Abstract
Understanding beta structures has been hindered by the challenge of designing well-folded, small β-sheet systems. Kier et al. developed a β-capping motif that helped to solve this problem, but with limitations. Due to the design of the Kier β-cap, the termini are not fully available for chain extension. Combining Coulombic side chain attractions with a Trp/Trp edge to face interaction produced a new capping motif that provided greater β-sheet stability, even in systems lacking a turn locus with a high propensity for chain direction reversal. This Coulombic cap was applied to a number of difficult systems with improvements in stability observed in all cases, affording an additional tool in the study of protein structure and folding.
Introduction
Compared to α-helicies,1–3 our knowledge about β-sheets is lacking. Information on the essential features of β-structures, including residue preferences within β-strands and the turns connecting them, should result in improved structure prediction based on protein sequences4–6. Also, the ability to design small stable β-strands is promising in the area of inhibiting7 and understanding protein aggregates8–9, which have β-sheet structures5. However, when removed from their protein frameworks, many protein β-hairpins have proven to be rather poorly folded10–14. As a result, re-designed hairpins have been used in many studies. Even in these cases, the formation of stable structures has been the exception12–13, 15–18 rather than the rule. There is still a need for design features that can stabilize β-hairpins with minimum change in the sequence. A number of design features have been explored as hairpin fold stabilizers: improved turn loci13,19–22, turn-flanking W/W pairs12–13, 23–26, and most recently the use of non-covalent interactions to stabilize the otherwise frayed ends of β-hairpins26–27. Previously, Kier et al. developed a β-capping motif that incorporated an acylated Trp at the N-terminus and a WTG-NH2 or WTGPK sequence at the C-terminus of hairpin structures. In addition to the quadrupolar Trp/Trp interaction, a number of hydrogen bonds between the residues of the two designed termini results in a highly structured β-cap, able to stabilize a wide range of β-hairpins. Although useful, the Kier β-cap requires specific terminal sequences and modifications, alkanoyl-W-hairpin-WTG. The present study was undertaken to develop β-capping units that could be applied to antiparallel β-sheets that do not link together the extreme termini of a sequence.
Herein we employ an alternative β-sheet model based on a disulfide dimer to explore alternative capping motifs. An earlier version of this motif (Cap-Strand-Cox-Strand-Cap)2 was reported in the original β-cap paper27. This “turnless” system allows for the investigation of the capping ability at the termini without complications from turn propensity effects on hairpin stability. The extent of β-strand association can be monitored from the diagnostic structuring chemical shifts observed at the HN and Hα protons in the β-strands28, with the capping interaction of the indole rings in the caps providing additional structuring shifts and a diagnostic exciton couplet in the circular dichroic (CD) spectra13, 25, 29.
Materials and Methods
Synthesis
All peptides were synthesized on either an Applied Biosystems 496 MOS, or a CEM Liberty Blue synthesizer, using standard Fmoc solid phase peptide synthesis as previously reported26. Preloaded Fmoc-protected Wang and unloaded Rink amide resins were used for synthesis. Peptides were cleaved from resin (0.1 mmol) using a cocktail of trifluoroacetic acid : triisopropylsilane : water (38:1:1, 9.5:0.25:0.25 ml) for 1.5 hr. The resin was filtered and washed with DCM, the resulting filtrate was concentrated in vacuo, crashed out and washed with cold (−20 °C) diethyl ether, giving the crude peptide. If disulfide formation was required, after drying the peptide, the pellet was dissolved in minimal amount of DMSO (0.5 ml), diluted with water (1 ml) and let stand for 24 hrs. at room temperature. After additionally diluting with water (3.5 ml) the solution was purified using RP-HPLC (Varian ProStar 220 HPLC, Agilent 21.2 × 50mm C18 column, 10 ml/min, Elutent A : Water with 0.1% TFA, Elutent B : Acetonitrile with 0.085% TFA), using a gradient of 10 – 50 %B over 19 min. Peaks were visualized at 215 and 280 nm with verification by mass spectrometry (Bruker Esquire ion trap with ESI ionization). The concentrated fractions were then lyophilized resulting in the purified peptide. C-termini amides were made using Rink amide Resin. N-termini were acetylated by adding DMF (3 ml), triethylamine (140 μl), acetic anhydride (95 μl) and shaking at room temp. for 1 hr, before cleaving from resin. If disulfide formation did not occur using the previous procedure, the dry peptide was dissolved in minimal amount of DMSO (0.5 ml) and diluted in 1 M HCl (1 ml) and left for 24 hrs. Covalently, dimers alone are formed, since only a single Cys residue is present in each monomer. The lack of a concentration dependence for both NMR shifts and the CD spectrum (see Supporting Information) indicates that other higher-order aggregate structures did not form.
NMR Spectroscopy
NMR samples were made at ~1 mM in 20 mM Potassium Phosphate buffer with 10% D2O and an internal standard of sodium 4,4-dimethyl-4-silapentane-1-sulfonate (DSS). Full 1H spectra assignments were made using peptide backbone connectivities determined by 2D TOCSY and NOESY experiments taken on either Bruker DRX 500 MHz, or AV 700 MHz spectrometers.
Structuring-induced chemical shift changes were analyzed as chemical shift deviations (CSDs, the observed shifts minus the coil reference values). The CSDs were calculated as previously reported13, 30 using the in-house version of CSDb algorithm which is also available on-line (andersenlab.chem.washington.edu/CSDb/about.php).
