Abstract
Purpose
Precision oncology develops and implements evidence-based personalized therapies that are based on specific genetic targets within each tumor. However, a major challenge that remains is the provision of a standardized, up-to-date, and evidenced-based precision medicine initiative across a geographic region.
Materials and Methods
We developed a statewide molecular tumor board that integrates academic and community oncology practices. The Precision Medicine Molecular Tumor Board (PMMTB) has three components: a biweekly Web-based teleconference tumor board meeting provided as a free clinical service, an observational research registry, and a monthly journal club to establish and revise evidence-based guidelines for off-label therapies. The PMMTB allows for flexible and rapid implementation of treatment, uniformity in practice, and the ability to track outcomes.
Results
We describe the implementation of the PMMTB and its first year of activity. Seventy-seven patient cases were presented, 48 were enrolled in a registry, and 38 had recommendations and clinical follow-up. The 38 subjects had diverse solid tumors (lung, 45%; GI, 21%; breast, 13%; other, 21%). Of these subjects, targeted therapy was recommended for 32 (84%). Clinical trials were identified for 24 subjects (63%), and nontrial targeted medicines for 16 (42%). Nine subjects (28%) received recommended therapy with a response rate of 17% (one of six) and a clinical benefit rate (partial response + stable disease) of 38% (three of eight). Although clinical trials often were identified, patients rarely enrolled.
Conclusion
The PMMTB provides a model for a regional molecular tumor board with clinical utility. This work highlights the need for outcome registries and improved access to clinical trials to pragmatically implement precision oncology.
INTRODUCTION
Precision oncology provides individualized treatment of patients on the basis of tumor genomic profiles. Today, next-generation sequencing assays are widely available and considered to be part of the standard of care in specific clinical situations.1,2 At the same time, precision medicine promises to match patients to gene-targeted treatments.3 However, many challenges exist in implementing precision medicine, including knowledge gaps, privacy, systems barriers, and reimbursement issues.4,5 The knowledge gaps arise from the extensive breadth and depth of knowledge required to effectively use genomic information, which is compounded by tumor complexity and the wide variation of genomic landscapes in tumors.6,7 To meet this challenge, knowledge in genomic technologies, cancer biology, human pathology, pharmacology, clinical patient care, and often detailed knowledge of gene structure and function are required.
Molecular tumor boards (MTBs) address gaps in knowledge and clinical utility by providing a forum for individuals with wide-ranging expertise to review patient medical histories and mutation profiles to guide patient-specific treatment options.8-12 In contrast to organ-specific tumor boards, MTBs consider patients with diverse histopathologies and focus on precision therapies directed at molecular targets. Although MTBs have been established at academic institutions, they are largely unavailable at nonacademic sites.
To expand access to MTBs, we have partnered with community practitioners at other health networks to develop an integrated MTB available to any provider in our geographic region, the Precision Medicine Molecular Tumor Board (PMMTB). The PMMTB provides a no-cost clinical service accessible to oncologists in the region through Web-based teleconference. In addition to its regional scope, the PMMTB is novel in its tripartite structure by separating clinical service, a registry protocol, and a journal club to establish parameters for evidence-based use of off-label therapies. Together, this structure ensures flexibility, standardization, and tracking of outcomes. In addition, the PMMTB provides a venue for education in molecular pathology and continuing medical education. In the first year of operation, 77 patient cases were presented in 23 meetings. We report PMMTB structure, operation, and clinical experience. The findings illustrate the value of a regional PMMTB and highlight the need to improve access to precision medicine clinical trials.
MATERIALS AND METHODS
The PMMTB Structure
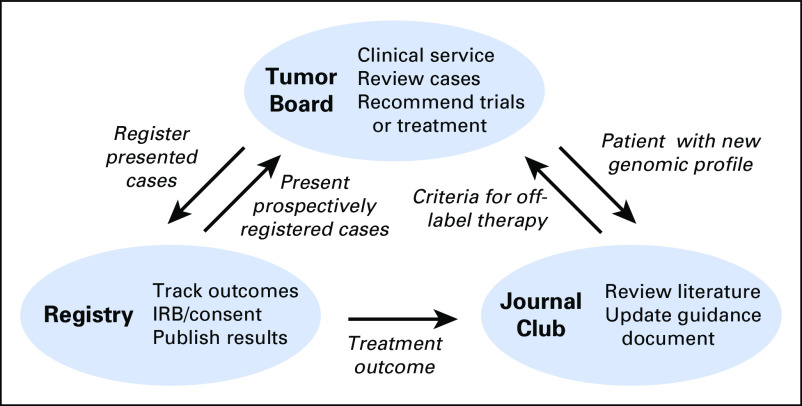
The Wisconsin PMMTB has a tripartite structure (Fig 1). The tumor board meeting is at the apex of this structure and is focused on reviewing clinical cases. Identified cases are submitted for review to the PMMTB coordinator, and deidentified for presentation to minimize the risk of breach of confidentiality. The primary mission of this clinical conference is service to patients. The second part of the structure is a registry that prospectively or retrospectively consents patients and collects data on patient outcomes. The goal is to enroll all patients whose cases are presented at the tumor board. Moreover, some patients are prospectively consented to the registry at the time that molecular testing and biopsy are performed; these patient cases are later presented at the tumor board. The third part of the structure is a monthly journal club meeting. Each journal club is structured as a 1-hour presentation where a volunteer advocate nominates a drug-biomarker pair. This advocate is responsible for circulating and presenting up to three citations that support the drug-biomarker pair. At the conclusion of the journal club, the tumor board decides whether to adopt or reject the proposed drug-biomarker pair and to update the guidance document that specifies when off-label therapies will be recommended. Journal clubs can be triggered by specific patient cases presented at the tumor board meeting. In addition, periodic review of cases in the tumor registry may reveal ineffective treatment approaches, which in turn will trigger a journal club to seek to retract a previously issued criterion in the guidance document.
