Abstract
Introduction.
An important factor for delayed healing of chronic wounds is the presence of bacteria. Quorum sensing (QS), a cell density-dependent signalling system, controls the production of many virulence factors and biofilm formation in Pseudomonas aeruginosa .
Aim.
Inhibition by sodium salicylate (NaSa) of QS-regulated virulence expression was evaluated in QS-characterized clinical wound isolates of P. aeruginosa, cultured in serum-containing medium.
Methodology.
Fourteen clinical P. aeruginosa strains from chronic wounds were evaluated for the production of QS signals and virulence factors. Inhibition of QS by NaSa in P. aeruginosa clinical strains, wild-type PAO1 and QS reporter strains was evaluated using in vitro assays for the production of biofilm, pyocyanin, siderophores, alkaline protease, elastase and stapholytic protease.
Results.
Six clinical strains secreted several QS-associated virulence factors and signal molecules and two were negative for all factors. Sub-inhibitory concentrations of NaSa downregulated the expression of the QS-related genes lasB, rhlA and pqsA and reduced the secretion of several virulence factors in PAO1 and clinical strains cultured in serum. Compared to serum-free media, the presence of serum increased the expression of QS genes and production of siderophores and pyocyanin but decreased biofilm formation.
Conclusions.
Pseudomonas aeruginosa from chronic wound infections showed different virulence properties. While very few strains showed no QS activity, approximately half were highly virulent and produced QS signals, suggesting that the targeting of QS is a viable and relevant strategy for infection control. NaSa showed activity as a QS-inhibitor by lowering the virulence phenotypes and QS signals at both transcriptional and extracellular levels.
Keywords: Pseudomonas aeruginosa, quorum sensing, wound infections, serum, sodium salicylate
Introduction
Chronic or hard-to-heal wounds have a significant impact on quality of life and are associated with a large health-economic burden (2–4 % of health care budgets) [1–3]. Between 1 and 1.5 % of the population in the industrialized world will have a hard-to-heal wound during their lifetime, and infection is an important complication. One reason for wound chronicity is the high prevalence of biofilms (in 80 % of chronic wounds) [4], which results in a drastic increase in bacterial tolerance towards antibiotics [5, 6]. Poor treatment outcome together with the rapid spread of antibiotic resistance, which is estimated to cause more yearly deaths than cancer by 2050 [7], stresses the need for alternative antimicrobial treatment strategies [3, 8].
Two of the most frequently isolated bacterial species in chronic wounds are Staphylococcus aureus and Pseudomonas aeruginosa , with respective prevalences ranging between 32–94 % and 17–52 % [9, 10]. Particularly, P. aeruginosa has been shown to destroy host tissues and is associated with stalled and deteriorating wounds [10–12]. The virulence of the infecting bacteria is believed to play an important role in wound chronicity [13]. Today, it is well established that quorum sensing (QS), a cell and signal density-dependent communication system used by a wide range of micro-organisms, regulates the synthesis of many virulence factors (for a recent review see Lee and Zhang [14]). Pseudomonas aeruginosa is globally regulated by at least three QS systems: two N-acyl-homoserine lactone (HSL)-mediated quorum-sensing systems, las and rhl, and the Pseudomonas quinolone signal (pqs) system (2-heptyl-3-hydroxy-4-quinolone, PQS). The HSL synthases LasI and RhlI mediate the synthesis of the autoinducers N-(3-oxo-dodecanoyl)-L-homoserine lactone (3-OC12-HSL) and N-butanoyl-L-homoserine lactone (C4-HSL), respectively.
The toxic effects of P. aeruginosa are largely mediated by QS-regulated virulence factors, including pyocyanin, elastase and alkaline protease. Pyocyanin is both toxic [15] and involved in biofilm architecture [16, 17]; elastase and alkaline protease degrade host tissues and are involved in iron acquisition from host proteins [18] and host immune evasion [19, 20]. Other virulence-associated factors are rhamnolipids, siderophores and stapholytic proteases. The siderophore pyoverdine plays an essential role in P. aeruginosa virulence by capturing iron and regulating the production of exotoxin A and an endoprotease [21]. Rhamnolipids are surfactants with haemolytic and necrotic activities [22], which influence the structure of biofilms [23], and like stapholytic protease [24], are toxic towards a variety of micro-organisms conferring upon P. aeruginosa a competitive advantage in the colonization of wound-tissue niches. QS has been found to influence biofilm formation in vitro [25, 26]. However, factors other than QS also play a role in biofilm formation since some QS-deficient clinical strains of P. aeruginosa were found to form biofilms in vivo [27] and in vitro with larger surface coverages than the fully QS-competent PAO1 [28]. All of these virulence factors, which are QS-dependent, are major contributors to the ability of this bacterium to cause disease.
Although there is evidence that P. aeruginosa clinical isolates from chronic wounds can produce QS signal molecules [13], there is less literature on the characterization of virulence factors. Since QS signals affect virulence-factor production and biofilm development [14], a wound-treatment concept based on QS inhibition would represent an attractive strategy for the prevention and/or treatment of wound infections. Salicylic acid, a metabolite of aspirin, has previously been evaluated as a quorum-sensing inhibitor (QSI) in vitro [29, 30]; however, its role as a potential anti-virulence compound to treat human infectious diseases remains to our best knowledge limited to keratitis caused by P. aeruginosa [31]. The poor solubility of salicylic acid led us to investigate the potential QS inhibitory effect of the sodium salt of salicylic acid, sodium salicylate (NaSa), which is several hundred-fold more soluble in water than salicylic acid [32, 33].
Screening and evaluation of QSIs are often carried out in simplified in vitro test systems based on the cultivation of bacteria in standard media, e.g. minimal medium or lysogeny broth (LB) [34–36]. Although of great importance in evaluating the QS pathways and potential QSIs, these standard media are far from in vivo wound conditions, which could result in the risk of limited compound efficacy when QSIs are tested in vivo and thus also clinically. A method that represents being one step closer to the evaluation of QSIs in a wound-relevant milieu is to supplement the test medium with serum to better simulate the characteristics of the wound exudate. The total protein content of chronic wound fluid has been determined to be approximately 50 % of the protein content found in serum [37, 38]. Serum and its abundant serum protein albumin are known to interact with and potentially decrease the bioavailability of a wide variety of compounds [39, 40]. Hence, including 50 % serum in the test medium when evaluating QSIs or when studying the phenotypic behaviour of wound pathogens in general increases the relevance of in vitro chronic wound models. Taken together, to evaluate the efficacy of potential QSI compounds in the treatment of chronic wounds, it is important to gain more knowledge on the virulence of wound clinical isolates and to use relevant media that resemble the wound bed environment.
The aim of this study was to first characterize 14 clinical P. aeruginosa strains isolated from chronic ulcers in terms of their ability to produce QS signal molecules and a range of virulence factors, and then to evaluate the effect of NaSa on the expression of QS genes and production of virulence factors in selected clinical and reference strains grown in serum-containing simulated wound fluid.
Results
Characterization of clinical strains isolated from chronic wounds
To add to the current understanding of QS in chronic ulcers and to test the NaSa treatment effect, 14 clinical P. aeruginosa strains isolated from chronic wounds were included in the study. All clinical strains were confirmed to belong to the P. aeruginosa species using the API kit (bioMérieux SA, Marcy-l’Etoile, France) based on 20 miniature biochemical tests to determine an analytical profile index (API) with scores ≥99.5 % (data not shown). All the strains, including the reference strain PAO1, were then characterized in terms of their ability to produce a number of QS-modulated virulence factors (Table 1), QS signals and biofilm (Table 2). Four clinical strains and PAO1 were positive for all seven tested virulence factors, and two additional strains were positive for five virulence factors. In contrast, two strains were completely negative for all virulence factors evaluated. Of the 14 clinical strains, pyocyanin and siderophores were found in eight strains, stapholytic protease and rhamnolipid in seven strains, alkaline protease in six strains, and elastase and swarming ability were detected in five strains.
Table 1.
Characterization of the ability of P. aeruginosa clinical isolates from chronic wounds to produce virulence factors. All assays were agar-plate-based except for rhamnolipid production, which was evaluated in a glycerol-based liquid medium. Serum-supplemented agar was used for characterization of siderophore production
|
Strain |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
PAO1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Pyocyanin |
+ |
− |
− |
+ |
+ |
+ |
− |
+ |
− |
− |
+ |
+ |
− |
+ |
+ |
|
Siderophores |
+ |
− |
+ |
+ |
+ |
+ |
− |
− |
− |
− |
+ |
+ |
+ |
− |
+ |
|
Alkaline protease |
+ |
− |
− |
+ |
+ |
+ |
− |
+ |
− |
− |
− |
+ |
− |
− |
+ |
|
Elastase |
+ |
− |
− |
+ |
+ |
+ |
− |
− |
− |
− |
− |
+ |
− |
− |
+ |
|
Stapholytic protease |
+ |
+ |
− |
− |
+ |
+ |
− |
+ |
− |
− |
− |
+ |
− |
+ |
+ |
|
Rhamnolipid |
+ |
− |
− |
+ |
+ |
+ |
+ |
+ |
− |
− |
− |
+ |
− |
− |
+ |
|
Swarming |
+ |
− |
− |
− |
+ |
+ |
− |
+ |
− |
− |
− |
+ |
− |
− |
+ |
Table 2.
