Abstract
Introduction.
Management of steroid-refractory ulcerative colitis has predominantly involved treatment with systemic cyclosporine A (CyA) and infliximab.
Aim.
The purpose of this study was to assess the effect of using a colon-targeted delivery system CyA formulation on the composition and functionality of the gut microbiota.
Methodology.
Ex vivo faecal fermentations from six healthy control subjects were treated with coated minispheres (SmPill) with (+) or without (−) CyA and compared with a non-treated control in a model colon system. In addition, the in vivo effect of the SmPill+CyA formulation was investigated by analysing the gut microbiota in faecal samples collected before the administration of SmPill+CyA and after 7 consecutive days of administration from eight healthy subjects who participated in a pilot study.
Results.
Analysis of faecal samples by 16S rRNA gene sequencing indicated little variation in the diversity or relative abundance of the microbiota composition before or after treatment with SmPill minispheres with or without CyA ex vivo or with CyA in vivo. Short-chain fatty acid profiles were evaluated using gas chromatography, showing an increase in the concentration of n-butyrate (P=0.02) and acetate (P=0.32) in the faecal fermented samples incubated in the presence of SmPill minispheres with or without CyA. This indicated that increased acetate and butyrate production was attributed to a component of the coated minispheres rather than an effect of CyA on the microbiota. Butyrate and acetate levels also increased significantly (P=0.05 for both) in the faecal samples of healthy individuals following 7 days’ treatment with SmPill+CyA in the pilot study.
Conclusion.
SmPill minispheres with or without CyA at the clinically relevant doses tested here have negligible direct effects on the gut microbiota composition. Butyrate and acetate production increased, however, in the presence of the beads in an ex vivo model system as well as in vivo in healthy subjects. Importantly, this study also demonstrates the relevance and value of using ex vivo colon models to predict the in vivo impact of colon-targeted drugs directly on the gut microbiota.
Keywords: Cyclosporine A, human microbiota, composition, functionality, 16S rRNA gene sequencing, short-chain fatty acids
Introduction
Perturbations in the microbial balance and host immune response in the gastrointestinal (GI) tract have been associated with the onset of inflammatory bowel disease (IBD), including ulcerative colitis (UC). Genetic predisposition in patients with IBD has been shown to cause a change in the immune response involved in the host’s interaction with bacteria in the gut [1]. Cyclosporine A (CyA) is a potent immunosuppressant approved for use in solid organ transplants to prevent rejection. It is a poorly soluble cyclic peptide and is subject to metabolism by various cytochrome P450 enzymes that are mainly expressed in the small intestine and liver [2]. It has been used off-label as a treatment for steroid-refractory UC for the past 20 years [3]. Other uses for CyA include treatment of autoimmune disorders such as rheumatoid arthritis, psoriasis and Crohn’s disease. The therapeutic guidelines recommend continuous i.v infusion of 2–4 mg/kg/day CyA followed by oral administration of 4–8 mg/kg/day for up to 3 months [4]. Recent studies comparing CyA and infliximab in patients with severe, i.v. steroid-refractory UC demonstrated that CyA’s efficacy is comparable to that of infliximab [5, 6]. The clear efficacy is counter-balanced by the risk of severe adverse events related to systemic exposure.
As systemic cyclosporine absorption from the colon is negligible [7], Sublimity Therapeutics developed a SmPill-enabled, colon-targeted, multiple minisphere formulation to deliver cyclosporine in a solubilized, and therefore active, form. An initial randomized controlled study in patients with mild–moderate UC demonstrated that 75 mg SmPill+CyA administered once daily for 4 weeks resulted in superior efficacy and comparable safety and tolerability when compared to placebo [8]. A follow-on study in pigs that were orally gavaged with ethylcellulose : pectin-based, SmPill-enabled CyA formulations demonstrated high levels of CyA in the colon accompanied with low systemic levels when compared to other available CyA formulations [9]. The pig model was used to monitor the release of CyA from the coated minispheres in the GI tract and to optimize the encapsulation design for increased efficiency. Based on this study, an ethylcellulose-based, SmPill-enabled CyA formulation was developed. In a pilot study in eight healthy subjects in which a range of optimized SmPill+CyA doses was evaluated for blood, colon luminal content and colon tissue concentrations, it was demonstrated that 75 mg ethylcellulose-coated, SmPill+CyA administered twice daily for 7 days resulted in colon luminal concentrations that translated to colon tissue concentrations comparable to those achieved following continuous i.v. infusion at the known-to-be-efficacious dose of 2 mg/kg/day (equivalent to 1 µM CyA). Importantly, systemic exposure following administration of the optimized SmPill+CyA across all doses tested was negligible. The CyA concentration in colon luminal contents sampled using flexible sigmoidoscopy in unprepared bowels was approximately 0.75 mg/ml [10]. With such low systemic exposure, it is not expected that SmPill+CyA will be associated with systemic adverse events, thus permitting long-term use of cyclosporine as a treatment for patients with UC. The purpose of this study was to evaluate the effect, if any, directly on the host microbiota within the human gut.
There is increasing interest in the effect of orally ingested drugs on the human gut microbiota. Drugs used for the treatment of IBD include sulfasalazine (SASP), 5-aminosalicyclic acid (5-ASA), corticosteroids, immunomodulatory drugs such as methotrexate (MTX), immunosuppressants such as calcineurin inhibitors [including cyclosporine (CyA) and tacrolimus], and anti-TNF-alpha antibodies such as infliximab. Sulfasalazine is a classic example of the interaction of the gut microbiota with orally administered drugs. Little is known about the interaction of most of these drugs with the gut microbiota. One of these drugs, however, sulfasalazine, is a well-known example of how bacteria can interact with the drug and change its function. Sulfasalazine is a prodrug made of sulfapyridine linked to 5-aminosalicyclic acid (5-ASA) through an azo bond. Sulfasalazine is partially absorbed in the jejunum after oral ingestion. The remaining drug passes to the colon. It is here that it is reduced by coliform bacteria through the bacterial azoreductase enzyme, yielding sulfapyridine and 5-aminosalicyclic acid. Aminosalicyclic acid is the active component that must be released from the compound in order to treat the inflammation [11].
