Abstract
Nearly all cellular processes are sensitive to mechanical inputs, and this plays a major role in diverse physiological processes. Mechanical stimuli are thought to be primarily detected through force-induced changes in protein structure. Approximately a decade ago, molecular tension sensors were created to measure forces across proteins within cells. Since then, an impressive assortment of sensors has been created and provided key insights into mechanotransduction, but comparisons of measurements between various sensors are challenging. In this review, we discuss the different types of molecular tension sensors, provide a system of classification based on their molecular-scale mechanical properties, and highlight how new applications of these sensors are enabling measurements beyond the magnitude of tensile load. We suggest that an expanded understanding of the functionality of these sensors, as well as integration with other techniques, will lead to consensus amongst measurements as well as critical insights into the underlying mechanisms of mechanotransduction.
2. Introduction
The mechanical nature of the cellular microenvironment is a key determinant of many developmental, physiological, and pathophysiological processes, and is an important variable in tissue engineering and regenerative medicine [1,2]. In vivo, a deformable extracellular matrix and neighboring cells provide mechanical support, while applied forces often stem from physiological processes, such as shear stresses on vessel walls due to blood flow or the forces generated as cells migrate through or over tissues [3,4]. Over the past several decades, mechanical stimuli have been shown to regulate a wide range of fundamental cellular processes, including adhesion, contraction, migration, polarity, differentiation, and growth [5]. However, despite their ubiquitous nature, the molecular mechanisms mediating cellular mechanosensitivity remain poorly understood.
Efforts to understand cellular mechanosensitivity have traditionally focused on events where mechanical stimuli are converted into biochemically detectable signals, a process referred to as mechanotransduction [6]. A major advance was the in vitro demonstration that pico-Newton (pN) forces, which can be generated by a relatively small number of molecular motors, sufficiently deform proteins to control specific protein-protein interactions [7]. Thus, a simple mechanism for mechanotransduction is that applied load alters protein structure, changing function [8]. While a variety of in vitro systems have demonstrated that force could affect protein-protein interactions, an inability to measure such small forces in cells prevented probing the existence of these phenomena beyond in vitro experiments [9].
Over the past decade, a variety of tools were designed to meet the challenge of measuring the mechanical forces experienced by specific proteins in living cells [10]. Collectively referred to as molecular tension sensors (MTS), these tools have been constructed using the various building blocks of molecular engineering, encompassing synthetic, protein-based, and DNA-based approaches. However, as with most fields undergoing a drastic expansion, discord has emerged. For example, estimates for the force supported by integrins vary by over two orders of magnitude [11]. Rather than a technical quagmire, we suggest that these discrepancies represent an incomplete understanding of critical functional aspects of MTSs, as well as key aspects of protein mechanosensitivity. In this article, we will first review the various classes of MTSs, highlighting key features of design and function. Next, we outline important functional considerations that may help explain the large variance in measurements, focusing on aspects of single molecule calibration as well as the effect of the cellular microenvironment. Then, we discuss the capability of MTSs to report more than tensile force magnitude. Finally, we discuss how this broadened view reconciles some disparate reports and provides new insights into the mechanisms mediating mechanotransduction.
3. The Rise of Molecular Tension Sensors
The principle underlying MTSs is the coupling of a mechanically-induced deformation to a change in optical signal (Fig. 1) [12]. Two physical phenomena involving distance-dependent transfer of energy from an excited donor fluorophore have been leveraged. In Forster Resonance Energy Transfer (FRET), energy is transferred from an excited fluorophore to a nearby fluorophore or quencher [13]. In nanometal surface energy transfer (NSET), energy is transferred to a gold nanoparticle [14]. An extension-sensing module can be created by placing a deformable domain between two entities capable of FRET/NSET and then incorporating this module into a load-bearing protein or complex. This was first done for alpha-actinin [15]. However, to move from an extension sensor to a tension sensor, the relationship between the applied load and FRET/NSET must be determined. Typically, the tools of single molecule force spectroscopy (SMFS) are used to create a calibration curve that relates FRET/NSET to applied force magnitude [13,16–18].
Figure 1:
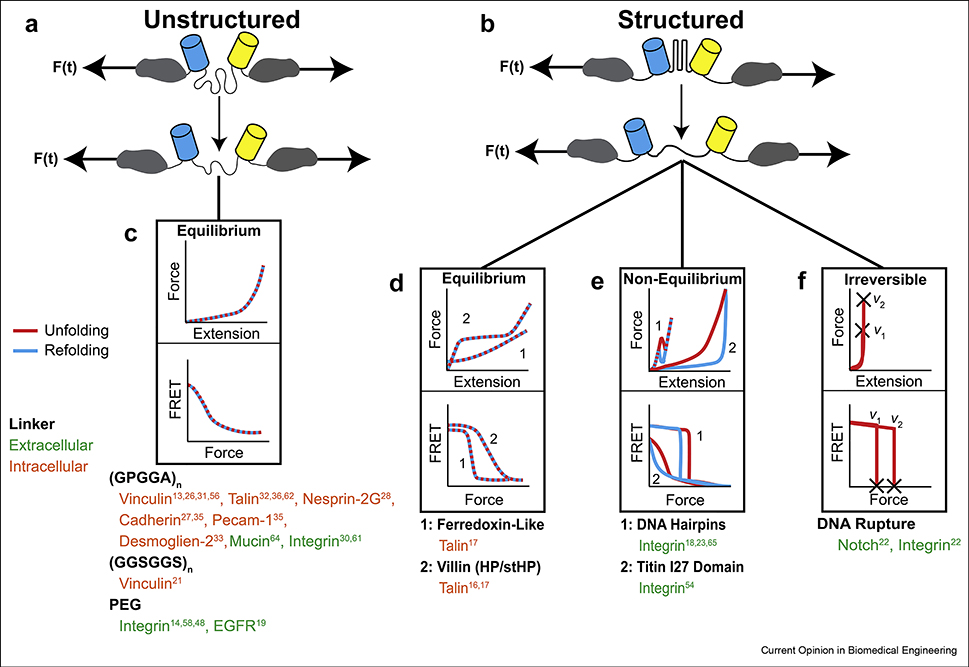
Molecular tension sensors can be classified based on linker mechanics. MTSs function by changes in the extension of a linker, inducing different FRET between two fluorophores flanking the linker. If the relationship between FRET and force is properly calibrated, the force across the linker can be determined. Broadly, linkers can be grouped into either (A) unstructured or (B) structured domains. (C) The force-extension relationship for unstructured linkers can be described according to the worm-like chain model, resulting in analog, monotonic FRET-force curves. The FRET-force relationship for structured domains depends on whether or not they extend at equilibrium. (D) For equilibrium linkers, the transition between states (e.g., folded or unfolded) happens much more quickly than changes in load and thus their response to load is not dependent on loading rate, duration, or history. Note that the unfolding and refolding curves overlay each other, indicating a lack of hysteresis. (E) However, if the transition rate is similar to changes in applied load, loading rate, duration, and history all affect the output of the sensor and they are not at equilibrium. In this case, the FRET-force curves are hysteretic. (F) Structured linkers can also undergo irreversible changes, which is an inherently non-equilibrium process, and thus depends on loading rate. The lists of MTS shown below each linker type are not comprehensive and represent specific examples we have chosen to focus on in this review.
