Abstract
Introduction.
Pneumocystis jirovecii pneumonia (PCP) is a severe disease affecting immunocompromised patients. Diagnosis is difficult due to the low sensitivity of direct examination and inability to grow the pathogen in culture. Quantitative PCR in bronchoalveolar lavage fluid (BAL) has high sensitivity, but limited specificity for distinguishing PCP from colonization.
Aim.
To assess the performance of an in-house quantitative PCR to discriminate between PCP and colonization.
Methodology.
This was a single-centre retrospective study including all patients with a positive PCR result for P. jirovecii in BAL between 2009 and 2017. Irrespective of PCR results, PCP was defined as the presence of host factors and clinical/radiological criteria consistent with PCP and (i) the presence of asci at direct examination of respiratory sample or (ii) anti-PCP treatment initiated with clinical response and absence of alternative diagnosis. Colonization was considered for cases who did not receive anti-PCP therapy with a favourable outcome or an alternative diagnosis. Cases who did not meet the above mentioned criteria were classified as ‘undetermined’.
Results.
Seventy-one patients with positive P. jirovecii PCR were included (90 % non-HIV patients). Cases were classified as follows: 37 PCP, 22 colonization and 12 undetermined. Quantitative PCR values in BAL were significantly higher in patients with PCP versus colonization or undetermined (P<0.0001). The cut-off of 5×103 copies/ml was able to discriminate PCP cases from colonization with 97 % sensitivity, 82 % specificity, 90 % positive predictive value and 95 % negative predictive value.
Conclusions.
Our quantitative PCR for P. jirovecii in BAL was reliable to distinguish PCP cases from colonization in this predominantly non-HIV population.
Keywords: pneumocystosis, molecular diagnosis, bronchoalveolar lavage fluid
Introduction
Pneumocystis jirovecii is an opportunistic fungus causing pneumonia (PCP), a severe lung disease in patients with T-lymphocyte mediated immune defect [1]. Historically, PCP was mainly observed among patients infected by the human immunodeficiency virus (HIV) with low CD4 count [2]. However, since the introduction of highly active antiretroviral therapy, the prevalence of the disease among HIV-infected patients has decreased in developed countries while the proportion of cases in non-HIV immunocompromised patients, such as haematologic cancer patients, has increased [3, 4]. Mortality rates are particularly high in this latter patients’ population (30–60 %) [3].
PCP is a life-threatening and rapidly progressive disease, which should be promptly detected [3]. The diagnosis of PCP is difficult because patients rarely produce sputum and invasive procedures, such as bronchoscopy, are often required to get respiratory specimens. Moreover, P. jirovecii cannot be isolated by routine culture methods and direct examination, albeit specific, has low sensitivity [5]. Molecular tools based on PCR are becoming a cornerstone for the diagnosis of PCP because of their high sensitivity for P. jirovecii detection [5–7]. While a negative PCR result is reliable to exclude the disease, a positive result could be difficult to interpret as it may reflect colonization instead of infection. Colonization, also referred to as asymptomatic carriage, corresponds to a situation in which the immune system is able to prevent the development of disease following recent exposure [8]. Because many pulmonary infections or non-infectious lung diseases may mimic PCP in non-HIV immunocompromised patients, the distinction between infection and colonization is challenging. In this study, we analysed the performance of an in-house quantitative P. jirovecii-specific PCR to discriminate PCP from colonization according to the fungal load in bronchoalveolar lavage fluid (BAL) in a predominantly non-HIV population.
