) is a well-known ligand-dependent nuclear receptor (NR) capable of activating or repressing thousands of genes in the human body.1 GR signaling is present in a variety of organ systems, including the nervous, visual, respiratory, reproductive, and epidermal systems, and regulates genes involved in metabolism and immune and inflammatory responses.2 Similar to other nuclear receptors, the presence of ligands disrupts the binding between the heat shock protein (HSP) and GR due to the higher affinity between GRs and the ligand; the ligand then binds to GR and the complex dimerizes and translocates into the nucleus.1 Activated GR binds to the glucocorticoid response element to regulate target genes, which then repress the inflammatory, allergic, and immune responses.3 Synthetic glucocorticoid-like substances, such as dexamethasone (Dex) and prednisolone, are used to treat inflammatory diseases. Such diseases include AMD (age-related macular degeneration), which is caused by an extruded substance known as drusen that accumulates due to retinal pigment epithelium (RPE) cell dysfunction,4–6 and pulmonary diseases, such as asthma.7 In addition, steroid hormones are used to treat rheumatic arthritis, ulcerative colitis, and organ transplant rejection.8 Although the GR pathway may have multiple functions, there is a common mechanism of activation in all cells. However, the type of genes regulated by GR differ from cell to cell.9–11 Given that multicellular organisms are composed of many types of cells that are functionally divergent, genes must be differentially activated based on the cell type.
In differentiated cells, the combinations of regulatory transcription factors required for the expression of individual genes vary across cell types. Although the mechanisms of cell type–specific gene expression are not yet clear, early studies have suggested that pioneer factors are important regulators of gene expression.12–14 Pioneer factors are distinguished from activators by their ability to bind to silent chromatin.15 The recruitment of transcription factors to specific genomic sequences within condensed chromatin is difficult. However, pioneer factors are able to open the binding site and facilitate the binding of proteins to DNA.13 Pioneer factors are able to regulate epigenetic factors by recruiting histone-modifying enzymes to either activate or repress gene expression, potentially facilitating cellular reprogramming, depending on the external conditions of the cell.16 The epigenetic effects of pioneer factors can be divided into histone modifications and DNA methylation. Changes to histone modifications affect gene expression by allowing activating or repressive modification enzymes to remain in place.17 In contrast, DNA methylation involves the binding of transcriptional corepressors to histones, which suppresses gene expression through H3K9 methylation.18,19
Hormone signaling induces chromatin remodeling and converts chromatin to an open state at certain previously inaccessible GR-binding sites.20,21 This suggests that prior to GR binding, pioneer factors remodel the chromatin structure to make it more accessible. FOXA1, a representative pioneer factor that binds to the nuclear receptors, interacts with the estrogen receptor (ER) and retinoic acid receptor (RAR), depending on the cell type.22,23 FOXA1 expression upregulates GR binding at the mouse mammary tumor virus (MMTV) promoter, a chromatin domain regulated by GR.24 Transcription factors, including GR, are recruited to the MMTV promoter in a FOXA1-dependent manner, leading to increased transcriptional activity.24 In contrast, recent studies have shown that hormone signal transduction through ER and GR promotes the restructuring of the FOXA1-chromatin complex. Another pioneer factor, C/EBPβ, initially binds GR-binding sites in mouse liver tissue to promote chromatin remodeling and maintain chromatin accessibility.25 C/EBPβ is required for the recruitment of GR and for additional chromatin remodeling at multiple GR-controlled de novo sites. On one hand, activating protein 1 (AP-1) acts as an essential pioneer factor to maintain chromatin in the open state at 40% of the GR responsive regions in mammary cells. On the other hand, GR recruits AP-1 to some binding sites in cell lines that express FOXA1.26,27
Here, we examined the differential patterns of GR-mediated gene expression in two different cell lines. Furthermore, we suggested that the mechanism underlying this cell type–specific gene expression might depend on the pioneer factor FOXA1. Based on this, we identified genes regulated by GR following treatment of two different cell lines, A549 and ARPE-19, with Dex, and examined how gene expression varied depending on FOXA1. Finally, we identified the factors that regulate cell type–specific gene regulation in A549 and ARPE-19 cells.