Drugs such as CyA, which have low permeability and poor solubility, have, by design – particularly when designed to be released in the colon – most contact with the host microbiota in the large intestine, as this is the site where both the human microbiota and the colon-targeted drug load is most concentrated. It is estimated that approximately 2000 bacterial species populate the lower region of the GI tract and it is here that the interplay with encapsulated drugs, prodrugs and their conjugated metabolites largely occurs [12]. The gut microbiota and the metabolites they produce significantly affect the health of the host. For instance, short-chain fatty acids (SCFAs) are end-products of the fermentation of undigested carbohydrates and protein by the gut microbiota in the large intestine and play a fundamental role in human health and disease [13]. The most dominant SCFAs (90–95 %) in the colon are butyric acid (butyrate), propionic acid and acetic acid. Butyrate is a major energy source for intestinal epithelial cells and plays a role in the maintenance of gut barrier function [14].
The aim of this study was to assess the effect of encapsulated CyA in SmPill minispheres on the diversity and relative abundance of the gut microbiota in an ex vivo colon model and in healthy subjects who participated in a 7-day pilot study. As such, we wanted to know whether there could be a microbiota effect that contributed to the in vivo efficacy of the treatment. We also investigated the production of SCFAs butyrate, propionate and acetate by the gut microbiota in the ex vivo model system in the presence of coated minispheres (SmPill) with (+) and without (−) CyA. We then assessed the effect of encapsulated CyA, using coated minispheres (SmPill)+CyA, on the levels of various metabolites and SCFAs in faecal samples from healthy human subjects, before and after participation in a 7-day pilot study. Ex vivo and in vivo results were compared to determine the efficacy of using an ex vivo colon model to study the effect of orally ingested drugs on the microbial communities of the GI tract.
Methods
Subject recruitment for ex vivo faecal fermentation study
Six healthy volunteers of varying ages (23–62 years) and genders (two male and four female) provided fresh faecal samples on the day of the experiment. Subjects were in good general health and had not taken antibiotics in the previous 6 months. This work was performed in compliance with APC Ethics Protocol 055.
Fermentation protocol
Fermentations were carried out using the MultiFors fermentation system (Infors UK), as follows: 20 % faecal slurry was prepared from faecal samples from the individual donors under anaerobic conditions by suspending the faecal samples in preboiled and anaerobically cooled 50 mM phosphate buffer, pH 7.0, containing 0.05 % cysteine (Sigma Aldrich, Wicklow, Ireland). One hundred and sixty millilitres of the faecal medium, prepared according to Fooks et al. [15], was added to each of three fermentation vessels. Anaerobic conditions were established by sparging the fermenters with oxygen-free nitrogen for at least 3 h prior to inoculation with 40 ml of the faecal slurry. The vessels contained (i) faecal medium, faecal slurry and 1.5 g coated SmPill minispheres (Sublimity Therapeutics Ltd, Dublin, Ireland) without CyA (minispheres −CyA), (ii) faecal medium, slurry and 1.5 g coated SmPill minispheres with CyA [minispheres +CyA (10 % w/w)] and (iii) faecal medium and slurry only (non-treatment control, NTC). Fermentations were conducted anaerobically at 37 °C for 24 h at pH 6.8, with stirring at 200 r.p.m. Samples were taken at 0, 5, 9, 20 and 24 h for CyA quantification and at 0 and 24 h for Miseq compositional sequencing.
Quantification of CyA by high-performance liquid chromatography (HPLC)
Samples from the faecal fermentations (2 ml) were centrifuged for 60 s at 21 000 g, filtered using a 0.45 µM filter and a 0.2 µM filter, and quantified using HPLC as detailed below. CyA (0.375 mg/ml) in 70 % acetonitrile was diluted 10-fold with 70 % acetonitrile to generate a working standard of 0.037 mg ml−1. The standard (50 µl) was injected onto a Proteo Jupiter (250×4.6 mm, 90 Å. 4 µm) RP-HPLC column (Phenomenex, Macclesfield, UK) with an 88–93 % acetonitrile 0.1 % triflouroacetic acid (TFA) gradient where buffer A is 0.1 % TFA and buffer B is 90 % acetonitrile, 0.1 % TFA. CyA elutes at 10 min or 90.5 % acetonitrile, which equates to 81.5 % acetonitrile. Each sample (50 µl) was run in triplicate and the CyA concentration (mg/ml) was calculated from the area of the standard.
Bacteroides strains and growth conditions
The growth of three Bacteroides sp. was tested in vitro in the presence of uncoated minispheres with (+) and without (−) CyA (0.75 mg/ml; equivalent to colon luminal concentration following administration of 75 mg SmPill+CyA twice daily for 7 days) and in growth medium alone. Minispheres that lacked the ethylcellulose and pectin polymer coating (referred to here as uncoated minispheres) were used to enable rapid release of the CyA to the growth medium. The Bacteroides strains ( Bacteroides distasonis , Bacteroides dorei and Bacteroides fragilis) were maintained long-term at −80 °C in 30 % glycerol/brain heart infusion (BHI) medium (Oxoid Ltd, Hampshire, UK) with 0.5 % (w/v) yeast extract (Oxoid Ltd, Hampshire, UK) (BHIYE). Prior to use, the strains were streaked onto trypticase soy agar (TSA; Becton, Dickinson and Co., Sparks, MD, USA) and incubated anaerobically at 37 °C for 48 h. Subsequently, one colony was inoculated into BHIYE broth and grown overnight anaerobically at 37 °C. SmPill uncoated minispheres +/−CyA were added to the BHIYE, such that the final concentration of CyA was 0.75 mg/ml. BHIYE without uncoated minispheres or CyA served as the negative controls. The medium was inoculated at 1 % with an overnight culture of each strain and incubated with shaking at 200 r.p.m. under anaerobic conditions at 37 °C for 48 h. After incubation a sample from the media was serially diluted and spread plated onto bile esculin agar (BEA) prior to incubation for 48 h anaerobically at 37 °C. After incubation the colonies were counted and the results expressed as colony forming units (c.f.u.)/ml.
