Abstract
Disulfide bonds between cysteine residues are essential to the structure and folding of many proteins. Yet their role in the design of structured peptides and proteins has frequently been limited to use as intra-chain covalent staples that reinforce existing structure or induce knot-like conformations. In beta hairpins, their placement at non-H-bonding positions across antiparallel strands has proven useful for achieving fully-folded positive controls.
Here we report a new class of designed beta sheet peptide dimers with strand-central disulfides as the key element. We have found that the mere presence of a disulfide bond near the middle of a short peptide chain is sufficient to nucleate some antiparallel β-sheet structure; addition of β capping units and other favorable cross-strand interactions yield hyperstable sheets. Strand-central cystines were found to be superior to the best designed reversing turns in terms of nucleating β-sheet structure formation. We have explored the limitations and possibilities of this technique (the use of disulfides as sheet nucleators) and we provide a set of rules and rationales for the application and further design of disulfide-tethered “turnless” β-sheets.
INTRODUCTION
Beta-hairpin peptides have proven useful as model systems for investigating the principles of protein folding and design, and as scaffolds for sidechain and loop display. However, outside the context of tertiary protein folds, β-hairpins must contain certain obligatory stabilizing features in order to maintain a well-populated folded state. Folding of isolated hairpins requires either a very tight nucleating turn or highly optimized β strands - typically with high-as-possible beta propensities and cross-strand Trp/Trp pairs. This limits the diversity of designable β-sheet structures.
Disulfide bonds can link adjacent antiparallel β-strands in proteins, but the dihedral angle preferences of disulfides constrain their use1–3. Specifically, disulfide bonds are only energetically favorable if the Cys residues are positioned at non-H-bonded (NHB) cross-strand sites.4–6 Outside of protein contexts, disulfides can be used to staple together otherwise poorly folded hairpins – though the same rule applies: the cyst(e)ine residues must be introduced at NHB sites. Properly positioned disulfide bonds have been used to cyclize peptide chains and create fully-folded controls for β-hairpin fold-population studies.5,7–9 They also stabilize and rigidify the natural β-hairpin motifs of antimicrobial peptides10–14, where multiple cross-strand disulfides are often found in a single β-hairpin. Of these examples, there are cases (e.g., protegrin13) where disulfides “rescue” beta hairpins which, due to their poor reversing turns or repulsive cross-strand Arg/Lys residues, are only poorly folded when the disulfides are reduced. This begs the question: if poor turns are allowed when this covalent strategy is used, are turns even necessary? Can small antiparallel β-sheets be brought together as structured disulfide dimers?
Before answering with the obvious “yes” and presenting our diverse array of turn-free β-sheet constructs, it is worth noting that disulfides have seen some use in stapling multi-strand β-sheet peptides together as symmetric dimers. For example, the Balaram group designed a C2-symmetric 8-stranded sheet held together by a central cross-strand NHB disulfide15 (PDB id # 1jy4, 1jy6). However, these previous examples consist of independently-folded β-sheets bound together as dimers, not natively-unfolded strands which form hyperstable two-stranded sheets on oxidative dimerization. Until recently, the closest thing to a designed, disulfide-nucleated β-sheet was the dimer reported by Cashman et al.16, which was only fold-stable in organic solvents. We recently reported17–18 two-strand oxidation-dependent homodimeric sheets, presenting them as ideal test systems for evaluating the extent of stabilization and end-fraying reduction resulting from our β-capping motifs. We have yet to give a detailed accounting of their strengths, limitations, and various forms. We now seek to present a breadth of examples of this useful class of peptides and lay down the rules for disulfide-mediated β-sheet formation.
METHODS
Synthesis
All peptides were synthesized on a CEM Liberty Blue synthesizer, using standard Fmoc solid phase peptide synthesis. Preloaded Fmoc-protected Wang and unloaded Rink amide resins were used for synthesis. Peptides were cleaved from resin (0.1 mmol) using a cocktail of trifluoroacetic acid : triisopropylsilane : water (38:1:1, 9.5:0.25:0.25 ml) for 1.5 hr. The resulting filtrate was concentrated in vacuo, crashed out and washed with cold (−20 °C) diethyl ether, giving the crude peptide. After drying the peptide, the pellet was dissolved in ~8mL water and purified using RP-HPLC (Varian ProStar 220 HPLC, Agilent 21.2 × 50mm C18 column, 10 ml/min, Elutent A : Water with 0.1% TFA, Elutent B : Acetonitrile with 0.085% TFA), using a gradient of 10 – 50 %B over 19 min. The resulting product was dried on a rotovap and oxidized via dissolution in a minimal amount of DMSO (0.5 ml), diluted with water (1 ml) and let stand for 24 hrs. at room temperature. After additionally diluting with water (3.5 ml) the solution was repurified via HPLC using the same method (to gauge the relative proportion, if any, of non-oxidized peaks. Oxidized peaks typically eluted before non-oxidized peaks, presumably due to folding and increased hydrophobic burial.) Peaks were visualized at 215 and 280 nm with verification by mass spectrometry (Bruker Esquire ion trap with ESI ionization). The concentrated fractions were then lyophilized resulting in the purified peptide. If disulfide formation did not proceed using the previous procedure, the dry peptide was dissolved in minimal amount of DMSO (0.5 ml) and diluted in 1 M HCl (1 ml) and left for 24 hrs. Higher-order assembly (D2-symmetric dimers of dimers) is a spontaneous process and not a result of covalent modifications or other chemical syntheses steps.
NMR Spectroscopy
NMR samples were made at ~1 mM in 20 mM Potassium Phosphate buffer with 10% D2O and an internal standard of sodium 4,4-dimethyl-4-silapentane-1-sulfonate (DSS). Full 1H spectra assignments were made using peptide backbone connectivities determined by 2D TOCSY and NOESY experiments taken on either Bruker DRX 500 MHz, or AV 700 MHz spectrometers.
Structuring-induced chemical shift changes were analyzed as chemical shift deviations (CSDs, the observed shifts minus the coil reference values) using the CSDb algorithm (andersenlab.chem.washington.edu/CSDb/about.php) as previously described.19–20 Converting CSDs to fold stability data (χF, the fraction folding) requires reference CSDs for the folded state. The CSDs of (CH3CH2CO-WTTVCIRKWTGPK-NH2)2, which is 98.5% folded based on H/D exchange data at 280 K17, served this function for most of the systems herein. More details appear in the Supporting Information.
CD spectroscopy
Stock solutions of approximately 200μM peptide concentration were prepared using 50mM aqueous pH 7.0 phosphate buffer. Accurate concentrations were determined by UV spectroscopy assuming the standard molar absorptivities for the Trp and Tyr residues present. The samples were typically diluted to obtain circa 30 μM polypeptide solutions, with spectra recorded on a Jasco J720 spectropolarimeter using 0.10 cm pathlength cells over a UV range of 190–270 nm as previously described.21–23 In melting studies, temperatures ranged from 5 to 95 °C in increments of 5° or 10°. For CD melting temperatures, the folded fraction (χF) was determined by defining the temperature-dependent CD signal of the unfolded and folded states and assuming a linear χF relationship for signals between the two lines. The CD spectrum of the unfolded state is expected to be sequence independent at 228nm, the position of the fold-diagnostic exciton couplet.
RESULTS
Single disulfide homodimers
It has long been known that disulfide bonds are favored at the non-H-bonded positions of antiparallel β-sheets.4–6 Until the present studies it had not been established that disulfide formation can nucleate antiparallel association of “turnless” strands into a β-sheet structure. In control peptides for some of our β-cap studies, we observed the nucleation of beta structure centered on a non-H-bonded cystine. This phenomenon was observed even for very small, symmetric peptide dimers with central cystine residues. For example, the oxidized disulfide form of (KKVCITT)2 displays chemical shift deviations (CSDs) from random coil norms that indicate antiparallel β-sheet formation with the folded state circa 65 % populated (χF ≈ 0.65) at 280K and pH 7, and χF ≈ 0.4 at pH 2. The pH effect suggests that the terminal salt bridges provide additional stabilization. When the beta strands flanking the disulfide bond are improved or lengthened, the resulting sheet structures can become remarkably stable, particularly with beta-capping motifs17,18,24 (Trp/Trp pairs at strand-terminal NHB positions and their supporting interactions) present. As dimeric structures the stabilizing caps appear twice in one β-sheet, and the net fold-stabilization they provide is doubled.
