Abstract
Alpelisib is a selective inhibitor of PI3Kα, shown to improve outcomes for PIK3CA mutant, hormone receptor positive (HR+) metastatic breast cancers (MBC) when combined with antiestrogen therapy. To uncover mechanisms of resistance, we conducted a detailed, longitudinal analysis of tumor and plasma circulating tumor DNA among such patients from a phase I/II trial combining alpelisib with an aromatase inhibitor (AI) (NCT01870505). The trial’s primary objective was to establish safety with maculopapular rash emerging as the most common grade 3 adverse event (33%). Among 44 evaluable patients, the observed clinical benefit rate was 52%. Correlating genetic alterations with outcome, we identified loss-of-function PTEN mutations in 25% of patients with resistance. ESR1 activating mutations also expanded in number and allele fraction during treatment and were associated with resistance. These data indicate that genomic alterations that mediate resistance to alpelisib or antiestrogen may promote disease progression and highlight PTEN loss as a recurrent mechanism of resistance to PI3Kα inhibition.
INTRODUCTION
Hormone receptor positive (HR+) breast cancer comprises roughly 75% of all cancers of the breast. Whilst many of these cancers can be cured through multi-modality therapy, there remain many deaths due to metastatic spread to distant organs1. These metastatic cancers are marked by their resilience in the face of potent targeted therapies and chemotherapies, with many tumors displaying an initial drug response followed by resistance2–4. Recently, genomic sequencing has identified recurrent, oncogenic alterations in HR+ metastatic breast cancer (MBC) with mutations in the catalytic alpha subunit of PI3K (PI3Kα, PIK3CA gene), in over 40% of cases5,6. This has raised hopes for more durable disease control through precise inhibition of this driver oncogene7,8.
Initial efforts to target mutant PI3Kα were met with challenges due to poor pharmacokinetic properties of the inhibitors9 or poor target selectivity from compounds that ultimately inhibited other PI3K family members10,11 or other proteins entirely12, limiting therapeutic index13,14. Moreover, the ability to durably inhibit tumor growth has been impaired by adaptive responses that lead to upregulation of additional oncogenic signals, particularly that of the estrogen receptor (ER)15–18. Indeed, genomic characterization of HR+ MBC has revealed the wide prevalence of ligand independent mutations in the ER gene (ESR1) further amplifying this problem3,4. More recently, selective inhibitors of PI3Kα, such as alpelisib (BYL719; Novartis Pharma AG) have been developed11,19 and combined with potent inhibitors of ER signaling. Although these drug combinations have demonstrated improved activity20,21, resistance remains ubiquitous. This was highlighted by the results of the SOLAR-1 Phase III study of alpelisib combined with fulvestrant in PIK3CA-mutated HR+ MBC that showed a markedly improved PFS over fulvestrant monotherapy but pervasive resistance nonetheless22.
To characterize the basis for such resistance to combination hormone plus oncoprotein targeted therapy, we conducted a detailed, longitudinal analysis of tumor and plasma circulating cell-free tumor DNA (ctDNA) among patients with HR+ MBC who participated in a phase I/II dose-escalation study of alpelisib in combination with letrozole or exemestane. Here, we report the primary results of the study and identify potential mechanisms of resistance through analyses of pre- and post-treatment samples.
RESULTS
Study population
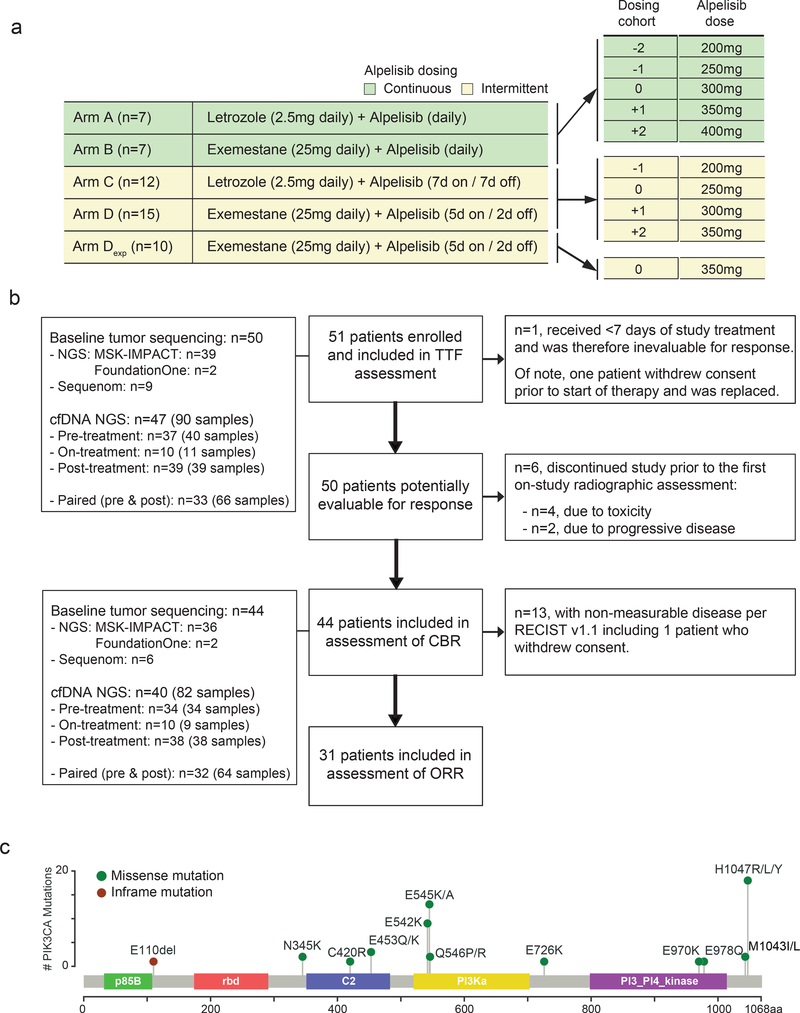
Fifty-one patients (Arm A: n=7, Arm B: n=7, Arm C: n=12, Arm D: n=25) were enrolled between June 2013 and September 2015 (Table 1, Fig. 1). Data were locked on April 30th, 2018. The median age of the cohort was 58 (range: 28–83). All patients had HR+, HER2 non-amplified MBC. 69% of the patients had visceral disease and 31% had bone-only disease; 88% were previously exposed to at least one line of endocrine therapy for metastatic disease (median 2 lines, range: 0–6) and 51% were previously exposed to at least one line of chemotherapy for metastatic disease (median 1 line, range: 0–9).Baseline PIK3CA mutational status of 50 patients were determined on pre-treatment tumor samples obtained for this study utilizing the MSK-IMPACT Next Generation Sequencing (NGS) assay (n=39 patients) or Foundation Medicine NGS assay (n=2 patients), or Sequenom mass spectrometry genotyping (n=9 patients; performed when tumor tissue did not meet the minimum quantity or quality for NGS). Additionally, ctDNA sequencing was performed utilizing a targeted 73-genepanel NGS panel on pre-, on-, and post-treatment (collected at the time of off-study) plasma samples from 47 patients (n=90 samples; including 32 paired pre- and post-treatment samples). Although the study did not mandate PIK3CA mutation on baseline tumor assessment, the majority of the patients were indeed selected based on having a hotspot activating mutation in PIK3CA at baseline (88%, n =44). The distribution of PIK3CA mutations in the cohort followed the anticipated pattern for HR+ breast cancer (Fig. 1c) and the most common mutations were in the kinase domain (including H1047R/L/Y) or helical domain (including E545K/A and E542K).
Table 1.
Patient characteristics
| Arm A | Arm B | Arm C | Arm D | All Arms | |
|---|---|---|---|---|---|
| (N=7) | (N=7) | (N=12) | (N=25) | (N=51) | |
| Age, median (range) | 55 (30–69) | 52 (37–69) | 58 (45–83) | 55 (28–78) | 55 (28–83) |
| ECOG PS | |||||
| 0, n | 4 | 6 | 3 | 14 | 27 |
| 1, n | 3 | 1 | 9 | 11 | 24 |
| Receptor Status | |||||
| ER+, n | 7 | 7 | 12 | 25 | 51 |
| PR+, n | 4 | 4 | 4 | 19 | 31 |
| HER2+, n | 0 | 0 | 0 | 0 | 0 |
| Histology | |||||
| Invasive ductal, n | 5 | 7 | 8 | 21 | 41 |
| Invasive lobular, n | 2 | 0 | 2 | 4 | 8 |
| Carcinoma NOS, n | 0 | 0 | 2 | 0 | 2 |
| Neoadjuvant or adjuvant chemotherapy, n | 5 (71%) | 5 (71%) | 7 (58%) | 13 (52%) | 30 (59%) |
| Adjuvant endocrine therapy, n | 5 (71%) | 5 (71%) | 10 (83%) | 17 (68%) | 37 (73%) |
| De novo metastatic, n | 2 (29%) | 2 (29%) | 2 (17%) | 8 (32%) | 14 (27%) |
| Total number of therapies in metastatic setting, median (range) | 3 (0–12) | 5 (1–10) | 4 (1–8) | 4 (0–10) | 4 (0–12) |
| Endocrine therapies, median (range) | 2 (0–4) | 2 (2–5) | 2 (1–3) | 2 (0–6) | 2 (0–6) |
| Chemotherapies, median (range) | 2 (0–9) | 3 (0–8) | 1 (0–5) | 1 (0–5) | 1 (0–9) |
| Endocrine Resistance, n | 6 (86%) | 7 (100 %) | 11 (92%) | 23 (92%) | 27 (53%) |
| Sites of metastasis | |||||
| Visceral disease, n | 4 | 5 | 9 | 17 | 35 |
| Bone only disease, n | 3 | 2 | 3 | 8 | 16 |
Fig. 1. Study design.
a) Treatment schema and dosing cohorts of the study arms. b) CONSORT diagram of patients included in the TTF, CBR and ORR evaluation. c) Distribution of PIK3CA mutations for the study cohort positioned by their amino acid coordinates across the protein domain based on the pre-treatment tumor sequencing results. Exp: expansion cohort, TFF: time to treatment failure, CBR: clinical benefit rate, ORR: objective response rate, NGS: next generation sequencing.