Participant recruitment for pilot study
Faecal samples were obtained from eight healthy volunteers (male/22–55) who participated in a pilot clinical study. The participants were administered CyA encapsulated in coated Smpill minispheres and faecal samples were taken at the beginning (day 0; before treatment) and at the end (day 7) of the trial. Samples were stored at −80 °C until required. Ethical approval was obtained by Sigmoid Pharma (SP) (protocol no: CYC 102).
Microbial DNA extraction, 16s rRNA gene amplification and Illumina MiSeq sequencing
In the ex vivo study, samples (1 ml) were taken from each fermentation vessel at 0 h and 24 h and snap frozen at −80 °C for DNA extraction. In the pilot study, 300–400 mg of each frozen faecal sample was taken for DNA extraction from samples taken before treatment (day 0) and after treatment (day 7). The faecal DNA MiniPrep kit (Zymo Research, Cambridge Bioscience, UK) was used for DNA extraction from all samples. The V3–V4 variable region of the 16S rRNA gene was amplified from faecal DNA using the 16S metagenomic sequencing library protocol (Illumina, Saffron Walden, UK). Samples were quantified and libraries were prepared for sequencing as described previously [16]. In brief, for the 16SrRNA amplicon sequencing, two PCR reactions were completed on the template DNA. Initially the DNA was amplified with primers specific to the V3–V4 region of the 16S rRNA gene, which also incorporates the Illumina overhang adaptor (forward primer 5′ TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG; reverse primer 5′ GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC). For the second PCR, two indexing primers (Illumina Nextera XT indexing primers, Illumina, Sweden) were added to allow for demultiplexing. Samples were run on the Illumina Miseq sequencer at the Teagasc Sequencing Centre, Moorepark, Fermoy, Co. Cork, Ireland using the Miseq 500 cycle v2 kit, following standard Illumina sequencing protocols.
Bioinformatic analysis of compositional sequencing data
Raw Illumina 250 bp paired-end sequence reads were merged using fast length adjustment of short reads to improve genome assemblies (FLASH) [17] and quality checked using the split_libraries script from the QIIME package [18]. Reads were then clustered into operational taxonomic units (OTUs) and chimeras were removed with the 64-bit version of USEARCH [19]. Subsequently OTUs were aligned and a phylogenetic tree was generated within QIIME [18]. The minimum level of sequence overlap used when merging the paired reads was 10 bp. Reads were filtered at >Q20 Phred score. A 97 % similarity threshold was used for clustering OTUs. Singletons and rare sequences were removed. Taxonomical assignments were reached using the silva 16S specific database (version 128) [20]. Alpha and beta diversity analysis was also implemented within QIIME, based on weighted and unweighted UniFrac distance matrices. Principal coordinate analysis (PCoA) plots were visualized using EMPeror −0.9.51 dev [21].
SCFA analysis of faecal fermentation samples
Gas chromatography with flame ionization detector (GC-FID) was used to determine SCFA content. A 100 mM SCFA stock solution (butyric acid, iso-butyric (ibutyric) acid, propionic acid and acetic acid) was prepared from reagents of the highest purity available (Sigma) with fresh deionized water. Two-ethyl-butyric acid (2-ETBA; Sigma) was used as the internal standard (IS). A 100 mM stock solution of 2-ETBA was prepared with formic acid. A 10 mM working solution of 2-ETBA and a 20 mM working solution of SCFA mix was made up with deionized milliQ water. Standard solutions containing 70.0, 60.0, 50.0, 40.0, 30.0, 20.0, 10·0, 8·0, 6·0, 4·0, 2·0, 1·0 and 0·5 mM of acetic acid, propionic acid, isobutyric acid and butyric acid were used for calibration of the instrument. Faecal fermentation samples that had been snap frozen at −80 °C were thawed and 2 ml of each sample was centrifuged at 16 000 g for 5 min. The supernatant was then filtered through a 0.2 µM filter. Thirty microlitres of working IS (10 mM) was added in triplicate to 1.5 ml Eppendorf tubes and 270 µl of filtered sample supernatant was then added to each tube. The tubes were vortexed and centrifuged at 16 000 g for 3 min. The supernatant was transferred to GC vials containing an insert and sealed. SCFA concentration was measured using a Varian 8400 GC flame ionization system, and fitted with a ZB-FFAP column (30 m×0·32 mm×0·25 mm) (Phenomenex, Macclesfield, UK) and a Restek Hydroguard deactivation guard column (5 m, 0.32 mm ID). Helium was used as the carrier gas at a flow rate of 1.3 ml/min. An initial oven temperature of 50 °C was held for 0.5 min, raised to 140 °C at a rate of 10 °C min−1, held for 0.5 min for a total of 10 min and finally raised to 240 °C at a rate of 20 °C/min and held for 12 min to give a total run time of 27 min. The temperature of the detector and injector was set at 300 °C. Peaks were integrated by using the Varian Star Chromatography Workstation version 6.0 software. Standards were included in each run to maintain calibration.
Metabolite analysis of faecal samples from the human pilot study
Preparation of faecal water
Four hundred milligrams of frozen faecal material was placed into a sterile 2 ml microcentrifuge tube. Eight hundred microlitres of sterile water was added to the 2 ml microcentrifuge tube, on ice. The sample was vortexed vigorously for 30 s to generate a slurry. Samples were then centrifuged at 12 000 g for 30 min at 4 °C. The supernatant was removed and transferred to a new 2 ml microcentrifuge tube. This step was then repeated. The supernatant was then centrifuged at 12 000 g for 30 min at 4 °C. The supernatant was removed again and transferred to a new 2 ml microcentrifuge tube. The supernatant was then centrifuged again at 12 000 g for 30 min at 4 °C. This supernatant was transferred to a 0.2 µM costar spin-x centrifuge tube (Sigma) and filtered by centrifugation at 12 000 g for 10 min at 4 °C. The filter was removed from the tube and the faecal water was stored at –20 °C for metabolomics testing. Samples were sent to MS-Omics ApS, Frederiksberg, Denmark for analysis of metabolites including the SCFA.
