Abstract
Systemic chemotherapy is identified as a curative approach to prolong the survival time of patients with colorectal cancer (CRC). Although great progress in therapeutic approaches has been achieved during the last decades, drug resistance still extensively persists and serves as a major hurdle to effective anticancer therapy for CRC. The mechanism of multidrug resistance remains unclear. Recently, mounting evidence suggests that a great number of microRNAs (miRNAs) may contribute to drug resistance in CRC. Certain of these miRNAs may thus be used as promising biomarkers for predicting drug response to chemotherapy or serve as potential targets to develop personalized therapy for patients with CRC. This review mainly summarizes recent advances in miRNAs and the molecular mechanisms underlying miRNA-mediated chemoresistance in CRC. We also discuss the potential role of drug resistance-related miRNAs as potential biomarkers (diagnostic and prognostic value) and envisage the future orientation and challenges in translating the findings on miRNA-mediated chemoresistance of CRC into clinical applications.
Keywords: biomarkers, colorectal cancer, drug resistance, miRNAs, targeted therapy
Introduction
Colorectal cancer (CRC) ranks the third for cancer incidence and fourth leading cause for cancer deaths worldwide.1 In 2018, there will be over 1.8 million new cases of CRC with approximately 881,000 deaths in the same year across the world, accounting for about 10% of cancer cases and deaths.2 CRC is a complex and heterogeneous disease with a multitude of genetic and epigenetic factors involved in the development of CRC.3 Despite great progress having been made in the detection, surgical resection, and adjuvant therapy of CRC, the mortality of CRC patient still remains relatively high. Therefore, it is imperative for researchers to elucidate the pathogenesis of CRC, further to develop more effective therapeutic measures and improve prognosis.
Chemotherapy is an available curative approach for CRC since most advanced patients cannot benefit from surgical resection or radiotherapy.4 However, the evolution of drug resistance contributes to therapy failure in CRC patients. Multidrug resistance (MDR), defined as the resistance of cancer cells to a variety of structurally and functionally unrelated drugs, can occur naturally or is acquired.5 The main mechanisms of intrinsic or acquired MDR include, but are not limited to, overexpression of MDR transporters, alterations in the regulation of cell cycle and checkpoints, dysregulation on the apoptotic and autophagy machinery, alteration in DNA repair and drug targets, enhancing drug metabolism, etc.6 In addition, cancer stem cells (CSCs) may also contribute to drug resistance.7 Nevertheless, the concrete mechanisms refer to MDR have not been fully clarified. For the sake of reversing chemoresistance, many researchers fling themselves into exploring the concrete mechanisms underlying drug resistance. More recently, accumulating evidence has reported that epigenetic modification such as miRNA dysregulation was significantly associated with the phenomenon of drug resistance.8,9
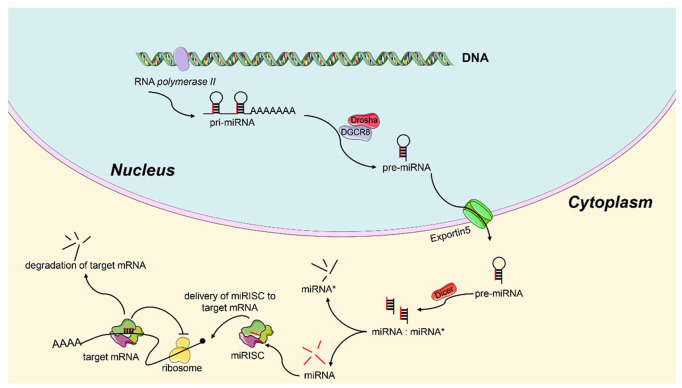
miRNAs, a subpopulation of noncoding RNAs, function as posttranscriptional regulatory elements consisting of 21–23 short nucleotides.10 The biogenesis mechanism of miRNAs has been well understood (Figure1).11 In brief, genes that encode miRNAs are transcribed primarily by RNA polymerase II in nucleus and create primary miRNA (pri-miRNA), which is subsequently processed by DiGeorge syndrome critical region 8 (DGCR8) protein associated with the Drosha ribonuclease III (Drosha) enzyme into a shorter stem-loop-structured precursor miRNA (pre-miRNA) (70–100 nucleotides).12 Then, the pre-miRNA is transported into cytoplasm by exportin-5. Next, the Dicer RNase III enzyme processes the pre-miRNA and produces the dsRNAs (18~25 nucleotides in length). One strand of dsRNAs, termed miRNA*, is degraded, another strand of the dsRNAs that serves as the guide strand is incorporated into RNA-induced silencing complex (RISC) and forms miRISC, which then targets the 3′-untranslated regions (UTRs) of its target mRNAs and regulates genes expression either by attenuating RNA translation or by promoting mRNA degradation.13,14 miRNAs play crucial roles in biological and pathological processes, encompassing metabolism, apoptosis, differentiation, cell proliferation, cell cycle as well as MDR, etc.15–17 miRNAs exhibit differential expression in many malignancies, including CRC,18 and function as either tumor suppressors or oncogenes.19 Emerging evidence shed light on the role of miRNAs in drug resistance of CRC and revealed some critical mechanisms involved in MDR.20 Aberrant expression of miRNAs can affect the expression and function of MDR-related mRNAs and proteins, further influencing the chemosensitivity of CRC cells. For example, miR-24, the first identification of chemoresistance-related miRNA, could contribute to methotrexate resistance via binding site polymorphism in the dihydrofolate reductase gene.21 In addition, forced expression of miR-24 can also reverse paclitaxel (PTX) resistance of breast cancer by targeting ATP binding cassette subfamily B Member 9 (ABCB9), indicating that miR-24 may serve as a promising biomarker to predict drug response and an effective target for cancer treatment.22 Therefore, miRNAs may serve as novel promising biomarkers and therapeutic targets in reversing chemoresistance of CRC.
Figure 1.
Schematic diagram of the biogenesis of miRNAs.
In the nucleus, the pri-miRNA transcript is firstly produced by RNA polymerase II. Then, the pri-miRNA is processed by the Drosha associates with DGCR8 and becomes pre-miRNA. Next, the pre-miRNA is transported into the cytoplasm by Exportin5. In the cytoplasm, the pre-miRNA further processed by the Dicer RNase III enzyme to form a mature miRNA.
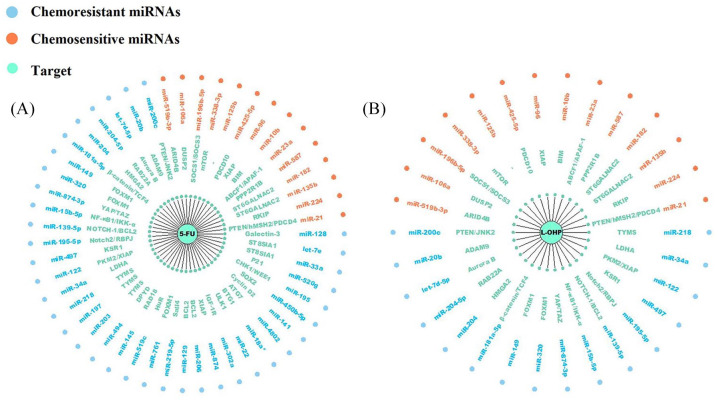
In the present review, a systematic electronic literature search of the Web of Science (http://apps.webofknowledge.com), PubMed (https://pubmed.ncbi.nlm.nih.gov), and EMBASE (http://www.embase.com) databases was conducted. Specific search strategies for each database were developed by using different combinations and variations of search terms, including ‘colorectal cancer’, ‘colon cancer’, ‘rectal cancer’, ‘drug resistance’, ‘chemoresistance’, ‘microRNA’, ‘miRNA’ and their variants. We summarize results from the published literatures on the role of miRNAs and their targets in drug-resistant CRC (Tables 1 and 2). Some of them even were regarded as potential biomarkers in CRC patients (Table 3). Furthermore, considering that 5-fluorouracil (5-FU) and oxaliplatin (L-OHP) are the most important first-line chemotherapeutic drugs for CRC, we summarize related miRNAs and their targets in Figure 2.
Table 1.
miRNAs mediate drug resistance via cellular signaling pathways in CRC.
| Mechanisms | Reported miRNA | Alteration | Validated miRNA targets | Corresponding drugs | Reference |
|---|---|---|---|---|---|
| PI3K/AKT signaling pathway | miR-17-5p | Up | PTEN | 5-FU, L-OHP and Irinotecan | Fang et al.23 |
| miR-21 | Up | PTEN | 5-FU | Si et al.24 | |
| miR-200c | Down | PTEN | 5-FU | Heydari et al.25 | |
| miR-7 | Up | EGFR and RAF-1 | Cetuximab | Suto et al.26 | |
| miR-20b | Down | ADAM9 | 5-FU | Fu et al.27 | |
| let-7 | Down | Ras | 5-FU | Salendo et al.28 | |
| miR-224 | Up | RKIP | 5-FU | Amankwatia et al.29 | |
| miR-204-5p | Down | RAB22A | L-OHP, 5-FU and CDDP | Yin et al.30 | |
| miR-204 | Down | HMGA2 | 5-FU | Wu et al.31 | |
| miR-194 | Down | VAPA | L-OHP, irinotecan | Chang et al.32 | |
| miR-135b and miR-182 | Up | ST6GALNAC2 | 5-FU | Liu et al.33 | |
| miR-587 | Up | PPP2R1B | 5-FU | Zhang et al.34 | |
| miR-199a-5p and miR-375 | Up | PHLPP1 | Cetuximab | Mussnich et al.35 | |
| Wnt/β-catenin signaling pathway | miR-181a-5p | Down | β-catenin/TCF4 | 5-FU | Han et al.36 |
| miR-149 | Down | FOXM1 | 5-FU | Liu et al.37 | |
| miR-320 | Down | FOXM1 | 5-FU, L-OHP | Wan et al.38 | |
| miR-203a-3p | Down | β-catenin, GRG5 | CDDP, PTX | Xiao et al.39 | |
| miR-224 | Up | GSK-3beta | ADM | Liang et al.40 | |
| miR-199a/b | Up | GSK-3β | CDDP | Chen et al.41 | |
| miR-100 | Up | DKK1 and ZNRF3 | Cetuximab | Lu et al.42 | |
| miR-125b | Up | ZNRF3, RNF43, DKK3 and APC2 | Cetuximab | Lu et al.42 | |
| TGF-β signaling pathway | miR-19b-3p | Up | SMAD4 | L-OHP | Jiang et al.43 |
| miR-34a | Down | SMAD4 | L-OHP | Sun et al.44 | |
| Hippo signaling pathway | miR-874-3p | Down | YAP and TAZ | 5-FU | Que et al.45 |
| NF-κB signaling pathway | miR-15b-5p | Down | NF-κB1, IKK-α | 5-FU | Zhao et al.46 |
| Notch signaling pathway | miR-139-5p | Down | NOTCH-1 | 5-FU | Liu et al.47 |
| miR-195-5p | Down | Notch2 and RBPJ | 5-FU | Xu et al.48 | |
| Raf/MEK/ERK signaling pathway | miR-497 | Down | KSR1 | 5-FU | Wang et al.49 |
ADM, adriamycin; AKT, Ser/Thr kinase; CDDP, cisplatin; CRC, colorectal cancer; 5-FU, 5-fluorouracil; L-OHP, oxaliplatin; miRNAs, microRNAs; NF-κB, nuclear factor kappa B; PI3K, phosphatidylinositol 3-kinase; PTX, paclitaxel; TGF-β, transforming growth factor beta.
Table 2.
Summary of reported drug resistance-related miRNAs and their target genes in CRC.
| Mechanisms | Reported miRNA | Alteration | Validated miRNA targets | Corresponding drugs | Ref. | |
|---|---|---|---|---|---|---|
| Aberrant metabolism | glucose metabolism | miR-122 | Down | PKM2 | 5-FU | He et al.50 |
| miR-34a | Down | LDHA | 5-FU | Li et al.51 | ||
| TS and dihydropyrimidine dehydrogenase | miR-218 | Down | TYMS | 5-FU | Li et al.52 | |
| miR-197 | Down | TYMS | 5-FU | Sun et al.53 | ||
| miR-203 | Down | TYMS | 5-FU | Li et al.54 | ||
| miR-494 | Down | DPYD | 5-FU | Chai et al.55 | ||
| DNA damage repair and MMR | miR-145 | Down | RAD18 | 5-FU | Liu et al.56 | |
| miR-21 | Up | hMSH2 | 5-FU | Deng et al.57 | ||
| miR-203 | Up | ATM | L-OHP | Zhou et al.58 | ||
| ABC transporters family | miR200c | Down | JNK2 | L-OHP,5-FU, CDDP, VCR | Sui et al.59 | |
| miR-302c-5p | Down | ABCB1 | L-OHP | Ghanbarian et al.60 | ||
| miR-451 | Down | ABCB1 | irinotecan | Bitarte et al.61 | ||
| miR-519c | Down | HuR | 5-FU, irinotecan | To et al.62 | ||
| miR-21 | Up | PDCD4 | 5-FU | Wu et al.63 | ||
| miR-23a | Up | ABCF1 | 5-FU | Li et al.64 | ||
| miR-761 | Down | FOXM1 | 5-FU | Cao et al.65 | ||
| miR-522 | Down | ABCB5 | DOX | Yang et al.66 | ||
| miR-506 | Down | MDR1/P-gp | L-OHP | Zhou et al.67 | ||
| miR-219-5p | Down | Sall4 | L-OHP,5-FU | Cheng et al.68 | ||
| miR-297 | Down | MRP-2 | L-OHP, VCR | Xu et al.69 | ||
| miR-34a | Down | OAZ2 | L-OHP | Li et al.70 | ||
| Apoptosis-related pathway | BCL-2 family | miR-1915 | Down | BCL2 | L-OHP | Xu et al.71 |
| miR-139-5p | Down | BCL2 | L-OHP,5-FU | Li et al.72 | ||
| miR-129 | Down | BCL2 | 5-FU | Karaayvaz et al.73 | ||
| miR-206 | Down | BCL2 | 5-FU | Meng et al.74 | ||
| miR-20a | Up | ASK1 | CDDP | Zhang et al.75 | ||
| miR-195 | Down | BCL2L2 | DOX | Qu et al.76 | ||
| miR-20a | Up | BID | TRAIL | Huang et al.77 | ||
| miR-10b | Up | BIM | 5-FU | Nishida et al.78 | ||
| XIAP | miR-874 | Down | XIAP | 5-FU | Han et al.79 | |
| miR-122 | Down | XIAP | L-OHP | Hua et al.80 | ||
| miR-96 | Up | XIAP | 5-FU | Kim et al.81 | ||
| IGF1R pathway | miR-302a | Down | IGF-1R | 5-FU | Liu et al.82 | |
| miR-143 | Down | IGF-1R | L-OHP | Qian et al.83 | ||
| Other apoptosis-related pathway | miR-27a | Up | Apaf-1 | TRAIL | Zhang et al.84 | |
| miR-23a | Up | APAF-1 | 5-FU | Shang et al.85 | ||
| miR-128 | Down | SIRT1 | TRAIL | Lian et al.86 | ||
| miR-425-5p | Up | PDCD10 | L-OHP,5-FU | Zhang et al.87 | ||
| miR-153 | Up | FOXO3a | L-OHP | Zhang et al.88 | ||
| miR-503-5p | Up | PUMA | L-OHP | Xu et al.89 | ||
| Autophagy-related pathway | miR-218 | Down | YEATS4 | L-OHP | Fu et al.90 | |
| miR-22 | Down | ULK1 | 5-FU | Zhang et al.91 | ||
| miR-125b | Up | - | 5-FU | Yu et al.92 | ||
| miR-409-3p | Down | Beclin-1 | L-OHP | Tan et al.93 | ||
| miR-338-3p | Up | mTOR | 5-FU | Han et al.94 | ||
| miR-18a* and | Down | BTG1 | L-OHP,5-FU | Yu et al.95 | ||
| miR-4802 | Down | ATG7 | ||||
| miR-34a | Down | Smad4 | L-OHP | Sun et al.44 | ||
| Cancer stem cells | miR-141 | Down | Cyclin D2 | 5-FU | Ye et al.96 | |
| miR-196b-5p | Up | SOCS1, SOCS3 | 5-FU | Ren et al.97 | ||
| miR-450b-5p | Down | SOX2 | 5-FU | Jin et al.98 | ||
| miR-106a | Up | DUSP2 | 5-FU | Qin et al.99 | ||
| miR-125a/b | Down | ALDH1, Mcl1 | PTX | Chen et al.100 | ||
| EMT | miR-223 | Up | FBXW7 | DOX | Ding et al.101 | |
| miR-514b-3p | Down | - | CDDP, Irinotecan | Ren et al.102 | ||
| miR-200c | Down | SUZ12 | L-OHP | Tanaka et al.103 | ||
| Cell cycle | miR-195 | Down | CHK1 and WEE1 | 5-FU | Kim et al.104 | |
| miR-520g | Down | P21 | 5-FU | Zhang et al.105 | ||
| Other chemoresistance-related miRNAs | miR-505 | Up | RASSF8 | Methotrexate | Chen et al.106 | |
| miR-519b-3p | Up | ARID4B | Cape,5-FU, L-OHP | Luo et al.107 | ||
| miR-124 | Down | SGK1 | DOX | Zhu et al.108 | ||
| miR-340 | Down | RLIP76 | L-OHP | Zhang et al.109 | ||
| miR-134 | Up | CREB1 | L-OHP | Ye et al.110 | ||
| let-7d-5p | Down | Aurora B | Trifluridine | Tsunekuni et al.111 | ||
| miR-33a/let-7e | Down | ST8SIA1 | 5-FU | Shan et al.112 | ||
| miR-128 | Down | Galectin-3 | 5-FU, L-OHP | Lu et al.113 | ||
| miR-200b-3p | Down | PRDX2 | L-OHP | Lv et al.114 | ||
| miR-186 | Down | CPEB2 | Methotrexate | Li et al.115 | ||
| miR-137 | Down | YBX1 | L-OHP | Guo et al.116 | ||
| miR-203 | Down | SIK2 | Taxol | Liu et al.117 | ||
| miR-492 | Down | CD147 | L-OHP | Peng et al.118 |
ABC, ATP binding cassette; BCL-2, B-cell lymphoma-2; CDDP, cisplatin; CRC, colorectal cancer; DOX, doxorubicin; EMT, epithelial–mesenchymal transition; 5-FU, 5-fluorouracil; IGF1R, insulin-like growth factor 1 receptor; L-OHP, oxaliplatin; miRNAs, microRNAs; MMR, mismatch repair; TS, thymidylate synthase; VCR, vincristine; XIAP, X-linked inhibitor of apoptosis protein.
