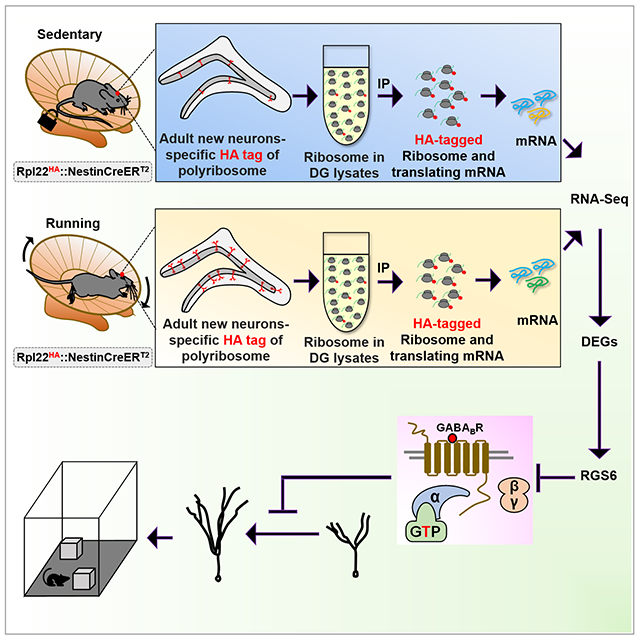
SUMMARY
Voluntary running enhances adult hippocampal neurogenesis, with consequences for hippocampal dependent learning ability and mood regulation. However, the underlying mechanism remains unclear. Here, we show that voluntary running induces unique and dynamic gene expression changes specifically within the adult-born hippocampal neurons, with significant impact on genes involved in neuronal maturation and human diseases. We identify the regulator of G protein signaling 6 (RGS6) as a key factor that mediates running impact on adult-born neurons. RGS6 overexpression mimics the positive effects of voluntary running on morphological and physiological maturation of adult new neurons and reduced sensitivity of adult-born neurons to inhibitory effect of GABAB receptor activation. Knocking down RGS6 abolishes running-enhanced neuronal maturation and hippocampal neurogenesis-dependent learning and anxiolytic effect. Our study provides the data resource unveiling the genome-wide intrinsic molecular changes in adult-born hippocampal neurons that contribute to voluntary running-induced neurogenesis.
Graphical Abstract
INTRODUCTION
Adult mammalian neurogenesis persists at a low level in the dentate gyrus (DG) where radial glial-like neural stem cells (NSCs) give rise to immediate progenitors (IPCs) that subsequently differentiate into new DG neurons integrating into the hippocampal circuity (Kempermann et al., 2015). Although the extent of adult neurogenesis in humans remains unresolved, mounting evidence support important roles of adult hippocampal neurogenesis in neuroplasticity, brain homeostasis, and behavioral adaptations (Deng et al., 2010; Hochgerner et al., 2018; Kempermann et al., 2015). Reduced adult hippocampal neurogenesis has been shown in neurodevelopmental and neurodegenerative diseases as well as normal aging, which may contribute to decline in cognitive functions (Christian et al., 2014; Thompson et al., 2008; Winner and Winkler, 2015). Therefore enhancing adult neurogenesis has been considered as a potential therapeutic method (Yun et al., 2016). Adult neurogenesis is sensitive to external stimuli and experiences (Aimone et al., 2010; Eisinger and Zhao, 2018). Among the positive stimuli, voluntary running is one of the most effective methods for promoting adult hippocampal neurogenesis. Since its first discovery (Kempermann et al., 1997; van Praag et al., 1999), extensive experimental evidence has supported that voluntary running significantly elevates adult hippocampal neurogenesis and improves hippocampus-dependent learning and memory (reviewed by(Eisinger and Zhao, 2018; Vivar et al., 2013). At the cellular level, voluntary running promotes adult NSC activation (Dong et al., 2019), increases NSC and IPC proliferation (van Praag et al., 1999), elevates immature neuron survival (Snyder et al., 2009), accelerates morphological maturation (Eadie et al., 2005; Sah et al., 2017; Steib et al., 2014), and enhances integration of new neurons (Deshpande et al., 2013; Piatti et al., 2011; Trinchero et al., 2017; Vivar et al., 2016). In fact, voluntary running is one of the most effective methods for reversing negative impact of aging and neurodegeneration on both adult neurogenesis and cognitive impairment (Berchtold et al., 2019; Trinchero et al., 2017). However, the mechanism underlying the effect of voluntary running on adult hippocampal neurogenesis remains poorly understood.
Both candidate gene approaches and more recently genome-wide transcriptomic methods have been used to interrogate the hippocampal or DG tissues of running animals and several genes have been identified, for example, brain derived neurotrophic factor (Bdnf) [reviewed by Eisinger and Zhao (Eisinger and Zhao, 2018)]. Although exercise-activated C-FOS positive cells has been isolated for transcriptomic analysis (Chatzi et al., 2019), no study has illustrated genome-wide expression profiles within adult newborn neurons in response to voluntary running. Because adult new neurons constitute an extremely small portion of the entire cell populations of the DG tissue that contains mostly mature neurons and glia as well as many other cell types (Hochgerner et al., 2018), the differentially expressed genes identified in the hippocampal or DG tissues may not represent those differentially expressed genes in the new neurons. The small number of adult newborn neurons presents a major challenge for genome-wide assessment specifically in these cells.
In this study, we showed that adult-born neurons mounted dynamic translational changes in response to voluntary running, with significant impact on genes involved in neuronal maturation and human diseases. We found that G protein signaling 6 (RGS6) is a key mediator of running-induced neurogenesis. Overexpression of RGS6 in adult new neurons increased morphological and electrophysiological maturation and reduced their sensitivity to inhibitory effect of GABAB receptor activation, mimicking the effects of running. Knocking down RGS6 in adult new neurons abolished running-enhanced neuronal maturation and hippocampal neurogenesis-dependent learning and anxiolytic effect. Our study has provided a genome-wide view of intrinsic molecular changes in the adult newborn neurons and unveiled mechanisms underlying voluntary running-enhanced neurogenesis.
RESULTS
Adult-born new neurons mount unique and dynamic translational responses to voluntary running
Voluntary running leads to increased cell proliferation in the subgranular zone (SGZ) of the DG (van Praag et al., 1999). To validate our running platform, we analyzed mice that have been running for 7-days and observed a significant increase (1.6-fold) in the number of BrdU-labeled cells in the running mice compared to controls (Figure S1A–C). A major challenge in studying gene expression in adult-born new DG neurons is the extremely small number of these new cells among vast mature cell populations. Fluorescent activated cell sorting used by others (Bracko et al., 2012) and us (Gao et al., 2017) is disruptive to live cells (Lacar et al., 2016). We therefore chose a conditional ribosome tagging (RiboTag) technology by generating inducible adult new neuron-specific ribosome-tagging mice (Rpl22HA:Nes-CreERT2; termed Nes-HA) through crossing inducible Nes-CreERT2 driver mice (Lagace et al., 2007) and conditional ribosome tagging mice (Rpl22HA) (Sanz et al., 2009). Tamoxifen (Tam) injection of Nes-HA mice leads to HA tagging of ribosomes specifically in NESTIN-expressing NSCs and their differentiated progenies (Figure 1A). To investigate translational responses to running in adult-born new neurons at different stages of neuronal development, the mice were exposed to wheels for 3 different time periods representing early, middle, and late stages of developmental maturation of adult new neurons (Kempermann et al., 2015): 11, 18 and 27 days (Figure 1B). We first assessed whether specificity of ribosome tagging. As expected, nearly all HA+ cells were located in the SGZ of the DG (Figure 1C, Figure S1). In both the 11-days and 18-days sedentary mice, around 80% HA+ cells expressed immature neuron marker Doublecortin (DCX, Figure 1D, Figure S1D–H). The percentage of DCX-positive (DCX+) cells among HA+ cells dropped to ~65% but the percentage of HA+ cells expressing mature neuron marker NeuN (NeuN+HA+) increased to ~80% in 27-days sedentary mice (Figure S1G, H), which is expected as a result of continuous maturation of HA+ cells. In the 18-days sedentary mice, 84.7 ± 0.7% HA+ cells were DCX+, 10.4 ± 0.7% were DCX−NeuN+, 3.5 ± 1.5% were DCX−MCM2+ cells, and only 1.4 ± 0.9% were other cell types (Figure 1C–D). Thus, most of the HA+DCX− cells were either mature neurons or dividing progenitors. Similar cell fate results were obtained in HA+ cells of running mice (Figure S1E–H). Therefore, Tam injection into Nes-HA mice leads to HA tagging ribosome (RiboTag) specifically in adult NSCs and their differentiated progenies in both running and sedentary mice, and the majority of cells labeled at all three time points were DCX+ neurons undergoing neuronal maturation.
Figure 1. Adult-born hippocampal neurons mount unique and dynamic translational responses to voluntary running.
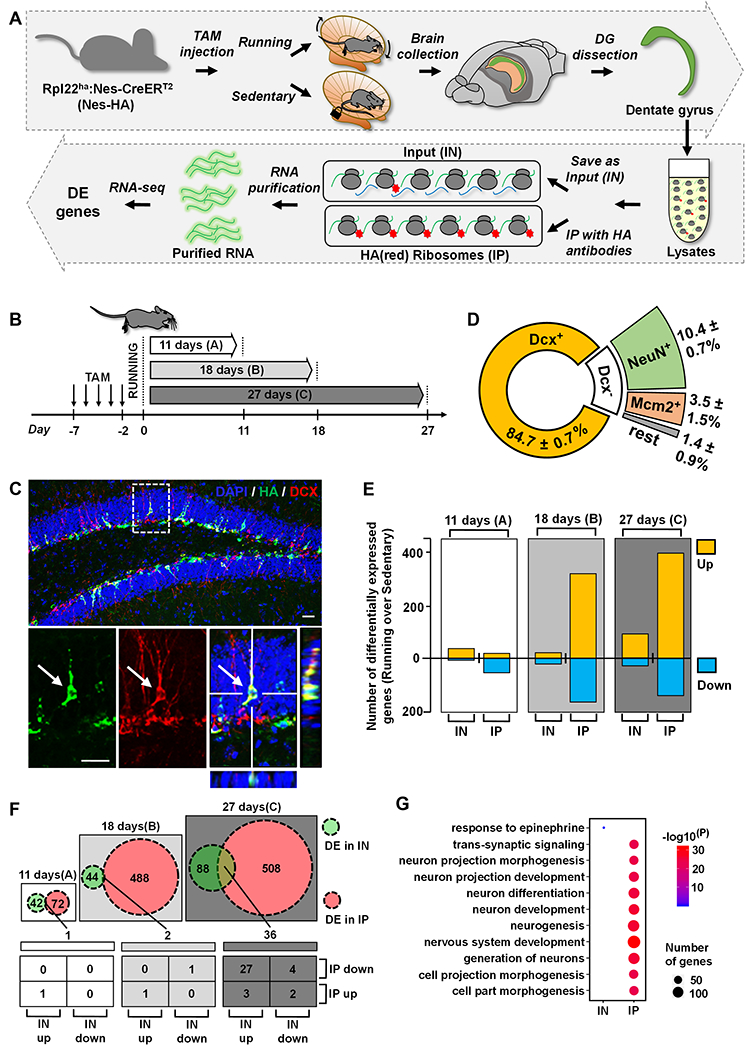
(A) A schematic drawing of the experimental design for translational profiling of adult new neurons in running and sedentary mice using RiboTag-Seq.
(B) Experimental scheme for RiboTag-Seq of mice subjected to running and sedentary treatments for three different time periods.
(C) Confocal images showing that most HA-positive (HA+, green) cells are Doublecortin-positive (DCX+, red) in 18-days sedentary mice. Scale bar, 50μm.
(D) Pie chart depicting the percentages of DCX+, NeuN+ and MCM2+ in HA+ cells in 18-days sedentary mice.
(E) Bar chart depicting numbers of upregulated or downregulated genes in the IN and IP samples in running mice.
(F) Numbers of shared differentially expressed genes (DEGs) in the IN and IP samples in running mice.
(G) GO analysis (top biological process GO terms) of the unique DEGs in the IN and IP in 27-days samples (the “88 genes” (IN) and “508 genes” (IP) in figure F).
We dissected DG tissues from another cohort of mice to perform RiboTag immunoprecipitation (IP) of HA-tagged ribosomes together with their associated translating mRNAs. The IP mRNAs, as well as input (IN) mRNAs, were then identified by RNA-sequencing (RiboTag-seq) (Figure 1A). There was no strong batch effect among both IP and IN samples (Figure S2A–B). We then performed PCA analysis of all 12 IP samples. The samples from 18-days and 27-days, but not 11-days, running mice can be successfully separated from corresponding sedentary mice (Figure S2C). Differentially expressed genes (DEGs) were successfully identified in running mice using embedded EBseq in RSEM with a cutoff of 0.05 of “PPEE” (posterior probability estimated by EBSeq that a gene/transcript is equally expressed) (Table S1). Bdnf showed higher expression in the running mice compared to control sedentary mice in both IP and IN samples whereas Calb2 (Calretinin) showed higher expression in only IP samples (Figure S2D–E), consistent with published literature (Brandt et al., 2003).
