Abstract
Background
The prevalence of allergy and other common chronic diseases is higher in developed than developing countries, and higher in urban than rural regions. Urbanization through its modification of environmental microbiomes may play a predominant role in the development of these conditions. However, no studies have been conducted to compare the microbiome in house dust among areas with different urbanization levels.
Methods
House dust from Xinxiang rural area (XR, n = 74), Xinxiang urban area (XU, n = 33), and Zhengzhou urban area (ZU, n = 32) in central China, and from Australia (AU, n = 58 [with pets AUP, n = 15, without pets AUNP, n = 43]) were collected during a summer season in China and Australia. High-throughput sequencing of 16S rDNA was employed to profile house dust bacterial communities.
Results
Settled dust collected in China was dominant with 2 bacterial phyla: Proteobacteria and Actinobacteria, while floor dust collected in Australia had a higher proportion of phylum Proteobacteria, Firmicutes, and Actinobacteria. XR dust samples presented higher bacterial richness and diversity compared with XU or ZU samples. Urbanization level (r2 = 0.741 P < 0.001) had a significant correlation with the distribution of house dust bacterial community. At the genus level, there was a positive correlation (r coefficient > 0.5) between urbanization level and bacterial genera Streptococcus, Bartonella, Staphylococcus, Pseudomonas, Acinetobacter, Bacteroides, Corynebacterium_1,and Enhydrobacter and a negative correlation (r coefficient < −0.5) with Rhodanobacter.
Conclusion
There was a significant difference in house dust microbiota among different urbanization areas. The areas with a lower urbanization level presented higher dust-borne bacterial richness and diversity. Modern urbanization has a significant influence on the bacterial microbiome profiles of indoor dust.
Keywords: House dust bacteria, Occupants, Pet, Level of urbanization
Introduction
With the development of urbanization, people spend most of their time indoors,1 which means that the time we interact with the indoor microbiome, mainly dust-borne microbes, is increasing. House dust is a complex mixture of inorganic and organic materials with microbes in abundance. It has been estimated that up to 500–1000 different species can be present in house dust.2 Dust-borne microbes and their products suspend in the air and produce a significant indoor exposure by inhaling.2 Environmental exposure to certain microorganisms that is potentially harmful to health as pathogens, such as antimicrobial-resistant Staphylococcus aureus,3 has been found to be responsible for the observed inverse association with atopic diseases such as asthma and hay fever.4,5 Microbial communities might even be beneficial to us in terms of ancillary mucosal barrier function, metabolism, and interaction with host immune response.6, 7, 8
We have seen a steep increase in allergic and other chronic inflammatory diseases in Western developed countries during the second half of the 20th century in marked contrast with the low prevalence in developing countries.9, 10, 11 The increased allergic disease expression has also been observed in residents living in urban compared to rural regions.12 These epidemiological evidences consistently indicate that modern urbanization plays a predominant role in the development of chronic conditions including allergic diseases. Reduced environmental microbial stimulation with better sanitation and living conditions is suggested to be at the origin of the increased prevalence of allergic diseases in Westernized countries, which is consistent with the "hygiene hypothesis".4,13,14 Furthermore, lower prevalence of allergic diseases in farming environments is possibly due to more exposure to a range of fungi and bacteria.4,5,15 In addition, reduced biodiversity that is associated with the environmental microbiome from soil and air has been implicated in adverse health outcomes.16 Given the substantial importance of the microbiome environments on health, it is surprising that there are sparse studies investigating the composition and diversity of environmental microbiomes, specifically in relation to different levels of urbanization in developing and developed countries as herein reported.
In the present study, we collected house dust samples in central China from 2 urban cities (Zhengzhou and Xinxiang) with different classifications of urbanization level, and a rural area (Xinxiang), as well as in Perth, the capital city of Western Australia. We aimed to compare the composition and diversity of the bacterial microbiome in house dust between rural and urban areas in central China, as well as between a developed country (Australia) and a developing country (China). We also investigated the effect of occupants and pets (dogs and cats) on the house dust microbiome.
Materials and methods
Study areas
House settled dust samples were collected from 3 areas with incremental levels of urbanization in central China (Xinxiang rural [XR] area, Xinxiang urban [XU] area and Zhengzhou urban [ZU] area), and floor dust samples from houses in Perth, the capital city of Western Australia [AU]. Zhengzhou is the provincial capital city of Henan province and it is a new first-tier city, while Xinxiang is a third-tier city in China. These study areas in China are geographically close with similar climate. The sampling houses from Zhengzhou and Xinxiang urban areas are in the high-tech zone, with a fast developing phase. The sampling houses selected from Xinxiang rural area are in the traditional farming area. The Globalization and World Cities Research Network in 2018 (GaWC 2018)17 classified the world cities into “Alpha”, “Beta” and “Gamma” tiers, based upon their international connectedness. Perth, the capital city of Western Australia is a Beta world city, Zhengzhou is a Gamma world city, and Xinxiang has no specific classification in GaWC 2018. We classified urbanization level into 0 (XR), 1 (XU), 2 (ZU) and 3 (AU).
Samples collection
Two types of house dust samples were collected: settled dust and floor dust. Settled dust was collected with a cotton swab, which had been disinfected by ultraviolet in the biosafety cabinet, on the top of a smooth surface, mainly on a cabinet. Floor dust was collected with a domestic vacuum machine by house keepers. And samples from Australia were transported to China in ambient room temperature, after about 10 h flights. A previous study showed that transport of dust samples at ambient indoor temperature within 12 h would not alter dust significantly.18 Samples were stored in −80 °C freezers until the analysis. In each sample, about 250 mg dust was collected. The dust samples were transferred directly into aluminum foil envelopes. Settled dust samples were collected in the rural (XR, n = 74) and urban areas (XU, n = 33; ZU, n = 32) in central China. Floor dust samples were collected in Perth city, Western Australia (AU, n = 58). Additionally, we divided AU samples into 2 groups: Australia sampling houses with pets (AUP); and Australia sampling houses without pets (AUNP). The samples from China were collected in July and August 2017, and those from Australia were collected in December 2017 and January 2018. China and Australia are located in the northern and southern hemispheres, respectively, and we matched the sampling season (summer) in the 2 countries. The average indoor temperature was 27 °C, and average indoor relative humidity was 42%. The occupant number in all sampling houses was recorded.
DNA preparation and sequencing
Bacterial DNA was extracted from 250 mg dried fine dust samples using the cetyltrimethylammonium bromide (CTAB) method. The concentration and purity of DNA were determined through electrophoresis on 1% agarose gel, and subsequently, the DNA was diluted with sterile water to a concentration of 1 ng/μL. For sequencing analyses, the V4 region of the 16s rRNA gene19, 20, 21 was amplified through polymerase chain reaction (PCR; 98 °C for 1min, followed by 30 cycles at 98 °C for 10s, 50 °C for 30s, and 72 °C for 30s, with a final extension at 72 °C for 5min) that used the primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGT WTCTAAT-3′). PCR reactions were performed in a 30-μL reaction solution containing 2X Phusion® High-Fidelity PCR Master Mix (New England Biolabs), 0.2 μM forward and reverse primers, and approximately 10 ng of template DNA (according to the DNA concentration). PCR products were mixed with the same volume of 1 × loading buffer and were detected through electrophoresis on 2% agarose gel. A Gene JET™ Gel Extraction Kit (Thermo Scientific) was used to purify PCR amplification products. High-throughput sequencing of 16s rRNA was conducted on the Ion S5™ XL platform.