Fraction folded (χF) calculations for peptides 2–11 were based on the CSDs of peptide 2, which were assumed to be 99% folded at 280 K, based on the observed melting of the CSDs and the agreement of these with the CSDs of (CH3CH2CO-WTTVCIRKWTGPK-NH2)2 , which was 98.5% folded based on H/D exchange data at 280 K27.
Results
Peptide system design.
We decided to employ a system consisting of two β-strands linked by a central disulfide bond, rather than a hairpin model, to eliminate possible effects of a turn on β-strand formation and alignment. Kier et al.27 reported that (CH3CH2CO-W-TTVCIRK-WTGPK-NH2)2 formed a remarkably stable β-sheet as an example of the application of the alkanoyl-W // WTG-NH2 type of β-cap. This model had a χF = 0.985 at 280 K based on amide NH exchange protection factors, which was reduced to χF = 0.72 (ΔΔGU = +7.54 kJ/mol) upon N-terminal deacylation. The N-propanoyl species displayed the usual diagnostics for an EtF (Edge to Face) W/W interaction with the N-terminal Trp as the face species: in the edge-indole (W9), Hε3 was 2.01 ppm upfield, compared to random coil values, due to the ring current from the face indole. As noted in the introduction, our aim was to replace this cap with units that did not need to be the extreme N- and C-terminus of a peptide sequence. We reasoned that Coulombic attraction between oppositely charged side-chains, possibly reinforced by an interaction between the chain terminal –NH3+ and –CO2− groups, might serve to stabilize the capping W/W interaction as well. When we prepared (R-WTTVCIRKW-E)2 this proved to be the case: the W10ε3 CSD (Chemical Shift Deviation) was −2.47 ppm at pH 8 and backbone site proton CSDs indicated that this system was comparably well-folded as the Kier cap system (1). The key chemical shift comparisons appear below.
The shifts for the edge-Trp in the coulombic cap, 2, indicated a more rigidly defined EtF geometry, compared to 1, with Hε3 and Hζ3 of the C-terminal Trp experiencing larger upfield shifts (Table 1). At pH 2, the folded structure appeared to be somewhat de-stabilized as indicated by decreased CSDs at T2HN, RHα and within the edge-Trp.
Table 1:
X-WTTVCIRKW-Z peptide shifts as the disulfide dimer, shown as CSDs (ppm)
| X- WTTVCIRKW-Z | β-Strand | C-terminal Trp | |||||||
| X / Z | pH | T2HN | KHN | RHα | Hε3 | Hβ3 | Hδ1 | Hζ3 | |
| 1 | CH3CH2CO / WTGPK | 3.5 | 1.31 | 1.08 | 0.69 | −2.01 | −1.31 | −0.58 | −0.43 |
| 2 | Arg / Glu | 8 | 1.42 | 1.08 | 1.06 | −2.47 | −1.22 | −0.34 | −0.62 |
| Arg / Glu | 2 | 1.07 | 1.09 | 0.95 | −2.39 | −1.18 | −0.38 | −0.57 | |
Samples in 20 mM potassium phosphate buffer at 300K. CSD = Chemical shift deviation, see Methods.
Since it would be difficult to quantitate fold stability improvements in this exceptionally well-folded system, we turned to a somewhat less stable disulfide system, (XWTTHCHRKWZ)2, (Figure 2) to examine the effects of alternative ending units, X and Z. The inclusion of His residues into the strands of peptide 2 should lower the intrinsic β-propensity, particularly at lower pH when His becomes a polar rather than hydrophobic residue. In addition, protonation of the His-Cys-His core of this β-sheet should result in substantial cross-strand Coulombic repulsion. This design element was confirmed by the ΔΔGU values associated with a pH change from 8 to 2 observed for (R-WTTVCIRKW-E)2 (1.78 kJ/mol) and peptide 3 = (R-WTTHCHRKW-E)2 (7.15 kJ/mol). The CSDs that were employed for fold-population measures were: W2Hα, T3HN, T4Hα, R8Hα, and K9HN . The CSDs of 3 (Figure 3) at pH 8 were equated with a fraction folded (χF) of 0.99 at 280 K based on the melting curves of the CSDs.