Dose escalation and MTD
Dose-limiting toxicities (DLTs) were defined for all the patients excluding the Arm D Expansion cohort (n=10 patients) based on NCI Common Toxicity Criteria for Adverse Events version 4.0 (Supplementary Table 1) and are shown in Table 2. On Arm A Cohort 0 (alpelisib 300mg daily + letrozole), 2 of 3 patients experienced DLTs, and 1 DLT (maculopapular rash) was observed among 3 patients treated with dose-reduced 250mg continuous dosing. Given the high frequency of grade 3 (G3) rash in the continuous dosing arms (50%, 7 of 14 patients), we halted accrual to Arms A and B and opted to explore two alternative intermittent dosing schedules. In murine xenograft breast cancer models, PI3K inhibition given on discontinuous schedules of administration has shown equal or improved antitumor activity compared to continuous dosing schedules23. On the continuous dosing schedule in our trial, 300mg of alpelisib (75% of single-agent MTD) was difficult to tolerate due to maculopapular rash, requiring breaks in therapy and the initiation of oral and topical steroids. Although the study arms were not designed to be compared for toxicity rate, the DLT rate appeared to be lower in patients enrolled on the intermittent arms (15% [4 of 27 patients] in the continuous arms vs. 64% [9 of 14 patients] in the intermittent arms) as was the overall incidence of rash (35% [13 of 37 patients, including Arm D expansion] all-grade on intermittent dosing arms vs. 64% [9 of 14 patients] on continuous, Supplementary Table 2).
Table 2.
Dose limiting toxicity
| Dose-limiting toxicity (Grade) | Arm A Daily (n=7) | Arm B Daily (n=7) | Arm C 7d on, 7d off (n=12) | Arm D 5d on, 2d off (n=15) | All Arms§ (n=41) |
||||
|---|---|---|---|---|---|---|---|---|---|
| 250mg n=3 | 300mg n=4 | 300mg n=7 | 250mg n=6 | 300mg n=6 | 250mg n=3 | 300mg n=6 | 350mg n=6 | ||
| Maculopapular rash | 1 (G3) | 2 (G3) | 4 (G3) | 0 | 1 (G3) | 0 | 1 (G3) | 1 (G3) | 10 (G3) |
| Hyperglycemia | 0 | 0 | 1 (G3) | 0 | 0 | 0 | 0 | 0 | 1 (G3) |
| Hypotension | 0 | 0 | 0 | 0 | 1 (G3) | 0 | 0 | 0 | 1 (G3) |
| Abdominal pain | 0 | 1 (G3)* | 0 | 0 | 0 | 0 | 0 | 0 | 1 (G3) |
G3 abdominal pain constituted DLT as it warranted hospitalization.
The Arm D expansion cohort (b=10) was not included in the DLT assessment.
On Arm C (letrozole + alpelisib, 7 days on followed by 7 days off), no DLTs were observed in patients treated with 250mg, whereas 2 of 6 patients experienced DLTs upon escalation of the alpelisib does to 300mg. Three additional patients were then enrolled at 250mg with no DLTs observed. Establishing 250mg as the MTD on Arm C would have required enrollment of 3 additional patients at this dose; however, accrual onto Arm C was halted without MTD determination as other clinical trials21 had established 300mg continuous dosing as the recommended phase II dose in combination with letrozole.
On Arm D (exemestane + alpelisib, 5 days on followed by 2 days off), no DLTs were seen at 250mg (Cohort 0), and 1 of 6 patients experienced G3 rash at 300mg dosing. The dose was then escalated to 350mg (highest dose for which escalation was planned), with 1 of 6 patients experiencing G3 rash. Thus, the MTD of alpelisib 5 days on, 2 days off with daily exemestane was determined to be 350mg. We expanded Arm D given the lower rate of rash in this arm and accrued an additional 10 patients at the 350mg dose level.
Safety Findings
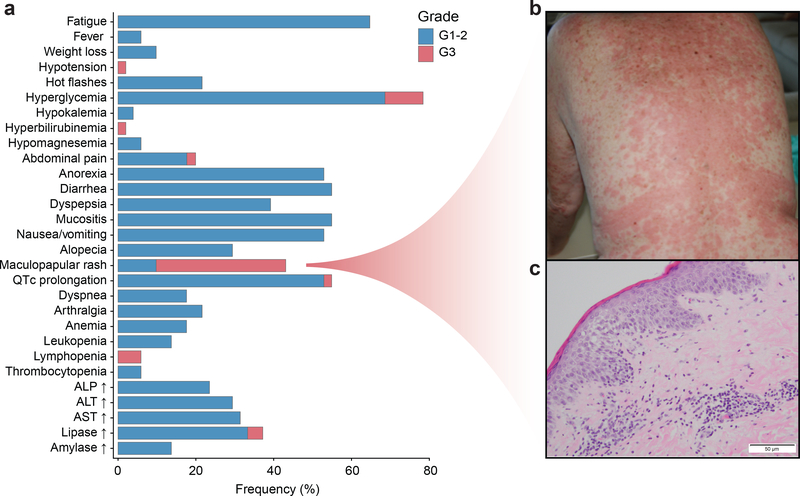
Adverse events (AEs) are presented in Fig. 2a and Supplementary Table 2. Most common AEs (all grades, arms, dosing cohorts) included: hyperglycemia (78%), fatigue (65%), mucositis (55%), QTc prolongation (55%), diarrhea (55%), nausea/vomiting (53%), and maculopapular rash (43%). The majority of AEs were grade 1 or 2. Maculopapular rash was the most common G3 AE and occurred in 17 patients (33%). In general, the toxicities, particularly gastrointestinal and dermatologic, were more prominent and appeared to be dose dependent with the continuous dosing schedule (Supplementary Table 2). Toxicities warranting study discontinuation included maculopapular rash in 5 patients (12%) and G1 pneumonitis and G3 hypotension in 1 patient (2%) each. No grade 4 toxicities were observed, and no deaths were attributable to the study drugs. Two deaths occurred within 30 days of end of study treatment due to rapidly progressive disease.
Fig. 2. Safety and adverse events for the study.
a) Treatment associated adverse events for 51 patients included in the study. b) Presentation of maculopapular rash in the trunk. c) Skin histology demonstrating a superficial lympho-histiocytic infiltrate with eosinophils.
We noted a relatively predictable timeline for onset of the maculopapular rash (Fig. 2b). The rash was characterized by maculopapules affecting the trunk, occasionally associated with pruritus. The rash generally arose within 9–15 days of treatment initiation, and the absence of hypersensitivity with interrupted dosing suggested that the underlying mechanism was a delayed, type IV hypersensitivity reaction. The rash responded in all patients to alpelisib interruption plus administration of oral antihistamines and/or topical or oral steroids. Pathologic evaluation of the rash revealed a lymphohistiocytic infiltrate with eosinophils in the dermis consistent with drug reaction (Fig. 2c). Notably, a higher incidence of rash was observed in this study as compared to prior studies of alpelisib19–22,24 and the frequent presence of eosinophilia suggests that the co-administration of an AI may contribute to cutaneous and/or pharmacodynamic differences, compared to single agent or combination with fulvestrant.
Response evaluation
Median time to treatment failure (TTF, defined as the time from on-study date to off-study date for any reason) for all 51 patients was 14 weeks (range: 2 – 169) (Fig. 3a). Median TTF by arm was: Arm A, 21 weeks (range: 2 – 169); Arm B, 8 weeks (range: 2 – 29); Arm C, 12 weeks (range: 3 – 72); Arm D, 17 weeks (range: 3 – 150). The reason for study discontinuation was progressive disease in 41 patients (Arm A: 4, Arm B: 5, Arm C: 11, Arm D: 21); toxicity in 7 patients (Arm A: 1, Arm B: 2, Arm C: 1, Arm D: 3); and withdrawal of consent unrelated to toxicity in 3 patients (Arm A: 2, Arm D: 1).
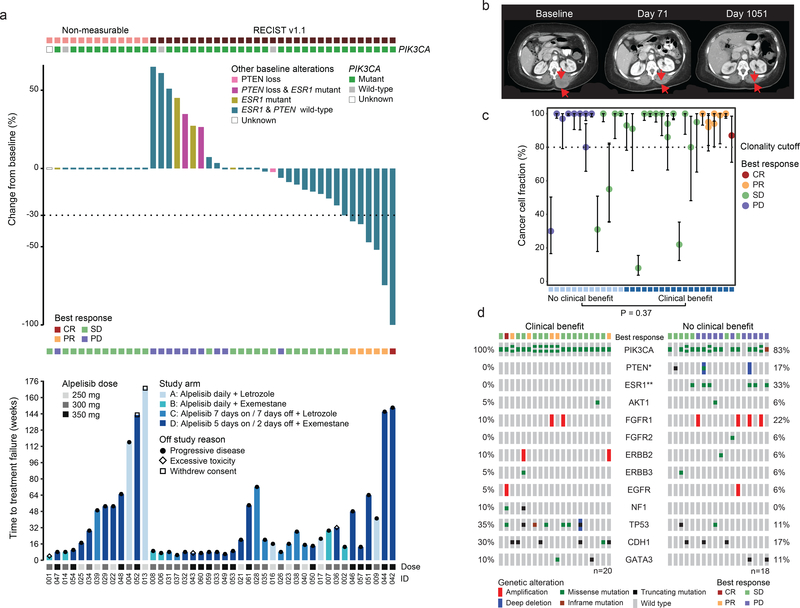
Fig. 3. Treatment outcome and response for 44 evaluable patients.
a) The top section shows the PIK3CA mutation status of the baseline tumors. The top barplot represents the percent best change from baseline in the target lesions assessed by the RECIST version 1.1 criteria. The bars are color coded based on the tumor tissue baseline PTEN or ESR1 mutation status. The middle section shows best overall response. The bottom barplot shows progression free survival color coded by the study arms. The lower section indicates the dose of alpelisib. b) Baseline, on- treatment and post-progression CT scan of the patient with a sustained complete response in the paravertebral mass. c) Cancer cell fractions (CCF) with binomial exact 95% confidence intervals calculated around the point estimate of CCF of PIK3CA mutations in 32 patients with tumor sequencing data sufficient for clonality analysis. Patients are ordered by increasing clinical benefit and weeks on therapy. P value is calculated based on two-sided Fisher’s exact test of clonality (clonal vs subclonal) and clinical benefit. d) Pre-treatment tumor tissue genomic alterations and clinical benefit in 38 evaluable patients with available next-generation sequencing results (one patient with evaluable genomic data was clinically unevaluable). PTEN loss of function alterations and ESR1 activating mutations were observed exclusively in baseline tumor tissue samples of patients with no clinical benefit. P values are based on two-sided Fisher’s exact test; *p= 0.0967, **p= 0.0067, the rest not significant.
