Abstract
The bacterial cell has less internal structure and genetic complexity than cells of eukaryotic organisms, yet it is a highly organized system that uses both temporal and spatial cues to drive its cell cycle. Key insights into bacterial regulatory programs that orchestrate cell cycle progression have come from studies of Caulobacter crescentus, a bacterium that divides asymmetrically. Three global regulatory proteins cycle out of phase with one another and drive cell cycle progression by directly controlling the expression of 200 cell-cycle-regulated genes. Exploration of this system provided insights into the evolution of regulatory circuits and the plasticity of circuit structure. The temporal expression of the modular subsystems that implement the cell cycle and asymmetric cell division is also coordinated by differential DNA methylation, regulated proteolysis, and phosphorylation signaling cascades. This control system structure has parallels to eukaryotic cell cycle control architecture. Remarkably, the transcriptional circuitry is dependent on three-dimensional dynamic deployment of key regulatory and signaling proteins. In addition, dynamically localized DNA-binding proteins ensure that DNA segregation is coupled to the timing and cellular position of the cytokinetic ring. Comparison to other organisms reveals conservation of cell cycle regulatory logic, even if regulatory proteins, themselves, are not conserved.
Cell duplication, whether in a mammal with complex organ systems or in a single-celled bacterium, must use rigorous cell cycle regulation to ensure that the cell is ready before proceeding from one step to the next. Premature entry into DNA synthesis (S) phase or exit from mitosis (M) could have drastic consequences, notably fatal damage to the genome. Once initiated, improper execution of physical aspects of the cell cycle such as chromosome segregation and cytokinesis is equally dangerous. Thus, the cell has robust mechanisms to assure fidelity of every step of the cell duplication process. The cell cycle of the aquatic α-proteobacterium Caulobacter crescentus (hereafter called Caulobacter) has been extensively studied at both the system and molecular levels.
Caulobacter has a life cycle characterized by precise developmental transitions and asymmetric cell division (Fig. 1A). Each division produces two cell types: a motile swarmer cell and a sessile stalked cell. After cytokinesis, a newborn swarmer cell is in the equivalent of G1 phase of the cell cycle, unable to replicate its chromosome (Degnen and Newton 1972). The swarmer cell has a period of motility and then differentiates into a stalked cell identical to its stalked sibling. In this process, it sheds its polar flagellum and builds a stalk at the same site while simultaneously initiating replication of the chromosome from a single origin of replication (Cori). Replicated portions of the chromosome are then segregated to opposite ends of the cell as replication proceeds (Viollier et al. 2004). While this is occurring, a flagellum is constructed at the pole opposite the stalk. Constriction of the cell envelope at the incipient division site begins before completion of chromosome segregation (Jensen 2006). The cytoplasm is divided into two distinct compartments shortly after the duplicated chromosomes are fully segregated (Judd et al. 2003; Jensen 2006). The differentiation of the two daughter cells is triggered by this compartmentalization event, as the genetic programs in each of the compartments immediately diverge. About 20 minutes later, the daughter cells separate to yield a newborn swarmer cell that undergoes an obligate G1 phase before becoming a stalked cell and the stalked cell that immediately reenters S phase.
Figure 1.
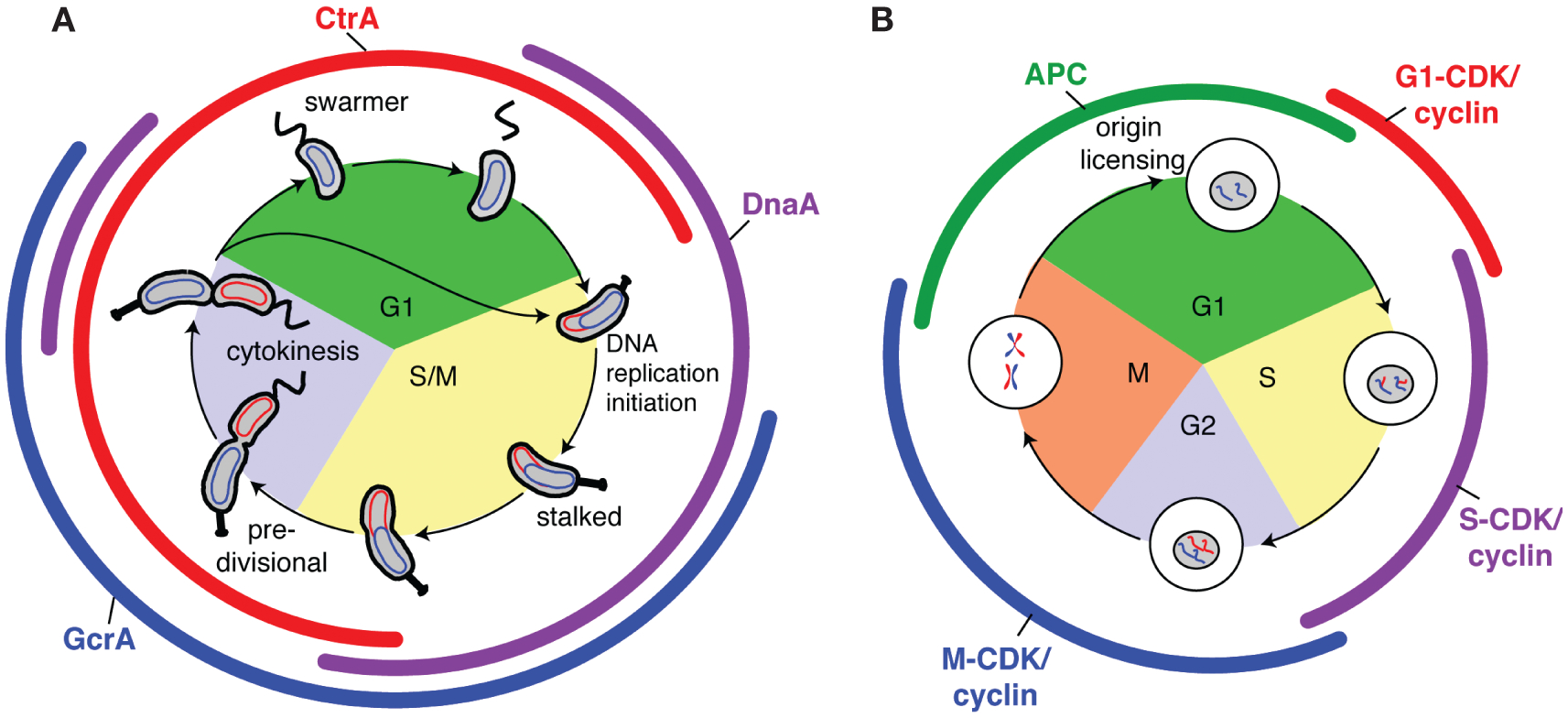
Replication initiation is limited to once per cell cycle by oscillating master regulators in both prokaryotes and eukaryotes. (A) Caulobacter cell cycle. The swarmer cell sheds its polar flagellum, and in the presence of low CtrA (red) and high DnaA (purple), DNA replication can initiate in the new stalked cell. (Colored arcs) Presence of the indicated master regulator protein, (curved ellipses) circular chromosome. (B) Generalized eukaryotic cell cycle. The APC is active from the metaphase-to-anaphase transition in late mitosis through most of G1 phase, keeping CDK–cyclin activity low and allowing loading of pre-RC complexes and origin licensing. When S-phase CDK cyclins are activated at the onset of S phase, licensed origins fire simultaneously.
