Abstract
PURPOSE
Guidelines advocate molecular profiling in the evaluation of advanced non–small-cell lung cancer (NSCLC) and support the use of plasma circulating tumor DNA (ctDNA)-based profiling for patients with insufficient tissue. Thorough prospective clinical validation studies of next-generation sequencing (NGS)-based ctDNA assays are lacking. We report the multicentered prospective clinical validation of the InVision ctDNA assay in patients with advanced untreated NSCLC.
METHODS
A total of 264 patients with untreated advanced NSCLC were prospectively recruited, and their plasma was analyzed using a ctDNA NGS assay for detection of genomic alterations in 36 commonly mutated genes. Tumor tissue was available in 178 patients for molecular profiling for comparison with plasma profiling. The remaining 86 patients were included to compare ctDNA profiles in patients with and without tissue for profiling.
RESULTS
Concordance of InVisionFirst with matched tissue profiling was 97.8%, with 82.9% positive predictive value, 98.5% negative predictive value, 70.6% sensitivity, and 99.2% specificity. Considering specific alterations in eight genes that most influence patient management, the positive predictive value was 97.8%, with 97.1% negative predictive value, 73.9% sensitivity, and 99.8% specificity. Across the entire study, 48 patients with actionable alterations were identified by ctDNA testing compared with only 38 by tissue testing. ctDNA NGS reported either an actionable alteration or an alteration generally considered mutually exclusive for such actionable changes in 53% of patients.
CONCLUSION
The liquid biopsy NGS assay demonstrated excellent concordance with tissue profiling in this multicenter, prospective, clinical validation study, with sensitivity and specificity equivalent to Food and Drug Administration–approved single-gene ctDNA assays. The use of plasma-based molecular profiling using NGS led to the detection of 26% more actionable alterations compared with standard-of-care tissue testing in this study.
INTRODUCTION
Non–small-cell lung cancer (NSCLC) accounts for more than 85% of lung cancer1; the majority of patients present with advanced-stage disease and are treated with systemic therapies. Great strides have been made in the development of therapies for such patients, including targeted therapies and immunotherapy. Targeted therapies require identification of specific molecular alterations in the cancer,2 and guidelines recommend broad genomic profiling to assess for therapeutic targets. However, the use of such comprehensive testing is still limited, often because of inadequate tumor tissue in many patients, given the high tissue demands of comprehensive genomic profiling (CGP) testing. A recent review of more than 800 patients from routine US community oncology practices revealed that only 59% of patients were profiled for two of the best known genomic alterations (epidermal growth factor receptor [EGFR] mutations and ALK fusions), and only 8% received CGP covering all the recommended alterations.3 Repeat biopsies are costly and often result in patient discomfort, and many patients may experience complications.4 A recent US Medicare-based analysis demonstrated that the average cost of a transthoracic biopsy was $14,587 once treatment of complications was included.5
Plasma-based assays for molecular profiling of tumor mutations through circulating tumor DNA (ctDNA) offer the potential to overcome difficulties associated with tissue-based CGP.6 These less-invasive liquid biopsies are now entering routine clinical practice, with recent National Comprehensive Cancer Network guidelines recommending their use in patients with NSCLC when tissue biopsy is not available.7
CONTEXT
Key Objective
In this report, the authors examine the application of plasma-based comprehensive genomic profiling (CGP) in untreated, newly diagnosed, advanced-stage non–small-cell lung cancer (NSCLC) compared with CGP using biopsy tissue in a prospective clinical study.
Knowledge Generated
Amplicon-based next-generation sequencing (NGS) using plasma was shown to provide a greater number of clinically actionable results than CGP via tissue in a clinically relevant population. Alterations detected by plasma NGS results were shown to be accurate, and the report suggests that the use of plasma testing for CGP can be considered as a viable alternative to biopsy tissue testing.
Relevance
Using amplicon-based plasma circulating tumor DNA profiling will help oncologists appropriately identify more patients for targeted therapy compared with tissue testing alone. This report supports the use of plasma NGS for providing CGP in clinical practice, particularly in those patients with inadequate tissue available for broad CGP testing or for whom tissue testing is unfeasible.
To enable routine clinical use of such assays, robust validation needs to be undertaken. Given differences in ctDNA levels across tumor types and stages of disease,8 characterizing performance in the intended-use setting is important to allow clinicians to understand assay performance in clinical use. The documentation of performance in well-designed prospective studies of patients who have not been previously profiled allows for a true characterization of assay performance in the real-world setting and comparison between patients who undergo tissue-based CGP and those who do not. To date, such validation studies have not been reported in patients with advanced untreated NSCLC, with many concordance studies being undertaken in archival sample sets that have already been characterized or only address orthogonal testing of patients who have previously been tested by the assay under scrutiny.9 Such studies make it difficult to interpret the actual assay performance, particularly sensitivity.
We report the prospective clinical validation of the InVisionFirst-Lung assay (Inivata, Research Triangle Park, NC) in its intended-use population. This plasma-based CGP assay can identify genomic alterations in 36 commonly mutated genes (Appendix Fig A1). It has undergone extensive analytical validation, demonstrating excellent sensitivity for identification of mutations, detecting 100% of single nucleotide variants (SNVs) with a variant allele fraction (VAF) of 0.5%, and 89% of SNVs in a VAF range of 0.13% to 0.16%.10
METHODS
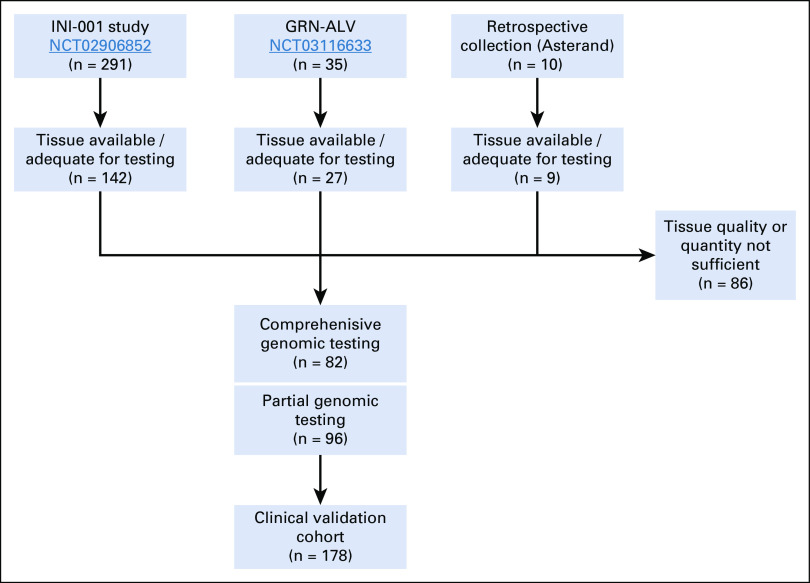
This prospective analysis combined patients with advanced, nonsquamous NSCLC recruited in two prospective US clinical studies (INI-001 [ClinicalTrials.gov identifier: NCT02906852] and GRN-ALV [ClinicalTrials.gov identifier: NCT03116633]) and samples obtained from a commercial biobank (Asterand, Detroit, MI; Appendix Fig A2). All enrolled patients were included if they met the following criteria: written informed consent; ages 18 years or older; stage IIIB/IV NSCLC; had not received therapy for advanced NSCLC; blood for plasma ctDNA analysis collected within 12 weeks of NSCLC tissue biopsy; and no anticancer therapy between the tissue and plasma collection.
