Abstract
Collagen, the most abundant protein in mammals, possesses notable cohesion and elasticity properties and efficiently induces tissue regeneration. The Gly-Pro-Hyp canonical tripeptide repeating unit of the collagen superhelix has been well-characterized. However, to date, the shortest tripeptide repeat demonstrated to attain a helical conformation contained 3–10 peptide repeats. Here, taking a minimalistic approach, we studied a single repeating unit of collagen in its protected form, Fmoc-Gly-Pro-Hyp. The peptide formed single crystals displaying left-handed polyproline II superhelical packing, as in the native collagen single strand. The crystalline assemblies also display head-to-tail H-bond interactions and an “aromatic zipper” arrangement at the molecular interface. The coassembly of this tripeptide, with Fmoc-Phe-Phe, a well-studied dipeptide hydrogelator, produced twisted helical fibrils with a polyproline II conformation and improved hydrogel mechanical rigidity. The design of these peptides illustrates the possibility to assemble superhelical nanostructures from minimal collagen-inspired peptides with their potential use as functional motifs to introduce a polyproline II conformation into hybrid hydrogel assemblies.
Keywords: collagen-inspired, polyproline II helix, single crystal, coassembly, hydrogel
Collagen is the most abundant protein in the human body, endowed with a range of important properties including cohesion, elasticity, and regeneration of connective tissues.1,2 Native collagen is composed of extended left-handed polyproline II triple helix polypeptide chains, arranged to form a right-handed superhelix.2,3 The triple helical domains of collagens are rich in glycine (Gly), proline (Pro), and hydroxyproline (Hyp), thus polypeptides of Gly-Aaa-Aaa sequences that fold into triple helical structures have been extensively studied. The structural analysis of poly(Pro-Pro-Gly), reported by Yonath and Traub in 1969, closely resembles the basic characteristics of the collagen structure, as previously proposed by the Rich and Crick model of collagen II.4,5 X-ray diffraction analysis of (Pro-Pro-Gly)10 peptide crystals subsequently reported by Okuyama et al. demonstrated clear evidence of helical symmetry.6,7 Many sequential polytripeptides and oligopeptides composed of collagen-like Gly-Aaa-Aaa sequences have subsequently been widely reported. Some exhibited self-assembly capabilities and thus demonstrated potential use for collagen mimetic peptides in biomedical applications.8−12 However, to date, the shortest Gly-Aaa-Aaa tripeptide repeats shown to attain a helical conformation have contained 3–10 peptide repeats. These results imply that identifying the minimal peptide sequence able to adopt a polyproline II helical conformation is of great interest.
Supramolecular nanoassemblies designed and fabricated with noncoded amino acids offer a wide range of chemical diversity, biocompatibility, and biodegradability but are often limited by the chemical nature of the building blocks, resulting in mechanically inferior materials. Designing short collagen mimetic oligopeptides with the aim of developing functional supramolecular assemblies often depends on a trial-and-error approach of shuffling the positions of proline and hydroxyproline while keeping the Gly-Aaa-Aaa tripeptide backbone intact.8−12 Several studies have reported that tripeptides with proline or hydroxyproline in the backbone have the highest propensity to aggregate and could even achieve a superhelical assembly with mechanical rigidity comparable to that of collagen fibers.13,14 However, these short peptide assemblies did not possess the characteristic canonical polyproline II architecture of collagen single strands.
Protein and peptide-based hydrogels possess unique biofunctionality, biocompatibility, and biodegradability that make them suitable for a wide range of applications. Among others, these include their use as scaffolds, for wound healing and tissue engineering, encapsulation and slow release of drugs and biomolecules, serving as templates for nanofabrication, and as catalysts for organic reactions.15−21 A prominent example is fluorenylmethoxycarbonyl-Phe-Phe (Fmoc-Phe-Phe), which forms a rigid biocompatible self-assembled scaffold, consisting of a fibrous network that is stable across a broad range of pH conditions and temperatures.22,23 Furthermore, coassembly of two different building blocks into one ordered structure demonstrated improvements in the mechanical properties, stability, and biofunctionality as compared to hydrogels assembled from each of the individual components.24−31
This study describes the self-assembly of Fmoc-Gly-Pro-Hyp, a single collagen tripeptide repeating unit with N-terminus modification. The X-ray single-crystal structure of the peptide demonstrated left-handed polyproline II superhelical packing with head-to-tail H-bond interactions and an “aromatic zipper” arrangement at the molecular interface. Moreover, the coassembly of Fmoc-Gly-Pro-Hyp with the Fmoc-Phe-Phe peptide formed a hybrid hydrogel with distinctive properties, different from those of the Fmoc-Phe-Phe hydrogel. Microscopic and spectroscopic analysis demonstrated that the coassembled hydrogel forms twisted fibrils with polyproline II helicity to produce improved mechanical properties. Thus, incorporation of Fmoc-Gly-Pro-Hyp induced structural and conformational changes, resulting in a hybrid hydrogel displaying collagen polyproline II features. Thus, we achieve a polyproline II conformation by a simple coassembly of a noncollagenous hydrogel building block. These findings can be utilized to develop potential structural biomaterials with desirable properties.
Results and Discussion
Design of a Collagen-Inspired Peptide
The high abundance of the Gly-Pro-Hyp triplet in native collagen sequences led us to explore the ability of this short tripeptide to self-assemble (Scheme 1). However, the tripeptide is highly soluble in water and does not form crystals or self-assemble to form ordered nanostructures. Inspired by the high propensity of Fmoc-protected single amino acids and short peptides to self-assemble as a result of π-stacking interactions between the Fmoc moieties,22,23,32 we further modified the tripeptide to produce Fmoc-Gly-Pro-Hyp (Scheme 1).
Scheme 1. Minimalistic Approach Designed to Develop Collagen-Mimicking Scaffolds.
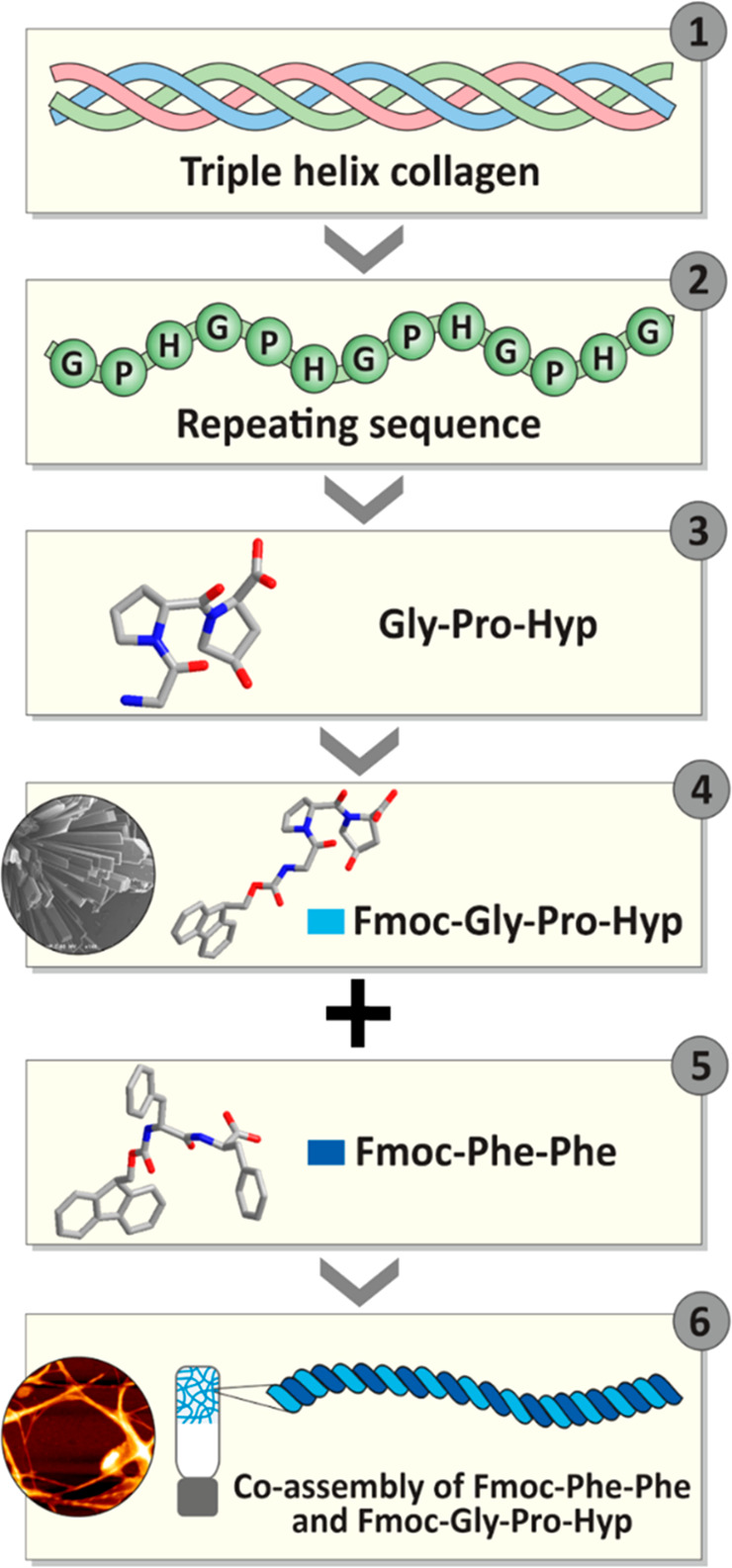
Schematic illustration of the structural design of a collagen-mimicking minimal tripeptide and its coassembly to form a rigid hydrogel with a twisted architecture showing (1) collagen triplehelix, (2) tripeptide repeating sequence in each strand of the triple helix, (3) minimal repeating sequence Gly-Pro-Hyp in a collagen helix, (4) a single crystal unit of Fmoc-modified Gly-Pro-Hyp revealing the presence of a polyproline helix II conformation, (5) Fmoc-Phe-Phe, the rigid hydrogelator, coassembled with Fmoc-Gly-Pro-Hyp, (6) development of a coassembled hybrid hydrogel, Fmoc-Phe-Phe:Fmoc-Gly-Pro-Hyp, displaying a polyproline helix II conformation and twisted helical fibrils.