Calibrated sensors can be classified by the nature of the extensible linker, which can be broadly described as either unstructured or structured (Fig. 1A,B). The original calibrated sensors were based on unstructured polypeptides (GPGGA)n or synthetic polymers (polyethylene glycol, PEG) (Fig. 1A) [13,19]. For unstructured linkers, the mechanical properties (Fig. 1C) can be quantitatively described using the worm-like chain model (WLC), which models the resistance to deformation as a reduction in entropy [20]. Sensors using unstructured linkers have analog, monotonic force-extension curves. Thus, subject to limits set by the temporal and ensemble averaging that occurs during imaging as well as the force dynamic range (the range of detectable forces) of the linker, these sensors report the exact load across the sensor. Importantly, given the wealth of theoretical and experimental characterization at the single-molecule level, the mechanical response of unstructured polypeptide linkers can be rationally tuned by controlling linker stiffness and length [19,21]. Recently, using different combinations of FRET-pairs, linker stiffnesses, and linker lengths, the force dynamic range of 1080 unique unstructured polypeptide MTSs were computationally predicted [21]. This resource enables the rational design of unstructured MTS through prediction of both the force dynamic range as well as the optimal FRET dynamic range. This tunability of the sensor properties is a main advantage of using unstructured linkers. Predictions suggest that sensors with different force dynamic ranges spanning 1–15 pN can be created with this approach. However, as extension of the linker begins instantaneously with the application of load, the force dynamic range and sensitivity (change in FRET per force) are coupled, resulting in sensors with high force dynamic ranges having smaller sensitivities.
To create MTSs with both high force dynamic ranges and sufficiently high sensitivities, a new class of extensible domains was needed, so researchers developed MTSs with structured domains as the deformable element. These domains are relatively inextensible at low forces, allowing the deformation and resulting change in FRET signal to be shifted to higher forces. (Fig. 1B). Key examples include ultrafast folding protein domains found in villin and ferredoxin (Fig. 1D), DNA hairpins (Fig. 1E), and DNA duplexes (tension gauge tethers, TGTs) (Fig. 1F) [17,22,23]. The mechanical response of these linkers can be conceptualized as two- or multi-state systems [24]. At low forces the linker is largely inextensible, and resistance to deformation mostly comes from enthalpic effects associated with non-covalent bond breaking. Under sufficient load, the linker experiences a transition to the unfolded or ruptured state (resulting in a rapid increase in extension) and adopts a largely unstructured conformation where mechanical resistance is due to entropic effects and well-described by the WLC model [16]. Structured sensors can undergo sharp transitions, displaying an approximately digital response. Digital sensors do not have a 1:1 relationship between FRET/NSET and force magnitude, limiting the accuracy of the force measurement away from the transition state. However, this functionality can provide accurate measurements of when the load experienced by the protein exceeds the transition threshold. Notably, the ability to compare areas of higher and lower tension (with respect to the transition threshold) is often what is most applicable to studies in cell or molecular biology. MTSs with structured domains, which often have a higher signal to noise ratio than unstructured sensors, may be particularly advantageous in these studies.
4. With Many Tension Sensors Comes Many Tension Measurements
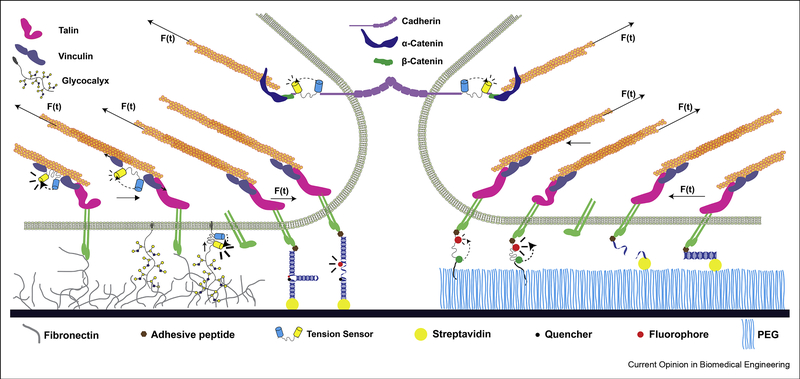
MTSs have been used to study the loads experienced by structural or adhesive proteins in both intra- and extracellular contexts (Fig. 2). Genetically-encoded sensors, which leverage cell machinery to produce sensors from recombinant DNA, are uniquely suited to probe the molecular forces within cells [25]. These sensors have been applied to proteins within focal adhesions, cell-cell contacts, and the nuclear membrane (Fig. 2) [16,17,26–33]. DNA-based and synthetic sensors enable greater design freedom, such as the incorporation of organic fluorophores, but are limited to extracellular applications due to challenges in delivery and/or limited stability inside cells. These sensors have been used to study forces experienced by cellular receptors, including integrins (Fig. 2) [18,22,23,34].
Figure 2:
Molecular tension sensors have been employed to study the load across a wide variety of mechanosensitive proteins. An unstructured linker flanked by two fluorescent proteins was originally genetically-encoded into vinculin and used to measure vinculin tension within focal adhesion [13]. The same linker has subsequently been applied to many different proteins, including the intracellular domain of cadherins [27,35], integrins [30,61], talin [32,36,62], PECAM [35], Nesprin-2G [28], and others [33,64]. Sensors with structured linkers were developed and applied to talin [16,17,62], integrins [18,22,23,54,65], and growth factor receptors [22]. These sensors include ultrafast folding structured protein domains to study load in talin (not shown), DNA hairpins which undergo an unfolding transition and rapidly reduce the quenching of a fluorophore at higher forces (shown here presenting an adhesive ligand), and DNA rupture probes, which separate irreversibly at higher force (also shown presenting an adhesive ligand). The spacing and relative size of each component was chosen to highlight MTS implementation and is not meant to perfectly capture real-world conditions (e.g., the glycocalyx is only shown in one region for clarity).
As diverse sensors have been developed, disparate measurements of the forces experienced by similar/identical proteins have emerged. Various cadherins have been reported to be under either constitutive loading or to only bear load within junctions [27,35]. The loads across talin have been reported to be both less than 6 pN or between 7–10 pN, with a small subset greater than 10 pN [17,36]. Perhaps more strikingly, tension measurements across individual ligated integrins range from 1 to over 100 pN [11]. The origins of these discrepancies are not immediately clear, but two obvious sources are the accuracy and appropriateness of the sensor calibrations, as well as differences between experimental conditions, including the choice of the sensor type.
As calibrations are used to convert extension (measured by NSET/FRET) into force, errors in calibration can directly account for discrepancies in reported force magnitude. Typically, calibration involves the use of SMFS techniques to directly assess the force-extension properties of the linkers. For the calibrations to be applicable to MTSs, at least three conditions must be met. Specifically, the calibration measurements must be conducted at equilibrium, in the correct environment, and account for the effects of load on each component of the sensor.
In all sensor designs to date, it has been assumed that changes in FRET/NSET are solely due to force magnitude. This requires that the extension/transition of the sensors occur at equilibrium (Fig. 3A). In non-equilibrium transitions, the apparent change in FRET/NSET will be dependent on the loading rate and history. The key aspects can be understood through a thermodynamic two-state model [24] (Fig. 3A). If the unfolding and refolding rates are fast in comparison to experimental time scales, then the system will sample all states and be in equilibrium. Thus, transitions will be induced quickly when the critical force is applied and the change in optical signal will occur at a single force magnitude. Sensors based on unstructured domains or ultrafast folding domains exhibit sufficiently rapid kinetics to undergo equilibrium transitions at all plausible loading rates within cells and the vast majority of SMFS systems [13,37].
