Abstract
Background
Tissue-specific fat deposition is regulated by a series of complex regulatory mechanisms. Reports indicate that epigenetic regulators, such as circular RNAs (circRNAs), are crucial in diseases progression, animal development, metabolism, and adipogenesis. In this study, to assess the functional roles of circRNAs in adipogenesis and tissue-specific fat deposition, we comprehensively analyzed the Ribo-Zero RNA-Seq and miRNAs data during chicken intramuscular and abdominal adipogenic differentiation.
Results
circRNAs and miRNAs profiles during chicken adipogenic differentiation were found in adipocytes derived from various adipose tissues. It was also discovered that high levels of downregulated miRNAs potentially promote adipogenesis by activating their target genes which are associated with fatty acid metabolism and adipogenic differentiation. Through analysis of the correlation between the expression levels of circRNAs and adipogenic genes, as well as the dynamic expression patterns of circRNAs during adipogenic differentiation, several candidate circRNAs were identified. Moreover, competing endogenous RNA (ceRNAs) networks were constructed during chicken intramuscular and abdominal adipogenesis by combining miRNAs with mRNAs data. Several candidate circRNAs potentially influence adipogenesis by regulating miRNAs via PPAR and fatty acid metabolism-related pathways were identified, such as circLCLAT1, circFNDC3AL, circCLEC19A and circARMH1.
Conclusion
In conclusion, our findings reveal that circRNAs and the circRNA-miRNAs-mRNAs-ceRNAs network may play important roles in chicken adipocytes differentiation and tissue-specific fat deposition.
Keywords: Chicken, circRNAs, ceRNA, Intramuscular fat, Abdominal fat, Adipogenic differentiation
Background
Different resources and organizational microenvironment dictate various physiological and biochemical characteristics of adipose tissues [1–3]. Since preadipocytes derived from various tissues exhibit distinct proliferation speed and adipogenic potential, it is believed that transcriptional and post-transcriptional regulation mechanisms cause stage- and tissue-specific fat deposition in animals [4–6].
An increasing number of studies have shown that non-coding RNAs regulates various normal and pathological processes, such as microRNAs (miRNAs) [7–10] and long non-coding RNAs (lncRNAs) [11–13]. Although recent studies on the role of miRNAs and lncRNAs in adipogenesis have reached a mature stage, information on the functional roles of other classes of non-coding RNAs, such as circular RNAs (circRNAs) remains scanty. circRNAs are a class of non-coding RNAs generated by alternative splicing exhibiting weak protein-coding potential [14, 15]. Increasing evidence indicates that circRNAs promote various biological processes, such as tummorigenesis [16–18], animal development [19, 20] and cell differentiation [21, 22]. circHECTD1 promotes gastric cancer progression by targeting miR-1256 through activation of the β-catenin/c-Myc pathway [23]. circ-ZNF609, when translated to a functional petide, regulates myogenesis in humans and mice [24]. circArhgap5–2 plays a crucial role in adipogenesis and obesity [25]. However, circRNAs related to poultry adipogenesis and tissues-specific fat deposition remain poorly studied.
In this study, comprehensive circRNAs atlas during chicken adipogenic differentiation across various tissues-derived preadipocytes were provided. Future, we identified differentially expressed (DE) circRNAs, miRNAs and their dynamic expression patterns during adipocytes differentiation. Then, circRNAs and RNA-Seq data were combined to construct circRNAs-miRNAs-mRNAs ceRNA network involved in chicken adipogenesis. This study provides an in-depth understanding of epigenetic underlying regulation mechanisms of adipogenesis and tissues-specific fat deposition in birds.
Results
Overview of circRNAs data by Ribo-zero RNA-Seq in chicken preadipocytes and adipocytes
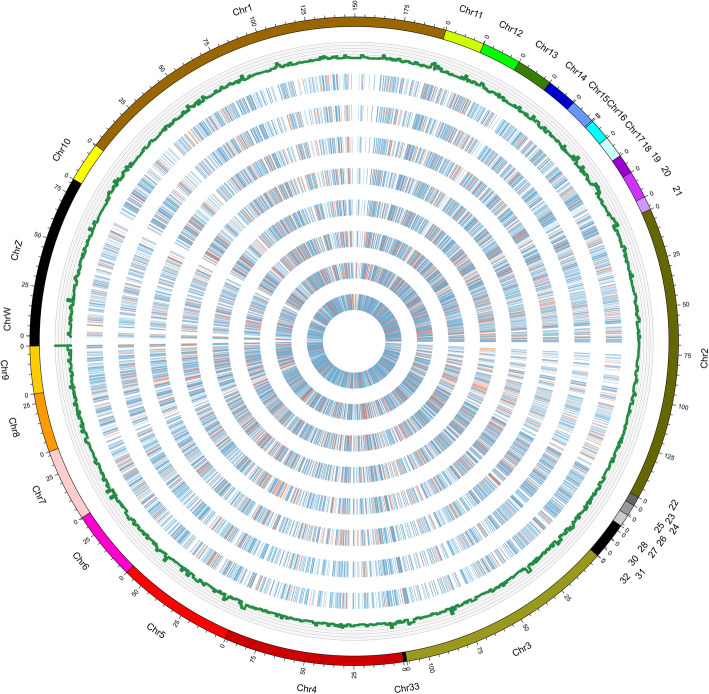
Here, we analyzed 960 million raw paired-end (PE) reads generated by Ribo-Zero RNA-Seq data. After filtering with low-quality reads, we retained a total of 910 million clean reads. Unique clean reads of 88–94% were mapped to the chicken genome 5 (galGal5) (Table 1). It was found that circRNAs were distributed in most of the chicken chromosomes. The expression levels of circRNAs in different tissue-derived adipocytes from the adipose tissue were identified then analyzed to find a better understanding of the cirRNAs in chicken adipocytes. As shown in Fig. 1a, intramuscular preadipocytes (IM_Pre) and adipocytes (IM_Ad), abdominal preadipocytes (Ab_Pre), and adipocytes (Ab_Ad) were fell into two clusters, suggesting that the differences within the adipose tissues are potentially larger than that of adipocytes. Moreover, the Pearson correlation coefficient of circRNA expression between the cell samples within groups waved from 0.86 to 0.93, suggesting a reliable consistency of cell samples within groups. The bar plot showed that no apparent difference in global expression levels of circRNAs across different groups (Fig. 1b). To classify the length distribution of circRNAs, the resource of all circRNAs in different gene regions were compared, including exon, intergenic, and intron regions. Eventually, 787, 710, 980, 1083, 1406, 1426, 1447 and 1004 circRNAs were identified in eight groups respectively (Fig. 1c). Results showed that the majority of circRNAs (65%) were generated from exon regions, followed by intergenic regions (27%) and intron regions (8%) (Fig. 1d). As shown in Fig. 1d, most of the intersect circRNAs (60%) shared from different software were 200–1000 nt in length. Moreover, a considerable number of circRNAs which longer than 3000 nt were identified in the present study (Fig. 1d). Besides, most of circRNAs (94%) were found more than two exons (Fig. 1e). The circRNAs were distributed across 31 chromosomes (chromosomes 1–28, 33, Z, and W) of chicken with chromosomes 1, 2, 3 and 21 carrying the largest share (Fig. 1f, Fig. 2).