Crystallization and Structural Characterization of Fmoc-Gly-Pro-Hyp
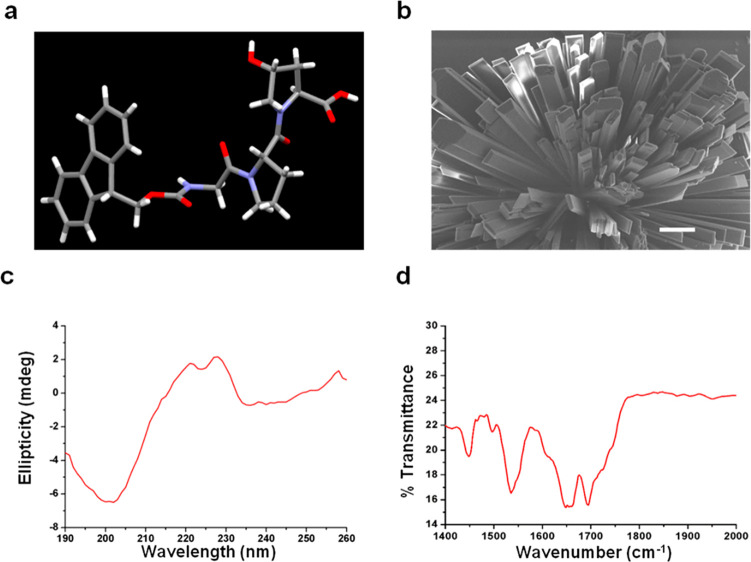
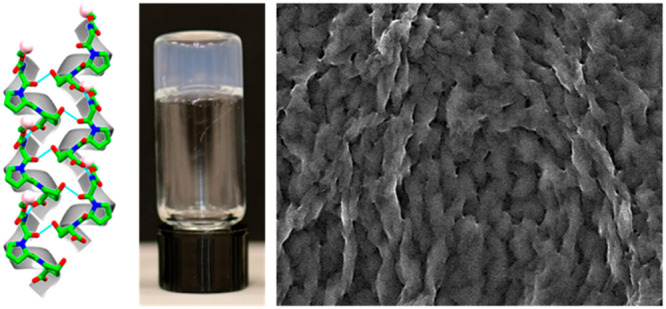
The Fmoc-Gly-Pro-Hyp formed crystals in a 2:1 MeOH/water solvent mixture at room temperature (Figure 1a,b). X-ray analysis of the molecular orientation and packing of the crystal structure of a single crystal of Fmoc-Gly-Pro-Hyp (Figure S1a) revealed one tripeptide molecule per asymmetric unit (Figure 1a and Table S1), with a trans orientation of all three peptide bonds (Figure 1a). The crystal displayed an orthorhombic space group P212121 with a = 9.4887 Å, b = 9.8639 Å, c = 26.3497 Å, and α = β = γ = 90°. The torsion angles calculated around the Fmoc-Gly peptide residue gave values for the φ1 and ψ1 angles of −69.21 and +136.24°, followed by −66.2 and +150.3° for the Gly-Pro residue (Figure 1a). These results suggest the presence of a left-handed polyproline II or collagen single-strand helix conformation, as observed in the Ramachandran plot (Figure S1b).
Figure 1.
Single-crystal of Fmoc-Gly-Pro-Hyp in MeOH/water solvent. (a) Peptide asymmetric unit. (b) Scanning electron microscopy image of the Fmoc-Gly-Pro-Hyp crystal. Scale bar: 10 μm. (c) Circular dichroism spectrum of the Fmoc-Gly-Pro-Hyp crystal suspension. (d) Fourier transform infrared spectrum of the Fmoc-Gly-Pro-Hyp crystal suspension.
Polyproline II helices have been reported to exhibit distinct spectral features in circular dichroism (CD) spectroscopy, with a negative peak around 200 nm and a positive peak around 220–230 nm.2,10 CD investigation of the secondary structure of the Fmoc-Gly-Pro-Hyp crystal suspension indeed revealed a negative maximum at ∼202 nm and a positive broad peak in the range of 220–230 nm (Figure 1c),2,10 confirming a polyproline II helical structural arrangement. Fourier transform infrared (FTIR) analysis displayed an amide I band at ∼1650 cm–1 and a shoulder at 1692 cm–1, findings in accordance with the presence of a predominantly polyproline II helical conformation, supported by a β-turn and carbamate moiety (Figure 1d).
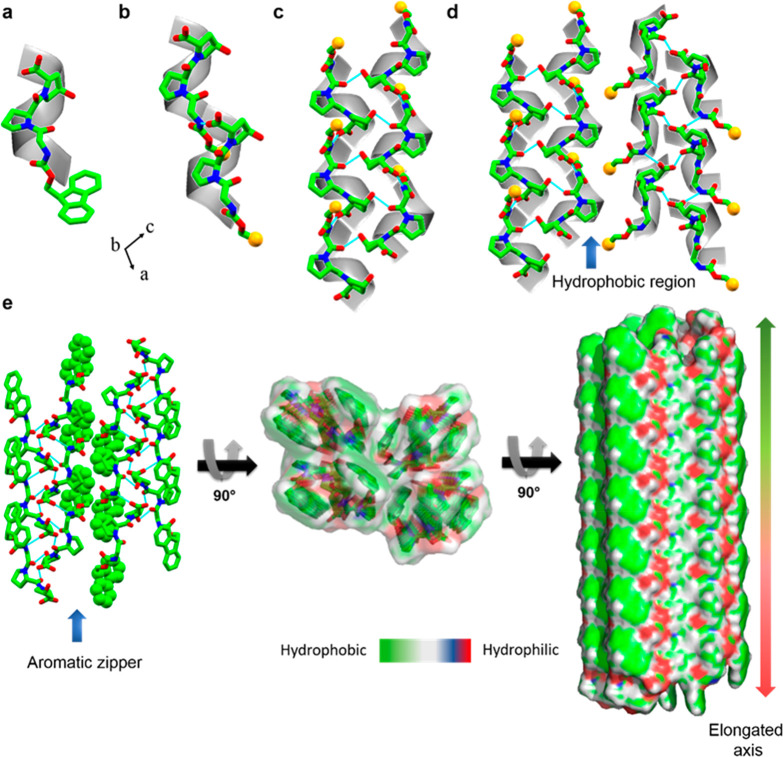
The molecular packing and higher-order organization of Fmoc-Gly-Pro-Hyp were assessed in a single crystal structure. Two types of H-bonds dictated the overall molecular arrangement: head-to-tail and side-by-side stacking. In the crystallographic a-direction, individual helical subunits (Figure 2a) were connected through head-to-tail H-bonding between the −COOH group of the terminal Hyp residue of one molecule and the −NH group of the Gly residue of the next molecule (N1–H1···O5),14,33 thus producing a single helical strand (Figure 2b). Proline rings generally adopt either a down puckering (Cγ-endo) or an up-puckering (Cγ-exo) conformation,34,35 whereas Hyp always shows a strong propensity to adopt an up-puckering conformation in the collagen triple helix structure.35,36 In the Fmoc-Gly-Pro-Hyp structure, both the Pro and Hyp residues adopted an up-puckering conformation with average χ1 (N–Cα–Cβ–Cγ) values of −31.41 and −30.20, respectively, in line with the preference for a collagen-like structure. Individual helical strands were connected to two similar adjacent helical strands through H-bond donation via the central amide groups, as shown from two different viewpoints in Figure 2c and Figures S2 and S3. These two interactions were mediated by one amide carbonyl group connecting with the −COOH group of the next molecule (O4···H6–O6) and the other interacting with the −OH part of the Hyp residue of another molecule (O3···H7–O7). Thus, the properties required to interact with adjacent helixes in order to fabricate a higher-order organization remain conserved in the short tripeptide module.36,37 Notably, this packing pattern is quite different from the sheet-like structure observed for the tripeptide, Boc-Pro-Hyp-Gly-OBzl, the terminal-protected other minimal repeating unit found in collagen.38 The differences in the molecular organization probably result from the major contribution of different terminal-protected groups. The asymmetric unit of Fmoc-Gly-Pro-Hyp does not include solvent molecules, with the −OH group playing a different role from that in previously reported structures.39,40 In the b-direction, the trimeric units interact through similar H-bonds to produce helical sheets and thus facilitate the lateral growth of the structure (Figure S3). In the crystallographic ac-plane, the helical sheets are arranged such that the centers of the sheets are dominated by hydrophilic moieties, whereas the surfaces are decorated with the hydrophobic Fmoc (Figure 2d).
