Abstract
DNA re-replication leads to genomic instability and has been implicated in the pathology of a variety of human cancers. Eukaryotic DNA replication is tightly controlled to ensure it occurs only once during each cell cycle. Geminin is a critical component of this control, it prevents DNA re-replication from occurring during S, G2 and early M phases by preventing MCM helicases from forming pre-replication complexes. Geminin is targeted for degradation by the anaphase promoting complex (APC/C) from anaphase through G1 phase, however, accumulating evidence indicates that Geminin is downregulated in late S phase due to an unknown mechanism. Here, we used a high throughput screen (HTS) to identify microRNAs that can induce excess DNA replication and found that miR-571 could reduce the protein level of Geminin in late S phase independent of the APC/C. Furthermore, miR-571 regulated efficient DNA replication and S phase cell cycle progression. Strikingly, c-Myc suppressed miR-571 expression by binding directly to the miR-571 promoter. At the beginning of S phase, Cdk2 phosphorylated c-Myc at Serine 62, promoting its association with the miR-571 promoter region. Collectively, we identify miR-571 as the first miRNA that prevents aberrant DNA replication, and the Cdk2-c-Myc-miR-571 axis as a new pathway for regulating DNA replication, cell cycle and genomic stability in cancer cells.
Keywords: Geminin, miR-571, Cdk2, MYC, DNA replication
INTRODUCTION
DNA re-replication leads to genomic instability and has been implicated in the pathology of a variety of human cancers. To maintain genomic stability, eukaryotic cells have evolved multiple mechanisms that ensure nuclear DNA is precisely replicated once and only once each time a cell divides. Overriding these mechanisms results in aberrant forms of DNA replication that lead to genomic instability (1,2). Of these mechanisms, the Geminin-mediated pathway helps to prevent licensing of DNA replication origins during the S, G2 and early M phases (3). Geminin is a 25 kDa protein in the metazoan that localizes to the nucleus where it binds specifically to Cdt1, a component of the pre-replication complex, and prevents recruitment of the MCM replicative helicase to DNA replication origins (4,5). We and others have demonstrated that down-regulation of Geminin results in DNA re-replication, DNA damage and apoptosis in both human and Drosophila cells, in which licensing of replication origins is not adequately regulated (6–10). Geminin protein is regulated in the cell cycle so that the Geminin level is low during G1 phase when pre-replication complexes are assembled at replication origins and high during S, G2 and early M phases to prevent assembly of new pre-replication complexes whose activation would cause DNA re-replication (4). When nuclear DNA replication is complete, Geminin protein is ubiquitinated by the anaphase promoting complex (APC/C) at the end of mitosis and throughout the subsequent G1 phase and degraded by the 26S proteasome (4,5).
The APC/C is activated during the transition from metaphase to anaphase, and then becomes inactive during late G1 phase due to phosphorylation of its substrate recognition subunit Cdh1 by a G1/S specific CDK activity until the next metaphase (11). Thus, S, G2 and early M phases are characterized by the absence of APC/C activity, which allows Geminin protein levels to increase during this period of cell division. However, the level of Geminin expression actually begins decreasing in late S phase and throughout G2/M phase(4). Thus, the question arises as to how Geminin expression is down-regulated in late S phase with the absence of APC/C activity. Is it possible that an as-yet unidentified APC/C-independent pathway exists to regulate Geminin expression?
Pre-replication complexes can only be assembled from anaphase through G1 phase when CDK activity is low. Cdk2/Cyclin E activity increases at the G1/S boundary where it activates the MCM helicase to begin DNA unwinding, and high levels of Cdk2/Cyclin A activity persist throughout S and G2 phase where it prevents DNA re-replication by phosphorylating specific proteins required for origin licensing (1,3,12). However, the mechanism by which Cdk2 prevents DNA re-replication and the identity of its inhibitory targets are not fully understood. Cdk2/Cyclin E also regulates c-Myc-mediated transcription during cellular senescence by phosphorylating the c-Myc protein at Serine 62 (Ser62) (13), suggesting that Cdk2/Cyclin E may regulate DNA replication by controlling c-Myc activity.
In fact, multiple studies have implicated c-Myc protein in the regulation of DNA replication at both transcriptional and non-transcriptional levels (14). For example, c-Myc can interact directly with pre-replication complexes (pre-RCs), localize to early sites of DNA synthesis, and promote replication origin activity (15–17). c-Myc could also regulate DNA replication by controlling expression of Cdt1 (18).
MiR-571 has been identified in multiple cancers, including melanoma, colorectal cancer, hepatocellular carcinoma, etc. (19–21), however, the biological function of miR-571 remains unknown. In this study, for the first time we used an unbiased high throughput screen (HTS) to identify miR-571 that induced DNA re-replication by down-regulating the level of Geminin mRNA. Inhibition of miR-571 expression decreased association of MCM proteins with chromatin at late G1 phase and delayed S phase progression, whereas forced expression of miR-571 increased association of MCM proteins with chromatin at late G1 phase, an event accompanied by DNA re-replication. Subsequent analyses revealed a novel pathway, in which Cdk2 phosphorylated c-Myc at Ser 62 during S phase, which promoted its binding to the miR-571 promoter and reduced miR-571 expression. This discovery marks the first time a miRNA regulates initiation of DNA replication, and elucidates a novel pathway in which Cdk2-c-Myc regulates expression of a specific miRNA that helps to maintain genomic stability during the transition from DNA replication to mitosis.
MATERIALS AND METHODS
High-throughput screen of microRNA induces excess of DNA replication
HCT116 cells were transfected with miRNA mimics library (Qiagen) as previously described (22). Briefly, 50nM individual miRNA mimic from human miRNA mimic (Qiagen) library was incubated with reaction buffer in 384 well plates for 1h, and then 1000 cells were seeded to each well and transfection was fulfilled to proceed for 72h as manufacturer instructions. After treatment for 72hr, cells were stained with Hoechst 33342 to visualize nuclear DNA and relative DNA content of each cell was quantified using an ImageXpress Micro System. The fraction of cells with more than 4N DNA content was selected as positive reads for further analysis.
Cell culture and Cell Synchronization
The human HCT116 and U2OS cells were purchased from ATCC. Cells were routinely tested for Mycoplasma and genetic identity of the cell lines were validated by short tandem repeat (STR) profiling (ATCC). Each cell lines were used within 10 passage after thawing. All cells were cultured at 37 °C with 5% CO2 in DMEM medium. Cells were synchronized in metaphase by culturing them with 40 ng/ml nocodazole for 16 hr, and the mitotic arrested cells were harvested by shaking off the dish. Harvested cells were washed in PBS and then released into nocodazole-free medium. To synchronize cells in G1 phase, cells were cultured in medium with L-mimosine (final concentration, 300 μM) for 24 hr.
Ligation reaction and PCR assay
DNA Oligos were designed to complementary almost half of 5’ and 3’ arm of the miR-571 sequence. The probe A that cover 5’ half of the miR-571 was phosphorylated that allows ligation to probe B. The sequence of each probe is listed in the Supplementary Table S1. Synthetic miRNAs (10nM) or total RNA from cells (1ug) were incubated with 0.1uM of each probe in 20ul ligation reaction (50mM Tris-HCl, 10mM MgCl2, 5mM DTT, 0.1mM spermidine, 0.1mM EDTA, 5 μM ATP, 10% PEG4000 and 1.5 μl T4 DNA ligase at 42°C for 2h) as previously described. The whole reaction was then inactivated at 80°C for 15 min. Primers for PCR amplification were as below, Forward: 5’-TCACCGACTGCCCATAGAGAGG-3’, Reverse: 5’-CCCTGCGTGTCTCCGACTCAG-3’. PCR products were fractionated using 15% polyacrylamide/TBE gels, stained with ethidium bromide, and pictured by UV camera.
RNA sequence
U2OS cells transfected with 50nM miR-Ctr, miR-571 mimic or 50nM of miR-571 inhibitor for 48 hr. Total RNA was extracted and prepared for cDNA libraries within Illumina TruSeq Stranded mRNA sample preparation kit (Illumina # RS-122–2103). All transcripts showed a false discovery rate (FDR) value < 0.5. Fold change more than 2.5 fold in miR-571 mimic group or more than 0.5 fold in inh-miR-571 group were selected as differentially expressed genes compared with cells treated with miR-Ctr or inh-miR-Ctr, respectively. Sequence data in this study have been deposited into GEO database with the accession codes GSE118998. All other remaining data are available in Supplementary files.
Chromatin Immuno-Precipitation (ChIP) and Electrophoretic mobility shift assay (EMSA)
Chromatin immunoprecipitation was carried out as previously described (23). Briefly, the cells were fixed with 1% paraformaldehyde for 24 hr and then sonicated to produce 200–1000 bp DNA fragments. The anti-Myc antibody and control mouse IgG were used for ChIP assays. The precipitated DNA fragments were quantitated by real-time PCR and normalized against the Input. The real-time PCR primers were as below: primer 1 (F: AGGATTCCAGGGTCTCACAGT, R: CAGTACCTGCCTCATGGAGT), primer 2 (F: TGGAGGATTCCAGGGTCTCAC, R: GCAGGATTGCACTGTGGCTAAG), primer 3 (F: GCCACAGTGCAATCCTGCAT, R: GGCGACCCGTTGAGCC), primer 4 (F: GAGAGATGATTCGCGAGCCT, R: AGTGCGCTGATTGGATACGG). DNA mobility shift assay was performed within LightShift® Chemiluminescent EMSA Kit as manufacturer instructions (ThermoScientific, Cat #20148). Briefly, four independent double-stranded DNA probes with 3’-end-labeled with biotin (Supplementary Table S2) were designed, annealed and purified on a Sephadex G-25 column. The probes were then incubated with recombinant c-Myc protein (expressed from E.coli and purified using Glutathione Agarose) for 2h at 37°C. For antibody super-shift analysis, DNA-protein complex was incubated with c-Myc antibody overnight at 4°C. For competition assays, 100-fold excess amount of unlabeled competitor was premixed with the biotin-labeled probes for 2h before adding the binding mixture reactions. DNA-protein complexed were then resolved on 15% non-denature PAGE and transferred to nylon membrane, UV-crosslinked, probed with streptavidin-HRP conjugate and incubated with the substrate, followed by Western blotting.
