SUMMARY
tRNA-derived small RNAs (tsRNAs) have been implicated in many cellular processes, yet the detailed mechanisms are not well defined. We previously found that the 3′ end of Leu-CAG tRNA-derived small RNA (LeuCAG3′tsRNA) regulates ribosome biogenesis in humans by maintaining ribosomal protein S28 (RPS28) levels. The tsRNA binds to coding (CDS) and non-coding 3′ UTR sequence in the RPS28 mRNA, altering its secondary structure and enhancing its translation. Here we report that the functional 3′ UTR target site is present in primates while the CDS target site is present in many vertebrates. We establish that this tsRNA also regulates mouse Rps28 translation by interacting with the CDS target site. We further establish that the change in mRNA translation occurred at a post-initiation step in both species. Overall, our results suggest that LeuCAG3′tsRNA might maintain ribosome biogenesis through a conserved gene regulatory mechanism in vertebrates.
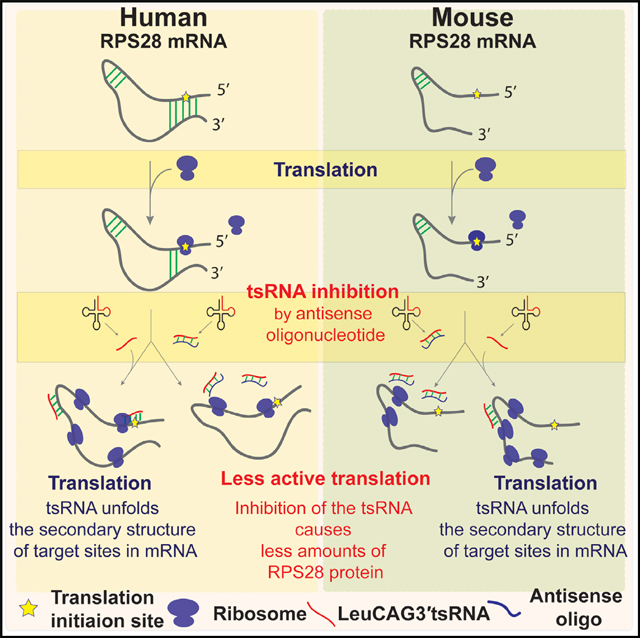
In Brief
Kim et al. determined that the LeuCAG3’tsRNA target site in the RPS28 coding sequence (CDS) is conserved in vertebrates and established that the tsRNA regulation of RPS28 mRNA translation is conserved between humans and mice. Their results suggest that the tsRNA-regulated mRNA translation might be a conserved process.
Graphical Abstract
Graphical abstract
INTRODUCTION
Historically, the central dogma has been that tRNAs recognize the mRNA triplet sequence on a ribosome to deliver the appropriate amino acid to a growing polypeptide chain. There is a growing appreciation that mature tRNAs alter gene expression in a more complex manner (Schimmel, 2018). tRNAs are differentially expressed in various cancers, tissues, and developmental stages. Each tRNA has an average of eleven to thirteen post-transcriptional modifications, which can affect tRNA folding and function. The tRNA-interacting enzymes potentially add complexity to their various functions. Furthermore, there is increasing evidence that tRNA-derived small RNAs (tsRNAs) (Haussecker et al., 2010), also called tRNA fragments (tRFs) (Lee et al., 2009), affect many cellular processes such as cell proliferation, apoptosis, global translation inhibition, epigenetic inheritance, and neuronal function (Kumar et al., 2016a; Schimmel, 2018). To date, more than six subtypes have been identified based on their cleavage site and length. The longer forms, 30- to 40- nt tsRNAs, are produced by angiogenin-mediated cleavage in the anti-codon loop of mature tRNA and are called tiRNA (tRNA-derived stress-induced RNA) (Yamasaki et al., 2009). The shorter forms, 18- to 26-nt tsRNAs, are somewhat similar to microRNAs (miRNAs) in terms of their length. However, they are not processed by Dicer and the microprocessor complex required for microRNA biogenesis (Haussecker et al., 2010; Kumar et al., 2014; Lee et al., 2009; Li et al., 2012).
One of the known roles for tsRNAs is to regulate mRNA translation by non-canonical mechanisms. In mammalian cells, the 5′ tiRNAs represses global translation by displacing translation eukaryotic initiation factor eIF4A and eIF4G from mRNAs (Guzzi et al., 2018; Ivanov et al., 2011). tiRNA-regulated translation is also observed in other organisms, including Haloferax volcanii and Trypanosoma brucei. However, the mechanisms of regulation differ among species (Fricker et al., 2019; Gebetsberger et al., 2012).
Distinct from the global translation-inhibitory effects of tsRNAs or tiRNAs, we recently discovered that a specific small non-coding RNA derived from the 3′ end of the Leu-CAG tRNA (LeuCAG3′tsRNA) maintains the translation of RPS28 (ribosomal protein S28) mRNAs and ultimately the number of ribosomes (Figure S1A) (Kim et al., 2017). RPS28 is a component of the 40S ribosome and is essential for the biogenesis of 18S rRNA (Robledo et al., 2008). Inhibition of LeuCAG3′tsRNA reduces RPS28 mRNA translation, resulting in reduced 18S rRNA processing and lower numbers of 40S ribosomal subunits. LeuCAG3′tsRNA inhibition leads to apoptosis in human cancer cells and an orthotopic hepatocellular carcinoma (HCC) patient-derived xenograft (PDX) model in mice (Kim et al., 2017; Slack, 2018).
Mechanistically, the LeuCAG3′tsRNA binds to two target sites in human RPS28 mRNA and disrupts the secondary structure of both target sites: target A in the coding sequence (CDS) and target B in the 3′ UTR enhancing mRNA translation (Figures S1A and S1B). Target A in the CDS forms a local hairpin structure, while target B in the 3′ UTR forms a duplex with a 20-nt region that straddles the translation initiation site (TIS) (Figure S1B) (Kim et al., 2017). This made it difficult to determine the step at which RPS28 mRNA translation was regulated. To establish the mechanism by which this tsRNA enhances the translation of its target mRNAs, we sought to predict the LeuCAG3′tsRNA target sites in RPS28 mRNAs across various vertebrate species and use this information to delineate the process by which the non-coding RNA regulates translation.
RESULTS
Target Site Conservation in the RPS28 mRNA for Vertebrate Species
We determined 22 nt of the 3′ end of the Leu-CAG tRNA in 44 vertebrate species from the Genomic tRNA Database (http://gtrnadb2009.ucsc.edu) and calculated the genetic distances (p distances) between species (Nei and Kumar, 2000). The p distance (0.198) was low, indicating that the LeuCAG3′tsRNA sequence was nearly identical across the tested species. This result was also confirmed by the 95%–100% identity of the mature LeuCAG3′tsRNA for thirteen representative vertebrate species in which the LeuCAG3′tsRNA sequence was identical or differed by just 1 nt (Figure S1C).
