Abstract
The cortex organizes sensory information to enable discrimination and generalization1–4. Systematic representations of chemical odor space have not been described in olfactory cortex, and so it remains unclear how odor relationships are encoded to place chemically distinct but similar odors, like lemon and orange, into perceptual categories, like citrus5–7. Here we demonstrate that both the piriform cortex (PCx) and its sensory inputs from the olfactory bulb represent chemical odor relationships through correlated patterns of activity. However, cortical odor codes differ from those in the bulb: cortex more strongly clusters together representations for related odors, selectively rewrites pairwise odor relationships, and better matches odor perception. The bulb-to-cortex transformation depends upon the associative network originating within PCx, and can be reshaped by passive odor experience. Thus, cortex actively builds a structured representation of chemical odor space that highlights odor relationships; this representation is similar across individuals but remains plastic, suggesting a means through which the olfactory system can assign related odor cues to common and yet personalized percepts.
In olfaction, perception depends upon chemistry8. Chemically-related odors evoke similar percepts within and across individuals, suggesting that cortex harbors a conserved mapping from chemical to neural space that organizes information about odor relationships to ultimately support perception6,7. Odors are detected by broadly tuned receptors expressed by olfactory sensory neurons, whose axons project to the olfactory bulb (OB)9,10. Within the mouse OB, these axons are organized into thousands of discrete and spatially organized information channels called glomeruli, each of which represents the tuning properties of an individual odor receptor11. Odor information is reformatted by OB circuits before being transmitted to cortex; it is not clear whether or to what degree this peripheral transformation preserves information about odor chemical relationships12–14.
The main recipient of OB afferents is the piriform cortex (PCx)1; axons from OB projection neurons are broadly dispersed across the entire surface of the PCx, and individual PCx neurons respond to multiple, chemically-distinct odorants15–17. These observations suggest that neurons in PCx randomly sample sensory inputs from the OB18,19. Consistent with this possibility, individual odors activate ensembles of spatially-distributed PCx neurons that lack apparent topographical organization with respect to chemical space18,20,21. Feed-forward random network models (which posit stochastic connectivity between OB glomeruli and PCx neurons) predict that PCx odor representations should be pervasively decorrelated relative to the bulb, but that PCx should maintain the pairwise odor relationships present in the OB; these models further argue that cortical codes for odor relationships should be invariant across individuals, as peripheral representations of chemical relationships are largely determined by odor receptor specificity, which is encoded in the genome19,22–24.
However, in addition to receiving inputs from the OB, PCx neurons are linked through a dense web of excitatory interconnections, suggesting that olfactory cortex may act as an auto-associative network1,25. Such networks use Hebbian mechanisms to construct cell assemblies that encode information about stimulus relationships (like feature similarity or temporal coincidence) through correlated activity. In the case of PCx, auto-associative mechanisms are predicted to both increase generalization across chemically similar odors, and to render cortical odor representations sensitive to passive odor experience, thereby reshaping pairwise odor relationships inherited from OB inputs. While PCx exhibits characteristics consistent with both random and auto-associative networks, it remains unclear whether cortex systematically encodes information about odor chemical relationships; whether any such representation preserves or reshapes odor relational information conveyed by the OB; or whether cortical odor representations are primarily decorrelated (thereby favoring odor discrimination as predicted by random network models) or correlated (thereby favoring odor generalization as predicted by auto-associative models).
To address these questions, we used multiphoton microscopy in mice expressing GCaMP6s within PCx to assess neural activity both in the input-dominated PCx layer 2 (L2), and in the more associational layer 3 (L3, whose odor responses have not yet been described)26 (Extended Data Fig. 1). We took advantage of a library of odor descriptors that quantifies thousands of physiochemical features, such as molecular weight, polarizability and hydrophobicity5,27, to rationally design three sets of 22 odors each: a “global” odor set, which included structurally diverse odorants well separated in odor space; a “clustered” odor set divided into six odor subsets, each of which shared functional groups and other structural features; and a “tiled” odor set, in which the carbon chain length of a ketone, an ester, an aldehyde and an acid was incrementally varied (Fig. 1a and Extended Data Fig. 1, see Methods). Although each odor set captured progressively less chemical variance, by construction individual odors in the clustered set (within each of the six subsets) were most closely related, while odors were separated at intermediate scales in the tiled set. We noted that under anesthesia odor responses in L3 (and to a lesser extent L2) were attenuated or absent; recordings were therefore performed during wakefulness, a state in which L3 neurons were significantly more active (Extended Data Fig. 2, see Methods).
Figure 1. Systematically probing relationships between odor chemistry and cortical odor representations.

a, Global, clustered and tiled odor sets (see Extended Data Fig. 1e for odor identities and structures), depicted in principal component space (see Methods). Color indicates functional group associated with each odor. Variance of full odor space (gray dots) spanned by each odor set is indicated.
b, Example single neuron responses for the clustered odor set, representing the trial-averaged response of single neurons (rows) across 22 odors (columns). Rows are sorted using hierarchical clustering, with PCx L2 and L3 rasters sorted independently (see Methods).
c, Pairwise odor distances (Pearson’s correlation) for all odor sets based on chemical descriptors (see Methods). Rows and columns represent individual odors sorted using hierarchical clustering (ordering identical to that in Extended Data Fig. 1e). Color bars indicate functional groups associated with each odor.
d, Pairwise odor distances based on pooled neural population responses in PCx L2 and L3 (see Methods), sorted as in c. Pearson’s correlation coefficient between the chemical and neural distance matrices reported below each matrix (global: p<10−7; clustered: p<10−16; tiled: p<10−18); rs (shuffle) obtained by independently permuting odor labels for each neuron. Blue boxes highlight ketone-ester and ketone-acid relationships between chemistry and PCx L3.
e, UMAP embeddings of cortical responses to the tiled odor set. Each dot represents a population response for one odor presentation (7 per odor), color-coded as in d.
f, Fraction of total variance in each mouse (L3 activity) attributable to shared across-animal structure determined by distance covariance analysis (see Methods).
g, k-nearest-neighbor classification of odor identity in a held-out mouse using odor distances from other mice. Data are bootstrap mean ± SEM; grey bars indicate shuffle control on odor labels (see Methods). (Accuracy is greater in PCx. global: p<10−3; clustered: p<10−60; tiled: p<10−22, Wilcoxon Rank Sum, two-sided). b, d-g, are based on all responsive neurons (see Methods) pooled by layer across mice (n mice, neurons (L2/L3) for global: 3, (854/616), clustered: 3, (867/488), tiled: 3, (427/334); see Methods for subject-specific statistics).
All odors evoked selective excitation and suppression, with PCx L3 responses being denser, broader and more reliable than those in L2 (Extended Data Fig. 3). Odors evoked more correlated activity across the population of PCx neurons (i.e., ensemble correlations) than was expected by chance, with greater correlations observed in L3 compared to L2 (Fig. 1b, Extended Data Fig. 3). These findings raised the possibility that correlated odor-evoked responses among PCx ensembles systematically reflect chemical relationships among odor stimuli. To explore this possibility, correlation distance matrices were generated for each odor set based upon the physiochemical descriptors that characterize each odorant (Fig. 1c, see Methods). Odors in the global set were the least chemically correlated with each other, while odors in the clustered odor set exhibited substantial block diagonal structure, consistent with subsets of odors sharing key chemical attributes. Because molecules in the tiled set are related along two chemical axes (for example, heptanone and octanone differ by one carbon atom while heptanone and pentyl acetate differ by one oxygen atom), the matrix describing these odors exhibited periodic on- and off-diagonal structure.
Visual comparison and quantification demonstrated that odor chemistry and neural responses were only weakly related in the global odor set; in contrast, cortical odor responses maintained the block diagonal physiochemical correlation structure apparent in the clustered odor set, demonstrating that at close chemical distances PCx represents odor chemical relationships (Fig. 1d). Interestingly, neural responses to the tiled odor set (in which odor relationships are organized at intermediate chemical distances) reflected on-diagonal chemical relationships, but did not uniformly encode off-diagonal relationships. For example, cortex appeared to emphasize chemical similarities between ketones and esters, while de-emphasizing chemical similarities between ketones and acids (see highlighted blue boxes in Fig. 1d). Structured chemical-neural relationships were apparent on a trial-by-trial basis, and persisted for several seconds after odor offset; as has been observed previously under anesthesia, no spatial ordering of neurons was observed with respect to odor chemistry during wakefulness, consistent with response correlations alone conveying information about odor relationships (Extended Data Fig. 4)18,20.
Both UMAP embeddings and manifold alignment revealed that cortical odor relationships were similar across mice (Figs. 1e–f); indeed, information about pairwise cortical odor distances derived from one mouse could be used to predict the identity of a held-out odorant based upon odor distances measured in a different mouse, with better performance observed in L3 than L2 (Fig. 1g, see Methods). Lasso optimization was used to identify chemical features relevant to driving neural responses in each of the odor sets; identified descriptors captured physiochemical features such as molecular weight, electronegativity, polarizability, and hydrophobicity, suggesting that ensemble-level odor representations are driven by diverse aspects of odor chemistry (SI Table 1). Identified features that predicted neural activity for each odor set also improved the correspondence between all the other odor sets and their associated neural activity, demonstrating that information about odor chemistry gleaned from one experiment can be used to predict cortical responses in a different experiment carried out using a separate set of odorants (Extended Data Fig. 5).
The selective differences between odor chemical relationships and cortical activity apparent in the tiled odor experiment could reflect correlation structure present in OB inputs to PCx (consistent with feed-forward random network models), or instead could be generated by cortex (consistent with auto-associative models). However, to date it has not been possible to quantify odor-evoked responses across the complete array of OB glomeruli, preventing characterization of correlations in bulb inputs to PCx. We therefore introduced synaptically-targeted GCaMP6s into projection neurons spanning the OB, and imaged odor evoked activity in boutons in PCx layer 1a, where they synapse with L2 and L3 neurons; because the axons and boutons of all OB glomeruli are spatially distributed across the PCx15,16, each cortical field of view effectively samples glomeruli from the entire bulb (see Methods, Extended Data Fig. 6).
Odors from the tiled odor set evoked both excitation and suppression in boutons, whose responses were similar across mice (Extended Data Fig. 7). Correlation distance matrices revealed that bouton responses reflected information about odor chemical relationships (Fig. 2a); in addition, identification of physiochemical features that optimized the observed chemical-bouton relationships improved predictions of bouton responses to held-out odors as well as predictions of cortical responses to the tiled odor set (Extended Data Fig. 5b, SI Table 1). Thus — like cortex — OB projection neuron boutons encode information about odor relationships and chemistry.
Figure 2. Correlation structure differs in olfactory bulb and cortex.
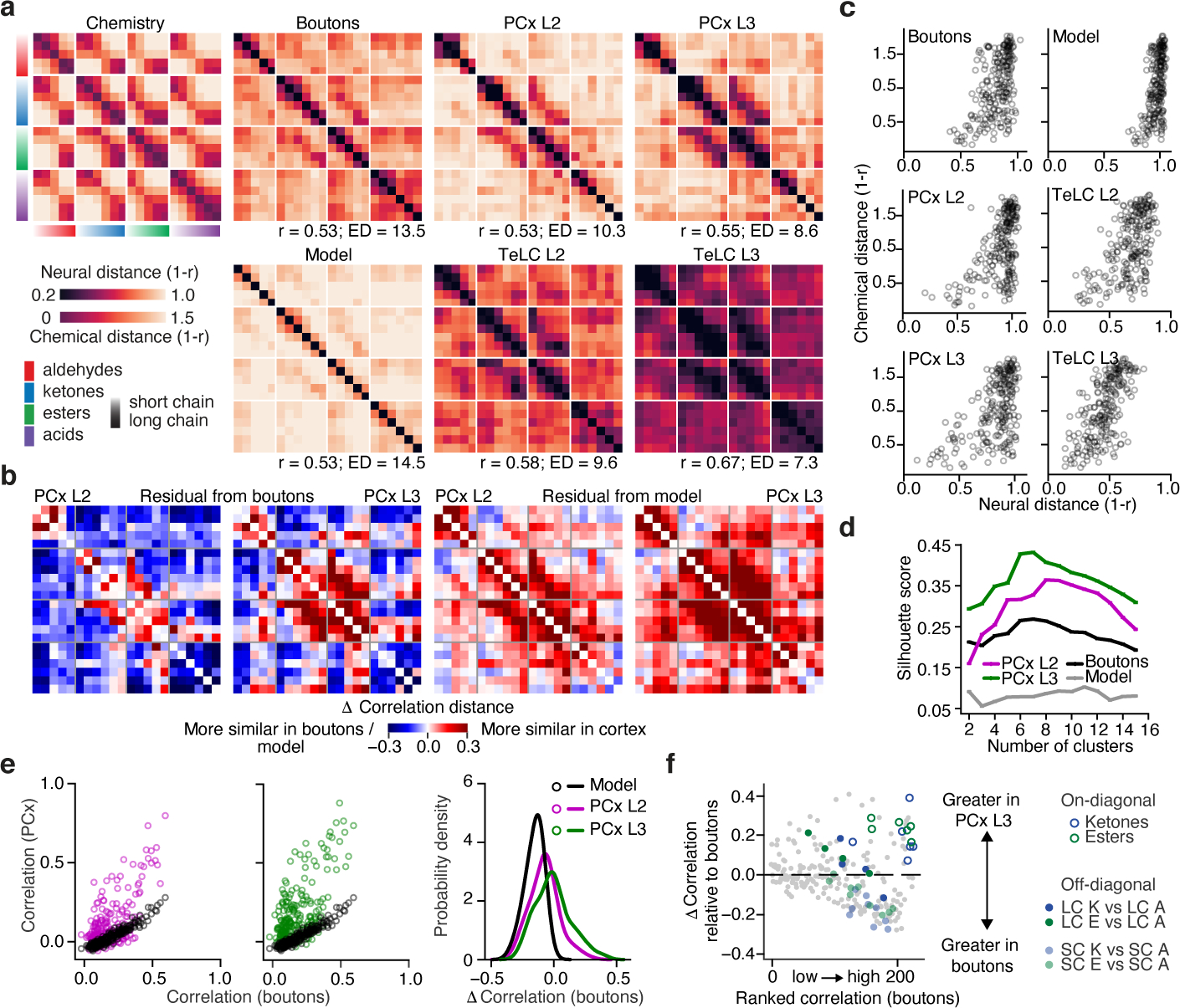
a, Correlation distance matrices for the tiled odor set across all conditions. (Top left), Distances obtained using chemical descriptors. (Right), Distances based on odor responses. Odor sorting as in Fig 1c. R values indicate Pearson’s correlation with odor chemistry (Boutons: p<10−17; PCx L2: p<10−17; PCx L3: p<10−19; Model: p<10−17; TeLC L2: p<10−21; TeLC L3: p<10−32; Shuffled Pearson’s r = 0.0 ± 0.063 SD, 1000 permutations on odor label). ED = effective dimensionality (see Methods).
b, (Left), Difference between PCx and bouton distances in a. (Right), Difference between PCx and random network model distances in a (see Methods).
c, Pairwise odor correlation distances based on neural responses plotted against corresponding chemical distances.
d, Silhoutte scores for clustered population responses (based upon Euclidean distances and grouped via k-means clustering) over a range of cluster sizes. Higher values indicate better clustering (see Methods).
e, Pairwise odor correlations in boutons and PCx predicted by the feed-forward random network model (see Methods) compared to observed correlations in PCx L2 and L3. (Right), Probability density distribution of differences between cortical (PCx L2 and L3) and input (boutons) pairwise odor correlations, superimposed on the distribution expected with the model (model vs L3: p<10−33, vs L2: p<10−17, Kolmagorov-Smirnov test).
f, Difference in pairwise odor correlations between PCx L3 and boutons (grey dots). Positive values indicate greater correlation in cortex. Odor pairs are ranked along the x-axis from least to highest correlation in the bouton data. Short-chain (SC) and long-chain (LC) comparisons between ketones (K), esters (E), and aldehydes (A) are color-coded as in legend.
