Abstract
Most tumor-targeted drug delivery systems must overcome a large variety of physiological barriers before reaching the tumor site and diffuse through the tight network of tumor cells. Many studies focus on optimizing the first part, the accumulation of drug carriers at the tumor site, ignoring the penetration efficiency, i.e., a measure of the ability of a drug delivery system to overcome tumor surface adherence and uptake. We used three-dimensional (3D) tumor spheroids in combination with light-sheet fluorescence microscopy in a head-to-head comparison of a variety of commonly used lipid-based nanoparticles, including liposomes, PEGylated liposomes, lipoplexes, and reconstituted high-density lipoproteins (rHDL). Whilst PEGylation of liposomes only had minor effects on the penetration efficiency, we show that lipoplexes are mainly associated with the periphery of tumor spheroids, possibly due to their positive surface charge, leading to fusion with the cells at the spheroid surface or aggregation. Surprisingly, the rHDL showed significantly higher penetration efficiency and high accumulation inside the spheroid. While these findings indeed could be relevant when designing novel drug delivery systems based on lipid-based nanoparticles, we stress that the used platform and the detailed image analysis are a versatile tool for in vitro studies of the penetration efficiency of nanoparticles in tumors.
1. Introduction
Over the last decades, nanoparticles (NPs) have been established as promising drug carriers for cancer therapy.1−3 The advantages of using NPs as drug carriers include specific targeting, enhanced drug stability, and accumulation in the tumor tissue relative to free drugs.4,5 Especially lipid-based NPs are attractive due to their high biocompatibility compared to other nanosized drug carriers, e.g., gold NPs or quantum dots.2,3 Most often, clinically approved lipid-based drug delivery systems rely on passive accumulation at the tumor site.3,4,6
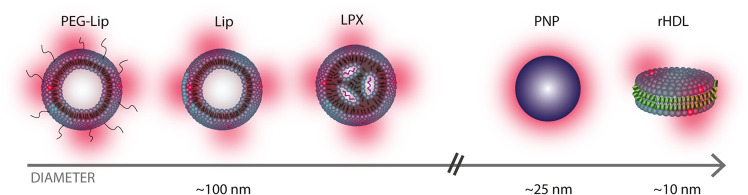
The majority of studies on NP drug delivery focus on the accumulation of NPs in the tumor tissue (through biodistribution studies) and are less concerned with the penetration profile of the NPs into the tumor. We emphasize that the penetration efficiency of the NPs is an important parameter as the therapeutic efficiency could be limited when the drug is only delivered to the outermost cells of the tumor.7,8 Whilst it is well established that NP penetration depends on the physicochemical properties of the NPs, e.g., size,9−12 a detailed head-to-head comparison of the penetration of a broad selection of lipid-based NPs is still wanted to learn which kind of lipid-based NPs to use for a certain drug candidate. Hence, this study provides a comparative investigation of the penetration efficiencies of liposomes (Lip), PEGylated liposomes (PEG-Lip), reconstituted high-density lipoproteins (rHDL), and lipoplexes (LPX), which have all reached clinical trials.2,13,14 As a non-lipid control, we used polystyrene nanoparticles (PNP) (Figure 1). Briefly, liposomes (Lip and PEG-Lip) are some of the most commonly used drug carriers. They are spherical vesicles comprising a lipid bilayer and an aqueous core, while surface grafting of polyethylene glycol (PEG) is frequently used to increase their blood circulation time.2,15,16 LPX are complexes of nucleic acids that are stabilized by cationic lipids13 and are usually used for gene-targeting applications.2 The rHDL are biomimetic particles similar to the endogenous high-density lipoproteins (HDL), a natural lipid transporter in the body;14 hence, rHDL are promising drug delivery carriers for various applications.17 There are several variants of the rHDL/HDL-mimicking particles,14,18 and in this study, we used the ∼10 nm-sized discoidal rHDL consisting of a lipid bilayer stabilized at the edge by two apolipoproteins A-I (apoA-I).
Figure 1.
Overview of the NPs in this study. The sketch of the lipid-based NPs used in this study: PEGylated liposomes (PEG-Lip), liposomes (Lip), lipoplexes (LPX), and reconstituted high-density lipoproteins (rHDL), consisting of lipid bilayers (blue), aqueous cores (white), fluorescent markers (red), nucleic acids (purple), and apoA-I proteins (green). For comparison, we also used a non-lipid particle (PNP).
The lipid-based NPs used here differ in several aspects like (i) size, (ii) surface charge, (iii) surface chemistry, and (iv) targeting mechanisms. For the latter, only rHDL are believed to be actively targeting as they are recognized by the scavenger receptor class B type 1 (SR-BI) that is overexpressed in many types of cancer cells,14,19 including the U87-MG brain cancer cells used in this study.20
To investigate the NP uptake and penetration, we used an in vitro three-dimensional (3D) tumor spheroid model,21,22 which is a well-established drug screening system.23−25 Even though this simple system is grown from monocultures, it still preserves the essential features present in cancer tumors in vivo, i.e., rapid proliferation at the surface and slow metabolism, or necrosis, in the center of spheroids.26 The majority of studies of NP distributions in tumor spheroids use tumor spheroid fixation,27−35 often in combination with histological sectioning.7,36−39 Nevertheless, there are also reports of NP penetration in live tumor spheroids using confocal-scanning microscopy.40−50 Live imaging of NP-treated tumor spheroids ensures that the studied NPs preserve localization, stability, and emission wavelength,51,52 as indicated for semiconducting NPs.53 However, the high scattering and absorption of a live tissue54 limits the imaging depth. Thus, to overcome these constraints, we used an Airy beam light-sheet microscope55,56 that allows for optical sectioning with little bleaching or photodamage.57 With this setup, we combine deep imaging of NPs with single-cell resolution of tumor spheroids of hundreds of micrometers.27,58
This study is a head-to-head comparison of the penetration efficiencies of several lipid-based NPs commonly used for drug delivery. We use a 3D tumor spheroid model in combination with optical sectioning and a custom-made analysis procedure, and we find that clustering on the tumor spheroid surface and penetration efficiency vary significantly among the tested lipid-based NP types. This in vitro platform can easily be adapted to study the penetration of a large variety of fluorescently labeled NPs. Hence, we provide an important tool to screen and optimize the penetration efficiency of NPs before conducting more complex in vivo studies.
2. Materials and Methods
2.1. Preparation of NPs
2.1.1. Materials
The lipids 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), cholesterol, 1,2-dioleoyl-3-trimethylammonium-propane (chloride salt) (DOTAP), and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[amino(polyethylene glycol)-2000] (ammonium salt) (DOPE-PEG2k) were supplied from Avanti Polar Lipids. The lipophilic carbocyanine dye 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI) was supplied from Thermo Fisher Scientific. The CpG ODN 2395 was supplied from Invivogen. The stealth liposome mix (consisting of hydrogenated soy l-α-phosphatidylcholine (HSPC), cholesterol, and DSPE-PEG2k in a weight ratio of 3:1:1) was acquired from Lipoid GmbH (Germany). Reagents for the buffers used for preparation of the NPs (phosphate-buffered saline (PBS), sodium chloride, sodium citrate, and Hepes, glucose, and sodium cholate hydrate), 99.9% absolute ethanol, and tert-Butanol were all supplied from Sigma-Aldrich (Denmark).
2.1.2. Liposomes/PEGylated Liposomes
Lip were prepared with DPPC/cholesterol/DiI in a molar ratio of (61.3:38.2:0.5), while PEG-Lip were prepared from the stealth liposome mix HSPC/cholesterol/DSPE-PEG (56.6:38.2:5.2) supplied with 0.5 mol % DiI. Initially, the lipids were dissolved in tert-butanol/MilliQ (9:1), snap-frozen in liquid nitrogen, and freeze-dried overnight using a Scanvac Coolsafe lyophilizer (Labogene, Denmark). The dry lipids were rehydrated in PBS to a total lipid concentration of 10 mM and put under 65 °C heating and magnetic stirring for 1 h. The size of the liposomes was controlled by extruding 21 times through Whatman filters (GE Healthcare, U.K.) with a pore size of 100 nm using a Mini-Extruder from Avanti Polar Lipids. The liposome suspension was transferred to a glass vial and stored at 4 °C.