Methods
Study design and patients
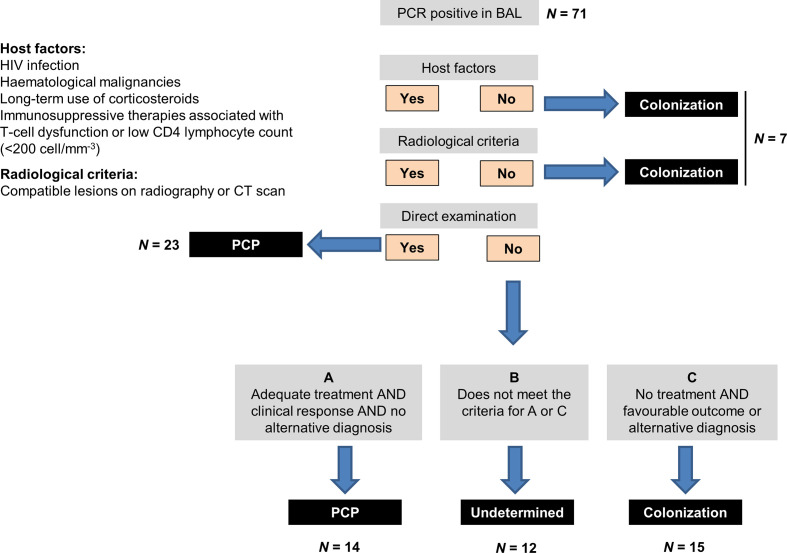
This was a retrospective study performed at the University Hospital of Lausanne (Switzerland) from October 2007 (date of introduction of PCP quantitative PCR in clinical routine) to December 2017. All patients with a positive P. jirovecii PCR in a BAL were enrolled. Clinical data were collected in medical records including patients’ underlying conditions, clinical and radiological presentation of pneumonia, microbiological results of BAL, therapeutic management of PCP and 12 week overall mortality. Two investigators (T.P. and A.K.) analysed the clinical data independently and without knowledge of the PCP PCR quantitative results. Irrespective of the diagnosis of PCP stated in medical records, cases were classified according to the algorithm of Fig. 1. PCP diagnosis was made in the presence of all the following criteria: (i) presence of at least one host criterion as defined in Fig. 1, (ii) clinical signs of respiratory infection with radiologic features consistent with PCP and (iii) presence of asci at direct examination by silver staining or introduction of antimicrobial therapy with anti-PCP activity with complete or partial improvement during the course of therapy in the absence of any alternative diagnosis. Patients were considered as ‘colonized’ if host criteria or clinical features of PCP were absent or if they did not receive anti-PCP therapy with a subsequent favourable outcome or a documented alternative diagnosis of respiratory infection or disease. All cases who did not fulfill the criteria of PCP or colonization were classified as ‘undetermined’. Discordant classification of cases was resolved by a consensus between F.L., T.P. and A.K.
Fig. 1.
Algorithm for classification of cases as PCP, colonization or undetermined. BAL, bronchoalveolar lavage fluid.
Diagnostic methods
Direct examination of P. jirovecii asci in the BAL sample was performed by the rapid methenamine silver-staining method [9]. The real-time quantitative PCR was run on our automated molecular diagnostic platform, as previously described [10]. The PCP PCR targets the mitochondrial 26S rDNA. The primers and protocol used in this study have been described elsewhere [11]. In brief, the BAL was centrifuged 30 min at 3000 g and the pellet was resuspended in remaining 2 ml of liquid. Then, 200 µl was used for the extraction procedure. The Kit DNA Process Control (Roche Molecular Systems) was used as extraction control. Amplification and detection were performed on an Applied Biosystems Quantstudio 7 Real-Time PCR System, using a TaqMan Fast Advanced Master Mix. In order to quantify the fungal load, plasmid-positive control dilutions corresponding to 104, 105 and 106 copies/ml were added to each run. A standard dilution curve using the cycle threshold (Ct) value of the known range of concentrations allowed the quantification of the fungal load of the original specimen in copies/ml. Plasmid positive controls are constructed according to our targeted PCR and produced by RD-Biotech, France.
PCP management
Antimicrobial therapy of PCP in our institution follows international recommendations [12]. The recommended regimen is trimethoprim-sulfamethoxazole (TMP-SMX) at 15 mg/kg/day of the trimethoprim component divided in three daily doses (orally or intravenously) for 3 weeks. For patients with TMP-SMX allergy, treatment consist of atovaquone 750 mg twice daily for mild disease or primaquine 30 mg once daily and clindamycin 600–900 mg three times daily for moderate/severe cases. Adjunctive corticosteroids are given in case of hypoxemia (PaO2<70 mmHg).
Statistical analyses
Quantitative PCR results were compared between the three groups (PCP, colonization, undetermined) using the non-parametric one-way ANOVA (Kruskal–Wallis) test with P-value considered as significant if ≤0.05. The performance of PCR to discriminate PCP from colonization (after exclusion of undetermined cases) among positive results was tested for different thresholds and expressed in terms of sensitivity, specificity, positive and negative predictive values, positive and negative likelihood ratios.
Ethical statement
This study was approved by Swissethics (commission of the ‘Canton de Vaud’, project number: 2018–01220) for retrospective use of clinical data with waiver of patients’ informed consent.