Characterization of clinical P. aeruginosa strains from chronic wounds according to their production of the QS signals C4-HSL, 3-OC12-HSL and PQS and biofilm formation on polystyrene. Supernatants from 24-h-old cultures of each strain in 50 % serum in saline were used for QS-signal analysis using fluorescent reporter strains. Biofilm formation was evaluated on polystyrene surfaces after 48 h culture in 50 % serum in saline using the Calgary biofilm device
|
Strain |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
13 |
14 |
PAO1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
C4-HSL |
+ |
− |
− |
+ |
+ |
+ |
− |
− |
+ |
− |
+ |
+ |
+ |
− |
+ |
|
3-OC12-HSL |
+ |
− |
− |
+ |
+ |
+ |
+ |
− |
− |
− |
+ |
+ |
+ |
− |
+ |
|
PQS |
+ |
− |
− |
+ |
+ |
+ |
− |
− |
+ |
− |
+ |
+ |
− |
− |
+ |
|
Biofilm OD520 (STDEV) |
1.2 (0.1) |
1.4 (0.4) |
1.1 (0.6) |
1.8 (0.2) |
2.4 (0.2) |
2.0 (0.7) |
1.6 (0.9) |
1.1 (0.5) |
1.2 (0.5) |
1.0 (0.4) |
0.9 (0.1) |
0.9 (0.4) |
1.0 (0.2) |
0.8 (0.2) |
2.0 (0.9) |
Regarding QS signal production, six out of 14 strains were positive for all three QS signals, nine strains and PAO1 were positive for ≥1 QS signal, while in five strains, none of the signals were detected (limits of detection: 0.39 nM, 0.39 nM and 19.5 nM for C4-HSL, 3-OC12-HSL and PQS, respectively) (Table 2). Concerning their abilities to form biofilm on polystyrene pegs, no clear connection was observed between signal/virulence-factor production and biofilm formation. Three strains (strains 4, 5 and 6) resulted in the highest OD values (similar biofilm biomass as PAO1); the same strains also produced all three QS signals and ≥5 virulence factors. Among the strains with the lowest biofilm biomass, strains 11 and 12 produced the three QS signals, and strain 13 produced two QS signals. In contrast, strains 2 and 7 were moderate biofilm producers with low production of virulence factors.
Effect of NaSa on QS modulation in PAO1
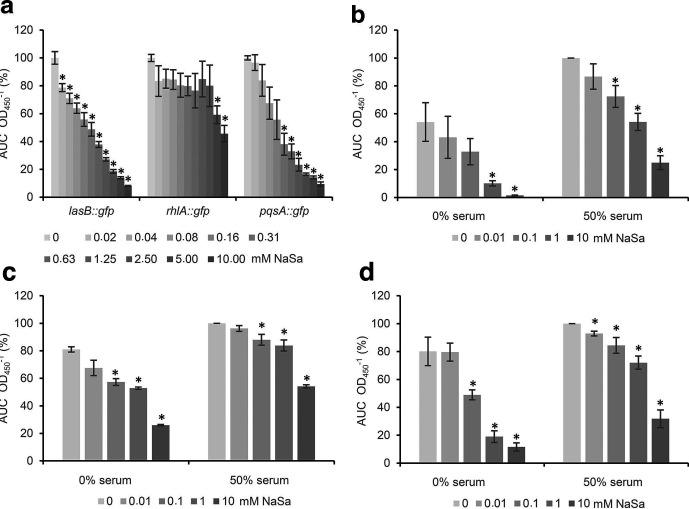
First, the QS modulatory effect of NaSa was evaluated using PAO1-based QS-reporter strains cultured in a commonly used minimal medium [34, 41]. Fluorescent signals from green fluorescent protein (GFP) were acquired and normalized by OD to account for any NaSa-dependent differences in growth rate (Fig. S1, available in the online version of this article). The highest concentration of NaSa tested was below the MIC for all strains (Table S1, available in the online version of this article). Treatment with up to 10 mM NaSa resulted in an overall downregulation of the las, rhl and pqs QS systems in a concentration-dependent manner, as measured by the three reporter strains (Fig. 1a). NaSa treatment was more effective against the las (lasB::gfp) and pqs (pqsA::gfp) systems than the rhl (rhlA::gfp) system. The highest dose of NaSa (10 mM) inhibited the expression levels of lasB by 12-fold, of pqsA by 11-fold and of rhlA by 2-fold. NaSa concentrations of 0.02 mM (for lasB), 0.08 mM (for pqsA) and 5 mM (for rhlA) resulted in a statistically significant reduction in QS expression compared to the control.
Fig. 1.
Inhibition of QS by sodium salicylate (NaSa) in P. aeruginosa . (a) Concentration-dependent effect of NaSa (0–10 mM) in AB medium on the gene expression of lasB, rhlA and pqsA evaluated with the reporter strains PAO1 lasB::gfp, rhlA::gfp and pqsA::gfp. Effect of NaSa treatment (0–10 mM) in AB media with or without 50 % serum on the expression of (b) lasB::gfp, (c) rhlA::gfp and (d) pqsA::gfp. For each group, the mean area under the curve (AUC) was calculated from each fluorescence intensity curve (baseline- and blank-adjusted) and normalized by OD from three independent experiments. Data in (a) are expressed as % of untreated control in each respective group (100 % corresponds to 4.6×107, 3.6×107 and 5.4×106 fluorescence units for lasB::gfp, rhlA::gfp and pqsA::gfp, respectively). Data in (b–d) are expressed as % of the untreated 50 % serum sample (100 % corresponds to 1.2×108, 7.0×107 and 6.8×106 fluorescence units for lasB::gfp, rhlA::gfp and pqsA::gfp, respectively). Error bars represent ±sd, N=3. * Indicates a statistically significant difference compared to untreated 0 mM NaSa group (in the 0 and 50% serum groups, respectively) with a P-value <0.05 based on one-way ANOVA Dunnett's post hoc test.
Next, to investigate the effect of NaSa on QS in serum-containing media, a series of experiments were performed using the same reporter strains. NaSa significantly reduced the gene expression of the three QS reporter strains, both in the absence and presence of up to 50 % serum, in a concentration-dependent manner (Fig. 1b–d). In 50 % serum, 10 mM NaSa reduced the gene expression of lasB, rhlA and pqsA genes by four-, two- and three-fold, respectively (Fig. 1b–d). In the control group (non-NaSa treated), the addition of 50 % serum resulted in an upregulation in gene expression by 1.85-fold for lasB, 1.2-fold for rhlA and 1.2-fold for pqsA compared to minimal media (no serum) (Fig. 1b–d).
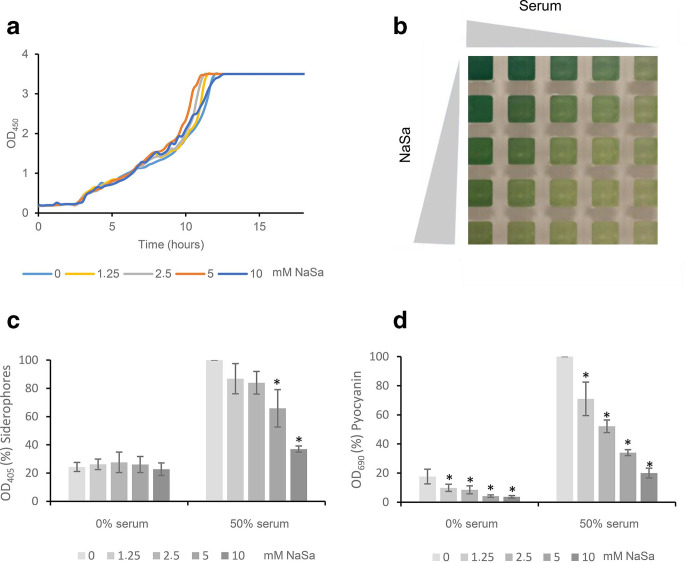
To investigate whether NaSa treatment also influences the synthesis of the actual virulence factors controlled by QS in serum-containing media, wild-type P. aeruginosa PAO1 was cultured in minimal AB medium supplemented with increasing concentrations of NaSa and serum using a checkerboard approach. NaSa did not affect the growth rate of PAO1 when grown in 50 % serum (Fig. 2a). In minimal media, NaSa reduced the growth rate (Fig. S2, available in the online version of this article), although after 24 h incubation, there was no difference in c.f.u. counts (approx. 2×109 c.f.u. ml−1) between the bacterial cultures taken from the four extreme conditions (NaSa/serum: +/-, -/+, +/+, -/-) represented by the four corners of the checkerboard assay (Fig. 2b).
Fig. 2.
Effect of sodium salicylate (NaSa) on siderophore and pyocyanin production in P. aeruginosa PAO1. (a) Growth curves of PAO1 with increasing concentrations of NaSa (0–10 mM) in 50 % serum in AB medium. (b) Visual appearance of supernatants. Effect of NaSa (0–10 mM) and serum (0 and 50 %) on (c) siderophores and (d) pyocyanin production after 3 days of growth. Note: 100% corresponds to OD 6.85 and 1.91 for siderophores and pyocyanin, respectively. Each bar represents the average data from three independent experiments. Error bars represent ±sd. * Indicates a statistically significant difference compared to the untreated 0 mM NaSa group (in the 0 and 50% serum groups, respectively) with a P-value <0.05 based on one-way ANOVA Dunnett's post hoc test.
NaSa treatment of PAO1 in medium supplemented with 50 % serum resulted in a linear decrease in the production of pyocyanin (five-fold decrease for 10 mM NaSa, R2=0.82, P<0.05) and siderophores (2.7-fold decrease for 10 mM NaSa, R2=0.99, P<0.001) compared to the control (Fig. 2b–d). The lowest concentration of NaSa tested (1.25 mM) in 50 % serum was enough to significantly reduce pyocyanin production compared to the control, while 5 mM NaSa was required to significantly reduce siderophore production.
In control wells (not NaSa treated), increasing concentrations of serum resulted in a linear increase of siderophore (R2=0.99, P<0.01) and pyocyanin (R2=0.93, P<0.01) production (Fig. S3c, d, available in the online version of this article) after 3 days of culture. In 50 % serum, the production of both virulence factors increased approximately six-fold compared to those detected with serum-free media.
A similar effect of serum was observed in the clinical isolates, where 50 % serum significantly increased the secretion of pyocyanin (median 4.3-fold increase) in 6 of 14 strains (Fig. S4a, available in the online version of this article) and siderophores (median 15.7-fold increase) in 11 of 14 strains (Fig. S4b).
In the majority of the strains, 11 out of 14, serum-supplemented culture medium resulted in a significant decrease in biofilm formation on polystyrene surfaces after 24 h (Fig. S4c), with an average reduction of 59 % in biofilm biomass.