Forty-four patients were evaluable for response per protocol (received at least 1 week of therapy and had on-study imaging; Fig. 1b) and were included in the assessment of clinical benefit rate (clinical benefit was predefined for this study as complete response, partial response of stable disease ≥ 16 weeks). The overall CBR was 52% (23/44 patients, 95% CI: 37%, 68%). Of the 44 evaluable patients, 31 had measurable disease and were included in the assessment of observed response rate (ORR, Fig. 3a). The ORR was 19% (6/31 patients, 95% CI: 7%, 37%), with 5 partial responses (Arm A: 1; Arm D: 4) and one complete response (Arm D, Fig. 3b).
In terms of the continuous (Arms A and B) versus intermittent arms (Arms C and D) of the study: TTF, CBR, and ORR were 11 weeks (range: 2 – 169), 45% (5/11 patients, 95% CI: 17%, 77%), and 14% (1/7 patients, 95% CI: 0, 58%) on the continuous arms and 15 weeks (range: 3 – 150), 55% (11/33 patients, 95% CI: 36%, 72%), and 21% (5/24 patients, 95% CI: 7%, 42%) on the intermittent arms (Fig. 3a).
Clinical benefit was only observed in patients whose tumors harbored PIK3CA mutations (CBR of 57.5%, 23/40 patients, 95% CI: 41%, 73%), with none of the evaluable PIK3CA-wild type patients demonstrating clinical benefit (Fig. 3a, 0/6 patients, 95% CI: 0, 46%; p = 0.025). These results, although based on small numbers, are consistent with those reported in other trials of PI3K inhibitors in HR+ MBC25.
Beyond PIK3CA mutational status, we sought to determine if other features of the baseline tumor mutational profile might be associated with clinical benefit. We restricted this analysis to the evaluable patients whose cancers underwent tissue-based NGS genomic profiling (n=38, excluding one patient with evaluable genomic data that was clinically unevaluable). We first assessed whether clinical benefit was associated with clonality of the PIK3CA mutation by inferring the cancer cell fraction (CCF, the percentage of cancer cells within the tumor harboring a given mutation)26 of the PIK3CA mutations from the sequencing data (Fig. 3c). The majority of the PIK3CA mutations (70%, 31/39 mutations in 32 evaluable pre-treatment tumors) were clonal (bioinfomatically inferred to be present in all of the sequenced cancer cells). Three of five patients whose tumors harbored only subclonal PIK3CA mutations had no clinical benefit (23% [3/13] subclonal with no clinical benefit and 11% [2/19] subclonal with clinical benefit). The association between the PIK3CA mutation clonality (clonal vs subclonal) was inconclusive due to lack of adequate statistical power (p = 0.37).
We next sought to assess whether any co-occurring alterations were associated with clinical benefit or intrinsic resistance, focusing on recurrent variants with established oncogenic potential in breast cancer27 (Fig. 3d). Activating ESR1 mutations that confer AI resistance (D538G, n=3; Y537S, n=3) were found in the baseline tumor samples of six patients where no clinical benefit was observed and were not detected in the baseline samples of any patients who experienced clinical benefit. ESR1 mutations were thus significantly associated with lack of clinical benefit (p = 0.0067).
In addition, despite the fact that mutations in PIK3CA and PTEN are mutually exclusive in primary breast cancer6, we had previously identified an individual patient where distinct PTEN loss-of-function mutations were associated with progression on alpelisib monotherapy28.In this trial, we identified two patients whose tumors harbored concurrent clonal activating PIK3CA mutations and PTEN loss (#032 and #060). Both of these patients derived no clinical benefit from alpelisib plus AI as both had progressive disease on their first radiographic evaluations on week 8 (Fig. 3d).
ctDNA and tissue analyses of pre- and post-progression samples
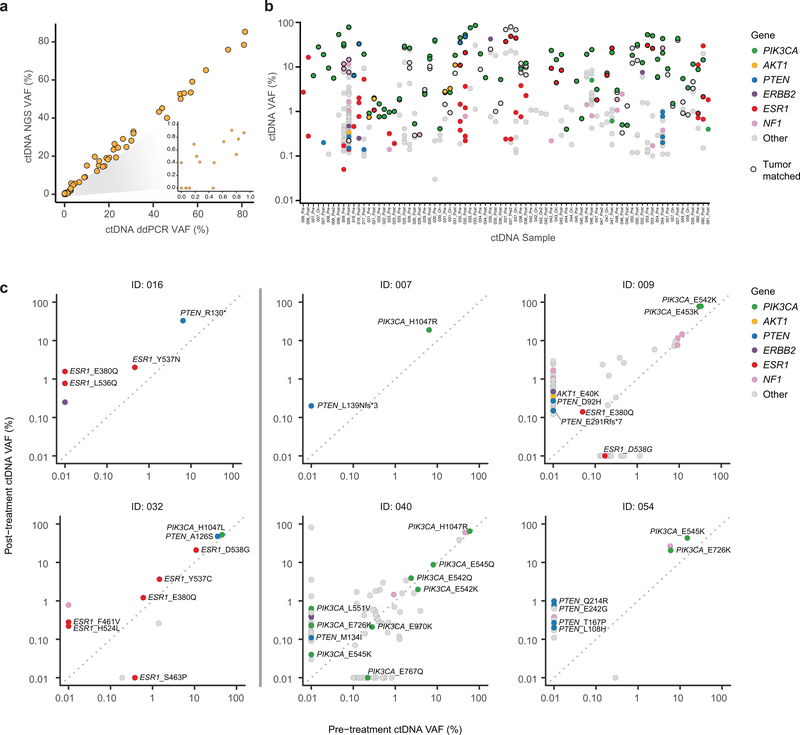
In breast cancer, spatial and temporal intra-tumor heterogeneity is frequently observed, particularly in advanced disease28–30. Individual needle biopsies may therefore not represent the full repertoire of tumor somatic alterations that influence clinical outcomes28–30. ctDNA can potentially provide a more comprehensive picture of the tumor genome as ctDNA is derived from the pool of tumor sites shedding DNA31–33. Hence, we utilized serial blood samples collected at baseline and after progression to characterize genomic alterations in ctDNA potentially affecting treatment response. Paired pre- and post-treatment ctDNA samples from 32 patients were sufficient for ultradeep targeted sequencing34,35. The ctDNA NGS analysis demonstrated performance characteristics necessary for detection of mutations near the molecular limits with a performance comparable to that of digital droplet PCR on the same ctDNA specimens (ddPCR; Extended Data Fig. 1a). Of note, in 89% (34/38) of patients, the PIK3CA mutations identified in the pre-treatment tumors were also identified in the ctDNA samples (Extended Data Fig. 1b).
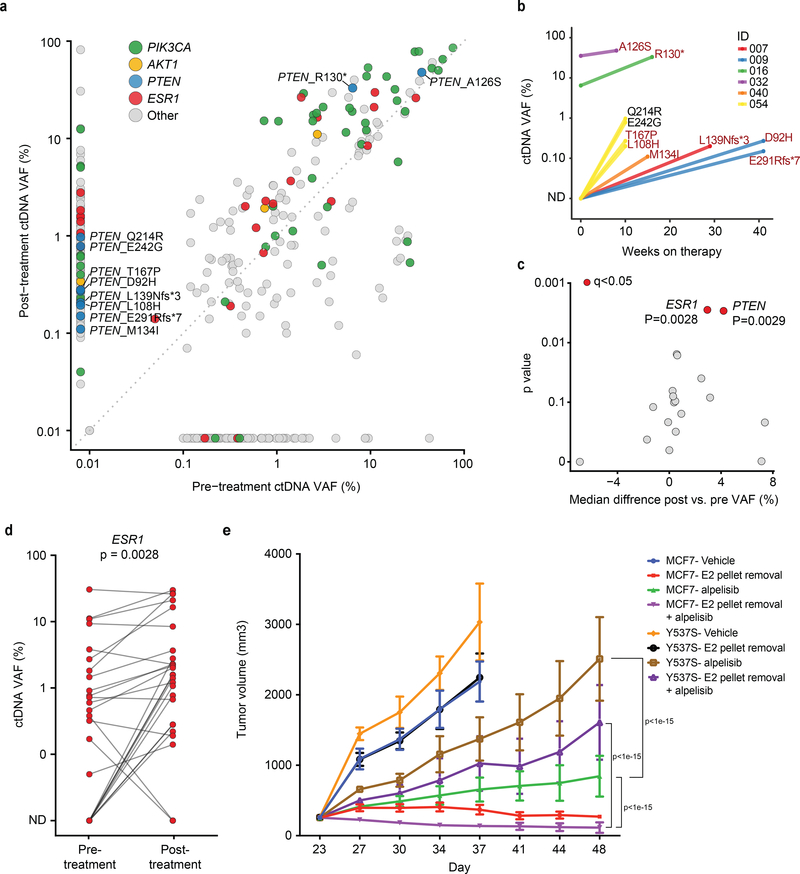
To more comprehensively assess whether de novo or acquired PTEN loss might contribute to resistance to the alpelisib plus AI combination, we interrogated the ctDNA samples for PTEN mutations and copy number alterations. We identified PTEN alterations in 25% of patients (8/32, Fig. 4a, Extended Data Fig. 1c). This included three patients with loss-of-function alterations in the pre-treatment samples (A126S36, R130*, and a homozygous deletion that was also confirmed in the pre-treatment tumor tissue) who had rapid progression of disease upon commencing alpelisib plus AI therapy (Figs. 4a–4b). As well, our analysis revealed PTEN loss-of-function mutations in post-treatment specimens from five patients (including two truncating mutations, four previously identified loss-of-function missense mutations36–38, and a homozygous deletion), all of whom had no evidence of these alterations in their pre-treatment ctDNA or tumor tissue specimens (Figs. 3d, 4a–4b). These data reveal that genomic alterations causing loss of PTEN function may be identified in the population of HR+ PIK3CA mutant patients receiving alpelisib therapy.
Fig. 4. ctDNA analyses of pre- and post-progression samples.
a) Comparison of variant allele fraction (VAF) of mutations detected in the pre-treatment and post-treatment ctDNA samples of 32 patients with evaluable paired ctDNA specimens (n=64). The colors of the circles indicate mutated gene, PIK3CA, AKT1, PTEN, ESR1, and others. b) Change in VAF of PTEN mutations (Y-axis) and time to treatment failure (X-axis). The colors of the lines indicate each individual patient. The PTEN mutations are labelled next to the circles and the previously identified loss-of-function mutations are indicated (shown in crimson). c) The median of pairwise change between the pre- and the post-treatment VAFs of mutations in each gene (including all the genes with a minimum frequency of 5 mutations in the cohort and excluding PIK3CA) for 32 patients with evaluable paired ctDNA specimens (n=64). The q values are calculated based on Benjamini and Hochberg method correction of two-sided Wilcoxon signed rank test p values. ESR1 and PTEN were the two significant genes (both q=0.0278) d) Change in pre- and post-treatment VAF of individual ESR1 mutations identified in ctDNA. The p value is calculated based on two-sided Wilcoxon signed rank test. The analysis includes 26 ESR1 mutations identified in 12 patients e) Proliferation of ESR1 Y537S mutant and parental MCF7 tumors treated with vehicle or 25mg/kg alpelisib with or without estradiol pellet. The error bars represent standard errors and the center represent the mean. Each experiment includes 10 animals. P values are two-sided and are based on a 2-way ANOVA test with Šidák correction for multiple comparisons.