Statistical analysis
Non-parametric Kruskal–Wallis and Mann–Whitney U tests were performed in SPSS (version 24) to identify significantly different bacterial taxa in the different groups. Statistical significance was accepted as P≤0.05, with adjusted measures. To control for multiple testing, false discovery rate (FDR)-adjusted q-values (q≤0.05) were calculated for comparisons of taxa using the Benjamini–Hochberg method [22] in GraphPad Prism 6.04. To minimize the number of null hypotheses that had to be corrected for, OTUs that were present in at least 50 % of samples with a minimum of 0.1 % relative abundance were included in the analysis. Univariate analysis of variance (ANOVA) and post-hoc Tukey tests were performed using SPSS to determine the significant differences between the groups for alpha diversity, SCFA and metabolites. Statistical significance was accepted as P≤0.05. Box plots were created using GraphPad Prism 6.04.
Results
The overall aim of this study was to investigate whether cyclosporine had any effect on the human gut microbiota. To achieve this, encapsulated cyclosporine was tested both ex vivo and in vivo.
Measurement of CyA release from coated minispheres during the ex vivo fermentation
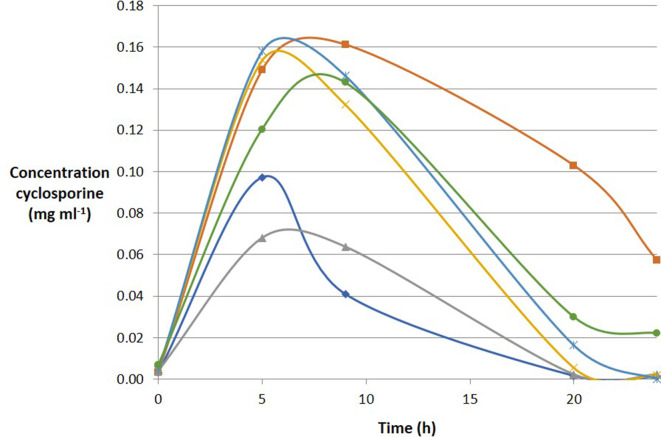
The concentration of CyA released from the coated minispheres, in the presence of a faecal inoculum prepared from each of six healthy donors, was measured over 24 h in the fermentation system (Fig. 1). The results showed that the release of CyA from the coated minispheres varied with the donor inoculum, but the peak detection time across the six donors ranged between 5 and 9 h.
Fig. 1.
Release of cyclosporine A (CyA) from SmPill minispheres over time during 24 h fermentation in an ex vivo model. Each colour represents an individual faecal sample.
The effect of CyA and coated minispheres on the faecal microbiota in the ex vivo model
Following total DNA extraction from faecal samples taken at 0 and 24 h during the ex vivo fermentation process, the V3–V4 16S rRNA gene amplicons were generated and sequenced using the Illumina MiSeq. A total of 9 747 625 reads passing the quality filter were obtained with an average of 270 767 reads per sample. To determine if alterations in microbial diversity occurred in the presence of coated minispheres, +/−CyA, diversity, richness and coverage estimations were calculated for each dataset (alpha diversity) (Table S1, available in the online version of this article). No significant difference in alpha diversity occurred between the three groups across six subjects. This was determined using Chao 1 (P=0.56), Simpson’s diversity index (P=0.23) and Shannon index measurements (P=0.38). Beta-diversity, a term for the comparison of samples based on the similarity of their OTUs, was also measured. Beta-diversity was estimated using distance matrices built from weighted and unweighted UniFrac distances. PCoA was performed on the distance matrices from the unweighted UniFrac distance matrices. The data show that the samples cluster by subject, not by treatment (Fig. S1). Sequence analysis revealed that Firmicutes (62 %), Bacteroidetes (30 %), Actinobacteria (4 %) Proteobacteria (3 %) and Cyanobacteria (2 %) were the dominant phyla across the six subjects at 0 h. Following 24 h fermentation, the treatment groups +/−CyA and NTC samples shared common phyla. No significant effect of minispheres +/−CyA on the composition of the gut microbiota was observed after incubation for 24 h (Fig. S2).
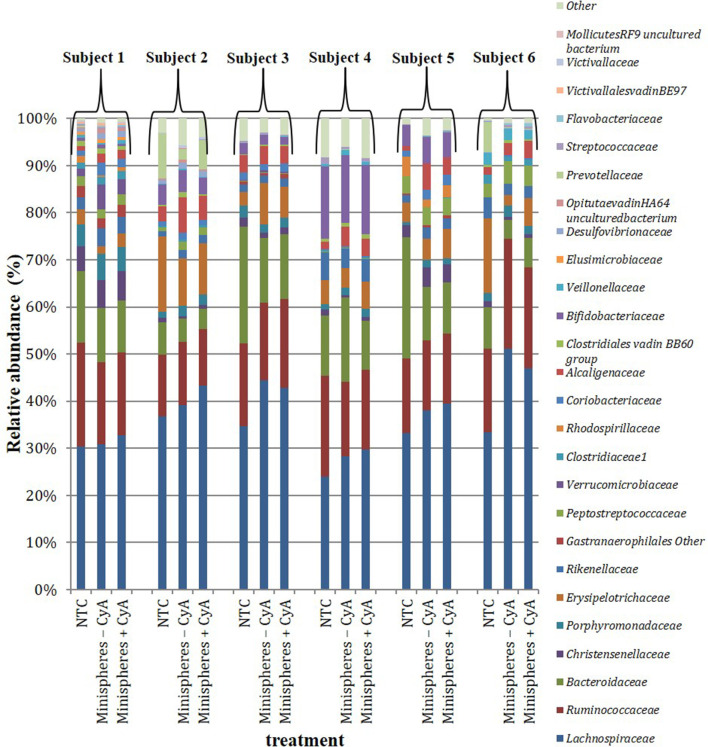
Seventy-six family-level taxa were identified, of which 44 were present in at least 1 subject, with a minimum relative abundance of 0.1 % or greater after 24 h across the 3 treatment groups. The most dominant families detected across all subjects and treatment groups at 24 h fermentation were Lachnospiraceae , Ruminococcaceae and Bacteroidaceae (Fig. 2). Treatment with minispheres +/−CyA did not have any effect on the relative abundance of any of the families present, compared to the NTC, and there were no significant differences observed between the three treatment groups. Two hundred and forty-two genera were identified, of which 133 were present in at least 1 subject with a minimum relative abundance of 0.1 % or greater after 24 h across the 3 treatment groups. The most dominant genera detected across all subjects were Bacteroides , Blautia , Faecalibacterium and Lachnospiraceae (Fig. S3). While there was a slight decrease in the relative abundance of Bacteroides after treatment with minispheres +/−CyA, this was not significant. There was, however, a slight decrease in Bacteroidetes phyla (not significant) in four out of six subjects in the groups with minispheres +/−CyA (median 16%) compared to the NTC (median 22%) (Fig. 3a). Since the phylum Bacteroidetes was the only one to be decreased in the presence of the minispheres +/−CyA compared to the NTC, the growth of three pure cultures of Bacteroides sp. that were representative of the phylum Bacteroidetes was tested in vitro in the presence of uncoated minispheres +/−CyA (0.75 mg/ml) and growth medium alone. The impact on cell viability of the minispheres +/−CyA varied between species. While there was no impact on the cell viability of B. fragilis after 48 h incubation, there was a 2-log reduction in cell viability for B. dorei in the presence of minispheres +/−CyA, indicating that it was the minispheres that were impacting on the viability of B. dorea and not the CyA. In contrast, there was a 1-log reduction in B. distasonis when incubated at 37 °C for 48 h in the presence of minispheres +CyA, but the minispheres alone did not have any additional effect (Table S6).