Global regulatory paradigms governing the Caulobacter cell cycle have emerged from the analysis of specific cell-cycle-dependent events such as DNA replication initiation and cell division site selection that parallel those in other bacteria, as well as in eukaryotes. Here, we describe key events in the Caulobacter cell cycle and highlight conserved regulatory themes that reflect constraints guiding the evolution of cell cycle control mechanisms in diverse organisms.
ENTRY INTO S PHASE
Master Regulators Provide a Cell Cycle Timing Mechanism and Regulate Entry into S Phase
Whole-genome microarray studies on synchronized populations of Caulobacter revealed a transcriptional cascade governing cell cycle progression, wherein expression of functional gene modules (e.g., replication initiation, flagellum biogenesis, chemotaxis apparatus assembly, and cell division) is sequentially activated just in time to accomplish their cellular functions (Laub et al. 2000). Central to this genetic circuitry is a set of three master regulators (DnaA, CtrA, and GcrA) that together affect expression of ~200 cell-cycle-regulated genes (Laub et al. 2002; Holtzendorff et al. 2004; Hottes et al. 2005). The protein levels of these master regulators oscillate out of phase with one another (Fig. 1A) (Collier et al. 2006) in a manner similar to the oscillations of cell cycle regulatory cyclin-dependent kinase (CDK)–cyclin complexes and the anaphase promoting complex (APC) in eukaryotes (Fig. 1B). Each master regulator controls expression of the next. A fourth protein, the CcrM DNA methyltransferase, completes the closed cell cycle control circuit (Fig. 2) (Collier et al. 2007). The robustness of the circuit is increased by posttranscriptional regulation of the activities of the master regulators, including phosphorylation, proteolysis, and dynamic spatial positioning of the phosphosignaling proteins and proteases. This tightly integrated spatial and temporal choreography of regulatory factors advances the cell cycle while simultaneously driving Caulobacter’s asymmetric development.
Figure 2.
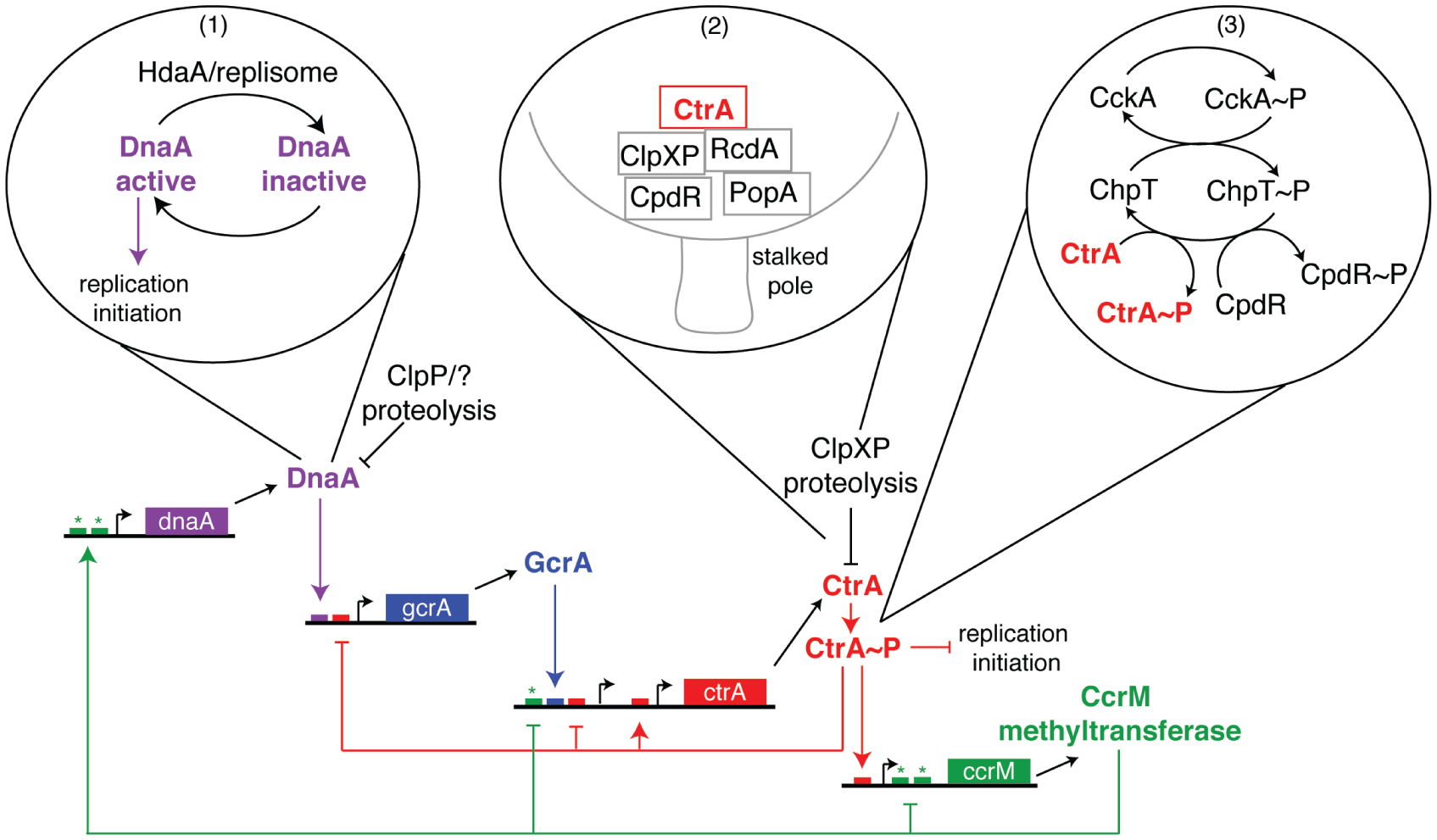
The Caulobacter core cell cycle transcriptional control circuit includes posttranslational regulation of master regulators. Each master regulator activates transcription of the next and, in the case of CtrA and CcrM, inhibits transcription of the previous master regulator in the cascade. Promoters and genes encoding the master regulators are depicted with regulatory and/or binding motifs (small boxes) indicated. The color of the regulatory motif correlates with the master regulator that governs it (i.e., small red boxes represent CtrA-binding sites, etc.). (Asterisks) CcrM methylation sites, (bubbles) posttranslational regulation of DnaA and CtrA. (1) DnaA is inactivated after HdaA joins the replisome upon initiation of replication. (2) CtrA is localized to the stalked pole, by the concerted action of RcdA, PopA, ClpXP, and CpdR, where it is subjected to ClpXP-mediated proteolysis. (3) CtrA is phosphorylated and activated by the same CckA-ChpT phosphorelay that phosphorylates and inactivates CpdR.