Apart from new mutations, selective enrichment of certain mutant alleles may be associated with a role in therapy resistance. To assess for this, we compared variant allele frequencies (VAFs) in post- and pre-treatment ctDNA samples (Fig. 4c). In addition to the expected enrichment in VAFs of mutant PTEN, we also observed a significant increase in the VAFs of hotspot, activating ESR1 mutations (Figs. 4c–4d). Unlike with PTEN, we did see instances where ESR1 mutation VAF only slightly increased or even decreased, but across the cohort we saw substantial increases as the more common pattern. Given prior work establishing the necessity of effective ER blockade during PI3K inhibition in HR+ breast cancer, we sought to determine whether such AI-resistant ESR1 mutations might indeed contribute to tumor progression in these cases. To address this, we turned to experimental models and constructed isogenic ER+ breast cancer cell lines harboring wild type (WT) ESR1 or the Y537S mutation through CRISPR/Cas9 knock in. Treatment of xenograft tumors from these models revealed that, compared to ESR1 WT tumors, ESR1 mutant-expressing tumors more rapidly progressed on the combination of alpelisib and estrogen deprivation (Fig. 4e). These data imply that genomic alterations promoting resistance to the hormone component of this combination therapy can significantly contribute to disease progression. Taken together, PTEN loss and ESR1 activating mutations were present in nearly half (15/32) of the patients on this study and their presence was associated with progression on therapy.
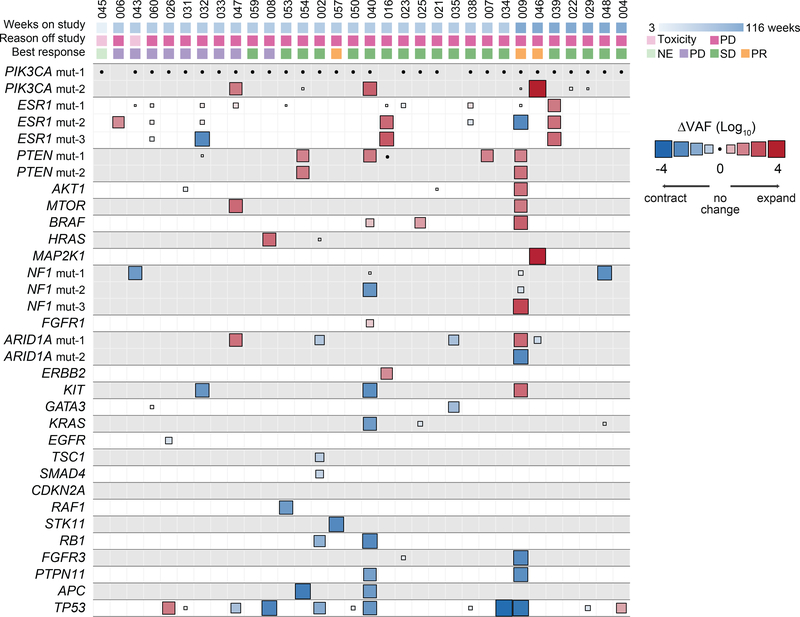
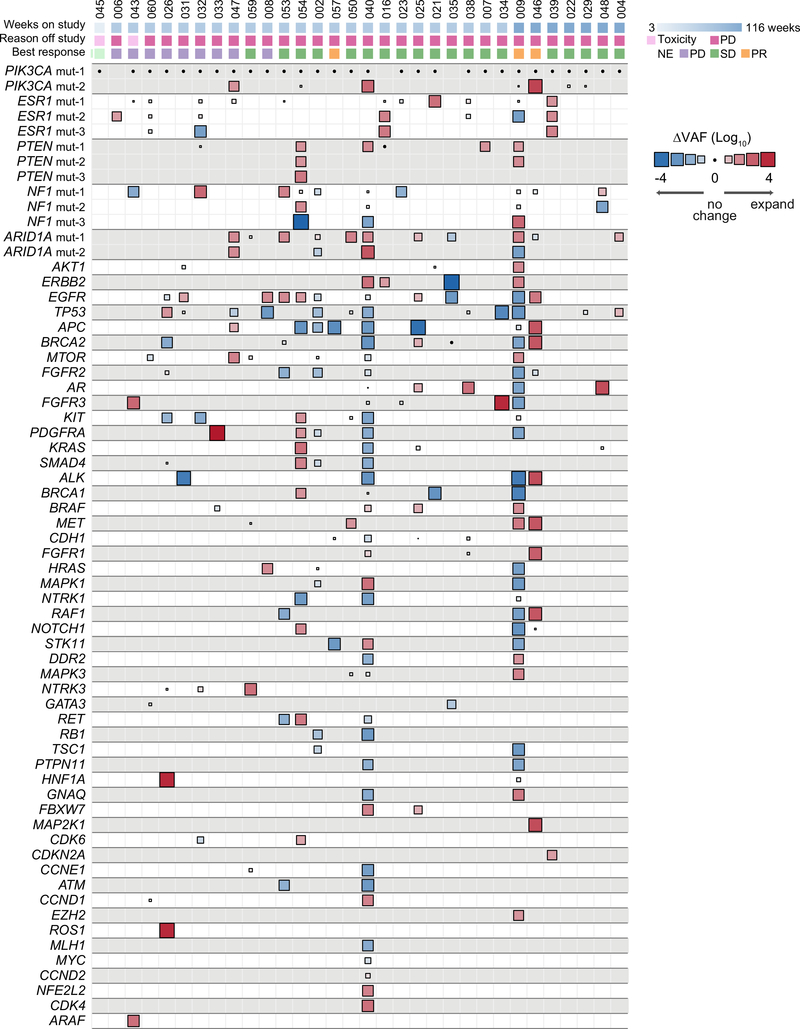
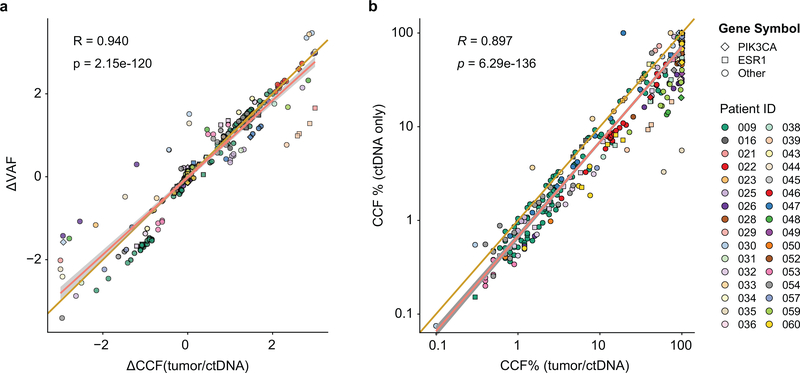
Beyond the associations between PTEN loss and ESR1 mutations and treatment resistance, we sought to determine if other genomic alterations of potential significance were acquired under the selective pressure of therapy. Previous work has demonstrated that changes in VAFs of the mutations in ctDNA can be used to ascertain disease burden and tumor evolution39–42. In order to identify mutations that expanded or contracted during therapy, we calculated a normalized ΔVAF which measures the change in VAF of mutations between pre- and post-treatment samples relative to the change in ctDNA fraction as a proxy of change in disease burden. For this analysis, we first calculated the difference in ctDNA VAF of mutations comparing the pre-treatment vs. post-treatment ctDNA samples. We then calculated the expected change in ctDNA VAF based on that of clonal mutations (usually PIK3CA) present in the pre- and post-treatment ctDNA samples. In doing so, we identified mutations whose VAF increased or decreased above what was expected given the change in VAF of PIK3CA reflective of change in disease burden (Fig. 5, Extended Data Figs. 2a and 3, Methods). As expected, our analysis revealed no disproportionate changes in the normalized ΔVAF for most PIK3CA mutations (Fig. 5) supporting our assumption of clonality of PIK3CA (present in all tumor cells, Fig. 3c). In almost all patients there was evidence of disease evolution based on selective expansion or contraction of the mutational repertoire (Extended Data Fig. 3). We then restricted our analysis to known pathogenic alterations27 (Fig. 5) and identified disproportionate expansions of multiple alterations involving genes downstream of the PI3K pathway (such as AKT1 and MTOR) and MAPK pathway alterations (including ERBB2, HRAS, NF1, BRAF, and MAP2K1). Each of these alterations has been previously linked to the pathogenesis or treatment resistance of HR+ MBC5,43. To support our finding, we performed a more stringent analysis on the subset of patients with available matched tumor sequencing and utilized the allele-specific copy number data from the tumors and the estimated ctDNA fractions (i.e. purity, Extended Data Fig. 2b, Methods) to determine the differences in ctDNA CCFs (ΔCCF) comparing the post-treatment and pre-treatment samples. The corresponding ΔCCF measuring changes in CCFs of mutations in ctDNA highly correlated with the normalized ΔVAF in line with our previous results (Extended Data Fig. 4a). We additionally estimated the CCF of ctDNA mutations using an orthogonal method solely based on ctDNA sequencing results44,45 and the results were concordant with our CCF estimates (Extended Data Fig. 4b).
Fig. 5. Change in variant allele fraction (ΔVAF) of known pathogenic mutations in post- vs pre-treatment ctDNA specimens.
Heatmap of change in VAF comparing the post-treatment with pre-treatment ctDNA of 32 evaluable patients (n=64 samples) normalized according to the change in ctDNA fraction as a proxy of change in disease burden. The size of the boxes represents the relative change and the color gradient of the boxes represent increase or decrease in ΔVAF. The top section shows the time to treatment failure (weeks), reason off study, and the best response on therapy. Multiple mutations in a same gene are indicated, for example patient #47 had two different PIK3CA mutations with one mutation having no change whilst the other one expanded in the post-treatment sample (positive ΔVAF). PR: partial response, SD: stable disease, PD: progressive disease, NE: not evaluable for response.
Taken together, our findings indicate that resistance to alpelisib in combination with aromatase inhibition is a convergent phenotype, and that genetic alterations in components of either the ERα or PI3K pathway may be sufficient to promote tumor progression.