Like (KKVCITT)2, peptide (KWRCIWD)2 is a seven residue peptide dimerized by a central cystine. However, it incorporates a version of the Coulombic β-cap18 at both ends of the homodimer, and the beta sheet structure it adopts is much more stable (ΔΔGU > 15 kJ/mol). It is effectively 100% folded at 280K and has a melting point of 70 °C based on the temperature dependent circular dichroism (CD) spectrum (see Supporting Information, figure S7, for full CD melt). This is remarkable for a peptide synthesized with just seven natural amino acids. The two β-caps impart not only great fold-stability but also provide useful spectroscopic diagnostics of fold formation: a high amplitude exciton couplet in the CD (max: 228 nm) and an Hε3 proton for the “edge” Trp shifted upfield > 2.2 ppm (at 100% folding). Some related examples of hyperstable beta sheet dimers are collected in Table 1.
Table1.
Cys-containing peptides displaying disulfide-mediated β-sheet formation.
| Sequence | χF | TM (CD) |
|---|---|---|
| VCI | 0.20 | |
| KKTCTTT | 0.52 | |
| KKVCITT | 0.72 | |
| PWVCKHT | 0.88 | |
| KWRCIWD | ≥0.98 | 69 |
| HWVCIWR | ≥0.98 | 87 |
| KWRTVCIRTWE | ≥0.98 a | 75 |
| KWRTIKVCITKRTWE | 0.99 | |
| Pr-WVCKWTGPK-NH2 | 0.97 | 72 |
| WVCKWTGPK-NH2 | 0.17 | |
| Pr-WTTVCIRKWTGP-NH2 | 0.985 b | 81 |
| WTTVCIRKWTGP-NH2 | 0.74 b | |
| Bz-WHTHCIRTWTGP-NH2 | 0.89 | |
| Bz-WITKCIRKWTGKK | 0.94 | 78 |
| KWTTHCHRKWT | 0.96 | |
| KWTTHCHRKTW | 0.06 | |
| KWTTHCHRKWA | 0.93 | 70 |
| KWTTHCHRKWA REDUCED | 0.00 |
Pr- : N-terminal propanoyl, Bz- : N-terminal benzoyl. All values are for the oxidized species, except as noted. The fraction folded values are based on NMR data for pH 6 – 8 at 280K. All reduced species show no evidence, whatsoever, of folding by NMR and CD spectroscopy. Complete NMR chemical shift data can be found in the Supporting Information for peptides KWRCIWD and HWVCIWR (table S2) and KWTTHCHRKXX (the last four entries of this table; found in table S6.)
. From reference 18
. From reference 17; determined with precision by amide H/D exchange methods.
The fold stability data in Table 1 indicates that the β capping strategy is orthogonal to disulfide dimerization; these techniques work together well, but don’t require each other. For example, compare Peptide Pr-WTTVCIRTWTGP-NH2 vs +WTTVCIRTWTGP-NH2; the latter has disabled β-caps, as it lacks the essential N-terminal alkanoyl function17. Though it loses circa 12 kJ/mol of stability it remains 70% folded at 280K. More dramatic is the case of peptide (KWTTHCHRKWT)2 which is stabilized by superior, hydrophilic caps. It is ≥96% folded at 280K (pH 8.0) while its isomer with C-terminal W and T residues swapped (thus abolishing the beta caps) is almost entirely unstructured under the same conditions – presumably due to the poor β-propensity of the disulfide-flanking histidine residues.
Our hyperstable constructs take two forms, (alkanoyl group)-W(X)n1C(X)n2WTG… (designated as a hydrophobic cap) and ZW(X)n1C(X)n2WZ (the hydrophilic cap, where Z can be any residue18). The alkanoyl group can be virtually any small molecule24, and X can be any residue(s) other than proline. However, n1 must equal n2, and the value must be an odd number. This is necessary to maintain the disulfide at the central position of the β-sheet, and keep it in-register with the capping tryptophan residues. The disulfide and both cross-strand Trp/Trp pairs must be at NHB positions; as a result, constructs of the form ZW(X)C(X)WZ (hydrophilic cap) are limited to β-sheet lengths that satisfy a 3+4n formula - 7, 11, 15, 19, etc. are possible strand lengths.
These peptides are contiguous β-sheets, analogous to long hairpins but with no turn at either end. The longer β-sheet constructs of this form have some local distortion (a change in the twist) at the disulfide, as evidenced by a slight anomaly in the CSD patterns (figure 5 provides the most extreme example), but overall a defined β sheet structure is maintained. See figure 1.
Figure 5.
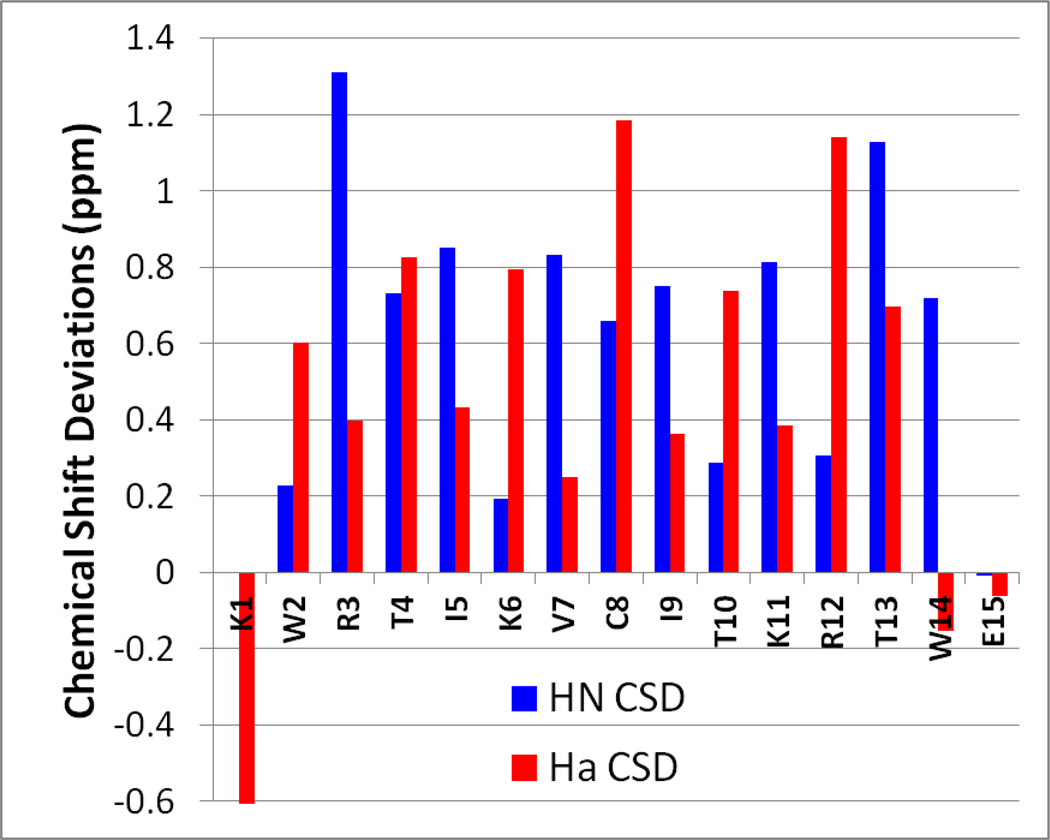
HN CSDs for peptide (RWKCKCKCKWE)2. The β-sheet is highly distorted, and this manifests as an inverted pattern of HN CSDs: larger CSDs are observed for the outwardly-directed non-H-bonded positions (the disulfide positions at C4, C6, C8) rather than the H-bonded positions (especially the presumably-H-bonded K5 and K7 sites).
