Abstract
The detection and quantification of biomarkers have numerous applications in biological research and medicine. The most widely used methods to detect nucleic acids require amplification via the polymerase chain reaction (PCR). However, errors arising from the imperfect copying fidelity of DNA polymerases, limited specificity of primers, and heat-induced damage reduce the specificity of PCR-based methods, particularly for single-nucleotide variants. Furthermore, not all analytes can be amplified efficiently. While amplification-free methods avoid these pitfalls, the specificity of most such methods is strictly constrained by probe binding thermodynamics, which for example hampers detection of rare somatic mutations. In contrast, single-molecule recognition through equilibrium Poisson sampling (SiMREPS) provides ultraspecific detection with single-molecule and single-nucleotide sensitivity by monitoring the repetitive interactions of a fluorescent probe with surface-immobilized targets. In this review, we discuss SiMREPS in comparison with other analytical approaches, and describe its utility in quantifying a range of nucleic acids and other analytes.
Keywords: Disease biomarker, single-molecule fluorescence detection, polymerase chain reaction, kinetic fingerprinting, single-nucleotide variant, single-nucleotide polymorphism
1. Introduction
1.1. Amplification-based nucleic acid detection and its limitations
Detection and/or quantification of nucleic acid biomarker sequences is crucial for applications as diverse as prenatal diagnostics [1], gene expression analysis [2], identification of genetic risk factors in disease [3], detection of sequence polymorphisms [4], quantification of telomerase activity [5], detection of pathogens [6], and identification of genetically-modified organisms [7]. Highly specific detection of low-abundance single-nucleotide variants (SNVs) in a large excess of wild-type sequences is increasingly important in clinical diagnostics of cancer and other diseases [8, 9]. Several techniques, most of which rely on the specific hybridization between the target nucleic acid molecule and one or more sequence-specific probes, have been developed to detect such low-abundance SNVs.
The most widely used methods to detect nucleic acids are based on PCR [10]. Quantitative PCR (qPCR)-based assays exploit the differential hybridization efficiency of primers and probes to detect homologous nucleic acid sequences [11]. In contrast, next-generation sequencing (NGS) technologies digitally tabulate the sequence of many individual nucleic acid fragments, offering the unique ability to detect nucleic acid sequences within heterogeneous mixtures without prior knowledge [12]; however, to detect rare SNVs, NGS typically depends on high sequencing depth, at considerable increase in cost, and/or targeted PCR of specific genetic loci.
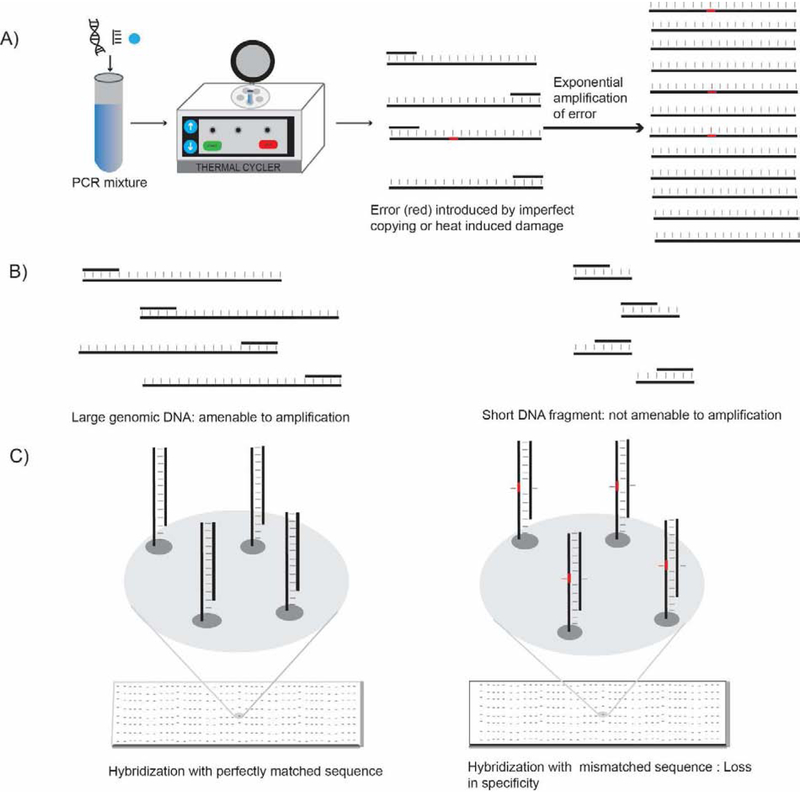
Two crucial performance parameters of any method for the detection and/or quantification of nucleic acids are sensitivity and specificity. By virtue of amplification, qPCR and droplet digital PCR (ddPCR) can achieve detection limits in the low attomolar range (2–5 copies/μL) [13]. However, PCR amplification introduces errors caused by heat-induced chemical damage (i.e. nucleotide deamination or oxidation) and imperfect copying fidelity of DNA polymerases [14–17] (Fig. 1A). Such errors can be exponentially amplified during thermal cycling in PCR. Thus, the necessity of amplification in PCR-based methods imposes limits on the specificity of nucleic acid detection, particularly for SNVs. While tagging individual molecular copies of nucleic acid molecules with unique identifier sequences (UIDs) prior to amplification for NGS can help to distinguish genuine SNVs from amplification artifacts [18], amplification-based methods still suffer from several other limitations, including:
PCR amplification of heterogeneous nucleic acid mixtures can give rise to differential amplification, affecting the relative abundance of sequences in the final amplified products and introducing systematic bias in quantification [19].
Contaminants in clinical samples such as heparin and heme can inhibit the activity of polymerases and ligases, necessitating additional purification steps prior to amplification [20, 21].
Direct amplification of epigenetic modifications, of short or fragmented nucleic acids, or of non-nucleic acid analytes, is difficult or impossible (Fig. 1B).
Adding UIDs to enable rare allele detection using NGS approaches increases the required depth of sequencing several-fold, significantly increasing the overall cost.
Fig. 1.
Shortcomings of PCR-based and hybridization-based methods for detecting nucleic acids. (A) Generation and exponential amplification of erroneous sequences causes false positive signal in PCR-based assays. (B) Short DNA or RNA fragments (<30 bp) are typically not amenable to amplification in PCR-based assays. (C) Low specificity of hybridization-based techniques arises from significant nonspecific binding of probes to non-cognate sequences.
1.2. Amplification-free nucleic acid detection methods and their limitations
To alleviate the limitations introduced by amplification while preserving high analytical sensitivity, several amplification-free single-molecule nucleic acid detection methods have been developed. One such method, termed associating and dissociating nanodimer analysis (ADNA), detects the change in plasmonic coupling-based scattering intensity due to DNA-assisted, dynamic association and dissociation of nanoparticles on a lipid micropattern [22]. The nCounter (NanoString) platform utilizes molecular barcodes and single-molecule imaging for the direct hybridization and detection of nucleic acids immobilized on an imaging surface [23]. Single-molecule arrays (Simoa), on the other hand, isolate single nucleic acid molecules in femtolitre-sized reaction wells and detect them by quantifying the fluorescent product generated by a signal-amplifying enzyme within each well containing at least one target molecule, analogous to ddPCR [24].
While these methods avoid amplification-related copying errors and permit detection of targets that cannot be efficiently amplified by PCR, they suffer from different sets of challenges. First, they generally cannot detect SNVs at relative abundances below ~0.1% (and sometimes 1–10%) due to a finite thermodynamic discrimination factor between closely related sequences [25] (Fig. 1C). This fundamental limitation of thermodynamic specificity is given by the parameter , where ΔΔG0 is the difference in the Gibbs free energy of hybridization of a particular detection probe to a target sequence and to a related but spurious target sequence. Depending on the specific sequence, Qmax,therm for SNVs can be as high as 104 or as low as ~20 [25].
Second, amplification-free methods rarely achieve the ability to detect single molecules due to their inability to completely suppress nonspecific binding of probes (e.g., fluorescent probes or signal-generating enzymes) to the detection surface. The lower limit of detection of target nucleic acids using these amplification-free methods ranges from 22 aM – 200 fM, with detection limits in the fM range significantly more common (Table 1) [22, 24, 26, 27].
Table 1.
Nucleic acid detection technologies and their analytical performance.