Sequencing analysis
Single-end reads were assigned to DNA samples by identifying their unique barcode; the raw reads were then truncated by cutting off the barcode and primer sequence by using QIIME using QIIME V1.9.1 (http://qiime.org/scripts/split_libraries_fastq.html) according to: (1) read-quality score not less than 19; (2) setting length not to fall below 3bp; (3) consecutive high quality base over 75%. Chimera sequences were removed with the usearch61 algorithm (http://qiime.org/scripts/identify_chimeric_seqs.html).22 Quality filtering on the raw reads was performed under specific filtering conditions to obtain the high-quality clean reads according to the Cutadapt23 quality controlled process. The reads were compared with the reference database using UCHIME algorithm24 to detect chimera sequences, and then the chimera sequences were removed.25 Then the Clean Reads finally obtained. Sequence analysis was performed by Uparse software (Uparse v7.0.1001),26 and sequences with a similarity greater than 97% were assigned to the same operational taxonomic units (OTUs) according to the SILVA reference database.27 Representative sequences for each OTU were screened for further annotation. The α-diversity including Chao 1 and Shannon indices describes the number of taxa in a single sample. QIIME (Version 1.9.1) was used to calculate the Chao1 estimator to estimate the species richness, the Shannon index to estimate community diversity as well as the Pielou index to estimate community evenness. The difference of these indices between groups was compared using a Wilcoxon rank sum test. The β-diversity analysis indicates the extent of similarity between microbial communities among samples. The distances/similarities between microbial communities were calculated using weighted or unweighted UniFrac. Principal coordinate analysis (PCoA) plots were created using the Brady-Curtis measure in QIIME to check the differences and variations in bacterial communities and correlated the similarity of different microbial communities in house dust samples. The difference of relative abundance between groups was compared using Kruskal-Wallis tests. Tax4Fun was employed based on SILVA database to predict the functional profiles of microbial communities of house dust samples. The estimated abundances from the Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology groups were compared between XR, XU and ZU groups, respectively, and the difference of the pathway between groups, using independent sample t-test. P values were adjusted for multiple testing using false discovery rate (FDR) control.
The canonical correspondence analysis (CCA) was performed to examine the relationships between urbanization level and other environmental factors (household occupant number, the number and duration of pets kept in the house) and bacterial community composition. In CCA biplot, the length of the arrow represents the correlation between environmental factors and the distribution of the bacterial community; the longer the arrow, the greater the correlation. EnvFit analysis was used to determine significant covariates and the significance values were estimated on the basis of 10,000 permutations tests. The corresponding value of CCA1 and CCA2 is the cosine of the angle between the arrow of environmental factor and the sorting axis, indicating the correlation between environmental factor and the sorting axis; r2 represents the determinant coefficient of the environmental factor on the distribution of species. Furthermore, Spearman's rank correlation analysis was used to examine the correlations between urbanization level and house dust bacterial diversity. R (version 2.15.3) was used to draw all diagrams and analyze the microbiome data. Bioinformatics analysis was conducted by Beijing Novogene Bioinformatics Technology Co., Ltd., under the authors' supervision.
Results
Housing characteristics
One hundred and ninety-seven houses were included in this study. XR had the highest occupant number (5.7 ± 1.8) and the lowest urbanization level (0), while AU had the lowest occupant number (AUP 3.2 ± 1.6, AUNP 3.2 ± 1.2) and the highest urbanization level (3). In AU sampling houses, the average pet number was 1.3, and the average years of pet kept in the house was 6.4 years. No pets were recorded in any of the sampling houses in China.
House dust bacterial composition
OTUs were single rarefied to get even depths of 64,269 for settled dust and 68,594 for floor dust, respectively. Approximately 52 bacterial phyla and 1152 bacterial genera were detected from the settled dust samples; 57 bacterial phyla and 1343 bacterial genera were detected from the floor dust samples. Phylum-level taxonomical assignment showed that Proteobacteria and Actinobacteria were dominant in settled dust samples: XR (47.62%, 15.17%), XU (46.91%, 18.77%) and ZU samples (40.84%, 19.04%). Proteobacteria (37.65%), Firmicutes (20.50%) and Actinobacteria (19.07%) were the most abundant phyla in floor dust samples collected in Australia. At the genus level, unidentified_Mitochondria (5.72%), Sphingomonas (8.26%), unidentified_Chloroplast (16.28%) and Staphylococcus (8.80%) was the most abundant genus in XR, XU, ZU and AU samples, respectively (Table 1).
Table 1.
Relative abundance of house dust bacteria at the phylum and genus levels in 4 areas.
| Phylum | Relative abundance (%) |
XR-AU |
XU-AU |
ZU-AU |
XR-XU |
XR-ZU |
XU-ZU |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| XR | XU | ZU | AU | q | q | q | q | q | q | |
| Proteobacteria | 47.62 | 46.91 | 40.84 | 37.65 | 0.000 | 0.008 | 0.118 | 0.114 | 0.020 | 0.144 |
| Firmicutes | 7.11 | 16.64 | 12.73 | 20.50 | 0.000 | 0.184 | 0.004 | 0.000 | 0.003 | 0.173 |
| Cyanobacteria | 5.52 | 1.53 | 17.26 | 9.50 | 0.000 | 0.000 | 0.045 | 0.000 | 0.000 | 0.000 |
