Abstract
Introduction.
Ankylosing spondylitis (AS) is a systemic progressive disease with an unknown etiology that may be related to the gut microbiome. Therefore, a more thorough understanding of its pathogenesis is necessary for directing future therapy.
Aim.
We aimed to determine the differences in intestinal microbial composition between healthy individuals and patients with AS who received and who did not receive treatment interventions. In parallel, the pathology of AS in each patient was analysed to better understand the link between AS treatment and the intestinal microbiota of the patients.
Methodology.
Sixty-six faecal DNA samples, including 37 from healthy controls (HCs), 11 from patients with untreated AS (NM), 7 from patients treated with nonsteroidal anti-inflammatory drugs (e.g. celecoxib; WM) and 11 from patients treated with Chinese herbal medicine (CHM), such as the Bushen–Qiangdu–Zhilv decoction, were collected and used in the drug effect analysis. All samples were sequenced using Illumina HiSeq 4000 and the microbial composition was determined.
Results.
Four species were enriched in the patients with AS: Flavonifractor plautii , Oscillibacter , Parabacteroides distasonis and Bacteroides nordii (HC vs. NM, P<0.05); only F. plautii was found to be significantly changed in the NM-HC comparison. No additional species were found in the HC vs. CHM analysis, which indicated a beneficial effect of CHM in removing the other three strains. F. plautii was found to be significantly increased in the comparison between the HC and WM groups, along with four other species ( Clostridium bolteae , Clostridiales bacterium 1_7_47FAA, C. asparagiforme and C. hathewayi ). The patients with AS harboured more bacterial species associated with carbohydrate metabolism and glycan biosynthesis in their faeces. They also had bacterial profiles less able to biodegrade xenobiotics or synthesize and transport vitamins.
Conclusion.
The gut microbiota of the patients with AS varied from that of the HCs, and the treatment had an impact on this divergence. Our data provide insight that could guide improvements in AS treatment.
Keywords: Ankylosing spondylitis, Pathogenesis, Drug effect, Flavonifractor plautii, Heparan sulfate degradation
Introduction
Ankylosing spondylitis (AS) is the most serious and atypical form of spondyloarthritis. Interestingly, inflammatory bowel disease and subclinical intestinal inflammation are often observed during the progression of AS [1].
To date, a spectrum of human host genetic factors has been implicated in the pathogenesis of AS. Over 40 host genetic variations have been identified as having an influence on the development and progression of AS [2]. Among these host genetic variations, human leukocyte antigen (HLA)-B27 has been verified to contribute to the risk for AS. HLA B27 (subtypes B*2701–2759) is a class I surface antigen encoded by the B locus in the major histocompatibility complex (MHC) on chromosome 6 and presents antigenic peptides (derived from self and non-self-antigens) to T cells, along with many other HLA-B (MHC, class I, B) alleles and non-MHC loci [3–5]. These AS-susceptibility genes do not seem to be directly involved in the ankylosing process [6], creating the need to further investigate the underlying mechanisms of AS. Given that AS shares a similar genetic background with inflammatory bowel disease [7], researchers consequently proposed that there may be a possibility that AS is a microbiome-driven disease. In support of this, both animal studies [8] and oral/gut microbiome studies have revealed several mechanisms that can explain the potential role of the microbiome in the etiology of AS. These include pathological alterations, such as changes in intestinal permeability, stimulation of the immune response and molecular mimicry [9]. Klebsiella species have been specifically implicated in many studies as being potentially associated with the pathogenesis of AS [10]. Recently, a metagenomic study conducted on a Chinese population found an alteration in the intestinal microbiome in patients with AS [11]. That study revealed that the enrichment of Actinobacteria in patients with AS may be correlated with IκBα, a member of an inhibitory family of proteins that bind to nuclear factor-κB transcription factors, the ubiquitination module and activation of nuclear factor-κB signalling in the immune and inflammatory signalling cascades. Bifidobacterium spp., Prevotella melaninogenica , Prevotella copri and Prevotella sp. C561 were also proposed as potential biomarkers in the pathogenesis or development of AS.
According to international guidelines, the currently recommended treatment for AS is a combination of physical rehabilitation and therapy against the inflammatory protein tumour necrosis factor alpha [12]. In China, traditional herbal medicine is also often used. Both strategies still primarily focus on relieving pain and slowing disease progression, without completely controlling or treating the disease; in some cases, the currently available treatments may simply have no effect. Therefore, a thorough investigation into disease etiology and the influence that current treatments have on pathological progression is necessary. Such further investigation is likely to aid the development of novel treatment options.
In the current study, we conducted a metagenomic analysis of 113 patients with AS and 37 healthy controls (HCs). Our objective was to identify the differences in the gut microbiota makeup between healthy individuals and patients with AS who received and who did not receive therapeutic intervention.
Methods
Patient recruitment and sample collection
Patients with AS were recruited from The Second Affiliated Hospital, Guangzhou University of Chinese Medicine, Guangzhou, PR China. The inclusion criteria were as follows: AS confirmed according to the modified New York criteria [13], patient age between 18 and 60 years, and normal values on recent screenings for liver and kidney function and routine blood tests. The patients with a Bath Ankylosing Spondylitis Disease Activity Index (BASDAI) of ≥4 were assigned to the high AS disease activity subgroup (AC) and those with a BASDAI of <4 to the low AS disease activity subgroup (UN) [14]. Individuals were excluded from the study if they had received antibiotic therapy in the preceding month. Faecal samples were collected, and DNA extraction was performed as previously described [15].