| Technology | Quantitative | Amplification | Sensitivity (dynamic range) | Single-Nucleotide Specificity | Multiplexing ability | Instrumentation | Reagents |
|---|---|---|---|---|---|---|---|
| qRT-PCR[13, 27, 30–32] | Yes | Amplification-based | zM to fM | >95% | Limited | Real-time PCR instrument (e.g., ThermoFisher QuantStudio. Bio-Rad CFX96) | Extraction kits, primers, fluorogenic probes, thermostable polymerase, dNTPs |
| ddPCR [27, 33] | Yes | Amplification-based | zM to fM | 99.99% | Limited | Droplet generator + droplet reader (e.g., Bio-Rad QX100), thermal cycler | Extraction kits, droplet generation reagents, primers, fluorogenic probes, thermostable polymerase |
| Illumina sequencing (TruSeq) [27] | Yes | Amplification-based | zM to fM | >99% | High | Illumina sequencer (e.g., MiSeq, HiSeq, NovaSeq), thermal cycler | Extraction kits, library preparation kits, fluorescent dNTPs, thermostable polymerase, flow cells |
| Microarray[27, 31] | Yes | Amplification-free | nM to mM | 50–80% | High | Microarray plate scanner (e.g., AffyMetrix GeneChip HT) | Extraction and PCR/labeling kits, microarray chips |
| ADNA [22] | Yes | Amplification-free | aM to fM | Not available | Not available | Dark field microscope (e.g., Zeiss Axiovert 200M) | Probe-conjugated nanoparticles, patterned lipid arrays |
| NanoString [27] | Yes | Amplification-free | fM to nM | 90–95% | High | nCounter system (e.g., nCounter MAX) | Flow cells, capture probes, fluorescently barcoded affinity probes |
| Simoa [24] | Yes | Amplification-free | fM to pM | >95% | Limited | Simoa analyzer (e.g., HD-1, SR-X) | Probe-conjugated microparticles, droplet generation reagents, primary and secondary detection probes, enzymatic signal developing reagents |
| Time gated FRET ligation [26] | Yes | Amplification-free | pM to nM | Not available | Limited | Fluorescence plate reader (e.g., ThermoFisher KRYPTOR) | FRET probes, adaptor probes, DNA or RNA ligase |
| SiMREPS [17] | Yes | Amplification-free | fM to pM | ≥99.99999% | Limited | TIRF microscope (e.g., Olympus IX83, Oxford NanoImager) | Flow cells, capture probes, fluorescent probes |
1.3. Single-molecule kinetic fingerprinting: an ultraspecific amplification-free method for nucleic acid detection
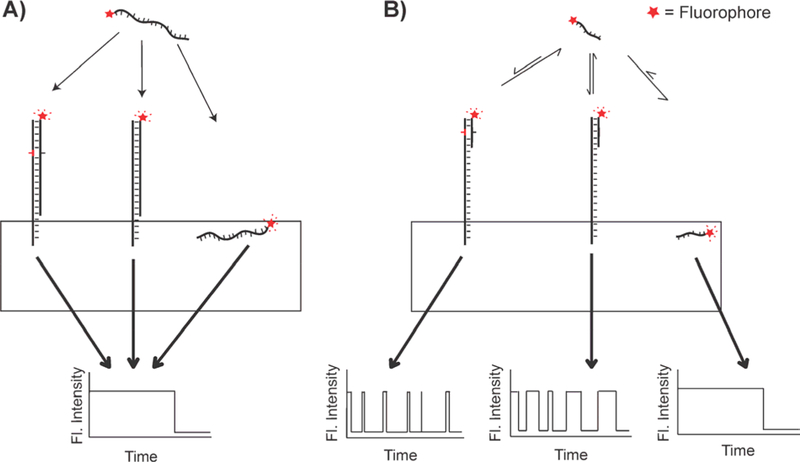
To detect nucleic acids without amplification and without being bound by the thermodynamic limits of specificity, a new approach termed single molecule recognition through equilibrium Poisson sampling (SiMREPS) has been developed [17, 28, 29]. In SiMREPS, instead of detecting the total fluorescence originating from the (irreversible) binding of a fluorescently labeled probe to the target nucleic acid (Fig. 2A), the repeated transient interaction of a fluorescent probe with the surface-bound target generates a time-dependent signal of fluctuations in fluorescence (Fig. 2B), or “kinetic fingerprint,” that is highly sensitive to even small differences in free energy of binding to the probe. This characterization in SiMREPS of the interaction kinetics over many cycles of probe-target binding thus provides exquisite discrimination between the specific binding of probes to the target analyte and non-specific binding to the detection surface or other non-target species, overcoming the challenge of other amplification-free single-molecule detection methods to achieve single-molecule sensitivity combined with high specificity for SNVs [17, 28, 29].
Fig. 2.
Kinetic fingerprinting with transient probing improves specificity. (A) The irreversible interaction between a fluorophore-labeled probe and a surface-bound target molecule is indistinguishable from nonspecific binding to spurious sequences on the surface, or to the surface itself. (B) The reversible interaction between a short fluorophore-labeled oligonucleotide probe and a surface bound target generates a kinetic fingerprint (fluorescence intensity versus time trace) that can be distinguished from nonspecific binding.
As the binding interaction of a fluorescent probe to a target nucleic acid can be modeled by Poisson statistics, the number of probe binding and dissociation events observed for each target molecule (Nb+d) will, on average, increase linearly with observation time. Thus, given an adequate number of binding events, probes with finite thermodynamic discrimination can be used to detect a target nucleic acid with arbitrarily high specificity. Using kinetic information, SiMREPS thus realizes the amplification-free detection of single target DNA molecules with a specificity of 99.99999% (Table 1) and an apparent discrimination factor (Qapp >106), exceeding Qmax,therm by more than two orders of magnitude [17], making the technique of great potential utility in clinical diagnostic applications, particularly settings that require specific detection of rare SNVs.
In this review, we will describe both theoretical and practical considerations for the ultraspecific detection of biomarkers using SiMREPS. We will also discuss the progress made towards the application of SiMREPS to the detection of various nucleic acid and non-nucleic acid analytes, and conclude by suggesting several improvements in performance and analyte scope that may be forthcoming in the near future.
2. Experimental considerations for single-molecule kinetic fingerprinting by SiMREPS
SiMREPS achieves extremely high specificity in single-molecule detection by monitoring the repeated binding of fluorescent probes to individual target molecules over time [29], which typically requires the target molecules to be captured at or near the surface of a microscope slide or coverslip for time-resolved imaging. This kinetic fingerprint consists of a number of metrics that are further elaborated below, and provides the ability to distinguish even nucleic acids that differ by only a single nucleotide [17, 29].
2.1. Analyte scope
Since SiMREPS does not rely on any nucleic acid-specific chemistry or enzymatic manipulation (e.g., PCR), it should be possible to quantify a broad range of analytes by SiMREPS. In principle, any analyte that can be immobilized at a surface – preferably via a specific interaction – and remain free to transiently recruit fluorescent detection probes from solution can be detected by SiMREPS. While the assays reviewed here are limited to short nucleic acids like miRNA, ~22–160 bp ssDNA or dsDNA, and small molecules like adenosine, acetamiprid and the toxin PCB-77 [28, 34], it is likely that SiMREPS will be adapted to the quantification of other classes of analyte in the future.
2.2. Sample Preparation
Sample preparation for SiMREPS consists primarily of preparation of a microscope slide or coverslip, surface capture of the analyte, and then introduction of an imaging solution containing a fluorescent probe (FP) for kinetic fingerprinting by single molecule fluorescence microscopy [28]. The first consideration is the type of slide and sample cell design to use for imaging. This is determined by two main factors: the sensitivity required, and the type of microscopy used. Single-molecule imaging is usually performed with total internal reflection fluorescence microscopy (TIRFM) [35, 36], where coherent excitation light is directed onto the microscope slide or coverslip at an angle shallow enough to result in complete reflection of the light from the glass-solution interface. Although the light is completely reflected, a so-called evanescent wave is generated at that interface that penetrates a very short distance (~100 nm) into the solution, which permits the efficient excitation of only those fluorophores that are at or near the surface of the slide. This approach greatly facilitates single-molecule detection by reducing background fluorescence from the bulk of the sample, which would otherwise overwhelm the relatively low-intensity emission from single fluorescent dye molecules. This single-molecule sensitivity is critical for SiMREPS, which requires separately characterizing the kinetics of probe binding to individual target molecules at equilibrium. The two most common approaches used are prism-type TIRF (pTIRF) and objective-type TIRF (oTIRF), and further discussion of these approaches can be found elsewhere [37, 38].
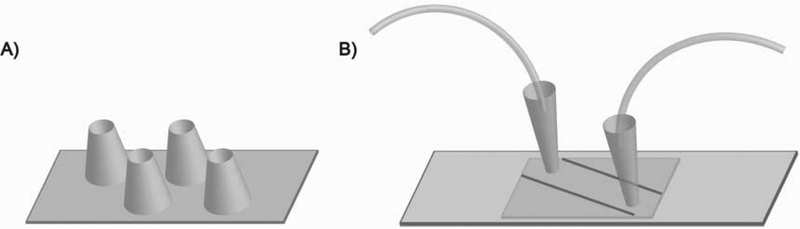
The two most common types of sample cell usable for SiMREPS are the slide-coverslip sandwich flow cell and the coverslip with cylindrical wells (Fig. 3). While the flow cell is usable on both pTIRF and oTIRF microscopes, the cylindrical wells are only usable on oTIRF microscopes due to their height. However, the cylindrical wells are preferred for high-sensitivity applications because they permit the capture of a larger number of molecules over a smaller area of the coverslip. A more detailed discussion can be found elsewhere [28].
Fig. 3.
Typical sample cell designs used in SiMREPS. (A) The coverslip with cylindrical wells uses a cleaned coverslip with cut pipette tips glued into position as wells for introduction of sample and imaging buffer. This design does not permit placement of a prism on top, and is thus only compatible with oTIRF. It generally provides much higher sensitivity because of the smaller surface area over which molecules are distributed. (B) The slide-coverslip sandwich flow cell uses a cleaned slide with two holes that is attached to a coverslip via double-sided tape and sealed around the edges with fast-curing epoxy adhesive, forming a channel for sample delivery. Cut pipette tips are placed into the holes and glued into place to provide an inlet and outlet for sample and imaging solutions. Tygon tubing is placed into the pipette tips and sealed with epoxy glue to allow injection of sample with a syringe. This design provides generally lower sensitivity, but is compatible with both oTIRF and pTIRF.
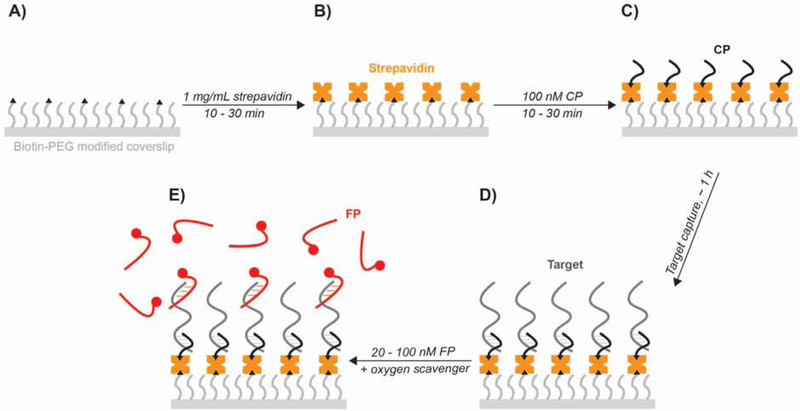
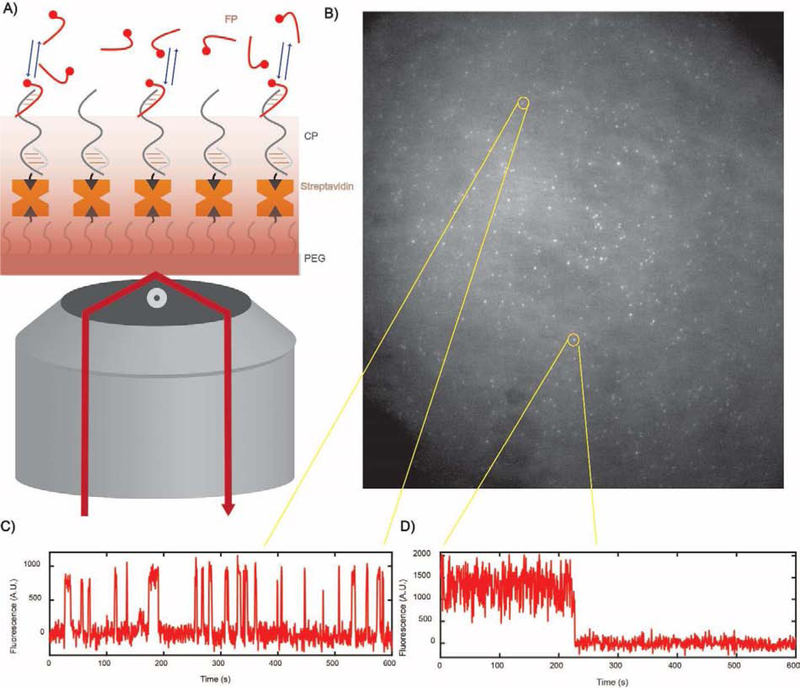
Prior to an experiment, the surface of the slide or coverslip is first stringently cleaned and functionalized with a biotin-containing moiety such as biotinylated polyethylene glycol (biotin-PEG) or biotinylated bovine serum albumin (bBSA). A multivalent avidin family molecule such as streptavidin is then added and bound to the biotinylated surface, providing an anchor point for the binding of a biotinylated capture probe (CP) that is complementary to the target (Fig. 4). The CP is then added to the sample cell for immobilization; optimal incubation times and concentration of the CP will vary depending on system-specific factors such as the accessibility of the biotin in the CP, but typically 100 nM of CP is incubated for 10–30 min.