Effect of NaSa on selected virulence factors in strains isolated from wounds
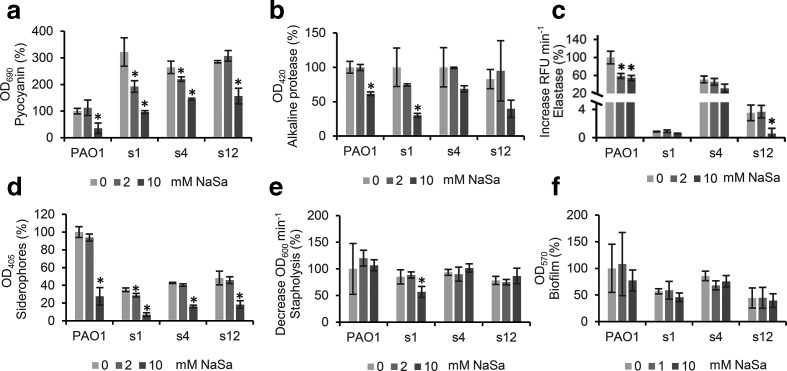
To investigate the effect of NaSa treatment on clinical isolates, a subset of three highly virulent strains (1, 4 and 12), positive for all investigated QS signals and virulence factors, were treated with 0, 2 or 10 mM NaSa in 50% serum-supplemented medium. These concentrations were chosen based on pyocyanin inhibition in pilot experiments (data not shown). While 2 mM NaSa significantly reduced pyocyanin production in two clinical isolates (strain 1: 40% and strain 4: 16% reduction) (Fig. 3a), 10 mM NaSa was required to significantly reduce alkaline protease activity (strain 1 : 69 % reduction), elastase activity (strain 12 : 83 % reduction) and siderophore production (strain 1 : 80%, strain 4 : 62 % and strain 12 : 62 % reduction) (Fig. 3b–d). Biofilm formation was not affected by NaSa treatment at these concentrations and stapholytic activity was only affected in strain 1 (Fig. 3e, f).
Fig. 3.
Effect of sodium salicylate (NaSa) on virulence of clinical P. aeruginosa strains from chronic wounds (strain 1, 4, 12) and PAO1 after 24 h culture in LB supplemented with 50 % serum and treated with 0 (light grey), 2 (mid grey) or 10 (dark grey) mM NaSa. (a) Pyocyanin production, (b) alkaline protease activity, (c) elastase activity, (d) siderophore production, (e) stapholytic activity and (f) biofilm formation. Pyocyanin production for PAO1 was evaluated after 48 h due to low production after 24 h. Biofilm formation was evaluated in 50 % serum in saline. Data are expressed as % activity or production relative to untreated PAO1 (100 % correspond to increase in fluorescence intensity per min of 9333 for elastase, decrease in OD per min of 0.0012 for stapholysis and to OD readings of 0.53, 0.42, 0.73 and 2.0 for pyocyanin, alkaline protease, siderophores and biofilm, respectively). Error bars represent ±sd, N=3. * Indicates a statistically significant difference compared to the untreated 0 mM NaSa group with a P-value <0.05 using one-way ANOVA Dunnett's post hoc test.
Discussion
Although some studies include clinical strains, the vast majority of research on QS in P. aeruginosa related to clinical issues is carried out using laboratory strains and various reporter strains [42–47]. As suggested by Soukarieh et al. [48], two important drawbacks for the further clinical development of QSIs are uncertainty in regard to anti-virulence profiles and efficacy towards P. aeruginosa clinical isolates, as well as the lack of standardization of methods used to assess the compounds. A recent study by Cornforth et al. [49] further highlights the need of proper evaluation and evidence-based selection of infection models. The authors propose a quantitative framework for model evaluation based on comparing transcriptome data acquired from a target system, such as clinical samples, resulting in an accuracy score [49].
The literature on P. aeruginosa QS from a clinical perspective has largely focused on cystic fibrosis [44, 47, 50, 51], whereas studies regarding the QS of clinical P. aeruginosa isolates from chronic wounds are limited. To increase the current knowledge, this study included 14 clinical strains of P. aeruginosa isolated from patients suffering from infected chronic ulcers and evaluated their ability to produce QS signals and selected virulence factors. PAO1 was included as a virulent reference strain. Although the strain collection was relatively small, this study demonstrates that six out of 14 strains showed a highly virulent phenotype, being positive for at least five out of seven investigated virulence factors. These results are in line with a previous study on chronic wounds where 56 % of the isolates produced two of three investigated virulence factors [46]. In addition, eight of the 14 strains produced at least two QS signals. This high prevalence of signal production was accompanied by increased virulence, as five of the six highly virulent strains were positive for all three QS signals, suggesting that an anti-QS strategy could be a viable treatment option for P. aeruginosa strains that cause wound infections.
According to the methodology used in this study, some of the clinical strains do not appear to produce any QS signals but still produce virulence factors (strains 8 and 14), while others produced two QS signals but only ≤1 virulence factor (strains 9 and 13). However, the apparent absence of signals in some strains does not necessarily mean that the strains are signal negative. Signal detection could be influenced by methodological limitations regarding signal stability, sampling time, and the limit of detection of the analytical technique. In fact, the lactone ring of AHL signals undergoes lactonolysis in a pH-dependent manner [52], resulting in signal inactivation. Furthermore, the presence of serum could inactivate as well as degrade signals [53, 54], making the addition of serum relevant to evaluate the potential effect of compounds. Moreover, sampling time also matters [55] and should thus be taken into account. In pilot experiments, different time-points were evaluated to choose the time-point with the highest detection value for some of the factors studied (data not shown). Furthermore, the observation that some of the strains were positive for QS signals but negative for virulence-factor production could be due to mutations in QS receptors. In particular, the LasR receptor is prone to mutation [56], which has been demonstrated in clinical isolates from cystic fibrosis [47] and intubated patients [45]. Hence, non-virulent strains could during establishment of an infection have been virulent, but at some point lost this phenotype. Similarly, strains negative in signal and virulence-factor production in vitro could potentially be virulent clinically in close proximity of QS signal-producing strains, since QS signals can be shared between strains [57]. Considering all the above aspects, the in vivo pathogenicity of P. aeruginosa is multifactorial and dependent on the infectious dose, virulence characteristics of the strains, the ability to produce, sense and respond to QS signals, as well as the host immune response. In this study, any potential relationship between the strains’ virulence and the wound healing outcome could not be studied, since clinical outcome data on the patients were not available.
As further confirmed in this study, P. aeruginosa in chronic wounds often expresses QS signals and virulence factors [13, 46], which to a large extent are QS-regulated [14]. Inhibition of QS has been proposed as an alternative treatment strategy for chronic infections [58] and has generated much research interest in the last decades, especially in light of the increasing global threat of antibiotic resistance. Treatment with a QSI could result in reduced expression of virulence factors, aiding the host immune system to clear the infecting pathogen. NaSa is the sodium salt of salicylic acid, a metabolite of the anti-inflammatory and analgesic drug aspirin (acetylsalicylic acid; ASA). This study reports that NaSa reduces QS gene expression in a physiologically relevant milieu, resulting in attenuated secretion of several virulence factors, which could be beneficial clinically. NaSa partially inhibited all three QS systems las, rhl and pqs at the gene expression level in a concentration-dependent manner, using reporter strains for lasB, rhlA and pqsA grown in medium with and without 50 % serum. Furthermore, a concentration-dependent effect could be demonstrated for the production of virulence factors, as NaSa significantly reduced the secretion of pyocyanin, siderophores, alkaline protease and elastase. A few previous in vitro studies have demonstrated the effect of ASA and salicylic acid on QS and/or the production of virulence factors in both Gram-negative and Gram-positive bacteria, e.g. those concerning P. aeruginosa [29, 30, 59, 60], S. aureus [61–63] and S. epidermidis [64]. Interestingly, a clinical study showed that orally administered aspirin reduced the healing time of wounds associated with chronic venous insufficiency, although the mechanism of action was not elucidated [65]. Since NaSa is closely related to ASA and salicylic acid, and because ASA is readily deacetylated in vivo [66], it is likely that NaSa could have similar effects. The in vitro studies performed on P. aeruginosa [29, 30, 59, 60] are all based on laboratory reference strains and two corneal isolates cultured in minimal medium or LB, highlighting the need and relevance of proving treatment efficacy with multiple clinical isolates and serum-supplemented culture conditions to closer resemble the wound environment. Mimicking the conditions where the treatment will be applied, e.g. the wound environment, potentially increases the chance of identifying active compounds with greater potential to be effective in vivo and, ultimately, in a clinical setting.
To further evaluate the potential of NaSa as a QSI, its effect was also assessed in three highly virulent clinical strains grown in serum-supplemented media. NaSa decreased the production of four of the five virulence factors tested, indicating that the substance is also effective on clinical P . aeruginosa strains. NaSa did not significantly affect the stapholytic activity, except for strain 1. Further, NaSa did not display any effect on the amount of biofilm biomass formed by any of the clinical wound isolates (only three shown here) or the PAO1 strain, when biofilms were grown on the surface of the Calgary biofilm device under dynamic conditions (Fig. 3f).
A drawback of the widely used microtitre plate assay for biofilm quantification using crystal violet is that bacterial aggregates formed in the bulk solution are not considered [67], and it could still be possible that NaSa may affect biofilm aggregation in suspension. It is well established that biofilm is formed in chronic wounds [4, 68] and that aggregated bacteria are abundant in the bulk solution of bacterial cultures [69, 70]. Further studies in our lab will evaluate the effect of NaSa in a three-dimensional biofilm model [67], using confocal laser scanning microscopy and labelling of biofilm matrix components, to evaluate how biofilm composition and architecture may be influenced by NaSa.
Previously, it was shown that ASA decreased biofilm formation on polystyrene in serum-free medium, although a high concentration of 34 mM was required [59]. Similarly, medium with 0.11 mM ASA was shown to reduce the total biofilm biomass produced in a biofilm flow-chamber system using minimal media supplemented with glucose [30]. However, since this is based on subjecting the bacterial cells to a constant flow of ASA-containing medium, it is difficult to determine the total amount of ASA that the cells were exposed to.
Including serum in the test medium drastically reduced P. aeruginosa biofilm formation on polystyrene pegs for the majority of clinical isolates after 24 h under dynamic conditions. Serum has previously been shown to reduce early biofilm formation by P. aeruginosa on plastic surfaces, possibly by increasing twitching motility [71, 72] or because of the low levels of free iron caused by iron-binding proteins such as lactoferrin [73–75].