Materials and methods
Cell culture
Human RPE cells (ARPE-19) purchased from the American Type Culture Collection (Manassas, VA, USA) were grown in Dulbecco’s modified Eagle’s medium F-12 obtained from Welgene (Daegu, Korea) at 37°C under 5% CO2 conditions. A549 cells were obtained from the Korean Cell Line Bank (KCLB, Seoul, Korea) and maintained in Roswell Park Memorial Institute (RPMI) 1640 medium with 2 mM L-glutamine and 10% fetal bovine serum (FBS) at 37°C under 5% CO2 conditions.
mRNA sequencing
Total RNA was extracted and an mRNA sequencing (mRNA-seq) library was prepared using the TruSeq Stranded mRNA kit (Illumina, San Diego, CA, USA). Polyadenylated mRNAs were purified using poly-T oligo-coupled magnetic beads. mRNAs were then fragmented using divalent cations under elevated temperature conditions. The fragmented RNA was subsequently used for first- and second-strand cDNA synthesis using reverse transcriptase with random primers and DNA polymerase I. These cDNA fragments were purified and enriched by polymerase chain reaction (PCR) to create a cDNA library. Each constructed library was sequenced using an Illumina NextSeq500 instrument (Illumina). The original image data were converted into sequence data and stored in the FASTQ format. Genes showing an absolute fold change of at least 2 and with a false discovery rate (FDR) < 0.05 between the groups were considered to be differentially expressed. Gene set enrichment was evaluated using EnrichR.28
RNA interference
Small interfering RNA (siRNA) experiments were performed according to previously published methods.29 Transfection of APRE-19 cells was performed with Oligofectamine (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The following siRNA sequences were used: siFOXA1(1): 5′-GAGAGAAAAAAUCAACAGCdTdT- 3′ (sense) and 5′-GCUGUUGAUUUUUUCUCUCdTdT-3′ (anti-sense); siFOXA1(2): 5′-GCGAAGUUUAAUGAUCCACdTdT-3′ (sense) and 5′-GUGGAUCAUUAAACUUCGCdTdT-3′ (anti-sense); siAP-1(1): 5′-CGGACCUUAUGGCUACAGUdTdT-3′ (sense) and 5′-ACUGUAGCCAUAAGGUCCGdTdT-3′ (anti-sense); siAP-1(2): 5′-GGCAUGUGCUGUGAUCAUUdTdT-3′ (sense) and 5′-AAUGAUCACAGCACAUGCCdTdT-3′ (anti-sense); and siNS: 5′- UUCUCCGAACGUGUCACGUdTdT-3′ (sense) and 5′-ACGUGACACGUUCGGAGAAdTdT-3′ (anti-sense).
Quantitative RT-PCR (RT-qPCR)
ARPE-19 and A549 cells were treated with Dex (100 nM) for 16 or 24 h; then, the total RNA was extracted using Trizol (Invitrogen, Carlsbad, CA, USA) and reverse transcribed (RT) using an iScript cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA, USA) in a total volume of 20 μL. The RT product was used for qPCR analysis with specific primers. The RT-qPCR primer sequences are listed in Table 1.
Table 1.
Primers used for RT-qPCR experiments.
| Name | Forward | Reverse |
|---|---|---|
| ENACα | AACGGTCTGTCCCTGATGCT | TTGGTGCAGTCGCCATAATC |
| 18S | GAGGATGAGGTGGAACGTGT | TCTTCAGTCGCTCCAGGTCT |
| FKBP5 | AGGCTGCAAGACTGCAGATC | CTTGCCCATTGCTTTATTGG |
| GILZ | AGATCGAACAGGCCATGGAT | TTACACCGCAGAACCACCAG |
ENACα: epithelial sodium channel; FKBP5: FK506 binding protein 5; GILZ: glucocorticoid-induced leucine zipper.
Statistical analysis
The P-values for pathway analysis (Kyoto Encyclopedia of Genes and Genomes (KEGG)) were obtained using Fisher’s extract test, which is a proportion test that assumes a binomial distribution and independence for the probability of any gene to belong to any set.28 Statistical analysis of the RT-qPCR results was performed using a Student’s two-tailed t-test; *P < 0.05 represents a significant difference (vs vehicle-treated control, n = 3).