Figure 2.
Single-crystal structure of Fmoc-Gly-Pro-Hyp. (a) Asymmetric unit. (b) Head-to-tail H-bonded elongation of a single helical chain along the crystallographic a-direction. (c) Side-by-side H-bond connection of a single helical chain with two nearby chains as viewed in the ac-plane. (d) Hydrophobic interaction between two nearby pairs of helical sheets. (e) Dry aromatic “zipper-like” stabilization of a pair of sheets (left), a top view (middle), and the formation of the elongated structure via alternative hydrophobic and hydrophilic interactions (right). For clear visualization, the peptide helix is superimposed over an ideal helical model in a–d. For clarity, the Fmoc group has been replaced by a yellow sphere in b–d. Nitrogen and oxygen heteroatoms are shown in blue and red, respectively.
Two adjacent helical sheets are separated from each other by a hydrophobic region. The dry aromatic “zipper-like” interactions between the Fmoc groups of two nearby sheets stabilize the higher-order arrangement and fabricate a layer-by-layer structure (Figure 2d,e). Moreover, the lack of solvent layers between the helical sheets means that the structure can grow along the a and b-axes in both directions because the stacking distances are comparable. The elongation along the helical axis is represented in Figure 2e, with the hydrophobic and hydrophilic regions marked by different colors.
Formation of Hybrid Hydrogels
The Fmoc-Phe-Phe peptide has previously been reported to form a fibrous hydrogel under physiological conditions.22,23 We have recently demonstrated the ability of various Fmoc-protected amino acids and biopolymers to coassemble with Fmoc-Phe-Phe to produce hybrid hydrogels with adjustable mechanical properties and with higher stability and better biofunctionality than any of the individual component building blocks.25,26,41−44 Motivated by our previous work, here, we coassembled the Fmoc-Gly-Pro-Hyp peptide with Fmoc-Phe-Phe to form a hydrogel that combines the properties of the two peptides (Scheme 1).
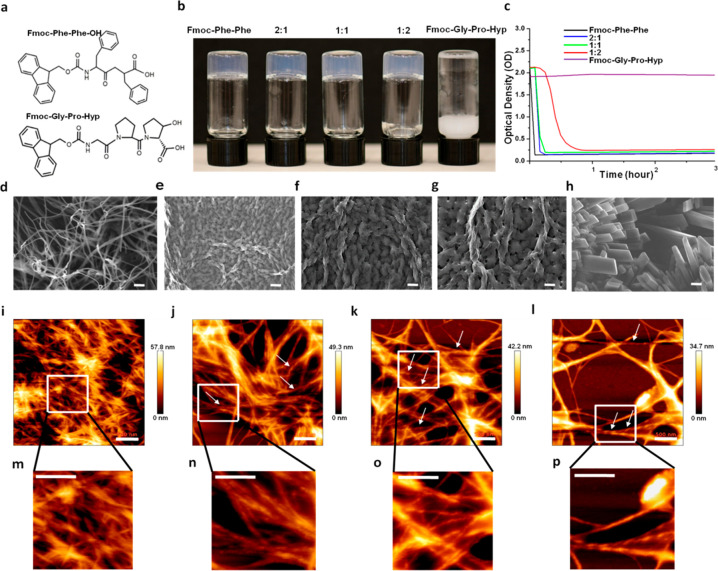
Fmoc-Phe-Phe and Fmoc-Gly-Pro-Hyp (Figure 3a) were mixed 2:1, 1:1, and 1:2 by the solvent switch method in 5% DMSO solution in water. Inverted vials containing the resultant hybrid and the individual components are shown in Figure 3b, demonstrating the formation of stable transparent hydrogels. The gelation time decreased in a concentration-dependent manner as the Fmoc-Phe-Phe concentration increased. To quantify the kinetics of the coassembly process, we monitored the absorbance spectra of the different hybrid hydrogels over time at 400 nm. Whereas the absorbance of the Fmoc-Phe-Phe solution decreased within 5 min, to reach an optical density (OD) of 0.125, the hybrid hydrogels showed slower kinetic profiles, with the 2:1 and 1:1 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp hybrids forming a clear hydrogel and reaching OD values of 0.125 and 0.186 after 10 and ∼15 min, respectively (Figure 3c). In contrast, the 1:2 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp hybrid required a significantly longer time of ∼75 min to form a clear hydrogel and to reach an OD of 0.26 (Figure 3c). Pristine Fmoc-Gly-Pro-Hyp did not form a gel under these conditions, and samples remained turbid until the appearance of white clusters that precipitated from the solution after 1 day. Scanning electron microscopy (SEM) and powder X-ray diffraction (PXRD) indicated these clusters to be Fmoc-Gly-Pro-Hyp crystals as they displayed a peak pattern similar to that of Fmoc-Gly-Pro-Hyp crystals obtained from the MeOH/water solvent (Figure S4a). Crystals observed in the 1:2 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp hybrid hydrogel after 5 days also displayed features similar to those of Fmoc-Gly-Pro-Hyp crystals as analyzed by SEM and PXRD (Figure S4b,c). It can be assumed that excess Fmoc-Gly-Pro-Hyp building blocks that did not coassemble with Fmoc-Phe-Phe were deposited as crystals within the hydrogel.
Figure 3.
Coassembled hydrogel of Fmoc-Phe-Phe and Fmoc-Gly-Pro-Hyp. (a) Molecular structure of the two Fmoc-Phe-Phe building blocks. (b) Inverted vials of the pristine and hybrid hydrogels. (c) Turbidity changes in the hydrogels over time (absorbance at 400 nm). SEM images of the studied hydrogels. (d) Fmoc-Phe-Phe, (e) 2:1 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp, (f) 1:1 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp, (g) 1:2 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp, (h) Fmoc-Gly-Pro-Hyp. Scale bar is 10 μm. Atomic force microscopy images of the studied hydrogels. (i) Fmoc-Phe-Phe, (j) 2:1 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp, (k) 1:1 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp, (l) 1:2 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp. Scale bar is 500 nm. Magnified insets of the fibrils in (m) Fmoc-Phe-Phe and twisted fibrils in (n) 2:1 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp, (o) 1:1 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp, and (p) 1:2 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp. Scale bar is 500 nm.
Ultrastructural Analysis of the Hybrid Hydrogels
SEM analysis was also used to examine the ultrastructural morphology of the hybrid hydrogels. Pristine Fmoc-Phe-Phe hydrogels, as well as 2:1, 1:1, and 1:2 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp hybrid hydrogels, were observed to comprise several micrometer long entangled fibrils (Figure 3d–g). Interestingly, the helical fibrils of all three hybrid hydrogels were thicker in diameter than the Fmoc-Phe-Phe fibrils, which formed long nonhelical structures (Figure 3d), whereas pristine Fmoc-Gly-Pro-Hyp formed crystals in flower-like clusters in a spherical arrangement (Figure 3h and Figure S4a). The helical fibrils observed in the hybrid hydrogels were further analyzed by atomic force microscopy (AFM). Whereas Fmoc-Phe-Phe formed several micrometer long nonhelical entangled fibrils, ∼10–20 nm in diameter (Figure 3i and Figure S5a), the three hybrid hydrogels contained several micrometer long twisted helical fibrils, ∼30–40 nm in diameter, as shown in the histogram plots of the fibrils (Figure 3j–l and Figure S5b–d). The formation of a bundled entangled network may explain the thicker fibrils observed by SEM in the hybrid hydrogels. Fibrils in the hybrid hydrogel displayed a morphology different than those in the individual Fmoc-Phe-Phe and Fmoc-Gly-Pro-Hyp components. We hypothesize that the presence of the Fmoc moiety in the two peptides facilitates π-stacking interactions and plays a crucial role in producing and stabilizing a coassembled hybrid material with the characteristic twisted fibrillar features.