Overall, the IP samples (new neurons) had more DEGs than IN samples (DG tissue). Both IP and IN had more upregulated genes compared to down regulated genes in both the 18-days and the 27-days time points (Figure 1E). Interestingly, there were only 1 and 2 DEGs shared between IP and IN samples at the11-days and 18-days time points, respectively. In contrast, 36 DEGs were shared between the IP and IN samples at the 27-days time point (Figure 1F)(Table S1). The shared DEGs at 11-days, Mylk3 (myosin light chain kinase 3) was upregulated in both IP and IN. Among the 2 shared DEGs at 18-days, Ttr (transthyretin) was upregulated in both IP and IN, whereas Csmd3 (CUB and Sushi multiple domains 3) was downregulated in both IP and IN samples. To our surprise, in 27-days samples, 29 out of 36 DEGs showed opposite directions of changes in IN vs IP, with 2 genes upregulated in IP but downregulated in the IN and 27 genes downregulated in IP but upregulated in the IN (Figure 1F), suggesting that new neurons responded differently to running compared to other mature DG cells. We performed GO analysis (Table S2) of the unique DEGs in IN (88 DEGs in figure 1F) and IP (508 DEGs in figure 1F) samples separately, the 88 genes DEGs in IN were associated with only one biological process GO term, “response to epinephrine cellular” (Figure 1G). In contrast, the 508 unique DEGs in IP were related to many neurogenesis and neuronal maturation-related biological process GO terms (Figure 1G) . We also performed pathway analysis of the 29 DEGs that showed opposite directions in IP vs IN in 27 days samples and identified cytosolic calcium ion concentration as one of 7 biological processes and membrane as the only cellular component, suggesting possible changes in neuronal activities in running mice (Table S2). In summary, these results demonstrate that adult born new neurons mount distinct gene expression response to voluntary running compare to majority of other cells residing in the DG.
Voluntary running leads to dynamic gene expression changes in adult-born new neurons across stages of neurogenesis.
Single nuclei RNA sequencing has identified transcriptional signatures of adult new DG cells over a 14-day period post-BrdU labeling (Habib et al., 2016). To interrogate the neurogenic stages of our IP samples, we utilized the data from Habib et al (Habib et al., 2016) to create a developmental trajectory using pseudotime analysis by Monocle (Habib et al., 2016; Trapnell et al., 2014). Using DEGs between running and sedentary as transcriptional signatures, we recovered the developmental trajectory of adult-born DG neurons over 14 days (Habib et al., 2016) and mapped gene expression profiles of our IP samples onto this trajectory (Figure 2A). The sedentary samples and 11-day running samples were clustered together with immature cells on the trajectory, implying that neither the sedentary condition nor short period running elicit significant impact on neuronal maturation process. On the other hand, the 18- and 27- days running samples were separated from other samples and mapped to more mature neurons. This suggests that the effects of running appear after a longer period of voluntary running and new neurons in running mice mature faster compared to those in the control sedentary mice.
Figure 2. Adult New Neurons Exhibit Dynamic Gene Express Changes Across Stages of Neurogenesis in Response to Running.
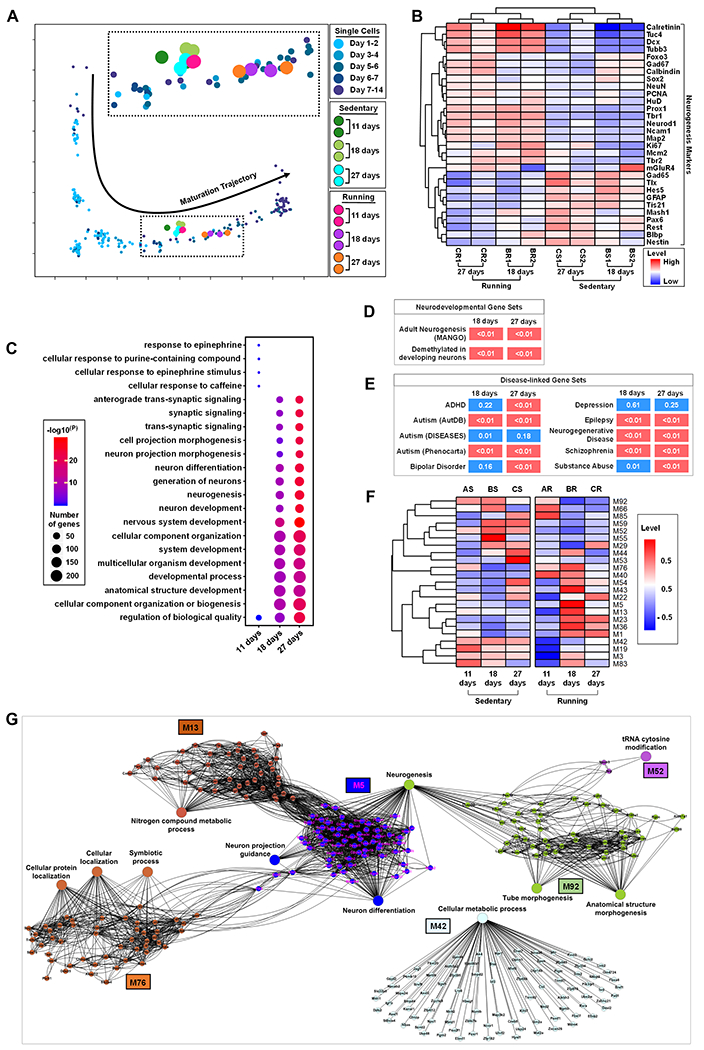
(A) Mapping of IP samples from running and sedentary mice in neuronal maturation single cell trajectory.
(B) Heatmap of 30 known neurogenesis related markers in 18-days and 27-days IP samples in sedentary and running mice.
(C) GO analysis of DEGs in 11-days, 18-days and 27-days running mice.
(D) Enrichment of 18-days and 27-days DEGs that are implicated in adult hippocampal neurogenesis and DNA demethylation in neurons during postnatal cortical development.
(E) Enrichment heatmap for disease-related genes in 18-days and 27-days DEGs. Diseases are represented by rows and DEGs are represented by columns. P values are shown in each cell and are color coded (<0.01 (red) and >0.01 or =0.01 (blue)). The Autism1 gene set was curated by Phenocarta, Autism2 by AutDB, and Autism3 by the DISEASES database. All other gene sets are from Phenocarta.
(F) Top 23 most different eigengene modules in sedentary and running mice.
(G) Gene network of the top 23 eigengene module genes in (F).
To further validate this observation, we assessed the expression patterns of 30 genes that that have been widely used to define the developmental stages of adult neurogenesis. Although there were fewer DEGs at the 11-days time point, running groups could be separated from the sedentary groups, with 18 neurogenesis early stage associated genes (Figure S2H), including striking upregulation of Calb2 (Figure S2F–G), an immature neuron maker shown to be upregulated in response to running (Brandt et al., 2003; Gregoire et al., 2014). For the 18-days and 27-days time points, running groups were clearly separated from the sedentary groups and different time points were also separated from each other (Figure 2B). The genes corresponding to both immature neurons (e.g. Calb2, Dcx, NeuroD1, and Tbr2) and mature neurons (e.g. HuD, Map2, Prox1) exhibited higher translational levels in running groups compared to controls. Whereas genes corresponding to immature NSCs and IPCs (Nestin, Pax6, and Tlx) displayed lower translational levels in running group compared to controls.
We performed GO analysis of DEGs (Table S2) and compiled the top ten biological process GO terms from each time point (Figure 2D). Interestingly, 4 out of top 5 biology process GO terms in 11-day running condition were specific to 11-days condition, such as response to epinephrine (Figure 2C). On the other hand, both the 18- and 27-days running time points had top biological process GO terms related to neuronal differentiation and maturation-related neurogenesis processes (Figure 2C). In addition, the DEGs in the 18-days and 27-days time points were significantly enriched with genes regulating adult hippocampal neurogenesis [MANGO: (Overall et al., 2012)] and genes exhibiting dynamic DNA demethylation in cortical neurons during postnatal neuronal maturation (Lister et al., 2013) (Figure 2D). We have shown that genes upregulated in more mature DCX-expressing cells in the adult DG are significantly enriched for genes involved in autism-related disorders (Gao et al., 2017). Using a similar analysis, we found that the DEGs in both 18- and 27-days time points were significantly enriched for genes associated with both neurodevelopmental and neurodegenerative disorders, such as autism, schizophrenia and substance abuse (Figure 2E). Therefore the new neurons mount dynamic and development-stage specific gene expression changes in response to voluntary running and many of these genes are important for neuronal development and functions.
Next, we asked whether gene expression changes were dynamic across time points. We performed gene co-expression network analysis using WGCNA and clustered genes into co-expression modules (Langfelder and Horvath, 2008). The genes with similar temporal expression dynamics were clustered together in the same module (e.g., select modules in Figure 2F, Figure S2I). In total, we identified 92 modules (Figure S2I) and 23 modules which exhibited most different expression dynamics between running and sedentary controls, as shown by modular eigengenes (Figure 2F). For example, some modules (e.g.M53, M59, M85) showed dramatic upregulation in the sedentary mice but down regulation in running mice from 11-days to 27-days. In contrast, some modules (e.g.M1, M23, M36) showed dramatic upregulation in the running mice but not in the sedentary mice from 11-days to 27-days (Figure 2F). These results demonstrate dynamic changes of gene expression during maturation that are differentially affected in running and sedentary mice.
To further identify the biological functions related to running-specific expression dynamics, we also identified enriched pathways in gene co-expression modules (Table S3). Several running-specific modules have enriched neurogenesis-related biological processes, for example, module M5 genes were enriched with neuron differentiation, neurogenesis and neuron projection guidance, and M92 genes were enriched with neurogenesis (Figure 2G). A number of biological processes including cellular protein localization, symbiotic process and cellular localization were associate with enriched genes in M76. In addition, M13 and M42 module genes were associated with biological processes nitrogen compound metabolic process and cellular metabolic process respectively. These results revealed dynamic regulations of biological processes during maturation of the adult-born new neurons and differential dynamic changes in running and sedentary mice. In summary, adult-born DG new neurons exhibit intrinsic dramatic changes in response to running and these changes correspond to an accelerated maturation.
Running elevates RGS6 expression in adult newborn neurons
Several genes exhibited significant changes in adult newborn neurons across all three time points (Figure 3A). The top downregulated gene was Abca8a, a mouse ortholog genes of human ABCA8 (ATP Binding Cassette Subfamily A Member 8). Abca8a was downregulated in both IP and IN. The top upregulated gene was Rgs6 which was specifically upregulated in IP, but not in the IN of running mice (Figure 3B–C), suggesting that RGS6 might mediate adult-born new neurons-specific response to running. We therefore decided to further assess the roles of RGS6 upregulation in running-induced neurogenesis. We first assessed the expression patterns of RGS6 during adult neurogenesis. We found that RGS6 protein was low in radial glia-like (GFAP+NESTIN+) NSCs and (NESTIN+ or Ki67+) IPCs , but was expressed in both DCX+ immature neurons and NeuN+ mature neurons (Figure S3A–D), suggesting a functional role of RGS6 in neurons (Figure S3C–D). Indeed RGS6 protein levels were upregulated in HA+ new neurons in 18-day running mice compared to controls (Figure 3D–E). RGS6 belongs to the R7-Rgs subfamily of RGS proteins that accelerate the GTPase activity of G(i/o)-class of Gα to terminate signaling cascades of G protein-coupled receptors (GPCRs) (Ross and Wilkie, 2000). RGS6 is enriched in both hippocampal and cortical neurons (Stewart et al., 2014) and has been shown to modulate GABAB receptor signaling (Maity et al., 2012) that inhibits adult neurogenesis (Giachino et al., 2014). However, whether RGS6 plays a role in any of the biological processes in adult-born new neurons has never been investigated.
Figure 3. Adult newborn neurons in running mice have increased RGS6 levels.
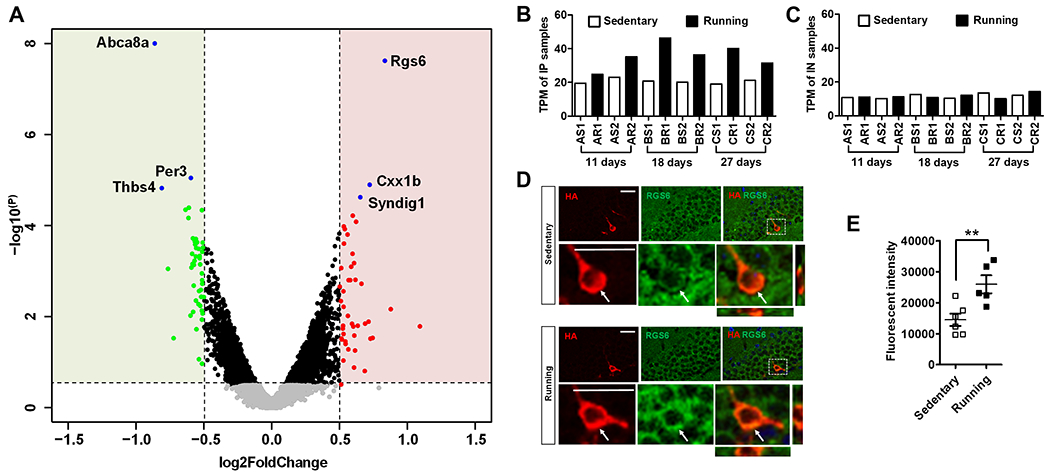
(A) A volcano plot showing DEGs between running and sedentary groups. Genes with more than 1.5 fold change (>1.5 or < −1.5-fold) and p-value (<0.05) were indicated with color (green, down regulated; red, upregulated in running).
(B-C) RNA-Seq TPM values showed Rgs6 levels were higher increased in the IP (B), but not in the IN samples (C) in running mice compared to sedentary mice.