The availability of a large number of bacterial genome sequences combined with systems-wide approaches to the identification of the genetic circuitry controlling the bacterial cell cycle has allowed analysis of the evolution of regulatory networks and the degree of plasticity of regulatory network structure (McAdams et al. 2004). An example of the plasticity of regulatory networks comes from the pathway controlled by the CtrA global regulator that directly activates 95 genes. Although the CtrA protein and multiple elements of the complex circuitry that controls the timing of CtrA expression and activation have been conserved through evolution of the α-class of proteobacteria, the portfolio of subsystem functions controlled by CtrA in each bacterial species differs to reflect their specific environmental niche (Bellefontaine et al. 2002; McAdams et al. 2004). Nevertheless, this analysis suggests that an intact genetic circuit can act as an evolutionary unit.
In addition to acting as transcription factors, DnaA and CtrA act coordinately to regulate entry into S phase. DnaA, an AAA+ ATPase, is a broadly conserved replication initiator in bacteria that binds to the replication origin (Cori) and locally unwinds the DNA to allow loading of the replication machinery (Mott and Berger 2007). Conversely, CtrA acts as a repressor of replication initiation by binding to five sites in Cori (Quon et al. 1998). The cell can only enter S phase when active CtrA levels are low and active DnaA levels are high. These conditions exist only once per cell cycle in Caulobacter: in stalked cells, either at the swarmer-to-stalked cell transition or in the nascent stalked cell compartment of late-predivisional cells (Fig. 1A).
Multiple Modes of DnaA and CtrA Regulation: Phosphorylation, Localization, and Proteolysis
CtrA is only active in its phosphorylated form (CtrA~P) (Domian et al. 1997). Its phosphorylation state is controlled by a phosphorelay from the CckA histidine kinase through the ChpT phosphotransferase (Fig. 2) (Jacobs et al. 1999; Biondi et al. 2006). CtrA~P is present in swarmer cells and in predivisional cells, but it is redundantly inactivated by both dephosphorylation and proteolysis at the swarmer-to-stalked cell transition and in the stalked compartment of late-predivisional cells (Domian et al. 1997). Inactivation of CtrA is critical for licensing DNA replication initiation in newborn stalked cells; expression of a constitutively active, stable mutant of CtrA causes cell cycle arrest in G1 (Domian et al. 1997).
To enter S phase, CtrA must be cleared from the cell. It is degraded by the essential ATP-dependent ClpXP protease complex (Jenal and Fuchs 1998), which proteolyzes CtrA in vitro in the absence of accessory factors (Chien et al. 2007). Remarkably, however, CtrA degradation in vivo requires not only that ClpXP be present, but that both ClpXP and its CtrA substrate be localized to the stalked pole (Ryan et al. 2002; McGrath et al. 2006). This colo-calization is achieved through the action of the CpdR phosphoprotein, which localizes ClpXP to the pole (Iniesta et al. 2006), and the combined actions of the RcdA localization factor (McGrath et al. 2006) and the PopA cyclic di-GMP effector protein (Duerig et al. 2009) that together bring the CtrA substrate to polar ClpXP (Fig. 2). The response regulator CpdR only localizes to the stalked pole and recruits ClpXP in its unphosphorylated form. The timing of CtrA degradation is therefore controlled in part by the phosphorylation state of CpdR, which changes as the cell cycle progresses. CpdR is phosphorylated in swarmer and early-predivisional cells, but it is dephosphorylated at the swarmer-to-stalked transition and in the stalked compartment of late-predivisional cells, just in time to promote clearance of CtrA (Iniesta et al. 2006). Interestingly, CpdR is phosphorylated (thereby preventing its polar localization) by the same phosphorelay that activates CtrA, the CckA-ChpT pathway (Biondi et al. 2006). Thus, the pathway that activates CtrA by phosphorylation also prevents its degradation by inactivating the CpdR polar localization factor, providing robust control of entry into S phase.
Caulobacter DnaA is also posttranscriptionally regulated. At least two mechanisms are likely to be involved in DnaA inactivation to prevent early entry into S phase or overinitiation of replication (Fig. 2). First, the DnaA protein is relatively unstable, with a half-life of about one-third of the cell cycle (Gorbatyuk and Marczynski 2005). Its short half-life combined with cell-cycle-regulated transcription results in high DnaA levels in stalked cells and early-predivisional cells and low levels of DnaA in swarmer and late-predivisional cells (Gorbatyuk and Marczynski 2005; Collier et al. 2006). The second mode of inactivation of DnaA uses the HdaA protein, which binds to the replisome upon replication initiation and inactivates DnaA, rendering any remaining DnaA protein incapable of reinitiating replication (J Collier and L Shapiro, in press).
This multilayered and tightly regulated control of the initiation of DNA replication ensures that Caulobacter replicates its chromosome only once per cell cycle. This is in contrast to Escherichia coli, where overlapping rounds of replication can take place in each cell cycle when grown in rich media. These contrasting modes of regulation of DNA replication reflect the different niches that the two species occupy. E. coli is adapted to achieve very rapid growth under intermittent high nutrient conditions in the mammalian gut. Caulobacter, on the other hand, is adapted to survive under the low nutrient conditions found in lakes and streams.
Preventing Improper Replication Initiation in Other Bacteria
Replication of the genome is central to the cell cycle, and it is perhaps not surprising that a factor essential for initiation of replication, DnaA, is highly conserved in eubacteria. The activity of DnaA in Caulobacter (J Collier and L Shapiro, in press) and E. coli (Kato and Katayama 2001) is restricted after initiation by HdaA and Hda, respectively. Hda acts by promoting conversion of active ATP-DnaA to inactive ADP-DnaA in E. coli. Given the conservation of the AAA+ ATPase activity of DnaA, it is likely that the control of replication via regulation of the ATPase activity of the initiator protein is broadly used. Indeed, the activities of eukaryotic origin recognition complex (ORC) AAA+ ATPase subunits and the initiator protein Cdc6 are regulated by modulation of their nucleotide-bound states as well (Bell and Dutta 2002).
Unlike DnaA, CtrA is not found outside the α-proteobacteria, and even in closely related Caulobacter species, the number and position of CtrA-binding sites near the origin of replication differ considerably (Shaheen et al. 2009). However, restricting access to DnaA-binding sites is a commonly used mechanism for preventing improper initiation of replication. In E. coli, for example, reinitiation is restricted in part by binding of SeqA to hemimethylated sites on newly replicated origins and transiently preventing DnaA binding (Slater et al. 1995). Another inhibitor that is remarkably similar to CtrA is Spo0A from the spore-forming Gram-positive bacterium Bacillus subtilis. Like CtrA, Spo0A is a response regulator and transcription factor that acts as a master regulator for entry into sporulation only in its phosphorylated form (Piggot and Hilbert 2004). In addition to its transcription-related activities, Spo0A also binds to sites in the origin overlapping DnaA-binding sites and prevents replication initiation once sporulation has commenced (Castilla-Llorente et al. 2006). In the case of both CtrA and Spo0A, it appears that the cell has adapted an existing transcription factor to couple developmentally regulated changes in gene expression to duplication of the genome. This strategy might be readily evolved, because it requires only a series of base pair changes in the region around the origin of replication to introduce a new binding site for a pre-existing DNA-binding protein.
