Abstract
The autophagic pathway involves the encapsulation of substrates in double-membraned vesicles, which are subsequently delivered to the lysosome for enzymatic degradation and recycling of metabolic precursors. Autophagy is a major cellular defense against oxidative stress, or related conditions that cause accumulation of damaged proteins or organelles. Selective forms of autophagy can maintain organelle populations or remove aggregated proteins. Dysregulation of redox homeostasis under pathological conditions results in excessive generation of reactive oxygen species (ROS), leading to oxidative stress and the associated oxidative damage of cellular components. Accumulating evidence indicates that autophagy is necessary to maintain redox homeostasis. ROS activates autophagy, which facilitates cellular adaptation and diminishes oxidative damage by degrading and recycling intracellular damaged macromolecules and dysfunctional organelles. The cellular responses triggered by oxidative stress include the altered regulation of signaling pathways that culminate in the regulation of autophagy. Current research suggests a central role for autophagy as a mammalian oxidative stress response and its interrelationship to other stress defense systems. Altered autophagy phenotypes have been observed in lung diseases such as chronic obstructive lung disease, acute lung injury, cystic fibrosis, idiopathic pulmonary fibrosis, and pulmonary arterial hypertension, and asthma. Understanding the mechanisms by which ROS regulate autophagy will provide novel therapeutic targets for lung diseases. This review highlights our current understanding on the interplay between ROS and autophagy in the development of pulmonary disease.
Keywords: Autophagy, Reactive oxygen species, Mitophagy, Oxidative stress, Mitochondria, Pulmonary disease
1. Introduction
Autophagy (“self-eating” in Greek) is a cellular degradation and recycling process for cytosolic macromolecules and damaged organelles [1,2]. Autophagy is highly conserved in all eukaryotes, and it is required to maintain cellular homeostasis. Thus, almost all types of cells have basal levels of autophagy. Under normal conditions, autophagy regulates physiological functions in which cellular components have to be degraded, building blocks have to be formed, and the cell has to respond to stress [2]. Autophagy is also highly inducible by environmental insults and alterations in the autophagic cycle rate (flux) is commonly observed in response to stress [3]. In most cases, induction of autophagy in response to stress acts as a cytoprotective mechanism. However, excessive self-degradation can lead to cell death [[4], [5], [6]].
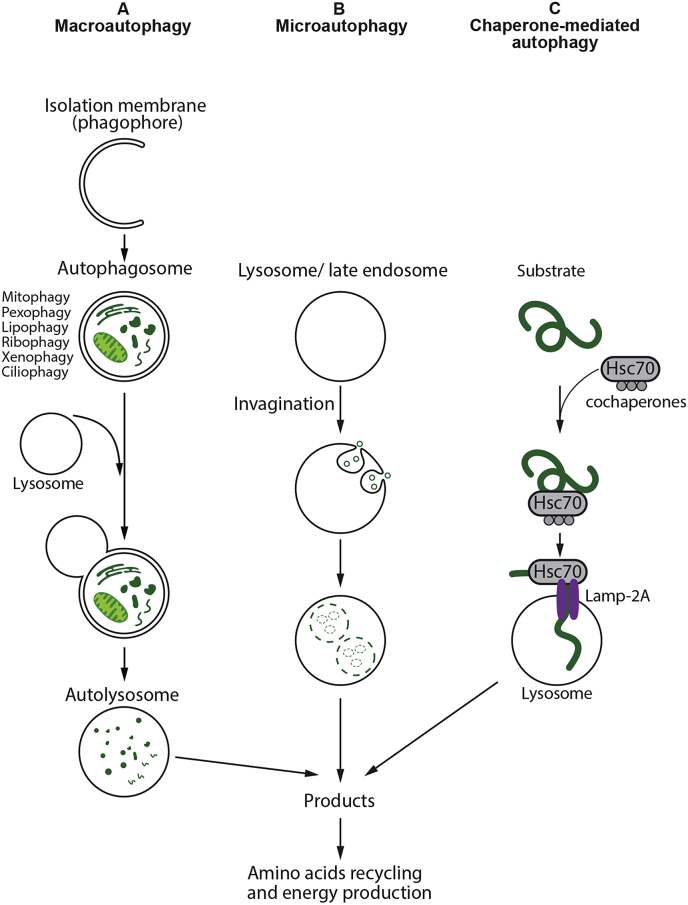
In mammalian cells, there are three primary types of autophagy: macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA). All the three pathways deliver their cargo to the lysosome for degradation and recycling (Fig. 1) but are mechanistically different from one another [7,8]. Macroautophagy (hereafter referred to as autophagy) has been the most investigated and characterized mechanism of autophagy [9]. It is induced by several stimuli, such as nutrient deprivation and other cellular stressors [10]. In contrast to microautophagy and CMA, macroautophagy involves de novo synthesis of double-membrane vesicles, autophagosomes that are used to sequester cargo and subsequently transport it to the lysosome [7,11]. Microautophagy does not require autophagosomes but involves the direct engulfment of the cargo by the invagination of the lysosomal membrane [12,13]. Lysosomal enzymes directly access the cargo by degeneration of the inner membrane [5,6,13]. Recent studies have demonstrated that a microautophagy-like process called endosomal microautophagy can also transport soluble cytosolic proteins to the vesicles of late endosomal multivesicular bodies [14,15]. Unlike microautophagy and macroautophagy, which both nonspecifically engulf bulk cytoplasm, CMA is highly specific, and has so far only been described in mammalian cells. CMA differs from microautophagy in that it does not use membranous structures to sequester cargo, but instead involves the chaperone protein Hsc70 to identify proteins with a consensus KFERQ [16]. These proteins are recognized by the lysosome-associated membrane protein type 2a (LAMP2a) receptor, a major regulator of CMA, to transport them into the lysosomes for degradation [17]. CMA degrades a wide range of substrate proteins, including certain glycolytic enzymes, transcription factors and their inhibitors, calcium and lipid binding proteins, proteasome subunits, and proteins involved in vesicular trafficking [18].
Fig. 1.
Different types of autophagy. (A) Macroautophagy: A cytosolic substrates or “cargo” is enclosed by an isolation membrane (also called phagophore) to form a double-membrane vesicles, termed autophagosomes. The outer membrane of the autophagosome fuses with the lysosome, and the internal material is degraded in the autolysosome. Macroautophagy can also target selective cargo for degradation such as organelles, proteins, microbes, and RNA. (B) Microautophagy: Small cytosolic components are directly engulfed into the lysosome or late endosomes by membrane invagination. (C) Chaperone-mediated autophagy: Substrate proteins containing a KFERQ-like motifs are recognized by chaperone Hsc70 and co-chaperones and translocated into the lysosomal lumen in a process that requires the lysosomal receptor protein LAMP-2A. After all three types of autophagy, the resultant degradation products can be used for different purposes, such as new protein synthesis or energy production.
While nonspecific autophagy can be induced in response to nutrient or energy deprivation to enable cell survival, growing evidence suggests that autophagy has a more selective role in the delivery of a wide range of specific cytoplasmic cargo to the lysosome for degradation [19,20]. Selective autophagy processes are named after the specific targets, for example: “mitophagy” refers to the selective removal of mitochondria by autophagy [21,22]. Other forms of selective autophagy have been identified to engage in organelle turnover such as peroxisomes (pexophagy) [23], endoplasmic reticulum (ER)-phagy [24], portions of the nucleus (nucleophagy) [25], and ribosomes (ribophagy) [26]. Selective autophagy also has the capacity to clear lipids (lipophagy) [27], RNA (Rnautophagy) [28], microorganisms (xenophagy) [29], protein aggregates (aggrephagy) [30], cilia components (ciliophagy) [31], ferritin (ferritinophagy) [32] and more recently, inflammasome complexes (inflammasomophagy) [33].
Interestingly, autophagy is regulated by reactive oxygen species (ROS) and oxidative stress [34,35]. Although the specific interactions between autophagy and oxidative stress have not been elucidated fully, it is known that autophagy can be modulated by oxidative stress, generally enhancing its induction [[36], [37], [38]]. Therefore, careful regulation of ROS and its interplay with autophagic degradation pathways is crucial for cellular homeostasis. It is also well established that both ROS and autophagy are strongly associated with pulmonary diseases, but clarifying the functional relationship of the two mechanisms has proven to be difficult due to their dual role in many disease processes. In this review, we will discuss the role of autophagy and its association with oxidative stress–mediated ROS signaling in the development and progression of pulmonary disease.
2. Molecular regulation of autophagy
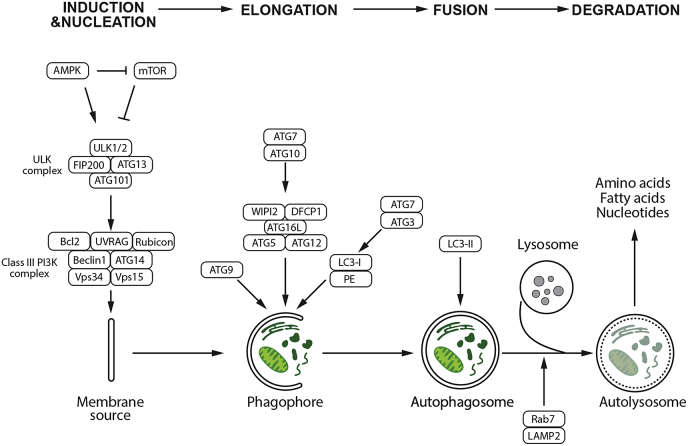
The autophagy regulatory system consists of over 30 autophagy-related gene (ATG) products that are homologues of similar proteins (Atg) originally identified in yeast [39]. The whole process of autophagy involves a set of 15–20 core conserved ATG genes. ATGs are implicated in all the different steps of the autophagic process, which can be divided into initiation, nucleation, elongation, fusion, and degradation (Fig. 2). Induction of autophagy can be triggered by several intracellular and extracellular stimuli, e.g. nutrient starvation including depletion of total amino acids [40], inhibitors of mTOR such as rapamycin [41], hypoxia [42], and oxidative stress [36]. Under autophagy-inducing conditions, the initiation of autophagy requires the formation of the Unc‐51 like autophagy activating kinase 1 or 2 (ULK1/2) complex at the phagophore assembly site (PAS). In humans, this complex includes the ATG scaffolds ATG13, ATG101, and FIP200 [43] (Fig. 2). The activity of ULK1 kinase complex can be regulated by both mammalian target of rapamycin complex 1 (mTORC1) and AMP-activated protein kinase (AMPK). Growth factors and amino acids stimulate mTORC1 and negatively regulate autophagy by directly phosphorylating ULK1 and inhibiting its activity [44,45]. Upon mTOR inhibition by starvation or rapamycin, ULK1 and ULK2 are activated and ATG13 and FIP200 are phosphorylated, which is an essential step for autophagy activity [[46], [47], [48]] (Fig. 2). Autophagy is also regulated by depletion of cellular energy through increased activity of AMPK. AMPK inactivates mTORC1 and phosphorylates and activates ULK1 to promote autophagy resulting in the degradation of cellular components to generate ATP [44,45,49]. The formation of ULK1/2 complex triggers the initiation of a double-membrane autophagosome formation. In mammals, however, the origin of the autophagosomal membrane is debated. Several studies suggest that endoplasmic reticulum (ER)-associated structures called omegasomes may contribute as initiation sites in mammals [50,51], other sources of membrane include ER-Golgi intermediate compartments [52], ER-mitochondria junctions [53], mitochondria [54], Golgi-endosomal membranes [55], and the plasma membrane [56].
Fig. 2.
Molecular regulation of autophagy. Autophagy consists of several stages: induction, nucleation, elongation, fusion to lysosome, and degradation. Under nutrient-rich conditions, activated mTOR depresses the protein kinase autophagy regulatory complex that includes ULK1/2, ATG13, FIP200, and ATG101; mTOR inhibits autophagy by inactivating the complex kinase-kinase ULK1/2; upon mTOR inactivation due to the lack of nutrients, growth factor deprivation, or other types of stress, autophagy is induced. The initial step requires the activity of the PI3K-III/Beclin-1 complex, which activate phagophore formation. ATG7 and ATG10 help ATG16L form a complex with ATG5 and ATG12, which lipidates LC3-I into LC3-II. ATG7, and ATG3 mediate LC3-II conjugating to PE. LC3-II remains associated with the maturing autophagosome The phagophore recruits cargo and closes to form the autophagosome, which fuses with a lysosome to form the autophagolysosome with the help of LAMP2 and Rab7, leading to degradation of its content.
Following the initiation step of autophagy, the nucleation stage involves the class III phosphatidylinositol 3-kinase (PI3K) complex consisting of Beclin 1, VPS34, VPS15, and ATG14 [57]. In mammalian cells, Beclin1 is a central component of PI3K complex. It interacts with a multitude of proteins including ATG14, UVRAG, Rubicon, and Bcl-2 to regulate the size and number of autophagosomes [[58], [59], [60]]. Activation of the Beclin 1 is responsible for the generation of phosphatidyl-inositol 3-phosphate (PtdIns3P), which is required for the formation of the autophagosome [61]. PtdIns3P recruits accessory protein factors that include the double FYVE-containing protein-1 (DFCP1) and WD-repeat protein interacting with phosphoinositide (WIPI) proteins, to mediate the initial stages of autophagosome formation including the recruitment of ATG16L and subsequently, the ATG5-ATG12 conjugation system to the PAS [62]. In addition, ATG9-containing vesicles generated by the secretory pathway are recruited to the PAS allowing the delivery of additional lipids and proteins contributing to membrane expansion [63]. Recent studies suggest that AMPK-dependent ULK1 phosphorylation regulates the trafficking of ATG9 [57]. Non-canonical, Beclin 1-independent autophagy has also been reported [64,65].
After the activation of PtdIns3K complex, the phagophore expands and elongates by membrane addition, which is accomplished by two essential ubiquitin‐like (Ubl) conjugation systems, which involve the Ubl proteins ATG12 and microtubule‐associated protein light chain 3 (LC3) [66,67]. The covalent conjugation of ATG12 to ATG5 involves ATG7 and ATG10 and is organized into a complex by a non-covalent association with ATG16L. This complex is essential for the elongation of the pre-autophagosomal membrane but dissociates from fully formed autophagosomes. The ATG12-ATG5-ATG16L complex localizes to the isolation membrane throughout its elongation process and serves as a ligase to promote LC3 conjugation with phosphatidylethanolamine (PE) [68]. The conjugation of PE to soluble LC3 (LC3-I) is mediated by the sequential action of the protease ATG4 (the sole protease in the ATG family), ATG7, and ATG3. Lipidation of LC3 facilitates its interaction with the forming autophagosomal membrane which facilitates cargo recruitment to autophagosomes, and allows expansion and closure of the isolation membrane to occur [69]. The lipidated form of LC3 (LC3-II) is specifically targeted to the elongating autophagosome and remains on autophagosomes until their fusion with lysosomes [70] (Fig. 2). LC3-II localized at the cytoplasmic face of autolysosomes is delipidated by ATG4 and recycled, while LC3-II found on the internal surface of autophagosomes is degraded in the autolysosomes [3]. Since LC3-II localizes to the autophagosome membranes, it is commonly used as an autophagy marker.
When initiation, nucleation, and elongation have completed, the expanding membrane closes around its cargo to form a complete autophagosome which fuses with lysosomes to form the autolysosome. The PtdIns3K complex, can activate the GTPase Rab7, which promotes fusion with lysosomes [71]. In addition, LAMP-2A, a lysosomal membrane protein, is required for the fusion of autophagosomes with lysosomes. Once fusion is complete, the autolysosome contents are degraded and the resulting molecules are released back into the cytosol as new energy sources for cell survival. In addition to providing new building blocks and energy sources, autophagy also helps to remove damaged organelles such as mitochondria as another important mechanism for cell survival [72].
3. The relationship between reactive oxygen species and autophagy
Dysregulation of ROS due to increased ROS generation and/or decreased local antioxidant defenses results in oxidative stress leading to oxidative damage of proteins, lipids, and nucleic acids [73,74]. Therefore, a balance between the generation and elimination of ROS is necessary for avoiding cell damage and this is critical for human health [75]. In biologic systems, ROS are generated by enzymatic and non-enzymatic redox reactions during cellular aerobic metabolism under both normal and pathological conditions, or upon exposure to environmental or xenobiotic agents. In general, ROS are comprised of oxygen free radicals such as superoxide (O2•−) and hydroxyl radical (OH•), as well as non-radical derivatives of oxygen, such as hydrogen peroxide (H2O2). These species are endogenously generated by several cell organelles including peroxisomes and mitochondria where approximately 90% of ROS (mtROS) are derived from the respiratory chain of the inner mitochondrial membrane [76,77]. Under physiological conditions, mitochondria produce very low levels of ROS. High ROS flux leads to irreversible alteration of target macromolecules contributing to the biological damage inside the cell associated with a number of physiological conditions, such as aging and senescence, as well as pathological states, such as cancer and neurodegeneration. Conversely, basal or physiological levels of ROS play an important homeostatic role in regulating cell proliferation, growth factor signaling, immune responses, differentiation, and autophagy [78,79]. To maintain cellular redox homeostasis, mammalian cells produce antioxidant enzymes and non-enzyme agents including superoxide dismutase (SOD), catalase, peroxidases, glutathione, and thioredoxin, which antagonize and detoxify ROS [80,81]. ROS-mediated oxidization of macromolecules and organelles or mild oxidative stress activate the autophagy pathway to eliminate damaged macromolecules and protein aggregates to maintain cellular homeostasis. Autophagy is one of the most important biological responses regulated by ROS and oxidative stress. Alterations in cellular redox state could be both, a trigger for and a regulator of autophagy, in contrast autophagy could be used by cells as a mechanism for regulating redox metabolism under various stress conditions [82,83]. Autophagy also regulate levels of ROS by other pathways such as the CMA-, p62 delivery-, mitophagy-, and pexophagy-pathways.