Furthermore, in control samples without NaSa, supplementing the medium with increasing concentrations of serum resulted in increased QS activation, most notably in the lasB::gfp and pqsA::gfp reporter strains. Serum also increased pyocyanin and siderophore production in P. aeruginosa clinical strains and PAO1. These results agree with a previous study in which serum was shown to increase the expression of the lasI, rhlA and pqsA genes in P. aeruginosa, accompanied by increased virulence-factor production in late stage cultures (16 h) [76]. It was proposed that in the late phase, serum enhances vfr expression, resulting in increased levels of the transcription modulator Vfr, which activates the lasR gene. This leads to increased 3-OC12-HSL production via enhanced lasI transcription. Additionally, LasR activates rhlR/rhlI transcription, increasing C4-HSL levels, and LasR enhances mvfR expression, inducing the transcription of the pqsA-E operon leading to higher levels of PQS. Since Vfr is a positive regulator of lasR- and rhlR transcription [77, 78], it could also explain the increased levels of siderophores and pyocyanin in the presence of serum, although vfr expression was not evaluated in the present study. In contrast, albumin, a major component of serum, has been shown to decrease las- and rhl-associated QS, but not that associated with pqs, by binding QS signal molecules [53], indicating that serum components other than albumin also play a role in las and rhl. The serum-dependent increased production of siderophores could also be due to changes in iron concentrations. Studies have shown that low levels of free iron, as in serum due to iron-binding proteins such as transferrin [79, 80], induce siderophore production. It has also been shown that serum increases the virulence of the problematic wound pathogen S. aureus , further highlighting the relevance of including serum when studying wound infection in vitro [81].
In the present study, a trend was observed in which the inclusion of serum in the test medium decreased the potency of NaSa treatment compared to that achieved in serum-free medium, although only the las system was significantly reduced with 10 mM NaSa treatment. It was previously shown that serum components such as albumin interact with a wide variety of pharmaceuticals [40], e.g. aspirin [82, 83]. Importantly, the present results show that NaSa was still effective in medium supplemented with 50 % serum. This indicates that NaSa could potentially be effective as a QSI in the wound environment where serum is a major component.
In conclusion, this study shows that NaSa significantly decreases the gene expression of the las, rhl and pqs QS systems and attenuates the production of pyocyanin, siderophores, alkaline protease and elastase in P. aeruginosa wound strains and PAO1 cultured in simulated wound fluid. Serum alone increased QS gene expression of the las system and production of pyocyanin and siderophores, while biofilm formation was reduced, stressing the importance of including serum in vitro when appropriate.
The characterization performed on the clinical isolates from chronic wounds demonstrated that approximately half of the strains were highly virulent and expressed several QS signals, highlighting the relevance of using QSIs such as NaSa in the clinical setting as an alternative to conventional antibiotics. The observed QS inhibitory effect of NaSa will be further evaluated in vivo.
Methods
Bacterial strains, culture conditions and chemicals
Fourteen P. aeruginosa clinical isolates from chronic leg ulcers, kindly provided by Professor Artur Schmidtchen (Department of Clinical Sciences, Lund University, Sweden) were used in this study. The mutants and reporter strains used were kindly provided by Professor Thomas Bjarnsholt (Costerton Biofilm Center, University of Copenhagen, Denmark) and Professor Yang Liang (Southern University of Science and Technology, China). All chemicals used were purchased from Sigma-Aldrich (USA) if not stated otherwise.
All clinical strains were propagated on 5 % (w/v) horse blood Columbia agar (Medium Department, Clinical Microbiology Lab, Sahlgrenska University Hospital, Sweden). QS reporter strains were cultured in minimal medium (AB medium) consisting of nine parts B-medium (1 mM MgCl2, 0.1 mM CaCl2, 0.01 mM FeCl3) and one part A10-buffer [20 g l−1 (NH4)2SO4 (Merck, USA), 60 g l−1 Na2HPO4×2H2O, 30 g l−1 KH2PO4 (Acros Organics, USA) and 30 g l−1 NaCl] supplemented with 0.5 % (w/v) casamino acids (BD, USA), 0.1 % (w/v) glucose (Merck, USA), and 0.025 % (w/v) thiamine. The AB medium for QS signal- and QS interference reporter strains was supplemented with 10 µg ml−1 gentamicin and 100 µg ml−1 ampicillin or 60 µg ml−1 gentamicin, respectively. All serum used was of foetal calf origin (non-heat inactivated, HyClone, GE Healthcare Life Sciences, USA). The strains used are listed in Table 3.
Table 3.
List of strains used in this study
|
Strain name |
Characteristic |
Reference |
|---|---|---|
|
From 1 to 14 |
Clinical wound isolates of P. aeruginosa |
[96] |
|
PAO1 |
Reference strain of P. aeruginosa, Pseudomonas Genetic Stock Center (http:// www.pseudomonas.med.ecu.edu, strain PAO0001) |
|
|
MH155 |
3-OC12-HSL signal reporter. Escherichia coli MT102 with lasB::gfp(ASV) |
[34] |
|
MH2O5 |
C4-HSL signal reporter. E. coli MT102 with ahyR-ahyI::gfp(ASV) |
[91] |
|
∆pqsC PpqsA::gfp(ASV) |
PQS signal reporter. PAO1 ∆pqsC with pqsA::gfp(ASV) |
[97] |
|
PAO1 PlasB::gfp(ASV) |
las reporter. PAO1 with a mini-Tn5 insert containing a translational fusion of the LasR-regulated lasB promoter and a gene coding for an unstable version of green fluorescent protein (GFP), gfp(ASV) |
[34] |
|
PAO1 rhlA::gfp(ASV) |
rhl reporter. PAO1 carrying a pMHRA plasmid containing RhlR-regulated rhlA::gfp(ASV) translational fusion inserted into the vector pMH391 |
[30] |
|
PAO1 pqsA::gfp |
pqs reporter. PAO1 carrying the plasmid pAC37 containing a transcriptional fusion of the pqs-regulated pqsA promoter and gfp(ASV) |
[97] |
|
MH694 |
PAO1 ∆lasI. QS-deficient mutant with a deletion of lasI gene |
[98] |
|
MH698 |
PAO1 ∆rhlI. QS-deficient mutant with a deletion of rhlI gene |
[35] |
|
∆pqsC |
PAO1 ∆pqsC. QS-deficient mutant with a deletion of pqsC gene |
[99] |
Characterization of P. aeruginosa clinical strains from chronic wounds
(i) Virulence characterization
Pyocyanin and siderophore production
Overnight cultures of the clinical strains and PAO1 on blood agar were diluted in saline [0.9 % (w/v) NaCl] until OD546=0.1 was reached, which corresponds to 108 c.f.u. ml−1, and then 100 µl was added to Kings agar A in 24-well plates and 5 µl was added to Pseudomonas F-agar in 9 cm diameter petri dishes for detection of pyocyanin and siderophore (pyoverdine and pyochelin) production [84], respectively. Plates were incubated for 24 h (siderophores) and 48 h (pyocyanin) at 35 °C in a humidified incubator. Visual detection of green colouration in the agar indicated that a strain was positive for pyocyanin production. Fluorescent colonies under UV light were considered positive for siderophore production [85]. At least two of three independent experiments needed to be positive for a strain to be considered a pyocyanin or siderophore producer.
Alkaline protease and elastase
The presence of alkaline protease and elastase was detected using modified agar-based assays [86, 87]. Overnight cultures of the clinical isolates and PAO1 on blood agar were diluted to OD546=0.1 in saline, and 5 µl were plated on agar plates containing 1.5 % (w/v) tryptic soy agar (TSA, Merck, USA) in 100 mM Tris adjusted to pH=10 and supplemented with 2 % (w/v) skimmed milk powder (Scharlau, Spain) for alkaline protease determination or on plates containing 1.5 % (w/v) TSA and 0.3 % (w/v) insoluble elastin from bovine neck ligaments for elastase determination. The plates were incubated at 35 °C for up to 48 h in a humidified incubator. Strains showing visual signs of degraded milk proteins (24 h) or elastin (48 h) around the colonies were considered positive for alkaline protease and elastase, respectively. Strains with at least two of three positive independent trial results were considered alkaline protease or elastase producers.
Swarming
Overnight cultures of the clinical isolates and PAO1 on blood agar were diluted to OD546=0.1 in saline, and 2 µl spots were plated on agar containing 1.5 % (w/v) TSA and 2 % (w/v) skimmed milk powder. Swarming behaviour was observed during a period of 48 h of incubation in a humidified incubator at 35 °C. The experiment was repeated three times.
Rhamnolipids
Rhamnolipid production was evaluated in a glycerol-based medium [88]. Overnight cultures of the clinical isolates and PAO1 on blood agar were diluted to OD546=0.1 in saline. A total of 100 µl of the OD suspension was added to 3 ml medium containing 42 g l−1 glycerol, 1.4 g l−1 NaNO3, 7 g l−1 K2HPO4, 3 g l−1 KH2PO4 and 0.2 g l−1 MgSO4×7 H2O. Rhamnolipids were quantified based on the orcinol assay [89]. In brief, after 6 days of culture at 35 °C and 100 r.p.m., the supernatants were collected by centrifugation at 14 000 g for 10 min. A volume of 300 µl of supernatant was added to 1.7 ml diethyl ether (VWR, USA), vortexed and centrifuged using a bench centrifuge (iFuge, Neuation, India) at 2000 g for 1 min. The diethyl ether fraction was collected and evaporated overnight prior to the addition of 300 µl dH2O. After vortexing, 100 µl of the rhamnolipid-water solution was added to 900 µl 0.19 % orcinol (w/w) in 53 % (w/w) H2SO4. The mixture was incubated at 80 °C for 30 min before cooling down at RT. Of this mixture, 200 µl was added to the wells of a 96-well plate (Nunc, Denmark), and the OD was read at 421 nm using a plate reader (FluostarOmega, BMG LABTECH, Germany). Test medium without bacteria, using the same sample preparation, was used as a blank. A strain was considered a rhamnolipid producer if the average OD from three independent experiments differed from that of the blank with statistical significance.
Stapholytic activity
LasA protease activity was measured by determining the ability of P. aeruginosa culture supernatants to lyse boiled Staphylococcus aureus cells [24]. Staphylococcus aureus ATCC 25923 was grown in 5 ml TSB at 35 °C and 100 r.p.m. in a shaking incubator (KS4000i, IKA, Germany). The cells were harvested by centrifugation at 4000 g for 10 min and added to 30 ml of 10 mM Na2PO4 (dH2O) containing 1.5 % (w/v) TSA. The mixture was boiled for 15 min and added to two sterile petri dishes (9 cm ø). After solidification, 5 µl of bacterial suspension of OD546=0.1 in saline of each clinical isolate and PAO1 was spotted on each agar plate. The plates were incubated upside down at 35 °C in a humidified incubator for 24 h. Clear zones in the agar around the colonies in at least two out of three independent experiments indicated that a strain was positive for stapholytic activity.