Figure 3:
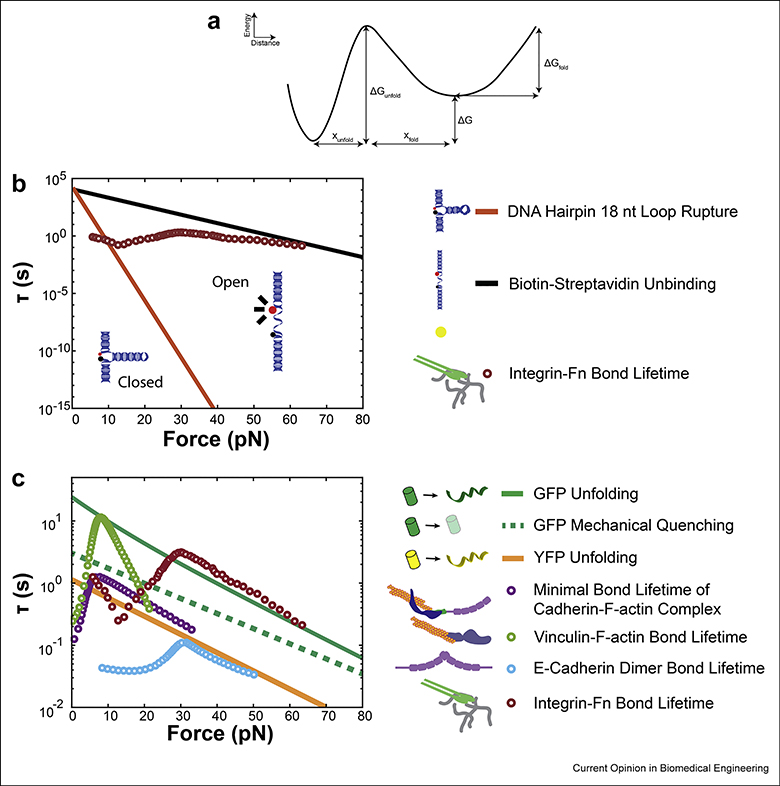
An order of magnitude comparison between two-state transition kinetics for common sensor components (lines) and the measured bond lifetimes of mechanosensitive proteins via single molecule force spectroscopy (SMFS, circles). A: A schematic of the energy landscape for a two-state transition. The wells represent the folded or unfolded states, with a transition distance (x, the increase in extension following force-induced unfolding) and activation energy (G) between the two. A detailed theoretical description can be found in [24]. B: Competing folded lifetimes for different MTS components influences force reporting. DNA hairpins undergo a rapid decrease in folded lifetime (τ, in seconds) as force is increased [41]. Biotin-streptavidin has a much longer folded lifetime at high forces [45]. An overlay of bond lifetime for integrins bound to fibronectin [46] shows that, for this example DNA hairpin there is a range of low forces where the integrin would unbind the ligand before sensor unfolding, a range of moderate forces where the sensor would unfold first, and a range of high forces where biotin may dissociate from streptavidin before integrin unbinding. C: Genetically-encoded linkers use fluorescent proteins (FPs) which bear the same force as the linker. FPs can experience mechanical quenching or unfolding. A plot of the lifetimes for GFP unfolding [51] and mechanical quenching [52] shows that unfolding occurs at a higher force than quenching. An overlay of bond lifetimes for commonly-studied mechanosensitive proteins [46,66–68] shows that many are capable (in single molecule experiments) of maintaining longer bond lifetimes than the lifetime for GFP quenching and, in the case of integrin ligation, may also be able to unfold GFP. This is corroborated by experimental data using cells [54]. We also provide an estimate of YFP unfolding lifetime based on data extracted from literature [50] and fit to a two-state transition model using methodology described in [79,80]. According to these data, YFP folded lifetime is much lower than GFP at physiologic forces, and importantly, is well below the regime of bond lifetime for actin-bound vinculin and cadherin observed in literature. This has implications for the selection and use of FPs in genetically-encoded sensors.
Non-equilibrium transitions occur when all of the states cannot be sampled. In this scenario, the system will exhibit kinetic or history-dependent effects [24]. In the context of force-induced changes in protein conformation, this occurs in two contexts. The first is in irreversible systems, when either unfolding or refolding rates drops to zero. Sensors that undergo irreversible transitions are clearly not at equilibrium, and the effects of loading history must be considered in the use and analysis of these sensors. The transitions induced by TGTs are irreversible, as the DNA strand containing the adhesive ligand unbinds and is then internalized by the cell [22]. Therefore, TGT rupture forces will depend on loading conditions. For instance, as TGTs unbinding is irreversible and mediated by non-covalent interactions, these domains will eventually rupture under any load given sufficient time [38,39]. This substantially limits the interpretability of these sensors as well as the biological phenomena they are capable of studying. Non-equilibrium effects can also be observed if loading happens at rates that approach the unfolding and refolding rates, and thus non-equilibrium effects are more likely to occur as the loading rate increases. In SMFS experiments, hysteresis has been observed for DNA structures that can undergo reversible transitions, such as hairpins, at relatively low loading rates (down to 30 nm/s) [40,41]. Determining if non-equilibrium transitions occur during MTS experiments requires knowledge of pertinent loading rates inside cells. However, the loading conditions imposed by cells on proteins are largely uncharacterized and very challenging to probe. One measurement has been made by direct, two-color single molecule imaging of the termini of talin dimers. These estimates revealed the loading to be approximately 10–60 nm/s with high spatial and temporal variation [42]. Together, the current data demonstrate that DNA-based sensors could potentially be undergoing non-equilibrium transitions when used as extracellular MTSs, and this may affect the ability of these sensors to accurately report force magnitude [40].
Second, sensors must be calibrated in an appropriate environment. This is particularly relevant for genetically-encoded sensors, which are intended to be used within cells but are typically calibrated using SMFS techniques in vitro. Therefore, differences in pH, salt concentration, protein concentration (which could induce molecular crowding effects), as well as the concentration of ATP (which is a biological hydrotrope and affects the solubility of disordered proteins [43]) could lead to differences in the behavior of the extensible domains. This has been observed for unstructured polypeptides, as distinct mechanical properties were noted between in vitro solutions and living cells [21]. This is likely due to differences in pH, salt concentration, and/or ATP, as crowding effects are proportional to linker length, and primarily affect longer linkers (greater than 70 amino acids), rather than the 30–50 amino acid linkers used to create unstructured extensible domains for MTSs [44]. To bypass these issues, a modeling-based approach utilizing linker mechanical properties measured in cells has been developed to predict the force dynamic range of these sensors, bypassing a need for in vitro calibration and removing a potential source of measurement discrepancies [21]. As the mechanical response of genetically-encoded structured linkers cannot be predicted a priori from models, this class of sensor still requires in vitro calibration with SMFS techniques. The effect of environmental conditions on genetically-encoded structured linkers is currently unknown and ensuring the direct applicability of in vitro calibration for use in cellulo or in vivo is a major goal of current work.