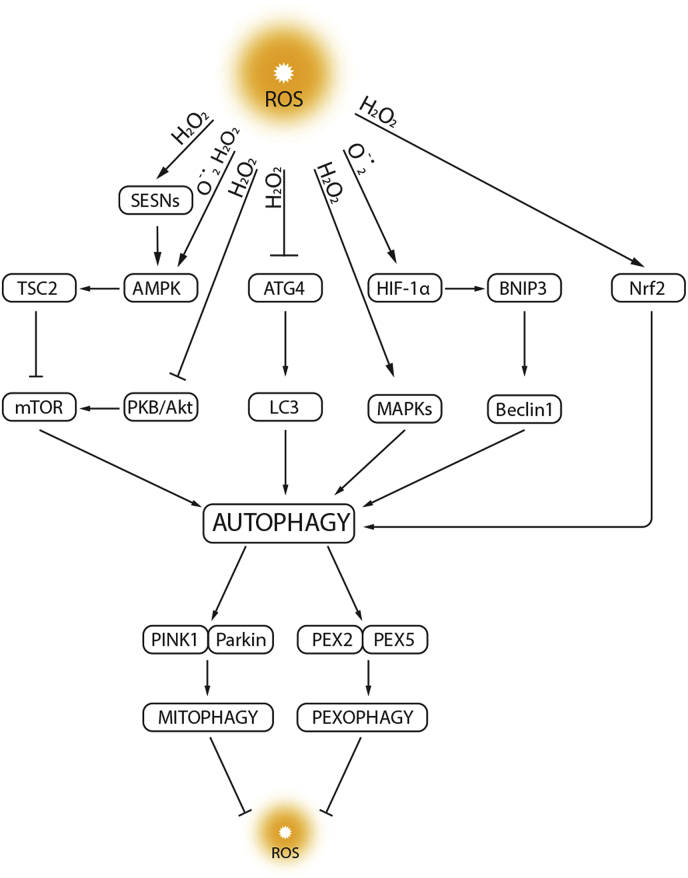
There is a growing consensus that the oxidative stress associated with increases in reactive oxygen species (ROS) and autophagy are intricately connected (Fig. 3). Autophagy can be regarded as a secondary defense to oxidative stress, by virtue of its ability to facilitate the turnover of damaged substrates which may accumulate during this process. ROS participate in the interplay between autophagy and apoptosis by ROS ability to mediate the redox signaling pathways [37,84]. This interaction triggers an autophagy response, which further induces oxidative stress when the autophagy function is disrupted. Under starvation conditions, ROS originating from mitochondria and NADPH oxidases activate autophagy to provide protection from nutrient starvation via the autophagic elimination of damaged organelles [85]. Moreover, autophagy is also a mechanism for the removal of damaged and dysfunctional mitochondria (mitophagy) that themselves are a significant source of oxidative stress. The removal of damaged mitochondria by mitophagy plays important role in cellular antioxidant defenses, by preventing pathological mtROS generation [86]. The regulation of the mammalian antioxidant response also overlaps with that of autophagy. Autophagy is crucial in ROS generation and scavenging, which is achieved by the release and activation of nuclear factor erythroid 2-related factor 2, Nrf2 [87]. Ubiquitin-binding protein p62 enhances Keap-1-Nrf2 dissociation and facilitates degradation of Keap1 by p62-dependent autophagy, further stimulating the antioxidant response [88,89]. In addition, sestrins (SESNs), proteins involved in protecting cells from oxidative stress also promote p62-dependent autophagic degradation of Keap-1 [90]. Therefore, there is a close relationship between oxidative stress and autophagy where oxidative stress can induce autophagy through various mechanisms and vice-versa [36,91]. The following sections describe in more detail the complex interplay between ROS and autophagy.
Fig. 3.
Relationship between autophagy and ROS. ROS act as signaling molecules in the regulation of autophagy by targeting autophagy genes (e.g., ATG4), transcription factors (e.g., hypoxia-inducible factor HIF-1α), and signal transduction systems (e.g., mTOR and MAPKs). In contrast, autophagy inhibits ROS accumulation through the elimination of damaged mitochondria (mitophagy) or peroxisomes (pexophagy). In response to abundant ROS, released Nrf2 is activated and stimulates transcription of antioxidant genes. Moreover, sestrins (SESNs) promote antioxidant adaptive responses via mTOR signaling inhibition. LC3, microtubule-associated protein 1 light chain 3; TSC2, tuberous sclerosis protein 2; BNIP3, BCL2/adenovirus E1B 19 kDa protein-interacting protein 3; PKB, protein kinase B; Nrf2, nuclear factor E2-related factor 2; PINK1, PTEN-induced putative kinase 1; PEX2, peroxisomal biogenesis factor 2; PEX5, peroxisomal biogenesis factor 5. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.1. Role of free radicals in autophagy regulation
There are several major sources of free radicals in cells including NADPH oxidase (NOX), xanthine oxidase, the mitochondrial electron transport chain (ETC), and nitric oxide synthase (NOS). Further, there are a number of free radicals produced within the cell including superoxide (O2·−) the hydroxyl radical (OH.), nitric oxide (NO), and peroxynitrite formed from the interaction of NO with O2·− and H2O2. There is a significant amount of literature that demonstrates the crosstalk between free radical generating systems and autophagy (Fig. 3 and Table 1). The NOX family consists of at least 7 members: NOX1-5 and DUOX-1 and -2 [92]. There is a significant amount of evidence that NOX isoforms are involved in the regulation of autophagy. However, the role of NOX in autophagy regulation appears to be dependent on the disease pathology. For example, increases in NOX1 (and NOX4) have been shown to be involved in increase in autophagy associated with HI injury [93], in transport stress in the pig jejunum [94], and in the decrease in angiogenesis associated with persistent pulmonary hypertension of the newborn (PPHN) [95]. Further, in the latter study a positive feedback relationship between autophagy and NOX activity was identified [95]. Conversely, and increase in NOX1 (and NOX4) expression in Retinal neovascularization (RNV) NOX1 (and NOX4) is associated with decreased autophagy flux [96]. Similarly, in high fat-fed mice, increased NOX2 activity appears to result in impaired lysosomal acidification and reduced autophagic flux [97]. However, the increase in autophagy associated with Parkinson's disease is associated with increases in multiple NOX isoforms: NOX1, NOX2, and NOX4 and these increases occur in both microglia and neurons within the substantia nigra of the brain [98]. Multiple NOX isoforms (NOX2, NOX4, and NOX5 but not NOX1) are also associated with the activation of autophagy associated with starvation stress in retinal pigment epithelial cells (RPE) [99]. In cultured rat cardiomyocytes, removing extracellular glucose triggers an increase in ROS that was dependent on NOX4 activity within the endoplasmic reticulum [100]. This NOX4-dependent increase in ROS induced a corresponding increase in autophagic flux [100]. In addition, NOX4-derived ROS appeared to trigger a transcriptional program mediated through a pathway involving protein kinase RNA-activated-like ER kinase (PERK) [101]. Although there is no evidence for NOX3 in regulating autophagy, DUOX has been shown to be activated downstream of autophagy in Drosophila [102,103]. Like NOX, XO appears to be able to be regulated by autophagy [104,105] and to regulate autophagy [106] although there is significantly less literature on this area which is a significant limitation in the field that needs to be addressed. Starvation-induced autophagy has been shown to depend on mitochondrial O2·− formation, which also regulates adenosine monophosphate-dependent protein kinase (AMPK) activation [107]. Moreover, amino acid deprivation induces the formation of H2O2 in mitochondria in a PI3KCIII-dependent manner, and this has also been linked to the induction of autophagy in response to starvation [108]. In addition, H2O2 can diffuse into the lysosome, particularly in neuronal cells, possibly undergoing Fenton like reactions to generate highly reactive OH. [109]. OH. is capable of reacting with the multitude of hydrolases in the lysosome, decreasing their ability to degrade molecules delivered to the lysosome. Hypoxia, or reduced pO2, also results in impaired mitochondrial electron transport chain activity, increased mitochondrial O2·− production, and mitochondrial dysfunction [110]. Under oxidative conditions Atg4 is oxidized and inactive, which allows lipidation of LC3 and the subsequent initiation of autophagy, revealing that the modulation of the action of Atg4 on LC3 can be controlled by H2O2 [34]. Other evidence suggests that H2O2 induces autophagy through a Beclin-1 dependent pathway that is associated with autophagic cell death [35].
Table 1.
Autophagy-Free radical cross-talk.
| Source of free redical | Regulator of autophagy | Regulated by Autophsy |
|---|---|---|
| NOX1 | ✓ | ✓ |
| NOX2 | ✓ | ✓ |
| NOX3 | ? | ? |
| NOX4 | ✓ | ✓ |
| NOX5 | ✓ | ✓ |
| DUOX1 | ✓ | ✓ |
| DUOX2 | ? | ? |
| Xanthine oxidase(XO) | ✓ | ✓ |
| mitochondrion | ✓ | ✓ |
| NOS | ✓ | ? |
There is also a significant literature demonstrating the involvement of NO in the regulation of autophagy. This appears to be linked to the role of NO in regulating apoptosis. However, conflicting roles of NO in regulating apoptosis and autophagy have been reported. For example, the induction of autophagy by NO is intimately involved in the survival of human dental pulp cells (HDPCs) against NO-induced apoptosis [111] and induces autophagy but not apoptosis in K562 cells [112]. Conversely, in acute liver failure, NO promotes apoptosis through autophagy inhibition in hepatic stellate cells [113]. A similar NO-mediated inhibition of autophagy is observed in hepatocellular carcinoma [114]. Interestingly in plants, there appears to be a yin-yang relationship between NO and ROS in regulating autophagy with NO suppressing, and ROS activating, autophagy [115]. As a number of vascular diseases are associated with a loss of NO signaling this could be another mechanism by which autophagy is stimulated during the disease process. One of the major sources of peroxynitrite is uncoupled NOS in which there is an uncoupling between the oxidation of NADH and the generation of NO [116]. Prevailing, though limited, data indicate that peroxynitrite is a regulator of autophagy. For example, Wogonin stimulates apoptosis in gastric cancer cells through increases in peroxynitrite generation [117] and peroxynitrite is involved in the excessive mitophagy associated with ischemia-reperfusion injury [118]. Peroxynitrite interacts with exposed tyrosine residues in certain proteins to form a covalent modification called a nitrotyrosine residue. However, at present only the transient receptor potential melastatin-related 2 (TRPM2) channel has been identified as a peroxynitrite target that stimulates autophagy [119]. It is critical to determine if there are other protein targets within the autophagy pathway susceptible to protein nitration and determine how the introduction of nitrotyrosine residues alters autophagy activity in different disease states.
3.2. The regulation of autophagy by ROS
Autophagy is activated in response to oxidative stress to protect the cells from apoptosis [120], whereas autophagy inhibition causes accumulation of oxidative stress damage and cell death [121]. Increased production of ROS activates the HIF-1α transcription factor, p53, FOXO3, and Nrf2. These transcription factors then induce the transcription of BNIP3 and NIX, TIGAR, LC3 and p62 [121]. Studies using nutrient deprivation have identified increases in O2•− [85] or H2O2 [108] as early inducers of autophagy [36]. In mammalian cells, H2O2 induces autophagy, via increased Beclin 1 expression and mTOR inhibition, and inhibiting autophagy stimulates apoptosis [122]. Increased generation of mitochondrial H2O2 oxidizes and inactivates a critical cysteine residue (Cys78) on ATG4 catalytic site [123]. Oxidized ATG4 is then capable of promoting lipidation of LC3 for autophagy initiation, followed by autophagosome formation. It was recently demonstrated that the vasoconstrictor effect of angiotensin II (Ang II) is associated with the induction of autophagy, suggesting that Ang II can stimulate the elongation of the phagophore via the development of LC3-containing autophagic vesicles and increases in ATG12–ATG5, ATG7 and ATG4 [124]. In human glomerular mesangial cells, Ang II-induced oxidative stress promotes autophagy via mTORC1 signaling causing early senescence [125]. In addition, Li et al. [126] found that the growth inhibitory effect of N-Benzoyl-O-d-phenylalanyl-d-phenylalaninol (BBP) involves induction of the autophagy in MCF-7 cells, which is modulated by ATG4 upregulation that requires increased ROS production.
Studies have shown that ROS regulate autophagy via mTOR-dependent pathways in the cytoplasm [37,127]. ROS can activate autophagy by inhibiting the PI3K-Akt-mTOR pathway or by activating AMPK to inhibit the mTOR signaling pathway [128]. Akt-mTOR activity is inhibited by elevated levels of ROS which oxidize phosphatase and tensin homologue (PTEN) [129]. Similarly, excess H2O2 can induce autophagy in an AMPK-dependent manner and this is accompanied by decreased mTORC1 activity [18]. Thioredoxin-interacting protein (TXNIP), has also been shown to regulate autophagy in a rat model of diabetic nephropathy again through the mTOR signaling pathway [130]. In addition, MAPK-p38 are responsive to the kinase apoptosis-signal kinase-1 (ASK-1), which is maintained in an inactive state until it is release by ROS-mediated oxidation of TXNIP [131]. It is also well established that SESNs are involved in regulating mTOR signaling by activating AMPK [82,132]. After exposure to cellular stress, increased levels of SESN2 can activate AMPK, resulting in the negative regulation of mTOR [132]. Morsch and colleagues [133] observed reduced SESN2 protein levels and AMPK phosphorylation, and increased mTOR phosphorylation in mice exposed to cigarette smoking for 45 days. Furthermore, mTORC2 activity is also sensitive to the redox state [134]. ROS and starvation combine increase mTORC2 signaling, leading to long-term cell cycle arrest, senescence, or differentiation [135] (Fig. 3).
Hypoxia, which is often seen in lung diseases, due to reduced lung function in these diseases is a major stimulus for the induction of autophagy [136]. Hypoxia can result in impaired mitochondrial electron transport chain activity, increased mitochondrial O2·− production, and mitochondrial dysfunction [[136], [137], [138], [139]]. Depending upon the degree of severity and duration of oxygen deprivation, hypoxia triggers different pathways of autophagy. Moderate and chronic hypoxia trigger HIF-1α as well as PKCδ–JNK1-mediated pathways to induce autophagy [42]. In moderate hypoxia, ROS can activate Beclin 1 dependent autophagy, as BNIP3, a protein that is upregulated by HIF-1α, prevents Bcl-2-mediated inhibition of Beclin 1 and free Beclin 1 induces autophagy [136]. Deletion of BECN1 or BNIP3 promotes hypoxia-induced cell death. It has been shown that stabilization of HIF-1α under hypoxia is dependent on the PI3K-Akt pathway [140]. Moreover, ROS accumulation mediated by hypoxia exposure triggers the activation of p38 and JNK-MAPK and stimulates BNIP3-induced autophagy [141]. Conversely, severe oxygen fluctuations induce autophagy via a HIF-1α-independent pathway [142] and unfolded protein response (UPR) [143]. In primary human lung vascular endothelial and smooth muscle-cells, hypoxia induces the expression of BECN1, causes LC3 activation, increases autophagosome formation, and stimulates autophagic flux resulting in protection against pulmonary vascular disease in mice [144].
Several studies have shown that ROS regulate RhoA GTPase activity, and RhoA, in turn, controls cellular redox balance by modulating the enzymes involved in ROS generation [[145], [146], [147]]. Autophagy has been shown to target and degrade active RhoA [148]. Intriguingly, RhoA can repress signaling through mTORC1, thus enhancing autophagy in a potential negative feedback loop [149]. When exposure to ROS becomes chronic, a long-term response is generated via the activation of specific transcriptional networks including the transcription factor and tumor suppressor, p53 [150]. p53-induced glycolysis and apoptosis regulator (TIGAR) as a p53 target interacts with hexokinase II (HKII), which modulates the glycolysis pathway, upregulating NADPH production and reducing mtROS levels [151,152]. Inhibition of TIGAR induces mtROS production and autophagy, whereas overexpression of TIGAR alleviates autophagy [108]. Moreover, p53-induced SESN1 and SESN2 increases autophagy via the activation of AMPK and the subsequent inhibition of mTORC1 [153]. In contrast, the sulfhydryl group-containing ubiquitin-like enzymes Atg3, Atg7, and Atg10 are redox-sensitive in a manner similar to the E1 and E2 enzymes of the ubiquitin system, and can undergo reversible oxidation and inactivation in the presence of ROS, resulting in inhibition of autophagy [154]. ROS can also modulate autophagy indirectly by affecting the activity of the master autophagy regulator, mTORC1 rather than the Atg proteins themselves [155]. Studies have shown that mTORC1 activity increases in the presence of oxidizing agents, such as diamide or the cysteine oxidant, phenylarsine oxide (PAO) [156,157]. ROS-induced inactivation of PTEN can also result in increased mTORC1 activity via the PI3K–Akt pathway, thus suppressing autophagy [158]. Thus, ROS can negatively regulate mTORC1 dependent autophagy via the oxidation, and inactivation, of PTEN. Moreover, this relationship between activated NOX2 and impaired autophagy has also observed in mouse models of Duchenne's muscular dystrophy [159]. Finally, it has been reported that CFTR deficiency causes defective autophagic flux in both human airway epithelial cells and nasal biopsies from CF patients, leading to the formation of aggresome through the production of ROS, upregulation of tissue transglutaminase (TG2), and subsequent accumulation of p62 [160]. Since autophagy is one of the primary mechanisms by which cells moderate ROS, autophagy impairment can generate a vicious feed-forward loop, where inability to remove elevated ROS levels leads to further ROS production, which may trigger cell death, and lead to disease progression.
3.3. Autophagy regulates ROS formation
Autophagy is a potential survival mechanism during oxidative stress by removing damaged cellular components [36,91]. Importantly, the removal of damaged mitochondria by mitophagy plays a crucial role in cellular antioxidant defense, by maintaining mitochondrial quality control and preventing pathological mtROS generation [86]. This represents a mechanism of negative feedback regulation by which autophagy eliminates the source of oxidative stress and protects the cell from oxidative injury. The beneficial role of autophagy in oxidative stress has been highlighted in genetic models of defective autophagy, which manifest with increased sensitivity to oxidative stress. In cells with defective autophagy proteins such as ATG5, Beclin 1, and ATG7, intracellular ROS levels increase and cells accumulate enlarged and defective mitochondria [161,162]. Thus, mitophagy plays an essential role in removal of damaged mitochondria and defects in mitophagy inhibits the selective degradation of defective mitochondria leading to subsequent cellular damage [163,164].
Mitochondria are highly dynamic organelles that maintain a stable quantity and quality through constant fusion, fission, biogenesis and mitophagy, which are the essential components of mitochondrial quality control. Mitochondria form a network of tubes through the fusion of inner and outer membranes for information exchange and mitochondrial DNA (mtDNA) damage repair [165]. Damaged mtDNA will be separated into mitochondria with low membrane potential during fission, which later will be recognized by mitophagy-related proteins and selectively cleared by the mitophagy pathway [86]. The mitophagy mediated clearance of damaged mitochondria is countered by mitochondrial biogenesis [166]. Mitophagy selectively degrades mitochondria through PINK1-Parkin and BNIP3-NIX-FUNDC1 pathways. PINK1 and Parkin appear to function as the first step of a signaling pathway that activates mitochondrial quality control pathways in response to mitochondrial damage [167]. Mitophagy can be triggered by a number of stimuli including mitochondrial depolarization, hypoxia and metabolic stress. Numerous studies suggest that the loss of mitochondrial potential (Δψm) serves as a signal for mitophagy [22,168,169]. The mitochondrial uncoupling agent carbonyl cyanide m-chlorophenyl hydrazone (CCCP), which induces mitochondrial depolarization, promotes PINK1-dependent mitophagy [168,169]. In mammalian cells, mitophagy starts with the accumulation of PTEN-putative kinase 1 (PINK1) on the depolarized mitochondria which recruits E3 ubiquitin ligase Parkin [170]. PINK1 activates Parkin by phosphorylation and active Parkin ubiquitinates mitochondrial surface proteins, and the ubiquitinated mitochondria are then delivered to the autophagosome via p62, facilitating autophagic removal of the damaged mitochondria [171]. Parkin ubiquitin targets on the mitochondria, include mitofusins (Mfn1 and Mfn2) and the mitochondrial outer membrane–voltage-dependent anion channel 1 (VDAC1). Recent studies have identified a role for ROS in promoting the translocation of Parkin to mitochondria [172,173]. Mitophagy is also activated by hypoxia through the induction of adaptor proteins: BNIP3, NIX, and FUNDC1. These proteins are localized in the outer mitochondrial membrane and contain an LC3-interacting motif to promote the recruitment of the autophagic machinery [136]. The expression of BNIP3 and NIX is increased by hypoxia through HIF-1α whereas FUNDC1 is dephosphorylated in hypoxia by Src kinase inactivation, increasing its affinity for LC3 [136,174]. Defects in BNIP3-dependent mitophagy promote tumor progression and metastasis via the upregulation of ROS, leading to enhanced HIF-1α signaling, glycolysis and angiogenesis [175]. In addition to BNIP3, ULK1 also regulates mitophagy via the mitochondrial translocation where it can then phosphorylate FUNDC1 on outer mitochondrial membrane [176]. There are also interactions between mitophagy and mitochondrial dynamics. It is now widely recognized that mitochondrial fission precedes mitophagy. Mild oxidative stress triggers mitophagy, which has been shown to be dependent on the mitochondrial fission factor dynamin-related protein (Drp) 1 [177,178]. This cooperativity is further enhanced in hypoxia with FUNDC1 recruiting Drp1 leading to mitochondrial fission and mitophagy [176]. The recognition of injured mitochondria for degradation through mitophagy is also regulated by mitochondrial membrane composition. Under normal conditions, cardiolipins, specific mitochondrial phospholipids, are mostly located in the inner mitochondrial membrane. Cardiolipin externalization to the outer mitochondrial membrane leads to its interaction with LC3 producing an elimination signal for mitophagy [179]. The hexameric intermembrane space protein, NDPK-D, has been recently identified as the translocation system for cardiolipin externalization [180].