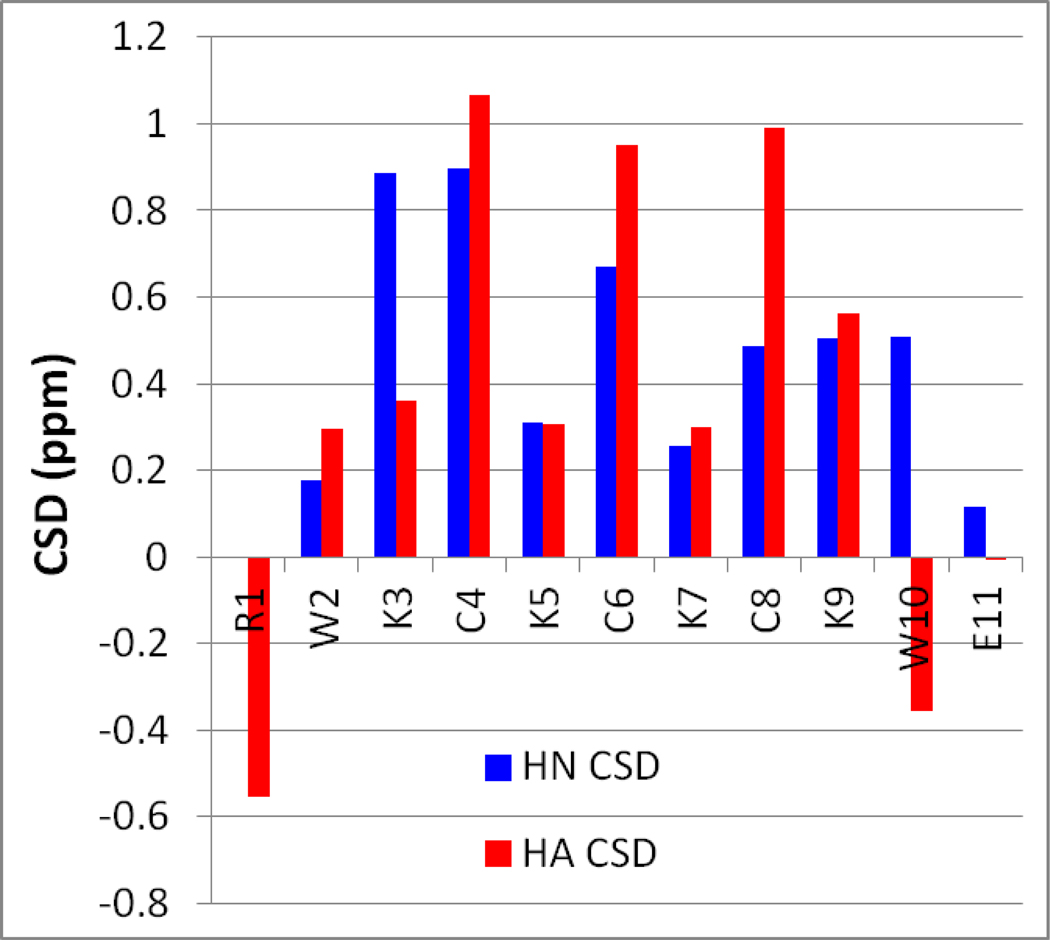
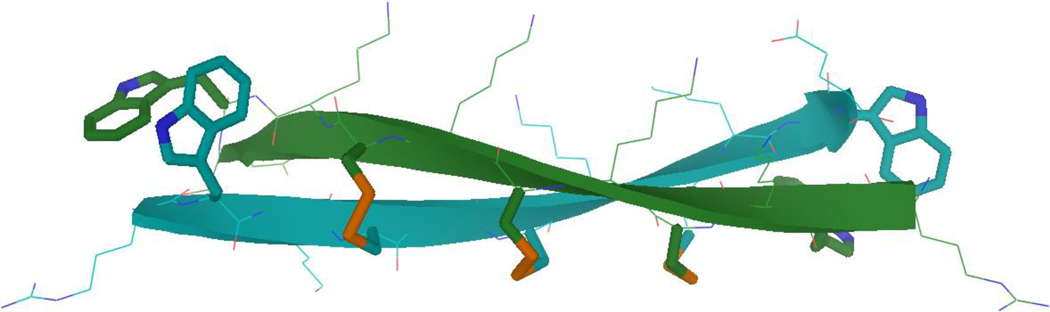
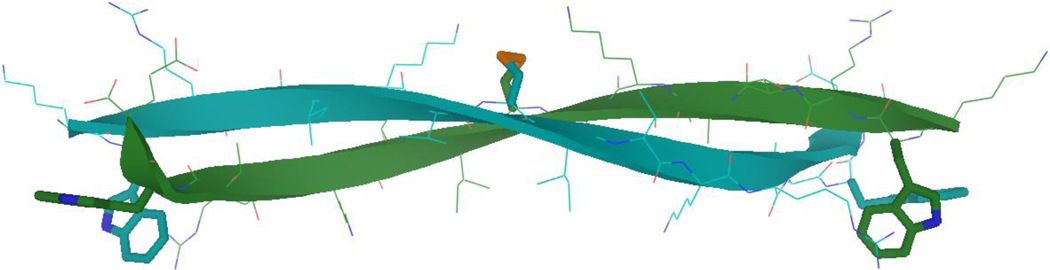
Figure 1.(a).
Chemical shift deviations (CSDs) of a long disulfide dimer, (KWRTIKVCITKRTWE)2. The expected β-sheet periodicity is observed: inward pointing Hα and HN protons are strongly shifted downfield from expected-coil values (0.7–1.3 ppm), while outward-pointing protons are less shifted, at 0.3–0.5 ppm.
Comparing the beta sheet nucleation strengths of disulfides versus an optimized turn sequence.
Previous work aimed at designing small, stable β-sheet constructs typically employed turn sequences including D-AAs to favor chain reversal. Among these, the heterochiral diproline unit (D-Pro L-Pro25–27) is probably the most effective. This sequence favors a type II’ β-turn and provides exceptional entropy-based stabilization for β hairpins; only a handful of exotic turn-nucleating small molecules are slightly better.28–29 All turn sequences composed only of L-AAs and glycine are significantly poorer. We have now established that a strand-central cystine residue has a higher β-structure nucleating propensity than the heterochiral diproline unit (Table 2).
Table 2. Peptides with identical strands (KWRWRTR and RKIKRWY) but different connecting units.
(All peptides containing Cys are oxidized, with disulfide bonds between strands.)
| Name | Sequence | description | TM (°C, CD) | 300 K χF (NMR) |
|---|---|---|---|---|
| C | KWRWRTR--C--RKIKRWY | (homodimer) | 44 | 0.95 |
| YWRKIKR--C--RTRWRWK | ||||
| CIC | KWRWRTR-CIC-RKIKRWY | (homodimer) | 58 | 0.96 |
| YWRKIKR-CIC-RTRWRWK | ||||
| pP | KWRWRTR-p | (hairpin monomer) | 36 | 0.88 |
| YWRKIKR-P | ||||
| Half | KWRWRTR--CR | (hairpin-like heterodimer) | 24 | 0.66 |
| YWRKIKR--CR |
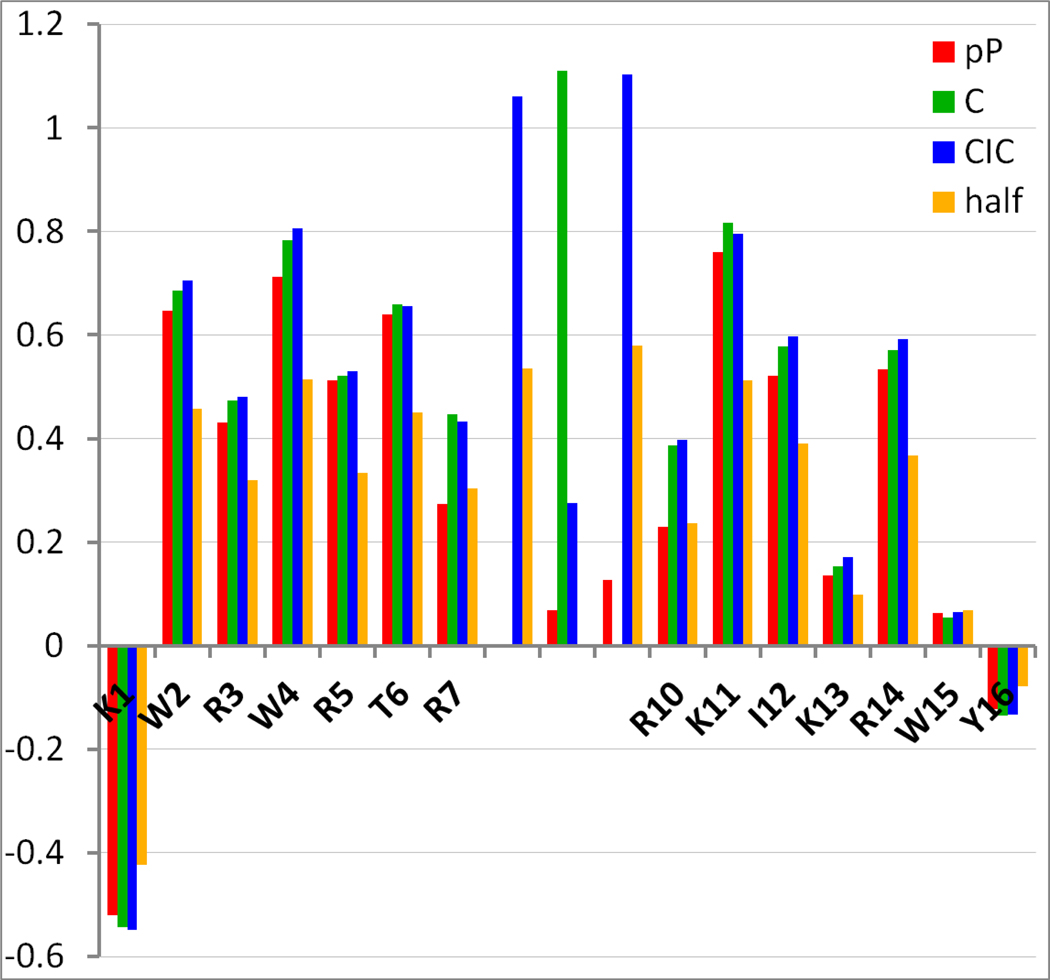
The comparison begins with peptides KWRWRTR-C-RKIKRWY and KWRWRTR-pP-RKIKRWY, identical strands with different linkers. The strands are favorably capped but the residues have only middling β-propensities; in addition, they are exceptionally rich in Arg and Lys, with two repulsive cross-strand R/R interactions per strand-pairing. This serves to alleviate any potential solubility problems, and reduces the fold populations to measurable values despite the presence of the β-cap (which provides the spectroscopic diagnostics for measuring χF ). Incidentally, these constructs strongly resemble antimicrobial peptides and RNA-binding peptides30 in residue composition.