2.1.3. Lipoplexes
LPX were prepared by the ethanol-injection method. They were formulated with cholesterol/DOTAP/DOPE-PEG2k/DiI (49.5:49:1:0.5) and CpG ODN 2395 (single-stranded synthetic DNA) as the encapsulated nucleic acid. Initially, the lipids were dissolved in 99.9% absolute ethanol, and CpG was diluted in a citrate buffer, pH 5. The lipid solution was mixed into the CpG solution under vigorous vortexing, yielding a DOTAP/CpG (8:1). Immediately after mixing, the mixture was diluted by adding twice the volume of buffered saline (0.3 M sodium chloride, 20 mM sodium citrate, pH 6.0). To remove excess ethanol, the solution was then dialyzed for 24 h using cassettes with 3.5k MWCO (Thermo Fisher Scientific) against 300 mM Hepes and 5% glucose buffer (pH 7.4) with two buffer exchanges. After dialysis, the LPX solution was transferred to amicon spin filters (MWCO 10 000) in 15 mL tubes and centrifuged three times at 4000g and 4 °C for 10 min. Between each cycle, the volume was replenished by adding 300 mM Hepes and 5% glucose buffer (pH 7.4) to ensure the removal of unbound CpG. After washing, the LPX solution (10 mM total lipid concentration) was transferred to a new vial and allowed to equilibrate at room temperature for 30 min. The LPX were stored at 4 °C and used within 4 weeks after preparation.
2.1.4. rHDL
The lipids DPPC/DiI (99.5:0.5) were dissolved in tert-butanol/MilliQ (9:1) before freeze-drying overnight. ApoA-I (purified from human plasma as described elsewhere19) dissolved in PBS containing 20 mM cholate was added to the lipid mixture such that it yielded a protein/lipid molar ratio of 100 and a total lipid concentration of 5 mM. The solution was heated to 41 °C for approximately 1 h to ensure that the lipids were dissolved. Subsequently, Bio-Beads from Bio-Rad (Denmark) were added to remove the cholate and induce the self-assembly of the rHDL. The sample was incubated overnight with the Bio-Beads that were removed afterward by spinning the liquid from a 2 mL Eppendorf tube with a perforated hole into a falcon tube. The sample was stored at 4 °C.
2.2. Characterization of the NPs
All lipid-based NPs were fluorescently labeled using 0.5 mol % DiI fluorophore (549/565 nm). They were diluted in PBS, such that they contained a similar amount of fluorophores, as estimated by absorbance (NanoDrop 2000/2000c Spectrophotometer, Thermo Fisher Scientific), (Supporting Information Figure S2), and were further diluted 10× in an imaging medium (see Section 2.4 ) when added to the spheroids.
The fluorescently labeled (580/605 nm) anionic polystyrene carboxylate-modified nanoparticles (Thermo Fisher, Lot No. = 1922891) were sonicated for 10 min and were diluted in an imaging medium (see Section 2.4 section) to a final concentration of 1.7 × 1011 particles/mL.
The hydrodynamic diameters of the NPs were measured by dynamic light scattering (DLS) on a ZetaSizer Nano ZS (Malvern Instruments, U.K.) over an average of three runs of 10–15 cycles. The results were estimated by number [%]. The ζ-potential was obtained by the same instrument using a universal “Dip” Cell (Malvern Panalytical, U.K.) and was estimated with an average of three runs with an automatically determined number of cycles (10–100 cycles). Before the ζ-potential measurements, the samples were diluted 25× in a buffer containing 10 mM Hepes and 5% glucose (pH 7.4). For both the size and ζ-potential measurements, a distribution was obtained from each run/cycle with mean and standard deviation (SD). In Table 1, we present the average of the means and average of the SD from the three runs.
Table 1. Characterization of the NPs Used in the Studya.
| Lip | PEG-Lip | LPX | rHDL | PNP | |
|---|---|---|---|---|---|
| size (nm) | 114 ± 35.9 | 112 ± 25.9 | 77.2 ± 28.7 | 8.70 ± 1.70 | 24.1 ± 6.77 |
| ζ-potential (mV) | 0.827 ± 6.24 | –8.83 ± 9.43 | 9.96 ± 4.43 | –9.39 ± 5.27 | –31.0 ± 20.1 |
The mean size is based on the number (%) from dynamic light scattering measurements. Data represent mean ± SD.
2.3. Cell Culture
Gliosblastoma multiforme U87-MG cells were cultured in high-glucose Dulbecco’s-modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% (100 units/mL penicillin and 100 μg/mL streptomycin), all from Gibco.
2.3.1. Gravitation-Assisted Spheroid Formation
Five hundred cells were seeded in each round-bottom ultralow attachment of 96 wells (Corning Costar) in a volume of 100 μL DMEM and incubated for 72 h (37° C, 5% CO2, and 100% humidity). After 72 h, the spheroids reached a radius of 134 ± 10 μm (mean ± SD, N = 19).
2.4. Sample Preparation
For imaging, the culture medium was replaced by phenol red-free DMEM (FluoroBrite, Gibco) without serum to minimize the noise. Fully formed spheroids were rinsed three times with imaging medium and were treated with nanoparticles in a fresh imaging medium for 5 h. After 3 h of incubation, the spheroids were nuclear-labeled by supplementing the wells with a cell-permeable nucleic acid stain (SYTO13, Invitrogen; 488/509 nm). Although the nuclear stain is a high-performance dye, it causes phototoxicity within 3 h,59 which restricts live-cell imaging. Spheroids were washed three times in PBS (Gibco) and were then immersed in a fresh imaging medium. The spheroids were embedded in 1% low-melt agarose (NuSieve GTG Agarose; Lonza, Rockland, ME), were transferred separately in fluorinated ethylene propylene (FEP) tubes (inner diameter 0.8 mm, outer diameter 1.6 mm) (Bohlender), and were mounted on the microscope.
2.5. Live Spheroid Imaging
We used an Aurora airy light-sheet microscope (M Squared) with a M2 filter cube with one objective for illumination (UMPlanFL N 10×/0.30 W) and one for detection (UMPlanFL N 20×/0.50 W), each with a 3.5 mm working distance. The light sheet was equipped with a blue (488 nm) and a yellow (570 nm) laser. The light sheet allowed deep sample penetration and high axial resolution over an extended field view (600 mm × 600 mm). The emitted light was collected through a band-pass filter (520–540 nm) and low-pass filter (>570 nm) for the 488 and 561 nm laser beams, respectively. Two stacks of images were recorded sequentially, one for the nuclear stain and one for the nanoparticle stain in steps of 0.4 μm in the axial direction using the build Aurora Alpha acquisition software. The laser intensity and exposure were kept constant among experiments, which were performed in room temperature with minimized background light.
2.6. Image Analysis
To measure the fluorescence intensity versus depth of the spheroid, we analyzed the cross-sections and quantified the intensity with a combination of Fiji plugins and custom-made Matlab routines. As our spheroids on average were 300 μm in diameter and the step size in the axial direction was 0.4 μm, the measured intensities of our cross-sections was averaged over 100 frames (which corresponds to 40 μm). We estimated the variation of the radius over this band to be less than the variations in Rθ in the imaging plane. The obtained average cross-sections were analyzed to obtain the mean intensities as in eq 1.
2.6.1. Statistics
Unless otherwise stated, data are shown as mean ± standard deviation (SD) or SD of the mean (SEM), N ≥ 3. To assess statistical significance, analysis of variance (ANOVA, one-way) was performed for comparison of means, followed by Tukey’s post hoc test. Upon comparison to the α level of 5%, a p-value of <0.05 was considered statistically significant.