| Actinobacteria | 15.17 | 18.77 | 19.04 | 19.07 | 0.000 | 0.351 | 0.553 | 0.015 | 0.039 | 0.823 |
| Chloroflexi | 0.56 | 1.34 | 0.68 | 0.69 | 0.000 | 0.008 | 0.125 | 0.000 | 0.280 | 0.438 |
| Bacteroidetes | 9.46 | 10.58 | 5.54 | 7.49 | 0.000 | 0.004 | 0.016 | 0.151 | 0.000 | 0.000 |
| Gemmatimonadetes | 5.61 | 0.52 | 0.42 | 0.49 | 0.000 | 0.184 | 0.008 | 0.000 | 0.000 | 0.005 |
| Acidobacteria | 2.55 | 0.45 | 0.76 | 0.92 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.027 |
| Verrucomicrobia | 3.34 | 0.21 | 0.15 | 0.33 | 0.000 | 0.011 | 0.001 | 0.000 | 0.000 | 0.288 |
| Deinococcus-Thermus | 0.41 | 1.06 | 0.62 | 0.86 | 0.000 | 0.404 | 0.040 | 0.000 | 0.048 | 0.148 |
| Genus | ||||||||||
| unidentified_Mitochondria | 5.72 | 1.60 | 5.80 | 1.64 | 0.100 | 0.010 | 0.000 | 0.124 | 0.001 | 0.000 |
| Rickettsiella | 0.00 | 0.04 | 0.00 | 1.06 | 0.000 | 0.172 | 0.000 | 0.000 | 0.005 | 0.000 |
| unidentified_Chloroplast | 4.86 | 1.20 | 16.28 | 5.38 | 0.089 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| Bifidobacterium | 0.23 | 2.19 | 0.24 | 0.29 | 0.000 | 0.000 | 0.315 | 0.000 | 0.003 | 0.007 |
| Bartonella | 0.04 | 0.37 | 0.16 | 1.98 | 0.000 | 0.006 | 0.001 | 0.001 | 0.004 | 0.645 |
| Staphylococcus | 0.43 | 2.06 | 1.37 | 8.80 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.325 |
| Pseudomonas | 0.41 | 1.54 | 1.01 | 3.49 | 0.000 | 0.015 | 0.000 | 0.000 | 0.000 | 0.110 |
| Acinetobacter | 0.65 | 3.06 | 2.66 | 3.57 | 0.000 | 0.204 | 0.063 | 0.000 | 0.000 | 0.364 |
| Phyllobacterium | 0.89 | 0.35 | 4.20 | 0.38 | 0.090 | 0.279 | 0.000 | 0.046 | 0.000 | 0.000 |
| Sphingomonas | 5.34 | 8.26 | 5.28 | 2.01 | 0.000 | 0.000 | 0.000 | 0.004 | 0.286 | 0.130 |
| Bacteroides | 0.13 | 0.64 | 0.39 | 1.04 | 0.000 | 0.231 | 0.015 | 0.000 | 0.001 | 0.259 |
| Rubellimicrobium | 1.48 | 3.87 | 2.29 | 0.47 | 0.129 | 0.000 | 0.000 | 0.000 | 0.000 | 0.063 |
| Exiguobacterium | 0.25 | 1.17 | 0.36 | 0.18 | 0.237 | 0.000 | 0.000 | 0.003 | 0.007 | 0.808 |
| Lactobacillus | 0.29 | 0.81 | 1.40 | 1.35 | 0.000 | 0.088 | 0.205 | 0.000 | 0.000 | 0.658 |
| Corynebacterium_1 | 0.43 | 1.70 | 1.10 | 2.97 | 0.000 | 0.031 | 0.000 | 0.000 | 0.000 | 0.351 |
| Rhodanobacter | 5.37 | 0.01 | 0.01 | 0.01 | 0.000 | 0.073 | 0.100 | 0.000 | 0.000 | 0.824 |
| Enhydrobacter | 0.08 | 1.73 | 0.58 | 0.83 | 0.000 | 0.030 | 0.174 | 0.000 | 0.000 | 0.023 |
| Micrococcus | 0.20 | 1.18 | 0.89 | 0.61 | 0.000 | 0.017 | 0.327 | 0.000 | 0.000 | 0.207 |
| Streptococcus | 0.36 | 0.89 | 0.87 | 2.04 | 0.000 | 0.007 | 0.030 | 0.000 | 0.000 | 0.654 |
| Bacillus | 0.61 | 1.58 | 0.87 | 0.68 | 0.032 | 0.000 | 0.003 | 0.000 | 0.002 | 0.046 |
| Massilia | 0.79 | 1.83 | 0.74 | 0.95 | 0.003 | 0.066 | 0.174 | 0.000 | 0.173 | 0.067 |
| Blastococcus | 0.50 | 0.65 | 0.94 | 1.04 | 0.000 | 0.003 | 0.178 | 0.063 | 0.000 | 0.009 |
| Ralstonia | 0.30 | 0.05 | 2.76 | 0.12 | 0.200 | 0.010 | 0.000 | 0.086 | 0.000 | 0.000 |
| Paracoccus | 0.87 | 2.90 | 1.23 | 0.77 | 0.074 | 0.000 | 0.016 | 0.000 | 0.010 | 0.006 |
| Kocuria | 1.35 | 1.42 | 2.98 | 1.11 | 0.024 | 0.032 | 0.000 | 0.015 | 0.000 | 0.013 |
| Chroococcidiopsis | 0.34 | 0.10 | 0.38 | 1.08 | 0.000 | 0.000 | 0.004 | 0.117 | 0.002 | 0.000 |
| Hymenobacter | 0.49 | 1.85 | 0.89 | 0.73 | 0.000 | 0.001 | 0.297 | 0.000 | 0.001 | 0.071 |
| Pedobacter | 1.08 | 1.55 | 0.58 | 0.33 | 0.000 | 0.000 | 0.029 | 0.045 | 0.000 | 0.007 |
| Craurococcus | 0.17 | 0.18 | 0.42 | 1.03 | 0.000 | 0.000 | 0.000 | 0.199 | 0.000 | 0.008 |
| Gemmatimonas | 1.52 | 0.07 | 0.02 | 0.04 | 0.000 | 0.064 | 0.054 | 0.000 | 0.000 | 0.001 |
| unidentified_Gemmatimonadaceae | 1.41 | 0.00 | 0.04 | 0.04 | 0.000 | 0.163 | 0.480 | 0.000 | 0.000 | 0.262 |
| Reyranella | 1.17 | 0.02 | 0.02 | 0.03 | 0.000 | 0.154 | 0.249 | 0.000 | 0.000 | 0.443 |
Note: The q is a FDR-corrected probability (p value) to control for multiple testing. “XR”, “XU” and “ZU” are abbreviations for Xinxiang rural area, Xinxiang urban area and Zhengzhou urban area, respectively. “AU” stands for the sampling area in Australia. Phyla and genus which are significantly different between different samples are annotated with a black bar accompanying the p-value of significance in difference
After adjusting for multiple comparisons, significant differences were found in bacterial composition among areas of different urbanization levels, and bacteria phylum and genus showed significant differences between areas with a larger distance in urbanization levels (Table 1). Between AUP and AUNP groups (Table 2), Proteobacteria, Actinobacteria and Cyanobacteria showed significant differences, but after FDR control for multiple corrections, there were no significant difference in these taxa between AUP and AUNP groups. At the genus level, Corynebacterium_1 (Phylum Actinobacteria) was present in a significantly lower frequency in AUP dust than AUNP dust with a FDR-controlled significance of P = 0.032. Table 3 shows the results of Spearman's correlation between the urbanization level and relative abundance of individual bacteria at the phylum and genus levels. Almost all house dust bacteria with a relative abundance >1% had a significant correlation with the urbanization level, presenting an either positive or negative trend. Three of the 10 bacterial phyla (30%) and 9 of the 32 bacterial genera (28%) had a correlation coefficient of >0.5 or < -0.5. At the phylum level, urbanization increased the habitation of Firmicutes (r = 0.507, q < 0.0001) and decreased the abundance of Gemmatimonadetes (r = −0.610, q < 0.0001) and Verrucomicrobia (r = −0.515, q < 0.0001). For bacterial genera, the positive correction (r > 0.5, q < 0.0001) with urbanization level was significant for Streptococcus, Bartonella, Staphylococcus, Pseudomonas, Acinetobacter, Bacteroides, Corynebacterium_1 and Enhydrobacter. In contrast, Rhodanobacter had a significant negative correlation (r = −0.715, q < 0.0001) with urbanization level.
Table 2.
