Abstract
Monitoring blood coagulation in response to an anticoagulant (heparin) and its reversal agent (protamine) is essential during and after surgery, especially with cardiopulmonary bypass (CPB). A current clinical standard is the use of activated clotting time (ACT), where the mechanical movement of a plunger through a whole blood-filled channel is monitored to evaluate the endpoint time of coagulation. As a rapid, simple, low-volume, and cost-effective alternative, we have developed a paper microfluidic assay and Raspberry Pi-based device with the aim of quantifying the extent of blood coagulation in response to varying doses of heparin and protamine. The flow rate of blood through the paper microfluidic channel is automatically monitored using Python-coded edge detection algorithm. For each set of assay, 8 μL of fresh human whole blood (untreated and undiluted) from human subjects is loaded onto each of 8 sample pads, which have been preloaded with varying amounts of heparin or protamine. Total assay time is 3-5 minutes including the time for sample loading and incubation.
Keywords: blood coagulation, heparin, paper microfluidics, protamine, Raspberry Pi
I. Introduction
BLOOD coagulation is the process by which fibrin clots form in blood. Monitoring coagulation is essential for patient safety in numerous clinical situations. Of particular interest, is monitoring blood coagulation while a patient is on cardiopulmonary bypass (CPB). If blood coagulates while a patient is on CPB, clots can be sent from the bypass machine into the bloodstream of the patient, which can cause serious complications such as stroke or embolism. The use of CPB requires that a patient’s blood be in an optimally anticoagulated state, and upon conclusion of CPB blood must be returned to homeostatic conditions [1].
To prevent coagulation from occurring, an anticoagulant, typically sodium heparin, is intravenously administered to patients. Heparin must be administered to a patient in a high enough dosage to prevent any clot formation for the duration of a surgery that requires CPB [2], [3]. Reaction and metabolic rates to a dose of heparin are significantly varied by individuals and the nature of treatments, leading to varied plasma heparin concentrations even when administered in identical dosages [4], [5].
Following administration of heparin to a patient, this anticoagulant must be reversed when anticoagulation is no longer necessary (i.e. when CPB is no longer necessary in this instance). To reverse the anticoagulant effects of heparin, a reversal agent, protamine sulfate, is intravenously administered to patients. Protamine reverses the actions of heparin by binding to the heparin molecules in the blood. Adverse side effects, such as anaphylaxis, hypotension, pulmonary hypertension, and platelet activation/dysfunction are known to occur following administration of protamine [6]. The risk of these adverse effects increase with excessive dosages of protamine [6]. Protamine dosage have previously been determined either empirically from heparin dosage administered or on patient weight [6], which do not account for individualized differences in interactions with heparin and protamine. For these reasons, a method to determine patient-specific dosing of protamine would be highly beneficial to reverse the remaining heparin in a patient’s blood exactly, without adding excess protamine to the blood. Pediatric patients are at a particular risk when administered heparin or protamine while on CPB or extracorporeal membrane oxygenation (ECMO). Pediatric patients have immature coagulation systems and are affected significantly by hemodilution [7], [8].
The most common method for monitoring blood coagulation during surgery is the use of activated clotting time (ACT) [1], [2]. ACT measures the time it takes for blood to clot after being exposed to an activator such as celite, kaolin, or glass beads. Before heparin is administered during CPB, the baseline ACT is determined. The patient is then administered a dosage of heparin that is typically determined based on their weight [6]. The ACT is again tested after the administration of heparin. If the ACT is not within the time range deemed safe at the particular hospital or by the surgeon, more heparin is administered until the ACT is within this safe range. Typically, the safe range of ACT is a value of over 400 s [9]. Throughout surgery, the ACT is tested approximately every 30 minutes to ensure that it remains within the safe range, or if more heparin needs to be administered to the patient. When CPB is no longer necessary, protamine will be administered to a patient, again based on weight or the dosage of heparin administered. The ACT is tested again until the patient is back to the initial baseline value. Surprisingly, many of the common practices related to CPB are still based on clinical experience rather than a strong foundation in evidence, as many factors still need to be studied [10].
It is well known that heparin concentrations within a patient do not correlate to ACT values [4]-[6]. Since metabolic rates of heparin (and protamine) are varied by individuals and treatment methods, a dosage of heparin administered is not indicative of the final plasma heparin concentration. Another concern with ACT is the wide range of ACT machines available and the inconsistencies between them [11]. There are large variations reported between identical samples tested repeatedly using one single machine, as well as between a newer and older model of the same brand and model of ACT machine [12].
Direct information on dosages of heparin and protamine can be provided via titration-based method. HMS Plus from Medtronic Cardiac Surgery is a good example [5]. Studies have shown that the use of the HMS results in fewer complications, fewer blood products needed, and less time in the operating room (OR) after protamine being administered in pediatric patients (a population of particular concern) [5].
Most point-of-care blood coagulation monitoring devices, including ACT equipment and HMS Plus, measure the coagulation endpoints in fresh whole blood samples in an electromechanical manner. In ACT Plus from Medtronic, for example, a plunger-flag assembly falls through the whole blood sample in each cartridge channel, which is monitored by a photo-optical system [13]. With fibrin clot formation, it falls slower than with unclotted sample. Such endpoint time will be recorded as ACT. While they have been successfully used in clinical settings, there still exist substantial rooms for improvement. Firstly, they measure the endpoint time of coagulation, which can be substantially shortened by evaluating coagulation in situ. Secondly, the mechanical system (e.g. plunger-flag) is rather complicated, which may increase the device and supplies cost and/or compromise assay reproducibility. Simpler mechanism could improve sensitivity and reproducibility, or at least maintain comparable sensitivity with shorter assay time. Thirdly, they require substantial volume of blood – ACT Plus requires 0.2 mL per channel and HMS Plus requires a total of at least 2.4 mL [5], [13]. Reduced volume requirement would certainly be an advantage, especially when repeated testing is necessary [1]. Finally and most importantly, the costs of many existing devices are substantial: ACT Plus device costs ~$4,400 and HMS Plus device costs ~$26,000. In addition, custom syringes and cartridges are necessary for each use, and substantial training time is also necessary [14].