DNA and metagenomic libraries for paired-end sequencing were constructed and sequenced in Illumina HiSeq 4000 in BGI Shenzhen, and the paired-end 150 sequencing strategy was used. An initial quality control was conducted to discard reads with adaptor contamination and low quality using the SOAPnuke filterMeta [16] (1.5.0, -Q 2 L 15 N 3 P 0 -q 20 l 30 R 0.5 C 0). The remaining reads were filtered to eliminate human DNA based on hg19 using Soap2 [17] (2.22, -M 4 m 400 -x 600 r 1 v 7 l 30 n 5 c 0.9). On average, 10.62 Gb of high-quality data was generated for each of the 150 samples.
Construction of genes and Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology (KO) profiles
For each disease dataset, in the construction of a gene profile, high-quality reads from each sample were aligned against the gene catalogue [18] using SOAP2 (17) with the criterion of identity ≥90 % (2.22, -m 226,-x 426,-r 2,-l 30, -M 4, S, -p 8, -v 5, -c 0.95). Sequence-based gene abundance profiling was performed as described previously [18]. On the basis that genes with exceedingly low frequencies among the samples might not appropriately reflect the actual conditions of the samples, genes that were present in less than 8 of the 150 samples were discarded. The KO was calculated from the relative abundance of their respective genes with annotations, as described previously [19].
Construction of species profiles
For each sample, MetaPhlAn2 [20] was used for profiling the composition of the microbial communities from the metagenomic shotgun sequencing data. MetaPhlAn relies on unique clade-specific marker genes identified from ∼17 000 reference genomes (∼13 500 bacterial and archaeal,~3500 viral and ~110 eukaryotic).
Permutational multivariate ANOVA (PERMANOVA)
We used the vegan package of the computational program R to perform PERMANOVA on the species abundance profiles [21]. This was conducted to assess the effects of each of the factors listed in Table S1 (available in the online version of this article) using the Bray–Curtis distance method and 9999 permutations.
Pathway analysis
The KO profiles that appeared in less than 40 % of the samples were first ruled out in the following analysis. The one-tailed Wilcoxon rank-sum test was performed on the filtered KO profiles with Benjamin–Hochberg multiple test adjustment to determine the differentially enriched KO modules between the HCs and patients with AS. The reporter score [22] was then calculated for each of the identified KO terms to quantify the enrichment of specific modules. The modules with a reporter score of >1.65 were considered as significantly differentiated modules. The KEGG was subsequently used to annotate the acquired distinct modules to the pathway level.
Sample classifier
We used the random forest model [22] (R 3.4.1, randomForest 4.6–14 package) with a tenfold cross-validation (CV) repeated ten times to construct the sample classifier. The input species profiles were omitted prior to this analysis if their frequency in all the 150 samples was lower than 20 %. For the training stage, 70 % of the 150 samples were randomly selected with a seed of 12 345. We then computed the CV error curves for each trial of the tenfold CV and calculated the sum of the average of the CV errors and the mean standard deviations for variable selection. Species with mean CV errors lower than the threshold were retained to construct the optimal training set. The probability of the patient group was calculated using this set, and a receiver-operating curve [23] was developed (R 3.0.2, pROC3 package) to assess the efficiency of the classification. The remaining 30 % of the 150 samples were then used for the testing stage, and the prediction error was determined.
Accession codes
The metagenomic sequencing data for all samples have been deposited in the European Bioinformatic Institute database under accession code PRJEB28545. The methods, associated references, any supplementary information and source data files are available in the online version of this paper.
Results
Dysbiosis of the gut microbiota in AS and the associated treatment type
According to the treatment type administered to the patients, we compartmentalized AS into three subgroups: NM (untreated), WM (treated with nonsteroidal anti-inflammatory drugs) and Chinese herbal medicine (CHM; treated with traditional CHM).
Deep metagenome sequencing on the faecal DNA samples collected from 150 Chinese individuals (113 patients and 37 controls) was performed, generating a mean of 11.45 Gb of raw paired-end reads for each sample. An average of 10.62 Gb of high-quality reads was free from human DNA and adaptor contamination. The mean comparison rate of the clean dataset summarized from all of the 150 samples to the 11.4M gene catalogue was 73.02 %, covering ~2.1 million genes (Fig. S1). The AS status had a strong effect on the gut microbiota (lowest P-value in PERMANOVA, Table S1). To delineate the features of the AS-associated gut microbiome (Fig. S2), MetaPhlAn2 [24] was used to analyse the microbial community.