Figure 2:
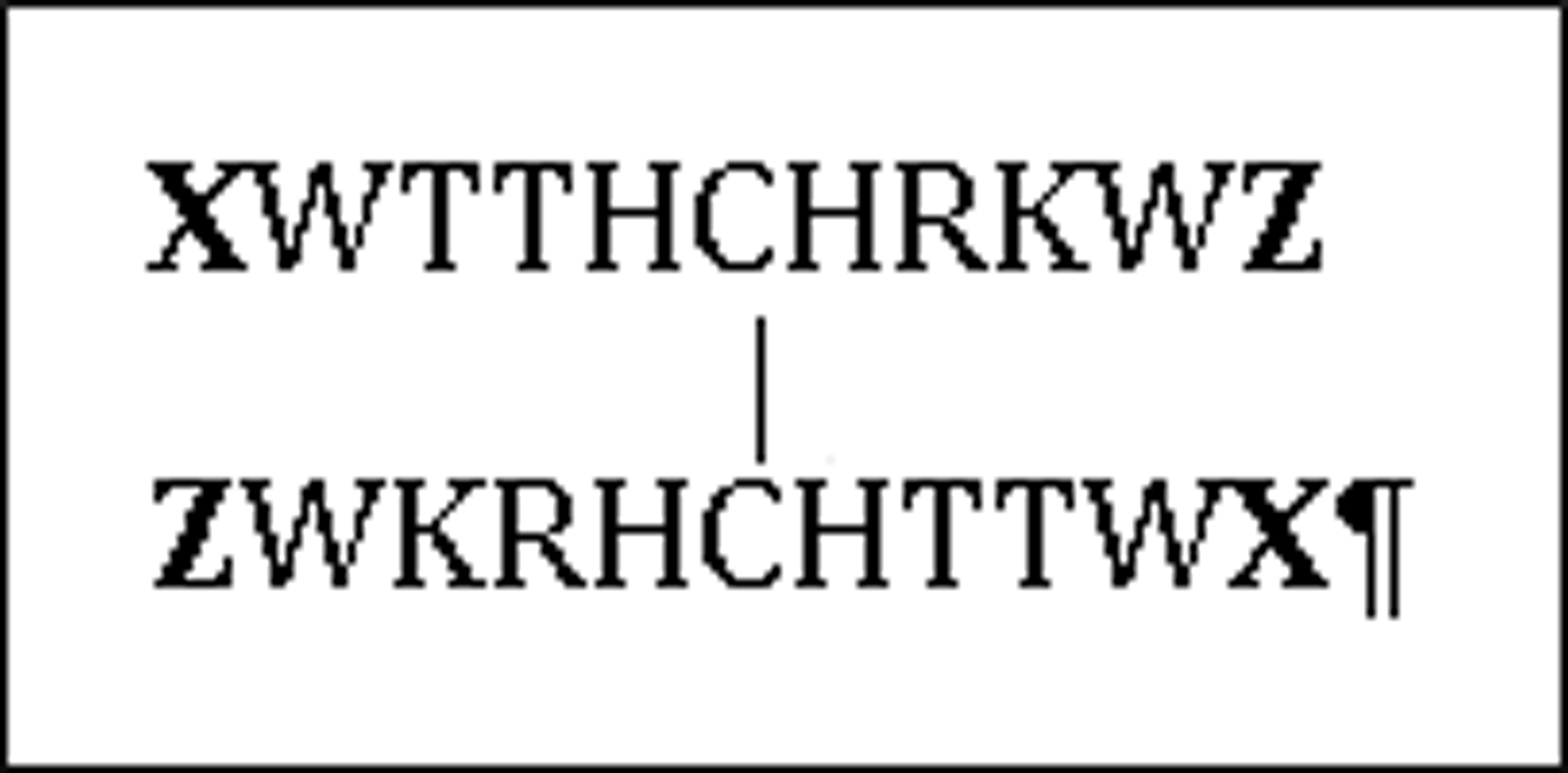
Disulfide Dimer
Figure 3:
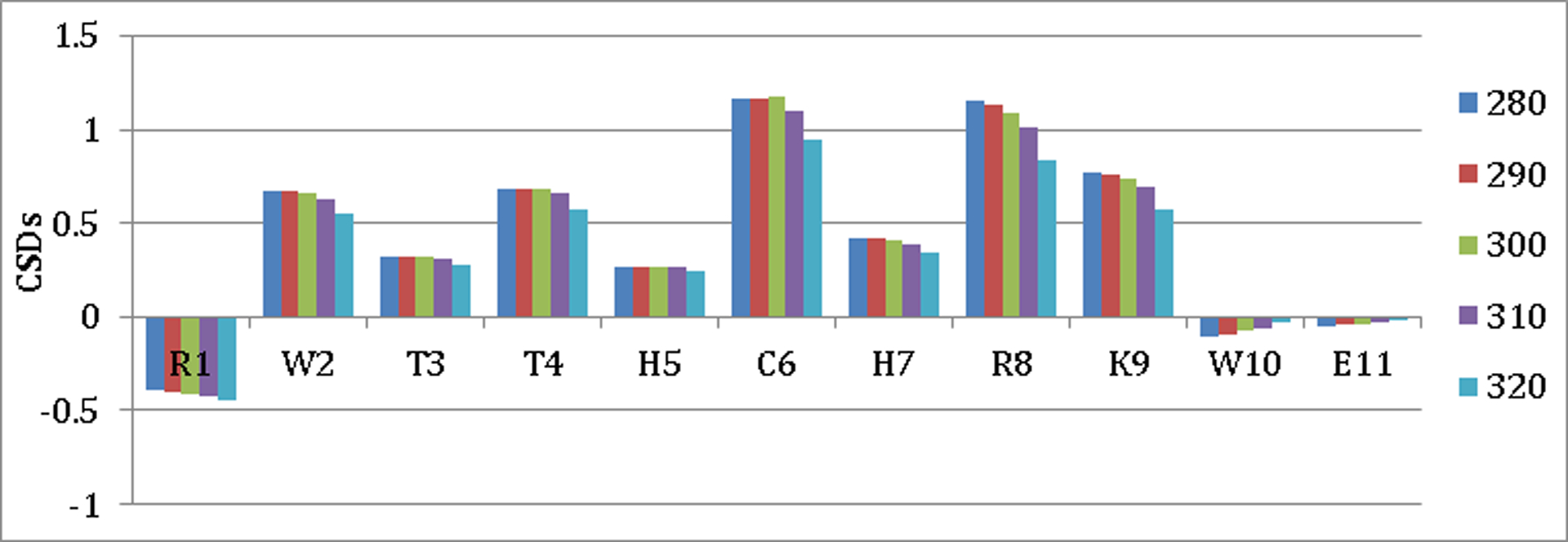
Chemical Shift Melting Profile For Hα Sites Along the Sequence of Peptide 3
The most diagnostic CSDs were also identical (± 2%) to those observed for the (CH3CH2CO-W-TTVCIRK-WTGPK-NH2)2 species previously examined27 by backbone NH exchange methods.