Statistical analysis
GraphPad Prism 5.0 software was used for all data analysis. Data were represented as mean ± SEM. Statistical analysis was performed using one-way ANOVA or Student’s t test. p≤0.05 was considered significant.
RESULTS
Identification of miR-571 that causes DNA re-replication through a high-throughput screen (HTS)
We initially developed a HTS assay to identify genes that prevent excess DNA replication, such as DNA re-replication (24). The same assay was then used to screen a library of miRNA mimics for miRNAs that induce excess DNA replication. Specifically, HCT116 cells were transfected with miRNA mimics library (Qiagen). After treatment with miRNA mimics for 72hr, cells were stained with Hoechst 33342 to visualize nuclear DNA using an ImageXpress Micro System (Molecular Devices) (24). Through this HTS, we identified a total of 11 miRNAs that induced excess DNA replication (Supplementary Table S3). We were particularly interested in the identification of miRNAs that regulate DNA re-replication. Since Geminin is critical for preventing DNA re-replication, we assumed that some of these miRNAs may regulate DNA re-replication by targeting Geminin. Therefore, we used the miRNA prediction program, Targetscan to identify those miRNAs that contained the seed sequence for binding to 3’-UTR region of Geminin mRNA (25). Interestingly, we found that miR-571 was predicted with seed sequences that could potentially bind to the 3’-UTR of Geminin mRNA at 88–94 sites (Supplementary Fig. S1A and Supplementary Table S4).
To determine whether or not miR-571 indeed targets Geminin, we constructed pGL3 luciferase reporter vectors containing the wild type Geminin 3’-UTR (pGL3-Gem 3’-UTR WT) or mutant Geminin 3’-UTR (pGL3-Gem 3’-UTR Mut) with mutation at miR-571 seed sequence (88–94 sites, as shown in Supplementary Fig. S1A). With these vectors, we then examined whether miR-571 binds to Geminin 3’-UTR by measuring relative luciferase activities. HCT-116 cells were co-transfected with pGL3 empty vector, pGL3-Gem 3’-UTR WT or pGL3-Gem 3’-UTR Mut together with a miRNA control (miR-Ctr) or miR-571 mimic (miR-571) (Fig. 1A). Compared to miR-Ctr, miR-571 significantly reduced the relative luciferase activity of WT Geminin 3’-UTR but not the mutant Geminin 3’-UTR (Fig. 1A), indicating that the sequence at 88–94 of 3’-UTR is the potential binding site for miR-571.
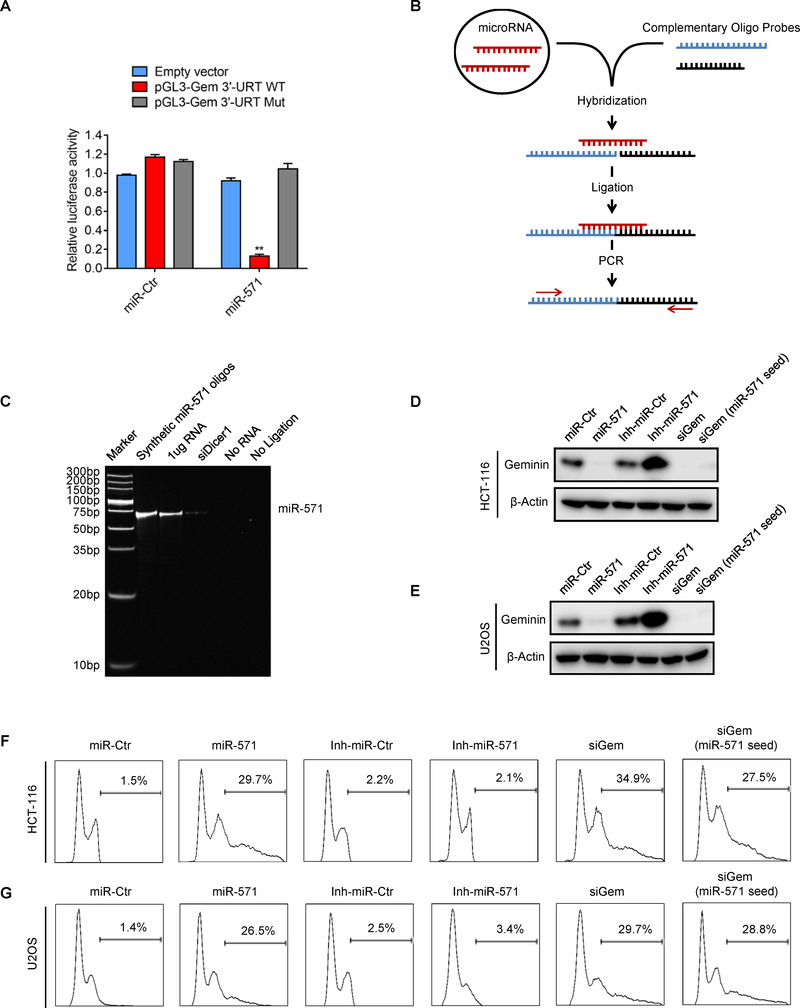
Figure 1. Identification of miR-571 that induces DNA re-replication.
(A) The relative luciferase activity in HCT-116 cells co-transfected with pGL3 empty vector, pGL3 vectors carrying wild type (pGL3-Gem 3’-UTR WT) or mutant Geminin 3’-UTR (pGL3-Gem 3’-UTR Mut) and miR-Ctr or miR-571. Relative luciferase activity was measured and normalized by firefly to Renilla ratio (Data represent mean ± SEM from three independent experiments, **p≤0.01). (B) Schematic depiction of the ligation-mediated PCR assay. miR-571 are hybridized to the half-complementary probe A and B in reaction buffer. Then all hybridized probes are joined by the T4 DNA ligase and amplified by PCR. (C) The ligated-based PCR products of miR-571 in HCT-116 cells are visualized by 15% polyacrylamide/TBE gels. (D and E) Immunoblot analysis of Geminin in HCT-116 (D) and U2OS cells (E) with indicated treatment after 48 hr. (F and G) Cells treated in D and E were collected for FACS analysis. The percentage of cells with re-replicated DNA was indicated (20000 events were counted for measure DNA over-replication ratio in each sample). Representative from three biological replicates was shown.
To determine whether or not miR-571 is a bona fide microRNA expressed in cells, we used a size-coded ligation-mediated polymerase chain reaction (SL-PCR) method, which could discriminate highly related miRNAs and detects mature miRNAs inclusive or exclusive of precursors(26). The SL-PCR is based on size-coded DNA probe hybridization in solution, followed-by ligation, PCR amplification and gel fractionation (26). Briefly, two DNA probes containing a half complementary sequence of miR-571 were incubated with synthetic miR-571 or total RNA extracted from cells for hybridization, followed by ligation and PCR amplification (Fig. 1B). As shown in Fig. 1C and Supplementary Fig. S1B, RNA extracted from HCT116 or U2OS cells generated a specific PCR product as predicted size (64bp, probe A + probe B), the same as the synthetic miR-571. However, RNA from cells treated with siDicer1 failed to generate PCR products since Dicer1 is critical to process microRNA precursors into mature microRNAs. As predicted, reaction without RNA or ligation enzyme failed to generate PCR product. Furthermore, we also used miScript SYBR Green PCR assay to detect miR-571 expression. Specifically, cDNA was synthesized and then followed by qPCR using miR-571-specific DNA primers for amplification. miR-571 was detected in control cells, which was significantly reduced in cells treated with siDicer1 and the inhibitor of miR-571 (Inh-miR-571) (Supplementary Fig. S1C–D). Together, these data strongly suggest that miR-571 is a bona fide microRNA expressed in HCT116 and U2OS cells.
To study the biological function of miR-571, we ectopically expressed miR-571 mimic or inhibitor in HCT-116 or U2OS cells. Interestingly, forced expression of miR-571 reduced both Geminin mRNA and protein level as well as induced DNA re-replication, which was also observed in cells transfected with siRNA against Geminin (siGem, targeting Geminin open reading frame) in both cells (Fig. 1D–G and Supplementary Fig. S1E–G). To confirm that miR-571 truly sufficiently induces DNA re-replication by targeting Geminin, we designed a siRNA (siGem miR-571 seed) containing 7-base miR-571 seed sequence to mimic the specific miR-571 targeting site in Geminin 3’-UTR region. Consistently, this specific siRNA notably reduced Geminin protein and mRNA level, as well as induced excess DNA replication in HCT-116 and U2OS cells (Fig. 1D–G and Supplementary Fig. S1E–G). Meanwhile, ectopic expression of miR-571 induced DNA damage as indicated by increased phosphorylation of Chk1 at Ser317, cell cycle arrest in G2/M phase as indicated by increased cyclin A levels, activation of G2/M phase checkpoint as indicated by increased phosphorylation of Cdc2 at Tyr15, and increasedγ-H2AX foci formation (Supplementary Fig. S1H–K). Moreover, ectopic expression of miR-571 significantly reduced cell viability as indicated by colony formation (Supplementary Fig. S1L–M). Since ectopic expression of miR-571 activates G2/M phase checkpoint, we assumed that inhibition of G2/M checkpoint by caffeine allows cells to enter mitosis and thereby apoptosis. Indeed, miR-571 or siGem alone did not induce apoptosis, but addition of caffeine dramatically induced apoptosis in cells treated with miR-571 or siGem (Supplementary Fig. S1N). Ectopic expression of miR-571 did not induce expression of mitotic marker p-H3 (S10), which was significantly elevated in nocodazole treated cells (Supplementary Fig. S1O–S1P), indicating that miR-571 treated cells do not arrest at mitosis.