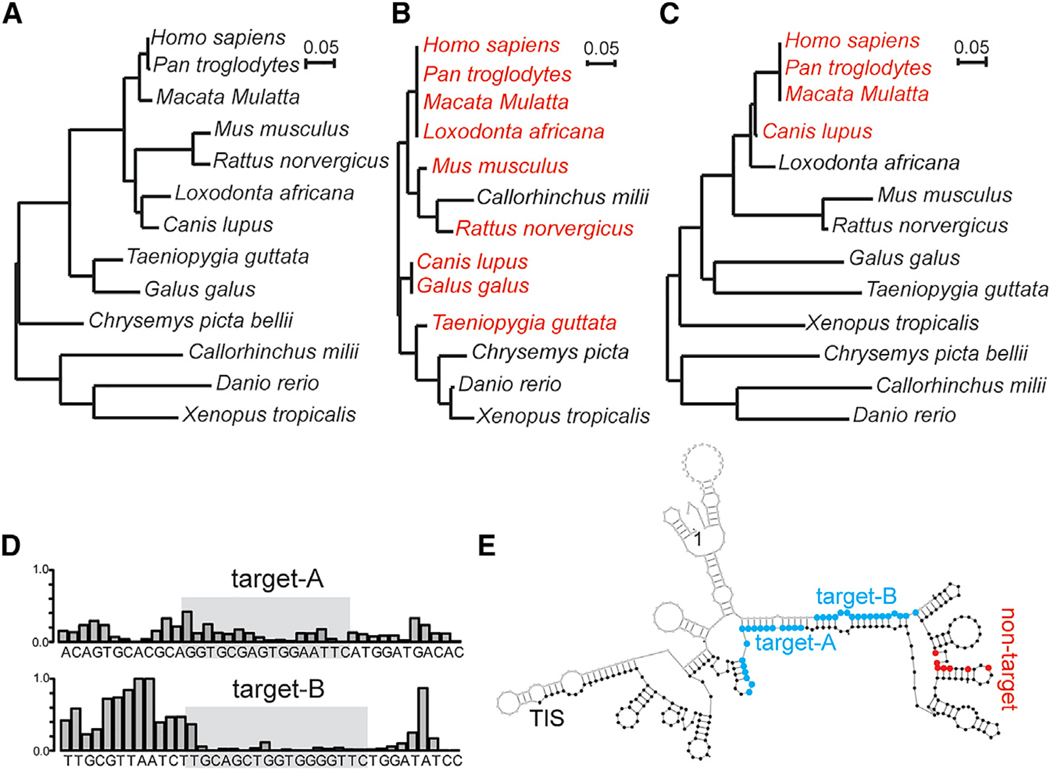
To determine the conservation of the LeuCAG3′tsRNA target sites in RPS28 mRNA for vertebrates, we built a phylogenetic tree of full-length RPS28 mRNA sequences from thirteen representative vertebrate species (Figure 1A) and predicted the potential targets of LeuCAG3′tsRNA in the RPS28 mRNA based on the intermolecular minimal free energy (m.f.e.) (Figure S1D). We found two major target sites, one in the CDS and the other in the 3′ UTR, from the many vertebrate species and generated the phylogenetic trees for both potential target sites (Figures 1B and 1C). The predicted nucleotide sequence making up the target A site in the CDS is nearly identical in mammals and birds (Figures 1B and S1D). We also examined the target A site in 100 vertebrate species by comparing an average of phyloP conservation scores of 22-nt sliding windows across the entire RPS28 CDS (Figure S1E). This analysis showed that the average conservation score of a group of seven 22-nucleotide windows spanning the target A site was ranked second among all seven grouped 22-nt windows spanning the entire RPS28 CDS, suggesting selective pressure to preserve the conserved tsRNA target site for 100 vertebrate species.
Figure 1. The Target A Site of Leu-CAG3′tsRNA in the RPS28 CDS Region Is Conserved in Mammals and Birds, and the Two Target Sites in Mouse Rps28 mRNA Have a Double-Stranded Secondary Structure.
(A–C) Phylogenetic trees of 13 vertebrate species based upon their RPS28 mRNA sequence (A), target A site sequence in the RPS28 CDS region (B), and target B site sequence in the RPS28 30 UTR (C). The branch length is proportional to the number of changes that have occurred in each species. The species that have predicted human LeuCAG3′tsRNA target sites are colored red (B and C).
(D) The icSHAPE data track the LeuCAG3′tsRNA binding sites in the mouse Rps28 mRNA. The icSHAPE data are scaled from 0 (no reactivity; double-strandedness) to 1 (maximum reactivity; single-strandedness). The gray box represents the target site. The complete icSHAPE data for mouse Rps28 mRNA are in Table S2.
(E) Schematic of the mouse Rps28 mRNA (NM_001355384.1) secondary structure predicted based on the icSHAPE analysis. Blue, the potential binding sites of the LeuCAG3′tsRNA; red, the modified nucleotide of the non-target Rps28 mutant; TIS, translation initiation site.
In contrast to the target A site, the target B site in the 3′ UTR is present only in some mammals, such as non-human primates and dogs (Figures 1C and S1D). We next asked whether target B in 3′ UTR forms a double-stranded secondary structure with the translation initiation site (TIS) in the chimpanzee, rhesus monkey, and dog (Figures S1F–S1H) like the human (Figure S1B), using the RNAfold program (Hofacker and Stadler, 2006). The chimpanzee and rhesus monkey, but not the dog, were predicted to have the correct target B secondary structure to interact with the TIS (Figures S1F–S1H). This structure prediction suggests that only primates might have a functional target B site regulating RPS28 mRNA translation. The lack of the functional target B site in non-primate species raised the question of whether LeuCAG3′tsRNA enhances RPS28 mRNA translation and, if yes, whether it does so by solely unfolding the target A site in the CDS.
Both Target Sites of LeuCAG3′tsRNA in Mouse RPS28 mRNA Are Double Stranded
We elected to investigate tsRNA-mediated Rps28 mRNA translation in the mouse. There are two Rps28 isoforms that differ by 4 nt in the 3′ UTR. Only transcript 2 (NM_001355384.1) is transcribed in the liver (Table S1) (Valdmanis et al., 2016) and is the most abundant in other tissues (Brawand et al., 2011); thus, it was selected for further study.
The tsRNA-target A and tsRNA-target B m.f.e. in mice were –23.7 and –24.6 kcal/mol, respectively (Figure S1I), suggesting that the tsRNA might bind to both regions of the mouse Rps28 mRNA transcript 2. Both targets have a double-stranded confirmation similar to that of the human RPS28 mRNA based on icSHAPE (in vivo click selective 2′-hydroxyl acylation and profiling experiment) data (Figure 1D; Table S2) (Spitale et al., 2015). However, icSHAPE-based or computational structure modeling predicted that the detailed secondary structure models of the tsRNA target sites differed between mouse and human mRNAs (Figures 1E, S1B, and S1J). Specifically, the mouse target B site, in contrast to the human site, does not form a duplex within the region straddling the translation initiation site (TIS) (Figures 1E, S1B, and S1J).
Therefore, our results suggest both the target A and target B sites in the expressed mouse Rps28 mRNA isoform exist as double-stranded forms in cells, although the nature of the double-stranded regions in the target B sites differs between mouse and human.
Mouse LeuCAG3′tsRNA Is Required for 18S rRNA Processing
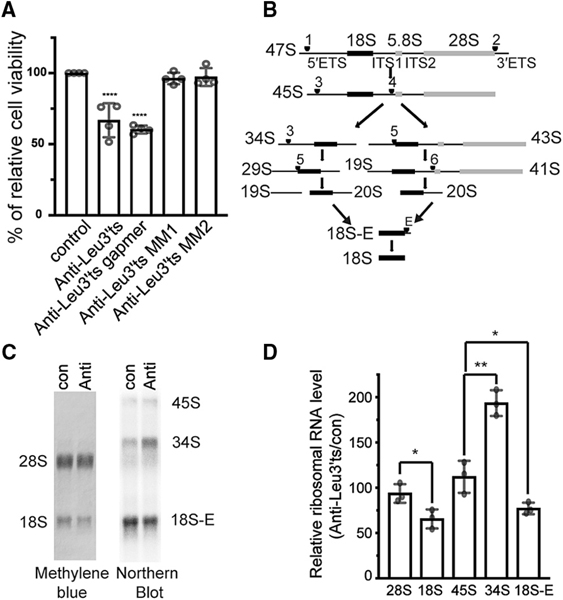
Decreasing the RPS28 protein by inhibiting the LeuCAG3′tsRNA impairs the 18S rRNA processing pathway in human cells, ultimately reducing the viability of human cancer cells (Kim et al., 2017). The LeuCAG3′tsRNA sequence is identical between mouse and human and is expressed at similar levels in HeLa (human cervical cancer) cells and Hepa 1–6 cells (mouse hepatoma cells) (Figure S2A). We next confirmed that as in the human (Kim et al., 2017), the anti-Leu3′ts ASO blocked mouse LeuCAG3′tsRNA, but not the cognate mature tRNA, while the scrambled and two 2-nt mismatched ASOs did not affect the tsRNA concentrations (Figure S2B). Similar to human cells (Kim et al., 2017), inhibition of the LeuCAG3′tsRNA significantly reduced Hepa 1–6 cell viability to 66.8% ± 9.3% compared with control (con) cells (Figure 2A).