DISCUSSION
To understand the spatial and temporal distribution of discrete genomic alterations that mediate resistance to targeted therapy in breast cancer, we undertook analyses of plasma and tumors from a clinical trial population receiving a combination of PI3Kα and aromatase inhibitors. The results revealed the immense challenge for successful targeted therapy in this disease context as oncogenic drivers appeared to arise and promote resistance: (1) from multiple different oncogenic pathways, (2) subclonally and often originating from different locations, and (3) rapidly, often during the first few months of therapy. This biology of widespread and progressive accumulation of oncogenic alterations likely engenders the substantial interpatient variability in duration of response to targeted therapy observed in this disease.
The clinical utility of combined inhibition of ER and PI3Kα was established in the SOLAR-1 trial where the addition of alpelisib to fulvestrant led to a five-month increase in PFS22. However, despite the select population enrolled to this study (second line, PIK3CA-mutant HR+ MBC), there was a wide range of clinical benefit observed. Our phase I/II study of alpelisib plus antiestrogen therapy was conducted in a heavily pre-treated but comparable population and likewise showed a wide range of clinical benefit prompting our investigation into the basis for therapy resistance.
To ascertain the basis for the initial or eventual disease progression, we analyzed a combination of pre-treatment tumor biopsies, pre-treatment ctDNA samples, and post-treatment ctDNA samples from the trial. As depicted in Fig. 3a, several cancers were intrinsically resistant manifesting progressive disease after ~2 months, however sequencing of pre-treatment tumor biopsies could only provide insight on resistance for a subset (18%). Among these cases, tumor biopsy or ctDNA identified a preexisting loss of function PTEN alteration in 3 instances. These data on PTEN loss are consistent with preclinical studies showing that PTEN loss promotes PI3Kα-independent, AKT activation46,47. The findings establish that concurrent activating PIK3CA mutation and PTEN loss may indeed be present in a meaningful portion of patients in line to receive alpelisib-based therapy and justify more comprehensive genomic approaches than merely testing PIK3CA status.
Given the previous findings of spatial genomic heterogeneity in HR+ MBC48–50, we turned to ctDNA as a method for investigating the contribution of other genomic alterations that might not be present at the specific site that was sampled by needle biopsy. Interestingly, these data highlighted alterations that promoted resistance to the hormone portion of the combination in the form of activating mutations in ESR1. Here, the data confirmed prior observations on the wide prevalence of subclonal alterations in ESR1 that promote resistance to aromatase inhibitors3,4. Based on the potent cross-talk between PI3K and ER signaling15–18, we speculated that these mutations might ultimately promote resistance to the alpelisib plus aromatase inhibitor combination. Preclinical studies (Fig. 4d) demonstrated that ESR1 mutations helped to accelerate tumor progression on the combination, perhaps explaining mutation enrichment among pre-treatment ctDNA samples of patients who did not benefit. Taken together, these data reveal that (1) resistance to combination therapy may manifest through genomic alterations that promote resistance to either component of therapy, and (2) sampling a single site of a highly heterogeneous disease like HR+ MBC may result in an inadequate biomarker that precludes more robust patient selection strategies. For instance, excluding tumors with ESR1 mutations or PTEN loss might yield meaningful improvements in the response rates observed. Patients with these specific alterations might be considered for trials of oral SERDs (ESR1) and/or AKT inhibitors (PTEN).
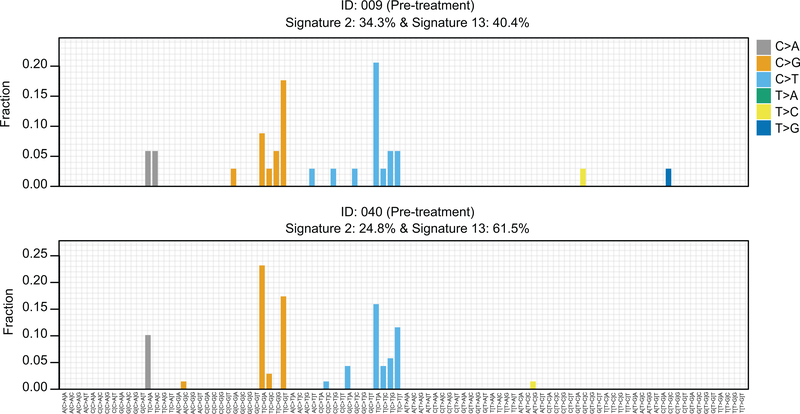
Apart from the patients with de novo PTEN or ESR1 mutation, we also noted several patients who had initial signs of clinical benefit prior to the disease subsequently progressing. For such cases, we sought to understand whether additional genomic alterations might have contributed to treatment resistance. Once again, the data pointed to selection for mutations in PTEN and ESR1 (Fig. 5). Additionally, we identified pathogenic alterations involving the downstream PI3K pathway (i.e. AKT1, MTOR) or MAPK pathway (i.e. ERBB2, HRAS, NF1, BRAF, and MAP2K1) that were selectively expanded upon therapy. Perhaps most surprising was the frequency and chronology of the appearance of these alterations. These data represent some of the first exploiting highly sensitive ctDNA NGS technologies31,51–53 to broadly survey for the acquisition of pathogenic genomic alterations in a focused clinical trial population. Together with reports from individual cases4,40,54, the data here imply HR+ MBC to be a highly evolvable disease state – able to adapt to combination therapy with precise pathway-intrinsic alterations in a very short time frame. The data also raise questions about the underlying mutational processes present in HR+ MBC that might enable such evolvability. Indeed, whilst the assays employed here were not sufficient to distinguish mutational signatures, we did find that 3/4 patients with acquired PTEN mutations had evidence of a hypermutator phenotype with two cases demonstrating mutational signature related to the activity of the APOBEC cytidine deaminases (Extended Data Fig. 5).
The findings presented here have several limitations in attempting to fully characterize the activity of the combination of alpelisib and aromatase inhibition. First, the study population was small, and the study lacked comparator groups such as aromatase inhibitor monotherapy. Second, the tools employed for defining genomic mechanisms of resistance were not comprehensive and changes in gene expression were not analyzed as part of this study. Despite these limitations, the findings are tightly linked to a wealth of biologic results from laboratory models on PTEN and ESR1 and provide strong clinical support of those preclinical observations.
Going forward in HR+ MBC, deployment of biomarkers and therapeutic strategies taking into consideration heterogeneity and evolvability are likely to be essential tools for ultimately overcoming drug resistance.
METHODS
Clinical trial eligibility
Eligible patients were women ≥18 years of age with histologically confirmed, measurable or non-measurable, metastatic or locally advanced unresectable HR+ breast cancer and either stable or progressive disease on letrozole or exemestane. HER2-amplified disease was permitted. Patients had Eastern Cooperative Oncology Group (ECOG) performance status ≤1 and were postmenopausal. Ovarian suppression with an LHRH-agonist was allowed. There were no limits on number of prior chemotherapy or endocrine therapies; prior everolimus was permitted. Patients were required to be >2 weeks from prior radiation, >4 weeks from major surgery, and >3 weeks from prior antineoplastic systemic therapy except the study AI. Initially, patients were required to be on AI alone for 4 weeks, at which point alpelisib was added. In January 2014, this AI-only lead-in period was reduced to 2 weeks.
Patients were required to have adequate hepatic, renal, and bone marrow function. Eligible patients were willing and able to comply with required study-related procedures including mandatory pre- and on-treatment biopsies, and then after a protocol amendment, optional post-progression biopsies. Exclusion criteria included prior treatment with a PI3K inhibitor; insulin-dependent diabetes; central nervous system metastases that were unstable, untreated, or treated ≤4 weeks from start of study treatment; rapidly extensive visceral disease; and baseline corrected QT interval >480msec.
Prior to study enrollment, all patients provided written informed consent according to institutional guidelines. The study protocol was reviewed and approved by the Institutional Review Board of MSKCC. Additional informed consent for taking and publishing photographs was obtained from the patient with maculopapular rash whose photograph was presented in Fig. 2b.
Study design
This was a single-institution, investigator-initiated, phase I/II dose-escalation trial. The study is registered under the clinicaltrials.gov identifier NCT01870505. The primary objective was to determine the MTD. Secondary exploratory objectives were to assess efficacy including overall response rate (ORR), 16-week clinical benefit rate (CBR) and time to treatment failure (TTF), as well as genomic characterization of pre-treatment biopsies to clarify predictors of sensitivity or resistance to treatment, and assessment of on-treatment samples (collected on C1 days 7–14) to assess pharmacodynamics of therapy.
Patients were initially accrued onto one of two treatment arms: Arm A with daily alpelisib and letrozole, or Arm B with daily alpelisib and exemestane. In January 2014, accrual to these arms was halted due to toxicity, predominantly maculopapular rash, limiting dose escalation on continuous Arms A and B. The study was amended to include 2 additional arms with intermittently dosed alpelisib (7 days on followed by 7 days off) plus letrozole (Arm C) or alpelisib (5 days on followed by 2 days off) plus exemestane (Arm D). Treatment including dose and schedule on all arms is shown in Fig. 1a. We hypothesized that intermittent dosing may mitigate adverse events without loss of therapeutic activity based on preclinical data demonstrating persistent activity with a PI3K inhibitor administered on intermittent rather than continuous schedules23.
Dosing proceeded through a standard 3+3 dose escalation/de-escalation design, with 28-day cycles. Patients were asked to take their study drugs concomitantly and to record administration in a pill diary. During cycle 1, a 7-day treatment hold was permitted for any reason; however, no dose reductions were permitted except in patients who experienced DLTs. After completion of the DLT period, up to 2 intrapatient dose reductions were allowed for toxicity. Intrapatient dose escalations were not permitted. Patients were treated until disease progression, unacceptable toxicity, or withdrawal of consent. All patients were required to undergo a pre-treatment tumor biopsy within 4 weeks prior to study treatment for genomic profiling and an on-treatment biopsy within 2 weeks of start of therapy for pharmacodynamics studies.
Safety and radiographic assessments
Clinical and laboratory assessments were performed at screening; cycle 1, days 1, 8 and 15; cycle 2, days 1 and 15, and monthly thereafter. Patients were evaluable for toxicity if they had received 1 dose of alpelisib with an AI. Toxicity was graded according to the National Cancer Institute Common Terminology Criteria for Adverse Events (CTCAE), version 4.0. A dose-limiting toxicity (DLT) was an adverse event (AE) or abnormal laboratory value that met protocol-specified criteria for severity, and that was at least possibly related to the study medication occurring during cycle 1. A confirmatory safety assessment was performed at the end of Cycle 2.