Taken together, via a HTS we have identified a previously uncharacterized miR-571, which could induce DNA re-replication by targeting Geminin 3’-UTR region.
miR-571 specifically targets Geminin mRNA to induce DNA re-replication
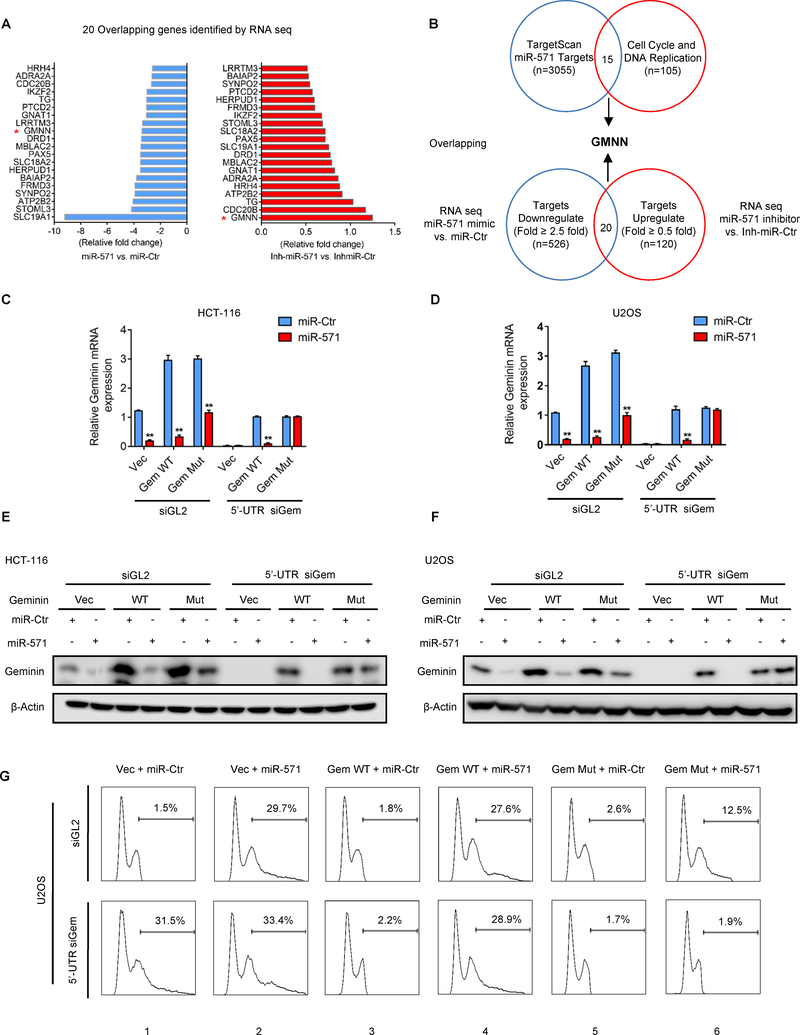
To rule out the possibility that miR-571 may target other genes rather than Geminin to induce DNA re-replication, we conducted RNA-Seq analyses of U2OS cells transfected with miR-Ctr versus miR-571, or inh-miR-Ctr versus inh-miR-571. The analyses revealed a total 20 genes, which are simultaneously downregulated by miR-571 and upregulated by inh-miR-571 (Fig. 2A). We then examined all miR-571 targets (n=3055) identified by TargetScan and identified a total of 15 genes that regulate cell cycle or DNA replication (n=105, Supplementary Table S5) (Supplementary Fig. S2A). Comparing these two gene sets, we found that Geminin (GMNN) is the only one overlapping gene, suggesting that miR-571 may primarily target Geminin to induce DNA re-replication (Fig. 2B).
Figure 2. miR-571 directly targets Geminin mRNA to induce DNA re-replication.
(A) RNA-Seq analyses to identify potential targeting genes by miR-571. Fold changes of genes potentially targeted by miR-571 in U2OS cells. Negative fold changes (left, blue) indicate genes downregulated by miR-571, positive fold changes (right, red) indicate genes upregulated by inhibitor of miR-571 (Inh-miR-571). (B) Identification of 15 genes that regulate the cell cycle & DNA replication and are predicted to be targeted by miR-571 using Targetscan. This gene set was compared with miR-571 targeting genes discovered by RNA-Seq to identify Geminin that is targeted by miR-571 and regulates DNA replication. (C-G) HCT116 cells and U2OS cells co-transfected with siGL2 or 5’-UTR siGem plus control, pIRES2-Gem WT or Mut vectors were treated with miR-Ctr or miR-571. 48 hr after transfection, Geminin mRNA levels (C and D) were detected by qPCR and protein levels (E and F) were analyzed by Western blot. U2OS cells treated as above were harvested for FACS analysis. The percentage of cells with re-replicated DNA was indicated. Representative from three biological replicates was shown.
If miR-571 induces DNA re-replication by targeting Geminin, we expect that ectopic expression of Geminin from cDNA (pEBG-Geminin) should be able to suppress DNA re-replication induced by miR-571 since miR-571 cannot target Geminin cDNA. Indeed, ectopic expression of Geminin from cDNA suppressed DNA re-replication caused by miR-571 expression in both HCT116 and U2OS cells (Supplementary Fig. S2B–D). Consistently, ectopic expression of Geminin reduced the giant nuclei formation by miR-571 mimic transfection (Supplementary Fig. S2E–F). These results strongly suggest that miR-571-induced DNA re-replication is dependent on Geminin expression.
To further confirm that miR-571 specifically targets Geminin to induce DNA re-replication, we constructed vectors containing Geminin cDNA together with the wild type Geminin 3’-UTR (pIRES2-Gem WT) or mutant Geminin 3’-UTR containing mutated miR-571 seed sequence (pIRES2-Gem Mut). We next co-expressed miR-571 mimic with control vector, pIRES2-Gem WT or pIRES2-Gem Mut in both HCT-116 and U2OS cells, in which endogenous Geminin was downregulated by siRNA targeting Geminin 5’-UTR, which should have an effect on the ectopic expression of pIRES2-Gem WT but not on pIRES2-Gem Mut. siRNA targeting Geminin 5’-UTR significantly reduced Geminin mRNA and protein level in both cells (Fig. 2C–F). Interestingly, expression of miR-571 reduced expression of WT but not mutant Geminin (Fig. 2C–F), suggesting that miR-571 specifically targets Geminin 3’-UTR at the site of 88–94. Consistently, in both HCT116 and U2OS cells, expression of WT or mutant Geminin dramatically suppressed DNA re-replication caused by siGem (compare columns 3 and 5 with 1 in Fig. 2G and Supplementary Fig. S2G–I), whereas co-expression of miR-571 with mutant but not WT Geminin greatly suppressed DNA re-replication (compare column 4 with 6 in Fig. 2G and Supplementary Fig. S2G–I), suggesting that miR-571 regulates DNA re-replication by specifically targeting Geminin 3’-UTR.
Two concerted pathways regulate Geminin expression
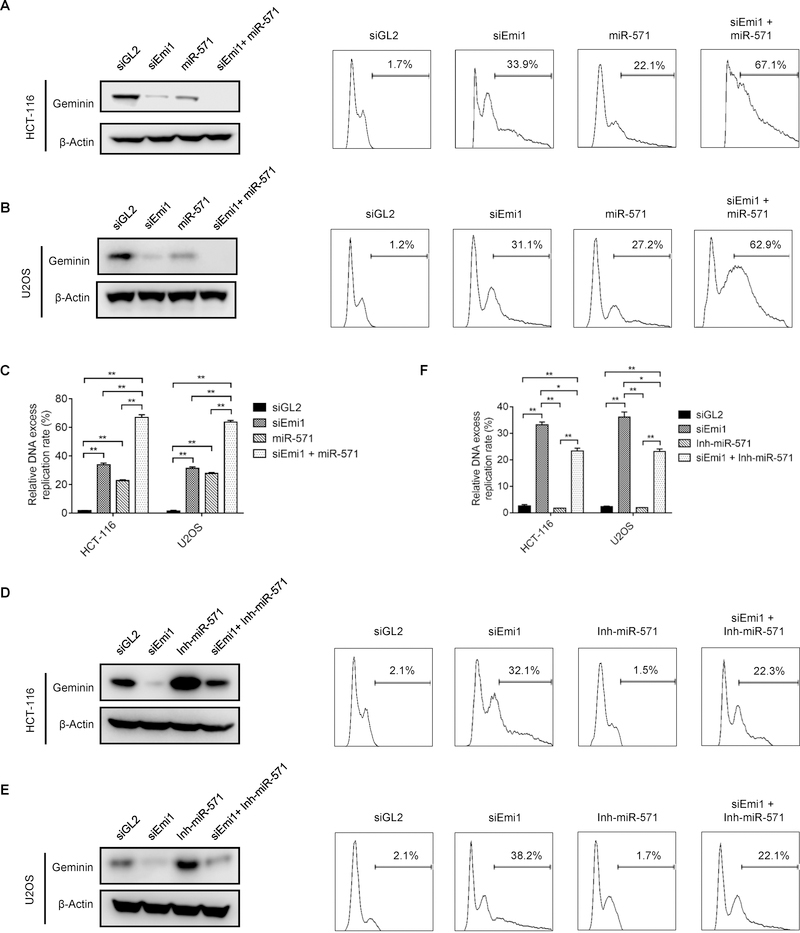
Geminin protein level has been proved to be down-regulated by APC/C-mediated degradation as cells transit from metaphase to anaphase during mitosis (5,27). Emi1 inhibits APC/C activity during S, G2 and early M phases, thereby preventing the APC/C from mediating degradation of geminin, cyclin A and other proteins (28). Thus, depletion of Emi1 causes DNA re-replication (27).