Figure 2. LeuCAG3′tsRNA Is Essential and Required for 18S rRNA Processing in Mouse Cells.
(A)Inhibition of LeuCAG3′tsRNA impairs Hepa 1–6 cell viability. Three days post-transfection, a MTS assay was performed (n = 4 independent experiments). Anti-Leu3′ts, antisense oligonucleotide to LeuCAG3′tsRNA; anti-Leu3′ts gapmer, antisense oligonucleotide that induces RNase H activity to cleave LeuCAG3′tsRNA; anti-Leu3′ts MM and MM2, two 2-nt mismatched oligonucleotides to LeuCAG3′tsRNA.
(B) Pre-rRNA processing pathways in mouse cells based on prior studies (Bowman et al., 1981; Kent et al., 2009). The 47S primary transcript is processed and categorized as 50 external transcribed spacers (5′ ETSs), 18S rRNA, internal transcribed spacer 1 (ITS1), 5.8S rRNA, internal transcribed spacer 2 (ITS2), 28S rRNA, and 3′ external transcribed spacers (3′ ETSs). There are two alternative processing pathways. Inhibition of LeuCAG3′tsRNA inhibits processing from the pre-34S to the pre-20S form depicted in pathway A. Arrowhead and number indicate cleavage sites.
(C and D) Inhibition of the LeuCAG3′tsRNA suppresses 5′ ETS processing in 18S rRNA biogenesis in Hepa 1–6 cells. Methylene blue staining (28S and 18S rRNA) and northern hybridization (45S, 34S, and 18S-E pre-rRNA) were performed with total RNA from Hepa 1–6 cells 24 h post-transfection (n = 3 in- dependent experiments). A representative image is shown in (C). Relative mature and pre-rRNA levels are shown in (D). The rRNA level from anti-Leu3′ts-transfected cells was normalized to that from con (control).
The mean is shown in (A) and (D). Error bar, SD. *p < 0.05, **p < 0.005, ****p < 0.0001 by one-way ANOVA (A) and two-tailed t test (D). Anti, anti-LeuCAG3′tsRNA; con, control.
As in the human, we ruled out direct binding of the ASO with rRNA and the Rps28 mRNA (Figures S2C and S2D) (Kim et al., 2017). In addition, the specificity of the anti-Leu3′ts ASO that binds to sequester the target RNA was confirmed using an anti-Leu3′ts gapmer ASO that induces RNase H-mediated cleavage of their target RNAs (Figure 2A) (Jepsen et al., 2004; Kim et al., 2017).
As before, the inhibition of LeuCAG3′tsRNA significantly decreased the 18S rRNA level to 65.5% ± 10.5% compared with control cells. In agreement with previous findings (Kim et al., 2017), the 28S rRNA was not significantly reduced (93.7% ± 10.3%), confirming that the LeuCAG3′tsRNA specifically affects the 18S, but not the 28S, rRNA abundance (Figures 2C and 2D). To determine the step of action on 18S rRNA processing, we measured the relative abundance of different 18S pre-rRNAs by northern hybridization (Figures 2B–2D). Inhibition of the LeuCAG3′tsRNA resulted in the accumulation of the 34S pre-rRNA (equivalent to human 30S pre-rRNA) to a level of 193.5% ± 14.3% compared with the level in control cells, while the 18S-E pre-rRNA level decreased to 77.1% ± 6.3% (Figures 2B–2D). However, the 45S primary transcript only slightly increased to 112.1% ± 71% (Figure 2B–2D). Altogether, these results suggest that the LeuCAG3′tsRNA does not affect rRNA transcription but is required for the processing of the 34S intermediate RNA, as was the case in human (Kim et al., 2017). In addition, in both human (Kim et al., 2017; Robledo et al., 2008) and mouse cells, RNAi knockdown of RPS28 mRNA resulted in a similar reduction in 5′ external transcribed spacer (ETS) rRNA processing (Figures S2E and S2F), suggesting that the RPS28 protein level plays a similar role in ribosome biogenesis in both species.
LeuCAG3′tsRNA Regulates Mouse Rps28 mRNA Translation
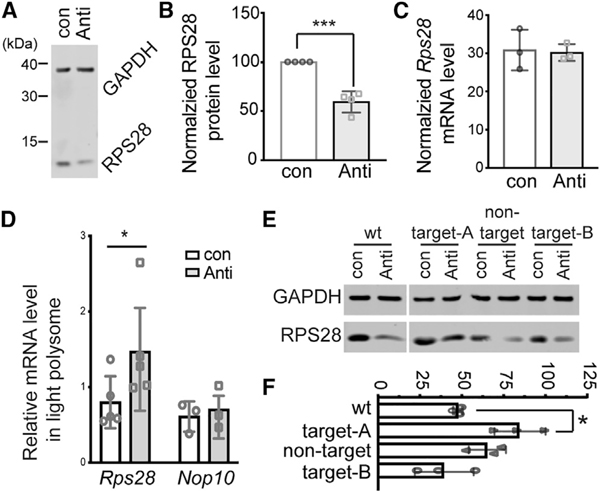
The RPS28 protein levels (Figures 3A and 3B) and mRNA levels (Figure 3C) were quantified by western blot and real-time PCR, respectively, after the ASO-mediated inhibition of Le-uCAG3′tsRNA. Reduction of the active LeuCAG3′tsRNA resulted in a decrease in the RPS28 protein while the mRNA concentration was unchanged, consistent with an effect on translation (Figures 3A–3C). To establish this, we performed sucrose gradient fractionation in LeuCAG3′tsRNA-inhibited cells (Figure S3). As observed in human cells, inhibition of the LeuCAG3′tsRNA caused a decrease in the 18S rRNA level (Figures 2C and 2D), likely resulting in the reduced 40S ribosomal subunit concentration and subsequent lowering of the 80S monosomes (Figure S3A). Next, to determine the polysomal distribution of Rps28 and two control mRNAs (Nop10 and Gapdh), we extracted total RNA from each fraction across the gradient for northern blot quantification (Figures S3B and S3C). Mouse Rps28 and Nop10 coding sequences are 210 and 195 nt, respectively, making their maximal ribosome density similar. Mouse Rps28 mRNA was most highly represented in fraction 11, indicative of three to four ribosomes per mRNA. Inhibition of LeuCAG3′tsRNA significantly shifted the Rps28 mRNA into the lighter fractions 8–10, corresponding to two to three ribosomes per mRNA (p = 0.0492 in fraction 10, p = 0.0155 in fraction 11), while the Nop10 and Gapdh mRNA were primarily found in fractions 11 and 14, respectively, and were not different in both anti-Leu3′ts ASO-and control ASO-treated cells (Figures S3B and S3C). There was no significant change in Rps28 mRNA concentrations in fractions 6 and 7, where the initiating ribosomes (monosomes) co-migrate. This suggested that translational regulation occurred at a step other than initiation.
Figure 3. LeuCAG3′tsRNA Is Required for Mouse Rps28 mRNA Translation through Base Pairing with the Target A Site in the CDS.
(A) Representative western blotting image of mouse RPS28 protein from Hepa 1–6 cells 24 h post-transfection (n = 4 independent experiments). GAPDH, loading control.
(B) Average value of four independent experiments shown in (A). The RPS28 protein level was normalized to the Gapdh protein level.
(C) Rps28 mRNA concentration was determined by real-time PCR 24 h post- transfection and normalized to the Gapdh mRNA level (n = 3 independent experiments).(D) The amount of specific mRNAs in the light polysome (fractions 8 to 10) was normalized to the amount in the heavy polysome (fractions 11 to 13) 24 h post-transfection of ASOs (con, control; anti, anti-Leu30ts) shown in Figure S3B (n = 3 independent experiments).
(E) A representative western blot result from co-transfection of ASOs and the Rps28 WT or mutant plasmids (n = 3 independent experiments). The altered target sites or non-target site are indicated in Figure 1E.
(F) The mean value of three independent experiments in (E). The relative RPS28 protein level from each sample was normalized to the GAPDH protein level; subsequently, the calculated relative RPS28 protein level in the anti-Leu30ts- transfected cells was normalized to the control transfected cells.