Fig 1.
Tripartite structure of the Precision Medicine Molecular Tumor Board, including the tumor board, registry, and journal club, and interactions among the components. IRB, institutional review board.
Tumor Board
The first part of the PMMTB structure consists of a twice-monthly tumor board convened through Web-based teleconference. Participants are experts in diverse fields that span basic cancer biology, medical/clinical genetics, molecular pathology, surgical pathology/cytopathology, pharmacology, medical oncology, and radiation oncology. At meetings, clinical cases of metastatic/incurable cancer are presented with next-generation sequencing results followed by discussion directed at optimal clinical trials or targeted drugs. Patient cases are submitted to the coordinator at least 7 days before the meeting with a brief clinical history and factors that affect clinical trial eligibility. Molecular test results from a Clinical Laboratory Improvement Amendments–certified laboratory are included with the case submission information.
Up to six cases are discussed per 1-hour meeting; when additional cases are submitted, review is either postponed or performed administratively by the PMMTB chairs. At the meeting, each case is presented by the treating physician or designee followed by review of histopathology and genomic results. The molecular pathology team reviews alterations and assesses their analytic validity, that is, whether a particular mutation is likely to alter gene function on the basis of site and allele frequency or whether an amplicon encompasses many reported genes. The tumor board prioritizes alterations relevant to current and emerging treatment options. Clinical significance of alterations are systematically discussed in the context of cancer databases (Cancer Gene Census/COSMIC [Catalogue of Somatic Mutations in Cancer]), The Cancer Genome Atlas, and human polymorphism databases (dbSNP, 1000 Genomes Project, ESP6500) along with knowledge tools (cBioPortal, My Cancer Genome, targeted cancer care, personalized cancer therapy).13-18 The board identifies and discusses possible clinical trials and off-label targeted therapies. Clinical trials are identified by using ClinicalTrials.gov and other resources. Off-label therapies are recommended per separate guidance document (see Journal Club). If deemed valuable, the PMMTB may also suggest genetic counseling and additional tumor testing. Final recommendations are provided in the form of a formal letter to the submitting physician, with acceptance at the discretion of the treating clinician (Data Supplement).
Journal Club
The PMMTB considers targeted Food and Drug Administration–approved drugs that are not labeled for use in that particular patient’s tumor type according to a guidance document. This guidance document lists specific or general drug-biomarker pairs deemed acceptable for recommending off-label targeted therapies (Data Supplement). The guidance document includes, for example, the targeting of oncogenes with known activating mutations by using drugs that directly inhibit their activity, combination therapies for BRAF mutations, and specific criteria for using genomic decisions for immune checkpoint inhibitors. The guidance document is dynamic and expands or contracts on the basis of new evidence presented at the monthly journal club meeting. During the meeting, up to three studies, including human and preclinical evidence, are provided and reviewed; members of the PMMTB decide whether to amend the guidance document, to provisionally amend it (if human data are extremely limited), or to reject it. Occasionally, a case presented in the PMMTB provides the impetus for convening a journal club meeting. If the guidance document is amended, the case is reviewed again at the next PMMTB meeting, and an updated recommendation letter is written. In this manner, all decisions to recommend off-label therapies are based on careful, but timely review of evidence.
Registry
Patients are enrolled in a registry to allow for the tracking of outcomes. The registry protocol was approved by our institutional review board (University of Wisconsin [UW] HS-IRB Approval #2015-1370) and was conducted in accordance with the Declaration of Helsinki. Although initially envisioned as prospective, regulatory delays resulted in a mixture of prospective and retrospective enrollment. To enroll, subjects provided written informed consent, except in the case of deceased individuals. Registry subjects are patients ≥ 18 years of age with histologically confirmed or suspected malignancy who had or were planning genetic testing of their tumor. All Clinical Laboratory Improvement Amendments–certified genomic tumor profiling assays are allowed, including commercial tests, tests performed at outside academic facilities, and tests performed at the UW Collaborative Genomics Core.
Subjects registered and presented at the PMMTB were followed for acceptance of recommendations, treatment, and disease response or progression. Data elements, including demographic information, prior therapies, genomic information, and pathology were obtained from subject electronic health records at the time of study entry. PMMTB recommendations were recorded after the meetings. Subjects were followed for study end points, including acceptance of PMMTB recommendations, barriers to following recommendations, and response to therapy. The primary objective of this study was to assess the clinical utility of the PMMTB, defined as the frequency of recommendation acceptance. Descriptive summaries of response to therapy, disease status, and demographic information are provided. To evaluate response, central radiologic review of tumor responses was made using Response Evaluation Criteria in Solid Tumors (RECIST) 1.1 for patients with measurable tumors. For nonmeasurable tumors, stable disease versus partial response was reported on the basis of radiologic follow-up and clinical assessment of the treating physician at the follow-up study performed 2 to 4 months after baseline.