Results
Differential gene expression mediated by GR in ARPE-19 and A549 cells
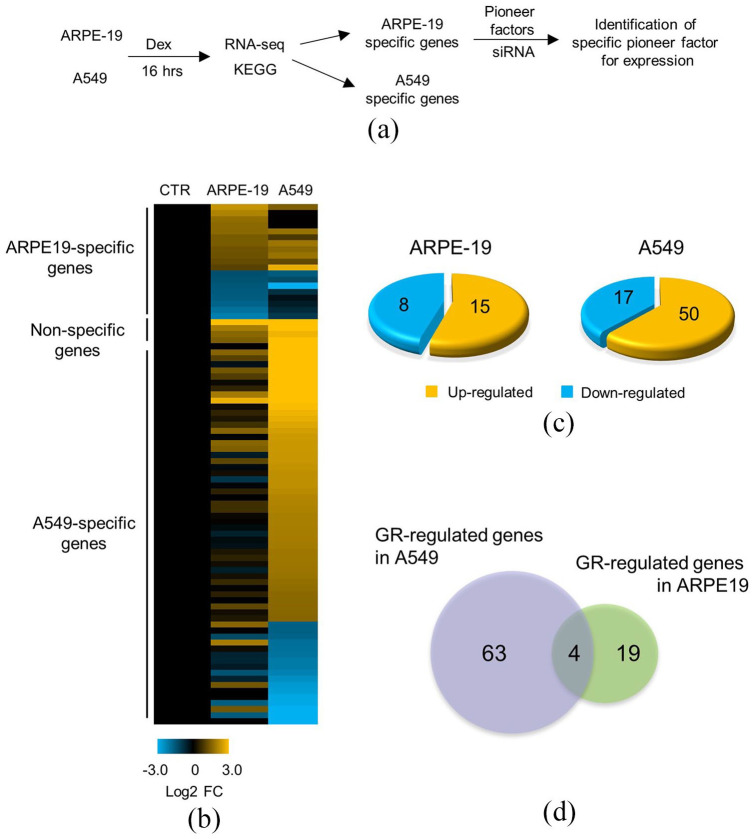
To explore the differing patterns of GR-mediated gene expression in two GR-responsive cell lines, ARPE-19 and A549, transcriptional analysis was performed on these cells following their treatment with Dex (Figure 1(a)). Eighty-six genes were differentially expressed between the two cell lines (Figure 1(b)). There were four genes that showed altered expression following Dex treatment, when compared to the control, in both cell lines. In ARPE-19 cells, the expression of 15 and 8 genes was increased and decreased, respectively, compared to the control (Figure 1(c), left panel). In A549 cells, 50 and 17 genes were upregulated and downregulated, respectively, compared to the control (Figure 1(c), right panel). RNA-seq analysis revealed that there were 19 ARPE-specific Dex-regulated genes. Moreover, while 63 genes were found to be A549-specific, four genes showed significantly different expression in both cell lines (Figure 1(d)). Based on the RNA-seq analysis, we confirmed the presence of cell-specific genes regulated by GR in ARPE-19 and A549 cells. These results suggest that the GR induced gene expression is cell type-specific.
Figure 1.
Differential gene expression induced by GR in ARPE-19 and A549 cells: (a) experimental setup for the identification of cell type–specific genes. (b) Heat map generated from RNA-seq analysis of ARPE-19 and A549 cells treated with Dex (100 nM) for 16 h, showing the differential expression of GR-responsive genes between the two groups. Values represent the log2 fold change (FC) relative to the vehicle-treated control (CTR). GR target genes are divided into three groups: ARPE-19-specific genes, A549-specific genes, and genes regulated by GR in both cell lines (non-specific genes). (c) The number of genes that were upregulated or downregulated by GR in ARPE-19 or A549 cells. (d) Venn diagram showing the number of genes that are ARPE-19-specific, A549-specific, and non-specific.