Fig. 2.
Relative abundance (%) of the bacterial families across treatment groups in faecal samples from six individual subjects (subjects 1–6) following 24 h fermentation in an ex vivo colon model. Groups: non-treatment control (NTC); minispheres without (−) CyA; minispheres with (+) CyA.
Fig. 3.
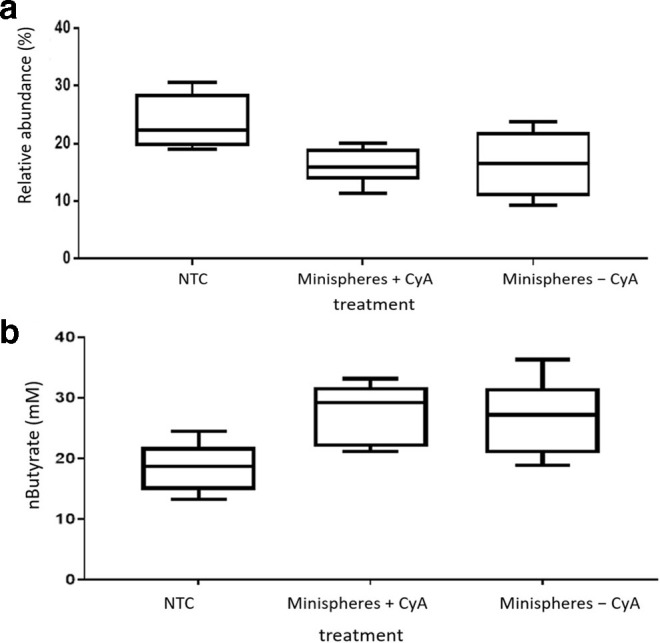
(a) Relative abundance in operational taxonomic units (OTU)s (%) of the phylum Bacteroidetes from each treatment group following 24 h fermentation in an ex vivo model. Groups: non-treatment control (NTC); minispheres with (+) CyA; minispheres without (−) CyA. (b) Butyrate concentration in samples from the treatment groups following 24 h fermentation in an ex vivo colon model. Groups: non-treatment control (NTC); minispheres +CyA and minispheres −CyA (P=0.018).
The effect of CyA and coated minispheres on SCFA production in an ex vivo colon model
SCFA production is an important beneficial function of the microbiota and therefore we investigated whether CyA and minispheres had an impact on SCFA levels. Butyrate, propionate and acetate levels were assessed as they are the most abundant SCFAs found in the colon. The concentration of n-butyrate was significantly higher in samples with coated minispheres +/−CyA compared to the NTC group (P=0.018), as shown in Fig. 3b. The data indicate that increased n-butyrate production is due to a component of the coated minisphere rather than the effect of CyA on the microbiota. There were increased acetate levels in the samples with minispheres (+/−CyA); however, the increase was not statistically significant. There was no significant change in the levels of isobutyrate or propionate between the three treatment groups (Table S2).
The effect of CyA encapsulated in coated minispheres (SmPill) on the faecal microbiota in healthy subjects participating in a 7-day pilot trial
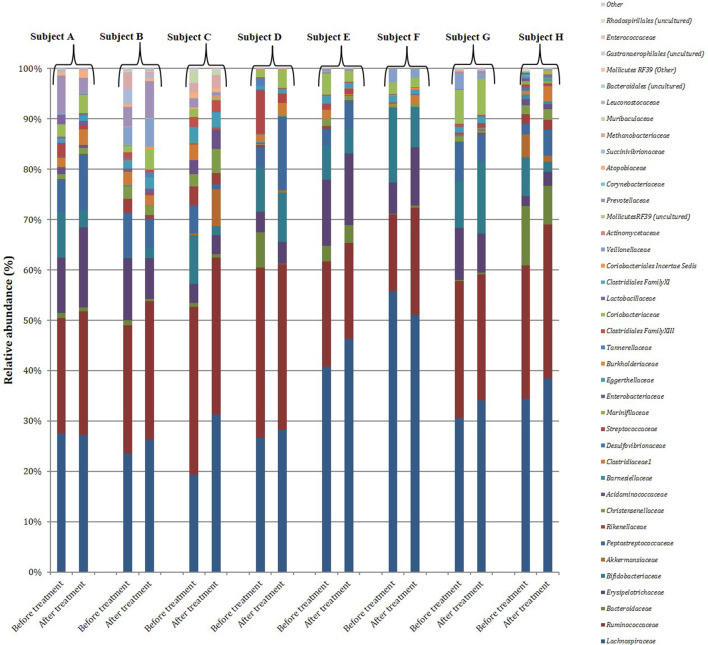
Faecal samples were collected from eight healthy subjects who participated in the 7-day pilot study before and after SmPill+CyA administration. Following total metagenomic DNA extraction from the 16 faecal samples, the V3–V4 16S rRNA gene amplicons were generated and sequenced using the Illumina MiSeq. A total of 4 481 418 reads passing the quality filter were obtained with an average of 280 089 reads per sample. Diversity, richness and coverage estimations were calculated for each dataset to determine if alterations in microbial diversity occurred in the presence of SmPill+CyA. No significant difference in alpha diversity occurred between the before and after treatment samples across eight subjects (Table S3). This was determined using Chao 1 (P=0.99), Simpson’s diversity index (P=0.84) and Shannon index measurements (P=0.96). There was no significant difference in beta diversity on the PCoA plots before and after treatment with SmPill+CyA (P=0.95). The data show that the samples clustered by subject and not by treatment. As in the faecal ferment samples, the most dominant phyla were Firmicutes , Bacteroidetes and Actinobacteria across all eight subjects before and after treatment. There was no significant difference in the distribution of any of the phyla before and after treatment with SmPill+CyA after 7 days (Fig. S4). Seventy-nine family-level taxa were identified, of which 39 were present in at least 1 sample with a minimum relative abundance of 0.1 % or greater. The most prevalent families detected across all subjects were Lachnospiraceae, Ruminococcaceae, Bacteroidaceae Bifidobacteriaceae and Erysipelotrichaceae (Fig. 4). There was no significant difference in the relative abundance observed before and after treatment in any of the families present.One hundred and thirty-four genus-level taxa were identified out of a total of 257 genera that were present in at least 1 sample with a minimum relative abundance of 0.1 % or greater. The most dominant genera across all subjects were Blautia, Faecalibacterium, Lachnospiraceae other, Bifidobacterium , Agathobacter , Romboutsia and Bacteroides (Fig. S5). There were no significant differences in the relative abundance of any of these before and after 7 days’ treatment with SmPill+CyA.