Secondary Structure Analysis of the Hybrid Hydrogels
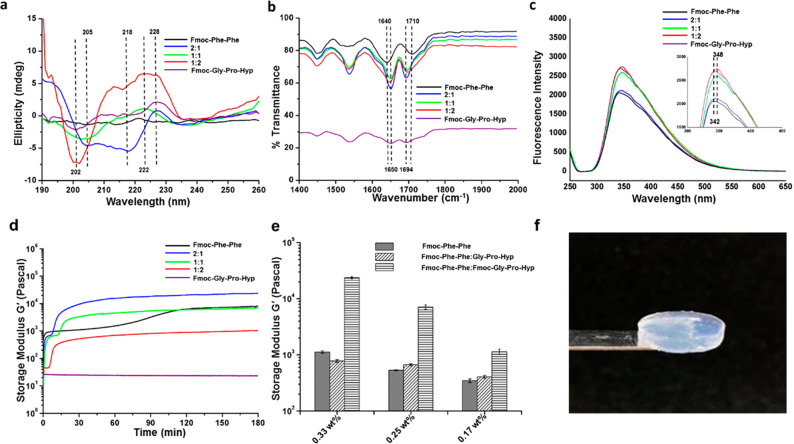
The secondary structure of the helical fibrils observed in the coassembled gels was characterized by CD spectroscopy. The 2:1 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp exhibited an α-helical pattern with negative peaks at 204 and 218 nm, both shifted by 4 nm from the typical α-helical peaks of 208 and 222 nm, respectively (Figure 4a). As the proportion of Fmoc-Gly-Pro-Hyp increased in the hybrid hydrogels, the CD spectroscopic patterns differed even more from a typical α-helix. Well-defined negative peaks at ∼200 nm and a positive peak at ∼220–230 nm were observed for the 1:1 and 1:2 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp hybrids, which is a characteristic pattern previously reported for the polyproline II helix conformation (Figure 4a).2,45,46 The pure Fmoc-Phe-Phe gel showed no such helical pattern, although pure Fmoc-Gly-Pro-Hyp in DMSO/water did exhibit the polyproline II helix pattern (Figure 4a). Thus, CD spectroscopic analysis confirms that the hybrid hydrogels attain a secondary helical structure that changes from an α-helix to a typical polyproline-II-like pattern as the fraction of Fmoc-Gly-Pro-Hyp increases, probably reflecting the contribution of Fmoc-Gly-Pro-Hyp to the secondary structure. We studied the thermal transition of the hybrid hydrogels from 5 to 90 °C by CD spectroscopy by monitoring the change in the polyproline II conformation positive peak at 227 nm (Figure S6). The 2:1 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp hybrid showed no change in the peak intensity over the temperature range, whereas for the 1:1 and 1:2 hybrids, the 227 nm peak showed a transition from positive to negative intensity at 35 and 42 °C, respectively, in further agreement with the polyproline II pattern in the hybrids.2,46,47
Figure 4.
Physicochemical characterization of the hydrogels. (a) CD spectra of the hydrogels. (b) FTIR spectra of the hydrogels. (c) Fluorescence emission spectra of the fluorenyl moiety of the multicomponent hydrogels at an excitation wavelength of 280 nm 3 h after gel formation (inset shows 6 nm shift in fluorescence spectra). (d) In situ time sweep oscillation measurements of hydrogel formation by pure Fmoc-Phe-Phe, 2:1, 1:1, and 1:2 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp, and Fmoc-Gly-Pro-Hyp. (e) Comparative study of the end point mechanical rigidity of the different types of hybrid hydrogels obtained from three repeats of experiment. (f) 2:1 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp self-supporting rigid hydrogel.
Following the CD spectroscopic analysis, we used FTIR to expand our understanding of the secondary structure of the hydrogels. The amide I region of the FTIR transmittance spectra of all hybrid hydrogels showed a distinct peak at ∼1650 cm–1 and a sharp peak at 1694 cm–1, suggesting that the hybrid hydrogels are rich in α-helix or polyproline II conformations (Figure 4b).48,49 In contrast, the spectra of pure Fmoc-Phe-Phe revealed a β-sheet-rich structure with peaks at 1640 cm–1 and a hump at 1690–1710 cm–1, reflecting the π–π stacking interactions between the Fmoc moieties and the presence of a carbamate moiety (Figure 4b).25,49 FTIR analysis of pure Fmoc-Gly-Pro-Hyp crystals indicated a polyproline II conformation, with a peak at 1650 cm–1. Thus, CD and FTIR analyses confirm the polyproline II helix conformation of the hybrid hydrogels as a result of the Fmoc-Gly-Pro-Hyp contribution. CD and FTIR analysis were also used to compare the secondary structures of type I collagen and the coassembled hydrogels (Figure S7). The CD spectrum of collagen showed the characteristic negative maximum at ∼202 nm and a positive broad peak in the range of 220–230 nm, indicative of a polyproline II helical conformation. The amide I band at ∼1650 cm–1 in FTIR analysis further implied the presence of a predominantly polyproline II helical arrangement in collagen. The results from the designed coassembled hydrogels are therefore very similar to those produced by the polyproline II conformation in collagen.
Spectroscopic and Rheological Characterization
The coassembled hybrid hydrogels were also analyzed by fluorescence spectroscopy. Photoluminescence analysis of the hydrogel formation was performed at two different time points, at the initiation of gelation (time point 0) and 180 min after completion of the process. At time point 0, all of the hybrid hydrogels showed a characteristic peak at ∼342 nm, which is typical of the fluorenyl peak (Figure S8). Interestingly, after 3 h, we could observe a ∼6 nm red shift in the peak maxima of all hybrid hydrogels, from 342 to 348 nm, which was not observed in the pristine Fmoc-Phe-Phe hydrogel or in the Fmoc-Gly-Pro-Hyp crystal solution (Figure 4c). This small red shift suggests additional stacking between the fluorenyl moieties, possibly as a result of the coassembly.50
The mechanical properties of the hybrid hydrogels were characterized by rheological analysis. Dynamic strain sweep (at 1 Hz frequency) over a range of 0.01–100% strain showed a wide linear viscoelastic region (LVR) in all hybrid hydrogels, indicating stable gel formation (Figure S9). All of the hybrid hydrogels showed an LVR of up to 10% strain (Figure S9). Frequency sweep analysis for the coassembled 2:1, 1:1, and 1:2 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp hydrogels was performed at the LVR from oscillatory sweep, using a frequency range of 0.1–100 Hz (Figure S10). Time sweep oscillatory measurements over 3 h, performed at a fixed strain of 0.1% and frequency of 1 Hz (Figure 4d), produced end point storage modulus values of ∼24,000, ∼7000, and ∼1100 Pa for the 2:1, 1:1, and 1:2 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp hybrid hydrogels, respectively, compared to the ∼8000 Pa value for the pristine Fmoc-Phe-Phe hydrogel. This suggests that the rigidity of the hybrid hydrogels decreases as the proportion of Fmoc-Gly-Pro-Hyp increases (Figure 4d). The initial rise in the rheological curves suggests the process of gel formation. However, the point where the gelation process and rigidification are complete, that is, the time in which the storage modulus G′ reaches its plateau, is a much longer process (∼45 to 60 min) for all hybrid hydrogels. The low storage modulus obtained for the 1:2 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp hybrid is in accordance with the SEM and AFM analyses, showing a less entangled fibrillary network in this material. Thus, the coassembly of 2:1 Fmoc-Phe-Phe and Fmoc-Gly-Pro-Hyp gives rise to a rigid hybrid hydrogel, with a storage modulus that is ∼3-fold higher than that of the pristine Fmoc-Phe-Phe gel at the same concentration (24,000 versus 8000 Pa). In order to demonstrate that the improved mechanical rigidity of the multicomponent hydrogel can be attributed solely to the different concentrations of Fmoc-Phe-Phe in the hybrid hydrogels, we performed time sweep analyses of Fmoc-Phe-Phe hydrogels with different compositions. The results gave G′ values of ∼1100, ∼650, and ∼400 Pa for the pure 0.33, 0.25, and 0.17 wt % Fmoc-Phe-Phe hydrogels, respectively. These values are considerably lower than those obtained from their hybrids with Fmoc-Gly-Pro-Hyp (Figures S11 and 4e) and emphasize the importance of stacking interactions between the Fmoc moieties of Fmoc-Phe-Phe and Fmoc-Gly-Pro-Hyp in the improved rigidity of the self-supporting coassembled hybrid hydrogels (Figure 4f).