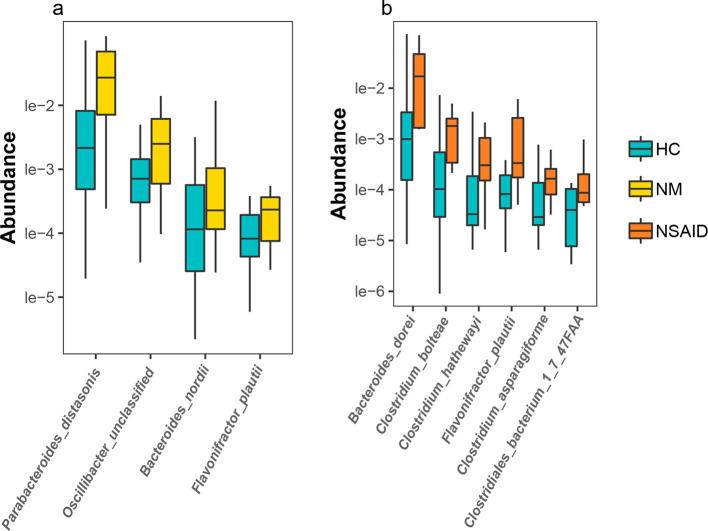
A comparison of the species profiles between the HC and NM groups provided information on the pathologically associated changes in the species composition, independent of the effect of treatment. We found four species that were significantly enriched in the NM group: Flavonifractor plautii , Oscillibacter unclassified (an unclassified species in the Oscillibacter genus), Parabacteroides distasonis and Bacteroides nordii (HC vs. NM, Wilcoxon test: P<0.05; (Figs 1a and S3).
Fig. 1.
Species that were differentially abundant in the HCs and patients with AS who received and who did not receive treatment. (a) Species whose abundance significantly increased in the patients with untreated AS (NM) were discovered on the basis of the species-abundance profiles. (b) The species whose abundance significantly increased in the patients with AS treated with nonsteroidal anti-inflammatory drugs (WM). Both the NM and WM groups were significantly different from the HC group (Kruskal–Wallis test). Yellow, orange and blue represent the NM, WM and HC groups, respectively. AS, ankylosing spondylitis; HC, healthy control; NSAID, nonsteroidal anti-inflammatory drug.
Similarly, comparing the HC and WM groups helped identify the effective mechanism associated with the standard treatments of AS. The abundance of three species ( B. dorei , Clostridium asparagiforme and C. bolteae ) increased in the patients administered with nonsteroidal anti-inflammatory drugs (WM group). These differentially abundant species in the WM group were mainly opportunistically pathogenic bacteria (HC vs. WM, Wilcoxon test: P<0.05, Fig. 1b), which may represent a pathological challenge to the patients’ health. We found some similar changes in the WM group whereby the abundance of F. plautii and Oscillibacter was still significantly higher in the WM group than in the HC group. With the exception of F. plautii , none of the other HC-NM differential species had significantly higher abundance in the CHM group. Therefore, to some extent, CHM may be beneficial for the gut microbiome, given that it did not increase the abundance of the opportunistic pathogenic bacteria as in the WM group. This may suggest that CHM helped maintain a more balanced and normal gut microbiome than no treatment and treatment with nonsteroidal anti-inflammatory drugs.
Functional discrepancies in AS
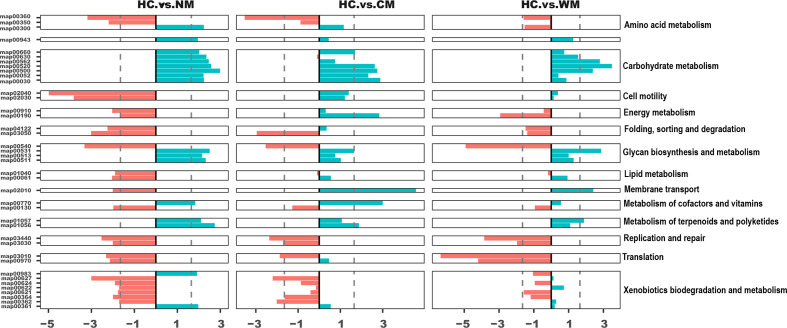
Some of the pathways were consistently enriched in the healthy cohort, including ‘energy metabolism’, ‘folding, sorting and degradation’, ‘replication and repair’, ‘translation’, ‘xenobiotics biodegradation’ and ‘metabolism’ (Fig. 2, reporter score ≤−1.65, HC vs. NM). However, the secondary function pathways significantly increased in the patients with AS were primarily involved in carbohydrate metabolism and glycan biosynthesis and metabolism (Figs 2 and 3).
Fig. 2.
Kyoto Encyclopedia of Genes and Genomes pathways enriched in the NM, CN, WM and HC groups. Bar plots represent the reporter scores of the pathways between two groups. The reporter score was presented by dividing the patients into three AS subgroups (NM, CN and WM) and the HC group (<−1.65, enriched in the former; >1.65, enriched in the latter). AS, ankylosing spondylitis.
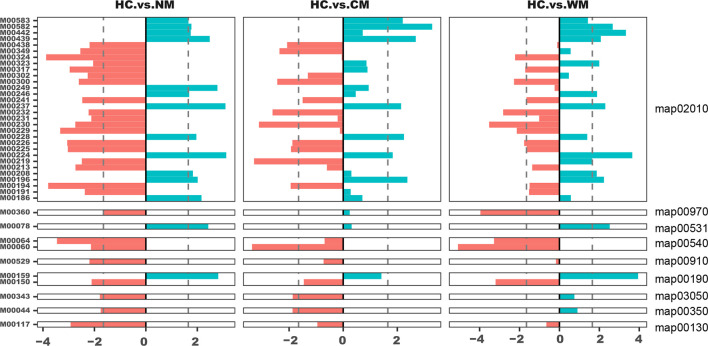
Fig. 3.
Kyoto Encyclopedia of Genes and Genomes modules corresponding to the differential pathways enriched in the NM, CN, WM and HC groups. Bar plots represent the z-scores of the modules between two groups. The reporter score was presented by dividing the patients into three AS subgroups (NM, CN and WM) and the HC group (<−1.65, enriched in the former; >1.65, enriched in the latter). AS, ankylosing spondylitis.