The primary aim was to examine concordance of ctDNA and tissue profiling. All patients meeting the inclusion criteria were eligible regardless of tissue availability to allow the comparison of ctDNA profiles in patients with and without tissue for profiling. All studies were undertaken within recognized ethical principles established in International Conference on Harmonisation Good Clinical Practice and the Declaration of Helsinki and were subject to institutional review board/ethical review and approval.
ctDNA, Tissue Analysis, and Concordance Analysis
Blood was collected into Streck-DNA tubes (Streck, La Vista, NE) and shipped to the Inivata Clinical Laboratory Improvement Amendments–accredited laboratory (Research Triangle Park, NC) for InVision ctDNA analysis. Full assay details have been described previously.10,11 Briefly, blood was processed to plasma by centrifugation. After plasma extraction, plasma was stored at −80°C according to validated specifications until analysis in batch. DNA was extracted from plasma using the QIAamp Circulating Nucleic Acid kit (Qiagen, Santa Clarita, CA). After quality control, sequencing libraries were prepared using a two-step amplification process, and libraries were sequenced by Illumina (San Diego, CA) NextSEquation 500. Sequencing data were analyzed using the Inivata analytical pipeline to identify genomic alterations (Appendix Fig A1).
Where sufficient tissue was available, CGP was performed in a Clinical Laboratory Improvement Amendments–certified laboratory (Caris Life Science, Irving, TX). Direct sequence analysis was performed on genomic DNA isolated from formalin-fixed paraffin-embedded tumor samples using the Illumina NextSeq platform. An Agilent (Santa Clara, CA ) custom-designed SureSelect XT assay was used to enrich 592 whole-gene targets, and all variants reported were detected with more than 99% confidence. Fusion analysis was performed on isolated mRNA using the ArcherDx (Boulder, CO) FusionPlex Solid Tumor Panel and the Illumina MiSeq. When patients had insufficient tissue for CGP, tissue molecular testing was allowed per the treating institutions’ routine pathways. Where both central CGP and local data were available, the centralized CGP data were used for concordance analysis (Appendix Table A1).
Both InVisionFirst and tissue analysis were performed blinded. Calls made in either ctDNA or tissue in genomic regions that were not covered by testing in the other were excluded from concordance analysis. The calling nomenclature for all identified mutations was reviewed along with underlying sequencing data where present to ensure that mutations were named consistently, and all calls were correctly classified for concordance.
Droplet Digital Polymerase Chain Reaction Validation Data
Thirty-one patients from the INI-001 study also underwent plasma droplet digital polymerase chain reaction (ddPCR) testing for key genomic alterations (KRAS G12C/G12D/G12V, EGFR exon19del/T790M/L858R, BRAF V600E, ALK/ROS1 fusions) via a commercial assay provider (GeneStrat; Biodesix, Boulder, CO) as part of the clinical site’s routine standard of care for their patients. Blood samples were collected and shipped according to the test specifications. Analysis was completed by Biodesix before any knowledge of tissue or ctDNA testing results. Inivata was blinded to the GeneStrat test results.
Statistical Analysis
Prospective validation was performed by combining blinded data from two prospective studies and a cohort of commercially available samples to increase sample size. A sensitivity analysis was included to ensure no bias was introduced. No formal samples size estimates were performed.
Before analysis, a core gene variant panel was defined for clinically relevant gene mutation hotspots as EGFR exons 18-21, BRAF V600, MET exon 14, ERBB2 ins 20, KRAS, and ALK and ROS structural variants on the basis of recent recommendations of ASCO and International Association for the Study of Lung Cancer biomarker guidelines when NGS panels are used for molecular profiling.12,13 On the basis of the emerging clinical interest, STK1114 was also included in the core gene panel.
All analyses were performed using R version 3.2.5. In the concordance analysis, the data were summarized using a 2-×-2 table (Table 1), referring to tissue as the standard.
TABLE 1.
The 2-×2 Table: Tissue as the Testing Standard
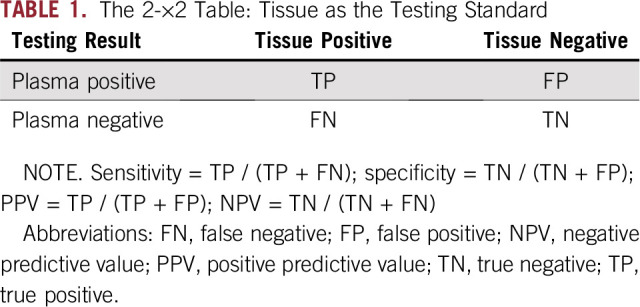
Utility of InVisionFirst testing was performed by assessing the number of patients who failed tissue analysis or for whom the analysis was not performed, specifically, for actionable mutations conferring sensitivity to approved or experimental therapies and correlation of the detection of actionable mutations in tissue versus blood.