Fig. 4.
Relative abundance (%) of the bacterial families identified in human faecal samples from eight subjects (A–H), before and after treatment with CyA encapsulated into Smpill minispheres.
Effect of CyA encapsulated in coated minispheres (SmPill) on the levels of metabolites in faecal samples from healthy subjects participating in a 7-day pilot study
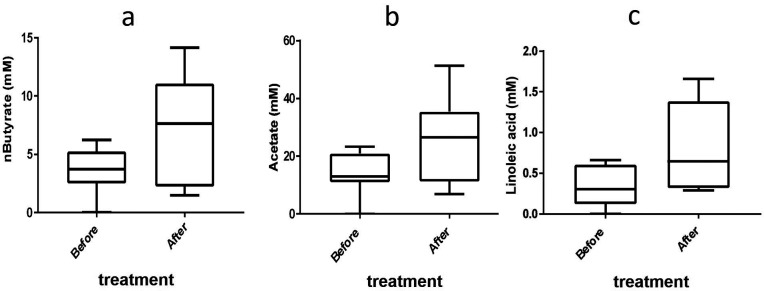
In vivo, the levels of n-butyrate, i-butyrate, propionate and acetate in the faecal samples of eight subjects were assessed before the start of the trial and after 7 days’ treatment. The levels of i-valerate, hexanoate and pentanoate were also measured (Table S4). Butyrate (P=0.05) and acetate (P=0.048) were shown to increase significantly after 7 days’ treatment with SmPill+CyA (See Fig. 5a, b). Thirty-three additional metabolites and amino acids commonly found in human faecal samples were identified (Table S5). There was a significant increase in the concentration of linoleic acid after 7 days’ treatment (P=0.048) (Fig. 5c) .
Fig. 5.
Effect of SmPill+CyA on (a) butyrate levels (P=0.05), (b) acetate levels (P=0.05) and (c) linoleic acid levels (P=0.048) in human faecal samples from subjects participating in a pilot study in which they were administered SmPill+CyA for 7 days. Samples were taken before and after treatment.
Discussion
The liver is the primary site of xenobiotic metabolism and biotransformation, but the way in which drugs are metabolized can vary between individuals. Drugs with narrow therapeutic windows and subject to extensive and variable enzymatic metabolism, such as CyA, often require careful systemic drug concentration monitoring to ensure that patient-specific dosing regimens are implemented [23]. Metabolism of the drug starts at the intestinal level and can have a significant impact on drug bioavailability, efficacy and safety, given that the intestinal microbiota can contribute to xenobiotic metabolism [12]. In addition, xenobiotic exposure has the potential to alter the composition of the gut microbiota [12, 24]. The interaction between drugs and the gut microbiota in the intestine can affect the rate of release of the drug and absorption or uptake in the body.
There is an increasing demand to develop novel methods to formulate poorly soluble drugs such that they withstand release in the upper GI tract and are released in solubilized form within specific regions of the intestine, such as the colon. This is particularly relevant for drugs that specifically target IBD. Therefore, the preferred outcome for a drug destined for release in the lower GI tract is to successfully treat the condition whilst having a beneficial effect or at the least a minimal impact on the gut microbes. This study therefore concentrated on determining the effect of SmPill+/−CyA on the gut microbiota in an ex vivo model of the distal colon that had been inoculated with stool samples from healthy adults. Previous work by our laboratory using the same ex vivo colon model with a similar number of subjects has shown the impact that antimicrobials such as vancomycin and metronidazole have on the gut microbiota diversity and abundance [25]. In addition, we measured the effect of a treatment-appropriate SmPill+CyA dosing regimen (75 mg BID) on the gut microbiota in eight healthy subjects who participated in a 7-day pilot study. While the number of subjects used in both the in vivo and ex vivo studies were limited in these pilot trials, we believe that overall we do see very good diversity in microbiota composition and enough so to evaluate whether cyclosporine would have any effect of magnitude. Indeed the results from the in vivo study supported the findings of the ex vivo work.
The rate of CyA release over time between donor inocula in the fermentation vessels was found to vary. This may be a consequence of CyA adhering to or entering the bacteria in the biomass that is removed before HPLC quantification, or may be due to CyA being metabolized differently depending on the bacterial composition of the inoculum. CyA has been shown to be highly stable in a colonic model [26] due to its unique cyclic structure, N-methylation and high lipophilicity [27, 28] making it less prone to metabolism by colonic bacteria. The CyA concentration in colon luminal contents sampled using flexible sigmoidoscopy in unprepared bowels of healthy subjects who were administered 75 mg twice daily for 7 consecutive days was approximately 0.75 mg/ml. The concentration range that we anticipated to measure therefore was between 0.05 and 0.75 mg/ml, representing levels that the bacteria would be exposed to in the human gut following administration of SmPill+CyA. Although the concentration of CyA released varied between subjects, sufficient levels of CyA were measured across all vessels to obtain clinically relevant colonic tissue concentrations.