RESULTS
Participant and Disease Characteristics
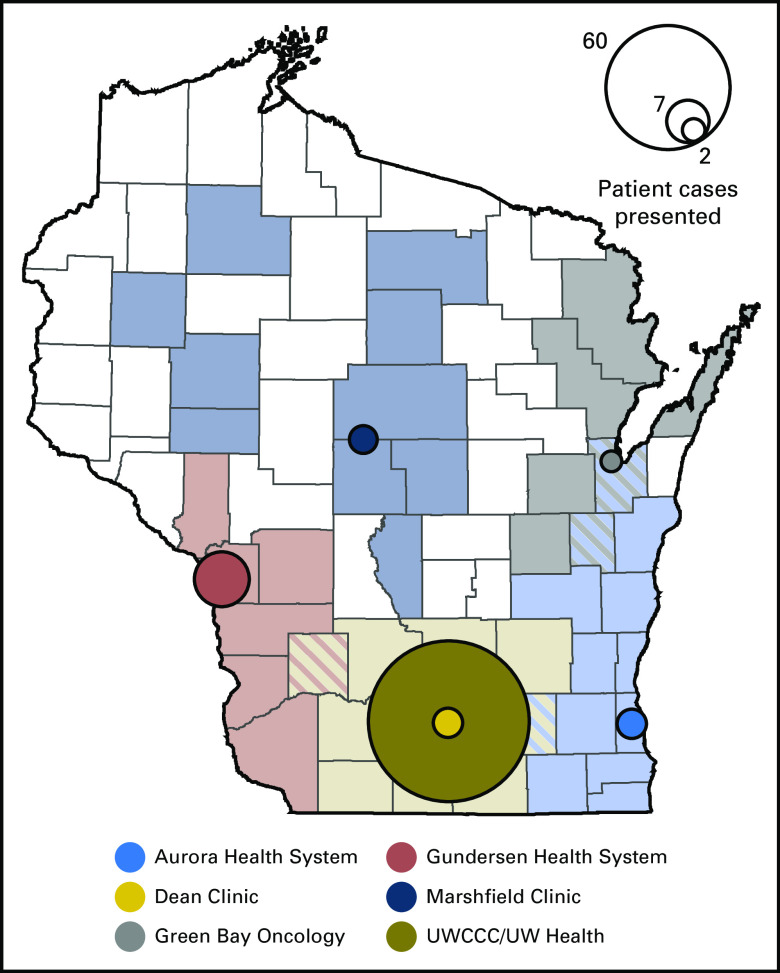
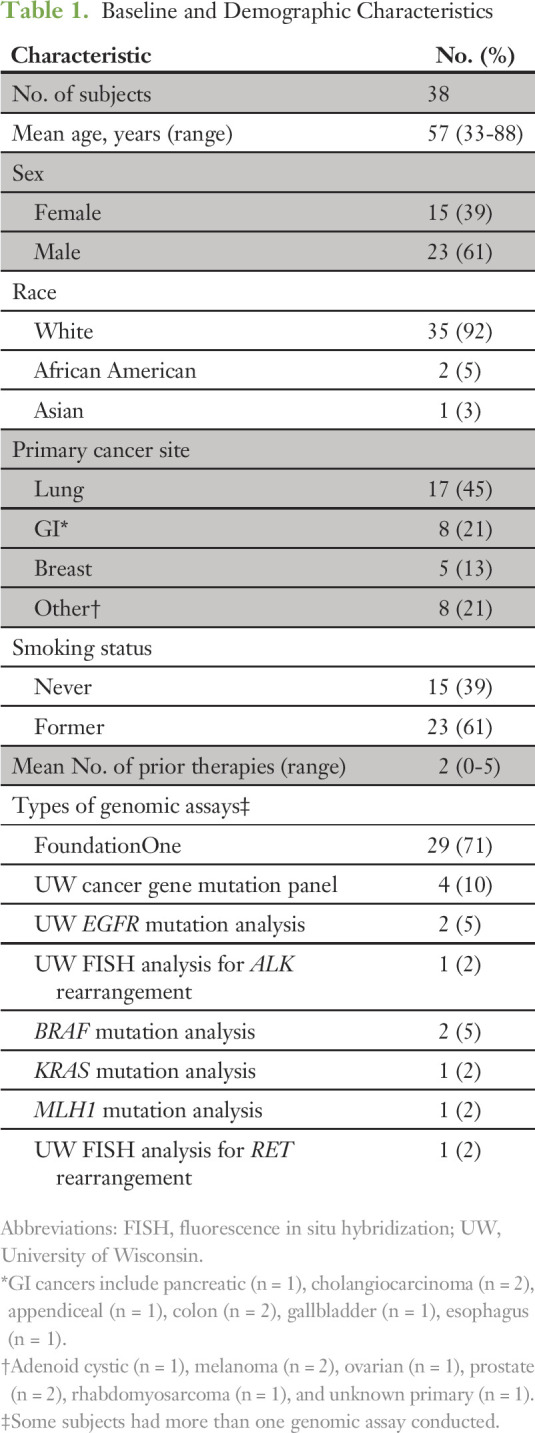
Because registry consent was not a prerequisite to PMMTB presentation, overlap was incomplete between patient cases presented and subjects enrolled. Seventy-seven patient cases were submitted to the tumor board between September 17, 2015, and September 1, 2016: 74 were presented to the board, and three were reviewed administratively. Patients were presented from six distinct health systems that cover regions that span most of the state (Fig 2). Forty-eight patients were enrolled in the registry protocol. Four registered patients who had planned genomic testing were not presented at the tumor board because molecular analysis of their tumor was not complete (eg, insufficient tissue). Of the 44 patient cases presented, 38 were deemed to have adequate follow-up by September 1, 2016, to report results here. Table 1 includes demographics and other baseline characteristics for these 38 patients. The mean age was 57 years, with a majority of patients being male (61%) and white (92%). Lung, GI, and breast cancers were the most common tumor types. Patients were previously treated with a mean of approximately two prior regimens. The most common genomic test used was the FoundationOne (Foundation Medicine, Cambridge, MA) assay, followed by the UW Collaborative Genomics Core 50-gene panel.