The mean is indicated in (B)–(D) and (F). Error bar, SD. *p < 0.05, ***p < 0.0005 by two-tailed t test.
To quantify how much Rps28 mRNA shifted from the heavier fractions (11 to 13) to lighter fractions (8 to 10), we calculated the relative Rps28 mRNA abundance in the lighter compared with the heavier polysomal fraction from the data shown in Figure S3B. The normalized light fraction contained 0.8 ± 0.34 of the Rps28 mRNA in wild-type (WT) cells, but this was increased to 1.47 ± 0.68 (p = 0.0179) when the LeuCAG3′tsRNA was inhibited. The relative amount of normalized Nop10 mRNA contained in the lighter fraction was marginally increased from 0.61 ± 0.20 to 0.71 ± 0.28 when the tsRNA was reduced (Figure 3D). These results confirmed that that LeuCAG3′tsRNA regulates Rps28 mRNA translation in both mouse and human cells.
LeuCAG3′tsRNA-Regulated Mouse Rps28 mRNA Translation Depends on a Target Site in the CDS
The mouse Rps28 mRNA has two potential LeuCAG3′tsRNA target sites; target A in the CDS and target B in the 3′ UTR (Figure 1E). The target A site sequence is almost identical between mouse and human, but the target B site is not (Figure S1D). In addition, the target B site does not form a duplex with the region straddling the TIS(Figures 1E and S1J). To establish whether the LeuCAG3′tsRNA modulates Rps28 mRNA translation with the potential target sites in mouse cells, we constructed expression plasmids containing either a wild type (WT) or various Rps28 mutations (target A, target B, and non-target), which alters mRNA nucleotides, but not the amino acid sequence (Figure 1E). To avoid codon biases that might affect expression, we replaced the codon sequence with those that have comparable codon usage in the Rps28 mutants. The target A and B mutants were predicted to disrupt the tsRNA- Rps28 mRNA interaction and therefore abolish the tsRNA regulation of mRNA translation, while the non-target mutants were not expected to affect the tsRNA-mediated translational regulation.
We co-transfected each plasmid with either control (con) or anti-Leu3′ts (anti) ASOs in Hepa 1–6 cells and examined protein expression by western blot analysis (Figures 3E and 3F). Compared with RPS28 protein expression from control cells, the anti-Leu3′ts ASO-mediated inhibition of LeuCAG3′tsRNA reduced the RPS28 protein concentrations from the wild-type and non-target mutant mRNAs to 47.6% ± 2.7% and 64.8% ± 11.1%, respectively, while the protein level derived from the target A mutant (the conserved target) mRNA was minimally decreased to 84.0% ± 15.2% (Figures 3E and 3F). Unlike the target A mutant, the target B mutant (the non-conserved target) had levels of RPS28 protein 39.1% ± 17.6% of wild-type (Figures 3E and 3F) after tsRNA inhibition, suggesting that the target B site was not active in mouse cells.
Altogether, these results suggest that mouse LeuCAG3′tsRNA regulates mouse Rps28 mRNA translation primarily using the more conserved target site (target A) in the CDS, while the less conserved sequence (target B) in the 3′ UTR is not used as a regulatory site
LeuCAG3′tsRNA Regulates RPS28 mRNA Translation at a Post-initiation Step in Humans and Mice
The human LeuCAG3′tsRNA unfolds the secondary structure at both the TIS and the coding sequence (Figure S1B) (Kim et al., 2017), while the mouse Rps28 mRNA TIS is not functionally active (Figures 1E, 3E, and 3F). As a result, we could not predict whether the tsRNA-based translational enhancement was more likely to occur at the step of initiation or post-initiation and whether these mechanisms were the same or different in humans and mice.
To establish whether a translational initiation block can be discriminated with sucrose gradient fractionation, we used small interfering RNAs (siRNAs) to knock down of RPL7 and RPL35A, which inhibits the production of the 60S subunit and hence the formation of the 80S monosome (Figure S4A). We quantified mRNAs in the various polysomal fractions after sucrose gradient fractionation and found the RPS28 and RPS13 mRNAs stall near the 40S ribosomal free subunit (fraction 4), indicating a block in forming new 80S initiation monosome complexes (Figure S4A).
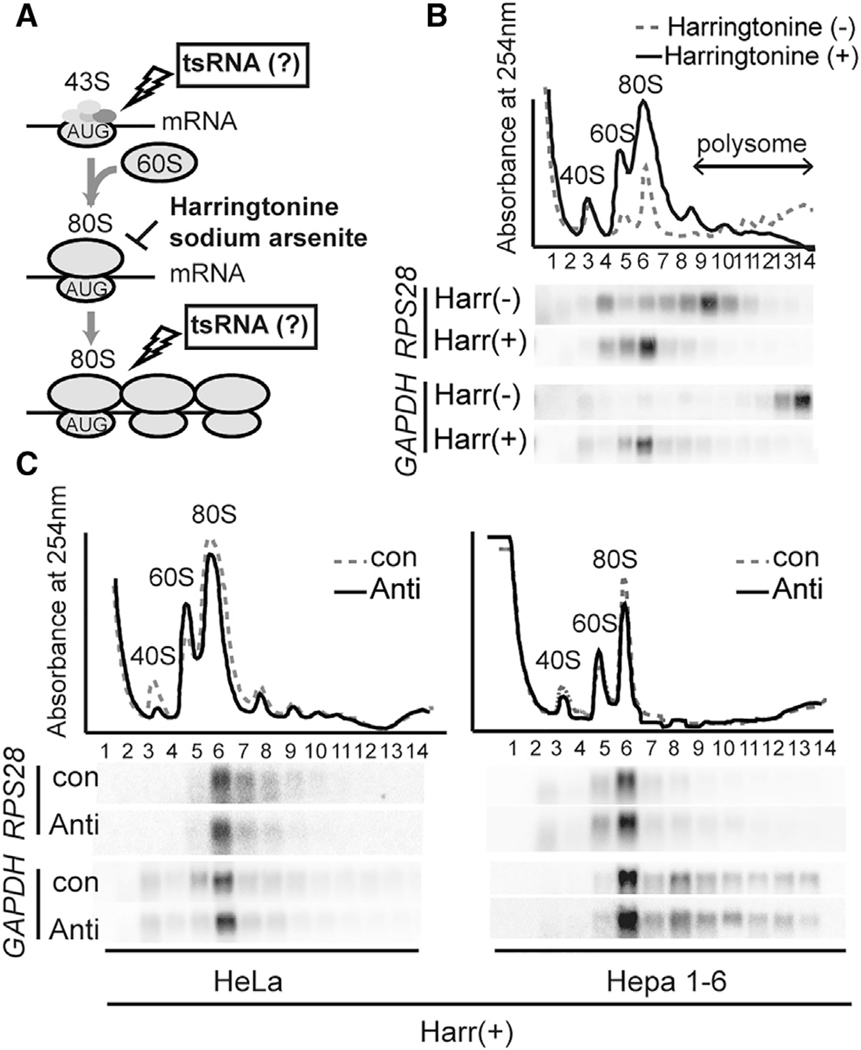
We next treated HeLa and Hepa 1–6 cells with either harringtonine (Figures 4A and 4B) or sodium arsenite (Figures 4A and S4B) and compared RPS28 and GAPDH mRNA sedimentation using sucrose gradient fractionation. Harringtonine prevents the first peptide bond formation (Fresno et al., 1977; Ingolia et al., 2012), and sodium arsenite reduces eukaryotic initiation factor 4E protein levels (Othumpangat et al., 2005) and/or induces the phosphorylation of eukaryotic initiation factor 2α (Kim et al., 2011). Both drugs result in the freezing of the mRNA-ribosome complexes after formation of an 80S monosome while allowing mRNAs in the polysomal fraction to run off (Figures 4B and S4B). Thus, if RPS28 mRNA translation was being suppressed at the level of post-initiation, the RPS28 mRNA would co-sediment with the 80S monosome, whereas if initiation is affected, the RPS28 mRNA would accumulate in the non-polysomal fractions lighter than the 80S monosome (e.g., 40S ribosomal free subunit) (Figures 4A and S4A).