3. Results and Discussion
To study the penetration efficiency of the NPs, we used a brain cancer tumor spheroid model grown from a seed of (∼200 cells/well) U87-MG cells in round-bottom ultralow attachment wells. While negatively charged coating impedes cell attachment to the wells, gravitation assists agglomeration (Figure 2A).60,61 After the initial spheroid formation, the growth rate was estimated to be ∼40 μm/day under our experimental conditions. Spheroids were harvested after 3 days, where they had reached a final radius of 134 ± 10 μm (mean ± SD, N = 19) and a density of about 3 × 10–4 cells/μ m3. All spheroids were confirmed to attain similar morphological characteristics, e.g., ellipticity of 0.5 ± 0.1 (Figure S3). In general, cancer spheroids have an outermost layer of proliferating cells, an intermediate quiescent region, and sometimes even a necrotic core.26 The cancer spheroids used in this study, however, are reported to have high viability even in the core region.62
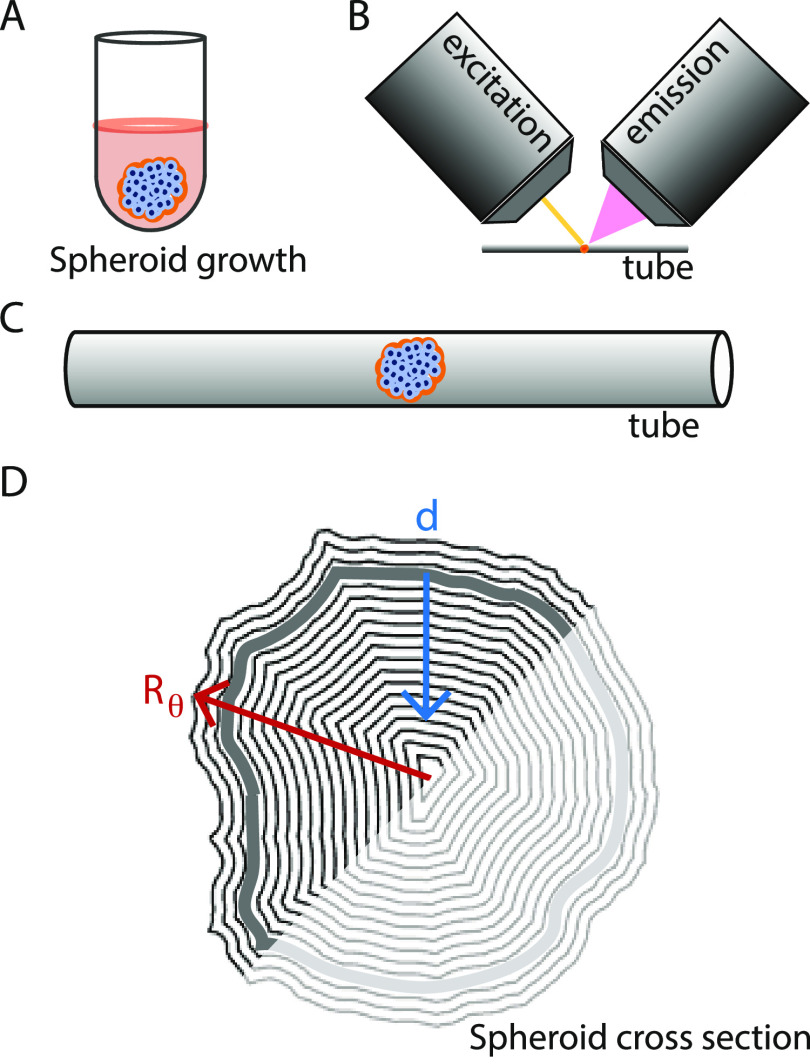
Figure 2.
Overview of the experimental setup. (A) Illustration of gravitation-assisted tumor spheroid growth. (B) Sketch of a spheroid mounted for imaging in an Airy beam light-sheet microscope, with one objective for the (yellow) light sheet (excitation) and one for collection of the emitted light (emission). (C) Close-up of a spheroid mounted in a tube for imaging. (D) Example of a spheroid’s cross-section mask, where the radial vector from the origin goes to the mask’s boundary; Rθ varies in length with respect to the polar angle θ. The mask boundary was slightly larger than the spheroid’s surface (gray contour) and the blue vector indicates the tumor depth, d. We restricted our analysis to the half-spheroid closest to the light-sheet objective (excitation).
3.1. Preparation and Characterization of the NPs
The lipid-based NPs (Figure 1) were prepared using standard methods as described in detail elsewhere for liposomes,63 LPX,64 and rHDL.19 Briefly, Lip and PEG-Lip were formed by hydration of freeze-dried lipids, followed by extrusion through a 100 nm filter. LPX were prepared by the ethanol-injection technique, where lipids dissolved in ethanol were rapidly injected into an aqueous buffer containing the nucleic acids CpG. rHDL were prepared by the detergent depletion method, where lipids were dissolved with detergents that were subsequently depleted using Bio-Beads in the presence of the apolipoprotein A-I (apoA-I) to induce the self-assembly of rHDL. The NPs’ size and surface charge, i.e., ζ-potential, are listed in Table 1.
The distribution and penetration of the lipid-based NPs were studied by detection of the DiI in the NPs. Besides being used as the fluorescent label, the lipid-conjugated DiI can also be considered as a model of lipid-conjugated drugs incorporated into the NPs. We confirmed that DiI was stably incorporated into the lipid-based NPs (Supporting Information Figure S1), as consistent with previous studies using similar types of fluorophores.15,19 We emphasize that all of the lipid-based NPs were prepared with equal mol% of fluorophores (0.5% DiI), and that the concentration of the NPs was adjusted such that the amount of fluorophores (based on absorbance) was equal in each sample (Supporting Information Figure S2). Thus, we assume that the lipid concentration was the same in each sample. Note that due to the different sizes of the lipid-based NPs, this implies that the particle concentration varied between samples.
3.2. Distribution of NPs in Tumor Spheroids
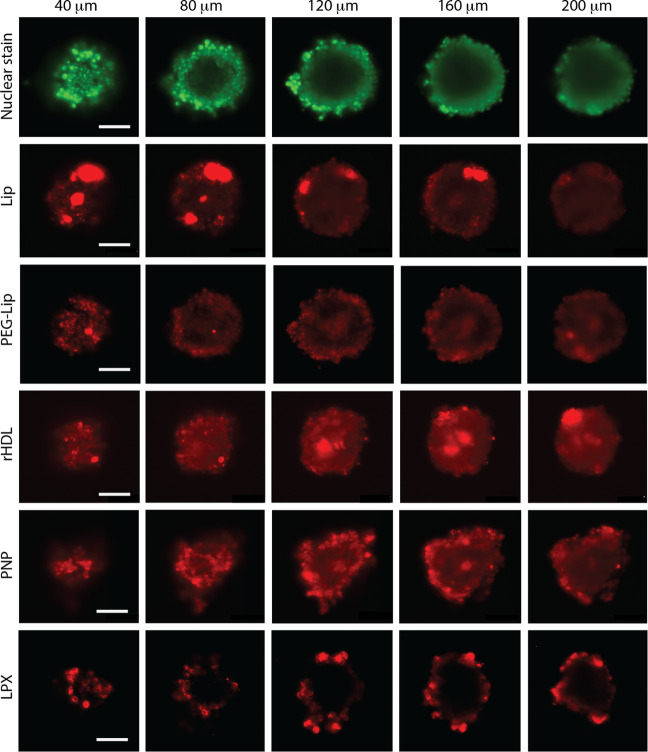
NP-treated tumor spheroids were optically sectioned (Figure 2B,C) in 400 nm steps to measure the fluorescence intensity on planar cross-sections of the spheroids. Notably, the intensity of the nuclear stain gradually decreased toward the center of the spheroids, which is likely explained by the diffusion gradient7 and effects from attenuation of the exciting/emitting light at increasing depths.57 Interestingly, the cross-sections (Figure 3) show a clear difference in the distribution of the different NPs in the tumor spheroids. While spheroids treated with Lip, PEG-Lip, rHDL, and PNP exhibited fluorescence intensity distributed across the whole cross-section (though with varying intensity), LPX accumulated primarily at the spheroid surface. Areas with augmented fluorescence were observed for rHDL, Lip, and LPX, and while these areas were confined to the spheroids’ surface for LPX and Lip, they also appeared deeper in the spheroids for rHDL. The augmented fluorescence likely corresponds to either aggregation of particles or high local cellular uptake of NPs. In particular, LPX, due to positive surface charges, might effectively fuse with the outermost cells or aggregate in the culture medium (at 37 °C), since LPX is considered relatively unstable without PEGylation.65 Aggregation of LPX would likely render their size larger than the pores in the tumors, thereby impeding penetration, while adherence to the surface of the cells or cellular uptake prevents aggregated LPX from being washed out before imaging. Similarly, Lip are considered more prone to aggregation than PEG-Lip.2,66,67 However, in contrast to the initially smaller LPX, the neutral surface charge of the Lip possibly allowed them to penetrate deeper into the tumor due to inferior NP-cell interactions, before aggregation or cellular uptake. On the other hand, rHDL showed enhanced accumulation at much greater penetration depths. This could in part be ascribed to the smaller size of rHDL, allowing them to pass more easily through the interstitial space between the cells, like in the case of PNP.
Figure 3.
Cross-sections of U87-MG tumor spheroids treated with NPs. The pseudocolored images are representative examples of optical sections from spheroids at different depths in the axial direction (40–200 μm). The spheroids were all nuclear stained (green) and treated with NPs (red): Lip, PEG-Lip, rHDL, PNP, or LPX, for 5 h. The images have uneven contrast settings between the different NPs, and the scale bars correspond to 100 μm.
3.3. NP Tumor Spheroid Penetration
The imaging of a dense spheroid is in brief limited by two conditions: (i) the attenuation of the fluorescence emission and (ii) the attenuation of the exciting laser through the spheroid. To balance (i), we did not use the cross-section from the spheroid equator but instead a cross-section closer to the imaging objective (emission Figure 2B) at a depth of 100–140 μm. Therefore, the measured intensities were averaged over 40 μm, which corresponds to 100 frames (Supporting Information Figure S4). Furthermore, to redeem the effect of (ii), we restricted our analysis to the half of the cross-section closest to the exciting laser (Figure 2D) and to a maximum depth of 80 μm.