Relative abundance of house dust bacteria at the phylum and genus levels between Australian houses with pets (AUP) and Australian houses without pets (AUNP).
| Phylum | Relative abundance (%) |
AUP-AUNP |
||
|---|---|---|---|---|
| AUNP | AUP | p | q | |
| Proteobacteria | 35.78 | 43.06 | 0.040 | 0.122 |
| Firmicutes | 20.85 | 19.46 | 0.223 | 0.143 |
| Actinobacteria | 20.09 | 16.17 | 0.035 | 0.122 |
| Cyanobacteria | 10.09 | 7.80 | 0.038 | 0.122 |
| Bacteroidetes |
7.07 |
8.71 |
0.079 |
0.126 |
| Genus |
AUNP |
AUP |
p |
q |
| Rickettsiella | 0.10 | 3.83 | 0.035 | 0.307 |
| Bartonella | 2.55 | 0.34 | 0.026 | 0.276 |
| Pseudomonas | 3.02 | 4.83 | 0.958 | 0.751 |
| unidentified_Mitochondria | 1.61 | 1.70 | 1.000 | 0.751 |
| Acinetobacter | 3.20 | 4.66 | 0.552 | 0.610 |
| Stenotrophomonas | 0.39 | 1.14 | 0.824 | 0.700 |
| Sphingomonas | 1.87 | 2.42 | 0.817 | 0.698 |
| Craurococcus | 1.02 | 1.04 | 0.051 | 0.371 |
| Staphylococcus | 10.11 | 5.04 | 0.002 | 0.107 |
| Lactococcus | 0.57 | 2.13 | 0.158 | 0.511 |
| Faecalibacterium | 0.23 | 1.30 | 0.025 | 0.276 |
| Lactobacillus | 1.57 | 0.72 | 0.166 | 0.515 |
| Streptococcus | 2.04 | 2.04 | 0.619 | 0.629 |
| unidentified_Chloroplast | 5.81 | 4.12 | 0.111 | 0.420 |
| Chroococcidiopsis | 1.09 | 1.03 | 0.019 | 0.251 |
| Bacteroides | 0.53 | 2.51 | 0.004 | 0.127 |
| Corynebacterium_1 | 3.59 | 1.22 | 0.000 | 0.032 |
| Blastococcus | 1.16 | 0.67 | 0.389 | 0.610 |
| Kocuria | 1.26 | 0.67 | 0.018 | 0.248 |
P: p value; q: q value denotes the p value after adjusting for multiple tests. Phyla and genus which are significantly different between different samples are annotated with a black bar accompanying the p-value of significance in difference
Table 3.
The correlation between urbanization level (URB) and bacteria richness for bacteria with a relative abundance >1% at the phylum and genus levels.
| Phylum | URB |
||
|---|---|---|---|
| P | Q | r | |
| Proteobacteria | <0.0001 | <0.0001 | −0.364 |
| Firmicutes | <0.0001 | <0.0001 | 0.507 |
| Cyanobacteria | <0.0001 | <0.0001 | 0.33 |
| Actinobacteria | 0.0003 | 0.0004 | 0.255 |
| Chloroflexi | 0.0078 | 0.0102 | −0.189 |
| Bacteroidetes | <0.0001 | <0.0001 | −0.281 |
| Deinococcus-Thermus | <0.0001 | <0.0001 | 0.378 |
| Gemmatimonadetes | <0.0001 | <0.0001 | −0.61 |
| Acidobacteria | 0.0003 | 0.0005 | −0.253 |
| Verrucomicrobia | <0.0001 | <0.0001 | −0.515 |
| Genus | |||
| unidentified_Mitochondria | 0.0712 | 0.0809 | 0.128 |
| Lactobacillus | <0.0001 | <0.0001 | 0.483 |
| Streptococcus | <0.0001 | <0.0001 | 0.534 |
| Rickettsiella | <0.0001 | <0.0001 | 0.403 |
| unidentified_Chloroplast | 0.0392 | 0.0547 | 0.147 |
| Bifidobacterium | <0.0001 | <0.0001 | 0.29 |
| Bartonella | <0.0001 | <0.0001 | 0.549 |
| Staphylococcus | <0.0001 | <0.0001 | 0.723 |
| Pseudomonas | <0.0001 | <0.0001 | 0.621 |
| Acinetobacter | <0.0001 | <0.0001 | 0.529 |
| Phyllobacterium | 0.8427 | 0.8485 | 0.014 |
| Sphingomonas | <0.0001 | <0.0001 | −0.499 |
| Bacteroides | <0.0001 | <0.0001 | 0.504 |
| Rubellimicrobium | 0.3876 | 0.407 | −0.062 |
| Exiguobacterium | 0.8484 | 0.8485 | −0.014 |
| Corynebacterium_1 | <0.0001 | <0.0001 | 0.557 |
| Rhodanobacter | <0.0001 | <0.0001 | −0.715 |
| Enhydrobacter | <0.0001 | <0.0001 | 0.567 |
| Micrococcus | <0.0001 | <0.0001 | 0.405 |
| Massilia | 0.0171 | 0.0218 | 0.17 |
| Bacillus | 0.1608 | 0.1777 | 0.1 |
| Blastococcus | <0.0001 | <0.0001 | 0.346 |
| Paracoccus | 0.3711 | 0.3996 | 0.064 |
| Ralstonia | 0.035 | 0.0419 | 0.15 |
| Kocuria | 0.0291 | 0.0359 | 0.156 |
| Chroococcidiopsis | <0.0001 | <0.0001 | 0.483 |
| Hymenobacter | 0.0006 | 0.0008 | 0.244 |
| Craurococcus | <0.0001 | <0.0001 | 0.602 |
| Peobacter | <0.0001 | <0.0001 | −0.595 |
| Gemmatimonas | <0.0001 | <0.0001 | −0.712 |
| Unidentified_Gemmatimonadaceae | <0.0001 | <0.0001 | −0.682 |
| Reyranella | <0.0001 | <0.0001 | −0.581 |
Note: q is a p value after false discovery rate correction to control for multiple testing (number of multiple comparisons: n = 42); r is a Spearman correlation coefficient; the absolute value of r coefficient >0.5 is highlighted. Phyla and genus which are significantly associated with urbanization are annotated with a black bar accompanying the p-value of significance
House dust bacterial diversity
Table 4 shows α-diversity indices of house dust bacterial community in the settled dust and floor dust samples for each group. The bacterial α-diversity comparisons between different sample groups in China and between Australian floor dust samples collected from houses with and without pets are shown in Table 5. We did not calculate the statistics between settled dust and floor dust samples as the sample method would significantly influence the bacterial diversity. Excluding AU floor dust samples, XR group had the highest bacteria richness and diversity at levels significantly higher than ZU group. ZU samples had the lowest Shannon index at a level significantly lower than XR (P = 0.007) and XU (P = 0.011) groups. For community evenness, ZU samples were significantly lower than that of XU samples. There was no significant difference in Chao1 richness, Shannon's indices and Pielou's indices between AUP and AUNP samples. Additionally, bacterial community richness, evenness and diversity in dust from household with different kind of pets also been described, and there were no significant difference between them (Table 4, Table 5).
Table 4.