Fig. 4.
Preparing the sample for imaging. (A) The slide or coverslip is functionalized with a biotin-containing moiety such as biotin-PEG or biotin-BSA. (B) An avidin family protein such as streptavidin is added to provide an anchor point for CP immobilization. (C) The CP is added and allowed to bind to streptavidin to permit capture of the target. (D) The target is added and incubated to allow capture at the surface. (E) The FP is added in an imaging solution containing an oxygen scavenger system to permit single-molecule kinetic fingerprinting through repetitive binding of the FP to the target.
In most cases where an assay with single-molecule sensitivity is desired, analyte concentrations will rarely exceed the low picomolar range; if this is the case, it is unlikely any sample dilution will be required. Furthermore, super-resolution analysis methods (see Section 5.1) can accommodate concentrations varying from ~1 fM to ~100 pM without varying dilution factors. However, for analytes that are routinely present at concentrations above 100 pM, prior dilution of the sample may be required to ensure that analytes are captured at surface densities that may be resolved with wide-field fluorescence methods. Standard curves will provide guidance as to the dynamic range for a given analyte and matrix, and whether any dilution is required for a given sample type.
Next, the target is incubated in the sample cell for approximately 1 h to allow binding to the CP. To inhibit degradation of nucleic acid analytes or sensors by nucleases, and to liberate any nucleic acid analytes from interfering binding partners, prior sample preparation steps are often necessary prior to this capture step, particularly for samples containing minimally processed biofluids [39]. Since many nucleases require divalent cations (i.e., Mg2+, Ca2+) as cofactors, the addition of a chelating agent such as EDTA is useful to prevent degradation of nucleic acids [40]. For direct capture of nucleic acids from crude biofluids such as cell extracts or serum, a pre-incubation step in ~2% (w/v) sodium dodecyl sulfate (SDS) and 0.16 U/μL of proteinase K (New England BioLabs, Inc.) has been used to liberate nucleic acids from any protein binding partners as well as to inactivate any nucleases present in the sample [29].
Another important consideration in the application of SiMREPS to the detection of nucleic acid analytes is to avoid the formation of duplexes or secondary structures in the analytes that would inhibit binding of the CP or FP. Therefore, a short denaturation period by heating and sequestering interfering sequences is often employed before immobilizing analytes to the surface. This is especially important for long nucleic acid analytes that have a higher propensity to form secondary structures, and for double-stranded nucleic acids.
After a final wash with PBS to remove excess, fluorescent probe (FP) complementary to a different sequence on the target is introduced in an imaging solution containing an oxygen scavenger system (OSS). An OSS is used in order to reduce the rate of oxygen-induced photobleaching of the FP, thus ensuring more accurate and reproducible measurement of the FP-target interaction kinetics. In addition, a triplet state quencher is often added to prevent blinking of the FP due to the adoption of dark triplet states [41]. The most commonly used OSS is a mixture of protocatechuate-3,4-dioxygenase (PCD), protocatechuic acid (PCA), and Trolox, wherein PCD catalyzes the oxygen-consuming conversion of PCA to 3-carboxy-cis,cis-muconic acid, and Trolox functions as a triplet state quencher [42, 43]. Additional details on this protocol can be found here [28]. Once these preparations are done, imaging may begin.
2.3. Data Acquisition and Processing
Data acquisition is performed on a pTIRF or oTIRF microscope, using an excitation intensity of 10–100 W/cm2 at a wavelength near the excitation maximum of the fluorescent dye employed (for example, a 640-nm continuous-wave laser with an output power of 25–100 mW is typically used to excite Cy5 or Alexa Fluor 647). Fluorophores excited toward the red end of the visible spectrum are particularly attractive for biological samples because of the relatively little autofluorescence of most biological samples in this range of wavelengths. Wide-field detection is usually accomplished using an electron-multiplying charge-coupled device (EMCCD) or scientific complementary metal-oxide-semiconductor (sCMOS) camera with a frame exposure time of 100–500 ms, or the fastest frame rate that provides sufficient signal-to-noise for accurate determination of kinetics (typically a signal/noise ratio of 3–10 for single fluorophores). The optimal excitation intensity and camera exposure time depend on probe concentration as well as the molar extinction coefficient, quantum yield, and photostability of the chosen fluorophore.
For each sample analyzed, data are collected from multiple (≥3) fields of view, resulting in multiple movies recording the repeated binding of single FP molecules to the imaging surface (Fig. 5A,B). The number of fields of view needed depends on the desired sensitivity and the type of sample cell used, and the acquisition time per field of view depends on how much time is needed to generate a fingerprint that is distinguishable from background and non-target molecules based on the metrics elaborated below. In prior studies, a recording time of 5–10 min per field of view was typical, and three fields of view were usually sufficient for limits of detection of ~1 fM [17, 29].
Fig. 5.
SiMREPS data acquisition. (A) FP-target binding and dissociation occurs over the observed time window and the resulting changes in fluorescent signal are observed via TIRFM. (B) A single-frame snapshot showing the locations of single bound FP molecules in a field of view under the microscope. (C) A molecule of target is present at this location, as confirmed by the repetitive binding-dissociation pattern of the FP that constitutes a kinetic fingerprint. (D) Nonspecific binding at other locations gives rise to a smaller number of binding-dissociation transitions and/or different kinetics that are easily distinguished so that only genuine target molecules are counted.
After movies are collected, they are subjected to further data processing to determine which fluorescent signals originate from FP binding to the target (as opposed to nonspecific binding to the surface or other surface-bound molecules). To accomplish this, MATLAB scripts are used to (1) identify regions of the image with frequent fluctuations in fluorescence intensity, (2) calculate intensity versus time trajectories (or “traces”) for each candidate region of the image, (3) perform hidden Markov modeling (HMM) to establish the number of transitions between bound and unbound states (Nb+d) as well as the lifetime in each state (τbound and τunbound), and (4) use filtering criteria outlined below to differentiate true positives from background [28]. Typical filtering criteria include: minimum signal-to-noise ratio, minimum intensity difference between bound and unbound state, minimum number of binding and dissociation events (Nb+d) per trace, minimum and maximum values for the median lifetimes in the bound (τbound,median) and unbound (τunbound,median) states in each trace. The appropriate thresholds for these values must be determined empirically by comparing positive and negative control experiments. To obtain the greatest difference between kinetic fingerprints generated by the target and nonspecific binding, optimizations can be made to the probe design or experimental conditions as outlined below. On a typical modern laptop, analysis of a 10-min movie requires 2–5 min of total processing time, depending on the overall density of probe binding events.
3. Theoretical considerations for ultraspecific detection with SiMREPS
3.1. Statistical properties of kinetic fingerprints and optimization of data acquisition time
If the lifetimes of the FP-bound and FP-unbound states are similar (τbound ≈ τunbound), the number of binding and dissociation events Nb+d is well modeled by a Poisson distribution [28]. This is because the repetitive binding of the FP constitutes a series of discrete events in which the average time between binding and dissociation events is known, but the precise time of each event is random—a behavior exemplary of a Poisson process. Additionally, the waiting periods between the binding and dissociation events—i.e., the dwell times in the bound and unbound states—can be idealized as gamma-distributed variables, since they are averages over many exponentially distributed random events [44]. These statistical distributions aid in understanding how the SiMREPS approach achieves high specificity through kinetic analysis, and can help estimate the appropriate kinetic behavior and dwell times required to effectively discriminate between specific and nonspecific binding, or between similar analytes such as an SNV and wild-type sequence. Namely, these statistical distributions illustrate how Nb+d, τbound, and τunbound become more precisely determined at the single-molecule level as observation time increases.
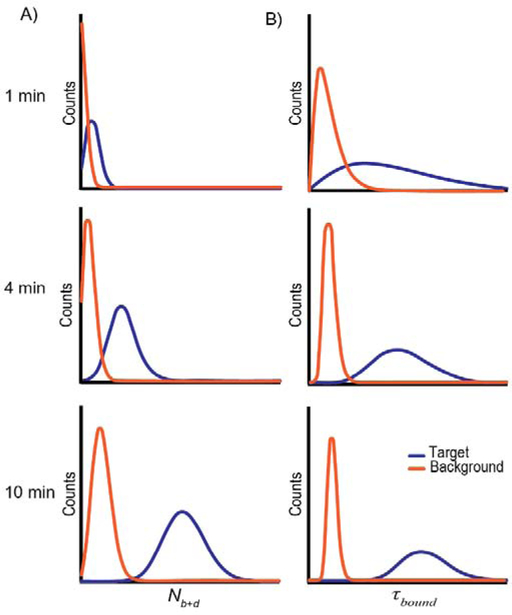
Modeling Nb+d as a Poisson process, the standard deviation is expected to scale only as , predicting that increasing acquisition time will yield an increasing separation of distributions for distinct processes—such as specific and nonspecific binding—allowing for increasing specificity as the acquisition time is increased (Fig. 6A) [28]. Consistent with this expectation, it has been shown experimentally that as acquisition time increases in SiMREPS measurements, the signal and background peaks become better resolved [29]. Johnson-Buck et al. provide a guideline to estimate the minimum observation period required to achieve a desired level of discrimination based on the apparent FP-target binding and dissociation rate constants (Kon and Koff) as well as the ratio between specific and nonspecific binding rates [28]. The statistical properties of Nb+d thus make it useful for distinguishing between the specific binding to the analyte and nonspecific binding to other features on the imaging surface.