The KO term ‘starch and sucrose metabolism’ (map00500) was found to be consistently enriched in the patients with AS, regardless of whether the patients received treatment. This abnormal enrichment indicated an increase in bacteria that mainly feed on starch. The metabolism of starch and sucrose could produce a diversity of saccharides that can potentially elicit an increased inflammatory response. This could then partially explain the inflammatory symptoms observed in patients with AS. According to the BMI density plot, we observed a significantly lower BMI in the patients with AS than in the HCs (Wilcoxon test: P<0.05; Fig. S4). In addition, we found that the ability of the patients to synthesize lipids through their microbiota was down-regulated (map00061 and map01040, reporter score of <−1.65). This observation could partially explain the somatotype emaciation of patients with AS.
Another pathway that we found to be highly enriched in the intestines of the patients with AS was glycosaminoglycan degradation (map00531), specifically heparan sulfate degradation (M00078). Bao et al. [25] previously showed that the binding of heparan sulfate to several major molecules, including l-selectin, chemokines and integrins, was involved in lymphocyte homing in vitro. Lymphocyte homing is one of the most important immune activities of the intestinal mucosal immune system. Further, it plays a role in the occurrence of acute/chronic inflammation in the intestinal mucosa. Thus, excessive degradation of heparan sulfate in AS could diminish its effect on chemokine concentration and impede lymphocyte homing.
We also noted that the ability to degrade dioxin (map00621) was impaired in AS (Fig. 2). Previously, it has been suggested that exotic pollutants can cause an inflammatory host-cell response [26]. Therefore, with the declining ability to deal with xenobiotics (e.g. dioxin), the intestinal environment could be exposed to accumulating toxins. This would then stimulate the gut epithelial cells to produce cytokine IL-8, leading to a pro-inflammatory state in the intestines [26]. Notably, this impairment was less evident in the patients who received treatment, which may indicate that medical therapy helps relieve inflammation by adjusting the unbalanced dioxin degradation capacity.
Patients with AS have a general tendency to develop osteoporosis or low bone mineral density. Studies have proven that vitamins, such as vitamin K, vitamin D3 and vitamin B, can effectively prevent osteoporosis or increase bone density [27]. Accordingly, among our identified differential modules, the vitamin B12 transport system (M00241) was found to be decreased in AS. Even in moderate cases, a deficit of vitamin B12 can result in neurological or haematological abnormalities [28] and impact the generation of osteoclasts indirectly [29]. Hence, the gradual deterioration and disequilibrium of the gut microbiota in AS disease progression could weaken the gut’s capacity for vitamin biosynthesis and transportation. This may consequently cause the previously observed bone-related complications.
Microbiota composition predicts the disease stage
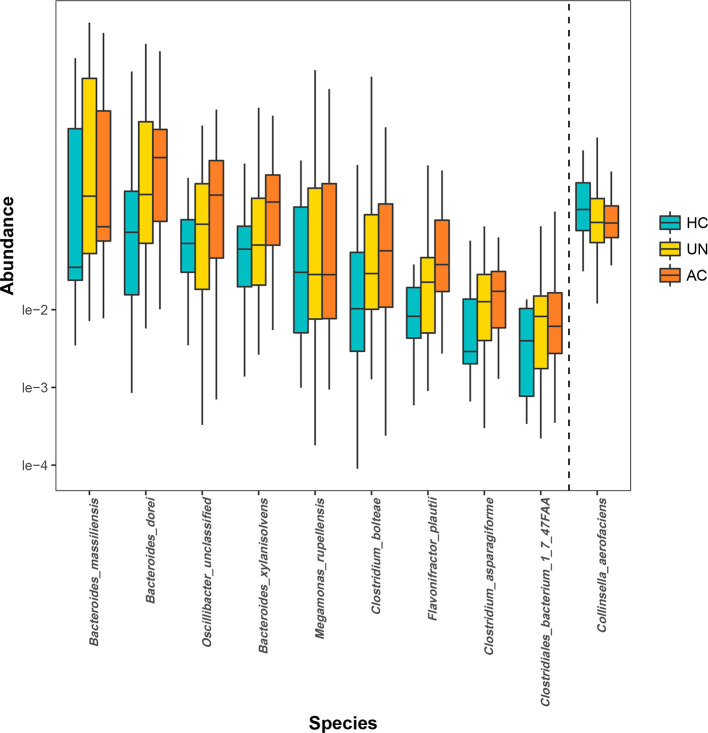
We compared the discrepancy in the intestinal microbiota between the HCs and patients with two stages of AS determined using the BASDAI criteria. For the entire course of the disease, B. dorei, B. massiliensis, C. asparagiforme, C. bolteae and F. plautii were found to be significantly enriched in the patients with AS, and their abundance increased further as the disease severity progressed. By comparison, the differences in the abundance of some species (i.e. B. xylanisolvens, Clostridiales bacterium 1_7_47FAA, Megamonas rupellensis and Oscillibacter ) only became significant in the more severe disease stages. Only one bacterium, Collinsella aerofaciens , was enriched in the HCs (Kruskal–Wallis test: P<0.05; Fig. 4).
Fig. 4.