Biofilm formation
Biofilm formation was studied using the MBEC P and G Assay (Calgary biofilm device, Innovotech, Canada) and crystal violet [90]. Overnight cultures of the clinical isolates and PAO1 on blood agar were diluted to OD546=0.1 in saline supplemented with 50 % (v/v) serum. A volume of 5 µl of each respective strain was added to 145 µl 50 % (v/v) serum-supplemented saline (2×107 c.f.u ml−1 final concentration) in the MBEC 96-well plate with polystyrene pegs on the lid. Medium without cells was used as blank. After 48 h incubation at 35 °C and 100 r.p.m. in a humidified shaking incubator, the lid was transferred to a new 96-well plate containing 170 µl per well of 0.1 % (w/v) crystal violet solution and incubated at RT for 15 min. Unbound dye and loosely attached bacterial cells were repeatedly washed off in a water bath until no more dye was visible in the eluent. The lid was air-dried for 30 min before the pegs were immersed in 200 µl 96 % (w/w) ethanol for elution of the dye. After 30 min, the OD was measured at 590 nm. The data are presented as blank-corrected average OD values from three independent experiments, each with three technical replicates.
(ii) QS signal characterization
QS signal measurements were based on protocols described previously for C4-HSL [91], 3-OC12-HSL [34] and PQS [92]. Overnight cultures of the clinical isolates on blood agar were diluted to OD546=0.2 in 50 % (v/v) serum in saline and were further diluted 1 : 10 in the same medium. A total of 10 ml of bacterial suspension was added to 50 ml Falcon tubes and cultured at 35 °C and 100 r.p.m. for 24 h. Supernatants were collected by centrifugation at 4000 g for 10 min, sterile filtered and frozen at −80 °C until further analysis. PAO1 was used as a positive control, and mutants negative for signal production were used as negative controls (PAO1 ∆lasI, PAO1 ∆rhlI and PAO1 ∆pqsC). Overnight cultures of the signal reporter strains MH155, MH205 and ∆pqsC PpqsA::gfp(ASV) were diluted to OD546=0.1 in AB medium supplemented with 10 µg ml−1 gentamicin and 100 µg ml−1 ampicillin. Purified signals [N-(3-oxo-dodecanoyl)-L-homoserine lactone ‘3-OC12-HSL’], [N-butanoyl-L-homoserine lactone ‘C4-HSL’ and 2-heptyl-3-hydroxy-4(1 H)-quinolone ‘PQS’] from frozen DMSO stock solutions (1 mg ml−1 for 3-OC12-HSL and C4-HSL or 20 mM for PQS) were serially diluted in 50 % (v/v) serum in saline from 20 µM to 0.2 nM, with pure medium used as a blank. Next, 25 µl of supernatant or purified signal was mixed with 25 µl of each respective signal reporter strain in a black 384-well plate (Greiner Bio-one, Austria) with a clear bottom. The plate was placed in a plate reader, and kinetic measurements of the absorbance (450 nm) and fluorescence (bottom reading, excitation 530/20 and emission at 485/30 filters) were recorded every 15 min for 20 h at 35 °C. The area under the curve (AUC) data of baseline- and blank-adjusted fluorescence intensity (FI) (OD normalized) from three independent experiments, with at least two technical replicates, were used to characterize the clinical strains in terms of signal production. As a cut-off value for signal production characterization, strains with AUC data within the standard curve and above that of the signal deficient mutant were considered positive for QS signal production.
Effect of NaSa on PAO1 QS activation and virulence-factor production in minimal media with/without serum supplementation
(i) MIC of NaSa
To ensure that the NaSa concentrations did not inhibit growth, the MIC of the clinical isolates, PAO1 and the QS reporter strains was determined in 50 % (v/v) serum in saline. Colonies from overnight cultures on blood agar plates were diluted to OD546=0.1 and further diluted 1 : 1000 in 50 % (w/v) serum to approximately 105 c.f.u ml−1. In a flat-bottomed 96-well plate (Nunc, Denmark), 150 µl inoculum was mixed 1 : 1 with different concentrations of NaSa in 50 % (w/v) serum in saline. Final concentrations of NaSa ranged between 2 and 250 mM using two-fold dilution steps. The plate was incubated for 24 h at 35 °C under static conditions. Next, the OD600 was measured, and concentrations of NaSa resulting in OD600<0.01 were used to define the MIC. At least two of the three trials needed to match for the results to be considered valid.
(ii) Effect of NaSa on QS activation in minimal media
To evaluate the effect of NaSa on QS activation, reporter strains were used as described previously [41] with modifications. In brief, overnight cultures of the reporter strains lasB::gfp(ASV), rhlA::gfp(ASV) and pqsA::gfp(ASV) grown in TSB supplemented with 60 µg ml−1 gentamicin were washed twice in AB medium by 5 min centrifugation at 4000 g, diluted to OD546=0.1 and further diluted 1 : 10 in AB medium. NaSa was diluted to 10 mM in the reporter-strain-containing AB medium and further two-fold diluted in same medium to 20 µM. A volume of 50 µl was added to the wells of a black 384-well plate with a clear bottom. Absorbance at 450 nm and bottom-read FI at an excitation of 530/20 nm and an emission of 485/30 nm were recorded every 15 min for 20 h at 35 °C in a plate reader. For each experimental group, the average AUC from triplicate wells was calculated from the FI curves (baseline- and blank-adjusted) and normalized by the OD. Average data from three independent experiments are presented.
(iii) Effect of NaSa on QS inhibition and virulence factor production in serum-supplemented minimal media
A checkerboard approach was used to investigate the effect of NaSa and serum on QS gene expression and virulence-factor production. In a 96-well plate, serum was added to the first column of five rows and serially diluted column-wise (1 : 2, four times) in AB medium; the fifth column contained only AB medium. Similarly, 20 mM NaSa in AB medium was added to the first row of five columns and serially diluted row-wise (1 : 10, four times); the fifth row contained only AB medium. Next, the two dilution series were mixed 1 : 1 with each other, creating a pattern with 25 unique serum-NaSa combinations. A volume of 45 µl of each combination was added to black 384-well plates with a clear bottom. Overnight cultures in 3 ml TSB of PAO1 lasB::gfp(ASV), PAO1 rhlA::gfp(ASV) or PAO1 pqsA::gfp(ASV) were washed twice in saline by centrifugation at 4 000 g for 5 min and resuspended in AB medium to an OD546 of 0.1. In total, 5 µl of the bacterial suspensions were added to the 45 µl checkerboard wells. Absorbance at 450 nm and bottom-read FI at an excitation of 530/20 nm and emission of 485/30 nm were recorded every 15 min during a 20 h incubation at 35 °C in a plate reader. All medium used contained 60 µg ml−1 gentamicin. The average AUC of baseline-adjusted and OD-normalized FI curves from three independent experiments was calculated. Growth curves of the reporter strains were obtained to ensure that the chosen serum and NaSa levels did not significantly inhibit bacterial growth (Fig. S1, available in the online version of this article).
The same checkerboard approach was used with P. aeruginosa PAO1, using the same medium but without antibiotics. After 72 h of incubation at 35 °C in a humidified incubator, cultures were centrifuged at 14 000 g for 10 min, and 50 µl supernatants were transferred to a clear 384-well plate (Nunc, Denmark). The OD was measured at 405 and 690 nm for indirect quantification of siderophores and pyocyanin production, respectively [93, 94]. For samples resulting in OD readings above the limit of detection of the plate reader, the well volumes were reduced and subsequent OD readings were pathway-corrected. Average data from three independent experiments are presented.
Effect of NaSa on clinical P. aeruginosa strains from chronic wounds
Overnight cultures of strains 1, 4, 12 and PAO1 on blood agar plates were diluted to OD546=0.1 in LB supplemented with 50 % (w/v) serum and NaSa (0, 2 or 10 mM final concentrations) and cultured in a 24-well microtiter plate (Nunc, Denmark) at 35 °C and 100 r.p.m. for 24 h and 48 h. Cell-free LB with 50 % (w/v) serum served as a blank sample. After 24 h and 48 h, 500 µl of bacterial suspension from each well was transferred to an Eppendorf tube and centrifuged at 14 000 g for 10 min. Next, the production of the following virulence factors was evaluated.
(i) Pyocyanin and siderophore production
A volume of 300 µl of the supernatant was transferred to a 96-well microtitre plate (Nunc, Denmark). The presence of pyocyanin and siderophores was evaluated after 48 h of growth using a plate reader by determining their blank-corrected ODs at 405 and 690 nm, respectively. Average data from three independent experiments were calculated.
(ii) Alkaline protease activity
Alkaline protease activity was evaluated in an azocasein-based assay [95] after 24 h of growth. Altogether, 100 µl of supernatant was added to 400 µl 1 % (w/v) azocasein solution in 100 mM TRIS (pH 10) and incubated at 35 °C and 100 r.p.m. for 30 min before 500 µl 10 % (w/v) trichloroacetic acid was added to stop the degradation of azocasein. The tubes were centrifuged at 14 000 g for 10 min before the supernatant was mixed with 1 M NaOH in a 1 : 1 ratio. Of the mixture, 100 µl was further diluted 1 : 1 in 1 M NaOH and transferred to a 384-well microtitre plate, and OD was recorded at 420 nm using a plate reader. Data were blank-subtracted against cell-free LB supplemented with 50 % (v/v) serum. Average data from three independent experiments were calculated.
(iii) Elastase protease activity
Elastase activity was measured after 24 h of growth. 5-FAM fluorophore-labelled elastin (Anaspec, USA) was diluted to 1 mg ml−1 in dH2O and further diluted 1 : 30 in PBS. Then, 25 µl elastin solution was added to 25 µl supernatant diluted 1 : 10, 1 : 100 and 1 : 1000 in a black 384-well plate. Porcine pancreas elastase was diluted two-fold in PBS from 50 U ml−1 to 0.7 U ml−1 to serve as the standard curve and added to the plate in triplicate. Pure PBS served as the blank. FI was recorded every 90 s in a plate reader at 35 °C using an excitation of 530/20 nm and emission of 485/30 nm. The average blank-corrected increase in relative FI per min (r.f.u. min−1) from three independent experiments was calculated. Samples were chosen from dilutions resulting in data within the linear part of the standard curve, and the results were corrected for any dilutions.