Fig. 6.
Predicted statistical properties of SiMREPS kinetic fingerprints as a function of observation time. (A) Increasing the acquisition time from 1 min to 10 min yields an increase in separation of the background from the target distribution of Nb+d. (B) Increasing the observation time results in observation of more dwell times and a larger shape parameter of the gamma-distributed estimates of bound- or unbound-state lifetime, resulting in a more precise determination of τbound and/or τunbound, and allowing for more complete separation between target and background.
In some circumstances, Nb+d may be similar for specific and off-target binding, such as in the discrimination between two closely related nucleic acid sequences. In such cases, the lifetimes in the bound and unbound states (particularly τbound, which is highly sensitive to the number of complementary base pairs between the FP and target) are more useful in distinguishing between specific and nonspecific binding. The idealization as a gamma distribution predicts that τbound and τunbound will become more precisely determined as the number of dwell times observed—corresponding to the so-called shape parameter of the gamma distribution—increases (Fig. 6B). For example, the miRNAs let-7a and let-7c, which differ by only one nucleotide, can be distinguished by their differing τbound values after observing approximately 15 events per target molecule [28]. This statistical property gives SiMREPS the capability to reliably distinguish specific from background binding as well as to discriminate between a wild-type and mutant sequence with even a single nucleotide substitution. With optimal probe design that positions the mutation near the middle of the FP-binding sequence, Hayward et al. used this property to detect the drug-resistance-conferring cancer mutation EGFR T790M, a single C-to-T substitution, with an estimated specificity of 99.99999% [17].
It is important to stress that, as with other approaches that do not employ sequencing, SiMREPS can only be used to detect known mutations. However, unlike many other techniques (such as qPCR and digital PCR), SiMREPS probes bind the target sequence reversibly, which in principle permits the same population of surface-captured analytes to be assayed for several different mutations by sequentially introducing a series of different fluorescent probes specific to each mutation of interest.
3.2. Optimization of fluorescent probes
Based on the above considerations, to ensure adequate specificity for distinguishing similar sequences, the probes utilized must result in the observation of many cycles of transient binding to each copy of the immobilized target, in as short an interval of time as the detection apparatus permits. To facilitate such observation, the analyte is immobilized by a biotinylated capture probe (CP) that binds irreversibly to the target. Additionally, given the necessity of observing reversible binding of the fluorescent probe (FP) for many cycles, the melting temperature (Tm) of the fluorescent probe and target should be near the temperature at which the assay is performed [28]. Near the Tm, the dissociation rate is highly sensitive to the number of complementary base pairs between a probe and target [45], making the number of base pairs with the FP an especially important parameter to optimize. Additionally, the C:G/A:T base pair ratio represents a significant parameter to optimize given its relationship to duplex stability. Finally, the position of a mismatch between the FP and a spurious target molecule has a strong impact on the ability to discriminate between SNVs; generally, positioning such mismatches toward the interior of the duplex results in greater destabilization than placing them near the 3′- or 5′-end of the probe [46], resulting in better mismatch discrimination.
The number of base pairs in the FP-target duplex strongly influences the stability due to the incorporation of 2–3 inter-strand hydrogen bonds as well as additional π-π stacking interactions upon the addition of each base pair [47], making the length of the FP-target interaction an adjustable parameter to exploit in tuning kinetics for SiMREPS. For very short duplexes with a Tm near room temperature (~6–9 base pairs), the kinetics of dissociation exhibit an exponential dependence upon the number of base pairs, with a dissociation lifetime on the order of milliseconds to seconds [45]. To ensure that kinetics are matched to the typical frame rate of high-sensitivity single-molecule fluorescence measurements (2–10 Hz), the FPs used in SiMREPS typically make 8–9 base pairs with the target. Empirically, we have found the best performance for FPs with GC content of 25–50%, with ~9 base pairs to RNA targets, and ~8 base pairs to DNA targets. However, FPs with >50% GC content can exhibit appropriate kinetics for SiMREPS with minor adjustments such as the addition of one or more mismatches [28], a reduction in the length of the duplex, or the use of denaturants or higher temperatures during imaging (see below).
3.3. Optimization of imaging conditions
Reducing the number of base pairs in the FP-target duplex will accelerate dissociation kinetics, but at the expense of specificity. To avoid this tradeoff, one can instead destabilize the interaction using higher observation temperatures or by adding a low concentration of a chemical denaturant of nucleic acids, such as formamide, to the imaging buffer. Formamide aids in lowering the melting temperature of DNAs by 2.4–2.9°C/(mol L−1) of formamide, contingent on base pairing ratios and state of hydration [48].
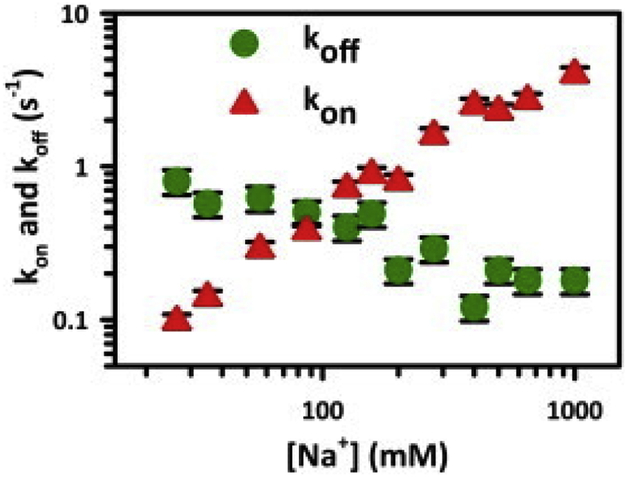
In addition, due to the important role cations play in stabilizing the interactions between two highly negatively charged nucleic acid strands, ionic strength is an important parameter to optimize in order to obtain optimal FP kinetics. Since the negatively charged phosphates tend to repel each other as well as the negative charges on the opposite strand of a DNA duplex, an increase in cation concentration in the buffer stabilizes DNA duplexes by screening electrostatic repulsion between the opposing strands, stabilizing the duplex. Dupuis et al. have experimentally shown that the rate constant of association of two short complementary DNA strands increases about 40-fold as the sodium ion concentration is increased from 20 mM to 1 M, while the dissociation rate constant decreases by less than tenfold over the same range (Fig. 7) [45]. Thus, a larger number of binding events can be observed in relatively high monovalent cation concentrations (i.e., >500 mM). This fact has been exploited in SiMREPS measurements, in which a buffer of high ionic strength (e.g., 4× PBS) is typically used to obtain rapid kinetics of binding of the FP to the target, which in turn allows for the observation of many binding and dissociation cycles in an observation period of (at most) ~10 min.
Fig. 7.
Duplex binding and dissociation rates for the 8 bp construct as a function of [Na+]. An imaging buffer containing 4X PBS (monovalent cation concentration ~650 mM) is utilized in SiMREPS experiments to obtain appropriate kinetics for SiMREPS. Reprinted with permission from ref. [45]. Copyright 2013 Cell Press.
Due to the presence of a large excess of FP over the surface-bound target, the FP exhibits pseudo-first-order kinetics of binding to the target that is proportional to the concentration of FP in the imaging buffer. Thus, the concentration of the FP in the imaging buffer directly influences the frequency of binding and, hence, how rapidly a SiMREPS fingerprint can be acquired. The optimal range under typical pTIRF and oTIRF observation conditions has been empirically found to be in the range of 25–50 nM FP. Lower concentrations than 25 nM will reduce frequency of FP binding, thus lengthening the required acquisition time for a kinetic fingerprint, while higher concentrations than 50 nM yield higher levels of background fluorescence from freely diffusing probes near the surface, reducing the signal-to-noise ratio for single fluorophore detection and thus reducing the accuracy of kinetic analysis [28].
Finally, if single-base specificity is needed, it is important to consider the optimal position of the mismatch within a duplex. Cisse et al. found that the Kd of a 9-bp DNA duplex varied by approximately three orders of magnitude depending on the location of a single-base mismatch within the duplex [46]. In particular, the strongest effect on Kd, kon, and koff was observed if the mismatch was close to the center of the duplex rather than in one of the first two or last two base pairs [46]. Consistent with this observation, the best single-base selectivity in SiMREPS assays has been observed when the mismatch is positioned toward the middle of the FP-target duplex (as in a previously published T790M assay [17]) rather than towards one end of the duplex (as by necessity in a let-7a/let-7c assay [28]).
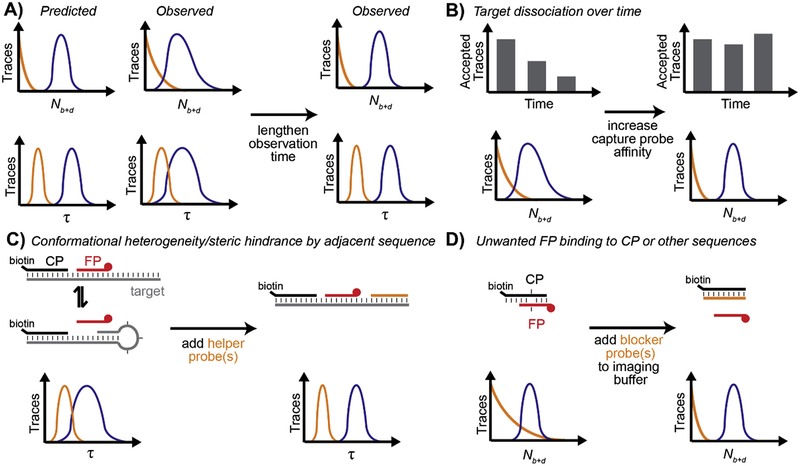
4. Deviations from ideal SiMREPS behavior
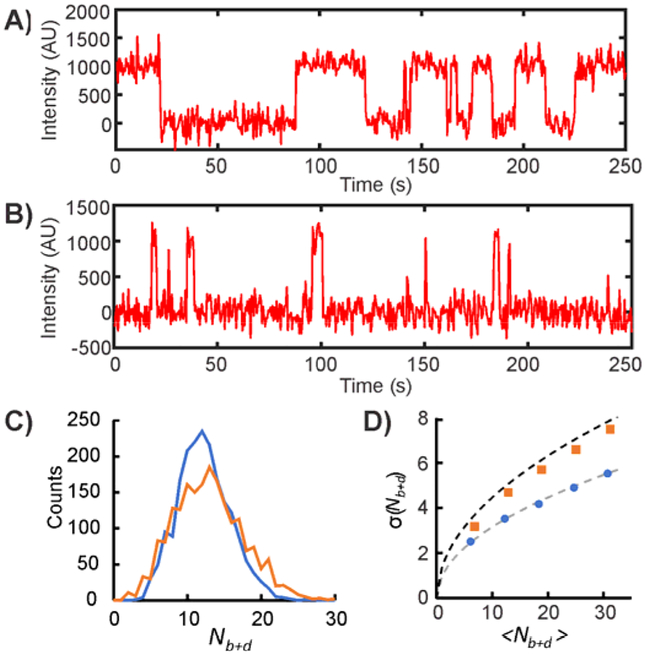
As described in section 3, binding of SiMREPS FPs to surface-bound targets is predicted to yield Poisson-distributed Nb+d values and gamma-distributed τ values. However, several factors can result in deviations from this idealized behavior. First, if τbound >> τunbound or τbound << τunbound, the FP binding and dissociation events will become increasingly correlated in time (i.e., a binding event will always follow soon after a dissociation event, or vice-versa). As a result, the Nb+d distribution will broaden somewhat relative to the predicted Poisson distribution; in the extreme case where τbound is infinitesimal compared to τunbound (or vice versa), there will be perfect temporal correlation between each pair of consecutive binding and dissociation events, and Nb+d/2 will become Poisson-distributed rather than Nb+d (Fig. 8). While such an assay still exhibits the key characteristic of SiMREPS—improved specific/nonspecific signal discrimination with increasing observation time—the separation of signal and background peaks requires a longer observation time when the lifetimes in the bound and unbound states are very different. Alternatively, the FP design or imaging conditions can be adjusted to ensure that τbound ≈ τunbound (see sections 3.2–3.3).