Table 1.
An overall review of the circRNAs data in the present study
| BMK-ID | Total Reads | Mapped Reads | Uniq Map Reads | Reads Map to ‘+’ | Reads Map to ‘-’ | GC(%) | Q30(%) |
|---|---|---|---|---|---|---|---|
| AbAd1 | 75,595,006 | 75,557,008 (99.95%) | 70,918,567 (93.81%) | 37,002,982 (48.95%) | 37,013,048 (48.96%) | 46.67 | 90.8 |
| AbAd2 | 126,110,450 | 125,959,112 (99.88%) | 118,894,690 (94.28%) | 61,664,929 (48.90%) | 61,680,043 (48.91%) | 46.03 | 91.1 |
| AbPre1 | 114,650,344 | 114,569,360 (99.93%) | 107,125,637 (93.44%) | 55,851,419 (48.71%) | 55,649,631 (48.54%) | 46.85 | 90.61 |
| AbPre2 | 119,804,518 | 119,721,318 (99.93%) | 111,531,581 (93.09%) | 58,436,387 (48.78%) | 58,456,954 (48.79%) | 47.1 | 91.15 |
| IMAd1 | 100,975,170 | 100,800,650 (99.83%) | 91,276,252 (90.39%) | 49,182,680 (48.71%) | 48,704,390 (48.23%) | 48 | 91.27 |
| IMAd2 | 107,859,100 | 107,660,296 (99.82%) | 96,647,866 (89.61%) | 52,391,877 (48.57%) | 52,116,610 (48.32%) | 48.6 | 90.86 |
| IMPre1 | 120,678,792 | 120,540,008 (99.88%) | 111,158,225 (92.11%) | 58,476,590 (48.46%) | 58,500,070 (48.48%) | 48.8 | 90.49 |
| IMPre2 | 96,725,572 | 96,635,868 (99.91%) | 85,825,333 (88.73%) | 47,124,244 (48.72%) | 47,116,981 (48.71%) | 48.85 | 91.49 |
Fig. 1.
The characteristics of circRNAs in chicken adipocytes. a The correlations analysis among various samples. b The total expression levels of circRNAs in different groups. c The numbers of identified circRNAs in various samples. d The length and genome region distribution of all circRNAs. e The exon number distribution of circRNAs in chicken adipocytes. f The chromosome distribution of all circRNAs
Fig. 2.
The circos plot of circRNAs in chicken adipocytes. The ring from the outer layer to the inner layer represent represents AbPre1, AbPre1, AbAd1, AbAd2, IMPre1, IMPre2, IMAd1and IMAd2 groups, respectively
Identification of differentially expressed (DE) circRNAs during adipogenic differentiation across various groups
To explore functional roles of circRNAs in tissues-specific adipogenic differentiation, we analyzed and compared the expression levels of circRNAs between different groups. Eventually, we identified 17, 14, 20, and 9 differentially expressed (DE) circRNAs across different groups (IMPre_vs_IMAd, AbPre_vs_AbAd, AbPre_vs_IMPre and AbAd_vs_IMAb) (Fig. 3, Table 2). Most of DE circRNAs were specificially found in preadipocytes or adipocytes. In addition, several DE circRNAs showed different expression patterns during adipogenic differentiation between AbF and IMF groups (Table S1). Among them, 7:22323550|22,327,655 was significantly downregulated in mature adipocytes when compared with preadipocytes in both IMF and AbF groups. Z:78636651|78,640,537 (circCDKN2A) was considerably downregulated in AbAd group, while upregulated in the IMAd group. Additionally, 7:22754779|22,756,470 (circTNS1) was only found in intramuscular fat preadipocytes and adipocytes, 21:6563897|6,564,555 (circHSPG2), 3:66699169|66,707,019 (circSLC22A16), and 16:242878|309,387 (circKIFC1L) were specifically expressed in intramuscular adipocytes. 3:79378778|79,415,580 (circBCKDHB), 28:1095851|1,096,498 and 1:83410785|83,437,136 were specifically expressed in abdominal adipocytes.
Fig. 3.
The identification of DE circRNAs across various groups. a Cluster analysis of DE circRNAs in different groups. b Volcano plots of DE circRNAs across various comparisons
Table 2.
The statistics of DE circRNAs across various comparisons
| Group | ALL_DE circRNAs | Up_regulated | down_regulated |
|---|---|---|---|
| AbAd_vs_IMAd | 9 | 8 | 1 |
| AbPre_vs_AbAd | 14 | 2 | 12 |
| AbPre_vs_IMPre | 20 | 9 | 11 |
| IMPre_vs_IMAd | 17 | 10 | 7 |
Functional characterization of DE circRNAs
To investigate the potential functions of DE circRNAs, GO terms analysis of host genes of DE circRNAs were performed. We found that the molecular function (MF) of gene ontology (GO) analysis during adipogenesis were primarily enriched in lipid binding, phospholipase activity, and lipase activity, whereas cell component (CC) were mainly enriched in regulation of autophagy, glycerophospholipid biosynthetic process, phosphatidylinositol-mediated signaling and inositol lipid-mediated signaling (Fig. 4).
Fig. 4.