DLTs comprised any grade (G) 4 adverse events (AEs), persistent or clinically significant G3 AEs, and G2 AEs deemed intolerable or leading to dose interruption of greater than 7 days (Supplementary Table 1). G3 hyperglycemia was a DLT if it did not resolve to G0 within 14 days of anti-diabetic treatment initiation. Toxicities were managed as deemed appropriate by treating physicians with guidance of protocol-specified recommendations. Toxicity data was recorded per institutional practice in the Clinical Research Database.
Radiographic disease evaluation with a CT scan performed at baseline (within 4 weeks of study start), every 8 weeks for the first 16 weeks, every 12 weeks thereafter, and at end of study. A bone scan was performed at baseline and as per investigator discretion thereafter. Patients were evaluable for response if they had received at least one week of study treatment. A study radiologist (B.C.) assessed all imaging according to the Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1.
Sequencing
Tumor tissue sequencing
DNA was extracted from formalin-fixed paraffin embedded (FFPE) pretreatment tumor biopsy samples using the QIAamp DNA Micro Kit (Qiagen). Mononuclear cells from peripheral blood were used to extract patient-matched normal DNA. Methods for tumor sequencing by MSK-IMPACT have been published previously55–57. In addition to the gene-level CNA calls generated by the clinical laboratory pipeline, for additional genome-wide copy number analyses we determined total and allele-specific copy number genome-wide using FACETS algorithm described previously58. To obtain allele-specific copy number calls, we use a Gaussian-non-central χ2 mixture model. Tumor purity, ploidy and clonal heterogeneity are factored in the model to obtain accurate ASCN output and facilitate the identification of subclonal events. We perform joint segmentation of total and allelic copy ratio to identify regions of copy number alterations, and estimate tumor purity, ploidy and integer copy number calls as described in Shen and Seshan58. The FACETS output was then used to estimate cancer cell fraction26 (CCF i.e. variant allele frequency corrected for purity, ploidy and focal copy number changes) for somatic mutation detected in the tumor biopsy. Binomial exact confidence interval (CI) was calculated around the point estimate of CCF. Each mutation was then classified into clonal and subclonal status. Mutations with lower bound of 95% CI ≥ 75% were classified as clonal. Mutations with CCF ≥ 80% and lower bound of 95% CI below 75% were classified as likely clonal. Mutations with CCF < 80% and lower bound of 95% CI <75% were classified as subclonal.
In case of MSK-IMPACT test failure due to technical issues or insufficient analyte, alternative sequencing was performed using Sequenom mass spectrometry genotyping for specific mutations in 8 genes (AKT1, BRAF, EGFR, ERBB2, KRAS, MAP2K1, NRAS, and PIK3CA). This assay was able to detect common PIK3CA mutations in residues R88, N345, C520, E542, E545, M1043, and H1047. Two patients underwent extramural next generation sequencing (NGS) and their raw sequencing data was not available for analysis.
Cell-free DNA extraction, library preparation, and sequencing
All plasma cell-free DNA (cfDNA) extraction, processing, and sequencing was performed in a CLIA-certified, CAP-accredited laboratory (Guardant Health, Inc.) as previously described34,35 utilizing a 73-gene targeted assay, which could detect SNVs and small indels in all exons of PIK3CA. Briefly, cfDNA was extracted from the entire plasma aliquot prepared from a single 10ml tube described above (QIAmp Circulating Nucleic Acid Kit, Qiagen, Inc). 5–30ng of extracted cfDNA was labeled with non-random oligonucleotide barcodes (IDT, Inc.) and used to prepare sequencing libraries, which were then enriched by hybrid capture (Agilent Technologies, Inc.), pooled, and sequenced by paired-end synthesis (NextSeq 500 and/or HiSeq 2500, Illumina, Inc.). Separate sequencing controls were utilized for SNVs and CNAs/fusions/indels (CFI). The SNV control comprised a mixture of healthy donor cfDNA pooled to target germline SNVs to 0.5%, 2.5%, and 6% allelic fraction. The CFI control comprised cell lines with known CNAs, fusions, and indels diluted into healthy donor cfDNA.
Bioinformatics analysis and variant detection
As previously described34,35 base call files generated by Illumina’s RTA software (v2.12) were de-multiplexed using bcl2fastq (v2.19) and processed with a custom pipeline for molecule barcode detection, sequencing adapter trimming, and base quality trimming (discarding bases below Q20 at the ends of the reads). Processed reads were then aligned to the human reference genome hg19 using BWA-MEM (arXiv:1303.3997v2) and used to build double-stranded consensus representations of original unique cfDNA molecules using both the inferred molecular barcodes and read start/stop positions. SNVs were detected by comparing read and consensus molecule characteristics to sequencing platform- and position-specific reference error noise profiles determined independently for each position in the panel by sequencing a training set of 62 healthy donors on both the NextSeq 500 and HiSeq 2500. Observed positional SNV error profiles were used to define calling cut-offs for SNV detection with respect to the number and characteristics of variant molecules, which differed by position but were most commonly ≥2 unique molecules, which in an average sample (~5,000 unique molecule coverage), the corresponds to a detection limit of ~0.04% VAF. Indel detection used two methods. For short (<50–70bp) indels, a generative background noise model was constructed to account for PCR artifacts arising frequently in homopolymeric or repetitive contexts, allowing for strand-specific and late PCR errors. Detection was then determined by the likelihood ratio score for observed feature weighted variant molecule support versus background noise distribution. Detection of indels >>50bp relies on secondary analysis of soft-clipped reads using methods described in the fusion section below and is only performed to detect specific genomic events (e.g. MET exon 14-skipping deletions). Reporting thresholds were event-specific as determined by performance in training samples but were most commonly at least one unique molecule for clinically actionable indels, which in an average sample corresponds to a detection limit of ~0.02% VAF.
To detect CNAs, probe-level unique molecule coverage was normalized for overall unique molecule throughput (the equivalent of library size), probe efficiency, GC content, and signal saturation and summarized at the gene level. CNA determinations were based on training set-established decision thresholds for both absolute copy number deviation from per-sample diploid baseline and deviation from the baseline variation of probe-level normalized signal in the context of background variation within each sample’s own diploid baseline. Per-sample relative tumor burden was determined by normalization to the mutational burden expected for tumor type and ctDNA fraction and reported as a z-score.
ctDNA fraction and cancer cell fraction assessment
ctDNA fraction for each plasma sample was estimated from clonal biopsy-matched mutations. CCF estimates for somatic mutations detected in the matched tumor biopsy sample. Allele specific copy number (ASCN) was estimated using the FACETS algorithm58. The FACETS output was then used to estimate CCD for somatic mutation detected in the matched tumor biopsy. Binomial exact confidence interval (CI) was calculated around the point estimate of CCF. Each biopsy-matched mutation was then classified into clonal and subclonal status. Mutations with lower bound of 95% CI ≥ 75% were classified as clonal. Mutations with CCF ≥ 80% and lower bound of 95% CI below 75% were classified as likely clonal. Mutations with CCF < 80% and lower bound of 95% CI <75% were classified as subclonal. The biopsy-matched clonal mutations were then used to estimate the plasma sample ctDNA fraction based on the plasma variant allele frequency. The cancer cell fraction of detected ctDNA mutations was estimated by maximum likelihood26 for patients with available matched tumor sequencing using the VAF observed in cfDNA and assuming similar integer level total and allelic copy numbers in the matched pre-treatment tumor biopsy.
Additionally, as an orthogonal approach to estimate CCF of ctDNA mutations, we utilized a ctDNA only approach based on the VAFs by normalizing against a log2 transformation of the reported copy numbers on a per gene basis44,45.
Mutational signatures from hypermutated patients
The contributions of different mutation signatures were identified for each sample according to distribution of the six substitution classes (C>A, C>G, C>T, T>A, T>C, T>G) and the bases immediately 5′ and 3′ of the mutated base, producing 96 possible mutation subtypes using deconstructSigs 59. For analyses in the manuscript, we focused on six signatures: (1) aging (signature 1 and 5), (2) APOBEC (signatures 2 and 13), (3) homologous recombination repair deficiency (HRD, signature 3), (4) MMR (signatures 6, 15, 20 and 26), (5) smoking (signature 4), and (6) POLE (signature 10).
cfDNA digital droplet PCR
Digital droplet PCR (ddPCR) was performed on PIK3CA H1047R, H1047L, H1047Y, E545K, E542K, C420R, N345K in all pre-, on- and post-treatment plasma specimens. The assays were designed and ordered through Biorad. Cycling conditions were tested to ensure optimal annealing/extension temperature as well as optimal separation of positive from empty droplets. All reactions were performed on a QX200 ddPCR system (Biorad). Each sample was evaluated in technical duplicates. PCR reactions contains primers and probes, DNA and digital PCR Supermix for probes (no dUTP). Reactions were partitioned into a median of ~16,000 droplets per well using the QX200 droplet generator. Emulsified PCRs were run on a 96-well thermal cycler using cycling conditions identified during the optimization step (95°C 10’;40 cycles of 94°C 30” 55°C 1’, 98°C 10’, 4°C hold). Plates were read and analyzed with the QuantaSoft software to assess the number of droplets positive for mutant DNA, wild-type DNA, both, or neither. Empirical sensitivity of 0.03% was assessed from total droplet counts across all metastatic sites and assessed mutations and we require that greater than 3 mutant droplets exist before a site is confirmed mutated.
Generation of ESR1 Y537S knock in MCF7 cell line
MCF7 cells were transfected with a single-guide RNA Cas9 vector and a repair template cloned into pUC vector at a 1:1 ratio using Xtremegene 9 HP (Roche). The repair template contained homology regions for incorporation of the Y537S mutation via homologous directed repair (HDR). The media was changed to estrogen deprived media, 48h after transfection, to select for HDR events for a period of 2 weeks. As a Puromycin resistance gene was encoded between the homology regions of the repair template, puromycin was also used for selection of HDR events after estrogen deprived media selection for another 2 weeks. After both selections were completed, single-cell clones were generated and screened with ddPCR for evidence of Y537S mutation. The clones positive for Y537S mutation were further screened for their proliferation rates, ERE-luciferase activity, induction of ER target genes in the absence of estrogen and fulvestrant sensitivity. Clones with estrogen independent activity level in all aspects as mentioned above were then sent for MSK-IMPACT analysis to make sure that they have no off-target mutations in the major oncogenes and tumor suppressors.