(iv) Stapholytic activity
Staphylococcus aureus ATCC 25923 was cultured, harvested and boiled as described above, but without the addition of TSA. The boiled cells were diluted in 10 mM Na2PO4 (dH2O) until OD521=0.1. Overall, 180 µl of S. aureus solution was added to a 96-well plate (Nunc, Denmark) prior to the transfer of 20 µl supernatant from 24 h old cultures to the wells. The OD at 600 nm was recorded every 30 s for 30 min in a plate reader at 35 °C. The average decrease in the blank-corrected OD per min during the first 10 min was calculated from three independent experiments.
(v) Biofilm formation
Biofilm formation was evaluated as described previously using strains 1, 4, 12 and PAO1 cultured in saline supplemented with 50 % (v/v) serum and 0, 1 or 10 mM NaSa. The data are presented as blank-corrected average OD values from three independent experiments, each with three technical replicates.
Effect of serum on selected virulence factors in clinical P. aeruginosa strains from chronic wounds
The effect of serum on the production of siderophores and pyocyanin was investigated in LB with and without the addition of 50 % (v/v) serum after 24 and 48 h cultures, respectively. Overnight cultures of strain 1–14 and PAO1 in TSB were washed twice by centrifugation at 4000 g for 10 min and resuspended in saline to OD546=0.1. In total, 5 µl of these bacterial suspensions were added to 195 µl LB or LB supplemented with 50 % (v/v) serum in 96-well microtitre plates. After incubation at 35 °C in a humidified incubator, 50 µl of bacterial supernatants of each sample were collected by centrifugation at 4000 g for 10 min and transferred to a 384-well plate (Nunc Denmark). Siderophore and pyocyanin production was evaluated by absorbance measurements at 405 nm and 690 nm, respectively. The experiment was repeated three times. Biofilm formation was studied as described before, but using LB with or without 50 % (v/v) serum as media; N=6.
Statistics
One-way ANOVA followed by two-sided Dunnett’s post hoc tests was performed to evaluate significant differences between the control (no treatment) and each of the different concentrations of NaSa and between the control (no serum) and each of the different concentrations of serum in medium with regard to the expression of lasB::gfp, rhlA::gfp and pqsA::gfp genes, production of siderophores, pyocyanin, elastase, alkaline protease, stapholytic protease and biofilm. Two-sided Student’s t-tests were performed to calculate differences in pyocyanin, siderophores and biofilm production between the control (no serum) and the serum-containing medium as well as for the production of rhamnolipids between the strains and the blank. Statistical analyses were performed using SPSS Statistics 21 (IBM Corporation, USA) with a significance level set at P<0.05.
Supplementary Data
Funding information
This study was financially supported by the Swedish Foundation for Strategic Research (SSF; RMA15-0110 2016), Mölnlycke Health Care AB (Sweden), the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 754412 (MoRE2020 - Region Västra Götaland), CARe - Centre for Antibiotic Resistance Research at University of Gothenburg, the Handlanden Hjalmar Svensson Foundation, the Adlerbertska Foundation and the Doctor Felix Neubergh Foundation.
Acknowledgements
The authors would like to express their gratitude to Professor Artur Schmidtchen (Department of Clinical Sciences, Lund University, Sweden) for sharing the clinical strains [1–14] used in this study and to Professor Thomas Bjarnsholt (Costerton Biofilm Center, University of Copenhagen, Denmark) and Professor Yang Liang (Southern University of Science and Technology, China) for kindly providing us with the mutant and reporter strains. Maria Hoffman is thanked for her technical assistance with the biochemical identification of the clinical strains. We thank Professor Peter Thomsen for valuable discussions on this work.
Author contributions
E.G., S.A., M.W. and M.T. planned the work and interpreted the results. E.G. conducted the experiments, data analysis and prepared the first manuscript draft. S.A., M.W. and M.T. reviewed the manuscript.
Conflicts of interest
E.G. and S.A. are employees of Mölnlycke Health Care AB. The other authors declare no conflict of interest.
Footnotes
Abbreviations: AHL, N-Acyl homoserine lactone; API, analytical profile index; ASA, acetylsalicylic acid; AUC, area under the curve; c.f.u., colony forming units; C4-HSL, N-butanoyl-L-homoserine lactone; FI, fluorescence intensity; GFP, green fluorescent protein; LB, lysogeny broth; MIC, minimum inhibitory concentration; NaSa, sodium salicylate; 3-OC12-HSL, N-(3-oxo-dodecanoyl)-L-homoserine lactone; OD, optical density; PQS, Pseudomonas quinolone signal; QS, quorum sensing; QSI, quorum-sensing inhibitor; r.f.u., relative fluorescence units; RT, room temperature; SWF, simulated wound fluid; TSA, tryptic soy agar; TSB, tryptic soy broth.
Four supplementary figures and one supplementary table are available with the online version of this article.
References
- 1.Phillips CJ, Humphreys I, Fletcher J, Harding K, Chamberlain G, et al. Estimating the costs associated with the management of patients with chronic wounds using linked routine data. Int Wound J. 2016;13:1193–1197. doi: 10.1111/iwj.12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green J, Jester R, McKinley R, Pooler A. The impact of chronic venous leg ulcers: a systematic review. J Wound Care. 2014;23:601–612. doi: 10.12968/jowc.2014.23.12.601. [DOI] [PubMed] [Google Scholar]
- 3.Gottrup F, Apelqvist J, Bjarnsholt T, Bjansholt T, Cooper R, et al. EWMA document: antimicrobials and non-healing wounds. Evidence, controversies and suggestions. J Wound Care. 2013;22:S1–89. doi: 10.12968/jowc.2013.22.Sup5.S1. [DOI] [PubMed] [Google Scholar]
- 4.Malone M, Bjarnsholt T, McBain AJ, James GA, Stoodley P, et al. The prevalence of biofilms in chronic wounds: a systematic review and meta-analysis of published data. J Wound Care. 2017;26:20–25. doi: 10.12968/jowc.2017.26.1.20. [DOI] [PubMed] [Google Scholar]
- 5.Sanchez CJ, Mende K, Beckius ML, Akers KS, Romano DR, et al. Biofilm formation by clinical isolates and the implications in chronic infections. BMC Infect Dis. 2013;13:47. doi: 10.1186/1471-2334-13-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. International Journal of Antimicrobial Agents. 2010;35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 7.Sugden R, Kelly R, Davies S. Combatting antimicrobial resistance globally. Nature Microbiology. 2016;1:16187. doi: 10.1038/nmicrobiol.2016.187. [DOI] [PubMed] [Google Scholar]
- 8.Hoiby N, Ciofu O, Johansen HK, Song ZJ, Moser C, et al. The clinical impact of bacterial biofilms. International Journal of Oral Science. 2011;3:55–65. doi: 10.4248/IJOS11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolcott RD, Hanson JD, Rees EJ, Koenig LD, Phillips CD, et al. Analysis of the chronic wound microbiota of 2,963 patients by 16S rDNA pyrosequencing. Wound Repair Regen. 2016;24:163–174. doi: 10.1111/wrr.12370. [DOI] [PubMed] [Google Scholar]
- 10.Gjodsbol K, Christensen JJ, Karlsmark T, Jorgensen B, Klein BM, et al. Multiple bacterial species reside in chronic wounds: a longitudinal study. Int Wound J. 2006;3:225–231. doi: 10.1111/j.1742-481X.2006.00159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halbert AR, Stacey MC, Rohr JB, Jopp-McKay A. The effect of bacterial colonization on venous ulcer healing. The Australasian Journal of Dermatology. 1992;33:75–80. doi: 10.1111/j.1440-0960.1992.tb00083.x. [DOI] [PubMed] [Google Scholar]
- 12.Madsen SM, Westh H, Danielsen L, Rosdahl VT. Bacterial colonization and healing of venous leg ulcers. APMIS: acta pathologica, microbiologica, et immunologica. Scandinavica. 1996;104:895–899. doi: 10.1111/j.1699-0463.1996.tb04955.x. [DOI] [PubMed] [Google Scholar]
- 13.Rickard AH, Colacino KR, Manton KM, Morton RI, Pulcini E, et al. Production of cell-cell signalling molecules by bacteria isolated from human chronic wounds. J Appl Microbiol. 2010;108:1509–1522. doi: 10.1111/j.1365-2672.2009.04554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J, Zhang L. The hierarchy quorum sensing network in Pseudomonas aeruginosa . Protein & cell. 2015;6:26–41. doi: 10.1007/s13238-014-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allen L, Dockrell DH, Pattery T, Lee DG, Cornelis P, et al. Pyocyanin production by Pseudomonas aeruginosa induces neutrophil apoptosis and impairs neutrophil-mediated host defenses in vivo. The Journal of Immunology. 2005;174:3643–3649. doi: 10.4049/jimmunol.174.6.3643. [DOI] [PubMed] [Google Scholar]
- 16.Ramos I, Dietrich LEP, Price-Whelan A, Newman DK. Phenazines affect biofilm formation by Pseudomonas aeruginosa in similar ways at various scales. Res Microbiol. 2010;161:187–191. doi: 10.1016/j.resmic.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das T, Kutty SK, Kumar N, Manefield M. Pyocyanin facilitates extracellular DNA binding to Pseudomonas aeruginosa influencing cell surface properties and aggregation. PloS one. 2013;8:e58299. doi: 10.1371/journal.pone.0058299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolz C, Hohloch K, Ocaktan A, Poole K, Evans RW, et al. Iron release from transferrin by pyoverdin and elastase from Pseudomonas aeruginosa . Infect Immun. 1994;62:4021–4027. doi: 10.1128/iai.62.9.4021-4027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laarman AJ, Bardoel BW, Ruyken M, Fernie J, Milder FJ, et al. Pseudomonas aeruginosa alkaline protease blocks complement activation via the classical and lectin pathways. Journal of immunology. 2012;188:386–393. doi: 10.4049/jimmunol.1102162. [DOI] [PubMed] [Google Scholar]