Species that were differentially abundant in the HC group and the two BASDAI groups. Species whose abundance significantly increased (left side of the dashed line) and decreased (right side of the dashed line) in the patients with AS were discovered on the basis of the species-abundance profiles. Both the AC group and UN group were significantly different from the HC group (Kruskal–Wallis test). Orange, yellow and blue correspond to the AC, UN and HC groups, respectively. HC, healthy control; AS, ankylosing spondylitis; BASDAI, Bath Ankylosing Spondylitis Disease Activity Index.
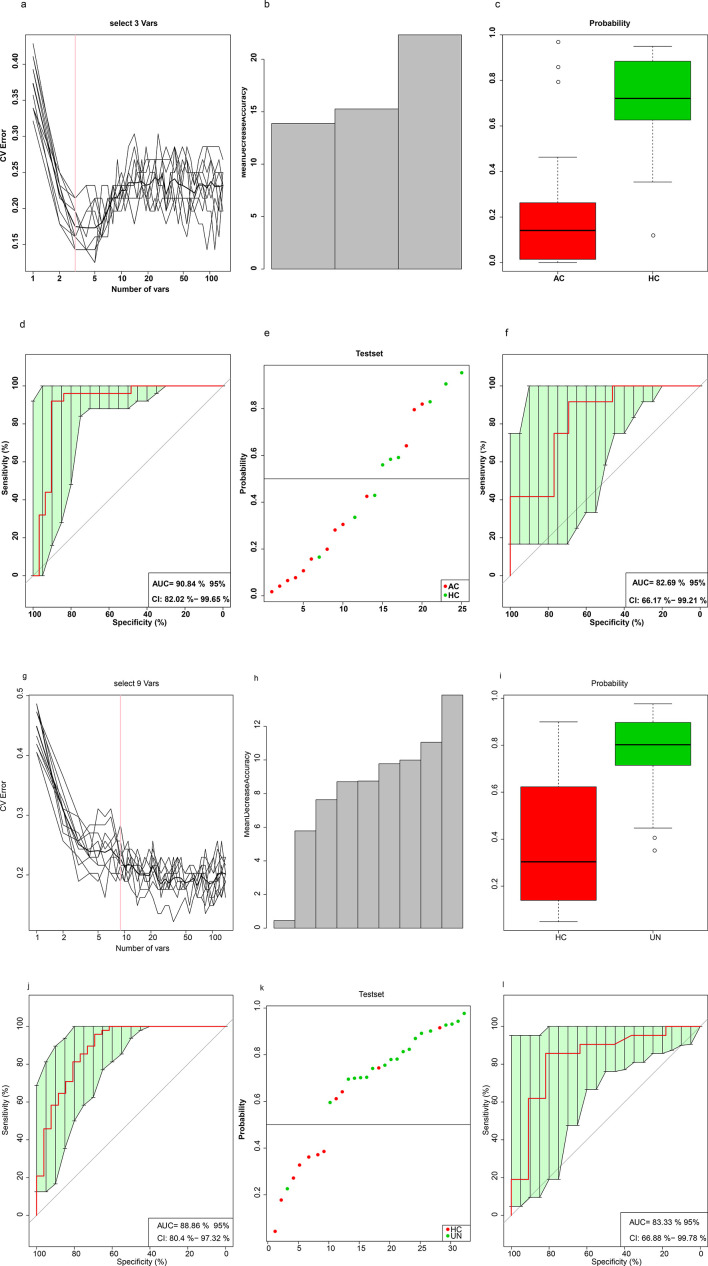
Ten trials of tenfold CV were separately performed on the training set of 70 % of randomly selected AC-HC (AC=31, NHC=25) and UN-HC (UN=48, HC=26) paired samples to obtain an optimal marker set for the training model (Fig. 5a–c, g–i). The receiver-operating curve was plotted to assess the classification efficiency (Fig. 5d–f, j–l). We identified three species ( B. massiliensis , O. unclassified and Anaerotruncus colihominis ) that can be used to distinguish the AC group from the HC group. Additionally, we found nine species (Enterococcus faecium, C. bolteae, Alistipes senegalensis, B. faecis, Bacillus cereus thuringiensis, B. dorei, B. massiliensis, C. asparagiforme and Sutterella wadsworthensis ) that could distinguish between the UN and HC groups. There were two species shared between these two sets of classifications ( B. massiliensis and O. unclassified). The abundance of these two species was also found to be significantly increased in the patients from both the AS course groups in the earlier rank sum test.
Fig. 5.