Our work is aimed at rapid, simple, low-volume, and cost-effecctive assay method of evaluating blood coagulation in response to varying concentrations of heparin and protamine. Towards this end, we replaced the cartridge-based channels with paper microfluidic channels. Whole blood samples traveled through paper fibers spontaneously via capillary action, also known as wicking. The lengths of such blood flow were measured over time in situ(= flow rates), before the endpoint of coagulation, as long as the data showed statistical significance. This would substantially reduce the assay time. In addition, the assay was greatly simplified since the flow was spontaneous and no complicated mechanical action was necessary. Volume could also be reduced substantially, as whole blood flowed through very thin paper fibers with submicron-scale pores. Cartridges were replaced with simple wax-printed paper strips and the photo-optical detection system with Raspberry Pi and Pi camera, thus significantly lowering the device cost.
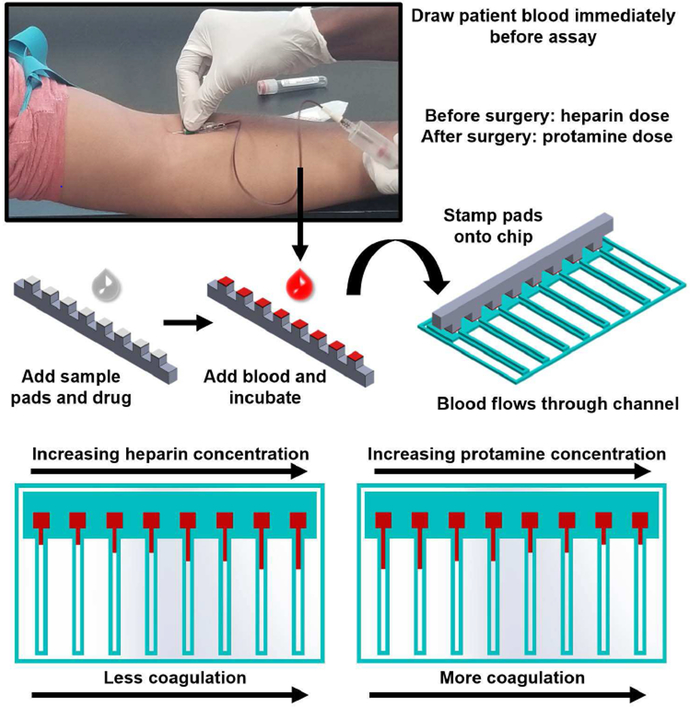
While blood analyses have extensively been studied on silicone-based microfluidic devices [15], [16], blood coagulation have not extensively been studied on such microfluidic platforms. A small number of recent publications are focused on evaluating the time for blood to coagulate, essentially duplicating ACT on microfluidic platforms [17]-[19]. In addition, these recent studies have demonstrated neither the use of fresh human blood immediately after withdrawal nor multiplexed assays with varying heparin and protamine concentrations. In this work, 8 assays were simultaneously performed on an 8 channel paper microfluidic chip to test for 4 different heparin or protamine concentrations in duplicates, with blood withdrawn from human subjects at the start of the assay. The flow length and subsequent flow rate of whole blood through paper fibers were monitored in situ, as compared to the endpoint coagulation time in other methods. Each of the 8 channels has a sample size of just 8 μL of human whole blood from human subjects; no dilution, processing, or reagents are required. Further, we have developed a Raspberry Pi-based device with Python-coded edge detection algorithm that automatically captures images of the paper microfluidic chip over the course of 2-3 minutes, automatically analyzes the flow rate of blood, and displays a result to the user. Fig. 1 outlines the assay process from drawing blood to displaying results.
Fig. 1.
Assay procedure and paper microfluidic chip. Blood is drawn from a volunteer immediately prior to assays, acting as a substitute for a patient’s blood during surgery. A 3D printed stamper block was designed to hold the glass fiber sample pads, where varying concentrations of heparin or protamine solutions (8 μL) are preloaded and allowed to dry fully at room temperature. 8 μL of undiluted and untreated human whole blood is loaded onto the sample pads using an 8 channel multipipette. The blood is incubated on the stamper block to allow sufficient time for the blood to interact with the preloaded drug (2 minutes for heparin and 1 minute for protamine). The sample pads are then stamped onto the loading area of the paper microfluidic chip, which starts the simultaneous blood flow over eight channels, initiating the assay. The paper microfluidic chips are then imaged by a Raspberry Pi camera and images are analyzed by a Raspberry Pi 3 with results displayed to the user on a touchscreen display.
II. Materials and Method
A. Study Design
Study groups in this controlled laboratory experiment include heparin sodium and protamine sulfate in varied concentrations added to human whole blood. Samples were randomly assigned a drug, and dosage and researchers were not blinded.
B. Blood Collection
Human whole blood was collected from volunteers according to protocol 1612094853 approved by the University of Arizona’s institutional review board (IRB). ASCP (American Society for Clinical Pathology) board-certified medical laboratory scientist withdrew the blood samples from the volunteers. No personal identifying information was collected. The total number of subjects was identified as 9. Blood was collected immediately before assays were conducted. For testing of heparin dosage, blood was used immediately following blood draw with no anticoagulants added. For testing of protamine dosage, 3 USP/mL of heparin was added to the blood during blood draw. Blood volume added to each of 8 sample pads was 8 μL.
C. Paper Microfluidic Chip Fabrication
Nitrocellulose paper with a lateral flow rate of 75 s per 4 cm (Millipore Sigma, Billerica, MA, USA) was patterned via wax printing using a Xerox ColorQube 8580 solid ink printer. After the pattern was printed on the nitrocellulose paper, chips were melted at about 130°C for 2-3 minutes, to allow for the wax to flow down into the paper fibers. The hydrophobic wax barrier was designed as (0, 255, 255) RGB intensity values, resulting in a teal color, used to distinguish blood (red) versus the wax barrier. The wax channels have a 4 mm × 4 mm sample pad loading area and channel (30 mm L × 1 mm W) that also acts as a wicking pad due to its extended length. Each paper microfluidic chip contains 8 channels, to allow for simultaneous testing of duplicates of each of four drug concentrations (Fig. 1). G041 glass fiber conjugate pad sheets (Millipore Sigma, Billerica, MA, USA) were mechanically cut into 4 mm × 4 mm squares and placed on a 3D printed stamper to act as sample pads (Fig. 1). Glass fibers acted as an activator for blood coagulation, similar to glass beads [1], [2].