The greater capping effectiveness of R-W- // -W-E versus Ac-W- // -WTG-NH2 was readily observed, using the same diagnostic CSDs, with this new set of strands.
The R//E terminated peptide dimer was significantly more stable at both high and low pH (ΔΔGU = 6.76, 4.84 kJ/mol at 300 K, pH 8, pH 2 respectively). As expected, fold stability was significantly reduced upon acidification, but several design features could contribute to this.
A significant portion of the de-stabilization at pH 2 seen in the comparison above was assumed to be due to His protonation, which would decrease its β-propensity and introduce repulsive Coulombic interactions in the folded dimer state. A series of mutations in the (XWTTHCHRKWZ)2 motif and further pH studies were employed to elucidate the contributions of the distinct individual interactions within the capping unit to fold stabilization and the His protonation effect on β-sheet stability (Table 3).
Table 3:
Mutations of the Coulombic Capping Interaction of (XWTTHCHRKWZ)2
| pH 2 | pH 5 | pH 8 | |||||||||
| X---Z | 280 | 300 | 320 | 280 | 300 | 320 | 280 | 300 | 320 | ||
| 3 | R---E | χF | 0.76 | 0.60 | 0.32 | 0.84 | - | - | 0.99 | 0.97 | 0.78 |
| 5 | K---E | χF | 0.66 | 0.48 | 0.27 | 0.83 | 0.77 | 0.55 | 0.98 | 0.94 | 0.63 |
| 6 | K---T | χF | 0.78 | 0.62 | 0.35 | 0.85 | 0.79 | 0.55 | 0.96 | 0.87 | 0.35 |
| 7 | K---A | χF | 0.63 | 0.28 | 0.15 | 0.78 | 0.60 | 0.33 | 0.93 | 0.83 | 0.56 |
| 8 | K---A-NH2 | χF | - | - | - | 0.61 | 0.43 | − | 0.90 | 0.80 | 0.62 |
Mutations were made at the N and C terminal positions (X, and Z respectively) of peptide 3. At the high pH, the N-terminal Lys species, 5, was found to be as stable as the Arg species, 2, but displayed lesser stability (ΔΔGU = 1.21 kJ/mol) at pH 2. The stability enhancement associated with both the C-terminal side-chain charge and that of the C-terminus of the peptide were examined with further mutations (species 6, 7, and 8). The E→T substitution was relatively well tolerated, but additional melting was evident at both pH 5 and 8. Advantages of the terminal side-chain Coulombic attraction can be seen by mutations at the C-terminus. The comparison between species 5 and 7 provides the best measures of the effect of an Ala to Glu side-chain substitution: at 300 K the ΔΔGU is −2.91, −2.27, and −2.16 for pHs 8, 5, and 2 respectively. The changes at the lower pHs are likely due to the increased β-propensity of the Glu residue compared to Ala, while protonated at pH 2. To investigate the stability gained from the C-terminal carboxylate of the peptide backbone, amidation of the C-terminus of 7 was performed. The most dramatic loss of stability was noted at pH 5: ΔΔGU = 1.71 kJ/mol at 300K.
Further studies aimed to ascertain the limitations associated with the X and Z terminal groups as β-sheet assisting elements. A potential advantage of this Coulombic capping strategy compared to the original β-cap would be a greater range of substitution and chain extension options at the termini. With the N-terminus acetylated and very specific units at the C terminus in the previous systems, this cap can only be used at the extreme termini of a peptide or protein sequence.
To investigate this, residue extensions were added to the termini of peptide 5 (Table 4). The evidence suggests that only one of the termini is fully compatible with strand extension. With the addition of Thr residues to both termini, 9, the capping ability is effectively destroyed; however, when a Gly spacer is added between the cap and terminal threonines, 10, the cap’s effectiveness is regained at all pHs. To determine which termini the cap can tolerate residue extensions, this spacer was asymmetrically place on the C or N-terminus (11 and 12 respectively). With a Gly spacer placed only at the C-terminus, 11, the peptide is almost completely unfolded, but when this spacer is placed on the N-terminus, 12, the folded state is regained, χF = 0.90 (pH 8, 300 K). Molecular modeling of peptides 11 and 12 show residues on the N-terminus of the cap disrupting the Trp/Trp interaction by crowding the C-terminal Trp which is the edge ring in the EtF interaction (Supporting information Figures S8–S9).