- 20.van der Plas MJ, Bhongir RK, Kjellstrom S, Siller H, Kasetty G, et al. Pseudomonas aeruginosa elastase cleaves a C-terminal peptide from human thrombin that inhibits host inflammatory responses. Nat Commun. 2016;7:11567. doi: 10.1038/ncomms11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamont IL, Beare PA, Ochsner U, Vasil AI, Vasil ML. Siderophore-Mediated signaling regulates virulence factor production in Pseudomonasaeruginosa . Proc Natl Acad Sci U S A. 2002;99:7072–7077. doi: 10.1073/pnas.092016999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen PO, Bjarnsholt T, Phipps R, Rasmussen TB, Calum H, et al. Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa . Microbiology. 2007;153:1329–1338. doi: 10.1099/mic.0.2006/003863-0. [DOI] [PubMed] [Google Scholar]
- 23.Davey ME, Caiazza NC, O'Toole GA. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J Bacteriol. 2003;185:1027–1036. doi: 10.1128/JB.185.3.1027-1036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kessler E, Safrin M, Olson JC, Ohman DE. Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J Biol Chem. 1993;268:7503–7508. [PubMed] [Google Scholar]
- 25.Davies DG. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 26.Passos da Silva D, Schofield MC, Parsek MR, Tseng BS. An update on the Sociomicrobiology of quorum sensing in gram-negative biofilm development. Pathogens. 2017;6 doi: 10.3390/pathogens6040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaber JA, Triffo WJ, Suh SJ, Oliver JW, Hastert MC, et al. Pseudomonas aeruginosa forms biofilms in acute infection independent of cell-to-cell signaling. Infect Immun. 2007;75:3715–3721. doi: 10.1128/IAI.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaber JA, Hammond A, Carty NL, Williams SC, Colmer-Hamood JA, et al. Diversity of biofilms produced by quorum-sensing-deficient clinical isolates of Pseudomonas aeruginosa. J Med Microbiol. 2007;56:738–748. doi: 10.1099/jmm.0.47031-0. [DOI] [PubMed] [Google Scholar]
- 29.Prithiviraj B, Bais HP, Weir T, Suresh B, Najarro EH, et al. Down regulation of virulence factors of Pseudomonas aeruginosa by salicylic acid attenuates its virulence on Arabidopsis thaliana and Caenorhabditis elegans . Infect Immun. 2005;73:5319–5328. doi: 10.1128/IAI.73.9.5319-5328.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang L, Rybtke MT, Jakobsen TH, Hentzer M, Bjarnsholt T, et al. Computer-Aided identification of recognized drugs as Pseudomonas aeruginosa quorum-sensing inhibitors. Antimicrob Agents Chemother. 2009;53:2432–2443. doi: 10.1128/AAC.01283-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bandara M, Sankaridurg P, Zhu H, Hume E, Willcox M. Effect of salicylic acid on the membrane proteome and virulence of Pseudomonas aeruginosa. Invest Ophthalmol Vis Sci. 2016;57:1213–1220. doi: 10.1167/iovs.15-18990. [DOI] [PubMed] [Google Scholar]
- 32.Yalkowsky SH, Banerjee S. Aqueous Solubility: Methods of Estimation for Organic Compounds. New York: Marcel Dekker; 1992. [Google Scholar]
- 33.WHO The International Pharmacopoeia. World Health Organization; 2006. [Google Scholar]
- 34.Hentzer M, Riedel K, Rasmussen TB, Heydorn A, Andersen JB, et al. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology. 2002;148:87–102. doi: 10.1099/00221287-148-1-87. [DOI] [PubMed] [Google Scholar]
- 35.Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. The EMBO journal. 2003;22:3803–3815. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang K, Zhang Y, Yu M, Shi X, Coenye T, et al. Evaluation of a new high-throughput method for identifying quorum quenching bacteria. Sci Rep. 2013;3:2935. doi: 10.1038/srep02935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aiba-Kojima E, Tsuno NH, Inoue K, Matsumoto D, Shigeura T, et al. Characterization of wound drainage fluids as a source of soluble factors associated with wound healing: comparison with platelet-rich plasma and potential use in cell culture. Wound Repair Regen. 2007;15:511–520. doi: 10.1111/j.1524-475X.2007.00259.x. [DOI] [PubMed] [Google Scholar]
- 38.Tarnuzzer RW, Schultz GS. Biochemical analysis of acute and chronic wound environments. Wound Repair Regen. 1996;4:321–325. doi: 10.1046/j.1524-475X.1996.40307.x. [DOI] [PubMed] [Google Scholar]
- 39.Trainor GL. The importance of plasma protein binding in drug discovery. Expert opinion on drug discovery. 2007;2:51–64. doi: 10.1517/17460441.2.1.51. [DOI] [PubMed] [Google Scholar]
- 40.Ghuman J, Zunszain PA, Petitpas I, Bhattacharya AA, Otagiri M, et al. Structural basis of the drug-binding specificity of human serum albumin. Journal of molecular biology. 2005;353:38–52. doi: 10.1016/j.jmb.2005.07.075. [DOI] [PubMed] [Google Scholar]
- 41.Rasmussen TB, Bjarnsholt T, Skindersoe ME, Hentzer M, Kristoffersen P, et al. Screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J Bacteriol. 2005;187:1799–1814. doi: 10.1128/JB.187.5.1799-1814.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rumbaugh KP, Hamood AN, Griswold JA. Analysis of Pseudomonas aeruginosa clinical isolates for possible variations within the virulence genes exotoxin A and exoenzyme S. The Journal of surgical research. 1999;82:95–105. doi: 10.1006/jsre.1998.5523. [DOI] [PubMed] [Google Scholar]
- 43.Li Y, HP Q, Liu JL, Wan HY. Correlation between group behavior and quorum sensing in Pseudomonas aeruginosa isolated from patients with hospital-acquired pneumonia. Journal of thoracic disease. 2014;6:810–817. doi: 10.3978/j.issn.2072-1439.2014.03.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le Berre R, Nguyen S, Nowak E, Kipnis E, Pierre M, et al. Quorum-Sensing activity and related virulence factor expression in clinically pathogenic isolates of Pseudomonas aeruginosa . Clinical Microbiology and Infection: the Official Publication of the European Society of Clinical Microbiology and Infectious Diseases. 2008;14:337–343. doi: 10.1111/j.1469-0691.2007.01925.x. [DOI] [PubMed] [Google Scholar]
- 45.Kohler T, Buckling A, van Delden C. Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc Natl Acad Sci U S A. 2009;106:6339–6344. doi: 10.1073/pnas.0811741106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guendouze A, Plener L, Bzdrenga J, Jacquet P, Remy B, et al. Effect of quorum quenching lactonase in clinical isolates of Pseudomonas aeruginosa and comparison with quorum sensing inhibitors. Front Microbiol. 2017;8:227. doi: 10.3389/fmicb.2017.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bjarnsholt T, Jensen PO, Jakobsen TH, Phipps R, Nielsen AK, et al. Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PloS one. 2010;5:e10115. doi: 10.1371/journal.pone.0010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soukarieh F, Williams P, Stocks MJ, Camara M. Pseudomonas aeruginosa quorum sensing systems as drug discovery targets: current position and future perspectives. Journal of medicinal chemistry. 2018 doi: 10.1021/acs.jmedchem.8b00540. [DOI] [PubMed] [Google Scholar]
- 49.Cornforth DM, Diggle FL, Melvin JA, Bomberger JM, Whiteley M. Quantitative framework for model evaluation in microbiology research using Pseudomonas aeruginosa and cystic fibrosis infection as a test case. mBio. 2020;11 doi: 10.1128/mBio.03042-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erickson DL, Endersby R, Kirkham A, Stuber K, Vollman DD, et al. Pseudomonas aeruginosa quorum-sensing systems may control virulence factor expression in the lungs of patients with cystic fibrosis. Infect Immun. 2002;70:1783–1790. doi: 10.1128/IAI.70.4.1783-1790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, et al. Quorum-Sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 52.Yates EA, Philipp B, Buckley C, Atkinson S, Chhabra SR, et al. N-Acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa . Infect Immun. 2002;70:5635–5646. doi: 10.1128/IAI.70.10.5635-5646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith AC, Rice A, Sutton B, Gabrilska R, Wessel AK, et al. Albumin inhibits Pseudomonas aeruginosa quorum sensing and alters polymicrobial interactions. Infect Immun. 2017;85 doi: 10.1128/IAI.00116-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang F, Wang LH, Wang J, Dong YH, JY H, et al. Quorum quenching enzyme activity is widely conserved in the sera of mammalian species. FEBS letters. 2005;579:3713–3717. doi: 10.1016/j.febslet.2005.05.060. [DOI] [PubMed] [Google Scholar]
- 55.Dekimpe V, Deziel E. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiology. 2009;155:712–723. doi: 10.1099/mic.0.022764-0. [DOI] [PubMed] [Google Scholar]
- 56.Sandoz KM, Mitzimberg SM, Schuster M. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc Natl Acad Sci U S A. 2007;104:15876–15881. doi: 10.1073/pnas.0705653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mund A, Diggle SP, Harrison F. The fitness of Pseudomonas aeruginosa quorum sensing signal cheats is influenced by the diffusivity of the environment. mBio. 2017;8 doi: 10.1128/mBio.00353-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hentzer M, Givskov M. Pharmacological inhibition of quorum sensing for the treatment of chronic bacterial infections. J Clin Invest. 2003;112:1300–1307. doi: 10.1172/JCI20074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.El-Mowafy SA, Abd El Galil KH, El-Messery SM, Shaaban MI. Aspirin is an efficient inhibitor of quorum sensing, virulence and toxins in Pseudomonas aeruginosa . Microb Pathog. 2014;74:25–32. doi: 10.1016/j.micpath.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 60.Bandara MB, Zhu H, Sankaridurg PR, Willcox MD. Salicylic acid reduces the production of several potential virulence factors of Pseudomonas aeruginosa associated with microbial keratitis. Invest Ophthalmol Vis Sci. 2006;47:4453–4460. doi: 10.1167/iovs.06-0288. [DOI] [PubMed] [Google Scholar]