Figure 4. LeuCAG3′tsRNA Regulates Both Human and Mouse RPS28 Translation in the Elongation Phase.
(A) A schematic representation of the possible translation steps affected by the LeuCAG3′tsRNA using harringtonine (B and C) and sodium arsenite (Figures S4B and S4C). If the tsRNA affects 80S complex formation, the harringtonine or sodium arsenite treatments stall RPS28 mRNA near the 40S subunit (Figure S4A). If the tsRNA affects the step after 80S complex formation, the harringtonine or sodium arsenite treatments stall RPS28 mRNA on the 80S complex (Figures 4B and S4B).
(B) Harringtonine treatment in HeLa cells (n = 2 independent experiments).
(C) Harringtonine treatment post-transfection in HeLa (left) and Hepa 1–6 (right) cells (n = 2 independent experiments). The polysome profile (top) and northern hybridization (bottom) were analyzed. The polysome profile indicates the position of 40S and 60S ribosomal free subunits, monosomes, and polysomes on each designated fraction.
Harringtonine (+) and Harr(+), treatment with harringtonine; Harringtonine (–) and Harr(−), no treatment with harringtonine; con, control; anti, anti-Leu3′ts.
As expected, RPS28 and GAPDH mRNAs that normally migrated in fractions 9 and 14, respectively, accumulated on the 80S monosome with harringtonine or sodium arsenite (Figures 4B and S4B). Inhibition of LeuCAG3′tsRNA did not change the accumulation of both mRNAs on the 80S monosome complex in cells treated with harringtonine or sodium arsenite (Figures 4C and S4C). Upon treatment with sodium arsenite, the presence of GAPDH mRNA at fraction 3 (43S pre-initiation) suggests that this drug also affected formation of the GAPDH mRNA-80S monosome complex (Figures S4B and S4C). These results strongly suggest that the LeuCAG3′tsRNA regulates RPS28 mRNA at the level of post-translation initiation in both species.
To support our model, we also determined whether the LeuCAG3′tsRNA is associated with polysomes (Figure S4D). We determined the migration of a mRNA, two microRNAs, a tRNA, and two tsRNAs in the polysomal fractions of a sucrose gradient. The GAPDH mRNA, miR-92a, Let-7, and mature Leu-CAG tRNAs co-migrated with heavy polysomes (fractions 12–14). Thirty-two percent of the LeuCAG3′tsRNAs migrated with fractions 9–14 (polysomes). In contrast, the LeuCAG5′tsRNA was primarily found in lighter fractions, which contain the free ribonucleoproteins (RNPs) and 40S ribosomal free subunits (fractions 1–3), showing they were not associated with the polysomes. To exclude the binding of LeuCAG3′tsRNAs with other non-polysomal complexes in the gradient, we treated the cells with puromycin, a drug that mimics charged tRNAs and terminates polypeptide chain elongation and release of the polysomes into 40S and 60S subunits. This also resulted in the release of the mature Leu-CAG tRNAs and LeuCAG3′tsRNA, specifying their association with polysomes in cells (Figure S4D). Altogether, our results strongly support that the LeuCAG3′tsRNA regulates RPS28 mRNA translation with the target A site at the level of post-initiation in humans and mice. Based on sequence and structure similarities of the target A site, it is highly likely that this mode of regulation is conserved for mammals and possibly vertebrates.
DISCUSSION
Ribosome biogenesis is a complex yet precisely regulated cellular process. Dysregulation of ribosome biogenesis or abnormal expression of ribosomal proteins (RPs) is associated with disease states such as Treacher Collins syndrome (TCS), Shwachman-Bodian-Diamond syndrome (SBDS), dyskeratosis congenita, 5q– syndrome, and Diamond-Blackfan anemia (DBA) (Freed et al., 2010; Sulima et al., 2017). In fact, a RPS28 mutation in TCS/MFD (mandibulofacial dysostosis) and DBA patients has been reported (Gripp et al., 2014). In addition, augmented ribosome biogenesis plays a role in various malignant processes (Bywater et al., 2013; Pelletier et al., 2018).
The importance of the RPS28 protein in translation can be inferred from its localization to the head of the small ribosomal subunit, where it contacts the 18S rRNA and mRNA near or in the exit E site (Fortier et al., 2015; Pisarev et al., 2008; Robledo et al., 2008). A ribosome that lacks RPS28 might have a detrimental effect on translation, making cell death a preferred outcome in humans and mice. As a result, the precise regulatory processes have likely evolved to regulate the production of this protein. It has been well documented that a decrease in specific ribosomal proteins (RPs) downregulates other RP levels and rRNA processing (Robledo et al., 2008), suggesting that multiple regulatory mechanisms could be in play to fine-tune the production of a subset or all ribosomal proteins.
Here, we predicted and then experimentally were able to demonstrate that the LeuCAG3′tsRNA-regulated RPS28 mRNA translation in both mouse and human cells. Based on sequence similarities, we predict that similar mechanisms might be operative in other vertebrates, but this will require additional experimentation.
We also demonstrated that the tsRNA regulated translation at the post-initiation step in humans and mice. The most likely post- initiation step regulated by this tsRNA is elongation. While translational initiation is well established for regulating the rates of translation (Kudla et al., 2009; Salis et al., 2009), there is growing evidence suggesting elongation can also play important roles in regulating protein synthesis (Chu et al., 2014; Firczuk et al., 2013). So far, many factors have been identified to affect translation elongation-GC (guanine-cytosine) content, length and structure of the 5′ UTR, codon optimization and/or rare codons, miRNA targets, and secondary structure. Our finding adds to a growing layer of regulatory processes likely required to maintain precise protein concentrations in cells.
The generation and function of multiple types of tsRNAs remain largely unknown. Moreover, quantifying their concentration within cells and tissues is complicated, because most tRNAs have an average of 11–13 modifications per gene (Phizicky and Hopper, 2010). For example, sequencing paradigms often identify 18-nt 3′ tsRNAs rather than the more predominant 22-nt isoform detected by northern hybridization (Kim et al., 2017; Kumar et al., 2015; Li et al., 2012). This is due to the presence of the N1-methyl-adenosine modification conserved at the 58th nt in the TΨC loop (Saikia et al., 2010), which inhibits reverse transcriptase (Findeiss et al., 2011; Renda et al., 2001), a required step for high-throughput sequencing. Even though AlkB-facilitated RNA de-methylation sequencing was recently developed (Cozen et al., 2015; Liu et al., 2016), it warrants more precise sequencing to overcome various tRNA modifications.
Emerging data revealed that the tRNA modifications affect tRNA stability and function, as well as tsRNA expression profiles. m5C38 (m5C at the C38 position) of Asp, Gly, and Val tRNAs is modified by DNMT2, a multi-substrate tRNA methyltransferase (Lyko, 2018). Disruption of Dnmt2-regulated m5C38 also altered the secondary structure of tsRNAs and their stability against RNase degradation (Zhang et al., 2018). Pseudouridine (Ψ) is one of the most abundant modifications in the RNA world (Charette and Gray, 2000). PUS7-mediated Ψ of 5′ tsRNA in human embryonic stem cells activates 5′ terminal oligoguanine (TOG)-containing, tsRNA-mediated global translation inhibition (Guzzi et al., 2018). The 22-nt 3′ tsRNAs also contain modifications, including m1A58 and Ψ, suggesting that these modifications might affect the biogenesis and/or structure and/or function, including the binding of the LeuCAG3′tsRNA with its target. In addition, it is not yet known whether the 3′ end is amino-acylated, a parameter that may affect the biogenesis and/or specific function. All of these findings warrant more investigation on the modification and biogenesis of 3′ tsRNAs.
There are still many unresolved questions about how these tsRNAs regulate translation. First, a potential seed region might be important in the interaction of the tsRNA and target mRNA. Indeed, the last 3 nt of the 3′ canonical end (CCA) of LeuCAG3′tsRNA do not bind to the Rps28 mRNA (Figure S1I), whereas the importance of the 3′ end of other non-3′ tsRNAs for the interaction with specific targets is noted (Wang et al., 2013; Zhou et al., 2017). Thus, the lack of the CCA interaction with the target might allow greater functional diversity across all 3′ tsRNAs. Second, the importance of the location of the anti-target site and the structure or sequence of the surrounding regions is not known. Lastly, how tsRNA binding influences other parameters known to influence translation is unclear.