The peroxisome is another organelle involved in ROS-induced autophagy. Peroxisomes are highly dynamic organelles that play an important role in fatty acid oxidation reactions. Peroxisomes serve dual functions in both the generation and elimination of ROS. Autophagic removal of damaged peroxisomes (pexophagy) is an important mechanism to maintain the correct redox balance in the cell [23]. Additionally, peroxisomal protein import defects have been shown to increase the rate of pexophagy [181]. Pexophagy relies on cytosolic adaptor proteins such as p62 or NBR1, which recognize ubiquitinated cargo proteins and direct them to the autophagosome [182]. Increase in ROS can stimulate pexophagy via the activation of ATM (ATM serine/threonine kinase), which phosphorylates PEX5, leading to its ubiquitination [183] (Fig. 3). ATM can also be directly activated by ROS through the formation of a disulfide-crosslinked dimer [184]. The loss of the mitochondrial chaperone, HSPA9, also leads to enhanced peroxisomal degradation by pexophagy [185]. The removal of damaged peroxisomes helps to maintain a healthy redox balance and serves as another example of how organelles signal distress through pathways that involve ROS-dependent autophagic stimulation.
The oxidation of proteins causes the exposure of hydrophobic moieties to the surface via partial unfolding and these sequences can then be targets of proteasomal degradation [186,187]. Ubiquitin-independent degradation by the 20S proteasome seems to mediate the degradation of the majority of oxidized proteins [188]. Mild oxidative stress has also been reported to increase substrate availability and up-regulate the ubiquitin-conjugation systems. In contrast, extensive oxidative stress appears to inhibit the ubiquitination system, decreasing the levels of ubiquitin conjugates and inducing the accumulation of oxidatively damaged proteins and increasing protein aggregation [189]. Covalent cross-links, disulfide bonds, hydrophobic interactions, and heavily oxidized stable proteins aggregates are not suitable for proteasomal degradation. Recent evidence suggests that autophagy plays a major role in the degradation of oxidized protein aggregates via their incomplete degradation within the lysosomal compartment. Finally, CMA has also been demonstrated to be involved in the turnover of oxidized proteins [190]. As previously described, CMA is the type of autophagy wherein a particular pool of substrate proteins is recognized by a chaperone–co-chaperone complex, with this complex delivering them to lysosome for degradation. Several components of the lysosomal translocation complex, such as the chaperoning activities of Hsp70 and Hsc70, as well as transcriptional up-regulation of LAMP-2A, are induced under oxidative stress [190,191]. Protein oxidation causes partial unfolding, not only exposing hidden recognition motifs for Hsc70 binding but also facilitating translocation to the lysosomal lumen. The process of protein unfolding can also expose hidden CMA-targeting motifs, which may facilitate their recognition by the cytosolic chaperone complex [190]. Another possibility is that oxidation of certain residues creates a previously non-existing KFERQ-like motif [121]. Exposure of CMA-incompetent cells to oxidant factors results in more severely compromised cell viability than in cells with preserved CMA function [192]. The impairment of CMA also up-regulates macroautophagy in order to maintain normal rates of long-lived protein degradation. Interestingly, up-regulation of macroautophagy is unable to compensate for the increased sensitivity of CMA-deficient mouse fibroblasts to oxidative stress [192]. Furthermore, inhibition of CMA stimulates accumulation of GAPDH, AKT1 phosphorylation, generation of ROS, and cellular apoptosis [193]. Significantly, a deletion of LAMP2, a major regulator of CMA, impairs mitochondrial health and increases ROS-induced cell death [194].
3.4. Autophagy and the antioxidant response
Numerous studies suggest a cross talk between antioxidant enzyme upregulation following oxidative stress and enhanced autophagy [36,195,196]. The oxidative stress response transcription factor, Nuclear factor erythroid 2-related factor 2 (Nrf2) is a master regulator of several genes encoding antioxidant and detoxifying enzymes that are required to maintain cellular antioxidant defenses [197,198]. Nrf2 function in controlling the transcription of antioxidant genes is mediated through interaction with the antioxidant-response element (ARE) [199] or electrophile response element (EpRE) [200]. Under normal growth conditions, Nrf2 levels in the cell are kept low due to proteasomal degradation by the Kelch-like ECH-associated protein 1 (Keap1) that functions as an adapter protein of the Cul3-ubiquitin E3 ligase complex, which ubiquitinates Nrf2 [197]. In response to oxidative stress, Nrf2 dissociates from its cytoplasmic inhibitor Keap1 and the released Nrf2 translocates to the nucleus where it binds to AREs in the promoter regions of antioxidant genes [197,198,201]. A number of oxidative agents modify cysteine residues on Keap1, resulting in conformational changes that prevent the degradation of newly synthesized Nrf2. This results in a rapid accumulation of Nrf2 in the cell. In addition to regulation of antioxidant genes, Nrf2 can regulate the transcription of p62, a protein involved in cargo recognition and autophagy. In this process, p62 interacts with the Nrf2-binding site on Keap1, and mTORC1-dependent phosphorylation of p62 significantly increases the binding affinity of p62 for Keap1, allowing release of Nrf2 and subsequent transcriptional activation of Nrf2 target genes [202]. Several studies have demonstrated that the Keap1-p62 complex formed in this process is recruited to the autophagosome by LC3, and subsequently degraded by autophagy [[201], [202], [203]] (Fig. 3). Nrf2 induces p62 gene expression in response to oxidative stress, leading to a further increase in Nrf2 [87,201]. Therefore, p62 participates in a positive-feedback loop to maintain the Nrf2 anti-oxidant effect through increased autophagy [201]. Nrf2−/− mice display increased oxidative stress, impaired antioxidant responses and decreased cardiac autophagy after high intensity exercise [204]. Thus, the disruption of the Nrf2-Keap1 pathway can suppress autophagy while autophagy deficiency leads to the overexpression of the Nrf2-Keap1 pathway. The regulation and co-ordination of Keap1–Nrf2 and the autophagy pathway are essential for cell growth, differentiation, redox signaling and survival, has been addressed in several studies [205,206]. A recent investigation showed that during prolonged exposure of oxidative stress, using tert-butyl hydroperoxide (TBHP), Nrf2 negatively regulates autophagy through the down-regulation of AMPK [207]. A recent report described a novel pathway for mitophagy where p62 translocates Keap1 to the mitochondria to ubiquitinate this organelle and trigger mitophagy [208]. Nrf2 can also regulate PINK1 expression during oxidative stress [209], highlighting the complex links between Nrf2 and mitophagy. Accordingly, Keap1 inhibitors trigger mitophagy [210]. Therefore, the p62 positive-feedback loop likely protects against oxidative stress-dependent cell death by increasing mitophagy. As Nrf2 can also regulate mitochondrial biogenesis [211], it can be considered a central regulator of mitochondrial homeostasis.
The expression of sestrins or SESNs is induced by oxidative stress as a cellular antioxidant response. SESNs promote an antioxidant adaptive response in cells by activating transcription factors, such as p53, Nrf2, AP-1, and FoxOs. Increasing evidence indicates that SESNs are positive regulators of autophagy in the face of diverse environmental stresses [[212], [213], [214]]. Although, there are some distinct differences in the functional biology of the various SESNs, they all converge to trigger activation of anti-oxidant factors and promote autophagy. SESNs have been shown to prevent ROS accumulation via inhibition of mTORC1 or stimulation of the Nrf2 [215]. Moreover, given that activation of mTORC1 increases the abundance of SESNs, SESNs are also negative feedback regulators of mTORC1 signaling. Studies indicate that SESN-dependent AMPK induction is important for mTORC1 suppression in diverse cellular contexts [216,217] (Fig. 3). SESN1 is induced by H2O2 in a p53-dependent manner, whereas the induction of SESN2 by oxidative stress is only partially dependent on p53 activation [218]. Overexpression of SESN1 or SESN2 induces autophagic degradation of Keap1 and increased Nrf2 activity [212]. Coimmunoprecipitation analyses have revealed that SESN2 interacts directly with p62 and promotes the autophagic degradation of p62-dependent targets, including Keap1, thus upregulating the transcription of antioxidant genes [219] and the depletion of SESN2 results in the accumulation of ROS [220]. SESN2 is also a substrate of ULK1 and its phosphorylation mediates mTORC1 inhibition [221], highlighting the complex crosstalk between the sestrin family and autophagy in regulating cellular redox metabolism. Beyond that, mitochondrial HKII shares an extensive relationship with autophagy and redox homeostasis. HKII induces the inactivation of mTORC1 and creates a preventive antioxidant defense by decreasing release of ROS [222].
Taken together, intracellular oxidative stress significantly regulates autophagy. Meanwhile, autophagy also regulates ROS levels in cells via the mitophagy-, pexophagy-, proteasomal-, and CMA-pathways. In addition, autophagy can directly regulate antioxidant pathways (i.e., Nrf2 and SESNs pathways) to modulate redox homeostasis and cell survival.
4. Autophagy and ROS in pulmonary diseases
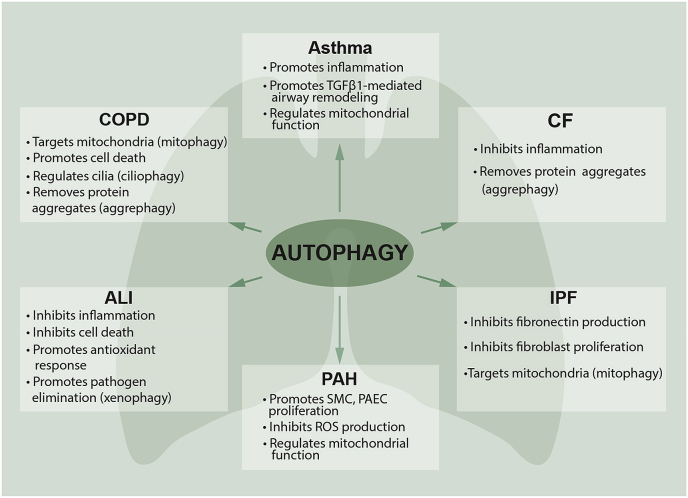
Autophagy plays a crucial role in lung homeostasis through several mechanisms, including the preservation of mitochondrial homeostasis and the removal of misfolded proteins [3,6]. As diverse pathophysiological roles of autophagy have been revealed, a number of studies exploring the roles of autophagy in pulmonary disease also have been reported [223]. Redox homeostasis has been shown to be altered in a variety of pulmonary diseases in both in vitro and experimental animal models [224]. In this section, we will discuss the role of ROS and imbalances in lung redox homeostasis signaling pathways in which autophagy is implicated in experimental pulmonary disease models and patients with pulmonary pathogenesis of chronic obstructive pulmonary disease, acute lung injury and sepsis, cystic fibrosis, idiopathic pulmonary fibrosis, pulmonary arterial hypertension, and asthma (Fig. 4).
Fig. 4.
Role of autophagy in pulmonary diseases. Autophagy plays a complex role in a growing number of pulmonary diseases. Both protective and pathogenic roles for autophagy have been proposed in these diseases. Specialized functions of autophagy (selective autophagy, such as mitophagy or aggrephagy) may directly contribute to the regulation of pathogenesis in pulmonary diseases. The upregulation of autophagy may initially act as a pro-survival mechanism responsible for the clearance of damaged proteins or organelles. In the chronic lung diseases, loss of autophagy drives lung inflammation and injury, suggesting autophagy plays a protective role. However, under certain circumstances, this naturally homeostatic cellular process may become overwhelmed and ultimately become unable to deal with excessive autophagic targets leading to cell death. COPD, chronic obstructive pulmonary disease; ALI, acute lung injury; CF, cystic fibrosis; IPF, idiopathic pulmonary fibrosis; PAH, pulmonary arterial hypertension.
4.1. Chronic obstructive pulmonary disease (COPD)
Chronic obstructive pulmonary disease (COPD) is a respiratory condition caused by direct and long-time exposure to toxic particles or gases that triggers airway or alveolar abnormalities, and typically presents with persistent respiratory symptoms and airflow limitations [225]. COPD includes both chronic bronchitis and emphysema. One of the major risk factors for COPD is cigarette smoke (CS). Epithelial cells lining the airways and alveoli are the primary targets of inhaled CS. Immune system dysregulation in response to the inhalation of CS drives prolonged and exaggerated inflammation in the lung, which contributes to the development of COPD. Other environmental factors include passive smoking, air pollution, and workplace exposure to dust, smoke or fumes [226,227]. COPD pathogenesis has been linked to excessive increases in autophagy and mitophagy (Fig. 4) which leads to epithelial programmed cell death leading to pulmonary emphysema [[228], [229], [230]]. Polymorphisms in the autophagy gene ATG16L have been shown to be a significant genetic risk factor for susceptibility to COPD, and autophagy is also increased in lung epithelium in patients with a genetic variant of emphysema, α1-antitrypsin deficiency (α1-AT) [231]. CS has been shown to produce abnormal autophagy and mitophagy leading to bronchial programmed cell death via apoptosis [228,230]. Further, impaired airway clearance caused by ciliophagy-regulated cilia shortening may prevent the elimination of pathogens from the airways, resulting in recurrent respiratory infections that exacerbate COPD.
In vitro studies using cultured epithelial cells subjected to aqueous cigarette smoke extract (CSE) have revealed increases in autophagosome formation and accumulation of LC3-II. Conversely, the inhibition of autophagy by silencing LC3B protects epithelial cells from CSE-induced apoptosis [228]. Additionally, expression of the active form of LC3, LC3-II, as well as ATG4b, ATG5, ATG12, and ATG7 levels are significantly increased in COPD lungs [232]. Alterations in mTOR signaling has also been linked to CS-induced COPD/emphysema. Pre-clinical studies using both human lung cells and tissues have shown that autophagy mediates accumulation of aggresome-bodies primarily comprised of aggregated (misfolded or damaged) proteins triggers chronic inflammatory-apoptotic responses leading to senescence and emphysema progression [[233], [234], [235]]. Acute CS exposure only initiates moderate changes in autophagy, but with chronic exposures autophagy-flux is impaired [233,236]. CS-induced mitochondrial-dysfunction and lack of mitophagy also combine to induce cellular senescence and the progression of COPD. Recent studies have shown that oxidative stress can accelerate aging by depleting stem cells, accumulation of dysfunctional mitochondria, and decline in autophagy, all of which generate additional oxidative stress [237].
Current treatment measures for COPD include administration of bronchodilators, inhaled or oral steroid medications, antibiotics, anti-inflammation drugs and anti-oxidants but these therapies do little to prevent the progression of the disease [238]. Therefore, enhancing mitophagy in human lung cells could prevent the accumulation of damaged mitochondria and the associated cellular senescence, and presents a potential therapeutic strategy in COPD [239].
4.2. Acute lung injury (ALI) and ventilator-induced lung injury (VILI)
Acute lung injury (ALI) and it more severe form acute respiratory distress syndrome (ARDS) is a life-threatening syndrome characterized by uncontrolled inflammation, oxidative injury, alveolar edema, and alveolar-capillary barrier disruption, leading to respiratory failure [240]. Autophagy is induced in response to diverse stimuli of ALI, including bacterial infection, lipopolysaccharide (LPS) exposure, sepsis, trauma, hyperoxia, pharmaceutical exposure, and mechanical ventilation [241]. One of the major inducers of autophagy in ALI is increased oxidative stress, which has been shown to promote autophagy in a number of lung cell lines and animal models of lung injury [[242], [243], [244]]. However, the role of autophagy in ALI still remains controversial (Fig. 4) with studies suggesting both protective and deleterious effects of autophagy on ALI development [[245], [246], [247]]. A protective role for autophagy in ALI comes from studies showing that the loss of key autophagy genes, such as ATG7, ATG5, and ATG4b or autophagy inhibition with chloroquine, significantly increases the development of ALI in mice suggesting autophagy may have protective effects against the progression of ALI [241,244,248,249]. Thus, inhibiting mTOR activity, with rapamycin increases autophagy and reduces both LPS-induced pro-inflammatory cytokine production and lung injury [250,251]. Moreover, mTOR knockdown significantly attenuates LPS-induced airway inflammation and injury, indicating that autophagy functions as a protective mechanism in ALI [252].
Additionally, autophagy plays significant role in preventing sepsis. In a cecal ligation and puncture (CLP) model of polymicrobial sepsis-induced ALI autophagy proteins are up-regulated in multiple organs, including heart, lungs, liver, and kidneys [[253], [254], [255], [256]]. Zhao et al [256] has demonstrated that there were significant increases in the autophagy markers LC3-II and Beclin 1, and lysosome-related protein Rab7 and LAMP2 in ALI induced by sepsis. Genetic depletion of autophagy genes such as LC3B, BECN1 increases inflammation, organ injury, and mortality in septic mice induced by CLP [251,257]. Conversely, rapamycin or LC3 overexpression reduces inflammatory response and apoptotic cell death and improves the survival in mice subjected to CLP [254,258]. In the endotoxic shock model, macrophages lacking LC3 or Beclin 1 demonstrated increased caspase-1 activation, the accumulation of depolarized mitochondria, increased mtROS, and the secretion of IL-1β and IL-18 [251]. Another pathway through which autophagy may be protective in ALI is associated with activation of Nrf2-regulated antioxidant responses to control oxidative stress and pulmonary injury [[259], [260], [261]]. Activation of Nrf2 with tert-butylhydroquinone (tBHQ) reduced LPS-induced lung injury in mice by promoting macrophages M2 polarization, increased tissue repair and reduced ROS production [261]. The lung injury associated with prolonged hyperoxia (elevated pO2) exposure also increases in the overall expression of LC3 marker in both the mouse lung through elevations in ROS [262] and in human bronchial epithelial cells [263] further emphasizing the complex interplay between autophagy/mitophagy and ROS in ALI. Pharmacological Nrf2 activation has been previously explored as a strategy to prevent and treat hyperoxia-induced ALI [264].
In contrast, several studies have shown that autophagy may have a detrimental role in ALI pathogenesis in certain circumstances [245,247,265,266]. For instance, inhibition of autophagy ameliorates acute lung injury caused by mechanical ventilation [245,247]. Mechanical ventilation, while a life-saving intervention, can expose the lung to excessive mechanical stress that exacerbates the existing injury in established ARDS, a process known as ventilator-induced lung injury (VILI). autophagy and oxidative stress contribute to the development of VILI. A growing body of evidence indicates that mechanical ventilation causes massive generation of mitochondrial ROS following mechanical stretch [[267], [268], [269]] and further promotes ROS generation from inflammatory cells [270]. LC3 expression increased in lung macrophages and genetic inhibition of autophagy abolished mechanical ventilation-induced NLRP3 inflammasome activation and subsequent release of IL-1β and IL-18 [247]. Similarly, ATG4b knockout mice also exhibited decreased NF-κB activation and alleviated inflammatory injury in response to VILI [245]. It has recently been reported that H2S (hydrogen sulfide) treatment could attenuate VILI through ROS formation inhibiting autophagy via the ROS/AMPK/mTOR pathway [[271], [272], [273]]. Subsequent studies have been performed to evaluate the effects of Nrf2 activation in the protection against VILI. Sodium sulfide protects against VILI by upregulating Nrf2 target genes involved in the restoration of redox balance [274]. Therefore, pharmacologically induced autophagy, and the subsequent activation of Nrf2, may constitute a good approach for lung protection during mechanical ventilation.