Peptide “C” was significantly more stable than “pP”, despite having one less residue. Thus, when designing stable β-sheets, a single disulfide is superior to the best known hairpin-nucleating turn sequence from peptide engineering efforts. Admittedly, it is not a perfect comparison for determining raw hairpin nucleation potential, as disulfides can nucleate beta sheet formation in two directions and form a single contiguous sheet double the length of an analogous monomeric hairpin. If considered as separate sheets, the two halves on either side of the central disulfide are cooperative in their folding; the folding of one half nucleates the other. Therefore, for a direct comparison, the disulfide-bound heterodimer comprised of KWRWRTRCR and RCRKIKRWY was synthesized and characterized. This species (listed as “Half” in Table 2) was found to form a reasonably stable hairpin, though not quite so stable as the pP variant (Tm circa 24 °C, vs 36 °C for pP.) CD data and the method for determining fraction folded appear in the Supporting Information.
Paired disulfides as beta-structure nucleators
As previously noted, antiparallel β-sheets with disulfide-linked, capped β-strands are limited in their diversity and application by the requirement that all of the W,C,W residues must be at NHB strand positions: many strand lengths and compositions are precluded. Some of these limitations can be lifted by using a CXC-sequence rather than a single disulfide. The data in Table 2 shows that this can provide even greater β-structure stabilization. Prior studies have established that peptides with CXC sequences can form both antiparallel and parallel homodimeric sheet structures on oxidation and that homodimers are strongly favored over intramolecular disulfide formation31.
In our studies, this simple two-disulfide i+2 motif is more redox-stable than isolated cystines, largely orthogonal to single-disulfide oxidation, and a better β-sheet nucleator. No parallel dimers were observed for peptide “CIC” (Table 2), likely because β-caps do not work on parallel β-sheets, and antiparallel sheets are generally more stable than their parallel counterparts. It also oxidized much more rapidly than its single-cyst(e)ine analog.
The strong preference for disulfide formation across CXC units leading to self-associated dimers, versus slower disulfide formation for other cysteine residues opens up convenient biotechnology applications. Specifically, functionalization of free cysteine residues can be carried out on a structured β-sheet held together by a set of i+2 disulfides. For example, we were able to purify RWCTKCICIRKWE as the partially-oxidized dimer after 14 hours of air exposure in neutral buffer. Higher order assembly on complete oxidation resulted in a complex mix of soluble, high-molecular-weight oligomers.
The structure-nucleating efficiency of alternative disulfide locations: heterodimers.
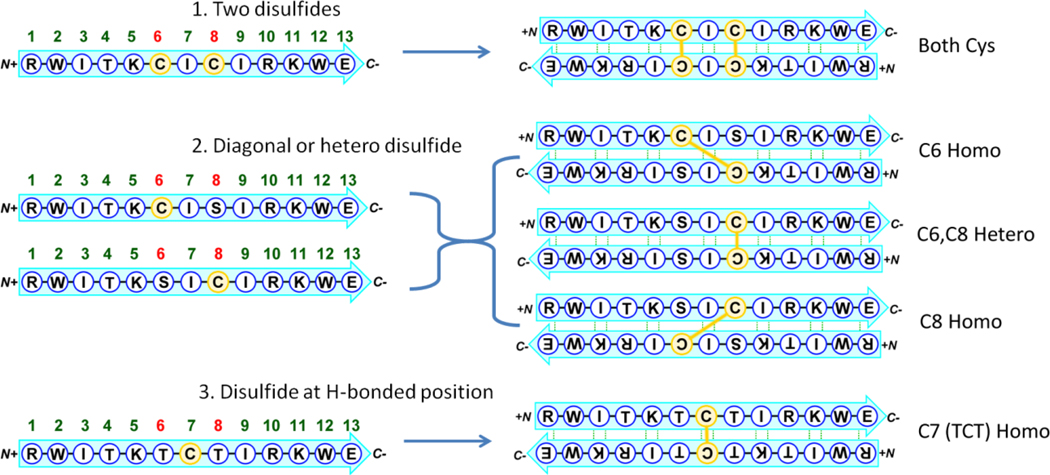
There is no inherent limitation of disulfide-mediated β sheets formation to homodimers. Heterodimers should also work, although there are clearly yield considerations when both homo- and hetero-dimers could form. To examine the effects of alternative disulfide linkage effects on β-sheet formation, we replaced individual cysteine residues of fully-folded (when oxidized) parent peptide RWITKCICIRKWE with serine, to investigate heterodimers with off-center disulfide linkages (Figure 3).
Figure 3.
Schematic illustrating the assembly and disulfide positions of the dimeric β-sheet peptides in Table 3.
As it turned out, yields for the oxidative disulfide-formation approached 100% heterodimer, with little interference from homo-dimers with diagonal disulfide linkages. Of note, the heterodimer allows formation of both a NHB cross-strand disulfide and two β-caps. The resulting off-center dimer was fully folded, with a TM of circa 95 °C (as observed via CD). Surprisingly, the chemical shifts were nearly identical for the two strands; there were slight differences for the asymmetric central region, but perfect peak overlap for the terminal (capping) residues.
We also investigated homodimers of these two single-disulfide species, to explore the fold-stabilizing capacity of disulfides bridging either of the non-equivalent diagonal positions. The natural twist of isolated β-sheets staggers the sidechains and favors one diagonal interaction over the other for disulfide formation. For right-handed, twisted β-sheets, sidechains at NHB positions can make close contacts with opposite-strand sidechains in the direction of their own strand’s C-terminus (sufficient, if not optimal, for disulfide formation) but not with sidechains toward their strand’s N-terminus – these are too distant for disulfide formation, in beta sheets of standard geometry. Thus, a diagonal disulfide between C6 and C6’ would be compatible with the register of a doubly-capped, stable β-sheet – while a potential disulfide between C8 and C8’ would not. There is some literature evidence for β-sheet stabilization by a diagonal disulfide (e.g., antimicrobial peptide androctonin,32 PDB id# 1CZ6) but only near the ends (rather than the middle) of the associated antiparallel β-strands.
We also investigated a nearly-identical sequence with the Cys residue at an H-bonded position. (H-bonded cysteines are technically close enough to their cross-strand symmetry pair to form a disulfide bond, but this is disfavorable due to steric features and rotamer preferences.) We changed the Cys-flanking residues to threonines as a compromise between isoleucine and serine.
As expected, the stability order was “Both Cys” (two cross-strand NHB disulfides) > C6,C8 heterodimer (one cross-strand NHB disulfide, off-center) > C6 homodimer (favored-diagonal disulfide) > C7 homodimer (cross-strand HB disulfide) > C8 homodimer (disfavored-diagonal disulfide). In all cases, strand register was fixed by the beta caps, not the disulfides. This series of peptides clearly illustrates that cross-strand disulfides are considerably more favorable than alternatives, yet even strongly-disfavored disulfides (e.g., C7 homo peptide’s HB-position disulfide) allow themselves to adopt strained conformations rather than alter the register as defined by the strong (but non-covalent) β-capping interactions. The capacity of cross-strand Trp/Trp interactions to “outcompete” disulfides as fold-stabilizers and strand register anchors of beta sheet structures has been observed previously.33
Deviation from canonical antiparallel β-sheet structure are observed for disulfide-dimerized strands.
