Abstract
The human gut microbiome is considered critical for establishing and maintaining intestinal function and homeostasis throughout life. Evidence for bidirectional communication with the immune and nervous systems has spawned interest in the microbiome as a key factor for human and animal health. Consequently, appreciation of the microbiome as a target of xenobiotics, including environmental pollutants such as heavy metals, has risen steadily because disruption of a healthy microbiome (dysbiosis) has been linked to unfavorable health outcomes. Thus, toxicology must consider toxicant effects on the host’s microbiome as an integral part of the holobiont. We discuss current findings on the impact of toxic metals on the composition, diversity, and function of the gut microbiome as well as the modulation of metal toxicity by the microbiome. Present limitations and future needs in elucidating microbiome-metal interactions and the potential of harnessing beneficial traits of the microbiota to counteract metal toxicity are also considered.
Keywords: Gut microbiome, microbiota, dysbiosis, heavy metal, toxicity, 16S rRNA, metagenomics, arsenic, cadmium, chromium, mercury, lead
Graphical Abstract

1. Introduction
A well-known quote from Paracelsus (1493 – 1541) states the founding principle of modern toxicology: “Solely the dose determines that a thing is not a poison” [1]. “Die wichtigsten Dinge des Lebens spielen sich zwischen Anfang und Ende des Verdauungskanals ab” [The most important things in life take place between the beginning and end of the digestive tract] is another quote ascribed to this ground-breaking physician and alchemist. These quotes combined are the basis of this brief review on the interactions of metals with the gut microbiome – the importance of dose in the toxicology of metals and our emerging comprehension how crucial the trillions of microorganisms in the gut are for human health and disease. As the most densely colonized site in the human body, the gastrointestinal tract (GIT) is inhabited by microbes in numbers ranging from 101–103 CFU/ml in the stomach to 1011–1012 CFU/ml in the colon, while overall more than 1,100 known bacterial species were identified in a recent culturomics and metagenomics study [2]. Bacteria represent the vast majority of the intestinal microorganisms, while archaea, fungi, and protozoa are considered minor components of a healthy gut microbiota. Recent studies on the intestinal virome have estimated that the number of bacteriophage particles in the gut reaches, if not surpasses, the number of bacteria – appreciation of the potential impact of the virome on the microbiome-host interaction is just emerging [3–6].
The crucial role of the microbiome in development, function and homeostasis of the GIT, as well as its integration with the host immune and nervous systems [7–11], raises many questions on how this “organ” within an organ interacts with ingested xenobiotics and how the outcome of these interactions might affect the individual host [12–15]. While the human gut microbiotas exhibit commonalities in structure and metabolic activity, the uniqueness of individual microbiomes, especially at the species and strain levels, should be an important factor in the assessment of toxicity of any xenobiotic.
As gut microbiomes potentially are exposed to myriads of xenobiotics such as oral therapeutic drugs, drugs of misuse, and environmental chemicals and pollutants, we will focus this critical review on recent insights on the effects of a selection of heavy metals on the gut microbiome and conversely, potential actions of gut microbes affecting the toxicity of metals. Most of these deliberations stem from findings in experimental animal research, because the literature on the interrelationships of toxic metals and the intestinal microbiome in humans is still relatively scarce.
2. Metals change the microbiota
Physiologically essential metals (e.g., Mg, Mn, Fe, Co, Ni, Cu) at high concentrations and some non-essential metals such as mercury, silver, and lead at much lower concentrations are toxic to many microorganisms. While these antimicrobial effects may be advantageous for growth suppression or killing of pathogens (for review [16]), especially when these are multi-drug resistant, the indiscriminate nature of metal toxicity likely will also harm commensal and beneficial microbes in complex communities such as the gut microbiota. Metal toxicity results from oxidative stress via generation of reactive oxygen species (ROS) and depletion of antioxidants, protein dysfunction via oxidation, structural damage and interference with catalysis, and from metal-induced damage to biological membranes. Furthermore, antimicrobial activity may also be based on disturbances of gene expression and DNA damage (genotoxicity) [16].
Exposure of the gut microbiota to toxic metals is likely to exert disparate effects on the resident species, depending on intestinal location (e.g., stomach, jejunum, ileum, cecum, or colon; lumen vs. mucus layer), microenvironmental conditions such as pH, oxygen availability and redox potential as well as the abundance of susceptible/resistant strains and overall diversity and metabolic activity of the local microbial community. In addition, a wide range of host factors including nutrition, sex, age, and immune status may influence the microbiome-metal interaction. In the following, we discuss a selection of the most toxic metals with emphasis on arsenic, whose compounds are arguably the most studied in context with the gut microbiome.