However, odor responses in boutons and cortex exhibited distinct patterns of correlation with respect to chemistry, with the greatest differences observed in PCx L3 (Fig. 2a–b). While average levels of correlated activity were similar in boutons and cortex, the distributions of these correlations differed, with bouton representations exhibiting higher effective dimensionality; in contrast, PCx L3 odor responses were more clustered, more selectively structured, and exhibited both lower effective dimensionality and a wider dynamic range for representing close chemical relationships (Figs. 2a, 2c–e, Extended Data Fig. 7). The presence of these structured correlations in part reflected increased grouping of closely-related odorants, as representations for odors nearest each other in chemical space (i.e., on-diagonal correlation matrix relationships) were more clustered in cortex than in boutons (Figs. 2a, 2f). One exception to this trend were acids, which as a class were correlated in the OB but relatively decorrelated in cortex (Fig. 2a).
Odor relationships were also reshaped in cortex compared to those in odor chemistry and boutons. UMAP embeddings demonstrated that boutons largely organize odor information along a single axis that emphasizes chain length (again, with the exception of acids; see Methods); in contrast, odor information in PCx L3 was largely organized in two dimensions based on functional group (Fig. 3a). Similar functional group-based reorganization was observed via hierarchical clustering (Fig. 3b). Lasso optimization confirmed that boutons and cortex differentially weight chemical features related to chain length and functional group (Extended Data Fig. 5c).
Figure 3. Cortical odor responses reformat odor relationships inherited from the OB.
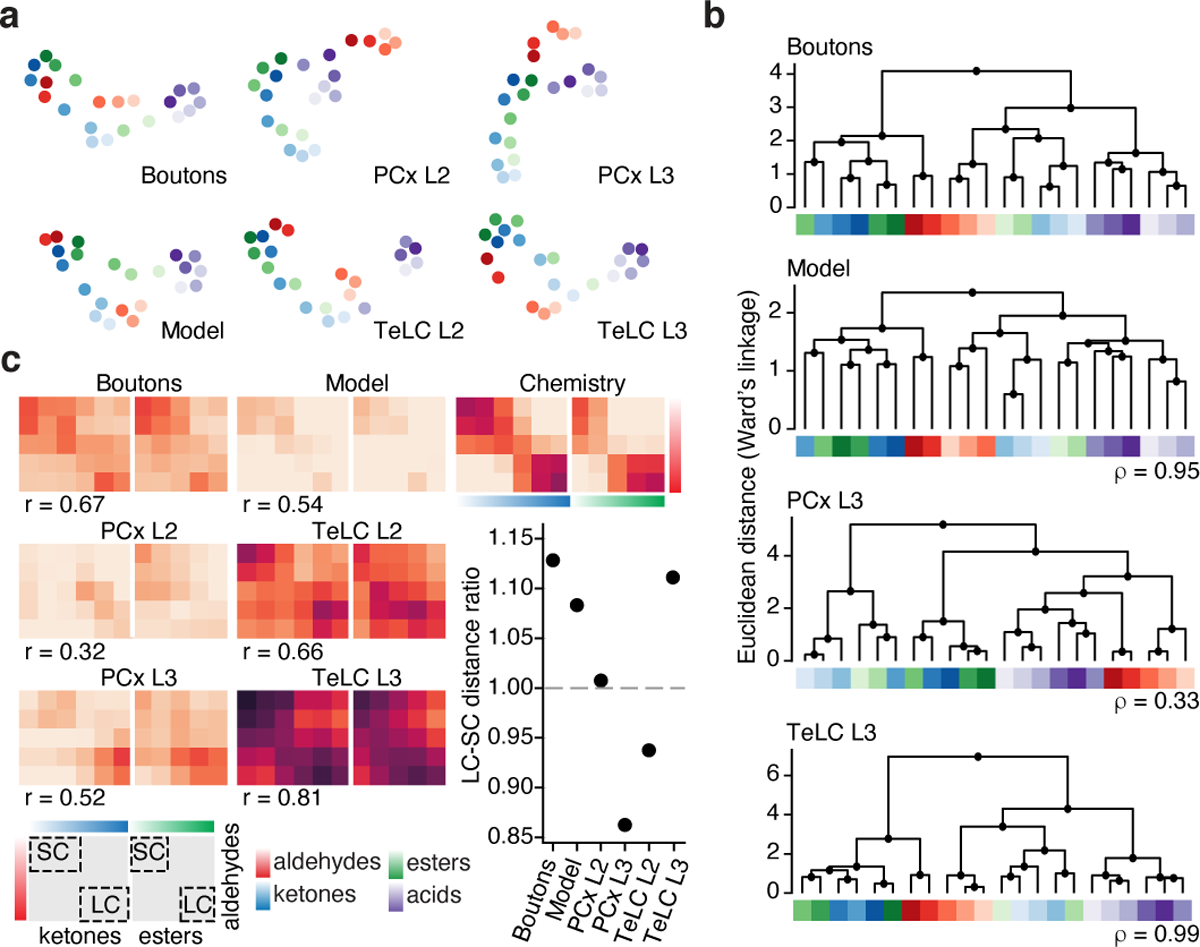
a, UMAP embeddings for all experimental conditions. Note that UMAP emphasizes relationships rather than distances, so these embeddings are similarly scaled (see Fig. 2a, Methods).
b, Hierarchical clustering of neural population responses; ρ values indicate clustering similarity to OB boutons (Spearman correlation on cophenetic distances between boutons and the other datasets).
c, Pullouts from panels in Fig. 2a depicting conserved and rearranged odor relationships between aldehydes, ketones and esters; inset: ratio of correlations between long-chain (LC) and short-chain (SC) comparisons (each dot indicates mean across odor pairs); r values indicate Pearson’s correlation to odor chemistry. For a,b, color code as in c.
Moreover, several pairwise odor relationships were reorganized in cortex on the basis of both chain length and function group. For example, in chemical space short-chain and long-chain odors with different functional groups were similarly cross-correlated; in boutons correlations between short-chain aldehydes and esters were emphasized while those between long-chains were diminished; and, in PCx L3 the opposite pattern was observed, with long-chain aldehydes and esters exhibiting stronger correlations and short-chains exhibiting weaker correlations (Fig. 3c). Chain-length dependent cortical reshaping of odor relationships was also apparent between aldehydes and ketones.
These differences in correlation structure suggest that PCx and boutons differentially encode information about odor identity and odor relationships. Linear decoders based upon cortical responses (particularly from PCx L3) were worse than OB-based decoders at predicting odor identity on each trial, consistent with bouton odor responses having a higher dimensionality (Fig. 4a). In contrast, cortex (particularly PCx L3) was on average better at encoding information about odor relationships (Fig. 4b); notably, however, OB was better at generalizing across short-chain ketones, aldehydes and esters while cortex was better at generalizing across the corresponding long-chains, consistent with the observed differences in correlation structure for these odor classes (Fig. 3c, 4c).
Figure 4. Cortical odor representations generalize across odors, are consistent with perception, and can be modified by experience.

a, (Left) Schematic depicting a linear SVM classifier trained to identify an odor associated with a held-out neural population response on a trial-by-trial basis. (Right) Decoding accuracy plotted against neural/bouton populations of different sizes.
b, Decoding analysis to quantify odor generalization; each line represents classifier confusion between any odor and all other odors, rank ordered by the degree of confusion.
c, Decoding accuracy of SVM classifiers predicting whether a held-out odor is a short-chain (SC) or long-chain (LC) molecule. The acid block was excluded for this analysis. Data are bootstrapped mean ± SEM across held-out odors and neural/bouton ensembles, (For a-c: tiled odor set, 22 odors; number of mice, neurons/boutons for PCx L2/L3 same as Fig. 1b, d–g, for boutons, 6 mice /3160 boutons. For b,c: 300 units, 100 bootstraps. See Methods for all decoding analyses).
d, (Left), Pairwise neural and behavioral odor distances from a cross-habituation assay for the tiled odor set (see Extended Fig. 8); ρ is Spearman correlation coefficient. Black line indicates regression fit (mean ± 95th % CI, 1000 bootstraps). Black circles are mean ± SEM across mice (n>3 for each comparison). (Right), Coefficient of determination (R2) based on short-chain:short-chain (SC) and long-chain:long-chain (LC) or all comparisons (median ± 66th % CI indicated, 1000 bootstraps. N=26 odor triplets; 122 mice across all conditions, see Methods for behavioral distance and odor identities).
e, (Left), Probability density estimates of cell-wise class preference index for naïve and passive odor exposure conditions, for neurons responding to at least one short-chain (SC) ketone or aldehyde (see Methods). (Right), Example z-scored fluorescence (and preference index) from neurons tuned to either SC ketones (cell 1), SC aldehydes (cell 2), or both (cell 3). Grey bars indicate odor onset.
f, Pairwise odor distances in PCx L3 from odor-naïve and odor-exposed animals. Passive exposure to the target mixture (short-chain (SC) ketones (K) and aldehydes (A)) specifically increased similarity between ketones and aldehydes, but not between control odor pairs SC K vs long-chain (LC) A and SC esters (E) vs SC acids (Ac) (see Extended Data Fig 10; *: p<0.002; n.s. middle: p=0.62; n.s. right: p=0.45, independent t-test, 2-tailed; number of mice / neurons for naïve: 3/334; exposed: 3/742).
Given these differences in information content, we assessed whether bulbar or cortical odor codes more closely correspond to perceptual odor relationships by measuring the innate perceptual similarity of odor pairs through a cross-habituation assay (Extended Data Fig. 8)28. Perceptual odor relationships better matched odor responses in PCx L3 than those in OB or PCx L2 (Fig. 4d); this closer correspondence to PCx L3 was particularly apparent for the short-chain:short-chain and long-chain:long-chain comparisons whose neural correlation patterns were inverted in bulb and cortex (Fig. 3c).
The transformation between bulb and cortex appears to reflect the combined influence of random network-type connectivity (which maintains odor relationships) and an auto-associative mechanism (which generally clusters and selectively reshapes odor relationships). To directly assess the contribution of random network-type connectivity to the observed cortical odor responses, bouton odor responses were passed through a previously-established feed-forward model in which simulated PCx neurons stochastically sample from multiple glomerular inputs19 (see Methods). Consistent with previous reports, the model predicted decorrelated cortical odor representations whose pairwise relationships were preserved relative to boutons (Fig. 2a). While cortical responses were in part consistent with model output — as many pairwise odor relationships were preserved — the model failed to capture the strong correlation structure present in cortex or the selective rewriting of pairwise odor relationships (Figs. 2a–e, 3).
To evaluate the relative influence of auto-associative mechanisms on cortical odor representations, we used an AAV-based method to express tetanus toxin light chain (TeLC) within PCx neurons; this approach blocks synaptic transmission and causes PCx to behave as if it largely receives feed-forward inputs29 (Extended Data Fig. 9). After attenuation of the associative network, single neuron odor tuning broadened, response densities rose, and odor correlations increased, consistent with known network-dependent recruitment of inhibition29 (Extended Data Fig. 9). Critically, after TeLC infection, cortical odor relationships more closely resembled those present in odor chemistry and OB axonal boutons as assessed via correlation matrices, UMAP clustering and hierarchical clustering; for example, the cortical restructuring of short-chain and long-chain odor relationships was abolished, as was the decorrelation among acids (Figs. 2a–c, 3).
Auto-associative networks are predicted to influence correlations among odor representations to reflect the coincidence of stimuli in the world; while reward-based experiments have revealed task-dependent changes in cortical odor relationships30,31, it has not yet been demonstrated that cortical odor correlations are sensitive to passive odor experience1,25. We therefore repeatedly exposed mice to a mixture of short-chain aldehydes and ketones (whose PCx L3 representations are relatively decorrelated, see Fig. 2a) before assessing cortical responses to the tiled odor set. Mixture experience specifically increased the cortical correlation between individual aldehydes and ketones and recruited single neuron tuning curves that reflected generalized responses to these specific odor classes (Fig. 4e–f, Extended Data Fig. 10). These observations demonstrate that cortical odor relationships can adapt to the statistics of the experienced odor environment.
Discussion
The olfactory system must synthesize information about chemical features to generate organized odor representations that support discrimination and generalization. Here we show that both OB boutons and cortex explicitly represent odor chemical relationships. The observation that many pairwise odor relationships are encoded similarly in these two brain areas is consistent with random connectivity models, which propose that PCx neurons stochastically sample glomeruli to generate a systematic population-level representation of odor chemical space19.
However, cortex differs from the bulb in two key respects. First, PCx better clusters odor representations, enabling it to preferentially signify odor relationships. Second, cortex reconfigures information about odor relationships inherited from the bulb — the cortex does not simply pool and normalize its inputs, but instead, in a network-dependent manner, actively builds an odor space to emphasize certain odor relationships and de-emphasize others; this re-writing is sensitive to odor exposure, which can recruit new single neuron tuning properties and modify odor relationships. The olfactory system, therefore, transforms a chemical feature space into a cortical space that represents stimulus relationships through correlated activity; the structure of this space reflects information inherited from the sensory periphery, the transformation imposed by cortical circuits, and the effects of sensory experience. The cortical grouping of representations for both structurally and temporally related odors suggests a mechanism for generalization across natural odor sources, which tend to emit related odor chemicals32. Future in vivo experiments will be required to understand how the instrinsic properties of PCx neurons and the associative network, which targets both excitatory pyramidal and inhibitory neurons, collaborate to transform odor representations.