GO analysis of host genes of DE circRNAs across various groups. a IMAd vs IMPre. b AbAd vs AbPre. c AbAd vs IMAd. d IMPre vs AbPre
Validation of circRNAs
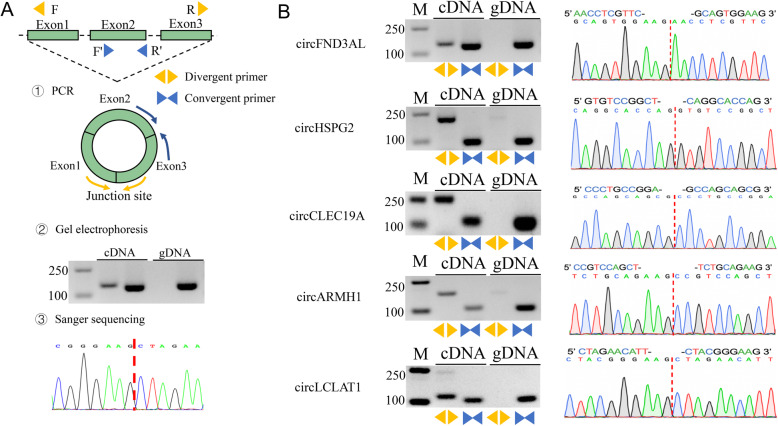
To confirm the circRNAs in chicken adipocytes, five DE circRNAs including circFNDC3AL, circHSPG2, circCLEC19A, circARMH1, and circLCLAT1 were randomly selected for validation using PCR and Sanger sequencing (Fig. 5a). Divergent and convergent primers were designed to amplify circRNAs back-spliced junction (BSJ) site and linear mRNAs. As showed in Fig. 5b, BSJ sites of circRNAs were amplified and validated by sanger sequencing. The amplified PCR products using convergent primers of circRNAs were distinct both in cDNA and gDNA samples, whereas the divergent primers only amplified the cDNA samples. PCR products amplified using the divergent primers were further identified through Sanger sequencing (Fig. 5b). The sequencing results corresponded to Ribo-zero RNA-Seq data, which suggested that the circRNAs data is reliable.
Fig. 5.
The validation of circRNAs in chicken adipocytes. a The validation flow of circRNAs in the present study. b The results of gel electrophoresis (left panel) and Sanger sequencing (right panel) of circRNAs. M, DL2000 DNA maker. The red dotted line represents junction sites
Expression characteristics of miRNAs in chicken intramuscular and abdominal adipogenesis
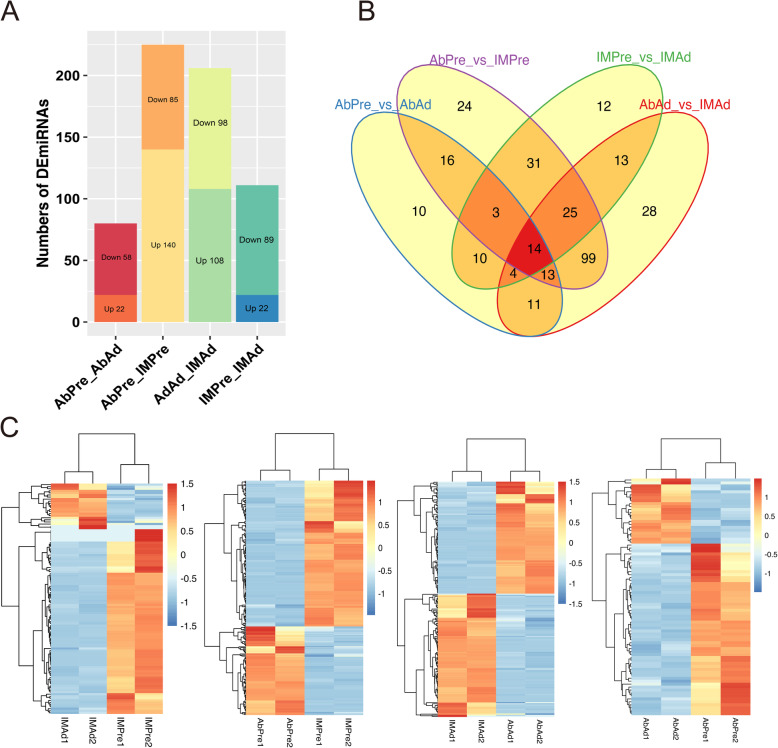
To comprehensively analyze the characteristics of miRNAs during tissue-specific adipogenesis, miRNAs expression levels were compared across various groups. A total of 80 DE miRNAs (58 upregulated and 22 downregulated) were identified between AbPre and AbAd groups, 225 DE miRNAs (85 downregulated and 140 DE miRNAs upregulated) between AbPre and IMPre groups, 206 DE miRNAs (98 downregulated and 108 upregulated) between AbAd and IMAd groups and 111 DE miRNAs (89 upregulated and 22 upregulated) between IMPre and IMAd groups respectively (Fig. 6a) (Table S2). Among them, 81 (66 up-regulated, 15 down-regulated) and 50 (34 up-regulated, 16 down-regulated) specific-DE miRNAs were identified in IM and Ab groups during adipogenic differentiation respectively (Fig. 6b). Moreover, it was observed that most of DE miRNAs were significantly downregulated in mature adipocytes, including miR-130b-5p, miR-148a-5p, miR-15c-5p, miR-16c-5p, miR-30a-3p (Fold change > 2, Q_value < 0.001). However, miR-146b-5p was significantly upregulated in mature adipocytes compared to preadipocytes (Fold change > 4, Q_value < 0.001). The heatmaps of DE miRNAs across various groups are displayed in Fig. 6c.
Fig. 6.
The characteristics of miRNAs during adipogenesis in different groups. a The numbers of DE miRNAs across various groups. b The Venn plot of DE miRNAs across various groups. c Heatmaps of DE miRNAs between different groups (IMAd vs IMPre, AbPre vs IMPre, IMAd vs AbAd, AbAd vs AbPre)
Construction of circRNAs-miRNAs-mRNAs network
Notably, 31 DE miRNAs were shared between IM and Ab groups during adipogenesis, including miR-15c-5p, miR-206, miR-148a-5p, miR-128-1-5p, miR-30a-3p, miR-27b-5p, miR-92-5p and three novel miRNAs (not shown) (Fig. 7a). To further investigate the functional roles of miRNAs in chicken adipogenesis, the target genes of DE miRNAs were predicted through bioinformatics analyses. KEGG pathway analysis revealed that their target genes were highly enriched in the PPAR signaling pathway, metabolic pathways, fatty acid metabolism pathway (Fig. 7b). Further, through integrating analysis of DE cirRNAs, miRNAs and mRNAs data, we constructed a ceRNAs network during chicken adipogenesis (Fig. 7c). It was noted that circFNDC3AL, circHSPG2, circLCLAT1 and circCLEC19A were the hub cirRNAs of the ceRNAs network (Fig. 7c). Furthermore, KEGG pathway analysis showed that the ceRNAs network regulates target genes mainly via fatty acid metabolism, focal adhesion and ECM-receptor interaction pathways (Fig. 7d).
Fig. 7.