Results
A total of 71 patients with a positive PCP PCR result in a BAL sample were included in the analysis. As shown in Fig. 1 and 37 cases were classified as PCP according to the above-mentioned criteria, of which 23 (62 %) had proven infection on the basis of a positive direct examination for asci in BAL. In 22 cases, PCP was excluded because of lack of host or clinical/radiological criteria or because patients did not receive anti-PCP therapy or had an alternative diagnosis of respiratory disease. The remaining 12 cases were classified as undetermined. Characteristics of patients are described for the three groups in Table 1. PCP cases were mainly non-HIV patients (81 %) with haematologic cancer (51 %) or solid tumours (19 %). All patients classified as PCP or undetermined cases received anti-PCP therapy (mainly with co-trimoxazole), while 18 % of the cases classified as colonization were treated. Anti-PCP therapy was already ongoing at the time of BAL in nine (17 %) of the treated patients (median of 1 day before BAL sampling, range 1–4 days).
Table 1.
Characteristics of patients with a positive P. jirovecii PCR
|
PCP (N=37) |
Undetermined (N=12) |
Colonization (N=22) |
|
|---|---|---|---|
|
Demographic data |
|||
|
Female / Male |
22 (59) / 15 (41) |
5 (42) / 7 (58) |
16 (73) / 6 (27) |
|
Age |
59 (2–78) |
64 (43–80) |
61 (27–81) |
|
Underlying diseases |
|||
|
HIV infection |
7 (19) |
0 (0) |
0 (0) |
|
Haematologic cancer |
19 (51) |
4 (33) |
11 (50) |
|
Acute leukemia |
6 (16) |
0 (0) |
3 (14) |
|
Lymphoproliferative disorder |
6 (16) |
3 (25) |
6 (27) |
|
Other haematologic cancer* |
7 (19) |
1 (8) |
2 (9) |
|
Solid tumour |
7 (19) |
5 (42) |
6 (27) |
|
Other† |
4 (11) |
3 (25) |
3 (14) |
|
Immunosuppression |
|||
|
Neutropenia (PMN<500/mm3) |
7 (19) |
0 (0) |
4 (18) |
|
Lymphocyte CD4<200/ mm3 |
18 (49) |
6 (50) |
8 (36) |
|
Corticosteroids>14 days |
17 (46) |
9 (75) |
5 (23) |
|
Other immunosuppressive drugs |
19 (51) |
7 (58) |
12 (55) |
|
Anti-PCP prophylaxis |
4 (11) |
0 (0) |
2 (9) |
|
Clinical/radiological features |
|||
|
Hypoxemia‡ |
31 (84) |
9 (75) |
11 (50) |
|
Bilateral lung lesions |
35 (95) |
11 (92) |
20 (91) |
|
Ground-glass opacity (chest CT) |
30 (81) |
9 (75) |
17 (77) |
|
Anti-PCP therapy |
|||
|
No treatment |
0 (0) |
0 (0) |
18 (82) |
|
TMP-SMX |
37 (100) |
10 (83) |
4 (18) |
|
Atovaquone |
0 (0) |
2 (17) |
0 (0) |
|
Adjunctive corticosteroids |
33 (89) |
10 (83) |
4 (18) |
|
Mortality at 12 weeks |
10 (27) |
4 (33) |
7 (32) |
Values are absolute numbers (percentage) for proportions and median (range) for continuous variables.
*Other haematologic cancer: multiple myeloma, myelodysplastic syndrome or myeloproliferative disorder
†Auto-immune disorders or solid-organ transplantation.
‡Room air PaO2 <70 mmHg or O2 saturation <92 %.
PCP, P. jirovecii pneumonia; HIV, human immunodeficiency virus; PMN, polymorphonuclear neutrophils; TMP-SMX, trimethoprim-sulfamethoxazole.
The quantitative results of PCP PCR were significantly higher among PCP cases compared to both undetermined and colonized cases (Fig. 2). The performance of the quantitative PCR to discriminate PCP from colonization for different thresholds is shown in Table 2. The optimal result was obtained for a cut-off of 5×103 copies/ml with a sensitivity, specificity, positive and negative predictive value of 97, 82, 90 and 95 %, respectively. A 100 % sensitivity and negative predictive value were obtained for the cut-off of 103 copies/ml, while a cut-off of 5×105 copies/ml was associated with 100 % specificity and positive predictive value.
Fig. 2.