Fig 2.
Catchment of health systems included in the Precision Medicine Molecular Tumor Board by county. Each circle identifies the central location of a health system in the catchment area. The area of the circle is proportional to the number of patient cases presented from that health system. UWCCC, University of Wisconsin Carbone Cancer Center.
Table 1.
Baseline and Demographic Characteristics
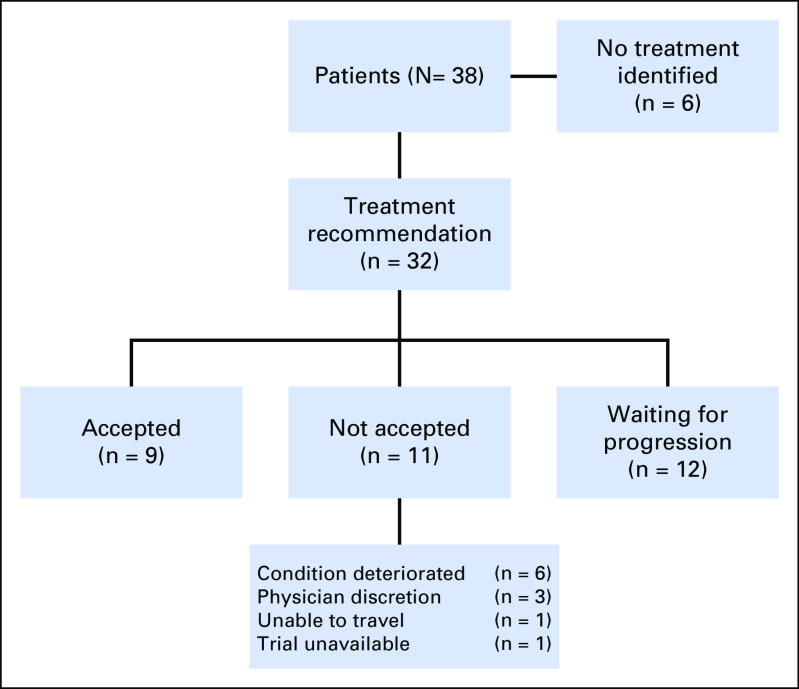
PMMTB Recommendations and Acceptance
Figure 3 depicts recommendations and acceptance of the 38 patient cases. An actionable target was defined as one that allowed identification of a molecular-targeted clinical trial or off-trial treatment that is based on the PMMTB guidance document. Of the cases presented to the PMMTB, an actionable target was found in 32 (84%). Treatment was accepted for nine (28%) of these 32 and not accepted for 11 (34%), and the remainder of patients continued on standard-of-care therapy, pending progression. The most common reason for not accepting the recommended treatment was clinical deterioration of the patient, but this was not likely due to PMMTB delays because the time from submission to presentation averaged 13.5 days.
Fig 3.
Identification and acceptance of Precision Medicine Molecular Tumor Board recommendations.
Because multiple treatment options often were identified, we tracked ones that were implemented by the treating physician (Table 2). The most commonly accepted recommendation was off-trial treatment with a targeted drug. In general, we found that patients were able to obtain these drugs when recommended by the PMMTB, which highlights the access that physicians and patients have to off-label therapies. The PMMTB may have facilitated this access by establishing and implementing a uniform set of criteria and recommendations provided as an official letter (Data Supplement).
Table 2.
Acceptance of Recommendations by Type (N = 38)*
Although clinical trials were preferentially recommended, they were rarely accepted. Among trials available within the state, only one of 14 potential patients enrolled. For clinical trials available outside the state, no patients enrolled. The major reason was that patients with metastatic cancer often were unable or unwilling to travel long distances for investigational therapy. Within the state, restrictive and often unanticipated eligibility criteria prohibited enrollment. For example, one patient was ineligible because of a recent amendment that precluded any history of drug allergy. Molecular-targeted basket studies were largely unavailable in the region partly because of a national hold placed on the National Cancer Institute Molecular Analysis for Therapy Choice (NCI-MATCH) trial during most of the period covered. These findings highlight poor access to clinical trials of precision medicines and the need for local availability of clinical trials with minimal eligibility restrictions and fast startup.