In nearly all our analyses the correlation structure of L2 odor representations was intermediate between that observed in boutons and L3, which may reflect relative differences in the prominence of bulb inputs in L2 and associational connectivity in L31,26. Because the network that interconnects PCx neurons also sends centrifugal projections to the OB, it is likely that under normal circumstances this network influences both bulb and cortical representations of odor relationships12,29,33,34. Although the TeLC experiment demonstrates that network activity originating in PCx is required for the bulb-cortex transformation, PCx is recurrently connected to an array of higher olfactory centers that may also play a role in shaping odor relationships2. Importantly, neural representations in PCx L3 more closely match perception than those present in bulbar inputs, suggesting a functional hierarchy among interconnected olfactory brain areas.
Relationships among cortical odor representations depend upon chemical distances, such that at close distances information about chemical relationships is largely maintained, at large distances cortex decorrelates odor representations, and at the intermediate distances captured by the tiled odor set the olfactory system sculpts relational representations for odors in a manner that respects but reshapes chemical relationships. Our findings with the clustered odor set are reminiscent of prior work demonstrating that similar odor mixtures recruit overlapping ensembles of PCx neurons, although in those experiments chemical distances were not quantified3. Although here we take advantage of functional groups and chain lengths to systematically alter odor distances at intermediate scales, many distinct chemical features differentially contribute to odor representations in both the bulb and cortex4. The finding that treating odor chemicals as buckets of physiochemical features — rather than organizing information about chemistry along arbitrary dimensions — identifies structured chemical-neural-perceptual relationships is consistent with the longstanding model that the odor receptor repertoire broadly samples chemical feature space5,10,27.
The relational information present in PCx cannot, in and of itself, assign a given odorant to its unique odor quality: the mapping observed here potentially explains why lemon characteristically smells similar to orange, but fails to explain why lemon smells like lemon. In particular, it is unclear how cortical information about odor relationships might be aligned to enable lemon odor to evoke a similar percept across individuals. We propose that relational information in PCx (and possibly other olfactory areas) is translated into invariant information about odor quality by using universal points of reference, much like a compass can be used to orient a paper map to the cardinal directions19. These points of reference may arise from invariant properties of specific odor receptors, or from hardwired circuits in olfactory areas like the accessory olfactory nucleus or the cortical amygdala15,16,35. Alternatively, reference points could be learned from shared experience; in this model exposure to stereotyped odors (e.g., amniotic fluid, mother’s milk, feces, urine, food) or common objects (like actual lemons) could orient chemical-neural mappings along (largely) invariant axes across individuals. Additional work aimed at understanding the interaction between fixed and flexible features of olfactory circuitry will be required for a full account of the relationship between chemistry, experience and perception.
Methods
Ethical compliance
All experimental procedures were approved by the Harvard Medical School Institutional Animal Care and Use Committee (protocol number 04930) and were performed in compliance with the ethical regulations of Harvard University as well as the Guide for Animal Care and Use of Laboratory Animals.
Animals
Acute imaging of piriform cortex (PCx) was performed in 8–16 week old C57/BL6J (Jackson Laboratories) male mice. Imaging of cortical neurons was performed in mice harboring the Vgat-ires-Cre knock-in allele (Jackson Stock No. 028862) and the ROSA26-LSL-TdTomato cre reporter allele (Jackson Stock No. 007914); imaging of boutons was performed in mice harboring the Tbx21-Cre allele (Jackson Stock No. 024507, gift of C. Dulac). TeLC-dependent elimination of cortical excitatory transmission and subsequent imaging was performed in Emx1-IRES-Cre animals (Jackson Stock No. 005628). Male mice were group-housed prior to viral delivery of GCaMP6s and singly-housed for 1–3 weeks post-injection.
Viral constructs
To generate pAAV-hSyn-FLEX-TeLC-P2A-dTom, pAAV-hSyn-FLEX-TeLC-P2A-EYFP (a gift from Bernardo Sabatini) was digested with AscI and NheI to remove EYFP. A gene fragment (synthesized by IDT) containing dTomato with the SV40 nuclear localization signal was cloned into the TeLC backbone via isothermal assembly (NEB HIFI E2621). AAVDJ-hSyn-FLEX-TeLC-P2A-dTom virus was produced by Vigene Biosciences, with a titer of 1.5 × 1013 GC/mL. AAV PHP.eB hSynapsin1-FLEx-axon-GCaMP6s virus was produced as previously described36. In brief, HEK293T cells were co-transfected with pAAV-hSynapsin1-FLEx-axon-GCaMP6s (a gift from Lin Tian, Addgene plasmid #112010), PHP.eB rep-cap (a gift from the Viviana Gradinaru Lab/Clover center), and Ad helper plasmids. After 5 days, viral particles were harvested and then purified via iodixanol gradient ultracentrifugation. The virus was titered via qPCR; the titer of all batches was between 2–3 × 1013 VG/mL.
Stereotaxic viral delivery
Vgat-ires-Cre; ROSA26-LSL-TdTomato male mice were injected with an AAV expressing the genetically encoded activity indicator GCaMP6s (AAV1.CAG.GCaMP6s.WPRE.SV40, Penn Vector Core). Injections were targeted to posterior PCx using Allen Brain Atlas coordinates: ML: −4.2, AP: −1.09, DV: −4.25, from the dura. FLEX-TeLC-P2A-dTom was targeted to anterior and posterior PCx of both hemispheres of Emx1-IRES-Cre animals at AP: 0.39 and −1.0; ML: −3.51 and −4.2; DV: −4.4 and −4.1. To uniformly infect olfactory bulb projection neurons, AAV PHP.eB FLEX-axon-GCaMP was delivered intravenously via retro-orbital injections in Tbx21-Cre transgenic mice (Jackson Stock No. 024507), which express Cre recombinase in OB projection neurons.
Full-titer viruses (500 – 1000 nl) were delivered to cortex at 1 nl/second using a Nanoject II dispensing pump (Drummond Scientific). GCaMP6s-injected animals were imaged 1–3 weeks after delivery. In TeLC experiments, GCaMP6s was injected at ML: −4.2, AP: −1.09, DV: −4.25, from the dura, 2–3 weeks after TeLC delivery. Animals were imaged 3–5 weeks post-TeLC delivery. Imaging of OB projection neuron axon terminals in layer 1a of PCx was performed 3–5 weeks after retro-orbital injection. To assess the influence of passive odor exposure on cortical representations (see Passive odor exposures, below), odor exposure to mixtures was initiated 1–2 weeks after viral delivery; as with imaging in odor-naïve animals, imaging in odor-exposed animals was performed 3–4 weeks after injection.
Uniform infection of L2 and L3 cortical neurons by AAV-TeLC across the rostro-caudal extent of PCx was confirmed histologically. To validate TeLC dependent inhibition of cortical excitatory synaptic transmission, odor-evoked single-unit activity was compared between control and infected mice as previously described37.
Surgical approach and craniotomy
We developed a surgical preparation compatible with PCx imaging during wakefulness and semi-paralysis; this preparation is similar to those used in the past to explore in vivo neural responses without the use of general anesthesia (but with effective analgesia) during the experiment38–40. Prior to exposing PCx, mice were anesthetized with isoflurane, and head-fixed with dental cement to a rotating headpost. The PCx was then accessed from the ventral surface of the mouse skull through surgical resection of the zygoma, the mandible, and associated musculature and fascia. A 2mm craniotomy overlying the PCx was made using a dental drill and secured with a custom-shaped cranial window.
To ensure that the animal was free of pain and discomfort during wakefulness and semi-paralysis, full hemifacial analgesia was provided by performing a complete trigeminal nerve block. This procedure is designed to abolish sensation around the surgical exposure, as well as the ipsilateral oro-facial region encompassing the entire dorsoventral extent of the head and extending from the nostril to the neck. The junction of the four branches of the trigeminal ganglion was readily identified at the external pterygoid ridge, which was rendered accessible when the mandible was removed. A 1–5 μl, 0.2–1mg/kg dose of bupivacaine was injected directly into the stalk of the trigeminal nerve bundle using a calibrated micropipette mounted on a micro-manipulator. By infusing bupivacaine solution proximally to the trigeminal ganglion, distribution along all trigeminal branches, including the mandibular, ophthalmic, and infraorbital branches, was ensured. To verify that the block infiltrated the entire nerve bundle, each injection was supplemented with DiI, a contrast agent used to identify myelinated nerve fibers, due to its lipophilic, infiltrating nature41. By including DiI in the block solution and monitoring its diffusion through the nerve adjacent to the injection site, proper micropipette placement directly inside the nerve bundle was confirmed. Successful DiI injections were characterized by uniform distribution of dye through the trigeminal bundle proximal to the injection site. In cases of insufficient labelling of the nerve bundle, multiple injections were administered until the entire nerve was labelled by visual inspection. This procedure was extensively evaluated through measurements of heart rate (which revealed no signs of distress, see Extended Data Fig. 2)42, and by systematically probing the depth of analgesia through the use of needle pricks along the entire dorso-ventral and rostro-caudal portion of the head ipsilateral to the injection site.
Upon completion of the surgical exposure, induction of analgesia, installation of paralytic infusion and retro-nasal sniffing lines, as well as placement of the EEG electrode (see Semi-Paralysis, Artificial Sniffing, EEG for Assessing Brain State sections below), isoflurane anesthesia was discontinued and mice were transferred to the imaging set-up equipped with custom-built sniff generator, oxygen respirator, as well as a peristaltic pump for paralytic infusion.
Semi-paralysis
Mice were provided with a continuous infusion of a low dose of the muscle relaxant pancuronium bromide into the jugular vein during the imaging phase of the experiment39. After calibration, the final dose and infusion speeds were chosen to be 0.024 ng/kg/10 min. At this dose, mice experience a loss of righting reflex, but maintain diaphragmatic contractions and toe-pinch reflexes. Because this dose was chosen as to minimize paralysis (which is not required for analgesia), if movement was observed in the experiment intermittent pushes of pancuronium were provided to ensure motion-free imaging. We refer to this preparation as being in the condition of “wakefulness” (see EEG section below) as opposed to “awake” given that the mice are incapable of gross movements and cannot actively sample odors because of the ventilator (see Artificial sniffing below).
Artificial sniffing
In order to control for potential differences in odor coding due to changes in odor sampling, the mouse’s sniffing rhythm was replaced with an 8 Hz fixed inspiration/expiration cycle synchronized in time to odor presentation; this cycle rate mimics known sniffing rates during active odor sampling43,44. We adopted a previously developed method in which a cannula was placed into the nasopharynx via the trachea and subsequently attached to a solenoid valve which draws air bidirectionally across the nasal epithelium45. The tube was secured to the trachea with a pair of nylon sutures and doused with silicone elastomer for further stability46. The distal portion of the tube was then coupled to a computer driven solenoid valve and a vacuum line, providing 50 ms pulses of suction every 75 ms at a flow rate of 100 ml/min.
EEG for assessing brain state
Anesthesia is thought to induce a brain-state similar to slow wave sleep that is characterized by large-amplitude fluctuations in the 0.5–4 Hz range and the absence of high-frequency activity from 40–100 Hz, which is typically present during wakefulness or behavioral engagement. Because power in the slow and fast frequency bands of the EEG is anti-correlated across these brain-states, their ratio has been traditionally used to assign an absolute value to the arousal state of the animal47,48. To compare changes in brain state between wakefulness and anesthesia and associated changes in odor representation, after completion of awake imaging, some mice were subsequently anesthetized with a ketamine/medetomidine dose of 50mg/kg + 0.5mg/kg IP, and the imaging session was immediately repeated. For all experiments, EEG activity was recorded using a silver wire inserted into the dorsal anterior PCx via a 0.5mm craniotomy. A grounding wire was placed into the contralateral cerebellum. This signal was amplified using an AM Systems 1800 amplifier and digitized with a National Instruments PXie-6341 acquisition card. Signals were detrended and bandpassed (0.5–500 Hz) prior to computing the EEG power ratio.
Odor space design
A major goal of this study was to rationally design odor sets such that chemical similarities and differences between odorants in each odor panel could be explicitly titrated. As has been done previously, to describe odor space we took advantage of 2584 molecules commonly used in the flavors and perfume industries from http://www.thegoodscentscompany.com/49. A large fraction of these molecules are odorous, in that they are < 300 daltons in size and sufficiently hydrophilic and volatile to readily access the environment of the nasal epithelium. This collection contains structurally diverse molecules that vary in carbon chain length, weight, polarizability, hydrophobicity, cyclicity, branching, constituent functional groups, and other chemical attributes. To characterize the physiochemical features of the odors within this large odor collection, we took advantage of a database of 3705 statistical metrics designed to quantify different molecular physiochemical properties (Dragon, KODE Inc), including those related to molecular weight, volume, ionization potential, etc. Using this descriptor database, each of the 2584 collected molecules were represented by a vector containing 3705 values (where each value is a quantitative description of a specific physiochemical feature), thereby constituting an odor chemical space where odor similarity can be expressed as the Euclidean, cosine or correlation distance between any two molecules50,51. Of these features, 2522 are quantified in the Dragon database as continuous variables (e.g., molecular weight) and 985 were binarized or categorical (e.g., the presence or absence of a N atom). The collection of these inter-odor distances (which holistically capture the quantified physiochemical differences between each odor pair) can be converted into correlation distance matrices (see Chemical and activity distances, below).
Odor selection
Three (“global,” “clustered,” and “tiled”) distinct odor sets were identified, each consisting of 22 odorants. The global odor set contained structurally diverse molecules that span the entire odor space. The clustered odor set consisted of 6 groups of 3–4 molecules, where all the odors within each group share a chemical functional group (as well as other common chemical features); these groups were designed such that the odors that belong to each group were maximally separated from the odors belonging to all other groups. The tiled odor set included closely-related aliphatic molecules that systematically varied along two dimensions; the first was the number of carbon atoms in the chain, and the second was the particular functional group attached to the carbon chain (i.e., aldehydes, esters, ketones and acids, all of which are related to each other). Odor selection for the first two odor sets was performed with stochastic optimization (see Simulated Annealing) to prevent human-induced biases in odor set design. The cost function for the global odor set was designed to maximize separation between all 22 odors (by maximizing the minimum pairwise distance among selected odors). For the clustered odor set, within-group similarity and between-group dissimilarity was maximized by using the silhouette coefficient as the cost function.
Odor delivery
A 23-valve olfactometer that can deliver up to 22 odorants was used to present odors (Island Motion, Tappan, NY). The 23rd valve was used to deliver a blank stimulus (no odor) between odor presentations. Custom Arduino software was used to control valve opening and closing, thereby enabling switching between odor vials and the blank vial. This software also controlled the output of two mass flow controllers (MFC). The first MFC delivered a constant carrier flow at 0.8 L/min of purified air into a common channel; the second MFC supplied a constant flow at 0.2 L/min of clean air that was injected into an odor vial (see below) and then merged with the carrier flow 1 inch in front of the mouse’s nose. A larger exhaust fan drew air from the imaging cage that enclosed the rig to prevent cross-contamination.