Confirmation of Fmoc-Phe-Phe and Fmoc-Gly-Pro-Hyp Coassembly
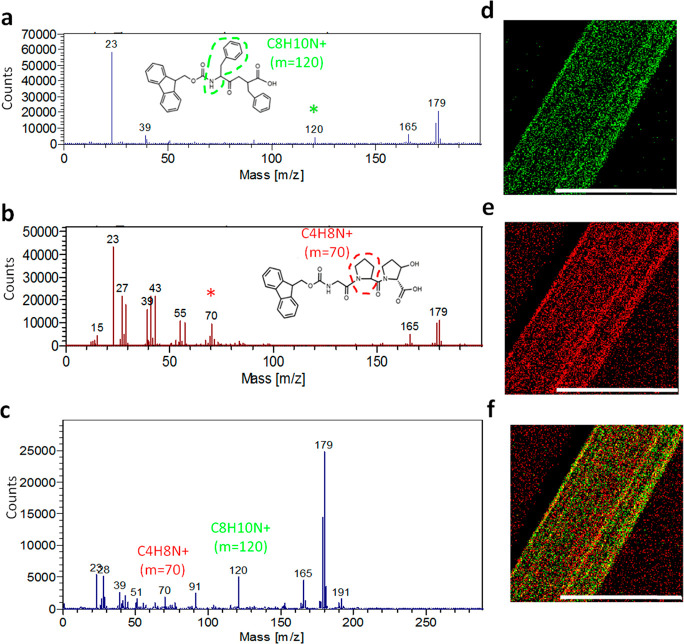
The coassembly of Fmoc-Phe-Phe and Fmoc-Gly-Pro-Hyp was further analyzed by chemical identification using ToF-SIMS (Figure 5).29,51 The positive secondary ion C8H10N+ 120 m/z was indicative of the presence of Fmoc-Phe-Phe (Figure 5a,d), whereas the positive ion C4H8N+ 70 m/z was chosen for Fmoc-Gly-Pro-Hyp (Figure 5b,e). ToF-SIMS analysis of the chemical composition of the thin dried bundled fibrils formed by the coassembled 1:1 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp hydrogel revealed the presence of peaks at both 120 and 70 m/z, confirming the presence of both moieties (Figure 5c). Chemical ion mapping was performed by assigning different colors to selected specific ions from the mass spectra and observing their location over a precise area of the fibrils. The significant presence of yellow dots over the mapped area confirms the colocalization of both C8H10N+ (green dots) and C4H8N+ (red dots) at the same point (Figure 5f). These results also confirm that the 1:1 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp hydrogel indeed comprises of a coassembly of the two peptide molecules.
Figure 5.
ToF-SIMS analysis of the chemical composition of the 1:1 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp hybrid hydrogel. (a–c) ToF-SIMS mass spectrometry analysis. (d–f) Chemical ion maps. (a,d) Fmoc-Phe-Phe, (b,e) Fmoc-Gly-Pro-Hyp, (c,f) 1:1 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp hybrid hydrogel. Scale bar represents 100 μm.
Formation of Fmoc-Phe-Phe/Gly-Pro-Hyp Hybrid Hydrogels
We developed hybrid hydrogels by mixing the nonprotected Gly-Pro-Hyp and Fmoc-Phe-Phe in the same ratios used for Fmoc-Phe-Phe/Gly-Pro-Hyp (Figure S12a). All combinations formed a clear gel, as shown in Figure S11b, and their ultrastructure was characterized by SEM and AFM (Figure S11). All three hybrid hydrogels exhibited long nonhelical fibrils (Figure S11c–h). PXRD analysis indicates that Gly-Pro-Hyp alone does not form crystals in DMSO/water or MeOH/water solvents (Figure S13). The FTIR spectra of the hybrid hydrogels showed an extended band in the range of 1600–1700 cm–1 with peak maxima at 1640 cm–1, suggesting a β-sheet structure as the major component (Figure S14). Photoluminescence analysis of all the Fmoc-Phe-Phe/Gly-Pro-Hyp hybrid hydrogels at 0 and 180 min of gelation did not reveal any spectral shift (Figure S15). Interestingly, the storage modulus values of the 2:1, 1:1, and 1:2 hybrid hydrogels (∼800, ∼700, and ∼450 Pa, respectively) are similar to those of pristine Fmoc-Phe-Phe at the same concentration (Figure 4c and Figure S11b).
We also analyzed the 1:1 Fmoc-Phe-Phe/Gly-Pro-Hyp hydrogel by ToF-SIMS. The positive secondary ion C14H11+ 179 m/z was characteristic of the Fmoc-Phe-Phe hydrogel, and the positive ion C4H8N+ 70 m/z was chosen for Gly-Pro-Hyp, as shown in Figure S15. In the chemical mapping, C14H11+ 179 m/z (green dots) and C4H8N+ 70 m/z (red dots) showed distinct, non-overlapping distributions, indicating that Gly-Pro-Hyp does not coassemble with Fmoc-Phe-Phe (Figure S16). Taken together, these results strongly suggest that Gly-Pro-Hyp does not coassemble with Fmoc-Phe-Phe and support the finding that the coassembly of Fmoc-Gly-Pro-Hyp with Fmoc-Phe-Phe is primarily driven by aromatic interactions mediated by the Fmoc groups. These give rise to the formation of a hybrid hydrogel with distinct features, including twisted helical fibrils with a polyproline II conformation, as found in a single helical strand of collagen.
Conclusion
Here, we describe the design and development of a Fmoc-modified form of a tripeptide module that is common in collagen. Single-crystal X-ray structural analysis of the peptide demonstrated superhelical packing with a polyproline II helical structure. The Fmoc modification facilitated the coassembly of this tripeptide, Fmoc-Gly-Pro-Hyp, with a well-established dipeptide hydrogelator, Fmoc-Phe-Phe, to produce a hybrid hydrogel comprising twisted helical fibrils. These display the characteristic polyproline II conformation with improved mechanical rigidity. This demonstration of a collagen-inspired polyproline II structural arrangement achieved by such a short peptide sequence is highly interesting. Moreover, the ability to introduce the polyproline II conformation, characteristic of native single strand, collagen into a hybrid hydrogel by a simple coassembly approach has enormous potential in the design of materials including scaffolds for tissue regeneration.
Materials and Methods
Materials
Lyophilized peptides Fmoc-Phe-Phe-OH (Fmoc-Phe-Phe), Fmoc-Gly-Pro-Hyp-OH (Fmoc-Gly-Pro-Hyp), and free H-Gly-Pro-Hyp-OH were purchased from Bachem (Budendorf, Switzerland). Bovine collagen type I was purchased from Sigma-Aldrich.
Crystal Preparation and Data Collection
Crystals used for data collection were grown using the slow evaporation method. The Fmoc-Gly-Pro-Hyp peptide was first dissolved in MeOH/DMSO at a concentration of 5 mg/mL, and 50 μL aliquots were deposited into a series of 8 × 40 mm vessels. Water was added dropwise until the solution became faintly turbid. At this point, each tube was sealed with parafilm, into which a needle was used to prick three small holes. The samples were left on the bench at room temperature, and crystals grew within 7–8 days. For data collection, crystals were coated in Paratone oil (Hampton Research), mounted on a MiTeGencryo-loop, and flash-frozen in liquid nitrogen. Crystal data were obtained for Fmoc-Gly-Pro-Hyp at 100 K on a RigakuXtaLabPro diffractometer equipped with λ(Cu Kα) = 1.54184 Å radiation and a Dectris Pilatus3R 200 K-A detector. The data were processed with CrysAlisPro programs (RigakuOD). The structure was solved by direct methods with SHELXT-2016/4 and refined with full-matrix least-squares refinement based on F2 with SHELXL-2016/4. The crystallographic data have been deposited in the Cambridge Crystallographic Data Centre (CCDC) under no. 1962894 and are presented in Supporting Information Table S1.
Single and Hybrid Hydrogel Preparation
Peptide hydrogels were prepared at a concentration of 5 mg/mL. Fmoc-Phe-Phe, Fmoc-Gly-Pro-Hyp, or Gly-Pro-Hyp powder was dissolved in dimethyl sulfoxide (DMSO) at a concentration of 100 mg/mL until a transparent solution was obtained. Single peptide hydrogels were prepared by adding 50 μL of the DMSO stock solutions to 950 μL of double distilled water under a vortex. Fmoc-Phe-Phe/Gly-Pro-Hyp and Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp coassembly solutions were prepared by combining the two peptides stock solutions at molar ratios of 2:1 (33.4 and 16.6 μL), 1:1 (25 and 25 μL), and 1:2 (16.6 and 33.4 μL), respectively. The coassembled solutions were then diluted in 950 μL of double distilled water under a vortex to a final concentration of 5 mg/mL.
Absorbance Kinetics of the Formation of Gels
Samples of 150 μL of each hydrogel were transferred into a 96-well plate. Absorbance at 400 nm was measured every 5 min using a TECAN Infinite M200PRO plate reader for a total of 24 h.