Activation of cell type–specific signaling pathways by GR
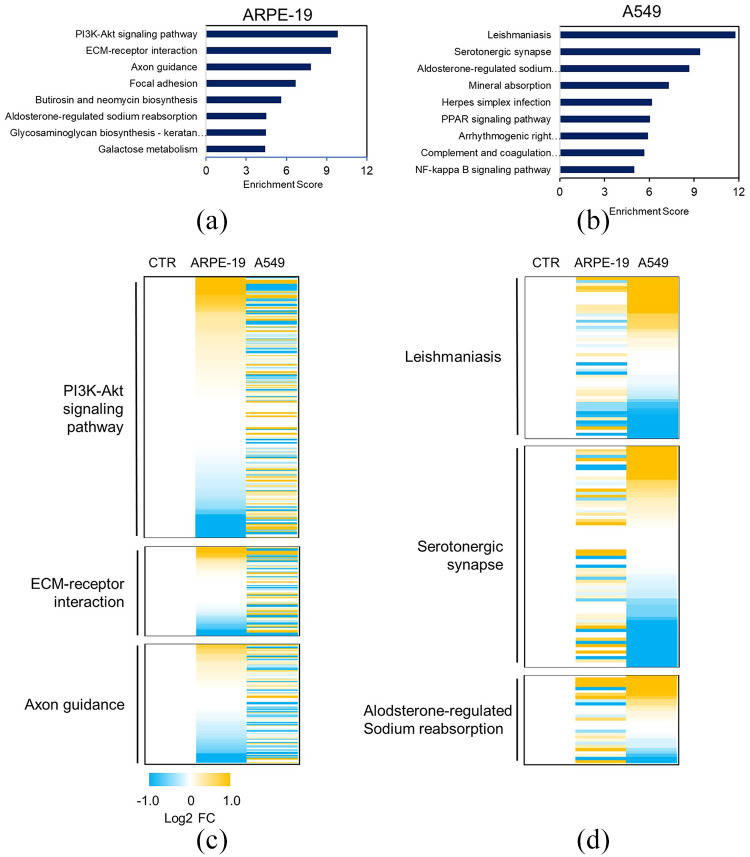
To confirm the expression pattern of the cell type–specific genes, we performed pathway analysis using genes regulated by GR in A549 and ARPE-19 cells. Pathway analysis (KEGG) was performed using EnrichR and revealed that GR was associated with different pathways in ARPE-19 and A549 cells. Specifically, genes that were differentially regulated by GR in ARPE-19 cells were enriched for the PI3K-Akt signaling pathway (P = 0.0076), extracellular matrix (ECM)–receptor interaction (P = 0.004), and axon guidance (P = 0.010) (Figure 2(a) and Supplemental Figure S1). In contrast, the genes that were differentially expressed following Dex treatment in A549 cells were enriched for leishmaniosis (P = 0.0019), serotonergic synapse (P = 0.0063), and aldosterone-regulated sodium reabsorption (P = 0.0076) (Figure 2(b)). From the KEGG pathway analysis, we selected a list of genes related to the three pathways with the highest enrichment scores from the RNA-seq results from each cell line. A heat map was generated to represent the significantly different gene expression patterns of the two cell lines after Dex treatment (Figure 2(c) and (d)). This suggests that different sets of genes are expressed in a cell type–specific manner after Dex treatment, which could be mediated by different mechanisms.
Figure 2.
Activation of cell type–specific signaling pathways by GR. (a, b) Among the GR-responsive genes, ARPE-19-specific and A549-specific genes were categorized according to gene function via KEGG pathway analysis. Statistically significant pathways (P < 0.05) are listed. (c, d) Heat map showing the expression level of genes belonging to major pathways identified through KEGG analysis of GR target genes in ARPE-19 and A549 cells.
Identification of cell type–specific genes regulated by GR
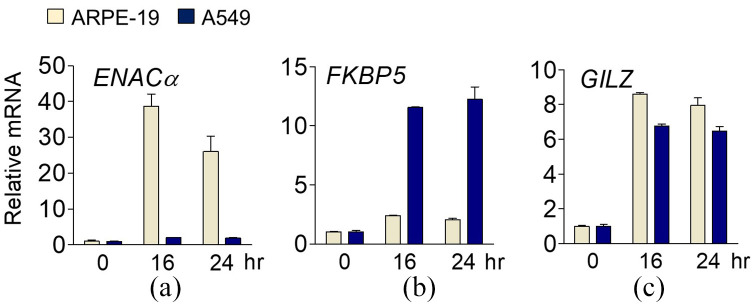
After identifying the presence of cell type–specific genes using transcriptome analysis, RT-qPCR was used to validate the genes selectively expressed in A549 and ARPE-19 cells, along with the genes regulated by GR in both cell types. After 16 and 24 h of Dex treatment in ARPE-19 cells, epithelial sodium channel (ENACα) expression increased by 38.6-fold and 26-fold, respectively, compared to the vehicle-treated control. In A549 cells, there was only a 2-fold and 1.9-fold increase in ENACα expression after 16 and 24 h of Dex treatment, respectively (Figure 3(a) and Supplemental Figure S2). Thus, ENACα was defined as an ARPE-19-specific GR target gene. In contrast, FK506 binding protein 5 (FKBP5) showed opposite expression patterns in both cell lines. ARPE-19 cells treated with Dex for 16 and 24 h exhibited 2.4-fold and 2.1-fold higher FKBP5 expression, respectively, compared to the control group. In contrast, the increase in FKBP5 expression in the A549 cells was 11.6-fold and 12.3-fold after 16 and 24 h of Dex treatment, respectively (Figure 3(b)). This suggests that FKBP5 is A549-specific. Unlike the above two cases, the expression of glucocorticoid-induced leucine zipper (GILZ) by Dex was significantly increased in both cell lines (Figure 3(c)).