From the masks of the averaged cross-sections, we defined the spheroid surface (d = 0 μm) as the maximum intensity depth of the nuclear stain. As the surface of the spheroid is rough, the radius of the spheroid, Rθ, changes in respect to the polar angle, θ (Figure 2D). Therefore, the intensity at a specific depth, d, of the averaged cross-section was quantified as follows68
| 1 |
for the M pairs of pixels, (xi,yi), that fulfill the requirement
| 2 |
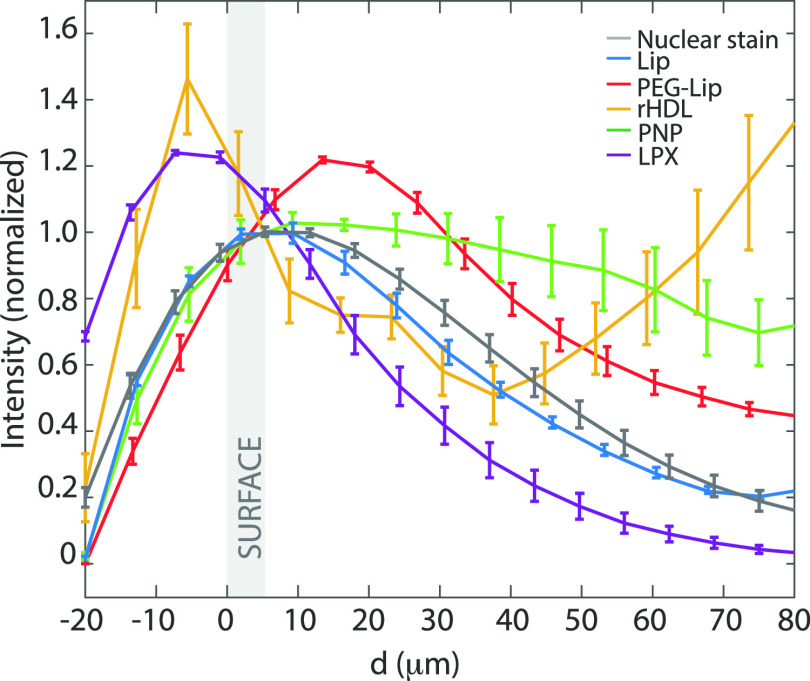
We found the normalized intensity profiles ⟨I⟩ averaged over all N spheroids of the nuclear stain (gray) and the investigated NPs as a function of the spheroid depth d (Figure 4). Regardless of the natural variations among the spheroids, the penetration profiles were specific for each type of NPs. Consistent with the cross-section images (Figure 3), we observed a gradual decrease in the nuclear stain’s intensity over d. Even though individual nuclei cannot be distinguished at penetration depths greater than ∼50 μm, it has been previously shown that U87-MG spheroids of similar sizes have uniform densities.62 Furthermore, we observed that both rHDL and LPX exhibited high surface adhesion. However, LPX remained at the spheroid rim, while all of the other NPs penetrated more effectively toward the center of the tumor spheroid.
Figure 4.
Penetration of NPs in U87-MG tumor spheroids. The normalized average intensity of NP fluorescence versus tumor spheroid depth, d. The intensity was averaged over all treated spheroids and normalized with the surface intensity (the surface was defined from the nuclear stain). All spheroids were exposed to both nuclear stain (gray, N = 19) and NPs: Lip (blue, N = 3), PEG-Lip (red, N = 5), rHDL (yellow, N = 4), PNP (green, N = 4), and LPX (purple, N = 3). The shaded area corresponds to the tumor spheroid surface, and error bars correspond to one SEM.
The penetration depth can be interpreted by the declination of the surface intensity by half d1/2 for the averaged profile. This measure confirmed the limited penetration depth of LPX, d1/2 = 26 μm, in contrast to the larger penetration depths of Lip, d1/2 = 39 μm, and PEG-Lip, d1/2 = 67 μm. The larger penetration depth of PEG-Lip relative to Lip could be attributed to reduced interactions with the cells due the PEGylation, thus allowing for increased penetration into the tumor spheroid, as also reported elsewhere for PEGylated NPs.7 Though the d1/2 indeed is a descriptive measure, it is not instrumental for the analysis of penetration profiles that differ from an exponential decay. For instance, rHDL had a unique secondary peak (d = 80 μm). Furthermore, PNP did not reduce the fluorescence intensity by half at the measured depths, although the intensity gradually decreased after the first surface cell layers.
The up-concentration in depth observed for rHDL might be explained by aggregation. Although rHDL are relatively stable,69,70 rHDL aggregation might occur due to varying environmental conditions toward the spheroid core. For example, the spheroid core is slightly more acidic relative to the surface due to the increased lactate production from the oxygen-deficient cells,71 and the acidic conditions could potentially cause the HDL-associated apoA-I to unfold.72 Interestingly, the PNP (∼24 nm) did not accumulate toward the spheroids’ center, though they seemingly also diffused throughout the tumor spheroids. This further confirms that the rHDL distribution in the spheroids is controlled by additional parameters besides size.
3.4. Penetration Efficiency
To further quantify the ability of the various NPs to penetrate the tumor spheroid, we defined the penetration efficiency as the following ratio
| 3 |
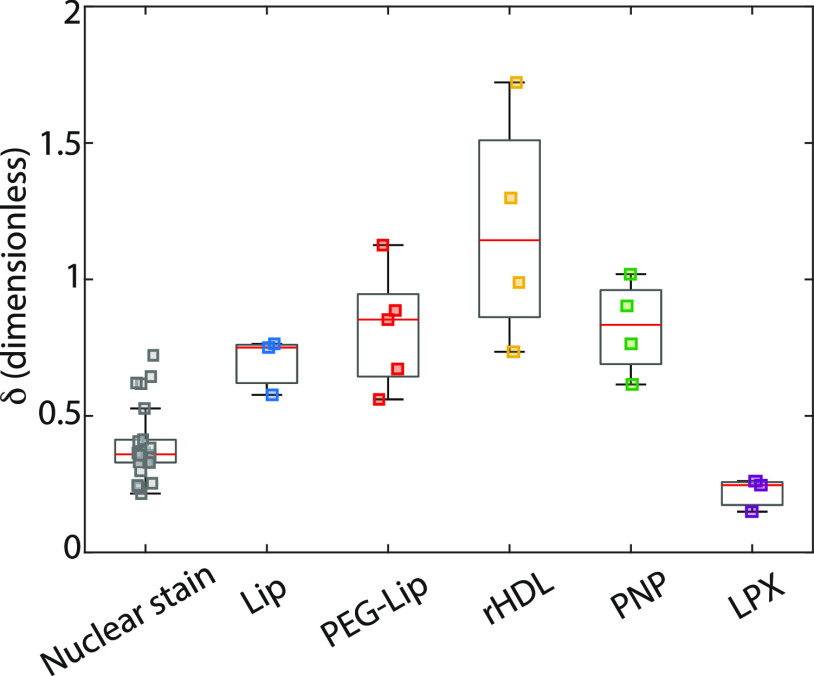
where N is the number of spheroids, ⟨I⟩out is the average NP fluorescence at shallow depths (0 μm < d < 20 μm), and ⟨I⟩in is the average intensity deeper in the spheroids (60 μm < d < 80 μm). A uniform distribution of NPs within the spheroid would yield a δ close to 1 convoluted with an exponential-like decay due to the attenuation of the laser light, etc. As the penetration efficiency δ of the nuclear stain reflects the diffusion gradient in combination with the attenuating factors, it can be used to assess the homogeneity between tumor spheroids. The consistent δ values for the nuclear stain (Figure 5) confirmed that the tumor spheroids were uniform in morphology and size. The penetration efficiency of the NPs (Figure 5) generally followed the trends observed from the cross-sections (Figure 3) and penetration profiles (Figure 4). Only LPX had lower δ than the nuclear stain and significantly lower than Lip (p < 0.04), PEG-Lip (p = 0.001), and PNP (p = 0.002), since they were mainly associated or were taken up by the outermost cells. The rHDL showed the highest penetration efficiency δ, which was significantly larger than Lip (p = 0.02), nuclear stain, and LPX (p < 0.0001, both). This is likely due to a combination of their small size, allowing them to easily diffuse through intercellular spaces of the tumor spheroids, and the surprising augmented accumulation of rHDL deeper in the spheroid.
Figure 5.
Boxplot of the penetration efficiency, δ, of nanoparticles in U87-MG tumor spheroids. The penetration efficiency is the ratio between the average fluorescence of the outermost band (0–20 μm depth) and an inner band (60–80 μm depth).