2.1. Arsenic
Arsenic is a highly relevant environmental toxicant found in food (e.g., rice, fish, seafood) and drinking water supplies. Arsenic exposure can lead to carcinogenesis and other adverse health outcomes affecting various organ systems [17]. Speciation of arsenic is crucial for health risk assessment and the potential for alteration of bioavailability through interactions with gut microbes. For example, water sources are almost exclusively contaminated with inorganic arsenic (arsenate, arsenite), while food may contain inorganic and organic species of arsenic. The trivalent arsenite and especially its methylated species (e.g., monomethylarsonous acid) are more toxic than the respective pentavalent states.
Richardson et al. [18] assessed the acute effects of toxic metal exposures on rat gut microbiotas by administering five different metals, each administered at a specific range of three different doses, for five consecutive days. Rats received daily oral gavages of sodium arsenite, cadmium chloride, cobalt chloride, sodium dichromate, or nickel chloride. Fecal samples for microbiota analyses were collected prior to the first administration and 24 hours after the fifth dosing [18]. Sequencing of 16S rRNA genes and computational prediction of microbial gene content (Phylogenetic investigation of communities by reconstruction of unobserved states; PICRUSt [19]) were used to characterize the early and metal-specific perturbations in the rat gut microbiota instigated by these toxicants with significant environmental and human health impacts (e.g., all are considered carcinogens [20,21]). Arsenic, cadmium, and nickel altered bacterial composition and diversity significantly and in a dose-dependent manner, while chromium and cobalt had weaker impacts on the microbiota, albeit still appeared to affect host physiology (e.g., causing weight loss) [18]. Importantly, the response to the metals were not uniform, showing specific changes in the microbiota depending on the administered compound. For example, the Bacteroidetes family S24–7 [22] was dramatically reduced by nickel, while other Bacteroidetes and Proteobacteria increased. The PICRUSt analyses showed increases of iron-importing gene functions in the nickel and arsenic-treated samples, which could be related to selection of bacteria capable of utilizing these genes to moderate the toxic metal effects.
Comparison of these results with other studies that employed different metal exposure regimens and dosages in other animal models illustrates how deeply experimental factors influence study outcomes.
Lu et al. [23] reported that arsenic exposure in drinking water (10 ppm arsenic for 4 weeks) alters the composition (e.g., decreases in some Firmicutes families) and metabolic profiles (e.g., alterations in indole metabolites, bile acid profiles) of the gut microbiota of female C57BL/6 mice. In another study with lower, environmentally relevant dosage of arsenic (100 ppb, 13 weeks), dysbiosis with alterations in composition and diversity of microbiota was accompanied by metagenomic changes in carbohydrate metabolism, short chain fatty acid synthesis and starch utilization systems [24]. Furthermore, arsenic increased oxidative stress indicators and DNA repair genes. Of particular concern is the observed enrichment of multidrug resistance and conjugative transposon genes in the arsenic-exposed animals, which could indicate that heavy metals promote the spread of multidrug resistance via horizontal gene transfer in the gut.
Sex-specific responses to arsenic exposure were explored by Chi and coworkers [25] who found not only differences in the resultant fecal microbiota compositions of male and female mice, but also clear distinctions in functional profiles as determined by metagenomics sequencing. Interestingly, sex-specific effects of arsenic exposure also were found in 6-week-old human infants who were part of the New Hampshire Birth Cohort Study [26]. Despite the relatively low arsenic exposure levels in this cohort, significant associations of elevated urinary arsenic levels with stool microbiome composition (e.g., reduced abundance of Bifidobacterium, Bacteroides, and Lactobacillus) were found in formula-fed male infants, but not in female formula-fed infants or breast-fed infants of both sexes.
The fecal microbiotas from a cohort of Bangladeshi children, who were exposed to low and high arsenic levels during prenatal development and early life, revealed higher abundance of Proteobacteria, in particular Gammaproteobacteria, in children with high exposure [27]. Concomitantly, virulence and multidrug resistance genes were enriched after high exposure; especially E. coli strains with arsenic resistance genes (ArsB, ArsC) were increased.
Gokulan et al. [28] investigated the impact of single and short-term repeated (8 days) arsenite exposure on gut microbiome composition as well as intestinal immune status in adult and juvenile CD-1 mice. Dose, duration of exposure, and developmental status of the animals effected distinct changes in bacterial recovery and microbiota composition. Repeated exposure increased the abundance of bacteria harboring arsenic resistance genes and induced arsenite methylation for detoxification by the host. Furthermore, reduction of bacteria involved in protein to butyrate conversion as well as indications of host immune modulation by arsenic exposure were revealed. Single doses of arsenite in juvenile mice elicited distinct bacterial populations, which illustrates how early-life arsenic exposure may have long-term consequences for development of a healthy gut microbiota.