Table 3.
Summary of key drug resistance-related miRNAs which are identified as biomarkers (diagnostic and prognostic value) in CRC.
| miRNA | Alteration | Potential values | Ref |
|---|---|---|---|
| miR-17-5p | Up | High level of miR-17-5p predicts poorer survival and poor response to chemotherapy | Fang et al.23 |
| miR-7 | Down | Low level of miR-7 predicts poorer survival | Suto et al.26 |
| miR-204-5p | Down | Low level of miR-204-5p predicts poorer survival | Yin et al.30 |
| miR-182 | Up | diagnostic marker for CRC | Liu et al.33 |
| miR-199a/b | Up | High level of miR-199a/b predicts poorer survival | Chen et al.41 |
| miR-19b-3p | Up | High level of miR-19b-3p predicts poorer survival | Jiang et al.43 |
| miR-218 | Down | Low level of miR-218 predicts poorer survival and poor response to chemotherapy | Li et al.52 |
| miR-1290 | Up | High level of miR-1290 predicts poorer survival | Ye et al.119 |
| miR-506 | Down | Low level of miR-506 predicts poorer survival | Zhou et al.67 |
| miR-10b | Up | High level of miR-10b predicts poorer survival | Nishida et al.78 |
| miR-143 | Down | diagnostic marker for CRC; Low level of miR-143 predicts poor response to chemotherapy | Qian et al.83 |
| miR-338-3p | Up | High level of miR-338-3p predicts poorer survival | Han et al.94 |
| miR-196b-5p | Up | diagnostic marker for CRC | Ren et al.97 |
| miR-128 | Down | Low level of miR-128 predicts poorer survival | Lu et al.113 |
| miR-1914* /1915 | Down | diagnostic marker for CRC | Hu et al.120 |
| miR-21 | Up | High level of miR-21 predicts poorer survival and poor response to chemotherapy | Kulda et al.121 |
CRC, colorectal cancer; miRNAs, microRNAs.
Figure 2.
(A) Modulation of 5-FU chemoresensitivity by chemoresistant miRNAs and chemosensitive miRNAs in CRC; (B) Modulation of L-OHP chemoresensitivity by chemoresistant miRNAs and chemosensitive miRNAs in CRC.
CRC, colorectal cancer; 5-FU, 5-fluorouracil; L-OHP, oxaliplatin.
miRNA-mediated signaling pathways in drug resistance of CRC
MDR refers to resistance to a variety of anticancer drugs, and poses a great challenge to cancer treatment. Mounting evidence indicates that several signaling pathways, such as the phosphatidylinositol 3-kinase (PI3K)/Ser/Thr kinase (AKT) signaling pathway and Wnt/β-catenin signaling pathway, contribute to chemoresistance in CRC. Some miRNAs have been shown to affect the function/expression of various components of these pathways, thereby changing the chemosensitivity of CRC cells. The related cellular signaling pathways through which miRNAs modulate efficacy of chemotherapy regimens of CRC are summarized in the following (Table 1).
PI3K/AKT signaling pathway
The PI3K/Akt signaling pathway confers drug resistance to various malignancies including CRC.122 The activity of the PI3K/AKT signaling pathway is regulated by various molecules, including phosphatase and tensin homolog (PTEN), epidermal growth factor receptor (EGFR), high mobility group protein A2 (HMGA2), and protein phosphatase 2 scaffold subunit A β (PPP2R1B), RAS, etc.123
PTEN functions as a lipid phosphatase to antagonize the PI3K/Akt pathway via dephosphorylating phosphatidylinositol 3,4,5-trisphosphate (PIP3). The decreased expression level of PTEN correlated with drug resistance to conventional therapy.124 PTEN mRNA is a direct target of miRNA-17-5p, which is significantly upregulated in CRC patients. Enhanced expression of miR-17-5p contributes to the development of resistance to chemotherapeutic treatments (5-FU, L-OHP and Irinotecan) and correlates with distant metastases and poor clinical stages in CRC patients. Either antisense oligo against miR-17-5p or reconstruction of PTEN expression could effectively sensitize CRC cells to cytotoxic drug, inducing increased cellular apoptosis and cell death.23 miR-21 also interacts with PTEN and is overexpressed in many malignancies, including CRC.24 PTEN expression was downregulated in miR-21 overexpressing xenograft. Up-take of difluorinated curcumin (CDF), a novel nontoxic analog of the dietary ingredient curcumin,125 has been shown to repressed miR-21expression and restored PTEN levels with subsequent reduction in Akt phosphorylation, contributing to inhibit the growth of 5-FU + L-OHP resistant colon cancer HCT116 and HT-29 cells. In short, Dysregulated of PTEN-Akt signaling mediated by miR-21 could be normalized by CDF in chemo-resistant CRC cells.126 Of note, decreased expression of miR-200c in CRC can promote 5-FU resistance by inversely regulation of PTEN expression and caspase-3 activity and subsequently inhibiting the AKT signaling pathway.25
One class of upstream regulators of the PI3K/AKT pathway is EGFR, whose phosphorylation leads to the activation of PI3K/AKT pathway and extracellular signal-regulated kinase (ERK).127 A recent study showed that 3′-UTRs of EGFR mRNA are direct functional targets of miR-7, the expression level of which was higher in CRC tissues than in adjacent normal tissues. miR-7 can promote cell proliferation and induce resistance to cetuximab in CRC cells via targeting EGFR. Therefore, miR-7 may serve as a promising biomarker for targeted therapy in CRC patients who exhibit resistance to EGFR-directed antibodies.26 Interestingly, a disintegrin and metalloprotease 9 (ADAM9), involving in EGFR-AKT signaling and regulating cell proliferation, is a direct target of miR-20b. miR-20b expression was decreased in the 5-FU-resistant colon cancer tissues and cells and was inversely correlated with ADAM9 and EGFR expression. miR-20b can reverse 5-FU resistance of CRC both in vitro and in vivo through suppressing the ADAM9/EGFR signaling pathway.27
Another PI3K/AKT signaling pathway upstream is RAS, which also modulates drug resistance of CRC.128 Ras activity was found to be increased via the inhibition of let-7, which targeted the 3′-untranslated region of Ras proteins and enhanced 5-FU resistance of CRC.28 In addition, the let-7 family also contributed to enhance sensitivity of metastatic colorectal cancer (mCRC) to anti-EGFR agents by post-transcriptionally downregulating KRAS.129 Indeed, higher Let-7a levels in KRAS-mutated mCRC patients treated with salvage cetuximab plus irinotecan correlated with favorable survival outcomes.130 Let-7b and let-7e were downregulated in HCT-116 cells (with mutated KRAS) following cetuximab treatment. All of these miRNAs were identified as potential biomarkers to predict cetuximab resistance in KRAS-mutated CRC.131 Interestingly, miR-146b-3p and miR-486-5p were also found to be up-regulated in KRAS-mutated CRC patients compared with wild-type KRAS,131 and KRAS mutations were considered as negative predictor of anti-EGFR therapy response in CRC,132 suggesting that these miRNAs may serve as predictive biomarkers of response to cetuximab in CRC. A recent study showed that mCRC patients who did not benefit from cetuximab treatment exhibited higher miR-31-5p/3p expression in comparison with responders, and miR-31-5p/3p were significantly associated with time to progression (TTP) in RAS wild-type (wt-RAS) mCRC patients who received cetuximab therapy.133 Thus miR-31-5p/3p could be used as a promisingly predictive biomarkers of response to cetuximab in wt-RAS mCRC patients. It is notable that miR-17-3p was upregulated in HCT-116 (with mutated KRAS) cells and downregulated in Caco-2 (with wild-type KRAS) cells after cetuximab treatment, suggesting that it may be used as potential predictive markers of cetuximab resistance in KRAS-mutated CRC patients.131 One study demonstrated that miR-224 made CRC cells exhibit a poor response to 5-FU-based chemotherapy via the RAS/PI3K/AKT signaling pathway.29 In addition, RAB22A, a member of the RAS oncogene family, is also involved in MDR of CRC. RAB22A is a direct functional target of miR-204-5p, which was downregulated in CRC tissues. Functional analyses revealed that restoring miR-204-5p expression significantly represses cell proliferation and invasion both in vitro and in vivo and enhances the sensitivity of CRC cells to 5-FU, L-OHP, and cisplatin (CDDP). Mechanistic investigations showed that restoring miR-204-5p expression inhibited colorectal cancer invasion and migration and increased CRC sensitivity to chemotherapy via directly targeting RAB22A.30
HMGA2, a substantiated activator of PI3K/AKT signaling pathway, plays a vital role in regulating cell proliferation and differentiation, and its overexpression was identified as a poor prognostic factor for colon cancer.134 A cross-section study showed that HMGA2 expression was associated with Dukes stages and metastasis of CRC patients.135 miR-204 is significantly downregulated in CRC, can directly target HMGA2 mRNA. miR-204/HMGA2 axis mediated the resistance of CRC cells to 5-Fu through activation of the PI3K/AKT pathway.31 Conversely, HMGA2 can bind to the promoters of miR-194 loci. Overexpression HMGA2 attenuated miR-194 expression and promoted cell growth, migration and drug resistance, whereas restoring miR-194 expression could compromise the previously mentioned biological activities.32
The PI3K/AKT signaling pathway is also associated with the sialyltransferases (STs). Aberrant expression of ST6GALNAC2 has been identified to correlate to proliferative potential and invasive property of cancer via the PI3K/AKT pathway. It was reported that the sialylation related gene ST6GALNAC2 was a direct target of miR-135b and miR-182, expression of which were remarkably elevated in CRC samples and cell lines. Forced expression miR-135b or miR-182 induced the resistance of HCT-8 and LoVo cell lines to 5-FU and promoted cell proliferation in vitro and in vivo, whereas ST6GALNAC2 silencing attenuated the previously mentioned effects. Inhibition of miR-135b or miR-182 may function as potential therapy targets to enhance the chemosensitivity to 5-FU via activation of PI3K/AKT pathway.33
PPP2R1B, a regulatory subunit of the PP2A complex, was identified as a negative regulator of the PI3K/AKT signaling pathway.136 Oncogenic miRNA, miR-587, was upregulated in 5-FU-resistant CRC patients, and its expression inversely correlated to PPP2R1B expression. Knockdown of PPP2R1B expression mediated by miR-587 leads to AKT phosphorylation, which gives rise to elevated XIAP expression and promotes 5-FU resistance. Of note, a specific and robust AKT inhibitor, MK2206 could attenuate miR-587-induced 5-FU resistance.34
Another negative regulator of the PI3K/AKT signaling pathway PHLPP1, is classified as a member of a phosphatase family.137 PHLPP1 expression was found to be downregulated in CRC cells after transfecting miR-199a-5p and miR-375, which exhibited decreased expression in colon cancer cells. Of note, miR-375 directly targets PHLPP1 mRNA and degrades it, whereas miR-199-5p functions only at the translational level. Up-regulation of miR-199a-5p and miR-375 led to PHLPP1 degradation and enhanced phospho-AKT levels, resulting in significant chemoresistance of CRC cells to cetuximab treatment. Conversely, restoration of PHLPP1 expression via silencing of the same miRNAs was able to reduces phospho-AKT levels, thus resensitizing CRC cells to cetuximab.35
Wnt/β-catenin signaling pathway
Wnt/β-catenin signaling plays fundamental roles in biological processes such as tissue regeneration and morphogenesis.138 Dysregulation of the Wnt/β-catenin pathway was identified as a central oncogenic driver in CRC via enhancing Wnt target genes, including Myc, cyclin D1, and so on.139,140 These genes correlate with cell proliferation and survival, and thus might be involved in the development of CRC.141 Emerging evidence indicated that miRNAs can affect chemotherapy resistance via the Wnt/β-catenin signaling pathway.142 The related miRNAs are summarized in the following.