In summary, autophagy and mitophagy can play protective or detrimental roles in ALI, depending on the underlying cause of the lung injury, on the cell types, on the stage of lung injury (early phase or recovery phase), and at which step autophagy is inhibited.
4.3. Cystic fibrosis (CF)
Cystic fibrosis (CF) is caused by mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene, leading to misfolding of the CFTR protein and damage to the lung among other organs [[275], [276], [277]]. The misfolding of CFTR leads to increased ROS, reduced autophagy, accumulation of misfolded proteins in the airway epithelia of CF patients and inflammation [160,278] (Fig. 4). Blocking the expression of mutant CFTR with Spautin-1 and homocysteine restores autophagic activity in CF [279,280]. Compounds such as VX-809 and VX-661 aim to restore proper CFTR folding reestablishing normal patterns of autophagy [277,281]. These findings demonstrate the importance of mutations in CFTR in the disruption of the autophagic process. Increased ROS in the airway epithelia could be the result of reduced levels of the antioxidant GSH in CFTR mutants, as demonstrated by lower GSH levels in CFTR-knockout mice and abnormal GSH efflux in CF [[282], [283], [284]]. Additionally, some patients with CF have increased levels of γ-glutamyltransferase (GGT), which catabolizes GSH to glutamate, that can exert ROS generation and inflammation in the airways [285,286]. Indeed, treatment with a ROS scavenger, such as N-acetylcysteine (NAC), or the synthetic superoxide dismutase (SOD) EUK 134 restores autophagic function [160]. ROS have also been shown to disrupt autophagy in CF by regulating the SUMOylation of TG2 leading to its activation and the crosslinking and sequestration of Beclin 1 with the PI3K complex III to form aggresomes. Inhibition of TG2 with the proteostasis regulator cysteamine can restore the autophagic function and reduce inflammation in the airway epithelium [160,287]. Autophagy is also restored by directly promoting the expression of autophagy related protein, Beclin 1 [160]. Demethylation of the ATG12 promoter region with the natural product epigallocatechin-3-gallate (EGCG) increases ATG12 expression, and improves both autophagy and the immune response in CF [288]. Autophagy is also restored when the mTOR pathway is inhibited, suggesting that regulation of the mTOR pathway represents another avenue to restore autophagy in CF [289]. Together, these studies suggest that autophagy is a survival mechanism in the pathogenesis of CF, and pharmacological induction of autophagy might be a promising strategy to delay CF progression.
4.4. Idiopathic pulmonary fibrosis (IPF)
A growing body of evidence supports an important role of autophagy in idiopathic pulmonary fibrosis (IPF) [[290], [291], [292]] (Fig. 4). IPF is a chronic progressive lung disease characterized by scar tissue formation, lung function impairment and respiratory failure. Although IPF etiology is unclear, a number of major risk factors have been described including genetics, CS, air pollution, and aging [293]. The prevailing literature suggests that aging mitochondria and impaired autophagy/mitophagy are involved in IPF pathogenesis [[294], [295], [296], [297]]. Excessive ROS generation produced by activation of several NOX isoforms induces aberrant redox signaling and disease progression in IPF (reviewed in Ref. [298]). Cross-talk between activated NOX isoforms and mitochondria also affects mitochondrial function and mtROS generation [299]. TGFβ1-mediated activation of NOX4 is a major contributor to IPF development. NOX4 is widely distributed in different types of vascular cells and is the only NOX isoform which is up-regulated by TGFβ. Recent studies demonstrated a functional link between TGFβ-induced NOX4 levels and autophagy rate in bleomycin-exposed mice and fibroblasts derived from IPF patients [300]. NOX4 inhibition by metformin [301], NOX4 degradation by azithromycin [300] or miRNA-directed downregulation of TGFβR2/NOX4 [302] reverse or prevent bleomycin-induced pulmonary fibrosis in animals. The oxidative stress induced by NOX4 induces cell senescence resulting in a pro-apoptotic phenotype in alveolar epithelial cells (AEC) and an anti-apoptotic phenotype in senescent myofibroblasts leading to increased extracellular matrix (ECM) deposition and fibrotic foci formation. AEC senescence is accompanied by the loss of PTEN, and the activation of the NF-κB pathway [303]. Interestingly, fibroblasts cultured in the supernatants collected from senescent AEC also exhibit increased collagen deposition [303]. Mitochondria-targeted antioxidants reverse fibroblast senescence suggesting mtROS contribute to this process [304]. The analysis of lung tissue samples obtained from IPF patients shows an accumulation of p62 and low levels of LC3-II, and TGF-β1 treatment of lung fibroblasts also downregulates autophagy [305]. These data indicate that the molecular mechanisms underlying IPF development include impaired autophagy. Furthermore, stimulating autophagy with rapamycin decreases the expression of fibronectin and α-smooth muscle actin in fibroblasts in vitro and exerts an anti-fibrotic effect in the bleomycin model in vivo [305]. Detailed studies of autophagy in IPF have revealed that the accumulation of dysfunctional mitochondria is associated with Parkin [306] and PINK1 deficiency which, in turn, depends on a decrease in ATF3 transcription factor activity [294,307]. PINK1-deficient mice are prone to lung fibrosis and this is characterized by the accumulation of dysfunctional mitochondria [294]. The anti-fibrotic agent, pirfenidone, has been shown to improve autophagy/mitophagy by enhancing PARK2 expression and by decreasing mtROS levels and attenuating fibrosis in bleomycin-exposed mice [308]. The decrease in mitophagy in IPF also leads to extracellular release of mitochondrial damage associated molecular patterns (mtDAMPs), such as mtDNA, which induces pro-inflammatory cell signaling via TLR9 [309]. These can be been used as a plasma marker of severe IPF in patients [310]. [310]. Molecular mechanisms associated with aging (such as genetic instability, epigenetic changes, aging-related histone PTMs etc.) may also contribute to mitochondria dysfunction and IPF pathogenesis [311]. Mucin 5B overexpression has been shown to enhance lung fibrosis in mice [312], suggesting a cross-talk between autophagy and secretory mechanisms. Further, increased expression of mucin 5B in the lungs of IPF patients has been recently linked to MUC5B promoter polymorphisms and to alterations in its epigenetic regulation by DNA methylation) [313]. Interestingly, in contrast to other hyperproliferative/fibrotic pulmonary diseases (such as PAH and asthma), autophagy impairment is a hallmark of IPF [[294], [295], [296], [297]] and also characteristic for CF (see Section 4.3). Indeed, the systematic analysis of data obtained for various pulmonary diseases suggests that both an impaired and hyper-activated autophagy can have a negative impact on cell homeostasis and contribute to the development of disease pathology.
4.5. Pulmonary arterial hypertension (PAH)
Pulmonary artery hypertension (PAH) is a chronic disease characterized by abnormal proliferation of vascular wall cells (including pulmonary artery endothelial/PAEC, smooth muscle cells/PASMC, and fibroblasts), vascular remodeling and resistance, a decreased vessel lumen and high blood pressure followed by a fatal right heart failure. An excessive generation of ROS and reactive nitrogen species (RNS) along with mitochondrial dysfunction are characteristics for PAH and have been well-documented [269,314,315]. Moreover, in vitro and in vivo experiments with the drugs aimed at scavenging ROS/RNS or improving mitochondrial function have identified their central role in PAH development [[316], [317], [318]]. However, oxidative stress can induce autophagy [319] and increased autophagy may be important for PAH development (Fig. 4). Since autophagy is responsible for degradation of defective organelles including mitochondria, it has become clear that this may be involved in modulation of PAH progression and have a therapeutic potential. Indeed, recent studies of animal models of PAH, have investigated autophagy markers such as LC3, Beclin 1, p62 etc., and have concluded that autophagy is accelerated in the cells or lung tissue isolated from experimental models of PAH including mice exposed to chronic hypoxia, or rats treated with monocrotaline- or sugen/hypoxia-treated rats [144,320,321]. Autophagy also appears to be enhanced in lung tissue samples obtained from PAH patients [144]. A number of publications have demonstrated that increased autophagy promotes the development of PAH and is involved in the development of the hyperproliferative/anti-apoptotic phenotype in PAEC and PASMC. As a note, this phenotype is a cellular signature of PAH pathogenesis [322,323]. In concordance with these findings, several autophagy inhibitors have been tested in animal models and were able to attenuate PAH development. Lysosomal inhibitors, chloroquine and hydroxychloroquine, which inhibit autophagy by preventing autophagosome maturation, were reported to diminish proliferation of SMC and reduce PAH pathogenesis in monocrotaline-treated rats [320] and hypoxia-exposed mice [324]. Zhang and colleagues reported hypoxia-induced proliferation of PASMC can be reversed by another inhibitor of autophagy, 3-MA [323]. Hypoxia-induced activation of AMP-activated protein kinase (AMPKα-1) induces autophagy and is critical for human PASMC survival [325]. And vice-versa, inhibition of AMPK or activation of mTOR pathway can inhibit autophagy and have a negative effect on PAH progression [323,325]. Interestingly, the dietary flavonoid quercetin, induces apoptosis and potentiates hypoxia-induced autophagy in PASMCs through a FoxO1-SENS3-mTOR pathway [322,326]. Furthermore, PASMCs treated with autophagic inhibitors are more susceptible to quercetin-induced apoptosis, suggesting that combining quercetin and autophagy inhibitors could be a novel therapeutic strategy for treating hypoxia-associated PH.
4.6. Asthma
Asthma is a complex respiratory illness characterized by chronic airway inflammation and hyperreactivity, bronchoconstriction, mucus overproduction, and airway wall remodeling [327]. A number of studies have demonstrated the central role of oxidative stress in the pathogenesis of asthma [328,329], with increased levels of ROS in sputum and breath condensates positively correlating with disease severity [330,331]. Autophagy plays an important role in asthma being involved in several key features of asthma pathogenesis, including eosinophilic airway inflammation [332], airway hyperresponsiveness [333], and airway remodeling [334] (Fig. 4). Increased autophagy levels have been detected in sputum granulocytes, neutrophils, peripheral blood eosinophils and peripheral blood cells of patients with asthma [333,335,336]. There are also more double-membrane autophagosomes in fibroblasts and epithelial cells in bronchial biopsy tissue from asthmatic patients than from a healthy subjects [335]. Genetic mutations in autophagy genes are also associated with asthma with single nucleotide polymorphisms (SNPs) in the autophagy gene, Atg5 correlating with reduced lung function [337] and promotion of airway remodeling in patients with severe asthma [335,337]. Further, SNPs in Atg5 and Atg7 are associated with the development of childhood asthma [333,338]. The activation of autophagy also correlates with the production of neutrophil extracellular traps (NETs) in neutrophils [339] and genetic polymorphisms of ATG5 and ATG7 contribute to neutrophilic airway inflammation in the pathogenesis of adult asthma [340]. Activation of autophagy in epithelial cell culture model systems exposed to allergens or other antigens, is also detrimental and promotes disease progression [341,342]. Genetic depletion of ATG5 and ATG14 or pharmacological inhibition of autophagy with 3-MA or bafilomycin A1 (Baf-A1) in epithelial cells treated with IL13 results in less mucus secretion and less CCL26 (eosinophil chemokine) production, demonstrating that autophagy is involved in the activation of the Th2 response in asthma [343].
There is a strong correlation between autophagy activation and asthma airway remodeling with higher expression of Beclin 1 and ATG5 along with reduced p62 in asthmatics compared with non-asthmatics [334]. Autophagy is also a regulator of TGF-β1-induced fibrogenesis [344] and autophagy inhibition using ATG5 deletion or treatment with Baf-A1 or 3-MA decreases the fibrotic effect of TGF-β1 [344]. Furthermore, mitochondrial dysfunction and elevated ROS production are also associated with asthma [[345], [346], [347]]. For example, fine particulate matter (PM2.5) increases ROS production, mitochondrial lipid peroxidation, disrupts mitochondrial dynamics and causes mitochondrial damage, which triggers the mitophagy via the PINK/Parkin pathway [348]. Thus, mitophagy may be critical in environmental pollutant-induced injury by removing damaged mitochondria and reducing excessive ROS. Moreover, genetic polymorphisms in the ATG5 gene are associated with autophagy and asthma. This finding, coupled with the fact that autophagy and mitophagy are enhanced in the asthmatic airway, suggests that autophagic pathway modulation has the potential to be a new therapeutic target in asthma treatment.
5. Therapeutic implications of autophagy dysregulation
Autophagy can both promote and perturb the pathogenesis of pulmonary diseases (Fig. 4). In some lung diseases, such as sepsis, autophagy appears to provide protection against bacterial infection and allow an inflammatory response. In the case of exposure to cigarette smoke, autophagy aggravates and promotes disease progression for many human lung diseases. To date the array of autophagy modulating therapeutics is mostly limited to experimental compounds. Currently, only a few compounds that can modulate autophagy have been evaluated for clinical use, including both activators of autophagy e.g., the mTOR inhibitor, rapamycin and inhibitors of autophagy e.g., chloroquine/hydroxychloroquine. Chloroquine/hydroxychloroquine has been explored as a cancer therapeutic [349,350]. However, the effectiveness of these compounds in a is limited by their off-target effects unrelated to the autophagy pathway. Thus, the pleiotropic effects of these compounds make it difficult to assign therapeutic effects directly to autophagy inhibition rather than other endpoints. However, the targeting of autophagy pathways might prove more beneficial than direct scavenging of ROS, which while capable of inhibiting damage, might also diminish the normal signals that act to augment autophagy. Thus, autophagy or mitophagy activators have the potential to prove to be significantly more efficacious that antioxidant therapies. Additional candidate therapeutic compounds that modulate autophagy have been described, including a Tat-Beclin 1 peptide [351], inhibitors of histone deacetylases [352], activators of the AMPK pathway [353], and Vitamin D [354]. Given the important roles of autophagy in various lung diseases (Fig. 4), utilizing autophagy modulators could also improve the therapeutic effects of the current interventions in various lung diseases when used in combination.
Autophagy has also been shown to be induced by, and to protect against, the oxidative stress induced by numerous drugs used in cancer treatment. Various anti-cancer treatments have been shown to activate ROS-induced autophagy which in turn leads to either the development of cell drug resistance or to the induction of autophagy and/or apoptosis. Considering the complexity of cellular effects observed during cancer therapy, several treatments have been proposed, predicated on the importance of ROS as well as autophagy or apoptosis in the disease pathogenesis. Autophagy can be induced by, and to protect against, the oxidative stress induced by numerous drugs used in cancer treatment. Various anti-cancer treatments activate ROS-induced autophagy which in turn leads to either the development of drug resistance or to the induction of autophagy and/or apoptosis. Considering the complexity of cellular effects observed during cancer therapy, several treatments have been proposed, to take advantage of the importance of ROS, autophagy, and apoptosis in the disease pathogenesis. Thus, cancer therapy strategies are now trying to exploit ROS and autophagy to achieve cytotoxicity [355]. Some chemotherapy drugs are known to induce oxidative stress leading to autophagic cell death, such as arsenic trioxide (As2O3) and 2-methoxyestradiol (2-ME). As2O3 can induce apoptotic and autophagic cell death in many types of cancer cells [356] while ROS formation is essential for As2O3 cytotoxicity [357]. Similarly, 2-ME increases ROS levels by inhibiting complex I and mitochondrial SOD [358]. ROS generation induces both apoptosis and autophagy; thus combining ROS and autophagy inducing agents could produce synergistic tumor killing effects [355,359]. Therefore, it is likely that in the future, treatment strategies will be based on cell characteristics such as basal autophagy levels, beclin-1 expression, ROS levels, and apoptosis resistance. Targeting mitophagy alone also has therapeutic potential. For example, rapamycin and metformin, as general autophagy-inducing drugs, have been shown to attenuate AMPK and mTOR activity, preserving energy metabolism by regulating mitophagy and mitochondrial biogenesis [360]. In addition, several naturally occurring compounds, such as spermidine, resveratrol, urolithin A and certain antibiotics, have been shown to maintain mitochondrial integrity through the induction of mitophagy and the promotion of mitochondrial biogenesis [361]. Thus, although we are still some way from utilizing autophagy modulator therapies clinically, it is clear that oxidative stress and autophagy are key elements in the pathological progression of many pulmonary (and other) diseases, and that they interact in intricate and important ways (Fig. 3). A comprehensive clinical approach that considers the complex interplay between these important processes will be essential for successful therapeutic interventions in humans.
6. Conclusions and future perspectives
One of the most important biological responses in the cell that is regulated by ROS and oxidative stress is autophagy [6]. Autophagy provides a protective or adaptive function during the pathogenesis of diseases that involve components of metabolic or mitochondrial dysfunction, protein aggregation, inflammation, and oxidative stress. Thus, the interplay between ROS and autophagy is critical for cellular homeostasis and survival. Autophagy is now recognized to exert key functions in the pathogenesis of various human diseases [224] and multiple studies have indicated that the interplay between ROS and autophagy is highly relevant in the development of many pulmonary diseases (Fig. 4).
Oxidative stress is a complex phenomenon with important physiological and pathophysiological implications in many lung disorders. Our lack of a through basic understanding of when oxidative stress is a major driver of disease, coupled with an absence of standardized biomarkers that can easily define clinical or functional outcomes, likely plays a significant role in the lack of success in a number of antioxidant therapy trials. In the future oxidative stress will need to be framed in the context of individual (phenotypical and genetic) risk factors and environmental exposures, to determine which individuals are most likely to benefit from a particular therapy. This personalized medicine approach will necessitate the development of clinically relevant biomarkers. For example, the rational selection of patients entering clinical trials with autophagy inhibitors would require a robust set of autophagy-related biomarkers that can be followed longitudinally and allow the rapid determination of likely clinical outcome. However, even if a robust biomarker is identified, a number of other issues will need to be overcome such as differences in staining techniques, scoring systems and cut-off points, sensitivity due to limited sample sizes, and the need for independent validation processes. One of the best studied autophagy-related proteins, LC3B, has long served as an autophagy marker in multiple in vitro assays. The association of high LC3B expression with aggressive disease and poor outcomes has been repeatedly reported in various cancers [[362], [363], [364]]. However, an increase or decrease in the expression levels of autophagosomal markers such as LC3B is difficult to monitor over time, especially in specific tissues and thus may not directly correlate with autophagic activity. Identifying appropriate biomarkers that will allow changes in autophagy to be accurately measured is a major challenge for clinical studies using autophagy modulators. In this context, novel biomarkers indirectly related to autophagic activity, such as autophagy products rather than the autophagy machinery, should be explored. Due to the dynamic nature of autophagy and the complexity of its regulation, simultaneous assessment of several autophagy markers will likely be required to assess autophagy status. However, many autophagy proteins have been shown to function in other cellular processes, including cell survival and apoptosis, modulation of cellular trafficking, protein secretion, cell signaling, transcription, translation and membrane reorganization [365]. Each protein known to be involved in the autophagy machinery, when evaluated separately from other autophagy proteins, could have an independent biomarker potential in pulmonary diseases. Thus, caution will be required when interpreting the results from studies that report association of disease expression of autophagy related proteins with clinicopathological characteristics and patient outcomes.