D. Heparin and Protamine Solutions
Sodium heparin (Sagent, Schaumburg, IL, USA) was diluted in Plasma-Lyte A to final concentrations of 0 USP/mL, 2 USP/mL, 4 USP/mL, and 6 USP/mL. 0 USP/mL of heparin provides the baseline of the patient without any anticoagulant. 2 USP/mL of heparin is used as a starting point to get an ACT of 300 s, where surgeons can safely begin cannulation. Between 2 and 4 USP/mL of heparin is a standard dosage of heparin to result in a patient being within the safe range of ACT (>400 s). 6 USP/mL is included as a high dose to evaluate those patients who are less sensitive to heparin and require a higher dosage. Protamine sulfate (Fresenius Kabi, Lake Zurich, IL, USA) was diluted in Plasma-Lyte A to final concentrations of 0 mg/mL, 2 mg/mL, 4 mg/mL, and 6 mg/mL. Typical protamine dosages range from 0.5-4 mg protamine per unit of heparin, with the variation often being attributed to the institution and the specific procedure. Again, 6 mg/mL of protamine was included as a high dose to evaluate those patients who are less sensitive to heparin or have excess heparin in their system. Sample pads were placed onto a 3D printed stamper, which allowed for simultaneous and consistent stamping of sample pads onto the loading area of the paper microfluidic chip (Fig. 1). 8 μL of each drug solution was loaded onto a single sample pad and allowed to dry fully at room temperature (about 25-35 minutes). Once dry, these sample pads were considered to be pre-loaded with heparin or protamine (relative to the start of the assay).
E. Optimization of Incubation Time
For each of the two drugs used, the incubation time was optimized by testing a wide range of incubation times from 0 to 10 minutes. The optimal incubation time was determined based on reproducibility of the assay and a lack of clot formation in the sample pads.
F. Assay Procedure
Prior to assays, heparin or protamine was preloaded onto sample pads that were placed on stamper blocks and allowed to dry fully. 8 μL of human whole blood was added to each of 8 sample pads following the blood collection procedure (Fig. 1). Sample pads preloaded with heparin were incubated with human whole blood at room temperature for 2 minutes, while those with protamine for 1 minute. The sample pads’ pore sizes were substantially bigger than those of the paper and the blood samples were fully absorbed and remained there in liquid form. Following the incubation period of the blood and the preloaded drug, sample pads were stamped onto the loading area of the paper microfluidic chip (Fig. 1). The assay monitoring device was started as sample pads were loaded onto the paper microfluidic chip, initiating the flow of blood through the paper microfluidic chip. For all samples a baseline flow length/rate was determined by the flow rate of an individual’s blood with no heparin and no protamine added to the blood, and used to obtain normalized flow lengths. The baseline flow lengths/rates at 180 s were used for all time points, so that the time-dependent increase could be monitored.
G. Assay Monitoring Device
The assay monitoring device consists of a Raspberry Pi 3, 7” touchscreen display, and Raspberry Pi camera module (Fig. 2). The program to capture and analyze images of the paper microfluidic chip throughout the assay is coded in Python. Upon initiation of the assay, the monitoring device captures images of the chips at time points of 10 s, 20 s, 30 s, 40 s, 50 s, 60 s, 120 s, and 180 s from the initiation of the assay. Images were saved to be later analyzed manually for initial optimization, and eventually with automated image analysis methods coded in Python.
Fig. 2.
Monitoring and analysis device. Users are able to interface with the device through the 7” touchscreen display shown in the top view, and results are displayed to the user. The Raspberry Pi camera, shown in the bottom view, is positioned above the paper microfluidic chip to capture images of a paper microfluidic chip automatically at set time points. The Raspberry Pi 3 inside of the device is coded in Python and is used to automatically capture images, analyze images, and display the results of the assay.
H. Sample Size
Each single assay consists of 8 channels, with 4 drug concentrations tested, meaning that each drug concentration was tested in duplicate for a single blood sample. Data shown is the compilation of the assays from 9 subjects, each duplicated, resulting in n = 18 for a given drug concentration at each time point. Assays were considered a failure if a known issue occurred during the assay (such as sample pads being stamped on late or sample pads being stamped partially outside of the sample pad loading area). Individual channels were considered a failure if a known issue occurred during the assay (such as wax blocking a single channel or a single sample pad not adhering to the loading area of the paper microfluidic chip). Averages of these compiled data sets are presented with error bars representing standard error of the mean.
I. Manual Data Analysis
Images were analyzed for length of flow at each known time point, resulting in information on the flow rate throughout the assay. The length of flow was measured using ImageJ (U.S. National Institutes of Health, Bethesda, MD, USA) from the base of the sample pad to the end of the blood flow in the channel. The tilt and rotation of chips within each image was corrected for using a correction factor based on the known consistent length of each wax printed hydrophobic channel barrier. Within each assay, data was normalized by dividing by the average of the 0 USP/mL heparin or 0 mg/mL protamine concentration, at given time point.