miR-181a-5p expression was decreased significantly in CRC. Enhanced expression of miR-181a-5p was associated with increased sensitivity of CRC cells to 5-FU treatment. Mechanically, β-catenin and TCF4 were direct targets of miR-181a-5p. Overexpression of miR-181a-5p could regulate the chemoresistance of CRC via modulating the activity of Wnt/β-catenin signaling.36
Another downregulated miRNA in 5-FU-resistant CRC cells was miR-149. Restoration of miR-149 could sensitize 5-FU-resistant CRC cells to 5-FU by increasing 5-FU-inducing apoptosis, while miR-149 silencing exhibits the opposite effect. Mechanistic investigations revealed that Forkhead box protein M1 (FOXM1) is a target of miR-149 in 5-FU-resistant CRC cells. FOXM1 could modulate β-catenin nuclear localization and regulate the Wnt/β-catenin signaling pathway.143 Knockdown of FOXM1 expression could mimic the effect of miR-149 overexpression on the 5-FU resistance of 5-FU-resistant CRC cells. Taken together, upregulation of miR-149 could reverse the 5-FU resistance of CRC cells via inhibition of Wnt/β-catenin signaling pathway by targeting FOXM1, targeting miR-149/FOXM1 signaling may be a promising therapeutic strategy for 5-FU-resistant CRC patients.37 Interestingly, another down-regulated miRNA in colon cancer, namely miR-320, can also directly bind to FOXM1, resulting in decreased expression of Wnt/β-catenin signing pathway associated molecules, including β-catenin, c-myc, and cyclin D1. Enhanced expression of miR-320 can inhibit CRC cell proliferation, invasion and increase sensitivity of CRC to 5-Fu and L-OHP by targeting FOXM1.38 The miR-320–FOXM1 axis may widen our horizon of knowledge about the concrete mechanisms of chemo-resistance for CRC, and the re-expression of miR-320 may provide a new therapeutic target for CRC treatment.
miR-203a-3p acts as a tumor suppressor in CRC. Ectopic expression of miR-203a-3p is critical for cell proliferation repression and chemoresistance reduction. β-catenin and GRG5, the downstream genes of Wnt/β-catenin signaling pathway, were identified as direct targets of miR-203a-3p. Overexpression of miR-203a-3p sensitized CRC cells to CDDP and PTX via inhibition of the Wnt/β-catenin signing pathway by targeting β-catenin and GRG5.39 miR-203a-3p-mediated Wnt/β-catenin signaling pathway may provide new insights into the mechanisms of drug resistance, and offer potential strategies for the treatment of CRC.
Enhanced expression of miR-224 is associated with adriamycin (ADM) resistance of CRC cells. Glycogen synthase kinase-3β (GSK-3β) is a target of miR-224. GSK-3β is able to phosphorylate and degrade β-catenin, and further inhibit the Wnt/β-catenin signal pathway. Suppression of miR-224 expression up-regulated GSK-3β expression, inhibited Wnt/β-catenin signal pathway activity and survivin expression, as well as reduced ADM resistance of CRC SW480 cells.40 GSK-3β was also identified as the direct target of miR-199a/b. Enhanced expression of miR-199a/b was detected in colorectal cancer stem cells (CCSCs) and correlated with CDDP resistance. miR-199a/b regulated Wnt/β-catenin pathway by targeting Gsk3β in CCSCs, contributing to the resistance of CDDP in CRC.41
Up-regulation of miR-100 and miR-125b were detected in CRC tissues and cell lines with cetuximab resistance. miR-100 and miR-125b coordinately drive cetuximab resistance by targeting five negative regulators of Wnt signaling, of which DKK1 and ZNRF3 were targets of miR-100, ZNRF3, RNF43, DKK3 and APC2 were targets of miR-125b. Targeting Wnt signaling can attenuate this mode of cetuximab resistance.42
TGF-β signaling pathway
Accumulating evidence has reported that the activation of the transforming growth factor (TGF)-β signaling pathway could promote tumorigenesis, including metastasis and chemoresistance.144 As a key mediator of the TGF-β signaling pathway, SMAD4 plays a pivotal role in promoting apoptosis and suppressing tumor progression. SMAD4 deficiency is associated with poor response to chemotherapeutic drugs and worse prognosis of patients with colon cancer.145 A study conducted by Jiang et al. reported that SMAD4 is a direct target of miR-19b-3p. High expression of miR-19b-3p promoted proliferation and mediated resistance to L-OHP-based chemotherapy via targeting SMAD4.43 In addition, SMAD4 is also identified as a direct target of miR-34a, which expression was inversely associated with TGF-β and SMAD4 in the blood samples of CRC patients following L-OHP treatment. miR-34a mediated the resistance of CRC patients to L-OHP by directly targeting SMAD4 through the TGF-β/SMAD4 pathway.44
Other signaling pathways and transcription factors
Apart from the previously mentioned cellular signaling pathways, aberrant activation or inactivation of the Hippo, NF-κB, Notch, Raf/MEK/ERK signing pathway, etc., have been reported to be implicated in chemotherapeutic resistance of CRC.146
The transcriptional co-activators YAP and TAZ of the Hippo signaling pathway play a fundamental role in the development and progression of CRC,147 were identified as direct targets of miR-874-3p, which was significantly downregulated in CRC. Enhanced expression of miR-874-3p can decrease the mitochondrial potential and increase the apoptosis ratio of CRC cells following 5-FU treatment in vitro and attenuated the chemoresistance of CRC cells treated with 5-FU in vivo, whereas knockdown of miR-874-3p exhibited opposite effects. Mechanically, miR-874-3p directly targeted YAP and TAZ, leading to the downregulation of Hippo signing pathway downstream genes, including BCL2L1, CTGF and cyclin A, and further attenuating 5-FU resistance of CRC cells.45
miR-15b-5p is regarded as a NF-κB-dependent gene, and its decreased expression was detected in CRC tissues and cell lines. Enhanced expression of miR-15b-5p increased cellular apoptosis rate of CRC cells treated with 5-FU and sensitized CRC to 5-FU. Mechanistic investigations indicated that miR-15b-5p modulates therapeutic effects via influencing the activity of NF-κB signaling pathway by negatively regulating the expression of NF-κB1 and IKK-α.46 Given this insight into the miRNA regulation of the drug-resistant phenotype, targeting miR-15b-5p might be a promising alternative for 5-FU-resistant CRC patients.
Dysregulation of Notch signaling pathway contributed to CRC progression, metastasis and inhibition of apoptosis. It was reported that enforced expression of miR-139-5p drove 5-FU sensitivity of CRC cells via regulating Notch signaling pathway. Mechanistic investigations revealed that miR-139-5p exerted its therapeutic effects by targeting NOTCH1 and influencing the expression of MRP-1 and BCL-2. Knockdown NOTCH-1 expression exerted an effect similar to miR-139-5p overexpression, whereas up-regulation of NOTCH-1 promoted the drug-resistant phenotype.47 Similar results have been duplicated in other studies.48 Therefore, miR-139-5p has the potential to enrich standard therapeutic approaches to CRC. The other two Notch signaling proteins Notch2 and RBPJ, which played key roles in CRC cell stemness and chemoresistance, and their 3′UTR possessed binding sites of miR-195-5p. Functional assays further confirmed that miR-195-5p could inhibit tumor sphere formation and increase CRC cells apoptosis following 5-FU treatment by targeting Notch2 and RBPJ.148 miR-195-5p could serve as a promising therapeutic target for CRC treatment.
Furthermore, Raf/MEK/ERK signaling pathway has effects on induction of drug resistance as well.149 Kinase suppressor of ras 1 (KSR1) acts as a molecular scaffold for the Raf/MEK/ERK phosphorylation cascade, binding to Raf, MEK, and ERK and further positively inducing ERK activation.150 KSR1 was found to be a direct target of miR-497, which was downregulated in CRC tissues. Forced expression of miR-497 inhibited malignant phenotypes and increased chemosensitivity of CRC cells to 5-FU treatment via attenuating KSR1 protein expression and ERK activation, whereas KSR1 overexpression abrogated the previously mentioned effects.49
Specific mechanisms of drug resistance mediated by miRNA in CRC
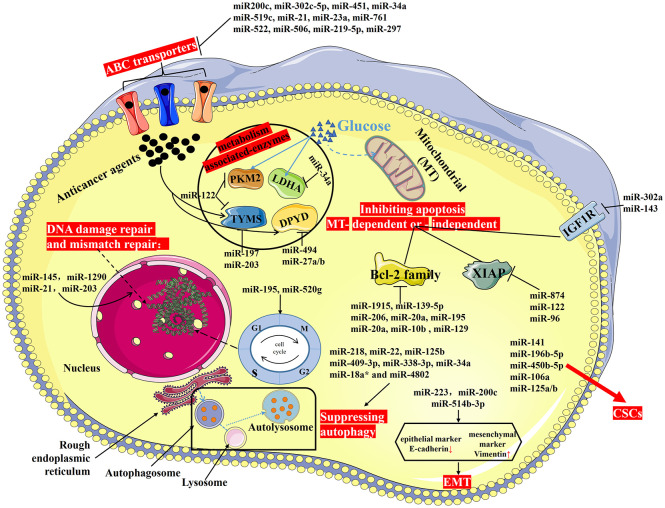
Emerging studies have demonstrated that drug resistance of CRC is attributed to various mechanisms including aberrant metabolism, enhanced DNA damage repair, dysregulation of ATP-binding cassette transporters (ABC transporters) activity, resistance to apoptosis, induction of autophagy, regulating properties of cancer stem cells, promoting epithelial-mesenchymal transition (EMT), alternations of cell cycle and checkpoints and so forth. We discuss the role of miRNAs in modulating efficacy of anticancer drugs through the mentioned mechanisms in CRC as follow (Figure 3; Table 2).
Figure 3.
Drug resistance (or MDR) of CRC. Drug resistance of CRC is attributed to various mechanisms including aberrant metabolism, enhanced DNA damage repair, dysregulation of ABC transporter activity, resistance to apoptosis, induction of autophagy, regulating properties of CSCs, promoting EMT, alternations of cell cycle and checkpoints, and so forth.
ABC, ATP-binding cassette; CRC, colorectal cancer; CSCs, cancer stem cells; EMT, epithelial–mesenchymal transition; MDR, multidrug resistance.
Aberrant metabolism
Abnormal metabolism, such as enhanced glycolysis and reduced mitochondrial oxidative phosphorylation, is characterized as a hallmark of cancer cells.151 Mounting evidence has shown that dysregulated cellular metabolism may also drive chemoresistance.152
Pyruvate kinase type M2 (PKM2) is considered to be one of key regulatory enzymes in glucose metabolism, catalyzing the final step of glycolysis and facilitating lactate production in cancer cells.153 Luciferase reporter assay showed that the 3′UTR region of PKM2 could interact with miR122, which was downregulated in 5-FU-resistant CRC cells. miR-122 overexpression sensitized CRC cells to 5-FU in vitro and in vivo through the inhibition of PKM2.50
In addition, another glucose metabolism-associated enzyme – lactate dehydrogenase A (LDHA), an isoenzyme of LDH enzymes – catalyzes the conversion of pyruvate to lactate during the glycolytic process and plays a pivotal role in the growth, invasion, and metastasis of various tumors including CRC.154 LDHA was found to be a direct target of miR-34a. Enhanced expression of miR-34a decreased the expression of LDHA through directly binding to its 3′UTR, resulting in the re-sensitization of 5-FU-resistant CRC cells to 5-FU treatment.51
Furthermore, it is well known that thymidylate synthase (TYMS), a critical enzyme of 5-FU metabolism, is significantly associated with the response to 5-FU-based therapy. One study has shown that overexpression of miR-122 re-sensitized 5-FU-resistant CRC cells to the drug through inhibition of TYMS expression. Of note, miR-218 was significantly downregulated in CRC tissues, and re-expression of miR-218 inhibited cell proliferation, promoted apoptosis and caused cell cycle arrest by suppressing BIRC5 expression.52 In addition, luciferase assay and western blot analysis confirmed that miR-197 can bind directly to TYMS and downregulate its expression. Upregulation of miR-197 increases the sensitivity of CRC cells to the cytotoxic effect of 5-FU by suppressing TYMS expression.53 Furthermore, miR-203 can also directly bind to the site of 3′UTR of TYMS mRNA and suppress its expression both in vitro and in vivo. Cytotoxicity assay confirmed that miR-203 overexpression enhanced 5-FU chemosensitivity in CRC cells via regulating TYMS expression. Knockdown of TYMS expression increased 5-FU chemosensitivity, similar to the effects of miR-203 overexpression.54 Taken together, downregulation of TYMS induced by miRNAs may provide a promising approach to overcome 5-FU resistance for CRC patients.
Apart from TYMS, dihydropyrimidine dehydrogenase (DPYD) also functions as a regulatory enzyme in the 5-FU catabolic pathway.155 3′UTR of DPYD could interact with miR-494, which was downregulated in 5-FU-resistant CRC cells. High miR-494 expression promoted apoptosis of CRC cells in vitro and repressed xenograft tumors growth in vivo in the presence of 5-Fu via directly suppressing DPYD expression, whereas upregulation of DPYD could abrogate miR-494-mediated 5-Fu sensitivity regulation.55 In silico analysis indicated that the 3′UTR of DPYD also has two conserved recognition sites interacting with miR-27a and miR-27b. Ectopic expression of miR-27a and miR-27b could sensitize CRC cells to 5-FU by directly targeting DPYD.156
MiRNAs and regulation of DNA damage repair and mismatch repair
Cell-inherent DNA repair pathways could abrogate the DNA-damaging effect of cytotoxic agents. Though the effects of DNA damage repair on drug resistance is not well appreciated, certain miRNAs have been identified to play a key role in DNA damage repair-mediated drug resistance. Tumor-suppressor miR-145 enhanced re-sensitivity of CRC cells to 5-FU in vitro and in vivo by downregulating the expression of RAD18, which is a DNA damage-activated E3 ubiquitin ligase and plays an important role in DNA damage repair.56
The mismatch repair (MMR) system is critical for the recognition and repair of DNA base mismatches. Dysregulation expression of MMR proteins induced deficient MMR (dMMR). Interestingly, miR-1290 was positively associated with dMMR status. Human mutS homolog 2 (hMSH2), a core mismatch repair (MMR) protein, was shown to be a direct target of miR-1290. Forced expression of miR-1290 promoted the resistance of CRC cells to 5-FU by directly targeting hMSH2.119 In addition, miR-21 also negatively regulated hMSH2 expression at transcriptional and protein level.57 High level of miR-21 expression significantly promoted cell proliferation and reduces 5-FU-induced G2/M arrest and apoptosis via downregulating hMSH2 and hMSH6. Furthermore, miR-21 overexpression attenuated the therapeutic efficacy of 5-FU in xenograft tumors.157 In silico analysis indicated that ataxia telangiectasia mutated (ATM) kinase – a primary mediator of the DNA damage response – was a potential target of miR-203. Upregulation of miR-203 expression is detected in L-OHP-resistant CRC cells. miR-203 silencing sensitized chemo-resistant CRC cells to L-OHP through regulating ATM protein expression.58
MiRNAs and regulation of ABC transporters
ATP-binding cassette (ABC) transporters) are members of P-type membrane ATPases that are involved in MDR.158 To date, 48 ATP-binding cassettes (ABC) transporters have been identified in the human genome, subdivided into seven families (A–G). ABC transporters facilitate the efflux of excessive intracellular drugs, thus giving rise to a significant impairment of chemotherapeutic effects. Growing evidence indicates that miRNAs play a pivotal role in regulating ABC transporters, contributing to the sensitivity or resistance to anticancer drugs.