The diverse roles of autophagy in lung disease pathogenesis might also be due to the different types of lung diseases, the diverse stressors for the etiology, the various cell types in the lung, and the different mechanisms underlying disease initiation and progression. In addition, the analytical methods, experimental approaches, reagents, and models with different cells and animals all contribute to variations between laboratories. Further, it still remains unclear how cells orchestrate nonselective autophagy and selective autophagy during disease initiation and progression, and whether nonselective autophagy cross-talks with selective autophagy [246]. The lack of autophagy inhibitors to specifically target nonselective autophagy and selective autophagy in a given lung cell type also remains a major challenge for therapeutic intervention. Furthermore, some of studies have only examined LC3 accumulation which might be a result of the activation or inhibition of autophagic flux–mediated degradation. Therefore, an increased understanding of the complex role of autophagy in the pathogenesis of disease will facilitate targeting this pathway for therapeutic gain in specific diseases. Thus, we will need to define the various pathways of selective autophagy and their relevance to disease pathogenesis in acute detail to allow the development of appropriate therapeutic agents designed for different diseases.
In conclusion, in this review, we have provided an overview of cellular autophagy, and then focused on the interrelation between ROS and autophagy. ROS-induced autophagy could be either a cellular protective mechanism that alleviates oxidative stress, or a destructive process. Autophagic degradation of Keap1 activates Nrf2 and protects against oxidative stress, while mitophagy and pexophagy regulates oxidative stress by degradation of ROS-producing organelles. Furthermore, macroautophagy and CMA play a major role in the degradation of oxidized proteins and aggregates. Finally, we discussed the interplay of ROS and autophagy in lung diseases including COPD, ALI, CF, IPF, PAH, and asthma. ROS-autophagy interaction plays a critical and complex role in the pathogenesis of these lung diseases. However, further investigation is needed to determine the underlying mechanisms and signaling pathways of the specific molecular interactions between ROS and autophagy. The appropriate adjustment of autophagy will be crucial for the development of new therapeutic strategies for chronic pathologies involving oxidative stress. Therefore, a new avenue for lung disease treatment might include combinations of antioxidants or autophagy inhibitors/activators.
Declaration of competing interest
The authors have no conflict of interest to declare related to this review article.
Acknowledgements
This research was supported in part by HL60190 (SMB), HL137282 (SMB/JRF), HL134610 (SMB), HL142212 (SMB/EZ), HL146369 (SMB), and HL061284 (JRF) all from the National Institutes of Health.
Abbreviations
- 3-MA
3-methyladenine
- ALI
acute lung injury
- AMPK
AMP-dependent protein kinase
- Ang II
angiotensin II
- AP-1
activator protein 1
- ARDS
acute respiratory distress syndrome
- ARE
antioxidant response element
- Atg
autophagy related gene
- Bcl2
B-cell lymphoma 2
- Bnip3
Bcl-2/adenovirus E1B 19-kDa-interacting protein 3
- CCCP
carbonyl cyanide m-chlorophenyl hydrazine
- CF
cystic fibrosis
- CFTR
cystic fibrosis transmembrane conductance regulator
- CMA –
chaperone mediated autophagy
- CO
carbon monoxide
- COPD
chronic obstructive pulmonary disease
- CS
cigarette smoke
- CSE
cigarette smoke extract
- DRP1
dynamin-related protein 1
- EpRE
electrophile response element
- FIP200
FAK-family interacting protein of 200 kDa
- FoxO
forkhead BoxO
- FUNDC1 –
FUN14 domain-containing protein 1
- GSH
reduced glutathione
- GTPase
guanosine triphosphatase
- HIF-1α
hypoxia-induced factor-1α
- HKII
hexokinase II;
- HLMVEC
human lung microvascular
- H2O2
hydrogen peroxide
- HSPA9
heat Shock Protein Family A (Hsp70) Member 9
- IPF
idiopathic pulmonary fibrosis IPS;
- JAK2
Janus kinase 2
- JNK
c-Jun-NH2-terminal kinase
- Keap1
Kelch-like ECH-associated protein-1
- LC3B
microtubule-associated protein-1 light chain3B
- LPS
lipopolysaccharide
- Mfn
mitofusin
- mtDNA
mitochondrial DNA
- mtROS
mitochondrial-derived reactive oxygen species
- mTOR
mechanistic target of rapamycin
- mTORC1
mTOR complex 1
- NOX
NADPH oxidase
- Nrf2
nuclear factor erythroid 2-related factor 2
- O2•−
superoxide anion
- PAH
pulmonary arterial hypertension
- PASMC
pulmonary artery smooth muscles cells
- PI3K
phosphoinositide 3-kinase
- PI3P
binding autophagy-linked FYVE domain protein AMPK
- PINK1
PTEN-induced putative kinase 1
- p62/SQSTM1
62-kDa autophagic substrate protein, sequestosome-1
- PTEN
phosphatase and tensin homologue
- RhoA
Ras homologous GTP-binding protein
- ROS
reactive oxygen species
- SMC
smooth muscle cells
- SOD
superoxide dismutase
- STAT1
signal transducer and activator of transcription 1
- TG2
transglutaminase
- TGF-β1
transforming growth factor β1
- TIGAR
P53-inducible glycolysis and apoptosis regulator
- TNFα
tumor necrosis factor α
- TSC2
tuberous sclerosis 2
- TXNIP
thioredoxin-interacting protein
- ULK1
upstream kinase UNC51-like kinase 1
- UVRAG
ultraviolet radiation resistance-associated gene protein
- VDAC1
voltage-dependent anion-selective channel 1
- VILI
ventilator-induced lung injury
- VPS34
vacuolar protein sorting 34
- Δψm
mitochondrial membrane potential
References
- 1.Klionsky D.J., Emr S.D. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290(5497):1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N., Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 3.Kroemer G., Marino G., Levine B. Autophagy and the integrated stress response. Mol. Cell. 2010;40(2):280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doherty J., Baehrecke E.H. Life, death and autophagy. Nat. Cell Biol. 2018;20(10):1110–1117. doi: 10.1038/s41556-018-0201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galluzzi L., Baehrecke E.H., Ballabio A., Boya P., Bravo-San Pedro J.M., Cecconi F., Choi A.M., Chu C.T., Codogno P., Colombo M.I., Cuervo A.M., Debnath J., Deretic V., Dikic I., Eskelinen E.L., Fimia G.M., Fulda S., Gewirtz D.A., Green D.R., Hansen M., Harper J.W., Jaattela M., Johansen T., Juhasz G., Kimmelman A.C., Kraft C., Ktistakis N.T., Kumar S., Levine B., Lopez-Otin C., Madeo F., Martens S., Martinez J., Melendez A., Mizushima N., Munz C., Murphy L.O., Penninger J.M., Piacentini M., Reggiori F., Rubinsztein D.C., Ryan K.M., Santambrogio L., Scorrano L., Simon A.K., Simon H.U., Simonsen A., Tavernarakis N., Tooze S.A., Yoshimori T., Yuan J., Yue Z., Zhong Q., Kroemer G. Molecular definitions of autophagy and related processes. EMBO J. 2017;36(13):1811–1836. doi: 10.15252/embj.201796697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizushima N. A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 2018;20(5):521–527. doi: 10.1038/s41556-018-0092-5. [DOI] [PubMed] [Google Scholar]
- 7.Yang Z., Klionsky D.J. Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010;22(2):124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ravikumar B., Sarkar S., Davies J.E., Futter M., Garcia-Arencibia M., Green-Thompson Z.W., Jimenez-Sanchez M., Korolchuk V.I., Lichtenberg M., Luo S., Massey D.C., Menzies F.M., Moreau K., Narayanan U., Renna M., Siddiqi F.H., Underwood B.R., Winslow A.R., Rubinsztein D.C. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol. Rev. 2010;90(4):1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 9.Juhasz G., Neufeld T.P. Autophagy: a forty-year search for a missing membrane source. PLoS Biol. 2006;4(2):e36. doi: 10.1371/journal.pbio.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galluzzi L., Pietrocola F., Levine B., Kroemer G. Metabolic control of autophagy. Cell. 2014;159(6):1263–1276. doi: 10.1016/j.cell.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yorimitsu T., Klionsky D.J. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12(Suppl 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W.W., Li J., Bao J.K. Microautophagy: lesser-known self-eating. Cell. Mol. Life Sci. 2012;69(7):1125–1136. doi: 10.1007/s00018-011-0865-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mijaljica D., Prescott M., Devenish R.J. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy. 2011;7(7):673–682. doi: 10.4161/auto.7.7.14733. [DOI] [PubMed] [Google Scholar]
- 14.Sahu R., Kaushik S., Clement C.C., Cannizzo E.S., Scharf B., Follenzi A., Potolicchio I., Nieves E., Cuervo A.M., Santambrogio L. Microautophagy of cytosolic proteins by late endosomes. Dev. Cell. 2011;20(1):131–139. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakai M., Araki N., Ogawa K. Lysosomal movements during heterophagy and autophagy: with special reference to nematolysosome and wrapping lysosome. J. Electron. Microsc. Tech. 1989;12(2):101–131. doi: 10.1002/jemt.1060120206. [DOI] [PubMed] [Google Scholar]
- 16.Dice J.F. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem. Sci. 1990;15(8):305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- 17.Kaushik S., Cuervo A.M. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 2018;19(6):365–381. doi: 10.1038/s41580-018-0001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orenstein S.J., Cuervo A.M. Chaperone-mediated autophagy: molecular mechanisms and physiological relevance. Semin. Cell Dev. Biol. 2010;21(7):719–726. doi: 10.1016/j.semcdb.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farre J.C., Subramani S. Mechanistic insights into selective autophagy pathways: lessons from yeast. Nat. Rev. Mol. Cell Biol. 2016;17(9):537–552. doi: 10.1038/nrm.2016.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gatica D., Lahiri V., Klionsky D.J. Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 2018;20(3):233–242. doi: 10.1038/s41556-018-0037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirota Y., Aoki Y., Kanki T. [Mitophagy: selective degradation of mitochondria by autophagy] Seikagaku. 2011;83(2):126–130. [PubMed] [Google Scholar]
- 22.Kim I., Rodriguez-Enriquez S., Lemasters J.J. Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 2007;462(2):245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho D.H., Kim Y.S., Jo D.S., Choe S.K., Jo E.K. Pexophagy: molecular mechanisms and implications for health and diseases. Mol. Cell. 2018;41(1):55–64. doi: 10.14348/molcells.2018.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cebollero E., Reggiori F., Kraft C. Reticulophagy and ribophagy: regulated degradation of protein production factories. Int J Cell Biol. 2012;2012:182834. doi: 10.1155/2012/182834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park Y.E., Hayashi Y.K., Bonne G., Arimura T., Noguchi S., Nonaka I., Nishino I. Autophagic degradation of nuclear components in mammalian cells. Autophagy. 2009;5(6):795–804. doi: 10.4161/auto.8901. [DOI] [PubMed] [Google Scholar]
- 26.Baltanas F.C., Casafont I., Weruaga E., Alonso J.R., Berciano M.T., Lafarga M. Nucleolar disruption and cajal body disassembly are nuclear hallmarks of DNA damage-induced neurodegeneration in purkinje cells. Brain Pathol. 2011;21(4):374–388. doi: 10.1111/j.1750-3639.2010.00461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A.M., Czaja M.J. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujiwara Y., Furuta A., Kikuchi H., Aizawa S., Hatanaka Y., Konya C., Uchida K., Yoshimura A., Tamai Y., Wada K., Kabuta T. Discovery of a novel type of autophagy targeting RNA. Autophagy. 2013;9(3):403–409. doi: 10.4161/auto.23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120(2):159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 30.Overbye A., Fengsrud M., Seglen P.O. Proteomic analysis of membrane-associated proteins from rat liver autophagosomes. Autophagy. 2007;3(4):300–322. doi: 10.4161/auto.3910. [DOI] [PubMed] [Google Scholar]
- 31.Lam H.C., Cloonan S.M., Bhashyam A.R., Haspel J.A., Singh A., Sathirapongsasuti J.F., Cervo M., Yao H., Chung A.L., Mizumura K., An C.H., Shan B., Franks J.M., Haley K.J., Owen C.A., Tesfaigzi Y., Washko G.R., Quackenbush J., Silverman E.K., Rahman I., Kim H.P., Mahmood A., Biswal S.S., Ryter S.W., Choi A.M. Histone deacetylase 6-mediated selective autophagy regulates COPD-associated cilia dysfunction. J. Clin. Invest. 2013;123(12):5212–5230. doi: 10.1172/JCI69636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mancias J.D., Wang X., Gygi S.P., Harper J.W., Kimmelman A.C. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509(7498):105–109. doi: 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evans T.D., Sergin I., Zhang X., Razani B. Target acquired: selective autophagy in cardiometabolic disease. Sci. Signal. 2017;10(468) doi: 10.1126/scisignal.aag2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sies H., Berndt C., Jones D.P. Oxidative stress. Annu. Rev. Biochem. 2017;86:715–748. doi: 10.1146/annurev-biochem-061516-045037. [DOI] [PubMed] [Google Scholar]
- 35.Szumiel I. Autophagy, reactive oxygen species and the fate of mammalian cells. Free Radic. Res. 2011;45(3):253–265. doi: 10.3109/10715762.2010.525233. [DOI] [PubMed] [Google Scholar]
- 36.Filomeni G., De Zio D., Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 2015;22(3):377–388. doi: 10.1038/cdd.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scherz-Shouval R., Elazar Z. Regulation of autophagy by ROS: physiology and pathology. Trends Biochem. Sci. 2011;36(1):30–38. doi: 10.1016/j.tibs.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Levonen A.L., Hill B.G., Kansanen E., Zhang J., Darley-Usmar V.M. Redox regulation of antioxidants, autophagy, and the response to stress: implications for electrophile therapeutics. Free Radic. Biol. Med. 2014;71:196–207. doi: 10.1016/j.freeradbiomed.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He C., Klionsky D.J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levine B., Klionsky D.J. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6(4):463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 41.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21(22):2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 42.Mazure N.M., Pouyssegur J. Hypoxia-induced autophagy: cell death or cell survival? Curr. Opin. Cell Biol. 2010;22(2):177–180. doi: 10.1016/j.ceb.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 43.Corona Velazquez A.F., Jackson W.T. So many roads: the multifaceted regulation of autophagy induction. Mol. Cell Biol. 2018;38(21) doi: 10.1128/MCB.00303-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Egan D., Kim J., Shaw R.J., Guan K.L. The autophagy initiating kinase ULK1 is regulated via opposing phosphorylation by AMPK and mTOR. Autophagy. 2011;7(6):643–644. doi: 10.4161/auto.7.6.15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim J., Kundu M., Viollet B., Guan K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S., Natsume T., Takehana K., Yamada N., Guan J.L., Oshiro N., Mizushima N. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 2009;20(7):1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hurley J.H., Young L.N. Mechanisms of autophagy initiation. Annu. Rev. Biochem. 2017;86:225–244. doi: 10.1146/annurev-biochem-061516-044820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 2010;22(2):132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 49.Loffler A.S., Alers S., Dieterle A.M., Keppeler H., Franz-Wachtel M., Kundu M., Campbell D.G., Wesselborg S., Alessi D.R., Stork B. Ulk1-mediated phosphorylation of AMPK constitutes a negative regulatory feedback loop. Autophagy. 2011;7(7):696–706. doi: 10.4161/auto.7.7.15451. [DOI] [PubMed] [Google Scholar]
- 50.Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 2009;11(12):1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 51.Yla-Anttila P., Vihinen H., Jokitalo E., Eskelinen E.L. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5(8):1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 52.Ge L., Melville D., Zhang M., Schekman R. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife. 2013;2 doi: 10.7554/eLife.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., Hiraoka Y., Amano A., Yoshimori T. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495(7441):389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 54.Hailey D.W., Rambold A.S., Satpute-Krishnan P., Mitra K., Sougrat R., Kim P.K., Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141(4):656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohashi Y., Munro S. Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol. Biol. Cell. 2010;21(22):3998–4008. doi: 10.1091/mbc.E10-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ravikumar B., Moreau K., Jahreiss L., Puri C., Rubinsztein D.C. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 2010;12(8):747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mack H.I., Zheng B., Asara J.M., Thomas S.M. AMPK-dependent phosphorylation of ULK1 regulates ATG9 localization. Autophagy. 2012;8(8):1197–1214. doi: 10.4161/auto.20586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Itakura E., Kishi C., Inoue K., Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell. 2008;19(12):5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fogel A.I., Dlouhy B.J., Wang C., Ryu S.W., Neutzner A., Hasson S.A., Sideris D.P., Abeliovich H., Youle R.J. Role of membrane association and Atg14-dependent phosphorylation in beclin-1-mediated autophagy. Mol. Cell Biol. 2013;33(18):3675–3688. doi: 10.1128/MCB.00079-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li X., He L., Che K.H., Funderburk S.F., Pan L., Pan N., Zhang M., Yue Z., Zhao Y. Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat. Commun. 2012;3:662. doi: 10.1038/ncomms1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burman C., Ktistakis N.T. Regulation of autophagy by phosphatidylinositol 3-phosphate. FEBS Lett. 2010;584(7):1302–1312. doi: 10.1016/j.febslet.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 62.Proikas-Cezanne T., Takacs Z., Donnes P., Kohlbacher O. WIPI proteins: essential PtdIns3P effectors at the nascent autophagosome. J. Cell Sci. 2015;128(2):207–217. doi: 10.1242/jcs.146258. [DOI] [PubMed] [Google Scholar]
- 63.Noda T. Autophagy in the context of the cellular membrane-trafficking system: the enigma of Atg9 vesicles. Biochem. Soc. Trans. 2017;45(6):1323–1331. doi: 10.1042/BST20170128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chu C.T., Zhu J., Dagda R. Beclin 1-independent pathway of damage-induced mitophagy and autophagic stress: implications for neurodegeneration and cell death. Autophagy. 2007;3(6):663–666. doi: 10.4161/auto.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Codogno P., Mehrpour M., Proikas-Cezanne T. Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat. Rev. Mol. Cell Biol. 2011;13(1):7–12. doi: 10.1038/nrm3249. [DOI] [PubMed] [Google Scholar]