J. Automated Data Analysis
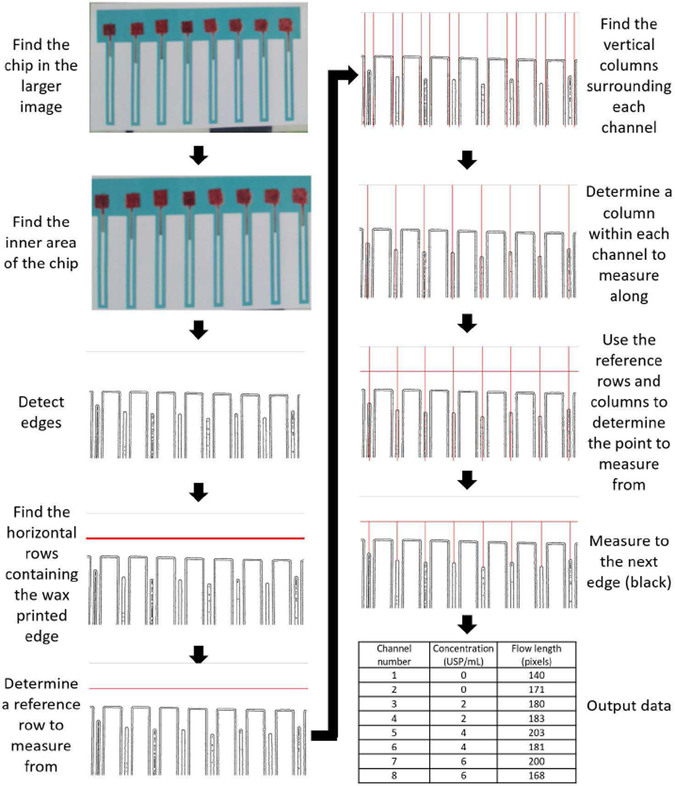
The automated analysis is coded in Python using OpenCV and NumPy. Contour detection is used initially, followed by edge detection. The initial image is converted to a binary image in this manner. From a binary image, the edge of the wax printed chip is detected and serves as a reference point. The rows and columns (i.e., pixel coordinates values) with respect to the reference line were evaluated for the beginning and the end of blood flow within each microfluidic channel, and the distance between these two points was evaluated as the flow length. Fig. 3 illustrates the automatic data analysis procedure.
Fig. 3.
Automated image analysis procedure. The automated image analysis program finds the paper microfluidic chip within a larger image, then carries out contour detection and edge detection algorithms to find the wax printed borders and the edge of blood flow. Each channel is found and a horizontal reference point is determined based on the wax printed borders. The distance from the reference point to the edge of blood flow is measured for each channel.
K. Statistics
Student’s t-test was performed to determine differences in the lengths of blood flow between drug concentrations. ANOVA was performed for each individual at different time point. A p-value of less than or equal to 0.05 was considered significant.
III. Results
A. Device
The assay monitoring and analysis device consists of a Raspberry Pi 3, coded in Python, a 7” touchscreen display, and a Raspberry Pi camera module. These components are housed in a 3D printed enclosure (Fig. 2). The device successfully captures and saves images of the paper microfluidic chip used for this assay at set time points of 10 s, 20 s, 30 s, 40 s, 50 s, 60 s, 120 s, and 180 s. These images have text overlay detailing the drug being tested, concentration, and the time point.
Initially, manual image analyses were performed and the flow rates (or the lengths of flow at each time point) were evaluated. For all assays, automated image analyses were also performed, and the averaged lengths of flow at each time point along with overall standard errors were collected.
B. Incubation Time Optimization
0 minute incubation time does not allow sufficient time for the drug to act on the blood, and therefore was excluded. Incubation times of 5-10 minutes resulted in blood clotting and drying on the sample pads, and were therefore excluded. It was found that the optimal incubation times (for reproducibility of the assay and to prevent drying) were 2 minutes for blood samples with heparin and 1 minute for blood samples with protamine.
C. Assay Time Optimization
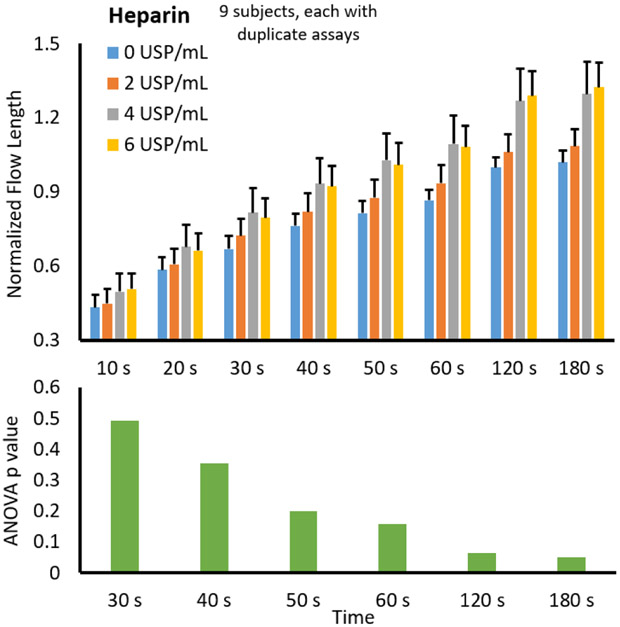
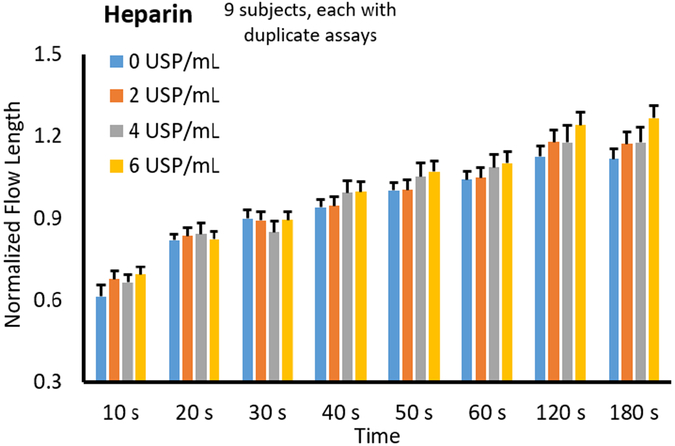
The normalized flow lengths from 9 different subjects were averaged and plotted against the varying heparin concentrations from 0 USP/mL to 6 USP/mL. All data were normalized with the baseline flow rate with 0 USP/mL at 180 s. Between 2 and 4 USP/mL of heparin is a standard dosage of heparin to typically result in the safe range of ACT. 6 USP/mL is included as a high dose for the patients who are less sensitive to heparin. Overall trends at each time point for varied preloaded heparin concentrations are shown in Fig. 4. It was expected that as heparin concentration increased, flow rate (or length of flow at each time point) would increase due to the anticoagulant effects of heparin. Fig. 4 shows that the data followed this expected trend. ANOVA was performed for each data set at different time points (data sets at 10 s and 20 s were excluded as the blood did not flow sufficiently before 30 s on several occasions). The resulting p values are shown below the main plot, indicating that the significance (p ≤ 0.05) could be achieved at 120 s and 180s. With the aim of reducing assay time, the optimum assay was determined to be 120 s.