Fig. 8.
Impact of relative bound and unbound state lifetimes on the distribution of Nb+d. (A,B) Simulated intensity versus time traces for probes with equal (τbound = τunbound = 20 s) and very different (τbound = 2 s, τunbound = 38 s) lifetimes in the bound and unbound states. (C) Nb+d distributions for 2,000 simulated intensity versus time traces with τbound = τunbound = 20 s (blue curve) or τbound = 2 s, τunbound = 38 s (orange curve). The distribution becomes broader if τbound ≠ τunbound due to strong temporal correlation between binding and dissociation events. (D) Standard deviation in the Nb+d values of 2,000 simulated intensity versus time traces as a function of average Nb+d value, which increases with increasing observation time. When τbound = τunbound (blue circles), σ(Nb+d) increases as (gray dashed line), consistent with expectations for a Poisson process. However, when τbound ≠ τunbound (orange squares), σ(Nb+d) increases more rapidly due to temporal correlation of binding and dissociation events. In the limit where either τbound or τunbound is infinitesimal, σ(Nb+d) increases as (black dashed line) – consistent with Nb+d/2 being Poisson-distributed.
Furthermore, the Poisson and gamma distribution idealizations assume population homogeneity: that is, all surface-bound targets interact with the FP according to the same rate constants, and these rate constants are invariant with time. In reality, single-molecule kinetic measurements almost always exhibit some degree of static or dynamic heterogeneity [49]. In the context of a SiMREPS measurement, heterogeneous kinetics can be caused by factors such as:
Variable microenvironment in the vicinity of the captured target molecule on the surface, such as other nearby surface-bound molecules or poorly passivated regions of the coverslip, resulting in persistent differences in probe binding kinetics;
Dissociation or binding of the target to the surface partway through the measurement, resulting in segments of the movie in which no FP binding events are observed for a given target molecule;
Conformational changes intrinsic to the target molecule (e.g., secondary structure formation) that affect FP binding and/or dissociation kinetics;
Uneven illumination across the field of view, resulting in lower signal-to-noise ratios (generally toward the edges of the field of view) and potential for missed binding events;
Imperfections in fitting algorithms that occasionally miss binding events.
Because of factors such as those listed above, the experimentally observed distributions of Nb+d and τ are often somewhat broader than those predicted by theory (Fig. 9A). This is not necessarily an impediment to SiMREPS measurement, but it means that empirical calibration of kinetic filtering parameters and observation time is usually necessary for optimal assay performance. For instance, if broadening of the distribution of Nb+d or τ makes it challenging to distinguish between specific and nonspecific signal, a slightly longer observation time (combined with an appropriate change in the Nb+d threshold) usually suffices to improve the separation between signal and background peaks.
Fig. 9.
Deviations from ideal SiMREPS behavior and suggested solutions. (A) Broadening of the Nb+d and/or τ distributions is frequently observed experimentally, resulting in less complete separation between specific (purple) and nonspecific (orange) binding distributions than theory predicts. This broadening can often be offset by a slight (e.g. < 2-fold) increase in the observation time to increase the number of FP binding events observed per target molecule. (B) If significant target dissociation occurs within the observation window, the Nb+d distribution will appear broader than expected. Increasing the CP’s affinity for the target may reduce the dissociation rate and tighten the distribution. (C) Secondary structure and other steric hindrance by adjacent sequence can broaden the apparent bound- or unbound-state lifetime distribution. In such cases, inclusion of one or more helper probes to block adjacent sequence can make the kinetics more homogeneous. (D) FPs may exhibit unwanted interactions with other sequences, particularly if other sequences are present at large surface densities or contain modifications such as LNAs that increase the affinity of base-pairing interactions. Even low-affinity interactions can interfere with measurements if they are numerous enough. In such cases, supplementing the imaging buffer with one or more blocker probes specific to the interfering sequence can improve the separation of signal and background peaks.
In contrast, dissociation of the target from the surface can be assessed by performing multiple consecutive measurements on the same field of view, and determining whether the number of molecules detected by SiMREPS is systematically decreasing over time (Fig. 9B). If this is the case, it may be necessary to redesign the CP so that it interacts more strongly with the target—for instance, by adding nucleotides or locked nucleic acid (LNA) [50] modifications to the CP to increase its affinity for the target sequence—or to stabilize the interaction by increasing the ionic strength of the imaging buffer (provided that this does not interfere with the transient binding behavior of the FP).
If no dissociation of the target is evident over time, but there are still conspicuous gaps (lacking FP binding) in the fluorescence versus time traces or a surprising degree of heterogeneity between different molecules, variable secondary structure of the target may be interfering with the FP interaction. The likelihood of this can be assessed through the use of secondary structure prediction software such as NUPACK [51] or mfold [52]. If this analysis yields a clear hypothesis regarding secondary structure interference, it may be helpful to test the hypothesis by designing and including “helper” probes that irreversibly bind to the target molecule in a manner that prevents the interfering structure from forming (Fig. 9C). Note that even if no specific secondary structure forms, a long single-stranded segment of RNA or DNA adjacent to the probe binding site can still provide steric hindrance to FP binding. In such circumstances, it may still be beneficial to include one or more “helper” probes that bind to the target in a position adjacent to the FP binding site, but separated from it by 2–3 nucleotides to prevent stacking interactions between the helper probe and the FP and associated changes in kinetics.
Although SiMREPS measurements are completely insensitive to a modest amount of nonspecific FP binding, nonspecific binding can be more of a problem if it begins to interfere with the detection of specific binding signal. For instance, sometimes a large number of very weak interactions between the FP and CP, or between the FP and a very abundant but spurious target (e.g., a mutant probe interacting briefly with a wild-type sequence) can result in enough nonspecific FP binding that it interferes with observation of the specific binding signal (Fig. 9D). If this occurs, a competitor probe that blocks the off-target interaction can be added to the imaging solution to prevent the interfering binding from occurring. For instance, a wild-type-specific competitor probe has been used to prevent nonspecific binding to the wild-type EGFR sequence in an assay for the point mutation T790M, improving the ability to detect mutant DNA at a relative abundance as low as 1:106 [17]. A blocker probe that binds to unoccupied CPs and prevents any unwanted base pairing between the FP and CP was also found to be helpful in the same study [17]. Note that the wild-type competitor can also bind to the target (mutant) DNA sequence, albeit with lower affinity; it is therefore important to use a concentration of the competitor that is just high enough to reduce nonspecific binding of the FP to the wild-type sequence, but no higher. A good rule-of-thumb is that the competitor should be at a sufficiently high concentration that the wild-type sequence is occupied by the competitor more than 50% of the time, but the mutant/target sequence is occupied by the competitor less than 50% of the time.
In rare cases, specific chemical damage can result in false positives. For example, heat-induced deamination or oxidation of cytosine can result in false positives for C>T mutations such as T790M, since deamination of cytosine yields uracil (U), which will almost perfectly resemble the mutant (T) sequence [14, 16]. For any assay distinguishing C and T nucleotides (particularly when false positives for the T-containing sequence are to be avoided), we therefore recommend avoiding excessive heating and, in the case of assays for DNA targets, we recommend treating the sample with repair enzymes such as uracil DNA glycosylase (UDG), formamidopyrimidine-DNA glycosylase (FpG), and endonuclease VIII following any heating steps to cleave deaminated or partly oxidized cytosine from the DNA strands. Both of these strategies were crucial to obtaining a specificity of 99.99999% for T790M by SiMREPS [17].
Sometimes nonspecific binding signal is caused by binding to the coverslip surface itself, rather than unintended nucleic acid sequences. This may lead to artifacts caused by photoblinking of stably bound probes, or nonspecific repeated binding in the same location. If this occurs, passivation may be improved by performing PEG functionalization of the coverslip surface for a longer period of time or by a pre-incubation step in TWEEN 20 [53].
Errors in the kinetic analysis will occasionally occur with any fitting algorithm, but can be minimized by choosing an appropriate number of minimum fitting iterations or improving the signal-to-noise of the raw data (e.g., by increasing the excitation intensity in the TIRF measurement). In addition, future improvements may be possible through the use of machine learning methods that assess features of the intensity versus time signal in a manner not confined to a Markov model with discrete states. For instance, we sometimes observe deviations from two-state Markovian behavior, particularly in background binding profiles. If a probe binds irreversibly to the glass coverslip and then undergoes rapid blinking, repeated quenching/dequenching, or other erratic intensity changes, the resulting signal can confuse the current analysis method that assumes two-state intensity behavior, resulting in false positives despite obvious differences from true positives that a human user would have noticed. Smarter algorithms with more free parameters, including deep learning methods, may be helpful in reducing the error rate of SiMREPS by correctly classifying a larger range of behaviors that might be mis-classified using traditional fitting methods.