High-Resolution Scanning Electron Microscopy
Samples of Fmoc-Phe-Phe, Fmoc-Gly-Pro-Hyp, Gly-Pro-Hyp, and the coassemblies were placed on glass slides and left to air-dry under ambient conditions. Samples were then coated with Au for conductance and viewed using a scanning electron microscope (JEOL, Tokyo, Japan) operating at 20 kV.
Atomic Force Microscopy
Single peptide and coassembled hydrogels were prepared at a concentration of 5 mg/mL. AFM images were obtained by depositing 5 μL solutions of the hydrogels onto freshly cleaved V1 grade mica (Ted Pella) immediately after preparation. The samples were allowed to dry under ambient conditions for 24 h. The samples were imaged using AFM (JPK Instruments AG) performed with Nano Wizard 3 with 5 N/m spring constant tips and a resonance frequency of ∼150 kHz in soft tapping mode. AFM analysis was performed on three different areas for each sample, and the fibril diameter was measured using the ImageJ program choosing a population of 10 fibrils from each area. The images were processed and analyzed by JPK Data Processing software.
Circular Dichroism Spectroscopy
Single peptide suspension, coassembled hydrogels, prepared at concentrations of 5 mg/mL, and bovine collagen solution type I from Sigma were characterized by CD spectroscopy. CD spectra were collected using a Chirascan spectrometer (Applied Photophysics) fitted with a Peltier temperature controller set to 25 °C and quartz cuvettes with an optical path length of 0.1 mm (Hellma Analytics). Absorbance was kept within the linear range of the instrument during measurements. Data acquisition was performed in steps of 1 nm in a wavelength range of 190–260 nm with a spectral bandwidth of 1.0 nm and an averaging time of 3 s. The spectra of each sample were collected three times and averaged. The baseline was similarly recorded for the DMSO/water solvent (5%) and subtracted from the sample spectra. Data processing was performed using the Pro-Data Viewer software (Applied Photophysics). CD spectra of Fmoc-Gly-Pro-Hyp solution in 2:1 MeOH/water solvent were similarly acquired with a baseline correction of the 2:1 MeOH/water solvent and subsequently subtracted from the sample data. For temperature scan CD analysis, coassembled hydrogels were prepared at concentrations of 5 mg/mL, and using quartz cuvettes with an optical path length of 0.1 mm, the change in the 227 nm peak was observed over a temperature range of 5–90 °C.
Fourier Transform Infrared Spectroscopy
Single peptide, coassembled hydrogels, prepared at concentrations of 5 mg/mL, and bovine collagen solution type I from Sigma were characterized by FTIR spectroscopy. FTIR spectroscopy was performed using a portion of pre-prepared samples of gels and hybrids, prepared as described above, 4 days after the initiation of assembly. The samples prepared in 5% DMSO/water by combining the two peptide stock solutions at molar ratios of 2:1 (33.4 and 16.6 μL), 1:1 (25 and 25 μL), and 1:2 (16.6 and 33.4 μL), respectively, for Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp and Fmoc-Phe-Phe/Gly-Pro-Hyp coassembled gels; Fmoc-Phe-Phe (5 mg/mL), Gly-Pro-Hyp (5 mg/mL), Fmoc-Gly-Pro-Hyp (5 mg/mL in DMSO/water solvent), and Fmoc-Gly-Pro-Hyp (5 mg/mL) in 2:1 MeOH/water solvent were deposited on disposable KBr IR sample cards (Sigma-Aldrich, Israel), which were then allowed to dry under vacuum. Transmission infrared spectra were collected using a Nexus 470 FTIR spectrometer (Nicolet, Offenbach, Germany) with a deuterated triglycine sulfate detector. Measurements were performed using the atmospheric suppression mode by averaging 64 scans at 4 cm–1 resolution.
Fluorescence Spectroscopy
The emission spectra of the gels were recorded using a Horiba JobinYvon FL3-11 fluorimeter (Horiba JobinYvon, NJ, USA). A quartz cuvette with an optical path length of 1 cm was used. The gels were assembled within the cuvette, and the spectrum was collected. The experiments were carried out using an excitation wavelength of 280 and 5 nm excitation and emission slits.
Rheology Analysis
Rheological analysis was performed using an AR-G2 rheometer (TA Instruments, USA). A sample of approximately 250 μL of freshly prepared hydrogel sample was placed on a flat-plate geometry with a 20 mm diameter with a solvent trap. A gap distance of 0.6 mm was used, and time sweep oscillatory tests were conducted at room temperature. In order to determine the linear viscoelastic region at which the time sweep oscillatory tests were performed, oscillatory strain (0.01–100%) and frequency sweeps (0.1–100 Hz) were conducted 1 h after sample placement (soak time). G′ and G″, the storage and loss moduli, respectively, were obtained at 1 Hz oscillation and 0.1% strain deformation for each sample. Results are reported as the end point storage modulus values of the hydrogels with standard deviations obtained from triplicate repeats of each measurement.
ToF-SIMS Analysis
A thin fibril of 1:1 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp hybrid hydrogel was prepared between two thin capillaries and dried. The dried fibril was deposited on a silicon wafer and analyzed by a PHI model 2100 TRIFT II ToF-SIMS instrument. The system used a pulsed primary ion beam to desorb and ionize species from the amino acids’ surface. The resulting secondary ions were accelerated into a mass spectrometer, where they were mass analyzed by measuring their time-of-flight from the sample surface to the detector. In addition, an image was generated by rostering a finely focused beam across the sample surface. Due to the parallel detection nature of ToF-SIMS, the entire mass spectrum was acquired from every pixel in the image. The ions related to 120 and 70 m/z were used to identify and evaluate the ionic image of the coassembled 1:1 Fmoc-Phe-Phe/Fmoc-Gly-Pro-Hyp hybrid hydrogel. The ions related to 179 and 70 m/z were used to identify and evaluate the ionic image of the coassembled 1:1 Fmoc-Phe-Phe/Gly-Pro-Hyp hybrid hydrogel. The mass spectrum and the secondary ion images were then used to determine the composition and distribution of sample surface constituents.
Powder X-ray Diffraction
Single peptide solutions and coassembled hydrogels were prepared at a concentration of 5 mg/mL. Samples (200 μL) were deposited on glass slides immediately after preparation and allowed to dry under ambient conditions for 2 days. The X-ray diffraction pattern was collected using a Bruker’s D8 Discover diffractometer; the used setup was a q:q Bragg–Brentano geometry, the source was copper anode, and the detector was a LYNXEYE XE linear detector. The diffraction patterns were collected between 4 and 40°2q with a step of 0.02°2q for 1 s per step.
Acknowledgments
This research was partially supported by the Israel Science Foundation (Grant No. 1732/17) (L.A.A.) and the Ministry of Science, Technology & Space, Israel (L.A.A.). M.G. and S.B. thank Tel Aviv University for a postdoctoral fellowship. We thank Davide Levy, Gal Radovsky, and Alexander Gladkikh for help with PXRD, AFM, and Tof-SIMS analysis, respectively, and the Chaoul Center for Nanoscale Systems of Tel Aviv University for the use of instruments and staff assistance. We thank Sharon Tsach for graphical assistance and the members of the Adler-Abramovich group for helpful discussions.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.0c03085.