Monomolecular odors were diluted in di-propylene glycol (DPG) according to individual vapor pressures obtained from www.thegoodscentscompany.com, to give a nominal concentration of 500 ppm. This vapor-phase concentration was further diluted 1:5 by the carrier airflow to yield 100 ppm at the exit port. Odor presentations lasted for two seconds and were interleaved by 30 seconds of blank (DPG) delivery. The order of presentation of odors was pseudo-randomized for each experiment, such that on any given trial, odors were presented once in no predictable order. Each odor was presented 7–10 times in each experiment.
Two-photon calcium imaging
High-speed volumetric imaging was performed using a 16kHz resonant galvo - regular galvo pair (Cambridge Technologies) housed in a custom-designed microscopy rig equipped with 2-inch optics. Acquisition was performed with a large working distance Nikon 16x objective (N16XLWD-PF, 0.8 NA, 3mm WD) mounted on a high-speed piezo actuator (nPoint 400). A Chameleon laser (Coherent) tuned to 930 nm delivered 50–120 mW of excitation power at the front end of the objective. Emitted fluorescence was detected using Hamamatsu H10770PA-40 PMTs. Scanimage 5 was used for hardware control and data acquisition.
For imaging neuronal cell bodies, acquisition volumes spanned 210 μm in the Z axis across PCx L2 and L3. Volumes were split into 6 optical slices each spanning 35 μm of cortex. Volumes were positioned such that 2 slices resided in L2 and 4 slices resided in L3. This allowed us to monitor similarly sized populations of neurons in L2 and L3 given the approximately 3-fold lower cell density of L3 in posterior PCx52. We typically discarded a single optical slice that spanned the boundary between layers to avoid any cross-contamination between layers. For axonal imaging, a single plane was acquired in PCx L1 at 60 Hz and subsequently downsampled by averaging to match the neural acquisition rate.
Note that our imaging fields are in the most anterior portion of posterior PCx. The degree of associational connectivity is known to systematically vary across the anterior-posterior axis of the PCx, with the least associational and most feed-forward connectivity anteriorly53,54. We chose to image in the “middle” of the PCx both because of anatomical constraints in our imaging field in the 2P configuration, and to ensure the representations we probed would include both feed-forward and associational connectivity. We would expect that if we imaged more anteriorly, we would observe representations that were progressively more “bulb”-like (given the relative predominance of inputs), and conversely that posterior PCx would deviate more strongly from the bulb (given the relative predominance of associational connectivity); because of surgical constraints, addressing this possibility will require the future development of alternative means of accessing anterior and posterior PCx both pre- and post-synaptically.
Data inclusion criteria
For experiments involving the global, clustered and tiled odor sets in odor-naïve animals, data was analyzed from 3 animals per odor set. For the passive odor exposure experiment, data was analyzed from 3 animals. For bouton imaging, data was analyzed from 6 animals. All animals that satisfied the following pre-determined criteria were included in the study: imaging volumes spanning both piriform cortical layers 2 and 3 (layer 1a for boutons) could be imaged continuously for the duration of the experiment; in each cortical layer, at least 150 GCaMP6s-labelled neurons could be identified (500 axonal boutons for axonal imaging); odor-evoked activity persisted over the course of the entire imaging session; and field-of-view drift and motion artifacts could be fully corrected with post-hoc image registration. Given the nature of this population imaging study, study sample size was not pre-determined, the experiments were not randomized, and the investigators were not blinded to study conditions.
Signal extraction
ROI detection, segmentation, and extraction of fluorescence signal was performed using the Suite2p software55. This package implements image registration, neuropil fluorescence correction and fluorescence source detection from spatially overlapping ROIs. To accommodate differences in ROI size between axonal boutons and somata, the expected ROI size paramater was set to five microns for axonal boutons and 12 microns for somata.
auROC-based detection of odor responses
Analysis was only performed on neurons that responded, in a statistically significant manner, to at least one odorant. To identify such neurons, we computed the area-under-the-receiver-operator-curve (auROC) statistic for each cell-odor pair. The auROC metric represents the probability that a neuron’s response, chosen at random from all presentations of the same odor, will be ranked higher than a randomly chosen sham response obtained using baseline activity. A value of 0.5 indicates no difference between a neuron’s activity during baseline and odor presentation. A value of 1 indicates a perfectly distinguishable excitatory response, while a value of 0 indicates a perfectly distinguishable suppressed response. For a single neuron and all presentations of a single odorant, the classifier was provided with the mean fluorescence obtained from 2-second time windows immediately flanking odor onset. A null distribution of auROC values for each cell-odor pair was constructed by randomly permuting the identity of the odor and baseline periods on each presentation. This procedure was repeated 1000 times. The actual auROC value was deemed significant if it resided outside the 1–99th percentile of the null distribution. Neurons that did not display a significant response to any odors, according to auROC analysis, were excluded from all subsequent analysis. Of those neurons imaged, the fraction of retained neurons (and the absolute number of neurons in each dataset) were: global L2 = 854 neurons, 82 percent; global L3 = 616 neurons, 89 percent; clustered L2 = 867 neurons, 87 percent; clustered L3 = 488 neurons, 85 percent; tiled L2 = 427 neurons, 59 percent; tiled L2 = 334 neurons, 52 percent; TeLC tiled L2 = 435 neurons, 51 percent; TeLC tiled L3 = 590 neurons, 68 percent; boutons tiled = 3160 boutons, 68 percent. Note that the number of neurons deemed responsive by auROC analysis is proportional to the extent to which each odor set captured chemical diversity, with the greatest number of responsive neurons observed in the global odor set, and the fewest observed in the tiled odor set. This distribution of responsive neurons (between 51 and 89 percent, depending upon the chemical diversity in each odor set) is consistent with prior work characterizing response densities and tuning breadths in PCx.
Gaussian mixture model for response type clustering
Cell-odor response type clustering was performed for visualization purposes only. Trial-averaged response time-courses spanning the odor presentation period were dimensionally reduced by PCA to capture 90% of the variance in the data and served as the input to a gaussian-mixture model, with the optimal number of clusters was assessed using the Bayesian Information Criterion.
Lifetime and population sparseness
Lifetime sparseness is a metric reflecting the tuning breadths of individual neurons, with neurons specifically tuned to small numbers of stimuli exhibiting a lifetime sparseness of close to 1; population sparseness is a metric reflecting the density of responses among a population of neurons to a set of stimuli, with less dense responses (i.e., fewer neurons or boutons responding to a stimulus set) exhibiting a population sparseness of close to 1. To determine a neuron’s odor-selectivity the lifetime sparseness metric was computed according to56:
where rj is the positive odor-evoked change in fluorescence to an odor j relative to baseline and averaged over multiple odor presentations, and N is the number of odors (22 in all odor sets). Inhibitory responses were zeroed (for this analysis only). Lifetime sparseness reflects the kurtosis of a neuron’s tuning profile and ranges from 0 to 1. Highly peaked, narrow tuning profiles yield values close to 1 and represent neurons that respond strongly and selectively to few odors. Values close to 0 indicate equal responsiveness to a large fraction of the odor set. Population sparseness for each odor was calculated using the same formula used for lifetime sparseness, but in this case, j indexes a neuron instead of an odor.
Signal and ensemble correlations
The extent to which any two neurons have similar odor preferences can be assessed by computing the Pearson’s product moment correlation between their trial-averaged odor response profiles (tuning curves). This is typically referred to as a “signal” correlation. Each neuron’s tuning curve was represented as a vector containing N elements, where N is the number of odors in the stimulus set. Each entry in this vector corresponds to the odor-evoked change in fluorescence relative to baseline and averaged over all presentations. For each neuron, responses across odors were z-scored. Populations were defined as all neurons that responded to at least one odor according to the auROC analysis.
We also wished to compute the similarity in odor responses exhibited by different odor-evoked ensembles of PCx neurons or boutons. We refer to this herein as “ensemble” correlation. In these analyses, we computed the Pearson’s correlation between the population responses to every pair of odors in our panel. Each odor’s population response was represented as a vector containing N elements, where N is the number of neurons/boutons in the population. Each entry in this vector represents a single neuron’s/ bouton’s trial-averaged response to the corresponding odor. For each neuron/bouton, responses across odors were z-scored. Populations were defined as all neurons/boutons that responded to at least one odor according to the auROC analysis.
Chemical and activity distance
Pair-wise odor distances (1- Pearson’s r) in neural activity space were computed between odor vector pairs where each matched vector entry corresponded to a single neuron’s trial-averaged response to the corresponding odor. This procedure was applied to neural populations from individual mice or to pseudo-populations of neurons built by pooling all responsive neurons from all mice for each layer and odor set. In chemical space, correlation distances between odor pairs were computed identically, except each vector entry (matched across odors) represented the odor-specific value assigned by a physiochemical descriptor. For presentation purposes, distance matrices were sorted using hierarchical clustering. For the global odor set, all odors were sorted collectively. For the clustered odor set, odors were sorted within each functional class first followed by sorting on functional classes. For the tiled odor set, functional group classes were sorted, but odors within each class were ordered according to increasing chain length. Row and column ordering of all activity and chemistry distance matrices is preserved across figures. Note that for all correlation analyses, both inhibitory and excitatory responses were included.
UMAP Embedding
For visualizing odor relationships in neural data, population responses were embedded in 2 dimensions using UMAP (uniform manifold approximation and projection for dimensionality reduction)57. Selection of optimal embedding settings was accomplished by minimizing the mean-squared error between correlation distance matrices built from data projected on the UMAP dimensions and those corresponding to the input data. Simulated cortical responses from the feed-forward models were processed in a similar manner. Because UMAP imposes an arbitrary rotation on projected data, each embedding was aligned to a reference using the orthogonal Procrustes transformation. For embeddings of pseudopopulation data for the tiled odor set across boutons as well as neural and simulated cortical data, bouton data served as the reference. For aligning embeddings obtained from individual mice, the orthogonal Procrustes transform (rotation and reflection only) was performed iteratively across animals in a pairwise manner58. Note that these embeddings are meant to visualize odor relationships (and are complemented by quantitative metrics); pairwise relationships cannot be changed by any of the rotations used herein to align embeddings to each other.
Distance-based nearest neighbor decoding and classification
To test whether PCx odor relationships are invariant across individuals, we asked whether we could identify any given odor from one mouse based upon the pairwise odor relationships observed in other mice. To decode odor identity based on odor relationships, nearest neighbor classifiers were trained on odor distances from 2 animals and tested on odor distances obtained from a held-out animal, such that each animal was tested once. For each such classifier, 100 bootstrap iterations were performed. For each iteration, correlation distance matrices were constructed using a randomly sampled neural ensemble containing 50 neurons. In each condition, each distance matrix represented all pairwise correlations between trial-averaged population responses (see Chemical and activity distance, above), such that any given training or testing odor was represented as a vector containing 21 pairwise distances. For a single run of the classifier, reported accuracy represents the fraction of odors that were correctly identified.
Distance covariance analysis
Distance covariance analysis (DCA) belongs to a set of statistical methods that seek to identify shared dimensions of variability between two different datasets. DCA – an extension of canonical correlation analysis (CCA) – was developed for identifying related dimensions of activity across two or more populations of neurons59. To measure the similarity of cortical odor relationships between individual mice, DCA was performed on response data (neurons by odor-trial) from all individual animals exposed to the same set of odors (code supplied with the reference). The output consisted of a set of orthogonal dimensions (one set per animal) and associated DCA statistics ranked from highest to lowest contribution to common activity between individuals. Each dimension was evaluated for significance by permutation testing. The null distribution of DCA statistics was constructed by shuffling the sequence of odor responses across all neurons in each animal in order to destroy between but not within-animal relationships. A dimension was deemed significant if its associated statistic was higher than the 95th percentile of the null distribution built from 100 permutations. Three to six dimensions were typically retained. Because DCA is not deterministic, this procedure was subjected to 100 independent restarts. The reported results correspond to the best modeling run. The fraction of an individual’s neural variance that could be explained by the shared embedding was determined by calculating the total accounted variance after regressing each neuron’s activity on the set of DCA dimensions.
Silhouette Coefficient
The degree of clustering in odor correlation distance matrices was evaluated using k-means clustering and the Silhouette coefficient. Correlation distances were computed between trial-averaged population responses to all 22 odors in the tiled odor set. Correlation distance matrices containing all pairwise odor distances were projected onto 21 principal components and subjected to k-means clustering. For this set of labelled data, the silhouette coefficient assigns a single value ranging from −1 (overlapping diffuse clusters) to 1 (compact, well-separated clusters) that represents the average silhouette score computed on an odor-by-odor basis: for an odor i, its score – Si, is defined as (bi-wi) / max(wi, bi) where wi is the average Euclidean distance between odor i and all other odors with the same class label, and bi is the average distance between odor i and all odors in the next nearest class. Qualitatively similar results between experimental conditions were obtained by running k-means and cluster evaluation directly on full population response data or by obtaining k-means labels from full population data and computing the silhouette coefficient using these labels on PCA embeddings of correlation distance matrices.
Effective dimension
Effective dimension (ED) of a population of neurons, a quantity reflecting the number of principal components required to capture the odor-evoked neural variance, was defined as previously described60,61. Briefly, for each experimental condition and for model cortical activity, ED was quantified from trial-averaged population responses (neural data only) after mean-centering units across odors. ED reflecting variance in similar ensemble sizes were calculated as averages across randomly chosen, 300-unit ensembles (100 bootstraps).
Hierarchical clustering
Dendrograms depicting the reconfiguration of odor relationships across boutons, PCx, TeLC, and the feed-forward random connectivity model PCx were constructed directly from correlation distance matrices associated with each experiment. First, each correlation matrix was projected, using PCA, onto K dimensions where K is the number of dimensions required to explain 95% of the variance in correlation distances. The resulting embedding expresses each odor’s contributions to the prominent similarity/dissimilarity modes in the original correlation distance matrix. Dendrograms were built by hierarchically clustering this data using Euclidean distance and Ward’s linkage. Clustering similarity between dendrogram pairs was assessed using the Cophenetic correlation coefficient after topologically aligning each pair.
Feed-forward connectivity model
To assess whether the observed cortical odor responses are expected under a random feed-forward model we simulated the OB-PCx network using a previously-established model62. In our implementation of this model (which hews as closely as possible to the published model), the OB and PCx layer contained 1000 and 100,000 units respectively, feed-forward connections were assigned randomly and can be either excitatory or inhibitory, and each cortical neuron was “innervated” by a random 20% of excitatory OB units and a random 40% of inhibitory units. Excitatory and inhibitory connection weights were set to 1 and −0.5 respectively, providing each PCx unit with balanced E/I innervation. Odor evoked activity in the OB layer was simulated from a log-normal multivariate distribution defined by population-mean response amplitudes and covariance obtained from bouton activity. Model PCx neurons linearly summed their inputs and are zero-rectified. The odor-average response fraction of model PCx units was adjusted to 8% to match the fraction of excitatory responses observed in PCx (detected by auROC analysis) on average across all odors and subjects.