Replication Licensing in Eukaryotes
The basic requirement for high DnaA and low CtraA activities to allow replication initiation in Caulobacter is strikingly similar to the mode of origin licensing in eukaryotes. There, licensing occurs before entry into S phase, when APC levels are high and CDK–cyclin levels are low (Fig. 1B) (for review, see Diffley 2004; Arias and Walter 2007). Only under those conditions can prereplication complexes (pre-RCs) load at chromosomal origins. Pre-RCs consist of the ORC, the MCM2–7 presumptive helicase, and two additional factors, Cdc6 and Cdt1, each of which is essential for origin licensing. Upon inactivation of the APC and activation of S-phase CDKs, origins that have been preloaded with pre-RCs can fire simultaneously for a single round of replication. Reinitiation is prevented because CDK levels are high from S phase until late M phase, disallowing assembly of additional pre-RCs at origins until the next late M and G1 phases. Unlike CtrA and DnaA in Caulobacter, however, the APC and CDKs exert their effects on replication licensing indirectly.
As described above, CtrA activity is regulated redundantly by phosphorylation–dephosphorylation and synthesis–proteolysis, which also relies on precise subcellular protein localization. Similar mechanisms regulate components of the pre-RC in yeasts and metazoans (for review, see Diffley 2004; Arias and Walter 2007). In the budding yeast Saccharomyces cerevisiae, for example, CDK phosphorylation of pre-RC components can target them for ubiquitin-mediated proteolysis, affect their subcellular localization, or regulate their enzymatic activities. In addition to CDK-mediated inactivation of pre-RC components, in metazoans the APC mediates destruction of the Cdt1 inhibitor geminin in M and G1 phases, allowing pre-RC assembly precisely in that window of time. Variations on these regulatory events, with CDK-mediated phosphorylation inactivating components of the pre-RC and the APC inactivating CDKs, are found in all eukaryotes that have been studied.
The requirement in many species to limit replication initiation to once per cell cycle has led to the independent evolution of redundant, and remarkably similar, regulatory mechanisms in diverse organisms. The fundamental paradigm of oscillation between two mutually exclusive states as a component of control of replication initiation (e.g., high DnaA–low CtrA in Caulobacter and high APC–low CDK in eukaryotes) appears across distant domains of life as the driver behind origin licensing and replication initiation. The combination of this oscillation with the additional elements of spatial and temporal regulation via phosphorylation, proteolysis, and localization of key components produces a rigorous system with numerous fail-safes to restrict DNA replication initiation until all conditions necessary for successful completion are satisfied.
AFTER ENTRY INTO S PHASE: CHROMOSOME SEGREGATION AND ORGANIZATION
Timing of Replication and Segregation: All Things Are Not Equal
As we have noted, striking parallels exist in the logic, if not the specific proteins, that drives cell cycle progression in eukaryotes and bacteria. However, there are notable differences. One significant difference is the strict temporal separation of replication and segregation in eukaryotes (Philpott and Yew 2008); these processes are concurrent in Caulobacter and other bacteria (Viollier et al. 2004; Thanbichler and Shapiro 2006a).
The ability to synchronize Caulobacter cells enabled Viollier et al. (2004) to demonstrate concurrent replication and segregation directly by visualizing segregation in live cells. Using 10 separate strains, they showed that segregation of genomic loci follows a strict order that corresponds with the order of replication. Furthermore, comparing the timing of replication with that of segregation demonstrated that segregation does not wait for replication to finish. It is important to note that this distinction, with prokaryotes initiating segregation of loci immediately after they are replicated, is not absolute. An exception occurs in E. coli, where an origin-proximal portion of the genome undergoes “sister chromatid cohesion” for a short period of time and is segregated as a single unit (Bates and Kleckner 2005; Espéli et al. 2008). Once this origin macrodomain has segregated, however, the rest of the E. coli chromosome undergoes concurrent replication and segregation (Nielsen et al. 2006a).
In stark contrast to bacterial systems, eukaryotic entry into mitosis can only occur after all DNA replication is completed. In the presence of unreplicated DNA, the checkpoint inducer protein Chk1 initiates a phosphorylation signaling cascade that results in inactivation of mitotic CDK–cyclin and cell cycle arrest (Dasso and Newport 1990; Kumagai et al. 1998), thus assuring that eukaryotic DNA replication is completed before initiating mitosis.
Chromosome Segregation Is an Active Process
In addition to differences in the relative timing of segregation with respect to replication, a second major difference between the eukaryotic and prokaryotic cell cycles is commonly cited, namely, eukaryotes have a dedicated machinery that separates the chromosomes, whereas bacteria do not. However, in recent years, this view has been challenged.
In 1964, Jacob et al. published a possible mode of chromosome segregation in bacteria based on the hypothesis that the origin of replication is anchored to the cell envelope (Jacob et al. 1964). If, after replication, the cell wall grows in between two chromosome attachment sites, this model predicts that DNA separation could happen passively as a result of cell growth. This first and very influential model of DNA segregation was the accepted paradigm for a considerable time. With the advent of methods to visualize DNA loci in live bacterial cells using fluorescent fusions to sequence-specific DNA-binding proteins (Robinett et al. 1996; Straight et al. 1996; Webb et al. 1997; Nielsen et al. 2006b), however, several groups demonstrated that the movement of labeled DNA loci is too fast to be accounted for by cell growth (Mohl and Gober 1997; Webb et al. 1997; Viollier et al. 2004; Fogel and Waldor 2006), thus challenging the Jacob model. Furthermore, several species of bacteria have proteins that specifically affect the localization of segments of the chromosome (Ben-Yehuda et al. 2003; Yamaichi and Niki 2004; Gitai et al. 2005; Fogel and Waldor 2006; Bowman et al. 2008; Ebersbach et al. 2008), and in Caulobacter, the existence a centromere, i.e., a site of force exertion on the DNA during segregation, has been demonstrated (Toro et al. 2008). This centromere, the widely conserved parS sequence, is the only demonstrated exception to the rule of sequential segregation mentioned above. By necessity, the site of force exertion must move ahead of all other sites, and parS was shown to be invariantly the first locus segregated, even when the genomic distance, and therefore time of duplication, between the replication origin and parS was increased considerably. Interestingly, Caulobacter parS is situated very close to the origin of replication, and a comparative genomics study found that this arrangement is true of most bacteria (Livny et al. 2007). This suggests that concurrent replication and segregation may require the centromere to be placed near the origin of replication.
In short, at least some bacteria actively segregate their chromosomes and have a functional equivalent of a mitotic spindle. The detailed molecular mechanism that drives this movement is still unknown, but the ParA/ParB/parS partitioning system is likely to be involved (Fogel and Waldor 2006; Toro et al. 2008). In several bacterial species, however, the ParA/ParB/parS system is not essential, and some species (e.g., E. coli) do not have it at all, so bacteria must have evolved several independent DNA segregation systems.