Species-abundance profiles distinguished the two BASDAI patient subgroups from the HCs. (a) Distribution of ten trials of tenfold CV error in the random forest classification of the AC group as the input species number increased. The training model was constructed using species-abundance profiles (pre-filtered by a cutoff of no less than 20 % frequency in all 150 samples) in a randomly selected 70 % of AC (NAC=31) and HC samples (NHC=25) with a random seed of 12 345. The black curve indicates the ten trials of CV. The pink line denotes the number of species used in the optimal set (n=3). (b) Mean decreased accuracy. (c) Box-and-whisker plot for the probability of the AC group in the CV training set according to the model in (a). (d) Receiver-operating curve (ROC) for the training set (n=56). The area under the receiver-operating curve (AUC) was 90.84 % with a 95 % confidence interval (CI) of 82.02–99.65 %. (e) Classification of the test set using the remaining 15 AC (red) and 11 HC (green) samples. (f) ROC for the test set (n=26). The AUC was 82.69%, and the 95 % CI was 66.17 %–99.21 %. (g–l) Training and testing of the model that classified the UN group from the controls, performed as in parts a–f of this figure. The AUC for the training set (NUN=48, NHC=26) was 88.65%, and the 95 % CI was 80.4 %–97.32 %; the AUC for the test set (NUN=21, NHC=11) was 83.33%, and the 95 % CI was 66.88 %–99.78 %. HC, healthy control; CV, cross-validation; BASDAI, Bath Ankylosing Spondylitis Disease Activity Index; ROC, receiver-operating curve; AUC, area under the receiver-operating curve; CI, confidence interval
Discussion
The comparison among the HC, NM, CHM and WM groups revealed several potential bacterial species that may participate in the pathogenesis of AS. For example, P. distasonis , which was significantly enriched in the NM group compared with that in the HC group, was recently studied in a mono-colonization study [30]. In the comparison in germ-free mice, P. distasonis could significantly induce regulatory T cells as well as elevate the concentrations of succinate and three short-chain fatty acids (SCFA), i.e. butyrate, acetate and propionate. Given that both regulatory T cells [22] and short-chain fatty acids [31] are known to contribute to the equilibrium of the gut environment and immune homeostasis, the enrichment of P. distasonis detected in the NM group could possibly be responsible for the elevated inflammatory response noted in patients with AS.
Another interesting bacterium observed was C. bolteae . The abundance of C. bolteae was highly increased in the WM vs. HC comparison (Fig. 1b). The C. bolteae strain is antibiotic-resistant based on previous reports and has fimbriae that enable it to adhere to intestinal epithelial cells [32, 33]. Owing to its ability to adhere, this bacterium is able to alter the intestinal barrier and may subsequently cause abdominal infection. C. bolteae may produce a conserved specific cell-wall capsular polysaccharide composed of repeating disaccharide blocks of rhamnose and mannose units. The capsular polysaccharide contributes towards immune evasion and is consequently a primary virulence factor. Given that AS predisposes patients to diarrhea and this bacterium was also enriched in patients after receiving treatment, we suggest that some medications could possibly be compounding the pathological changes in the patients’ gut microbiota.
Conversely, the presence of B. dorei in the treated groups may be an indicator of the relieving effects of medical treatments. One experimental study exploring various co-cultures of bacteria on two common polysaccharide substrates (i.e. inulin and xylan) revealed that only the combination of B. dorei and C. symbiosum incubated with xylan could produce a sufficient concentration of butyrate to relieve inflammation in cellular models [34]. Further, C. symbiosum alone or in combination with other bacteria was not able to achieve this effect. The anti-inflammatory effect of high concentrations of butyrate is thought to be mediated by a decrease in the expression of IL-8 in the epithelial HT-29 cells. Furthermore, the metabolites of B. dorei may also bind directly to and activate the farnesoid X receptor, which could help ameliorate intestinal inflammation [35]. However, another study found that C. asparagiforme could produce acetate and lactate through glucose fermentation [36], and these fermentation products would then be used by some other specific bacteria to produce butyrate [34]. Therefore, both B. dorei and C. asparagiforme were suggested to promote butyrate production, which would indirectly help relieve intestinal inflammation.
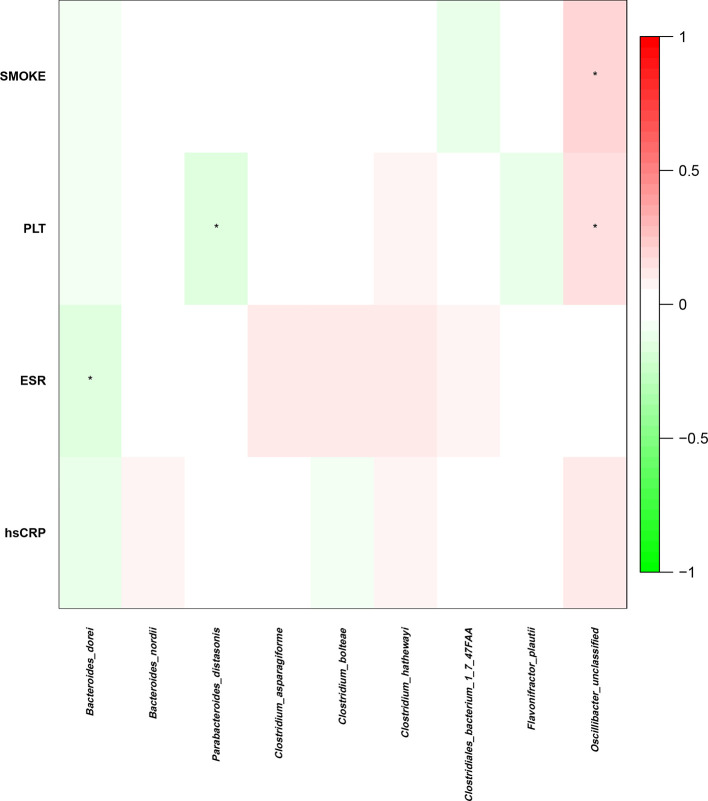
To identify the influence of these species on AS further, we calculated their correlation with the clinical data (Fig. 6). According to the heatmap, there was a negative correlation between the abundance of P. distasonis and the platelet counts in the NM group. The platelet count is a biomarker of inflammation severity, with higher counts indicating more severe inflammation. This negative correlation could indicate that patients with more P. distasonis could have a milder inflammatory response, implying a possible beneficial function of this bacterium. Nevertheless, future studies are needed to validate these findings.