In the first year of the PMMTB, journal clubs primarily were used to expand the guidance document (Data Supplement). Although no specific guidelines were specified for adopting a drug-biomarker pair, the accepted changes were made when early clinical data suggested objective response rates that exceed approximately 15% or if the mutation was extremely rare and strong preclinical or early clinical data were available. Conditions were added, for example, to include olaparib for BRCA1/2 mutant cancers and pembrolizumab for tumors with mismatch repair deficiency confirmed by immunohistochemistry or microsatellite testing. Journal clubs also were used as educational sessions for trainees. Some journal clubs were selected on the basis of a case presented at the prior PMMTB. For example, vandetanib was adopted provisionally for the treatment of tumors with fumarate hydratase loss-of-function mutations, which led to an updated recommendation issued at the next PMMTB meeting. In short, the PMMTB Journal Club proved effective at striking a balance between rigor and flexibility. On the basis of the updated guidelines, three cases were reviewed again.
The genomic profiles and recommendations of the PMMTB are displayed in Figure 4. In this diagram, some samples were tested for large gene sets (FoundationOne, including gray boxes for nonaltered genes) and others for limited gene sets through the UW Collaborative Genomics Core 50-gene panel or subset; genes not tested are depicted as white areas. Among the alterations found, the TP53 mutation was most common followed by deletion of CDKN2A/B and various EGFR mutations. However, tremendous diversity of mutational profiles was observed, even within a given tumor type. Additional genes identified in these subjects are listed in the Data Supplement. The PMMTB identified treatment options that targeted 18 distinct genes in these patients (red boxes). More treatment options were identified for tumors tested with comprehensive panels. With the inclusion of both trial and nontrial treatments, we identified a mean of 1.7 treatment options with the 405-gene panel versus 1.1 with a more-restrictive panel of ≤ 50 genes. The most commonly used treatment was everolimus for tumors with a mutation of MTOR, PIK3CA, or PIK3R1. For assessment of response, we excluded the patient who received osimertinib for lung cancer with EGFR T790M. Of the remaining eight patients who received the recommended therapy, the clinical benefit rate was 38% (three of eight) and the response rate was 17% (one of six) by central review. Three patients administered recommended therapy have continued treatment (patients 2, 9, and 12). An additional 13 patients received standard therapy, with clinicians planning to use PMMTB recommendations to guide subsequent therapy at disease progression. The PMMTB recommended genetic counseling for three patients, and two received this counseling.
Fig 4.
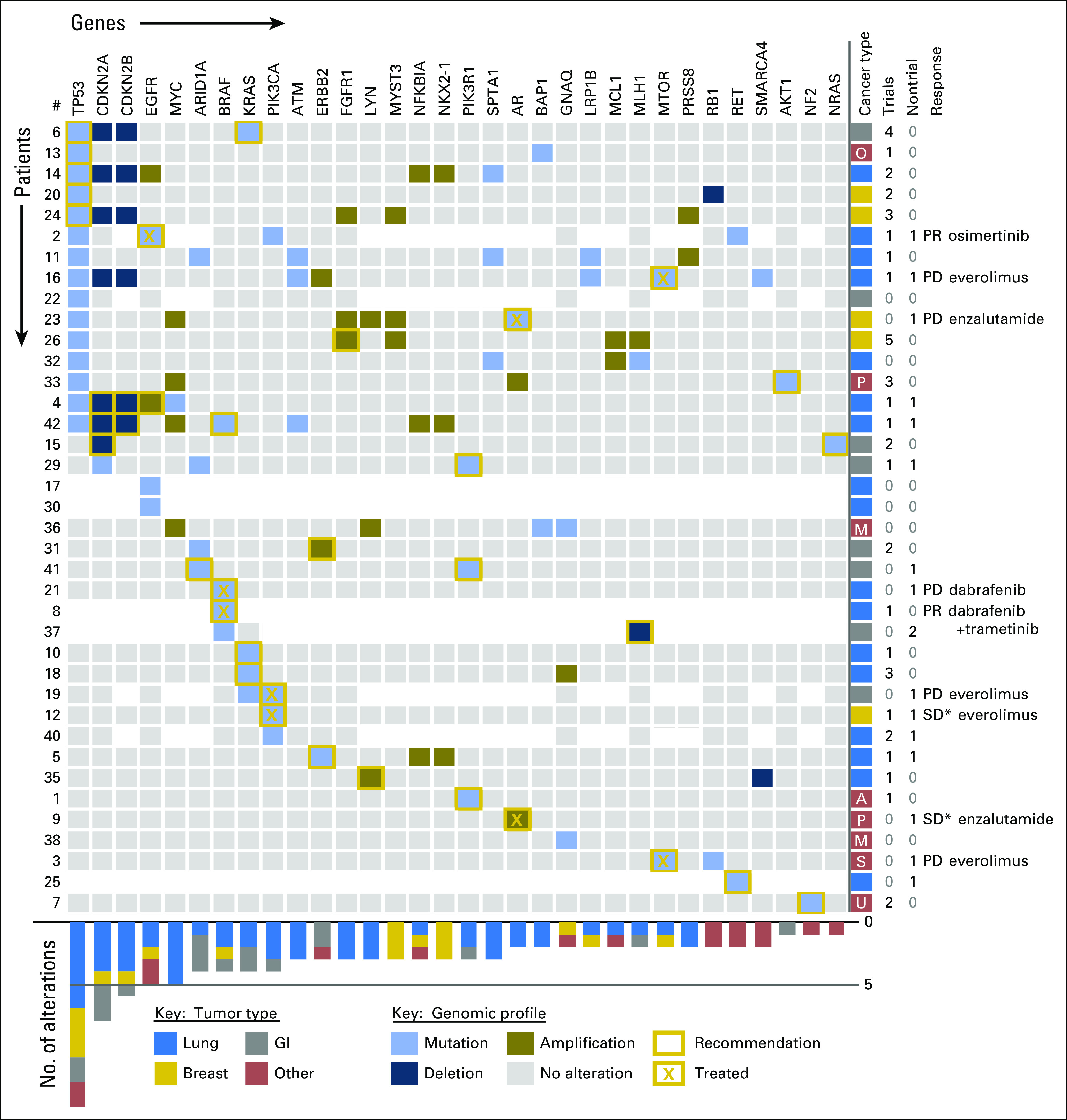
Genomic profiles, recommendations, treatment, and outcomes of the evaluated subjects. Genomic profiles are displayed for the 38 evaluable subjects. The genes with identified alterations are displayed as columns, with rows displaying the tumor profile for a given subject. Areas shown in white indicate genes that were not evaluated in that tumor. Gold boxes indicate genes that were considered actionable for targeted therapy, including clinical trials. An X within the box illustrates the accepted treatment recommendation. Cancer type by color code is shown on the right, as are the number of clinical trials and nontrial treatments recommended by the Precision Medicine Molecular Tumor Board, the response to therapy, and the treatment given. The chart on the bottom displays the frequency of alterations in each gene shown at the top by tumor type. Response Evaluation Criteria in Solid Tumors (RECIST 1.1) for measurable disease: PD, progressive disease; PR, partial response; SD, stable disease. (*) Nonmeasurable disease. A, adenoid cystic; M, melanoma; O, papillary serous ovary; P, prostate; S, alveolar rhabdomyosarcoma; U, carcinoma of unknown primary.
DISCUSSION