Permeability glycoprotein (P-gp), also known as ATP binding cassette subfamily B Member 1 (ABCB1), was the first identified ABC transporter that contributed to chemoresistance.159 For example, ABCB1/P-gp expression correlated inversely with miR-200c expression, which was downregulated in MDR colorectal cancer cells [L-OHP, 5-Fu, CDDP and vincristine (VCR)]. JNK2 was shown to be a direct target of miR-200c. miR-200c overexpression downregulated the levels of ABCB1/P-gp specifically via the JNK2-mediated JNK signaling pathway, resulting in increased sensitivity to anticancer drugs and inhibition of metastasis in vitro and in vivo.59 It was reported that the level of ABCB1 level was also inversely correlated with the level of miR-302c-5p level in CRC cells. Reduced miR-302c-5p expression was detected in L-OHP-resistant CRC cells. Enhanced miR-302c-5p expression could increase the sensitivity of CRC cells to L-OHP via regulating ABCB1 expression.60 In addition, miR-451 was also reported to regulate ABCB1 expression in drug-resistant CRC. miR-451 restoration significantly reduced the expression of ABCB1 and reversed drug resistance of CRC cells to irinotecan.61
Another ABC transporter that is significantly associated with MDR is ABCG2. It has been shown that miR-519c downregulated the expression of ABCG2 directly or indirectly via modulating mRNA binding protein HuR expression, causing increased sensitivity of CRC to 5-FU.62 A recent study revealed that miR-21 remarkably elevated the expression of ABC transporter ABCC5 and the stem cell marker CD44 via negatively regulating programmed cell death protein 4 (PDCD4) expression (a target gene of miR-21), resulting in increased 5-FU resistance to CRC. miR-21 silencing significantly reduced the IC50 of 5-FU in CRC cells and increased the apoptosis ratio.63 One study that focused on the role of miR-23a in chemosensitivity of microsatellite instability (MSI) CRC cells provided evidence that reduced miR-23a could increase 5-FU-induced apoptosis via directly targeting ABCF1.64 In addition, miR-761 overexpression increased the sensitivity of CRC cells to 5-FU and ectopic expression of miR-761 suppressed CR cell proliferation, invasion, cell cycle, and colony formation partly via targeting FOXM1 expression.65 FOXM1 silencing overcame 5-FU resistance of CRC cells through regulating ABCC10 expression.160 In the case of miR-522, the authors showed that miR-522 significantly inhibited cell survival and doxorubicin (DOX) resistance in CRC cells by directly targeting ABCB5.66
Furthermore, multidrug resistance-associated protein (MRP1) is generally referred to ABCC1, playing a pivotal role in the development of MDR of CRC cells against various chemotherapeutic agents.158 In L-OHP-resistant CRC cells, miR-506 upregulation accompanied with downregulated low MDR1/P-gp expression suppressed cell growth and elevated L-OHP-induced cell apoptosis. Mechanically, miR-506 overexpression reduced MDR1/P-gp expression in L-OHP-resistant CRC cells by inhibiting the Wnt/β-catenin signing pathway, enhancing the sensitivity of CRC cells to L-OHP.67 Similarly, ectopic enforced miR-219-5p expression decreased the level of P-gp and MRP1 by directly regulating Sall4 expression, resulting in reduced resistance of CRC to 5-FU and L-OHP. And ectopic expression of miR-219-5p also suppressed cell proliferation, migration, invasion and G0/G1 cell cycle arrest by targeting Sall4 in CRC.68 A recent study revealed that transfection of miR-297 mimics into L-OHP-resistant HCT116/L-OHP cells remarkably downregulated MDR-associated protein 2 (MRP2) expression, overexpression of which was related to platinum-drug resistance.161 Ectopic expression of miR-297 sensitizes MDR CRC cells to some anti-cancer drugs by directly inhibiting MRP-2 expression at the post-transcriptional level.69
In addition to the previously mentioned miRNAs, miR-34a has also been proven to involve in MDR of CRC through regulating the activation of ABC transporters and anti-apoptosis pathways. miR-34a overexpression reversed the resistance of L-OHP-resistant colon cancer to L-OHP-resistant colon cancer via inhibition of P-gp, MRP2, BCRP, and Bcl-2 by positively targeting the 3′UTR of ornithine decarboxylase antizyme 2 (OAZ2). Augmentation of OAZ2 expression potentiated the chemosensitivity of CRC cells to L-OHP.70
MiRNAs and apoptosis regulating genes
BCL-2 family
Apoptosis refers to the autonomous and orderly death of cells controlled by genes in order to maintain the homeostasis of the internal environment, and resistance to apoptosis is commonly characterized as a hallmark of cancer and limits the effectiveness of anti-cancer drug treatment in CRC.151 Drug resistance in CRC is significantly associated with dysregulation of apoptosis-related genes, of which the B-cell lymphoma-2 (BCL-2) family members, including the death antagonists (Bcl-2, Bcl-XL, Bcl-w, Mcl-1, and A1/Bfl1), death agonists (Bax, Bak, Bad, Bcl-xS, Bid, Bik, and Hrk), and the BCL-2 homology 3 (BH3)-only (BH3-only) proteins,162 which play important roles in inhibiting mitochondria-dependent intrinsic and extrinsic cell death pathways and in the development of drug resistance in CRC.163 Investigations have stated that many miRNAs regulate drug resistance by targeting apoptosis-related genes, particularly those of BCL-2 family members.
Decreased expression of miR-1915 was detected in L-OHP-resistant HCT116 cells (HCT116/L-OHP), and HCT116/L-OHP overexpressed Bcl-2 mRNA and protein in comparison with their parental cells. miR-1915 directly reduced Bcl-2 expression at the posttranscriptional level through binding to its 3′-UTR. Modulation of miR-1915 expression sensitized CRC cells to L-OHP-induced apoptosis via targeting Bcl-2.71 In addition, Bcl-2 is also a target of miR-139-5p, which exerted its inhibitory effects on CRC tumorigenesis, metastasis and drug sensitivity by the downregulation of its target Bcl-2. Of note, Bcl-2 overexpression reversed the inhibitory effects of miR-139-5p on progression and drug resistance in CRC cells.72 Karaayvaz et al. reported that miR-129 is downregulated in CRC, and its forced expression inhibited cell proliferation, caused cell-cycle arrest, and promoted apoptosis. Importantly, up-regulated miR-129 could sensitize CRC cells to 5-FU both in vitro and in vivo through triggering the intrinsic apoptotic pathway via inhibiting Bcl-2. In addition to inducing apoptosis by targeting Bcl2, miR-129 can also activate the intrinsic apoptotic pathway by cleavage of caspase-9 and caspase-3.73 Another study conducted by Meng et al. demonstrated that miR-206 can also target Bcl-2 to mediate 5-FU resistance, proliferation, and apoptosis in CRC.74 In addition, inhibition of Bcl-2 has been reported to enhance CDDP-induced apoptosis via the mitochondrial pathway.164 ASK1, a key mediator in the ROS-dependent cell death pathway, was identified as a target of miR-20a.165 Knockdown of miR-20a promoted the CDDP-induced production of ROS, which subsequently augmented ASK1 activation and further increased the cellular level of phosphorylated JNK in CRC cells. Activation of JNK signaling in CRC cells promoted mitochondrial apoptosis by inhibiting the anti-apoptotic function of Bcl-2.166 Taken together, knockdown of miR-20a reversed CDDP resistance in CRC cells through the ROS/ASK1/JNK pathway.75
miRNAs also regulate other members of BCL-2 family, contributing to drug resistance in CRC. In the case of miR-195, the authors demonstrated that blockage of miR-195 significantly enhanced DOX resistance in CRC cells through repression of BCL2L2 expression.76 Huang et al. found that high level of miR-20a was associated with TRAIL resistance in CRC. miR-20a silencing sensitized the CRC cells to TRAIL-induced cell death, whereas miR-20a mimics attenuated the cytotoxic effect of TRAIL. Mechanically, knockdown of miR-20a reversed TRAIL resistance on account of the dysfunction of mitochondria triggered by upregulation of BID.77 Another miRNA which confers resistance to 5-FU in CRC is oncogenic miR-10b. High level of miR-10b expression is correlated significantly with high incidence of lymphatic invasion and poor prognosis in CRC patients. BIM, a member of BH3-only proteins, was identified as a direct target of miR-10b. miR-10b modulates 5-FU resistance in CRC cells via directly inhibiting proapoptotic BIM.78
X-linked inhibitor of apoptosis protein pathway
X-linked inhibitor of apoptosis protein (XIAP), an important member of the IAP family proteins, has been found to inhibit the activities of caspase-3, -7 and -9, leading to inhibition of apoptosis.167 Accumulating evidence suggests that XIAP is upregulated and functions as an oncogene in multiple cancers, including CRC.168 Previous studies have shown that overexpression of XIAP promoted cell proliferation and inhibited apoptosis, as well as conferred resistance to chemotherapeutic agents in cancer cells.169 Evidence has shown that some miRNAs were involved in the formation of CRC MDR by regulating XIAP expression.
For instance, miR-874 functions as a tumor suppressor, is downregulated in CRC tissues and cell lines, and its expression is negatively correlated with TNM stage and lymph node metastasis. Ectopic expression of miR-874 inhibited cell proliferation and colony formation, enhanced cell apoptosis and sensitized CRC cells to 5-FU in vitro, as well as repressed tumor growth in vivo through inhibition of XIAP.79 Another study conducted by Hua et al. showed that miR-122 was downregulated in L-OHP-resistant CRC cells, and restoration of miR-122 can reverse L-OHP resistance in CRC by targeting XIAP.80 Furthermore, forced expression of miR-96 accelerated cell proliferation and sensitized CRC cells to 5-FU through indirect negative regulation of expression of XIAP in a three-dimensional (3D) tumor spheroid model of CRC.81
The insulin-like growth factor 1 receptor pathway
The insulin-like growth factor 1 receptor (IGF1R), a transmembrane protein, plays a pivotal role in activating certain downstream signaling pathways including PI3K/AKT pathway for regulating angiogenesis and tumorigenesis.170 Activation of IGF‑1R‑dependent pathways has also been considered as a critical step that confers chemotherapeutic agents resistance to CRC.171 In addition, miRNAs were recently reported to participate in IGF1R-mediated drug resistance in CRC.
IGF‑1R is identified as a direct target of miR-302a, which was significantly downregulated in CRC cells. In addition to IGF‑1R, miR-302a could target Akt and inhibit Akt signaling. Ectopic expression of miR‑302a enhancing 5‑FU‑induced cell death and viability inhibition by downregulating IGF‑1R expression and inactivating Akt signaling in CRC.82 Similarly, miR-143 was significantly downregulated both in CRC patients’ blood samples and tumor specimens, and its expression level was inversely correlated with IGF-1R expression. Importantly, miR-143 expression was remarkably associated with clinical stages and lymph node metastasis in CRC, upregulation of miR-143 inhibited cell proliferation, migration, tumor growth and angiogenesis and sensitized CRC cells to L-OHP treatment through inhibiting IGF-1R expression.83
Other apoptosis-related pathways
Apart from the apoptotic pathways mentioned previously, other networks also contribute to drug resistance in CRC via regulating apoptosis. For instance, the cytosolic protein apoptosis-activating factor-1 (APAF-1), also known as the human homolog of the Caenorhabditis elegans cell death protein CED-4, is required for mitochondrial-mediated apoptosis.172 Apaf-1 oligomerizes into the apoptosome upon binding to deoxyadenosine triphosphate and cytochrome c, subsequently recruiting and activating cell-killing caspases.173 Apaf-1 was found to be a target of microRNA-27a in CSCs that exhibited high level of miR-27a expression. miR-27a overexpression was strongly correlated with the resistance to TRAIL in CSCs. Knockdown of miRNA-27a rescued the expression level of Apaf-1, thus promoting the formation of the Apaf-1-caspase-9 complex and subsequently enhancing TRAIL-induced apoptosis in CSCs. In summary, miR-27a silencing has the potential to reverse the chemoresistance to TRAIL by promoting the formation of an Apaf-1-caspase-9 complex in CSCs.84 In addition, miR-23a can also bind directly to the 3′UTR of Apaf-1 and downregulate its expression. Knockdown of miR-23a re-sensitized CRC cells to 5-FU-induced apoptosis through the Apaf-1/caspase-9 apoptotic pathway.85 Furthermore, ectopic expression of miR-128 can also sensitize CRC cells to TRAIL-induced cytotoxicity by regulating apoptosis. Mechanistically, miR-128 can directly target sirtuin 1 (SIRT1) and suppress SIRT1 expression, which accelerated the production of reactive oxygen species (ROS) in CRC cells following TRAIL treatment. Elevated ROS expression subsequently increased death receptor 5 (DR5) expression, and thus enhanced TRAIL-induced apoptosis in CRC cells. Altogether, these results indicated that miR-128 sensitized CRC cells to TRAIL-induced apoptosis via targeting the SIRT1/ROS/DR5 pathway.86
Another apoptosis-related gene programmed cell death protein 10 (PDCD10) was identified as a direct target of miR-425-5p, which was up-regulated in chemo-resistant CRC cells as compared with isogenic parental cells. Knockdown of miR-425-5p sensitized CRC cells to 5-FU and L-OHP both in vitro and in vivo through inducing apoptotic cell death by modulating PDCD10 expression level.87
The forkhead transcription factor Forkhead box O3a (FOXO3a) plays a critical role in initiating apoptotic programs via upregulating proapoptotic genes such as Bim and PUMA.174 FOXO3a was identified as a target of miR-153. miR-153 overexpression reduced FOXO3a expression at transcriptional and protein level, inhibiting the apoptotic response to CDDP. Restoration of FOXO3a can reverse L-OHP resistance induced by miR-153 in CRC.88 Moreover, the proapoptotic gene PUMA is also a posttranscriptional repression target of miR-503-5p. Knockdown of miR-503-5p expression could reverse L-OHP resistance of CRC by modulating PUMA expression.89
MiRNAs and autophagy
Autophagy promotes the survival of tumor cells under therapeutic and metabolic stress,175 contributing to the development of acquired drug resistance.176 Mounting evidence revealed that miRNAs could mediated drug resistance via regulating autophagy by targeting autophagy-related molecules.177
Previously, researchers found that miR-218 was downregulated in CRC cells and regulated 5-FU resistance of CRC cells by modulation of apoptosis via targeting BIRC5.52 A recent study suggested that this miRNA could modulate L-OHP resistance of CRC cells via inhibiting cytoprotective autophagy by directly downregulating YEATS4 expression.90 miR-22 was another autophagy-related modulator that was downregulated in CRC. miR-22 overexpression was found to reverse 5-FU-induced chemoresistance through suppressing autophagy, as evidenced by enhancement of p62 (an autophagy marker, also known as SQSTM1) and reduced LC3-II (a hallmark protein for increased autophagy) expression. B-cell translocation gene 1 (BTG1) is identified as a direct target of miR-22 in regulating autophagy. Re-expression of BTG1 in miR-22-overexpressing CRC cells potentiated the formation of LC3-II and abrogated the effects of miR-22.91 Augmentation of 5-FU-induced cleavage of LC3-Ι into LC3-II was observed in the miR-125b mimics-transfected CRC cells, which exhibited poor response to 5-FU. In addition, xenograft tumor model with miR-125b overexpression also exhibited enhancement of cleaved LC3-II and autophagic proteins beclin-1 as well as elevated formation of autophagosomes following 5-FU treatment, indicating that miR-125b confers CRC cells to 5-FU resistance through inducing cell autophagy.92