Table 4:
Effects of residue extension at the termini of a Coulombic cap.
| pH 2 | pH 5 | pH 8 | |||||||||
| X---Z | 280 | 300 | 320 | 280 | 300 | 320 | 280 | 300 | 320 | ||
| 5 | K---E | χF | 0.66 | 0.48 | 0.27 | 0.83 | 0.77 | 0.55 | 0.98 | 0.94 | 0.63 |
| 9 | TT-K---E-TT | χF | - | - | - | 0.12 | 0.11 | 0.11 | - | - | - |
| 10 | TG-K---E-GT | χF | 0.77 | 0.55 | 0.24 | 0.81 | 0.66 | 0.34 | 0.92 | 0.82 | 0.37 |
| 11 | GT-K---E-GT | χF | - | - | - | - | - | - | 0.06 | 0.08 | 0.11 |
| 12 | TG-K---E-TG | χF | - | - | - | - | - | - | 0.92 | 0.90 | 0.62 |
To demonstrate the usefulness of the Coulombic cap, it was incorporated into a number of β-strand containing systems for a direct comparison with the original Kier β-cap, in these systems. Longer loops capped by an antiparallel β-sheet provide particularly telling examples (Table 5). In these systems, the β-turn of a hairpin is replaced by a longer unstructured, or partially-structured, loop. Folding to an antiparallel β-sheet (Fig. 4) requires a β-capping interaction. These long loop species serve as models of antiparallel sheets in proteins: where, in many cases, two associated β-strands are connected by a longer loops rather than a tight β-turn. Peptides 13–19 each contain two 7 residue capped β-strands with a 10 residue loop between them. The corresponding β-hairpins, with the loop replaced by GG or HG display χF values in excess of 0.97 at 300K. In all cases, replacing GG or HG with a longer sequence is destabilizing, but not to as great an extent as one might expect.
Table 5:
Capped peptide loop systems
| Fraction Fold (χF) | ||||
| Cap‐ β ‐Loop‐ β ‐Cap | 280 K | 300 K | 320 K | |
| 13 | RW- β -G4K2G4- β’ -WE | 0.84 | 0.75 | 0.51 |
| 14 | HW- β -G4K2G4- β’ -WE | 0.78 | 0.66 | 0.51 |
| 15 | Ac-W- β -G4K2G4- β’ -WTG-NH2 | 0.69 | 0.54 | 0.32 |
| 16 | RW- β* -WG4K2G4W- β# -WE | 0.97 | 0.97 | 0.95 |
| 17 | Ac-W- β* -WG4K2G4W- β# -WTG-NH2 | 0.93 | 0.96 | 0.92 |
| 18 | RW- β -G3IpGKG3- β’ -WE | 0.93 | 0.91 | 0.86 |
| 19 | Ac-W- β -G3IpGKG3- β’ -WTG-NH2 | 0.87 | 0.80 | 0.65 |
β = ITVTI , β’ = KKIRV , β* = ITVRI , β# = KTIRV, p = D-Pro
Figure 4:
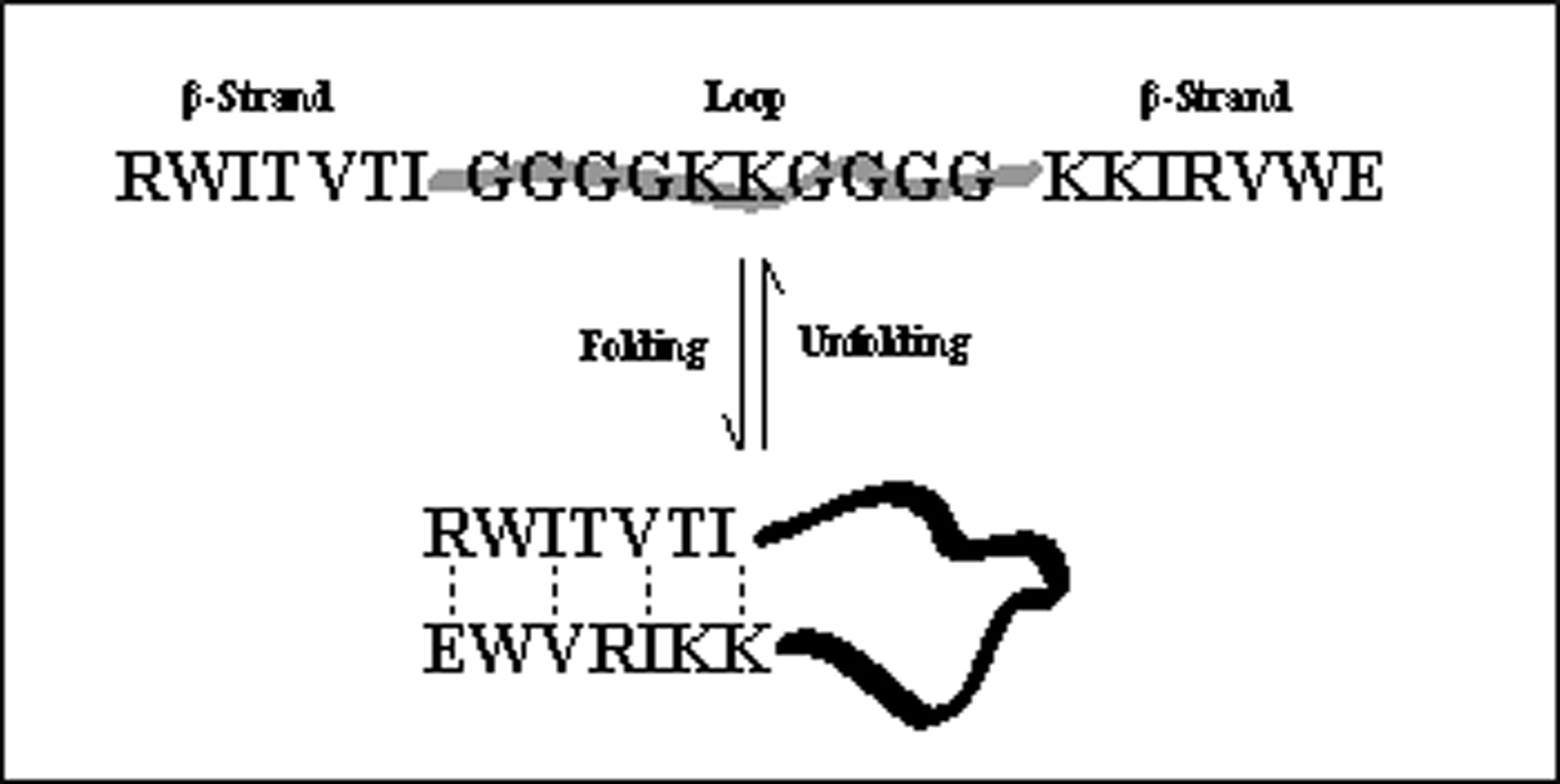
Folding of Looped systems
Peptides 13–15 contain a completely flexible 10 residue loop (-GGGGKKGGGG-). The RW/WE Coulombic cap of peptide 13 provides a gain in stability of ΔΔGU = +2.34 kJ/mol, at 300 K, compared to the standard β-cap, 15. When this same loop sequence is flanked with a Trp/Trp EtF interaction creating strands with a exceptionally high Tm , a moderate stability gain of ΔΔGU = −1.34 kJ/mol is still evident at 320 K. The loops in peptides 13-19 are, with the exception of peptides 18 and 19, unstructured. The loops in peptides 18 and 19 contain an IpGK sequence flanked by three glycines on each side. The IpGK sequence is one the best turn forming sequences known15, 19, 28, 30. The extent of [2:4]-turn formation can be assessed based on diagnostic CSDs within the “turn unit”: a downfield shift at the Ile-HN and upfield shifts at GHα3 and K-HN. These indicate circa 40% turn-formation in peptides 18 and 19 at 280K. This provides a significant increment of stability to the capped loop species. However, the incremental stability increase associated with replacing the Ac-W/WTG-NH2 cap with the RW/WE Coulombic cap is retained, particularly at the higher temperatures. The Coulombic cap maintains stability in the strands even at the higher temperature, while the turn within the loop melts out quickly, due to the segmental motion within the loop. In fact the strands of all the looped systems discussed, 13–19, remain more stable at high temperatures using the Coulombic cap.