5. Applications of SiMREPS assays
5.1. Analyte scope
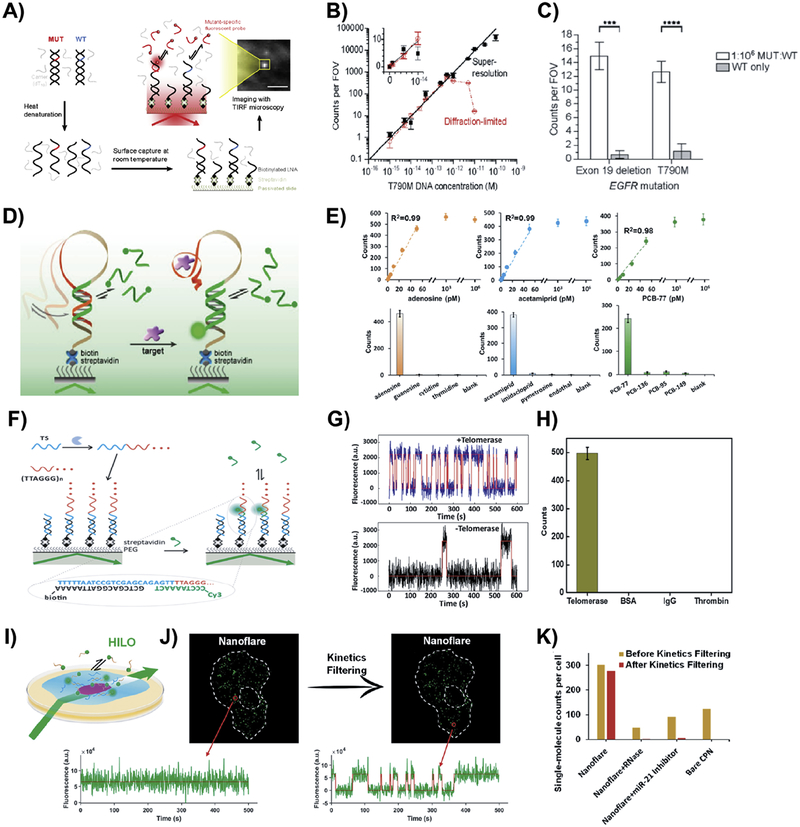
Due to the amplification-free and enzymatic reaction-free nature of SiMREPS, any analyte that can be immobilized on a surface and bound by a fluorescent affinity probe can, in principle, be detected by SiMREPS. Typically, analytes are immobilized or captured by surface probes via specific interactions such as hybridization of complementary nucleotides between analytes and probes. Freely diffusing fluorescent probes in the imaging buffer transiently bind to the surface-bound analytes, generating detectable single-molecule fingerprints for identification of the analytes. So far, several analytes including miRNAs (including let-7 family members differing by a single nucleobase) [29], DNA sequences with oncogenic point mutations [17, 54] and small molecules [34] have been successfully detected utilizing SiMREPS. Fig. 10 shows schematic diagrams and representative data from the application of SiMREPS to the detection of various analytes. Fig. 10A shows a schematic description of SiMREPS experiments for detecting oncogenic point mutations in DNA. Super-resolution imaging yielded a linear dynamic range of ~5 orders of magnitude (Fig. 10B), and single-nucleotide specificity was high enough to detect EGFR mutations in a 1 million-fold excess of wild-type EGFR (Fig. 10C).
Fig. 10.
Practical applications of SiMREPS. (A) Schematic depicting the use of SiMREPS for analysis of double-stranded DNA. (B) Standard curves from a SiMREPS assay for the cancer point mutation EGFR T790M using super-resolution and diffraction-limited analysis methods. (C) Comparison of accepted traces from low mutant allelic fraction (1:1 million) and wild-type-only conditions illustrating the high specificity of SiMREPS. (D) Schematic depiction of SiMREPS-based aptasensors for the detection of small molecules. (E) Standard curves and specificity of detection of different small molecules and spiked-in small molecule samples using SiMREPS-based aptasensors. (F) Schematic depicting the detection of telomerase activity on single substrate molecules. (G) Time traces of fluorescent probes in the presence and absence of telomerase. (H) High selectivity of the SiMREPS assay for the detection of telomerase. (I) Schematic of in situ detection of miRNAs using SiMREPS with nanoflare probes. (J) Kinetic behaviors of fluorescent probes undergoing specific binding to a single miRNA molecule and non-specific binding; (K) Quantification of the control experiments shown in (K) illustrating the importance of kinetic filtering. (A)-(C) Reprinted with permission from ref. [17]. Copyright 2018 Journal of the American Chemical Society. (D) and (E) Reprinted with permission from ref. [34]. Copyright 2019 Analytical Chemistry. (F)-(H) Reprinted with permission from ref. [57]. Copyright 2017 Analytical Chemistry. (I)-(K) Reprinted with permission from ref. [64]. Copyright 2019 Analytical Chemistry.
In the detection of small molecules, Weng et al. [34] performed SiMREPS with aptamers to overcome the hurdles of signal leakage by aptasensors in the absence of analytes, yielding a dramatic enhancement in sensitivity and specificity. Fig. 10E shows calibration curves (sensitivity) and selectivity for adenosine, acetamiprid and PCB-77 small molecules with a detection limit of 0.3 pM, 0.35 pM and 0.72 pM, respectively, which is about 1–3 orders of magnitude lower (i.e., more sensitive) than other aptasensors published recently [55, 56].
In addition to detecting a variety of analytes, SiMREPS can be used to monitor the activity of certain enzymes. For example, Su et al. [57] used SiMREPS to detect the activity of telomerase with high confidence, high sensitivity, high selectivity and zero background (Fig. 10F–H). In this case, the reaction products of telomerase were analyzed by transient binding of FPs complementary to the reaction product (the repeated sequence TTAGGG).
5.2. Detection of analytes in single cells
Single-molecule fluorescence in situ hybridization (smFISH) is an invaluable approach for the localization and quantification of nucleic acids at the single-cell level and at single-molecule resolution [58, 59]. In smFISH, a multitude of fluorescent dye-labeled oligonucleotide probes designed to be complementary to the target nucleic acids are introduced into fixed cells and allowed to hybridize to the analytes within the cell, and their accumulation on the target and location detected via fluorescence microscopy. The smFISH approach has been widely applied to the detection of RNA (especially mRNA) in single cells to better understand the regulation of gene expression [60, 61]. However, performing smFISH is often challenging due to 1) the low abundance of many analytes (i.e., many RNAs) in a single cell; 2) the limited sequence “real estate” on short target molecules to accumulate a detectable number of probes; and 3) the poor signal-to-noise ratio due to cellular autofluorescence and the inability to completely wash away unbound probes [62]. To combat these problems, either signal amplification or more complex fluorescent probe schemes are often employed in smFISH, which in turn makes the detection more complicated and tedious [58, 60, 63]. Furthermore, for short targets such as miRNAs, it is not possible to bind multiple fluorescent probes to the same target, making some of the usual strategies to improve signal-to-noise ineffective. However, as an amplification-free method with very low background, SiMREPS may provide a more general solution to the challenges of detecting single, low-abundance analytes in fixed cells and tissues. In principle, the additional time axis of the kinetic fingerprint allows for better separation between signal and background in analyte detection compared to conventional smFISH approaches. Recently, Li et al. [64] have used SiMREPS together with a novel type of probe fabricated on fluorescent polymer nanoparticles to detect cellular miRNA molecules in situ. Single-molecule sensitivity and single-mismatch specificity of detection of miRNA without any amplification steps were also demonstrated in vitro. In situ single-detection of miRNA in fixed cells was successfully performed (Fig. 10I–K).
6. Potential future improvements of SiMREPS
6.1. FRET-based SiMREPS
Originally inspired by the DNA-PAINT method [65], SiMREPS provides a means to detect various analytes with high accuracy and sensitivity. However, as with DNA-PAINT, a major drawback of SiMREPS is the relatively long acquisition times required. This is ultimately the result of the significant fluorescent background caused by freely diffusing fluorescent probes in the imaging buffer, even when TIRF or HILO [66] illumination is employed to reduce such background fluorescence. To minimize this background, a relatively low concentration of fluorescent probe (~ 25–50 nM) is often used, resulting in fairly slow kinetics of FP binding to the target and, hence, longer acquisition times to separate true positives from false positives.
Combining SiMREPS with smFRET might allow further reduction of background signal and, for enabling higher probe concentrations, faster data acquisition times. This principle has already been demonstrated for DNA-PAINT in the form of FRET-PAINT [67, 68]. In this approach, two fluorescent probes are present in the imaging buffer: one bearing a FRET donor, and the other a FRET acceptor. Only when the two fluorescent probes are simultaneously bound to the same docking strand can FRET signal be detected. The background induced by the freely diffusing fluorescent probes is therefore reduced by detecting the fluorescence of the acceptor probes, which will only be excited when simultaneously bound to the same docking strand as a donor probe. As a result of this lower background signal, higher concentrations of fluorescent probes can be used without compromising signal-to-noise, permitting shorter acquisition times. Moreover, incorporation of smFRET with SiMREPS may provide a strategy for multiplex detection of different analytes in the future. For instance, Deußner-Helfmann et al. [69] stated that, by combining smFRET with DNA-PAINT imaging, they were able to perform multiplex super-resolution imaging with low background. Multiplexing was achieved by either sequential imaging with different probe sets, or by designing different probe sets with distinguishable FRET efficiencies.
6.2. Increasing the sensitivity of SiMREPS
Compared to most other amplification-free techniques for detecting nucleic acid analytes, SiMREPS has far better sensitivity, with a typical limit of detection around 1 fM [17]. This limit of detection is still significantly higher than amplification-based PCR approaches (e.g., digital PCR), despite the fact that SiMREPS can detect single molecular copies of the analyte. This is a significant challenge to overcome in applications such as liquid biopsies involving circulating tumor DNA, where analyte concentrations may be in the attomolar or zeptomolar range. A key factor limiting the sensitivity of SiMREPS is the analyte/target capture step, in which analytes diffuse to the detection surface and bind to the CP. The hybridization reaction between the analyte and CP is much faster than the rate of diffusion of the analyte to the surface. Thus, to maximize the potential of SiMREPS for high-sensitivity detection, it is especially important to improve the mass transfer of analytes to the imaging surface. Previously, for hybridization-based detection approaches (i.e., DNA microarrays) researchers have applied electric fields to accelerate the transport of nucleic acids to the detection surface for faster hybridization [70, 71]. Similar application of electric fields in SiMREPS assays is also likely to be beneficial. Another route to increasing the sensitivity of SiMREPS might be to combine SiMREPS with a technique called isotachophoresis (ITP). ITP is an electrophoresis-based approach with the capability of pre-concentrating and separating charged species [72]. This technique has been employed to speed up microarray hybridization with an improvement in sensitivity of 8-fold for a 30-min assay [73]. Alternatively, pre-concentration of analytes using aqueous two phase systems (ATPS) [74] may also serve as a means to improve the sensitivity of SiMREPS. In this case, analytes such as nucleic acids pre-concentrated into the smaller of two or more phases, and can be brought close to the probe surface for more efficient capture. Another approach to improving the sensitivity of SiMREPS may be to use magnetic nanoparticles for analyte capture [75]. In this approach, instead of waiting for analytes to diffuse to the detection surface, CP-modified magnetic nanoparticles capture the analytes in bulk suspension and then are rapidly pulled to the detection surface by applying a magnetic field.