The construction of circRNAs-miRNAs-mRNAs ceRNAs networks during chicken adipogenesis. a Heatmap of 28 DE miRNAs across various groups. b The pathway enrichment analysis of target genes of miRNAs. c The circRNAs-media ceRNAs networks during chicken adipogenesis. The cube represents circRNAs, circles represent miRNAs and arrows represent genes. Sea-blue color represents a downregulated trend, while dark-red and brown-yellow color represents an upregulated trend. d The pathway enrichment analysis of downstream genes in ceRNAs networks
Candidate circRNAs related to adipogenic differentiation
Further, correlation analysis between circRNAs and their target genes based on the ceRNAs network was performed to investigate the crucial roles of circRNAs in the adipogenesis of chicken. As shown in Fig. 8a, the expression levels of circFNDC3AL and circHSPG2 were significantly and positively related to PPAR signaling pathway genes in intramuscular adipogenesis (r > 0.90, p < 0.01), such as peroxisome proliferator-activated receptor γ (PPARγ), fatty acid binding protein 4 (FABP4), perilipin 2 (PLIN2), CD36 molecule (CD36). In contrast, the expression level of circLCLAT1 was significantly and negatively related to PPAR signaling pathway genes and positively related to lysocardiolipin acyltransferase 1 (LCLAT1) and fatty acid desaturase 2 (FADS2) (r > 0.95, p < 0.01) (Fig. 8a). Moreover, the expression levels of circARMH1 were significantly and negatively related to those of genes in abdominal adipocytes (r > 0.92, p < 0.01), while circCLEC19A was significantly and positively related to those of genes (r > 0.90, p < 0.01) (Fig. 8b). To further investigate the potential roles of circRNAs in the fat tissue-specific deposition, the expression of cirRNAs was analyzed in different tissues. Our results demonstrated that the expression of these four candidate circRNAs was highly expressed in the abdominal fat tissue of chicken (Fig. 8c). In addition, the dynamistic expression patterns of candidate circRNAs were detected during tissue-specific adipogenetic differentiation. Our results showed that the expression levels of circFNDC3AL were decreased after 4 days of induction, while dramatically upregulated in the late stage of intramuscular adipogenesis (Fig. 8d). The expression levels of circCLEC19A were significantly increased during the late stage of abdominal adipogenesis (Fig. 8 d). In contrast, the expression levels of circCLEC19A were dramatically decreased after 2 days’ of adipogenetic induction, while slightly increased in the late stage of intramuscular adipogenesis. In addition, the expression levels of circARMH1 were decreased during the abdominal adipogenesis (Fig. 8d). Interestingly, we noticed that chicken circLCLAT1 was highly conservative among humans, mice and pigs (Fig. S1), implying that circLCLAT1 potentially regulates animal adipogenesis or lipid metabolism.
Fig. 8.
Candidate circRNAs associated with tissue-specific adipogenesis. a The Pearson correlations between the expression levels of circFNDC3AL, circHSPG2, circLCLAT1, (b) circARMH1, and circCLEC19A and genes associated with adipogenesis and lipid metabolism, respectively. c The relative expression levels of candidate circRNAs in five chicken tissues. d The dynamical expression patterns of candidate circRNAs during IM and Ab adipogenic differentiation, respectively (n = 3, Mean ± S.E.M). *p < 0.05, **p < 0.01
Validation of circRNA-miRNA-mRNAs ceRNA networks
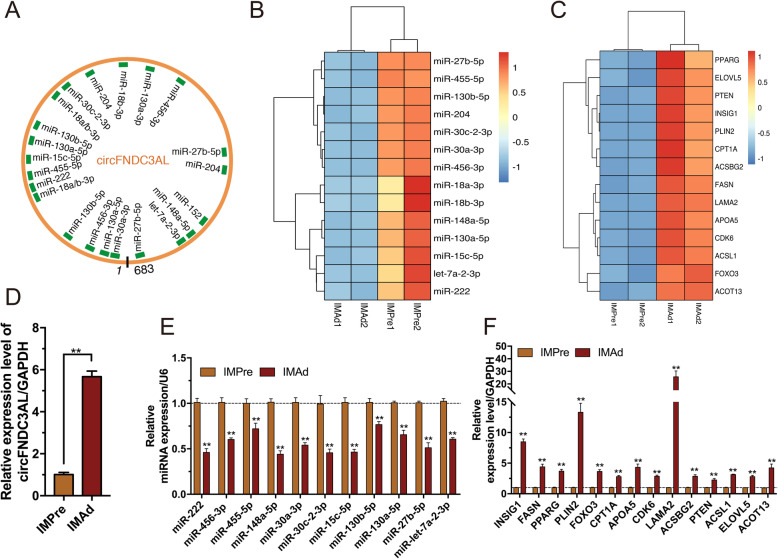
To verify the circRNA-miRNA-mRNAs regulation networks during intramuscular adipogenic differentiation, we analyzed their expression levels in intramuscular preadipocytes and mature adipocytes. Bioinformatics analysis revealed that cirFNDC3AL exhibit many MREs (miRNAs response elements), such as miR-130a-5p, miR-15c-5p, miR-18a/b-3p (Fig. 9a). Results from RNA-Seq and RT-qPCR showed that cirFNDC3AL and its downstream genes were significantly upregulated in differentiated-mature intramuscular adipocytes, compared to intramuscular preadipocytes (Fig. 9c, d, f). On the other hand, several adipogenic differentiation-related miRNAs were significantly downregulated in adipocytes, compared to intramuscular preadipocytes, including miR-222, miR-456-3p, miR-15C-5p, miR-130b-5p, miR-130a-5p (Fig. 9b, e). Additionally, it was worth noting that the target genes of circFNDC3AL were mainly associated with lipid metabolism and adipogenesis, such as PPARG, PLIN2, CPT1A, INSIG1. In addition, bioinformatic analysis and qRT-PCR results found that miR-146b-5p and miR-147 potentially regulate RUNXT1 and FADS2, while miR-34a-5p and miR-30e-5p might regulate PDGFRA, IGF2BP3, FNDC3B, MYH9 (Fig. S2). circLCLAT1 potentially influence chicken adipogenesis by sponging these miRNAs, thus regulating target genes. The findings suggested that circLCLAT1-miRNAs (miR-146b-5p, miR-147, miR-30e-5p, and miR-34a-5p)-mRNAs pathway showed opposite expression patterns during intramuscular adipogenic differentiation (Fig. 10). CircLCLAT1 and its downstream genes were significantly downregulated in differentiated-mature intramuscular adipocytes (Fig. 10c, d and f), while miR-146b-5p and miR-147 were significantly upregulated in mature adipocytes compared to preadipocytes (Fig. 10b, e).