The α-diversity indexes (mean ± standard deviation).
| Group | Observed OTUs | Chao1 richness estimate | Shannon's index | Pielou's index | |
|---|---|---|---|---|---|
| Settled dust | XR | 60557.77 ± 8551.25 | 2990 ± 55 | 8.90 ± 0.06 | 0.74 ± 0.06 |
| XU | 64477.09 ± 4912.36 | 2777 ± 114 | 8.70 ± 0.22 | 0.74 ± 0.09 | |
| ZU | 72638.28 ± 4092.33 | 2688 ± 100 | 8.33 ± 0.21 | 0.68 ± 0.14 | |
| Floor dust | AUNP | 68033.09 ± 7468.36 | 3247 ± 124 | 8.72 ± 0.16 | 0.72 ± 0.09 |
| AUP | 68183.67 ± 8026.00 | 3356 ± 201 | 8.70 ± 0.38 | 0.72 ± 0.11 | |
| AUP-DOG | 2939.78 ± 773.83 | 3363.01 ± 892.86 | 8.57 ± 1.65 | 0.72 ± 0.11 | |
| AUP-CAT | 2590.50 ± 578.44 | 2927.42 ± 654.06 | 8.35 ± 1.37 | 0.72 ± 0.09 | |
| AUP-OTHERS | 3076.50 ± 0.71 | 3605.22 ± 24.61 | 9.03 ± 0.13 | 0.74 ± 0.02 |
Note: “XR”, “XU”, “ZU” and “AU” represents samples in Xinxiang rural area, Xinxiang urban area, Zhengzhou urban area and Australia area, respectively. “AUP” and “AUNP” stands for the samples from house with pets and without pets, respectively. “AUP-DOG”, “AUP-CAT”, and “AUP-OTHERS” means samples from house with dog, cat and other pets respectively
Table 5.
The α-diversity indexes comparison between sample groups.
| Groups |
P.value |
|||
|---|---|---|---|---|
| Observed OTUs | Chao1 richness estimate | Shannon's index | Pielou's index | |
| XR-XU | 0.592 | 0.295 | 0.78 | 0.28 |
| XR-ZU | <0.0001 | 0.016 | 0.007 | 0.649 |
| XU-ZU | <0.0001 | 0.238 | 0.011 | 0.04 |
| AUNP-AUP | 0.582 | 0.296 | 0.673 | 0.852 |
| AUNP-AUPCAT | 0.3848 | 0.5756 | 0.5455 | 0.909 |
| AUNP-AUPDOG | 0.5233 | 0.3003 | 0.9819 | 0.781 |
| AUNP-AUPOTHERS | 0.3886 | 0.1467 | 0.7954 | 0.474 |
| AUPCAT-AUPDOG | 0.2536 | 0.8829 | 0.5892 | 0.758 |
| AUPCAT-AUPOTHERS | 0.2149 | 0.3774 | 0.5611 | 0.643 |
| AUPDOG-AUPOTHERS | 0.617 | 0.3869 | 0.8187 | 0.814 |
Note: “XR”, “XU”, “ZU” and “AU” represents samples in Xinxiang rural area, Xinxiang urban area, Zhengzhou urban area and Australia area, respectively. “AUP” and “AUNP” stands for the samples from house with pets and without pets, respectively. “AUP-DOG”, “AUP-CAT”, and “AUP-OTHERS” means samples from house with dog, cat and other pets respectively. The alpha indices which are significantly different between different samples are annotated with a black bar accompanying the p-value of significance in difference
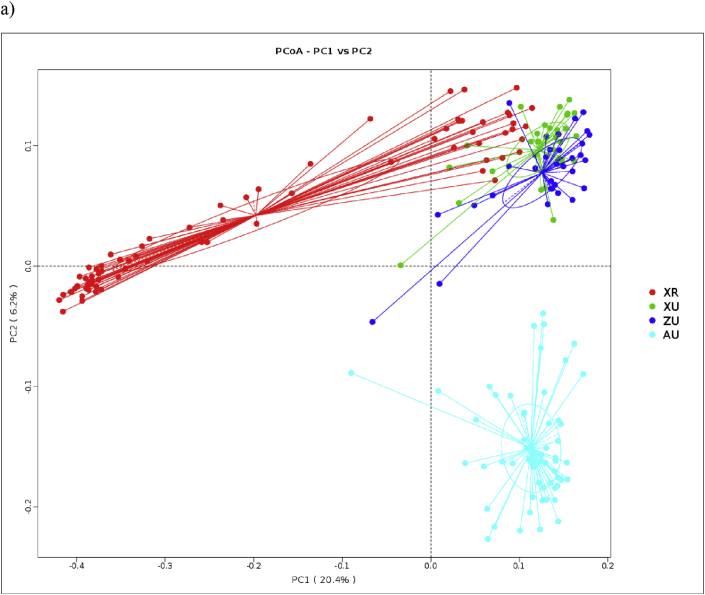
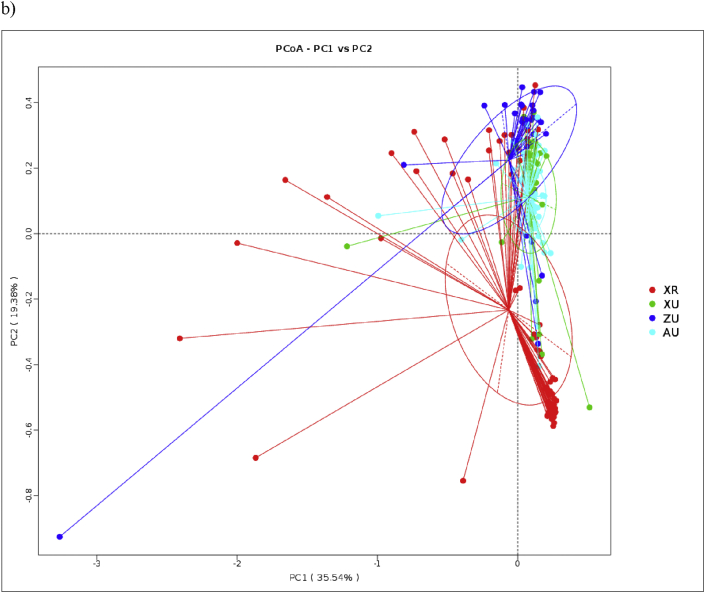
PCoA analysis of bacterial communities (Fig. 1) showed that settled dust was clustered together for the XU and ZU samples, suggesting that house dust bacterial community of urban dust samples in XU and ZU groups had higher similarity. XR samples scattered widely indicating the large variations of bacterial community among the rural dust samples. Floor dust samples were clustered together, which is also evident that bacterial community in AU group structure is similar. The Adonis statistical model analyses disclosed significant differences in the house dust bacterial communities between the different areas (Table 6).
Fig. 1.
The beta diversity of house dust samples among different urbanization level areas: Beta diversities for dust samples are presented by a) unweighted and b) weighted principal component analyses (PCoAs). Samples from different areas are shown in different colors, and the x- and y-axes are the 2 major principle components. The colors green, blue, red and cyan correspond to the samples from Xinxiang urban area, Zhengzhou urban area, Xinxiang rural area and Australia area, respectively. The numbers next to PCO 1 and PCO 2 explain the percentages of community variations. “XR”, “XU”, “ZU” and “AU” stands for Xinxiang rural area, Xinxiang urban area, Zhengzhou urban area and Australia area, respectively
Table 6.