(D) Sample confocal images used for quantitative analysis in F, showing RGS6 (green) and HA (red) immunoreactivities in the DG of sedentary and running Nes-HA mice. Scale bar, 25μm.
(E) Quantitative results of RGS6 protein levels in the HA+ cells in 18-days sedentary (n=6) and running (n=5) mice (**, p < 0.01; t-test).
Enhancing RGS6 expression in adult-born new neurons promotes neuronal maturation
To determine the potential neurogenic functions of elevated RGS6 in adult born neurons, we performed GO analysis of all the genes within the eigengenes-module M5 that contains Rgs6 (Table S3). Interestingly, the most significant biological processes generated from M5 module genes were related to neuronal maturation (Figure 4A). Since running enhances morphological maturation in running mice (Steib et al., 2014), we hypothesized that increased levels of RGS6 in new neurons played a role in running-enhanced neuronal maturation.
Figure 4. Enhancing RGS6 levels in adult-born neurons promotes neuronal maturation.
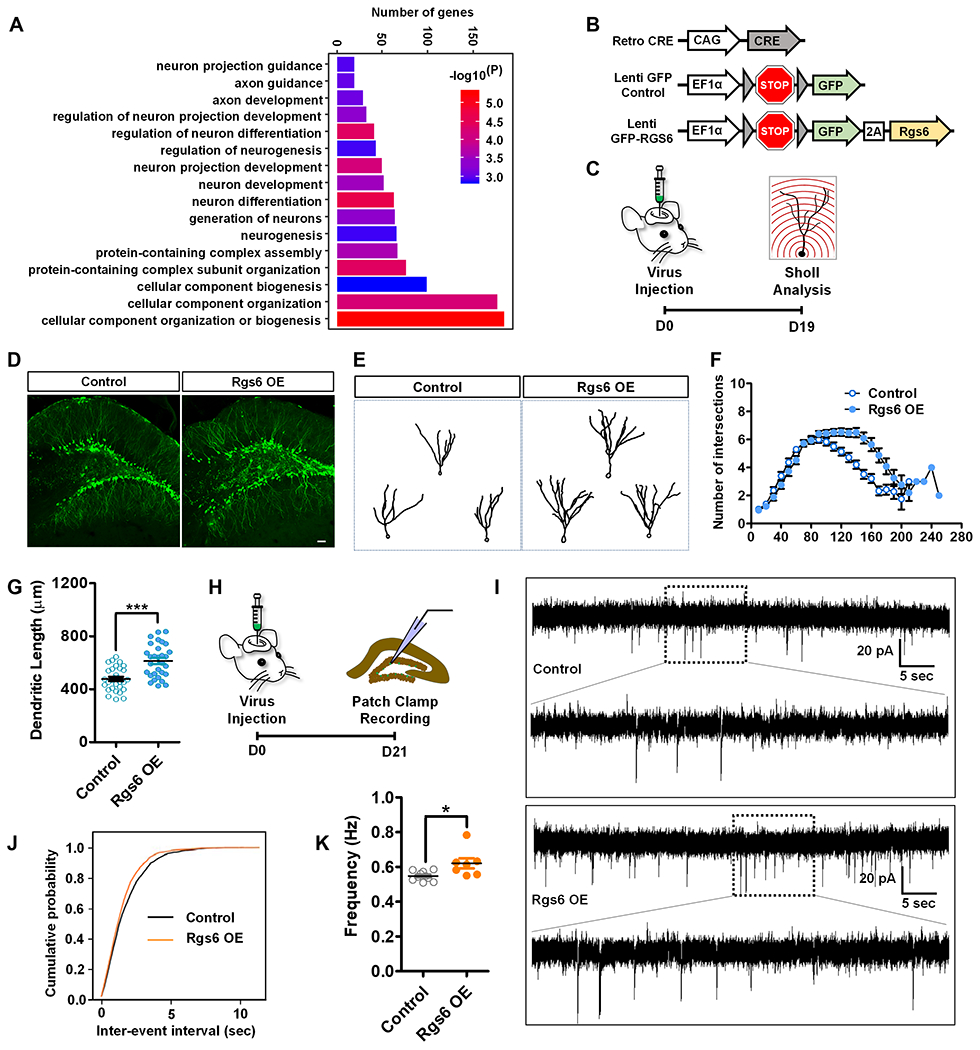
(A) GO analysis of Rgs6 (M5) module genes.
(B) A schematic illustration of viral vectors used for in vivo targeting of adult new neurons: retroviral vector expressing Cre recombinase (Retro-CRE), lentiviral vector with CRE-dependent expression of GFP (Lenti-GFP-Control), and lentiviral vector with CRE-dependent expression of GFP-RGS6 (Lenti-GFP-RGS6).
(C) Timeline of in vivo viral targeting for morphological analysis of adult-born new neurons.
(D) Sample confocal images of virus-infected adult new neurons. Scale bar, 50μm.
(E) Sample traces of GFP-expressing neurons.
(F) Overexpression of RGS6 increases the dendritic complexity compared to controls (F1,60= 26.357, p < 0.001, MANOVA).
(G) Exogenous expression of RGS6 increases dendritic length of adult new neurons compared to controls (p<0.0001, t-test).
(H) Timeline of in vivo viral targeting for electrophysiological analysis of adult-born new neurons.
(I) Representative traces of mEPSCs recorded from GFP+ new neurons in acute hippocampal slices derived from Control and RGS6 OE mice 3 weeks after retroviral injection.
(J-K) Cumulative probability distribution (Control, n= 10 cells; RGS6 OE, n= 7 cells.) and average frequency (P=0.0154, on two tailed t-test). *P< 0.05, **P< 0.01, ***P< 0.001.
There are multiple RGS6 protein isoforms resulted from alternative splicing. Although the antibody we used (Figure 3D) recognized all major RGS6 isoforms, only one Rgs6 isoform, equivalent to the human alpha2 long RGS6 isoform (RGS6Lα2; AH011570.2) (Chatterjee et al., 2003), was highly expressed in adult-born neurons and was also differentially expressed between running and sedentary conditions (Figure S4A). We next overexpressed RGS6Lα2 in adult new neurons using a lentivirus vectors expressing express either RGS6 (GFP-RGS6) or GFP alone (Control) in a Cre-dependent manner, together with retrovirus expressing Cre into the adult DG (Figure 4B, C and Figure S4B–C). Only dividing adult NSC or IPCs could be targeted by retrovirus. RGS6 overexpression did not significantly impact NPC proliferation (Figure S5). (Figure 4D–E). We found that at 19 days after viral injection newborn neurons expressing GFP-RGS6 showed increased dendritic complexity and total dendritic length compared to neurons expressing GFP alone (Figure 4D–G). Therefore increased RGS6 levels in adult-born new neurons of sedentary mice leads to enhanced dendritic maturation.
Adult new neurons exhibit electrophysiological changes reflecting maturation stages (Ge et al., 2006; Trinchero et al., 2019). We performed whole-cell patch clamp recording at 3 weeks post-viral injection (Figure 4H). New neurons infected with GFP-RGS6 virus exhibited increased frequency of miniature excitatory postsynaptic currents (mEPSCs) compared to new neurons infected with GFP virus (Figure 4I–K). Taken together, increased expression levels of RGS6 in adult-born neurons in sedentary mice mimics the effect of running on neuronal maturation in running mice.
Enhancing RGS6 expression in adult-born neurons reduces their sensitivity to GABAB receptor activation
RGS6 accelerates the GTPase activity of G(i/o) to terminate GPCR signaling (Ross and Wilkie, 2000). In fact, “regulation of GTPase activity” and “GTPase binding” GO terms were identified in 508 unique DEGs of 27-days IP samples but not IN samples (Table S2). Prior studies have shown that RGS6 modulates signaling by the GABAB receptor (Maity et al., 2012). We found that GABA related GO terms were identified in 508 unique DEGs 27-days IP samples but not in IN samples (Table S2). GABAB receptor activation is known to inhibit maturation of adult hippocampal new neurons (Giachino et al., 2014). We hypothesized that running-induced upregulation of RGS6 in adult-born immature neurons render them less sensitive to GABAB receptor activation (Figure 5A). Downstream effects of GABAB receptor activation include activation of G protein-gated inwardly rectifying K+ channels (GIRKs) and inhibition of voltage-gated Ca2+ channels (VGCCs) (Hollinger and Hepler, 2002; Padgett and Slesinger, 2010). Since immature adult-born neurons initially lack functional GIRK signaling (Gonzalez et al., 2018), we assessed the strength of GABAB receptor-mediated suppression of VGCCs by the GABAB receptor agonist baclofen. We first tested whether voluntary running altered baclofen-induced suppression of VGCCs, using Ascl1-CreERT2::Rosa26-STOP-TdTomato (Ascl1-tdT) mice to identify adult-born neurons (Yang et al., 2015) (Figure S6). After tamoxifen induction, Ascl1-tdT mice were exposed to running wheels or locked wheels for 22-26 days and then we recorded pharmacologically-isolated Ca2+ currents in tdT+ adult-born neurons (Figure 5B). In sedentary mice, baclofen (30 μM) reduced the peak Ca2+ current to 41 ± 8% of control (Figure 5C). On the contrary, in running mice baclofen did not significantly reduce peak Ca2+ currents (81 ± 10% of control, Figure 5D). This result suggests that running renders adult new neurons less sensitive to GABAB-receptor mediated suppression of neuronal activation.
Figure 5. Both Voluntary Running and Elevating RGS6 Expression Levels in Adult-born Neurons Reduces Their Sensitivity to GABAB Receptor-Mediated inhibition.
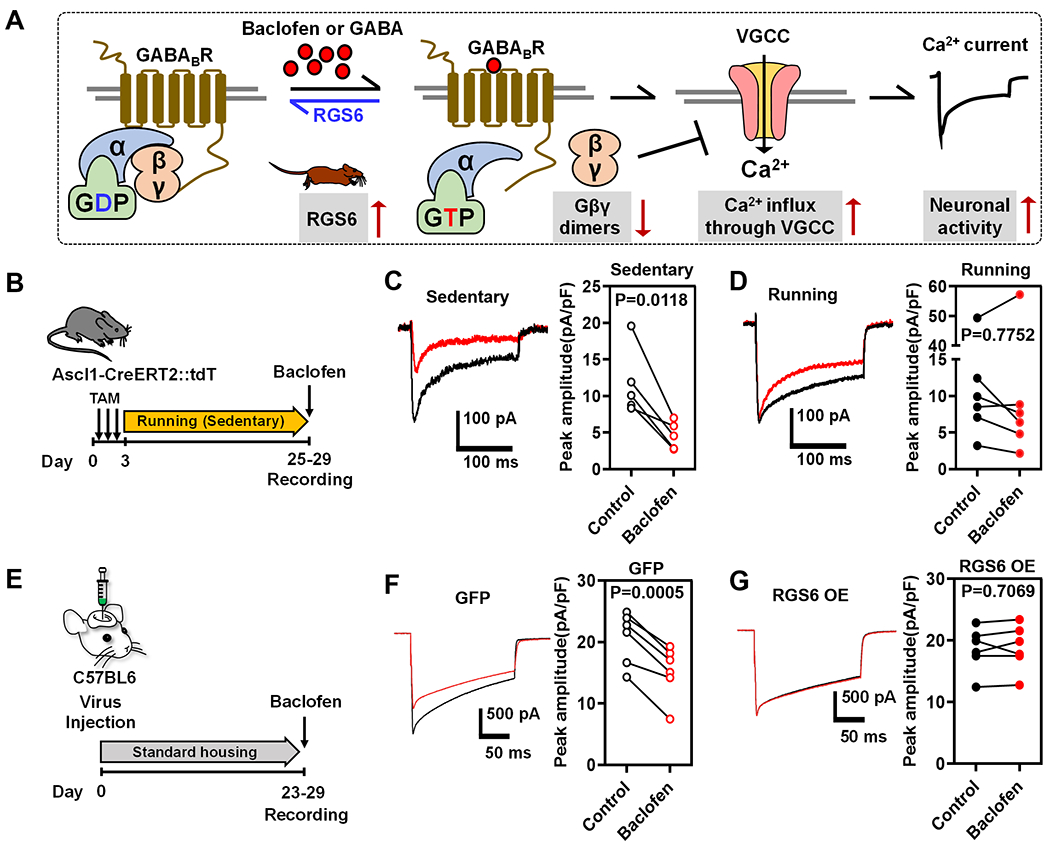
(A) Hypothetic model for the role of RGS6 in reduced sensitivity to GABAB receptor signaling in running mice. GABAB receptor activation by GABA or baclofen leads to activation of Gα and dissociation of Gβγ subunits from trimeric G protein complex. Gβγ is inhibitory to voltage-gated calcium channel (VGCC) in adult new neurons. Elevated RGS6 accelerates GTP hydrolysis therefore terminates GABAB receptor activation and relieves the inhibitory effects on VGCC, rendering adult new neurons less sensitive to GABA or baclofen-activated GABAB signaling.
(B) Experimental timeline for testing the efficiency of the GABAB agonist baclofen on suppression of Ca2+ currents in immature GCs in sedentary and running mice.
(C-D) Average traces (left) of baclofen-induced (30 μM, red) suppression of peak Ca2+ currents recorded in immature neurons from sedentary (C) and running (D) mice. In sedentary mice (C, right), baclofen reduced the peak current from 11.7 ± 2.3 pA/pF to 4.6 ± 0.9 pA/pF (n=5, P=0.01, paired t-test), whereas baclofen had no significant effect on currents in running mice (15.1 ± 7 pA/pF to 14.5 ± 8.6 pA/pF, n=6, P=0.8, paired t-test)(D, right).