To evaluate if NPs were taken up by the cells or simply adhered to their surface, we imaged the surface cells of the spheroid individually (Figure 6). As these images show single sheets in the imaging stacks (0.4 μm thickness) inside the more than 10 μm cells, the NP fluorescence (red) measured in proximity to the nucleus (green) is likely to be internalized NPs. These results are indicative of a view where all NPs were internalized (passively or actively) but to a greater extent for rHDL and LPX compared to Lip and PEG-Lip. Evidently, LPX, rHDL, and Lip were not only taken up by the cells but they also formed aggregations that adhered to the spheroid surface (Figure 3). The effect of cell internalization on NP penetration in tumors is disputed, as it has been found to both reduce penetration due to depletion7 and to facilitate penetration by transcellular transport.73
Figure 6.
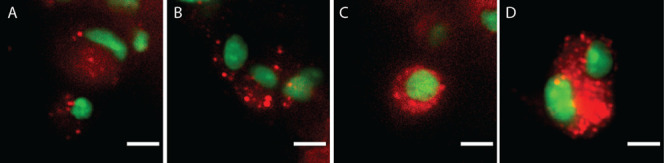
Uptake of NPs by spheroid surface cells. Representative optical sections of cells on the surface. Overlay of pseudocolored images of nuclear-stained cells (green) on the spheroids’ surface, treated with the various lipid-based NPs (red): PEG-Lip (A), Lip (B), rHDL (C), and LPX (D). Intensity and contrast levels were kept constant, and scale bars correspond to 10 μm.
Interestingly, Lip and PEG-Lip show similar uptake by outermost cells (Figure 6A,B) and have similar δ (p = 0.95), though the d1/2 indicated that PEG-Lip penetrated deeper into the spheroid. This can in part be explained by the measured intensity peak of PEG-Lip just beneath the tumor spheroid surface (Figure 4). Hence, the PEG-Lip apparently overcame the adherence to the spheroid surface and penetrated the first cell layers efficiently, while only a limited amount reached deeper into the spheroid. We speculate that their PEGylation reduces the interactions between PEG-Lip and the cell membrane74 to allow rapid diffusion through the spheroid,7,42,75 in contrast to the less negatively charged/non-PEGylated Lip and to the slightly positively charged LPX.
3.5. Effect of the NP Design on the Penetration Efficiency
The penetration efficiency is an interplay between the NP design, tumor morphology, and cell internalization sensitivity.9,12 We found that LPX exhibited a very different penetration efficiency than Lip and PEG-Lip despite their similar size (∼100 nm). This might be attributed to the positive surface charge of LPX causing fusion-based uptake by the outermost cells (Figure 6) and lower stability of the LPX. However, the importance of the surface charge has also been observed in other studies where charged NPs were shown to penetrate less efficiently than neutral NPs.49,76,77
The small differences in the sizes of rHDL and PNP (∼10 nm versus ∼24 nm) might contribute to the slightly different penetration efficiencies, as shown for other NPs.9,78 We suggest, however, that the great accumulation of rHDL inside the spheroid is attributed to effects from the different environments at the spheroid core relative to the surface, e.g., the slightly acidic environment inside the spheroid might cause rHDL aggregation or alter the interactions between the rHDL/apoA-I and the extracellular matrix and/or cells.79 Furthermore, viability of the cells might vary toward the core, thus potentially affecting the NP penetration. Interestingly, Lee et al.11 showed decreased penetration depth for NPs with targeting ligands, as the NP binding and uptake by the outermost cells inhibited further NP penetration into the spheroid. As SR-BI is expressed by the cells, the rHDL could, in principle, be actively taken up by all cells in the spheroids, thus resulting in likewise decreased accumulation toward the tumor center due to enhanced clearance by the outermost cells, compared to similar-sized nontargeting NPs. In contrast, Tang et al.30 suggested that SR-BI receptor-mediated cell uptake of HDL-mimicking NPs in the core of the tumor spheroid is important for enhanced penetration of the NPs. They based this claim on the observation that PEGylation of the HDL-mimicking NPs decreased the penetration into the tumor spheroid, which they attribute to SR-BI interactions being diminished by PEG. However, we emphasize that SR-BI is presumably expressed on all of the cells in the tumor spheroid, and we therefore suggest that additional parameters contribute to the observed increased rHDL accumulation toward the spheroid core.
The penetration efficiency of NPs is an important evaluation measure of therapeutic delivery systems that aim at distributing drugs uniformly in the tumor and must be considered together with the equally important biodistribution of NPs, which determines how much of the intravenously administered NPs end up in in vivo tumor tissue compared to other organs.10 More specifically, PEGylation of liposomes is believed to be very important for long circulation times and results in high accumulation in the tumor due to the expected enhanced permeability and the retention (EPR) effect.3,16 However, we emphasize that the relevance of the EPR effect is disputed80 and that the exact relation between circulation and biodistribution is under much debate (chapter 5 in ref (81)). We observed only slight (but not significant) difference in the penetration efficiency between Lip and PEG-Lip. In accordance with other in vitro studies, we observed higher penetration efficiency of the smaller NPs (rHDL and PNP).82 In contrast in vivo, ∼100 nm is believed to be the optimal size for uptake in tumors through the EPR effect.83 Despite their small size, intravenously injected rHDL are reported to yield relatively high tumor accumulation, often attributed to their SR-BI targeting feature.30,84−86 Our results suggest that the rHDL can be retained in the tumor, possibly as a result of the naturally occurring gradients in the cancer spheroid; hence, this might add to the explanation of the enhanced tumor accumulation of rHDL. Consequently, we highlight that the amount of NPs distributed in the tumor tissue is an interplay between NP biodistribution and penetration efficiency.
4. Conclusions
We performed a head-to-head comparison of the penetration efficiencies of lipid-based NPs that varied in size, surface charge, surface chemistry, stability, and targeting ability. Besides the direct comparison between the selected lipid-based NPs, our findings shed light on important and relevant effects on the penetration of NPs into tumors. While we only observed a limited effect on the penetration efficiency from PEGylation of liposomes, we did observe an effect of the NPs’ stability and surface charge, as indicated by the limited penetration efficiency of LPX compared to Lip/PEG-Lip. We also observed a high penetration efficiency for rHDL and speculated that rHDL’s unique penetration profile is attributed to a combination of their small size and the effects associated with the spheroid environment. It could be relevant in future experiments to thoroughly investigate the mechanisms that drive the different penetration efficiencies of the NPs.
The use of light-sheet microscopy and the detailed image analysis can be easily adapted to screen the penetration efficiency of any fluorescent NPs in 3D tumor spheroids. Therefore, the testing of NPs is only limited by the build-in optics of the light-sheet microscope, i.e., lasers and detectors. Thus, we present a powerful tool and analysis method for the in vitro evaluation of the penetration efficiency of NPs, a measure that can be highly relevant for the therapeutic effect of the corresponding drug delivery system.
Acknowledgments
L.J. acknowledges funding from the Danish National Research Council (DNRF116) and fruitful discussions with Lene Oddershede, Pand Petra Hammerlik.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c02879.