The mammalian homologue of yeast autophagy‑related gene 6,also known as Beclin‑1, contributes to the formation of autophagosome.178 Beclin‑1 was found to be a direct target of miR-409-3p, which was significantly downregulated in CRC cells. Importantly, the miR-409-3p expression levels were negatively associated with resistance to L-OHP in CRC. miR-409-3p overexpression sensitized CRC cells to L-OHP by suppressing Beclin-1-mediated autophagy.93
Another autophagy marker, the mammalian target of rapamycin (mTOR) has been reported to negatively regulate autophagy and correlates with 5-FU-induced apoptosis.179 miR-338-3p can bind to the 3′UTR of mTOR, inhibiting its expression and further activating autophagy in 5-FU-treated CRC cells. miR-338-3p-mTOR-autophagy attenuated 5-FU induced apoptosis and promoted 5-FU resistance.94
Of note, Fusobacterium nucleatum confers CRC resistance to L-OHP and 5-FU by activating the autophagy pathway, and the autophagy elements ULK1 and ATG7 participate in the F. nucleatum-mediated chemoresistance in CRC cells. miR-18a* and miR-4802, which were significantly downregulated in the CRC patients administrating a high amount of F. nucleatum, could reverse F. nucleatum-mediated chemoresistance by blocking F. nucleatum-induced autophagy activation via regulating the expression of autophagy elements ULK1 and ATG7 respectively.95
Furthermore, macroautophagy is a critical regulator of L-OHP resistance in CRC.180 CRC patients treated with L-OHP-based combination chemotherapy exhibited downregulation of miR-34a and upregulation of TGF-β/Smad4, whereas miR-34a mimics reduced expression of LC3-II, beclin-1, SMAD4 and TGF-β. The miR-34a can target SMAD4 and suppress macroautophagy, mediating resistance to L-OHP in CRC patients following L-OHP treatment.44
MiRNAs and CSCs
CRC cancer stem cells (CSCs) are characterized by their ability to self-renew, proliferate indefinitely and differentiate into cancer cells, and have been reported to mediate the development of chemoresistance.181 Studies have shown that miRNAs could modulate drug resistance by regulating the properties of CSCs.182
miR-141 expression was decreased in CSCs as compared with differentiated CRC cells. Cyclin D2, a member of the D-type cyclin protein family that regulates cell cycle progression, was identified as a novel target gene of miR-141 and an important regulator of self-renewal of human embryonic stem cells.183 miR-141 affects the maintenance of stemness in CSCs via targeting cyclin D2, thereby enhancing 5-FU and L-OHP susceptibility in CRC.96
High expression of miR-196b-5p was detected in CRC patients with poor response to 5-FU treatment. Silencing miR-196b-5p represses the stem-cell-like phenotype and re-sensitizes CRC cells to 5-FU in vitro and in vivo. Mechanically, miR-196b-5p enhances stemness and chemoresistance of CRC cells to 5-FU via activating signal transducer and activator of transcription 3 (STAT3) signaling pathway by negatively regulating the expression of cytokine signaling 1 (SOCS1) and SOCS3.97 Conversely, miR-450b-5p overexpression inhibited stemness and re-sensitizes CRC cells to 5-FU by targeting the transcription factor SOX2, which is essential for maintaining CSCs properties.98
Dual-specificity phosphatases 2 (DUSP2) suppressed CRC cell stemness and its downregulation was significantly associated with chemoresistance.184 Dual luciferase assay revealed that DUSP2 was a direct target of miR-106a.The levels of miR-106a and DUSP2 were inversely correlated in CRC tissues. miR-106a can enhance cell stemness and resistance of CRC cells to 5-FU by negatively regulating DUSP2 expression.99
Furthermore, aldehyde dehydrogenase (ALDH1) A3 and Mcl1(a member of the prosurvival BCL2 family) played key roles in promoting survival of CSCs and were identified as targets of miR-125a/b, downregulation of which was responsible for chemoresistance to PTX in colon cancer cells.185,186 miR-125a/b upregulation overcame chemoresistance in PTX-resistant colon cancer cells in vitro and in vivo by downregulating ALDH1A3 and Mcl1 expression, subsequently enhancing cell apoptosis and inhibiting cell survival and tumor growth.100
Epithelial-mesenchymal transition
The epithelial-mesenchymal transition (EMT) plays a critical role in cancer invasion and metastasis, and has been proved to contribute to the development of chemoresistance.187 Ding et al. proposed that upregulation of miR-223 promotes the DOX resistance of CRC cells via regulating EMT by targeting a tumor suppressor F-box and WD repeat domain containing 7 (FBXW7), as evidenced by downregulation of the epithelial marker E-cadherin and upregulation of the mesenchymal marker Vimentin.101 Conversely, overexpression of miR-514b-3p suppressed EMT process by upregulation of the epithelial marker(E-cadherin and CLDN-1) and downregulation of the mesenchymal marker (fibronectin-1 and vimentin) in both mRNA and protein levels, further accelerating cell death in CRC cells treated with CDDP or Irinotecan.102
In addition, zinc finger E-box-binding homeobox 1 (ZEB1),a transcription factor that can bind to the E-box elements of CDH1 (encoding E-cadherin), can cause EMT by downregulating the expression of E-cadherin.188 ZEB1 was found to be a direct target of miR-200c, which was significantly downregulated in L-OHP-resistant CRC cells. miR-200c plays a role in mediating selective resistance to L-OHP in CRC cells through regulation of EMT.103
Regulations on cell cycle and checkpoint
CRC MDR is attributed partly to altering cell cycle and checkpoint.189 miRNAs mediating chemoresistance in CRC by impacting cell cycle and checkpoint were also identified. miR-195 regulates chemoresistance by alleviating G2/M phase arrest induced by 5-FU partially through inhibiting the expression of check point kinase 1 (CHK1) and G2 check point kinase WEE1 in CRC. CHK1 and WEE1, known to play critical roles in cell cycle regulation, were identified as direct targets of miR-195. miR-195 silencing sensitized CRC cells to 5-FU by downregulating CHK1 and WEE1.104 In addition, a novel p53/miR-520g/p21 signaling axis identified by Zhang et al. played a critical role in the response of CRC cells to chemotherapy chemotherapeutic agents including 5-FU and L-OHP. P53 inhibited miR-520g expression, whereas deletion of p53 up-regulated miR-520g expression. MiR-520g mediated drug resistance through downregulating p21 expression, a major cycle regulator that is required for 5-FU-induced apoptosis.105 Restoration of p21 enhanced 5-FU-induced apoptosis in miR-520g-expressing CRC cells.
Other chemoresistance-related miRNAs
Besides the examples mentioned previously, a great number of miRNAs as well as their associated molecular targets have been identified (Table 2), whereas detailed mechanisms and intracellular pathways of these miRNAs in regulation of chemosensitivity in CRC remain largely unclear. Further prospective studies are warranted to explore the concrete mechanisms and related pathways by which miRNAs modulate the MDR of CRC.
Drug resistance-related miRNAs as biomarkers in CRC
Although great progress has been made in the treatment of CRC in the past decades, including colorectal resection, perioperative management, adjuvant chemotherapy, neoadjuvant chemotherapy, and radiotherapy, CRC patients still present with a poor prognosis on account of there being no effective approach for early detection and prognostic evaluation in current clinical practice. Therefore, it is imperative to identify novel biological markers for CRC patients (especially those who exhibit poor response to chemotherapy) to improve the early diagnosis rate and develop individual therapies. Several studies have indicated that miRNAs are involved in modulating MDR and might serve as promising diagnostic or prognostic biomarkers of CRC.
For example, a variety of miRNAs that are released into the bloodstream, such as miR-182 and miR-143,33,83 had high sensitivity and specificity for detecting CRC patients amongst cancer-free controls. In addition, Kaplan–Meier survival curves and univariate and multivariate survival analysis, as well as Cox proportional hazards risk analysis, showed that elevated expression of miR-182 was associated with poor OS in patients with CRC, indicating that miR-182 could serve as an independent and adverse prognostic biomarker for CRC patients.33 Moreover, the plasma miR-1914* and -1915 also play a role in the diagnosis of CRC.120
Notably, recent studies demonstrated that exosome-derived miRNAs were identified as highly stable and noninvasive biomarkers, exhibiting great potential to detect CRC. For instance, miR-196b-5p is dramatically upregulated in the serum exosomes in CRC patients and positively correlated with T stage and M-category, indicating exosomal 196b-5p may serve as a valuable serum biomarker for the diagnosis of CRC.97
Kaplan–Meier analysis showed that a group of drug resistance-related miRNAs, including miR-17-5p, miR-199a/b, miR-10b, miR-21, miR-1290, miR-19b-3p, and miR-338-3p were upregulated in CRC and associated with poor prognosis,23,41,43,78,94,119,121 while CRC patients with the low expression of miR-7, miR-218, miR-204-5p, miR-506 or miR-128 had worse outcomes.26,30,52,67,113 In addition, multivariate analysis indicated that miR-204-5p, miR-7, miR-19b-3p and miR-10b were independent prognostic factors for CRC survival (Table 3).26,30,43,78 Of note, miR-224 overexpression not only promotes cell proliferation, migration and invasion, but also correlates with a high risk of relapse in CRC.190
Since miRNAs are not prone to degradation and could be easily isolated and detected in paraffin-embedded CRC tissue using quantitative real-time PCR or in situ hybridization, certain drug resistance-related miRNAs are identified as promising predictors of chemotherapy response in CRC. For instance, aberrant expression of miR-17-5p,23 miR-497,49 miR-197,53 miR-21,57 miR-203,58 miR-519c,62 miR-218,90 miR-22,91 miR-450b-5p,98 miR-143,191 miR-519b-3p were demonstrated to be potential biomarkers to predict drug efficacy in CRC.107 Some of them, like miR-17-5p,23 miR-203,58 miR-519c,62 and miR-21890, were proven to have potential to mediate MDR in CRC.
A growing body of studies on miRNAs shed new light on the concrete mechanisms underlying drug resistance in CRC and provide an experimental basis for the clinical development of novel and advantageous targets for reversing CRC MDR. The previously mentioned studies on the role of miRNAs in drug resistance of CRC demonstrated that some miRNAs can simultaneously regulate various MDR-related genes, such as miR-21,126 miR-200c,25 and miR-125b,42 etc., could be candidates for therapeutic biomarkers of CRC patients with MDR. In addition, different miRNAs could target the same MDR-related gene, suggesting that combining a panel of miRNAs may present a promising alternative tool for overcoming MDR.
Conclusion
miRNAs have been established as a means of inducing CRC sensitivity or resistance to anticancer drugs. Identification of MDR-related miRNAs and their targets is the key to understanding the molecular mechanisms of CRC chemoresistance and to designing novel effective therapeutic targets. In addition, combining miRNAs with existing chemotherapeutic agents may be a promising therapeutic approach to maximize therapeutic effect and improve clinical outcomes in CRC patients. Moreover, it is plausible that targeting multiple MDR-associated pathways to reverse miRNAs-mediated CRC chemoresistance in future clinical practice on account of the complicated interactions among these miRNAs and target molecules.
The combination of immunotherapy and traditional therapies has shown great potential in the treatment of tumors.192 Interestingly, miRNAs play a pivotal role in immune cell development, differentiation and regulation, as well as in innate and adaptive immune response.193,194 Of note, a recent study reported that the miRNA-200/ZEB1 axis could not only modulate tumor cell PD-L1 expression and intratumoral immunosuppression,195 but was also implicated in regulating drug resistance of CRC via induction of EMT,103 highlighting the roles of miRNAs in modulating the sensitivity of immunotherapy and in reversing MDR in tumors. Taken together, an miRNA-based strategy combined with traditional chemotherapy and immunotherapy has a bright prospect in prolonging the survival of CRC patients. Further prospective studies are warranted to explore this field.
However, there have been no studies on the safety and efficacy of miRNA-based treatment for CRC to date. Furthermore, several obstacles, such as variability in patient characteristics and lack of standardized methods for miRNA detection, hinder the translation of promising findings into clinical application. Further investigations are anticipated to engage in large clinical trials to explore the underlying mechanisms in CRC and to validate the therapeutic value, as well as prognostic and predictive potential, of these MDR-related miRNAs.
Footnotes
Conflict of interest statement: The authors declare that there is no conflict of interest.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported in part by grant from the Scientific Foundation of Shaanxi Province (Grant number 2019ZDLSF01-02-01, 2018SF-240), grant from the State Key Laboratory of Cancer Biology (Grant number CBSKL2014Z13) and grant from the National Clinical Research Center for Digestive Diseases (Grant number 2015BAI13B07). It was not supported by any private or public company or organization.
ORCID iD: Liu Hong  https://orcid.org/0000-0002-8276-2345
https://orcid.org/0000-0002-8276-2345
Contributor Information
Lili Duan, State Key Laboratory of Cancer Biology, National Clinical Research Center for Digestive Diseases, and Xijing Hospital of Digestive Diseases, Fourth Military Medical University, Xi’an, China.
Wanli Yang, State Key Laboratory of Cancer Biology, National Clinical Research Center for Digestive Diseases, and Xijing Hospital of Digestive Diseases, Fourth Military Medical University, Xi’an, China.
Weibo Feng, State Key Laboratory of Cancer Biology, National Clinical Research Center for Digestive Diseases, and Xijing Hospital of Digestive Diseases, Fourth Military Medical University, Xi’an, China.
Lu Cao, Department of Biomedical Engineering, Fourth Military Medical University, Xi’an, China.
Xiaoqian Wang, State Key Laboratory of Cancer Biology, National Clinical Research Center for Digestive Diseases, and Xijing Hospital of Digestive Diseases, Fourth Military Medical University, Xi’an, China.
Liaoran Niu, State Key Laboratory of Cancer Biology, National Clinical Research Center for Digestive Diseases, and Xijing Hospital of Digestive Diseases, Fourth Military Medical University, Xi’an, China.
Yiding Li, State Key Laboratory of Cancer Biology, National Clinical Research Center for Digestive Diseases, and Xijing Hospital of Digestive Diseases, Fourth Military Medical University, Xi’an, China.
Wei Zhou, State Key Laboratory of Cancer Biology, National Clinical Research Center for Digestive Diseases, and Xijing Hospital of Digestive Diseases, Fourth Military Medical University, Xi’an, China.
Yujie Zhang, State Key Laboratory of Cancer Biology, National Clinical Research Center for Digestive Diseases, and Xijing Hospital of Digestive Diseases, Fourth Military Medical University, Xi’an, China.