- 61.Sedlacek M, Gemery JM, Cheung AL, Bayer AS, Remillard BD. Aspirin treatment is associated with a significantly decreased risk of Staphylococcus aureus bacteremia in hemodialysis patients with tunneled catheters. American Journal of Kidney Diseases: the Official Journal of the National Kidney Foundation. 2007;49:401–408. doi: 10.1053/j.ajkd.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 62.Kupferwasser LI, Yeaman MR, Nast CC, Kupferwasser D, Xiong YQ, et al. Salicylic acid attenuates virulence in endovascular infections by targeting global regulatory pathways in Staphylococcus aureus. J Clin Invest. 2003;112:222–233. doi: 10.1172/JCI16876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alvarez LP, Barbagelata MS, Gordiola M, Cheung AL, Sordelli DO, et al. Salicylic acid diminishes Staphylococcus aureus capsular polysaccharide type 5 expression. Infect Immun. 2010;78:1339–1344. doi: 10.1128/IAI.00245-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Muller E, Al-Attar J, Wolff AG, Farber BF. Mechanism of salicylate-mediated inhibition of biofilm in Staphylococcus epidermidis . J Infect Dis. 1998;177:501–503. doi: 10.1086/517386. [DOI] [PubMed] [Google Scholar]
- 65.del Rio Sola ML, Antonio J, Fajardo G, Vaquero Puerta C. Influence of aspirin therapy in the ulcer associated with chronic venous insufficiency. Ann Vasc Surg. 2012;26:620–629. doi: 10.1016/j.avsg.2011.02.051. [DOI] [PubMed] [Google Scholar]
- 66.Bakar SK, Niazi S. Stability of aspirin in different media. Journal of pharmaceutical sciences. 1983;72:1024–1026. doi: 10.1002/jps.2600720914. [DOI] [PubMed] [Google Scholar]
- 67.Werthen M, Henriksson L, Jensen PO, Sternberg C, Givskov M, et al. An in vitro model of bacterial infections in wounds and other soft tissues. APMIS : acta pathologica, microbiologica, et immunologica. Scandinavica. 2010;118:156–164. doi: 10.1111/j.1600-0463.2009.02580.x. [DOI] [PubMed] [Google Scholar]
- 68.Kirketerp-Moller K, Jensen PO, Fazli M, Madsen KG, Pedersen J, et al. Distribution, organization, and ecology of bacteria in chronic wounds. Journal of clinical microbiology. 2008;46:2717–2722. doi: 10.1128/JCM.00501-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alhede M, Kragh KN, Qvortrup K, Allesen-Holm M, van Gennip M, et al. Phenotypes of non-attached Pseudomonas aeruginosa aggregates resemble surface attached biofilm. PloS one. 2011;6:e27943. doi: 10.1371/journal.pone.0027943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schleheck D, Barraud N, Klebensberger J, Webb JS, McDougald D, et al. Pseudomonas aeruginosa PAO1 preferentially grows as aggregates in liquid batch cultures and disperses upon starvation. PloS one. 2009;4:e5513. doi: 10.1371/journal.pone.0005513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hammond A, Dertien J, Colmer-Hamood JA, Griswold JA, Hamood AN. Serum inhibits P. aeruginosa biofilm formation on plastic surfaces and intravenous catheters. The Journal of Surgical Research. 2010;159:735–746. doi: 10.1016/j.jss.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 72.Ruhal R, Antti H, Rzhepishevska O, Boulanger N, Barbero DR, et al. A multivariate approach to correlate bacterial surface properties to biofilm formation by lipopolysaccharide mutants of Pseudomonas aeruginosa . Colloids and Surfaces B, Biointerfaces. 2015;127:182–191. doi: 10.1016/j.colsurfb.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 73.Singh PK, Parsek MR, Greenberg EP, Welsh MJ. A component of innate immunity prevents bacterial biofilm development. Nature. 2002;417:552–555. doi: 10.1038/417552a. [DOI] [PubMed] [Google Scholar]
- 74.Berlutti F, Morea C, Battistoni A, Sarli S, Cipriani P, et al. Iron availability influences aggregation, biofilm, adhesion and invasion of Pseudomonas aeruginosa and Burkholderia cenocepacia . International Journal of Immunopathology and Pharmacology. 2005;18:661–670. doi: 10.1177/039463200501800407. [DOI] [PubMed] [Google Scholar]
- 75.Banin E, Vasil ML, Greenberg EP. Iron and Pseudomonas aeruginosa biofilm formation. Proc Natl Acad Sci U S A. 2005;102:11076–11081. doi: 10.1073/pnas.0504266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kruczek C, Qaisar U, Colmer-Hamood JA, Hamood AN. Serum influences the expression of Pseudomonas aeruginosa quorum-sensing genes and QS-controlled virulence genes during early and late stages of growth. MicrobiologyOpen. 2014;3:64–79. doi: 10.1002/mbo3.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Albus AM, Pesci EC, Runyen-Janecky LJ, West SE, Iglewski BH. Vfr controls quorum sensing in Pseudomonas aeruginosa . J Bacteriol. 1997;179:3928–3935. doi: 10.1128/jb.179.12.3928-3935.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Croda-Garcia G, Grosso-Becerra V, Gonzalez-Valdez A, Servin-Gonzalez L, Soberon-Chavez G. Transcriptional regulation of Pseudomonas aeruginosa rhlR: role of the CRP orthologue Vfr (virulence factor regulator) and quorum-sensing regulators LasR and RhlR. Microbiology. 2011;157:2545–2555. doi: 10.1099/mic.0.050161-0. [DOI] [PubMed] [Google Scholar]
- 79.Burton MO, Campbell JJ, Eagles BA. The mineral requirements for pyocyanin production. Canadian Journal of Research. 1948;26:15–22. doi: 10.1139/cjr48c-002. [DOI] [PubMed] [Google Scholar]
- 80.Totter JR, Moseley FT. Influence of the concentration of iron on the production of fluorescin by Pseudomonas aeruginosa . J Bacteriol. 1953;65:45–47. doi: 10.1128/jb.65.1.45-47.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Oogai Y, Matsuo M, Hashimoto M, Kato F, Sugai M, et al. Expression of virulence factors by Staphylococcus aureus grown in serum. Appl Environ Microbiol. 2011;77:8097–8105. doi: 10.1128/AEM.05316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Omidvar Z, Asoodeh A, Chamani J. Studies on the antagonistic behavior between cyclophosphamide hydrochloride and aspirin with human serum albumin: time-resolved fluorescence spectroscopy and isothermal titration calorimetry. J Solution Chem. 2013;42:1005–1017. [Google Scholar]
- 83.Nafisi S, Bagheri Sadeghi G, PanahYab A. Interaction of aspirin and vitamin C with bovine serum albumin. J Photochem Photobiol B. 2011;105:198–202. doi: 10.1016/j.jphotobiol.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 84.King EO, Ward MK, Raney DE. Two simple media for the demonstration of pyocyanin and fluorescin. The Journal of Laboratory and Clinical Medicine. 1954;44:301–307. [PubMed] [Google Scholar]
- 85.Lamichhane JR, Varvaro L. A new medium for the detection of fluorescent pigment production by pseudomonads. Plant Pathol. 2013;62:624–632. [Google Scholar]
- 86.Rust L, Messing CR, Iglewski BH. Elastase assays. Methods in enzymology. 1994;235:554–562. doi: 10.1016/0076-6879(94)35170-8. [DOI] [PubMed] [Google Scholar]
- 87.Rajamani S, Hilda A. Plate assay to screen fungi for proteolytic activity. Current Science. 1987;56:1179–1181. [Google Scholar]
- 88.Dobler L, de Carvalho BR, Alves WS, Neves BC, Freire DMG, et al. Enhanced rhamnolipid production by Pseudomonas aeruginosa overexpressing estA in a simple medium. PloS one. 2017;12:e0183857. doi: 10.1371/journal.pone.0183857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koch AK, Kappeli O, Fiechter A, Reiser J. Hydrocarbon assimilation and biosurfactant production in Pseudomonas aeruginosa mutants. J Bacteriol. 1991;173:4212–4219. doi: 10.1128/jb.173.13.4212-4219.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.O'Toole GA. Microtiter dish biofilm formation assay. Journal of visualized experiments : JoVE. 2011 doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Garde C, Bjarnsholt T, Givskov M, Jakobsen TH, Hentzer M, et al. Quorum sensing regulation in Aeromonas hydrophila . Journal of molecular biology. 2010;396:849–857. doi: 10.1016/j.jmb.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 92.Seviour T, Hansen SH, Yang L, Yau YH, Wang VB, et al. Functional amyloids keep quorum-sensing molecules in check. J Biol Chem. 2015;290:6457–6469. doi: 10.1074/jbc.M114.613810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Martinez-Solano L, Macia MD, Fajardo A, Oliver A, Martinez JL. Chronic Pseudomonas aeruginosa infection in chronic obstructive pulmonary disease. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2008;47:1526–1533. doi: 10.1086/593186. [DOI] [PubMed] [Google Scholar]
- 94.Meyer JM, Halle F, Hohnadel D, Lemanceau P, Ratefiarivelo H. Siderophores of Pseudomonas–biological properties. In: Winkelmann G, Van der Helm D, Neilands JB, editors. Iron Transport in Microbes, Plants, and Animals. Weinheim Germany: VCH; 1987. pp. 189–205. [Google Scholar]
- 95.Charney J, Tomarelli RM. A colorimetric method for the determination of the proteolytic activity of duodenal juice. J Biol Chem. 1947;171:501–505. [PubMed] [Google Scholar]
- 96.Schmidtchen A, Wolff H, Hansson C. Differential proteinase expression by Pseudomonas aeruginosa derived from chronic leg ulcers. Acta Derm Venereol. 2001;81:406–409. doi: 10.1080/000155501317208336. [DOI] [PubMed] [Google Scholar]
- 97.Yang L, Barken KB, Skindersoe ME, Christensen AB, Givskov M, et al. Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa . Microbiology. 2007;153:1318–1328. doi: 10.1099/mic.0.2006/004911-0. [DOI] [PubMed] [Google Scholar]
- 98.Pearson JP, Pesci EC, Iglewski BH. Roles of Pseudomonas aeruginosa LAS and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang VB, Chua SL, Cao B, Seviour T, Nesatyy VJ, et al. Engineering PQS biosynthesis pathway for enhancement of bioelectricity production in Pseudomonas aeruginosa microbial fuel cells. PloS one. 2013;8:e63129. doi: 10.1371/journal.pone.0063129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.