Coulombic capping can also be used as a means to improve the stability of more typical β-hairpins and eliminate terminal fraying. Hairpin 20 is only moderately stable, with a χF of 0.30 in the β-strands and 0.42 when the turn region CSDs are employed to judge the extent of folding. When this hairpin is capped, 21, at the termini, a significant increase in folding is observed, χF=0.90 for the β-strands, and χF = 0.84 for the turn region, demonstrating that the resulting reduction in terminal fraying produced by the capping interaction contributes significantly to overall hairpin stability. In peptide 21, the W/W pair appears at the S±6 positions in the β-strands. When the capping interaction is moved closer to the turn, with the W/W pair at the S±4 positions (peptide 22), a more stable hairpin also results. The lower hairpin population of 22 versus 21, is attributed to the shorter β-strand length. When an S±4 W/W pair is added to peptide 20 without the strand truncation that places this pair within the β-cap, only a small gain of stability (ΔΔGU= −1.41 kJ/mol) results (peptide 23). The W/W pair is central within the β-strands and lacks the flanking ionized sidechains that complete the Coulombic capping unit.
We opted to confirm the capping effect with an S±4 W/W pair in another peptide series with an –NPATGK- turn (peptides 24-26). In this case we also examined the effect of chain extension (peptide 26) while retaining the RW/WE unit as a potential β-cap. Peptide 24 with the RW/WE unit as a terminal β-cap was more stable (ΔΔG = 5.6 kJ/mol) than the species with longer β-strands in which the Arg and Glu units could still provide a Coulombic interaction to favor hairpin formation but were separated from the Trp residues by two intervening residues. In agreement with CSD measure of fold population, the Trp/Trp exciton couplet (as measured by the [θ]228 maximum) is much reduced in peptide 25 versus 24: [θ]228= +154,000 and [θ]228=+344,000 deg. cm2 dmol−1, respectively. This couplet is diagnostic of the Trp/Trp EtF interaction25, 27, 30, 31.
An additional test of our Coulombic cap was to establish that it can stabilize a hairpin that has additional chain extensions at each end. Since data presented previously indicated that a glycine spacer is needed before the N-terminal RW capping unit, in peptide 26 we added an Ac-AAG unit to the terminus of peptide 24. The C-terminal extension was a TA-NH2 unit. Since the N and C termini were acetylated and amidated respectively, any stability gain versus peptide 25 could attribute solely to the hairpin capping interaction. Based on the CSDs that measure β-strand association, a non-chain-terminal RW/WE cap provides at least a 4.0 kJ/mol increment in hairpin fold stability. Evidence that 26 is capped similar to 24 can be observed by the exciton couplet of [θ]228=+300,000 deg. cm2 dmol−1, and the upfield shifted indole protons of Trp16 in 26, CSDs of Hε3= −1.564, Hζ3= −0.560, Hδ1= −0.258 ppm at 300 K. (CD comparisons of 24–26 shown in Supporting Information)
We provide one additional example of fold enhancement by an RW / WD Coulombic β-cap. The first hairpin of the Pin1 WW domain is known to be the fold nucleating feature of this protein motif32. We recently established that the isolated hairpin, the GWEKRM-SRSSGR-VYYFN-H sequence in the native form, by analogy to RWEKRM-SRSSGR-VYYFNS26, does not form a stable hairpin fold. As another application of the Coulombic cap, we prepared RWEKRM-SRSSGR-VYYFWD (corresponding to an NS to WD mutation at the C-terminus). This modification completes a Coulombic β-cap, providing the charged sidechain and the missing indole ring, and brought the hairpin population from < 0.30 to 0.83 (ΔΔGU better than −5.7 kJ/mol). The CSDs comparisons, including the native hairpin in the native structure appear in the Supporting Material.