7. Conclusions
Assays based on single-molecule kinetic fingerprinting with SiMREPS enable the detection of single analyte molecules without amplification and with essentially no background. The raw single-base selectivity of the technique is unparalleled by any other technique, and is sufficient to detect point mutations with an apparent discrimination factor of >1 million. In the future, it is likely that the scope of analytes assayed by this technique will broaden to include other classes of biomolecules for which there is no PCR-equivalent method to amplify the analyte prior to detection. For nucleic acid analytes, the greatest improvements will come from methods that increase the capture speed and efficiency, and permit rapid multiplexed measurements from the same sample.
Highlights.
SiMREPS achieves 99.99999% single-nucleotide specificity without PCR
Specificity of SiMREPS exceeds that of digital PCR by at least an order of magnitude
Observation time can be extended for theoretically unlimited specificity
Optimal performance is achieved with equally rapid probe binding and dissociation
Applications to date include miRNAs, cancer mutations, and small molecules
Acknowledgements
The authors acknowledge support from a Michigan Economic Development Corporation MTRAC for Life Sciences grant to M.T., N.G.W. and A.J-B.; pilot grants from the University of Michigan MCubed 2.0 program, the James Selleck Bower Permanently Endowed Innovative Promise Funds for Cancer Research as well as a Cellular Cancer Biology Imaging Research (CCBIR) Program Pilot Grant from the University of Michigan Rogel Cancer Center, and a Fast Forward GI Innovation Fund to M.T. and N.G.W.; and NIH grants R21 CA204560 and R33 CA229023 to M.T. and N.G.W. The authors also wish to thank the Single Molecule Analysis in Real-Time (SMART) Center of the University of Michigan, seeded by NSF MRI-R2-ID award DBI-0959823 to N.G.W., as well as J. D. Hoff for training, technical advice, and use of the objective-type TIRF microscope.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest
The University of Michigan has filed patent applications related to the single-molecule kinetic fingerprinting approach described herein, on which A.J.B, M.T. and N.G.W. are co-inventors. A.J.B., M.T., and N.G.W. are co-founders of a startup company, aLight Sciences LLC, which seeks to commercialize this technology.
References
- [1].Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR, Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood, Proc Natl Acad Sci U S A, 105 (2008) 16266–16271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Foulkes WD, Knoppers BM, Turnbull C, Population genetic testing for cancer susceptibility: founder mutations to genomes, Nat Rev Clin Oncol, 13 (2016) 41–54. [DOI] [PubMed] [Google Scholar]
- [3].Vasan RS, Biomarkers of cardiovascular disease: molecular basis and practical considerations, Circulation, 113 (2006) 2335–2362. [DOI] [PubMed] [Google Scholar]
- [4].Umek RM, Lin SS, Chen Yp Y, Irvine B, Paulluconi G, Chan V, Chong Y, Cheung L, Vielmetter J, Farkas DH, Bioelectronic detection of point mutations using discrimination of the H63D polymorphism of the Hfe gene as a model, Mol Diagn, 5 (2000) 321–328. [DOI] [PubMed] [Google Scholar]
- [5].Kim NW, Wu F, Advances in quantification and characterization of telomerase activity by the telomeric repeat amplification protocol (TRAP), Nucleic Acids Res, 25 (1997) 2595–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].van der Meide WF, Schoone GJ, Faber WR, Zeegelaar JE, de Vries HJ, Ozbel Y, Lai AFRF, Coelho LI, Kassi M, Schallig HD, Quantitative nucleic acid sequence-based assay as a new molecular tool for detection and quantification of Leishmania parasites in skin biopsy samples, J Clin Microbiol, 43 (2005) 5560–5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Liu GY, Isolation, sequence identification, and tissue expression profile of 3 novel porcine genes: NCF2, BCKDHB and BCKDHA, J Appl Genet, 50 (2009) 47–50. [DOI] [PubMed] [Google Scholar]
- [8].Diaz LA Jr., Bardelli A, Liquid biopsies: genotyping circulating tumor DNA, J Clin Oncol, 32 (2014) 579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rybicka M, Stalke P, Bielawski KP, Current molecular methods for the detection of hepatitis B virus quasispecies, Rev Med Virol, 26 (2016) 369–381. [DOI] [PubMed] [Google Scholar]
- [10].Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H, Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction, Cold Spring Harb Symp Quant Biol, 51 Pt 1 (1986) 263–273. [DOI] [PubMed] [Google Scholar]
- [11].Sreejith KR, Ooi CH, Jin J, Dao DV, Nguyen NT, Digital polymerase chain reaction technology - recent advances and future perspectives, Lab Chip, 18 (2018) 3717–3732. [DOI] [PubMed] [Google Scholar]
- [12].Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B, Detection and quantification of rare mutations with massively parallel sequencing, Proc Natl Acad Sci U S A, 108 (2011) 9530–9535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sofi Ibrahim M, Kulesh DA, Saleh SS, Damon IK, Esposito JJ, Schmaljohn AL, Jahrling PB, Real-time PCR assay to detect smallpox virus, J Clin Microbiol, 41 (2003) 3835–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chen G, Mosier S, Gocke CD, Lin MT, Eshleman JR, Cytosine deamination is a major cause of baseline noise in next-generation sequencing, Mol Diagn Ther, 18 (2014) 587–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chen L, Liu P, Evans TC Jr., Ettwiller LM, DNA damage is a pervasive cause of sequencing errors, directly confounding variant identification, Science, 355 (2017) 752–756. [DOI] [PubMed] [Google Scholar]
- [16].Potapov V, Ong JL, Examining Sources of Error in PCR by Single-Molecule Sequencing, PLoS One, 12 (2017) e0169774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hayward SL, Lund PE, Kang Q, Johnson-Buck A, Tewari M, Walter NG, Ultraspecific and Amplification-Free Quantification of Mutant DNA by Single-Molecule Kinetic Fingerprinting, J Am Chem Soc, 140 (2018) 11755–11762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Newman AM, Lovejoy AF, Klass DM, Kurtz DM, Chabon JJ, Scherer F, Stehr H, Liu CL, Bratman SV, Say C, Zhou L, Carter JN, West RB, Sledge GW, Shrager JB, Loo BW Jr., Neal JW, Wakelee HA, Diehn M, Alizadeh AA, Integrated digital error suppression for improved detection of circulating tumor DNA, Nat Biotechnol, 34 (2016) 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Giraldez MD, Spengler RM, Etheridge A, Godoy PM, Barczak AJ, Srinivasan S, De Hoff PL, Tanriverdi K, Courtright A, Lu S, Khoory J, Rubio R, Baxter D, Driedonks TAP, Buermans HPJ, Hoen ENMNT, Jiang H, Wang K, Ghiran I, Wang YE, Van Keuren-Jensen K, Freedman JE, Woodruff PG, Laurent LC, Erle DJ, Galas DJ, Tewari M, Comprehensive multi-center assessment of small RNA-seq methods for quantitative miRNA profiling (vol 36, pg 746, 2018), Nat Biotechnol, 36 (2018) 899–899. [DOI] [PubMed] [Google Scholar]
- [20].Akane A, Matsubara K, Nakamura H, Takahashi S, Kimura K, Identification of the Heme Compound Copurified with Deoxyribonucleic-Acid (DNA) from Bloodstains, a Major Inhibitor of Polymerase Chain-Reaction (Pcr) Amplification, J Forensic Sci, 39 (1994) 362–372. [PubMed] [Google Scholar]
- [21].Satsangi J, Jewell D, Welsh K, Bunce M, Bell J, Effect of Heparin on Polymerase Chain-Reaction, Lancet, 343 (1994) 1509–1510. [DOI] [PubMed] [Google Scholar]
- [22].Kim K, Oh JW, Lee YK, Son J, Nam JM, Associating and Dissociating Nanodimer Analysis for Quantifying Ultrasmall Amounts of DNA, Angew Chem Int Ed Engl, 56 (2017) 9877–9880. [DOI] [PubMed] [Google Scholar]
- [23].Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L, Dimitrov K, Direct multiplexed measurement of gene expression with color-coded probe pairs, Nat Biotechnol, 26 (2008) 317–325. [DOI] [PubMed] [Google Scholar]
- [24].Cohen L, Hartman MR, Amardey-Wellington A, Walt DR, Digital direct detection of microRNAs using single molecule arrays, Nucleic Acids Res, 45 (2017) e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhang DY, Chen SX, Yin P , Optimizing the specificity of nucleic acid hybridization, Nat Chem, 4 (2012) 208–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Jin Z, Geissler D, Qiu X, Wegner KD, Hildebrandt N, A Rapid, Amplification-Free, and Sensitive Diagnostic Assay for Single-Step Multiplexed Fluorescence Detection of MicroRNA, Angew Chem Int Ed Engl, 54 (2015) 10024–10029. [DOI] [PubMed] [Google Scholar]
- [27].Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, Chen C, Cheo D, D’Andrade P, DeMayo M, Dennis L, Derveaux S, Feng Y, Fulmer-Smentek S, Gerstmayer B, Gouffon J, Grimley C, Lader E, Lee KY, Luo S, Mouritzen P, Narayanan A, Patel S, Peiffer S, Ruberg S, Schroth G, Schuster D, Shaffer JM, Shelton EJ, Silveria S, Ulmanella U, Veeramachaneni V, Staedtler F, Peters T, Guettouche T, Wong L, Vandesompele J, Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study, Nat Methods, 11 (2014) 809–815. [DOI] [PubMed] [Google Scholar]
- [28].Johnson-Buck A, Li J, Tewari M, Walter NG, A guide to nucleic acid detection by single-molecule kinetic fingerprinting, Methods, 153 (2019) 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Johnson-Buck A, Su X, Giraldez MD, Zhao M, Tewari M, Walter NG, Kinetic fingerprinting to identify and count single nucleic acids, Nat Biotechnol, 33 (2015) 730–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cissell KA, Rahimi Y, Shrestha S, Hunt EA, Deo SK, Bioluminescence-based detection of microRNA, miR21 in breast cancer cells, Anal Chem, 80 (2008) 2319–2325. [DOI] [PubMed] [Google Scholar]