(E) Experimental timeline for testing the efficiency of baclofen on suppression of Ca2+ currents in immature GCs in GFP (GFP only Control) and RGS6 OE (RGS6 overexpression) mice.
(F-G) Left, examples of baclofen-induced (30 μM, red) suppression of peak Ca2+ currents recorded from control GFP immature GCs (F) or RGS6 OE immature GCs (G). Right, in control GFP immature GCs baclofen reduced the peak Ca2+ current from 20.7 ± 1.7 pA/pF to 15.2 ± 1.7 (n=6, P<0.001, paired t-test). In RGS6 OE immature GCs baclofen did not affect Ca2+ currents (18.6 ± 1.4 to 18.8 ± 1.5, n=6, P=0.7).
Next we assessed whether elevated RGS6 contributed to the running-induced reduction in GABAB receptor-mediated signaling using the same viral targeting strategy (Figure 5E). In mice expressing control GFP, baclofen (30 μM) uniformly reduced peak Ca2+ currents to 73 ± 5% of control (Figure 5F), whereas in mice targeted with RGS6 OE, baclofen did not significantly reduce peak Ca2+ currents (101 ± 3% of control, Figure 5G). Hence, elevating expression of RGS6 specifically in adult-born immature neurons rendered them less sensitive to GABAB receptor activation. Therefore both running mice with elevated RGS6 in adult neurons and targeted RGS6 overexpression in adult new neurons indicate that RGS6 overexpression suppresses GABAB-receptor mediated G protein signaling.
RGS6 is required for running-enhanced maturation of adult-born new neurons
We next assessed whether RGS6 is necessary for running-enhanced neuronal maturation. We targeted adult-born neurons using retrovirus expressing either control shRNA (shNC) or shRgs6 (Figure S7), as well as GFP. We first subjected shNC retrovirus injected mice to either voluntary running or sedentary conditions for 17 days (Figure 6A–B). GFP+ newborn neurons in running mice showed significant increase in dendritic complexity and total dendritic length (Figure 6C–F), consistent with literature (Steib et al., 2014). We then subjected shRgs6 injected mice with to either running or sedentary treatment for 17 days. Newborn neurons with RGS6 knockdown did not show significant differences in either dendritic complexity or dendritic length (Figure 6G–I) between running and sedentary conditions. Therefore RGS6 is a necessary mediator for running-induced neuronal dendritic maturation.
Figure 6. Knockdown of RGS6 in adult-born new neurons abolishes running effect on new neuron maturation.
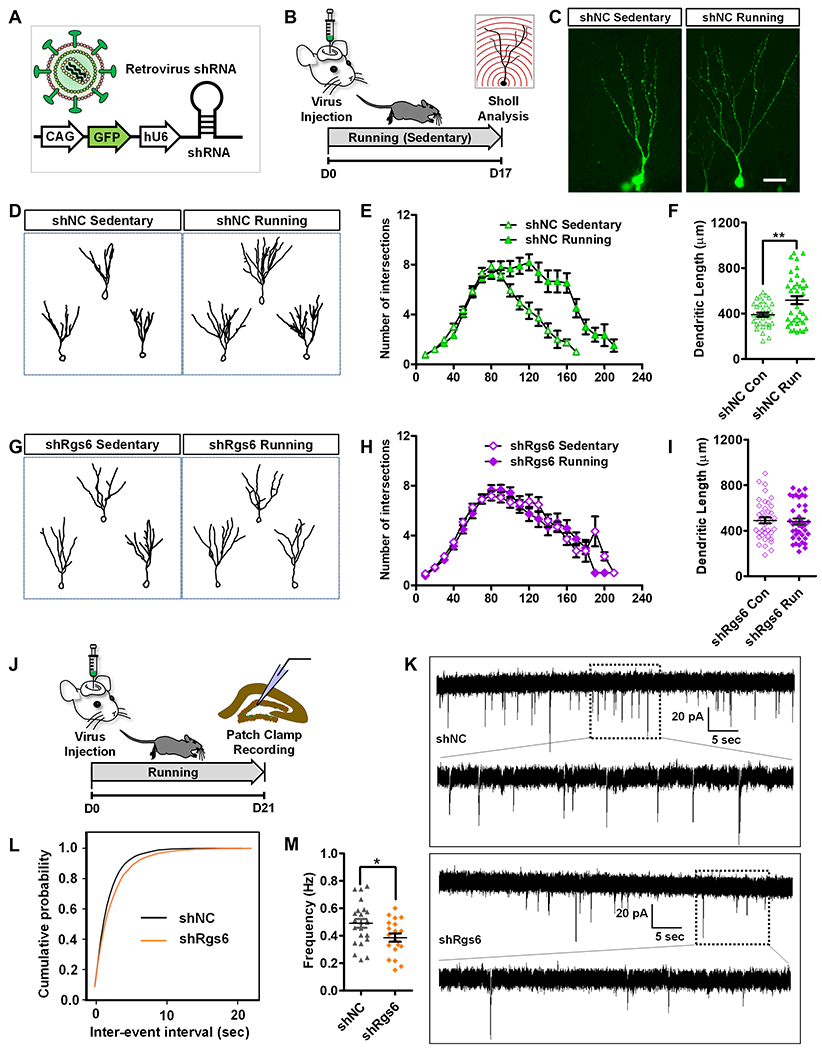
(A) A schematic illustration of retroviral vector expressing shRNA and GFP for targeting adult new neurons.
(B) Experimental timeline for accessing the effect of RGS6 knockdown on dendritic morphology of adult new neurons in running and sedentary mice.
(C) Sample confocal images of virus-infected and GFP+ neurons. Scale bar, 50μm.
(D) Sample traces of retrovirus shNC-infected neurons in running and sedentary mice.
(E) Running increases dendritic complexity of adult-born new neurons in mice injected with control shNC virus (F1,72=9.241, p < 0.01, MANOVA).
(F) Running increases the dendritic length of adult-born new neurons (p<0.01, t-test).
(G-I) RGS6 knockdown abolishes running-induced increased dendritic complexity (shRgs6-sedentary vs. shRgs6-running, F1,80 =0.23, p=0.633, MANOVA) and increased dendritic length (p>0.05, t-test).
(J) Experimental timeline for patch clamp recording of adult-born new neurons with RGS6 knockdown.
(K) Representative traces of mEPSCs recorded from GFP+ DG neurons in acute hippocampal slices derived from shNC and shRgs6 mice 3 weeks after retroviral injection.
(L-M) Cumulative probability distribution (shNC, n= 23 cells; shRgs6, n= 19 cells. t-test) and average frequency (P=0.0259, t-test). *P< 0.05, **P< 0.01, ***P< 0.001.
To further assess the role of RGS6 upregulation played in neuronal maturation, we performed whole-cell patch clamp recording of immature neurons infected with retroviral shRgs6 or shNC (shRNA control) in the running mice (Figure 6J). The frequency of mEPSCs recorded from RGS6 knockdown cells was significantly decreased compared with those in controls (Figure 6K–M). These results suggest that RGS6 knockdown in immature neurons abolishes the running effects of elevated ability to receive excitatory transmission which may result from dendritic maturation deficits. Therefore, RGS6 is essential for running effects on neuronal maturation.
RGS6 is required for running-enhanced hippocampal-dependent learning ability and anxiolytic effect
Voluntary running enhances both spatial and associative learning (reviewed by (Eisinger and Zhao, 2018; Vivar et al., 2013)), as well as exerts anxiolytic effect (Schoenfeld et al., 2016) in rodents. We first injected control lentivirus into the DG of adult mice, housed them with either running or locked wheels for 4 weeks, and subjected them to several tests that have been used to demonstrate running-induced behavioral changes, including novel location test for spatial learning (Snigdha et al., 2014), contextual fear conditioning forassociative learning (Lin et al., 2012), and elevated plus maze for anxiety (Schoenfeld et al., 2016). We found that running mice exhibited reduced anxiety (Figure 7B–C) and enhanced spatial and contextual learning, compared to sedentary mice (Figure 7E–F and Figure 7H–I), which was consistent with literature (Lin et al., 2012; Schoenfeld et al., 2016; Snigdha et al., 2014). We then injected another cohort of adult mice with virus expressing either shNC or shRgs6 and then subjected both groups of mice to running followed by the same behavioral tests. We found that shRgs6 virus-injected mice had higher levels of anxiety, reduced spatial and contextual learning performance compared to shNC virus-infected mice (Figure 7D, G and J). In fact, running mice injected with shRgs6 virus exhibited similar levels of learning ability as the sedentary mice injected with control shNC virus (Figure 7C–D, Figure 7F–G and Figure 7I–J). Therefore, RGS6 is required for running-induced adult-born new neuron maturation and hippocampal neurogenesis-dependent learning enhancement and anxiolytic effect.
Figure 7. RGS6 is Required for Running-induced Learning and Memory Enhancement and Anxiolytic Effect.
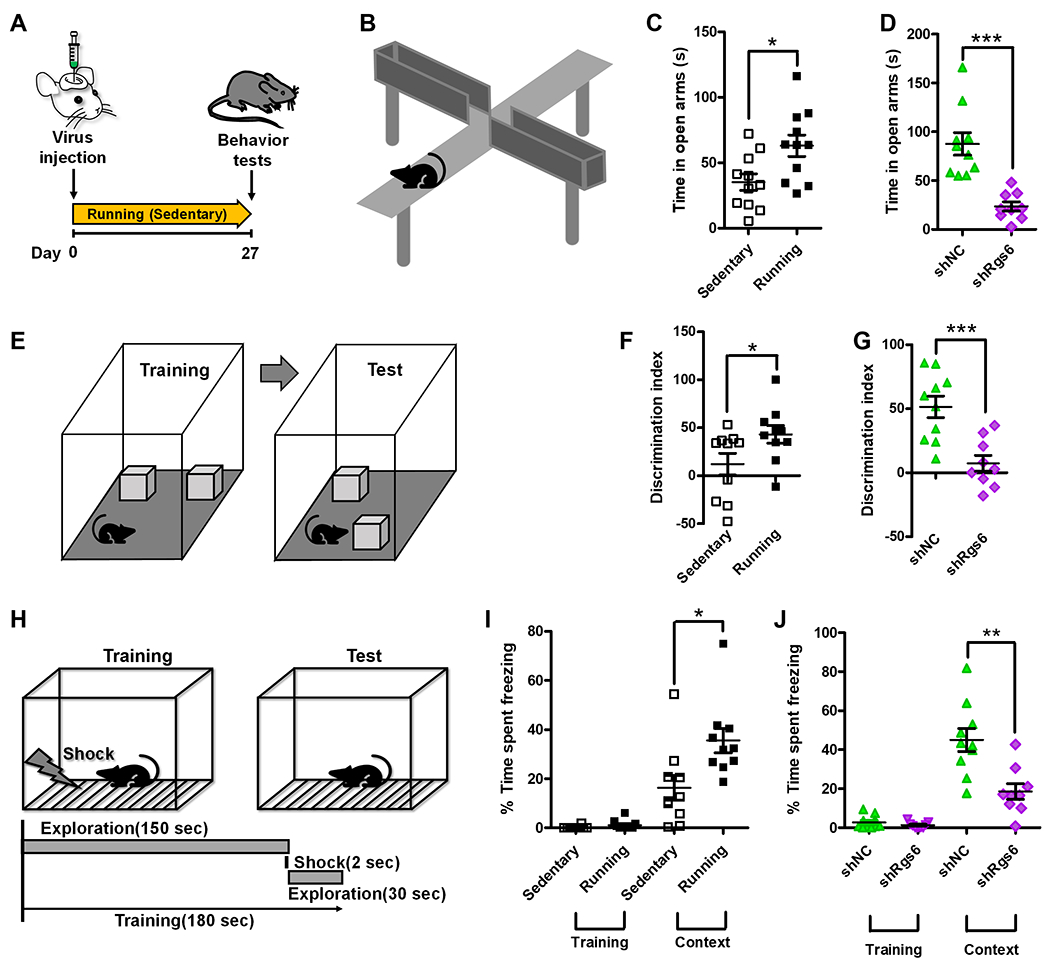
(A) Experimental timeline for assessing the effect of RGS6 knockdown on anxiety, spatial learning and memory in running and sedentary mice.
(B) A scheme cartoon illustrates elevated plus maze (EPM) test.
(C) Running mice exhibit reduced anxiety compared to sedentary mice as measured using an EPM test (P= 0.0141, Sedentary: 35.22 ± 6.25; Running: 63.02 ± 8.21).
(D) Knockdown of Rgs6 abolishes running-induced reduced anxiety in running mice (P= 0.0001, shNC: 87.60 ± 11.46; shRgs6: 23.51 ± 4.73).
(E) A scheme cartoon illustrates novel location test (NLT).
(F) Running mice exhibit improved spatial learning compared to sedentary mice measured by NLT (P= 0.0485, Sedentary: 12.07 ± 11.38; Running: 43.08 ± 9.233).
(G) Knockdown of RGS6 abolishes running-induced improvement on spatial learning in running mice (P= 0.0007, shNC: 51.46 ± 8.36; shRgs6: 7.27 ± 6.27).
(H) A scheme cartoon illustrates contextual fear conditioning test.
(I) Running mice exhibit improved contextual learning compared to sedentary mice measured by a contextual fear conditioning test (P= 0.014, Sedentary: 16.35 ± 5.05; Running: 35.54 ± 4.93).