Third, the effects of load on the different MTS components must be considered. This is relevant for the linker, the fluorescent proteins, and the linkages between components of the MTS. For extracellular MTSs, integrin to ligand, linker, and linker to substrate interfaces are all subject to force-sensitive unbinding/rupture kinetics at competing timescales. The effects are most pronounced in DNA TGTs, which will rupture under any applied load given sufficient loading time [38]. Although this precludes their ability to report force magnitude, computational analyses of TGTs suggest that placing two TGTs in series would enable the reporting of force history with high resolution [39]. Notably, the integrin-FN linkage is dynamic and force-sensitive. This leads to a complex relationship between integrin-FN unbinding and TGT rupture. Although this effect is not as pronounced due to the reversibility of hairpin rupture, similar effects can be observed with DNA hairpins. Additionally, the linkage between the MTS to the substrate is subject to force-dependent dynamics. Figure 3B shows an overlay of SMFS data for integrin-FN bond lifetime, DNA hairpin folded lifetime (τ(F)), and biotin-streptavidin bond lifetime (τ(F)). There are three key regimes. At low forces, the lifetime of the integrin-FN bond is shorter than the timescale for DNA hairpin unfolding. At intermediate forces, the integrin-FN bond lifetime is long enough to unfold the hairpin. At high forces, the integrin-FN bond is long enough to dissociate biotin from streptavidin (Fig. 3B) [41,45–47]. Notably, hairpins are assumed to function in the intermediate regime, where the hairpin is the shortest-lived component. Thus, large differences in reported force magnitude involving DNA hairpins could be compounded by these dynamic effects. Dissociation of non-covalent linkages between a complex and substrate has been observed during MTS use and in SMFS experiments probing the force-sensitive unbinding kinetics of FN-ligated integrins, where the antibody-Fc fragment interaction used to link probes to the surface dissociated before the integrin:FN bond [46,48]. This means that the lifetime of the integrin-FN linkage at high forces (Fig. 3B) may be underestimated. Therefore, as more diverse extracellular tension sensors are developed, the effect of load on each component in an MTS must be validated [48,49].
The same concept of force-sensitive components applies to genetically-encoded sensors. Genetically-encoded sensors use FPs to measure linker extension via FRET. SMFS experiments, as well as molecular dynamics simulations, have shown that load can have substantial effects on FP optical properties, including the loss of fluorescence via mechanical quenching at lower forces and complete denaturing of the protein at very high forces (Fig. 3C) [50–53]. These processes can also be described as two- or multi-state systems and are likely subject to non-equilibrium transitions which depend upon both the force magnitude and duration [51,52]. Thus, the effect of load on FPs is an important consideration when designing and interpreting MTSs. However, the relevance of this phenomenon for genetically-encoded MTSs has not yet been directly probed. To estimate the relative chance of FP mechanical quenching or unfolding during MTS use, we compared data (generated via SMFS experiments) on force-sensitive bond lifetimes of mechanosensitive proteins with GFP/YFP denaturing and GFP mechanical quenching lifetimes (τ(F), Fig. 3C). At certain forces, the lifetimes of some of the mechanical linkages, such as the integrin:FN catch bond, are longer than the timescales required to induce mechanical quenching or denaturation, suggesting that forces within the cell may affect the function of genetically-encoded MTSs. Notably, this interplay might be quite complex, as the lifetimes of the vinculin:actin catch bond can likely induce mechanical quenching, but are on the threshold for mechanical denaturation of GFP. An uncalibrated extracellular MTS has demonstrated irreversible GFP denaturation under mechanical load from integrins [54]. In contrast, SMFS experiments have suggested that the YPET-mCherry FRET pair did not denature in response to loads of 24 pN applied for 300 sec [16]. These data suggest that differences in the mechanical robustness of various FPs may be quite large, and the suitability of the FPs should be evaluated before use in genetically-encoded MTSs.
The technical details of MTS interpretation and implementation outlined above are likely to contribute to much, but not all, of the discrepancies in reported force magnitude. They show how some MTSs are not suited to solely report force magnitude but are sensitive to additional mechanical variables, such as loading rate or loading duration.
5. Learning Through Discrepancies: Moving Beyond Tension
Another reason for variation in measurements across various MTSs is due to relevant differences between biological systems. The forces generated by cells vary spatiotemporally in magnitude and duration, are regulated by cell signaling, propagate through a dynamic cytoskeleton, and change in response to microenvironmental cues [55]. This suggests that loads experienced by proteins are equally complex. Consistent with this idea, the molecular load of several structural proteins has been shown to spatially vary across the cell, within individual adhesion structures, and even within an individual protein [17,21,56,57]. Small variations in integrin load have been observed with modulated spacing of engineered ligands, and talin load is reduced on softer gels [17,58]. Notably, recent models of mechanotransduction have focused on the importance of loading rate for the key structural proteins in the cytoskeleton [59,60]. However, a common assumption when interpreting MTS data is that proteins are subject to static tensile loads and thus load magnitude is the sole key parameter. In this section, we first review recently developed techniques for probing diverse aspects of the mechanical loads experienced by proteins in living cells. In the next section, we then illustrate how sensors with different mechanical properties, as well as differences between measurements with various sensors, could illustrate key aspects of the loading, behavior, and function of mechanosensitive proteins.
A percentage of structural proteins within load-bearing structures do not bear substantial force, suggesting that the percentage of loaded molecules is a key variable. Notably, the percentage of loaded proteins can be calculated directly with single molecule imaging techniques (Fig 4A) [61]. Additionally, switch-like genetically-encoded MTSs that respond to small forces have been made to estimate the percentage of proteins subject to mechanical load [17]. A common finding in these systems is that a substantial portion of structural proteins within adhesive structures are not subject to mechanical load. For instance, it was estimated that as few as 40 – 70% of talin molecules (depending on where the sensor was inserted within talin) within an FA support significant load [17]. A similar trend was observed in Drosophila, as the percentage of loaded talin reduced as muscles developed [62]. Also, an unstructured extracellular MTS revealed that the majority of integrins experienced low forces, but could be loaded quickly in response to key stimuli [61]. In total, these data suggest that mechanical redundancy could be a common theme in load-bearing structures, and that cells may regulate molecular engagement as a part of mechanosensitive signaling in addition to regulating load. Furthermore, such mechanical redundancy could explain observations of low force magnitudes from ensemble measurements while single molecule studies yield much higher and more variable measurements.
Figure 4:
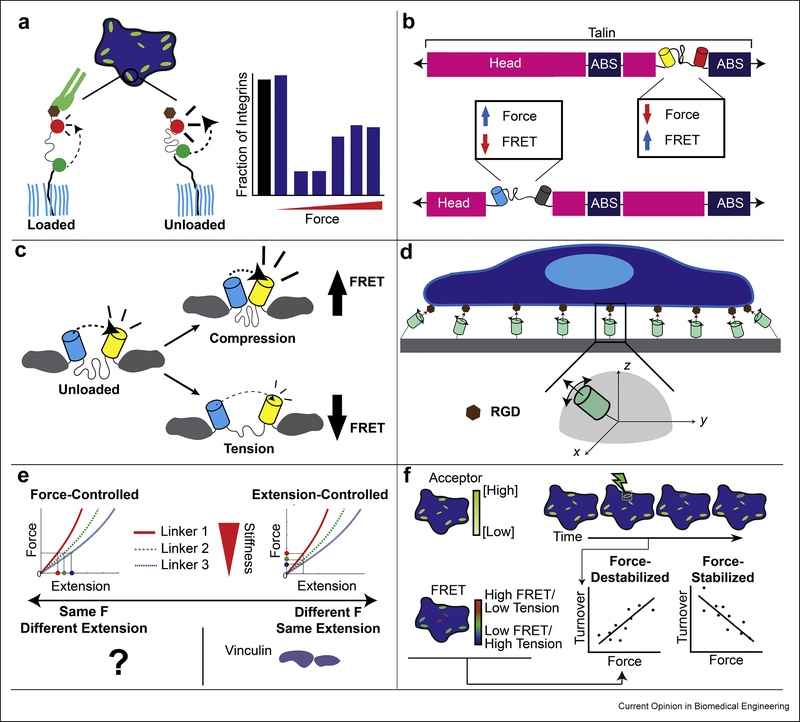
Newly designed molecular tension sensors and the combination of molecular tension sensors with other techniques enables the elucidation of more than just force. A: Different techniques (including single molecule fluorescence) can show the percentage of MTSs which are under load. A study of the distribution of integrin loading showed that, within an FA, the distribution is skewed and context-dependent [61]. B: Multiplexing MTSs in different regions within a protein has shown that force is not uniform across an entire protein. Two sensors inserted into different regions within talin showed that force was heterogeneously distributed. A sensor between the head and rod domain showed forces >7 pN, while a sensor between two actin binding sites showed forces >3 pN [17]. C: In addition to reporting tension, sensors can also report compression. Reports of compression within mucin [64] and vinculin (in cells under geometric constraint, [56]) have been made. D: Excitation-resolved fluorescence polarization microscopy, combined with careful sensor design, can report both the orientation and magnitude of load. A study of integrin force orientation showed that it was highly aligned within FAs [65]. E: Extensible linkers with different properties can differentiate between force and extension-based control of protein loading. Using 3 carefully designed sensors within vinculin showed that it was under extension, not force control [21]. F: Combining FRET on the vinculin MTS with FRAP on the acceptor fluorophore can show how protein dynamics and force are related: protein turnover can increase (force-destabilized) or decrease (force-stabilized) in response to force. A study of the force-sensitive dynamics of vinculin showed that they were fundamental to a variety of cell processes, including migration [26].