Disulfides are well-suited for non-H-bonded positions, but they do impose some additional structural constraints. While the examples in the previous section show they are not always capable of defining their strands’ register to achieve a non-H-bonded cross-strand disulfide, they are nonetheless capable of altering the twist and buckle of their beta sheet. The Varadarajan laboratory4,34 found that, even at their most-allowed cross-strand NHB positions, disulfide bonds lowered cyst(e)ine Cα distances and caused beta sheets to buckle inward on the disulfide-bearing face. This imposition is expected to be greater for a set of two or more i+2 disulfides.
Typical β-hairpins display a characteristic pattern of chemical shift deviations. Most notably, every second amide proton (the inward-pointing protons) exhibits far-downfield (circa 1 ppm) CSDs as a result of strong H-bonding to the cross-strand carbonyl, and an in-plane alignment between the β-strands. Disulfide-disrupted beta structure can be identified most readily by a disruption to this pattern of HN chemical shift deviations. The cyst(e)ine residues have higher than expected chemical shifts for non-H-bonding strand positions, and neighboring (H-bonded) residues are not as downfield as expected. H-bonds are presumably still present at these sites, but they may be weakened by strand buckling and/or other distortions required to accommodate the disulfide bond. We have observed that this buckling effect is subtle for lone disulfides and short sheets, but becomes increasingly obvious when: a) the length of the associated β-strands increases, b) the number of consecutive i+2 disulfides increases, or c) the neighboring residues (at H-bonded sites) are less bulky (not beta-branched). The Supporting Information provides a table of ratios between the HN CSDs for cysteine and its immediate neighbors; we view this ratio as a diagnostic for disulfide-induced β strand distortion. An extreme test-case, a de novo antimicrobial peptide with a triple-disulfide (RWKCKCKCKWE)2, (three consecutive i+2 disulfides, no beta-branched residues, moderately long sheet) has an inverted pattern of HN CSDs. The data is shown in figure 4.
Figure 4.
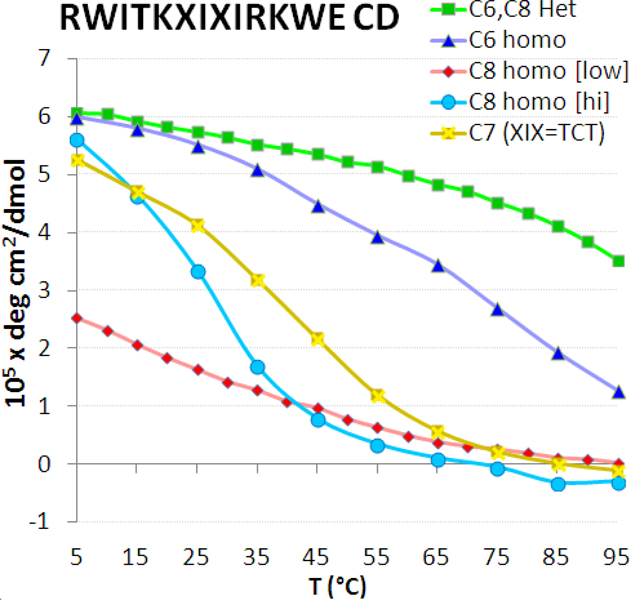
The CD melting curves (molar ellipticity measured at the 228nm maxima of the exciton couplet versus temperature) for the five peptides in Table 3. Peptide C8 homodimer was investigated at both high (1.5 mM) and low (30 μM) concentrations. For the corresponding NMR comparisons see Figure S6 in the Supporting Information.
Building on the core sheet: how to make four-stranded sheets
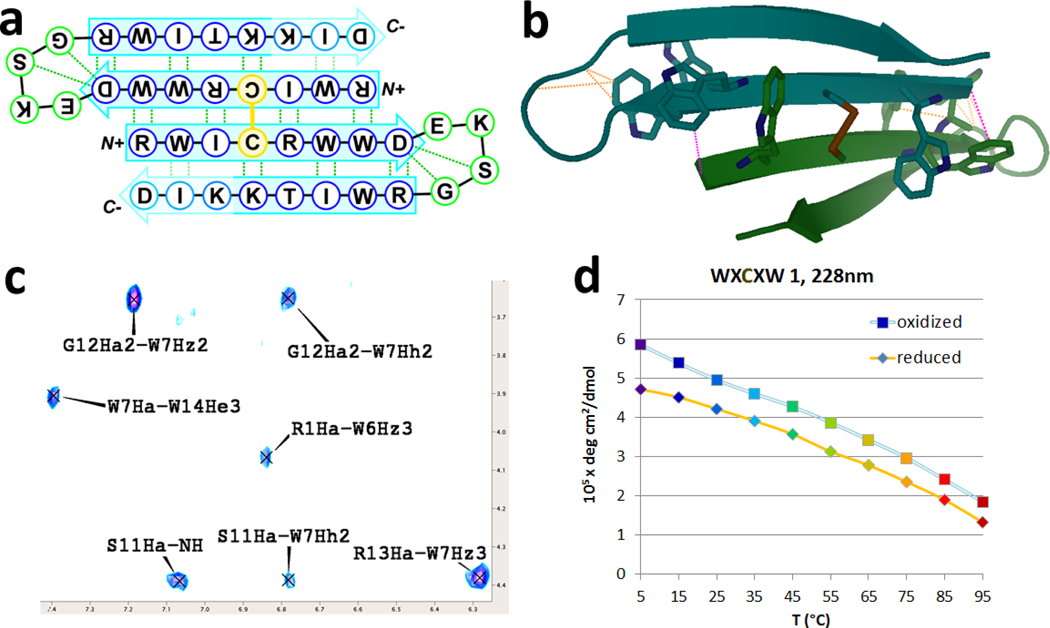
Disulfide-dimerized β-sheets would appear to be ideal cores for capture and docking of outer β-strands. Curiously, of our four attempted designs of four-stranded sheets (dimers of beta hairpins sewn together by a disulfide centered on one strand), all but one yielded a stable central core juxtaposed with an unstructured region. We had expected a strong templating effect from the fully-folded core strand, as is typical of 3-stranded sheets35,36. We now conclude that the distortion of the end-capped hub strands (e.g., WXCXW) precludes formation of all but the most favorable β-hairpin dimers. Data for the successful four-stranded sheet motif (sequence: (RWICRWWDEKSGRWITKKID)2 ) is shown in figure 6. Further examples appear in the Supporting Information.
Figure 6.
(a) Cartoon image illustrating beta-beta dimer topology. (b) Designed structure; validated by 2D NMR NOEs & CSDs. Selected, structurally diagnostic NOEs are marked as orange lines (intrachain) and purple lines (interchain). (c) A slice of the NOESY spectra showing the structurally-diagnostic contacts highlighted in the upper right panel, here as raw NOEs. (Complete NMR data appear in the Supporting Information). (d) CD-derived melting plot; intensity vs. temperature. (Trp/Trp exciton couplet maxima at 228nm.) The reduced form still adopts a fully-folded hairpin. NMR data (see Supporting Information) suggest that the reduced species remains monomeric, and that the intrachain Trp/Trp interaction (present in both oxidized dimer and reduced monomer) produces a stronger exciton couplet than the interchain Trp/Trp (oxidized dimer only) pair.
Disulfide stabilization of antiparallel β-sheet structures does not apply when the disulfide unit is not located in the β-strands
Disulfide bonds can impart β-sheet structure as well or better than the best turns, yet their use in this regard is highly context-dependent. The example of RWITKXIXIRKWE (table 3, figures 3 and 4) illustrates that, while cross-strand non-H-bonded positions are strongly preferred, this preference is not enough to shift the strand register required by beta caps.
Table 3.