Our observation shows that two species have the same function of LeuCAG3′tsRNA, which strongly increases the likelihood that other tsRNAs might disrupt or unwind secondary structures of other mRNAs during translation and that the tsRNAs may have co-evolved with their targets to fine-tune the production of specific proteins, many of which may be involved in protein synthesis. In addition, rRNA, ribosomal proteins (RPs), associated proteins, or modifying proteins may result in ribosome heterogeneity, which may in turn regulate specialized translation of specific transcripts, providing an additional layer to complex gene regulation during cell differentiation and organismal development (Genuth and Barna, 2018).
We demonstrated that the LeuCAG3′tsRNA enhances RPS28 protein synthesis in humans and mice, and more recently, Luo et al. (2018) found that non-3′ tsRNAs repress ribosomal protein expressions in Drosophila, suggesting that varied tsRNAs might be important de novo factors for regulating ribosome biogenesis in various species (Kim et al., 2017).
In addition, our results highlight a potential explanation for why at least some mRNA levels do not necessarily correlate with protein levels (Vogel and Marcotte, 2012; Wilhelm et al., 2014). Further delineating the biogenesis of the more than 150 unique 3′ tsRNAs in mammals, as well as their RNA targets, and their detailed mechanistic functions may reveal an overarching regulatory circuit for fine-tuning gene expression.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
This study did not generate new unique reagents. Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author, Mark A. Kay (markay@stanford.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines were grown at 37°C in 5% CO2 humidified incubators. HeLa (human cervical cancer) and Hepa 1–6 (mouse hepatoma) cells were grown in DMEM medium containing 10% heat-inactivated fetal bovine serum (FBS), 2mM L-glutamine, 100 U/ml penicillin, and 100U/ml streptomycin.
METHOD DETAILS
Transfection
30–60 nM of locked nucleic acid (LNA) mixmers and/or plasmids were transfected using Lipofectamine 2000 (Thermo Fisher) according to the manufacturer’s instructions. 700ng of the appropriate expression plasmid was transfected in 6 well dishes for RPS28 wt or mutant western blot analyses. Anti-sense oligonucleotides (DNA and LNA mixer) were synthesized by Exiqon. The siRPS28, siRPL7, siRPL35 and sicontrol were purchased from Dharmacon. DNA oligonucleotides were synthesized by IDT.
Oligonucleotides
All oligonucleotides used for cell viability experiments with inhibition of LeuCAG3′tsRNA and for detection of LeuCAG3′tsRNA and 18S rRNA precursors are listed in Key Resources Table. All PCR primers used for generation of northern probes to detect mRNAs are listed in Table S3.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-RPS28 | Aviva systems biology | Cat# ARP65601_P050 |
| Rabbit polyclonal anti-RPS7 | Bethyl laboratories | Cat# A300–740A; RRID:AB_533451 |
| Rabbit polyclonal anti-RPL35A | Bethyl laboratories | Cat# A305–106A; RRID:AB_2631501 |
| Mouse monoclonal anti-GAPDH | Thermo Fisher | Cat# AM4300; RRID:AB_437392 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Cycloheximide | Sigma-Aldrich | Cat# C4859–1ML |
| Sodium arsenite | Sigma-Aldrich | Cat# 1062771000 |
| Critical Commercial Assays | ||
| Cell 96 Aqueous One Solution Cell Proliferation Assay (MTS) | Promega | Cat# G3582 |
| Lipofectamine 2000 | Thermo Fisher | Cat# 11668019 |
| Superscript IV RT kit | Thermo Fisher | Cat# 12594100 |
| TRIZOL reagent | Thermo Fisher | Cat# 15596026 |
| Experimental Models: Cell Lines | ||
| Mouse: Hepa 1–6 cells | ATCC | CRL-1830 |
| Human: HeLa cells | ATCC | CCL-2 |
| Oligonucleotides | ||
| Anti-control: GtaCgCgGaaTaCTtC | Exiqon | N/A |
| Anti-Leu3ťs: tGTcAGgAgTggGaT | Exiqon | N/A |
| Anti-Leu3ťsMM: tCTcACgAgTggGaT | Exiqon | N/A |
| Anti-Leu3ťsMM2: tGTcAAgAcTggGaT | Exiqon | N/A |
| Northern probe for LeuCAG3ťsRNA: 5'-gtgtcagg agtgggattcg-3' | IDT | N/A |
| Northern probe for mouse ITS1: 5'-acgccgccgct cctccacagtctcccgtt-3' | IDT | N/A |
| Northern probe for mRNAs: See Table S3 | IDT | N/A |
| Oligonucleotides for site-directed mutagenesis: See Table S4 | IDT | N/A |
| Recombinant DNA | ||
| CMV promoter-mouseRPS28 wt | This paper | N/A |
| CMV promoter-mouseRPS28 target-A mutant | This paper | N/A |
| CMV promoter-mouseRPS28 target-B mutant | This paper | N/A |
| CMV promoter-mouseRPS28 non-target mutant | This paper | N/A |
| Software and Algorithms | ||
| RNA-hybrid program | Krüger and Rehmsmeier, 2006 | https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid |
| Mega software | Kumar et al., 2016b | https://www.megasoftware.net |
| MegAlign program | Burland, 2000 |
https://www.dnastar.com/software/ molecular-biology/ |
| PRISM 8.0 | N/A | https://www.graphpad.com/ |
| Integrative genomics viewer | Robinson et al., 2011 | https://software.broadinstitute.org/software/igv/ |
| Clustal Omega | Sievers et al., 2011 | https://www.ebi.ac.uk/Tools/msa/clustalo/ |
| TopHat2 (v.2.0.14) | Trapnell et al., 2009 | https://ccb.jhu.edu/software/tophat/index.shtml |
Plasmid constructs
The full-length mouse Rps28 gene was amplified from Hepa 1–6 cDNA with primers (5′-ctcgcgagagcgaaagtgag-3′ and 5′-taata taaatgctttatttaacagttgcag-3′) and was cloned into the pcDNA3.3 plasmid. Site-directed mutagenesis was performed with the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene) to generate point mutations or deletions in the recombinant Rps28 gene. All oligonucleotides for site directed mutagenesis are listed in Table S4. All plasmid clones were confirmed by DNA sequencing.
Western blotting
24 h post-transfection, cell lysates were prepared using 1X cell lysis buffer (Cell Signaling) with 1 mM PMSF (Cell Signaling). 10–15 ug of protein lysate was run on 4%–12% SDS-PAGE and transferred to Hybond-P or nitrocellulose membrane (GE Healthcare). The membrane was incubated for 20 min at room temperature (RT) in Blocking Buffer (LI-COR Biosciences), washed, and incubated overnight (O/N) at 4°C with anti-RPS28 (1:1000, Aviva systems biology, ARP65601_P050), anti-RPS7 (1:1000, Bethyl laboratories, A300–740A), anti-RPL35A (1:1000, Bethyl laboratories, A305–106A) or anti-GAPDH antibodies (1:5000, Life Technologies, clone 6C5). After washing and incubation for 2 h at RT with secondary antibody (1:10,000, Fisher, 92568071 and 92532210), the protein signal was detected using Odyssey CLx imaging system (LI-COR Biosciences) according to the manufacturer’s instructions.
Real time PCR
500 ng of total RNA was reverse transcribed with the superscript IV RT kit (Thermo Fisher) and subjected to gene expression analyses with gene-specific TaqMan probes (Mm99999915_g1 for GAPDH and Mm04203728_gH for mouse Rps28). Real time PCR was performed on a CFX384 Real-Time system (Bio-Rad).