Mitigation of acute arsenic toxicity (25 or 100 ppm inorganic sodium arsenate) by the microbiota was demonstrated by Coryell at al. [29] using antibiotic-treated, transgenic (arsenite methyltransferase As3mt detoxification enzyme knock-out), germ-free, and gnotobiotic mice. While microbiome disruption by antibiotic treatment increased arsenic bioaccumulation, germ-free status in concert with As3mt-deficiency was associated with high mortality after acute exposure. Interestingly, transplantation of human stool microbiota to the hypersensitive germ-free transgenic mice protected the recipients from arsenic-induced mortality. Moreover, gnotobiotic mouse experiments showed that Faecalibacterium prausnitzii, a bacterium commonly associated with healthy human microbiomes [30], provided at least partial protection against arsenic toxicity [29] – thus, specific microbiome manipulation may aid in prevention and treatment of arsenic poisoning.
2.2. Cadmium
Cadmium is another toxic metal with significant environmental impact [31]. For the general population, cadmium accumulated in food poses the main risk of exposure [31]. The GIT is a main target for cadmium toxicity [32]. Impairment of the gut barrier function in concert with Cd-induced changes in viability of components of the gut microbiota lead to increases in proinflammatory molecules (e.g., lipopolysaccharide, LPS) and may result in systemic inflammation. In addition to the aforementioned study by Richardson et al. [18], the impact of Cd toxicity on the intestinal microbiome of mice was investigated by several other research groups [33–35] who all reported significant alterations in bacterial communities; however, in detail, changes were disparate, even opposite, most likely due to differences in dosing, animal model, and sequencing methodology.
2.3. Lead
A similar situation is encountered with lead. Exposure to lead remains a public health issue, globally and in the U.S., as the water crisis in Flint, MI, has demonstrated [36–38]. A recent multi-omics study by Gao and coworkers [39] assessed the effects of lead on the gut microbiota composition, diversity, and metabolic activity (via whole-genome metagenomics sequencing and gas chromatography-mass spectrometry metabolomics) revealing that lead exposure altered the development of the gut microbiota and concomitantly affected multiple metabolic pathways, including some related to oxidative stress and detoxification. Such a multipronged approach certainly provides a more comprehensive insight into the impact of toxic metals on the gut microbiome.
2.4. Mercury
Elemental, inorganic and organic forms of mercury are global pollutants with disparate toxicity (Hg0 < inorganic Hg, mostly Hg2+ < organic Hg, mostly CH3Hg). As a potent neurotoxicant, methylmercury is most concerning, especially because of its tendency to bioaccumulate in fish relevant for dietary consumption [40]. A recent study by Rothenberg et al. [41] investigating potential correlations of the gut microbiota structure and metabolic activities with fetal methylmercury exposure in pregnant women revealed a significant correlation of 17 bacterial genera with mercury biomarkers. Dietary methylmercury also led to changes in the gut microbiome and metabolome of mice and larval fish [42].
3. Microbiotas change metals
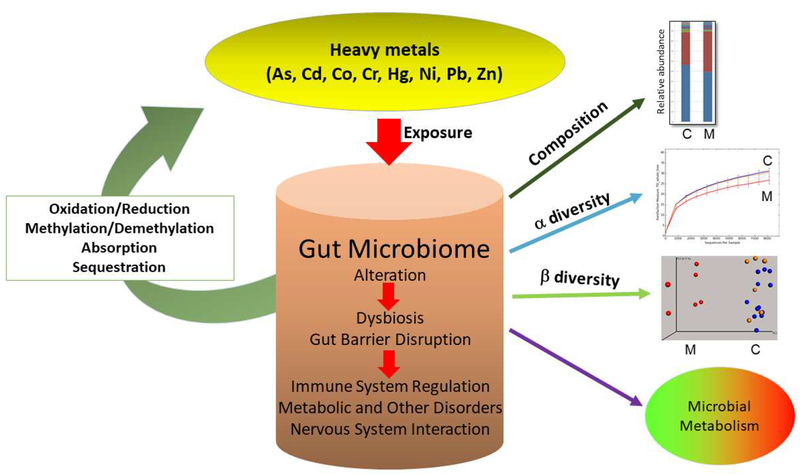
In addition to host metabolism, biotransformation by gut bacteria such as reduction, oxidation, methylations or demethylations may modulate metal toxicity (Fig. 1). For example, trivalent arsenite and especially its methylated species (e.g., monomethylarsonous acid) are more toxic than the respective pentavalent states. Both host cells and gut microbes can act on arsenic and transform it into less or more toxic forms (for review [43]. Other examples are the reduction of highly toxic chromate [Cr(VI)] to its less-toxic form [44–46] or the potential biotransformations of mercury [44,47]. The high neurotoxicity of organic mercury has drawn attention to microorganisms that can methylate mercury and concomitantly increase its bioavailability. The gene pair hgcAB¸ first described in two sulfate-reducing bacteria [48], has become a genetic marker used to screen for orthologous methylation genes in bacterial and archaeal genomes as well as many metagenomes [49–52]. While the evidence for effective mercury methylation in the vertebrate intestine remains scarce, potential detoxification reactions by the microbiota such demethylation of methylmercury or reduction of inorganic mercury to its least toxic elemental form (via activities encoded by bacterial mer operons) are actively investigated [47,51]. Another mechanism how the microbiota may interfere with metal toxicity is the binding of metals by intestinal microorganisms, which could aid in elimination of the toxicants from the GIT (Fig. 1). A recent study in pregnant women and children has provided evidence for such beneficial effects exerted by certain bacteria: a probiotic-supplemented yogurt reduced the bioaccumulation of arsenic and mercury [53]. In summary, the resident gut microbiota is likely to interfere with bioavailability and toxicity of metals. Consequently, the gut microbiome could have a substantial influence on an individual’s susceptibility to toxic metal exposure.