(J) RGS6 Knockdown abolishes running-induced improvement on contextual learning in running mice (P= 0.0021, shNC: 45.01 ± 5.85; shRgs6: 18.61 ± 4.04). *P< 0.05, **P< 0.01, ***P< 0.001. Data are presented as mean ± s.e.m.
DISCUSSION
We have discovered that, in response to voluntary running, adult-born new neurons in the DG mount dynamic translational changes that are distinct from those of other cell types within the DG tissue with genes important for neuronal maturation dramatically differentially expressed in new neurons but not in DG tissue. We demonstrated that Rgs6 has an important role in running-induced new neuron maturation and behavioral improvements. Therefore, our study has provided a genome-wide view of intrinsic molecular changes in the adult newborn neurons and unveil a molecular mechanism underlying voluntary running-induced neurogenesis.
Adult hippocampal neurogenesis is sensitive to external stimuli and experiences, a feature that is important for both understanding diseases and developing therapeutics. Some of the most effective positive stimuli for adult neurogenesis include voluntary running and enriched environment (Kempermann, 2015). Since the first publications on positive effects of running on adult neurogenesis (Kempermann et al., 1997; van Praag et al., 1999), significant interest and effort have been dedicated into the understanding the underlying molecular mechanisms. To date, at least ten research groups have used either microarray or RNA sequencing to interrogate the hippocampal or DG tissues in rodent models subjected to running wheels and post-mortem hippocampal tissues from humans who have been physically active for at least two decades (Berchtold et al., 2019; Chatzi et al., 2019; Eisinger and Zhao, 2018; Gregoire et al., 2018). However none of these studies has assessed genome-wide changes specifically in adult-born new neurons in response to voluntary running. Our study has therefore filled this gap of knowledge.
Although adult born immature neurons represent an extremely small portion of the total cell populations in the DG, therefore assessing intrinsic changes in adult neurons is difficult by using dissected hippocampal and DG tissues. A number of new methods have been applied to isolate new neurons from the adult DG including cell sorting used by several others and us (Bracko et al., 2012; Gao et al., 2017; Shin et al., 2015). However tissue dissociation may cause extensive cellular stress, especially to neurons (Habib et al., 2016). The adult new neuron-specific RiboTag-seq used here does not require dissociation of live cells therefore avoid confound of disruptive cell isolation. This method proves powerful in identifying intrinsic genome-wide molecular changes in adult new neurons, as well as other small number of cells in mature tissues. As a future direction, it would be informative to assess impact of enriched environment, aging, and disease mutations on translational responses of adult new neurons using this method and compare to the effect of running. A limitation of our method is that the HA-labeled cells contain adult born new cells that are not exactly at the same developmental stages. Single cell analysis of directly isolated adult Nestin+ NPCs (Shin et al., 2015) and DCX+ immature neurons (Gao et al., 2017) have been used to assess developmental progression of these new cells but tissue dissociation may not recover mRNA localized in dendrites. In addition, the read coverage of single cell analysis remain limited, prohibiting the identification of some low expressing regulators. RiboTag-seq at single cell levels has not yet been developed.
RGS6 belongs to a group of RGS protein that regulates G protein activity which in turn modulate the activity of neurotransmitter receptors and diverse signaling pathways including those associated with cell growth and differentiation (Hollinger and Hepler, 2002). Among RGS proteins, RGS6, 7, 11 share homology and targets (Anderson et al., 2009). However only RGS6 was upregulated in running mice. In addition, although both adult new neurons and mature neurons express RGS6, RGS6 upregulation was specific to adult-born new neuron. Furthermore, upregulation of RGS6 and knockdown of RGS6 in adult new neurons had significant impact on morphology, excitability of new neurons, which have been shown to significantly impact behaviors (Goncalves et al., 2016). Therefore the behavioral deficits we observed in running mice with RGS6 knockdown is likely, if not solely, resulted from RGS6 reduction in adult new neurons rather than other cell types in the DG. Our results also point to a possible explanation for the role of RGS6 in activity-dependent new neuron maturation. GABAergic mechanisms contribute to activity-dependent modulation of adult neurogenesis via both GABAA and GABAB receptors. RGS6 inhibits the GABAB-GIRK signaling pathway by accelerating deactivation of Gα and reducing the availability of free Gβγ (Hollinger and Hepler, 2002; Maity et al., 2012; Padgett and Slesinger, 2010). While GABAB-GIRK signaling is a well-known target of RGS6, the late development of functional GIRK channels during adult neurogenesis makes it unlikely to mediate RGS6 effects on running-induced maturation (Gonzalez et al., 2018). Regulation of VGCCs is more likely to mediate activity-dependent regulation at early stages of maturation, when depolarization generates Ca2+ influx that is important for many intracellular signaling pathways regulating neurodevelopment including dendrite arborization (Chevaleyre et al., 2002; Greer and Greenberg, 2008; Rajan and Cline, 1998; Wong and Ghosh, 2002). In adult-born neurons, Ca2+ influx generated by GABAA receptor depolarization is thought to promote activity-dependent development (Ge et al., 2007). Our data suggests that RGS6 could promote neuronal maturation by releasing the inhibitory effect of GABAB receptor signaling on voltage-activated Ca2+ influx (Figure 5), potentially providing a synergistic interaction with GABAA mediated depolarization. It would be interesting for future studies to investigate this interaction, as well as to investigate how running leads to elevated RGS6 in adult new neurons. In addition, our current investigation has focused on how upregulation of RGS6 in new neurons of running mice promoted neuronal maturation. It would be interesting to further investigate how RGS6 and specific isoforms of RGS6 regulate functions of neurons in both with or without external stimuli.
Because our IN data are essentially transcriptome of the DG tissues, we compared our 27-days IN data with the only DG RNA-seq dataset from 28-days running CD1 strain of mice (Gregoire et al., 2018). We found 25 of 124 of down-regulated and 18 of 124 of up-regulated our DEGs overlapped with published DEGs; however most (~80%) of our DEGs were not shared with the published DEGs. This difference could be due to different genetic background and tissue processing methods. In addition, we found that different sets of genes in IN samples responded to voluntary running. The genes upregulated in IN of 11-days running mice are associated with synaptic related biological process GO terms such as chemical synaptic transmission (Table S4). Since at 11-days, most new neurons are quite immature, these synaptic changes in the IN likely represent the early response of mature neurons. Interestingly, genes upregulated in IN of longer-period running (18-days and 27-days) do not seem to contribute to synaptic changes. Therefore our data demonstrate that mature DG neurons and adult-born new neurons mount different gene expression responses in response to voluntary running.
In conclusion, our study has provided the first genome-wide view of intrinsic molecular changes in the adult newborn neurons and that contribute to voluntary running-induced neurogenesis. The novel mediators we have identified such as RGS6 may have the potentials to serve as therapeutic targets for neurodegenerative diseases and aging.
STAR ★ METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Xinyu Zhao (Xinyu.zhao@wisc.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
All figures except for Figure 2A were generated from raw data or processed data. The data generated and/or analyzed during this study are available from the Lead Contact upon reasonable request. The next generation sequencing data have been submitted to GEO (GSE142678). Gene co-expression network analysis using WGCNA and clustered genes into co-expression modules (Langfelder and Horvath, 2008). The GO analysis performed using online g:Profiler (website: https://biit.cs.ut.ee/gprofiler/gost). The generation of the developmental trajectory using pseudotime analysis by Monocle (Trapnell et al., 2014).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice and animal husbandry
We performed all procedures involving live mice in accordance with the NIH Guide for the Care and Use of Laboratory Animals and the protocols approved by the University of Wisconsin-Madison and the University of Alabama at Birmingham Animal Care and Use Committee. We generated the inducible conditional ribosome tagging female mice (Tg(Nestin-cre/ERT2)::RPL22HA/WT, simplified as Nes-CreERT2::RiboTag) by crossing transgenic driver Tg(Nes-cre/ERT2) mice (Jackson Laboratory Stock No:016261)(Lagace et al., 2007) with RPL22HA/WT mice (Jackson Laboratory Stock No:029977) (Sanz et al., 2009). Nestin-GFP male mice were the same mouse line published previously (Jobe et al., 2017). Mice were kept in 12 hours light/12 dark cycle housing. Both the Tg(Nes-Cre/ERT2) and RPL22HA/WT mice were C57BL/6J genetic background. To induce recombination, mice (9–11 weeks old) received Tamoxifen (Sigma-Aldrich) daily for 5 days (160 mg kg–1 i.p., 40 mg ml–1 in 10% ethanol mixed with sunflower oil, Sigma-Aldrich). Running mice were individually housed in regular mouse cages that contain one running wheel with wireless monitor per cage (Med Associates inc). Sedentary mice were housed in identical cages as running mice except that the wheels were glued (blocked) and without wireless monitor (Med Associates, inc). To record from newly generated adult born neurons, we used Ascl1CreERT2 (Ascl1tm1(Cre/ERT2)Jejo/J) male and female mice, obtained from the Jackson Laboratory (Stock No:012882) crossed with CAGfloxStop-tdTomato (Ai14) (B6;129S6-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J) conditional reporter line (Jackson Laboratory Stock No:007914). Tamoxifen was delivered as described above. Functional and locked running wheels were attached to standard cages (Kaytee brand). The mice were kept in 12 hours light/12 dark cycle housing.
METHOD DETAILS
Tissue preparation, immunohistochemistry and confocal imaging
Brain tissue processing and histological analysis of mouse brains were performed as described in our publications(Gao et al., 2015; Gao et al., 2017). For in vivo cell proliferation analyses using BrdU labeling, mice were given two intraperitoneal injections of BrdU (100 mg/kg) 24 h before perfusion. At 11, 18 or 27 days after wheel running, mice were euthanized by intraperitoneal injection of sodium pentobarbital and then transcardially perfused with saline followed by 4% PFA. Brains were dissected out, post-fixed overnight in 4% PFA, and then equilibrated in 30% sucrose. Forty-μm brain sections were generated using a sliding microtone and stored in a −20°C freezer as floating sections in 96-well plates filled with cryoprotectant solution (glycerol, ethylene glycol, and 0.1M phosphate buffer, pH 7.4, 1:1:2 by volume). Immunohistology was performed as published previously(Gao et al., 2015; Gao et al., 2017). The tissue sections were pre-blocked with TBS++ (TBS containing 3% goat or donkey serum and 0.2% Triton X-100) for 1 h at room temperature, followed by incubation with primary antibodies diluted in TBS++ overnight in 4 ℃. After washing 3 times, secondary antibodies were incubated 1 h at room temperature. All sections were counterstained with a nuclear counter stain, DAPI (4’, 6-diamidine-2’-phenylindole dihydrochloride, 1:2000, Roche Applied Science, Indianapolis, IN). For RGS6 staining, before blocking, there was an antigen retrieval treatment with 10 mM sodium citrate (pH 9) for 30 min at 80 °C. For BrdU staining, we performed immunohistological analysis on 1-in-6 serial floating brain sections (240 mm apart).
The primary antibodies used were: Rabbit anti-RGS6 (1:200, antibodies-online Inc., #ABIN634403, Limerick, PA, USA), Mouse anti-HA.11 (1:1000, Covance (now Biolegend), # MMS-101R, Princeton, NJ, USA), Rabbit anti-MCM2 (1:1000, Cell Signaling Technology, Inc., #4007, Danvers, MA, USA), Goat anti-DCX (1:500, Santa Cruz Biotechnology, Inc., # SC-8066, Dallas, TX, USA), Rabbit anti-GFAP (1:2000, DAKO, #Z0334, Carpinteria, CA, USA), Rabbit anti-NeuN (1:3000, Cell Signaling Technology, Inc., #4007, Danvers, MA, USA), Rabbit anti-Phospho-CREB(Ser133) (1:800, Cell Signaling Technology, Inc., #9198S, Danvers, MA, USA). Fluorescent secondary antibodies used by 1:1000 dilution: Donkey anti-mouse 488 (A21202, Invitrogen), Donkey anti-mouse 568 (A10037, Invitrogen), Donkey anti-rabbit 488 (A21206, Invitrogen), Donkey anti-rabbit 647 (A31573, Invitrogen), Donkey anti-goat 568 (A11057, Invitrogen), Goat anti-mouse 488 (A11029, Invitrogen), Goat anti-rabbit 568 (A11036, Invitrogen), After staining, sections were mounted and maintained at 4oC in the dark until analysis. Z-stack confocal images were taken using Nikon A1 confocal microscope. The Z-stack images were analyzed using a Zeiss Apotome microscope equipped by StereoInvestigator and Microlucida software (MBF Biosciences,Inc.)
Plasmids
Retro-CAG-GFP-shRgs6 was cloned using Retro-shNC(Guo et al., 2015) as the backbone. Briefly, U6-shNC cassette was cloned into the backbone through HpaI/ClaI restriction sites. Retro-Cre were generated by replacing red fluorescent protein (RFP) coding sequence in the Retro-CAG-RFP (Zhao et al., 2006) with Cre coding sequence. Lenti-GFP-Control and Lenti-GFP-RGS6 were generated using lenti-CMV-GFP vector as a backbone and the CMV-GFP cassette were replaced with CAG-LoxP-STOP-LoxP-GFP and CAG-LoxP-STOP-LoxP-GFP-P2A-RGS6 correspondently. The Rgs6 shRNA sequences are: shRgs6-1: 5‘- CAAGTGAAGATTGACCGGAAA-‘3; shRgs6-2: 5‘-CGGCGTTTGAAGAATCCACAA -‘3; shRgs6-3: 5‘- GCTATGAGATAACCAGTCAAA -‘3.