- 66.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2001;2(3):211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 67.Schaaf M.B., Keulers T.G., Vooijs M.A., Rouschop K.M. LC3/GABARAP family proteins: autophagy-(un)related functions. Faseb. J. 2016;30(12):3961–3978. doi: 10.1096/fj.201600698R. [DOI] [PubMed] [Google Scholar]
- 68.He H., Dang Y., Dai F., Guo Z., Wu J., She X., Pei Y., Chen Y., Ling W., Wu C., Zhao S., Liu J.O., Yu L. Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J. Biol. Chem. 2003;278(31):29278–29287. doi: 10.1074/jbc.M303800200. [DOI] [PubMed] [Google Scholar]
- 69.Mizushima N., Yoshimori T., Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xie Y., Kang R., Sun X., Zhong M., Huang J., Klionsky D.J., Tang D. Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy. 2015;11(1):28–45. doi: 10.4161/15548627.2014.984267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liang C., Lee J.S., Inn K.S., Gack M.U., Li Q., Roberts E.A., Vergne I., Deretic V., Feng P., Akazawa C., Jung J.U. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 2008;10(7):776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Williams J.A., Ding W.X. Mechanisms, pathophysiological roles and methods for analyzing mitophagy - recent insights. Biol. Chem. 2018;399(2):147–178. doi: 10.1515/hsz-2017-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sies H., Cadenas E. Oxidative stress: damage to intact cells and organs. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1985;311(1152):617–631. doi: 10.1098/rstb.1985.0168. [DOI] [PubMed] [Google Scholar]
- 74.Sohal R.S., Allen R.G. Oxidative stress as a causal factor in differentiation and aging: a unifying hypothesis. Exp. Gerontol. 1990;25(6):499–522. doi: 10.1016/0531-5565(90)90017-v. [DOI] [PubMed] [Google Scholar]
- 75.Pizzino G., Irrera N., Cucinotta M., Pallio G., Mannino F., Arcoraci V., Squadrito F., Altavilla D., Bitto A. Oxidative stress: harms and benefits for human health. Oxid Med Cell Longev. 2017;2017:8416763. doi: 10.1155/2017/8416763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shadel G.S., Horvath T.L. Mitochondrial ROS signaling in organismal homeostasis. Cell. 2015;163(3):560–569. doi: 10.1016/j.cell.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Murphy M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417(1):1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reczek C.R., Chandel N.S. ROS-dependent signal transduction. Curr. Opin. Cell Biol. 2015;33:8–13. doi: 10.1016/j.ceb.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Holmstrom K.M., Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat. Rev. Mol. Cell Biol. 2014;15(6):411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 80.Netto L.E., Antunes F. The roles of peroxiredoxin and thioredoxin in hydrogen peroxide sensing and in signal transduction. Mol. Cell. 2016;39(1):65–71. doi: 10.14348/molcells.2016.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Venditti P., Di Stefano L., Di Meo S. Mitochondrial metabolism of reactive oxygen species. Mitochondrion. 2013;13(2):71–82. doi: 10.1016/j.mito.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 82.Cordani M., Sanchez-Alvarez M., Strippoli R., Bazhin A.V., Donadelli M. Sestrins at the interface of ROS control and autophagy regulation in health and disease. Oxid Med Cell Longev. 2019;2019:1283075. doi: 10.1155/2019/1283075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roca-Agujetas V., de Dios C., Leston L., Mari M., Morales A., Colell A. Recent insights into the mitochondrial role in autophagy and its regulation by oxidative stress. Oxid Med Cell Longev. 2019;2019:3809308. doi: 10.1155/2019/3809308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Circu M.L., Aw T.Y. Reactive oxygen species, cellular redox systems, and apoptosis. Free Radic. Biol. Med. 2010;48(6):749–762. doi: 10.1016/j.freeradbiomed.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen Y., Azad M.B., Gibson S.B. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 2009;16(7):1040–1052. doi: 10.1038/cdd.2009.49. [DOI] [PubMed] [Google Scholar]
- 86.Youle R.J., Narendra D.P. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 2011;12(1):9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y.S., Ueno I., Sakamoto A., Tong K.I., Kim M., Nishito Y., Iemura S., Natsume T., Ueno T., Kominami E., Motohashi H., Tanaka K., Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010;12(3):213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 88.Dodson M., Redmann M., Rajasekaran N.S., Darley-Usmar V., Zhang J. KEAP1-NRF2 signalling and autophagy in protection against oxidative and reductive proteotoxicity. Biochem. J. 2015;469(3):347–355. doi: 10.1042/BJ20150568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kageyama S., Saito T., Obata M., Koide R.H., Ichimura Y., Komatsu M. Negative regulation of the keap1-nrf2 pathway by a p62/Sqstm1 splicing variant. Mol. Cell Biol. 2018;38(7) doi: 10.1128/MCB.00642-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen S.D., Yang J.L., Lin T.K., Yang D.I. Emerging roles of sestrins in neurodegenerative diseases: counteracting oxidative stress and beyond. J. Clin. Med. 2019;8(7) doi: 10.3390/jcm8071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gao Q. Oxidative stress and autophagy. Adv. Exp. Med. Biol. 2019;1206:179–198. doi: 10.1007/978-981-15-0602-4_9. [DOI] [PubMed] [Google Scholar]
- 92.Panday A., Sahoo M.K., Osorio D., Batra S. NADPH oxidases: an overview from structure to innate immunity-associated pathologies. Cell. Mol. Immunol. 2015;12(1):5–23. doi: 10.1038/cmi.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cui Y., Wang Y., Li G., Ma W., Zhou X.S., Wang J., Liu B. The Nox1/Nox4 inhibitor attenuates acute lung injury induced by ischemia-reperfusion in mice. PloS One. 2018;13(12) doi: 10.1371/journal.pone.0209444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.He Y., Sang Z., Zhuo Y., Wang X., Guo Z., He L., Zeng C., Dai H. Transport stress induces pig jejunum tissue oxidative damage and results in autophagy/mitophagy activation. J. Anim. Physiol. Anim. Nutr. 2019;103(5):1521–1529. doi: 10.1111/jpn.13161. [DOI] [PubMed] [Google Scholar]
- 95.Teng R.J., Du J., Welak S., Guan T., Eis A., Shi Y., Konduri G.G. Cross talk between NADPH oxidase and autophagy in pulmonary artery endothelial cells with intrauterine persistent pulmonary hypertension. Am. J. Physiol. Lung Cell Mol. Physiol. 2012;302(7):L651–L663. doi: 10.1152/ajplung.00177.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang S., Ji L.Y., Li L., Li J.M. Oxidative stress, autophagy and pyroptosis in the neovascularization of oxygeninduced retinopathy in mice. Mol. Med. Rep. 2019;19(2):927–934. doi: 10.3892/mmr.2018.9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jaishy B., Zhang Q., Chung H.S., Riehle C., Soto J., Jenkins S., Abel P., Cowart L.A., Van Eyk J.E., Abel E.D. Lipid-induced NOX2 activation inhibits autophagic flux by impairing lysosomal enzyme activity. J. Lipid Res. 2015;56(3):546–561. doi: 10.1194/jlr.M055152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Belarbi K., Cuvelier E., Destee A., Gressier B., Chartier-Harlin M.C. NADPH oxidases in Parkinson's disease: a systematic review. Mol. Neurodegener. 2017;12(1):84. doi: 10.1186/s13024-017-0225-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang Y., Ren S., Gu Y., Wang J., Liu Z., Zhang Z. Taurine attenuates calpain-2 induction and a series of cell damage via suppression of NOX-derived ROS in ARPE-19 cells. Oxid Med Cell Longev. 2018;2018:4596746. doi: 10.1155/2018/4596746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sciarretta S., Zhai P., Shao D., Zablocki D., Nagarajan N., Terada L.S., Volpe M., Sadoshima J. Activation of NADPH oxidase 4 in the endoplasmic reticulum promotes cardiomyocyte autophagy and survival during energy stress through the protein kinase RNA-activated-like endoplasmic reticulum kinase/eukaryotic initiation factor 2alpha/activating transcription factor 4 pathway. Circ. Res. 2013;113(11):1253–1264. doi: 10.1161/CIRCRESAHA.113.301787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sciarretta S., Volpe M., Sadoshima J. NOX4 regulates autophagy during energy deprivation. Autophagy. 2014;10(4):699–701. doi: 10.4161/auto.27955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee K.A., Cho K.C., Kim B., Jang I.H., Nam K., Kwon Y.E., Kim M., Hyeon D.Y., Hwang D., Seol J.H., Lee W.J. Inflammation-modulated metabolic reprogramming is required for DUOX-dependent gut immunity in Drosophila. Cell Host Microbe. 2018;23(3):338–352. doi: 10.1016/j.chom.2018.01.011. e5. [DOI] [PubMed] [Google Scholar]
- 103.Lee K.A., Kim B., Bhin J., Kim D.H., You H., Kim E.K., Kim S.H., Ryu J.H., Hwang D., Lee W.J. Bacterial uracil modulates Drosophila DUOX-dependent gut immunity via Hedgehog-induced signaling endosomes. Cell Host Microbe. 2015;17(2):191–204. doi: 10.1016/j.chom.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 104.Huang C.C., Chen K.L., Cheung C.H.A., Chang J.Y. Autophagy induced by cathepsin S inhibition induces early ROS production, oxidative DNA damage, and cell death via xanthine oxidase. Free Radic. Biol. Med. 2013;65:1473–1486. doi: 10.1016/j.freeradbiomed.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 105.Huang C.C., Lee C.C., Lin H.H., Chen M.C., Lin C.C., Chang J.Y. Autophagy-regulated ROS from xanthine oxidase acts as an early effector for triggering late mitochondria-dependent apoptosis in cathepsin S-targeted tumor cells. PloS One. 2015;10(6) doi: 10.1371/journal.pone.0128045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Recuero M., Munive V.A., Sastre I., Aldudo J., Valdivieso F., Bullido M.J. A free radical-generating system regulates AbetaPP metabolism/processing: involvement of the ubiquitin/proteasome and autophagy/lysosome pathways. J Alzheimers Dis. 2013;34(3):637–647. doi: 10.3233/JAD-121510. [DOI] [PubMed] [Google Scholar]
- 107.Li L., Chen Y., Gibson S.B. Starvation-induced autophagy is regulated by mitochondrial reactive oxygen species leading to AMPK activation. Cell. Signal. 2013;25(1):50–65. doi: 10.1016/j.cellsig.2012.09.020. [DOI] [PubMed] [Google Scholar]
- 108.Scherz-Shouval R., Shvets E., Elazar Z. Oxidation as a post-translational modification that regulates autophagy. Autophagy. 2007;3(4):371–373. doi: 10.4161/auto.4214. [DOI] [PubMed] [Google Scholar]
- 109.Pivtoraiko V.N., Stone S.L., Roth K.A., Shacka J.J. Oxidative stress and autophagy in the regulation of lysosome-dependent neuron death. Antioxidants Redox Signal. 2009;11(3):481–496. doi: 10.1089/ars.2008.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang H., Bosch-Marce M., Shimoda L.A., Tan Y.S., Baek J.H., Wesley J.B., Gonzalez F.J., Semenza G.L. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J. Biol. Chem. 2008;283(16):10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 111.Park S.Y., Park M.Y., Park H.G., Lee K.J., Kook M.S., Kim W.J., Jung J.Y. Nitric oxide-induced autophagy and the activation of activated protein kinase pathway protect against apoptosis in human dental pulp cells. Int. Endod. J. 2017;50(3):260–270. doi: 10.1111/iej.12616. [DOI] [PubMed] [Google Scholar]
- 112.Datta S., Chakraborty S., Panja C., Ghosh S. Reactive nitrogen species control apoptosis and autophagy in K562 cells: implication of TAp73alpha induction in controlling autophagy. Free Radic. Res. 2018;52(4):491–506. doi: 10.1080/10715762.2018.1449210. [DOI] [PubMed] [Google Scholar]
- 113.Jin L., Gao H., Wang J., Yang S., Wang J., Liu J., Yang Y., Yan T., Chen T., Zhao Y., He Y. Role and regulation of autophagy and apoptosis by nitric oxide in hepatic stellate cells during acute liver failure. Liver Int. 2017;37(11):1651–1659. doi: 10.1111/liv.13476. [DOI] [PubMed] [Google Scholar]
- 114.Zhang X., Jin L., Tian Z., Wang J., Yang Y., Liu J., Chen Y., Hu C., Chen T., Zhao Y., He Y. Nitric oxide inhibits autophagy and promotes apoptosis in hepatocellular carcinoma. Canc. Sci. 2019;110(3):1054–1063. doi: 10.1111/cas.13945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sadhu A., Moriyasu Y., Acharya K., Bandyopadhyay M. Nitric oxide and ROS mediate autophagy and regulate Alternaria alternata toxin-induced cell death in tobacco BY-2 cells. Sci. Rep. 2019;9(1):8973. doi: 10.1038/s41598-019-45470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pacher P., Beckman J.S., Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol. Rev. 2007;87(1):315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hong Z.P., Wang L.G., Wang H.J., Ye W.F., Wang X.Z. Wogonin exacerbates the cytotoxic effect of oxaliplatin by inducing nitrosative stress and autophagy in human gastric cancer cells. Phytomedicine. 2018;39:168–175. doi: 10.1016/j.phymed.2017.12.019. [DOI] [PubMed] [Google Scholar]
- 118.Feng J., Chen X., Lu S., Li W., Yang D., Su W., Wang X., Shen J. Naringin attenuates cerebral ischemia-reperfusion injury through inhibiting peroxynitrite-mediated mitophagy activation. Mol. Neurobiol. 2018;55(12):9029–9042. doi: 10.1007/s12035-018-1027-7. [DOI] [PubMed] [Google Scholar]
- 119.Jiang Q., Gao Y., Wang C., Tao R., Wu Y., Zhan K., Liao M., Lu N., Lu Y., Wilcox C.S., Luo J., Jiang L.H., Yang W., Han F. Nitration of TRPM2 as a molecular switch induces autophagy during brain pericyte injury. Antioxidants Redox Signal. 2017;27(16):1297–1316. doi: 10.1089/ars.2016.6873. [DOI] [PubMed] [Google Scholar]
- 120.Mizushima N. Physiological functions of autophagy. Curr. Top. Microbiol. Immunol. 2009;335:71–84. doi: 10.1007/978-3-642-00302-8_3. [DOI] [PubMed] [Google Scholar]
- 121.Lee J., Giordano S., Zhang J. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem. J. 2012;441(2):523–540. doi: 10.1042/BJ20111451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang H., Kong X., Kang J., Su J., Li Y., Zhong J., Sun L. Oxidative stress induces parallel autophagy and mitochondria dysfunction in human glioma U251 cells. Toxicol. Sci. 2009;110(2):376–388. doi: 10.1093/toxsci/kfp101. [DOI] [PubMed] [Google Scholar]
- 123.Scherz-Shouval R., Shvets E., Fass E., Shorer H., Gil L., Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26(7):1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mondaca-Ruff D., Riquelme J.A., Quiroga C., Norambuena-Soto I., Sanhueza-Olivares F., Villar-Fincheira P., Hernandez-Diaz T., Cancino-Arenas N., San Martin A., Garcia L., Lavandero S., Chiong M. Angiotensin II-regulated autophagy is required for vascular smooth muscle cell hypertrophy. Front. Pharmacol. 2018;9:1553. doi: 10.3389/fphar.2018.01553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yang S., Sun D., Wang L., Wang X., Shi M., Jiang X., Gao X. The role of STAT3/mTOR-regulated autophagy in angiotensin II-induced senescence of human glomerular mesangial cells. Cell. Signal. 2019;53:327–338. doi: 10.1016/j.cellsig.2018.10.021. [DOI] [PubMed] [Google Scholar]
- 126.Li Y., Luo Q., Yuan L., Miao C., Mu X., Xiao W., Li J., Sun T., Ma E. JNK-dependent Atg4 upregulation mediates asperphenamate derivative BBP-induced autophagy in MCF-7 cells. Toxicol. Appl. Pharmacol. 2012;263(1):21–31. doi: 10.1016/j.taap.2012.05.018. [DOI] [PubMed] [Google Scholar]
- 127.Zhang L., Wang H., Xu J., Zhu J., Ding K. Inhibition of cathepsin S induces autophagy and apoptosis in human glioblastoma cell lines through ROS-mediated PI3K/AKT/mTOR/p70S6K and JNK signaling pathways. Toxicol. Lett. 2014;228(3):248–259. doi: 10.1016/j.toxlet.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 128.Zhang J., Kim J., Alexander A., Cai S., Tripathi D.N., Dere R., Tee A.R., Tait-Mulder J., Di Nardo A., Han J.M., Kwiatkowski E., Dunlop E.A., Dodd K.M., Folkerth R.D., Faust P.L., Kastan M.B., Sahin M., Walker C.L. A tuberous sclerosis complex signalling node at the peroxisome regulates mTORC1 and autophagy in response to ROS. Nat. Cell Biol. 2013;15(10):1186–1196. doi: 10.1038/ncb2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Salmeen A., Barford D. Functions and mechanisms of redox regulation of cysteine-based phosphatases. Antioxidants Redox Signal. 2005;7(5–6):560–577. doi: 10.1089/ars.2005.7.560. [DOI] [PubMed] [Google Scholar]
- 130.Huang C., Zhang Y., Kelly D.J., Tan C.Y., Gill A., Cheng D., Braet F., Park J.S., Sue C.M., Pollock C.A., Chen X.M. Thioredoxin interacting protein (TXNIP) regulates tubular autophagy and mitophagy in diabetic nephropathy through the mTOR signaling pathway. Sci. Rep. 2016;6:29196. doi: 10.1038/srep29196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hsieh C.C., Papaconstantinou J. Thioredoxin-ASK1 complex levels regulate ROS-mediated p38 MAPK pathway activity in livers of aged and long-lived Snell dwarf mice. Faseb. J. 2006;20(2):259–268. doi: 10.1096/fj.05-4376com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wei J.L., Fang M., Fu Z.X., Zhang S.R., Guo J.B., Wang R., Lv Z.B., Xiong Y.F. Sestrin 2 suppresses cells proliferation through AMPK/mTORC1 pathway activation in colorectal cancer. Oncotarget. 2017;8(30):49318–49328. doi: 10.18632/oncotarget.17595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Morsch A., Wisniewski E., Luciano T.F., Comin V.H., Silveira G.B., Marques S.O., Thirupathi A., Silveira Lock P.C., De Souza C.T. Cigarette smoke exposure induces ROS-mediated autophagy by regulating sestrin, AMPK, and mTOR level in mice. Redox Rep. 2019;24(1):27–33. doi: 10.1080/13510002.2019.1601448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nayak B.K., Feliers D., Sudarshan S., Friedrichs W.E., Day R.T., New D.D., Fitzgerald J.P., Eid A., Denapoli T., Parekh D.J., Gorin Y., Block K. Stabilization of HIF-2alpha through redox regulation of mTORC2 activation and initiation of mRNA translation. Oncogene. 2013;32(26):3147–3155. doi: 10.1038/onc.2012.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bernard M., Yang B., Migneault F., Turgeon J., Dieude M., Olivier M.A., Cardin G.B., El-Diwany M., Underwood K., Rodier F., Hebert M.J. Autophagy drives fibroblast senescence through MTORC2 regulation. Autophagy. 2020:1–13. doi: 10.1080/15548627.2020.1713640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bellot G., Garcia-Medina R., Gounon P., Chiche J., Roux D., Pouyssegur J., Mazure N.M. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol. Cell Biol. 2009;29(10):2570–2581. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chandel N.S., Maltepe E., Goldwasser E., Mathieu C.E., Simon M.C., Schumacker P.T. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc. Natl. Acad. Sci. U. S. A. 1998;95(20):11715–11720. doi: 10.1073/pnas.95.20.11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Brunelle J.K., Bell E.L., Quesada N.M., Vercauteren K., Tiranti V., Zeviani M., Scarpulla R.C., Chandel N.S. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metabol. 2005;1(6):409–414. doi: 10.1016/j.cmet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 139.Chandel N.S., McClintock D.S., Feliciano C.E., Wood T.M., Melendez J.A., Rodriguez A.M., Schumacker P.T. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. J. Biol. Chem. 2000;275(33):25130–25138. doi: 10.1074/jbc.M001914200. [DOI] [PubMed] [Google Scholar]