Precision oncology promises to markedly improve cancer therapy by targeting genes or pathways that are active in each individual cancer. However, implementation remains a challenge because of the depth and breadth of knowledge required to make effective decisions. To bridge this gap, MTBs that comprise experts with a diverse knowledge base have been established at academic institutions8-11 but remain largely inaccessible to community oncologists and their patients. We describe a regional partnership between academic and community practitioners to develop and implement the tripartite PMMTB. By separating the clinical service from the registry, we were able to rapidly launch and provide service across the region, although this method partially limited the number of reportable patients. Moreover, the PMMTB used a separate decision-making process (Journal Club) for standardizing recommendations for off-label therapy, which allowed the board to focus on clinical cases rather than on time-consuming excursions to the clinical/preclinical literature. Moreover, the monthly Journal Club allowed for a timely and careful review of conditions for which off-label therapies are reasonably expected to be effective.
Most previously described MTBs appear to be focused on a single institution or a single precision medicine clinic within the institution.8-12 Such centralization can standardize clinical practice and facilitate access to clinical trials. By contrast, we developed a decentralized approach that allows patients to be managed by their oncologist while accessing the expertise of colleagues and knowledge experts. However, some challenges come with this approach. For instance, the registry protocol for collecting clinical information needs to be opened and coordinated at multiple sites through a regional clinical trial network. Despite this limitation, the decentralized approach is expected to be superior in fostering regional collaboration and access of patients to precision medicine.
We found actionable mutations in 86% of the selected cohort. Although this number appears high, we note that the number of patients with actionable mutations depends on the particular definition of actionable as well as on the clinical trials and approved off-label drugs available at that time. In addition, this statistic can be biased because physicians are expected to select for submission patient cases with actionable alterations. Moreover, although we were able to identify molecular-based clinical trials for many patients, a number of these were inaccessible to patients unable to travel. If we limit the data by excluding out-of-state trials, the rate of actionable mutations becomes 63%, which is similar to the 71% reported by a another tumor board.9
An important finding of this study is that clinical trials often are unavailable for patients. Although this registry study was launched contemporaneously with NCI-MATCH, the expected competition for the two approaches was not realized partly because of the relative complexity of MATCH and the clinical hold on accruals. Moreover, few subjects enrolled in MATCH were treated. Thus, the PMMTB was found to complement rather than compete with this type of study. Given that patients generally are unwilling to travel outside the region, our focus will be to bring studies to Wisconsin rather than to identify trials elsewhere. In addition, the development of molecular-targeted studies in which eligibility restrictions do not seriously limit patient access to investigational therapy is important. Nevertheless, our experience and that of others suggest that off-trial therapies will remain an important component of an MTB.3 Thus, the tracking of patient outcomes is important to provide data on treatment outcomes by molecular profile.
On the basis of our experience and that reported by others,3,8-10 continued identification of Food and Drug Administration–approved therapies and maintenance of a registry that tracks patient outcomes will be important. Although cancer genomics data are publicly available for > 10,000 cases, these profiles have not yet been linked with detailed clinical data. We anticipate that the engagement of community practitioners and development of stronger regional networks to collect and organize genomic data and matched clinical outcomes will facilitate national efforts, such as CancerLinQ,19,20 the American Association for Cancer Research GENIE (Genomics Evidence Neoplasia Information Exchange) project,21 ORIEN (Oncology Research Information Exchange Network), commercial databases, and anticipated NCI/Moonshot data banks.