Production of lentivirus and retrovirus
Lentivirus and retrovirus production were performed using previous protocol (Gao et al., 2015; Gao et al., 2017) with modification. Briefly, lentiviral DNA was co-transfected with packaging plasmids pMDL, REV and pCMV-Vsvg into HEK293T cells using PEI method. Retroviral DNA was co-transfected with packaging plasmids pCMV-gag-pol and pCMV-Vsvg into HEK293T cells using PEI method. The viral transfer vector DNA and packaging plasmid DNA were transfected into 5X15 cm dishes of cultured HEK293T cells using PEI. The medium containing lentivirus was collected at 36, 60 and 84 hours post-transfection, pooled, filtered through a 0.2-μm filter, and concentrated using an ultracentrifuge at 19,000 rpm for 2 hours at 4°C using a SW32Ti rotor (Beckman). The virus was washed once and then resuspended in 100 μl PBS. We routinely obtained 1X109 infectious viral particles /ml for lentivirus and 1X108 infectious viral particles /ml for retrovirus.
In vivo retroviral grafting and morphological analysis of targeted neurons
In vivo virus grafting was performed as described (Gao et al., 2015). Briefly, 7-week-old male mice were anesthetized with isofluorane and placed in a stereotactic instrument (DAVID KOPF Instruments, Tujunga, CA). Microinjections were performed using custom-made injection 33-gauge needles (Hamilton, #776206, Reno, NV, USA) connected to a 10 μL syringe (Hamilton, #87930). Virus (1.25 μl with titer greater than 1X108/ml) was mixed and then stereotaxically injected into the dentate gyrus using the following coordinates relative to bregma, caudal: +2.0 mm; lateral: +/−1.6 mm; ventral: −1.9 mm. 17, 19 or 21 days post viral grafting, mice were perfused for different analysis. Mice were deeply anesthetized with pentobarbital and perfused with saline followed by 4% PFA. Morphological analyses of retroviral labeled new neurons were performed as described (Gao et al., 2015). Two hundred micrometer-thick floating brain sections containing eGFP were selected. For dendritic branching analysis, GFP-positive cells were imaged on an A1 confocal microscope. The dendrites and the cell body of single expression of GFP neurons were analyzed by Neurolucida software (Micro-BrightField, Burlington, Vermont, http://www.mbfbioscience.com/). At least 30 neurons per group were traced.
In vivo cell quantification
Quantification of BrdU+ cells in the adult DG was performed using an AxioImagerZ2 ApoTome microscope (Zeiss) equipped with motorized stage and operated by StereoInvestigator software (MBF Biosciences, Inc). The cell number was quantified using design-based stereology with the optical fractionator function of StereoInvestigator software (MBF Biosciences, Inc), as described (Li et al., 2016; Shen et al., 2019). Briefly, 1 in 6 serial 40μm brain sections starting at beginning of hippocampus (relative to bregma, −1.5 mm) to the end of hippocampus (relative to bregma, −3.5 mm) were selected for staining using an anti-BrdU antibody as described above. The thickness of the mounted sections were measured (around 30 um) and guard zones of 3 μm were set for each surface of the section. The contour of the DG was drawn with one cell body extra space along the hilar surface of the DG. The counting frame was set at 200.0 μm width (X) and 200.0 μm height (Y). The number of sampling sites were around 350 and the section evaluation interval was 6. The estimated CE (Cruz-Orive/Geiser) < 0.1. Approximately 10-100 cells were counted in each DG (one side). The total number of cells were calculated using StereoInvestigator using preset algorithm. The volume of analyzed region for each animal was obtained. The percentage of HA+ cells that were colabeled with other cell markers was analyzed as described previously (Li et al., 2016; Shen et al., 2019). Briefly, the z-stack images about 500-1200 BrdU+ cells were obtained using an AxioImagerZ2 ApoTome confocal microscope (Zeiss).
Immunofluorescent intensity quantification
The signal intensity of RGS6 in HA+ cells in the DG of Tamoxifen injected Nes-CreERT2;RiboTag mice was quantified using Image J software. The z-stack images were acquired using Nikon A1 confocal microscope at 20X magnification with 2 μm interval. A line was drawn through the soma without the cell nuclear of each HA+ cell and the intensity of RGS6 was measured using Image J. The background area was selected and measured with Image J as well. The RGS6 intensity values were calculated as: (RGS6 Area) * (RGS6 mean intensity – Background mean intensity). In total, 5 running mice (115 cells) and 6 sedentary mice (90 cells) were used for this quantification.
RiboTag-Seq and identification of differentially expressed genes (DEGs)
Dentate gyri were dissected from 2-4 Nes-HA mice from either running or sedentary conditions, pooled, and immediately homogenized in 1ml of homogenization buffer (50mM Tris [pH 7.4], 100mM KCl, 12mM MgCl2, 1% IGEPAL, 1mM DTT, 1x RNase inhibitor, 100μg/mL Cycloheximide, 1mg/mL Heparin) with 1X complete protease inhibitors (Roche). Nuclei and debris were pelleted at 10,000 X g for 10 min. An aliquot (20 μl) of lysate input was saved for extraction of input (IN) RNA. 3-6 μl monoclonal antibody against HA Tag (Covance catalog# MMS-101R) was incubated with supernatant at 4°C for 6 hrs and before adding Dynabeads (ThermoFisher). The antibody/Dynabeads conjugation was rotated at 4°C overnight and washed with high salt buffer (50mM Tris [pH 7.4], 300mM KCl, 12mM MgCl2, 1% IGEPAL, 1mM DTT, 100μg/mL Cycloheximide) to remove unbound proteins and RNA. Finally, Trizol was added to the beads and the supernatant was saved for RNA isolation. RNA from the RiboTag IP was isolated using the Direct-zol™ RNA MicroPrep Kit (Zymo Research Corporation. Irvine, CA, USA). Quality, size, and concentration of the isolated RNA were analyzed using Bioanalyser 2100 (RNA Pico Kit, Agilent). The libraries were then made using SMARTer® Stranded Total RNA-Seq Kit v2 (Takara Bio USA, Inc. Mountain View, CA) according to the user manual. The constructed libraries were analyzed with Qubit (ThermoFisher Scientific, Waltham, MA) and Bioanalyser 2100 (RNA Pico Kit, Agilent). Cluster generation and high-throughput sequencing were performed on a HiSeq 2500 (Illumina), using the single-end 100 bp protocol. RSEM was used to align read pairs to the mm10 transcriptome and estimate gene expression levels (Li and Dewey, 2011). Differentially expressed genes (DEGs) were successfully identified in running mice using embedded EBseq in RSEM with a cutoff of 0.05 of “PPEE” (posterior probability estimated by EBSeq that a gene/transcript is equally expressed). The next generation sequencing data have been submitted to GEO (GSE142678).
Single Cell trajectory by pseudotime analysis
We performed the pseudo-timing analysis for our IP dataset and the Div-Seq single cell dataset for neuronal development (270 cells) covering the stages of 1-2, 3-4, 5-6, 6-7, 7-14 day (Habib et al., 2016). In particular, we input gene expression data (TPM values) of these datasets to Monocle (Trapnell et al., 2014)for revealing the pseudo timing. The differentially expressed genes (N=460) between running and sedentary samples at Day-18 were used to construct the developmental trajectories (Figure 2a), representing pseudo-timing ordering between Div-Seq cells and our six IP samples (running & sedentary Day- 11, 18, 27).
Gene co-expression network and module analysis
We constructed the gene co-expression networks by connecting all possible gene pairs by edges with edge weights being the Pearson correlations of their time-series gene expression profiles across Day-11, Day-18 and Day-27 for running and sedentary datasets. The gene co-expression network was further clustered by WGCNA (weighted correlation network analysis), R package (Langfelder and Horvath, 2008) into the gene co-expression modules (minimum module size = 100 genes, scale-free fitting beta>0.8). The eigengenes of modules were calculated and provided by WGCNA as well. An eigengene is a vector with its elements representing the expression levels across time points, representing the most likely temporal gene expression changes across time points (temporal dynamic pattern) of its co-expression module.
Enrichment analysis
Enrichment analyses including GO, KEGG and REACTOME for differentially expressed genes or gene co-expression modules were performed in “g:GOST” fuctional profiling using g: Profiler (https://biit.cs.ut.ee/gprofiler/gost). Genes linked to adult hippocampal neurogenesis were downloaded from the Mammalian Adult Neurogenesis Gene Ontology (MANGO) database (http://mango.adult-neurogenesis.de/documents/annotations?show=20&expression=true) (Overall et al., 2012). Genes that are demethylated during development of the mammalian brain were provided by Lister et al (Lister et al., 2013). Disease-linked genes were curated from Phenocarta (https://pavlidislab.github.io/Gemma/phenocarta.html), the DISEASES database (http://diseases.jensenlab.org/Search) (Pletscher-Frankild et al., 2015), and AutDB (http://autism.mindspec.org/autdb/Welcome.do) (Basu et al., 2009). Overrepresentation of gene sets was statistically assessed within identified gene clusters against the transcriptome background in R using the Modular Single-set Enrichment Test (Eisinger et al., 2013).
RNA isolation, cDNA synthesis and qPCR
RNA was isolated from TRIzol samples using the TRIzol Reagent following the manufacturer’s instructions. Reverse transcription and Real-time PCR was performed using standard methods as described (Gao et al., 2015). The first-strand complementary DNA (cDNA) was generated by reverse transcription with Random 6 mers primer (Takara Bio USA, Inc. Mountain View, CA). To quantify the mRNA expression using real-time PCR, aliquots of first-stranded cDNA were amplified with gene-specific primers and Power SYBR Green PCR Master Mix (Bio-Rad) using a StepOne Real-Time PCR System (Applied Biosystems). The PCR reactions contained cDNA, Universal Master Mix (Applied Biosystems), and 10 mM of forward and reverse primers in a final reaction volume of 20 ml. The mRNA expression of different samples was calculated using 2-deltadelta Ct method (Livak and Schmittgen, 2001).
Electrophysiology
Three weeks after virus injection, mice were euthanized with CO2 and decapitated, followed by quick transfer of brain into ice-cold dissection solution comprising (mM) 93 NMDG, 93 HCl, 2.5 KCl, 1.2 NaH2PO4, 30 NaHCO3, 20 HEPES, 25 glucose, 5 sodium ascorbate, 2 Thiourea, 3 sodium pyruvate, 10 MgSO4, 0.5 CaCl2. Coronal slices (400 μm thick) were obtained and transferred to artificial cerebrospinal fluid (ACSF) containing (mm): 124 NaCl, 2.5 KCl, 2.5 CaCl2, 1.2 MgCl2, 1.25 NaH2PO4, 26 NaHCO3, and 15 glucose (equilibrated with 95% O2 and 5% CO2; pH 7.4). Slices were incubated at 34 °C for 10 min and stored at room temperature for 1 hour for recovery. Then slices were transferred to a submerged recording chamber (flow rate 2-2.5 ml/min at room temperature). For recording miniature excitatory postsynaptic currents (mEPSCs), pipettes were filled with (in mM): 140 K-gluconate, 7.5 KCl, 10 HEPES-K, 0.5 EGTA-K, 4 Mg-ATP and Li-GTP (pH 7.2). mEPSCs were recorded in the presence of 1 mM tetrodotoxin (TTX). Adult born neurons were identified by expression of GFP under epifluorescence using differential interference contrast video microscopy and recorded in voltage clamp mode (-70 mV). Whole-cell patch-clamp recordings were filtered at 2 kHz and digitalized at 10 kHz using Multiclamp 700B, Digidata 1440 A and pClamp 10.4 Software (Molecular Devices). Series resistance was typically within 10–30 MΩ. Miniature EPSC events were analyzed using Python. Threshold was typically kept at −5 pA and adjusted with respect to the baseline noise. Artifacts were screened through visual inspection and overlapping events (event frequency of ≥ 100 Hz) were discarded. Raw current traces of 300 s were analyzed for peak amplitude and frequency which were averaged for each cell and used for further statistical analysis.