Decoding analysis
Linear support vector machine (SVM) classifiers were trained to predict either odor identity or odor class (based on chain-length) in the tiled odor set on the basis of odor-evoked population activity.
For odor identity discrimination (Fig. 4a), all neurons/boutons that responded to at least one odor in the tiled odor set (according to auROC analysis) were pooled to build three pseudo-populations of neurons/boutons (boutons, L2, L3). Z-scored responses of a population of up to Nn randomly selected neurons/boutons (the maximum common number of neurons recorded across the layers) were then considered, given t presentations of j odors as a matrix X with Nn rows (neurons/boutons) and t × j columns (trials/instances × odors/classes). Each column of this matrix was thus a vector of Nn responses, one for each neuron/bouton in response to a given odor in each trial. Each decoding session started with a split of the matrix X into a training and test set: the training set included 0.9 × t randomly chosen trials for each class and the test set comprised the 0.1 × t held out trials for each class (i.e., a standard 9:1 training:testing split). This procedure, which is instantiated as part of the standard LIBSVM library (http://www.csie.ntu.edu.tw/~cjlin/libsvm/), allows us to use a binary classification algorithm (such as an SVM) to compare multiple classes. In any given experiment, the train-test procedure was iterated 100 times (with training and test data randomly chosen on each iteration) to cross-validate classifier performance. For differently sized subpopulations of neurons/boutons, a randomly selected subset of neurons/boutons was used for each cross-validation cycle, and at the end of this procedure the outcomes of each individual iteration were averaged to generate a measure of classification accuracy across all restarts; this is the overall measure that is reported in the main text (Fig 4a). The hyperplanes for each classifier were determined using the LIBSVM library with a linear kernel, the C-SVC algorithm, and cost c. Cost c is the only free parameter for a linear kernel, and it was found by a grid search on an initial dataset including 50 randomly chosen neurons/boutons from each dataset in order to maximize the accuracy of the decoder’s classification.
For Fig. 4b, classifiers were trained on 21 of 22 odors in the tiled odor set, with all trials associated with any training odor assigned to 1 of 21 classes. SVM class predictions for each held-out odor were converted to confusion probabilities (the probability that any given held-out odor is associated with any other of 21 odors) using the Python scikit-learn library63 implementation of Platt Scaling64. Class probabilities for each tested odor were rank-ordered prior to averaging across all odors.
For Fig. 4c, classifiers were trained to predict the class (either short-chain (SC) or long-chain (LC)) of a held-out odor after training on a single SC-LC odor pair. All odor trials were presented on each train-test iteration and accuracy was determined as the fraction of correctly labelled held-out trials. For each randomly chosen subpopulation of neurons, on each of 100 restarts, training and testing was performed on all possible SC-LC odor pairs and associated held-out odors. Cross-validated generalization accuracy corresponds to the average performance across all restarts and folds of the data. The SC class contained aldehydes: propanal, butanal, and pentanal; ketones: propanone, butanone, and pentanone; esters: ethyl and butyl acetate. The LC class contained aldehydes: heptanal and octanal; ketones: hexanone, heptanone and octanone; esters: pentyl and hexyl acetates. The acid block is excluded for this analysis.
Lasso optimization
For finding small, optimal combinations of Dragon descriptors that predict neural odor relationships, an L1-regularized optimization routine was designed to maximize the correlation between matched odor-pair distances across chemical and neural spaces. During each step of optimization (‘L-BFGS-B’ gradient descent), descriptor weights (0 bounded) were modified and chemical distances were recomputed. The optimization objective sought to minimize the residual sum of squares between the modified descriptor distances and corresponding neural odor distances. The Lasso component set the sparseness of the final solution and was selected for each model by cross-validation. Models were trained and validated on individual odor sets containing 22 molecules with 5-fold cross-validation (random splits) such that on any split of the data, 17*(17–1)/2 odor pairs made up the training set and 5*(22–5) odor pairs comprised the validation set. For assessing generalization to other odor sets, models were retrained with all 22 odors prior to testing. Since the global and clustered odor sets share odors, overlapping odors were removed from training when cross-applying to the held-out odor set. For within-odor set cross-validation and cross-application to the tiled odor set all odors were included. For Extended Data Fig. 5c, we sought to determine the relative contribution of the full set of descriptors belonging to the “molecular properties” block of the Dragon database to bouton/cortical relationships; for this analysis, optimization was performed without imposing L1-regularization.
Simulated Annealing
SA was used for odor set design, as well as for predictive modeling of neural odor relationships. Simulated annealing (SA) is a well-validated Monte Carlo sampling variant designed for stochastic optimization65. SA optimization works by slowly decreasing a pre-specified cost function over the course of sampling, thereby enabling good initial coverage of solution space and progressive convergence on a global optimum. All SA routines were implemented using the open-source Python package simanneal available at https://pypi.python.org/pypi/simanneal. For odor set selection, the number of features was reduced, via PCA, such that the transformed odor space accounted for 95% of the original variance. Optimization for the global and clustered odor sets (see Odor selection) was carried out using Euclidean distance in this reduced space.
In addition, our findings using Lasso optimization were verified using SA; here, the SA objective was designed to identify small sets of Dragon physiochemical features describing a set of molecules such that molecular distances in chemical space were maximally correlated with corresponding odor distances in neural space. Qualitatively similar findings using SA were observed as reported in the manuscript using Lasso optimization (data not shown).
Cross-habituation for assessing perceptual odor similarity
C57 males (5–6 week-old) obtained from Jackson labs were housed on a reverse light schedule for 48 hours before beginning behavioral experiments. Established procedures for assessing odor similarity were slightly modified66. Twenty-six pairwise comparisons were obtained, with 12 odorants serving as the first odorants in each comparison:
Butanal vs. pentanal or propyl acetate
Butanoic acid vs. heptanoic acid or butanone
Butanone vs. butanoic acid or propyl acetate or butanal
Heptanal vs. octanal, or pentyl acetate
Heptanone vs. pentyl acetate or heptanal or hexyl acetate or octanal
Hexyl acetate vs. pentyl acetate or octanone or octanal or octanoic acid
Octanal vs. butanal or heptanal or octanone
Octanoic acid vs. octanal
Octanone vs. octanoic acid
Pentyl acetate vs. hexyl acetate
Propanoic acid vs. butanoic acid
Propyl acetate vs. butanone or butanal or butanoic acid.
Short-chain pairwise odor comparisons, different functional groups:
(butanone, butanal)
(butanone, butanoic acid)*
(propyl acetate, butanone)*
(propyl acetate, butanal)*
(propyl acetate, butanoic acid)
Long-chain pairwise odor comparisons, different functional groups:
(heptanal, pentyl acetate)
(octanal, octanone)
(heptanone, pentyl acetate)
(heptanone, heptanal)
(heptanone, hexyl acetate)
(heptanone, octanal)
(octanone, octanoic acid)
(pentyl acetate, hexyl acetate)*
(hexyl acetate, octanone)
(hexyl acetate, octanal)
(hexyl acetate, octanoic acid)
(octanoic acid, octanal)
Remaining pairs:
(heptanal, octanal)
(octanal, butanal)
(butanal, pentanal)
(propionic acid, butanoic acid)
(butanoic acid, heptanoic acid)
Because three odors were presented to each mouse, two adjacent odor pairs were included in analysis from each animal. Presentation order effects were considered by swapping the order of any given triplet in different experiments. For some comparisons the same pair was presented at different positions in the triplet as indicated above with an asterisk. Investigation time was scored manually, using video footage obtained during each experiment. Scoring was done blinded to experimental conditions, and with no knowledge of odor identity. Odor investigation was defined as periods of orienting to the odor source on the half of the cage containing the odor source as well as by stereotyped bouts of sniffing and associated head-bobbing.
Perceptual similarity (behavioral distance) between 2 odors was defined as the difference in time spent investigating the first odor in a pair during its last presentation and the investigation time associated with the first presentation of the subsequent odor (see Extended Data Fig. 8). In Fig. 4d, Linear regression was used to relate behavorial to neural distance. Because behavioral distance increases monotonically but not necessarily linearly with neural distance, we also used Spearman ρ, which measures correlation based on ranks and is less restrictive than linear regression. Three to twelve mice were used for each pairwise comparison (eight mice on average per triplet experiment). Each mouse was used for a single set of odor comparisons.
Passive odor exposure
C57 males (5–6 week-old) were housed on a reverse light schedule for 48 hours prior to behavioral training. Group-housed mice were subjected to daily odor exposures for a period of two weeks. On each training session (30 minutes; 3 times per day, 14 consecutive days) mice were simultaneously presented with two short-chain aldehydes (propanal, butanal) and two short-chain ketones (propanone, butanone) for 1 minute separated by a 5-minute inter-stimulus-interval. Odor delivery was designed to closely approximate odor presentation during cortical imaging. Briefly, odors were delivered to the home cage using a custom-built olfactometer consisting of an activated-carbon purification unit, master air flow controllers, and a valve bank coupled to 4 odor vials and 1 blank vial. Flow rates for carrier and odor lines were set to 0.8 and 0.2 L/min respectively. Monomolecular odorants were diluted in di-propylene glycol (DPG) according to individual vapor pressures to yield a final concentration of 100 ppm at the output of the olfactometer. During odor delivery, odorants were mixed in air phase by simultaneous opening of all 4 valves. During the inter-stimulus-interval, air was passed through the blank odor vial containing only DPG. Mice were not subject to this protocol on the day of imaging.
Measuring changes in cortical odor representations after mixture exposure
Cortical representations of all 22 odors of the tiled odor set were obtained after cessation of behavioral training. Single neuron and population representations of the target aldehydes – propanal and butanal – and target ketones – propanone and butanone – were compared to data obtained from odor-naïve animals exposed to the tiled odor set. Off-target control comparisons were made between chain-length matched esters and acids (ethyl acetate, propyl acetate vs. propanoic acid, and butanoic acid) as well as between target ketones and off-target long-chain aldehydes (heptanal, octanal).
For comparing changes in response profiles of individual neurons across the target classes, a class preference index was assigned to each neuron using ROC binary classification. Each neuron’s trial-averaged responses were labelled as either aldehydes or ketones, resulting in a single auROC value reflecting the degree of discriminability between classes. The class preference index was obtained by rescaling auROC values from a range of 0 to 1 to a range of −1 to 1 with 0 representing low discriminability. Because the class preference index combines both the magnitude and frequency of responses, a neuron with weak preference for a single class could be strongly and uniformly responsive or non-responsive to the target odors. We therefore limited analysis to neurons exhibiting at least one response to any short-chain aldehyde or ketone of magnitude greater than 2 SD above the mean response across all 22 presented odors. Changes to odor similarity at the population level were assessed by comparing the correlation distance between odor pairs across classes. Because differences in the average magnitude of ensemble correlations (which are not relevant to the pairwise restructuring we focus on herein) may uniformly bias comparisons between experimental conditions, prior to computing pairwise correlation distances (see Chemical and activity distance), the trial-averaged tuning profile of each neuron was initially centered on its mean).
Descriptor relevance
The chemical descriptors in SI Table 1 identified by Lasso optimization afford an algorithmic representations of chemical structure. However, each descriptor incorporates some information about semantic molecular properties, such as molecular weight, electronegativity, polarizability, ionization potential, molecular volume, and hydrophobicity. Each descriptor’s relevance, obtained from the (Dragon, KODE Inc) website https://chm.kode-solutions.net/products_dragon_descriptors.php, is presented next to the descriptor name.
Statistical Tests
For comparing two normal independent distributions, the Student’s t-test (two-sided) was used. For comparing two independent distributions when normality cannot be assumed, significance was assessed by permutation testing or using the Wilcoxon Rank Sum test (two-sided). The Kolmogorov-Smirnov test was used to determine equivalence between two distributions. For testing significance of a single statistic against null distributions obtained by permutation, the true value was deemed significant if it resided outside the 5–95th percentile of the null statistic distribution. Error bars refer to either 95th CI, standard error of the mean, or SD, as indicated in the figure legends. For regression modeling, confidence intervals are computed over bootstraps (with replacement) of the data. For establishing correspondence between 2 distance matrices, Pearson’s product moment correlation was used on the upper diagonal of each set of measurements.
Extended Data
Extended Data Figure 1. Volumetric population imaging of PCx L2 and L3 during wakefulness using rationally designed odor sets.

a, (Left), Cartoon of volumetric multiphoton imaging approach to characterize odor responses in PCx in wakeful, semi-paralyzed mice (see Methods). (Right), Approximate position of an imaging volume (green dotted line) in a typical experiment superimposed on a Nissl-stained coronal section through PCx. Scanning volumes were oriented to acquire similarly-sized cortical populations in L2 and L3 (red dotted lines), despite decreased neuron density in L3 (see Methods). Imaging was performed in the most anterior portion of the posterior PCx.
b, Sample fields of view for a single imaging session. PCx L2 is depicted on top; PCx L3 on bottom. Segmentation masks associated with each layer are shown on the right.
c, Global, clustered, and tiled odor sets superimposed on the collection of odors constituting odor space as defined by principal components analysis (see Methods). Global odors are indicated by black dots; tiled and clustered odor sets via the indicated color code
d, Plot of the amount of molecular variance contributed by each additional principal component for each odor set in descriptor space; this analysis reveals that each odor set tiles odor space at a distinct level of resolution.
e, Molecular structures and associated PID signals of the odors comprising the global, clustered, and tiled odor sets. These PID traces are shown to illustrate the controlled kinetics of the olfactometer only; because detector reports depend upon ability of an odor to be photo-ionized, the relative amplitudes of the traces between odors are not meaningful. For example, heavy aliphatics elicit a minimal PID response because their photo-ionization energies lie outside the range of the detector; however, odors with low/absent PID traces still induced cortical activity in 5–20% of the imaged population, consistent with effective odor delivery. Five odors are shared between the global and clustered odor sets. These are indicated by bold lettering (and in c, as black circles with colored edges). Color code as in c.
Extended Data Figure 2. Odor responses in cortex are substantially altered by anesthesia.