Few trials have compared molecular-targeted therapies versus standard of care to determine the benefit of genome sequencing. The SHIVA trial randomly assigned patients to standard of care or to a molecular match. Forty percent with a match formed the experimental group. In this study, the molecular-targeted therapy provided no improvement in progression-free survival.22 However, the assignments in SHIVA may have been suboptimal, considered only three targetable pathways (hormone receptors, PI3K/AKT/mTOR, and RAF/MEK), prioritized expression of hormone receptors, assigned treatments on the basis of amplifications, and used single-agent targeted therapies. By contrast, other studies have reported improved outcomes and enhanced cost effectiveness through a precision medicine approach.12,23 Similarly, clinical benefit of adaptable use of nonstandard molecular-targeted therapy has been seen in lung cancer24,25 and by other MTBs.9,10,12 We observed a 13% objective response rate by standardized criteria and a clinical benefit rate of 38% (partial response plus stable disease). Although modest with a small sample, the observed rate compares favorably with the 5% response reported for unmatched phase I trials and the 4% to 16% response rate reported for second-line chemotherapy for colorectal cancer.26,27 Similarly, a retrospective analysis has found improved progression-free survival without increased cost when patients are treated with a precision medicine approach.23 Thus, our initial findings are consistent with these observations and can be improved by experience.
This work has some limitations. The study was observational with a small sample size and patient responses were assessed by their treating physicians as part of standard-of-care evaluations. In addition, the majority of patients presented were of advanced-stage disease and had limited standard-of-care treatment options, which suggests that these results may not be generalizable to patients earlier in the course of their disease. However, this study reflects actual practice in our region and forms the basis for our next steps. We will continue to use the journal club format to assess the usefulness of off-label treatment. Local access to molecularly targeted therapies is the most pressing need for our patients with advanced cancer.
In conclusion, we have developed and implemented a regional MTB that integrates clinical service, a registry, and a journal club. The strength of this approach is that it provides flexibility and rapid implementation. The principal weaknesses are that the registry is observational, largely retrospective, and does not include all patient cases presented at the tumor board. Our experience demonstrates that we can identify actionable mutations for a high proportion of patients and identify off-trial targeted therapies. The PMMTB demonstrates one venue to coordinate a precision medicine initiative across a region. We anticipate that the development of additional regional networks to implement coordinated data collection and reporting to national databases and to enhance access to clinical trials will be important.
Footnotes
See accompanying editorial doi:https://doi.org/10.1200/PO.16.00022
Supported by National Cancer Institute Grant No. P30 CA014520 to the University of Wisconsin Carbone Cancer Center.
AUTHOR CONTRIBUTIONS
Conception and design: Mark E. Burkard, Dustin A. Deming, Benjamin M. Parsons, Michael A. Thompson, Jennifer Laffin, Kristina A. Matkowskyj, William M. Rehrauer, Jill Kolesar
Administrative support: Mark E. Burkard, Marissa R. Schuh
Financial support: Mark E. Burkard
Provision of study materials or patients: Mark E. Burkard, Ticiana Leal, Nataliya Uboha, Ruth Warren, Kristina A. Matkowskyj, Darya G. Buehler
Collection and assembly of data: Mark E. Burkard, Marissa R. Schuh, Ticiana Leal, Ruth Warren, Mary S. Mably, Jennifer Laffin, Darya G. Buehler, William M. Rehrauer, Jill Kolesar
Data analysis and interpretation: Mark E. Burkard, Dustin A. Deming, Benjamin M. Parsons, Paraic A. Kenny, Ticiana Leal, Nataliya Uboha, Joshua M. Lang, Michael A. Thompson, Jordan Bauman, Mary S. Mably, Jennifer Laffin, Catherine R. Paschal, Angela M. Lager, Kristy Lee, Kristina A. Matkowskyj, William M. Rehrauer, Jill Kolesar
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Implementation and Clinical Utility of an Integrated Academic-Community Regional Molecular Tumor Board
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or po.ascopubs.org/site/ifc.