- 140.Mottet D., Dumont V., Deccache Y., Demazy C., Ninane N., Raes M., Michiels C. Regulation of hypoxia-inducible factor-1alpha protein level during hypoxic conditions by the phosphatidylinositol 3-kinase/Akt/glycogen synthase kinase 3beta pathway in HepG2 cells. J. Biol. Chem. 2003;278(33):31277–31285. doi: 10.1074/jbc.M300763200. [DOI] [PubMed] [Google Scholar]
- 141.Zhang J., Zhang C., Jiang X., Li L., Zhang D., Tang D., Yan T., Zhang Q., Yuan H., Jia J., Hu J., Zhang J., Huang Y. Involvement of autophagy in hypoxia-BNIP3 signaling to promote epidermal keratinocyte migration. Cell Death Dis. 2019;10(3):234. doi: 10.1038/s41419-019-1473-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 142.Papandreou I., Lim A.L., Laderoute K., Denko N.C. Hypoxia signals autophagy in tumor cells via AMPK activity, independent of HIF-1, BNIP3, and BNIP3L. Cell Death Differ. 2008;15(10):1572–1581. doi: 10.1038/cdd.2008.84. [DOI] [PubMed] [Google Scholar]
- 143.Rouschop K.M., van den Beucken T., Dubois L., Niessen H., Bussink J., Savelkouls K., Keulers T., Mujcic H., Landuyt W., Voncken J.W., Lambin P., van der Kogel A.J., Koritzinsky M., Wouters B.G. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J. Clin. Invest. 2010;120(1):127–141. doi: 10.1172/JCI40027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lee S.J., Smith A., Guo L., Alastalo T.P., Li M., Sawada H., Liu X., Chen Z.H., Ifedigbo E., Jin Y., Feghali-Bostwick C., Ryter S.W., Kim H.P., Rabinovitch M., Choi A.M. Autophagic protein LC3B confers resistance against hypoxia-induced pulmonary hypertension. Am. J. Respir. Crit. Care Med. 2011;183(5):649–658. doi: 10.1164/rccm.201005-0746OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ferro E., Goitre L., Retta S.F., Trabalzini L. The interplay between ROS and ras GTPases: physiological and pathological implications. J Signal Transduct. 2012;2012:365769. doi: 10.1155/2012/365769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Heo J., Raines K.W., Mocanu V., Campbell S.L. Redox regulation of RhoA. Biochemistry. 2006;45(48):14481–14489. doi: 10.1021/bi0610101. [DOI] [PubMed] [Google Scholar]
- 147.Jin L., Ying Z., Webb R.C. Activation of Rho/Rho kinase signaling pathway by reactive oxygen species in rat aorta. Am. J. Physiol. Heart Circ. Physiol. 2004;287(4):H1495–H1500. doi: 10.1152/ajpheart.01006.2003. [DOI] [PubMed] [Google Scholar]
- 148.Belaid A., Cerezo M., Chargui A., Corcelle-Termeau E., Pedeutour F., Giuliano S., Ilie M., Rubera I., Tauc M., Barale S., Bertolotto C., Brest P., Vouret-Craviari V., Klionsky D.J., Carle G.F., Hofman P., Mograbi B. Autophagy plays a critical role in the degradation of active RHOA, the control of cell cytokinesis, and genomic stability. Can. Res. 2013;73(14):4311–4322. doi: 10.1158/0008-5472.CAN-12-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Gordon B.S., Kazi A.A., Coleman C.S., Dennis M.D., Chau V., Jefferson L.S., Kimball S.R. RhoA modulates signaling through the mechanistic target of rapamycin complex 1 (mTORC1) in mammalian cells. Cell. Signal. 2014;26(3):461–467. doi: 10.1016/j.cellsig.2013.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fischer M. Census and evaluation of p53 target genes. Oncogene. 2017;36(28):3943–3956. doi: 10.1038/onc.2016.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bensaad K., Tsuruta A., Selak M.A., Vidal M.N., Nakano K., Bartrons R., Gottlieb E., Vousden K.H. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126(1):107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 152.Cheung E.C., Ludwig R.L., Vousden K.H. Mitochondrial localization of TIGAR under hypoxia stimulates HK2 and lowers ROS and cell death. Proc. Natl. Acad. Sci. U. S. A. 2012;109(50):20491–20496. doi: 10.1073/pnas.1206530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Budanov A.V., Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134(3):451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Filomeni G., Desideri E., Cardaci S., Rotilio G., Ciriolo M.R. Under the ROS...thiol network is the principal suspect for autophagy commitment. Autophagy. 2010;6(7):999–1005. doi: 10.4161/auto.6.7.12754. [DOI] [PubMed] [Google Scholar]
- 155.Zoncu R., Efeyan A., Sabatini D.M. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 2011;12(1):21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yoshida S., Hong S., Suzuki T., Nada S., Mannan A.M., Wang J., Okada M., Guan K.L., Inoki K. Redox regulates mammalian target of rapamycin complex 1 (mTORC1) activity by modulating the TSC1/TSC2-Rheb GTPase pathway. J. Biol. Chem. 2011;286(37):32651–32660. doi: 10.1074/jbc.M111.238014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sarbassov D.D., Sabatini D.M. Redox regulation of the nutrient-sensitive raptor-mTOR pathway and complex. J. Biol. Chem. 2005;280(47):39505–39509. doi: 10.1074/jbc.M506096200. [DOI] [PubMed] [Google Scholar]
- 158.Cao J., Schulte J., Knight A., Leslie N.R., Zagozdzon A., Bronson R., Manevich Y., Beeson C., Neumann C.A. Prdx1 inhibits tumorigenesis via regulating PTEN/AKT activity. EMBO J. 2009;28(10):1505–1517. doi: 10.1038/emboj.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pal R., Palmieri M., Loehr J.A., Li S., Abo-Zahrah R., Monroe T.O., Thakur P.B., Sardiello M., Rodney G.G. Src-dependent impairment of autophagy by oxidative stress in a mouse model of Duchenne muscular dystrophy. Nat. Commun. 2014;5:4425. doi: 10.1038/ncomms5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Luciani A., Villella V.R., Esposito S., Brunetti-Pierri N., Medina D., Settembre C., Gavina M., Pulze L., Giardino I., Pettoello-Mantovani M., D'Apolito M., Guido S., Masliah E., Spencer B., Quaratino S., Raia V., Ballabio A., Maiuri L. Defective CFTR induces aggresome formation and lung inflammation in cystic fibrosis through ROS-mediated autophagy inhibition. Nat. Cell Biol. 2010;12(9):863–875. doi: 10.1038/ncb2090. [DOI] [PubMed] [Google Scholar]
- 161.Wu J.J., Quijano C., Chen E., Liu H., Cao L., Fergusson M.M., Rovira, Gutkind S., Daniels M.P., Komatsu M., Finkel T. Mitochondrial dysfunction and oxidative stress mediate the physiological impairment induced by the disruption of autophagy. Aging (Albany NY) 2009;1(4):425–437. doi: 10.18632/aging.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tal M.C., Sasai M., Lee H.K., Yordy B., Shadel G.S., Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc. Natl. Acad. Sci. U. S. A. 2009;106(8):2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yang S., Xia C., Li S., Du L., Zhang L., Zhou R. Defective mitophagy driven by dysregulation of rheb and KIF5B contributes to mitochondrial reactive oxygen species (ROS)-induced nod-like receptor 3 (NLRP3) dependent proinflammatory response and aggravates lipotoxicity. Redox Biol. 2014;3:63–71. doi: 10.1016/j.redox.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kurihara Y., Kanki T., Aoki Y., Hirota Y., Saigusa T., Uchiumi T., Kang D. Mitophagy plays an essential role in reducing mitochondrial production of reactive oxygen species and mutation of mitochondrial DNA by maintaining mitochondrial quantity and quality in yeast. J. Biol. Chem. 2012;287(5):3265–3272. doi: 10.1074/jbc.M111.280156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Archer S.L. Mitochondrial dynamics--mitochondrial fission and fusion in human diseases. N. Engl. J. Med. 2013;369(23):2236–2251. doi: 10.1056/NEJMra1215233. [DOI] [PubMed] [Google Scholar]
- 166.Scarpulla R.C. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 2008;88(2):611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 167.Harper J.W., Ordureau A., Heo J.M. Building and decoding ubiquitin chains for mitophagy. Nat. Rev. Mol. Cell Biol. 2018;19(2):93–108. doi: 10.1038/nrm.2017.129. [DOI] [PubMed] [Google Scholar]
- 168.Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C.A., Sou Y.S., Saiki S., Kawajiri S., Sato F., Kimura M., Komatsu M., Hattori N., Tanaka K. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 2010;189(2):211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R.L., Kim J., May J., Tocilescu M.A., Liu W., Ko H.S., Magrane J., Moore D.J., Dawson V.L., Grailhe R., Dawson T.M., Li C., Tieu K., Przedborski S. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. U. S. A. 2010;107(1):378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wang Y., Nartiss Y., Steipe B., McQuibban G.A., Kim P.K. ROS-induced mitochondrial depolarization initiates PARK2/PARKIN-dependent mitochondrial degradation by autophagy. Autophagy. 2012;8(10):1462–1476. doi: 10.4161/auto.21211. [DOI] [PubMed] [Google Scholar]
- 171.Eiyama A., Okamoto K. PINK1/Parkin-mediated mitophagy in mammalian cells. Curr. Opin. Cell Biol. 2015;33:95–101. doi: 10.1016/j.ceb.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 172.Xiao B., Deng X., Lim G.G.Y., Xie S., Zhou Z.D., Lim K.L., Tan E.K. Superoxide drives progression of Parkin/PINK1-dependent mitophagy following translocation of Parkin to mitochondria. Cell Death Dis. 2017;8(10) doi: 10.1038/cddis.2017.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Xiao B., Goh J.Y., Xiao L., Xian H., Lim K.L., Liou Y.C. Reactive oxygen species trigger Parkin/PINK1 pathway-dependent mitophagy by inducing mitochondrial recruitment of Parkin. J. Biol. Chem. 2017;292(40):16697–16708. doi: 10.1074/jbc.M117.787739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sowter H.M., Ratcliffe P.J., Watson P., Greenberg A.H., Harris A.L. HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Can. Res. 2001;61(18):6669–6673. [PubMed] [Google Scholar]
- 175.Chourasia A.H., Tracy K., Frankenberger C., Boland M.L., Sharifi M.N., Drake L.E., Sachleben J.R., Asara J.M., Locasale J.W., Karczmar G.S., Macleod K.F. Mitophagy defects arising from BNip3 loss promote mammary tumor progression to metastasis. EMBO Rep. 2015;16(9):1145–1163. doi: 10.15252/embr.201540759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wu W., Tian W., Hu Z., Chen G., Huang L., Li W., Zhang X., Xue P., Zhou C., Liu L., Zhu Y., Zhang X., Li L., Zhang L., Sui S., Zhao B., Feng D. ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. 2014;15(5):566–575. doi: 10.1002/embr.201438501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Frank M., Duvezin-Caubet S., Koob S., Occhipinti A., Jagasia R., Petcherski A., Ruonala M.O., Priault M., Salin B., Reichert A.S. Mitophagy is triggered by mild oxidative stress in a mitochondrial fission dependent manner. Biochim. Biophys. Acta. 2012;1823(12):2297–2310. doi: 10.1016/j.bbamcr.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 178.Buhlman L., Damiano M., Bertolin G., Ferrando-Miguel R., Lombes A., Brice A., Corti O. Functional interplay between Parkin and Drp1 in mitochondrial fission and clearance. Biochim. Biophys. Acta. 2014;1843(9):2012–2026. doi: 10.1016/j.bbamcr.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 179.Chu C.T., Ji J., Dagda R.K., Jiang J.F., Tyurina Y.Y., Kapralov A.A., Tyurin V.A., Yanamala N., Shrivastava I.H., Mohammadyani D., Wang K.Z.Q., Zhu J., Klein-Seetharaman J., Balasubramanian K., Amoscato A.A., Borisenko G., Huang Z., Gusdon A.M., Cheikhi A., Steer E.K., Wang R., Baty C., Watkins S., Bahar I., Bayir H., Kagan V.E. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat. Cell Biol. 2013;15(10):1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kagan V.E., Jiang J., Huang Z., Tyurina Y.Y., Desbourdes C., Cottet-Rousselle C., Dar H.H., Verma M., Tyurin V.A., Kapralov A.A., Cheikhi A., Mao G., Stolz D., St Croix C.M., Watkins S., Shen Z., Li Y., Greenberg M.L., Tokarska-Schlattner M., Boissan M., Lacombe M.L., Epand R.M., Chu C.T., Mallampalli R.K., Bayir H., Schlattner U. NDPK-D (NM23-H4)-mediated externalization of cardiolipin enables elimination of depolarized mitochondria by mitophagy. Cell Death Differ. 2016;23(7):1140–1151. doi: 10.1038/cdd.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Nuttall J.M., Motley A.M., Hettema E.H. Deficiency of the exportomer components Pex1, Pex6, and Pex15 causes enhanced pexophagy in Saccharomyces cerevisiae. Autophagy. 2014;10(5):835–845. doi: 10.4161/auto.28259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Rogov V., Dotsch V., Johansen T., Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell. 2014;53(2):167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 183.Zhang J., Tripathi D.N., Jing J., Alexander A., Kim J., Powell R.T., Dere R., Tait-Mulder J., Lee J.H., Paull T.T., Pandita R.K., Charaka V.K., Pandita T.K., Kastan M.B., Walker C.L. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat. Cell Biol. 2015;17(10):1259–1269. doi: 10.1038/ncb3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Guo Z., Kozlov S., Lavin M.F., Person M.D., Paull T.T. ATM activation by oxidative stress. Science. 2010;330(6003):517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 185.Jo D.S., Park S.J., Kim A.K., Park N.Y., Kim J.B., Bae J.E., Park H.J., Shin J.H., Chang J.W., Kim P.K., Jung Y.K., Koh J.Y., Choe S.K., Lee K.S., Cho D.H. Loss of HSPA9 induces peroxisomal degradation by increasing pexophagy. Autophagy. 2020:1–15. doi: 10.1080/15548627.2020.1712812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Grimm S., Hohn A., Grune T. Oxidative protein damage and the proteasome. Amino Acids. 2012;42(1):23–38. doi: 10.1007/s00726-010-0646-8. [DOI] [PubMed] [Google Scholar]
- 187.Jung T., Grune T. The proteasome and its role in the degradation of oxidized proteins. IUBMB Life. 2008;60(11):743–752. doi: 10.1002/iub.114. [DOI] [PubMed] [Google Scholar]
- 188.Pickering A.M., Davies K.J. Degradation of damaged proteins: the main function of the 20S proteasome. Prog Mol Biol Transl Sci. 2012;109:227–248. doi: 10.1016/B978-0-12-397863-9.00006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kriegenburg F., Poulsen E.G., Koch A., Kruger E., Hartmann-Petersen R. Redox control of the ubiquitin-proteasome system: from molecular mechanisms to functional significance. Antioxidants Redox Signal. 2011;15(8):2265–2299. doi: 10.1089/ars.2010.3590. [DOI] [PubMed] [Google Scholar]
- 190.Kiffin R., Christian C., Knecht E., Cuervo A.M. Activation of chaperone-mediated autophagy during oxidative stress. Mol. Biol. Cell. 2004;15(11):4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Callahan M.K., Chaillot D., Jacquin C., Clark P.R., Menoret A. Differential acquisition of antigenic peptides by Hsp70 and Hsc70 under oxidative conditions. J. Biol. Chem. 2002;277(37):33604–33609. doi: 10.1074/jbc.M202890200. [DOI] [PubMed] [Google Scholar]
- 192.Massey A.C., Kaushik S., Sovak G., Kiffin R., Cuervo A.M. Consequences of the selective blockage of chaperone-mediated autophagy. Proc. Natl. Acad. Sci. U. S. A. 2006;103(15):5805–5810. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Saha T. LAMP2A overexpression in breast tumors promotes cancer cell survival via chaperone-mediated autophagy. Autophagy. 2012;8(11):1643–1656. doi: 10.4161/auto.21654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lee J.J., Ishihara K., Notomi S., Efstathiou N.E., Ueta T., Maidana D., Chen X., Iesato Y., Caligiana A., Vavvas D.G. Lysosome-associated membrane protein-2 deficiency increases the risk of reactive oxygen species-induced ferroptosis in retinal pigment epithelial cells. Biochem. Biophys. Res. Commun. 2020;521(2):414–419. doi: 10.1016/j.bbrc.2019.10.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Pajares M., Cuadrado A., Engedal N., Jirsova Z., Cahova M. The role of free radicals in autophagy regulation: implications for ageing. Oxid Med Cell Longev. 2018;2018:2450748. doi: 10.1155/2018/2450748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Pajares M., Jimenez-Moreno N., Garcia-Yague A.J., Escoll M., de Ceballos M.L., Van Leuven F., Rabano A., Yamamoto M., Rojo A.I., Cuadrado A. Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes. Autophagy. 2016;12(10):1902–1916. doi: 10.1080/15548627.2016.1208889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Suzuki T., Yamamoto M. Stress-sensing mechanisms and the physiological roles of the Keap1-Nrf2 system during cellular stress. J. Biol. Chem. 2017;292(41):16817–16824. doi: 10.1074/jbc.R117.800169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kobayashi A., Kang M.I., Watai Y., Tong K.I., Shibata T., Uchida K., Yamamoto M. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol. Cell Biol. 2006;26(1):221–229. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kaspar J.W., Niture S.K., Jaiswal A.K. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic. Biol. Med. 2009;47(9):1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Mitsuishi Y., Motohashi H., Yamamoto M. The Keap1-Nrf2 system in cancers: stress response and anabolic metabolism. Front Oncol. 2012;2:200. doi: 10.3389/fonc.2012.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Jain A., Lamark T., Sjottem E., Larsen K.B., Awuh J.A., Overvatn A., McMahon M., Hayes J.D., Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 2010;285(29):22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ichimura Y., Waguri S., Sou Y.S., Kageyama S., Hasegawa J., Ishimura R., Saito T., Yang Y., Kouno T., Fukutomi T., Hoshii T., Hirao A., Takagi K., Mizushima T., Motohashi H., Lee M.S., Yoshimori T., Tanaka K., Yamamoto M., Komatsu M. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol. Cell. 2013;51(5):618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 203.Taguchi K., Fujikawa N., Komatsu M., Ishii T., Unno M., Akaike T., Motohashi H., Yamamoto M. Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proc. Natl. Acad. Sci. U. S. A. 2012;109(34):13561–13566. doi: 10.1073/pnas.1121572109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kumar R.R., Narasimhan M., Shanmugam G., Hong J., Devarajan A., Palaniappan S., Zhang J., Halade G.V., Darley-Usmar V.M., Hoidal J.R., Rajasekaran N.S. Abrogation of Nrf2 impairs antioxidant signaling and promotes atrial hypertrophy in response to high-intensity exercise stress. J. Transl. Med. 2016;14:86. doi: 10.1186/s12967-016-0839-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wang Y., Mandal A.K., Son Y.O., Pratheeshkumar P., Wise J.T.F., Wang L., Zhang Z., Shi X., Chen Z. Roles of ROS, Nrf2, and autophagy in cadmium-carcinogenesis and its prevention by sulforaphane. Toxicol. Appl. Pharmacol. 