The Chromosome Is Structured within the Nucleus and Nucleoid
An exciting recent development in cell biology has been the realization that chromosomal DNA is nonrandomly organized. In the case of bacteria, it was first discovered in Caulobacter and B. subtilis that the origin and terminus of replication held fixed positions within the cell (Mohl and Gober 1997; Webb et al. 1997). Later, a comprehensive study of 112 different loci in Caulobacter demonstrated that the cellular position of any given locus is linearly correlated with its position on the chromosome (Viollier et al. 2004), a finding that is likely to be true of other bacteria (Teleman et al. 1998; Niki et al. 2000). Interestingly, whereas B. subtilis and Caulobacter arrange their chromosomes so that the origin of replication is at one pole and the terminus at the other, E. coli arranges the left and right replichores along this axis, instead (Wang et al. 2006). The reason for this difference is unknown; an explanation will require an understanding of the mechanism that controls chromosome orientation within the cell. Toward this end, two factors have been identified that control chromosomal anchoring to the cell wall, one in B. subtilis and the other in Caulobacter (Ben-Yehuda et al. 2003; Bowman et al. 2008; Ebersbach et al. 2008; Toro et al. 2008). The relevant questions now are(1) how is chromosomal anchoring regulated and (2) are there other anchoring points in the cell?
In Caulobacter, the linear arrangement of genetic loci within the cell, and the simultaneous replication and segregation of these loci, is a factor in the temporal regulation of transcription of the dnaA and ctrA master regulator genes that orchestrate cell cycle progression (Reisenauer and Shapiro 2002; Collier et al. 2007). Replication initiates on a fully methylated chromosome. As the replication fork proceeds, the DNA copies become hemimethylated and remain in that state until the completion of DNA replication, because the CcrM DNA methyltransferase, which methylates the chromosome, is only present for a short period of time after completion of replication. The dnaA gene, which is adjacent to Cori on the chromosome, is preferentially transcribed from a fully methylated promoter. When the replication fork passes through dnaA, the two copies of the dnaA gene become hemimethylated, decreasing the transcription of dnaA. The ctrA gene, on the other hand, is farther from Cori and is preferentially transcribed from a hemimethylated promoter. This assures that ctrA transcription is activated later than dnaA, and the timing of activation of both transcripts is thus linked to the passage of the replication fork.
The realization that chromosomes are nonrandomly organized within the nucleus of eukaryotic cells originated from microirradiation experiments on synchronized hamster cells (Zorn et al. 1979). These authors showed that irradiation of a small section of the nucleus resulted in large amounts of damage to a small number of chromosomes, rather than a small amount of damage to all. This suggested that chromosomes occupy a small fraction of the nucleus, termed a “chromosome territory.” Later experiments confirmed this interpretation (Cremer et al. 1993; Croft et al. 1999) and showed that different cell types arrange their chromosomes in different configurations. For example, mouse hepatocytes tend to keep chromosomes 5 and 6 together and chromosomes 12 and 15 apart, a situation that is reversed in lymphocytes (Parada et al. 2004). This observation is particularly interesting because mouse hepatomas frequently show translocations between chromosomes 5 and 6, whereas lymphomas tend to show translocations between 12 and 15. A second striking example is found in the organization of heterochromatin in rod photoreceptors of nocturnal mammals. The majority of eukaryotic cells examined thus far localize the bulk of the heterochromatin to the nuclear periphery. Rod photoreceptors in nocturnal mammals, however, are an exception to this rule, and computer simulations suggest that this inverted nuclear organization works as a lens that channels light more efficiently to the light-sensing segments of the rod cells (Solovei et al. 2009). This remarkable finding underscores the fact that DNA organization within the cell is under strong selection not only for regulatory efficiency, but also for the physical implications of its structure.
DNA Mobility within the Nucleus
Localization is not the only aspect of chromosome organization that is strictly controlled. In bacteria, the mobility of DNA segments is restricted to subcellular domains (Fiebig et al. 2006). The spatial organization of the chromosome thus appears to favor certain DNA–DNA interactions much more than others. In the case of Caulobacter and B. subtilis, interactions that are symmetric around the origin–terminus axis are favored, whereas E. coli favors interactions perpendicular to this axis. Comparative genomics as well as experimental manipulations have shown that there is strong selection for equal length along the two arms of the bacterial chromosome (Esnault et al. 2007). Hence, the orientation of the chromosome will determine the relative likelihood of neutral versus deleterious inversions. Taken together, these findings paint a picture in which the three-dimensional organization of the DNA within the nucleoid is highly regulated and has an important role in maintaining the structural and functional integrity of the genome.
DIRECTING THE DIVISION PLANE
MipZ Communicates Information from the Chromosomes and the Poles to the Cell Division Machinery in Caulobacter
The final stage of the cell cycle is the physical separation of the cell into two daughter cells, or cytokinesis. Selection of the site of division is tightly controlled to assure that each daughter receives a full, undamaged copy of the genome. Bacterial cell division is mediated by the concerted action of a molecular machine called the divisome (Harry et al. 2006). The first protein to localize to the incipient division plane is the tubulin homolog FtsZ, which polymerizes into a ring structure (the Z ring) that forms the basis for assembly of the remainder of the divisome. The subcellular position of FtsZ assembly thereby dictates the site of cell division. In all bacteria that have been characterized, positioning of the Z ring involves negative regulators of FtsZ assembly that prevent division everywhere except mid cell. In Caulobacter, the locations of the duplicated chromosomes are intimately linked to the placement of the Z ring through a Walker A cytoskeletal ATPase (WACA) called MipZ that is conserved in α-proteobacteria (Thanbichler and Shapiro 2006b).
MipZ interacts directly with FtsZ in vitro and stimulates its GTPase activity, thereby promoting depolymerization (Thanbichler and Shapiro 2006b). This activity is spatially controlled in vivo so that the highest concentration of MipZ activity is at the cell poles and the lowest is near mid cell. MipZ is localized to the cell poles by virtue of its interaction with the ParB partitioning protein (Thanbichler and Shapiro 2006b), which, in turn, binds to both the PopZ polar anchoring protein (Bowman et al. 2008; Ebersbach et al. 2008) and the parS centromere (Mohl and Gober 1997; Toro et al. 2008). Importantly, the site of MipZ localization therefore reflects both the location of the centromere(s) and the locations of the cell poles. These interactions, combined with the dynamics of the chromosome, lead to a defined spatial arrangement of the MipZ and FtsZ proteins over the course of the cell cycle (Fig. 3A) (Thanbichler and Shapiro 2006b). Before duplication and segregation of parS, MipZ (along with ParB/PopZ) is observed at the stalked pole and FtsZ is found in a focus at the opposite new pole. Upon segregation and bipolarization of the parS/ParB/MipZ/PopZ complex, FtsZ depolymerizes at the new pole and reassembles at the site of lowest MipZ concentration in the cell, roughly the middle. The accuracy of this system relies on two characteristics of the Caulobacter chromosome: (1) The parS centromeres are anchored at the cell poles after duplication and segregation and (2) the single chromosome is duplicated exactly once per cell cycle.
Figure 3.