Fig. 4.
Assay time optimization with heparin. The flow lengths of the blood incubated with varied heparin concentrations on the sample pads are plotted against the assay time. Manual image analyses were used. Averages from 9 different subjects, each duplicated using a new microfluidic chip each time. Error bars represent the standard errors of the mean. ANOVA was performed for each data set at different time points, e.g. 0, 2, 4 and 6 USP/mL heparin data at 120 s as one data set. Data sets at 10 s and 20 s were excluded as the blood did not flow sufficiently before 30 s on several occasions, leading to inconsistent results. The resulting p values are plotted against the assay time for heparin (bottom left) and protamine (bottom right).
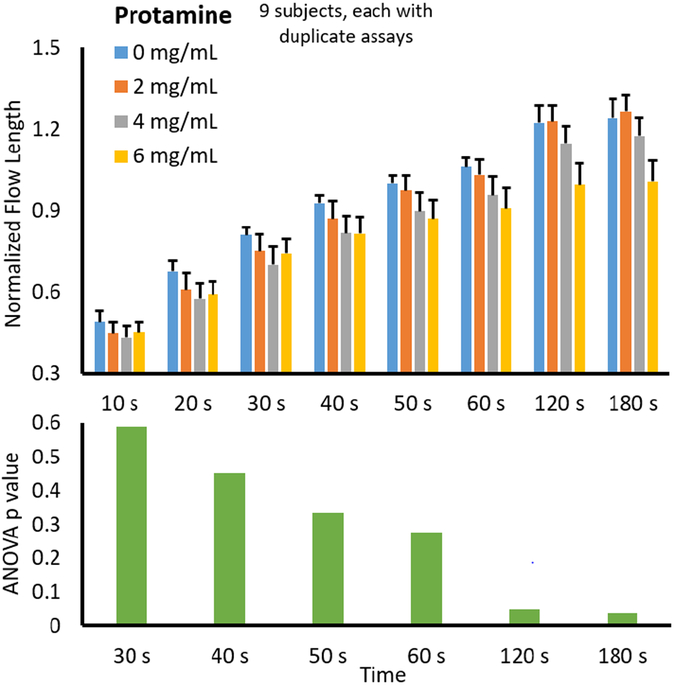
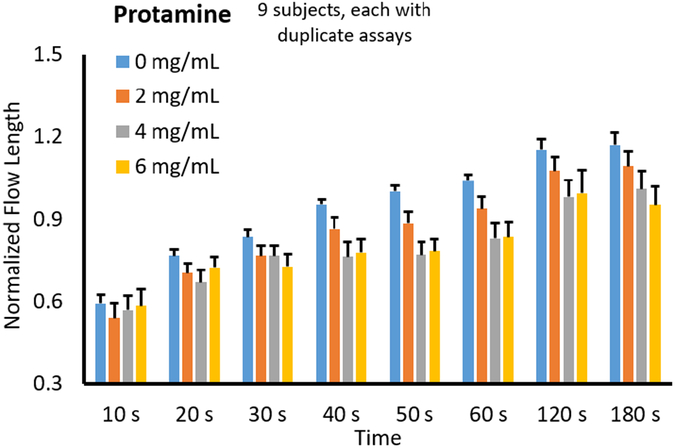
Additionally, the normalized flow lengths from 9 different subjects were averaged and plotted against the varying protamine concentrations from 0 mg/mL to 6 mg/mL. All data were normalized with the baseline flow rate with neither heparin nor protamine at 180 s. As all blood samples were added with heparin, the normalized flow length at 180 s was substantially higher than 1 with low protamine doses, while it approached to 1 with high protamine dose. Typical protamine dosages range from 0.5-4 mg protamine per unit of heparin.
Again, 6 mg/mL of protamine was included as a high dose for the patients who are less sensitive to protamine or have excess heparin in their system. Overall trends at each time point for varied preloaded protamine concentrations are shown in Fig. 5. It was expected that as protamine concentration increased, flow rate (or length of flow at each time point) would decrease due to protamine reversing the anticoagulant effects of heparin. Fig. 5 shows that the data follow this expected trend. ANOVA was performed for each data set at different time points (data sets at 10 s and 20 s were again excluded as the blood did not flow sufficiently before 30 s on several occasions). The resulting p values are shown below the main plot, indicating that the significance (p ≤ 0.05) could be achieved again at 120 s and 180s. With the aim of reducing assay time, the optimum assay was again determined to be 120 s.
Fig. 5.
Assay time optimization with protamine. All other experimental conditions are identical to those with heparin (Fig. 4).
D. Automated Image Analysis
To automatically analyze these images, OpenCV and NumPy were used in Python. The automated analysis code opens the captured images, finds the paper microfluidic chip, finds each of the 8 channels within the chip, finds the wax printed edge as a reference, and measures from the reference edge to the edge of blood flow (Fig. 3). The results are then automatically analyzed following the same normalization procedure outlined in the manual analysis section, and displayed to the user.
By analyzing the length of blood flow using this automated method, standard error for all 9 subjects was reduced by 47.2% for the average heparin dose response curves, and by 24.7% for the average protamine dose response curves by eliminating human error (Fig. 6 and Fig. 7).
Fig. 6.
Automated image analysis with heparin. The flow lengths of the blood incubated with varied heparin concentrations on the sample pads, following automated image analysis, were plotted against the assay time, showing the smaller error bars than those with manual analysis (Fig. 4). The standard errors of the averages from 9 different subjects, each duplicated, were significantly decreased using automated image analysis versus manual image analysis by eliminating human error.
Fig. 7.
Automated image analysis with protamine. All other experimental conditions are identical to those with heparin (Fig. 6).
Of the 560 data points for each of the two drugs tested, 95.2% and 85.2% were successfully analyzed retroactively for heparin and protamine dose response curves, respectively (Fig. 6 and Fig. 7). Fewer data points were analyzed for protamine dose response curves due to a failure to automatically analyze one complete chip. Ambient lighting conditions were not controlled at the time of capture, and this contributes heavily to the success of the automated analysis.