Fig. 6.
Relationships between the intestinal microbial species and clinical indices. Presentation of the correlation coefficient between four clinical indices and eight previously discovered bacterial species, which were differentially abundant in the patients with AS (NM and WM) and HCs (P<0.1; Spearman’s rank correlation was calculated for numerical data and polyserial correlation for ordinal data). PLT, platelet; ESR, erythrocyte sedimentation rate; hsCRP, high-sensitivity C-reactive protein.
Supplementary Data
Funding information
This study was supported by the National Natural Science Foundation of China (No. 81673898), the Science and Technology project of Guangdong Province (No. 2016A020226041), the Key Research Project of Guangzhou University of Chinese Medicine (No. XK2019021), the Science and Technology project of Guangzhou City (No. 201710010076), and grants from Guangdong Provincial Hospital of Chinese Medicine (No. YN2015MJ06, No. YN2018ML08 and No. YN2018ZD06).
Acknowledgements
The authors acknowledge the technical support for sampling and analytical measures on volunteers provided by The Second Affiliated Hospital, Guangzhou University of Chinese Medicine (Guangdong Provincial Hospital of Chinese Medicine).
Author contributions
R. H., F. L., X. T. and Z. Z. designed the study and managed the project. Y. Z., F. P., J. L., D. Q., Y. X., Y. L. and H. Y. collected the samples and computed the disease activity index. J. H., F. L., Z. Z. and Y. C., designed the analyses. F. L., Z. Z., Y. C., J. L., Y. T. and Y. S. analysed and interpreted the sequencing data. Y. C. and F. L. wrote the manuscript. Q. G., X. F., M. S., C. Z. and L. F., contributed to the text revision and data discussion. Y. H., R. H., and J. H., interpreted the data and revised the paper.
Conflicts of interest
All authors approved the submitted version and agree to be accountable for all aspects of the work. The authors declare that there are no conflicts of interest.
Ethical statement
This study was conducted in accordance with the recommendations of the International Committee of Medical Journal Editors and the Declaration of Helsinki principles. The protocol was approved by the Ethics Committee of Guangdong Provincial Hospital of Chinese Medicine. All subjects provided written informed consent to participate.
Footnotes
Abbreviations: AS, Ankylosing spondylitis; BASDAI, Bath Ankylosing Spondylitis Disease Activity Index; CHM, Chinese herbal medicine; CM, Chinese herbal medicine (CHM) treated AS patient; HC, Health Control; HLA-B27, human leukocyte antigen (HLA)-B27; MHC, major histocompatibility complex; NM, untreated Ankylosing spondylitis patients; NSAID, Nonsteroidal Antiinflammatory Drugs treated AS patients.
One supplementary table and four supplementary figures are available with the online version of this article.
References
- 1.Costello ME, Elewaut D, Kenna TJ, Brown MA, Microbes BMA. Microbes, the gut and ankylosing spondylitis. Arthritis Res Ther. 2013;15:214. doi: 10.1186/ar4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown MA, Kenna T, Wordsworth BP. Genetics of ankylosing spondylitis--insights into pathogenesis. Nat Rev Rheumatol. 2016;12:81–91. doi: 10.1038/nrrheum.2015.133. [DOI] [PubMed] [Google Scholar]
- 3.Kiltz U, Baraliakos X, Karakostas P, Igelmann M, Kalthoff L, et al. The degree of spinal inflammation is similar in patients with axial spondyloarthritis who report high or low levels of disease activity: a cohort study. Ann Rheum Dis. 2012;71:1207–1211. doi: 10.1136/annrheumdis-2011-200508. [DOI] [PubMed] [Google Scholar]
- 4.Braun J, Sieper J. Ankylosing spondylitis. The Lancet. 2007;369:1379–1390. doi: 10.1016/S0140-6736(07)60635-7. [DOI] [PubMed] [Google Scholar]
- 5.Evans DM, Spencer CCA, Pointon JJ, Su Z, Harvey D, et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet. 2011;43:761–767. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsui F, Tsui HW, Akram A, Haroon N, Inman R. The genetic basis of ankylosing spondylitis: new insights into disease pathogenesis. Appl Clin Genet. 2014;7:105–115. doi: 10.2147/TACG.S37325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenbaum JT, Lin P, Asquith M, Costello M-E, Kenna TJ, et al. Does the microbiome play a causal role in spondyloarthritis? Clin Rheumatol. 2014;33:763–767. doi: 10.1007/s10067-014-2664-5. [DOI] [PubMed] [Google Scholar]
- 8.Hammer RE, Maika SD, Richardson JA, Tang JP, Taurog JD. Spontaneous inflammatory disease in transgenic rats expressing HLA-B27 and human beta 2M: an animal model of HLA-B27-associated human disorders. Cell. 1990;63:1099–1112. doi: 10.1016/0092-8674(90)90512-D. [DOI] [PubMed] [Google Scholar]