a, (Left), EEG power spectral density plot from an individual subject depicting differences in cortical state between ketamine-medetomidine anesthesia and wakefulness (see Methods). Under anesthesia, the EEG signal is enriched in the delta band (0.5 – 4 Hz) at the expense of high frequency (40–100 Hz) gamma oscillations; in contrast, gamma frequency predominates over delta activity during wakfeulness. (Right) Summary of differences in EEG power content expressed as delta/gamma ratio during anesthesia and wakefulness averaged from 4 subjects. Error bars indicate SEM.
b, Comparison of the fraction of responsive neurons (obtained from the population of neurons that respond to at least 1 odor during the wakefulness, see Methods) to the tiled odor set in the same field of view (obtained from PCx L2 and PCx L3) during the awake state and under anesthesia. Responses are defined according to auROC analysis (see Methods). Each dot represents a single odor (L2: 504 neurons, L3: 418 neurons).
c, (Top), Black trace represents heart rate (average over 10 second, non-overlapping windows) recorded from an awake mouse in the home cage. Blue traces are example raw heart rate (HR) signal indicating the range of HR fluctuations observed during the awake state. The high variability in heart rates (which span ~350 to ~650 beats per minute) reflects ongoing behavior in the awake mouse. (Bottom), same as in the top panel, but for HR recorded during wakefulness and after induction of ketamine-medetomidine anesthesia (see Methods). Grey arrow indicates time of induction. Grey and Red rectangles and associated inset traces are 20-second segments of real-time heart-rate signal. During wakefulness, fluctuations in heart rate remain within a physiologically normal range of 300–500 beats per minute, without any detectible episodes of tachycardia (see Methods). Periodic dips in the recorded heart rate during wakefulness reflect moments when pharmacological agents are being administered, which briefly interrupts the heart rate monitor.
Extended Data Figure 3. PCx L3 neurons exhibit denser, broader and more reliable odor responses than neurons in PCx L2.
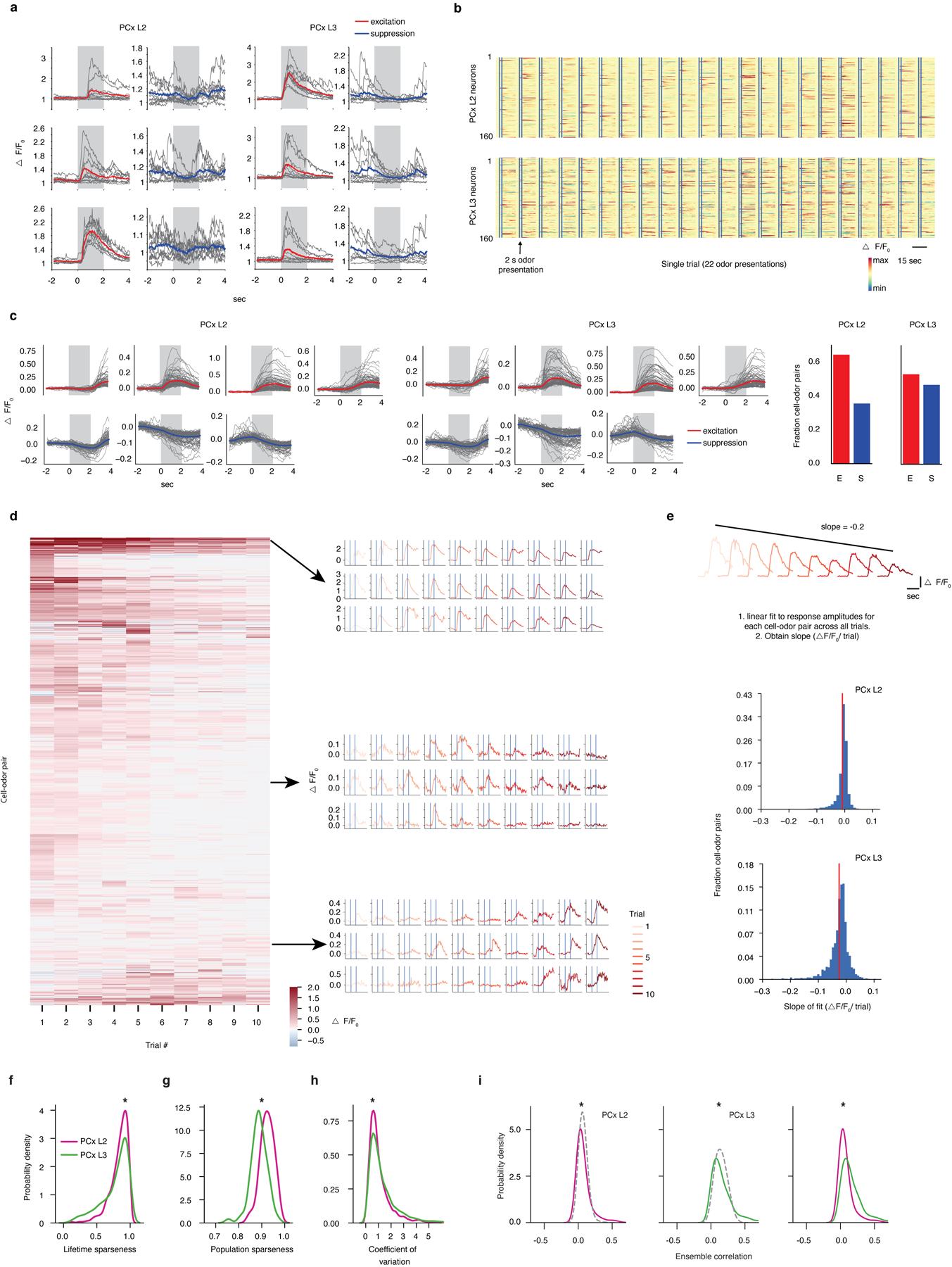
a, Examples of odor-evoked excitation and suppression in PCx. Each panel corresponds to a single cell-odor pair. Grey lines represent individual trials. Colored overlays represent trial-mean activity. Shaded grey rectangles delimit the odor presentation period.
b, Trial-averaged population response raster depicting odor-evoked activity in response to 22 odors (global odor set) across L2 and L3. Responses are ΔF/F0 with redder colors indicating excitatory transients and bluer colors indicating odor-evoked suppression. x-axis is time; double vertical bars delimit 2-second odor presentation periods.
c, Response types observed in L2 and L3 (clustered odor set). Individual panels correspond to clusters identified using a gaussian mixture model (see Methods). Grey traces correspond to trial-averaged cell-odor pairs. Colored overlays represent mean response time-course associated with each cluster. Right: fraction of all cell-odor pairs exhibiting excitation or suppression.
d, Response amplitudes of cell-odor pairs obtained from PCx L3 depicted on a trial-by-trial basis. Each row represents a given neuron’s response to 10 consecutive presentations of the same odor. Neurons are sorted hierarchically using average linkage and correlation distance. Despite the presence of some habituation in response to multiple presentations of the same odorant across the experiment, habituation does not appear uniform across the neural population nor does it appear to dominate neural responses to odors. Different groups of neurons were identified with maximal responses to an odor peaking at different times across the experiment; see examples depicted on the right. Each row of traces corresponds to a single cell-odor pair.
e, At the population level, odor responses do not uniformly habituate across the experiment. (Top), Cartoon depiction of procedure for determining change in response amplitude over the course of the experiment for a single cell odor pair. (Middle and Bottom), pooled data for all cell-odor pairs, sorted by layer. Red lines correspond to distribution means (clustered odor set).
f, Lifetime sparseness distributions (used to quantify tuning breadth, see Methods) in L2 and L3 across all experiments (1 = perfectly odor selective, 0 = completely non-selective, * = p<0.01, permutation test on layer label). Distributions are built using all responsive neurons (significant response to at least one odor by auROC analysis) pooled by layer across all experiments (here and throughout, global: n = 3 mice, L2 = 854 neurons, L3 = 616 neurons; clustered: n = 3 mice, L2 = 867 neurons, L3 = 488 neurons; tiled: n = 3 mice, L2 = 427 neurons, L3 = 334 neurons).
g, Population sparseness distributions (used to quantify response density, see Methods) in L2 and L3 (1 = few neurons active overall, 0 = all neurons active overall to an equal level. * = p<0.01, permutation test on layer label).
h, Probability density distributions of coefficient of variation for all significant cell-odor pairs identified with auROC analysis. (* = p<0.01, permutation test on layer label).
i, Probability density distributions of ensemble correlations (i.e., pairwise correlations between odor-evoked ensembles) between trial-averaged population odor responses in L2 (left) and L3 (middle). Dashed control curves indicate the distribution of ensemble correlations after shuffling odor labels independently across neurons. Ensemble correlations were determined independently for each animal, and subsequently pooled (* = p<0.01). L3 exhibits greater correlations at the population level than L2 (right) (* = p<0.01, permutation test on layer label).
Extended Data Figure 4. Cortical odor representations are stable from trial to trial and not chemotopically organized.

a, (Left), Pairwise odor chemical correlation matrices for the global, clustered and tiled odor sets. Rows and columns are sorted according to the chemical similarity between odors as assessed by hierarchical clustering (see Methods). (Middle and Right), Pairwise correlation distances of single-trial, population representations for odors in the global, clustered, and tiled odor experiments in PCx L2 and L3 (and boutons for the tiled odor set). Rows and columns are sorted according to the chemical similarity between odors as on (Left). Chemical color code (x and y axis labels of matrices, indicating functional group associated with each group of molecules) is shown in the legend. R values indicate Pearson’s correlation to odor chemistry.
b, (Top), Structured odor relationships persist from trial to trial over the course of the experiment. Blue line represents the similarity of two correlation distance matrices built from population responses obtained on consecutive trials. Grey dashed line indicates mean across all trial-pair comparisons (10 trials, 9 trial pairs; clustered odor set, L3). (Bottom), chemistry-based odor relationships correspond to matched cortical relationships obtained on a trial-by-trial basis. Dashed grey line represents the similarity of chemical and neural activity distances on a trial-by-trial basis.
c, Correspondence between odor structure in PCx L3 (clustered odor set) and odor chemistry using 3 different distance metrics (correlation distances, Euclidean distances and cosine distances). Distance matrices calculated from population activity are obtained using instantaneous ΔF/F0 over 130 ms increments (F0: baseline fluorescence averaged over a 1 second sliding window). Vertical lines delimit the 2 second odor presentation.
d, Odor chemical relationships emerge within a few hundred milliseconds after odor onset and persist for several seconds after odor offset (see Extended Data Fig. 1e for associated PID traces.
e, Example PCx L2 and L3 FOVs from a single animal with each responsive neuron colored according to its preferred odor in the clustered odor set. Neurons preferring odors belonging to different classes (legend) appear spatially intermingled in both L2 and L3.
f, Contour plots of pairwise signal correlations, plotted with respect to distance in L2 and L3 for the clustered and tiled experiments. Darker colors indicate increased density (see margin distributions). Pearson’s r is overlaid and indicates no spatial organization of odor representations in PCx.
Extended Data Figure 5. Lasso optimization identifies parsimonious sets of chemical descriptors that predict neural odor relationships.

a, (Left), Descriptors identified through training on one odor set also improve Pearson’s correlation (r) between corresponding chemical and neural distances for held-out sets of odors (G = global, C= clustered, T=tiled. A value of 1 in the matrix corresponds to no improvement from baseline Pearson’s r after optimization. Baseline chemical-neural correlation is 0.22 for global; 0.48 for clustered; 0.37 for tiled (see SI Table 1 for optimal descriptor sets). (Right), Reduction in mean-squared error (MSE) between chemical and neural odor pair distances for held-out odor sets (indicated below the x-axis) after training on a single odor set (indicated above). Note that the 5 odors in common between the global and clustered odor sets (names in bold case in Extended Data Fig. 2e) were discarded when evaluating performance on held-out data. The chemical features learned from the tiled odor set improved chemical-neural Pearson’s correlations in the clustered odor experiment but not the global odor experiment, consistent with the odors belonging to the tiled set covering only a limited region of chemical odor space (left). However, despite the limited chemical overlap between the tiled and global odor sets, training on the tiled odor set still improved the correspondence between odor chemistry and neural responses for the global odor set as assessed by a reduction in MSE (right).
b, Identifying a subset of chemical descriptors (from the original superset used to define odor space) using Lasso optimization on odor distances improves the correspondence to cortical activity (see Methods, SI Table 1). Training data was derived from the bouton dataset, and testing was performed for bouton responses to held-out odors within the tiled odor set, and also to cortical responses of the tiled odor set; Data is mean ± SEM over cross-validation folds.
c, The same procedure as in b was performed on a limited subset of 15 semantically-relevant descriptors that comprise the “molecular properties” block of the Dragon database; these descriptors include metrics that reflect molecular properties associated with functional groups (e.g. donor or acceptor atom surface area), molecular weight (e.g. van der Waals molecular volume) or a combination of both, like “hydrophilic factor,” and reflect the main axes of diversity in the tiled odor set. Most descriptors enriched in the olfactory bulb covary with molecular weight (red descriptors). Most descriptors enriched in PCx reflect the combined presence of a charged atom and variable number of carbon atoms along the aliphatic series of the tiled odor set (blue descriptors). Note that these descriptors differ from those identified when querying the entire Dragon set using Lasso optimization (SI Table 1), as this limited set of targeted descriptors (selected because their semantic meaning is transparent) may not afford optimal predictions over neural data.
Extended Data Figure 6. Functional imaging of OB axons in PCx via axonally-targeted gCaMP6s.