Estimation of the dynamics of the lipid-conjugated fluorophore label incorporated into the lipid-based NPs; determination of the dilution of NPs required to obtain an equal amount of fluorophore in the samples; 3D reconstruction, distribution of ellipticities and radii; optical cross-sections of a spheroid (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Wang A. Z.; Langer R.; Farokhzad O. C. Nanoparticle Delivery of Cancer Drugs. Annu. Rev. Med. 2012, 63, 185–198. 10.1146/annurev-med-040210-162544. [DOI] [PubMed] [Google Scholar]
- Puri A.; Loomis K.; Smith B.; Lee J. H.; Yavlovich A.; Heldman E.; Blumenthal R. Lipid-based nanoparticles as pharmaceutical drug carriers: From concepts to clinic. Crit. Rev. Ther. Drug Carrier Syst. 2009, 26, 523–580. 10.1615/CritRevTherDrugCarrierSyst.v26.i6.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yingchoncharoen P.; Kalinowski D. S.; Richardson D. R. Lipid-based drug delivery systems in cancer therapy: What is available and what is yet to come. Pharmacol. Rev. 2016, 68, 701–787. 10.1124/pr.115.012070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen T. M.; Cullis P. R. Drug Delivery Systems: Entering the Mainstream. Science 2004, 303, 1818–1822. 10.1126/science.1095833. [DOI] [PubMed] [Google Scholar]
- Senapati S.; Mahanta A. K.; Kumar S.; Maiti P. Controlled drug delivery vehicles for cancer treatment and their performance. Signal Transduction Targeted Ther. 2018, 3, 7 10.1038/s41392-017-0004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barenholz Y. Doxil - The first FDA-approved nano-drug: Lessons learned. J. Controlled Release 2012, 160, 117–134. 10.1016/j.jconrel.2012.03.020. [DOI] [PubMed] [Google Scholar]
- Tchoryk A.; Taresco V.; Argent R. H.; Ashford M.; Gellert P. R.; Stolnik S.; Grabowska A.; Garnett M. C. Penetration and Uptake of Nanoparticles in 3D Tumor Spheroids. Bioconjugate Chem. 2019, 30, 1371–1384. 10.1021/acs.bioconjchem.9b00136. [DOI] [PubMed] [Google Scholar]
- Li L.; Sun J.; He Z. Deep Penetration of Nanoparticulate Drug Delivery Systems into Tumors: Challenges and Solutions. Curr. Med. Chem. 2013, 20, 2881–2891. 10.2174/09298673113209990004. [DOI] [PubMed] [Google Scholar]
- Cabral H.; Matsumoto Y.; Mizuno K.; Chen Q.; Murakami M.; Kimura M.; Terada Y.; Kano M. R.; Miyazono K.; Uesaka M.; et al. Others, Accumulation of sub-100 nm polymeric micelles in poorly permeable tumours depends on size. Nat. Nanotechnol. 2011, 6, 815. 10.1038/nnano.2011.166. [DOI] [PubMed] [Google Scholar]
- Perrault S. D.; Walkey C.; Jennings T.; Fischer H. C.; Chan W. C. W. Mediating tumor targeting efficiency of nanoparticles through design. Nano Lett. 2009, 9, 1909–1915. 10.1021/nl900031y. [DOI] [PubMed] [Google Scholar]
- Lee H.; Fonge H.; Hoang B.; Reilly R. M.; Allen C. The effects of particle size and molecular targeting on the intratumoral and subcellular distribution of polymeric nanoparticles. Mol. Pharm. 2010, 7, 1195–1208. 10.1021/mp100038h. [DOI] [PubMed] [Google Scholar]
- Ernsting M. J.; Murakami M.; Roy A.; Li S.-D. Factors controlling the pharmacokinetics, biodistribution and intratumoral penetration of nanoparticles. J. Controlled Release 2013, 172, 782–794. 10.1016/j.jconrel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullis P. R.; Hope M. J. Lipid Nanoparticle Systems for Enabling Gene Therapies. Mol. Ther. 2017, 25, 1467–1475. 10.1016/j.ymthe.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen J. B. Evaluation of reconstituted high-density lipoprotein (rHDL) as a drug delivery platform-a detailed survey of rHDL particles ranging from biophysical properties to clinical implications. Nanomed.: Nanotechnol., Biol. Med. 2016, 12, 2161–2179. 10.1016/j.nano.2016.05.009. [DOI] [PubMed] [Google Scholar]
- Münter R.; Kristensen K.; Pedersbæk D.; Larsen J. B.; Simonsen J. B.; Andresen T. L. Dissociation of fluorescently labeled lipids from liposomes in biological environments challenges the interpretation of uptake studies. Nanoscale 2018, 10, 22720–22724. 10.1039/C8NR07755J. [DOI] [PubMed] [Google Scholar]
- Allen T. M. Liposomal Drug Formulations. Drugs 1998, 56, 747–756. 10.2165/00003495-199856050-00001. [DOI] [PubMed] [Google Scholar]
- Pedersbæk D.; Jønsson K.; Madsen D. V.; Weller S.; Bohn A. B.; Andresen T. L.; Simonsen J. B. A quantitative ex vivo study of the interactions between reconstituted high-density lipoproteins and human leukocytes. RSC Adv. 2020, 10, 3884–3894. 10.1039/C9RA08203D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon K. M.; Plebanek M. P.; Thaxton C. S. Properties of native high-density lipoproteins inspire synthesis of actively targeted in vivo siRNA delivery vehicles. Adv. Funct. Mater. 2016, 26, 7824–7835. 10.1002/adfm.201602600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersbæk D.; Kráemer M. K.; Kempen P. J.; Ashley J.; Braesch-Andersen S.; Andresen T. L.; Simonsen J. B. The composition of reconstituted high-density lipoproteins (rHDL) dictates the degree of rHDL cargo- and size-remodeling via direct interactions with endogenous lipoproteins. Bioconjugate Chem. 2019, 30, 2634–2646. 10.1021/acs.bioconjchem.9b00552. [DOI] [PubMed] [Google Scholar]
- Berney E.; Sabnis N.; Panchoo M.; Raut S.; Dickerman R.; Lacko A. G. The SR-B1 Receptor as a Potential Target for Treating Glioblastoma. J. Oncol. 2019, 2019, 1805841 10.1155/2019/1805841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y.; Pjesivac-Grbovic J.; Cantrell C.; Freyer J. P. A multiscale model for avascular tumor growth. Biophys. J. 2005, 89, 3884–3894. 10.1529/biophysj.105.060640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisboeck T. S.; Berens M. E.; Kansal A. R.; Torquato S.; Stemmer-Rachamimov A. O.; Chiocca E. A. Pattern of self-organization in tumour systems: complex growth dynamics in a novel brain tumour spheroid model. Cell Proliferation 2001, 34, 115–134. 10.1046/j.1365-2184.2001.00202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral R. L. F.; Miranda M.; Marcato P. D.; Swiech K. Comparative analysis of 3D bladder tumor spheroids obtained by forced floating and hanging drop methods for drug screening. Front. Physiol. 2017, 8, 605 10.3389/fphys.2017.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler F.; Obeïd P.; Rulina A. V.; Haguet V.; Gidrol X.; Balakirev M. Y. High-Content Monitoring of Drug Effects in a 3D Spheroid Model. Front. Oncol. 2017, 7, 293 10.3389/fonc.2017.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sant S.; Johnston P. A. The production of 3D tumor spheroids for cancer drug discovery. Drug Discovery Today: Technol. 2017, 23, 27–36. 10.1016/j.ddtec.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhaeuser F.; Menne H.; Dittfeld C.; West J.; Mueller-Klieser W.; Kunz-Schughart L. A. Multicellular tumor spheroids: an underestimated tool is catching up again. J. Biotechnol. 2010, 148, 3–15. 10.1016/j.jbiotec.2010.01.012. [DOI] [PubMed] [Google Scholar]
- Lazzari G.; Vinciguerra D.; Balasso A.; Nicolas V.; Goudin N.; Garfa-Traore M.; Fehér A.; Dinnyés A.; Nicolas J.; Couvreur P.; Mura S. Light sheet fluorescence microscopy versus confocal microscopy: in quest of a suitable tool to assess drug and nanomedicine penetration into multicellular tumor spheroids. Eur. J. Pharm. Biopharm. 2019, 142, 195–203. 10.1016/j.ejpb.2019.06.019. [DOI] [PubMed] [Google Scholar]
- Cutrona M. B.; Simpson J. C. A High-Throughput Automated Confocal Microscopy Platform for Quantitative Phenotyping of Nanoparticle Uptake and Transport in Spheroids. Small 2019, 15, 1902033 10.1002/smll.201902033. [DOI] [PubMed] [Google Scholar]