To explore the relationship between Emi1-mediated regulatory pathway and the ability of miR-571 to suppress Geminin protein levels, HCT116 and U2OS cells were transfected with Emi1/Fbxo5 siRNA (siEmi1), miR-571 mimic, or both. As shown in Fig. 3A–C, either siEmi1 or miR-571 mimic alone reduced Geminin protein levels, but co-transfection of both siEmi1 and miR-571 mimic reduced Geminin protein levels severely. Moreover, these decreases in Geminin levels corresponded to increases in the extent of DNA re-replication. SiEmi1 and miR-571 mimic individually induced DNA re-replication in 33.9% and 22.1% in HCT-116 and 31.1% and 27.2% in U2OS cells, respectively), but the effect of siEmi1 and miR-571 mimic together was additive, they induced DNA re-replication up to 67.1% in HCT-116 and 62.9% in U2OS cells. Conversely, co-transfection of siEmi1 and miR-571 inhibitor partially restored the level of Geminin protein and reduced the percentage of cells with re-replicated DNA by siEmi1 (Fig. 3D–F). These results together revealed that Geminin protein levels are regulated by two concerted pathways, miR-571-mediated suppression of Geminin mRNA translation and APC/C-mediated degradation of Geminin protein.
Figure 3. miR-571 and APC/C are two concerted pathways to regulate Geminin expression and DNA re-replication.
(A and B) HCT 116 cells (A) and U2OS cells (B) transfected with indicated siRNAs, miRNAs or both. 48 hr after transfection, Geminin protein levels were detected by Western blot (left panel) and DNA re-replication (right panel) were analyzed by FACS. (C) Quantification results of HCT-116 and U2OS cells treated in Fig. 3A and 3B (Data represent mean ± SE from three independent experiments, **p≤0.01). (D and E) HCT 116 cells (D) and U2OS cells (E) transfected with indicated siRNAs, miRNA inhibitors or both. 48 hr after transfection, Geminin protein levels (left panel) and DNA re-replication (right panel) were analyzed. (F) Quantification results of HCT-116 and U2OS cells treated in Fig. 3D and 3E (Data represent mean ± SE from three independent experiments, *p≤0.05, **p≤0.01).
miR-571 is critical for efficient DNA replication and S phase cell cycle progression
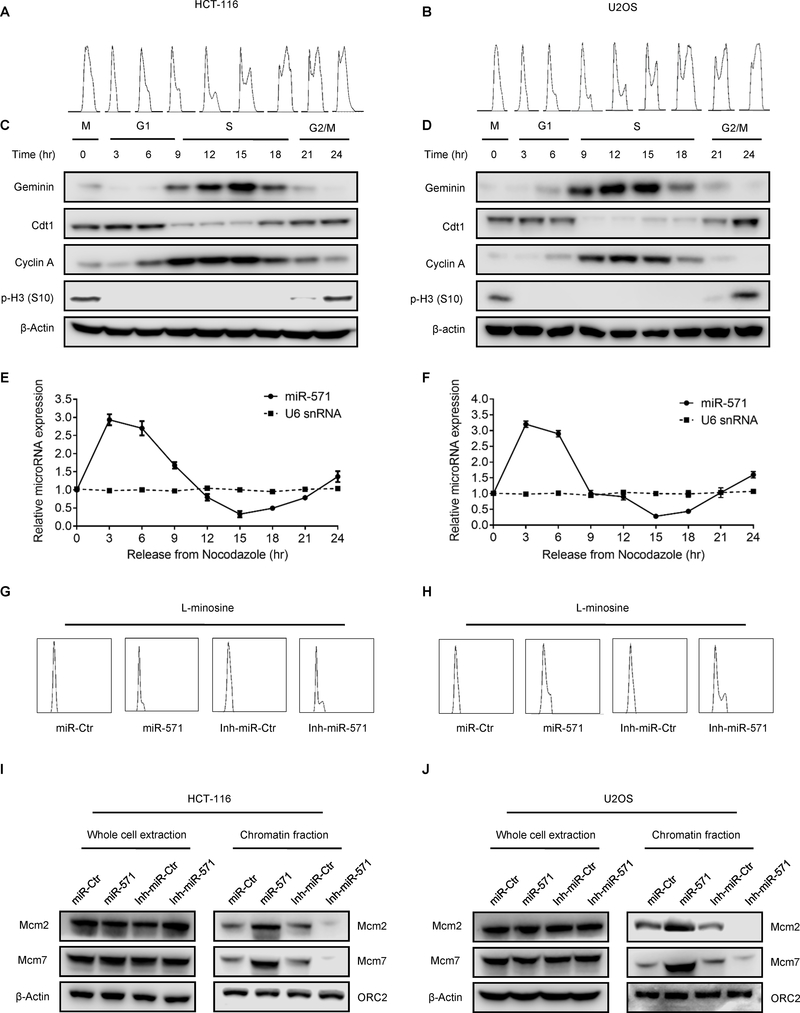
If endogenous miR-571 regulates Geminin expression during the cell cycle, then miR-571 levels should be inversely related to Geminin levels. To test this hypothesis, HCT116 and U2OS cells were synchronized in mitosis with nocodazole, and then released into nocodazole-free medium. FACS analysis confirmed that cells were synchronized in mitosis and re-entered G1 phase 3–6 hr after release from the nocodazole block, followed by S phase at 9–18 hr and G2/M phase at 21–24 hr (Fig. 4A–B). Cell cycle dependent proteins including Geminin, Cdt1, Cyclin A and pho-Histone H3 accumulated during specific cell cycle phases (Fig. 4C–D). Geminin level was disappeared when cells were released from mitosis into G1 phase, then its expression returned when cells entered S phase, where Geminin protein accumulated to its highest level and sharply decreased during late S phase to early G2 phase (Fig. 4C–D), as previously reported (4). In contrast, Cdt1 was accumulated in nocodazole arrested cells and remained throughout G1 phase until S phase, where its level was reduced. At G2/M phase, Cdt1 levels increased again for the next round Pre-RC complex formation (Fig. 4C–D). Cyclin A was present predominantly during S phase and then reduced in G2/M phases, where Cdk2/Cyclin A prevents re-licensing of replication origins and Cdk1/Cyclin A facilitates entrance into M phase. Phosphorylation of histone H3 (pH3) indicated that cells indeed entered mitosis (Fig. 4C–D). As predicted, qPCR results revealed that miR-571 levels increased as cells entered G1 phase, and gradually decreased as cells entered S phase, and then increased again as cells transited from S to G2/M phases, while the control U6 snRNA levels remained steady throughout the cell cycle (Fig. 4E–F). Clearly, miR-571 levels were inversely correlated to Geminin levels during cell cycle.
Figure 4. miR-571 dynamic expression in the cell cycle.
(A and B)HCT116 cells (A) and U2OS cells (B) were synchronized in mitosis with nocodazole and then were released for the indicated times. Cells were harvested and were analyzed by FACS. (C and D) Western blot analyses for indicated proteins in HCT-116 cells (C) and U2OS cells (D) following the release scheme described in A and B. (E and F) qPCR analysis for miR-571 and U6 snRNA in HCT-116 cells (E) and HCT-116 cells (F) following the release scheme described in A and B. (G and H) HCT116 cells (G) and U2OS cells (H) were synchronized at G1 phase by L-mimosine and then were harvested for FACS analysis. (I and J) HCT116 (I) and U2OS (J) synchronized at G1 phase were harvested for chromatin-binding assay. Whole cell and chromatin fractions were resolved on SDS-PAGE gel followed by Western blot for indicated proteins. Representative from three biological replicates was shown.
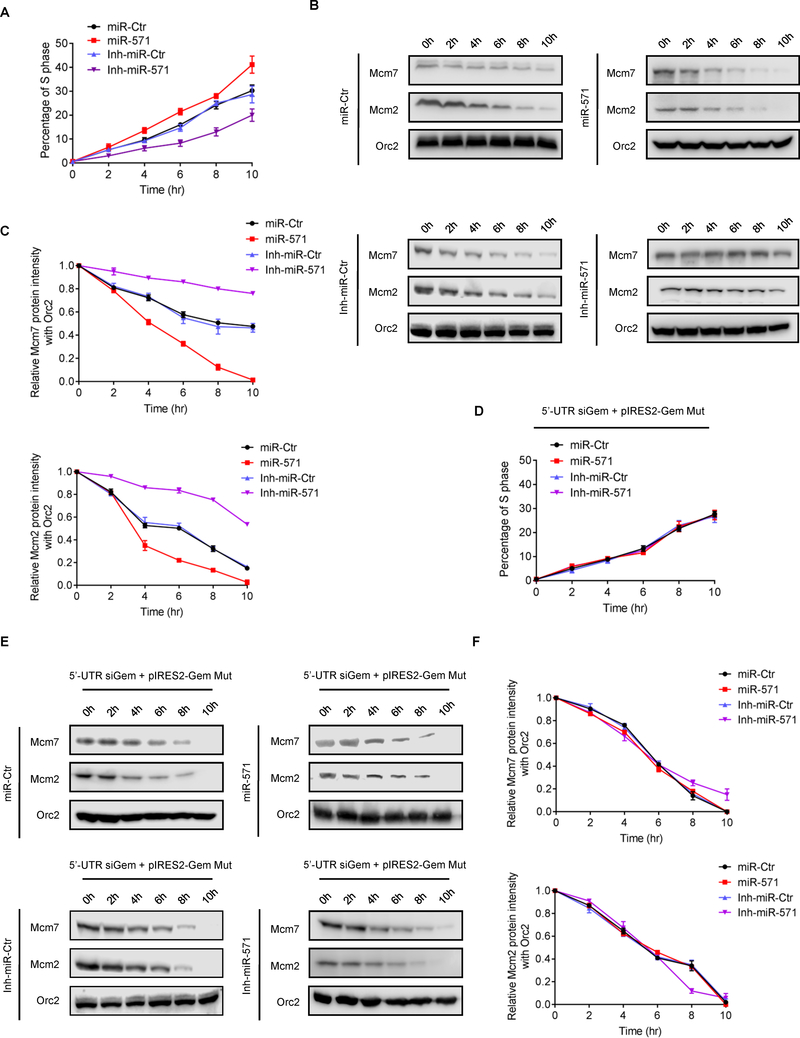
Since Geminin inhibits origin licensing by preventing the MCM helicase from loading onto replication origins for formation of pre-replicative complex (pre-RC) at late G1 phase (4), miR-517 would also be expected to interfere with association of MCM proteins with chromatin at late G1 phase. To test this hypothesis, HCT116 and U2OS cells synchronized at G1 phase by L-mimosine were transfected with miR-Ctr and miR-571 or Inh-miR-Ctr and Inh-miR-571, then the levels of Mcm2 and Mcm7 were examined in whole cell lysates and chromatin fraction from these cells (Fig. 4G–H and Supplementary Fig. S2J–K) (29). Neither miR-571 mimic nor miR-571 inhibitor affected the whole cellular levels of Mcm2 and Mcm7, but the levels of chromatin associated Mcm2 and Mcm7 were increased in G1 phase arrested cells with forced expression of miR-571, consistent with a role for miR-571 in regulating pre-RC assembly by inhibiting Geminin (Fig. 4I–J). To support this notion, inhibition of miR-571 expression significantly reduced the chromatin association of Mcm2 and Mcm7 in cells arrested in G1 phase (Fig. 4I–J).