For recording Ca2+ currents, Ascl1-CreERT2::Ai14 or virus-injected mice were anesthetized with 2,2,2-tribromoethanol (Avertin; Sigma-Aldrich), and perfused intracardially with ice-cold modified ACSF containing the following (in mM): 110 choline chloride, 25 D-glucose, 7 MgCl2, 2.5 KCl, 1.25 Na2PO4, 0.5 CaCl2, 1.3 Na-ascorbate, 3 Na-pyruvate, and 25 NaHCO3, bubbled with 95% O2/5% CO2. Brains were removed and horizontal slices (300 μm) from both hemispheres were cut using a vibratome (VT1200S, Leica Instruments) and transferred to a chamber containing recording ACSF (in mM) as follows: 125 NaCl, 2.5 KCl, 1.25 Na2PO4, 2 CaCl2, 1 MgCl2, 25 NaHCO3, and 25 D-glucose bubbled with 95% O2/5% CO2. Slices were incubated at 37°C for 30 min and then stored at room temperature for at least 30 min before mounted on a BX51WI Olympus microscope and superfused with ACSF solution at 30°C. Immature neurons were visually identified in the granule cell layer by fluorescence (TxRed or GFP-A filter sets, Leica) using custom Dodt gradient contrast videomicroscopy. Whole-cell calcium currents were recorded with fire-polished borosilicate glass electrodes (outer diameter, 1.5 mm, inner diameter, 0.86 mm; Sutter Instrument) with resistance of 5–7 MΩ when filled with the following intracellular solution (in mM): 122 CsMeSO3, 9 HEPES, 1.8 MgCl2, 4 Mg-ATP, 0.3 Na-GTP, 14 phosphocreatine, 0.45 EGTA, pH 7.3. Pipettes were mounted on the headstage (CV-7B) of a Multiclamp 700A amplifier (Molecular Devices) with cancellation of pipette capacitive transients. Currents were filtered at 2 kHz and sampled at 10 kHz. Calcium currents were isolated using NBQX (10 μM), R-CPP (5 μM), SR9553 (10 μM) and TTX (1 μM) with extracellular calcium in the ACSF exchanged for 2 mM BaCl2. The holding potential was set at −70 mV and 200 ms voltage steps to 0 mV were applied to evoke calcium currents every 20s. Capacitance and leak currents were subtracted using P/4 leak subtraction. Currents were analyzed with Axograph X (Axograph Scientific) or Clampfit (Molecular Devices). Peak amplitude was measured from an average of 5-10 currents in each condition and normalized to capacitance that was measured from a 10 mV voltage step. Recordings and analysis were performed blinded to condition. Drugs and chemicals were obtained from Sigma-Aldrich, Tocris Bioscience, or Ascent Scientific. Two-tailed paired or unpaired t-tests were used with significance at p < 0.05 (GraphPad Software). TdTomato fluorescence in fixed sections (50 μm) from Ascl1CreERT2/Ai14 mice was amplified with rabbit anti-DsRed polyclonal antibody (1:800, #632496, Takara) and goat anti-rabbit Alexa Fluor 568 (1:400, A-11011, Invitrogen). Sections were imaged with a water-immersion objective on a confocal microscope (10x, Olympus FV1200, z steps of 0.5 μm).
Behavioral analysis
Viral injected mice were subjected to either freely running wheel (Run) or locked wheels (sedentary controls) for 27 days before behavioral testing. All mice were subjected to open-field activity chamber test for assessing their general health and activity levels, both before and after the behavioral tests described below. The mice were always subjected to elevated plus maze first, then novel location text, and finally fear condition test.
Elevated plus-maze
Elevated plus-maze test was performed using the procedure described (Allan et al., 2008). Mice were placed in the center square (6X 6 cm) of a Plexiglas maze shaped in a cross that was elevated two feet above the ground and was located in a moderately lit, sound-attenuated room. The supports for the maze were made of clear Plexiglas and were positioned in the middle of the arms, so the mouse was unable to detect them. The maze had two open arms (30X6 cm each) that were made of clear Plexiglas floor with no wall. The closed arms were covered with black contact paper and had 6 cm high walls covered with black contact paper. An observer blind to genotype monitored behavior for 10 min. The time spent in the open arms and closed arms and the numbers of entries into each arm were recorded. The data were analyzed using one way ANOVA.
Novel location test
This test measures spatial memory through an evaluation of the ability of mice to recognize the new location of a familiar object with respect to spatial cues. The procedures of this test was performed as previously described (Shen et al., 2019). Briefly, mice were handled for approximately 5min per day for a maximum of 5 days prior to the experiment. All procedures were conducted during the light cycle of the animal between 9 a.m. and 6 p.m. Before the trial session, mice were brought into testing room and were allowed to acclimate for at least 30min. Testing consisted of five 6-min trials, with a 3-min intertrial interval between each trial. During the intertrial interval, the mouse was placed in a holding cage, which remained inside the testing room. In the first trial (pre-exposure), each mouse was placed individually into the center of the otherwise empty open arena (38.5 cm long×38.5 cm wide, and 25.5 cm high walls) for 6min. For the next three trials (sample trials 1–3), two identical objects were placed equidistantly from the arena wall in the corners against the wall with the colored decal. Objects were taped to the floor of the arena. Then, each mouse was placed individually into the center of the arena and allowed to explore for 6min. At the end of the trial, the mouse was removed and returned to the home cages for 3min. In the last trial (test), one of the objects was moved to a novel location, and the mouse was allowed to explore the objects for 6 min, and the total time spent exploring each object was measured. During the test phase, exploration time was defined as any investigative behavior (that is, head orientation, climbing on, sniffing occurring within<1.0 cm) or other motivated direct contact occurring with each object. To control for possible odor cues, objects were cleaned with 70% ethanol solution at the end of each trial and the floor of the arena wiped down to eliminate possible scent/trail markers. During the test phase, two objects were wiped down prior to testing so that the objects would all have the same odor. The discrimination index was calculated as the percentage of time spent investigating the object in the new location minus the percentage of time spent investigating the object in the old location: discrimination index = (novel location exploration time/total exploration time×100) − (old location exploration time/total exploration time×100). A higher discrimination index is considered to reflect greater memory retention for the novel location object. All experiments were videotaped and scored by scientists who were blinded to experimental conditions to ensure accuracy.
Fear conditioning
These tests were performed based on published method with modification (Li et al., 2009). Apparatus: Animals were placed into a Coulbourn Habitest™ fear conditioning system equipped with a stainless-steel grid floor for administration of a foot shock. Training: After 2.5 minutes of habituation in the conditioning apparatus, a 2-second electric foot shock of 0.7mA was presented. Mice were left in the conditioning apparatus for 30 more seconds, then were returned to their home cages. Testing: At 24 hours following training mice were tested for context freezing. For the context freezing, mice were placed into the conditioning context for 300 seconds and were observed for freezing behavior throughout the context testing period. Behavior was analyzed in a blinded fashion with regard to genotype and treatment. All training was videotaped for subsequent viewing, re-scoring and documentation. The trace conditioning apparatus was cleaned between each mouse with 70% isopropylalcohol.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
Statistical analysis was performed using ANOVA and Student’s t test, unless specified, with the aid of SPSS version 22 and GraphPad software. Two tailed and unpaired t-test was used to compare two conditions. Two-way ANOVA with Tukey’s post hoc analysis was used for analyzing multiple groups. One-Way ANOVA with Bonferroni post hoc test used for comparison among for in vivo dendritic analysis of different genotypes (GraphPad software 6). Scholl analysis was carried out using multivariate analysis of variance (MANOVA) using SPSS statistical software. All data were shown as mean with standard error of mean (mean ± SEM). Probabilities of p < 0.05 were considered as significant.
Supplementary Material
Table S1. DEGs in sedentary and running mice. Related to Figure 1
Table S3. GO of top 23 module-genes. Related to Figure 2
Table S4. GO analysis of upregulated IN genes. Related to Figure 1
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-RGS6 | antibodies-online Inc | #ABIN634403 |
| Mouse anti-HA.11 | Biolegend | # MMS-101R |
| Rabbit anti-MCM2 | Cell Signaling Technology | #4007 |
| Goat anti-DCX | Santa Cruz Biotechnology | # SC-8066 |
| Rabbit anti-GFAP | DAKO | #Z0334 |
| Rabbit anti-NeuN | Cell Signaling Technology | #4007 |
| Rabbit anti-DsRed | Takara | #632496 |
| Donkey anti-mouse 488 | Invitrogen | A21202 |
| Donkey anti-mouse 568 | Invitrogen | A10037 |
| Donkey anti-rabbit 488 | Invitrogen | A21206 |
| Donkey anti-rabbit 647 | Invitrogen | A31573 |
| Donkey anti-goat 568 | Invitrogen | A11057 |
| Goat anti-mouse 488 | Invitrogen | A11029 |
| Goat anti-rabbit 568 | Invitrogen | A11036 |
| Goat anti-rabbit 568 | Invitrogen | A11011 |
| Bacterial and Virus Strains | ||
| Lentivirus-STOP-RGS6-GFP | This paper | N/A |
| Lentivirus-STOP-GFP | This paper | N/A |
| Retrovirus-shRgs6-GFP | This paper | N/A |
| Retrovirus-shNC-GFP | (Guo et al., 2015) | N/A |
| Deposited Data | ||
| Source data associated with RiboTag sequencing | This paper | GEO: GSE142678 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | Jackson Laboratory | RRID:IMSR_JAX:000664 |
| Mouse: Tg(Nestin-cre/ERT2) | Jackson Laboratory | RRID:IMSR_JAX:016261 |
| Mouse: RPL22HA/WT | Jackson Laboratory | RRID:IMSR_JAX:029977 |
| Mouse: Ascl1tm1(Cre/ERT2)Jejo/J | Jackson Laboratory | RRID:IMSR_JAX:012882 |
| Mouse: B6;129S6-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | Jackson Laboratory | RRID:IMSR_JAX:007914 |
| Mouse: Nestin-GFP | Yamaguchi et al., 2000) | (Yamaguchi et al., 2000) |
| Oligonucleotides | ||
| Rgs6 Forward: GGATCGCTGATCCAGAAGAG | This study | N/A |
| Rgs6 Reverse: GGGGATTTTGGAGAGAAAGC | This study | N/A |
| Pgk1Forward: CTCCGCTTTCATGTAGAGGAAG | This study | N/A |
| Pgk1Reverse: GACATCTCCTAGTTTGGACAGTG | This study | N/A |
| Recombinant DNA | ||
| Retro-CAG-GFP-shRgs6 | This study | N/A |
| Retro-shNC | (Gao et al., 2015) | N/A |
| Retro-CAG-RFP | (Zhao et al., 2006) | N/A |
| Lenti-GFP-Control | This study | N/A |
| Lenti-GFP-RGS6 | This study | N/A |
| Software and Algorithms | ||
| Prism | GraphPad | https://www.graphpad.com; RRID:SCR_002798 |
| ImageJ | National Institute of Health | https://imagej.nih.gov/ij/docs/guide/user-guide.pdf |
| ANYmaze | Stoelting | https://www.stoeltingco.com/anymaze.html |
| RStudio | https://rstudio.com/ | |
| WGCNA | (Langfelder and Horvath, 2008) | |
| g:Profiler | https://biit.cs.ut.ee/gprofiler/gost | |
| Monocle | (Trapnell et al., 2014) | |
ACKNOWLEDGMENTS
We thank Y. Xing, R. Spitzer, D. Wagner, A. Meara, S. Pham, Y. Choi, K. Schoeller, J. Le, M. Syed, M. Seelig and Y. Sun for technical assistance, J. Pinnow, M. Eastwood, D. Bollig and K. Knobel at the Waisman IDD Model Core, S. Splinter-BonDurant at tUW-Madison Biotechnology Center for next generation sequencing services. This work was supported by grants from the National Institutes of Health (R01NS105200, R56MH113146, and R21NS095632 to X.Zhao, R01HD064743 to Q. Chang, R01NS064025 and R01NS105438 to L. Overstreet-Wadiche, R01AG067025 U01MH116492 to D. Wang, U54HD090256 to the Waisman Center). American Epilepsy Society postdoctoral fellowship (J.C. Gonzalez). UW Vilas Trust (Mid-Career Award), Wisconsin Alumni Research Foundation, and Jenni and Kyle Professorship (to X.Zhao).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Aimone JB, Deng W, and Gage FH (2010). Adult neurogenesis: integrating theories and separating functions. Trends in cognitive sciences 14, 325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan AM, Liang X, Luo Y, Pak C, Li X, Szulwach KE, Chen D, Jin P, and Zhao X (2008). The loss of methyl-CpG binding protein 1 leads to autism-like behavioral deficits. Human molecular genetics 17, 2047–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GR, Posokhova E, and Martemyanov KA (2009). The R7 RGS protein family: multi-subunit regulators of neuronal G protein signaling. Cell biochemistry and biophysics 54, 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu SN, Kollu R, and Banerjee-Basu S (2009). AutDB: a gene reference resource for autism research. Nucleic acids research 37, D832–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold NC, Prieto GA, Phelan M, Gillen DL, Baldi P, Bennett DA, Buchman AS, and Cotman CW (2019). Hippocampal gene expression patterns linked to late-life physical activity oppose age and AD-related transcriptional decline. Neurobiology of aging 78, 142–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracko O, Singer T, Aigner S, Knobloch M, Winner B, Ray J, Clemenson GD Jr., Suh H, Couillard-Despres S, Aigner L, et al. (2012). Gene expression profiling of neural stem cells and their neuronal progeny reveals IGF2 as a regulator of adult hippocampal neurogenesis. The Journal of neuroscience : the official journal of the Society for Neuroscience 32, 3376–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt MD, Jessberger S, Steiner B, Kronenberg G, Reuter K, Bick-Sander A, von der Behrens W, and Kempermann G (2003). Transient calretinin expression defines early postmitotic step of neuronal differentiation in adult hippocampal neurogenesis of mice. Molecular and cellular neurosciences 24, 603–613. [DOI] [PubMed] [Google Scholar]
- Chatterjee TK, Liu Z, and Fisher RA (2003). Human RGS6 gene structure, complex alternative splicing, and role of N terminus and G protein gamma-subunit-like (GGL) domain in subcellular localization of RGS6 splice variants. The Journal of biological chemistry 278, 30261–30271. [DOI] [PubMed] [Google Scholar]