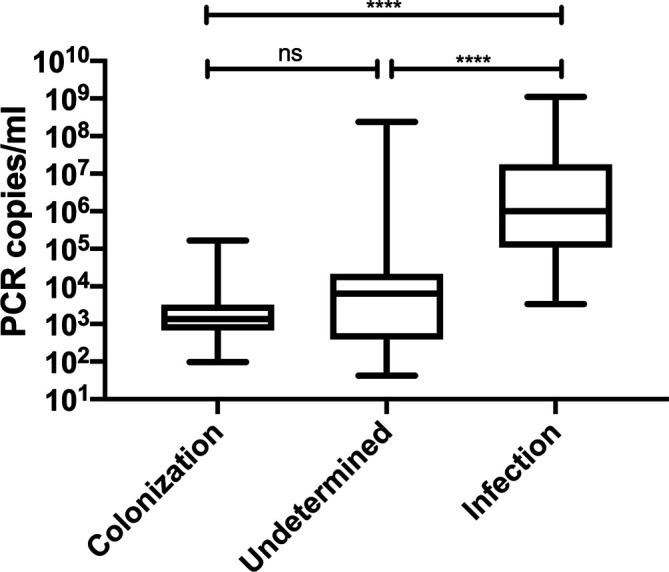
Comparison of P. jirovecii quantitative PCR results between PCP, colonization and undetermined cases. Analysis performed by non-parametric one-way ANOVA (Kruskal–Wallis) test with P-value considered as significant if ≤0.05. **** P value <0.0001.
Table 2.
Diagnostic performance of P. jirovecii PCR for the discrimination between PCP and colonization
|
Cut-off |
Sensitivity |
Specificity |
PPV |
NPV |
LR + |
LR - |
DOR |
|---|---|---|---|---|---|---|---|
|
102 |
1 |
0.05 |
0.64 |
1 |
1.05 |
0 |
NC |
|
5×102 |
1 |
0.23 |
0.69 |
1 |
1.29 |
0 |
NC |
|
103 |
1 |
0.36 |
0.73 |
1 |
1.57 |
0 |
NC |
|
5×103 |
0.97 |
0.82 |
0.90 |
0.95 |
5.35 |
0.03 |
162 |
|
104 |
0.95 |
0.82 |
0.90 |
0.90 |
5.20 |
0.07 |
78.75 |
|
5×104 |
0.81 |
0.91 |
0.94 |
0.74 |
8.92 |
0.21 |
42.86 |
|
105 |
0.76 |
0.95 |
0.97 |
0.70 |
16.65 |
0.25 |
65.33 |
|
5×105 |
0.62 |
1 |
1 |
0.61 |
NC |
0.38 |
NC |
|
106 |
0.51 |
1 |
1 |
0.55 |
NC |
0.49 |
NC |
|
5×106 |
0.30 |
1 |
1 |
0.46 |
NC |
0.70 |
NC |
|
107 |
0.30 |
1 |
1 |
0.46 |
NC |
0.70 |
NC |
DOR, diagnostic odds ratio; LR+, positive likelihood ratio; LR-, negative likelihood ratio; NC, not calculated; NPV, negative predictive value; PPV, positive predictive value.
Discussion
PCP is increasingly observed among non-HIV patients with different types and intensity of immunosuppression. Atypical clinical presentation and concomitant respiratory infections are frequent in this population making PCP diagnosis particularly challenging. Our analysis suggests an excellent ability of our quantitative PCR to discriminate PCP from colonization in a predominantly non-HIV population with a suggested cut-off of 5×103 copies/ml.
Some studies have previously assessed the performance of quantitative PCR cut-off values to distinguish PCP from colonization [13–18]. These studies (compared in Table 3) exhibit important heterogeneity regarding the type of population (proportion of HIV and non-HIV patients), the type of sample (BAL only or including induced sputum samples), the PCR target (mitochondrial rRNA, dihydropteroate synthase gene, major surface glycoprotein), the quantification method of fungal load (Ct cycles or copies per ml), the definition of PCP cases (i.e. clinical criteria to distinguish probable cases from colonization), and the expression of results. Similarly to our analysis, most studies demonstrate significant differences of mean fungal loads between the categories of proven PCP, versus probable/possible cases and colonization. However, the assessment of an acceptable fungal load cut-off for distinguishing PCP from colonization gives various results. While most studies suggest a ‘grey zone’ between cut-offs providing near 100 % positive/negative predictive values for PCP diagnosis, some of them found a large and overlapping distribution precluding the assessment of a reliable cut-off [15, 18]. Compared to previous analyses, our study has the highest number and proportion of haematologic cancer patients. Diagnosis of PCP is notoriously difficult in this population exposed to various types of lung injury including infectious causes and secondary effects of anti-cancer therapy [19].