- Darrigues E.; Nima Z. A.; Nedosekin D. A.; Watanabe F.; Alghazali K. M.; Zharov V. P.; Biris A. S. Tracking Gold Nanorods’ Interaction with Large 3D Pancreatic-Stromal Tumor Spheroids by Multimodal Imaging: Fluorescence, Photoacoustic, and Photothermal Microscopies. Sci. Rep. 2020, 10, 3362 10.1038/s41598-020-59226-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J.; Kuai R.; Yuan W.; Drake L.; Moon J. J.; Schwendeman A. Effect of size and pegylation of liposomes and peptide-based synthetic lipoproteins on tumor targeting. Nanomed.: Nanotechnol., Biol. Med. 2017, 13, 1869–1878. 10.1016/j.nano.2017.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Wang Y.; Yang D.; Huang W.; Hao P.; Feng S.; Appelhans D.; Zhang T.; Zan X. Shape effect of nanoparticles on tumor penetration in monolayers versus spheroids. Mol. Pharm. 2019, 16, 2902–2911. 10.1021/acs.molpharmaceut.9b00107. [DOI] [PubMed] [Google Scholar]
- Mollo V.; Scognamiglio P.; Marino A.; Ciofani G.; Santoro F. Probing the Ultrastructure of Spheroids and Their Uptake of Magnetic Nanoparticles by FIB–SEM. Adv. Mater. Technol. 2020, 5, 1900687 10.1002/admt.201900687. [DOI] [Google Scholar]
- Carver K.; Ming X.; Juliano R. L. Multicellular tumor spheroids as a model for assessing delivery of oligonucleotides in three dimensions. Mol. Ther. - Nucleic Acids 2014, 3, e153 10.1038/mtna.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Dakdouki M. H.; Puré E.; Huang X. Development of drug loaded nanoparticles for tumor targeting. Part 2: Enhancement of tumor penetration through receptor mediated transcytosis in 3D tumor models. Nanoscale 2013, 5, 3904–3911. 10.1039/c3nr90022c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal R.; Jurney P.; Raythatha M.; Singh V.; Sreenivasan S. V.; Shi L.; Roy K. Effect of shape, size, and aspect ratio on nanoparticle penetration and distribution inside solid tissues using 3D spheroid models. Adv. Healthcare Mater. 2015, 4, 2269–2280. 10.1002/adhm.201500441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa A.; Jakubowski N.; Koellensperger G.; Theiner S.; Schweikert A.; Flemig S.; Iwahata D.; Traub H.; Hirata T. Quantitative Imaging of Silver Nanoparticles and Essential Elements in Thin Sections of Fibroblast Multicellular Spheroids by High Resolution Laser Ablation Inductively Coupled Plasma Time-of-Flight Mass Spectrometry. Anal. Chem. 2019, 91, 10197–10203. 10.1021/acs.analchem.9b02239. [DOI] [PubMed] [Google Scholar]
- Fleddermann J.; Susewind J.; Peuschel H.; Koch M.; Tavernaro I.; Kraegeloh A. Distribution of SiO2 nanoparticles in 3D liver microtissues. Int. J. Nanomed. 2019, 14, 1411. 10.2147/IJN.S189888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durymanov M.; Kroll C.; Permyakova A.; Reineke J. Role of Endocytosis in Nanoparticle Penetration of 3D Pancreatic Cancer Spheroids. Mol. Pharmaceutics 2019, 16, 1074–1082. 10.1021/acs.molpharmaceut.8b01078. [DOI] [PubMed] [Google Scholar]
- Olsen T. R.; Mattix B.; Casco M.; Herbst A.; Williams C.; Tarasidis A.; Evans G.; Jenkins L.; McMahan C. L.; Simionescu D.; Visconti R. P.; Alexis F. Processing cellular spheroids for histological examination. J. Histotechnol. 2014, 37, 138–142. 10.1179/2046023614Y.0000000047. [DOI] [Google Scholar]
- Lu H.; Su J.; Mamdooh R.; Li Y.; Stenzel M. H. Cellular Uptake of Gold Nanoparticles and Their Movement in 3D Multicellular Tumor Spheroids: Effect of Molecular Weight and Grafting Density of Poly(2-hydroxyl ethyl acrylate). Macromol. Biosci. 2020, 20, 1900221 10.1002/mabi.201900221. [DOI] [PubMed] [Google Scholar]
- Priwitaningrum D. L.; Blondé J.-B. G.; Sridhar A.; van Baarlen J.; Hennink W. E.; Storm G.; Le Gac S.; Prakash J. Tumor stroma-containing 3D spheroid arrays: A tool to study nanoparticle penetration. J. Controlled Release 2016, 244, 257–268. 10.1016/j.jconrel.2016.09.004. [DOI] [PubMed] [Google Scholar]
- Huang K.; Boerhan R.; Liu C.; Jiang G. Nanoparticles penetrate into the multicellular spheroid-on-chip: effect of surface charge, protein corona, and exterior flow. Mol. Pharm. 2017, 14, 4618–4627. 10.1021/acs.molpharmaceut.7b00726. [DOI] [PubMed] [Google Scholar]
- Lee J.; Lilly G. D.; Doty R. C.; Podsiadlo P.; Kotov N. A. In vitro toxicity testing of nanoparticles in 3D cell culture. Small 2009, 5, 1213–1221. 10.1002/smll.200801788. [DOI] [PubMed] [Google Scholar]
- Leite P. E. C.; Pereira M. R.; Harris G.; Pamies D.; Dos Santos L. M. G.; Granjeiro J. M.; Hogberg H. T.; Hartung T.; Smirnova L. Suitability of 3D human brain spheroid models to distinguish toxic effects of gold and poly-lactic acid nanoparticles to assess biocompatibility for brain drug delivery. Part. Fibre Toxicol. 2019, 16, 22 10.1186/s12989-019-0307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X.; Xin H.; Gu J.; Xu X.; Xia W.; Chen S.; Xie Y.; Chen L.; Chen Y.; Sha X. Solid tumor penetration by integrin-mediated pegylated poly (trimethylene carbonate) nanoparticles loaded with paclitaxel. Biomaterials 2013, 34, 1739–1746. 10.1016/j.biomaterials.2012.11.016. [DOI] [PubMed] [Google Scholar]
- Kankala R. K.; Liu C.-G.; Yang D.-Y.; Wang S.-B.; Chen A.-Z. Ultrasmall platinum nanoparticles enable deep tumor penetration and synergistic therapeutic abilities through free radical species-assisted catalysis to combat cancer multidrug resistance. Chem. Eng. J. 2020, 383, 123138 10.1016/j.cej.2019.123138. [DOI] [Google Scholar]
- Skobeltsin A.; Farrakhova D.; Maklygina Y.; Romanishkin I.; Ryabova A.; Yakovets I.; Millard M.; Bolotine L.; Plyutinskaya A.; Karmakova T. 3D spheroid cultures for evaluation of nanophotosensitizers accumulation. J. Phys.: Conf. Ser. 2020, 1439, 012032 10.1088/1742-6596/1439/1/012032. [DOI] [Google Scholar]
- Mellor H.; Davies L.; et al. Optimising non-viral gene delivery in a tumour spheroid model. J. Gene Med. 2006, 8, 1160–1170. 10.1002/jgm.947. [DOI] [PubMed] [Google Scholar]
- Kostarelos K.; Emfietzoglou D.; Papakostas A.; Yang W. H.; Ballangrud A. A.; Sgouros G. Binding and interstitial penetration of liposomes within avascular tumor spheroids. Int. J. Cancer 2004, 112, 713–721. 10.1002/ijc.20457. [DOI] [PubMed] [Google Scholar]
- Kou L.; Yao Q.; Sivaprakasam S.; Luo Q.; Sun Y.; Fu Q.; He Z.; Sun J.; Ganapathy V. Dual targeting of l-carnitine-conjugated nanoparticles to OCTN2 and ATB{0,+} to deliver chemotherapeutic agents for colon cancer therapy. Drug Delivery 2017, 24, 1338–1349. 10.1080/10717544.2017.1377316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess M. W.; Pfaller K.; Ebner H. L.; Beer B.; Hekl D.; Seppi T.. 3D Versus 2D Cell Culture. In Methods in Cell Biology; Elsevier, 2010; Vol. 96, pp 649–670. [DOI] [PubMed] [Google Scholar]
- Kiernan J. A. Formaldehyde, Formalin, Paraformaldehyde And Glutaraldehyde: What They Are And What They Do. Microsc. Today 2000, 8, 8–13. 10.1017/S1551929500057060. [DOI] [Google Scholar]
- Williams Y.; Byrne S.; Bashir M.; Davies A.; Whelan A.; Gunko Y.; Kelleher D.; Volkov Y. Comparison of three cell fixation methods for high content analysis assays utilizing quantum dots. J. Microsc. 2008, 232, 91–98. 10.1111/j.1365-2818.2008.02083.x. [DOI] [PubMed] [Google Scholar]
- He Y.; Laugesen K.; Kamp D.; Sultan S. A.; Oddershede L. B.; Jauffred L. Effects and side effects of plasmonic photothermal therapy in brain tissue. Cancer Nanotechnol. 2019, 10, 8 10.1186/s12645-019-0053-0. [DOI] [Google Scholar]
- Tomer R.; Khairy K.; Amat F.; Keller P. J. Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy.. Nat. Methods 2012, 9, 755–63. 10.1038/nmeth.2062. [DOI] [PubMed] [Google Scholar]
- Vettenburg T.; Dalgarno H. I. C.; Nylk J.; Coll-Lladó C.; Ferrier D. E. K.; Čižmár T.; Gunn-Moore F. J.; Dholakia K. Light-sheet microscopy using an Airy beam. Nat. Methods 2014, 11, 541–544. 10.1038/nmeth.2922. [DOI] [PubMed] [Google Scholar]