In closing, a versatile new β-capping strategy has been characterized and applied to a number of peptide systems with incompletely formed antiparallel β-sheets. In all cases, the cap addition results in a fully-folded β-sheet, even without the use of unnatural sequences that favor turn-formation in designed β-hairpins. When comparisons are made to a previous β-capping system27 this new Coulombic capping motif provides more fold-stabilization and offers greater versatility of use. Unlike the previous β-capping unit, with the Coulombic caps reported herein, the N- and C-termini are open to modification or residue extension. However, the N-terminus does require a Gly spacer between the cap and any modification or chain extension. Hopefully the addition of this tool will help to expand the ability to design stable β-sheet models for the study of β-sheet folding rates and mechanisms.
Supplementary Material
Figure 1:
Tryptophan’s aromatic protons
Table 2:
Coulombic cap (2) compared to original β-Cap (3).
| X---Z | pH 2 | pH 8 | |||||||
| T | 280 | 300 | 320 | 280 | 300 | 320 | |||
| 3 | R---E | χF | 0.76 | 0.60 | 0.32 | 0.99 | 0.97 | 0.78 | |
| 4 | Ac-W---WTG-NH2 | χF | 0.39 | 0.16 | 0.05 | 0.83 | 0.64 | 0.49 | |
χF = Fraction folded
Table 6:
Capped β-Hairpins
| β-Strand-Turn- β-Strand | Strand χF |
Turn χF |
|
| 20 | KKLTVS-INGK-KITVSA | 0.3030 | 0.32 |
| 21 | KWLTVS-INGK-KITVWE | 0.90 | 0.84 |
| 22 | KWVS-INGK-KIWE | 0.62 | 0.81 |
| 23 | KKLWVS-INGK-KIWVSA | 0.4330 | 0.87 |
| 24 | RWTV-NPATGK-ITWE | 0.83 | 0.67 |
| 25 | RAVWTV-NPATGK-ITWVIE | 0.34 | 0.61 |
| 26 | Ac-AAGRWTV-NPATGK-ITWETA-NH2 | 0.72 | 0.63 |
The χF values were derived from the structuring shifts (CSDs) observed at 300 K.
Acknowledgements
The reported studies were supported by NIH (GM099889) and NSF (CHE-1152218) grants to N.H.A. Peptide 21 was prepared and characterized by Brice Jurban in the Andersen laboratory.
References
- (1).Serrano L, Fersht AR (1989) Capping and α-helix stability. Nature 342, 296–299. [DOI] [PubMed] [Google Scholar]
- (2).Pertukhov M, Tatsu Y, Tamaki K, Murase S, Uekawa H, Yoshikawa S, Serrano L, Yumoto N (2009) Design of Stable α-helices Using Global Sequence Optimization. J. Pept. Sci 15, 359–365. [DOI] [PubMed] [Google Scholar]
- (3).Andersen NH, Tong H (1997) Empirical Parameterization of a Model for Predicting Peptide Helix/Coil Equilibrium Populations. Protein Science, 6, 1920–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ruczinski I, Kooperberg C, Bonneau R, Baker D (2002) Distributions of Beta Sheets in Proteins With Application to Structure Prediction. Proteins: Struct., Funct., Genet 48, 85–97. [DOI] [PubMed] [Google Scholar]
- (5).Subramani A, Floudas CA (2014) β-sheet Topology Prediction with High Precision and Recall for β and Mixed α/β Proteins. PLoS one 7, e32461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ (1998) JPred: a Consensus Secondary Structure Prediction Server. Bioinformatics 14, 892–893 [DOI] [PubMed] [Google Scholar]
- (7).Huggins KN, Bisaglia M, Bubacco L, Tatarek-Nossol M, Kapurniotu A, Andersen NH (2011) Designed Hairpin Peptides Interfere with Amyloidogenesis Pathways: Fibril Formation and Cytotoxicity Inhibition, Interception of Preamyloid State. Biochemistry 50, 8202–8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Kar K, Hoop CL, Drombosky KW, Baker WA, Kodali R, Arduini I, van der Wel PC, Horne WS, Wetzel R (2013) β−Hairpin-Mediated Nucleation of Polyglutamine Amyloid Formation. J. Mol. Biol 425, 1183–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Chiti F and Dobson CM (2006) Protein Misfolding, Functional Amyloid, and Human Disease. Annu. Rev. Biochem 75, 333–366. [DOI] [PubMed] [Google Scholar]
- (10).Blanco FJ, Rivas G, Serrano L (1994) A Short Linear Peptide that Folds into a Native Stable β-hairpin in Aqueous Solution. Nature Struct. Biol 1, 584–590. [DOI] [PubMed] [Google Scholar]
- (11).Searle MS, Williams DH, Packman LC (1995) A Short Linear Peptide Derived from the N-terminal Sequence of Ubiquitin Folds into a Water-stable non-native β-hairpin. Nature Struct. Biol 2, 999–1006. [DOI] [PubMed] [Google Scholar]