Mark E. Burkard
Research Funding: Pfizer, AbbVie, Genentech
Consulting or Advisory Role: Pointcare Genomics
Dustin A. Deming
No relationship to disclose
Benjamin M. Parsons
Honoraria: Amgen, Celgene
Paraic A. Kenny
No relationship to disclose
Marissa R. Schuh
No relationship to disclose
Ticiana Leal
Consulting or Advisory Role: Genentech, Ariad
Nataliya Uboha
No relationship to disclose
Joshua M. Lang
Stock and Other Ownership Interests: Salus Discovery
Michael A. Thompson
No relationship to disclose
Ruth Warren
No relationship to disclose
Jordan Bauman
No relationship to disclose
Mary S. Mably
No relationship to disclose
Jennifer Laffin
No relationship to disclose
Catherine R. Paschal
No relationship to disclose
Angela M. Lager
No relationship to disclose
Kristy Lee
No relationship to disclose
Kristina A. Matkowskyj
No relationship to disclose
Darya G. Buehler
No relationship to disclose
William M. Rehrauer
No relationship to disclose
Jill Kolesar
No relationship to disclose
REFERENCES
- 1.Kruglyak KM, Lin E, Ong FS: Next-generation sequencing in precision oncology: Challenges and opportunities. Expert Rev Mol Diagn 14:635-637, 2014 [DOI] [PubMed] [Google Scholar]
- 2.Garraway LA, Verweij J, Ballman KV: Precision oncology: An overview. J Clin Oncol 31:1803-1805, 2013 [DOI] [PubMed] [Google Scholar]
- 3. Meric-Bernstam F, Brusco L, Shaw K, et al: Feasibility of large-scale genomic testing to facilitate enrollment onto genomically matched clinical trials. J Clin Oncol 33:2753-2762, 2015 doi: 10.1200/JCO.2014.60.4165. [DOI] [PMC free article] [PubMed]
- 4.Adams SA, Petersen C: Precision medicine: Opportunities, possibilities, and challenges for patients and providers. J Am Med Inform Assoc 23:787-790, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guttmacher AE, Porteous ME, McInerney JD: Educating health-care professionals about genetics and genomics. Nat Rev Genet 8:151-157, 2007 [DOI] [PubMed] [Google Scholar]
- 6. Vogelstein B, Papadopoulos N, Velculescu VE, et al: Cancer genome landscapes. Science 339:1546-1558, 2013. [DOI] [PMC free article] [PubMed]
- 7. Kandoth C, McLellan MD, Vandin F, et al: Mutational landscape and significance across 12 major cancer types. Nature 502:333-339, 2013. [DOI] [PMC free article] [PubMed]
- 8. Schwaederle M, Parker BA, Schwab RB, et al: Molecular tumor board: The University of California-San Diego Moores Cancer Center experience. Oncologist 19:631-636, 2014. [DOI] [PMC free article] [PubMed]
- 9. Tafe LJ, Gorlov IP, de Abreu FB, et al: Implementation of a molecular tumor board: The impact on treatment decisions for 35 patients evaluated at Dartmouth-Hitchcock Medical Center. Oncologist 20:1011-1018, 2015 doi: 10.1634/theoncologist.2015-0097. [DOI] [PMC free article] [PubMed]
- 10. doi: 10.1634/theoncologist.2016-0049. Hirshfield KM, Tolkunov D, Zhong H, et al: Clinical actionability of comprehensive genomic profiling for management of rare or refractory cancers. Oncologist 10.1634/theoncologist.2016-0049 [epub ahead of print on August 26, 2106] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ortiz MV, Kobos R, Walsh M, et al. : Integrating genomics into clinical pediatric oncology using the molecular tumor board at the Memorial Sloan Kettering Cancer Center. Pediatr Blood Cancer 63:1368-1374, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radovich M, Kiel PJ, Nance SM, et al. : Clinical benefit of a precision medicine based approach for guiding treatment of refractory cancers. Oncotarget 7:56491-56500, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forbes SA, Beare D, Gunasekaran P, et al. : COSMIC: Exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res 43:D805-D811, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Day IN: dbSNP in the detail and copy number complexities. Hum Mutat 31:2-4, 2010 [DOI] [PubMed] [Google Scholar]
- 15.Abecasis GR, Auton A, Brooks LD, et al. : An integrated map of genetic variation from 1,092 human genomes. Nature 491:56-65, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson A, Zeng J, Bailey AM, et al. : The right drugs at the right time for the right patient: The MD Anderson precision oncology decision support platform. Drug Discov Today 20:1433-1438, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao J, Aksoy BA, Dogrusoz U, et al. : Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6:pl1, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Micheel CM, Lovly CM, Levy MA: My cancer genome. Cancer Genet 207:289, 2014 [Google Scholar]
- 19.Schilsky RL, Michels DL, Kearbey AH, et al. : Building a rapid learning health care system for oncology: The regulatory framework of CancerLinQ. J Clin Oncol 32:2373-2379, 2014 [DOI] [PubMed] [Google Scholar]
- 20.Shah A, Stewart AK, Kolacevski A, et al. : Building a rapid learning health care system for oncology: Why CancerLinQ collects identifiable health information to achieve its vision. J Clin Oncol 34:756-763, 2016 [DOI] [PubMed] [Google Scholar]
- 21.Rose S: Huge data-sharing project launched. Cancer Discov 6:4-5, 2016 [DOI] [PubMed] [Google Scholar]
- 22.Le Tourneau C, Delord JP, Gonçalves A, et al. : Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): A multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol 16:1324-1334, 2015 [DOI] [PubMed] [Google Scholar]
- 23. doi: 10.1200/JOP.2016.011486. Haslem DS, Van Norman SB, Fulde G, et al: A retrospective analysis of precision medicine outcomes in patients with advanced cancer reveals improved progression-free survival without increased health care costs. J Oncol Pract 10.1200/JOP.2016.011486 [epub ahead of print on September 6, 2016] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaderbhai CG, Boidot R, Beltjens F, et al. : Use of dedicated gene panel sequencing using next generation sequencing to improve the personalized care of lung cancer. Oncotarget 7:24860-24870, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johnson BE, Kris MG, Berry LD, et al: A multicenter effort to identify driver mutations and employ targeted therapy in patients with lung adenocarcinomas: The Lung Cancer Mutation Consortium (LCMC). J Clin Oncol 31, 2013 (suppl; abstr 8019) [Google Scholar]
- 26.Tsimberidou A-M, Iskander NG, Hong DS, et al. : Personalized medicine in a phase I clinical trials program: The MD Anderson Cancer Center initiative. Clin Cancer Res 18:6373-6383, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tournigand C, André T, Achille E, et al. : FOLFIRI followed by FOLFOX6 or the reverse sequence in advanced colorectal cancer: A randomized GERCOR study. J Clin Oncol 22:229-237, 2004 [DOI] [PubMed] [Google Scholar]