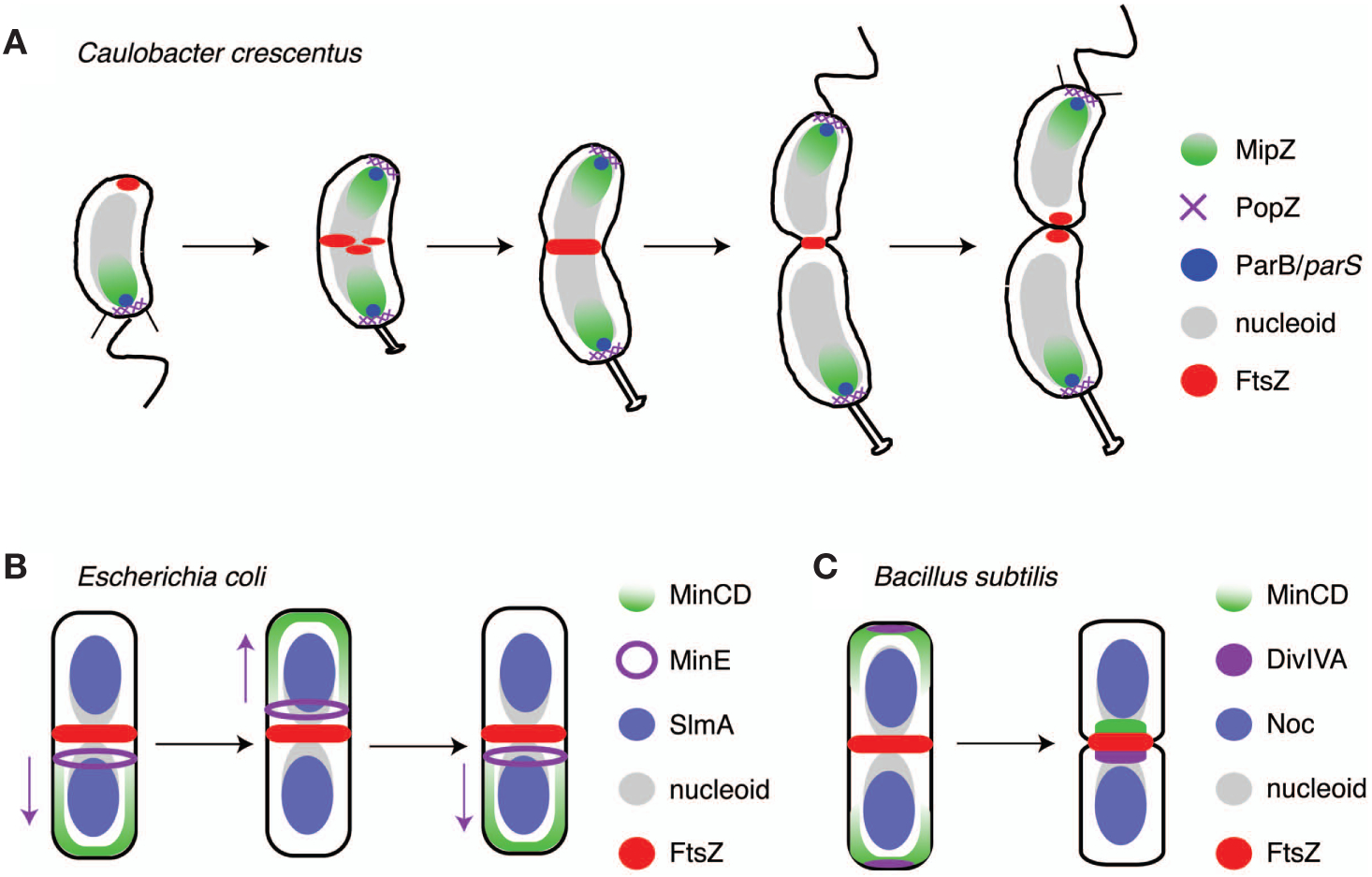
Placement of the division site integrates positional information from the poles and the chromosomes. (A) In Caulobacter, the MipZ complex is localized at the old pole and FtsZ is observed at the new pole. Upon segregation of parS, MipZ becomes bipolar, displacing FtsZ from the pole and targeting it to mid cell. (B) In E. coli, MinCD oscillates from pole to pole driven by MinE (which moves in the direction of the purple arrows). Additionally, SlmA on the segregated chromosomes inhibits FtsZ assembly over the bulk of the nucleoid. (C) B. subtilis also uses MinCD to inhibit polymerization of FtsZ next to newly formed poles but is localized by DivIVA. The activities of the Min system are complemented by Noc, which prevents Z-ring assembly over the chromosome(s).
Other Bacteria Use Separate Systems to Sense the Cell Poles and the Replicating Chromosome
Mechanisms of division site selection in E. coli and B. subtilis also reflect the positions of the cell poles and the segregated chromosomes. However, they achieve this using two separate mechanisms, each of which is distinct from MipZ (Fig. 3B,C). Polar assembly of FtsZ is inhibited in these organisms by MinC, a protein that directly interacts with FtsZ and antagonizes lateral interactions between protofilaments that are thought to be important for formation of a stable divisome (Dajkovic et al. 2008; Scheffers 2008). MinC localizes by interacting with the membrane-associated WACA MinD. In E. coli, MinCD oscillates from pole to pole driven by the action of a third protein, MinE (Fig. 3B) (Hu and Lutkenhaus 1999, 2001; Raskin and de Boer 1999a,b; Fu et al. 2001), whereas in B. subtilis, MinCD localizes by binding to DivIVA, a protein unrelated to MinE that localizes to the septum and to the new pole (Fig. 3C) (Marston et al. 1998). Unlike MipZ, MinCD is not essential in E. coli or B. subtilis because a second system is in place to inhibit FtsZ assembly by a nucleoid occlusion mechanism. Nucleoid occlusion functions in E. coli and B. subtilis through unrelated effectors that act as negative regulators of Z-ring assembly: SlmA in the former and Noc in the latter (Wu and Errington 2004; Bernhardt and de Boer 2005). In both cases, the nucleoid occlusion protein associates asymmetrically with the chromosome(s) and inhibits assembly of the FtsZ ring on the membrane around the nucleoid (Fig. 3B,C). This limits initiation of cell division to areas of low DNA concentration, either close to the poles (which is inhibited by MinCD and therefore disallowed) or between the chromosomes after segregation.
Ancestors to the α-proteobacteria probably originally possessed MinC and/or MinD and lost them, because these proteins are present in diverse species including other classes of proteobacteria, Gram-positive organisms, and even some chloroplasts. MipZ may have arisen originally as a gene duplication of another WACA protein such as ParA. It could then adapt to direct FtsZ localization while the mechanisms allowing polar positioning of parS were evolved, eliminating the need for MinCD. It has not been determined if MinCD artificially expressed in Caulobacter can oscillate from pole to pole, but we speculate that the distinct biochemical identities of the poles in Caulobacter might affect the efficiency of such a system. Moreover, the MipZ division plane positioning mechanism allows FtsZ to assemble over the bulk of the nucleoid early in the cell cycle, where it can direct both elongation of the cell before invagination at the division site (Aaron et al. 2007) and the process of division. A nucleoid occlusion system would eliminate the ability of FtsZ to direct the early phase of growth at mid cell, and this activity may have supplied selective pressure for eliminating nucleoid occlusion in Caulobacter or for preventing its inception. On the other hand, MipZ alone would not function efficiently in E. coli or B. subtilis because these organisms can have multiple rounds of chromosome replication occurring per cell division. In Caulobacter, the cell division regulatory mechanism may in turn select against mutations that alter positioning of the centromere or that allow reinitiation of DNA replication before division in α-proteobacteria, leading to stable coupling of chromosome dynamics and division site selection.