Jinqiang Liu, State Key Laboratory of Cancer Biology, National Clinical Research Center for Digestive Diseases, and Xijing Hospital of Digestive Diseases, Fourth Military Medical University, Xi’an, China.
Hongwei Zhang, State Key Laboratory of Cancer Biology, National Clinical Research Center for Digestive Diseases, and Xijing Hospital of Digestive Diseases, Fourth Military Medical University, Xi’an, China.
Qingchuan Zhao, State Key Laboratory of Cancer Biology, National Clinical Research Center for Digestive Diseases, and Xijing Hospital of Digestive Diseases, Fourth Military Medical University, Xi’an, China.
Liu Hong, State Key Laboratory of Cancer Biology, National Clinical Research Center for Digestive Diseases, and Xijing Hospital of Digestive Diseases, Fourth Military Medical University, Xi’an, 710032, China.
Daiming Fan, State Key Laboratory of Cancer Biology, National Clinical Research Center for Digestive Diseases, and Xijing Hospital of Digestive Diseases, Fourth Military Medical University, Xi’an, China.
References
- 1. Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87–108. [DOI] [PubMed] [Google Scholar]
- 2. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68: 394–424. [DOI] [PubMed] [Google Scholar]
- 3. Bae JM, Kim JH, Kang GH. Molecular subtypes of colorectal cancer and their clinicopathologic features, with an emphasis on the serrated neoplasia pathway. Arch Pathol Lab Med 2016; 140: 406–412. [DOI] [PubMed] [Google Scholar]
- 4. Geng F, Wang Z, Yin H, et al. Molecular targeted drugs and treatment of colorectal cancer: recent progress and future perspectives. Cancer Biother Radiopharm 2017; 32: 149–160. [DOI] [PubMed] [Google Scholar]
- 5. An X, Sarmiento C, Tan T, et al. Regulation of multidrug resistance by microRNAs in anti-cancer therapy. Acta Pharm Sin B 2017; 7: 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gong J, Jaiswal R, Mathys JM, et al. Microparticles and their emerging role in cancer multidrug resistance. Cancer Treat Rev 2012; 38: 226–234. [DOI] [PubMed] [Google Scholar]
- 7. Holohan C, Van Schaeybroeck S, Longley DB, et al. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer 2013; 13: 714–726. [DOI] [PubMed] [Google Scholar]
- 8. Bach DH, Hong JY, Park HJ, et al. The role of exosomes and miRNAs in drug-resistance of cancer cells. Int J Cancer 2017; 141: 220–230. [DOI] [PubMed] [Google Scholar]
- 9. Berman M, Mattheolabakis G, Suresh M, et al. Reversing epigenetic mechanisms of drug resistance in solid tumors using targeted microRNA delivery. Expert Opin Drug Deliv 2016; 13: 987–998. [DOI] [PubMed] [Google Scholar]
- 10. De Robertis M, Poeta ML, Signori E, et al. Current understanding and clinical utility of miRNAs regulation of colon cancer stem cells. Semin Cancer Biol 2018; 53: 232–247. [DOI] [PubMed] [Google Scholar]
- 11. Song JH, Meltzer SJ. MicroRNAs in pathogenesis, diagnosis, and treatment of gastroesophageal cancers. Gastroenterology 2012; 143: 35–47e32. [DOI] [PubMed] [Google Scholar]
- 12. Anfossi S, Babayan A, Pantel K, et al. Clinical utility of circulating non-coding RNAs - an update. Nat Rev Clin Oncol 2018; 15: 541–563. [DOI] [PubMed] [Google Scholar]
- 13. Lee H, Han S, Kwon CS, et al. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell 2016; 7: 100–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dangwal S, Thum T. MicroRNAs in platelet biogenesis and function. Thromb Haemost 2012; 108: 599–604. [DOI] [PubMed] [Google Scholar]
- 15. Zare M, Soleimani M, Akbarzadeh A, et al. A Novel protocol to differentiate induced pluripotent stem cells by neuronal microRNAs to provide a suitable cellular model. Chem Biol Drug Des 2015; 86: 232–238. [DOI] [PubMed] [Google Scholar]
- 16. Fallah P, Arefian E, Naderi M, et al. miR-146a and miR-150 promote the differentiation of CD133+ cells into T-lymphoid lineage. Mol Biol Rep 2013; 40: 4713–4719. [DOI] [PubMed] [Google Scholar]
- 17. Wu WK, Lee CW, Cho CH, et al. MicroRNA dysregulation in gastric cancer: a new player enters the game. Oncogene 2010; 29: 5761-5771. 2010/08/31. DOI: 10.1038/onc.2010.352. [DOI] [PubMed] [Google Scholar]
- 18. Wu WK, Law PT, Lee CW, et al. MicroRNA in colorectal cancer: from benchtop to bedside. Carcinogenesis 2011; 32: 247–253. [DOI] [PubMed] [Google Scholar]
- 19. Zhang B, Pan X, Cobb GP, et al. microRNAs as oncogenes and tumor suppressors. Dev Biol 2007; 302: 1–12. [DOI] [PubMed] [Google Scholar]
- 20. Zhang Y, Wang J. MicroRNAs are important regulators of drug resistance in colorectal cancer. Biol Chem 2017; 398: 929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mishra PJ, Humeniuk R, Mishra PJ, et al. A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc Natl Acad Sci U S A 2007; 104: 13513–13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gong JP, Yang L, Tang JW, et al. Overexpression of microRNA-24 increases the sensitivity to paclitaxel in drug-resistant breast carcinoma cell lines via targeting ABCB9. Oncol Lett 2016; 12: 3905–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fang L, Li H, Wang L, et al. MicroRNA-17-5p promotes chemotherapeutic drug resistance and tumour metastasis of colorectal cancer by repressing PTEN expression. Oncotarget 2014; 5: 2974–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Si ML, Zhu S, Wu H, et al. miR-21-mediated tumor growth. Oncogene 2007; 26: 2799–2803. [DOI] [PubMed] [Google Scholar]
- 25. Heydari K, Saidijam M, Sharifi MR, et al. The effect of miR-200c inhibition on chemosensitivity (5- FluoroUracil) in colorectal cancer. Pathol Oncol Res 2018; 24: 145–151. [DOI] [PubMed] [Google Scholar]
- 26. Suto T, Yokobori T, Yajima R, et al. MicroRNA-7 expression in colorectal cancer is associated with poor prognosis and regulates cetuximab sensitivity via EGFR regulation. Carcinogenesis 2015; 36: 338–345. [DOI] [PubMed] [Google Scholar]
- 27. Fu Q, Cheng J, Zhang J, et al. miR-20b reduces 5-FU resistance by suppressing the ADAM9/EGFR signaling pathway in colon cancer. Oncol Rep 2017; 37: 123–130. [DOI] [PubMed] [Google Scholar]
- 28. Salendo J, Spitzner M, Kramer F, et al. Identification of a microRNA expression signature for chemoradiosensitivity of colorectal cancer cells, involving miRNAs-320a, -224, -132 and let7g. Radiother Oncol 2013; 108: 451–457. [DOI] [PubMed] [Google Scholar]
- 29. Amankwatia EB, Chakravarty P, Carey FA, et al. MicroRNA-224 is associated with colorectal cancer progression and response to 5-fluorouracil-based chemotherapy by KRAS-dependent and -independent mechanisms. Br J Cancer 2015; 112: 1480–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yin Y, Zhang B, Wang W, et al. miR-204-5p inhibits proliferation and invasion and enhances chemotherapeutic sensitivity of colorectal cancer cells by downregulating RAB22A. Clin Cancer Res 2014; 20: 6187–6199. [DOI] [PubMed] [Google Scholar]
- 31. Wu H, Liang Y, Shen L, et al. MicroRNA-204 modulates colorectal cancer cell sensitivity in response to 5-fluorouracil-based treatment by targeting high mobility group protein A2. Biol Open 2016; 5: 563–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chang HY, Ye SP, Pan SL, et al. Overexpression of miR-194 reverses HMGA2-driven signatures in colorectal cancer. Theranostics 2017; 7: 3889–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu B, Liu Y, Zhao L, et al. Upregulation of microRNA-135b and microRNA-182 promotes chemoresistance of colorectal cancer by targeting ST6GALNAC2 via PI3K/AKT pathway. Mol Carcinog 2017; 56: 2669–2680. [DOI] [PubMed] [Google Scholar]
- 34. Zhang Y, Talmon G, Wang J. MicroRNA-587 antagonizes 5-FU-induced apoptosis and confers drug resistance by regulating PPP2R1B expression in colorectal cancer. Cell Death Dis 2015; 6: e1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mussnich P, Rosa R, Bianco R, et al. MiR-199a-5p and miR-375 affect colon cancer cell sensitivity to cetuximab by targeting PHLPP1. Expert Opin Ther Targets 2015; 19: 1017–1026. [DOI] [PubMed] [Google Scholar]
- 36. Han P, Li JW, Zhang BM, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol Cancer 2017; 16: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu X, Xie T, Mao X, et al. MicroRNA-149 increases the sensitivity of colorectal cancer cells to 5-fluorouracil by targeting forkhead box transcription factor FOXM1. Cell Physiol Biochem 2016; 39: 617–629. [DOI] [PubMed] [Google Scholar]
- 38. Wan LY, Deng J, Xiang XJ, et al. miR-320 enhances the sensitivity of human colon cancer cells to chemoradiotherapy in vitro by targeting FOXM1. Biochem Biophys Res Commun 2015; 457: 125–132. [DOI] [PubMed] [Google Scholar]
- 39. Xiao Z, Qu Z, Chen Z, et al. LncRNA HOTAIR is a prognostic biomarker for the proliferation and chemoresistance of colorectal cancer via MiR-203a-3p-mediated Wnt/ß-catenin signaling pathway. Cell Physiol Biochem 2018; 46: 1275–1285. [DOI] [PubMed] [Google Scholar]
- 40. Liang CQ, Fu YM, Liu ZY, et al. The effect of miR-224 down-regulation on SW80 cell proliferation and apoptosis and weakening of ADM drug resistance. Eur Rev Med Pharmacol Sci 2017; 21: 5008–5016. [PubMed] [Google Scholar]
- 41. Chen B, Zhang D, Kuai J, et al. Upregulation of miR-199a/b contributes to cisplatin resistance via Wnt/β-catenin-ABCG2 signaling pathway in ALDHA1(+) colorectal cancer stem cells. Tumour Biol 2017; 39: 1010428317715155. [DOI] [PubMed] [Google Scholar]
- 42. Lu Y, Zhao X, Liu Q, et al. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/β-catenin signaling. Nat Med 2017; 23: 1331–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jiang T, Ye L, Han Z, et al. miR-19b-3p promotes colon cancer proliferation and oxaliplatin-based chemoresistance by targeting SMAD4: validation by bioinformatics and experimental analyses. J Exp Clin Cancer Res 2017; 36: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sun C, Wang FJ, Zhang HG, et al. miR-34a mediates oxaliplatin resistance of colorectal cancer cells by inhibiting macroautophagy via transforming growth factor-β/Smad4 pathway. World J Gastroenterol 2017; 23: 1816–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Que K, Tong Y, Que G, et al. Downregulation of miR-874-3p promotes chemotherapeutic resistance in colorectal cancer via inactivation of the Hippo signaling pathway. Oncol Rep 2017; 38: 3376–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhao C, Zhao Q, Zhang C, et al. miR-15b-5p resensitizes colon cancer cells to 5-fluorouracil by promoting apoptosis via the NF-κB/XIAP axis. Sci Rep 2017; 7: 4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu H, Yin Y, Hu Y, et al. miR-139-5p sensitizes colorectal cancer cells to 5-fluorouracil by targeting NOTCH-1. Pathol Res Pract 2016; 212: 643–649. [DOI] [PubMed] [Google Scholar]
- 48. Xu K, Shen K, Liang X, et al. MiR-139-5p reverses CD44+/CD133+-associated multidrug resistance by downregulating NOTCH1 in colorectal carcinoma cells. Oncotarget 2016; 7: 75118–75129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang L, Jiang CF, Li DM, et al. MicroRNA-497 inhibits tumor growth and increases chemosensitivity to 5-fluorouracil treatment by targeting KSR1. Oncotarget 2016; 7: 2660–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. He J, Xie G, Tong J, et al. Overexpression of microRNA-122 re-sensitizes 5-FU-resistant colon cancer cells to 5-FU through the inhibition of PKM2 in vitro and in vivo. Cell Biochem Biophys 2014; 70: 1343–1350. [DOI] [PubMed] [Google Scholar]
- 51. Li X, Zhao H, Zhou X, et al. Inhibition of lactate dehydrogenase A by microRNA-34a resensitizes colon cancer cells to 5-fluorouracil. Mol Med Rep 2015; 11: 577–582. [DOI] [PubMed] [Google Scholar]
- 52. Li PL, Zhang X, Wang LL, et al. MicroRNA-218 is a prognostic indicator in colorectal cancer and enhances 5-fluorouracil-induced apoptosis by targeting BIRC5. Carcinogenesis 2015; 36: 1484–1493. [DOI] [PubMed] [Google Scholar]
- 53. Sun Z, Zhou N, Han Q, et al. MicroRNA-197 influences 5-fluorouracil resistance via thymidylate synthase in colorectal cancer. Clin Transl Oncol 2015; 17: 876–883. [DOI] [PubMed] [Google Scholar]
- 54. Li T, Gao F, Zhang XP. miR-203 enhances chemosensitivity to 5-fluorouracil by targeting thymidylate synthase in colorectal cancer. Oncol Rep 2015; 33: 607–614. [DOI] [PubMed] [Google Scholar]
- 55. Chai J, Dong W, Xie C, et al. MicroRNA-494 sensitizes colon cancer cells to fluorouracil through regulation of DPYD. IUBMB Life 2015; 67: 191–201. [DOI] [PubMed] [Google Scholar]
- 56. Liu RL, Dong Y, Deng YZ, et al. Tumor suppressor miR-145 reverses drug resistance by directly targeting DNA damage-related gene RAD18 in colorectal cancer. Tumour Biol 2015; 36: 5011–5019. [DOI] [PubMed] [Google Scholar]
- 57. Deng J, Lei W, Fu JC, et al. Targeting miR-21 enhances the sensitivity of human colon cancer HT-29 cells to chemoradiotherapy in vitro. Biochem Biophys Res Commun 2014; 443: 789–795. [DOI] [PubMed] [Google Scholar]
- 58. Zhou Y, Wan G, Spizzo R, et al. miR-203 induces oxaliplatin resistance in colorectal cancer cells by negatively regulating ATM kinase. Mol Oncol 2014; 8: 83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sui H, Cai GX, Pan SF, et al. miR200c attenuates P-gp-mediated MDR and metastasis by targeting JNK2/c-Jun signaling pathway in colorectal cancer. Mol Cancer Ther 2014; 13: 3137–3151. [DOI] [PubMed] [Google Scholar]
- 60. Ghanbarian M, Afgar A, Yadegarazari R, et al. Through oxaliplatin resistance induction in colorectal cancer cells, increasing ABCB1 level accompanies decreasing level of miR-302c-5p, miR-3664-5p and miR-129-5p. Biomed Pharmacother 2018; 108: 1070–1080. [DOI] [PubMed] [Google Scholar]
- 61. Bitarte N, Bandres E, Boni V, et al. MicroRNA-451 is involved in the self-renewal, tumorigenicity, and chemoresistance of colorectal cancer stem cells. Stem Cells 2011; 29: 1661–1671. [DOI] [PubMed] [Google Scholar]