Single-crystal XRD of a Fmoc-Gly-Pro-Hyp crystal; side-by-side H-bond interaction of a single crystal of Fmoc-Gly-Pro-Hyp; interaction of trimeric units through H-bonds in single-crystal Fmoc-Gly-Pro-Hyp; SEM images of Fmoc-Gly-Pro-Hyp crystals and XRD of coassembled hydrogel; hydrogel fibril diameter determination by histogram plots from an AFM image; thermal transition of circular dichroism analysis of the coassembled hydrogels; CD and FTIR spectra of collagen; fluorescence emission spectra of the multicomponent hydrogels; rheological frequency sweep analyses; rheological strain sweep analyses; in situ time sweep oscillation measurements of hydrogel formation; molecular structure of the two building blocks, Fmoc-Phe-Phe and Gly-Pro-Hyp; inverted vials of the single and hybrid hydrogels; SEM and AFM images of the studied hydrogels; PXRD characterization of dried Gly-Pro-Hyp in MeOH/water; FTIR spectra of the hydrogels formed by Fmoc-Phe-Phe and Gly-Pro-Hyp; fluorescence emission spectra of the hydrogels formed by Fmoc-Phe-Phe and Gly-Pro-Hyp; ToF-SIMS mass spectrometry analysis of the hydrogels formed by Fmoc-Phe-Phe and Gly-Pro-Hyp (PDF)
X-ray crystallographic data for Fmoc-Gly-Pro-Hyp (CIF)
Author Contributions
M.G. and L.A.-A. conceived the idea and designed the experiments. M.G. performed all of the experiments and crystallized the peptides. L.J.W.S. collected the single-crystal diffraction data and solved the crystal structures. M.G., S.B., and L.J.W.S. analyzed the single-crystal data. M.G. and S.S. performed absorbance and rheological characterizations. M.G., S.B., and L.A.-A. wrote the paper. All authors commented on the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Sarkar B.; O’Leary L. E. R.; Hartgerink J. D. Self-Assembly of Fiber-Forming Collagen Mimetic Peptides Controlled by Triple-Helical Nucleation. J. Am. Chem. Soc. 2014, 136, 14417–14424. 10.1021/ja504377s. [DOI] [PubMed] [Google Scholar]
- O’Leary L.; Fallas J.; Bakota E.; Kang M.; Hartgerink J. Multi-Hierarchical Self-Assembly of a Collagen Mimetic Peptide from Triple Helix to Nanofibre and Hydrogel. Nat. Chem. 2011, 3, 821–828. 10.1038/nchem.1123. [DOI] [PubMed] [Google Scholar]
- Ottl J.; Gabriel D.; Murphy G.; Knäuper V.; Tominaga Y.; Nagase H.; Kröger M.; Tschesche H.; Bode W.; Moroder L. Recognition and Catabolism of Synthetic Heterotrimeric Collagen Peptides by Matrix Metalloproteinases. Chem. Biol. 2000, 7, 119–132. 10.1016/S1074-5521(00)00077-6. [DOI] [PubMed] [Google Scholar]
- Yonath A.; Traub W. Polymers of Tripeptides as Collagen Models IV.? Structure Analysis of Poly (L-Prolyl-Glycyl-L-Proline). J. Mol. Biol. 1969, 43, 461–477. 10.1016/0022-2836(69)90352-0. [DOI] [PubMed] [Google Scholar]
- Rich A.; Crick F. The Molecular Structure of Collagen. J. Mol. Biol. 1961, 3, 483–506. 10.1016/S0022-2836(61)80016-8. [DOI] [PubMed] [Google Scholar]
- Okuyama K.; Okuyama K.; Arnott S.; Takayanagi M.; Kakudo M. Crystal and Molecular Structure of a Collagen-Like Polypeptide (Pro-Pro-Gly)10. J. Mol. Biol. 1981, 152, 427–443. 10.1016/0022-2836(81)90252-7. [DOI] [PubMed] [Google Scholar]
- Okuyama K.; Nagarajan V.; Kamitori S. 7/2-Helical Model for Collagen-Evidence from Model Peptides. Chem. Sci. 1999, 111, 19–34. [Google Scholar]
- Xu F.; Li J.; Jain V.; Tu R.; Huang Q.; Nanda V. Compositional Control of Higher Order Assembly Using Synthetic Collagen Peptides. J. Am. Chem. Soc. 2012, 134, 47–50. 10.1021/ja2077894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekkat A.; Poppleton E.; An B.; Visse R.; Nagase H.; Kaplan D. L.; Brodsky B.; Lin Y.-S. Effects of Flexibility of the α2 Chain of Type I Collagen on Collagenase Cleavage. J. Struct. Biol. 2018, 203, 247–254. 10.1016/j.jsb.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y.; Melacini G.; Taulane J.; Goodman M. Acetyl-Terminated and Template-Assembled Collagen-Based Polypeptides Composed of Gly-Pro-Hyp Sequences. 2. Synthesis and Conformational Analysis by Circular Dichroism, Ultraviolet Absorbance, and Optical Rotation. J. Am. Chem. Soc. 1996, 118, 10351–10358. 10.1021/ja961260c. [DOI] [Google Scholar]
- Melacini G.; Feng Y.; Goodman M. Acetyl-Terminated and Template-Assembled Collagen-Based Polypeptides Composed of Gly-Pro-Hyp Sequences. 3. Conformational Analysis by 1H-NMR and Molecular Modeling Studies. J. Am. Chem. Soc. 1996, 118, 10359–10364. 10.1021/ja9612615. [DOI] [Google Scholar]
- Jalan A.; Jochim K.; Hartgerink J. Rational Design of a Non-Canonical ″Sticky-Ended″ Collagen Triple Helix. J. Am. Chem. Soc. 2014, 136, 7535–7538. 10.1021/ja5001246. [DOI] [PubMed] [Google Scholar]
- Frederix P.; Scott G.; Abul-Haija Y.; Kalafatovic D.; Pappas C.; Javid N.; Hunt N.; Ulijn R.; Tuttle T. Exploring the Sequence Space for (Tri-)Peptide Self-Assembly to Design and Discover New Hydrogels. Nat. Chem. 2015, 7, 30–37. 10.1038/nchem.2122. [DOI] [PubMed] [Google Scholar]
- Bera S.; Mondal S.; Xue B.; Shimon L.; Cao Y.; Gazit E. Rigid Helical-Like Assemblies from a Self-Aggregating Tripeptide. Nat. Mater. 2019, 18, 503–509. 10.1038/s41563-019-0343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L.; Parker S. T.; Syoji D.; Wang X.; Lewis J. A.; Kaplan D. L. Direct-Write Assembly of 3D Silk/Hydroxyapatite Scaffolds for Bone Co-Cultures. Adv. Healthcare Mater. 2012, 1, 729–735. 10.1002/adhm.201200057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okesola B.; Mata A. Multicomponent Self-Assembly as a Tool to Harness New Properties from Peptides and Proteins in Material Design. Chem. Soc. Rev. 2018, 47, 3721–3736. 10.1039/C8CS00121A. [DOI] [PubMed] [Google Scholar]
- Frederix P.; Ulijn R.; Hunt N.; Tuttle T. Virtual Screening for Dipeptide Aggregation: Toward Predictive Tools for Peptide Self-Assembly. J. Phys. Chem. Lett. 2011, 2, 2380–2384. 10.1021/jz2010573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada A.; Levin A.; Toprakcioglu Z.; Shen Y.; Lutz-Bueno V.; Baumann K. N.; Mohammadi P.; Linder M. B.; Mezzenga R.; Knowles T. P. J. Modulating the Mechanical Performance of Macroscale Fibers Through Shear-Induced Alignment and Assembly of Protein Nanofibrils. Small 2020, 16, 1904190. 10.1002/smll.201904190. [DOI] [PubMed] [Google Scholar]
- Dasgupta A.; Mondal J.; Das D. Peptide Hydrogels. RSC Adv. 2013, 3, 9117–9149. 10.1039/c3ra40234g. [DOI] [Google Scholar]
- Redondo-Gómez C.; Abdouni Y.; Becer C.; Mata A. Self-Assembling Hydrogels Based on a Complementary Host-Guest Peptide Amphiphile Pair. Biomacromolecules 2019, 20, 2276–2285. 10.1021/acs.biomac.9b00224. [DOI] [PubMed] [Google Scholar]
- Toprakcioglu Z.; Challa P.; Xu C.; Knowles T. P. J. Label-Free Analysis of Protein Aggregation and Phase Behaviour. ACS Nano 2019, 13, 13940–13948. 10.1021/acsnano.9b05552. [DOI] [PubMed] [Google Scholar]
- Mahler A.; Reches M.; Rechter M.; Cohen S.; Gazit E. Rigid, Self-Assembled Hydrogel Composed of a Modified Aromatic Dipeptide. Adv. Mater. 2006, 18, 1365–1370. 10.1002/adma.200501765. [DOI] [Google Scholar]
- Jayawarna V.; Ali M.; Jowitt T.; Miller A.; Saiani A.; Gough J.; Ulijn R. Nanostructured Hydrogels for Three-Dimensional Cell Culture Through Self-Assembly of Fluorenylmethoxycarbonyl-Dipeptides. Adv. Mater. 2006, 18, 611–614. 10.1002/adma.200501522. [DOI] [Google Scholar]