RNA isolation and Northern blotting
Total RNA was isolated with TRIZOL reagent (Thermo Fisher) according to the manufacturer’s instructions. Total RNA was resolved by electrophoresis on 15% (w/v) polyacrylamide gel with 7 M urea for detection of small RNAs less than 200 bp or on 0.9% agarose denaturating gel for detection of large RNAs (> 200 bp) followed by transfer onto a Hybond-N+ nylon membrane (Amersham). P32-labeled oligonucleotides or amplified cDNA probes were hybridized to the membrane in PerfectHyb Plus hybridization buffer (Sigma).
Polysome gradient and RNA preparation
Polysome gradient and RNA preparation were performed as described previously (Kim et al., 2017). 24 h post-transfection, cells were treated with 100 mg/ml of cycloheximide (Sigma) for 3min, were washed with cold DPBS (Sigma) containing 100 mg/ml of cyclohexmide 2 times, and were lysed in buffer containing 15mM Tris–HCl (pH 7.5), 150mM KCl, 5mM MgCl2, 500u/ml RNasin (Promega), and 1% Triton X-100 for 10 m. The lysates were cleared by centrifugation at 8,500 g-force for 5 m. For dissociation of 40S and 60S ribosomal subunit from ribosome, 200 ug/ml of Puromycin (Sigma-Aldrich, MO) was treated for 30 m before harvesting cells. Inhibition of translation initiation was performed as described previously (Ingolia et al., 2012). Cells were incubated with the 2 mg/ml harringtonine (Abcam) for 2 m or 1mM sodium arsenite (Fluka) for 1 h, followed by the treatment of cycloheximide before harvesting cells. The cleared lysates were loaded onto 10%–50% sucrose gradients (15 mM Tris–HCl (pH 7.5), 150mM KCl, 5 mM MgCl2, 20u/ml SUPERaseIn (Thermo Fisher), and 100 mg/ml cycloheximide). Gradients were centrifuged at 35,000 rpm for 2 h 45 m in a SW41 rotor at 4°C and were collected into 14 tubes by pumping 70% sucrose into the bottom of the gradient and collecting from the top using a Teledyne Isco Foxy R1 Retriever/ UA-6 detector system with measurement of the absorbance at 254nm. Each obtained fractions were sequentially treated for 30 m at 37°C with 0.5 mg/ml proteinase K (New England Biolabs) in the presence of 5mM EDTA and 1% SDS. RNAs were extracted with an equal volume of phenol-chloroform-isoamylalcohol (25:24:1; Thermo Fisher), re-extracted with chloroform, and EtOH precipitation was performed.
The abundance of Rps28 transcript variants in mouse liver tissue
TopHat2 (v.2.0.14) (Trapnell et al., 2009) was used to align RNA sequence reads to the mouse mm9 genome. BAM files were visualized using the integrative genomics viewer (Robinson et al., 2011) and a Sashimi plot was generated to quantify the number of reads that mapped to splice junctions of each isoform of Rps28.
Structure probing of tsRNA targets and mouse Rps28 mRNA secondary structure prediction
The in vivo click selective 2′-hydroxyl acylation and profiling experiment (icSHAPE) generates the global view of RNA secondary structures in living cells for all four nucleotides. We retrieved RNA structure data from a previous study that probed nucleotide reactivity (i.e., single-strandedness) in mouse embryonic cells by using icSHAPE (Spitale et al., 2015). Mouse Rps28 mRNA sequences were downloaded from NCBI RefSeq database (O’Leary et al., 2016) and the secondary structure was predicted using RNAStructure or RNAfold software by using default parameters, with and without icSHAPE data as constraints, respectively (Hofacker and Stadler, 2006; Reuter and Mathews, 2010). The secondary structure was visualized and edited using the VARNA program (Darty et al., 2009). Sequencing depth for icSHAPE is not adequate to obtain. icSHAPE scores for all mRNAs or in some instances the complete sequence of an individual mRNA.
tsRNA target prediction
LeuCAG3′tsRNA sequences of various species were obtained from the tRNA database (GtRNAdb) (Chan and Lowe, 2016) and the RPS28 mRNA sequences were downloaded from the NCBI RefSeq database (O’Leary et al., 2016). The m.f.e. (minimal free energy) between the LeuCAG3′tsRNA and the binding site in the RPS28 mRNA in various species was predicted using a RNA-hybrid program (Krüger and Rehmsmeier, 2006). The potential target sites were predicted using a —20 kcal/mol of energy threshold and three nucleotides loop constraints. The tsRNA binding predictions do not take into account nucleotide modifications.
Genetic distances of LeuCAG3′tsRNA
The genetic distances of LeuCAG3′tsRNA from various species can be measured by computing the proportion of nucleotide differences between each pair of sequences. The p genetic distances of all LeuCAG3′tsRNA sequences across different 44 species were calculated using MEGA software (Kumar et al., 2016b).
Percent identity
Percent identity is a quantitative measurement of the similarity between each pair of sequences. Closely related species are expected to have a higher percent identity for a given sequence than distantly related species, and thus percent identity to a degree reflects relatedness. The percent identity of LeuCAG3′tsRNA sequences across various species was calculated using the MegAlign program from DNASTAR software package (Burland, 2000).
Sequence alignment of RPS28 sequences
RPS28 mRNA sequences across different species were downloaded in FASTA format from NCBI (O’Leary et al., 2016). Then sequence alignment were conducted using Clustal Omega (Sievers et al., 2011).
Conservation analysis of LeuCAG3′tsRNA target site
The phyloP conservation scores of RPS28 for 100 vertebrate species were downloaded from the UCSC Table Browser (Karolchik et al., 2004). The sites predicted to be conserved are assigned positive scores, while sites predicted to be tolerant to nucleotide changes are assigned negative scores. We calculated the average conservation score of each 22-nucleotides sliding window across the RPS28 CDS region and subsequently generated the conservation ranking for the LeuCAG3′tsRNA target site.
QUANTIFICATION AND STATISTICAL ANALYSIS
All data are presented as mean ± SD. Figure 2D (n = 3 independent experiments), Figure 3B (n = 4 independent experiments), Figure 3D (n = 3 independent experiments), and Figure 3F (n = 3 independent experiments) were analyzed by the two-tailed Student’s t test and Figure 2A (n = 4 independent experiments) was analyzed with one-way ANOVA using GraphPad Prism version 8.0.0 for Mac, GraphPad Software, San Diego, California USA, https://www.graphpad.com/.A P value of 0.05 or lower was considered significant.
Supplementary Material
Highlights.
LeuCAG3′tsRNA target site in the RPS28 coding sequence is conserved in vertebrates
LeuCAG3′tsRNA regulates RPS28 translation after the initiation step in humans and mice
LeuCAG3′tsRNA-regulated translation is conserved between humans and mice
tsRNA-regulated translational mechanism might be conserved among vertebrates
ACKNOWLEDGMENTS
This work was supported by grants to M.A.K. from the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (R01DK114483).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
DATA AND CODE AVAILABILITY
This study did not generate/analyze datasets/code.