Fig. 9.
circFNDC3AL regulated chicken adipogenesis via the ceRNA pathway. a The miRNAs binding sites of circFNDC3AL. b The expression patterns of candidate miRNAs and (c) their target genes during intramuscular adipogenesis in RNA-Seq data. The expression levels of (d) circFNDC3AL, (e) miRNAs and (f) targets genes before and after intramuscular adipogenic differentiation. (n = 3, Mean ± S.E.M) *p < 0.05, **p < 0.01
Fig. 10.
circLCLAT1 regulated chicken adipogenesis via the ceRNA pathway. a The miRNAs binding sites of circLCLAT1. b The expression patterns of candidate miRNAs and (c) their target genes during intramuscular adipogenesis in RNA-Seq data. The expression levels of (d) circLCLAT1, (e) miRNAs, and (f) targets genes before and after intramuscular adipogenic differentiation. (n = 3, Mean ± S.E.M) *p < 0.05, **p < 0.01
Discussion
Preadipocytes-derived from abdominal fat (AbF) have previously been reported to exhibit higher adipogenic differentiation potential than the ones derived from intramuscular fat (IMF) in chickens [26]. Adipogenesis is controlled by a series of complex regulatory mechanisms, including transcription level, post-transcription level, among others [27–29]. Noncoding RNAs have notable effects on adipogenesis and lipid metabolism [30, 31]. Several studies about noncoding RNAs on adipogenesis were geared towards mammals [11, 12, 30, 31]. However, information on circular RNAs (circRNAs) in poultry adipogenesis is limited.
In this work, we analyzed Ribo-zero RNA-Seq and corresponding miRNAs data during chicken tissue-specific adipogenic differentiation. Three software (CIRI2, CIRCexplore2 and find_circ) were used to identify circRNAs. Although fewer circRNAs were obtained with this way, it improved the accuracy rate of our findings. A total of 8843 circRNAs were identified in preadipocytes and adipocytes. This suggested that several circRNAs are expressed in chicken preadipocytes and adipocytes. Of note, the characteristics of circRNAs based on length, exon number, and genomic distribution in chicken were in line with previous reports [19, 32]. Previous studies revealed that the expression level of circRNAs was tissues- and stage-specific [33, 34]. Abdominal and intramuscular fat deposition resulted from a variety of complex molecular mechanisms. Notably, tissue-specific adipogenesis is potentially regulated by circRNAs [25]. In the present study, we identified DE circRNAs across various tissue and cell types. Also, previous reports identified several DE circRNAs in chicken, such as circCELC19A which was suggested to play functional roles in follicle development [32]. In addition, circFNDC3AL and circTNS1 were identified in embryonic muscle tissues of chicken [19].
Previously, we reported that miRNAs could be crucial regulators in chicken adipogenesis [8, 35]. Here, we systematically analyzed and compared the expression patterns of miRNAs during intramuscular and abdominal adipogenesis. It was revealed that several miRNAs were significantly downregulated in mature adipocytes. On the other hand, many genes associated with lipid metabolism and adipogenesis were upregulated during the adipogenic differentiation across various tissues, implying that the downregulated miRNAs potentially promote adipogenesis through their target genes in chicken.
Generally, it is believed that circRNAs could function as a molecular sponge of miRNAs thereby regulating their target genes [36]. By integrating the analysis of circRNAs, miRNAs, and mRNAs data, hub ceRNAs networks were constructed during chicken adipogenic differentiation. Notably, the hub genes in the ceRNAs network were enriched in fatty acid metabolism, PPAR signaling pathway, and p53 signaling pathway. Furthermore, the downstream genes in the ceRNA pathways were strongly related to candidate circRNAs in the present study (Fig. 7c and Fig. 8a), suggesting that these circRNAs might play functional roles during adipogenesis. RT-qPCR results of the circFNDC3AL-miRNAs-mRNAs pathway corroborated the results of bioinformatics analysis. Interestingly, fibronectin type III domain-containing protein 3B (FNDC3B) and 5 (FNDC5) regulate white fat browning and adipogenesis [37–40]. Besides, perlecan (HSPG2, Heparan Sulfate Proteoglycan 2) was found to regulate obesity and lipid deposition [41]. Thus, we speculated that circFNDC3AL and circHSPG2 potentially regulate chicken adipogenesis. However, in-depth studies on the functions of chicken circFNDC3AL and circHSPG2 on adipogenic differentiation are essential. Previous research reported that runt-related transcription factor 1 (RUNX1T1) [42, 43], myosin, heavy chain 9, non-muscle (MYH9), fatty acid desaturase 2 (FADS2), insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) were associated with adipogenesis and fat deposition [44–47]. We found that circLCLAT1 might influence adipogenic differentiation by regulating downstream genes (RUNX1T1, FADS2, MYH9, IGF2BP3, and PDGFRA) through four miRNAs (miR-34a-5p, miR-30e-5p, miR-146b-5p, and miR-147). Nonetheless, future in-depth studies should be undertaken to explore the molecular regulation mechanism of circFNDC3AL and circLCLAT1/miRNAs/mRNAs pathway in chicken adipogenesis.
Conclusions
In summary, we comprehensively identified expression profiles of circRNAs in adipocytes derived from various adipose tissues in chicken. By integrating circRNAs, miRNAs, and mRNAs data, we constructed ceRNAs networks that regulated chicken adipogenesis. Over all, this study offers insights into poultry tissue-specific adipogenesis in poultry and reveals novel clues to studying circRNAs on adipogenic differentiation and fat deposition in poultry.
Methods
Tissues and cell samples collection
The experimental birds were obtained from the Avian Farm of Henan Agriculture University (Zhengzhou, Henan, China). Here, five types of tissues (heart, liver, breast muscle, abdominal, and kidney tissues) were collected and stored at − 80 °C until use. Under deep anesthesia, the birds were euthanized by intramuscular injection of pentobarbital (Sigma, St. Louis, MO, USA) (40 mg/kg). Intramuscular and abdominal preadipocytes were isolated from the breast muscle and abdominal adipose tissue of 2 weeks old-chicken following the method described by Zhang et al [8, 26]. Pollution-free treatment of animal carcasses was performed after the experiment. In vitro adipogenic differentiation model of chicken preadipocytes constructed according to the method by Zhang et al [26]. After 80–90% confluence, cells were exposed to the MDIO differentiation medium. Cell samples were harvested at 0, 2, 4, 6, 8, and 10 days after adipogenic induction.
Data resources, annotation and quantification of circRNA
Ribo-Zero RNA-Seq and miRNAs data were downloaded from the NCBI database (No. PRJNA429489 and PRJNA453673). Four groups were included in the PRJNA429489 and PRJNA453673 program, including preadipocytes derived from chicken intramuscular fat (IM_Pre), abdominal fat (Ab_Pre) and differentiated-mature adipocytes (IM_Ad, Ab_Ad). Each group included two biological replicates. The paired-end (PE) reads were mapped to the Gallusgallus 5 genome (https://www.ncbi.nlm.nih.gov/genome/?term=chicken) using Bowtie2 (v2.2.9) with default parameters [48]. The output of Bowtie2 was scanned and analyzed by CIRI2 (v2.0.6) [49], find_circ (v1.0) [50] and CIRCexplore2 (v2.3.3) [51] with default parameters. The identification of circRNA was based on the intersection of the three software. TPM was applied to calculate the relative expression abundance of circRNA.