The p-values of β-diversity comparison between each 2 groups areas.
| β-diversity | Adonis |
|
|---|---|---|
| Unweighted UniFracPCoA | Weighted UniFracPCoA | |
| XR-XU | 0.001 | 0.001 |
| XR-ZU | 0.001 | 0.001 |
| XU-ZU | 0.001 | 0.001 |
| AUP-AUNP | 0.002 | 0.012 |
Note: “XR”, “XU”, “ZU” and “AU” represents samples in Xinxiang rural area, Xinxiang urban area, Zhengzhou urban area and Australia area, respectively. “AUP” and “AUNP” stands for the samples from house with pets and without pets, respectively
Functional characterization of the house dust bacterial community
The metabolism pathway was dominantly expressed for both settled dust and floor dust samples with a proportion of 0.47. At the first KEGG level, the metabolism pathway bacteria in ZU samples were significantly more abundant than XU (P = 0.001) and XR (P = 0.001) dust samples. At the third KEGG level, when compared with the other regions (XU, ZU, AU), XR dust were enriched in a substantially wider range of predicted KEGG pathways (Supplementary Figure 1), which indicates dust samples in XR rural area accommodate microbes with more diverse function relative to urban dust samples.
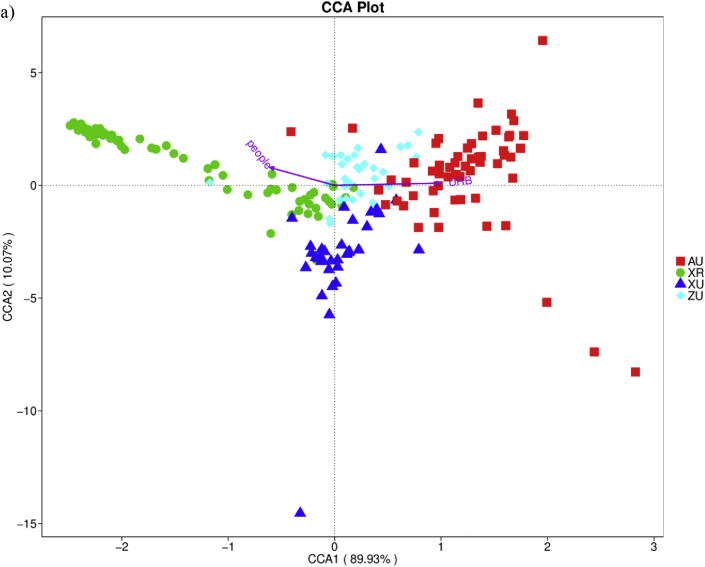
CCA analysis for urbanization level and house dust bacterial community
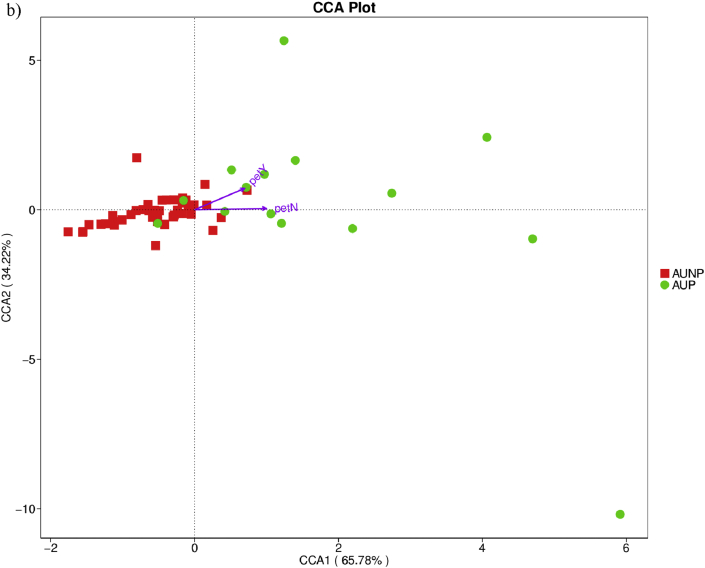
CCA was used to analyze the relationships between bacterial community composition and urbanization level and other environmental factors. The CCA biplot (Fig. 2a) distinctively exhibits the 4 dust sample groups with XR samples positioned on the left of the y axis, XU and ZU in the middle and AU samples on the right along the y axis. Urbanization level was a significant instrumental factor and positively correlated with the x and y axes, suggesting that urbanization level had a strong correlation with the distribution of house dust samples. The occupant number was also a significant factor that influenced the distribution of these samples in the CCA biplot. For AU samples only, the number of pets and the duration of pets kept in the house were significantly correlated with the distribution of floor dust samples (Fig. 2b). Additionally, "envfit" function with the CCA analysis was used to verify the relationships of these environmental factors with dust bacterial community composition (Supplementary Table 1). Urbanization level and occupant number variables explain 74% (r2 = 0.741 P < 0.001) and 31% (r2 = 0.311 P < 0.001) of the variation in the bacterial community composition, respectively.
Fig. 2.
Canonical correspondence analysis shows the relationships between environmental factors and the house dust bacterial community composition at the genus level: The length of the arrow represents the correlation between environmental factors and the distribution of the bacterial community; the longer the arrow, the greater the correlation. Acute angle shows a positive correlation and obtuse shows a negative correlation. People: the occupant number; URB: the urbanization level; petN: pet number and petY: the duration of pets kept in the house. “XR”, “XU”, “ZU” and “AU” stands for Xinxiang rural area, Xinxiang urban area, Zhengzhou urban area and Australia area, respectively. AUP: Australia sampling houses with pets; AUNP: Australia sampling houses without pets
Discussion
To the best of our knowledge, this present study is the first to compare the relative abundance and diversity of the house dust microbiome according to urbanization levels and country classification (developing vs. developed countries). We observed considerable differences in house dust microbiome profiles by urbanization level, with higher dust bacterial richness and diversity in rural than urban areas. Rural dust samples have inhabited microbes with more diverse function than urban dust samples. Most variations (74%) of the bacterial community composition could be explained by urbanization. Evidently, urbanization has significantly reshaped the indoor bacterial microbiome profiles in domestic environments where people spend most of their time. Many chronic diseases are more prevalent in developed countries and in people dwelling in urban areas. Given the continuing trend of people migrating from rural to urban areas and immigration from poor (developing) to rich (developed) countries, worldwide urbanization is leading to many health challenges. For example, asthma prevalence in adults is highest in developed and urbanized countries or areas, such as Australia (20.96%), and lowest in developing or rural areas, such as in China (0.19%).28 People from low-risk countries immigrating to high-risk countries experience a gradual increase in allergic diseases, which suggested that environmental factors including air pollution, allergens in the air, appear to play an important role in this process.29,30 Data from WHO showed that the concentration of fine particulate matter (PM2.5) was 7.19 μg/m3 in Australia and 49.16 μg/m3 in China.31 In addition, western environment and lifestyle have been associated with the development of allergic disease.32,33 Australia has the typical western lifestyle. Our previous study has showed that Chinese immigrants living in Australia for a longer period of time have increased allergic symptoms.34 Though China is a developing country with its own lifestyle, it has been experiencing great changes in home environments and lifestyle factors due to modernization and urbanization.35 Global urbanization is suspected as a cause for the allergy epidemic worldwide. Our study provides invaluable data to facilitate further research on the interrelationships of urbanization and environmental microbiome exposure as well as association with the development of common chronic diseases such as asthma and allergy.