In MTS studies, proteins and the sensing modules are often conceptualized as springs in series, where the force across the module and protein are the same. However, for complex proteins this simple assumption does not always hold, as demonstrated by the discovery that forces vary even within the same protein [17]. Through the clever design of multiple sensors using a combination of FPs and genetically-encoded quenchers, MTSs can be multiplexed at different points within a protein. Used in two positions within talin, these MTSs reported different tension, demonstrating that loads can vary across individual talin molecules (Fig. 4B) [17]. These differences are likely due to multiple vinculin and actin binding sites throughout talin, which could serve as different nodes for mechanotransduction. Therefore, discrepancies between MTSs for the same protein, with the tension sensing module located in different positions, could be indicative of multiple points of loading within the protein.
As force is a vector, its full description requires a magnitude and a direction. In many SMFS experiments the direction is implicitly known because loads are assumed to be applied along the axis of linker molecules. In cells, the direction of the applied load is unknown. Traction force experiments have revealed that cells can generate contractile shear stress as well as compressive normal forces on the surrounding microenvironment [63], demonstrating that the direction of the applied load is variable. MTSs based on unstructured domains essentially act like springs with a rest length. Thus, while tensile loads are detected through a reduction in FRET/NSET from the unloaded state, compression can be detected though an increase in FRET/NSET from the unloaded state (Fig. 4C). To date these measurements are qualitative as the sensors have not been calibrated for compressive loads, but both vinculin and mucin-1 have been shown to be subject to compressive loads [56,64]. Discrepancies involving FRET/NSET efficiencies higher than those in the unloaded state in unstructured sensors and no apparent load in structured sensors could be indicative of significant compressive loads. Furthermore, a key advance was the combination of MTSs with polarized light microscopy, enabling measurement of tensile load orientation (Fig. 4D) [65]. As the orientation and geometry of how a noncovalent linkage bears tension affects its bond strength and lifetimes [45], the ability to detect orientation may lead to significant advances in the understanding of integrin activation as well as the activation, or inhibition, of mechanosensitive signaling.
Alterations in the stiffness of the microenvironment lead to complex temporal changes in cell-generated stresses [55]. At the molecular level, the simplest possible explanation is that proteins involved in mechanosensing are regulated by either a specific force (force-control) or by a set extension (extension-control). Possibly inspired by SMFS force clamp experiments and classical ideas in cell biology like tensional homeostasis, force-control has been broadly assumed. Recently a suite of sensors with distinct force-extension curves were used to distinguish between force and extension-control paradigms in vinculin [21]. Surprisingly, extension control was observed (Fig. 4E). The generalizability of this result to other proteins is unknown and provides an interesting topic for further investigation. Discrepancies between loads reported by sensors with the same force dynamic range but different linker rigidities could be indicative of extension control or other more complex regulatory schemes. While the molecular basis of extension control is not known, simple physical arguments suggest it occurs in response to the loading of adhesive structures being subject to regulated displacements instead of forces [21]. Also, if mechanosensitive signaling is induced by the binding of a protein within a cryptic binding site, molecular extension might be a more pertinent regulating factor than force.
Very little is known about the loading rates, durations, and histories of proteins within or bound to a cell. SMFS experiments have established that protein-protein interactions can demonstrate both slip and catch behavior, where bond durations are reduced or increased respectively under load [46,66–68]. To probe the relationship between mechanical load and protein dynamics in living cells, a new technique combining FRET-based tension sensors with Fluorescence Recovery After Photobleaching (FRAP) was developed (FRET-FRAP) [26]. Utilizing this technique, the relationship between load, as determined by the tension sensor, and dynamics, as measured with FRAP, can be probed in cells (Fig. 4F) [26]. Vinculin SMFS experiments have shown that open vinculin and actin form a catch bond [66]. In cells, FRET-FRAP reported force-stabilized dynamics (higher force correlated with slower turnover), potentially consistent with catch bond behavior. Furthermore, forcing vinculin into a closed conformation, which also localizes to FAs, leads to force-destabilized dynamics (higher force correlated with faster turnover) [69]. The ability of vinculin to exhibit force-stabilized dynamics is required for cells to undergo migration through pores [70,71]. Discrepancies between tension sensors with similar force dynamic ranges but with either equilibrium or non-equilibrium transitions, could be indicative of differences in loading duration, rate, or history. Notably, the conformation (e.g. open versus closed) of vinculin is subject to biochemical regulation [69,71], Therefore, vinculin may act as a regulatable “mechanical switch,” allowing the same force magnitude to have different effects depending on the biochemical state within the cell. FRET-FRAP is an important step forward in the development of MTS-based techniques that probe the effects of force on protein function in living cells.
6. The Future of Molecular Tension Sensors
Molecular tension sensors have greatly expanded our understanding of mechanobiology. However, recent disparate results have caused confusion in the field. We suggest that these discrepancies are in part due to misinterpretation of MTSs, but mainly indicative of the complexity of the underlying mechanobiology. To date, integrins are the protein most studied with MTSs of different types and have the greatest disparity in measurements [11]. In hindsight, perhaps this should not be surprising. Integrins are complex molecules that are known to be sensitive to more than the magnitude of an applied force. For instance, the direction of the applied load as well as the loading history is thought to play a key role in the activation of integrins [72,73]. Once active, changes in force magnitude can further alter integrin conformation, affecting their affinity and enhancing their ability to support load through catch bond formation [46,47,73,74]. Also, as the transitions between the active states are thought to be rather slow, they may be non-equilibrium processes. Consistent with this idea, the loading history of the integrin affects its ability to become activated and its ability to bear load in complex waves, such as cyclic priming [47].
Integrins are known to be key mediators of mechanosensitive signaling and cell contractility regulation [75,76]. Thus, their loading could in turn activate cell signaling pathways that regulate bulk cell contractility, which in turn could influence integrin conformation, alterations in bound ligands, as well as local alignment of cytoskeleton structures and FA components [36,72–74,77,78]. Given this complex behavior, ability to respond to diverse cues, and a loading history dependence, it is not surprising that the magnitude of forces integrins are reported to support is widely variable. Additionally, some of this variability may have been introduced by the sensors themselves. While MTSs with linkers of different stiffnesses should not affect the reported force magnitude in a force-control regime, it would cause the cell to exert larger forces in an extension-control regime. Therefore, it is possible that the force discrepancies are at least partially induced by the introduction of stiffer sensors. Consistent with this idea, the largest measurements of integrin force are observed with sensors based on the unfolding of GFP or I27, which comprise the stiffest deformable elements used thus far [54].