Identification of differentially expressed (DE) circRNAs and host genes functional annotation
DE circRNAs were analyzed using the edgeR package [52], with the threshold of p-value < 0.05 and |fold change| ≥ 2. Host genes of circRNAs were used for predicting the potential functions of circRNAs. KOBAS 3.0 (http://kobas.cbi.pku.edu.cn/kobas3/?t=1) online software was used for Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses.
The conservative analysis of circRNAs
CircRNAs sequences of humans and mice were downloaded from the circBase database (http://www.circbase.org), porcine circRNAs sequences were download from pigcircNet database (http://lnc.rnanet.org/circ/). The sequences were compared using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) for multiple sequence alignment.
Genomic DNA (gDNA) and total RNA extraction, cDNA synthesis and quantitative real-time PCR (qRT-PCR)
Genomic DNA (gDNA) and total RNA were extracted from tissues/cell samples using animal genomic DNA kit (Tiangen, China) and RNAiso plus (Takara, Dalian, China) following to the manufacturers’ instructions respectively. cDNA was synthesized with a PrimeScriptTM RT reagent Kit with gDNA Eraser (Takara, Dalian, China). Primers used for qRT-PCR were designed by primer3plus (Supplementary Table S3). qRT-PCR was performed with the SYBR® Premix Ex TaqTM II kit (Takara) on a LightCycler® 96 Real-Time PCR system. GAPDH and U6 were used as the reference genes of circRNAs, mRNAs, and miRNAs, respectively. The 2−ΔΔCt method was used to calculate gene expression levels.
Validation of circRNAs
To validate the circRNAs related to chicken adipogenesis, PCR and Sanger sequencing assays were performed. Divergent primer was designed to amplify the junction sites of circRNA, whereas a convergent primer was designed to amplify the linear mRNA (Supplementary Table S3). Random primers were used for cDNA synthesis. Then, the cDNA of intramuscular and abdominal preadipocytes and adipocytes were mixed to a cDNA pool. The PCR products were sequenced by Sangon Biotech Co. Ltd. (Shanghai, China).
The integrative analysis of circRNAs, miRNAs, and mRNAs
Since circRNAs can act as sponges of miRNA, the potential miRNAs binding sites in chicken adipogenesis were analyzed. The four groups of circRNA data including IMPre, IMAd, AbPre, and AbAd were analyzed during chicken adipogenesis were analyzed, and the corresponding four groups of miRNA data during adipogenesis were descripted and analyzed in our previous studies [35, 53]. miRNAs with |fold change| > 2 and FDR < 0.05 were regarded as differentially expressed miRNAs. TargetScan (http://www.targetscan.org/vert_72/), miRanda (http://www.microrna.org/microrna/home.do), and RNAhybrid software (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid) were used to predict the miRNA sites in mRNAs and circRNAs with default parameters, and investigate putative interactions between miRNAs and mRNAs or circRNAs. Furthermore, the target genes with opposite expression trends to miRNAs were regarded as candidate target genes. Based on the co-expression of DE circRNAs and miRNAs (|Pearson’s correlation coefficient| > 0.8 and p < 0.05), the circRNAs-miRNAs-mRNAs ceRNA networks were constructed and visualized using Cytoscape (version 3.5.0) (http://www.cytoscape.org/) [54].
Statistical analysis
All statistical data were analyzed using SPSS 22.0 (SPSS Inc., Chicago, IL, USA). All results were presented as mean ± S.E.M, a two-tailed t-test to analyzed the data. p < 0.05 and ** p < 0.01 were considered significant and extremely significant respectively.
Supplementary information
Additional file 1: Table S1. The list of differentially expressed (DE) circRNAs across various groups. Table S2. The list of differentially expressed (DE) miRNAs across various groups. Table S3. The primers used for qRT-PCR in the present study.
Additional file 2 Figure S1. circLCLAT1 is a sequence-conservative circRNA. (A) Genome locate of chicken circLCLAT1. (B) Multiple sequence alignment analysis of circLCLAT1 among species. (C) The evolutionary tree of circLCLAT1 among species.
Additional file 3 Figure S2. The binding sites of some upregulated miRNAs (eg. miR-146b-5p, miR-147, miR-34a-5p, miR-30e-5p) in the 3’UTRs of potential target genes.
Acknowledgements
We would like to thank the Guirong Sun Ph.D. for providing cells and reagents for this research.
Abbreviations
- IMF
Intramuscular fat
- AbF
Abdominal fat
- circRNAs
Circular RNAs
- miRNAs
MicroRNAs
- MREs
MiRNAs response element
- DE
Differentially expressed
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- BSJ
Back-spliced junction
- FNDC3AL
Fibronectin type III domain containing 3A-like
- FNDC3B
Fibronectin type III domain-containing protein 3
- FNDC5
Fibronectin type III domain-containing protein 5
- TNS1
Tensin 1
- HSPG2
Heparan sulfate proteoglycan 2
- CLEC19A
C-type lectin domain containing 19A
- ARMH1
Armadillo like helical domain containing 1
- LCLAT1
Lysocardiolipin acyltransferase 1
- RUNX1T1
Runt-related transcription factor 1
- MYH9
Myosin, heavy chain 9, non-muscle
- FADS2
Fatty acid desaturase 2
- IGF2BP1
Insulin-like growth factor 2 mRNA-binding protein 1
- PPARγ
Peroxisome proliferator-activated receptor γ
- FABP4
Fatty acid binding protein 4
- PLIN2
Perilipin 2
- CD36
CD36 molecule
Authors’ contributions
ZL and SZ conceived the project, supervised the experiments and revised the manuscript. MZ and YH performed the experiments and analyzed data, MZ wrote the manuscript. YZ, XM and XA contributed the part of regents and data analysis. The final manuscript was approved by all authors.
Funding
No funding was obtained for this study.
Availability of data and materials
The sequencing data has been submitted to NCBI SRA under BioProject ID: PRJNA429489 and PRJNA453673.
Ethics approval and consent to participate
The animal experiments were performed with the permission of Avian Farm of Henan Agriculture University (Zhengzhou, Henan, China), and was approved by the Institutional Animal Care and Use Committee (IACUC) of Henan Agricultural University (11–0085). All effects were made to minimize birds suffering.
Consent for publication
Not applicable.
Competing interests
The authors declared no competing interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Meng Zhang, Email: zm19911213@gmail.com.
Yu Han, Email: hany18@gmail.com.