Table 3.
Comparative performance of quantitative P. jirovecii PCR in various studies
|
Study first author, year (reference) |
PCR target |
No. of PCP cases* HIV / HM / others [%] |
PCP classification† |
Results |
|---|---|---|---|---|
|
Alanio, 2011 [13] |
Mitochondrial large-subunit rRNA gene |
16 56/19/25 |
High vs low probability |
<120 TFEq/ml: 100 % NPV >1900 TFEq/ml: 100 % PPV |
|
Botterel, 2012 [14] |
Mitochondrial large-subunit rRNA gene |
17 47/35/18 |
Proven vs IFA-negative |
≤2.6 log10 copies/µl: 100 % NPV ≥4 log10 copies/µl: 100 % PPV |
|
Matsumura, 2016 [17] |
Dihydropteroate synthase gene |
53 9/17/74 |
(a) Proven vs colonization (b) proven/probable vs colonization |
(a) 1300 copies/ml: 100 % NPV, 79 % PPV (b) 340 copies/ml: 44 % NPV, 91 % PPV |
|
Robert-Gangneux, 2014 [18] |
Mitochondrial large-subunit rRNA gene |
35 12/37/51 |
Proven/probable vs possible vs colonization |
Significant differences of fungal load (Ct value between groups). Large distribution, no cut-off proposed. |
|
Maillet, 2014 [16] |
Major surface glycoprotein |
11 18/73/9 |
Proven/probable vs possible/colonization |
≤3160 copies/ml: 100 % NPV ≥31 600 copies/ml: 100 % PPV |
|
Fauchier. 2016 [15] |
Mitochondrial large-subunit rRNA gene |
73 41/26/33 |
Proven/probable vs colonization (no possible category) |
Ct 32 : 72 % sen, 75 % spe Ct >35: exclusion PCP (80%sen/63%spe) |
|
Kritikos, 2020 (present study) |
Mitochondrial 26S rDNA |
37 19/51/30 |
Proven/probable vs colonized (possible excluded) |
≤103 copies/ml: 100 % NPV 5×103 copies/ml: 95 % NPV, 90 % PPV ≥5×105 copies/ml: 100 % PPV |
*Patients with PCP classified as proven or probable.
†Nomenclature for PCP classification: proven: IFA positive, probable: IFA negative / PCR positive, with defined clinical criteria for PCP diagnosis, possible: IFA negative / PCR positive, without fulfilling all clinical criteria for PCP diagnosis. Colonization: IFA negative / PCR positive, and no PCP diagnosis retained on the basis of clinical assessment.
HIV, human immunodeficiency virus; HM, haematologic malignancies; IFA, immunofluorescence assay; NPV, negative predictive value; PCP, P. jirovecii pneumonia; Sen, sensitivity; Spe, specificity; TFEq, trophic form equivalents.
The absence of a reliable gold standard to define PCP infection is an important limitation. In this study, we used stringent criteria to define PCP with a majority of patients (62 %) with proven infection on the basis of a positive direct examination and the remaining cases having responded to anti-PCP therapy in the absence of alternative diagnosis. Nonetheless, a small proportion of cases were classified as undetermined and were excluded from the analysis. The median fungal burden among these patients was close to the cut-off, suggesting a grey zone in the interpretation. However, our results suggest a reliable and relatively narrow cut-off margin with values <103 and >5×103 copies/ml providing 100 and 90% negative and positive predictive values, respectively, which seems appropriate to trigger therapeutic decisions. Targeted treatment of PCP cases based on reliable PCR cut-offs may avoid unnecessary use of antimicrobial agents and their associated risks of toxicity or resistance.
Use of the 1,3-beta-d-glucan test in serum could be helpful for the assessment of PCP probability. Unfortunately, BDG results were not available for most cases of the present study, as the test was introduced in our institution only at the end of the study period. Another limitation consisted of the absence of negative controls. However, the excellent negative predictive value of PCP PCR (≥99 %) has already been established [5]. Accordingly, we did not observe any confirmed case of PCP with a negative PCR result during the study period. The actual challenge for PCP diagnosis relies on the discrimination between PCP and colonization among patients with a positive PCR result, which was the specific question addressed in the present study. Other roles of PCP PCR, for instance as a screening tool in the setting of nosocomial PCP outbreaks, could also be investigated in the future [20, 21].