Since many mechanosensitive processes depend on loading rate, duration, and history, sensors which are sensitive to these parameters may help to reveal important mechanisms of mechanotransduction. If the transition rates of non-equilibrium sensors were matched to those of key mechanosensitive proteins, changes in optical properties could indicate a conformational change in the protein of interest. For example, FP mechanical quenching/unfolding is a non-equilibrium process with different force-dependent lifetimes between variants (e.g., GFP vs. YFP, Fig 3C) [50–52]. FP variants with similar transition kinetics to a load-bearing protein of interest could be used to illuminate whether that protein has experienced a transition. Structured non-equilibrium sensors whose transition kinetics can be tuned, such as DNA hairpins, could be used in a similar manner. This approach could be applied in series with an unstructured MTS to add context to the force magnitude reported by the unstructured MTS. Therefore, non-equilibrium sensors with proper calibration may be well suited to fulfilling the original motivation of MTS design: understanding mechanotransduction.
Furthermore, the techniques currently being developed through the combination of MTSs with other technologies could be further improved through integration with single molecule imaging. This may enable further elucidation of the roles loading rate, duration, and history play in mechanotransduction. For instance, rate information is challenging to extract from ensemble average measurements of an unstructured linker. The use of time-resolved super-resolution microscopy to image protein extension at the single molecule level inside cells has enabled the measurement of extension rate and could be applied to unstructured MTSs to measure both the magnitude and rate of force application [42]. These measurements could also be coupled with non-equilibrium structured linkers to detect non-equilibrium transitions in protein conformation and probe how force magnitude, loading rate, and history drive changes in protein conformation. Additionally, the combination of single molecule imaging with multiple unstructured MTSs of different stiffnesses could be used to directly determine key scales in extension and force control. Approaches to measure force at the single molecule level could also be combined with particle tracking to provide direct measurements of the relationship between force and dynamics, potentially providing a bridge between the force stabilized/destabilized dynamics observed with FRET-FRAP and catch/slip bonds observed in SMFS experiments.
7. Conclusion
In summary, we conclude that the future for MTS is bright. An appreciation for the function of the various MTSs as well as the underlying mechanobiology will stimulate the design of sensors capable of reporting the plethora of mechanical cues experienced by cells and proteins beyond force magnitude, enabling further advancement towards a molecular scale understanding of mechanotransduction.
8. Acknowledgements
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number 5R01GM121739 and by a National Science Foundation CAREER award, both to Dr. Brenton Hoffman. The views presented here do not necessarily reflect those of the NSF or NIGMS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
9. References
- 1.Lampi MC, Reinhart-King CA, Targeting extracellular matrix stiffness to attenuate disease: From molecular mechanisms to clinical trials, Sci Transl Med. 10 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Matellan C, Del Rio Hernandez AE, Engineering the cellular mechanical microenvironment - from bulk mechanics to the nanoscale, J Cell Sci. 132 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Lintz M, Munoz A, Reinhart-King CA, The Mechanics of Single Cell and Collective Migration of Tumor Cells, J Biomech Eng. 139 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marsden AL, Truskey GA, The future of biomedical engineering – Vascular bioengineering, Current Opinion in Biomedical Engineering. 5 (2018) iii–v. [Google Scholar]
- 5.LaCroix AS, Rothenberg KE, Hoffman BD, Molecular-Scale Tools for Studying Mechanotransduction, In: Annual Review of Biomedical Engineering, Vol 17 Edited by Yarmush ML; 2015:287–316. [DOI] [PubMed] [Google Scholar]
- 6.Hoffman BD, Grashoff C, Schwartz MA, Dynamic molecular processes mediate cellular mechanotransduction, Nature. 475 (2011) 316–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP, Stretching single talin rod molecules activates vinculin binding, Science. 323 (2009) 638–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orr AW, Helmke BP, Blackman BR, Schwartz MA, Mechanisms of mechanotransduction, Dev Cell. 10 (2006) 11–20. [DOI] [PubMed] [Google Scholar]
- 9.Hu X, Margadant FM, Yao M, Sheetz MP, Molecular stretching modulates mechanosensing pathways, Protein Science. 26 (2017) 1337–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freikamp A, Mehlich A, Klingner C, Grashoff C, Investigating piconewton forces in cells by FRET-based molecular force microscopy, Journal of Structural Biology. 197 (2017) 37–42. [DOI] [PubMed] [Google Scholar]
- 11.Erickson HP, Protein unfolding under isometric tension-what force can integrins generate, and can it unfold FNIII domains?, Curr Opin Struct Biol. 42 (2017) 98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LaCroix AS, Rothenberg KE, Berginski ME, Urs AN, Hoffman BD, Construction, imaging, and analysis of FRET-based tension sensors in living cells, Methods Cell Biol. 125 (2015) 161–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T, et al. , Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics, Nature. 466 (2010) 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Yehl K, Narui Y, Salaita K, Tension Sensing Nanoparticles for Mechano-Imaging at the Living/Nonliving Interface, Journal of the American Chemical Society. 135 (2013) 5320–5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meng F, Suchyna TM, Sachs F, A fluorescence energy transfer-based mechanical stress sensor for specific proteins in situ, Febs Journal. 275 (2008) 3072–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Austen K, Ringer P, Mehlich A, Chrostek-Grashoff A, Kluger C, Klingner C, Sabass B, Zent R, Rief M, Grashoff C, Extracellular rigidity sensing by talin isoform-specific mechanical linkages, Nature Cell Biology. 17 (2015) 1597–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ringer P, Weissl A, Cost AL, Freikamp A, Sabass B, Mehlich A, Tramier M, Rief M, Grashoff C, Multiplexing molecular tension sensors reveals piconewton force gradient across talin-1, Nat Methods. 14 (2017) 1090–1096.** This paper details the development and multiplexing of structured tension sensors based on ultrafast folding protein domains. The sensors are applied to different regions within talin, and the authors show that loads are distinct in various regions of talin. They also show that not all talin molecules are loaded at once, consistent with the idea of mechanical redundancy in adhesion structures.
- 18.Blakely BL, Dumelin CE, Trappmann B, McGregor LM, Choi CK, Anthony PC, Duesterberg VK, Baker BM, Block SM, Liu DR, et al. , A DNA-based molecular probe for optically reporting cellular traction forces, Nature Methods. 11 (2014) 1229−+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stabley DR, Jurchenko C, Marshall SS, Salaita KS, Visualizing mechanical tension across membrane receptors with a fluorescent sensor, Nature Methods. 9 (2012) 64–U172. [DOI] [PubMed] [Google Scholar]
- 20.Becker NB, Rosa A, Everaers R, The radial distribution function of worm-like chains, Eur Phys J E Soft Matter. 32 (2010) 53–69. [DOI] [PubMed] [Google Scholar]
- 21.LaCroix AS, Lynch AD, Berginski ME, Hoffman BD, Tunable molecular tension sensors reveal extension-based control of vinculin loading, Elife. 7 (2018).** This paper details a method for calibrating unstructured polypeptide MTSs in cells and computationally predicts the behaviors of a library of sensors with different fluorescent proteins, stiffnesses, and lengths to enable the rational selection of sensors. It also demonstrates that the mechanical linker protein vinculin is subject to regulated extension, instead of the commonly assumed paradigm of regulated force.