- 9.Yang L, Wang L, Wang X, Xian C, Lu H. A possible role of intestinal microbiota in the pathogenesis of ankylosing spondylitis. Int J Mol Sci. 2016;17:2126. doi: 10.3390/ijms17122126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rashid T, Ebringer A. Autoimmunity in rheumatic diseases is induced by microbial infections via crossreactivity or molecular mimicry. Autoimmune Dis. 2012;2012:539282. doi: 10.1155/2012/539282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wen C, Zheng Z, Shao T, Liu L, Xie Z, et al. Quantitative metagenomics reveals unique gut microbiome biomarkers in ankylosing spondylitis. Genome Biol. 2017;18:142. doi: 10.1186/s13059-017-1271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lubrano E, Spadaro A, Amato G, Benucci M, Cavazzana I, et al. Tumour necrosis factor alpha inhibitor therapy and rehabilitation for the treatment of ankylosing spondylitis: a systematic review. Semin Arthritis Rheum. 2015;44:542–550. doi: 10.1016/j.semarthrit.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Goei The HS, Steven MM, van der Linden SM, Cats A. Evaluation of diagnostic criteria for ankylosing spondylitis: a comparison of the Rome, New York and modified New York criteria in patients with a positive clinical history screening test for ankylosing spondylitis. Rheumatology. 1985;24:242–249. doi: 10.1093/rheumatology/24.3.242. [DOI] [PubMed] [Google Scholar]
- 14.Garrett S, Jenkinson T, Kennedy LG, Whitelock H, Gaisford P, et al. A new approach to defining disease status in ankylosing spondylitis: the Bath ankylosing spondylitis disease activity index. J Rheumatol. 1994;21:2286–2291. [PubMed] [Google Scholar]
- 15.Qin J, Li Y, Cai Z, Li S, Zhu J, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y, Chen Y, Shi C, Huang Z, Zhang Y, et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience. 2018;7:1–6. doi: 10.1093/gigascience/gix120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li R, Yu C, Li Y, Lam TW, Yiu SM, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen HB, Almeida M, Juncker AS, Rasmussen S, Li J, et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol. 2014;32:822–828. doi: 10.1038/nbt.2939. [DOI] [PubMed] [Google Scholar]
- 19.Xie H, Guo R, Zhong H, Feng Q, Lan Z, et al. Shotgun Metagenomics of 250 adult twins reveals genetic and environmental impacts on the gut microbiome. Cell Systems. 2016;3:572–584. doi: 10.1016/j.cels.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Short DF, Zemel BS, Gilsanz V, Kalkwarf HJ, Lappe JM, et al. Fitting of bone mineral density with consideration of anthropometric parameters. Osteoporos Int. 2011;22:1047–1057. doi: 10.1007/s00198-010-1284-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patil KR, Nielsen J. Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc Natl Acad Sci USA. 2005;102:2685–2689. doi: 10.1073/pnas.0406811102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods. 2015;12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 25.Bao X, Moseman EA, Saito H, Petryanik B, Thiriot A, et al. Endothelial heparan sulfate controls chemokine presentation in recruitment of lymphocytes and dendritic cells to lymph nodes. Immunity. 2010;33:817–829. doi: 10.1016/j.immuni.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Defois C, Ratel J, Garrait G, Denis S, Le Goff O, et al. Food chemicals disrupt human gut microbiota activity and impact intestinal homeostasis as revealed by in vitro systems. Sci Rep. 2018;8:11006. doi: 10.1038/s41598-018-29376-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmer D, El Miedany Y. Tackling comorbidity associated with rheumatic diseases in standard practice. Br J Nurs. 2017;26:380–387. doi: 10.12968/bjon.2017.26.7.380. [DOI] [PubMed] [Google Scholar]
- 28.Shipton MJ, Thachil J. Vitamin B12 deficiency – A 21st century perspective. Clin Med. 2015;15:145–150. doi: 10.7861/clinmedicine.15-2-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai Z, Koh W-P. B-Vitamins and bone Health–A review of the current evidence. Nutrients. 2015;7:3322–3346. doi: 10.3390/nu7053322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med. 2014;6:220ra11. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, et al. The role of short-chain fatty acids in health and disease. Adv Immunol. 2014;121:91–119. doi: 10.1016/B978-0-12-800100-4.00003-9. [DOI] [PubMed] [Google Scholar]
- 32.Pequegnat B, Sagermann M, Valliani M, Toh M, Chow H, et al. A vaccine and diagnostic target for Clostridium bolteae, an autism-associated bacterium. Vaccine. 2013;31:2787–2790. doi: 10.1016/j.vaccine.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 33.Dehoux P, Marvaud JC, Abouelleil A, Earl AM, Lambert T, et al. Comparative genomics of Clostridium bolteae and Clostridium clostridioforme reveals species-specific genomic properties and numerous putative antibiotic resistance determinants. BMC Genomics. 2016;17:819. doi: 10.1186/s12864-016-3152-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomson P, Medina DA, Ortúzar V, Gotteland M, Garrido D. Anti-Inflammatory effect of microbial consortia during the utilization of dietary polysaccharides. Food Res Int. 2018;109:14–23. doi: 10.1016/j.foodres.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X, Osaka T, Tsuneda S. Bacterial metabolites directly modulate farnesoid X receptor activity. Nutr Metab. 2015;12:48. doi: 10.1186/s12986-015-0045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohan R, Namsolleck P, Lawson PA, Osterhoff M, Collins MD, et al. Clostridium asparagiforme sp. nov., isolated from a human faecal sample. Syst Appl Microbiol. 2006;29:292–299. doi: 10.1016/j.syapm.2005.11.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.