In addition, the automated analyses generated a general increasing trend in normalized flow length against heparin concentration, while the manual analyses did not – in fact, the manual analysis results with 4 and 6 USP/mL were similar to each. We have occasionally noticed a small extent of separation of plasma from red blood cells within paper channels, especially with high dose of heparin (4 and 6 USP/mL), which led to difficulty in manually identifying the flow lengths. With automated analyses, consistent and reproducible identification of flow front could be made, leading to smaller error bars and distinction between 4 and 6 USP/mL. Similar results were obtained with protamine experiments: the manual analysis results with low dose of protamine (0 and 2 mg/mL) were similar to each other (where the added heparin has not been reversed yet), while they were distinct with automated analyses.
E. Dose Responses
As the optimal assay time was determined to be 120 s, we can focus on the 120 s data set in Fig. 6 to evaluate the heparin dose responses. It shows a continuous increase over the heparin concentration; in fact, such linear increases can be observed in most other set time points. These results suggest that all heparin concentrations (2, 4, and 6 USP/mL) were effective in increasing the flow length, demonstrating their anti-coagulation capability.
Protamine dose responses can also be evaluated by examining the 120 s data set in Fig. 7. It shows a decrease up to 4 mg/mL, followed by an increase in 6 mg/mL, demonstrating their reversal effects only up to 4 mg/mL.
IV. Discussion
A. Concept of Research
Blood is a unique fluid due to its ability to clot readily. As blood clots form, the fluid becomes thicker and contains increasingly larger particles (blood clots). Such change will inevitably change the viscosity of blood. The capillary flow rate is reduced to the general Lucas-Washburn equation: l2/tα RγLG cos θ / 2μ, where l is flow length, t is time, R is capillary or pore radius, γLG is the surface tension between liquid (blood) and gas phases, θ is the blood contact angle over capillary or pore surface, and μ is dynamic viscosity [19]. As R cannot be varied and γLG and θ do not change significantly upon coagulation, the flow length (l) or rate (l2/t) depends almost solely on viscosity μ. Therefore, it is possible to use the flow length or rate as a measure of blood coagulation.
Based on these hypotheses, we designed and tested a paper microfluidic assay to monitor the flow rate of blood, and therefore the ability of the blood to coagulate, in response to varied concentrations of heparin or protamine added to the blood. The assay is preloaded with these drugs, to allow for a single step test. The glass fiber pad on a stamper acted as an activator for blood coagulation, similar to the use of glass beads as an activator in AcT [1], [2]. Based on the data shown in Fig. 4 and Fig. 5, it is clear that the results could be obtained 2 minutes after sample loading, as significant differences do not occur between 2 and 3 minutes (120 s and 180 s) of assay time. The total assay time would be 3-5 minutes including the time for sample loading and incubation. our assay time is significantly faster than the current gold standard of AcT, which can take up to 17 minutes; even a few minutes is often critical in a situation where a patient is on CPB.
The use of a stamper allowed us to provide a uniform pressure over 8 microfluidic channels in a single paper microfluidic chip, ensuring the reproducibility and uniformity within a single data set. However, the stamper’s pressure applied to paper might vary significantly over different paper microfluidic chips and subsequently different subjects. Normalizing the flow lengths by the baseline flow rate, as described in Materials and Method, addressed such variations, at least partially.
B. Assay Time and Sample Volume Requirement
Achieving and maintaining an appropriate inhibition of coagulation ability of the blood, utilizing heparin, is critical for patient safety while on CPB, as mentioned in the introduction. Following the administration of heparin, protamine needs to be administered in a dose that will fully and exactly reverse the heparin remaining in the patient’s blood. Protamine has been shown to have adverse effects when administered in excess. The assay we have demonstrated through this work has the potential to provide information to clinicians on exactly what dose of heparin and protamine will achieve these goals in a specific patient. Further, the assay demonstrated has the ability to provide this information in 3-5 minutes with only a few microliters of blood used per assay, which is beneficial in a surgical environment where both time and blood loss are major concerns.
Our assay has the distinct benefits over ACT that the assay time is 2 minutes for a simple coagulation test (vs. up to 1000 s or 17 minutes) and the blood volume is 8 μL per assay (vs. up to 20-30 μL per assay). Our assay has the distinct benefits over HMS that the blood volume is 8 μL per channel, resulting in 64 μL per assay for a full dose response curve, versus 2.4 mL of blood needed per assay for the HMS. The device is significantly more cost effective than the HMS, without requiring custom syringes or cartridges and extensive training. These benefits allow for answers significantly faster, which is critical in a surgical environment, and repeated testing to have extremely low impact on blood loss. Further, if increased heparin levels are required during surgery, ACT provides no direct information on this, while our assay provides specific information regarding patient response to a variety of dosages.
C. Future Impact
The assay created through this work has the potential to contribute to personalized medicine advancements by accounting for the response of an individual’s blood to a specified dosage of heparin or protamine.
If expanded to other blood-thinners, such as those taken daily by patients, our assay and monitoring device could potentially provide a patient and their physician with information on the proper dosage of the anticoagulant they are taking. Since only 8 μL of sample is needed for each channel, a simple finger-pricking device could be used rather than relying on a certified phlebotomist in a laboratory setting to monitor blood coagulation. Such an expansion could allow this technology to impact a patient in their daily life, rather than only in a surgical setting.
V. Conclusion
We have demonstrated a flow rate-based paper microfluidic assay for fresh, untreated, and undiluted human whole blood from human volunteers, that could quantify whole blood coagulation in a rapid, simple, low-volume, and cost-effective manner. The assay takes only 3-5 min including the time for sample loading and incubation, and requires only 8 μL of blood per channel. Our assay is monitored by a Raspberry Pi-based device, which provides a user-friendly interface and allows for immediate and automated analysis of results with Python-coded edge detection algorithm. Our device is able to provide information on patient-specific dosing of an anticoagulant and its reversal agent, unlike current standards that only tell an arbitrary time for blood to clot.