Figure 3.
Identification of cell type–specific genes regulated by GR. (a, c) Expression of GR target genes (ENACα, FKBP5, and GILZ) in ARPE-19 and A549 cells determined by RT-qPCR. Cells were treated with Dex (100 nM) for 16 or 24 h before being harvested. Total RNA was analyzed by RT-qPCR. Levels of all mRNAs were normalized to that of 18S rRNA.
FOXA1 acts as a pioneer factor for ARPE-19-specific ENACα expression
Among the genes identified by RT-qPCR, ENACα was specifically expressed in ARPE-19 cells. With regard to ENACα expression, we hypothesized that there might be an underlying mechanism responsible for differential gene regulation in ARPE-19 and A549 cells, and that this might be attributed to a pioneer factor. To investigate the pioneer factor potentially involved in ENACα expression induced by Dex in ARPE-19 cells, we observed changes in the Dex-induced expression of ENACα after knocking down FOXA1 or AP-1 in ARPE-19 cells.
ARPE-19 cells deficient in FOXA1 showed significantly reduced ENACα expression after treatment with Dex for 8 or 24 h compared to the untreated group (Figure 4(a)). In contrast, AP-1 knockdown in ARPE-19 cells resulted in no statistically significant change in ENACα expression (Figure 4(b)). These results suggest that FOXA1 plays a role as a pioneer factor to enable selective expression of ENACα in ARPE-19 cells, but not in A549 cells.
Figure 4.
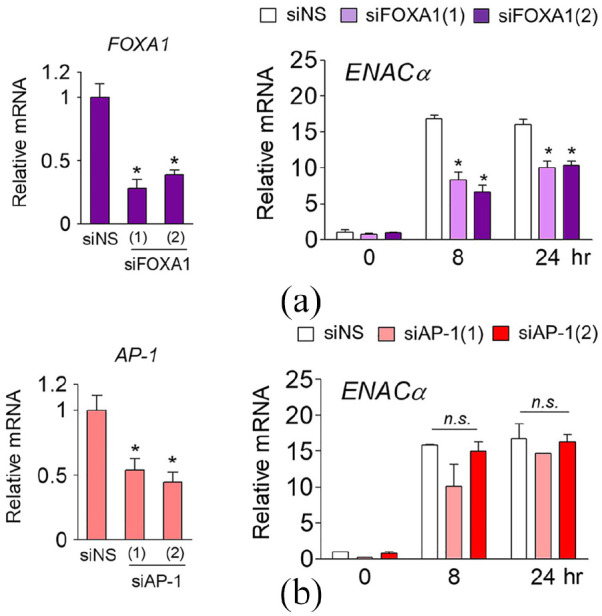
FOXA1 is a pioneer factor mediating ARPE-19-specific expression of ENACα (a) ARPE-19 cells were transfected with non-specific siRNA (siNS) or two siRNAs targeting different sites of the FOXA1 mRNA (siFOXA1(1) and siFOXA1(2)) to knock down FOXA1 (left panel). Cells were treated with Dex (100 nM) for 8 or 24 h before being harvested, and ENACα expression in ARPE-19 cells was assessed by RT-qPCR. Total RNA was analyzed by RT-qPCR. All mRNA levels were normalized to that of 18S rRNA (right panel). (b) The expression of ENACα in AP-1-depleted ARPE-19 cells (left panel) was assessed by RT-qPCR. Total RNA was analyzed by RT-qPCR. All mRNA levels were normalized to that of 18S rRNA (right panel).
n.s.: not significant.