- 62. To KK, Leung WW, Ng SS. Exploiting a novel miR-519c-HuR-ABCG2 regulatory pathway to overcome chemoresistance in colorectal cancer. Exp Cell Res 2015; 338: 222–231. [DOI] [PubMed] [Google Scholar]
- 63. Wu L, Li S, Peng R, et al. Drug resistance of colon cancer cells to 5-fluorouracil mediated by microRNA-21. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2015; 32: 620–624. [DOI] [PubMed] [Google Scholar]
- 64. Li X, Li X, Liao D, et al. Elevated microRNA-23a expression enhances the chemoresistance of colorectal cancer cells with microsatellite instability to 5-fluorouracil by directly targeting ABCF1. Curr Protein Pept Sci 2015; 16: 301–309. [DOI] [PubMed] [Google Scholar]
- 65. Cao S, Lin L, Xia X, et al. MicroRNA-761 promotes the sensitivity of colorectal cancer cells to 5-Fluorouracil through targeting FOXM1. Oncotarget 2018; 9: 321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yang G, Jiang O, Ling D, et al. MicroRNA-522 reverses drug resistance of doxorubicin-induced HT29 colon cancer cell by targeting ABCB5. Mol Med Rep 2015; 12: 3930–3936. [DOI] [PubMed] [Google Scholar]
- 67. Zhou H, Lin C, Zhang Y, et al. miR-506 enhances the sensitivity of human colorectal cancer cells to oxaliplatin by suppressing MDR1/P-gp expression. Cell Prolif 2017; 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cheng J, Deng R, Zhang P, et al. miR-219-5p plays a tumor suppressive role in colon cancer by targeting oncogene Sall4. Oncol Rep 2015; 34: 1923–1932. [DOI] [PubMed] [Google Scholar]
- 69. Xu K, Liang X, Shen K, et al. miR-297 modulates multidrug resistance in human colorectal carcinoma by down-regulating MRP-2. Biochem J 2012; 446: 291–300. [DOI] [PubMed] [Google Scholar]
- 70. Li Y, Gong P, Hou JX, et al. miR-34a regulates multidrug resistance via positively modulating OAZ2 signaling in colon cancer cells. J Immunol Res 2018; 2018: 7498514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Xu K, Liang X, Cui D, et al. miR-1915 inhibits Bcl-2 to modulate multidrug resistance by increasing drug-sensitivity in human colorectal carcinoma cells. Mol Carcinog 2013; 52: 70–78. [DOI] [PubMed] [Google Scholar]
- 72. Li Q, Liang X, Wang Y, et al. miR-139-5p inhibits the epithelial-mesenchymal transition and enhances the chemotherapeutic sensitivity of colorectal cancer cells by downregulating BCL2. Sci Rep 2016; 6: 27157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Karaayvaz M, Zhai H, Ju J. miR-129 promotes apoptosis and enhances chemosensitivity to 5-fluorouracil in colorectal cancer. Cell Death Dis 2013; 4: e659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Meng X, Fu R. miR-206 regulates 5-FU resistance by targeting Bcl-2 in colon cancer cells. Onco Targets Ther 2018; 11: 1757–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang L, He L, Zhang H, et al. Knockdown of MiR-20a enhances sensitivity of colorectal cancer cells to cisplatin by increasing ASK1 expression. Cell Physiol Biochem 2018; 47: 1432–1441. [DOI] [PubMed] [Google Scholar]
- 76. Qu J, Zhao L, Zhang P, et al. MicroRNA-195 chemosensitizes colon cancer cells to the chemotherapeutic drug doxorubicin by targeting the first binding site of BCL2L2 mRNA. J Cell Physiol 2015; 230: 535–545. [DOI] [PubMed] [Google Scholar]
- 77. Huang G, Chen X, Cai Y, et al. miR-20a-directed regulation of BID is associated with the TRAIL sensitivity in colorectal cancer. Oncol Rep 2017; 37: 571–578. [DOI] [PubMed] [Google Scholar]
- 78. Nishida N, Yamashita S, Mimori K. et al. MicroRNA-10b is a prognostic indicator in colorectal cancer and confers resistance to the chemotherapeutic agent 5-fluorouracil in colorectal cancer cells. Ann Surg Oncol 2012; 19: 3065–3071. [DOI] [PubMed] [Google Scholar]
- 79. Han J, Liu Z, Wang N, et al. MicroRNA-874 inhibits growth, induces apoptosis and reverses chemoresistance in colorectal cancer by targeting X-linked inhibitor of apoptosis protein. Oncol Rep 2016; 36: 542–550. [DOI] [PubMed] [Google Scholar]
- 80. Hua Y, Zhu Y, Zhang J, et al. miR-122 Targets X-linked inhibitor of apoptosis protein to sensitize oxaliplatin-resistant colorectal cancer cells to oxaliplatin-mediated cytotoxicity. Cell Physiol Biochem 2018; 51: 2148–2159. [DOI] [PubMed] [Google Scholar]
- 81. Kim SA, Kim I, Yoon SK, et al. Indirect modulation of sensitivity to 5-fluorouracil by microRNA-96 in human colorectal cancer cells. Arch Pharm Res 2015; 38: 239–248. [DOI] [PubMed] [Google Scholar]
- 82. Liu N, Li J, Zhao Z, et al. MicroRNA-302a enhances 5-fluorouracil-induced cell death in human colon cancer cells. Oncol Rep 2017; 37: 631–639. [DOI] [PubMed] [Google Scholar]
- 83. Qian X, Yu J, Yin Y, et al. MicroRNA-143 inhibits tumor growth and angiogenesis and sensitizes chemosensitivity to oxaliplatin in colorectal cancers. Cell Cycle 2013; 12: 1385–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhang R, Xu J, Zhao J, et al. Knockdown of miR-27a sensitizes colorectal cancer stem cells to TRAIL by promoting the formation of Apaf-1-caspase-9 complex. Oncotarget 2017; 8: 45213–45223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Shang J, Yang F, Wang Y, et al. MicroRNA-23a antisense enhances 5-fluorouracil chemosensitivity through APAF-1/caspase-9 apoptotic pathway in colorectal cancer cells. J Cell Biochem 2014; 115: 772–784. [DOI] [PubMed] [Google Scholar]
- 86. Lian B, Yang D, Liu Y, et al. miR-128 Targets the SIRT1/ROS/DR5 pathway to sensitize colorectal cancer to TRAIL-induced apoptosis. Cell Physiol Biochem 2018; 49: 2151–2162. [DOI] [PubMed] [Google Scholar]
- 87. Zhang Y, Hu X, Miao X, et al. MicroRNA-425-5p regulates chemoresistance in colorectal cancer cells via regulation of programmed cell death 10. J Cell Mol Med 2016; 20: 360–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhang L, Pickard K, Jenei V, et al. miR-153 supports colorectal cancer progression via pleiotropic effects that enhance invasion and chemotherapeutic resistance. Cancer Res 2013; 73: 6435–6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Xu K, Chen G, Qiu Y, et al. miR-503-5p confers drug resistance by targeting PUMA in colorectal carcinoma. Oncotarget 2017; 8: 21719–21732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Fu Q, Cheng J, Zhang J, et al. Downregulation of YEATS4 by miR-218 sensitizes colorectal cancer cells to L-OHP-induced cell apoptosis by inhibiting cytoprotective autophagy. Oncol Rep 2016; 36: 3682–3690. [DOI] [PubMed] [Google Scholar]
- 91. Zhang H, Tang J, Li C, et al. MiR-22 regulates 5-FU sensitivity by inhibiting autophagy and promoting apoptosis in colorectal cancer cells. Cancer Lett 2015; 356: 781–790. [DOI] [PubMed] [Google Scholar]
- 92. Yu X, Shi W, Zhang Y, et al. CXCL12/CXCR4 axis induced miR-125b promotes invasion and confers 5-fluorouracil resistance through enhancing autophagy in colorectal cancer. Sci Rep 2017; 7: 42226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Tan S, Shi H, Ba M, et al. miR-409-3p sensitizes colon cancer cells to oxaliplatin by inhibiting Beclin-1-mediated autophagy. Int J Mol Med 2016; 37: 1030–1038. [DOI] [PubMed] [Google Scholar]
- 94. Han J, Li J, Tang K, et al. miR-338-3p confers 5-fluorouracil resistance in p53 mutant colon cancer cells by targeting the mammalian target of rapamycin. Exp Cell Res 2017; 360: 328–336. [DOI] [PubMed] [Google Scholar]
- 95. Yu T, Guo F, Yu Y, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 2017; 170: 548–563e516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ye J, Wang Z, Zhao J, et al. MicroRNA-141 inhibits tumor growth and minimizes therapy resistance in colorectal cancer. Mol Med Rep 2017; 15: 1037–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ren D, Lin B, Zhang X, et al. Maintenance of cancer stemness by miR-196b-5p contributes to chemoresistance of colorectal cancer cells via activating STAT3 signaling pathway. Oncotarget 2017; 8: 49807–49823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Jin Y, Jiang Z, Guan X, et al. miR-450b-5p suppresses stemness and the development of chemoresistance by targeting SOX2 in colorectal cancer. DNA Cell Biol 2016; 35: 249–256. [DOI] [PubMed] [Google Scholar]
- 99. Qin Y, Chen X, Liu Z, et al. miR-106a reduces 5-fluorouracil (5-FU) sensitivity of colorectal cancer by targeting dual-specificity phosphatases 2 (DUSP2). Med Sci Monit 2018; 24: 4944–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Chen J, Chen Y, Chen Z. MiR-125a/b regulates the activation of cancer stem cells in paclitaxel-resistant colon cancer. Cancer Invest 2013; 31: 17–23. [DOI] [PubMed] [Google Scholar]
- 101. Ding J, Zhao Z, Song J, et al. MiR-223 promotes the doxorubicin resistance of colorectal cancer cells via regulating epithelial-mesenchymal transition by targeting FBXW7. Acta Biochim Biophys Sin (Shanghai) 2018; 50: 597–604. [DOI] [PubMed] [Google Scholar]
- 102. Ren LL, Yan TT, Shen CQ, et al. The distinct role of strand-specific miR-514b-3p and miR-514b-5p in colorectal cancer metastasis. Cell Death Dis 2018; 9: 687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Tanaka S, Hosokawa M, Yonezawa T, et al. Induction of epithelial-mesenchymal transition and down-regulation of miR-200c and miR-141 in oxaliplatin-resistant colorectal cancer cells. Biol Pharm Bull 2015; 38: 435–440. [DOI] [PubMed] [Google Scholar]
- 104. Kim C, Hong Y, Lee H, et al. MicroRNA-195 desensitizes HCT116 human colon cancer cells to 5-fluorouracil. Cancer Lett 2018; 412: 264–271. [DOI] [PubMed] [Google Scholar]
- 105. Zhang Y, Geng L, Talmon G, et al. MicroRNA-520g confers drug resistance by regulating p21 expression in colorectal cancer. J Biol Chem 2015; 290: 6215–6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Chen Y, Bian L, Zhang Y. MiR-505 mediates methotrexate resistance in colorectal cancer by targeting RASSF8. J Pharm Pharmacol 2018; 70: 937–951. [DOI] [PubMed] [Google Scholar]
- 107. Luo J, Liu L, Zhou N, et al. miR-519b-3p promotes responsiveness to preoperative chemoradiotherapy in rectal cancer patients by targeting ARID4B. Gene 2018; 655: 84–90. [DOI] [PubMed] [Google Scholar]
- 108. Zhu J, Zhang R, Yang D, et al. Knockdown of long non-coding RNA XIST inhibited doxorubicin resistance in colorectal cancer by upregulation of miR-124 and downregulation of SGK1. Cell Physiol Biochem 2018; 51: 113–128. [DOI] [PubMed] [Google Scholar]
- 109. Zhang LL, Xie FJ, Tang CH, et al. miR-340 suppresses tumor growth and enhances chemosensitivity of colorectal cancer by targeting RLIP76. Eur Rev Med Pharmacol Sci 2017; 21: 2875–2886. [PubMed] [Google Scholar]
- 110. Ye Q, Su L, Chen D, et al. Astragaloside IV induced miR-134 expression reduces EMT and increases chemotherapeutic sensitivity by suppressing CREB1 signaling in colorectal cancer cell line SW-480. Cell Physiol Biochem 2017; 43: 1617–1626. [DOI] [PubMed] [Google Scholar]
- 111. Tsunekuni K, Konno M, Asai A, et al. MicroRNA profiles involved in trifluridine resistance. Oncotarget 2017; 8: 53017–53027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Shan Y, Liu Y, Zhao L, et al. MicroRNA-33a and let-7e inhibit human colorectal cancer progression by targeting ST8SIA1. Int J Biochem Cell Biol 2017; 90: 48–58. [DOI] [PubMed] [Google Scholar]
- 113. Lu W, Wang J, Yang G, et al. Posttranscriptional regulation of Galectin-3 by miR-128 contributes to colorectal cancer progression. Oncotarget 2017; 8: 15242–15251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Lv Z, Wei J, You W, et al. Disruption of the c-Myc/miR-200b-3p/PRDX2 regulatory loop enhances tumor metastasis and chemotherapeutic resistance in colorectal cancer. J Transl Med 2017; 15: 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Li C, Gao Y, Li Y, et al. TUG1 mediates methotrexate resistance in colorectal cancer via miR-186/CPEB2 axis. Biochem Biophys Res Commun 2017; 491: 552–557. [DOI] [PubMed] [Google Scholar]
- 116. Guo Y, Pang Y, Gao X, et al. MicroRNA-137 chemosensitizes colon cancer cells to the chemotherapeutic drug oxaliplatin (OXA) by targeting YBX1. Cancer Biomark 2017; 18: 1–9. [DOI] [PubMed] [Google Scholar]
- 117. Liu Y, Gao S, Chen X, et al. Overexpression of miR-203 sensitizes paclitaxel (Taxol)-resistant colorectal cancer cells through targeting the salt-inducible kinase 2 (SIK2). Tumour Biol 2016; 37: 12231–12239. [DOI] [PubMed] [Google Scholar]
- 118. Peng L, Zhu H, Wang J, et al. MiR-492 is functionally involved in Oxaliplatin resistance in colon cancer cells LS174T via its regulating the expression of CD147. Mol Cell Biochem 2015; 405: 73–79. [DOI] [PubMed] [Google Scholar]
- 119. Ye L, Jiang T, Shao H, et al. miR-1290 Is a Biomarker in DNA-mismatch-repair-deficient colon cancer and promotes resistance to 5-fluorouracil by directly targeting hMSH2. Mol Ther Nucleic Acids 2017; 7: 453–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Hu J, Cai G, Xu Y, et al. The plasma microRNA miR-1914* and -1915 suppresses chemoresistant in colorectal cancer patients by down-regulating NFIX. Curr Mol Med 2016; 16: 70–82. [DOI] [PubMed] [Google Scholar]
- 121. Kulda V, Pesta M, Topolcan O, et al. Relevance of miR-21 and miR-143 expression in tissue samples of colorectal carcinoma and its liver metastases. Cancer Genet Cytogenet 2010; 200: 154–160. [DOI] [PubMed] [Google Scholar]
- 122. Wang ZH, Gao QY, Fang JY. Loss of PTEN expression as a predictor of resistance to anti-EGFR monoclonal therapy in metastatic colorectal cancer: evidence from retrospective studies. Cancer Chemother Pharmacol 2012; 69: 1647–1655. [DOI] [PubMed] [Google Scholar]
- 123. Danielsen SA, Eide PW, Nesbakken A, et al. Portrait of the PI3K/AKT pathway in colorectal cancer. Biochim Biophys Acta 2015; 1855: 104–121. [DOI] [PubMed] [Google Scholar]