We next examined whether or not miR-571 regulates S phase cell cycle progression. HCT116 and U2OS cells transfected with miR-Ctr, miR-571 or Inh-miR-Ctr, Inh-miR-571 were arrested in late G1 phase by L-mimosine and then released into cell cycle. We expected that overexpression of miR-571 could speed up S phase progression due to the accumulation of Mcm2/7 complex at G1 phase when Geminin level is reduced, therefore promoting more efficient DNA replication and faster S phase progression, whereas expression of miR-571 inhibitor should have the opposite effects. The results showed that cells were nicely synchronized at late G1 phase (Supplementary Fig. S3A–B). As expected, once released into S phase, cells transfected inh-miR-571 exhibited a slower progression through S phase than control treated cells, whereas miR-571 transfected cells exhibited an opposite S phase progression (Fig. 5A and Supplementary Fig. S3A–3B). Consistently, the chromatin associated Mcm2/7 rapidly decreased during S phase progression in cells transfected with miR-571 compared to cells transfected with miR-Ctr, indicating a faster DNA replication and S phase progression (Fig. 5B–C). Thus, miR-571 plays a critical role in the regulation of S phase DNA replication and cell cycle progression.
Figure 5. miR-571 regulates DNA replication and S phase progression.
(A) HCT116 cells transfected with miR-Ctr, miR-571 mimic, inh-miR-Ctr or Inh-miR-571 were synchronized at late G1 phase by L-mimosine. Cells were then released into L-mimosine-free medium and harvested at indicated time points for FACS analysis. The percentage of cell S phase population was indicated in Y-axis. Data represent means ± SEM from three independent experiments. The representative detailed cell cycle results were presented in Supplementary Fig. S 3A. (B) Cells treated as in A were analyzed by chromatin binding assays. Chromatin associated proteins were resolved on SDS-PAGE gel and Western blotted for indicated proteins. (C) Quantification of chromatin associated Mcm2/7 levels in cells treated in A. (D) HCT116 cells with downregulated Geminin by 5’-UTR siGem were co-transfected with pIRES2-Gem Mut and miR-Ctr, miR-571, inh-miR-Ctr or inh-miR-571. The transfected cells were then synchronized at late G1 phase by L-mimosine, followed by released into L-mimosine-free medium and harvested at indicated time points for FACS analysis. Data represent means ± SEM from three independent experiments. The detailed representative detailed cell cycle results were presented in Supplementary Fig. S 3B. (E) Cells treated as in D was analyzed by chromatin binding assays. Chromatin associated proteins were resolved on SDS-PAGE gel and Western blotted for indicated proteins. (F) Quantification of chromatin associated Mcm2/7 levels in cells treated in E. Representative from three biological replicates was shown.
To further examine the function of miR-571 in DNA replication and S phase cell cycle progression, miR-Ctr, miR-571, inh-miR-Ctr or inh-miR-571 were co-transfected with pIRES2-Gem Mut vector into HCT116 cells, in which endogenous Geminin was downregulated by 5’-UTR siGem. Transfected cells were arrested in late G1 phase with L-mimosine and then released into cell cycle. As expected, since miR-571 no longer targets mutant Geminin 3’-UTR region, pIRES2-Gem Mut expressed cells transfected with miR-571 exhibited a similar S phase progression and Mcm2/7 chromatin binding levels as cells treated with miR-Ctr, inh-miR-Ctr or inh-miR-571 (Fig. 5D, 5E–F and Supplementary Fig. S3C–D). These data suggest that miR-571 regulates DNA replication and S phase progress by specifically targeting Geminin mRNA.
c-Myc suppresses miR-571 expression by binding to the miR-571 promoter region
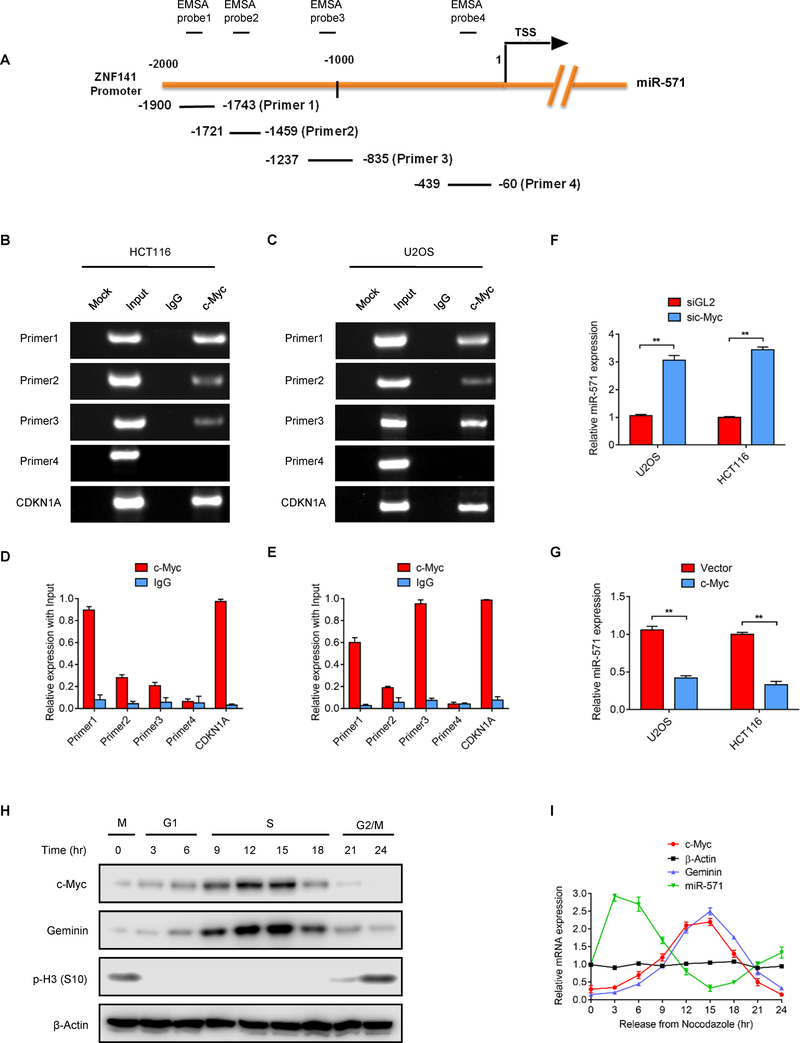
To determine how miR-571 expression might be regulated, the JASPAR database of experimentally defined binding sites for eukaryotic transcription factors were examined for transcription factors that might govern the expression of miR-571 directly (30). One promising candidate was c-Myc, a transcription factor that is not only implicated in regulating DNA replication, but also can bind to the promoter region of ZNF141, the host gene of miR-571. To explore whether or not c-Myc indeed binds to the promoter region of the miR-571 (ZNF141), chromatin immunoprecipitation (ChIP) assays with c-Myc associated DNA fragments were examined using four primers that covered approximately 2000bp of the ZNF141 promoter region (Fig. 6A). The ChIP results from both HCT116 and U2OS cells confirmed that c-Myc bound the region −835 to −1900 nucleotides before the start codon of miR-571 promoter. As a positive control, CDKN1A (p21) promoter region was detected from c-Myc ChIPs in both cells (Fig. 6B–6E). To further confirm the direct binding of c-Myc to the ZNF141 promoter region in vitro, we also performed Electrophoretic mobility shift assay (EMSA). Consistently, c-Myc specifically interacted with the EMSA probe 1,2 and 3 containing the −835 to −1900 region, but not probe 4 (Supplementary Fig. S4A). Moreover, addition of c-Myc antibody to the reaction buffer resulted in a super-shifted band, while incubation with unlabeled competitor abolished the shifted band in both HCT116 and U2OS cells. These results demonstrated that c-Myc directly binds to ZNF141 promoter region (−835 to −1900).
Figure 6. c-Myc directly binds to the promoter of miR-571.