CONCLUSIONS
We have described here the extraordinary lengths to which all cells go to preserve the integrity of their genomes. Intricate timing and synchronization mechanisms ensure accurate progression of the cell cycle into S phase only when necessary precursor processes are complete and nutrients are adequate. During and after replication, the chromosomes are highly organized and are actively shaped and moved during segregation. Finally, cell division occurs at a time and place in the cell that reflects constraints communicated from the replicated, segregated chromosomes. These general rules for preserving genome integrity are found throughout all domains of life, and the conservation of the regulatory logic and mechanisms underpinning cell cycle control in diverse organisms is truly remarkable.
In general, however, it is problematic that the great majority of our cell biological knowledge comes from a handful of organisms, particularly in the case of bacteria. The fact that E. coli and B. subtilis diverged about 1.5 billion years ago (Ochman and Wilson 1987) underscores the immense molecular variation that is contained in prokaryotes, but which is vastly underexploited. Exploration into other, divergent organisms will yield new insights into the conservation and evolution of the regulatory logic and genetic circuit design that drives cell cycle progression.
ACKNOWLEDGMENTS
Work in the L.S. and H.H.M. laboratories is supported in part by National Institutes of Health grants R01 GM51426 R24 and GM073011-04 (to L.S.) and Department of Energy grant DE-FG02-05ER64136 (to H.H.M. and L.S.). E.T. was funded by the Smith Stanford Graduate Fellowship. E.D.G. is a Helen Hay Whitney postdoctoral fellow.
REFERENCES
- Aaron M, Charbon G, Lam H, Schwartz H, Vollmer W, Jacobs-Wagner C. 2007. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol 64: 938–952. [DOI] [PubMed] [Google Scholar]
- Arias EE, Walter JC. 2007. Strength in numbers: Preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev 21: 497–518. [DOI] [PubMed] [Google Scholar]
- Bates D, Kleckner N. 2005. Chromosome and replisome dynamics in E. coli: Loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell 121: 899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Dutta A. 2002. DNA replication in eukaryotic cells. Annu Rev Biochem 71: 333–74. [DOI] [PubMed] [Google Scholar]
- Bellefontaine AF, Pierreux CE, Mertens P, Vandenhaute J, Letesson JJ, De Bolle X. 2002. Plasticity of a transcriptional regulation network among α-proteobacteria is supported by the identification of CtrA targets in Brucella abortus. Mol Microbiol 43: 945–960. [DOI] [PubMed] [Google Scholar]
- Ben-Yehuda S, Rudner DZ, Losick R. 2003. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299: 532–536. [DOI] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PA. 2005. SlmA, a nucleoid-associated,FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol Cell 18: 555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT. 2006. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature 444: 899–904. [DOI] [PubMed] [Google Scholar]
- Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, Downing KH, Moerner WE, Earnest T, Shapiro L. 2008. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell 134: 945–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilla-Llorente V, Munoz-Espin D, Villar L, Salas M, Meijer WJ. 2006. Spo0A, the key transcriptional regulator for entrance into sporulation, is an inhibitor of DNA replication. EMBO J 25: 3890–3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien P, Perchuk BS, Laub MT, Sauer RT, Baker TA. 2007. Direct and adaptor-mediated substrate recognition by an essential AAA+ protease. Proc Natl Acad Sci 104: 6590–6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier J, Shapiro L. 2009. Feedback control of DnaA-mediated replication initiation by replisome-associated HdaA protein in Caulobacter. J Bacteriol (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier J, Murray SR, Shapiro L. 2006. DnaA couples DNA replication and the expression of two cell cycle master regulators. EMBO J 25: 346–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier J, McAdams HH, Shapiro L. 2007. A DNA methylation ratchet governs progression through a bacterial cell cycle. Proc Natl Acad Sci 104: 17111–17116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Kurz A, Zirbel R, Dietzel S, Rinke B, Schröck E, Speicher MR, Mathieu U, Jauch A, Emmerich P, et al. 1993. Role of chromosome territories in the functional compartmentalization of the cell nucleus. Cold Spring Harbor Symp Quant Biol 58: 777–792. [DOI] [PubMed] [Google Scholar]
- Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA. 1999. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol 145: 1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajkovic A, Lan G, Sun SX, Wirtz D, Lutkenhaus J. 2008. MinC spatially controls bacterial cytokinesis by antagonizing the scaffolding function of FtsZ. Curr Biol 18: 235–244. [DOI] [PubMed] [Google Scholar]
- Dasso M, Newport JW. 1990. Completion of DNA replication is monitored by a feedback system that controls the initiation of mitosis in vitro: Studies in Xenopus. Cell 61: 811–823. [DOI] [PubMed] [Google Scholar]
- Degnen ST, Newton A. 1972. Chromosome replication during development in Caulobacter crescentus. J Mol Biol 64: 671–680. [DOI] [PubMed] [Google Scholar]
- Diffley JF. 2004. Regulation of early events in chromosome replication. Curr Biol 14: R778–R786. [DOI] [PubMed] [Google Scholar]
- Domian IJ, Quon KC, Shapiro L. 1997. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 90: 415–424. [DOI] [PubMed] [Google Scholar]
- Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. 2009. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev 23: 93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. 2008. A self-associating protein critical for chromosome attachment, division, and polar organization in Caulobacter. Cell 134: 956–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault E, Valens M, Espéli O, Boccard F. 2007. Chromosome structuring limits genome plasticity in Escherichia coli. PLoS Genet 3: e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espéli O, Mercier R, Boccard F. 2008. DNA dynamics vary according to macrodomain topography in the E. coli chromosome. Mol Microbiol 68: 1418–1427. [DOI] [PubMed] [Google Scholar]
- Fiebig A, Keren K, Theriot JA. 2006. Fine-scale time-lapse analysis of the biphasic, dynamic behaviour of the two Vibrio cholerae chromosomes. Mol Microbiol 60: 1164–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel MA, Waldor MK. 2006. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev 20: 3269–3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Shih YL, Zhang Y, Rothfield LI. 2001. The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia coli division cycle. Proc Natl Acad Sci 98: 980–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120: 329–341. [DOI] [PubMed] [Google Scholar]
- Gorbatyuk B, Marczynski GT. 2005. Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol Microbiol 55: 1233–1245. [DOI] [PubMed] [Google Scholar]
- Harry E, Monahan L, Thompson L. 2006. Bacterial cell division: The mechanism and its precison. Int Rev Cytol 253: 27–94. [DOI] [PubMed] [Google Scholar]
- Holtzendorff J, Hung D, Brende P, Reisenauer A, Viollier PH, McAdams HH, Shapiro L. 2004. Oscillating global regulators control the genetic circuit driving a bacterial cell cycle. Science 304: 983–987. [DOI] [PubMed] [Google Scholar]
- Hottes AK, Shapiro L, McAdams HH. 2005. DnaA coordinates replication initiation and cell cycle transcription in Caulobacter crescentus. Mol Microbiol 58: 1340–1353. [DOI] [PubMed] [Google Scholar]