RESULTS
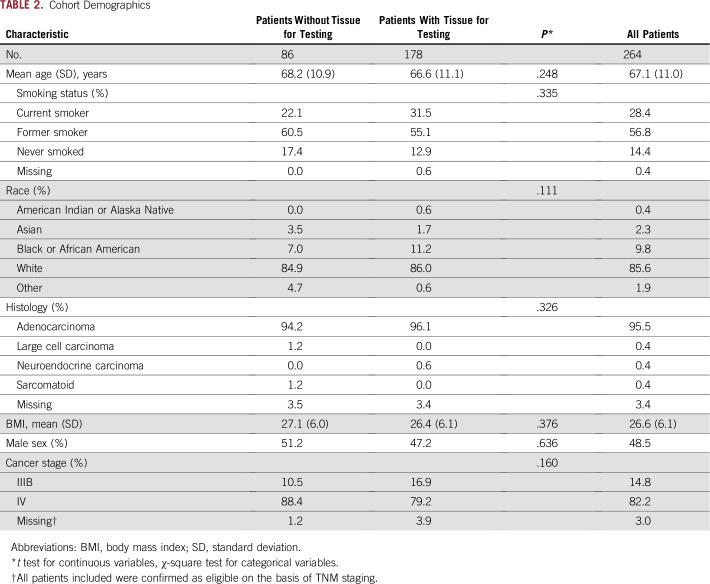
A total of 254 eligible patients were recruited across 41 centers in the prospective studies, with an additional 10 patients from retrospective collections, making a total of 264 patients analyzed. Baseline demographics for the cohort are listed in Table 2 and were consistent with expectations. Patients with and without tissue testing had similar demographics; no bias was observed.
TABLE 2.
Cohort Demographics
InVisionFirst ctDNA Profile
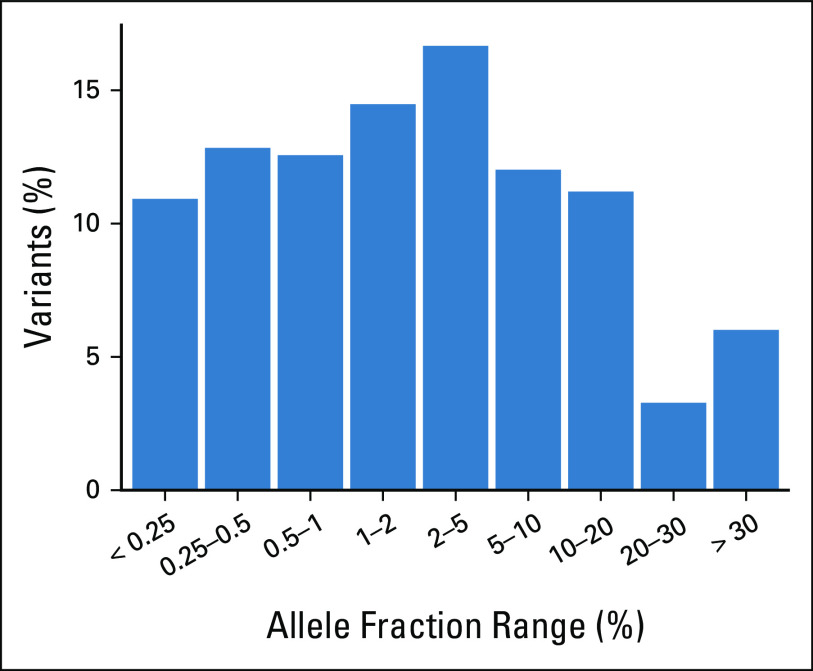
All patients were successfully tested for SNVs, indels, and amplifications. Testing for ALK/ROS1 fusions was successful in 252 patients (95.5%). Figure 1 shows the mutation profiles across all 264 patients. Overall, 204 patients (77.3%) had one or more alterations detected by ctDNA. The mean number of alterations identified per patient was 1.5. Of the SNVs and indels identified, 35.5% had an allele fraction lower than 1%, and 23.1% had an allele fraction lower than 0.5% (Appendix Fig A3).
FIG 1.
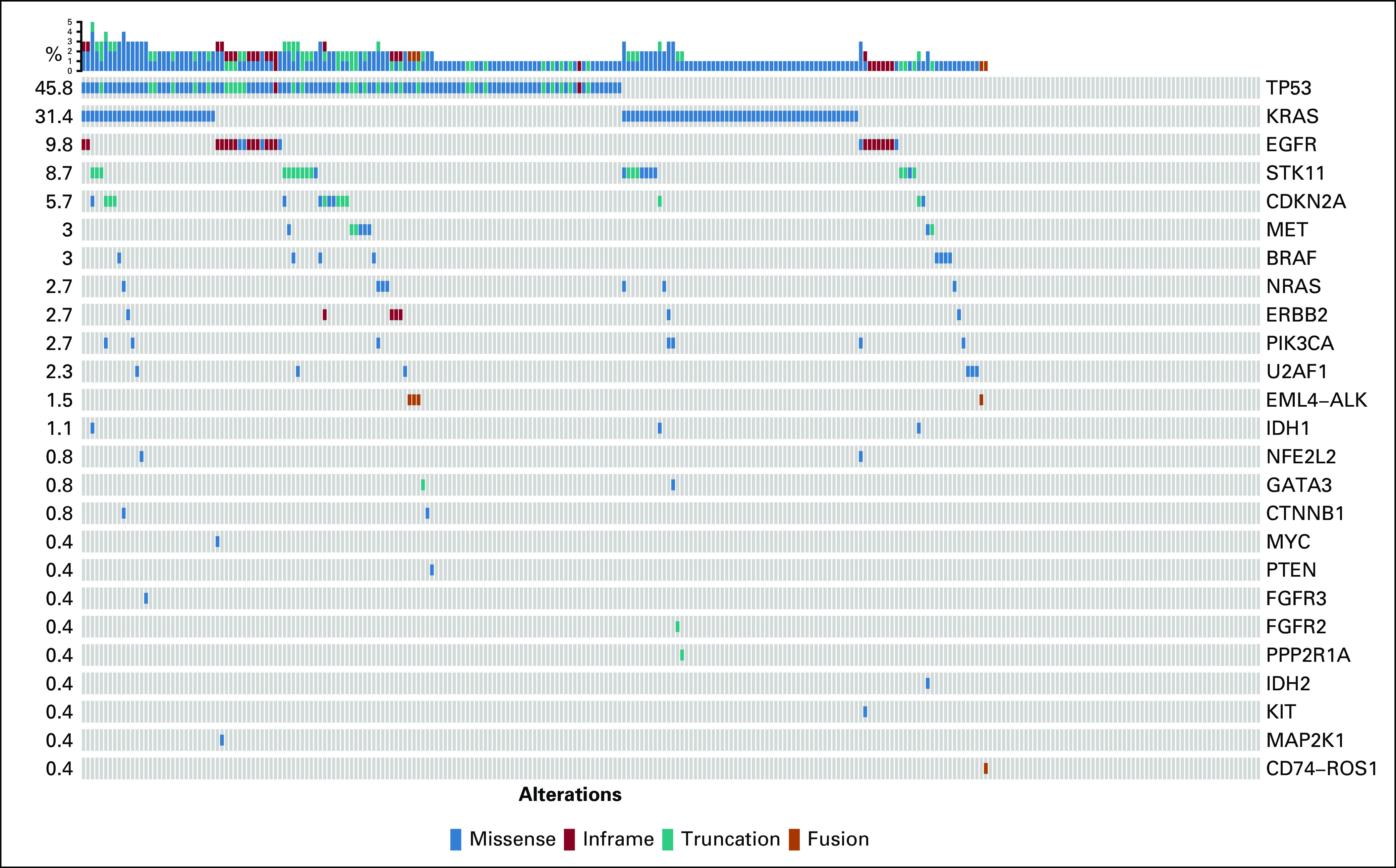
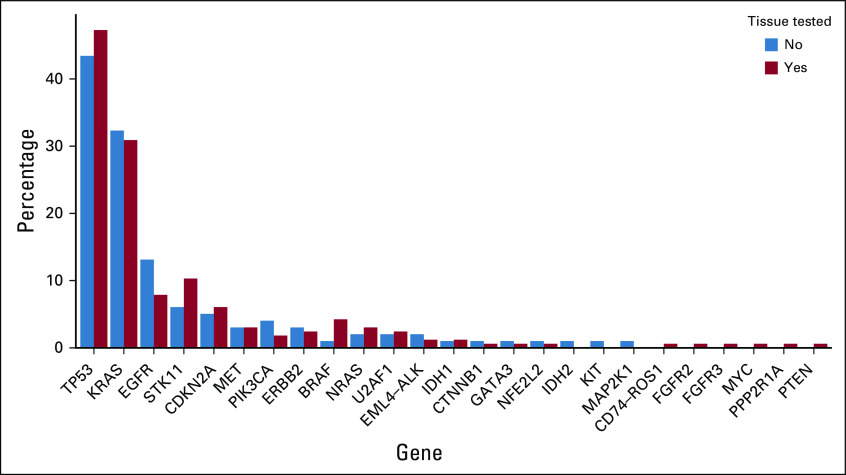
Single nucleotide variants, indels, and fusions identified in plasma in the full cohort of 264 patients. Rows refer to genes, and columns denote patients. Percentages refer to gene alteration incidence identified in this cohort.