- Andilla J.; Jorand R.; Olarte O. E.; Dufour A. C.; Cazales M.; Montagner Y. L. E.; Ceolato R.; Riviere N.; Olivo-Marin J.-C.; Loza-Alvarez P.; Lorenzo C. Imaging tissue-mimic with light sheet microscopy: A comparative guideline. Sci. Rep. 2017, 7, 44939 10.1038/srep44939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verveer P. J.; Swoger J.; Pampaloni F.; Greger K.; Marcello M.; Stelzer E. H. High-resolution three-dimensional imaging of large specimens with light sheet-based microscopy. Nat. Methods 2007, 4, 311–313. 10.1038/nmeth1017. [DOI] [PubMed] [Google Scholar]
- Hubbard K. S.; Gut I. M.; Scheeler S. M.; Lyman M. E.; McNutt P. M. Compatibility of SYTO 13 and Hoechst 33342 for longitudinal imaging of neuron viability and cell death. BMC Res. Notes 2012, 5, 437 10.1186/1756-0500-5-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhans S. A. Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front. Pharmacol. 2018, 9, 6 10.3389/fphar.2018.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinci M.; Gowan S.; Boxall F.; Patterson L.; Zimmermann M.; Court W.; Lomas C.; Mendiola M.; Hardisson D.; Eccles S. A. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012, 10, 29 10.1186/1741-7007-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell H. S.; Whittle I. R.; Walker M.; Leaver H. A.; Wharton S. B. The development of necrosis and apoptosis in glioma: Experimental findings using spheroid culture systems. Neuropathol. Appl. Neurobiol. 2001, 27, 291–304. 10.1046/j.0305-1846.2001.00319.x. [DOI] [PubMed] [Google Scholar]
- Münter R.; Kristensen K.; Pedersbæk D.; Andresen T. L.; Simonsen J. B.; Larsen J. B.. Nanotechnology Characterization Tools for Tissue Engineering and Medical Therapy; Springer: Berlin, Heidelberg, 2019; pp 333–359. [Google Scholar]
- Semple S. C.; Klimuk S. K.; Harasym T. O.; Dos Santos N.; Ansell S. M.; Wong K. F.; Maurer N.; Stark H.; Cullis P. R.; Hope M. J.; Scherrer P. Efficient encapsulation of antisense oligonucleotides in lipid vesicles using ionizable aminolipids: Formation of novel small multilamellar vesicle structures. Biochim. Biophys. Acta, Biomembr. 2001, 1510, 152–166. 10.1016/S0005-2736(00)00343-6. [DOI] [PubMed] [Google Scholar]
- Leung A. K.; Tam Y. Y. C.; Cullis P. R.. Advances in Genetics; Elsevier, 2014; Vol. 88, pp 71–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland J. W.; Hui C.; Cullis P. R.; Madden T. D. Poly(ethylene glycol)-lipid conjugates regulate the calcium-induced fusion of liposomes composed of phosphatidylethanolamine and phosphatidylserine. Biochemistry 1996, 35, 2618–2624. 10.1021/bi952000v. [DOI] [PubMed] [Google Scholar]
- Pattni B. S.; Chupin V. V.; Torchilin V. P. New developments in liposomal drug delivery. Chem. Rev. 2015, 115, 10938–10966. 10.1021/acs.chemrev.5b00046. [DOI] [PubMed] [Google Scholar]
- Bendix P. M.; Reihani S. N. S.; Oddershede L. B. Direct Measurements of Heating by Electromagnetically Trapped Gold Bilayers. ACS Nano 2010, 4, 2256–2262. 10.1021/nn901751w. [DOI] [PubMed] [Google Scholar]
- Cavigiolio G.; Shao B.; Geier E. G.; Ren G.; Heinecke J. W.; Oda M. N. The interplay between size, morphology, stability, and functionality of high-density lipoprotein subclasses. Biochemistry 2008, 47, 4770–4779. 10.1021/bi7023354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. S.; Kang Y.; Son K. H.; Choi S. M.; Kim K. Y. Manufacturing and shelf stability of reconstituted high-density lipoprotein for infusion therapy. Biotechnol. Bioprocess Eng. 2011, 16, 785–792. 10.1007/s12257-010-0421-4. [DOI] [Google Scholar]
- Nunes A. S.; Barros A. S.; Costa E. C.; Moreira A. F.; Correia I. J. 3D tumor spheroids as in vitro models to mimic in vivo human solid tumors resistance to therapeutic drugs. Biotechnol. Bioeng. 2019, 116, 206–226. 10.1002/bit.26845. [DOI] [PubMed] [Google Scholar]
- Nguyen S. D.; Öörni K.; Lee-Rueckert M.; Pihlajamaa T.; Metso J.; Jauhiainen M.; Kovanen P. T. Spontaneous remodeling of HDL particles at acidic pH enhances their capacity to induce cholesterol efflux from human macrophage foam cells. J. Lipid Res. 2012, 53, 2115–2125. 10.1194/jlr.M028118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H.; Stenzel M. H. Multicellular Tumor Spheroids (MCTS) as a 3D In Vitro Evaluation Tool of Nanoparticles. Small 2018, 14, 1702858 10.1002/smll.201702858. [DOI] [PubMed] [Google Scholar]
- Torchilin V. P.; Klibanov A. L.; Huang L.; O’Donnell S.; Nossiff N. D.; Khaw B. A. Targeted accumulation of polyethylene glycol-coated immunoliposomes in infarcted rabbit myocardium. FASEB J. 1992, 6, 2716–2719. 10.1096/fasebj.6.9.1612296. [DOI] [PubMed] [Google Scholar]
- Kim B.; Han G.; Toley B. J.; Kim C.-k.; Rotello V. M.; Forbes N. S. Tuning payload delivery in tumour cylindroids using gold nanoparticles. Nat. Nanotechnol. 2010, 5, 465. 10.1038/nnano.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieleg O.; Baumgärtel R. M.; Bausch A. R. Selective filtering of particles by the extracellular matrix: An electrostatic bandpass. Biophys. J. 2009, 97, 1569–1577. 10.1016/j.bpj.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T.; Koreeda N.; Yamashita F.; Takakura Y.; Hashida M. Effect of particle size and charge on the disposition of lipid carriers after intratumoral injection into tissue-isolated tumors. Pharm. Res. 1998, 15, 128–132. 10.1023/A:1011921324952. [DOI] [PubMed] [Google Scholar]
- Albanese A.; Lam A. K.; Sykes E. A.; Rocheleau J. V.; Chan W. C. W. Tumour-on-a-chip provides an optical window into nanoparticle tissue transport. Nat. Commun. 2013, 4, 2718 10.1038/ncomms3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramella N. A.; Rimoldi O. J.; Prieto E. D.; Schinella G. R.; Sanchez S. A.; Jaureguiberry M. S.; Vela M. E.; Ferreira S. T.; Tricerri M. A. Human apolipoprotein A-I-derived amyloid: Its association with atherosclerosis. PLoS One 2011, 6, e22532 10.1371/journal.pone.0022532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindhwani S.; Syed A. M.; Ngai J.; Kingston B. R.; Maiorino L.; Rothschild J.; MacMillan P.; Zhang Y.; Rajesh N. U.; Hoang T. The entry of nanoparticles into solid tumours. Nat. Mater. 2020, 19, 566–575. 10.1038/s41563-019-0566-2. [DOI] [PubMed] [Google Scholar]
- Jauffred L.; Samadi A.; Klingberg H.; Bendix P. M.; Oddershede L. B. Plasmonic Heating of Nanostructures. Chem. Rev. 2019, 119, 8087–8130. 10.1021/acs.chemrev.8b00738. [DOI] [PubMed] [Google Scholar]
- Schleh C.; Semmler-Behnke M.; Lipka J.; Wenk A.; Hirn S.; Schäffler M.; Schmid G.; Simon U.; Kreyling W. G. Size and surface charge of gold nanoparticles determine absorption across intestinal barriers and accumulation in secondary target organs after oral administration. Nanotoxicology 2012, 6, 36–46. 10.3109/17435390.2011.552811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.-D.; Huang L.; Nanoparticles B. Pharmacokinetics and biodistribution of nanoparticles. Mol. Pharm. 2008, 5, 496–504. 10.1021/mp800049w. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Chen J.; Ding L.; Jin H.; Lovell J. F.; Corbin I. R.; Cao W.; Lo P. C.; Yang M.; Tsao M. S.; Luo Q.; Zheng G. HDL-mimicking peptide-lipid nanoparticles with improved tumor targeting. Small 2010, 6, 430–437. 10.1002/smll.200901515. [DOI] [PubMed] [Google Scholar]
- Perez-Medina C.; Tang J.; Abdel-Atti D.; Hogstad B.; Merad M.; Fisher E. A.; Fayad Z. A.; Lewis J. S.; Mulder W. J. M.; Reiner T. PET Imaging of Tumor-Associated Macrophages with 89Zr-Labeled High-Density Lipoprotein Nanoparticles. J. Nucl. Med. 2015, 56, 1272–1277. 10.2967/jnumed.115.158956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuai R.; Subramanian C.; White P. T.; Timmermann B.; Moon J. J.; Cohen M.; Schwendeman A. Synthetic high-density lipoprotein nanodisks for targeted withalongolide delivery to adrenocortical carcinoma. Int. J. Nanomed. 2017, 12, 6581–6594. 10.2147/IJN.S140591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.