*P < 0.05.
Discussion
Most GR-binding sites are constitutively open and accessible. However, the binding of pioneer factors to specific sites on genomic loci causes local chromatin accessibility and promotes the recruitment of other transcription factors. FOXA1 has been studied as a pioneer factor that interacts with steroid receptors to bind to specific genomic loci. In particular, FOXA1 is the main factor interacting with ER during the development of breast cancer.23,30–32 FOXA1-mediated regulation of GR has been identified from the analysis of the MMTV promoter region, a chromatin domain regulated by GR. FOXA1 alters the chromatin structure, regardless of whether GR binds to MMTV.24 The binding of GR is enhanced by FOXA1, which optimizes the chromatin structure and mediates the binding of additional transcription factors associated with GR. In a FOXA1-dependent manner, FOXA1 and GR recruit related transcription factors, including NF1 and Oct1, leading to in an increase in transcriptional activity of the genes.24 The interaction between NRs and transcription factors can be either dependent or independent of DNA binding. Mechanisms independent of DNA binding include the binding between GR and AP-1, a pro-inflammatory transcription factor, which is mediated through protein–protein interactions in the absence of GR responsive element (GRE). AP-1 also maintains the accessible chromatin structure, allowing the selective access of GR to specific sites. The loss of AP-1 significantly reduces the binding of GR to the DNA.33,34 This could be due to the fact that GR must bind with AP-1 to bind the DNA or because of the increased chromatin accessibility established by the activation of AP-1. In addition, AP-1 is recruited to the TR responsive element (TRE) inflammatory gene to induce transcription and serves as a pioneer for the subsequent recruitment of GR.33,34 Regulators that involve in GR binding may be cell type-specific. The AP-1 motif was identified at the GR-binding site in hepatocytes, and the enrichment of motifs for other proteins (e.g. SP1 and forkhead motifs) was confirmed.35 These results suggest that the role of this factor in the binding of GR to the chromatin is to provide cell type–associated specificity. In this study, we demonstrated that different genes are expressed in different cell types in response to a common activator (GR), resulting in the activation of different biological pathways. In addition, various transcriptional mechanisms are involved in different cell types to control the expression of the same gene, suggesting that pioneer factors play an important role in these processes. One limitation of our study was that we used only two cell lines in our RNA-seq experiment. Follow-up studies can also aim to identify if there are additional patterns of GR-mediated gene expression in various tissue-specific cell types. Another limitation of this study is that the existence of intracellular signaling pathways coupled with epigenetic regulators (e.g. coactivators or corepressors) of cell type–specific gene expression was not considered. For example, receptor-mediated intracellular signaling cascades such as SGK1, ERK, and PKA have been reported as a mechanism regulating the expression of ENAC.36,37 Most recently, GR has been reported to activate transcription via the direct binding of a 1.3-kilobase portion of the ENACα gene with CARM1 and p300.38
Conclusion
Our findings indicate that the expression of ENACα in ARPE-19 cells is regulated via FOXA1 and provide insights into the molecular mechanisms of the cell type–specific expression of GR-regulated genes. We also highlight the potential for the treatment of clinical diseases (e.g. liddle syndrome, ulcerative colitis, and pulmonary edema) due to the imbalance of ENAC function.
Supplemental Material
Supplemental material, 2020_05_18_A549_ARPE19_Supple_Figures_R1 for Cell-specific expression of ENACα gene by FOXA1 in the glucocorticoid receptor pathway by Young Sun Chung, Hong Lan Jin and Kwang Won Jeong in International Journal of Immunopathology and Pharmacology
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was supported by the Basic Science Research Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education (2014R1A1A2056066) and by the Gachon University research fund of 2018 (GCU-2018-0665).
ORCID iD: Kwang Won Jeong  https://orcid.org/0000-0003-2258-8354
https://orcid.org/0000-0003-2258-8354
Supplemental material: Supplemental material for this article is available online.