RWITXIXIRKWE peptides: the effect of relative cyst(e)ine placement.
| Name | sequence | TM (CD) | 280 K χF (NMR) |
|---|---|---|---|
| C8 homodimer | RWITKSICIRKWE | 29* | 0.95* |
| (bad diagonal) | EWKRICISKTIWR | ||
| C6 homodimer | RWITKCISIRKWE | 72 | 0.98 |
| (good diagonal) | EWKRISICKTIWR | ||
| C6,C8 hetero | RWITKCISIRKWE | ~95 | 0.99 |
| (X-strand) | EWKRICISIRKWE | ||
| Both Cys | RWITKCICIRKWE | >100 | >0.99 |
| (2x X-strand) | EWKRICICKTIWR | ||
| C7 (TCT) homo | RWITKTCTIRKWE | 42 | 0.88 |
| (HB position) | EWKRITCTKTIWR |
At high concentration (1mM), as the tetramer. See part “Higher Order Oligomers” section (below) for details.
Further counterexamples help set the boundaries of the β-sheet-enforcing capabilities of disulfides. Peptide RWITVTIGGGGCGGGGKKIRVWE forms an antiparallel β-sheet structure (χF = 0.91 at 280K) even in its reduced form with the nine residue GGGGCGGGG sequence remaining a fully-flexible loop devoid of structure. While disulfide formation would not decrease the length of loop that would need to be conformationally searched to achieve β-strand association, it can effectively lower loop search times by increasing local concentrations of equivalent strands. Furthermore, if one hypothesizes that disulfides are exceptionally effective nucleators of β-sheet structures, one might expect the flexible loop to overcome the significant entropy barrier imposed by the eight glycine residues and form into a contiguous β-sheet, as shown below.
A RWITVTIGGGGCGGGGKKIRVWE B
B’ EWVRIKKGGGGCGGGGITVTIWR A’
As it turns out, the glycine loop region of both the reduced and oxidized forms of this peptide display nearly identical chemical shifts with negligible CSDs; this rules out the possibility shown immediately above. The β-sheet fold population of the oxidized species is somewhat higher (χF => 0.97, at 280K, vs. 0.91) but this may be nothing more than a local concentration effect which increases the folding rate. When oxidized, strand A can find both strand B and equivalent B’ nearby, with which to form a beta sheet. When reduced, strand A has only strand B to pair with. Due to the flexibility of the GGGGCGGGG segment, all combinations of AB, A’B, AB’, and A’B’ are almost perfectly equivalent. (See Supporting Information Figure S3 for a schematic of this peptide’s potential folding pathways.) Thus we can delineate the limitations of the disulfide strand-nucleation strategy: disulfides are not magic bullets that will generate local β-sheet structures where no propensity to form sheets is otherwise present. Also, the enhancement of association through a local concentration effect does not appear to be dramatic or essential in cases where folding is possible without dimerization.
Higher order oligomers (D2-symmetric dimer of dimers)
The use of disulfides to form stable β-sheets has an interesting consequence: highly charged, amphipathic β-sheets can be constructed with ease. Whereas rows of cross-strand arginines and lysines would normally repel each other and preclude folding, disulfides can force them into proximity. A secondary consequence is that beta sheets can have not only very high net charges, but can concentrate these charges onto one face of a sheet structure or hairpin, resulting in amphipathic peptides. This architecture is exploited by many natural antimicrobial peptides, where their high positive charge and lipophilic face enables them to disrupt bacterial membranes. (It helps that disulfides can, in themselves, form a hydrophobic face.13) This easy route to amphipathicity can also afford beta sheets capable of oligomerizing while remaining soluble.
Some of the peptides in this study were observed to form soluble oligomers – typically dimers-of-dimers – though most evidence was indirect. For example, triple-disulfide homodimer RWKCKCKCKWE has a hydrophilic face (R,K,K,K,K,E) and a hydrophobic face (W,C,C,C,W). Its severe line-broadening (for strand-central residues only) and certain NOEs on its hydrophobic face cannot be explained by the homodimer structure, suggesting the formation of a non-covalent dimer-of-dimers or some other soluble oligomeric form.
All short dimers of the form (XWXCXWX)2 are amphipathic, with a fully hydrophobic W,C,W face regardless of the identities of the other residues. This surface is capable of sticking to a copy of itself as a dimer-of-dimers. However, this second dimerization event is remarkably slow for a noncovalent interaction between small peptides. Two distinct, sharp sets of peaks corresponding to two beta sheets of slightly different structure are observed in NMR spectra. Simple models suggest these peaks represent disulfide-swapped structures, wherein the edge-to-edge cross-strand disulfide is exchanged for a pair of face-to-face disulfides in a D2-symmetric tetramer. Tellingly, NOEs are observed between opposite ends of individual indole rings for the isomer responsible for one of the sets of peaks. The set of peaks with the intra-indole NOE also sports higher overall NOE intensity (higher NOESY spectra quality) relative to its 1D peak intensities; this would be the result of the slower tumbling and increased rigidity of a dimer of dimers structure. A proposed model that satisfies the CSDs and NOEs is shown in figure 7.
Figure 7.
A proposed structure of the D2-symmetric tetramer form of KWRCIWD which reconciles the intra-indole Hδ1 –Hζ3 NOE observed. These are interchain NOEs between equivalent indoles stacking at the interface. This tetramer, with disulfides linking sheets face-to-face rather than edge-to-edge, results in efficient packing of the mostly-unchanged dimer units. Conformational exchange between this structure and the C2-symmetric dimer (with the typical edge-to-edge cross-strand disulfides) should be very slow at low temperatures, and this is indeed what we observe by NMR. Specifically, two roughly equal sets of peaks (likely corresponding to dimer and tetramer) are observed. See Supporting Information for CD and NMR data.
There was another case where evidence existed for a disulfide-swapped (face-to-face instead of edge-to-edge) dimer of dimers. We initially observed a discrepancy between CD and NMR data for the oxidized form of peptide C8 (RWITKSICIRKWE)n. The CD (at 30 μM) displayed an EtF W/W exciton couplet which decreased rapidly on warming, with an apparent melting point below the coldest assayed temperature (TM < 280 K), while NMR (measured at 1.5 mM) revealed a near-fully folded species at 280K. In addition, the NMR spectrum showed a regular CSD pattern suggestive of a canonical β-sheet with none of the serious twisting, buckling, or register shifting expected for a beta sheet with a strained, “backwards diagonal” disulfide bond.
Re-running the CD experiment in a 0.1mm CD cell using a portion of the original NMR sample yielded a CD spectra consistent with a high 280K fold population (χF = 0.95 by NMR) and more closely resembling the other peptides of the series (TM circa 29 °C; see figure 4). We propose an alternate disulfide arrangement; most likely a dimer of dimers with face-to-face disulfide bonds between strands as per the example shown in figure 7.
DISCUSSION
We have provided guidelines for designing stable, disulfide-central β-sheet homodimers, and illustrated their structural superiority to the best available hairpin models available. Heterodimers can be assembled using the same methods, which expands the potential applications of this strategy. Heterodimer synthesis requires the production of two discrete chains and possesses an innate yield penalty of up to 50% - but if the strands are designed to disfavor homodimerization, yields can approach 100% (as per the greatly favored C6/C8 heterodimer; see Figure 3). Two-disulfide …CXC… sequences oxidize faster and produce β-sheet dimers which are more stable (if also more distorted) compared to their single-disulfide analogues. The limited orthogonality between …CXC… and …C… allows for purification of partially oxidized products which can be labeled at the remaining free thiols.
Potential application of disulfide nucleated β-sheets: antimicrobials, hydrogels, etc.