In conclusion, quantitative PCP PCR results in BAL were reliable to distinguish infection from colonization in this predominantly non-HIV population and may be used to guide therapeutic decisions. Thresholds should be assessed for each individual in-house PCR method, as different values may be observed according to the molecular method and/or patients’ population. Development of standardized procedures and commercial kits for quantitative PCP PCR is warranted. Such efforts of standardization with comparison of PCP PCR performance across centres and between different assays are ongoing [22].
Funding information
This work was done as part of our routine practice without any specific funding.
Acknowledgements
We are grateful to Zahera Naseri, René Brouillet and all the technical staff of the Molecular Diagnostics Unit of the Institute of Microbiology of Lausanne University Hospital.
Author contributions
T.P. data collection, data analyses, draft of manuscript, A.K. data analyses, draft of manuscript, P.M.H. PCR design, review of manuscript, K.J. PCR design, data extraction, review of manuscript, F.L. data analyses, redaction of manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
This study was approved by Swissethics (commission of the ‘Canton de Vaud’, project number: 2018–01220) for retrospective use of clinical data with waiver of patients’ informed consent.
Footnotes
Abbreviations: BAL, Bronchoalveolar lavage; BDG, Beta-d-glucan; HIV, Human immunodefiency virus; PCP, Pneumocystis jirovecii pneumonia; TMP-SMX, Trimethoprim-sulfamethoxazole.
References
- 1.Calderón EJ, Gutiérrez-Rivero S, Durand-Joly I, Dei-Cas E. Pneumocystis infection in humans: diagnosis and treatment. Expert Rev Anti Infect Ther. 2010;8:683–701. doi: 10.1586/eri.10.42. [DOI] [PubMed] [Google Scholar]
- 2.Phair J, Muñoz A, Detels R, Kaslow R, Rinaldo C, et al. The risk of Pneumocystis carinii pneumonia among men infected with human immunodeficiency virus type 1. Multicenter AIDS cohort study Group. N Engl J Med. 1990;322:161–165. doi: 10.1056/NEJM199001183220304. [DOI] [PubMed] [Google Scholar]
- 3.Cordonnier C, Cesaro S, Maschmeyer G, Einsele H, Donnelly JP, et al. Pneumocystis jirovecii pneumonia: still a concern in patients with haematological malignancies and stem cell transplant recipients. J Antimicrob Chemother. 2016;71:2379–2385. doi: 10.1093/jac/dkw155. [DOI] [PubMed] [Google Scholar]
- 4.Maini R, Henderson KL, Sheridan EA, Lamagni T, Nichols G, et al. Increasing Pneumocystis pneumonia, England, UK, 2000-2010. Emerg Infect Dis. 2013;19:386–392. doi: 10.3201/eid1903.121151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White PL, Backx M, Barnes RA. Diagnosis and management of Pneumocystis jirovecii infection. Expert Rev Anti Infect Ther. 2017;15:435–447. doi: 10.1080/14787210.2017.1305887. [DOI] [PubMed] [Google Scholar]
- 6.Fan L-C, Lu H-W, Cheng K-B, Li H-P, Xu J-F. Evaluation of PCR in bronchoalveolar lavage fluid for diagnosis of Pneumocystis jirovecii pneumonia: a bivariate meta-analysis and systematic review. PLoS One. 2013;8:e73099. doi: 10.1371/journal.pone.0073099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu Y, Ling G, Qiang C, Ming Q, Wu C, et al. Pcr diagnosis of Pneumocystis pneumonia: a bivariate meta-analysis. J Clin Microbiol. 2011;49:4361–4363. doi: 10.1128/JCM.06066-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alanio A, Bretagne S. Pneumocystis jirovecii detection in asymptomatic patients: what does its natural history tell us? F1000Res. 2017;6:739. doi: 10.12688/f1000research.10619.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahan CT, Sale GE. Rapid methenamine silver stain for Pneumocystis and fungi. Arch Pathol Lab Med. 1978;102:351–352. [PubMed] [Google Scholar]
- 10.Greub G, Sahli R, Brouillet R, Jaton K. Ten years of R&D and full automation in molecular diagnosis. Future Microbiol. 2016;11:403–425. doi: 10.2217/fmb.15.152. [DOI] [PubMed] [Google Scholar]
