To the Editor:
Light-sheet microscopy is revolutionizing biology by enabling live in toto imaging of entire embryos or organs with minimal phototoxicity1. We present an open hardware and software platform for constructing a customizable microscope for selective-plane illumination microscopy (SPIM). The OpenSPIM platform is shared with the scientific community through a public website (http://openspim.org/), making light-sheet microscopy more accessible so that it can be optimized for various applications.
SPIM is ideally suited for capturing anatomy and patterns of gene activity in large developing specimens expressing fluorescent reporters2. However, because genomes contain thousands of genes and development can last for days, it is necessary to increase the throughput of SPIM acquisitions. We propose to do this by building arrays of affordable SPIM systems that allow the parallel imaging of many samples.
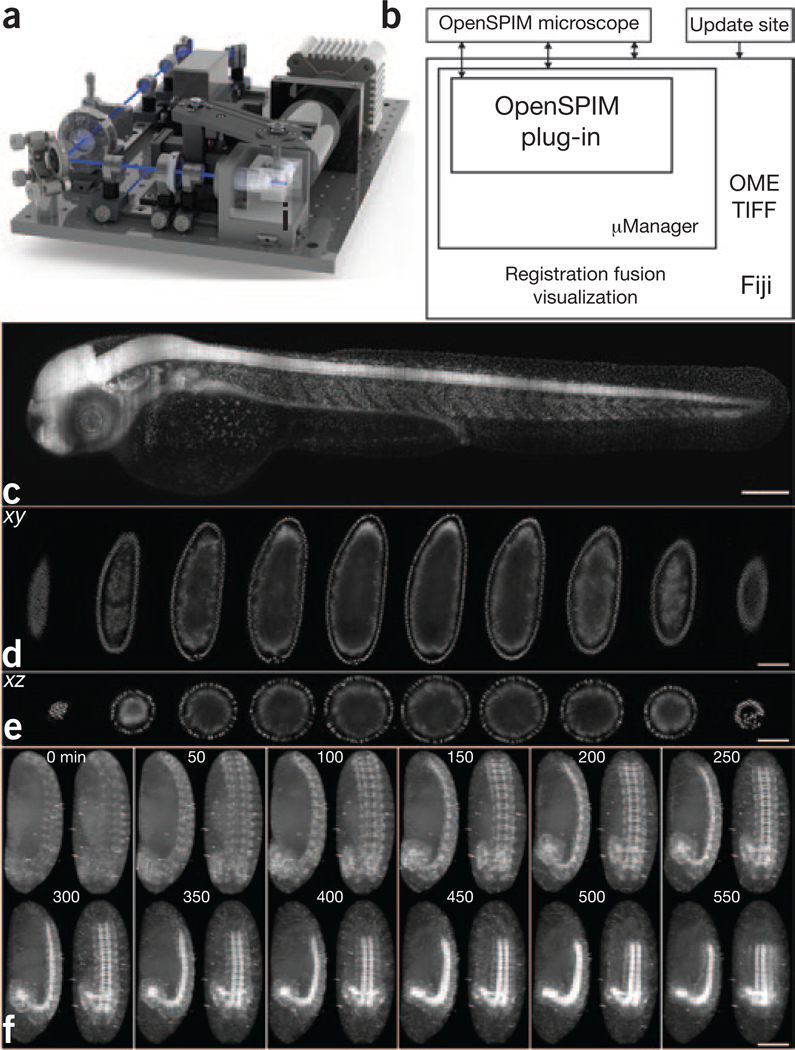
The initial version of the OpenSPIM setup implements a single-sided illumination and single-sided detection light-sheet arrangement (Fig. 1a and Supplementary Fig. 1), the most basic realization of the SPIM principle3. The microscope is controlled via a dedicated plug-in (Supplementary Fig. 2) extending the popular open-source microscopy control software μManager4 running inside Fiji5 on standard computer hardware. This architecture enables active feedback of on-the-fly image processing on image acquisition and facilitates reconstruction and analysis6 (Fig. 1b). To demonstrate the ability of OpenSPIM to image large specimens, we imaged a zebrafish embryo expressing histone H2A– EGFP by acquiring six overlapping fields of view (Fig. 1c and Supplementary Video 1). OpenSPIM can capture fast biological processes, such as the beating zebrafish heart, by recording a single plane at the maximum frame rate of the camera (Supplementary Video 2). For samples that strongly scatter light, such as Drosophila melanogaster embryos, OpenSPIM offers multiview acquisition (Fig. 1d,e and Supplementary Videos 3 and 4)—which, in combination with Fiji’s multiview registration6, can be used to record embryonic development (Supplementary Fig. 3 and Supplementary Video 5). Finally, the four-dimensional recording of the dynamics of gene expression pattern formation during development (Fig. 1f and Supplementary Video 6) is the representative application for which the parallelization offered by OpenSPIM provides the necessary imaging throughput.