- 22.Wang X, Ha T, Defining single molecular forces required to activate integrin and notch signaling, Science. 340 (2013) 991–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dutta PK, Zhang Y, Blanchard AT, Ge C, Rushdi M, Weiss K, Zhu C, Ke Y, Salaita K, Programmable Multivalent DNA-Origami Tension Probes for Reporting Cellular Traction Forces, Nano Lett. 18 (2018) 4803–4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rief M, Fernandez JM, Gaub HE, Elastically coupled two-level systems as a model for biopolymer extensibility, Physical Review Letters. 81 (1998) 4764–4767. [Google Scholar]
- 25.Freikamp A, Cost A-L, Grashoff C, The Piconewton Force Awakens: Quantifying Mechanics in Cells, Trends in Cell Biology. 26 (2016) 838–847. [DOI] [PubMed] [Google Scholar]
- 26.Rothenberg KE, Scott DW, Christoforou N, Hoffman BD, Vinculin Force-Sensitive Dynamics at Focal Adhesions Enable Effective Directed Cell Migration, Biophys J. 114 (2018) 1680–1694.**This paper details the combination of force measurements with FRAP to study force-activated protein dynamics in cells. Vinculin exhibits force-stabilized dynamics, consistent with the observation of catch-bond behavior at the single molecule level. This approach is one of the first to use molecular tension sensors to understand the effects of protein load on protein function in cells.
- 27.Borghi N, Sorokina M, Shcherbakova OG, Weis WI, Pruitt BL, Nelson WJ, Dunn AR, E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch, Proc Natl Acad Sci U S A. 109 (2012) 12568–12573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arsenovic Paul T, Ramachandran I, Bathula K, Zhu R, Narang Jiten D, Noll Natalie A, Lemmon Christopher A, Gundersen Gregg G, Conway Daniel E, Nesprin-2G, a Component of the Nuclear LINC Complex, Is Subject to Myosin-Dependent Tension, Biophysical Journal. 110 (2016) 34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang Y, Liu Z, Zhang Y, Galior K, Yang J, Salaita K, A General Approach for Generating Fluorescent Probes to Visualize Piconewton Forces at the Cell Surface, Journal of the American Chemical Society. 138 (2016) 2901–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morimatsu M, Mekhdjian AH, Chang AC, Tan SJ, Dunn AR, Visualizing the Interior Architecture of Focal Adhesions with High-Resolution Traction Maps, Nano Letters. 15 (2015) 2220–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brenner MD, Zhou R, Conway DE, Lanzano L, Gratton E, Schwartz MA, Ha T, Spider Silk Peptide Is a Compact, Linear Nanospring Ideal for Intracellular Tension Sensing, Nano Lett. 16 (2016) 2096–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar A, Ouyang M, Van den Dries K, McGhee EJ, Tanaka K, Anderson MD, Groisman A, Goult BT, Anderson KI, Schwartz MA, Talin tension sensor reveals novel features of focal adhesion force transmission and mechanosensitivity, Journal of Cell Biology. 213 (2016) 371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baddam S, Arsenovic P, Narayanan V, Duggan N, Mayer C, Newman S, Abutaleb D, Mohan A, Kowalczyk A, Conway D, et al. , The Desmosomal Cadherin Desmoglein-2 Experiences Mechanical Tension as Demonstrated by a FRET-Based Tension Biosensor Expressed in Living Cells, Cells. 7 (2018) 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Galior K, Ma VP, Salaita K, Molecular Tension Probes for Imaging Forces at the Cell Surface, Acc Chem Res. 50 (2017) 2915–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conway DE, Breckenridge MT, Hinde E, Gratton E, Chen CS, Schwartz MA, Fluid Shear Stress on Endothelial Cells Modulates Mechanical Tension across VE-Cadherin and PECAM-1, Current Biology. 23 (2013) 1024–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar A, Anderson KL, Swift MF, Hanein D, Volkmann N, Schwartz MA, Local Tension on Talin in Focal Adhesions Correlates with F-Actin Alignment at the Nanometer Scale, Biophys J. 115 (2018) 1569–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zoldak G, Stigler J, Pelz B, Li H, Rief M, Ultrafast folding kinetics and cooperativity of villin headpiece in single-molecule force spectroscopy, Proc Natl Acad Sci U S A. 110 (2013) 18156–18161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mosayebi M, Louis AA, Doye JP, Ouldridge TE, Force-Induced Rupture of a DNA Duplex: From Fundamentals to Force Sensors, ACS Nano. 9 (2015) 11993–12003. [DOI] [PubMed] [Google Scholar]
- 39.Murad Y, Li ITS, Quantifying Molecular Forces with Serially Connected Force Sensors, Biophys J. 116 (2019) 1282–1291.** The authors use a computational approach to predict that TGT rupture depends on force history and adhesion event density over multiple orders of magnitude, in addition to varying with different receptor-ligand pairs. Interestingly, they show that a system of two TGTs in series can reconstruct force history with high force resolution.
- 40.Alemany A, Ritort F, Force-Dependent Folding and Unfolding Kinetics in DNA Hairpins Reveals Transition-State Displacements along a Single Pathway, J Phys Chem Lett. 8 (2017) 895–900. [DOI] [PubMed] [Google Scholar]
- 41.Bercy M, Bockelmann U, Hairpins under tension: RNA versus DNA, Nucleic Acids Res. 43 (2015) 9928–9936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu X, Jing C, Xu X, Nakazawa N, Cornish VW, Margadant FM, Sheetz MP, Cooperative Vinculin Binding to Talin Mapped by Time-Resolved Super Resolution Microscopy, Nano Lett. 16 (2016) 4062–4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patel A, Malinovska L, Saha S, Wang J, Alberti S, Krishnan Y, Hyman AA, ATP as a biological hydrotrope, Science. 356 (2017) 753–756. [DOI] [PubMed] [Google Scholar]
- 44.Ohashi T, Galiacy SD, Briscoe G, Erickson HP, An experimental study of GFP-based FRET, with application to intrinsically unstructured proteins, Protein Sci. 16 (2007) 1429–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sedlak SM, Bauer MS, Kluger C, Schendel LC, Milles LF, Pippig DA, Gaub HE, Monodisperse measurement of the biotin-streptavidin interaction strength in a well-defined pulling geometry, PLoS One. 12 (2017) e0188722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kong F, Garcia AJ, Mould AP, Humphries MJ, Zhu C, Demonstration of catch bonds between an integrin and its ligand, J Cell Biol. 185 (2009) 1275–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong F, Li Z, Parks WM, Dumbauld DW, Garcia AJ, Mould AP, Humphries MJ, Zhu C, Cyclic mechanical reinforcement of integrin-ligand interactions, Mol Cell. 49 (2013) 1060–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jurchenko C, Chang Y, Narui Y, Zhang Y, Salaita KS, Integrin-Generated Forces Lead to Streptavidin-Biotin Unbinding in Cellular Adhesions, Biophysical Journal. 106 (2014) 1436–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X, Rahil Z, Li ITS, Chowdhury F, Leckband DE, Chemla YR, Ha T, Constructing modular and universal single molecule tension sensor using protein G to study mechano-sensitive receptors, Scientific Reports. 6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perez-Jimenez R, Garcia-Manyes S, Ainavarapu SR, Fernandez JM, Mechanical unfolding pathways of the enhanced yellow fluorescent protein revealed by single molecule force spectroscopy, J Biol Chem. 281 (2006) 40010–40014. [DOI] [PubMed] [Google Scholar]
- 51.Dietz H, Rief M, Exploring the energy landscape of GFP by single-molecule mechanical experiments, Proc Natl Acad Sci U S A. 101 (2004) 16192–16197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ganim Z, Rief M, Mechanically switching single-molecule fluorescence of GFP by unfolding and refolding, Proc Natl Acad Sci U S A. 114 (2017) 11052–11056.** This paper combines single molecule force measurements using optical tweezers with measurements of GFP fluorescence. They observe mechanical quenching of GFP, a process that occurs prior to unfolding, and show that fluorescence can be recovered upon refolding. This finding has large implications for the function of genetically-encoded molecular tension sensors.