- 11.Richard S, Almeida JMGCF, Cissé OH, Luraschi A, Nielsen O, et al. Functional and expression analyses of the Pneumocystis MAT genes suggest obligate sexuality through primary homothallism within host lungs. mBio. 2018;9:e02201-17. doi: 10.1128/mBio.02201-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maschmeyer G, Helweg-Larsen J, Pagano L, Robin C, Cordonnier C, et al. ECIL guidelines for treatment of Pneumocystis jirovecii pneumonia in non-HIV-infected haematology patients. J Antimicrob Chemother. 2016;71:2405–2413. doi: 10.1093/jac/dkw158. [DOI] [PubMed] [Google Scholar]
- 13.Alanio A, Desoubeaux G, Sarfati C, Hamane S, Bergeron A, et al. Real-Time PCR assay-based strategy for differentiation between active Pneumocystis jirovecii pneumonia and colonization in immunocompromised patients. Clin Microbiol Infect. 2011;17:1531–1537. doi: 10.1111/j.1469-0691.2010.03400.x. [DOI] [PubMed] [Google Scholar]
- 14.Botterel F, Cabaret O, Foulet F, Cordonnier C, Costa J-M, et al. Clinical significance of quantifying Pneumocystis jirovecii DNA by using real-time PCR in bronchoalveolar lavage fluid from immunocompromised patients. J Clin Microbiol. 2012;50:227–231. doi: 10.1128/JCM.06036-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fauchier T, Hasseine L, Gari-Toussaint M, Casanova V, Marty PM, et al. Detection of Pneumocystis jirovecii by quantitative PCR to differentiate colonization and pneumonia in immunocompromised HIV-positive and HIV-negative patients. J Clin Microbiol. 2016;54:1487–1495. doi: 10.1128/JCM.03174-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maillet M, Maubon D, Brion JP, François P, Molina L, et al. Pneumocystis jirovecii (Pj) quantitative PCR to differentiate Pj pneumonia from Pj colonization in immunocompromised patients. Eur J Clin Microbiol Infect Dis. 2014;33:331–336. doi: 10.1007/s10096-013-1960-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsumura Y, Ito Y, Iinuma Y, Yasuma K, Yamamoto M, et al. Quantitative real-time PCR and the (1→3)-β-D-glucan assay for differentiation between Pneumocystis jirovecii pneumonia and colonization. Clin Microbiol Infect. 2012;18:591–597. doi: 10.1111/j.1469-0691.2011.03605.x. [DOI] [PubMed] [Google Scholar]
- 18.Robert-Gangneux F, Belaz S, Revest M, Tattevin P, Jouneau S, et al. Diagnosis of Pneumocystis jirovecii pneumonia in immunocompromised patients by real-time PCR: a 4-year prospective study. J Clin Microbiol. 2014;52:3370–3376. doi: 10.1128/JCM.01480-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alanio A, Hauser PM, Lagrou K, Melchers WJG, Helweg-Larsen J, et al. ECIL guidelines for the diagnosis of Pneumocystis jirovecii pneumonia in patients with haematological malignancies and stem cell transplant recipients. J Antimicrob Chemother. 2016;71:2386–2396. doi: 10.1093/jac/dkw156. [DOI] [PubMed] [Google Scholar]
- 20.Nevez G, Le Gal S, Noel N, Wynckel A, Huguenin A, et al. Investigation of nosocomial Pneumocystis infections: usefulness of longitudinal screening of epidemic and post-epidemic Pneumocystis genotypes . J Hosp Infect. 2018;99:332–345. doi: 10.1016/j.jhin.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 21.Robin C, Alanio A, Gits-Muselli M, la Martire G, Schlemmer F, et al. Molecular demonstration of a Pneumocystis outbreak in stem cell transplant patients: evidence for transmission in the daycare center. Front Microbiol. 2017;8:700. doi: 10.3389/fmicb.2017.00700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gits-Muselli M, White PL, Mengoli C, Chen S, Crowley B, et al. The fungal PCR initiative's evaluation of in-house and commercial Pneumocystis jirovecii qPCR assays: toward a standard for a diagnostics assay. Med Mycol. 2019 doi: 10.1093/mmy/myz115. [DOI] [PubMed] [Google Scholar]