(A) Schematic of promoter region of miR-571 host gene ZNF141 and primers used for ChIP and EMSA assay. (B-E) ChIP assay to detect the association of c-Myc with the promoter region of the miR-571 host gene ZNF141. HCT116 and U2OS cells were harvested and cell lyses were immuno-precipitated with IgG or anti-c-Myc antibodies. c-Myc associated DNA was examined by Gel electrophoresis as in B or C and qPCR as in D or E. CDKN1A was used as the positive control. The PCR primers were indicated in A. (F) HCT116 and U2OS cells (G) transfected with indicated siRNAs or vectors were harvested to examine miR-571 expression level by qPCR 48 hr after transfection. Data represent means ± SEM from three independent experiments. **p≤0.01. (H) HCT116 cells treated as in Fig. 4A were harvested at indicated time points by Western blot for indicated proteins. (I) Relative mRNA expression levels of indicated genes and miR-571 were examined by qPCR in HCT116 cells as treated in H. Representative from three biological replicates was shown.
To explore how c-Myc regulates expression of miR-571, we either depleted c-Myc in HCT116 and U2OS cells by siRNA, or overexpressed c-Myc by ectopic expression of recombinant c-Myc protein. As shown in Fig. 6F–G, depletion of c-Myc significantly elevated miR-571 level, whereas overexpressed c-Myc markedly reduced miR-571 expression, indicating that c-Myc negatively regulates miR-571 expression (Fig. 6F–G). We next investigated whether c-Myc regulates miR-571 processing by examining the expression of pri-miR-571 and pre-miR-571. Consistently, inhibition c-Myc markedly increased pri/pre-miR-571 levels, while overexpression c-Myc notably decreased pri/pre-miR-571 expression in both HCT116 and U2OS cells (Supplementary Fig. S4B–E). As expected, qPCR analyses indicated that c-Myc also regulates the expression of miR-571 host gene ZNF141 (Supplementary Fig. S4F).
Having identified a direct regulation of c-Myc on miR-571 expression, we next assessed the correlation between expression levels of c-Myc and miR-571. To this end, dynamic expression of c-Myc protein levels during the cell cycle was examined. Although c-Myc protein was present throughout the cell cycle, it increased markedly during S phase, before subsiding when cells entered G2 phase (Fig. 6H–I). The changes of c-Myc protein levels coincided with down-regulation of miR-571 and up-regulation of Geminin expression (Fig. 6H–I), suggesting that c-Myc may regulate expression of Geminin.
c-Myc regulates Geminin expression through miR-571
To explore whether c-Myc directly regulates the transcription of Geminin, we constructed Geminin promoter region into pGL3 vector (pGL3-Gem Pro) for luciferase reporter analyses. Ectopic expression of c-Myc reduced the relative luciferase activity of CDKN1A (p21), indicating a direct interaction of c-Myc with the promoter region of CDKN1A (Supplementary Fig. S4G). However, ectopic expression of c-Myc had no effects on the luciferase activity of pGL3-Gem Pro (Supplementary Fig. S4G), suggesting that c-Myc does not bind to Geminin promoter. Consistently, depletion of c-Myc by siRNA increased the luciferase activity of pGL3-CDKN1A but not pGL3-Gem Pro (Supplementary Fig. S4H). These results demonstrated that c-Myc does not transcriptionally regulate Geminin level.
We next examined how c-Myc regulates Geminin expression. To this end, we either depleted c-Myc by siRNA or ectopically expressed c-Myc in U2OS cells. Significantly, depletion of c-Myc reduced the Geminin protein levels and transfection of miR-571 inhibitor partially restored the Geminin protein levels in c-Myc depleted cells (Supplementary Fig. S 5A). Depletion of c-Myc caused a G1 phase arrest but not a DNA re-replication (Supplementary Fig. S5B–C). This observation was discussed in the discussion part. Consistently, overexpression of c-Myc increased Geminin protein levels and partially restored Geminin protein levels that have been reduced by forced expression of miR-571 mimic (Supplementary Fig. S5D). Consistently, overexpression of c-Myc reduced the percentage of cells with DNA re-replication that was induced by forced expression of miR-571 mimic (Supplementary Fig. S5E–F). Thus, these results strongly demonstrate the role of c-Myc in the regulation of Geminin by targeting miR-571.
The depletion or overexpression of c-Myc may affect the cell cycle progression, thereby altering the expression level of Geminin. To rule out this possibility, HCT116 cells were synchronized at the G1/S border by L-mimosine. Under these conditions, depletion of c-Myc by siMyc decreased Geminin protein levels, while over-expression of c-Myc increased Geminin protein levels (Supplementary Fig. S5G). Furthermore, these changes were accompanied by corresponding changes in miR-571 levels (Supplementary Fig. S5H). Thus, c-Myc regulates Geminin expression independently of cell cycle progression.
Cdk2 inhibits miR-571 expression by phosphorylating c-Myc at Ser 62
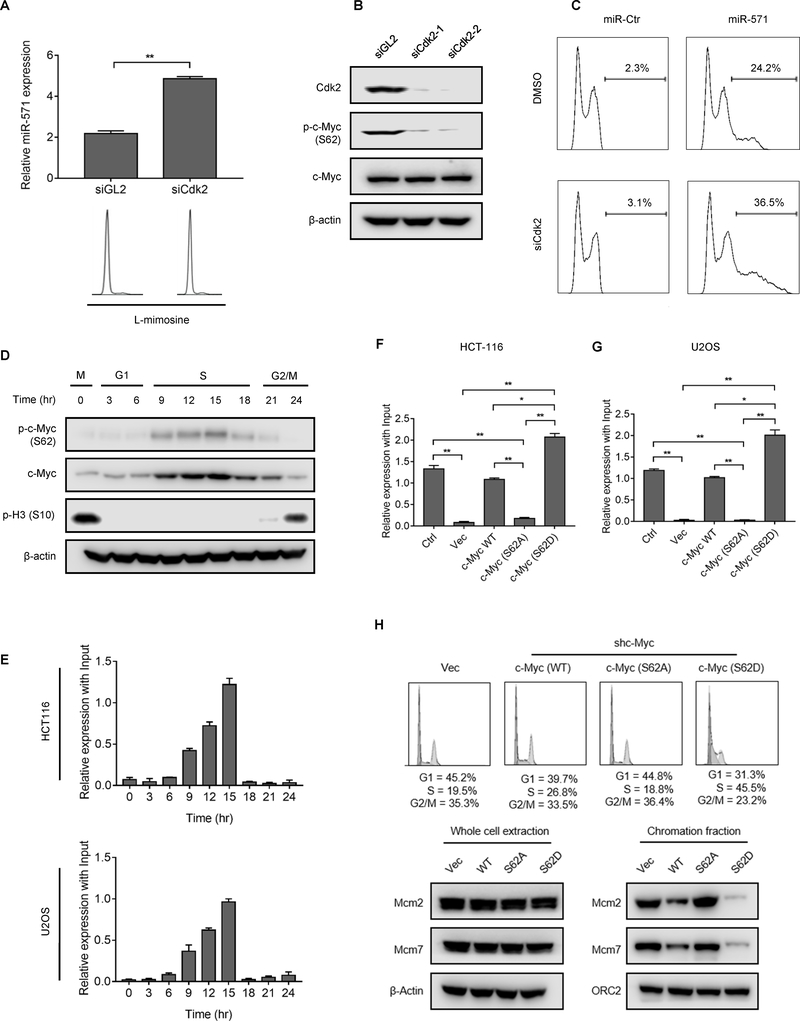
Cdk2 is an essential regulator of DNA replication and c-Myc activity (31). Since Cdk2 regulates c-Myc function in cell senescence by phosphorylating c-Myc at Serine 62 (Ser62) (13), we considered that Cdk2 might also regulate expression of miR-571 by phosphorylating c-Myc protein. To test this hypothesis, HCT116 cells synchronized at the G1/S boundary by L-mimosine were transfected with siGL2 or siCdk2. Depletion of Cdk2 resulted in a significant increase of miR-571 expression (Fig. 7A). The similar results were also observed in cells treated with CVT-313 (CV Therapeutics, Inc.) (Supplementary Fig. S6A), a selective inhibitor of Cdk2 activity (13). Moreover, depletion of Cdk2 reduced the level of phosphorylation c-Myc at Ser62 as indicated by western blot using an antibody that specifically recognized c-Myc protein phosphorylated at Ser62 (Fig. 7B). These data suggested that Cdk2 is required for the phosphorylation of c-Myc at Ser62. Consequently, Cdk2 inhibition by siRNA or CVT-313 promoted DNA re-replication in cells with forced expression of miR-571 (Fig. 7C and Supplementary Fig. S6B–D). Taken together, these data suggest that Cdk2-c-Myc axis plays an important role in the regulation of Geminin levels by targeting miR-571 during S phase.
Figure 7. Cdk2 regulates miR-571 expression and DNA replication by phosphorylating c-Myc at Ser62.
(A) HCT116 cells treated with siGL2 or siCdk2 were arrested in G1 phase by L-mimosine. Cells were then harvested to examine miR-571 expression by qPCR (upper) or for FACS analysis (lower). Data represent means ± SEM from three independent experiments.* p≤0.05; ** p≤0.01. (B) HCT116 cells treated with two independent Cdk2 siRNAs for 48 hr and then harvested for Western blot for indicated proteins. Ser62 indicated as S62. (C) HCT116 cells transfected with indicated miRNAs were treated with siGL2 or siCdk2 for 24 hr. Cells were then harvested for FACS analysis. (D) HCT116 cells treated as in Fig. 4A were harvested at indicated time points for Western blot for indicated proteins. (E) HCT116 and U2OS cells treated in D were immunoprecipitated with anti-phospho c-Myc (S62) antibody. p-c-Myc (S62) associated DNA at the promoter region of miR-571 was examined by qPCR. (F and G) HCT116 and U2OS cells transfected with indicated vectors were harvested for c-Myc ChIP assays to examine the association of c-Myc or c-Myc mutants (S62A, S62D) with the promoter region of miR-571. Data represent means ± SEM from three independent experiments. * p≤0.05, ** p≤0.01. (H) HCT116 cells with downregulated c-Myc by shc-Myc were transfected with vectors expressing indicated proteins and then were harvested for FACS analysis (upper) and Western blot (lower) for indicated proteins. The percentage of cell cycle stages was indicated. Representative from three biological replicates was shown.