Yanhui Zhai, Email: 524699311@qq.com.
Xiangfei Ma, Email: mxf1228@126.com.
Xinglan An, Email: anxinglan@jlu.edu.cn.
Sheng Zhang, Email: lncyzhangsheng@126.com.
Ziyi Li, Email: ziyi@jlu.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12864-020-07000-3.
References
- 1.Sun WX, Wang HH, Jiang BC, Zhao YY, Xie ZR, Xiong K, et al. Global comparison of gene expression between subcutaneous and intramuscular adipose tissue of mature Erhualian pig. Genet Mol Res. 2013;12(4):5085–5101. doi: 10.4238/2013.October.29.3. [DOI] [PubMed] [Google Scholar]
- 2.Huang WL, Zhang XX, Li A, Miao XY, et al. Identification of differentially expressed genes between subcutaneous and intramuscular adipose tissue of large White pig using RNA-seq. Yi Chuan. 2017;39(6):501–511. doi: 10.16288/j.yczz.17-038. [DOI] [PubMed] [Google Scholar]
- 3.Bong JJ, Cho KK, Baik M. Comparison of gene expression profiling between bovine subcutaneous and intramuscular adipose tissues by serial analysis of gene expression. Cell Biol Int. 2009;34(1):125–133. doi: 10.1042/CBI20090046. [DOI] [PubMed] [Google Scholar]
- 4.Lim YC, Chia SY, Jin S, Han W, Ding C, Sun L. Dynamic DNA methylation landscape defines brown and white cell specificity during adipogenesis. Mol Metab. 2016;5(10):1033–1041. doi: 10.1016/j.molmet.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao T, Liu L, Li H, Sun Y, Luo H, Li T, et al. Long noncoding RNA ADINR regulates Adipogenesis by transcriptionally activating C/EBPalpha. Stem Cell Reports. 2015;5(5):856–865. doi: 10.1016/j.stemcr.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hudson NJ, Reverter A, Griffiths WJ, Yutuc E, Wang Y, Jeanes A, et al. Gene expression identifies metabolic and functional differences between intramuscular and subcutaneous adipocytes in cattle. BMC Genomics. 2020;21(1):77. doi: 10.1186/s12864-020-6505-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan TT, Ren LL, Shen CQ, Wang ZH, Yu YN, Liang Q, et al. miR-508 defines the stem-like/mesenchymal subtype in colorectal cancer. Cancer Res. 2018;78(7):1751–1765. doi: 10.1158/0008-5472.CAN-17-2101. [DOI] [PubMed] [Google Scholar]
- 8.Zhang M, Li DH, Li F, Sun JW, Jiang RR, Li ZJ, et al. Integrated analysis of MiRNA and genes associated with meat quality reveals that Gga-MiR-140-5p affects intramuscular fat deposition in chickens. Cell Physiol Biochem. 2018;46(6):2421–2433. doi: 10.1159/000489649. [DOI] [PubMed] [Google Scholar]
- 9.Zhang M, Li CC, Li F, Li H, Liu XJ, Loor JJ, et al. Estrogen promotes hepatic synthesis of Long-chain polyunsaturated fatty acids by regulating ELOVL5 at post-transcriptional level in laying hens. Int J Mol Sci. 2017;18(7):1405. [DOI] [PMC free article] [PubMed]
- 10.Han H, Gu S, Chu W, Sun W, Wei W, Dang X, et al. miR-17-5p regulates differential expression of NCOA3 in pig intramuscular and subcutaneous adipose tissue. Lipids. 2017;52(11):939–949. doi: 10.1007/s11745-017-4288-4. [DOI] [PubMed] [Google Scholar]
- 11.Zhu M, Liu J, Xiao J, Yang L, Cai M, Shen H, et al. Lnc-mg is a long non-coding RNA that promotes myogenesis. Nat Commun. 2017;8:14718. doi: 10.1038/ncomms14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Wang Y, He X, Zhang S, Wang K, Wu H, et al. LncRNA TINCR/miR-31-5p/C/EBP-alpha feedback loop modulates the adipogenic differentiation process in human adipose tissue-derived mesenchymal stem cells. Stem Cell Res. 2018;32:35–42. doi: 10.1016/j.scr.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 13.Xu S, Wang P, Zhang J, Wu H, Sui S, Zhang J, et al. Ai-lncRNA EGOT enhancing autophagy sensitizes paclitaxel cytotoxicity via upregulation of ITPR1 expression by RNA-RNA and RNA-protein interactions in human cancer. Mol Cancer. 2019;18(1):89. doi: 10.1186/s12943-019-1017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869–881. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen S, Huang V, Xu X, Livingstone J, Soares F, Jeon J, et al. Widespread and functional RNA circularization in localized prostate cancer. Cell. 2019;176(4):831–843. doi: 10.1016/j.cell.2019.01.025. [DOI] [PubMed] [Google Scholar]
- 18.Ruan H, Xiang Y, Ko J, Li S, Jing Y, Zhu X, et al. Comprehensive characterization of circular RNAs in ~ 1000 human cancer cell lines. Genome Med. 2019;11(1):55. doi: 10.1186/s13073-019-0663-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ouyang H, Chen X, Wang Z, Yu J, Jia X, Li Z, et al. Circular RNAs are abundant and dynamically expressed during embryonic muscle development in chickens. DNA Res. 2018;25(1):71–86. doi: 10.1093/dnares/dsx039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei X, Li H, Yang J, Hao D, Dong D, Huang Y, et al. Circular RNA profiling reveals an abundant circLMO7 that regulates myoblasts differentiation and survival by sponging miR-378a-3p. Cell Death Dis. 2017;8(10):e3153. doi: 10.1038/cddis.2017.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Cao X, Dong D, Shen X, Cheng J, Jiang R, et al. Circular RNA TTN acts as a miR-432 sponge to facilitate proliferation and differentiation of myoblasts via the IGF2/PI3K/AKT signaling pathway. Mol Ther Nucleic Acids. 2019;18:966–980. doi: 10.1016/j.omtn.2019.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Yang J, Wei X, Song C, Dong D, Huang Y, et al. CircFUT10 reduces proliferation and facilitates differentiation of myoblasts by sponging miR-133a. J Cell Physiol. 2018;233(6):4643–4651. doi: 10.1002/jcp.26230. [DOI] [PubMed] [Google Scholar]
- 23.Cai J, Chen Z, Wang J, Wang J, Chen X, Liang L, et al. circHECTD1 facilitates glutaminolysis to promote gastric cancer progression by targeting miR-1256 and activating beta-catenin/c-Myc signaling. Cell Death Dis. 2019;10(8):576. doi: 10.1038/s41419-019-1814-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in Myogenesis. Mol Cell. 2017;66(1):22–37. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arcinas C, Tan W, Fang W, Desai TP, Teh DCS, Degirmenci U, et al. Adipose circular RNAs exhibit dynamic regulation in obesity and functional role in adipogenesis. Nat Metab. 2019;1(7):688–703. doi: 10.1038/s42255-019-0078-z. [DOI] [PubMed] [Google Scholar]