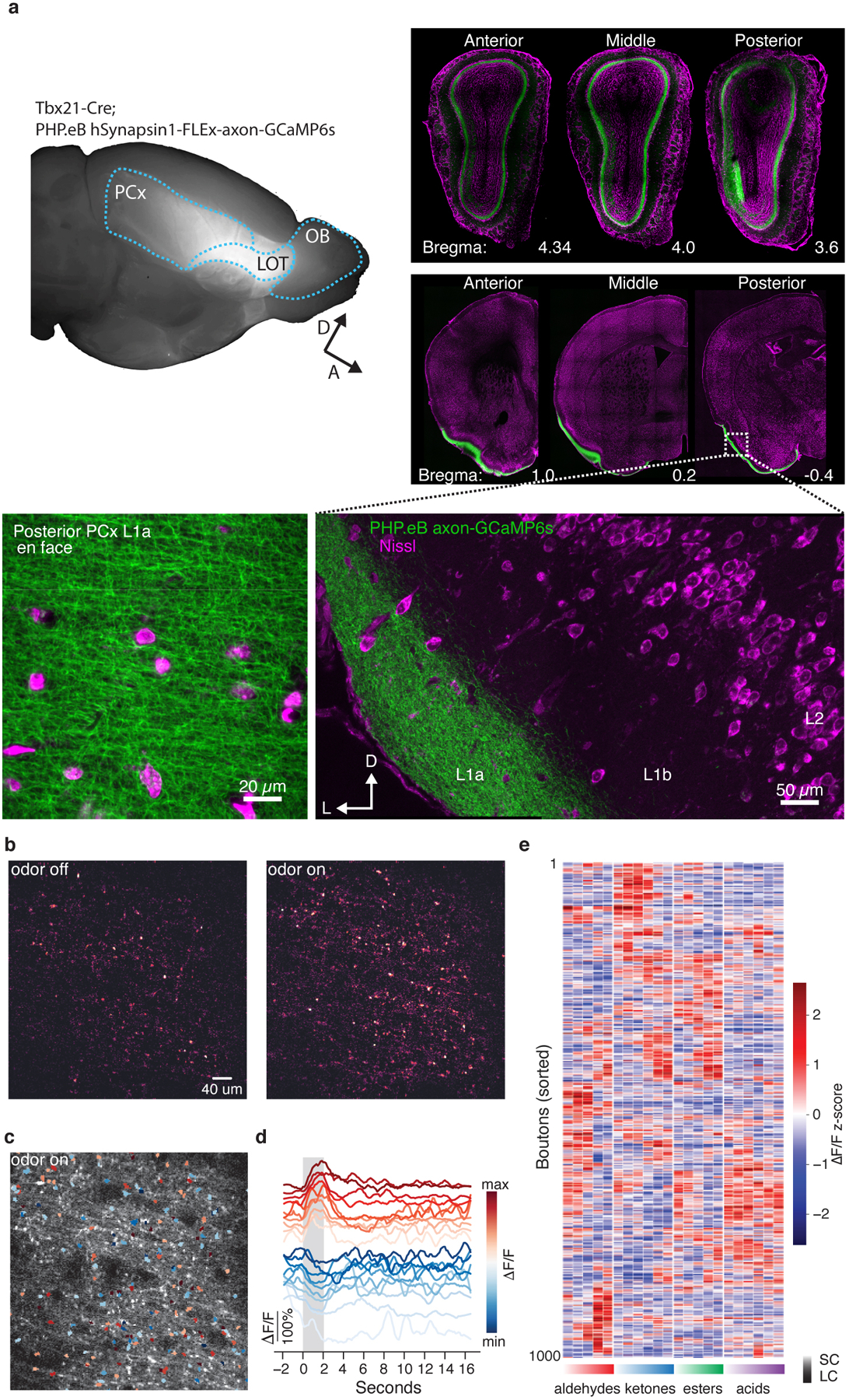
a, (Left), Whole-mount depicting Tbx21-Cre dependent expression of AAV PHP.eB hSynapsin1-FLEX-axon-GCaMP6s in OB projection neuron axons. GCaMP6s fluorescence is broadly distributed across piriform cortex. (Right), coronal sections depicting GCaMP6s signal (green) in the mitral cell layer across the entire anterior-posterior extent of the olfactory bulb and cortex. (Inset, bottom), GCaMP6s labelled axons shown coursing through PCx layer 1a. (Bottom left), En face image of layer 1a depicts dense and uniform distribution of axonal boutons.
b, Difference heatmap of a typical field-of-view (FOV) depicting baseline and odor-driven fluctuations in GCaMP6s signal. The strongest activation (light color) is associated with axonal boutons.
c, Time-averaged fluorescence signal of FOV in b. Overlay: segmented ROIs corresponding to axonal boutons depicting increases (red) or decreases (blue) in fluorescence, averaged over multiple presentations of a single odor from the tiled odor set.
d, Example average fluorescence from several boutons in a. Grey bar indicates odor delivery period, scale bar indicates response amplitude. For clarity, fluorescence time-courses for each example bouton are offset along the y-axis.
e, Example bouton responses for the tiled odor set. Each row represents the trial-averaged response of a single bouton for two seconds during and after odor exposere (columns) depicted as z-scored ΔF/F0; rows are sorted hierarchically using correlation distance and average linkage. The functional group and carbon chain-length associated with each odor are indicated below each column; light-to-saturated gradient indicates progression from short-chain (SC) to long-chain (LC) odors. Note that, as has been observed previously for OB projection neurons, boutons exhibit a substantial amount of odor-driven suppression.
Extended Data Figure 7. Bouton odor response properties.

a, Probability density distributions for boutons, PCx L2, and PCx L3 for signal correlations.
b, (Left), Same as in a, but for ensemble correlations. (Right), for the top 5% most similar odor pairs identified in boutons, correlation for the same odor pairs in PCx. Ensemble responses in both PCx L2 and PCx L3 exhibit stronger similarity than boutons.
c-d, Probability density distributions for boutons, PCx L2, and PCx L3, for lifetime and population sparseness.
e, Cumulative neural variance explained with increasing numbers of principal components, indicating relatively higher dimensionality in boutons compared to PCx (i.e., more uniform distribution of variance across principal components).
f, Probability density distributions for boutons, PCx L2, and PCx L3 for coefficient of variation representing trial-to-trial response variability across cell-odor pairs. These data demonstrate that observed odor responses in boutons are more reliable than similar responses in cortex. For a-f only the tiled odor set is used. For lifetime sparseness, 1 = perfectly odor selective, 0 = completely non-selective. For population sparseness, 1 = few neurons responsive, 0 = all neurons equally responsive. Distributions are built using all responsive neurons/boutons (significant response to at least one odor by auROC analysis; Boutons: 3160 ROIs across 6 subjects, PCx L2: 427 neurons across 3 subjects. PCx L3: 334 neurons across 3 subjects). (* indicates significant difference between boutons and either L2 or L3: a, vs L2 p<10−27; vs L3 p=0.02; b, vs L2 p<10−20; vs L3 p<0.005; c, vs L2 p<10−9; vs L3 p=0.93; d, vs L2 p<10−7 vs L3 p<10−4; f, vs L2: p<10−20; vs L3: p<10−23; two-sided Wilcoxon Rank Sum test for all comparisons).
g, Single-trial Z-scored ΔF/F0 for 1000 boutons recorded in PCx L1a during presentation of 22 odors belonging to the tiled odor set indicated by black lines. Redder colors indicating excitatory transients and bluer colors indicating odor-evoked suppression.
h, Response types observed in boutons (tiled odor set). Individual panels correspond to clusters identified using a gaussian mixture model (see Methods). Grey traces correspond to trial-averaged bouton-odor pairs. Colored overlays represent mean response time-course associated with each cluster. Blue vertical lines mark periods of odor presentation.
i, Fraction of total odor-driven bouton variance in each individual animal that can be attributed to the shared across-animal structure as quantified by distance covariance analysis (see Methods).
Extended Data Figure 8. Habituation-dishabituation test for assessing perceptual similarity of odor pairs.

a, (Left) Mice presented with novel odors exhibit investigation that diminishes over multiple consecutive presentations of the same odorant. Subsequent presentation of a perceptually different odor reinstates investigation while presentation of a similar odor has little effect. The extent to which two odorants are perceptually related is assessed by the magnitude of rekindled interest in the second odor after habituation has occurred to the first.
b, Investigation times for two different odor triplets. Data is mean ± SEM, (n=7 and n=8 mice, respectively). After habituation to heptanal, investigation of the closely related octanal (1-carbon difference) does not significantly increase. Presentation of butanal following habituation to octanal (4-carbon difference) induces greater investigation. For the second triplet, presentation of heptanal following habituation to heptanone (0-carbon difference, different functional group) induces greater investigation, while subsequent presentation of octanal following habituation to heptanal (1-carbon difference, same functional group), induces much less investigation.
Extended Data Figure 9. Inhibition of the associative network through cell-autonomous expression of tetanus toxin light chain in excitatory PCx neurons.
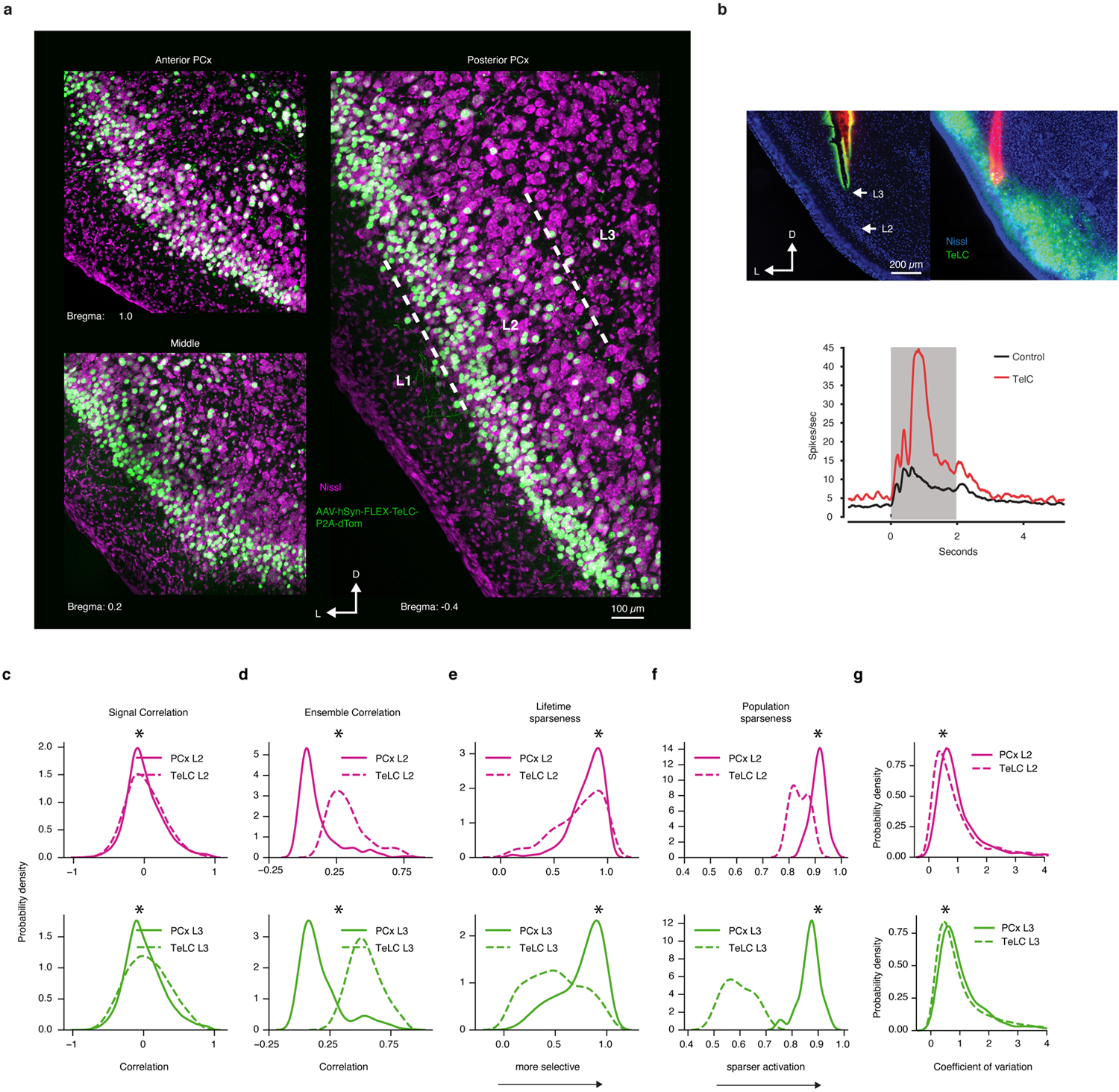
a, Uniform infection of excitatory pyramidal neurons in PCx L2 and L3 with AAV-hSyn-FLEX-TeLC-P2A-NLS-dTom in an Emx1-Cre mouse.
b, (Left), Coronal section through PCx indicating placement of recording electrode. (Right), Single-unit odor-evoked activity (grand-average of all excitatory responses deemed as significant by auROC analysis) in Emx1-cre mice expressing TeLC or wild-type controls. Disruption of cortical recurrent excitation enhances odor-evoked excitation, consistent with disruption of feedback inhibition. Grey bar indicates odor presentation (n = 121 cell-odor pairs from two Emx1-cre mice expressing TeLC; n = 229 cell-odor pairs from four mice).
c-g, Probability density distributions for the TeLC experiment for signal and ensemble correlations, lifetime and population sparseness, and coefficient of variation, constructed as in Extended Data Fig. 7, here only for the tiled odor set. For lifetime sparseness, 1 = perfectly odor selective, 0 = completely non-selective. For population sparseness, 1 = few neurons responsive, 0 = all neurons equally responsive. Distributions are built using all responsive neurons (significant response to at least one odor by auROC analysis; TeLC L2: 435 neurons across 3 subjects. TeLC L3: 590 neurons across 3 subjects. PCx L2: 427 neurons across 3 subjects. PCx L3: 334 neurons across 3 subjects). (* TeLC is significantly different from PCx L2 or L3: c, L2 p<10−8; L3 p<10−198; d, L2 p<10−46; L3 p<10−55; e, L2 p<10−05; L3 p<10−37; f, L2 p<10−7 L3 p<10−8; g, L2: p<10−10; L3: p<10−4; two-sided Wilcoxon Rank Sum test for all comparisons).
Extended Data Figure 10. Passive odor experience modifies odor relationships.
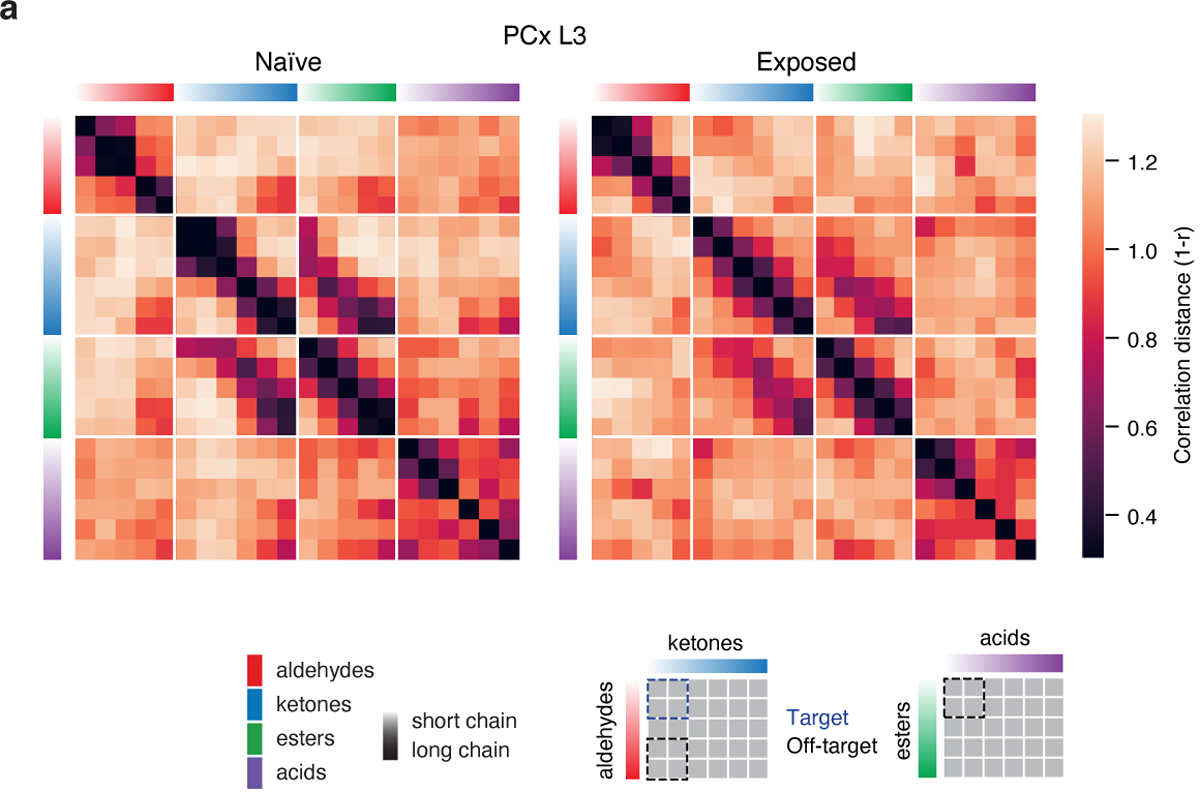
a, Correlation distance matrices for the tiled odor set obtained from odor-naïve (same data as in Figs 1–4) mice as well as mice passively exposed to a target mixture of two short-chain aldehydes and two short-chain ketones in the home cage (see Methods, Fig. 4 e–f). Passive experience with the mixture increases odor similarity specifically between mixture components (target comparisons indicated in the legend in blue), but not between target ketones and long-chain aldehydes or short-chain esters and short-chain acids with which mice had no prior experience (off-target comparisons indicated in legend in black, naïve: 334 neurons, n=3 mice; exposed: 742 neurons, n=3 mice).