Figure 1 |. OpenSPIM hardware, software and data.
(a) SolidWorks rendering of the OpenSPIM setup. (b) Schematic of the OpenSPIM steering software architecture. (c) A 2-d-old zebrafish larva expressing H2A-EGFP under the control of β-actin promoter Tg(Bactin:H2A-EGFP) was imaged as a set of six overlapping fields of view. (d,e) Lateral (d) and axial (e) planes through a blastoderm-stage Drosophila embryo expressing histone-YFP imaged from six views at 6-μm steps of the light sheet. The data were reconstructed using a bead-based registration algorithm and fused with multiview deconvolution in Fiji. (f) Drosophila embryos expressing Csp-sGFP protein fusion under native promoter control imaged from five views every 10 min. Maximum-intensity projections along the lateral and dorsal-ventral axes are shown for every fifth time point, highlighting the dynamic morphogenetic movement of the nervous system. Scale bars, 100 μm.
The unique feature of the OpenSPIM project is its emphasis on an open development process and community. We have established a wiki site that documents in great detail the building, alignment, software and operation of OpenSPIM. The wiki contains a comprehensive list of parts (http://openspim.org/Table_of_parts); detailed instructions for assembly supplemented with photographs, drawings and videos (http://openspim.org/Step_by_step_assembly); narrated tutorials for light-sheet alignment and sample preparation (http://openspim.org/Operation); and descriptions of the open-source software that are useful for users as well as developers (http://openspim.org/Category:Software).
The OpenSPIM system was intentionally designed to be simple, compact, modular and accessible. The current configuration of OpenSPIM (Supplementary Fig. 4a) can be extended to more advanced SPIM geometries (two-sided illumination and/or detection) (Supplementary Fig. 4b,c), and its modular properties will allow users to multiplex time-lapse acquisitions by building several setups operating in parallel (OpenSPIM farm; Supplementary Fig. 4d). Its simplicity makes it an excellent teaching tool for both applying and building SPIM technology. The combination of open-access hardware and open-source software creates a platform adapt-able for specific imaging tasks and a starting point for prototyping of new concepts. The OpenSPIM wiki invites interested researchers to build the setup and in turn contribute their knowledge and innovations back to the wiki site. We envision that this will nucleate an interdisciplinary community in which biologists interested in using SPIM will interact with technology developers and bring to life innovative ideas for addressing complex scientific questions.
Supplementary Material
ACKNOWLEDGMENTS
We thank V. Surendranath for help with photography, H. Bellen (Baylor College of Medicine, Texas) for the Csp-sGFP transgene, and S. Singh, Sonal and S. Simmert for seeding the wiki with material during the Dresden International PhD Program course. We thank B. Cox and G. Petry of the Morgridge Institute for Research for assistance with 3D printing and parts fabrication. J.S., L.S. and K.W.E. were supported by US National Institutes of Health grants RC2GM092519 and R01CA136590. S.P. was supported by the Human Frontier Science Program Postdoctoral Fellowship. P.T. and P.G.P. were supported by the European Research Council Community’s Seventh Framework Programme (FP7/2007-2013) grant agreement 260746.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Weber M & Huisken J. Curr. Opin. Genet. Dev. 21, 566–572 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Ejsmont RK, Sarov M, Winkler S, Lipinski KA & Tomancak P. Nat. Methods 6, 435–437 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Huisken J, Swoger J, Del Bene F, Wittbrodt J & Stelzer EH Science 305, 1007–1009 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Edelstein A, Amodaj N, Hoover K, Vale R & Stuurman N. Curr. Protoc. Mol. Biol. 92, 14–20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schindelin J et al. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Preibisch S, Saalfeld S, Schindelin J & Tomancak P. Nat. Methods 7, 418–419 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.