The predominant alterations identified were TP53 (47% of patients) and KRAS (32% of patients). Twenty-seven SNVs or indels in EGFR exon 18-21 were identified in 26 patients (10%). Gene fusions were identified in five patients (2%), including EML4-ALK in four patients and CD74-ROS1 in one patient. The pattern and frequency of genomic alterations were similar across patients with and without tissue (Appendix Fig A4).
Tissue Testing and Tissue-ctDNA Concordance
Of the 264 recruited patients, 178 had successful tissue testing for at least one genomic alteration. One hundred sixty-five patients (62.5%) were tested for point mutations/indels, and 159 (60.2%) were tested for ROS1 and/or ALK fusions. The most frequently tested gene in tissue was EGFR (164 patients; 62.1%). A total of 95 patients were tested for all eight of the key genes previously described, and 121 were tested for fusions in ALK and ROS1 and mutations in EGFR, MET, and BRAF (Appendix Table A1).
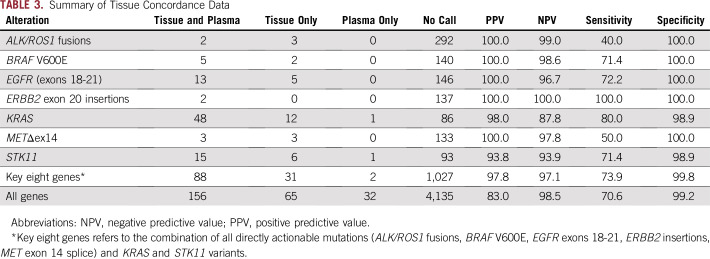
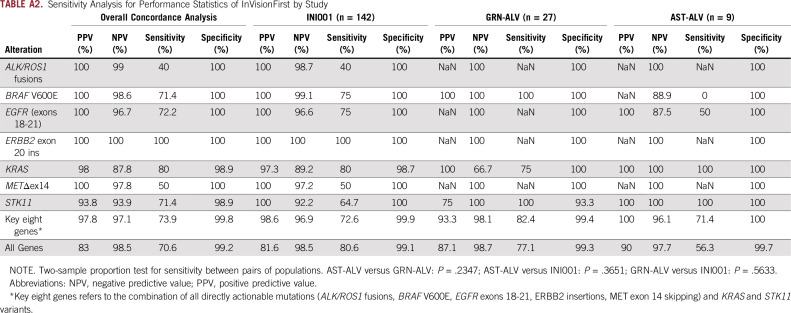
Considering tissue as the reference, the sensitivity of InVisionFirst across the entire panel was 70.6%. Considering only clinically actionable alterations in eight genes of most relevance, the sensitivity was 73.9%, with a positive predictive value (PPV) of 97.8% (Fig 2; Table 3; Appendix Table A2). The PPV was 100% when only considering the directly actionable variants (ALK/ROS1 fusions/EGFR exons18-21/ERBB2 insertions/ MET exon 14 skipping (METΔex14)/BRAF V600E).
FIG 2.