We previously reported an evident difference in the compositions of the oropharyngeal and gut microbiomes in Chinese children living in China and Australia.36 The exposure to different local dust bacterial microbiomes in Australia and China may contribute to the distinct separation of the human microbiome in the 2 populations. In an immigrant population, we recently found a marked shift in innate and adaptive immune responses for those with less than versus those with more than 5 years of residency in Australia.34,37 Hence, further investigation is required on the association of local environment microbiome exposure, population specific human microbiomes and their influences on immunological adaptation in immigrants on the time course of disease.
The composition of certain microbiome has markedly increased/decreased as the urbanization level increases. For example, the phylum Verrucomicrobia and Gemmatimonadetes that are mainly isolated from soil38, 39, 40 decreased with modern urbanization. Phylum Firmicutes, most of which are Gram-positive,41 including some notable pathogens such as Staphylococcus and Streptococcus, had a higher relative abundance in the higher urbanization level areas. Modern urbanization also significantly influenced the relative abundance of individual bacteria at the genus level. The bacterial diversity of house dust was higher in the rural area. Early life diverse microbial exposure has been consistently shown to protect against the development of atopic disease.42 In line with our research findings in Chinese immigrants, greater bacterial diversity was hypothesized in the farming area, and this hypothesis has been fulfilled in the present study. Consistent with our findings, Pakarinen et al also reported higher bacterial content and diversity, especially the animal-associated bacteria, in house dust from Russian Karelia where there is a low occurrence of atopy and atopic disease relative to Finnish Karelian area.43 Rural dust microbes had more diverse pathway function with low dominance for metabolism pathway, which may also be related to the protective effect of rural environments against allergy.44
Our study found that the type of pets did not have an influence on the house dust microbiome richness and diversity, likely because of the small sample size. Previous studies have shown that pet ownership, especially dogs, can increase the house dust diversity, and houses with a dog or cat had a significant influence on the type of bacteria in the house environment.45,46 The presence of pets has consistently been found to influence the house-associated microbial community.45,47 In Australian floor dust, we found that most of the bacterial genera that belong to Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes showed a significant difference between houses with and without dogs or cats, and these bacteria phylum were associated with the number and duration of pets kept in the house. This is consistent with a study that found 337 taxa belonging to phyla Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes were significantly higher in abundance in houses where pets were kept.45
A limitation of this study is that different sampling methods were used to collect dust samples in China and Australia. This is because we cannot collect sufficient amounts of settled dust in Australian houses using the same method as in China. Our original study design required the collection of 250 mg dust samples to guarantee the experimental quality. With this limitation, the bacterial diversity parameters in dust samples collected in China and Australia are not comparable. However, for the correlation between bacterial richness and urbanization level, the relative abundance of individual bacteria reported in this study is to some extent comparable in dust samples collected using the 2 sampling methods. In addition, we used the same protocol of sampling and laboratory analysis for the dust samples collected from rural and urban areas in China. It is consistently agreed that the environment plays a crucial role in the development of allergy and asthma. There are many environmental factors that can have an influence on both the environmental microbiome and development of allergic diseases. Specifically for domestic environments, species of pets, keeping pets indoors or outdoors, hygiene habits and lifestyle of the inhabitants, building materials and furniture and humidity are all important factors that affect the house dust microbiome and may also play a role in the development of allergic conditions. Our future research will investigate the complex interrelationship of environmental risk factors, environmental microbiomes, and allergic diseases.
Conclusions
We found evident differences in the composition of the house dust microbiome between areas by urbanization level. Areas with lower urbanization level had higher dust-borne bacterial richness and diversity. Modern urbanization was significantly correlated with the relative abundance of most house dust bacteria. Modern urbanization has remodeled the bacterial microbiome profiles of house dust in domestic environments.
Ethics approval and consent to participate
This study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of Xinxiang Medical University. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of Xinxiang Medical University for Human Studies (IRB number XY-HS04).
Consent for publication
All authors agreed to publish the work.
Competing interests
The authors declare that they have no competing interests.
Funding
This study was supported by the National Key Research and Development Program of China (2017YFD0400301) and the National Natural Science Foundation of China (81573112).
Footnotes
Full list of author information is available at the end of the article
Supplementary data to this article can be found online at https://doi.org/10.1016/j.waojou.2020.100452.
Contributor Information
Weidong Wu, Email: wdwu2013@126.com.
Guicheng Zhang, Email: brad.zhang@curtin.edu.au.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Klepeis N.E., Nelson W.C., Ott W.R. The National Human Activity Pattern Survey (NHAPS): a resource for assessing exposure to environmental pollutants. J Expo Anal Environ Epidemiol. 2000;11(3):231–252. doi: 10.1038/sj.jea.7500165. [DOI] [PubMed] [Google Scholar]
- 2.Rintala H., Pitkaranta M., Taubel M. Microbial communities associated with house dust. Adv Appl Microbiol. 2012;78:75–120. doi: 10.1016/B978-0-12-394805-2.00004-X. [DOI] [PubMed] [Google Scholar]
- 3.Ludden C., Cormican M., Austin B., Morris D. Rapid environmental contamination of a new nursing home with antimicrobial-resistant organisms preceding occupation by residents. J Hosp Infect. 2013;83(4):327–329. doi: 10.1016/j.jhin.2012.11.023. [DOI] [PubMed] [Google Scholar]
- 4.Ege M.J., Melanie M., Anne-Cécile N. Exposure to environmental microorganisms and childhood asthma. N Engl J Med. 2011;364(8):701–709. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- 5.Lynch S.V., Wood R.A., Homer B. Effects of early-life exposure to allergens and bacteria on recurrent wheeze and atopy in urban children. J Allergy Clin Immunol. 2014;134(3):593–601.e12. doi: 10.1016/j.jaci.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abreu N.A., Nagalingam N.A., Song Y. Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci Transl Med. 2012;4(151) doi: 10.1126/scitranslmed.3003783. 151ra124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tremaroli V., Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489(7415):242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 8.Hansen C.H.F., Nielsen D.S., Kverka M. Patterns of early gut colonization shape future immune responses of the host. Plos One. 2012;7(3) doi: 10.1371/journal.pone.0034043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hugg T., Ruotsalainen R., Jaakkola M.S., Pushkarev V., Jaakkola J.J.K. Comparison of allergic diseases, symptoms and respiratory infections between Finnish and Russian school children. Eur J Epidemiol. 2008;23(2):123–133. doi: 10.1007/s10654-007-9217-z. [DOI] [PubMed] [Google Scholar]