REFERENCES
- Bowman LH, Rabin B, and Schlessinger D. (1981). Multiple ribosomal RNA cleavage pathways in mammalian cells. Nucleic Acids Res. 9, 4951–4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D, Soumillon M, Necsulea A, Julien P, Csárdi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, et al. (2011). The evolution of gene expression levels in mammalian organs. Nature 478, 343–348. [DOI] [PubMed] [Google Scholar]
- Burland TG (2000). DNASTAR’s Lasergene sequence analysis software. Methods Mol. Biol 132, 71–91. [DOI] [PubMed] [Google Scholar]
- Bywater MJ, Pearson RB, McArthur GA, and Hannan RD (2013). Dys-regulation of the basal RNA polymerase transcription apparatus in cancer. Nat. Rev. Cancer 13, 299–314. [DOI] [PubMed] [Google Scholar]
- Chan PP, and Lowe TM (2016). GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44 (D1), D184–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charette M, and Gray MW (2000). Pseudouridine in RNA: what, where, how, and why. IUBMB Life 49, 341–351. [DOI] [PubMed] [Google Scholar]
- Chu D, Kazana E, Bellanger N, Singh T, Tuite MF, and von der Haar T. (2014). Translation elongation can control translation initiation on eukaryotic mRNAs. EMBO J. 33, 21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, and Lowe TM (2015). ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods 12, 879–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darty K, Denise A, and Ponty Y. (2009). VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 25, 1974–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findeiss S, Langenberger D, Stadler PF, and Hoffmann S. (2011). Traces of post-transcriptional RNA modifications in deep sequencing data. Biol. Chem 392, 305–313. [DOI] [PubMed] [Google Scholar]
- Firczuk H, Kannambath S, Pahle J, Claydon A, Beynon R, Duncan J, Westerhoff H, Mendes P, and McCarthy JE (2013). An in vivo control map for the eukaryotic mRNA translation machinery. Mol. Syst. Biol 9, 635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortier S, MacRae T, Bilodeau M, Sargeant T, and Sauvageau G. (2015). Haploinsufficiency screen highlights two distinct groups of ribosomal protein genes essential for embryonic stem cell fate. Proc. Natl. Acad. Sci. USA 112, 2127–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EF, Bleichert F, Dutca LM, and Baserga SJ (2010). When ribosomes go bad: diseases of ribosome biogenesis. Mol. Biosyst 6, 481–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fresno M, Jiménez A, and Vázquez D. (1977). Inhibition of translation in eukaryotic systems by harringtonine. Eur. J. Biochem 72, 323–330. [DOI] [PubMed] [Google Scholar]
- Fricker R, Brogli R, Luidalepp H, Wyss L, Fasnacht M, Joss O, Zywicki M, Helm M, Schneider A, Cristodero M, and Polacek N. (2019). A tRNA half modulates translation as stress response in Trypanosoma brucei. Nat. Commun 10, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebetsberger J, Zywicki M, Künzi A, and Polacek N. (2012). tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012, 260909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genuth NR, and Barna M. (2018). The Discovery of Ribosome Heterogeneity and Its Implications for Gene Regulation and Organismal Life. Mol. Cell 71, 364–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gripp KW, Curry C, Olney AH, Sandoval C, Fisher J, Chong JX, Pilchman L, Sahraoui R, Stabley DL, and Sol-Church K; UW Center for Mendelian Genomics (2014). Diamond-Blackfan anemia with mandibulofacial dystostosis is heterogeneous, including the novel DBA genes TSR2 and RPS28. Am. J. Med. Genet. A 164A, 2240–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzi N, Cieśla M, Ngoc PCT, Lang S, Arora S, Dimitriou M, Pimková K, Sommarin MNE, Munita R, Lubas M, et al. (2018). Pseudouridylation of tRNA-Derived Fragments Steers Translational Control in Stem Cells. Cell 173, 1204–1216.e26. [DOI] [PubMed] [Google Scholar]
- Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, and Kay MA (2010). Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16, 673–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL, and Stadler PF (2006). Memory efficient folding algorithms for circular RNA secondary structures. Bioinformatics 22, 1172–1176. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, and Weissman JS (2012). The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc 7, 1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P, Emara MM, Villen J, Gygi SP, and Anderson P. (2011). Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 43, 613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepsen JS, Sørensen MD, and Wengel J. (2004). Locked nucleic acid: a potent nucleic acid analog in therapeutics and biotechnology. Oligonucleotides 14, 130–146. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, and Kent WJ (2004). The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent T, Lapik YR, and Pestov DG (2009). The 5′ external transcribed spacer in mouse ribosomal RNA contains two cleavage sites. RNA 15, 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JHJ, Park SMS, Park JHJ, Keum SJS, and Jang SKS (2011). eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J. 30, 2454–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HK, Fuchs G, Wang S, Wei W, Zhang Y, Park H, Roy-Chaudhuri B, Li P, Xu J, Chu K, et al. (2017). A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature 552, 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger J, and Rehmsmeier M. (2006). RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 34, W451–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, and Plotkin JB (2009). Coding-sequence determinants of gene expression in Escherichia coli. Science 324, 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Anaya J, Mudunuri SB, and Dutta A. (2014). Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 12, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Mudunuri SB, Anaya J, and Dutta A. (2015). tRFdb: a database for transfer RNA fragments. Nucleic Acids Res. 43, D141–D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Kuscu C, and Dutta A. (2016a). Biogenesis and Function of Transfer RNA-Related Fragments (tRFs). Trends Biochem. Sci 41, 679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, and Tamura K. (2016b). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, and Dutta A. (2009). A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 23, 2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Ender C, Meister G, Moore PS, Chang Y, and John B. (2012). Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res. 40, 6787–6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J, Wang X, Hao Z, Dai Q, Zheng G, et al. (2016). ALKBH1-Mediated tRNA Demethylation Regulates Translation. Cell 167, 816–828.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, He F, Luo J, Dou S, Wang Y, Guo A, and Lu J. (2018). Drosophila tsRNAs preferentially suppress general translation machinery via antisense pairing and participate in cellular starvation response. Nucleic Acids Res. 46, 5250–5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F. (2018). The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet 19, 81–92. [DOI] [PubMed] [Google Scholar]
- Nei M, and Kumar S. (2000). Molecular Evolution and Phylogenetics (Oxford University Press; ). [Google Scholar]
- O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44 (D1), D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Othumpangat S, Kashon M, and Joseph P. (2005). Sodium arsenite-induced inhibition of eukaryotic translation initiation factor 4E (eIF4E) results in cytotoxicity and cell death. Mol. Cell. Biochem 279, 123–131. [DOI] [PubMed] [Google Scholar]
- Pelletier J, Thomas G, and Volarević S. (2018). Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat. Rev. Cancer 18, 51–63. [DOI] [PubMed] [Google Scholar]
- Phizicky EM, and Hopper AK (2010). tRNA biology charges to the front. Genes Dev. 24, 1832–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen CUT, and Pestova TV (2008). Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 27, 1609–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renda MJ, Rosenblatt JD, Klimatcheva E, Demeter LM, Bambara RA, and Planelles V. (2001). Mutation of the methylated tRNA(Lys)(3) residue A58 disrupts reverse transcription and inhibits replication of human immunodeficiency virus type 1. J. Virol 75, 9671–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter JS, and Mathews DH (2010). RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat. Biotechnol 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robledo S, Idol RA, Crimmins DL, Ladenson JH, Mason PJ, and Bessler M. (2008). The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA 14, 1918–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia M, Fu Y, Pavon-Eternod M, He C, and Pan T. (2010). Genomewide analysis of N1-methyl-adenosine modification in human tRNAs. RNA 16, 1317–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salis HM, Mirsky EA, and Voigt CA (2009). Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol 27, 946–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P. (2018). The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell Biol 19, 45–58. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack FJ (2018). Tackling Tumors with Small RNAs Derived from Transfer RNA. N. Engl. J. Med 378, 1842–1843. [DOI] [PubMed] [Google Scholar]
- Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung J-W, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, and Chang HY (2015). Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519, 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulima SO, Hofman IJF, De Keersmaecker K, and Dinman JD (2017). How Ribosomes Translate Cancer. Cancer Discov. 7, 1069–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, and Salzberg SL (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdmanis PN, Gu S, Chu K, Jin L, Zhang F, Munding EM, Zhang Y, Huang Y, Kutay H, Ghoshal K, et al. (2016). RNA interference-induced hepatotoxicity results from loss of the first synthesized isoform of microRNA-122 in mice. Nat. Med 22, 557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, and Marcotte EM (2012). Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet 13, 227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Lee I, Ren J, Ajay SS, Lee YS, and Bao X. (2013). Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol. Ther 21, 368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M, Schlegl J, Hahne H, Gholami AM, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, et al. (2014). Mass-spectrometry-based draft of the human proteome. Nature 509, 582–587. [DOI] [PubMed] [Google Scholar]
- Yamasaki S, Ivanov P, Hu G-F, and Anderson P. (2009). Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol 185, 35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang X, Shi J, Tuorto F, Li X, Liu Y, Liebers R, Zhang L, Qu Y, Qian J, et al. (2018). Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat. Cell Biol 20, 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu S, Chen Y, Fu Y, Silver AJ, Hill MS, Lee I, Lee YS, and Bao X. (2017). Identification of two novel functional tRNA-derived fragments induced in response to respiratory syncytial virus infection. J. Gen. Virol 98, 1600–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.