- [31].de Planell-Saguer M, Rodicio MC, Analytical aspects of microRNA in diagnostics: a review, Anal Chim Acta, 699 (2011) 134–152. [DOI] [PubMed] [Google Scholar]
- [32].de Planell-Saguer M, Rodicio MC, Detection methods for microRNAs in clinic practice, Clin Biochem, 46 (2013) 869–878. [DOI] [PubMed] [Google Scholar]
- [33].Zhang R, Chen B, Tong X, Wang Y, Wang C, Jin J, Tian P, Li W, Diagnostic accuracy of droplet digital PCR for detection of EGFR T790M mutation in circulating tumor DNA, Cancer Manag Res, 10 (2018) 1209–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Weng R, Lou S, Li L, Zhang Y, Qiu J, Su X, Qian Y, Walter NG, Single-Molecule Kinetic Fingerprinting for the Ultrasensitive Detection of Small Molecules with Aptasensors, Anal Chem, 91 (2019) 1424–1431. [DOI] [PubMed] [Google Scholar]
- [35].Wazawa T, Ueda M, Total internal reflection fluorescence microscopy in single molecule nanobioscience, Microscopy Techniques, 95 (2005) 77–106. [DOI] [PubMed] [Google Scholar]
- [36].Axelrod D, Thompson NL, Burghardt TP, Total Internal-Reflection Fluorescent Microscopy, Journal of Microscopy-Oxford, 129 (1983) 19–28. [DOI] [PubMed] [Google Scholar]
- [37].Walter NG, Huang CY, Manzo AJ, Sobhy MA, Do-it-yourself guide: how to use the modern single-molecule toolkit, Nat Methods, 5 (2008) 475–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Martin-Fernandez ML, Tynan CJ, Webb SED, A ‘pocket guide’ to total internal reflection fluorescence, Journal of Microscopy, 252 (2013) 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M, Circulating microRNAs as stable blood-based markers for cancer detection, P Natl Acad Sci USA, 105 (2008) 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Dominguez K, Ward WS, A novel nuclease activity that is activated by Ca(2+) chelated to EGTA, Syst Biol Reprod Med, 55 (2009) 193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ha TJ, Ting AY, Liang J, Deniz AA, Chemla DS, Schultz PG, Weiss S, Temporal fluctuations of fluorescence resonance energy transfer between two dyes conjugated to a single protein, Chemical Physics, 247 (1999) 107–118. [Google Scholar]
- [42].Rasnik I, McKinney SA, Ha T, Nonblinking and long-lasting single-molecule fluorescence imaging, Nat Methods, 3 (2006) 891–893. [DOI] [PubMed] [Google Scholar]
- [43].Aitken CE, Marshall RA, Puglisi JD, An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments, Biophys J, 94 (2008) 1826–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Floyd DL, Harrison SC, van Oijen AM, Analysis of kinetic intermediates in single-particle dwell-time distributions, Biophys J, 99 (2010) 360–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Dupuis NF, Holmstrom ED, Nesbitt DJ, Single Molecule Kinetics Reveal Cation Promoted DNA Duplex Formation through Ordering of Single Stranded Helices (vol 105, pg 756, 2013), Biophysical Journal, 106 (2014) 2737–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Cisse II, Kim H, Ha T, A rule of seven in Watson-Crick base-pairing of mismatched sequences, Nat Struct Mol Biol, 19 (2012) 623–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Yakovchuk P, Protozanova E, Frank-Kamenetskii MD, Base-stacking and base-pairing contributions into thermal stability of the DNA double helix (vol 34, pg 564, 2006), Nucleic Acids Research, 34 (2006) 1082–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Blake RD, Delcourt SG, Thermodynamic effects of formamide on DNA stability, Nucleic Acids Res, 24 (1996) 2095–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Marek MS, Johnson-Buck A, Walter NG, The shape-shifting quasispecies of RNA: one sequence, many functional folds, Phys Chem Chem Phys, 13 (2011) 11524–11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Singh SK, Nielsen P, Koshkin AA, Wengel J, LNA (locked nucleic acids): synthesis and high-affinity nucleic acid recognition, Chemical Communications, (1998) 455–456. [Google Scholar]
- [51].Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM, Pierce NA, NUPACK: Analysis and design of nucleic acid systems, J Comput Chem, 32 (2011) 170–173. [DOI] [PubMed] [Google Scholar]
- [52].Zuker M, Mfold web server for nucleic acid folding and hybridization prediction, Nucleic Acids Research, 31 (2003) 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Pan H, Xia Y, Qin M, Cao Y, Wang W, A simple procedure to improve the surface passivation for single molecule fluorescence studies, Phys Biol, 12 (2015) 045006. [DOI] [PubMed] [Google Scholar]
- [54].Su X, Li L, Wang S, Hao D, Wang L, Yu C, Single-Molecule Counting of Point Mutations by Transient DNA Binding, Sci Rep, 7 (2017) 43824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wang YM, Wu Z, Liu SJ, Chu X, Structure-Switching Aptamer Triggering Hybridization Chain Reaction on the Cell Surface for Activatable Theranostics, Anal Chem, 87 (2015) 6470–6474. [DOI] [PubMed] [Google Scholar]
- [56].S. H Long Feng, Wang Hongchen, Fluorescence resonance energy transfer based aptasensor for the sensitive and selective detection of 17β-estradiol using a quantum dot-bioconjugate as a nano-bioprobe, RSC Advances, 4 (2014) 2935–2941. [Google Scholar]
- [57].Su X, Li Z, Yan X, Wang L, Zhou X, Wei L, Xiao L, Yu C, Telomerase Activity Detection with Amplification-Free Single Molecule Stochastic Binding Assay, Anal Chem, 89 (2017) 3576–3582. [DOI] [PubMed] [Google Scholar]
- [58].Kwon S, Single-molecule fluorescence in situ hybridization: quantitative imaging of single RNA molecules, BMB Rep, 46 (2013) 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Cui C, Shu W, Li P, Fluorescence In situ Hybridization: Cell-Based Genetic Diagnostic and Research Applications, Front Cell Dev Biol, 4 (2016) 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Haimovich G, Gerst JE, Single-molecule Fluorescence in situ Hybridization (smFISH) for RNA Detection in Adherent Animal Cells, Bio-Protocol, 8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Xie F, Timme KA, Wood JR, Using Single Molecule mRNA Fluorescent in Situ Hybridization (RNA-FISH) to Quantify mRNAs in Individual Murine Oocytes and Embryos, Sci Rep, 8 (2018) 7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Moffitt JR, Hao J, Bambah-Mukku D, Lu T, Dulac C, Zhuang X, High-performance multiplexed fluorescence in situ hybridization in culture and tissue with matrix imprinting and clearing, Proc Natl Acad Sci U S A, 113 (2016) 14456–14461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wu C, Simonetti M, Rossell C, Mignardi M, Mirzazadeh R, Annaratone L, Marchio C, Sapino A, Bienko M, Crosetto N, Nilsson M, RollFISH achieves robust quantification of single-molecule RNA biomarkers in paraffin-embedded tumor tissue samples, Commun Biol, 1 (2018) 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Li L, Yu Y, Wang C, Han Q, Su X, Transient Hybridization Directed Nanoflare for Single-Molecule miRNA Imaging, Anal Chem, 91 (2019) 11122–11128. [DOI] [PubMed] [Google Scholar]
- [65].Jungmann R, Steinhauer C, Scheible M, Kuzyk A, Tinnefeld P, Simmel FC, Single-Molecule Kinetics and Super-Resolution Microscopy by Fluorescence Imaging of Transient Binding on DNA Origami, Nano Letters, 10 (2010) 4756–4761. [DOI] [PubMed] [Google Scholar]
- [66].Tokunaga M, Imamoto N, Sakata-Sogawa K, Highly inclined thin illumination enables clear single-molecule imaging in cells, Nat Methods, 5 (2008) 159–161. [DOI] [PubMed] [Google Scholar]
- [67].Auer A, Strauss MT, Schlichthaerle T, Jungmann R, Fast, Background-Free DNA-PAINT Imaging Using FRET-Based Probes, Nano Lett, 17 (2017) 6428–6434. [DOI] [PubMed] [Google Scholar]
- [68].Lee J, Park S, Kang W, Hohng S, Accelerated super-resolution imaging with FRET-PAINT, Mol Brain, 10 (2017) 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Deussner-Helfmann NS, Auer A, Strauss MT, Malkusch S, Dietz MS, Barth HD, Jungmann R, Heilemann M, Correlative Single-Molecule FRET and DNA-PAINT Imaging, Nano Lett, 18 (2018) 4626–4630. [DOI] [PubMed] [Google Scholar]
- [70].Tymoczko J, Schuhmann W, Gebala M, Electrical potential-assisted DNA hybridization. How to mitigate electrostatics for surface DNA hybridization, ACS Appl Mater Interfaces, 6 (2014) 21851–21858. [DOI] [PubMed] [Google Scholar]
- [71].Fixe F, Branz HM, Louro N, Chu V, Prazeres DM, Conde JP, Electric-field assisted immobilization and hybridization of DNA oligomers on thin-film microchips, Nanotechnology, 16 (2005) 2061–2071. [DOI] [PubMed] [Google Scholar]
- [72].Garcia-Schwarz G, Rogacs A, Bahga SS, Santiago JG, On-chip isotachophoresis for separation of ions and purification of nucleic acids, J Vis Exp, (2012) e3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Han CM, Katilius E, Santiago JG, Increasing hybridization rate and sensitivity of DNA microarrays using isotachophoresis, Lab Chip, 14 (2014) 2958–2967. [DOI] [PubMed] [Google Scholar]
- [74].Iqbal M, Tao Y, Xie S, Zhu Y, Chen D, Wang X, Huang L, Peng D, Sattar A, Shabbir MA, Hussain HI, Ahmed S, Yuan Z, Aqueous two-phase system (ATPS): an overview and advances in its applications, Biol Proced Online, 18 (2016) 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Bordelon H, Russ PK, Wright DW, Haselton FR, A magnetic bead-based method for concentrating DNA from human urine for downstream detection, PLoS One, 8 (2013) e68369. [DOI] [PMC free article] [PubMed] [Google Scholar]