References
- 1. Zheng Y, Murphy LC. (2016) Regulation of steroid hormone receptors and coregulators during the cell cycle highlights potential novel function in addition to roles as transcription factors. Nucl Recept Signal 14: e001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kadmiel M, Cidlowski JA. (2013) Glucocorticoid receptor signaling in health and disease. Trends Pharmacol Sci 34: 518–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Koenen M, Culemann S, Vettorazzi S, et al. (2018) Glucocorticoid receptor in stromal cells is essential for glucocorticoid-mediated suppression of inflammation in arthritis. Ann Rheum Dis 77(11): 1610–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Al-Zamil WM, Yassin SA. (2017) Recent developments in age-related macular degeneration: A review. Clin Interv Aging 12: 1313–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Galloway CA, Dalvi S, Hung SSC, et al. (2017) Drusen in patient-derived hiPSC-RPE models of macular dystrophies. Proc Natl Acad Sci U S A 114: E8214–E8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Diniz B, Rodger DC, Chavali VR, et al. (2015) Drusen and RPE atrophy automated quantification by optical coherence tomography in an elderly population. Eye 29: 272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ozturk Aktas O, Sekerel BE, Kalyoncu AF. (2017) Glucocorticoid sparing of benralizumab in asthma. New Engl J Med 377: 1204–1205. [DOI] [PubMed] [Google Scholar]
- 8. Hajialilo M, Ghorbanihaghjo A, Valaee L, et al. 2016. A double-blind randomized comparative study of triamcinolone hexacetonide and dexamethasone intra-articular injection for the treatment of knee joint arthritis in rheumatoid arthritis. Clin Rheumatol 35(12): 2887–2891. [DOI] [PubMed] [Google Scholar]
- 9. Wandler AM, Huang BJ, Craig JW, et al. (2020) Loss of glucocorticoid receptor expression mediates in vivo dexamethasone resistance in T-cell acute lymphoblastic leukemia. Leukemia. Epub ahead of print 17 February. DOI: 10.1038/s41375-020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Love MI, Huska MR, Jurk M, et al. (2017) Role of the chromatin landscape and sequence in determining cell type-specific genomic glucocorticoid receptor binding and gene regulation. Nucleic Acids Res 45: 1805–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Taves MD, Mittelstadt PR, Presman DM, et al. (2019) Single-cell resolution and quantitation of targeted glucocorticoid delivery in the thymus. Cell Rep 26: 3629–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jozwik KM, Carroll JS. (2012) Pioneer factors in hormone-dependent cancers. Nature 12: 381–385. [DOI] [PubMed] [Google Scholar]
- 13. Zaret KS, Carroll JS. (2011) Pioneer transcription factors: Establishing competence for gene expression. Genes Dev 25: 2227–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. El-Mayet FS, Sawant L, Thunuguntla P, et al. (2020) Two pioneer transcription factors, Kruppel-like transcription factor 4 and glucocorticoid receptor, cooperatively transactivate the bovine herpesvirus 1 ICP0 early promoter and stimulate productive infection. J Virol 94: e01670-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ye Z, Chen Z, Sunkel B, et al. (2016) Genome-wide analysis reveals positional-nucleosome-oriented binding pattern of pioneer factor FOXA1. Nucleic Acids Res 44: 7540–7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jiang G, Wang X, Sheng D, et al. (2019) Cooperativity of co-factor NR2F2 with Pioneer Factors GATA3, FOXA1 in promoting ERalpha function. Theranostics 9: 6501–6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang L, Tian S, Pei M, et al. (2019) Crosstalk between histone modification and DNA methylation orchestrates the epigenetic regulation of the costimulatory factors, Tim3 and galectin9, in cervical cancer. Oncol Rep 42(6): 2655–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sasidharan Nair V, El Salhat H, Taha RZ, et al. (2018) DNA methylation and repressive H3K9 and H3K27 trimethylation in the promoter regions of PD-1, CTLA-4, TIM-3, LAG-3, TIGIT, and PD-L1 genes in human primary breast cancer. Clin Epigenetics 10: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao Q, Zhang J, Chen R, et al. (2016) Dissecting the precise role of H3K9 methylation in crosstalk with DNA maintenance methylation in mammals. Nat Commun 7: 12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jing D, Huang Y, Liu X, et al. (2018) Lymphocyte-specific chromatin accessibility pre-determines glucocorticoid resistance in acute lymphoblastic leukemia. Cancer Cell 34: 906–921.e8. [DOI] [PubMed] [Google Scholar]