Fig. 1. Metal-Gut Microbiome Interactions.
Ingested toxic metals exposure can alter the composition (abundance of taxa), alpha and beta-diversity, and metabolic activities of resident microbiota in the gut. Dysbiosis and gut barrier disruption may activate the immune system, lead to metabolic and other disorders, and also could affect the bidirectional communication with the CNS (gut-brain axis). However, members of the gut microbiota could also modulate the toxicity of ingested metals via oxidation, reduction, methylation or demethylation reactions as well as binding and sequestration of metal species. M: Metal-exposed; C: control.
4. Challenges, Limitations, and Opportunities
Undoubtedly, the gut microbiome does have a profound impact on the toxicity of metals and their health effects on humans and animals. Current challenges and limitations in understanding the microbiome-metal interrelationship are posed by differences in experimental design (animal model, metal dosing, mode of exposure, sequencing technologies, data analytics, etc.), quality control (QC) measures, and by the sheer overwhelming complexity of the microbiome-host interactions. Each individual’s microbiome is unique and dynamic, constantly influenced by environmental, dietary, and biological factors.
At present, strong efforts are underway to make microbiome research more reliable and reproducible; for example, by using mock communities and spike-in controls for QC as well as standardized sampling procedures and data analysis pipelines [54,55]. As briefly summarized in this review, correlations of microbiome structure with metal toxicity are not enough - for elucidation of the mechanisms operating at the metal-microbiome-host interface, mechanistic confirmation using multiple omics such as metagenomics, transcriptomics, proteomics, and especially metabolomics will be necessary. Furthermore, the gut virome’s involvement needs to be evaluated.
Similarly, careful and considerate use of experimental models is required, but must be cognizant of the many potential pitfalls in design and transferability of results [56]. Alternative preclinical models, such as microbiome research in the highly social prairie voles (Microtus ochrogaster), can provide novel insights into the microbiome-gut-brain behavior axis [57,58]. Additionally, most animal studies to date have addressed single exposure, however, exposure to multiple metals or pollutants is frequent. Therefore, research studies with exposures to two or more toxicants must be conducted that are more realistic scenarios of exposure [59].
Environmental pollution by toxic metals is a global threat to human health and well-being. Therefore, well-designed surveillance studies are necessary to uncover, combat, and prevent human exposure. Microbiome research will continue to be essential for our understanding of toxicology and precision medicine. An individual’s microbiome must be considered in risk assessment and treatment of toxic metal exposure.
Heavy metal exposure can alter the composition of the intestinal microbiome
Heavy metals may affect the diversity of the gut microbiota
Metabolic activities of the gut microbiota may change during heavy metal exposure
Components of the gut microbiome can mitigate or exacerbate the toxicity of heavy metals
Acknowledgements
Research in the authors’ laboratory related to this work was supported partially by the National Institute of General Medical Sciences of the National Institutes of Health under award number R15GM110593 and Health Research Award project number HR13-013 from the Oklahoma Center for the Advancement of Science and Technology. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Footnotes
Conflicts of Interest
Nothing declared.
References
Papers of particular interest, published with the period of the review have been highlighted as:
• special interest
•• outstanding interest.