Animal Studies
Athymic nu/nu BALB/c female mice of 6–8 weeks old were obtained from Envigo and maintained in pressurized ventilated caging. All studies were performed in compliance with institutional guidelines under an Institutional Animal Care and Use Committee–approved protocol (MSKCC#12–10-016). The mice were first implanted subcutaneously with 0.18-mg sustained release 17β-estradiol pellets using a 10 g trocar into one flank and then injected with 1 × 107 cells suspended 1:1 (volume) with reconstituted basement membrane (Matrigel, Collaborative Research) on the opposite side 3 days afterward for the establishment of parental MCF7 and MCF7 Y537S xenograft tumors. When the tumors reached a size of ∼200 mm3, the mice bearing tumors from each cell line were randomized into 4 treatment groups, with either vehicle, estradiol pellet removed, 25 mg/kg of alpelisib via oral gavage once daily or estradiol pellet removed plus 25mg/kg of alpelisib. Tumor dimensions were measured with vernier calipers and tumor volumes calculated [π/6 × larger diameter × (smaller diameter)2]. In this study, there was no blinding of the investigator as randomization of animals was done. Based upon our previous work measuring the variability in size and growth of MCF7 xenografts, we estimated 10 mice/group would allow us to detect tumor size differences of >200 mm3.
Statistics and Reproducibility
The data included in the analysis reflect the data lock on April 31, 2018 from patients enrolled up to September 4, 2015. All patients who received at least one dose of alpelisib were included in the safety study. All the patients were off trial at the time of data analysis. Associations between genomic alterations and response were assessed using Fisher exact test or χ2 test (where appropriate). The exact confidence intervals were calculated for the clinical benefit rate. The difference in paired ctDNA post- and pre-treatment VAFs were calculated using Wilcoxon signed rank tests and the p-values were adjusted for multiple testing using the standard Benjamini-Hochberg multiple testing correction. The differences between xenograft growth curves were calculated based on 2-way ANOVA and Šidák correction for multiple comparisons. All statistical hypothesis tests were two-sided with α = 0.05 and carried out in R/Bioconductor.
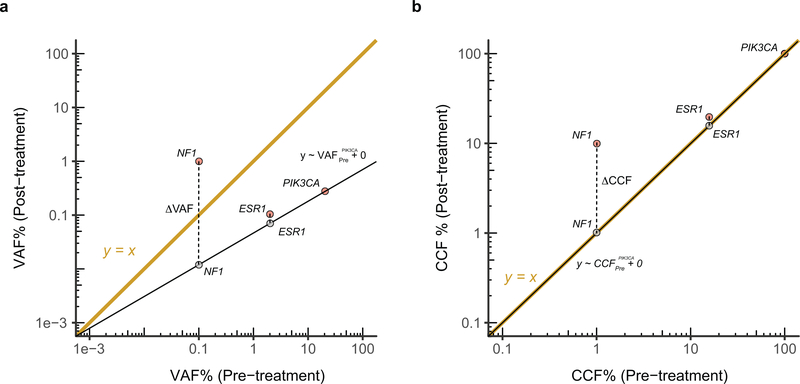
The ΔCCF or normalized ΔVAF quantify the additional change in CCF or VAF between pre- and post-treatment samples given the value expected from the difference in CCF of a canonical PIK3CA mutation i.e. E542K, E545K, H1047R, etc. (Extended Data Fig. 2), the latter being a surrogate of change in tumor burden. In case a patient did not have a canonical or any PIK3CA mutation, we used the mutation with highest VAF and is predicted to be fully clonal (CCF ~ 1.0) in pre-treatment tumor biopsy provided it is also present at high VAF and is fully clonal (CCF ~ 1.0) both in the pre- and post-treatment cfDNA sequencing. In the case of ΔVAF, we used a linear regression with zero y-intercept and compute the VAF as the Log10 difference between the observed VAF in the post-treatment cfDNA and the value predicted from the fitted line. In the case of ΔCCF, this amounts to the difference between the observed CCF in the post-treatment cfDNA and the line. However, in order to avoid numerical inconsistencies for the clonal PIK3CA mutation, we used a linear regression with zero y-intercept similar to that for the VAF. The normalized ΔVAF and ΔCCF thus computed express the ratio of observed and expected VAF or CCF, respectively.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1. Characterization of ctDNA variants.
a) Comparison of variant allele fraction (VAF) of PIK3CA mutations measured using the targeted ctDNA assay (y-axis) and ddPCR (x-axis). cfDNA samples extracted from 65 samples (35 patients) with canonical hotspot PIK3CA mutations were subjected to ddPCR. An aliquot of the same cfDNA isolate was used for targeted DNA assay (G360 assay, Guardant Health, CA). b) ctDNA VAFs of all the somatic variants detected in ctDNA restricted to 38 patients with available tumor next generation sequencing results. Colors indicate the altered gene and black borders indicate whether the alteration was detected in the tumor tissue. c) Comparison of VAF of mutations detected in the pre-treatment and post-treatment ctDNA samples of six patients with evaluable paired ctDNA specimens and PTEN loss-of-function mutations in either sample. The colors of the circles indicate mutated gene.
Extended Data Fig. 2. Normalized ΔVAF and ΔCCF model.
Toy model showing the calculation of a) normalized ΔVAF and b) ΔCCF for a fictitious pair of pre- and post-treatment ctDNA samples. Three mutations are shown; PIK3CA, ESR1 and NF1. The canonical PIK3CA mutation is expected to be clonal in the pre- and post-treatment ctDNA samples. The difference in VAF of PIK3CA between the pair is a surrogate of difference in purity and tumor burden. To quantify the change in VAF or CCF of other mutations in addition to what is expected from the difference in purity or tumor burden, we calculate the Log10 difference between the post-treatment cfDNA and the value obtained from the regression i.e. in the examples above, observed post-treatment NF1 VAF – expected post-treatment NF1 VAF. In both cases, the regression has zero y-intercept.
Extended Data Fig. 3. Change in variant allele fraction (ΔVAF) of all mutations in post- vs pre-treatment ctDNA specimens.
Heatmap of change in VAF comparing the post-treatment with pre-treatment ctDNA of 32 evaluable patients (n=64 samples) normalized according to the change in ctDNA fraction as a proxy of change in disease burden. The size of the boxes represents the relative change and the color gradient of the boxes represent increase or decrease in ΔVAF. The top section shows the time to treatment failure (weeks), reason off study, and the best response on therapy. Multiple mutations in a same gene are indicated, for example patient #47 had two different PIK3CA mutations with one mutation having no change whilst the other one expanded in the post-treatment sample (positive ΔVAF). PR: partial response, SD: stable disease, PD: progressive disease, NE: not evaluable for response.
Extended Data Fig. 4. Comparison of normalized change in ctDNA mutations.
a) Comparison of normalized change in ctDNA mutations (ΔVAF) with relative change in mutations cancer cell fractions (ΔCCF). The analysis includes 256 mutations. b) Comparison of estimated CCF based on combined tumor and ctDNA approach (Methods) with CCF estimated based on ctDNA-only approach54,55. The analysis includes 379 mutations. Pearson’s correlation coefficients (R) and two-sided p values are provided.
Extended Data Fig. 5. Mutational signatures in two hypermutated cases with acquired PTEN mutations.
96 base substitution profiles of pre-treatment ctDNA samples from the two hypermutated cases that eventually developed PTEN mutations under therapy showing dominant APOBEC signatures (Signatures 2 and 13).
Supplementary Material
Acknowledgements
This work was supported by Novartis Pharmaceuticals, and National Institutes of Health awards P30 CA008748, R01 CA190642 (M.S.), R01 CA234361 (D.B.S., S.C.), R01CA204999 (S.C.), the Breast Cancer Alliance Young Investigator Award (P.R.), Conquer Cancer Foundation Young Investigator Award (P.D.S.), the Breast Cancer Research Foundation (MS, JSR-F, SC), Damon Runyon Cancer Research Foundation (S.C.), Stand Up to Cancer (M.S., Cancer Drug Combination Convergence Team), the V Foundation (M.S.), the National Science Foundation (M.S.), the Geoffrey Beene Cancer Research Center, and a kind gift from Mrs. Barbara Smith.
Footnotes
Competing Interests
P.R. reports consulting or advisory role for Novartis, AstraZeneca, Foundation Medicine, and institutional research support from Illumina and GRAIL; M.N.D. is an employee of Eli Lilly, P.D.S. reports consulting with Tmnity and research funding from AstraZeneca; B.T.L. reports consulting/advisory board for Genentech, ThermoFisher Scientific, Guardant Health, Hengrui Therapeutics, Mersana Therapeutics, Biosceptre Australia and institutional research support from Illumina, GRAIL, Genentech, AstraZeneca; N.V. reports consulting or advisory role for Novartis; K.J. reports consulting or advisory role for ADC Therapeutics; AstraZeneca; Jounce therapeutics; Novartis; Pfizer; Spectrum; Taiho, research funding from ADC Therapeutics (Inst); Clovis Oncology (Inst); Debio (Inst); Genentech (Inst); Novartis (Inst); Novita (Inst); Pfizer (Inst) and other relationship with Jounce Therapeutics; Novartis; Pfizer; Taiho; A.C. reports being an advisory board member for Accurate Medical a Stockholder for Amgen; L.N. reports honoraria from Advanced Breast Cancer 4 International Consensus Conference; Bluprint Oncology Concepts; Celgene; Context Therapeutics; MCI Breast Cancer Symposium, consulting or advisory role for Advanced Breast Cancer International Consensus Conference; Bluprint Oncology Concepts; Celgene; Context Therapeutics; MCI Breast Cancer Symposium and travel, accommodations, expenses from Advanced Breast Cancer International Consensus Conference; Celgene; MCI Breast Cancer Symposium; A.S., R.J.N., J.I.O., and R.B.L. are employees and stockholders of Guardant Health; M.E.R. reports honoraria from AstraZeneca, consulting or advisory role for AstraZeneca; McKesson; Merck; Pfizer, research funding from Abbvie (Inst); AstraZeneca (Inst); InVitae (Inst); Medivation (Inst); Myriad Genetics (Inst); Tesaro (Inst), and travel, accommodations, expenses from AstraZeneca; M.E.L. reports serving as a consultant or speaking for Legacy Healthcare Services, Adgero Bio, Amryt, Celldex, Debiopharm, Galderma, Johnson & Johnson, Novocure, Lindi, Merck, Sharp and Dohme Corp, Helsinn Healthcare SA, Janssen Research & Development LLC, Menlo Therapeutics, Novartis, Roche, AbbVie, Boehringer Ingelheim, Allergan, Amgen, E. R. Squibb & Sons, LLC, EMD Serono, AstraZeneca, Genentech, Leo Pharma, Seattle Genetics, Bayer, Manner, SAS, Lutris, Pierre Fabre, Paxman Coolers, Adjucare, Dignitana, Biotechspert, Teva Mexico, Parexel, OnQuality, Novartis, Our Brain Bank, and Takeda Millenium; and receiving research funding from Veloce, US Biotest, Berg, BMS, Lutris, Paxman, and Novocure; J.S.R-F reports personal/consultancy fees from VolitionRx, Page.AI, Goldman Sachs, Grail, Ventana Medical Systems, Roche, Genentech and Invicro; D.B.S. received honoraria and consulted for Pfizer, Loxo, Vivideon, Illumina and Lilly Oncology; M.S. is in the Advisory Board of Bioscience Institute and Menarini Ricerche, received research funds from Puma Biotechnology, Daiichi Sankyo, Targimmune, Immunomedics and Menarini Ricerche, is a co-founder of Medendi Medical Travel and received honoraria from Menarini Ricerche and ADC; S.C. reports institutional research funding from Novartis, Eli Lilly, Sanofi, Daiichi Sankyo, Genentech and Ad hoc consulting for Novartis, Context Therapeutics, Sermonix, Eli Lilly, BMS, and Revolution Medicine. The other coauthors report no competing interests.