CyA encapsulated in SmPill did not affect the alpha or beta diversity of faecal microbiota in vivo in healthy human subjects administered the drug for 7 days or in the ex vivo fermentation conditions. In the ex vivo study, sequencing analysis revealed that the phylum Bacteroidetes was slightly decreased in the presence of minispheres +/−CyA when compared to the NTC. The exposure of three pure cultures of Bacteroides sp. to the minispheres +/−CyA revealed variable responses in that the minispheres: B. dorei showed reduced cell viability, while CyA decreased the viability of B. distasonis , but neither the minispheres nor the drug impacted on the viability of B. fragilis . In addition, CyA had no inhibitory effect on pure cultures of Lactobacillus or Bifidobacterium species that had been tested previously in our laboratory (data not shown). Therefore, the impact of SmPill+CyA on Bacteroidetes abundance levels is presumably influenced by the base levels and types of Bacteroidetes members in the microbiota of each individual prior to treatment.
Concentrations of the SCFAs butyrate, propionate and acetate were measured at 0 and 24 h in the ex vivo model. Levels of n-butyrate and acetate were shown to increase in the presence of the SmPill minispheres +/−CyA, indicating that a bead component, yet to be identified, was having an influence on the gut microbiota.
The coating on the minispheres is ethylcellulose based. Ethylcellulose is a plant-derived polysaccharide that can be broken down by microbial carbohydrate-degrading enzymes such as cellulases. Some cellulolytic strains that have been isolated and characterized from human faeces include Ruminococcus sp., Clostridium sp., Bacteroides sp. and Eubacterium sp. [29]. SCFAs are products of microbial fermentation of such indigestible carbohydrates in the large intestine. A high proportion of these SCFAs are absorbed by the host and are therefore an important energy source to the host. It is estimated that these SCFAs account for around 10 % of the calories obtained from the diet that otherwise would be excreted as undegraded substrate in the faeces [29, 30].
Other health benefits of SCFAs include maintenance of gut barrier function [14], protection against colorectal cancer (CRC) [31, 32] and protection against obesity and insulin resistance [33]. Butyrate reduces inflammation, inhibits tumour cell progression, induces apoptosis, promotes colon motility and increases visceral irrigation [31, 32]. The relative abundance of other butyrate-producing bacteria that have been suggested to be implicated in UC, such as Faecalibacterium prausnitzii and Roseburia sp., was also investigated in this study, but no significant changes were observed either in the ex vivo model or in vivo (data not shown). The increase in butyrate concentration could be viewed as a favourable outcome in terms of human health.
Conclusions
We observed that the concentration of encapsulated CyA in the SmPill formulation (likely to be encountered by the gut microbiota in vivo) did not impact on gut microbiota composition when tested in both an ex vivo colon model and an in vivo pilot study in healthy volunteers. CyA is an immunosuppressant and we cannot rule out that its impact on the host immune response could indirectly influence the gut microbiota. However, our study revealed that some component of the SmPill formulation (described previously) significantly increased the concentration of n-butyrate, a microbial metabolite with known positive attributes for host health.
An initial randomized, controlled study in patients with mild–moderate UC demonstrated that 75 mg SmPill+CyA administered once daily for 4 weeks resulted in superior efficacy and comparable safety and tolerability when compared to placebo.
The 7-day time period of the pilot study should have been sufficient to see changes in the macro-composition of the gut microbiota in this study (as we do observe with different antibiotics), but it does appear to be sufficient to alter the function of the microbes. We observed that the levels of acetate, butyrate and linoleic acid were all increased (approximately doubled) following SmPill treatment. The results from the compositional and SCFA analysis of the healthy human subjects correspond to the ex vivo faecal fermentation findings, suggesting that the ex vivo model is effective for studying the impact of orally ingested drugs on the gut microbiota. Moving forward with SmPill-encapsulated CyA, future studies with a larger cohort will investigate the efficacy, safety and tolerability across a range of doses in UC patients in clinical trials.
Supplementary Data
Funding information
This publication has emanated from research supported by a research centre grant from Science Foundation Ireland to APC Microbiome Ireland under grant number SFI/12/RC/2273 and a research grant from Sublimity Therapeutics Ltd (formerly known as Sigmoid Pharma Ltd).
Author contributions
C. O. R. performed the laboratory assays, data analysis and wrote the manuscript. O. O. S. performed the bioinformatic analysis. F. C. performed the 16 s rRNA gene sequencing. P. M. O. C. performed the HPLC analysis. I. C., R. P. R., F. S., M. C. R., C. H. and A. C. conceived and supervised the study.
Conflicts of interest
This work was supported by Science Foundation Ireland, APC Microbiome Ireland and Sublimity Therapeutics Ltd (formerly known as Sigmoid Pharma Ltd).
Ethical statement
The ex vivo studies were carried out in compliance with APC Ethics Protocol 055. The in vivo study was performed in compliance with the protocol CYC 102. Ethical approval was obtained by Sigmoid Pharma Ltd. Written informed consent was obtained from all participants.
Footnotes
Abbreviations: CRC, colorectal cancer; CyA, cyclosporine A; GI, gastrointestinal; HPLC, high-performance liquid chromatography; IBD, inflammatory bowel disease; NTC, non-treatment control; SCFA, short-chain fatty acids.
Five supplementary figures and six supplementary tables are available with the online version of this article.