- 1.Borzelleca JF: Paracelsus: herald of modern toxicology. Toxicol Sci 2000, 53:2–4; DOI: 10.1093/toxsci/53.1.2. [DOI] [PubMed] [Google Scholar]
- 2.Mailhe M, Ricaboni D, Vitton V, Gonzalez J-M, Bachar D, Dubourg G, Cadoret F, Robert C, Delerce J, Levasseur A, et al. : Repertoire of the gut microbiota from stomach to colon using culturomics and next-generation sequencing. BMC Microbiology 2018, 18:157; DOI: 10.1186/s12866-018-1304-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.•. Beller L, Matthijnssens J: What is (not) known about the dynamics of the human gut virome in health and disease. Curr Opin Virol 2019, 37:52–57; DOI: 10.1016/j.coviro.2019.05.013. [DOI] [PubMed] [Google Scholar]; A current review on the human gut virome discussing its complexity, interpersonal variation, and the dominance of temperate phages in the interactions with the bacteriome.
- 4.Dalmasso M, Hill C, Ross RP: Exploiting gut bacteriophages for human health. Trends Microbiol 2014, 22:399–405; DOI: 10.1016/j.tim.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Lepage P, Leclerc MC, Joossens M, Mondot S, Blottiere HM, Raes J, Ehrlich D, Dore J: A metagenomic insight into our gut’s microbiome. Gut 2013, 62:146–158; DOI: 10.1136/gutjnl-2011-301805. [DOI] [PubMed] [Google Scholar]
- 6.Mills S, Shanahan F, Stanton C, Hill C, Coffey A, Ross RP: Movers and shakers: influence of bacteriophages in shaping the mammalian gut microbiota. Gut Microbes 2013, 4:4–16; DOI: 10.4161/gmic.22371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.••. Fung TC, Olson CA, Hsiao EY: Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci 2017, 20:145–155; DOI: 10.1038/nn.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]; An exellent review on the interrelationships of the gut microbiome with the host immune system and nervous systems. The role of the gut-brain axis in brain development and behavior is discussed, as is the influcence of the microbiota in various diseases.
- 8.Gensollen T, Iyer SS, Kasper DL, Blumberg RS: How colonization by microbiota in early life shapes the immune system. Science 2016, 352:539–544; DOI: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.•. Abdel-Haq R, Schlachetzki JCM, Glass CK, Mazmanian SK: Microbiome-microglia connections via the gut-brain axis. J Exp Med 2019, 216:41–59; DOI: 10.1084/jem.20180794. [DOI] [PMC free article] [PubMed] [Google Scholar]; The role of microglia in the bidirectional gut-brain crosstalk and implications for the pathogenesis of diseases are discussed providing a mechanistic link between the microbiome and CNS.
- 10.•. Yoo BB, Mazmanian SK: The Enteric Network: Interactions between the Immune and Nervous Systems of the Gut. Immunity 2017, 46:910–926; DOI: 10.1016/j.immuni.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; Discussion of the enteric nervous system, the “second brain”, which is embedded in anatomy and physiology of the GIT and constitues a crucial interface between gut microbes, the immune system and the CNS.
- 11.Sharon G, Sampson TR, Geschwind DH, Mazmanian SK: The Central Nervous System and the Gut Microbiome. Cell 2016, 167:915–932; DOI: 10.1016/j.cell.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.••. Clarke G, Sandhu KV, Griffin BT, Dinan TG, Cryan JF, Hyland NP: Gut Reactions: Breaking Down Xenobiotic-Microbiome Interactions. Pharmacol Rev 2019, 71:198–224; DOI: 10.1124/pr.118.015768. [DOI] [PubMed] [Google Scholar]; An extensive review on the reciprocal relationships of the gut microbiome and xenobiotics.
- 13.Claus SP, Guillou H, Ellero-Simatos S: The gut microbiota: a major player in the toxicity of environmental pollutants? NPJ Biofilms Microbiomes 2016, 2:16003; DOI: 10.1038/npjbiofilms.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koppel N, Maini Rekdal V, Balskus EP: Chemical transformation of xenobiotics by the human gut microbiota. Science 2017, 356; DOI: 10.1126/science.aag2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.••. Spanogiannopoulos P, Bess EN, Carmody RN, Turnbaugh PJ: The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat Rev Microbiol 2016, 14:273–287; DOI: 10.1038/nrmicro.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]; An expert review on the underappreciated role of the “second genome”, the gut microbiome, in xenobiotic metabolism with a discussion of the great potential for development of novel precision therapeutics.
- 16.•. Lemire JA, Harrison JJ, Turner RJ: Antimicrobial activity of metals: mechanisms, molecular targets and applications. Nat Rev Microbiol 2013, 11:371–384; DOI: 10.1038/nrmicro3028. [DOI] [PubMed] [Google Scholar]; This review summarizes the antimicrobial activities of metals and discusses how to harness these activities for designing metal-based anti-microbials as alternatives to antibiotics.