To further explore the mechanistic insight of how Cdk2-c-Myc axis regulates miR-571 expression, we examined c-Myc phosphorylation at Ser62 in the cell cycle using HCT116 cells released from nocodozale block. Significantly, c-Myc phosphorylation at Ser62 was increased when cells entered into S phase at 9 hr and remained at high levels until cells entered G2/M phase (Fig. 7D). We next examined the association of phosphorylated c-Myc with miR-571 promoter in the cell cycle by ChIP assay. Strikingly, accumulation of phosphorylated c-Myc at the promoter regions of miR-571 was detected when cells entered S phase until the late S phase in both HCT116 and U2OS cells (Fig. 7E and Supplementary Fig. S6E–F). It appears that at S phase, c-Myc is phosphorylated at Ser62 and phosphorylated c-Myc prefers to bind the promoter region of miR-571.
We next generated stable U2OS and HCT116 cell lines in which endogenous c-Myc was down-regulated by constitutive expression of shc-Myc from lentivirus. These stable cells were then transfected with vectors expressing wild-type c-Myc, a non-phosphorylate c-Myc (S62A) mutant, or a phospho-mimetic c-Myc-S62D mutant. Phosphorylation of c-Myc at Ser62 was detected in both cells transfected with wild-type c-Myc or c-Myc (S62D), but not c-Myc (S62A) mutant (Supplementary Fig. S7A–B). ChIP assay indicated that c-Myc (S62D) exhibited a stronger association with the promoter region of miR-571 than wild type c-Myc (Fig. 7F–G). On the other hand, the association of c-Myc (S62A) with miR-571 promoter was significantly reduced compared with wild-type c-Myc (Fig. 7F–G and Supplementary Fig. S7C–D). Thus, phosphorylation of c-Myc at Ser62 is critical for its binding to the promoter region of miR-571.
To examine whether or not phosphorylation of c-Myc at Ser62 affects miR-571 expression, control vector (vec), wild-type c-Myc, c-Myc (S62A), or c-Myc (S62D) were overexpressed in U2OS cells whose endogenous c-Myc was down-regulated by lentivirus expressing shc-Myc. Compared with shRNA control treated cells (Ctrl), cells expressing shc-Myc strongly stimulated miR-571 expression and decreased Geminin protein level (Supplementary Fig. S7E–H, compare shCtr. with Vec). Expression of c-Myc or c-Myc (S62D) but not c-Myc (S62A) significantly reduced miR-571 expression and increased Geminin level in both HCT-116 and U2OS cells (Supplementary Fig. S7E–H). As expected, c-Myc (S62D) reduced more miR-571 levels than wild-type c-Myc (Supplementary Fig. S7E–H). These data strongly suggesting that the phosphorylation of c-Myc at Ser62 is critical for its regulation on miR-571 expression.
Given that Cdk2 regulates miR-571 expression by phosphorylating c-Myc at Ser62, we hypothesized that overexpression of c-Myc mutants may affect S phase progress due to its influence on miR-571 levels. Indeed, overexpression of WT c-Myc but not c-Myc (S62A) slightly increased S phase population (Fig. 7H and Supplementary Fig. 7I). However, over-expression of c-Myc (S62D) mutant significantly increased the fraction of cells in S phase from 19% to 44% (Fig. 7H), indicating an S phase arrest. Consistently, the chromatin associated Mcm2/7 levels were dramatically reduced in cells with expression of c-Myc (S62D) compared to cells with expression of c-Myc (S62A) or WT c-Myc (Fig. 7H), indicating a DNA replication defect. Taken together, these data demonstrate a critical role of Cdk2 in the regulation of DNA replication during S phase by phosphorylating c-Myc at Ser62.
DISCUSSION
Previous studies on the mitotic cell cycles of metazoan concluded that the level of Geminin protein was regulated post-translationally, primarily if not exclusively, by the APC/C proteasome pathway(5). From anaphase through G1 phase, Geminin protein is ubiquitinated by the APC/C and degraded by the 26S proteasome. This event coincides with licensing of DNA replication origins(4). During the transition from G1 to S phase, APC/C activity is inhibited specifically by the Emi1 protein (32). This event coincides with the activation of licensed replication origins and the appearance of Geminin protein (4). Emi1 gene transcription increases under the control of E2F transcription factors, and Emi1 protein is degraded in prophase after it is phosphorylated by Plk1 (33). APC/C activity is restored when metaphase is completed(34).
The results presented here demonstrate the existence of a second, previously undiscovered pathway that utilizes a unique microRNA to down-regulate translation of the Geminin gene to regulate DNA replication during the mitotic cell cycle (Supplementary Fig. S8A, schematic model). The Cdk2-c-Myc-miR-571 pathway operates in synchrony but independent with the APC/C pathway. It appears that suppression of miR-571 expression by c-Myc is dependent on Cdk2. During the transition from G1 to S phase, Cdk2 phosphorylates c-Myc protein at Ser62, thereby promoting its binding to the promoter of miR-571 and resulting in inhibition of miR-571 expression (Supplementary Fig. S8A). The importance of this pathway may appear in megakaryocytes and liver development. Megakaryocytes are terminally differentiated polyploid cells derived from hematopoietic stem cells in the bone marrow. Megakaryocytes undergo multiple rounds of replication (35). Serum levels of miR-571 closely correlate with liver cirrhosis (36), a disease associated with increased production of polyploid hepatocytes (37). Further studies are needed to investigate this functional link.
DNA re-replication is completely different from endoreduplication. Endoreduplication is replication of entire genome DNA in the absence of mitosis, while DNA re-replication occurs when one or more of the normal controls that prevent re-utilization of replication origins during S-phase is circumvented. Unlike endoreduplication, DNA re-replication is due to misfiring (re-firing) of replication origin during S phase, which leads to over-replication of partial genomic DNA, DNA damage, G2/M checkpoint activation, and is p53-independent. The detailed description of these two processes and difference was reviewed previously (38–40). To rule out the possibility that miR-571 induces DNA re-replication via mitotic failure, we conducted several experiments. First, cells were first arrested in mitosis by Nocodazole and arrested cells were then transfected with miR-Ctr or miR-571. In mitotic arrested cells, miR-571 did not induce DNA re-replication (Supplementary Fig. S8B–8E). Failure to induce DNA re-replication by miR-571 may be due to the lack of origin firing at mitosis. To support this notion, we released these cells into Nocodazole-free medium to allow cells to re-enter G1 and S phases. As expected, DNA re-replication was observed in miR-571 transfected cells but not in control treated cells (Supplementary Fig. S8F), suggesting that miR-571-induced DNA re-replication is dependent on origin firing at S phase. We assumed that once DNA replication origins already fire, addition of Nocodazole will not prevent DNA re-replication. Indeed, cells pre-transfected with miR-571 still exhibit DNA re-replication even when Nocodazole was added into medium to arrest cells at mitosis (Supplementary Fig.S8G–8H). In addition to above evidence, we have provided with several other strong evidence demonstrating that miR-571 regulates DNA re-replication but NOT endoreduplication. First, DNA damage was observed in cells with DNA re-replication by miR-571 (Supplementary Fig.1J–1K). Second, G2/M checkpoint was activated in cells with DNA re-replication by miR-571 arrest (Supplementary Fig.1H–1I, Supplementary Fig.1O–1P). Third, cells with DNA re-replication by miR-571 were not arrested in mitosis (Supplementary Fig.1O–1P). Fourth, DNA re-replication induced by miR-571 was seen in cells with wild type p53 (U2OS and HCT116 cells used in this study are p53 positive cells). Fifth, ectopic expression of geminin suppresses DNA re-replication induced by miR-571 (Figure 2G and Supplementary Fig.2G), suggesting miR-571 is involved in geminin-mediated DNA re-replication, a well-studied DNA re-replication process in human cells (6,7).
Cdk2 is a critical regulator of DNA replication in the metazoan. Cdk2/Cyclin E activity is required at the G1/S transition to convert pre-replication complexes into preinitiation complexes (3). Cdk2/Cyclin A activity inhibits re-initiation of DNA replication during S phase by phosphorylating Orc1, Cdc6 and Cdt1, critical components of the MCM helicase loader (1,3,12,41,42). The present study demonstrates that Cdk2 can also increase Geminin expression levels by the virtue of its ability to phosphorylate c-Myc, thereby preventing DNA re-replication by increasing Geminin expression during S phase. Cdk2 appears to inhibit DNA re-replication by targeting multiple replication factors to ensure DNA replicated once and only once per cell cycle. This study provides another example that cells have evolved multiple mechanisms to prevent DNA re-initiation after origin firing. Unexpectedly, depletion of c-Myc does not lead to DNA re-replication, instead it leads to G1 phase arrest (Supplementary Fig. S5B). We are not surprised by these results given that c-Myc regulates the expression of many genes that are essential for G1 to S phase transition and cell proliferation (43).
The results presented here reveal that c-Myc plays two important roles in nuclear DNA replication. c-Myc promotes DNA replication by regulating both transcription and translation of various cellular activities (14), and c-Myc prevents genomic instability by preventing DNA regions that have been replicated once from replicating a second time before mitosis is completed. Thus, it is not surprising that Myc is the most frequently amplified oncogene, and the elevated expression of c-Myc commonly observed in cancers correlates with aggressive malignancy and poor clinical prognosis (44,45). However, over-expression of c-Myc causes genome instability. For example, over-expression of c-Myc in Rat1 fibroblasts causes DNA amplification at specific regions (46). Ectopically over-expressed c-Myc cooperates with transforming growth factor-α to induce aneuploidy in the livers of transgenic mice (47), and c-Myc over-expression uncouples DNA replication from mitosis (48). These results raise the question as to how cancer cells with high levels of c-Myc avoid mitotic catastrophe. One explanation is that the ability of the Cdk2-c-Myc-miR-571 pathway to maintain genomic stability by preventing DNA re-replication counters the ability of c-Myc to drive cells into S phase. How these two opposing forces act to promote malignancy and metastasis remains to be investigated.