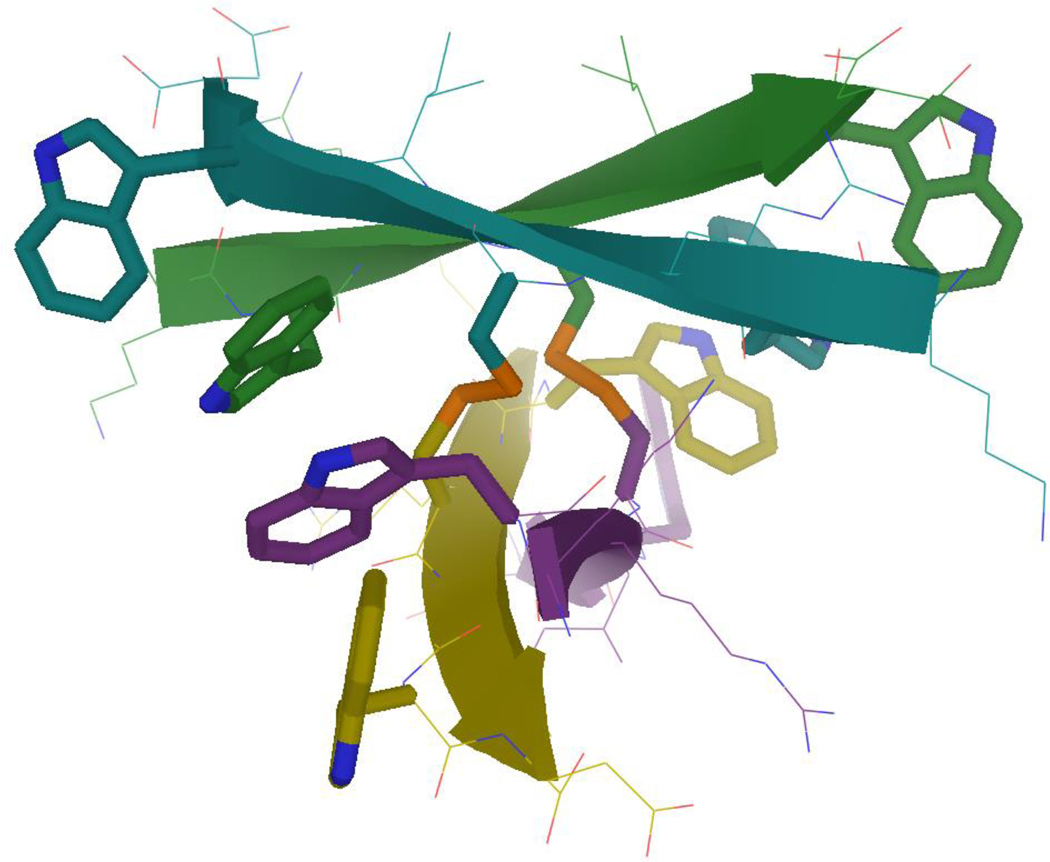
On oxidation, peptide RWKCKCKCKWE formed a single product: the expected antiparallel β-sheet. In addition to providing an example of a thoroughly disulfide-distorted β-sheet (figures 5 and 8), this rigid, amphipathic, highly cationic dimer proved to be a reasonably effective antimicrobial agent with an IC50 of 31 μM vs. E. coli (the details regarding this antibacterial and others will be presented elsewhere).
Figure 8.
Structure of the “lysine carpet” disulfide dimer (RWKCKCKCKWE)2, with the key disulfide and capping Trp residues highlighted. (Structure is supported by NMR data, including all expected cross-strand NOEs, but not explicitly generated via annealing to NOE distance constraints.)
Disulfide-rich peptides of this form can be used to make a carpet of any polar residue; e.g., a surface made entirely of histidine or aspartic acid sidechains, which have very strong cross-strand His+/His+ or Asp-/Asp- repulsion. A poly-disulfide sheet is the only way to force repulsive residues into a “beta carpet” at pH values where they are entirely charged. In addition to obvious antimicrobial (membrane disruptive) applications (as per the related but much more synthetically problematic theta-defensin13–14), this class of amphipathic peptides could be useful for catalysis37, metal binding, or selective detergents38, and already resembles a common hydrogel –forming structure consisting of alternating valine/lysine.39–44
Structurally rigid β-sheets with central disulfides have a range of other potential applications, such as minimalist scaffolds for pharmacophore display, rigid spacers, and building blocks for more complex structures. To further establish the precise structures of disulfide-bound homodimers and thus better apply them to these and other problems, we seek to establish crystal structures for one or more such constructs. While NMR can confirm β-sheet structure and register, it is not always enough to establish the twist of the hairpin, or the nature of any higher-order assembly that takes place (e.g., the precise conformation of any D2-symmetric tetramer).
Folding of Disulfide-Mediated Beta Sheet Dimers
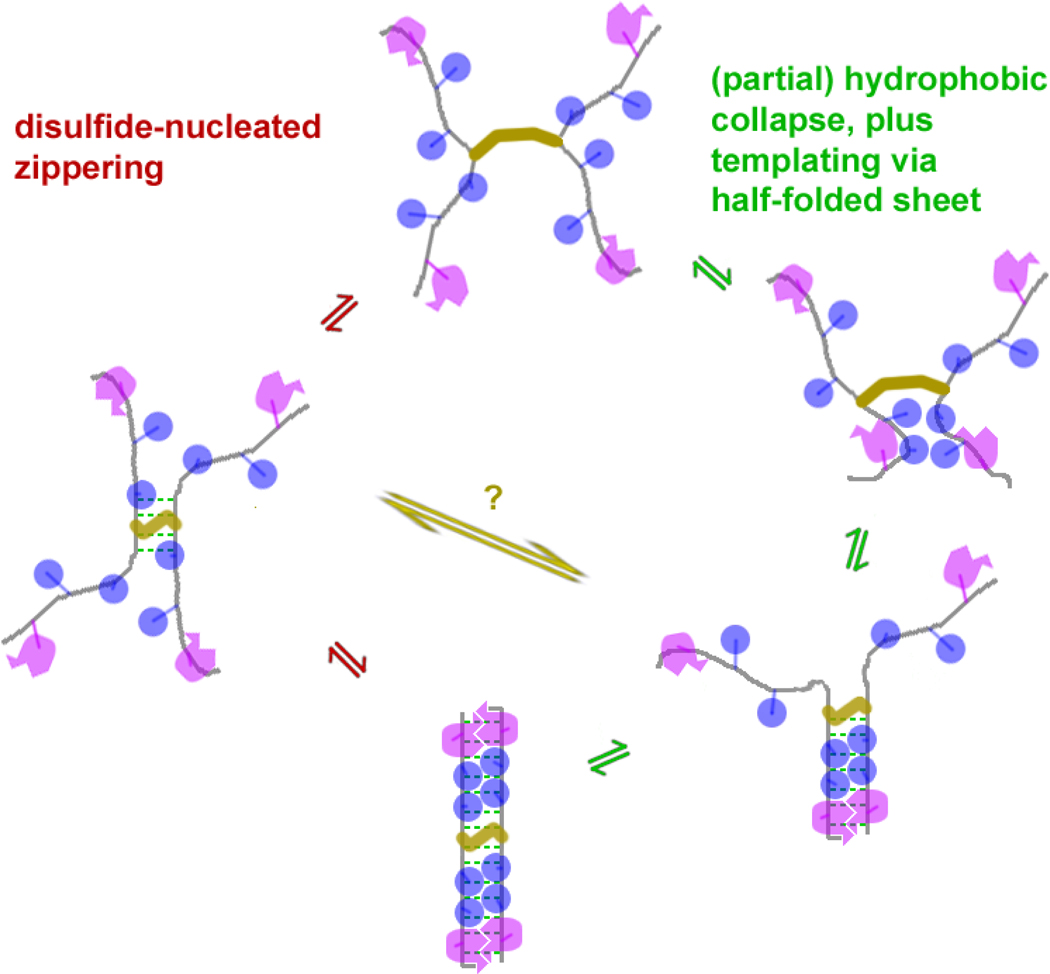
It is reasonable to assume that disulfide-dimer β-sheets fold via a cooperative mechanism, analogous to that of three-stranded sheets. (See figure 9.) Once one portion of the beta sheet has formed, a zippering or templating mechanism completes the other half. Theoretically, the folding rate in the forward direction should be faster than that of an equivalent monomeric hairpin, since there are two degenerate fold pathways. The unfolding rate may be slower as well, since a partially-unfolded sheet is poised to “zip up” and re-fold. β-Hairpins can be thought of as “internally cooperative” in the sense that a folded region (either the turn or a sequence-remote strand pair) can promote β-sheet folding in neighboring regions.45 Thus turnless β sheets with disulfides at their centers could be considered “cooperative” (folding of one section catalyzes the folding of another) despite the fact that they are comprised of a single contiguous secondary structure unit. As the inherent fold-pathway degeneracy makes it difficult to experimentally distinguish between potential folding mechanisms, molecular dynamics simulations of folding will be essential in analyzing these assumptions.
Figure 9.
Potential folding mechanisms for disulfide-mediated homodimers. These are analogous to related β hairpin folding mechanisms of: 1.turn nucleation plus zippering, and 2.hydrophobic collapse followed by packing optimization & H-bond formation. These folding mechanisms are not mutually exclusive, and even mixed mechanisms (e.g., via the yellow middle path) may be possible.
The ease of designing very stable disulfide-nucleated β-sheets suggests that the folding mechanisms of disulfide-rich proteins deserve a closer look. Once thought to be unfavorable1–3, we have firmly established that disulfides at cross-strand NHB positions in antiparallel β-sheets are not only favorable (as has since been determined, e.g., 4–6) but also more stabilizing for β-folds than the best reversing turns. Separate from their role as covalent staples, disulfide bonds have some propensity to form nascent β-sheets, to the benefit (via fold-nucleation) or detriment (via kinetic-trapping from off-path structures) of a protein’s native fold.