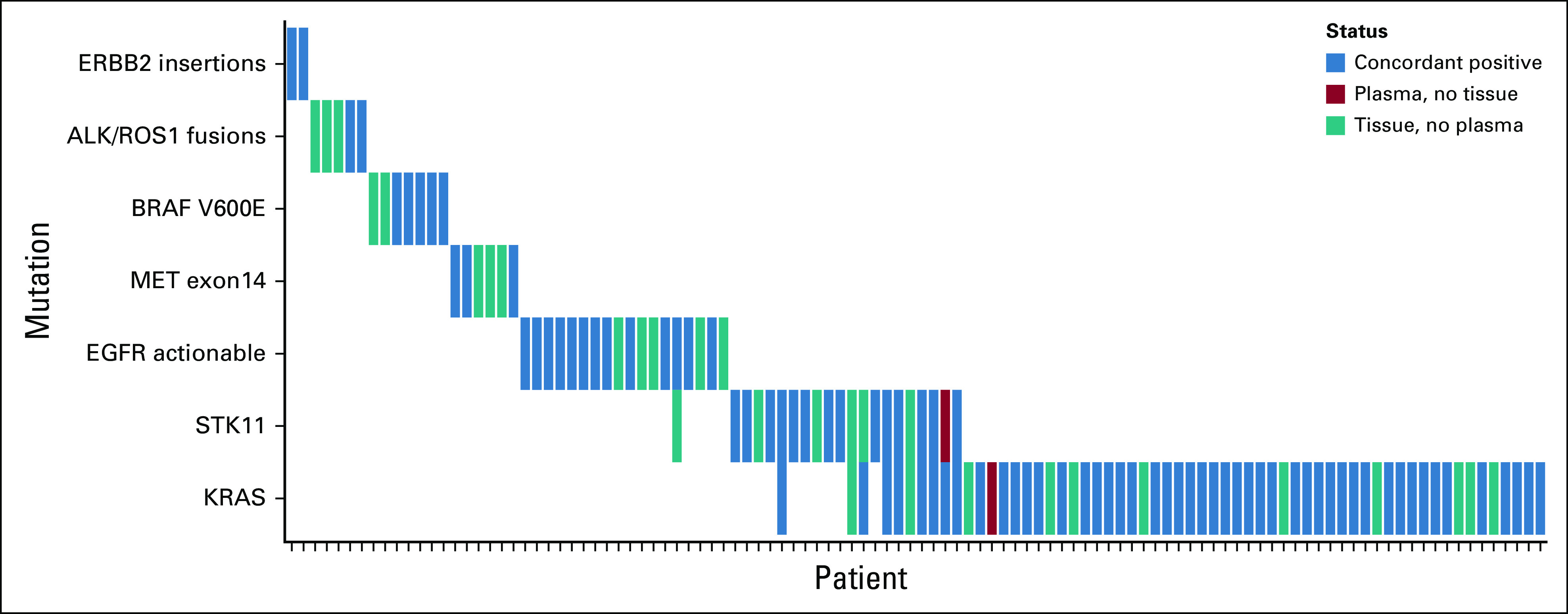
Concordance data for clinically relevant alterations detected in the eight key genes when both tissue and circulating tumor DNA testing was successful. EGFR, epidermal growth factor receptor.
TABLE 3.
Summary of Tissue Concordance Data
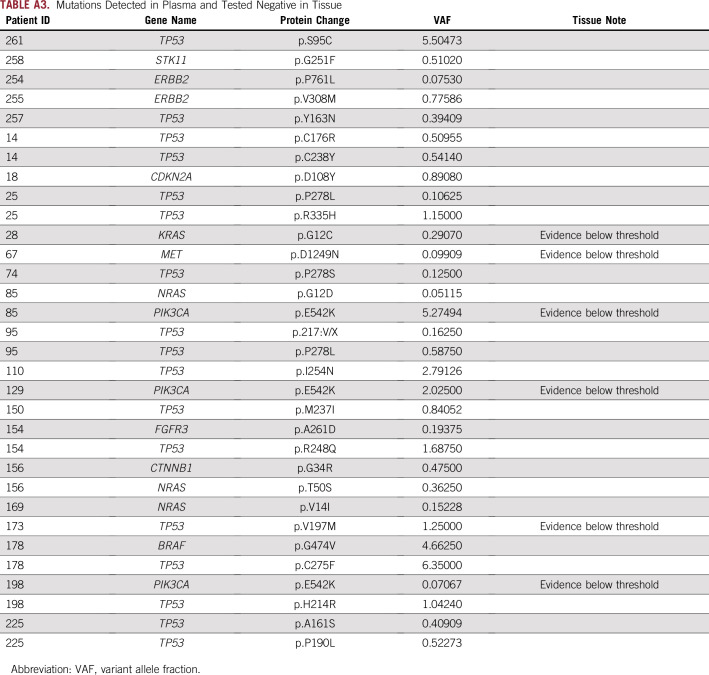
InVisionFirst detected 32 mutations in 23 patients that were not detected by tissue analysis (Appendix Table A3), including TP53 (17 mutations), PIK3CA (three mutations), NRAS (three mutations), and ERBB2 (two mutations). For 30 of these, read alignment data from tissue NGS were available. Review by the testing laboratory found evidence of the mutations below the standard calling threshold in six of the 30 mutations: PIK3CA E542K (three occurrences), KRAS G12C, MET D1249N, and TP53 V197M (Appendix Table A3).
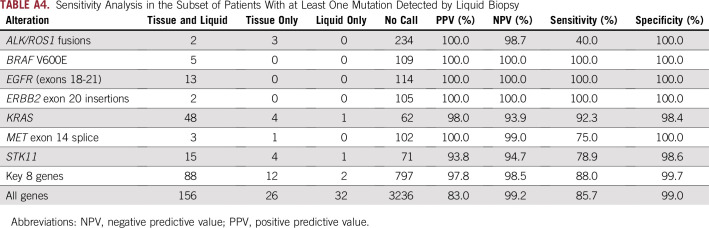
Two hundred four patients (77.27%) had at least one mutation detected by InVisionFirst (Appendix Table A4). In this cohort, the sensitivity was 88.0% for clinically relevant alterations in the key eight genes.
Utility Analysis
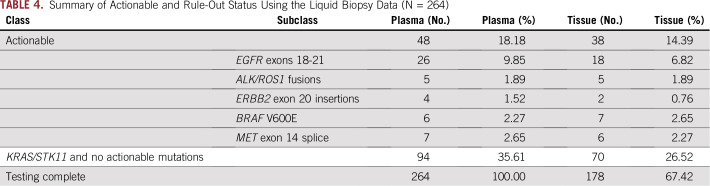
Tissue CGP was funded as part of the study and performed for all patients where sufficient tissue was available. Despite this, InVisionFirst testing resulted in a much higher rate of testing compared with tissue testing across the entire recruited population (Fig 3). Table 4 (Appendix Table A1) details clinically relevant alterations detected across all patients enrolled. Of 264 patients included, 48 patients qualified for a targeted treatment on the basis of InVisionFirst testing compared with 38 patients on the basis of tissue testing; 26% more identified actionable mutations by InVisionFirst (48 v 38). Forty-eight percent of the actionable alterations detected by InVisionFirst were in patients who had not been tested for that alteration in tissue because of incomplete tissue testing (insufficient or unavailable tissue).