- 26.Zhang M, Li F, Ma XF, Li WT, Jiang RR, Han RL, et al. Identification of differentially expressed genes and pathways between intramuscular and abdominal fat-derived preadipocyte differentiation of chickens in vitro. BMC Genomics. 2019;20(1):743. doi: 10.1186/s12864-019-6116-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karbiener M, Fischer C, Nowitsch S, Opriessnig P, Papak C, Ailhaud G, et al. microRNA miR-27b impairs human adipocyte differentiation and targets PPARgamma. Biochem Biophys Res Commun. 2009;390(2):247–251. doi: 10.1016/j.bbrc.2009.09.098. [DOI] [PubMed] [Google Scholar]
- 28.Loftus TM, Lane MD. Modulating the transcriptional control of adipogenesis. Curr Opin Genet Dev. 1997;7(5):603–608. doi: 10.1016/s0959-437x(97)80006-8. [DOI] [PubMed] [Google Scholar]
- 29.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79(7):1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 30.Romao JM, Jin W, Dodson MV, Hausman GJ, Moore SS, Guan LL. MicroRNA regulation in mammalian adipogenesis. Exp Biol Med (Maywood) 2011;236(9):997–1004. doi: 10.1258/ebm.2011.011101. [DOI] [PubMed] [Google Scholar]
- 31.Sun L, Goff LA, Trapnell C, Alexander R, Lo KA, Hacisuleyman E, et al. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci U S A. 2013;110(9):3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen M, Li T, Zhang G, Wu P, Chen F, Lou Q, et al. Dynamic expression and functional analysis of circRNA in granulosa cells during follicular development in chicken. BMC Genomics. 2019;20(1):96. doi: 10.1186/s12864-019-5462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73(5):1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 34.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun G, Li F, Ma X, Sun J, Jiang R, Tian Y, et al. Gga-miRNA-18b-3p inhibits intramuscular adipocytes differentiation in chicken by targeting the ACOT13 gene. Cells. 2019;8(6):556. [DOI] [PMC free article] [PubMed]
- 36.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bostrom P, Wu J, Jedrychowski MP, Korde A, Ye L, Lo JC, et al. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012;481(7382):463–468. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wrann CD, White JP, Salogiannnis J, Laznik-Bogoslavski D, Wu J, Ma D, et al. Exercise induces hippocampal BDNF through a PGC-1alpha/FNDC5 pathway. Cell Metab. 2013;18(5):649–659. doi: 10.1016/j.cmet.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lourenco MV, Frozza RL, de Freitas GB, Zhang H, Kincheski GC, Ribeiro FC, et al. Exercise-linked FNDC5/irisin rescues synaptic plasticity and memory defects in Alzheimer's models. Nat Med. 2019;25(1):165–175. doi: 10.1038/s41591-018-0275-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng Y, Li H, Li X, Yu S, Xiang H, Peng J, et al. MicroRNA-215 impairs adipocyte differentiation and co-represses FNDC3B and CTNNBIP1. Int J Biochem Cell Biol. 2016;79:104–112. doi: 10.1016/j.biocel.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 41.Yamashita Y, Nakada S, Yoshihara T, Nara T, Furuya N, Miida T, et al. Perlecan, a heparan sulfate proteoglycan, regulates systemic metabolism with dynamic changes in adipose tissue and skeletal muscle. Sci Rep. 2018;8(1):7766. doi: 10.1038/s41598-018-25635-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24(12):1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rochford JJ, Semple RK, Laudes M, Boyle KB, Christodoulides C, Mulligan C, et al. ETO/MTG8 is an inhibitor of C/EBPbeta activity and a regulator of early adipogenesis. Mol Cell Biol. 2004;24(22):9863–9872. doi: 10.1128/MCB.24.22.9863-9872.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang HY, Liu RR, Zhao GP, Li QH, Zheng MQ, Zhang JJ, et al. Integrated analysis of microRNA and mRNA expression profiles in abdominal adipose tissues in chickens. Sci Rep. 2015;5:16132. doi: 10.1038/srep16132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J, Ren X, Li L, Lu S, Chen T, Tan L, et al. Integrative analyses of mRNA expression profile reveal the involvement of IGF2BP1 in chicken Adipogenesis. Int J Mol Sci. 2019;20(12):2923. [DOI] [PMC free article] [PubMed]
- 46.Pfisterer SG, Gateva G, Horvath P, Pirhonen J, Salo VT, Karhinen L, et al. Role for formin-like 1-dependent acto-myosin assembly in lipid droplet dynamics and lipid storage. Nat Commun. 2017;8:14858. doi: 10.1038/ncomms14858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Romao JM, He ML, McAllister TA, Guan LL. Effect of age on bovine subcutaneous fat proteome. Molecular mechanisms of physiological variations during beef cattle growth. J Anim Sci. 2014;92(8):3316–3327. doi: 10.2527/jas.2013-7423. [DOI] [PubMed] [Google Scholar]
- 48.Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao Y, Zhang J, Zhao F. Circular RNA identification based on multiple seed matching. Brief Bioinform. 2017;19(5):803–810. doi: 10.1093/bib/bbx014. [DOI] [PubMed] [Google Scholar]
- 50.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 51.Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26(9):1277–87. [DOI] [PMC free article] [PubMed]
- 52.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England) 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma X, Sun J, Zhu S, Du Z, Li D, Li W, et al. MiRNAs and mRNAs analysis during abdominal Preadipocyte differentiation in chickens. Animals (Basel). 2020;10(3):468. [DOI] [PMC free article] [PubMed]
- 54.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. The list of differentially expressed (DE) circRNAs across various groups. Table S2. The list of differentially expressed (DE) miRNAs across various groups. Table S3. The primers used for qRT-PCR in the present study.
Additional file 2 Figure S1. circLCLAT1 is a sequence-conservative circRNA. (A) Genome locate of chicken circLCLAT1. (B) Multiple sequence alignment analysis of circLCLAT1 among species. (C) The evolutionary tree of circLCLAT1 among species.
Additional file 3 Figure S2. The binding sites of some upregulated miRNAs (eg. miR-146b-5p, miR-147, miR-34a-5p, miR-30e-5p) in the 3’UTRs of potential target genes.
Data Availability Statement
The sequencing data has been submitted to NCBI SRA under BioProject ID: PRJNA429489 and PRJNA453673.