Some limitations need to be overcome while advancing this work further. Automated image analysis was conducted retroactively on the data presented, therefore ambient lighting conditions were not controlled in each image captured. Future work should include controlled ambient lighting conditions to improve the success rate of the automated image analysis program. Future work would be aimed at decreasing the limit of detection, to improve the precision in dosing of each drug tested. Future work would also be aimed at determining the exact dosage that protamine has reversed the actions of heparin without being in excess of heparin, which could be accomplished by looking at lower and more precise dosages of protamine. Future work can also be aimed at determining the impact of varied protamine doses on blood with varied heparin concentrations, as the current work only evaluated the impact of varied protamine doses on blood with 3 USP/mL heparin. A key factor to consider going forward is the testing of blood from patients with coagulation disorders, such as hemophilia, which is problematic with currently monitoring methods such as ACT. After advancing this heparin/protamine assay, expansion to other anticoagulants would be considered.
Acknowledgment
The authors would like to thank Mr. Jacob Pfeiffer for his experimental assistance.
This paragraph of the first footnote will contain the date on which you submitted your paper for review. This work was supported by the Cardiovascular Biomedical Engineering Training Grant through the U.S. National Institutes of Health (NIH) under Grant T32HL007955.
Biography

Robin E. Sweeney was born in Albany, NY, USA in 1992. She received B.S. degrees in biology and biomathematics from The University of Scranton, Scranton, PA, USA in 2014, and Ph.D. degree in biomedical engineering from The University of Arizona, Tucson, AZ, USA 2017. Her research interests include the development of biosensors for medical monitoring and diagnostics. Dr. Sweeney is a recipient of the University of Arizona College of Engineering Graduate Fellowship, the Cardiovascular Biomedical Engineering Training Grant (U.S. National Institutes of Health, grant number T32HL007955), the Tony B. Travel Award, and the H.E. Carter Travel Award. Dr. Sweeney currently works at Unchained Labs in Pleasanton, CA, USA.

Vina Nguyen was born in Houston, Texas and received her B.S. degree in mathematical biology from University of Houston in 2013 and the M.S. degree in medical pharmacology at the University of Arizona College of Medicine in 2016 where she participated in the Biosensors Lab in Tucson, AZ, USA. After being board certified, she is currently a cardiovascular perfusionist for Pacific Life Lines, Inc. in the San Francisco Bay Area as well as an extracorporeal membrane oxygenation (ECMO) perfusionist at Stanford Hospital, Palo Alto, CA, USA.

Benjamin Alouidor received the B.S. degree in biotechnology from Pennsylvania State University, University Park, PA, USA, in 2013 and the M.S. degree in medical pharmacology – perfusion science from the University of Arizona, Tucson, AZ, USA, in 2018. Currently he is a practicing cardiovascular perfusionist at Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Alouidor was a recipient of both the American Society of Extracorporeal Technology Scientific Presentation Award and the International Society of Mechanical Circulatory Support Innovation Challenge Award in 2017.

Elizabeth Budiman is from the San Francisco Bay Area. She will receive a B.S. in biomedical engineering from the University of Arizona in 2019. Since 2015, she has been an undergraduate research assistant in the University of Arizona Biosensors Lab in Tucson, AZ, USA. In 2018, she was an intern for W. L. Gore & Associates. Her research interest has been in low-cost medical diagnostics.
Ms. Budiman is a three time recipient of the Spirit of Inquiry Research Grant from the University of Arizona Honors College.

Raymond K. Wong was born in Ithaca, NY but grew up in Malaysia. He returned to the US to earn his B.S. in bioengineering at Syracuse University. He then trained to be a clinical perfusionist and a clinical engineer while earning his M.S. and Ph.D. in physiological sciences at The University of Arizona, finishing in 2001. From 2001 until 2013, he practiced as a perfusionist at the Mayo Clinic Hospital, except for a two-year break from 2004-2006, when he worked in industry in product development and as a regulatory scientist. Since 2013, he has held appointments as an Assistant Professor of Pharmacology and as the Program Director of the Perfusion Sciences Graduate Program at the University of Arizona College of Medicine – Tucson. His research interests include topics in cardiopulmonary bypass, extracorporeal membrane oxygenation, mechanical circulatory assist devices, anticoagulation/hemostasis therapy and diagnostic testing and blood management in the cardiac surgery patient population. He is an associate editor of The Journal of Extra-Corporeal Technology and a member of both the American Academy of Cardiovascular Perfusion and the American Society of Extra-Corporeal Technology.

Jeong-Yeol Yoon was born in Seoul, Republic of Korea, and received his B.S., M.S. and Ph.D. degrees in chemical engineering from Yonsei University, Seoul, in 1992, 1994 and 1999, respectively, where he worked primarily on polymer colloids. He received his second Ph.D. degree in biomedical engineering from University of California, Los Angeles, CA, USA in 2004, working on lab-on-a-chip and biomaterials. He joined the faculty at the University of Arizona, Tucson, AZ, USA in August 2004 and currently Professor and Associate Department Head in biomedical engineering as well as biosystems engineering and chemistry & biochemistry. He is currently directing Biosensors Lab. He has published numerous journal articles in the field of biosensors, including IEEE Sensors Journal, Biosensors and Bioelectronics, Science Advances, Advanced Materials, Lab on a Chip, etc. He is also a sole author of Introduction to Biosensors: From Electric Circuits to Immunosensors(currently second edition; New York, NY, Springer, 2016). Dr. Yoon is a member of Institute of Biological Engineering (IBE), American Society of Agricultural and Biological Engineers (ASABE), and SPIE: The International Society for Optics and Photonics. He was President of IBE in the calendar year 2015. He currently serves as Editor-in-Chief for Journal of Biological Engineering (BMC), and Editorial Board Member for Scientific Reports (Nature Publishing Group).
Contributor Information
Robin E. Sweeney, Department of Biomedical Engineering, The University of Arizona, Tucson, AZ 85721, USA; Unchained Labs, Pleasanton, CA, USA.
Vina Nguyen, Perfusion Sciences Graduate Program, Department of Medical Pharmacology, The University of Arizona College of Medicine, Tucson, AZ 85721, USA; Pacific Life Lines, San Carlos, CA, USA.
Benjamin Alouidor, Perfusion Sciences Graduate Program, Department of Medical Pharmacology, The University of Arizona College of Medicine, Tucson, AZ 85721, USA; Cedars-Sinai Medical Center, Los Angeles, CA, USA..
Elizabeth Budiman, Department of Biomedical Engineering, The University of Arizona, Tucson, AZ 85721, USA.