- 21. Stavreva DA, Coulon A, Baek S, et al. (2015) Dynamics of chromatin accessibility and long-range interactions in response to glucocorticoid pulsing. Genome Res 25(6): 845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hurtado A, Holmes Ross-Innes CS, Schmidt D, et al. (2011) FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 43(1): 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takaku M, Grimm SA, De Kumar B, et al. (2020) Cancer-specific mutation of GATA3 disrupts the transcriptional regulatory network governed by Estrogen Receptor alpha, FOXA1 and GATA3. Nucleic Acids Res 48: 4756–4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Whirledge S, Kisanga EP, Taylor RN, et al. (2017) Pioneer factors FOXA1 and FOXA2 assist selective glucocorticoid receptor signaling in human endometrial cells. Endocrinology 158: 4076–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mihailidou C, Panagiotou C, Kiaris H, et al. (2016) Crosstalk between C/EBP homologous protein (CHOP) and glucocorticoid receptor in lung cancer. Mol Cell Endocrinol 436: 211–223. [DOI] [PubMed] [Google Scholar]
- 26. Swinstead EE, Miranda TB, Paakinaho V, et al. (2016) Steroid receptors reprogram FoxA1 occupancy through dynamic chromatin transitions. Cell 165: 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hirose I, Kanda A, Noda K, et al. (2019) Glucocor-ticoid receptor inhibits Muller glial galectin-1 expression via DUSP1-dependent and -independent deactivation of AP-1 signalling. J Cell Mol Med 23: 6785–6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kuleshov MV, Jones MR, Rouillard AD, et al. (2016) Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44: W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang L, Jeong KW. (2019) Flightless-I mediates the repression of estrogen receptor alpha target gene expression by the glucocorticoid receptor in MCF-7 cells. Endocrine J 66: 65–74. [DOI] [PubMed] [Google Scholar]
- 30. Jeong KW, Andreu-Vieyra C, You JS, et al. (2014) Establishment of active chromatin structure at enhancer elements by mixed-lineage leukemia 1 to initiate estrogen-dependent gene expression. Nucleic Acids Res 42(4): 2245–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jing X, Liang H, Hao C, et al. (2019) Analyses of an epigenetic switch involved in the activation of pioneer factor FOXA1 leading to the prognostic value of estrogen receptor and FOXA1 co-expression in breast cancer. Aging 11: 7442–7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kawase M, Toyama T, Takahashi S, et al. (2015) FOXA1 expression after neoadjuvant chemotherapy is a prognostic marker in estrogen receptor-positive breast cancer. Breast Cancer 22(3): 308–316. [DOI] [PubMed] [Google Scholar]
- 33. Li L, Liu C, Mao W, et al. (2019) Taurochenodeoxycholic acid inhibited AP-1 activation via stimulating glucocorticoid receptor. Molecules 24: 4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu L, Aleksandrowicz E, Schonsiegel F, et al. (2017) Dexamethasone mediates pancreatic cancer progression by glucocorticoid receptor, TGFbeta and JNK/AP-1. Cell Death Dis 8: e3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zou H, Jiang Z, Li R, et al. (2013) p53 cooperates with Sp1 to regulate breed-dependent expression of glucocorticoid receptor in the liver of preweaning piglets. PLoS ONE 8(8): e70494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. He J, Qi D, Tang XM, et al. (2019) Rosiglitazone promotes ENaC-mediated alveolar fluid clearance in acute lung injury through the PPARgamma/SGK1 signaling pathway. Cell Mol Biol Lett 24: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Niisato N, Ohta M, Eaton DC, et al. (2012) Hypotonic stress upregulates beta- and gamma-ENaC expression through suppression of ERK by inducing MKP-1. Am J Physiol Renal Physiol 303: F240–F252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bittencourt D, Wu DY, Jeong KW, et al. (2012) G9a functions as a molecular scaffold for assembly of transcriptional coactivators on a subset of glucocorticoid receptor target genes. Proc Natl Acad Sci U S A 109: 19673–19678. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, 2020_05_18_A549_ARPE19_Supple_Figures_R1 for Cell-specific expression of ENACα gene by FOXA1 in the glucocorticoid receptor pathway by Young Sun Chung, Hong Lan Jin and Kwang Won Jeong in International Journal of Immunopathology and Pharmacology