- 53.Saeger J, Hytonen VP, Klotzsch E, Vogel V, GFP’s Mechanical Intermediate States, Plos One. 7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Galior K, Liu Y, Yehl K, Vivek S, Salaita K, Titin-Based Nanoparticle Tension Sensors Map High-Magnitude Integrin Forces within Focal Adhesions, Nano Letters. 16 (2016) 341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lecuit T, Lenne PF, Munro E, Force generation, transmission, and integration during cell and tissue morphogenesis, Annu Rev Cell Dev Biol. 27 (2011) 157–184. [DOI] [PubMed] [Google Scholar]
- 56.Rothenberg KE, Neibart SS, Lacroix AS, Hoffman BD, Controlling Cell Geometry Affects the Spatial Distribution of Load Across Vinculin, Cellular and Molecular Bioengineering. 8 (2015) 364–382. [Google Scholar]
- 57.Sarangi BR, Gupta M, Doss BL, Tissot N, Lam F, Mege RM, Borghi N, Ladoux B, Coordination between Intra- and Extracellular Forces Regulates Focal Adhesion Dynamics, Nano Lett. 17 (2017) 399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Y, Medda R, Liu Z, Galior K, Yehl K, Spatz JP, Cavalcanti-Adam EA, Salaita K, Nanoparticle Tension Probes Patterned at the Nanoscale: Impact of Integrin Clustering on Force Transmission, Nano Letters. 14 (2014) 5539–5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Elosegui-Artola A, Trepat X, Roca-Cusachs P, Control of Mechanotransduction by Molecular Clutch Dynamics, Trends Cell Biol. 28 (2018) 356–367. [DOI] [PubMed] [Google Scholar]
- 60.Oria R, Wiegand T, Escribano J, Elosegui-Artola A, Uriarte JJ, Moreno-Pulido C, Platzman I, Delcanale P, Albertazzi L, Navajas D, et al. , Force loading explains spatial sensing of ligands by cells, Nature. 552 (2017) 219–224.** This paper shows that decreased ligand density results in FA growth on softer substrates but leads to FA collapse on stiffer substrates. Their finding is explained through the molecular clutch model and related to the ability of cells to redistribute force loading across clutches. This highlights the mechanical complexity of adhesion structures and suggests that a consideration of force magnitude alone is insufficient to understand adhesion mechanobiology.
- 61.Chang AC, Mekhdjian AH, Morimatsu M, Denisin AK, Pruitt BL, Dunn AR, Single Molecule Force Measurements in Living Cells Reveal a Minimally Tensioned Integrin State, Acs Nano. 10 (2016) 10745–10752.**The authors report single molecule measurements of integrin tension in living cells. The distribution of integrin load shows that most integrins experience a small load, with a smaller subpopulation experiencing larger loads. This observation is also consistent with mechanical redundancy, where adhesion structures have an excess of key constituents primed for loading.
- 62.Lemke SB, Weidemann T, Cost AL, Grashoff C, Schnorrer F, A small proportion of Talin molecules transmit forces at developing muscle attachments in vivo, PLoS Biol. 17 (2019) e3000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maskarinec SA, Franck C, Tirrell DA, Ravichandran G, Quantifying cellular traction forces in three dimensions., Proceedings of the National Academy of Sciences of the United States of America. 106 (2009) 22108–22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paszek MJ, DuFort CC, Rossier O, Bainer R, Mouw JK, Godula K, Hudak JE, Lakins JN, Wijekoon AC, Cassereau L, et al. , The cancer glycocalyx mechanically primes integrin-mediated growth and survival, Nature. 511 (2014) 319–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brockman JM, Blanchard AT, Pui-Yan VM, Derricotte WD, Zhang Y, Fay ME, Lam WA, Evangelista FA, Mattheyses AL, Salaita K, Mapping the 3D orientation of piconewton integrin traction forces, Nat Methods. 15 (2018) 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang DL, Bax NA, Buckley CD, Weis WI, Dunn AR, Vinculin forms a directionally asymmetric catch bond with F-actin, Science. 357 (2017) 703–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Buckley CD, Tan JL, Anderson KL, Hanein D, Volkmann N, Weis WI, Nelson WJ, Dunn AR, The minimal cadherin-catenin complex binds to actin filaments under force., Science. 346 (2014) 1254211–1254211– 1254218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rakshit S, Zhang Y, Manibog K, Shafraz O, Sivasankar S, Ideal, catch, and slip bonds in cadherin adhesion, Proc Natl Acad Sci U S A. 109 (2012) 18815–18820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Case LB, Baird MA, Shtengel G, Campbell SL, Hess HF, Davidson MW, Waterman CM, Molecular mechanism of vinculin activation and nanoscale spatial organization in focal adhesions, Nature Cell Biology. 17 (2015) 880–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bays JL, DeMali KA, Vinculin in cell-cell and cell-matrix adhesions, Cell Mol Life Sci. 74 (2017) 2999–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Auernheimer V, Lautscham LA, Leidenberger M, Friedrich O, Kappes B, Fabry B, Goldmann WH, Vinculin phosphorylation at residues Y100 and Y1065 is required for cellular force transmission, J Cell Sci. 128 (2015) 3435–3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nordenfelt P, Elliott HL, Springer TA, Coordinated integrin activation by actin-dependent force during T-cell migration, Nat Commun. 7 (2016) 13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li J, Springer TA, Integrin extension enables ultrasensitive regulation by cytoskeletal force, Proc Natl Acad Sci U S A. 114 (2017) 4685–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen Y, Lee H, Tong H, Schwartz M, Zhu C, Force regulated conformational change of integrin alpha(v)beta(3), Matrix Biology. 60–61 (2017) 70–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parsons JT, Horwitz AR, Schwartz MA, Cell adhesion: integrating cytoskeletal dynamics and cellular tension, Nat Rev Mol Cell Biol. 11 (2010) 633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Puklin-Faucher E, Sheetz MP, The mechanical integrin cycle, Journal of Cell Science. 122 (2009) 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Swaminathan V, Kalappurakkal JM, Mehta SB, Nordenfelt P, Moore TI, Koga N, Baker DA, Oldenbourg R, Tani T, Mayor S, et al. , Actin retrograde flow actively aligns and orients ligand-engaged integrins in focal adhesions, Proc Natl Acad Sci U S A. 114 (2017) 10648–10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Benito-Jardon M, Klapproth S, Gimeno-Lluch I, Petzold T, Bharadwaj M, Muller DJ, Zuchtriegel G, Reichel CA, Costell M, The fibronectin synergy site re-enforces cell adhesion and mediates a crosstalk between integrin classes, Elife. 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schlierf M, Li H, Fernandez JM, The unfolding kinetics of ubiquitin captured with single-molecule force-clamp techniques, Proc Natl Acad Sci U S A. 101 (2004) 7299–7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dudko OK, Hummer G, Szabo A, Theory, analysis, and interpretation of single-molecule force spectroscopy experiments, Proc Natl Acad Sci U S A. 105 (2008) 15755–15760. [DOI] [PMC free article] [PubMed] [Google Scholar]