Supplementary Material
Acknowledgements
We thank members of the Datta lab, Jonathan Pillow, Richard Axel, Larry Abbott, Ashok Litvin-Kumar, Carl Schoonover, Andrew Fink and Vanessa Ruta for helpful comments on the manuscript, Sigrid Knemeyer for technical illustrations, and Neha Bhagat for laboratory assistance. We thank Natasha Mathur for assistance with viral delivery of GCaMP6s. We thank Andrew Giessel for early development of tools for image acquisition and analysis. We thank Ofer Mazor and Pavel Gorelik from the Research Instrumentation Core Facility and Ludo Cacheux for engineering support. Core facility support is provided by NIH P30 grants HD18655 and NS072030. SRD is supported by fellowships from the Vallee Foundation, by grants RO11DC016222 and U19 NS112953 from the National Institutes of Health and by the Simons Collaboration on the Global Brain.
Footnotes
Authors declare no competing interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Data and code availability
All data and code will be posted to Github or made available upon reasonable request (www.github.com/dattalab).
References
- 1.Haberly LB Parallel-distributed processing in olfactory cortex: new insights from morphological and physiological analysis of neuronal circuitry. Chemical Senses 26, 551–576 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Courtiol E & Wilson DA The Olfactory Mosaic: Bringing an Olfactory Network Together for Odor Perception. Perception 46, 320–332, doi: 10.1177/0301006616663216 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes DC, Hofacer RD, Zaman AR, Rennaker RL & Wilson DA Olfactory perceptual stability and discrimination. Nature Neuroscience 11, 1378–1380, doi: 10.1038/nn.2217 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson DA & Sullivan RM Cortical Processing of Odor Objects. Neuron 72, 506–519, doi: 10.1016/j.neuron.2011.10.027 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haddad R et al. A metric for odorant comparison. Nature methods 5, 425–429, doi: 10.1038/nmeth.1197 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Dravnieks A Odor quality: semantically generated multidimensional profiles are stable. Science 218, 799–801 (1982). [DOI] [PubMed] [Google Scholar]
- 7.Schiffman SS Physicochemical correlates of olfactory quality. Science 185, 112–117 (1974). [DOI] [PubMed] [Google Scholar]
- 8.Amoore JE Stereochemical theory of olfaction. Nature 198, 271–272 (1963). [DOI] [PubMed] [Google Scholar]
- 9.Buck L & Axel R A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65, 175–187 (1991). [DOI] [PubMed] [Google Scholar]
- 10.Malnic B, Hirono J, Sato T & Buck LB Combinatorial receptor codes for odors. Cell 96, 713–723 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Mombaerts P et al. Visualizing an olfactory sensory map. Cell 87, 675–686 (1996). [DOI] [PubMed] [Google Scholar]
- 12.Otazu GH, Chae H, Davis MB & Albeanu DF Cortical Feedback Decorrelates Olfactory Bulb Output in Awake Mice. Neuron 86, 1461–1477, doi: 10.1016/j.neuron.2015.05.023 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedrich RW & Wiechert MT Neuronal circuits and computations: pattern decorrelation in the olfactory bulb. FEBS Lett 588, 2504–2513, doi: 10.1016/j.febslet.2014.05.055 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Chae H et al. Mosaic representations of odors in the input and output layers of the mouse olfactory bulb. Nature Publishing Group 22, 1306–1317, doi: 10.1038/s41593-019-0442-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sosulski DL, Lissitsyna Bloom M, Cutforth T, Axel R & Datta SR Distinct representations of olfactory information in different cortical centres. Nature 472, 213–216, doi: 10.1038/nature09868 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyamichi K et al. Cortical representations of olfactory input by trans-synaptic tracing. Nature 472, 191–196, doi: 10.1038/nature09714 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davison IG & Ehlers MD Neural circuit mechanisms for pattern detection and feature combination in olfactory cortex. Neuron 70, 82–94, doi: 10.1016/j.neuron.2011.02.047 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stettler DD & Axel R Representations of odor in the piriform cortex. Neuron 63, 854–864, doi: 10.1016/j.neuron.2009.09.005 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Schaffer ES et al. Odor Perception on the Two Sides of the Brain: Consistency Despite Randomness. Neuron, doi: 10.1016/j.neuron.2018.04.004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roland B, Deneux T, Franks KM, Bathellier B & Fleischmann A Odor identity coding by distributed ensembles of neurons in the mouse olfactory cortex. Elife 6, doi: 10.7554/eLife.26337 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iurilli G & Datta SR Population Coding in an Innately Relevant Olfactory Area. Neuron 93, 1180–1197 e1187, doi: 10.1016/j.neuron.2017.02.010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Babadi B & Sompolinsky H Sparseness and expansion in sensory representations. Neuron 83, 1213–1226, doi: 10.1016/j.neuron.2014.07.035 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Barak O, Rigotti M & Fusi S The sparseness of mixed selectivity neurons controls the generalization-discrimination trade-off. Journal of Neuroscience 33, 3844–3856 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dasgupta S, Stevens CF & Navlakha S A neural algorithm for a fundamental computing problem. Science 358, 793–796 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Haberly LB & Bower JM Olfactory cortex: model circuit for study of associative memory? Trends Neurosci 12, 258–264 (1989). [DOI] [PubMed] [Google Scholar]
- 26.Bekkers JM & Suzuki N Neurons and circuits for odor processing in the piriform cortex. Trends in neurosciences 36, 429–438, doi: 10.1016/j.tins.2013.04.005 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Saito H, Chi Q, Zhuang H, Matsunami H & Mainland JD Odor Coding by a Mammalian Receptor Repertoire. Science Signaling 2, ra9–ra9, doi: 10.1126/scisignal.2000016 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cleland TA, Morse A, Yue EL & Linster C Behavioral models of odor similarity. Behav Neurosci 116, 222–231 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Bolding KA & Franks KM Recurrent cortical circuits implement concentration-invariant odor coding. Science (New York, NY) 361, eaat6904, doi: 10.1126/science.aat6904 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chapuis J & Wilson DA Bidirectional plasticity of cortical pattern recognition and behavioral sensory acuity. Nature Publishing Group 15, 155–161, doi: 10.1038/nn.2966 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shakhawat AM, Harley CW & Yuan Q Arc visualization of odor objects reveals experience-dependent ensemble sharpening, separation, and merging in anterior piriform cortex in adult rat. Journal of Neuroscience 34, 10206–10210, doi: 10.1523/JNEUROSCI.1942-14.2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sell CS in Chemistry and the Sense of Smell Ch. 5, 237–296 (Wiley, 2014). [Google Scholar]
- 33.Diodato A et al. Molecular signatures of neural connectivity in the olfactory cortex. Nature communications 7, 12238, doi: 10.1038/ncomms12238 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyd AM, Kato HK, Komiyama T & Isaacson JS Broadcasting of cortical activity to the olfactory bulb. Cell Rep 10, 1032–1039, doi: 10.1016/j.celrep.2015.01.047 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schoenfeld TA & Macrides F Topographic organization of connections between the main olfactory bulb and pars externa of the anterior olfactory nucleus in the hamster. J Comp Neurol 227, 121–135 (1984). [DOI] [PubMed] [Google Scholar]
- 36.Challis RC et al. Systemic AAV vectors for widespread and targeted gene delivery in rodents. Nat Protoc 14, 379–414, doi: 10.1038/s41596-018-0097-3 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Bolding KA & Franks KM Recurrent cortical circuits implement concentration-invariant odor coding. Science (New York, NY) 361, doi: 10.1126/science.aat6904 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruno RM & Sakmann B Cortex is driven by weak but synchronously active thalamocortical synapses. Science 312, 1622–1627, doi: 10.1126/science.1124593 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Minamisawa G, Funayama K, Matsuki N & Ikegaya Y Intact internal dynamics of the neocortex in acutely paralyzed mice. J Physiol Sci 61, 343–348, doi: 10.1007/s12576-011-0155-x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simons DJ & Carvell GE Thalamocortical response transformation in the rat vibrissa/barrel system. Journal of neurophysiology 61, 311–330, doi: 10.1152/jn.1989.61.2.311 (1989). [DOI] [PubMed] [Google Scholar]
- 41.Maklad A, Quinn T & Fritzsch B Intracranial distribution of the sympathetic system in mice: DiI tracing and immunocytochemical labeling. The Anatomical Record, doi: 10.1002/ar.1083 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Doevendans PA, Daemen MJ, de Muinck ED & Smits JF Cardiovascular phenotyping in mice. Cardiovascular Research 39, 34–49, doi: 10.1016/s0008-6363(98)00073-x (1998). [DOI] [PubMed] [Google Scholar]
- 43.Carey RM & Wachowiak M Effect of Sniffing on the Temporal Structure of Mitral/Tufted Cell Output from the Olfactory Bulb. Journal of Neuroscience 31, 10615–10626, doi: 10.1523/JNEUROSCI.1805-11.2011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kepecs A & Uchida N The sniff as a unit of olfactory processing. Chemical senses, doi: 10.1093/chemse/bjj016 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Cheung M & Carey R A Method for Generating Natural and User-Defined Sniffing Patterns in Anesthetized or Reduced Preparations. Chemical senses, doi: 10.1093/chemse/bjn051 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moldestad O, Karlsen P, Molden S & Storm JF Tracheotomy improves experiment success rate in mice during urethane anesthesia and stereotaxic surgery. Journal of Neuroscience Methods 176, 57–62, doi: 10.1016/j.jneumeth.2008.08.015 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Ecker AS et al. State dependence of noise correlations in macaque primary visual cortex. Neuron 82, 235–248, doi: 10.1016/j.neuron.2014.02.006 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goard M & Dan Y Basal forebrain activation enhances cortical coding of natural scenes. Nat Neurosci 12, 1444–1449, doi: 10.1038/nn.2402 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saito H, Chi Q, Zhuang H, Matsunami H & Mainland JD Odor Coding by a Mammalian Receptor Repertoire. Science Signaling 2, doi: 10.1126/scisignal.2000016 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haddad R et al. A metric for odorant comparison. Nature methods 5, 425–429, doi: 10.1038/nmeth.1197 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Ma L et al. Distributed representation of chemical features and tunotopic organization of glomeruli in the mouse olfactory bulb. Proc Natl Acad Sci U S A 109, 5481–5486, doi: 10.1073/pnas.1117491109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Srinivasan S & Stevens C A quantitative description of the mouse piriform cortex. biorxiv.org, doi: 10.1101/099002 (2017). [DOI] [Google Scholar]
- 53.Hagiwara A, Pal SK, Sato TF, Wienisch M & Murthy VN Optophysiological analysis of associational circuits in the olfactory cortex. Frontiers in neural circuits 6, 18, doi: 10.3389/fncir.2012.00018 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luna VM & Morozov A Input-specific excitation of olfactory cortex microcircuits. Frontiers in neural circuits, doi: 10.3389/fncir.2012.00069 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pachitariu M, Stringer C, Dipoppa M & Schröder S Suite2p: beyond 10,000 neurons with standard two-photon microscopy. BioRxiv, doi: 10.1101/061507 (2017). [DOI] [Google Scholar]
- 56.Willmore B & Tolhurst DJ Characterizing the sparseness of neural codes. Network 12, 255–270, doi: 10.1088/0954-898x/12/3/302 (2001). [DOI] [PubMed] [Google Scholar]
- 57.McInnes L, Healy J & Melville J UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv e-prints, doi: arXiv:1802.03426v2 (2018). [Google Scholar]
- 58.Haxby JV et al. A common, high-dimensional model of the representational space in human ventral temporal cortex. Neuron 72, 404–416, doi: 10.1016/j.neuron.2011.08.026 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cowley B et al. in Artificial Intelligence and Statistics. 242–251.
- 60.Litwin-Kumar A, Harris KD, Axel R, Sompolinsky H & Abbott LF Optimal Degrees of Synaptic Connectivity. Neuron 93, 1153–1164 e1157, doi: 10.1016/j.neuron.2017.01.030 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abbott LF, Rajan K & Sompolinsky H Interactions between Intrinsic and Stimulus-Evoked Activity in Recurrent Neural Networks. arXiv e-prints, doi: arXiv:0912.3832v2 (2009). [Google Scholar]
- 62.Schaffer ES et al. Odor Perception on the Two Sides of the Brain: Consistency Despite Randomness. Neuron 98, 736–742 e733, doi: 10.1016/j.neuron.2018.04.004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pedregosa F et al. Scikit-learn: Machine Learning in Python. The Journal of Machine Learning Research 12, 2825–2830 (2011). [Google Scholar]
- 64.Platt J Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. Advances in large margin classifiers 10, 61–74 (1999). [Google Scholar]
- 65.Kirkpatrick S, Gelatt CD Jr. & Vecchi MP Optimization by simulated annealing. Science 220, 671–680, doi: 10.1126/science.220.4598.671 (1983). [DOI] [PubMed] [Google Scholar]
- 66.Wilson DA & Linster C Neurobiology of a simple memory. Journal of neurophysiology 100, 2–7, doi: 10.1152/jn.90479.2008 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