Data Availability
The assembled prospective somatic mutational data from ctDNA and tumors for the entire cohort have been deposited for visualization and download in the cBioPortal for Cancer Genomics (http://cbioportal.org/) under the following URL: https://www.cbioportal.org/study/summary?id=breast_alpelisib_2020
Source data for Figs. 1–5 and Extended Data Figs. 1,3–5 have been provided as the following Source Data files: Source_Data_Fig_1.xlsx; Source_Data_Fig_2.xlsx; Source_Data_Fig_3.xlsx; Source_Data_Fig_4.xlsx; Source_Data_Fig_5.xlsx; Source_Data_Extended_Data_Fig_1.xlsx; Source_Data_Extended_Data_Fig_3.xlsx; Source_Data_Extended_Data_Fig_4.xlsx; Source_Data_Extended_Data_Fig_5.xlsx.
All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Code Availability
The custom codes written to analyze the cfDNA data are available at https://github.com/ndbrown6/MSK-BYL-NC
REFERENCES
- 1.Siegel RL, Miller KD & Jemal A Cancer statistics, 2019. CA Cancer J Clin 69, 7–34 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Ma CX, Reinert T, Chmielewska I & Ellis MJ Mechanisms of aromatase inhibitor resistance. Nat Rev Cancer 15, 261–275 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Toy W, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nature genetics 45, 1439–1445 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schiavon G, et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med 7, 313ra182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Razavi P, et al. The Genomic Landscape of Endocrine-Resistant Advanced Breast Cancers. Cancer Cell 34, 427–438 e426 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Isakoff SJ, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res 65, 10992–11000 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Lauring J, Park BH & Wolff AC The phosphoinositide-3-kinase-Akt-mTOR pathway as a therapeutic target in breast cancer. J Natl Compr Canc Netw 11, 670–678 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlo MI, et al. A Phase Ib Study of BEZ235, a Dual Inhibitor of Phosphatidylinositol 3-Kinase (PI3K) and Mammalian Target of Rapamycin (mTOR), in Patients With Advanced Renal Cell Carcinoma. Oncologist 21, 787–788 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarker D, et al. First-in-human phase I study of pictilisib (GDC-0941), a potent pan-class I phosphatidylinositol-3-kinase (PI3K) inhibitor, in patients with advanced solid tumors. Clin Cancer Res 21, 77–86 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Juric D, et al. Phase I Dose-Escalation Study of Taselisib, an Oral PI3K Inhibitor, in Patients with Advanced Solid Tumors. Cancer Discov 7, 704–715 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bohnacker T, et al. Deconvolution of Buparlisib’s mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention. Nat Commun 8, 14683 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bendell JC, et al. Phase I, dose-escalation study of BKM120, an oral pan-Class I PI3K inhibitor, in patients with advanced solid tumors. J Clin Oncol 30, 282–290 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Baselga J, et al. Buparlisib plus fulvestrant versus placebo plus fulvestrant in postmenopausal, hormone receptor-positive, HER2-negative, advanced breast cancer (BELLE-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 18, 904–916 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosch A, et al. PI3K inhibition results in enhanced estrogen receptor function and dependence in hormone receptor-positive breast cancer. Sci Transl Med 7, 283ra251 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fox EM, Kuba MG, Miller TW, Davies BR & Arteaga CL Autocrine IGF-I/insulin receptor axis compensates for inhibition of AKT in ER-positive breast cancer cells with resistance to estrogen deprivation. Breast Cancer Res 15, R55 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toska E, et al. PI3K pathway regulates ER-dependent transcription in breast cancer through the epigenetic regulator KMT2D. Science 355, 1324–1330 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Creighton CJ, et al. Proteomic and transcriptomic profiling reveals a link between the PI3K pathway and lower estrogen-receptor (ER) levels and activity in ER+ breast cancer. Breast Cancer Res 12, R40 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juric D, et al. Phosphatidylinositol 3-Kinase alpha-Selective Inhibition With Alpelisib (BYL719) in PIK3CA-Altered Solid Tumors: Results From the First-in-Human Study. J Clin Oncol 36, 1291–1299 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juric D, et al. Alpelisib Plus Fulvestrant in PIK3CA-Altered and PIK3CA-Wild-Type Estrogen Receptor-Positive Advanced Breast Cancer: A Phase 1b Clinical Trial. JAMA Oncol, e184475 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayer IA, et al. A Phase Ib Study of Alpelisib (BYL719), a PI3Kalpha-specific Inhibitor, with Letrozole in ER+/HER2-Negative Metastatic Breast Cancer. Clin Cancer Res (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andre F, et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N Engl J Med 380, 1929–1940 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Will M, et al. Rapid induction of apoptosis by PI3K inhibitors is dependent upon their transient inhibition of RAS-ERK signaling. Cancer Discov 4, 334–347 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayer IA, et al. A Phase II Randomized Study of Neoadjuvant Letrozole Plus Alpelisib for Hormone Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Negative Breast Cancer (NEO-ORB). Clin Cancer Res 25, 2975–2987 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baselga J, et al. Phase III study of taselisib (GDC-0032)+ fulvestrant (FULV) v FULV in patients (pts) with estrogen receptor (ER)-positive, PIK3CA-mutant (MUT), locally advanced or metastatic breast cancer (MBC): Primary analysis from SANDPIPER. Journal of Clinical Oncology 36(2018). [Google Scholar]
- 26.Carter SL, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 30, 413–421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakravarty D, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol 2017(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Juric D, et al. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kalpha inhibitor. Nature 518, 240–244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yates LR, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med 21, 751–759 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng CKY, et al. Genetic Heterogeneity in Therapy-Naive Synchronous Primary Breast Cancers and Their Metastases. Clin Cancer Res 23, 4402–4415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murtaza M, et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun 6, 8760 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Mattos-Arruda L, et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO 25, 1729–1735 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Razavi P, et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Odegaard JI, et al. Validation of a Plasma-Based Comprehensive Cancer Genotyping Assay Utilizing Orthogonal Tissue- and Plasma-Based Methodologies. Clin Cancer Res 24, 3539–3549 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Lanman RB, et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PLoS One 10, e0140712 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez-Escudero I, et al. A comprehensive functional analysis of PTEN mutations: implications in tumor- and autism-related syndromes. Hum Mol Genet 20, 4132–4142 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Marsh DJ, et al. Mutation spectrum and genotype-phenotype analyses in Cowden disease and Bannayan-Zonana syndrome, two hamartoma syndromes with germline PTEN mutation. Hum Mol Genet 7, 507–515 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Han SY, et al. Functional evaluation of PTEN missense mutations using in vitro phosphoinositide phosphatase assay. Cancer Res 60, 3147–3151 (2000). [PubMed] [Google Scholar]
- 39.Diehl F, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 14, 985–990 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dawson SJ, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 368, 1199–1209 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Murtaza M, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497, 108–112 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Newman AM, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 20, 548–554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castel P & Scaltriti M Mechanisms of Resistance to PI3K and AKT Inhibitors in Resistance to Anti-Cancer Therapeutics Targeting Receptor Tyrosine Kinases and Downstream Pathways (eds. Yarden Y & Elkabets M) 117–146 (Springer International Publishing, Cham, 2018). [Google Scholar]
- 44.Zill OA, et al. The Landscape of Actionable Genomic Alterations in Cell-Free Circulating Tumor DNA from 21,807 Advanced Cancer Patients. Clin Cancer Res 24, 3528–3538 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Siravegna G, et al. Radiologic and Genomic Evolution of Individual Metastases during HER2 Blockade in Colorectal Cancer. Cancer Cell 34, 148–162 e147 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Jia S, et al. Essential roles of PI(3)K-p110beta in cell growth, metabolism and tumorigenesis. Nature 454, 776–779 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wee S, et al. PTEN-deficient cancers depend on PIK3CB. Proc Natl Acad Sci U S A 105, 13057–13062 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spoerke JM, et al. Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat Commun 7, 11579 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chandarlapaty S, et al. Prevalence of ESR1 Mutations in Cell-Free DNA and Outcomes in Metastatic Breast Cancer: A Secondary Analysis of the BOLERO-2 Clinical Trial. JAMA Oncol 2, 1310–1315 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ng CK, et al. Intra-tumor genetic heterogeneity and alternative driver genetic alterations in breast cancers with heterogeneous HER2 gene amplification. Genome Biol 16, 107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toledo RA, et al. Exome Sequencing of Plasma DNA Portrays the Mutation Landscape of Colorectal Cancer and Discovers Mutated VEGFR2 Receptors as Modulators of Antiangiogenic Therapies. Clin Cancer Res 24, 3550–3559 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Chicard M, et al. Whole-Exome Sequencing of Cell-Free DNA Reveals Temporo-spatial Heterogeneity and Identifies Treatment-Resistant Clones in Neuroblastoma. Clin Cancer Res 24, 939–949 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Frenel JS, et al. Serial Next-Generation Sequencing of Circulating Cell-Free DNA Evaluating Tumor Clone Response To Molecularly Targeted Drug Administration. Clin Cancer Res 21, 4586–4596 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keup C, et al. Targeted deep sequencing revealed variants in cell-free DNA of hormone receptor-positive metastatic breast cancer patients. Cell Mol Life Sci (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 55.Cheng DT, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 17, 251–264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng DT, et al. Comprehensive detection of germline variants by MSK-IMPACT, a clinical diagnostic platform for solid tumor molecular oncology and concurrent cancer predisposition testing. BMC Med Genomics 10, 33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zehir A, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 23, 703–713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shen R & Seshan VE FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 44, e131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rosenthal R, McGranahan N, Herrero J, Taylor BS & Swanton C DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol 17, 31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.