Supplementary Material
Figure 1.(b).
Example structure of the disulfide dimer from Figure 1a (KWRTIKVCITKRTWE)2, with the key disulfide and capping Trp residues highlighted as sticks. (This structure is supported by and consistent with NMR data, including all expected cross-strand NOEs, but not explicitly generated via annealing to NOE distance constraints.)
Figure 2.
NMR structuring shift comparisons (CSDs) for peptides C, pP, CIC, and “Half” (Table 2) at 300 K. The corresponding CD spectral comparisons appear in the Supporting Information.
ACKNOWLEDGEMENTS
This work was supported in large part by National Institutes of Health (NIH) Grant GM099889 (N.H.A., P.I.); partial salary support from an NSF grant (CHE-1152218) is also acknowledged.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Circular dichroism data, NMR data (as chemical shifts and chemical shift deviations), NOE-validated structures of (RWITKXXXIRKWE)n peptides, a table of the ratios of cystine HN : neighboring HN chemical shift deviations, schematic of the folding pathways available to long-loop hairpins dimerized by a cystine in the middle of a long flexible loop. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Richardson JS Adv. Protein Chem, 1981, 34, 167–339. [DOI] [PubMed] [Google Scholar]
- 2.Thornton JM J. Mol. Biol, 1981, 151, 261–287. [DOI] [PubMed] [Google Scholar]
- 3.Wouters MA; George RA; Haworth NL Curr. Protein. Pept. Sci. 2007, 8, 484–95. [DOI] [PubMed] [Google Scholar]
- 4.Indu S; Kochat V; Thakurela S; Ramakrishnan C; Varadarajan R. Prot. Struct. Funct. Bioinf. 2011, 79, 244–260. [DOI] [PubMed] [Google Scholar]
- 5.Wouters MA; Curmi PM. Proteins 1995, 22, 119–131. [DOI] [PubMed] [Google Scholar]
- 6.Santiveri CM; León E; Rico M; Jiménez MA Chem. Eur. J. 2008, 14, 488–499. [DOI] [PubMed] [Google Scholar]
- 7.Blandl T; Cochran AG; Skelton NJ Prot. Sci. 2003, 12, 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell SJ; Blandl T; Skelton NJ; Cochran AG J. Am. Chem. Soc. 2003. 125, 388–95. [DOI] [PubMed] [Google Scholar]
- 9.Pletneva EV; Laederach AT; Fulton DB; Kostić NM J. Am. Chem. Soc, 2001, 123, 6232–6245. [DOI] [PubMed] [Google Scholar]
- 10.Doherty T; Waring AJ; Hong M. Biochemistry 2008, 47, 1105–16. [DOI] [PubMed] [Google Scholar]
- 11.Matsuzaki K; Nakayama M; Fukui M; Otaka A; Funakoshi S; Fujii N; Bessho K; Miyajim K. Biochemistry 1993, 32, 11704–10. [DOI] [PubMed] [Google Scholar]
- 12.Lehrer RI; Cole AM; Selsted ME J. Biol. Chem. 2012, 287, 27014–27019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conibear AC; Rosengren KJ; Daly NL; Henriques ST; Craik DJ J. Biol. Chem. 2013, 288, 10830–10840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yasin B; Pang M; Turner JS; Cho Y; Dinh NN; Waring AJ; Lehrer RI; Wagar EA Eur. J. Clin. Microbiol. Infect. Dis. 2000, 19, 187–194. [DOI] [PubMed] [Google Scholar]
- 15.Venkatraman J; Nagana Gowda GA; Balaram PJ Am. Chem. Soc. 2002, 124, 4987–4994. [DOI] [PubMed] [Google Scholar]
- 16.Cashman TJ; Linton BR Org. Lett. 2007, 9, 5457–5460. [DOI] [PubMed] [Google Scholar]
- 17.Kier BL; Shu I; Eidenschink LA; Andersen NH Proc. Natl. Acad. Sci. USA, 2010, 107, 10466–10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson JM; Kier BL; Shcherbakov AA; Andersen NH FEBS Lett, 2014, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eidenschink LA; Kier BL; Huggins KNL; Andersen NH Proteins Struct. Funct. Bioinf. 2009, 75, 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fesinmeyer RM; Hudson FM; Andersen NH J. Am. Chem. Soc. 2004, 126, 7238. [DOI] [PubMed] [Google Scholar]
- 21.Kier BL; Andersen NH J. Am. Chem. Soc. 2008, 130, 14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersen NH; Olsen KA; Fesinmeyer RM; Tan X; Hudson FM; Eidenschink LA; Farazi SR J. Am. Chem. Soc. 2006, 128, 6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams DV; Byrne A; Stewart JM; Andersen NH Biochemistry, 2011, 50, 1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kier BL; Andersen NH J. Peptide Sci. 2014, 20, 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haque TS; Gellman SH J. Am. Chem. Soc. 1994, 116, 4105. [Google Scholar]
- 26.Haque TS; Gellman SH J. Am. Chem. Soc. 1997, 119, 2303. [Google Scholar]
- 27.Favre M; Moehle K; Jiang L; Pfeiffer B; Robinson JA J. Am. Chem. Soc. 1999, 121, 2679–2685. [Google Scholar]
- 28.Schneider JP; Kelly JW Chem. Rev. 1995, 95, 2169–2187. [Google Scholar]
- 29.Fuller AA; Du D; Liu F; Davoren JE; Bhabha G; Kroon G; Case DA; Dyson HJ; Powers ET; Wipf P; Gruebele M; Kelly JW Proc. Natl. Acad. Sci. U S A. 2009, 106, 11067–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davidson A; Patora-Komisarska K; Robinson JA; Varani G. Nucleic Acids Res. 2011, 39, 248–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu C; Leroux J-C; Gauthier MA Nature Chem, 2012, 4, 1044–1049. [DOI] [PubMed] [Google Scholar]
- 32.Mandard N; Sy D; Maufrais C; Bonmatin JM; Bulet P; Hetru C; Vovelle FJ Biomol. Struct. Dyn. 1999, 17, 367–380. [DOI] [PubMed] [Google Scholar]
- 33.Mirassou Y; Santiveri CM; Pérez de Vega MJ; González-Muñiz R; Jiménez MA ChemBioChem, 2009, 10, 902–910. [DOI] [PubMed] [Google Scholar]
- 34.Chakraborty K; Thakurela S; Pajapati RS; Indu S; Ali PSS; Ramakrishnan C and Varadarajan R. Biochemistry, 2005, 44, 14638–14646. [DOI] [PubMed] [Google Scholar]
- 35.Hudson FM; Andersen NH Biopolymers, 2006, 83, 424. [DOI] [PubMed] [Google Scholar]
- 36.Syud FA; Stanger HE; Schenck Mortell H; Espinosa JF; Fisk JD; Fry CG; Gellman SH J. Mol. Biol. 2003, 326, 553. [DOI] [PubMed] [Google Scholar]
- 37.Matsumoto M; Lee SL; Waters ML; Gagné MR J. Am. Chem. Soc. 2014, in press. [Google Scholar]
- 38.Bartocci S; Mazzier D; Moretto A; Mba M. Org. & Biomol. Chem. 2014, in press. [DOI] [PubMed] [Google Scholar]
- 39.Branco MC; Pochan DJ; Wagner NJ; Schneider JP Biomaterials 2009, 30, 1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deming TJ Soft Matter, 2005, 1, 28–35. [DOI] [PubMed] [Google Scholar]
- 41.Ozbas B,; Kretsinger JK; Rajagopal K; Schneider JP; Pochan DJ Macromolecules, 2004, 37, 7331–7337. [Google Scholar]
- 42.Kretsinger JK; Haines LA; Ozbas B; Pochan DJ; Schneider JP Biomaterials, 2005, 26, 5177–5186. [DOI] [PubMed] [Google Scholar]
- 43.Haines-Butterick L; Rajagopal K; Branco M; Salick D; Rughani R; Pilarz M; Lamm MS; Pochan DJ; Schneider JP Proc. Natl. Acad. Sci. U S A, 2007, 104, 7791–7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salick DA; Kretsinger JK; Pochan DJ; Schneider JP J. Am. Chem. Soc, 2007, 129, 14793–14799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stanger HE; Syud FA; Espinosa JF; Giriat I; Muir T; Gellman SH Proc. Natl. Acad. Sci. 2001, 98, 12015–12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.