- Hu Z, Lutkenhaus J. 1999. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol Microbiol 34: 82–90. [DOI] [PubMed] [Google Scholar]
- Hu Z, Lutkenhaus J. 2001. Topological regulation of cell division in E. coli. Spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol Cell 7: 1337–1343. [DOI] [PubMed] [Google Scholar]
- Iniesta AA, McGrath PT, Reisenauer A, McAdams HH, Shapiro L. 2006. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc Natl Acad Sci 103: 10935–10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Brenner S, Cuzin F. 1964. On the regulation of DNA replication in bacteria. Cold Spring Harbor Symp Quant Biol 28: 329–348. [Google Scholar]
- Jacobs C, Domian IJ, Maddock JR, Shapiro L. 1999. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell 97: 111–120. [DOI] [PubMed] [Google Scholar]
- Jenal U, Fuchs T. 1998. An essential protease involved in bacterial cell-cycle control. EMBO J 17: 5658–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB. 2006. Coordination between chromosome replication, segregation, and cell division in Caulobacter crescentus. J Bacteriol 188: 2244–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd EM, Ryan KR, Moerner WE, Shapiro L, McAdams HH. 2003. Fluorescence bleaching reveals asymmetric compartment formation prior to cell division in Caulobacter. Proc Natl Acad Sci 100: 8235–8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J, Katayama T. 2001. Had, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J 20: 4253–4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Guo Z, Emami KH, Wang SX, Dunphy WG. 1998. The Xenopus Chk1 protein kinase mediates a caffeine-sensitive pathway of checkpoint control in cell-free extracts. J Cell Biol 142: 1559–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L. 2000. Global analysis of the genetic network controlling a bacterial cell cycle. Science 290: 2144–2148. [DOI] [PubMed] [Google Scholar]
- Laub MT, Chen SL, Shapiro L, McAdams HH. 2002. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc Natl Acad Sci 99: 4632–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livny J, Yamaichi Y, Waldor MK. 2007. Distribution of centromere-like parS sites in bacteria: Insights from comparative genomics. J Bacteriol 189: 8693–8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston AL, Thomaides HB, Edwards DH, Sharpe ME, Errington J. 1998. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev 12: 3419–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdams HH, Srinivasan B, Arkin AP. 2004. The evolution of genetic regulatory systems in bacteria. Nat Rev Genet 5: 169–178. [DOI] [PubMed] [Google Scholar]
- McGrath PT, Iniesta AA, Ryan KR, Shapiro L, McAdams HH. 2006. A dynamically localized protease complex and a polar specificity factor control a cell cycle master regulator. Cell 124: 535–547. [DOI] [PubMed] [Google Scholar]
- Mohl DA, Gober JW. 1997. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell 88: 675–684. [DOI] [PubMed] [Google Scholar]
- Mott ML, Berger JM. 2007. DNA replication initiation: Mechanisms and regulation in bacteria. Nat Rev Microbiol 5: 343–354. [DOI] [PubMed] [Google Scholar]
- Nielsen HJ, Li Y, Youngren B, Hansen FG, Austin S. 2006a. Progressive segregation of the Escherichia coli chromosome. Mol Microbiol 61: 383–393. [DOI] [PubMed] [Google Scholar]
- Nielsen HJ, Ottesen JR, Youngren B, Austin SJ, Hansen FG. 2006b. The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol Microbiol 62: 331–338. [DOI] [PubMed] [Google Scholar]
- Niki H, Yamaichi Y, Hiraga S. 2000. Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev 14: 212–223. [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Wilson AC. 1987. Evolution in bacteria: Evidence for a universal substitution rate in cellular genomes. J Mol Evol 26: 74–86. [DOI] [PubMed] [Google Scholar]
- Parada LA, McQueen PG, Misteli T. 2004. Tissue-specific spatial organization of genomes. Genome Biol 5: R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott A, Yew PR. 2008. The Xenopus cell cycle: An overview. Mol Biotechnol 39: 9–19. [DOI] [PubMed] [Google Scholar]
- Piggot PJ, Hilbert DW. 2004. Sporulation of Bacillus subtilis. Curr Opin Microbiol 7: 579–586. [DOI] [PubMed] [Google Scholar]
- Quon KC, Yang B, Domian IJ, Shapiro L, Marczynski GT. 1998. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci 95: 120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin DM, de Boer PA. 1999a. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J Bacteriol 181: 6419–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin DM, de Boer PA. 1999b. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci 96: 4971–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisenauer A, Shapiro L. 2002. DNA methylation affects the cell cycle transcription of the CtrA global regulator in Caulobacter. EMBO J 21: 4969–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS. 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol 135: 1685–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KR, Judd EM, Shapiro L. 2002. The CtrA response regulator essential for Caulobacter crescentus cell-cycle progression requires a bipartite degradation signal for temporally controlled proteolysis. J Mol Biol 324: 443–455. [DOI] [PubMed] [Google Scholar]
- Scheffers DJ. 2008. The effect of MinC on FtsZ polymerization is pH dependent and can be counteracted by ZapA. FEBS Lett 582: 2601–2608. [DOI] [PubMed] [Google Scholar]
- Shaheen SM, Ouimet MC, Marczynski GT. 2009. Comparative analysis of Caulobacter chromosome replication origins. Microbiology 155: 1215–1225. [DOI] [PubMed] [Google Scholar]
- Slater S, Wold S, Lu M, Boye E, Skarstad K, Kleckner N. 1995. E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell 82: 927–936. [DOI] [PubMed] [Google Scholar]
- Solovei I, Kreysing M, Lanctot C, Kosem S, Peichl L, Cremer T,Guck J, Joffe B. 2009. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 137: 356–368. [DOI] [PubMed] [Google Scholar]
- Straight AF, Belmont AS, Robinett CC, Murray AW. 1996. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol 6: 1599–1608. [DOI] [PubMed] [Google Scholar]
- Teleman AA, Graumann PL, Lin DC, Grossman AD, Losick R. 1998. Chromosome arrangement within a bacterium. Curr Biol 8: 1102–1109. [DOI] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L. 2006a. Chromosome organization and segregation in bacteria. J Struct Biol 156: 292–303. [DOI] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L. 2006b. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126: 147–162. [DOI] [PubMed] [Google Scholar]
- Toro E, Hong SH, McAdams HH, Shapiro L. 2008. Caulobacter requires a dedicated mechanism to initiate chromosome segregation. Proc Natl Acad Sci 105: 15435–15440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L. 2004. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci 101: 9257–9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Liu X, Possoz C, Sherratt DJ. 2006. The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev 20: 1727–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CD, Teleman A, Gordon S, Straight A, Belmont A, Lin DC, Grossman AD, Wright A, Losick R. 1997. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell 88: 667–674. [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J. 2004. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117: 915–925. [DOI] [PubMed] [Google Scholar]
- Yamaichi Y, Niki H. 2004. migS, a cis-acting site that affects bipolar positioning of oriC on the Escherichia coli chromosome. EMBO J 23: 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn C, Cremer C, Cremer T, Zimmer J. 1979. Unscheduled DNA synthesis after partial UV irradiation of the cell nucleus. Distribution in interphase and metaphase. Exp Cell Res 124: 111–119. [DOI] [PubMed] [Google Scholar]