- 17.Naujokas MF, Anderson B, Ahsan H, Aposhian HV, Graziano JH, Thompson C, Suk WA: The broad scope of health effects from chronic arsenic exposure: update on a worldwide public health problem. Environ Health Perspect 2013, 121:295–302; DOI: 10.1289/ehp.1205875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.•. Richardson JB, Dancy BCR, Horton CL, Lee YS, Madejczyk MS, Xu ZZ, Ackermann G, Humphrey G, Palacios G, Knight R, et al. : Exposure to toxic metals triggers unique responses from the rat gut microbiota. Sci Rep 2018, 8:6578; DOI: 10.1038/s41598-018-24931-w. [DOI] [PMC free article] [PubMed] [Google Scholar]; A well-designed study on the immediate effects of exposure to several metals that reveals unique responses of the gut microbiome to the studied metals.
- 19.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, et al. : Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013, 31:814–821; DOI: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim HS, Kim YJ, Seo YR: An Overview of Carcinogenic Heavy Metal: Molecular Toxicity Mechanism and Prevention. J Cancer Prev 2015, 20:232–240; DOI: 10.15430/JCP.2015.20.4.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.NTP (National Toxicology Program): Report on Carcinogens, Fourteenth Edition Edited by. Research Triangle Park: U.S. Department of Health and Human Services; 2016 [Google Scholar]
- 22.Ormerod KL, Wood DL, Lachner N, Gellatly SL, Daly JN, Parsons JD, Dal’Molin CG, Palfreyman RW, Nielsen LK, Cooper MA, et al. : Genomic characterization of the uncultured Bacteroidales family S24–7 inhabiting the guts of homeothermic animals. Microbiome 2016, 4:36; DOI: 10.1186/s40168-016-0181-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu K, Abo RP, Schlieper KA, Graffam ME, Levine S, Wishnok JS, Swenberg JA, Tannenbaum SR, Fox JG: Arsenic exposure perturbs the gut microbiome and its metabolic profile in mice: an integrated metagenomics and metabolomics analysis. Environ Health Perspect 2014, 122:284–291; DOI: 10.1289/ehp.1307429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chi L, Bian X, Gao B, Tu P, Ru H, Lu K: The Effects of an Environmentally Relevant Level of Arsenic on the Gut Microbiome and Its Functional Metagenome. Toxicol Sci 2017, 160:193–204; DOI: 10.1093/toxsci/kfx174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chi L, Bian X, Gao B, Ru H, Tu P, Lu K: Sex-Specific Effects of Arsenic Exposure on the Trajectory and Function of the Gut Microbiome. Chem Res Toxicol 2016, 29:949–951; DOI: 10.1021/acs.chemrestox.6b00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.•. Hoen AG, Madan JC, Li Z, Coker M, Lundgren SN, Morrison HG, Palys T, Jackson BP, Sogin ML, Cottingham KL, et al. : Sex-specific associations of infants’ gut microbiome with arsenic exposure in a US population. Sci Rep 2018, 8:12627; DOI: 10.1038/s41598-018-30581-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors established sex-specific relationships between urinary arsenic concentration and infant stool microbiome composition during a critical window of infant development in formula-fed infants.
- 27.Dong X, Shulzhenko N, Lemaitre J, Greer RL, Peremyslova K, Quamruzzaman Q, Rahman M, Hasan OS, Joya SA, Golam M, et al. : Arsenic exposure and intestinal microbiota in children from Sirajdikhan, Bangladesh. PLoS One 2017, 12:e0188487; DOI: 10.1371/journal.pone.0188487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gokulan K, Arnold MG, Jensen J, Vanlandingham M, Twaddle NC, Doerge DR, Cerniglia CE, Khare S: Exposure to Arsenite in CD-1 Mice during Juvenile and Adult Stages: Effects on Intestinal Microbiota and Gut-Associated Immune Status. MBio 2018, 9; DOI: 10.1128/mBio.01418-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.•. Coryell M, McAlpine M, Pinkham NV, McDermott TR Walk ST: The gut microbiome is required for full protection against acute arsenic toxicity in mouse models. Nat Commun 2018, 9:5424; DOI: 10.1038/s41467-018-07803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; This elegant study demonstrates that the gut microbiome and even a single bacterial species, Faecalibacterium prausnitzii, exert protective effects against arsenic exposure.