- 10.James A.L., Knuiman M.W., Divitini M.L. Changes in the prevalence of asthma in adults since 1966: the Busselton health study. Eur Respir J. 2010;35(2):273–278. doi: 10.1183/09031936.00194308. [DOI] [PubMed] [Google Scholar]
- 11.Anandan C., Nurmatov U., Van Schayck O.C.P., Sheikh A. Is the prevalence of asthma declining? Systematic review of epidemiological studies. Allergy. 2010;65(2):152. doi: 10.1111/j.1398-9995.2009.02244.x. [DOI] [PubMed] [Google Scholar]
- 12.Viinanen A., Munhbayarlah S., Zevgee T. The protective effect of rural living against atopy in Mongolia. Allergy. 2007;62(3):272–280. doi: 10.1111/j.1398-9995.2007.01279.x. [DOI] [PubMed] [Google Scholar]
- 13.Rook G.A. Regulation of the immune system by biodiversity from the natural environment: An ecosystem service essential to health. Proc Natl Acad Sci USA. 2013;110(46):18360–18367. doi: 10.1073/pnas.1313731110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strachan D.P. Hay fever, hygiene, and household size. BMJ. 1989;299(6710):1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Mutius E., Vercelli D. Farm living: effects on childhood asthma and allergy. Nat Rev Immunol. 2010;10(12):861–868. doi: 10.1038/nri2871. [DOI] [PubMed] [Google Scholar]
- 16.Ilkka H., Leena V.H., Nanna F. Environmental biodiversity, human microbiota, and allergy are interrelated. Proc Natl Acad Sci USA. 2012;109(21):8334–8339. doi: 10.1073/pnas.1205624109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Globalization and World Cities Research Network. 2018. https://en.wikipedia.org/wiki/Globalization_and_World_Cities_Research_Network [cited 2020 2th January]; Available from: [Google Scholar]
- 18.Macher J.M. Review of methods to collect settled dust and isolate culturable microorganisms. Indoor Air. 2001;11(2):99–110. doi: 10.1034/j.1600-0668.2001.110204.x. [DOI] [PubMed] [Google Scholar]
- 19.Dannemiller K.C., Gent J.F., Leaderer B.P., Peccia J. Indoor microbial communities: Influence on asthma severity in atopic and nonatopic children. J Allergy Clin Immunol. 2016;138(1):76–83.e1. doi: 10.1016/j.jaci.2015.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dannemiller K.C., Gent J.F., Leaderer B.P., Peccia J. Influence of housing characteristics on bacterial and fungal communities in homes of asthmatic children. Indoor Air. 2016;26(2):179–192. doi: 10.1111/ina.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jayaprakash B., Adams R.I., Kirjavainen P. Indoor microbiota in severely moisture damaged homes and the impact of interventions. Microbiome. 2017;5(1):138. doi: 10.1186/s40168-017-0356-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso J.G., Kuczynski J., Stombaugh J. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10–12. [Google Scholar]
- 24.Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haas B.J., Gevers D., Earl A.M. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21(3):494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 27.Quast C., Pruesse E., Yilmaz P. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41(D1):D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.To T., Stanojevic S., Moores G. Global asthma prevalence in adults: findings from the cross-sectional world health survey. BMC Publ Health. 2012;12(1):204. doi: 10.1186/1471-2458-12-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H.-Y., Wong G.W.K., Chen Y.-Z. Prevalence of asthma among Chinese adolescents living in Canada and in China. Can Med Assoc J. 2008;179(11):1133–1142. doi: 10.1503/cmaj.071797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leung R.C., Carlin J.B., Burdon J.G., Czarny D. Asthma, allergy and atopy in Asian immigrants in Melbourne. Med J Aust. 1994;161(7):418–425. doi: 10.5694/j.1326-5377.1994.tb127522.x. [DOI] [PubMed] [Google Scholar]
- 31.WHO . 2020. Concentrations of fine particulate matter (PM2.5)https://www.who.int/data/gho/data/indicators/indicator-details/GHO/concentrations-of-fine-particulate-matter-(pm2-5) [cited 2020 3/15]; Available from: [Google Scholar]
- 32.Von Mutius E., Martinez F.D., Fritzsch C. Prevalence of asthma and atopy in two areas of West and East Germany. Am J Respir Crit Care Med. 1994;149(2 Pt 1):358–364. doi: 10.1164/ajrccm.149.2.8306030. [DOI] [PubMed] [Google Scholar]
- 33.Haahtela T., Laatikainen T., Alenius H. Hunt for the origin of allergy – comparing the Finnish and Russian Karelia. Clin Exp Allergy. 2015;45(5):891–901. doi: 10.1111/cea.12527. [DOI] [PubMed] [Google Scholar]
- 34.Saiganesh A., Hales B.J., Chen S. The Western environment reduces innate immune cytokine production in Chinese immigrants. J Allergy Clin Immunol. 2018;141(4):1504–1507.e3. doi: 10.1016/j.jaci.2017.11.031. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y., Mo J., Weschler C.J. Reducing health risks from indoor exposures in rapidly developing urban China. Environ Health Perspect. 2013;121(7):751–755. doi: 10.1289/ehp.1205983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo J., Lv Q., Ariff A. Western oropharyngeal and gut microbial profiles are associated with allergic conditions in Chinese immigrant children. World Allergy Org J. 2019;12(8):100051. doi: 10.1016/j.waojou.2019.100051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saiganesh A., Hales B.J., Li Y. A marked shift in innate and adaptive immune response in chinese immigrants living in a western environment. Allergy. 2018;73(10):2092. doi: 10.1111/all.13531. [DOI] [PubMed] [Google Scholar]
- 38.Fawaz M Naomi. 2013. Revealing the Ecological Role of Gemmatimonadetes Through Cultivation and Molecular Analysis of Agricultural Soils. [Google Scholar]
- 39.Parveen S., Suzana K., Davis K.E.R., Michelle S., Janssen P.H. Detection and cultivation of soil verrucomicrobia. Appl Environ Microbiol. 2005;71(12):8402. doi: 10.1128/AEM.71.12.8402-8410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thrash J.C., Coates J.D. 2015. Acidobacteria phyl. nov. Bergey;s Manual of Systematics of Archaea and Bacteria; pp. 1–5. [Google Scholar]
- 41.Konya T., Koster B., Maughan H. Associations between bacterial communities of house dust and infant gut. Environ Res. 2014;131:25–30. doi: 10.1016/j.envres.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 42.Beigelman A., Weinstock G.M., Bacharier L.B. The relationships between environmental bacterial exposure, airway bacterial colonization and asthma. Curr Opin Allergy Clin Immunol. 2014;14(2):137. doi: 10.1097/ACI.0000000000000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jaakko P., Anne H.R., Mirja S.S. Predominance of Gram-positive bacteria in house dust in the low-allergy risk Russian Karelia. Environ Microbiol. 2010;10(12):3317–3325. doi: 10.1111/j.1462-2920.2008.01723.x. [DOI] [PubMed] [Google Scholar]
- 44.Pérez-Losada M., Castro-Nallar E., Bendall M.L., Freishtat R.J., Crandall K.A. Dual transcriptomic profiling of host and microbiota during health and disease in pediatric asthma. Plos One. 2015;10(6):e0131819. doi: 10.1371/journal.pone.0131819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujimura K.E., Johnson C.C., Ownby D.R. Man’s best friend? The effect of pet ownership on house dust microbial communities. J Allergy Clin Immunol. 2010;126(2):410–412.e3. doi: 10.1016/j.jaci.2010.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barberan A., Dunn R.R., Reich B.J. The ecology of microscopic life in household dust. Proc Biol Sci. 2015;282(1814) doi: 10.1098/rspb.2015.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dunn R.R., Fierer N., Henley J.B., Leff J.W., Menninger H.L. Home life: factors structuring the bacterial diversity found within and between homes. Plos One. 2013;8(5):e64133. doi: 10.1371/journal.pone.0064133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.