References
- 1.Knights D, Lassen KG, Xavier RJ. Advances in inflammatory bowel disease pathogenesis: linking host genetics and the microbiome. Gut. 2013;62:1505–1510. doi: 10.1136/gutjnl-2012-303954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guada M, Beloqui A, Kumar MNVR, Préat V, Dios-Viéitez MDC, et al. Reformulating cyclosporine A (CsA): more than just a life cycle management strategy. J Control Release. 2016;225:269–282. doi: 10.1016/j.jconrel.2016.01.056. [DOI] [PubMed] [Google Scholar]
- 3.Sharkey L, Bredin F, Nightingale A, Parkes M. The use of cyclosporin A in acute steroid-refractory ulcerative colitis: long term outcomes. J Crohns Colitis. 2011;5:91–94. doi: 10.1016/j.crohns.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Ordás I, Eckmann L, Talamini M, Baumgart DC, Sandborn WJ. Ulcerative colitis. The Lancet. 2012;380:1606–1619. doi: 10.1016/S0140-6736(12)60150-0. [DOI] [PubMed] [Google Scholar]
- 5.Laharie D, Bourreille A, Branche J, Allez M, Bouhnik Y, et al. Ciclosporin versus infliximab in patients with severe ulcerative colitis refractory to intravenous steroids: a parallel, open-label randomised controlled trial. The Lancet. 2012;380:1909–1915. doi: 10.1016/S0140-6736(12)61084-8. [DOI] [PubMed] [Google Scholar]
- 6.Williams JG, Alam MF, Alrubaiy L, Arnott I, Clement C, et al. Infliximab versus ciclosporin for steroid-resistant acute severe ulcerative colitis (construct): a mixed methods, open-label, pragmatic randomised trial. Lancet Gastroenterol Hepatol. 2016;1:15–24. doi: 10.1016/S2468-1253(16)30003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drewe J, Beglinger C, Kissel T. The absorption site of cyclosporin in the human gastrointestinal tract. Br J Clin Pharmacol. 1992;33:39–43. doi: 10.1111/j.1365-2125.1992.tb03998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Donoghue DP, Bloom S, Coulter I. Colon targeted, low systemic absorption soluble ciclosporin in ulcerative colitis. Gut. 2013;62:A1.3–A2. doi: 10.1136/gutjnl-2013-305143.3. [DOI] [Google Scholar]
- 9.Keohane K, Rosa M, Coulter IS, Griffin BT. Enhanced colonic delivery of ciclosporin a self-emulsifying drug delivery system encapsulated in coated minispheres. Drug Dev Ind Pharm. 2016;42:245–253. doi: 10.3109/03639045.2015.1044905. [DOI] [PubMed] [Google Scholar]
- 10.Colombel J-F, Coulter IS, Hall J, Rosa M, McDonald B, et al. P053 targeted release oral cyclosporine formulation as a potential new therapy for ulcerative colitis. Gastroenterology. 2019;156:S37–S38. doi: 10.1053/j.gastro.2019.01.109. [DOI] [Google Scholar]
- 11.Peppercorn MA, Goldman P. The role of intestinal bacteria in the metabolism of salicylazosulfapyridine. J PharmacolExpTher. 1972;181:555. [PubMed] [Google Scholar]
- 12.Wilson ID, Nicholson JK. Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl Res. 2017;179:204–222. doi: 10.1016/j.trsl.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, et al. The role of short-chain fatty acids in health and disease. Advanced Immunology. 2014;121:91–119. doi: 10.1016/B978-0-12-800100-4.00003-9. [DOI] [PubMed] [Google Scholar]
- 14.Ríos-Covián D, Ruas-Madiedo P, Margolles A, Gueimonde M, de Los Reyes-Gavilán CG, et al. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol. 2016;7:185. doi: 10.3389/fmicb.2016.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fooks LJ, Gibson GR. Mixed culture fermentation studies on the effects of synbiotics on the human intestinal pathogens Campylobacter jejuni and Escherichia coli. Anaerobe. 2003;9:231–242. doi: 10.1016/S1075-9964(03)00043-X. [DOI] [PubMed] [Google Scholar]
- 16.Fouhy F, Deane J, Rea MC, O’Sullivan Órla, Ross RP, et al. The effects of freezing on faecal microbiota as determined using MiSeq sequencing and culture-based investigations. PLoS One. 2015;10:e0119355. doi: 10.1371/journal.pone.0119355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magoč T, Salzberg SL. Flash: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar RC. Search and clustering orders of magnitude faster than blast. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 20.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, et al. The Silva ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vázquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. Emperor: a tool for visualizing high-throughput microbial community data. Gigascience. 2013;2:16. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B. 1995;57:289–300. [Google Scholar]
- 23.Jorga A, Holt DW, Johnston A. Therapeutic drug monitoring of cyclosporine. Transplant Proc. 2004;36:S396–S403. doi: 10.1016/j.transproceed.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 24.Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rea MC, Dobson A, O'Sullivan O, Crispie F, Fouhy F, et al. Effect of broad- and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc Natl Acad Sci U S A. 2011;108:4639–4644. doi: 10.1073/pnas.1001224107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Yadav V, Smart AL, Tajiri S, Basit AW. Stability of peptide drugs in the colon. Eur J Pharm Sci. 2015;78:31–36. doi: 10.1016/j.ejps.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 27.Biron E, Chatterjee J, Ovadia O, Langenegger D, Brueggen J, et al. Improving oral bioavailability of peptides by multiple N-methylation: somatostatin analogues. Angew. Chem. Int. Ed. 2008;47:2595–2599. doi: 10.1002/anie.200705797. [DOI] [PubMed] [Google Scholar]
- 28.Haviv F, Fitzpatrick TD, Swenson RE, Nichols CJ, Mort NA, et al. Effect of N-methyl substitution of the peptide bonds in luteinizing hormone-releasing hormone agonists. J Med Chem. 1993;36:363–369. doi: 10.1021/jm00055a007. [DOI] [PubMed] [Google Scholar]
- 29.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012;3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNeil NI. The contribution of the large intestine to energy supplies in man. Am J Clin Nutr. 1984;39:338–342. doi: 10.1093/ajcn/39.2.338. [DOI] [PubMed] [Google Scholar]
- 31.Canani RB, Costanzo MD, Leone L, Pedata M, Meli R, et al. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. WJG. 2011;17:1519–1528. doi: 10.3748/wjg.v17.i12.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keku TO, Dulal S, Deveaux A, Jovov B, Han X. The gastrointestinal microbiota and colorectal cancer. Am J Physiol Gastrointest Liver Physiol. 2015;308:G351–G363. doi: 10.1152/ajpgi.00360.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao Z, Yin J, Zhang J, Ward RE, Martin RJ, et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009;58:1509–1517. doi: 10.2337/db08-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.