- 30.Lopez-Siles M, Duncan SH, Garcia-Gil LJ, Martinez-Medina M: Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J 2017, 11:841–852; DOI: 10.1038/ismej.2016.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rahman Z, Singh VP: The relative impact of toxic heavy metals (THMs) (arsenic (As), cadmium (Cd), chromium (Cr)(VI), mercury (Hg), and lead (Pb)) on the total environment: an overview. Environ Monit Assess 2019, 191:419; DOI: 10.1007/s10661-019-7528-7. [DOI] [PubMed] [Google Scholar]
- 32.Tinkov AA, Gritsenko VA, Skalnaya MG, Cherkasov SV, Aaseth J, Skalny AV: Gut as a target for cadmium toxicity. Environ Pollut 2018, 235:429–434; DOI: 10.1016/j.envpol.2017.12.114. [DOI] [PubMed] [Google Scholar]
- 33.Breton J, Massart S, Vandamme P, De Brandt E, Pot B, Foligne B: Ecotoxicology inside the gut: impact of heavy metals on the mouse microbiome. BMC Pharmacol Toxicol 2013, 14:62; DOI: 10.1186/2050-6511-14-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Li Y, Liu K, Shen J: Exposing to cadmium stress cause profound toxic effect on microbiota of the mice intestinal tract. PLoS One 2014, 9:e85323; DOI: 10.1371/journal.pone.0085323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S, Jin Y, Zeng Z, Liu Z, Fu Z: Subchronic Exposure of Mice to Cadmium Perturbs Their Hepatic Energy Metabolism and Gut Microbiome. Chem Res Toxicol 2015, 28:2000–2009; DOI: 10.1021/acs.chemrestox.5b00237. [DOI] [PubMed] [Google Scholar]
- 36.Laidlaw MA, Filippelli GM, Sadler RC, Gonzales CR, Ball AS, Mielke HW: Children’s Blood Lead Seasonality in Flint, Michigan (USA), and Soil-Sourced Lead Hazard Risks. Int J Environ Res Public Health 2016, 13:358; DOI: 10.3390/ijerph13040358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeWitt RD: Pediatric lead exposure and the water crisis in Flint, Michigan. JAAPA 2017, 30:43–46; DOI: 10.1097/01.JAA.0000511794.60054.eb. [DOI] [PubMed] [Google Scholar]
- 38.Gomez HF, Borgialli DA, Sharman M, Shah KK, Scolpino AJ, Oleske JM, Bogden JD: Analysis of blood lead levels of young children in Flint, Michigan before and during the 18-month switch to Flint River water. Clin Toxicol (Phila) 2019, 57:790–797; DOI: 10.1080/15563650.2018.1552003. [DOI] [PubMed] [Google Scholar]
- 39.Gao B, Chi L, Mahbub R, Bian X, Tu P, Ru H, Lu K: Multi-Omics Reveals that Lead Exposure Disturbs Gut Microbiome Development, Key Metabolites, and Metabolic Pathways. Chem Res Toxicol 2017, 30:996–1005; DOI: 10.1021/acs.chemrestox.6b00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Driscoll CT, Mason RP, Chan HM, Jacob DJ, Pirrone N: Mercury as a Global Pollutant: Sources, Pathways, and Effects. Environmental Science & Technology 2013, 47:4967–4983; DOI: 10.1021/es305071v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rothenberg SE, Keiser S, Ajami NJ, Wong MC, Gesell J, Petrosino JF, Johs A: The role of gut microbiota in fetal methylmercury exposure: Insights from a pilot study. Toxicol Lett 2016, 242:60–67; DOI: 10.1016/j.toxlet.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bridges KN, Zhang Y, Curran TE, Magnuson JT, Venables BJ, Durrer KE, Allen MS, Roberts AP: Alterations to the Intestinal Microbiome and Metabolome of Pimephales promelas and Mus musculus Following Exposure to Dietary Methylmercury. Environ Sci Technol 2018, 52:8774–8784; DOI: 10.1021/acs.est.8b01150. [DOI] [PubMed] [Google Scholar]
- 43.•. Roggenbeck BA, Leslie EM, Walk ST, Schmidt EE: Redox metabolism of ingested arsenic: Integrated activities of microbiome and host on toxicological outcomes. Current Opinion in Toxicology 2019, 13:90–98; DOI: 10.1016/j.cotox.2018.09.003. [DOI] [Google Scholar]; Detailed review on the interactions of arsenic with the microbiome and host summarizing current knowledge and unresolved aspects.