FIG 3.
The number of patients across the entire study successfully tested for key genomic alterations by InVisionFirst and tissue testing. EGFR, epidermal growth factor receptor.
TABLE 4.
Summary of Actionable and Rule-Out Status Using the Liquid Biopsy Data (N = 264)
Mutations in KRAS and STK11 are generally mutually exclusive with actionable driver mutations in untreated nonsquamous NSCLC,15-17 and their detection could provide additional confidence that patients without actionable alterations are true negative rather than false negative. Combining patients for whom InVisionFirst identified an actionable alteration (18.2% of the cohort) with patients in whom it did not identify those but identified KRAS and/or STK11 mutations, 142 patients (53.8% of the cohort) had an actionable alteration detected or ruled out. Of the KRAS/STK11 mutations detected in plasma in these patients, 90 had the same variant tested in tissue, and 88 of these were detected (97.8% PPV; Table 3). The two mutations not detected in tissue included a KRAS G12C, which was observed below the threshold for calling, and a mutation in STK11 detected in plasma in a patient who was also KRAS positive by both tissue and plasma. In the 96 patients where InVisionFirst identified mutations in KRAS and/or STK11, tissue data did not detect any actionable alterations.
Orthogonal Validation by ddPCR
Plasma orthogonal testing in 31 patients by GeneStrat (Biodesix, Boulder, CO) ddPCR revealed an overall concordance of alteration calls of 98.5% (275 of 279), with positive agreement of 87.5% and negative agreement of 98.9% when considering ddPCR as the reference. Eleven alterations were seen in this population. Discordance was observed in four alterations: EGFR exon19del (one patient) and KRAS G12C (two patients) were detected by InVisionFirst but not by ddPCR. In one patient, EGFR L858R was detected by ddPCR but not by InVisionFirst. Tissue was available in two of these patients and confirmed the presence of one KRAS G12C mutation and the EGFR L858R mutations. Finally, one KRAS G12A mutation was detected by both the InVisionFirst assay and tissue sequencing but was identified as KRAS G12D by the ddPCR assay. The ddPCR panel did not test for KRAS G12A but reported the sample as KRAS G12D nonetheless. Comparison of the variant allele frequency percentage of the ddPCR and InVisionFirst is not possible because the GeneStrat report only indicates whether a variant was detected or not.
DISCUSSION
Our data provide clinical validation of the InVisionFirst assay for molecular stratification of newly diagnosed patients with stage IIIb/IV NSCLC. This study goes beyond previously published concordance studies that compared results of plasma and tissue testing in selected subsets of samples and, to our knowledge, represents the first prospective validation of a ctDNA NGS platform for molecular stratification of patients with advanced untreated NSCLC.
Using tissue as the reference, concordance for the full 36 genes in the InVisionFirst panel with matched tissue profiling was 97.8%. Considering clinically actionable alterations in eight genes that can most influence routine clinical patient management, the PPV was 97.8%, negative predictive value was 97.1%, sensitivity was 73.9%, and specificity was 99.4%. Of all mutations detected in plasma, 23% had an allele fraction below 0.5%, highlighting the need for highly sensitive assays with strong performance at low allelic frequencies. The InVisionFirst assay has demonstrated excellent sensitivity in analytical validation studies,10 but despite this high level of sensitivity, approximately 23% of these newly diagnosed patients with stage IIIb/IV NSCLC had no mutations detected in ctDNA.
High sensitivity needs to be coupled with high specificity to ensure that false-positive results do not lead to inappropriate therapy. Across the full panel, the PPV was 83.0%, compared with 100% for actionable driver alterations only, the difference being a consequence of 32 nonactionable variants detected in plasma but not in tissue. In six of these patients, there was evidence for the variant below thresholds required for calling in tissue. Sixteen of the remaining 26 calls were TP53 variants. These may be subclonal events that only occur at low levels or may be completely absent from the biopsy site. The over-representation of TP53 in ctDNA may also be explained by clonal hematopoiesis.18 Of note, where tissue was available, all clinically actionable alterations detected by InVisionFirst in plasma were confirmed by tissue profiling. This provides reassurance of the high specificity of the assay and is supported by previous studies of InVisionFirst in NSCLC.19,20
In total, 18.2% of patients tested by InVisionFirst had an actionable change detected. An additional 35.6% were found to have a genomic alteration generally mutually exclusive with such actionable alterations. Therefore, 53.8% of patients had an informative result that could prevent the need for additional invasive biopsies. The strength of this rule-out classification was confirmed by the absence of any actionable alterations detected in available tissue in these patients.
Despite excitement regarding ctDNA NGS platforms, there are currently no robust studies in an equivalent clinical setting to provide comparisons across assays. Because ctDNA levels vary between patients at different stages of disease,8 sensitivity is affected by the population in the study. Compared with newly diagnosed patients studied here, the clinical sensitivity of InVisionFirst in previous studies was higher in the relapse setting, with 100% sensitivity (compared with tissue) reported for the EGFR driver mutation at relapse in 30 patients with tyrosine kinase inhibitor–treated NSCLC.20 Another assay was also reported to have sensitivities ranging from 35.7% to 90.3% in different disease settings.21-23 Taken together, these observations dictate that clinical validation of assays should be performed in unselected patients from the intended-use population with clinical characteristics consistent with the proposed clinical indication before clinical adoption.24
The clinical sensitivity of the InVisionFirst assay demonstrated here is comparable to published data on the Food and Drug Administration–approved Roche Molecular Systems (Pleasanton, CA) CobasV2 single-gene EGFR ctDNA assay.25 Such single-gene tests only identify the small subset of patients with mutations in those genes and are inconclusive for the great majority of patients who potentially require additional testing. The InVisionFirst assay provides data across a panel of genes and can provide a definitive result in more than 50% of patients through a rule-in/rule-out approach.