- Makam P.; Gazit E. Minimalistic Peptide Supramolecular Co-Assembly: Expanding the Conformational Space for Nanotechnology. Chem. Soc. Rev. 2018, 47, 3406–3420. 10.1039/C7CS00827A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halperin-Sternfeld M.; Ghosh M.; Sevostianov R.; Grigoriants I.; Adler-Abramovich L. Molecular Co-Assembly as a Strategy for Synergistic Improvement of the Mechanical Properties of Hydrogels. Chem. Commun. 2017, 53, 9586–9589. 10.1039/C7CC04187J. [DOI] [PubMed] [Google Scholar]
- Ghosh M.; Halperin-Sternfeld M.; Grigoriants I.; Lee J.; Nam K.; Adler-Abramovich L. Arginine-Presenting Peptide Hydrogels Decorated with Hydroxyapatite as Biomimetic Scaffolds for Bone Regeneration. Biomacromolecules 2017, 18, 3541–3550. 10.1021/acs.biomac.7b00876. [DOI] [PubMed] [Google Scholar]
- Diaferia C.; Morelli G.; Accardo A. Fmoc-Diphenylalanine as a Suitable Building Block for the Preparation of Hybrid Materials and Their Potential Applications. J. Mater. Chem. B 2019, 7, 5142–5155. 10.1039/C9TB01043B. [DOI] [PubMed] [Google Scholar]
- Helbing C.; Deckert-Gaudig T.; Firkowska-Boden I.; Wei G.; Deckert V.; Jandt K. D. Protein Handshake on the Nanoscale: How Albumin and Hemoglobin Self-Assemble into Nanohybrid Fibers. ACS Nano 2018, 12, 1211–1219. 10.1021/acsnano.7b07196. [DOI] [PubMed] [Google Scholar]
- Adler-Abramovich L.; Marco P.; Arnon Z.; Creasey R.; Michaels T.; Levin A.; Scurr D.; Roberts C.; Knowles T. P. J.; Tendler S.; Gazit E. Controlling the Physical Dimensions of Peptide Nanotubes by Supramolecular Polymer Coassembly. ACS Nano 2016, 10, 7436–7442. 10.1021/acsnano.6b01587. [DOI] [PubMed] [Google Scholar]
- Cross E.; Sproules S.; Schweins R.; Draper E.; Adams D. Controlled Tuning of the Properties in Optoelectronic Self-Sorted Gels. J. Am. Chem. Soc. 2018, 140, 8667–8670. 10.1021/jacs.8b05359. [DOI] [PubMed] [Google Scholar]
- Fuentes-Caparrós A.; De Paula Gómez-Franco F.; Dietrich B.; Wilson C.; Brasnett C.; Seddon A.; Adams D. Annealing Multicomponent Supramolecular Gels. Nanoscale 2019, 11, 3275–3280. 10.1039/C8NR09423C. [DOI] [PubMed] [Google Scholar]
- Ryan D.; Anderson S.; Nilsson B. The Influence of Side-Chain Halogenation on the Self-Assembly and Hydrogelation of Fmoc-Phenylalanine Derivatives. Soft Matter 2010, 6, 3220–3231. 10.1039/c0sm00018c. [DOI] [Google Scholar]
- Okuyama K.; Miyama K.; Morimoto T.; Masakiyo K.; Mizuno K.; Bächinger H. Stabilization of Triple-Helical Structures of Collagen Peptides Containing a Hyp-Thr-Gly, Hyp-Val-Gly, or Hyp-Ser-Gly Sequence. Biopolymers 2011, 95, 628–640. 10.1002/bip.21625. [DOI] [PubMed] [Google Scholar]
- Banwell E.; Abelardo E.; Adams D.; Birchall M.; Corrigan A.; Donald A.; Kirkland M.; Serpell L.; Butler M.; Woolfson D. Rational Design and Application of Responsive α-Helical Peptide Hydrogels. Nat. Mater. 2009, 8, 596–600. 10.1038/nmat2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedemeyer W.; Welker E.; Scheraga H. Proline Cis-Trans Isomerization and Protein Folding. Biochemistry 2002, 41, 14637–14644. 10.1021/bi020574b. [DOI] [PubMed] [Google Scholar]
- Okuyama K.; Miyama K.; Mizuno K.; Bächinger H. Crystal Structure of (Gly-Pro-Hyp)9: Implications for the Collagen Molecular Model. Biopolymers 2012, 97, 607–616. 10.1002/bip.22048. [DOI] [PubMed] [Google Scholar]
- Berisio R.; Vitagliano L.; Mazzarella L.; Zagari A. Crystal Structure of a Collagen-Like Polypeptide with Repeating Sequence Pro-Hyp-Gly at 1.4 Å Resolution: Implications for Collagen Hydration. Biopolymers 2000, 56, 8–13. . [DOI] [PubMed] [Google Scholar]
- Doi M.; Imori K.; Sakaguchi N.; Asano A. Boc-Pro-Hyp-Gly-OBzl and Boc-Ala-Hyp-Gly-OBzl, Two Repeating Triplets Found in Collagen. Acta Crystallogr., Sect. C: Cryst. Struct. Commun. 2006, 62, o577–o580. 10.1107/S0108270106028101. [DOI] [PubMed] [Google Scholar]
- Guerin S.; Syed T.; Thompson D. Deconstructing Collagen Piezoelectricity Using Alanine-Hydroxyproline-Glycine Building Blocks. Nanoscale 2018, 10, 9653–9663. 10.1039/C8NR01634H. [DOI] [PubMed] [Google Scholar]
- Berisio R.; Vitagliano L.; Mazzarella L.; Zagari A. Crystal Structure of a Collagen-Like Polypeptide with Repeating Sequence Pro-Hyp-Gly at 1.4 Å Resolution: Implications for Collagen Hydration. Biopolymers 2000, 56, 8–13. . [DOI] [PubMed] [Google Scholar]
- Schnaider L.; Ghosh M.; Bychenko D.; Grigoriants I.; Ya’Ari S.; ShalevAntsel T.; Matalon S.; Sarig R.; Brosh T.; Pilo R.; Gazit E.; Adler-Abramovich L. Enhanced NanoAssembly-Incorporated Antibacterial Composite Materials. ACS Appl. Mater. Interfaces 2019, 11, 21334–21342. 10.1021/acsami.9b02839. [DOI] [PubMed] [Google Scholar]
- Ghosh M.; Halperin-Sternfeld M.; Grinberg I.; Adler-Abramovich L. Injectable Alginate-Peptide Composite Hydrogel as aScaffold for Bone Tissue Regeneration. Nanomaterials 2019, 9, 497. 10.3390/nano9040497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv M.; Halperin-Sternfeld M.; Grigoriants I.; Buzhansky L.; Mironi-Harpaz I.; Seliktar D.; Einav S.; Nevo Z.; Adler-Abramovich L. Improving the Mechanical Rigidity of Hyaluronic Acid by Integration of a Supramolecular Peptide Matrix. ACS Appl. Mater. Interfaces 2018, 10, 41883–41891. 10.1021/acsami.8b08423. [DOI] [PubMed] [Google Scholar]
- Diaferia C.; Ghosh M.; Sibillano T.; Gallo E.; Stornaiuolo M.; Giannini C.; Morelli G.; Adler-Abramovich L.; Accardo A. Fmoc-FF and Hexapeptide-Based Multicomponent Hydrogels as Scaffold Materials. Soft Matter 2019, 15, 487–496. 10.1039/C8SM02366B. [DOI] [PubMed] [Google Scholar]
- Shi Z.; Olson A.; Rose G.; Baldwin R.; Kallenbach N. Polyproline II Structure in a Sequence of Seven Alanine Residues. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 9190–9195. 10.1073/pnas.112193999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowska J.; Rodziewicz-Motowidlo S.; Baginska K.; Vila J. A.; Liwo A.; Chmurzynski L.; Scheraga H. A. Polyproline II Conformation is One of Many Local Conformational States and Is Not an Overall Conformation of Unfolded Peptides and Proteins. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 1744–1749. 10.1073/pnas.0510549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei A. A.; Sternberg M. J. E.; Makarov A. A. Polyproline-II Helix in Proteins: Structure and Function. J. Mol. Biol. 2013, 425, 2100–2132. 10.1016/j.jmb.2013.03.018. [DOI] [PubMed] [Google Scholar]
- Fleming S.; Frederix P.; Ramos Sasselli I.; Hunt N.; Ulijn R.; Tuttle T. Assessing the Utility of Infrared Spectroscopy as a Structural Diagnostic Tool for β-Sheets in Self-Assembling Aromatic Peptide Amphiphiles. Langmuir 2013, 29, 9510–9515. 10.1021/la400994v. [DOI] [PubMed] [Google Scholar]
- Kubelka J.; Keiderling T. Differentiation of β-Sheet-Forming Structures: Ab Initio-Based Simulations of IR Absorption and Vibrational CD for Model Peptide and Protein β-Sheets. J. Am. Chem. Soc. 2001, 123, 12048–12058. 10.1021/ja0116627. [DOI] [PubMed] [Google Scholar]
- Tang C.; Ulijn R.; Saiani A. Effect of Glycine Substitution on Fmoc-Diphenylalanine Self-Assembly and Gelation Properties. Langmuir 2011, 27, 14438–14449. 10.1021/la202113j. [DOI] [PubMed] [Google Scholar]
- Bera S.; Mondal S.; Tang Y.; Jacoby G.; Arad E.; Guterman T.; Jelinek R.; Beck R.; Wei G.; Gazit E. Deciphering the Rules for Amino Acid Co-Assembly Based on Interlayer Distances. ACS Nano 2019, 13, 1703–1712. 10.1021/acsnano.8b07775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.