- 124. Nagata Y, Lan KH, Zhou X, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell 2004; 6: 117–127. [DOI] [PubMed] [Google Scholar]
- 125. Padhye S, Banerjee S, Chavan D, et al. Fluorocurcumins as cyclooxygenase-2 inhibitor: molecular docking, pharmacokinetics and tissue distribution in mice. Pharm Res 2009; 26: 2438–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Roy S, Yu Y, Padhye SB, et al. Difluorinated-curcumin (CDF) restores PTEN expression in colon cancer cells by down-regulating miR-21. PLoS One 2013; 8: e68543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Noordhuis MG, Eijsink JJ, Ten Hoor KA, et al. Expression of epidermal growth factor receptor (EGFR) and activated EGFR predict poor response to (chemo)radiation and survival in cervical cancer. Clin Cancer Res 2009; 15: 7389–7397. [DOI] [PubMed] [Google Scholar]
- 128. Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 2008; 26: 1626–1634. [DOI] [PubMed] [Google Scholar]
- 129. Fanale D, Castiglia M, Bazan V, et al. Involvement of non-coding RNAs in chemo- and radioresistance of colorectal cancer. Adv Exp Med Biol 2016; 937: 207–228. [DOI] [PubMed] [Google Scholar]
- 130. Ruzzo A, Graziano F, Vincenzi B, et al. High let-7a microRNA levels in KRAS-mutated colorectal carcinomas may rescue anti-EGFR therapy effects in patients with chemotherapy-refractory metastatic disease. Oncologist 2012; 17: 823–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Ragusa M, Majorana A, Statello L, et al. Specific alterations of microRNA transcriptome and global network structure in colorectal carcinoma after cetuximab treatment. Mol Cancer Ther 2010; 9: 3396–3409. [DOI] [PubMed] [Google Scholar]
- 132. Mlcochova J, Faltejskova P, Nemecek R, et al. MicroRNAs targeting EGFR signalling pathway in colorectal cancer. J Cancer Res Clin Oncol 2013; 139: 1615–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Mlcochova J, Faltejskova-Vychytilova P, Ferracin M, et al. MicroRNA expression profiling identifies miR-31-5p/3p as associated with time to progression in wild-type RAS metastatic colorectal cancer treated with cetuximab. Oncotarget 2015; 6: 38695–38704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer 2007; 7: 899–910. [DOI] [PubMed] [Google Scholar]
- 135. Li Y, He J, Zhong D, et al. High-mobility group box 1 protein activating nuclear factor-κB to upregulate vascular endothelial growth factor C is involved in lymphangiogenesis and lymphatic node metastasis in colon cancer. J Int Med Res 2015; 43: 494–505. [DOI] [PubMed] [Google Scholar]
- 136. Beaulieu JM, Sotnikova TD, Marion S, et al. An Akt/beta-arrestin 2/PP2A signaling complex mediates dopaminergic neurotransmission and behavior. Cell 2005; 122: 261–273. [DOI] [PubMed] [Google Scholar]
- 137. Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell 2005; 18: 13–24. [DOI] [PubMed] [Google Scholar]
- 138. Basu S, Haase G, Ben-Ze’ev A. Wnt signaling in cancer stem cells and colon cancer metastasis. F1000Res 2016; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 1999; 398: 422–426. [DOI] [PubMed] [Google Scholar]
- 140. You Z, Saims D, Chen S, et al. Wnt signaling promotes oncogenic transformation by inhibiting c-Myc-induced apoptosis. J Cell Biol 2002; 157: 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Morin PJ. beta-catenin signaling and cancer. Bioessays 1999; 21: 1021–1030. [DOI] [PubMed] [Google Scholar]
- 142. Rahmani F, Avan A, Hashemy SI, et al. Role of Wnt/β-catenin signaling regulatory microRNAs in the pathogenesis of colorectal cancer. J Cell Physiol 2018; 233: 811–817. [DOI] [PubMed] [Google Scholar]
- 143. Zhang N, Wei P, Gong A, et al. FoxM1 promotes β-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer Cell 2011; 20: 427–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Colak S, Ten Dijke P. Targeting TGF-β signaling in cancer. Trends Cancer 2017; 3: 56–71. [DOI] [PubMed] [Google Scholar]
- 145. Yan P, Klingbiel D, Saridaki Z, et al. Reduced expression of SMAD4 is associated with poor survival in colon cancer. Clin Cancer Res 2016; 22: 3037–3047. [DOI] [PubMed] [Google Scholar]
- 146. Martz CA, Ottina KA, Singleton KR, et al. Systematic identification of signaling pathways with potential to confer anticancer drug resistance. Sci Signal 2014; 7: ra121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Guo PD, Lu XX, Gan WJ, et al. RARγ downregulation contributes to colorectal tumorigenesis and metastasis by derepressing the hippo-yap pathway. Cancer Res 2016; 76: 3813–3825. [DOI] [PubMed] [Google Scholar]
- 148. Jin Y, Wang M, Hu H, et al. Overcoming stemness and chemoresistance in colorectal cancer through miR-195-5p-modulated inhibition of notch signaling. Int J Biol Macromol 2018; 117: 445–453. [DOI] [PubMed] [Google Scholar]
- 149. McCubrey JA, Steelman LS, Chappell WH, et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim Biophys Acta 2007; 1773: 1263–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Kortum RL, Lewis RE. The molecular scaffold KSR1 regulates the proliferative and oncogenic potential of cells. Mol Cell Biol 2004; 24: 4407–4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- 152. Rahman M, Hasan MR. Cancer metabolism and drug resistance. Metabolites 2015; 5: 571–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Wu S, Le H. Dual roles of PKM2 in cancer metabolism. Acta Biochim Biophys Sin 2013; 45: 27–35. [DOI] [PubMed] [Google Scholar]
- 154. Doherty JR, Cleveland JL. Targeting lactate metabolism for cancer therapeutics. J Clin Invest 2013; 123: 3685–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Mattison LK, Soong R, Diasio RB. Implications of dihydropyrimidine dehydrogenase on 5-fluorouracil pharmacogenetics and pharmacogenomics. Pharmacogenomics 2002; 3: 485–492. [DOI] [PubMed] [Google Scholar]
- 156. Offer SM, Butterfield GL, Jerde CR, et al. microRNAs miR-27a and miR-27b directly regulate liver dihydropyrimidine dehydrogenase expression through two conserved binding sites. Mol Cancer Ther 2014; 13: 742–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Valeri N, Gasparini P, Braconi C, et al. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2). Proc Natl Acad Sci U S A 2010; 107: 21098–21103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Kathawala RJ, Gupta P, Ashby CR, Jr., et al. The modulation of ABC transporter-mediated multidrug resistance in cancer: a review of the past decade. Drug Resist Updat 2015; 18: 1–17. [DOI] [PubMed] [Google Scholar]
- 159. Kartner N, Riordan JR, Ling V. Cell surface P-glycoprotein associated with multidrug resistance in mammalian cell lines. Science 1983; 221: 1285–1288. [DOI] [PubMed] [Google Scholar]
- 160. Xie T, Geng J, Wang Y, et al. FOXM1 evokes 5-fluorouracil resistance in colorectal cancer depending on ABCC10. Oncotarget 2017; 8: 8574–8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Haimeur A, Conseil G, Deeley RG, et al. The MRP-related and BCRP/ABCG2 multidrug resistance proteins: biology, substrate specificity and regulation. Curr Drug Metab 2004; 5: 21–53. [DOI] [PubMed] [Google Scholar]
- 162. Leibowitz B, Yu J. Mitochondrial signaling in cell death via the Bcl-2 family. Cancer Biol Ther 2010; 9: 417–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Kim R. Unknotting the roles of Bcl-2 and Bcl-xL in cell death. Biochem Biophys Res Commun 2005; 333: 336–343. [DOI] [PubMed] [Google Scholar]
- 164. Low SY, Tan BS, Choo HL, et al. Suppression of BCL-2 synergizes cisplatin sensitivity in nasopharyngeal carcinoma cells. Cancer Lett 2012; 314: 166–175. [DOI] [PubMed] [Google Scholar]
- 165. Liang T, Zhang X, Xue W, et al. Curcumin induced human gastric cancer BGC-823 cells apoptosis by ROS-mediated ASK1-MKK4-JNK stress signaling pathway. Int J Mol Sci 2014; 15: 15754–15765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Stander XX, Stander BA, Joubert AM. Synergistic anticancer potential of dichloroacetate and estradiol analogue exerting their effect via ROS-JNK-Bcl-2-mediated signalling pathways. Cell Physiol Biochem 2015; 35: 1499–1526. [DOI] [PubMed] [Google Scholar]
- 167. Salvesen GS, Duckett CS. IAP proteins: blocking the road to death’s door. Nat Rev Mol Cell Biol 2002; 3: 401–410. [DOI] [PubMed] [Google Scholar]
- 168. Xiang G, Wen X, Wang H, et al. Expression of X-linked inhibitor of apoptosis protein in human colorectal cancer and its correlation with prognosis. J Surg Oncol 2009; 100: 708–712. [DOI] [PubMed] [Google Scholar]
- 169. Deveraux QL, Reed JC. IAP family proteins-suppressors of apoptosis. Genes Dev 1999; 13: 239–252. [DOI] [PubMed] [Google Scholar]
- 170. Dong X, Javle M, Hess KR, et al. Insulin-like growth factor axis gene polymorphisms and clinical outcomes in pancreatic cancer. Gastroenterology 2010; 139: 464–473, 473.e461–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Dallas NA, Xia L, Fan F, et al. Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res 2009; 69: 1951–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Cecconi F, Alvarez-Bolado G, Meyer BI, et al. Apaf1 (CED-4 homolog) regulates programmed cell death in mammalian development. Cell 1998; 94: 727–737. [DOI] [PubMed] [Google Scholar]
- 173. Zhou M, Li Y, Hu Q, et al. Atomic structure of the apoptosome: mechanism of cytochrome c- and dATP-mediated activation of Apaf-1. Genes Dev 2015; 29: 2349–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. You H, Mak TW. Crosstalk between p53 and FOXO transcription factors. Cell Cycle 2005; 4: 37–38. [DOI] [PubMed] [Google Scholar]
- 175. Degenhardt K, Mathew R, Beaudoin B, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 2006; 10: 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Wei MF, Chen MW, Chen KC, et al. Autophagy promotes resistance to photodynamic therapy-induced apoptosis selectively in colorectal cancer stem-like cells. Autophagy 2014; 10: 1179–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Zhao J, Nie Y, Wang H, et al. MiR-181a suppresses autophagy and sensitizes gastric cancer cells to cisplatin. Gene 2016; 576: 828–833. [DOI] [PubMed] [Google Scholar]
- 178. Helgason GV, Holyoake TL, Ryan KM. Role of autophagy in cancer prevention, development and therapy. Essays Biochem 2013; 55: 133–151. [DOI] [PubMed] [Google Scholar]
- 179. Zhao S, Chen C, Wang S, et al. MHY1485 activates mTOR and protects osteoblasts from dexamethasone. Biochem Biophys Res Commun 2016; 481: 212–218. [DOI] [PubMed] [Google Scholar]
- 180. Yang AD, Fan F, Camp ER, et al. Chronic oxaliplatin resistance induces epithelial-to-mesenchymal transition in colorectal cancer cell lines. Clin Cancer Res 2006; 12: 4147–4153. [DOI] [PubMed] [Google Scholar]
- 181. Kozovska Z, Gabrisova V, Kucerova L. Colon cancer: cancer stem cells markers, drug resistance and treatment. Biomed Pharmacother 2014; 68: 911–916. [DOI] [PubMed] [Google Scholar]
- 182. Takahashi RU, Miyazaki H, Ochiya T. The role of microRNAs in the regulation of cancer stem cells. Front Genet 2014; 4: 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Becker KA, Ghule PN, Lian JB, et al. Cyclin D2 and the CDK substrate p220(NPAT) are required for self-renewal of human embryonic stem cells. J Cell Physiol 2010; 222: 456–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Hou PC, Li YH, Lin SC, et al. Hypoxia-induced downregulation of DUSP-2 phosphatase drives colon cancer stemness. Cancer Res 2017; 77: 4305–4316. [DOI] [PubMed] [Google Scholar]
- 185. Douville J, Beaulieu R, Balicki D. ALDH1 as a functional marker of cancer stem and progenitor cells. Stem Cells Dev 2009; 18: 17–25. [DOI] [PubMed] [Google Scholar]
- 186. Schwickart M, Huang X, Lill JR, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature 2010; 463: 103–107. [DOI] [PubMed] [Google Scholar]
- 187. Hu QD, Chen W, Yan TL, et al. NSC 74859 enhances doxorubicin cytotoxicity via inhibition of epithelial-mesenchymal transition in hepatocellular carcinoma cells. Cancer Lett 2012; 325: 207–213. [DOI] [PubMed] [Google Scholar]
- 188. Korpal M, Lee ES, Hu G, et al. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 2008; 283: 14910–14914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Shah MA, Schwartz GK. Cell cycle-mediated drug resistance: an emerging concept in cancer therapy. Clin Cancer Res 2001; 7: 2168–2181. [PubMed] [Google Scholar]
- 190. Mencia N, Selga E, Noé V, et al. Underexpression of miR-224 in methotrexate resistant human colon cancer cells. Biochem Pharmacol 2011; 82: 1572–1582. [DOI] [PubMed] [Google Scholar]
- 191. Simmer F, Venderbosch S, Dijkstra JR, et al. MicroRNA-143 is a putative predictive factor for the response to fluoropyrimidine-based chemotherapy in patients with metastatic colorectal cancer. Oncotarget 2015; 6: 22996–23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Langer CJ, Gadgeel SM, Borghaei H, et al. Carboplatin and pemetrexed with or without pembrolizumab for advanced, non-squamous non-small-cell lung cancer: a randomised, phase 2 cohort of the open-label KEYNOTE-021 study. Lancet Oncol 2016; 17: 1497–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Tili E, Michaille JJ, Croce CM. MicroRNAs play a central role in molecular dysfunctions linking inflammation with cancer. Immunol Rev 2013; 253: 167–184. [DOI] [PubMed] [Google Scholar]
- 194. Ueda R, Kohanbash G, Sasaki K, et al. Dicer-regulated microRNAs 222 and 339 promote resistance of cancer cells to cytotoxic T-lymphocytes by down-regulation of ICAM-1. Proc Natl Acad Sci U S A 2009; 106: 10746–10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Chen L, Gibbons DL, Goswami S, et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun 2014; 5: 5241. [DOI] [PMC free article] [PubMed] [Google Scholar]