- 44.Monachese M, Burton JP, Reid G: Bioremediation and tolerance of humans to heavy metals through microbial processes: a potential role for probiotics? Appl Environ Microbiol 2012, 78:6397–6404; DOI: 10.1128/AEM.01665-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu G, Xiao X, Feng P, Xie F, Yu Z, Yuan W, Liu P, Li X: Gut remediation: a potential approach to reducing chromium accumulation using Lactobacillus plantarum TW1–1. Sci Rep 2017, 7:15000; DOI: 10.1038/s41598-017-15216-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Younan S, Sakita GZ, Albuquerque TR, Keller R, Bremer-Neto H: Chromium(VI) bioremediation by probiotics. J Sci Food Agric 2016, 96:3977–3982; DOI: 10.1002/jsfa.7725. [DOI] [PubMed] [Google Scholar]
- 47.Li H, Lin X, Zhao J, Cui L, Wang L, Gao Y, Li B, Chen C, Li YF: Intestinal Methylation and Demethylation of Mercury. Bull Environ Contam Toxicol 2019, 102:597–604; DOI: 10.1007/s00128-018-2512-4. [DOI] [PubMed] [Google Scholar]
- 48.Parks JM, Johs A, Podar M, Bridou R, Hurt RA Jr., Smith SD, Tomanicek SJ, Qian Y, Brown SD, Brandt CC, et al. : The genetic basis for bacterial mercury methylation. Science 2013, 339:1332–1335; DOI: 10.1126/science.1230667. [DOI] [PubMed] [Google Scholar]
- 49.Gilmour CC, Bullock AL, McBurney A, Podar M, Elias DA: Robust Mercury Methylation across Diverse Methanogenic Archaea. MBio 2018, 9; DOI: 10.1128/mBio.02403-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gilmour CC, Podar M, Bullock AL, Graham AM, Brown SD, Somenahally AC, Johs A, Hurt RA Jr., Bailey KL, Elias DA: Mercury methylation by novel microorganisms from new environments. Environ Sci Technol 2013, 47:11810–11820; DOI: 10.1021/es403075t. [DOI] [PubMed] [Google Scholar]
- 51.Podar M, Gilmour CC, Brandt CC, Soren A, Brown SD, Crable BR, Palumbo AV, Somenahally AC, Elias DA: Global prevalence and distribution of genes and microorganisms involved in mercury methylation. Sci Adv 2015, 1:e1500675; DOI: 10.1126/sciadv.1500675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Christensen GA, Somenahally AC, Moberly JG, Miller CM, King AJ, Gilmour CC, Brown SD, Podar M, Brandt CC, Brooks SC, et al. : Carbon Amendments Alter Microbial Community Structure and Net Mercury Methylation Potential in Sediments. Appl Environ Microbiol 2018, 84; DOI: 10.1128/AEM.01049-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bisanz JE, Enos MK, Mwanga JR, Changalucha J, Burton JP, Gloor GB, Reid G: Randomized open-label pilot study of the influence of probiotics and the gut microbiome on toxic metal levels in Tanzanian pregnant women and school children. MBio 2014, 5:e01580–01514; DOI: 10.1128/mBio.01580-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morton JT, Marotz C, Washburne A, Silverman J, Zaramela LS, Edlund A, Zengler K, Knight R: Establishing microbial composition measurement standards with reference frames. Nat Commun 2019, 10:2719; DOI: 10.1038/s41467-019-10656-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sinha R, Abu-Ali G, Vogtmann E, Fodor AA, Ren B, Amir A, Schwager E, Crabtree J, Ma S, Abnet CC, et al. : Assessment of variation in microbial community amplicon sequencing by the Microbiome Quality Control (MBQC) project consortium. Nat Biotechnol 2017, 35:1077–1086; DOI: 10.1038/nbt.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCoy KD, Geuking MB, Ronchi F: Gut Microbiome Standardization in Control and Experimental Mice. Curr Protoc Immunol 2017, 117:23 21 21–23 21 13; DOI: 10.1002/cpim.25. [DOI] [PubMed] [Google Scholar]
- 57.Assefa S, Ahles K, Bigelow S, Curtis JT, Köhler GA: Lactobacilli with probiotic potential in the prairie vole (Microtus ochrogaster). Gut Pathogens 2015, 7:1–16; DOI: 10.1186/s13099-015-0082-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Curtis JT, Assefa S, Francis A, Kohler GA: Fecal microbiota in the female prairie vole (Microtus ochrogaster). PLoS One 2018, 13:e0190648; DOI: 10.1371/journal.pone.0190648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.••. Tsiaoussis J, Antoniou MN, Koliarakis I, Mesnage R, Vardavas CI, Izotov BN, Psaroulaki A, Tsatsakis A: Effects of single and combined toxic exposures on the gut microbiome: Current knowledge and future directions. Toxicol Lett 2019, 312:72–97; DOI: 10.1016/j.toxlet.2019.04.014. [DOI] [PubMed] [Google Scholar]; The review expertly discusses the importance of investigating the impacts on the gut microbiome of real-life, combined exposures with pollutants, which to date have not been studied in sufficient depth.