Raymond K. Wong, Perfusion Sciences Graduate Program, Department of Medical Pharmacology, The University of Arizona College of Medicine, Tucson, AZ 85721, USA
Jeong-Yeol Yoon, Department of Biomedical Engineering, The University of Arizona, Tucson, AZ 85721, USA`.
References
- [1].Ural K and Owen C, “Pro: the Hepcon HMS should be used instead of traditional activated clotting time (ACT) to dose heparin and protamine for cardiac surgery requiring cardiopulmonary bypass,” J. Cardiothorac. Vasc. Anesth, vol. 30, no. 6, pp. 1727–1729, 2016. [DOI] [PubMed] [Google Scholar]
- [2].Horton S and Augustin S, “Activated clotting time (ACT),” in Haemostasis: Methods and Protocols, New York, NY, USA: Humana Press, 2013, ch. 12, pp. 155–16. [DOI] [PubMed] [Google Scholar]
- [3].Paparella D, Brister SJ, and Buchanan MR, “Coagulation disorders of cardiopulmonary bypass: a review,” Intensive Care Med., vol. 30, pp. 1873–1881, 2004. [DOI] [PubMed] [Google Scholar]
- [4].Culliford AT, Gitel SN, Starr N, Thomas ST, Baumann FG, Wessler S, and Spencer FC, “Lack of correlation between activated clotting time and plasma heparin during cardiopulmonary bypass,” Ann. Surg, vol. 193, no. 1, pp. 105–111, 1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Machovec KA, Jooste EH, Walczak RJ, Homi HM, Jacquiss RDB, Lodge AJ, and Ames WA, “A change in anticoagulation monitoring improves safety, reduces transfusion, and reduces costs in infants on cardiopulmonary bypass,” Paediatr. Anesth, vol. 25, pp. 580–586, 2015. [DOI] [PubMed] [Google Scholar]
- [6].Cuenca JS, Diz PG, Sampedro FG, Zincke JMG, Acuna HR, and Fontanillo MMF, “Method to calculate protamine dose necessary for reversal of heparin as a function of activated clotting time in patients undergoing cardiac surgery,” J. Extra Corpor. Technol, vol. 45, pp. 235–241, 2013. [PMC free article] [PubMed] [Google Scholar]
- [7].Kern FH, Morana NJ, Sears JJ, and Hickey PR, “Coagulation defects in neonates during cardiopulmonary bypass,” Ann. Thorac. Surg., vol. 54, pp. 541–546, 1992. [DOI] [PubMed] [Google Scholar]
- [8].Baird CW, Zurakowski D, Robinson B, Gandhi S, Burdis-Koch L, Tamblyn J, Munoz R, Fortich K, and Pigula FA, “Anticoagulation and pediatric extracorporeal membrane oxygenation: Impact of activated clotting time and heparin dose on survival,” Ann. Thorac. Surg, vol. 83, pp. 912–920, 2007. [DOI] [PubMed] [Google Scholar]
- [9].Young JA, Kisker T, and Doty DB, “Adequate anticoagulation during cardiopulmonary bypass determined by activated clotting time and the appearance of fibrin monomer,” Ann. Thorac. Surg, vol. 26, no. 3, pp. 231–240, 1978. [DOI] [PubMed] [Google Scholar]
- [10].Hessel EA, “A brief history of cardiopulmonary bypass,” Semin. Cardiothorac. Vasc. Anesth, vol. 18, no. 2, pp. 87–100, 2014. [DOI] [PubMed] [Google Scholar]
- [11].Ojito JW, Hannan RL, Burgos MM, Lim H, Huynh M, Velis E, Arocha M, Tirotta CF, and Burke RP, “Comparison of point-of-care activated clotting time systems utilized in a single pediatric institution,” J. Extra Corpor. Technol, vol. 44, pp. 15–20, 2012. [PMC free article] [PubMed] [Google Scholar]
- [12].Dzemali O, Ganter MT, Zientara A, Graves K, Behr R, Genoni M, and Hofer CK, “Evaluation of a new sonoclot device for heparin management in cardiac surgery,” Clin. Appl. Thromb. Hemost, vol. 23, no. 1, pp. 20–26, 2017. [DOI] [PubMed] [Google Scholar]
- [13].Massachusetts General Hospital – Pathology Service, “Medtronic ACT Plus Procedure,” Boston, MA, USA: Massachusetts General Hospital, 2004. [Online]. Available: https://www.massgeneral.org/pathology/assets/pdf/Medtronic%20ACT%20Plus%20Procedure%20(LTR21325).pdf [Accessed: 10-Dec-2018]. [Google Scholar]
- [14].CAP TODAY, “Coagulation analyzers – point of care, self-monitoring,” Northfield, IL, USA: College of American Pathologists, 2011, pp. 28–36. [Google Scholar]
- [15].Toner M and Irimia D, “Blood-on-a-chip,” Annu. Rev. Biomed. Eng, vol. 7, pp. 77–103, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hou HW, Bhagay AAS, Lee WC, Huang S, Han J, and Lim CT, “Microfluidic devices for blood fractionation,” Micromachines, vol. 2, pp. 319–343, 2011. [Google Scholar]
- [17].Li H, Han D, Pauletti GM, and Steckl AJ, “Blood coagulation screening using a paper-based microfluidic lateral flow device,” Lab Chip, vol. 14, pp. 4035–4041, 2014. [DOI] [PubMed] [Google Scholar]
- [18].Dudek MM, Kent NJ, Gu P, Fan H, and Killard AJ, “Development of a fluorescent method for detecting the onset of coagulation in human plasma on microstructured lateral flow platforms,” Analyst, vol. 136, pp. 1816–1825, 2011. [DOI] [PubMed] [Google Scholar]
- [19].Hajjarian Z, Tripathi MM, and Nadkarni SK, “Optical thromboe-lastography to evaluate whole blood coagulation,” J. Biophotonics, vol. 8, no. 5, pp. 372–381, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Klug KE, Reynolds KA, and Yoon J-Y, “A capillary flow dynamics-based sensing modality for direct environmental pathogen monitoring,” Chem. Eur. J, vol. 24, pp. 6025–6029. [DOI] [PubMed] [Google Scholar]