- Chatzi C, Zhang Y, Hendricks WD, Chen Y, Schnell E, Goodman RH, and Westbrook GL (2019). Exercise-induced enhancement of synaptic function triggered by the inverse BAR protein, Mtss1L. eLife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevaleyre V, Moos FC, and Desarmenien MG (2002). Interplay between presynaptic and postsynaptic activities is required for dendritic plasticity and synaptogenesis in the supraoptic nucleus. The Journal of neuroscience : the official journal of the Society for Neuroscience 22, 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian KM, Song H, and Ming GL (2014). Functions and dysfunctions of adult hippocampal neurogenesis. Annual review of neuroscience 37, 243–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Aimone JB, and Gage FH (2010). New neurons and new memories: how does adult hippocampal neurogenesis affect learning and memory? Nature reviews Neuroscience 11, 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande A, Bergami M, Ghanem A, Conzelmann KK, Lepier A, Gotz M, and Berninger B (2013). Retrograde monosynaptic tracing reveals the temporal evolution of inputs onto new neurons in the adult dentate gyrus and olfactory bulb. Proceedings of the National Academy of Sciences of the United States of America 110, E1152–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Pan YB, Wu XR, He LN, Liu XD, Feng DF, Xu TL, Sun S, and Xu NJ (2019). A neuronal molecular switch through cell-cell contact that regulates quiescent neural stem cells. Science advances 5, eaav4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eadie BD, Redila VA, and Christie BR (2005). Voluntary exercise alters the cytoarchitecture of the adult dentate gyrus by increasing cellular proliferation, dendritic complexity, and spine density. The Journal of comparative neurology 486, 39–47. [DOI] [PubMed] [Google Scholar]
- Eisinger BE, Saul MC, Driessen TM, and Gammie SC (2013). Development of a versatile enrichment analysis tool reveals associations between the maternal brain and mental health disorders, including autism. BMC neuroscience 14, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger BE, and Zhao X (2018). Identifying molecular mediators of environmentally enhanced neurogenesis. Cell and tissue research 371, 7–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Su J, Guo W, Polich ED, Magyar DP, Xing Y, Li H, Smrt RD, Chang Q, and Zhao X (2015). Inhibition of miR-15a Promotes BDNF Expression and Rescues Dendritic Maturation Deficits in MeCP2-Deficient Neurons. Stem cells 33, 1618–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wang F, Eisinger BE, Kelnhofer LE, Jobe EM, and Zhao X (2017). Integrative Single-Cell Transcriptomics Reveals Molecular Networks Defining Neuronal Maturation During Postnatal Neurogenesis. Cerebral cortex 27, 2064–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S, Goh EL, Sailor KA, Kitabatake Y, Ming GL, and Song H (2006). GABA regulates synaptic integration of newly generated neurons in the adult brain. Nature 439, 589–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S, Pradhan DA, Ming GL, and Song H (2007). GABA sets the tempo for activity-dependent adult neurogenesis. Trends in neurosciences 30, 1–8. [DOI] [PubMed] [Google Scholar]
- Giachino C, Barz M, Tchorz JS, Tome M, Gassmann M, Bischofberger J, Bettler B, and Taylor V (2014). GABA suppresses neurogenesis in the adult hippocampus through GABAB receptors. Development 141, 83–90. [DOI] [PubMed] [Google Scholar]
- Goncalves JT, Schafer ST, and Gage FH (2016). Adult Neurogenesis in the Hippocampus: From Stem Cells to Behavior. Cell 167, 897–914. [DOI] [PubMed] [Google Scholar]
- Gonzalez JC, Epps SA, Markwardt SJ, Wadiche JI, and Overstreet-Wadiche L (2018). Constitutive and Synaptic Activation of GIRK Channels Differentiates Mature and Newborn Dentate Granule Cells. The Journal of neuroscience : the official journal of the Society for Neuroscience 38, 6513–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer PL, and Greenberg ME (2008). From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron 59, 846–860. [DOI] [PubMed] [Google Scholar]
- Gregoire CA, Bonenfant D, Le Nguyen A, Aumont A, and Fernandes KJ (2014). Untangling the influences of voluntary running, environmental complexity, social housing and stress on adult hippocampal neurogenesis. PloS one 9, e86237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire CA, Tobin S, Goldenstein BL, Samarut E, Leclerc A, Aumont A, Drapeau P, Fulton S, and Fernandes KJL (2018). RNA-Sequencing Reveals Unique Transcriptional Signatures of Running and Running-Independent Environmental Enrichment in the Adult Mouse Dentate Gyrus. Frontiers in molecular neuroscience 11, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Polich ED, Su J, Gao Y, Christopher DM, Allan AM, Wang M, Wang F, Wang G, and Zhao X (2015). Fragile X Proteins FMRP and FXR2P Control Synaptic GluA1 Expression and Neuronal Maturation via Distinct Mechanisms. Cell reports 11, 1651–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib N, Li Y, Heidenreich M, Swiech L, Avraham-Davidi I, Trombetta JJ, Hession C, Zhang F, and Regev A (2016). Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science 353, 925–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochgerner H, Zeisel A, Lonnerberg P, and Linnarsson S (2018). Conserved properties of dentate gyrus neurogenesis across postnatal development revealed by single-cell RNA sequencing. Nature neuroscience 21, 290–299. [DOI] [PubMed] [Google Scholar]
- Hollinger S, and Hepler JR (2002). Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacological reviews 54, 527–559. [DOI] [PubMed] [Google Scholar]
- Jobe EM, Gao Y, Eisinger BE, Mladucky JK, Giuliani CC, Kelnhofer LE, and Zhao X (2017). Methyl-CpG-Binding Protein MBD1 Regulates Neuronal Lineage Commitment through Maintaining Adult Neural Stem Cell Identity. The Journal of neuroscience : the official journal of the Society for Neuroscience 37, 523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G (2015). Activity Dependency and Aging in the Regulation of Adult Neurogenesis. Cold Spring Harbor perspectives in biology 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G, Kuhn HG, and Gage FH (1997). More hippocampal neurons in adult mice living in an enriched environment. Nature 386, 493–495. [DOI] [PubMed] [Google Scholar]
- Kempermann G, Song H, and Gage FH (2015). Neurogenesis in the Adult Hippocampus. Cold Spring Harbor perspectives in biology 7, a018812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacar B, Linker SB, Jaeger BN, Krishnaswami SR, Barron JJ, Kelder MJE, Parylak SL, Paquola ACM, Venepally P, Novotny M, et al. (2016). Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nature communications 7, 11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, and Horvath S (2008). WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li H, Liu X, Bao G, Tao Y, Wu Z, Xia P, Wu C, Li B, and Ma L (2009). Regulation of amygdalar PKA by beta-arrestin-2/phosphodiesterase-4 complex is critical for fear conditioning. Proceedings of the National Academy of Sciences of the United States of America 106, 21918–21923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Stockton ME, Bhuiyan I, Eisinger BE, Gao Y, Miller JL, Bhattacharyya A, and Zhao X (2016). MDM2 inhibition rescues neurogenic and cognitive deficits in a mouse model of fragile X syndrome. Science translational medicine 8, 336ra361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TW, Chen SJ, Huang TY, Chang CY, Chuang JI, Wu FS, Kuo YM, and Jen CJ (2012). Different types of exercise induce differential effects on neuronal adaptations and memory performance. Neurobiology of learning and memory 97, 140–147. [DOI] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. (2013). Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, and Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Maity B, Stewart A, Yang J, Loo L, Sheff D, Shepherd AJ, Mohapatra DP, and Fisher RA (2012). Regulator of G protein signaling 6 (RGS6) protein ensures coordination of motor movement by modulating GABAB receptor signaling. The Journal of biological chemistry 287, 4972–4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overall RW, Paszkowski-Rogacz M, and Kempermann G (2012). The mammalian adult neurogenesis gene ontology (MANGO) provides a structural framework for published information on genes regulating adult hippocampal neurogenesis. PloS one 7, e48527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett CL, and Slesinger PA (2010). GABAB receptor coupling to G-proteins and ion channels. Advances in pharmacology 58, 123–147. [DOI] [PubMed] [Google Scholar]
- Piatti VC, Davies-Sala MG, Esposito MS, Mongiat LA, Trinchero MF, and Schinder AF (2011). The timing for neuronal maturation in the adult hippocampus is modulated by local network activity. The Journal of neuroscience : the official journal of the Society for Neuroscience 31, 7715–7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletscher-Frankild S, Palleja A, Tsafou K, Binder JX, and Jensen LJ (2015). DISEASES: text mining and data integration of disease-gene associations. Methods 74, 83–89. [DOI] [PubMed] [Google Scholar]
- Rajan I, and Cline HT (1998). Glutamate receptor activity is required for normal development of tectal cell dendrites in vivo. The Journal of neuroscience : the official journal of the Society for Neuroscience 18, 7836–7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross EM, and Wilkie TM (2000). GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annual review of biochemistry 69, 795–827. [DOI] [PubMed] [Google Scholar]
- Sah N, Peterson BD, Lubejko ST, Vivar C, and van Praag H (2017). Running reorganizes the circuitry of one-week-old adult-born hippocampal neurons. Scientific reports 7, 10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, and Amieux PS (2009). Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proceedings of the National Academy of Sciences of the United States of America 106, 13939–13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfeld TJ, McCausland HC, Sonti AN, and Cameron HA (2016). Anxiolytic Actions of Exercise in Absence of New Neurons. Hippocampus 26, 1373–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M, Wang F, Li M, Sah N, Stockton ME, Tidei JJ, Gao Y, Korabelnikov T, Kannan S, Vevea JD, et al. (2019). Reduced mitochondrial fusion and Huntingtin levels contribute to impaired dendritic maturation and behavioral deficits in Fmr1-mutant mice. Nature neuroscience 22, 386–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Berg DA, Zhu Y, Shin JY, Song J, Bonaguidi MA, Enikolopov G, Nauen DW, Christian KM, Ming GL, et al. (2015). Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell stem cell 17, 360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snigdha S, de Rivera C, Milgram NW, and Cotman CW (2014). Exercise enhances memory consolidation in the aging brain. Frontiers in aging neuroscience 6, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder JS, Glover LR, Sanzone KM, Kamhi JF, and Cameron HA (2009). The effects of exercise and stress on the survival and maturation of adult-generated granule cells. Hippocampus 19, 898–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steib K, Schaffner I, Jagasia R, Ebert B, and Lie DC (2014). Mitochondria modify exercise-induced development of stem cell-derived neurons in the adult brain. The Journal of neuroscience : the official journal of the Society for Neuroscience 34, 6624–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart A, Maity B, Wunsch AM, Meng F, Wu Q, Wemmie JA, and Fisher RA (2014). Regulator of G-protein signaling 6 (RGS6) promotes anxiety and depression by attenuating serotonin-mediated activation of the 5-HT(1A) receptor-adenylyl cyclase axis. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 28, 1735–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A, Boekhoorn K, Van Dam AM, and Lucassen PJ (2008). Changes in adult neurogenesis in neurodegenerative diseases: cause or consequence? Genes, brain, and behavior 7 Suppl 1, 28–42. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature biotechnology 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinchero MF, Buttner KA, Sulkes Cuevas JN, Temprana SG, Fontanet PA, Monzon-Salinas MC, Ledda F, Paratcha G, and Schinder AF (2017). High Plasticity of New Granule Cells in the Aging Hippocampus. Cell reports 21, 1129–1139. [DOI] [PubMed] [Google Scholar]
- Trinchero MF, Herrero M, Monzon-Salinas MC, and Schinder AF (2019). Experience-Dependent Structural Plasticity of Adult-Born Neurons in the Aging Hippocampus. Frontiers in neuroscience 13, 739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Praag H, Kempermann G, and Gage FH (1999). Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nature neuroscience 2, 266–270. [DOI] [PubMed] [Google Scholar]
- Vivar C, Peterson BD, and van Praag H (2016). Running rewires the neuronal network of adult-born dentate granule cells. NeuroImage 131, 29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivar C, Potter MC, and van Praag H (2013). All about running: synaptic plasticity, growth factors and adult hippocampal neurogenesis. Current topics in behavioral neurosciences 15, 189–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winner B, and Winkler J (2015). Adult neurogenesis in neurodegenerative diseases. Cold Spring Harbor perspectives in biology 7, a021287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong RO, and Ghosh A (2002). Activity-dependent regulation of dendritic growth and patterning. Nature reviews Neuroscience 3, 803–812. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Saito H, Suzuki M, and Mori K (2000). Visualization of neurogenesis in the central nervous system using nestin promoter-GFP transgenic mice. Neuroreport 11, 1991–1996. [DOI] [PubMed] [Google Scholar]
- Yang SM, Alvarez DD, and Schinder AF (2015). Reliable Genetic Labeling of Adult-Born Dentate Granule Cells Using Ascl1 CreERT2 and Glast CreERT2 Murine Lines. The Journal of neuroscience : the official journal of the Society for Neuroscience 35, 15379–15390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun S, Reynolds RP, Masiulis I, and Eisch AJ (2016). Re-evaluating the link between neuropsychiatric disorders and dysregulated adult neurogenesis. Nature medicine 22, 1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Teng EM, Summers RG Jr., Ming GL, and Gage FH (2006). Distinct morphological stages of dentate granule neuron maturation in the adult mouse hippocampus. The Journal of neuroscience : the official journal of the Society for Neuroscience 26, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. DEGs in sedentary and running mice. Related to Figure 1
Table S3. GO of top 23 module-genes. Related to Figure 2
Table S4. GO analysis of upregulated IN genes. Related to Figure 1
Data Availability Statement
All figures except for Figure 2A were generated from raw data or processed data. The data generated and/or analyzed during this study are available from the Lead Contact upon reasonable request. The next generation sequencing data have been submitted to GEO (GSE142678). Gene co-expression network analysis using WGCNA and clustered genes into co-expression modules (Langfelder and Horvath, 2008). The GO analysis performed using online g:Profiler (website: https://biit.cs.ut.ee/gprofiler/gost). The generation of the developmental trajectory using pseudotime analysis by Monocle (Trapnell et al., 2014).