Tissue testing for the most common alterations was only successful in 62% of patients in the study, consistent with statistics reported in a recent study across community oncology institutions.3 Full CGP was successful in significantly fewer patients. Routine implementation of ctDNA testing by InVisionFirst could help to increase the proportion of patients eligible for targeted therapies. Within this study, InVisionFirst identified 48 actionable alterations compared with 38 that were detected by standard-of-care tissue testing supplemented by CGP. Nearly half of the alterations detected by InVisionFirst were in patients who were not profiled for the alteration because of limitations in tissue testing. This increased detection of actionable alterations would be delivered while reducing costs, patient discomfort, and complications associated with repeated invasive tissue biopsies.
Patients with advanced NSCLC progress rapidly, and the time taken to obtain results of molecular profiling is therefore paramount. Results for the InVisionFirst assay are now routinely available in 7 days from blood draw. The use of such testing early in the work-up of patients with advanced NSCLC may therefore enable earlier therapeutic intervention.
ACKNOWLEDGMENT
We thank the patients, their families, the investigators, and the study teams for their contributions to this work.
Appendix
FIG A1.
InVisionFirst-Lung liquid biopsy tumor profiling panel (InvCore v1.5). CNVs, copy number variations; SNVs, single nucleotide variants.
FIG A2.
Consortium diagram for prospective clinical validation of InVisionFirst-Lung assay.
FIG A3.
Distribution of allele fractions of indels and single nucleotide variants identified in plasma samples.
FIG A4.
Gene alteration frequencies detected in plasma defined by tissue testing status.
TABLE A1.
Summary of Liquid Biopsy and Tissue Molecular Profiling to Detect Clinically Relevant Mutations
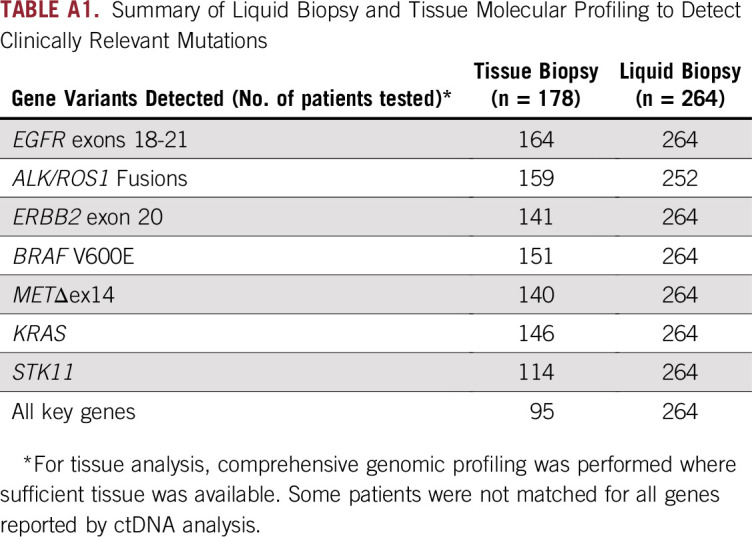
TABLE A2.
Sensitivity Analysis for Performance Statistics of InVisionFirst by Study
TABLE A3.
Mutations Detected in Plasma and Tested Negative in Tissue
TABLE A4.
Sensitivity Analysis in the Subset of Patients With at Least One Mutation Detected by Liquid Biopsy
Footnotes
Supported by Inivata.
Clinical trial information: INI-001 study: NCT02906852; GRN-ALV study: NCT03116633.
AUTHOR CONTRIBUTIONS
Conception and design: Michael A. Pritchett, D. Ross Camidge, Katherine Baker-Neblett, Nitzan Rosenfeld, Clive D. Morris, Ramaswamy Govindan
Financial support: Katherine Baker-Neblett
Administrative support: Katherine Baker-Neblett
Provision of study materials or patients: D. Ross Camidge, Jamil Khatri, Steven Boniol, Elke K. Friedman, Abderrahim Khomani, Katherine Baker-Neblett
Collection and assembly of data: Michael A. Pritchett, D. Ross Camidge, Manu Patel, Jamil Khatri, Steven Boniol, Elke K. Friedman, Samir Dalia, Katherine Baker-Neblett, Gregory R. Jones, Clive D. Morris, Ramaswamy Govindan
Data analysis and interpretation: Michael A. Pritchett, D. Ross Camidge, Abderrahim Khomani, Samir Dalia, Katherine Baker-Neblett, Vincent Plagnol, Karen D. Howarth, Gregory R. Jones, Clive D. Morris, Ramaswamy Govindan
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Michael A. Pritchett
Consulting or Advisory Role: Biodesix, AstraZeneca, Medtronic, BodyVision Medical
Speakers' Bureau: AstraZeneca, Medtronic, Boehringer Ingelheim, Actelion, United Therapeutics
D. Ross Camidge
Honoraria: Roche, G1 Therapeutics, Mersana, Takeda, AstraZeneca, Genoptix, Ignyta, Daiichi Sankyo, Hansoh, Lycera, Biothera, Revolution Medicines
Research Funding: Takeda
Elke K. Friedman
Leadership: HCA Healthcare
Stock and Other Ownership Interests: AbbVie (I)
Honoraria: Gilead Sciences
Consulting or Advisory Role: Gilead Sciences
Travel, Accommodations, Expenses: Gilead Sciences
Samir Dalia
Stock and Other Ownership Interests: 32nd Street Surgery Center (I)
Consulting or Advisory Role: New Century Health
Research Funding: Alnylam (Inst), Bristol-Myers Squibb (Inst), Calithera Biosciences (Inst), Helsinn Healthcare (Inst), Genentech (Inst), Celgene (Inst), Odonate Therapeutics (Inst)
Katherine Baker-Neblett
Employment: Inivata
Stock and Other Ownership Interests: GlaxoSmithKline
Travel, Accommodations, Expenses: Inivata
Vincent Plagnol
Employment: Inivata
Stock and Other Ownership Interests: Inivata
Patents, Royalties, Other Intellectual Property: Inivata patents
Karen D. Howarth
Employment: Inivata
Stock and Other Ownership Interests: Inivata
Research Funding: Inivata
Patents, Royalties, Other Intellectual Property: Patent pending
Gregory R. Jones
Employment: Inivata
Nitzan Rosenfeld
Employment: Storm Therapeutics (I)
Leadership: Inivata
Stock and Other Ownership Interests: Inivata, Mission Therapeutics (I)
Research Funding: AstraZeneca (Inst)
Patents, Royalties, Other Intellectual Property: Patents and patent applications relating to cancer classifications, detection, or analysis of microRNA and circulating tumor DNA, detection of rare sequence variants, applications in molecular diagnostics
Clive D. Morris
Employment: Inivata
Leadership: Inivata
Stock and Other Ownership Interests: Inivata
Ramaswamy Govindan
Honoraria: Genentech/AbbVie, AbbVie
Consulting or Advisory Role: GlaxoSmithKline, Genentech, AbbVie, Celgene, AstraZeneca/MedImmune, Inivata, Merck Serono, Pfizer, Bristol-Myers Squibb, EMD Serono, Eli Lilly, Ignyta, Nektar, Phillips Gilmore Oncology, Jounce Therapeutics
No other potential conflicts of interest were reported.