- (12).Cochran AG, Skelton NJ, Starovasnik MA (2001) Tryptophan zippers: Stable, monomeric β-hairpins. Proc. Natl. Acad. Sci 98, 5578–5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Fesinmeyer RM; Hudson FM; Andersen NH (2004) Enhanced Hairpin Stability through Loop Design: The Case of the Protein G B1 Domain Hairpin. J. Am. Chem. Soc 126, 7238. [DOI] [PubMed] [Google Scholar]
- (14).Olsen KA, Fesinmeyer RM, Stewart JM, Andersen NH (2005) Hairpin Folding Rates Reflect Mutations Within and Remote from the Turn Region. Proc. Natl. Acad. Sci 102, 15483–15487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Kier BL, Andersen NH (2008) Probing the Lower Size Limit for Protein-Like Fold Stability: Ten-Residue Microproteins With Specific, Rigid Structures in Water. J. Am. Chem. Soc 130, 14675–14683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Park JH, Waters ML (2013) Positional Effects of Click Cyclization on β-Hairpin Structure, Stability, and Function. Org. Biomol. Chem 11, 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Riemen AJ, Waters ML (2010) Positional Effects of Phosphoserine on β-Hairpin Stability. Org. Biomol. Chem 8, 5411–5417. [DOI] [PubMed] [Google Scholar]
- (18).Riemen AJ, Waters ML (2009) Design of Highly Stabilized β-Hairpin Peptides through Cation−π Interactions of Lysine and N-Methyllysine with an Aromatic Pocket. Biochemistry 48, 1525–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Haque TS, Gellman SH (1997) Insights into β-hairpin stability in aqueous solution from peptides with enforced type I′ and type II′ β-turns. J Am Chem Soc 119, 2303–2304. [Google Scholar]
- (20).Favre M, Moehle K, Jiang L, Pfeiffer B, Robinson J (1999) Structure Mimicry of Canonical Conformation in Antibody Hypervariable Loops Using Cyclic Peptides Containing a Heterochiral Diproline Template. J. Am. Chem. Soc 121, 2679–2685. [Google Scholar]
- (21).Karle IL, Awasthi SK, Balaram PA (1996) A designed β-hairpin peptide in crystals. Proc. Natl. Acad. Sci 93, 8189–8193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Syud FA, Espinosa JF, Gellman SH (1999) NMR-Based Quantification of β-Sheet Populations in Aqueous Solution through Use of Reference Peptides for the Folded and Unfolded States. J. Am. Chem. Soc 121, 11577–11578. [Google Scholar]
- (23).Santiveri CM, Jiménez MA (2010) Tryptophan Residues: Scarce in Proteins but Strong Stabilizers of β-Hairpin Peptides. Biopolymers 94, 779–790. [DOI] [PubMed] [Google Scholar]
- (24).Wu L, McElheny D, Takekiyo T, Keiderling TA (2010) Geometry and Efficacy of Cross-Strand Trp/Trp, Trp/Tyr, and Tyr/Tyr Aromatic Interaction in a β-Hairpin Peptide. Biochemistry 49, 4705–4714. [DOI] [PubMed] [Google Scholar]
- (25).Andersen NH, Olsen KA, Fesinmeyer RM, Tan X, Hudson FM, Eidenschink LA, Farazi SR (2006) Minimization and Optimization of Designed β-Hairpin Folds. J. Am. Chem. Soc 128, 6101–6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Kier BL, Anderson JM, Andersen NH (2014) Circular Permutation of a WW Domain: Folding Still Occurs after Excising the Turn of the Folding-Nucleating Hairpin. J. Am. Chem. Soc 136, 741–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Kier BL, Shu I, Eidenschink LA, Andersen NH (2010) Stabilizing Capping Motif for β-Hairpins and Sheets. Proc. Natl. Acad. Sci 23, 10466–10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Fesinmeyer RM, Hudson FM, Olsen KA, White GWN, Euser A, Andersen NH (2005) Chemical Shifts Provide Fold Populations and Register of β Hairpins and β Sheets. J. Biomol. NMR 33, 213–231. [DOI] [PubMed] [Google Scholar]
- (29).Wu L, McElheny D, Huang R, Keiderling TA (2009) Role of Tryptophan-Tryptophan Interactions in Trpzip β-Hairpin Formation, Structure, and Stability. Biochemistry 48, 10362–10371. [DOI] [PubMed] [Google Scholar]
- (30).Eidenschink L, Kier BL, Huggins KNL, Andersen NH (2008) Very Short Peptides with Stable Folds: Building on the Interrelationship of Trp/Trp. Trp/cation, and Trp/backbone-amide Interaction Geometries. Proteins 75, 308–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Scian M, Shu I, Olsen HA, Hassam K, Andersen NH (2013) Mutational Effects on the Folding Dynamics of a Minimized Hairpin. Biochemistry 52, 2556–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Jäger M, Nguyen H, Crane JC, Kelly JW, Gruebele M (2001) The Folding Mechanism of a β-Sheet: The WW Domain. J. Mol. Biol 311, 373–393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