Supplementary Material
Significance.
Findings identify a novel regulatory mechanism that is critical for maintaining genome integrity by regulating DNA replication and cell cycle progression.
ACKNOWLEDGMENTS
This work was partially supported by funding from the National Institutes of Health (CA177898 and CA184717 to W.Z.). W.Z. was supported by a Research Scholar Grant (RSG-13-214-01-DMC) from the American Cancer Society, and a grant from the McCormick Genomic and Proteomic Center. This work was partially supported by National Institute of Child Health and Human Development and a NIH Director’s Challenge grant awarded to M.L.D. We thank Dr. Scott Martin for help on HTS assay.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict interest
REFERENCES
- 1.Machida YJ, Hamlin JL, Dutta A. Right place, right time, and only once: replication initiation in metazoans. Cell 2005;123:13–24 [DOI] [PubMed] [Google Scholar]
- 2.Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nature reviews Molecular cell biology 2005;6:476–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annual Review of Biochemistry 2002;71:333–74 [DOI] [PubMed] [Google Scholar]
- 4.Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 2000;290:2309–12 [DOI] [PubMed] [Google Scholar]
- 5.McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 1998;93:1043–53 [DOI] [PubMed] [Google Scholar]
- 6.Zhu W, Chen Y, Dutta A. Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Molecular and cellular biology 2004;24:7140–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melixetian M, Ballabeni A, Masiero L, Gasparini P, Zamponi R, Bartek J, et al. Loss of Geminin induces rereplication in the presence of functional p53. J Cell Biol 2004;165:473–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abbas T, Keaton MA, Dutta A. Genomic Instability in Cancer. Cold Spring Harbor perspectives in medicine 2013;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mihaylov IS, Kondo T, Jones L, Ryzhikov S, Tanaka J, Zheng JY, et al. Control of DNA replication and chromosome ploidy by geminin and cyclin A. Molecular and cellular biology 2002;22:1868–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGarry TJ. Geminin deficiency causes a Chk1-dependent G2 arrest in Xenopus. Molecular biology of the cell 2002;13:3662–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Visintin R, Prinz S, Amon A. CDC20 and CDH1: A family of substrate-specific activators of APC-dependent proteolysis. Science 1997;278:460–3 [DOI] [PubMed] [Google Scholar]
- 12.Moritani M, Ishimi Y. Inhibition of DNA binding of MCM2–7 complex by phosphorylation with cyclin-dependent kinases. Journal of biochemistry 2013;154:363–72 [DOI] [PubMed] [Google Scholar]
- 13.Hydbring P, Bahram F, Su YT, Tronnersjo S, Hogstrand K, von der Lehr N, et al. Phosphorylation by Cdk2 is required for Myc to repress Ras-induced senescence in cotransformation. P Natl Acad Sci USA 2010;107:58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dominguez-Sola D, Gautier J. MYC and the control of DNA replication. Cold Spring Harbor perspectives in medicine 2014;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, et al. Non-transcriptional control of DNA replication by c-Myc. Nature 2007;448:445–51 [DOI] [PubMed] [Google Scholar]
- 16.Sankar N, Kadeppagari RK, Thimmapaya B. c-Myc-induced aberrant DNA synthesis and activation of DNA damage response in p300 knockdown cells. J Biol Chem 2009;284:15193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srinivasan SV, Dominguez-Sola D, Wang LC, Hyrien O, Gautier J. Cdc45 is a critical effector of myc-dependent DNA replication stress. Cell reports 2013;3:1629–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valovka T, Schonfeld M, Raffeiner P, Breuker K, Dunzendorfer-Matt T, Hartl M, et al. Transcriptional control of DNA replication licensing by Myc. Scientific reports 2013;3:3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Latchana N, DiVincenzo MJ, Regan K, Abrams Z, Zhang XL, Jacob NK, et al. Alterations in patient plasma microRNA expression profiles following resection of metastatic melanoma. J Surg Oncol 2018;118:501–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim JG, Chae YS, Lee SJ, Kang BW, Park JY, Lee EJ, et al. Genetic variation in microRNA-binding site and prognosis of patients with colorectal cancer. J Cancer Res Clin 2015;141:35–41 [DOI] [PubMed] [Google Scholar]
- 21.Yu F, Shen XY, Fan L, Yu ZC. Genome-wide analysis of genetic variations assisted by Ingenuity Pathway Analysis to comprehensively investigate potential genetic targets associated with the progression of hepatocellular carcinoma. Eur Rev Med Pharmaco 2014;18:2102–8 [PubMed] [Google Scholar]
- 22.Zhu W, Lee CY, Johnson RL, Wichterman J, Huang R, DePamphilis ML. An image-based, high-throughput screening assay for molecules that induce excess DNA replication in human cancer cells. Molecular cancer research : MCR 2011;9:294–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Im H, Grass JA, Johnson KD, Boyer ME, Wu J, Bresnick EH. Measurement of protein-DNA interactions in vivo by chromatin immunoprecipitation. Methods Mol Biol 2004;284:129–46 [DOI] [PubMed] [Google Scholar]
- 24.Lee CY, Johnson RL, Wichterman-Kouznetsova J, Guha R, Ferrer M, Tuzmen P, et al. High-throughput screening for genes that prevent excess DNA replication in human cells and for molecules that inhibit them. Methods 2012;57:234–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Molecular cell 2007;27:91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arefian E, Kiani J, Soleimani M, Shariati SAM, Aghaee-Bakhtiari SH, Atashi A, et al. Analysis of microRNA signatures using size-coded ligation-mediated PCR. Nucleic acids research 2011;39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Machida YJ, Dutta A. The APC/C inhibitor, Emi1, is essential for prevention of rereplication. Genes Dev 2007;21:184–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reimann JDR, Freed E, Hsu JY, Kramer ER, Peters JM, Jackson PK. Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell 2001;105:645–55 [DOI] [PubMed] [Google Scholar]
- 29.Li YM, Xiao HJ, de Renty C, Jaramillo-Lambert A, Han ZY, DePamphilis ML, et al. The Involvement of Acidic Nucleoplasmic DNA-binding Protein (And-1) in the Regulation of Prereplicative Complex (pre-RC) Assembly in Human Cells. Journal of Biological Chemistry 2012;287:42469–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic acids research 2004;32:D91–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bretones G, Delgado MD, Leon J. Myc and cell cycle control. Bba-Gene Regul Mech 2015;1849:506–16 [DOI] [PubMed] [Google Scholar]
- 32.Chang LF, Zhang ZG, Yang J, McLaughlin SH, Barford D. Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature 2015;522:450–+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu JY, Reimann JD, Sorensen CS, Lukas J, Jackson PK. E2F-dependent accumulation of hEmi1 regulates S phase entry by inhibiting APC(Cdh1). Nature cell biology 2002;4:358–66 [DOI] [PubMed] [Google Scholar]
- 34.Castro A, Bernis C, Vigneron S, Labbe JC, Lorca T. The anaphase-promoting complex: a key factor in the regulation of cell cycle. Oncogene 2005;24:314–25 [DOI] [PubMed] [Google Scholar]
- 35.Trakala M, Rodriguez-Acebes S, Maroto M, Symonds CE, Santamaria D, Ortega S, et al. Functional Reprogramming of Polyploidization in Megakaryocytes. Dev Cell 2015;32:155–67 [DOI] [PubMed] [Google Scholar]
- 36.Roderburg C, Mollnow T, Bongaerts B, Elfimova N, Cardenas DV, Berger K, et al. Micro-RNA Profiling in Human Serum Reveals Compartment-Specific Roles of miR-571 and miR-652 in Liver Cirrhosis. Plos One 2012;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gentric G, Desdouets C. Polyploidization in liver tissue. Am J Pathol 2014;184:322–31 [DOI] [PubMed] [Google Scholar]
- 38.Hook SS, Lin JJ, Dutta A. Mechanisms to control rereplication and implications for cancer. Curr Opin Cell Biol 2007;19:663–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ullah Z, Lee CY, Lilly MA, DePamphilis ML. Developmentally programmed endoreduplication in animals. Cell cycle 2009;8:1501–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu WG, Dutta A. Activation of Fanconi anemia pathway in cells with re-replicated DNA. Cell cycle 2006;5:2306–9 [DOI] [PubMed] [Google Scholar]
- 41.Elsasser S, Chi Y, Yang P, Campbell JL. Phosphorylation controls timing of Cdc6p destruction: A biochemical analysis. Molecular biology of the cell 1999;10:3263–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jallepalli PV, Brown GW, Muzi-Falconi M, Tien D, Kelly TJ. Regulation of the replication initiator protein p65cdc18 by CDK phosphorylation. Genes Dev 1997;11:2767–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nasi S, Ciarapica R, Jucker R, Rosati J, Soucek L. Making decisions through Myc. Febs Letters 2001;490:153–62 [DOI] [PubMed] [Google Scholar]
- 44.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010;463:899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene 1999;18:3004–16 [DOI] [PubMed] [Google Scholar]
- 46.Mai S, Hanley-Hyde J, Fluri M. c-Myc overexpression associated DHFR gene amplification in hamster, rat, mouse and human cell lines. Oncogene 1996;12:277–88 [PubMed] [Google Scholar]
- 47.Sargent LM, Sanderson ND, Thorgeirsson SS. Ploidy and karyotypic alterations associated with early events in the development of hepatocarcinogenesis in transgenic mice harboring c-myc and transforming growth factor alpha transgenes. Cancer research 1996;56:2137–42 [PubMed] [Google Scholar]
- 48.Li Q, Dang CV. c-Myc overexpression uncouples DNA replication from mitosis. Molecular and cellular biology 1999;19:5339–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.