REFERENCES
- 1.Molina JR, Yang P, Cassivi SD, et al. Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Noone AM, Howlader N, Krapcho M et al. SEER cancer statistics review, 1975-2015. https://seer.cancer.gov/csr/1975_2015.
- 3.Gutierrez ME, Choi K, Lanman RB, et al. Genomic profiling of advanced non-small cell lung cancer in community settings: Gaps and opportunities. Clin Lung Cancer. 2017;18:651–659. doi: 10.1016/j.cllc.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Heerink WJ, de Bock GH, de Jonge GJ, et al. Complication rates of CT-guided transthoracic lung biopsy: Meta-analysis. Eur Radiol. 2017;27:138–148. doi: 10.1007/s00330-016-4357-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lokhandwala T, Bittoni MA, Dann RA, et al. Costs of diagnostic assessment for lung cancer: A Medicare claims analysis. Clin Lung Cancer. 2017;18:e27–e34. doi: 10.1016/j.cllc.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Rolfo C, Mack PC, Scagliotti GV, et al. Liquid biopsy for advanced non-small cell lung cancer (NSCLC): A statement paper from the IASLC. J Thorac Oncol. 2018;13:1248–1268. doi: 10.1016/j.jtho.2018.05.030. [DOI] [PubMed] [Google Scholar]
- 7.National Comprehensive Cancer Network NCCN Clinical Practice Guidelines in Oncology. Version 3.2019. Non-Small Cell Lung Cancer. https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf [Google Scholar]
- 8. Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 6:224ra24-224ra24, 2014. [DOI] [PMC free article] [PubMed]
- 9.Odegaard JI, Vincent JJ, Mortimer S, et al. Validation of a plasma-based comprehensive cancer genotyping assay utilizing orthogonal tissue- and plasma-based methodologies. Clin Cancer Res. 2018;24:3539–3549. doi: 10.1158/1078-0432.CCR-17-3831. [DOI] [PubMed] [Google Scholar]
- 10.Plagnol V, Woodhouse S, Howarth K, et al. Analytical validation of a next generation sequencing liquid biopsy assay for high sensitivity broad molecular profiling. PLoS One. 2018;13:e0193802. doi: 10.1371/journal.pone.0193802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gale D, Lawson ARJ, Howarth K, et al. Development of a highly sensitive liquid biopsy platform to detect clinically-relevant cancer mutations at low allele fractions in cell-free DNA. PLoS One. 2018;13:e0194630. doi: 10.1371/journal.pone.0194630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalemkerian GP, Narula N, Kennedy EB, et al. Molecular testing guideline for the selection of patients with lung cancer for treatment with targeted tyrosine kinase inhibitors: American Society of Clinical Oncology endorsement of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology clinical practice guideline update. J Clin Oncol. 2018;36:911–919. doi: 10.1200/JCO.2017.76.7293. [DOI] [PubMed] [Google Scholar]
- 13.Lindeman NI, Cagle PT, Aisner DL, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: Guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Thorac Oncol. 2018;13:323–358. doi: 10.1016/j.jtho.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Skoulidis F, Goldberg ME, Greenawalt DM, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 2018;8:822–835. doi: 10.1158/2159-8290.CD-18-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koivunen JP, Kim J, Lee J, et al. Mutations in the LKB1 tumour suppressor are frequently detected in tumours from Caucasian but not Asian lung cancer patients. Br J Cancer. 2008;99:245–252. doi: 10.1038/sj.bjc.6604469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tam IYS, Chung LP, Suen WS, et al. Distinct epidermal growth factor receptor and KRAS mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res. 2006;12:1647–1653. doi: 10.1158/1078-0432.CCR-05-1981. [DOI] [PubMed] [Google Scholar]
- 18.Hu Y, Ulrich BC, Supplee J, et al. False positive plasma genotyping due to clonal hematopoiesis. Clin Cancer Res. 2018;24:4437–4443. doi: 10.1158/1078-0432.CCR-18-0143. [DOI] [PubMed] [Google Scholar]
- 19.Remon J, Caramella C, Jovelet C, et al. Osimertinib benefit in EGFR-mutant NSCLC patients with T790M-mutation detected by circulating tumour DNA. Ann Oncol. 2017;28:784–790. doi: 10.1093/annonc/mdx017. [DOI] [PubMed] [Google Scholar]
- 20.Guibert N, Hu Y, Feeney N, et al. Amplicon-based next-generation sequencing of plasma cell-free DNA for detection of driver and resistance mutations in advanced non-small cell lung cancer. Ann Oncol. 2018;29:1049–1055. doi: 10.1093/annonc/mdy005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chae YK, Davis AA, Carneiro BA, et al. Concordance between genomic alterations assessed by next-generation sequencing in tumor tissue or circulating cell-free DNA. Oncotarget. 2016;7:65364–65373. doi: 10.18632/oncotarget.11692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zill OA, Greene C, Sebisanovic D, et al. Cell-free DNA next-generation sequencing in pancreatobiliary carcinomas. Cancer Discov. 2015;5:1040–1048. doi: 10.1158/2159-8290.CD-15-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chae YK, Davis AA, Jain S, et al. Concordance of genomic alterations by next-generation sequencing in tumor tissue versus circulating tumor DNA in breast cancer. Mol Cancer Ther. 2017;16:1412–1420. doi: 10.1158/1535-7163.MCT-17-0061. [DOI] [PubMed] [Google Scholar]
- 24.Merker JD, Oxnard GR, Compton C, et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. J Clin Oncol. 2018;36:1631. doi: 10.1200/JCO.2017.76.8671. [DOI] [PubMed] [Google Scholar]
- 25. Roche Molecular Systems: Cobas EGFR Mutation Test v2. https://diagnostics.roche.com/global/en/products/params/cobas-egfr-mutation-test-v2.html.