2018;353:23–30. doi: 10.1016/j.taap.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Son Y.O., Pratheeshkumar P., Divya S.P., Zhang Z., Shi X. Nuclear factor erythroid 2-related factor 2 enhances carcinogenesis by suppressing apoptosis and promoting autophagy in nickel-transformed cells. J. Biol. Chem. 2017;292(20):8315–8330. doi: 10.1074/jbc.M116.773986. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 207.Kosztelnik M., Kurucz A., Papp D., Jones E., Sigmond T., Barna J., Traka M.H., Lorincz T., Szarka A., Banhegyi G., Vellai T., Korcsmaros T., Kapuy O. Suppression of AMPK/aak-2 by NRF2/SKN-1 down-regulates autophagy during prolonged oxidative stress. Faseb. J. 2019;33(2):2372–2387. doi: 10.1096/fj.201800565RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Yamada T., Murata D., Adachi Y., Itoh K., Kameoka S., Igarashi A., Kato T., Araki Y., Huganir R.L., Dawson T.M., Yanagawa T., Okamoto K., Iijima M., Sesaki H. Mitochondrial stasis reveals p62-mediated ubiquitination in parkin-independent mitophagy and mitigates nonalcoholic fatty liver disease. Cell Metabol. 2018;28(4):588–604 e5. doi: 10.1016/j.cmet.2018.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Murata H., Takamatsu H., Liu S., Kataoka K., Huh N.H., Sakaguchi M. NRF2 regulates PINK1 expression under oxidative stress conditions. PloS One. 2015;10(11) doi: 10.1371/journal.pone.0142438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Georgakopoulos N.D., Frison M., Alvarez M.S., Bertrand H., Wells G., Campanella M. Reversible Keap1 inhibitors are preferential pharmacological tools to modulate cellular mitophagy. Sci. Rep. 2017;7(1):10303. doi: 10.1038/s41598-017-07679-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Gureev A.P., Shaforostova E.A., Popov V.N. Regulation of mitochondrial biogenesis as a way for active longevity: interaction between the Nrf2 and PGC-1alpha signaling pathways. Front. Genet. 2019;10:435. doi: 10.3389/fgene.2019.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Bae S.H., Sung S.H., Oh S.Y., Lim J.M., Lee S.K., Park Y.N., Lee H.E., Kang D., Rhee S.G. Sestrins activate Nrf2 by promoting p62-dependent autophagic degradation of Keap1 and prevent oxidative liver damage. Cell Metabol. 2013;17(1):73–84. doi: 10.1016/j.cmet.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 213.Hou Y.S., Guan J.J., Xu H.D., Wu F., Sheng R., Qin Z.H. Sestrin2 protects dopaminergic cells against rotenone toxicity through AMPK-dependent autophagy activation. Mol. Cell Biol. 2015;35(16):2740–2751. doi: 10.1128/MCB.00285-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Maiuri M.C., Malik S.A., Morselli E., Kepp O., Criollo A., Mouchel P.L., Carnuccio R., Kroemer G. Stimulation of autophagy by the p53 target gene Sestrin2. Cell Cycle. 2009;8(10):1571–1576. doi: 10.4161/cc.8.10.8498. [DOI] [PubMed] [Google Scholar]
- 215.Sanchez-Alvarez M., Strippoli R., Donadelli M., Bazhin A.V., Cordani M. Sestrins as a therapeutic bridge between ROS and autophagy in cancer. Cancers (Basel) 2019;11(10) doi: 10.3390/cancers11101415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Morrison A., Chen L., Wang J., Zhang M., Yang H., Ma Y., Budanov A., Lee J.H., Karin M., Li J. Sestrin2 promotes LKB1-mediated AMPK activation in the ischemic heart. Faseb. J. 2015;29(2):408–417. doi: 10.1096/fj.14-258814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ben-Sahra I., Dirat B., Laurent K., Puissant A., Auberger P., Budanov A., Tanti J.F., Bost F. Sestrin2 integrates Akt and mTOR signaling to protect cells against energetic stress-induced death. Cell Death Differ. 2013;20(4):611–619. doi: 10.1038/cdd.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Sablina A.A., Budanov A.V., Ilyinskaya G.V., Agapova L.S., Kravchenko J.E., Chumakov P.M. The antioxidant function of the p53 tumor suppressor. Nat. Med. 2005;11(12):1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Rhee S.G., Bae S.H. The antioxidant function of sestrins is mediated by promotion of autophagic degradation of Keap1 and Nrf2 activation and by inhibition of mTORC1. Free Radic. Biol. Med. 2015;88(Pt B):205–211. doi: 10.1016/j.freeradbiomed.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 220.Budanov A.V., Sablina A.A., Feinstein E., Koonin E.V., Chumakov P.M. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304(5670):596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- 221.Kimball S.R., Gordon B.S., Moyer J.E., Dennis M.D., Jefferson L.S. Leucine induced dephosphorylation of Sestrin2 promotes mTORC1 activation. Cell. Signal. 2016;28(8):896–906. doi: 10.1016/j.cellsig.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Roberts D.J., Tan-Sah V.P., Ding E.Y., Smith J.M., Miyamoto S. Hexokinase-II positively regulates glucose starvation-induced autophagy through TORC1 inhibition. Mol. Cell. 2014;53(4):521–533. doi: 10.1016/j.molcel.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Chichger H., Rounds S., Harrington E.O. Endosomes and autophagy: regulators of pulmonary endothelial cell homeostasis in health and disease. Antioxidants Redox Signal. 2019;31(13):994–1008. doi: 10.1089/ars.2019.7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Ryter S.W., Bhatia D., Choi M.E. Autophagy: a lysosome-dependent process with implications in cellular redox homeostasis and human disease. Antioxidants Redox Signal. 2019;30(1):138–159. doi: 10.1089/ars.2018.7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Rodriguez-Roisin R., Rabe K.F., Vestbo J., Vogelmeier C., Agusti A. Global initiative for chronic obstructive lung disease (GOLD) 20th anniversary: a brief history of time. Eur. Respir. J. 2017;50(1) doi: 10.1183/13993003.00671-2017. 00671-2017. [DOI] [PubMed] [Google Scholar]
- 226.Rabe K.F., Hurd S., Anzueto A., Barnes P.J., Buist S.A., Calverley P., Fukuchi Y., Jenkins C., Rodriguez-Roisin R., van Weel C., Zielinski J. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am. J. Respir. Crit. Care Med. 2007;176(6):532–555. doi: 10.1164/rccm.200703-456SO. [DOI] [PubMed] [Google Scholar]
- 227.Strzelak A., Ratajczak A., Adamiec A., Feleszko W. Tobacco smoke induces and alters immune responses in the lung triggering inflammation, allergy, asthma and other lung diseases: a mechanistic review. Int. J. Environ. Res. Publ. Health. 2018;15(5) doi: 10.3390/ijerph15051033. (pii) (2018) ijerph15051033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Chen Z.-H., Lam H.C., Jin Y., Kim H.-P., Cao J., Lee S.-J., Ifedigbo E., Parameswaran H., Ryter S.W., Choi A.M.K. Autophagy protein microtubule-associated protein 1 light chain-3B (LC3B) activates extrinsic apoptosis during cigarette smoke-induced emphysema. Proc. Natl. Acad. Sci. Unit. States Am. 2010;107(44):18880–18885. doi: 10.1073/pnas.1005574107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Gegg M.E., Cooper J.M., Chau K.Y., Rojo M., Schapira A.H., Taanman J.W. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum. Mol. Genet. 2010;19(24):4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Mizumura K., Cloonan S.M., Nakahira K., Bhashyam A.R., Cervo M., Kitada T., Glass K., Owen C.A., Mahmood A., Washko G.R., Hashimoto S., Ryter S.W., Choi A.M. Mitophagy-dependent necroptosis contributes to the pathogenesis of COPD. J. Clin. Invest. 2014;124(9):3987–4003. doi: 10.1172/JCI74985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Chen C.Z., Ou C.Y., Wang R.H., Lee C.H., Lin C.C., Chang H.Y., Hsiue T.R. Association of Egr-1 and autophagy-related gene polymorphism in men with chronic obstructive pulmonary disease. J. Formos. Med. Assoc. 2015;114(8):750–755. doi: 10.1016/j.jfma.2013.07.015. [DOI] [PubMed] [Google Scholar]
- 232.Chen Z.H., Kim H.P., Sciurba F.C., Lee S.J., Feghali-Bostwick C., Stolz D.B., Dhir R., Landreneau R.J., Schuchert M.J., Yousem S.A., Nakahira K., Pilewski J.M., Lee J.S., Zhang Y., Ryter S.W., Choi A.M. Egr-1 regulates autophagy in cigarette smoke-induced chronic obstructive pulmonary disease. PloS One. 2008;3(10) doi: 10.1371/journal.pone.0003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Bodas M., Silverberg D., Walworth K., Brucia K., Vij N. Augmentation of S-nitrosoglutathione controls cigarette smoke-induced inflammatory-oxidative stress and chronic obstructive pulmonary disease-emphysema pathogenesis by restoring cystic fibrosis transmembrane conductance regulator function. Antioxidants Redox Signal. 2017;27(7):433–451. doi: 10.1089/ars.2016.6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Shivalingappa P.C., Hole R., Westphal C.V., Vij N. Airway exposure to E-cigarette vapors impairs autophagy and induces aggresome formation. Antioxidants Redox Signal. 2016;24(4):186–204. doi: 10.1089/ars.2015.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Tran I., Ji C., Ni I., Min T., Tang D., Vij N. Role of cigarette smoke-induced aggresome formation in chronic obstructive pulmonary disease-emphysema pathogenesis. Am. J. Respir. Cell Mol. Biol. 2015;53(2):159–173. doi: 10.1165/rcmb.2014-0107OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Bodas M., Patel N., Silverberg D., Walworth K., Vij N. Master autophagy regulator transcription factor EB regulates cigarette smoke-induced autophagy impairment and chronic obstructive pulmonary disease-emphysema pathogenesis. Antioxidants Redox Signal. 2017;27(3):150–167. doi: 10.1089/ars.2016.6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Mercado N., Ito K., Barnes P.J. Accelerated ageing of the lung in COPD: new concepts. Thorax. 2015;70(5):482–489. doi: 10.1136/thoraxjnl-2014-206084. [DOI] [PubMed] [Google Scholar]
- 238.Bernardo I., Bozinovski S., Vlahos R. Targeting oxidant-dependent mechanisms for the treatment of COPD and its comorbidities. Pharmacol. Ther. 2015;155:60–79. doi: 10.1016/j.pharmthera.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 239.Ahmad T., Sundar I.K., Lerner C.A., Gerloff J., Tormos A.M., Yao H., Rahman I. Impaired mitophagy leads to cigarette smoke stress-induced cellular senescence: implications for chronic obstructive pulmonary disease. Faseb. J. 2015;29(7):2912–2929. doi: 10.1096/fj.14-268276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Beasley M.B. The pathologist's approach to acute lung injury. Arch. Pathol. Lab Med. 2010;134(5):719–727. doi: 10.5858/134.5.719. [DOI] [PubMed] [Google Scholar]
- 241.Li Z.Y., Wu Y.F., Xu X.C., Zhou J.S., Wang Y., Shen H.H., Chen Z.H. Autophagy as a double-edged sword in pulmonary epithelial injury: a review and perspective. Am. J. Physiol. Lung Cell Mol. Physiol. 2017;313(2):L207–L217. doi: 10.1152/ajplung.00562.2016. [DOI] [PubMed] [Google Scholar]
- 242.Chen Q., Yu S., Zhang K., Zhang Z., Li C., Gao B., Zhang W., Wang Y. Exogenous H2S inhibits autophagy in unilateral ureteral obstruction mouse renal tubule cells by regulating the ROS-AMPK signaling pathway. Cell. Physiol. Biochem. 2018;49(6):2200–2213. doi: 10.1159/000493824. [DOI] [PubMed] [Google Scholar]
- 243.Malaviya R., Laskin J.D., Laskin D.L. Oxidative stress-induced autophagy: role in pulmonary toxicity. Toxicol. Appl. Pharmacol. 2014;275(2):145–151. doi: 10.1016/j.taap.2013.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Zhang D., Zhou J., Ye L.C., Li J., Wu Z., Li Y., Li C. Autophagy maintains the integrity of endothelial barrier in LPS-induced lung injury. J. Cell. Physiol. 2018;233(1):688–698. doi: 10.1002/jcp.25928. [DOI] [PubMed] [Google Scholar]
- 245.Lopez-Alonso I., Aguirre A., Gonzalez-Lopez A., Fernandez A.F., Amado-Rodriguez L., Astudillo A., Batalla-Solis E., Albaiceta G.M. Impairment of autophagy decreases ventilator-induced lung injury by blockade of the NF-kappaB pathway. Am. J. Physiol. Lung Cell Mol. Physiol. 2013;304(12):L844–L852. doi: 10.1152/ajplung.00422.2012. [DOI] [PubMed] [Google Scholar]
- 246.Mizumura K., Cloonan S., Choi M.E., Hashimoto S., Nakahira K., Ryter S.W., Choi A.M. Autophagy: friend or foe in lung disease? Ann Am Thorac Soc. 2016;13(Suppl 1):S40–S47. doi: 10.1513/AnnalsATS.201507-450MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Zhang Y., Liu G., Dull R.O., Schwartz D.E., Hu G. Autophagy in pulmonary macrophages mediates lung inflammatory injury via NLRP3 inflammasome activation during mechanical ventilation. Am. J. Physiol. Lung Cell Mol. Physiol. 2014;307(2):L173–L185. doi: 10.1152/ajplung.00083.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Aguirre A., López-Alonso I., González-López A., Amado-Rodríguez L., Batalla-Solís E., Astudillo A., Blázquez-Prieto J., Fernández A.F., Galván J.A., Dos Santos C.C., Albaiceta G.M. Defective autophagy impairs ATF3 activity and worsens lung injury during. endotoxemia. 2014;92(6):665–676. doi: 10.1007/s00109-014-1132-7. [DOI] [PubMed] [Google Scholar]
- 249.Saitoh T., Fujita N., Jang M.H., Uematsu S., Yang B.-G., Satoh T., Omori H., Noda T., Yamamoto N., Komatsu M., Tanaka K., Kawai T., Tsujimura T., Takeuchi O., Yoshimori T., Akira S. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 2008;456(7219):264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 250.Lorne E., Zhao X., Zmijewski J.W., Liu G., Park Y.-J., Tsuruta Y., Abraham E. Participation of mammalian target of rapamycin complex 1 in toll-like receptor 2– and 4–induced neutrophil activation and acute lung. Injury. 2009;41(2):237–245. doi: 10.1165/rcmb.2008-0290OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Nakahira K., Haspel J.A., Rathinam V.A., Lee S.J., Dolinay T., Lam H.C., Englert J.A., Rabinovitch M., Cernadas M., Kim H.P., Fitzgerald K.A., Ryter S.W., Choi A.M. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 2011;12(3):222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Hu Y., Lou J., Mao Y.Y., Lai T.W., Liu L.Y., Zhu C., Zhang C., Liu J., Li Y.Y., Zhang F., Li W., Ying S.M., Chen Z.H., Shen H.H. Activation of MTOR in pulmonary epithelium promotes LPS-induced acute lung injury. Autophagy. 2016;12(12):2286–2299. doi: 10.1080/15548627.2016.1230584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Chien W.S., Chen Y.H., Chiang P.C., Hsiao H.W., Chuang S.M., Lue S.I., Hsu C. Suppression of autophagy in rat liver at late stage of polymicrobial sepsis. Shock. 2011;35(5):506–511. doi: 10.1097/SHK.0b013e31820b2f05. [DOI] [PubMed] [Google Scholar]
- 254.Lo S., Yuan S.S., Hsu C., Cheng Y.J., Chang Y.F., Hsueh H.W., Lee P.H., Hsieh Y.C. Lc3 over-expression improves survival and attenuates lung injury through increasing autophagosomal clearance in septic mice. Ann. Surg. 2013;257(2):352–363. doi: 10.1097/SLA.0b013e318269d0e2. [DOI] [PubMed] [Google Scholar]
- 255.Takahashi W., Watanabe E., Fujimura L., Watanabe-Takano H., Yoshidome H., Swanson P.E., Tokuhisa T., Oda S., Hatano M. Kinetics and protective role of autophagy in a mouse cecal ligation and puncture-induced sepsis. Crit. Care. 2013;17(4):R160. doi: 10.1186/cc12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Zhao H., Chen H., Xiaoyin M., Yang G., Hu Y., Xie K., Yu Y. Autophagy activation improves lung injury and inflammation in sepsis. Inflammation. 2019;42(2):426–439. doi: 10.1007/s10753-018-00952-5. [DOI] [PubMed] [Google Scholar]
- 257.Carchman E.H., Rao J., Loughran P.A., Rosengart M.R., Zuckerbraun B.S. Heme oxygenase-1-mediated autophagy protects against hepatocyte cell death and hepatic injury from infection/sepsis in mice. Hepatology. 2011;53(6):2053–2062. doi: 10.1002/hep.24324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Yen Y.T., Yang H.R., Lo H.C., Hsieh Y.C., Tsai S.C., Hong C.W., Hsieh C.H. Enhancing autophagy with activated protein C and rapamycin protects against sepsis-induced acute lung injury. Surgery. 2013;153(5):689–698. doi: 10.1016/j.surg.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 259.Chan K., Kan Y.W. Nrf2 is essential for protection against acute pulmonary injury in mice. Proc. Natl. Acad. Sci. U. S. A. 1999;96(22):12731–12736. doi: 10.1073/pnas.96.22.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Rojo de la Vega M., Dodson M., Gross C., Mansour H.M., Lantz R.C., Chapman E., Wang T., Black S.M., Garcia J.G., Zhang D.D. Role of Nrf2 and autophagy in acute lung injury. Curr Pharmacol Rep. 2016;2(2):91–101. doi: 10.1007/s40495-016-0053-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Wei J., Chen G., Shi X., Zhou H., Liu M., Chen Y., Feng D., Zhang P., Wu L., Lv X. Nrf2 activation protects against intratracheal LPS induced mouse/murine acute respiratory distress syndrome by regulating macrophage polarization. Biochem. Biophys. Res. Commun. 2018;500(3):790–796. doi: 10.1016/j.bbrc.2018.04.161. [DOI] [PubMed] [Google Scholar]
- 262.Tanaka A., Jin Y., Lee S.J., Zhang M., Kim H.P., Stolz D.B., Ryter S.W., Choi A.M. Hyperoxia-induced LC3B interacts with the Fas apoptotic pathway in epithelial cell death. Am. J. Respir. Cell Mol. Biol. 2012;46(4):507–514. doi: 10.1165/rcmb.2009-0415OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Wang X., Wang Y., Kim H.P., Nakahira K., Ryter S.W., Choi A.M. Carbon monoxide protects against hyperoxia-induced endothelial cell apoptosis by inhibiting reactive oxygen species formation. J. Biol. Chem. 2007;282(3):1718–1726. doi: 10.1074/jbc.M607610200. [DOI] [PubMed] [Google Scholar]
- 264.Reddy N.M., Suryanaraya V., Yates M.S., Kleeberger S.R., Hassoun P.M., Yamamoto M., Liby K.T., Sporn M.B., Kensler T.W., Reddy S.P. The triterpenoid CDDO-imidazolide confers potent protection against hyperoxic acute lung injury in mice. Am. J. Respir. Crit. Care Med. 2009;180(9):867–874. doi: 10.1164/rccm.200905-0670OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Hu R., Chen Z.F., Yan J., Li Q.F., Huang Y., Xu H., Zhang X., Jiang H. Complement C5a exacerbates acute lung injury induced through autophagy-mediated alveolar macrophage apoptosis. Cell Death Dis. 2014;5 doi: 10.1038/cddis.2014.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Xu D., Chen B., Gu J., Chen L., Belguise K., Wang X., Yi B., Lu K. Inhibition of autophagy ameliorates pulmonary microvascular dilation and PMVECs excessive proliferation in rat experimental hepatopulmonary syndrome. Sci. Rep. 2016;6:30833. doi: 10.1038/srep30833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Martinez-Caro L., Nin N., Sanchez-Rodriguez C., Ferruelo A., El Assar M., de Paula M., Fernandez-Segoviano P., Esteban A., Lorente J.A. Inhibition of nitro-oxidative stress attenuates pulmonary and systemic injury induced by high-tidal volume mechanical ventilation. Shock. 2015;44(1):36–43. doi: 10.1097/SHK.0000000000000381. [DOI] [PubMed] [Google Scholar]
- 268.Wu J., Yan Z., Schwartz D.E., Yu J., Malik A.B., Hu G. Activation of NLRP3 inflammasome in alveolar macrophages contributes to mechanical stretch-induced lung inflammation and injury. J. Immunol. 2013;190(7):3590–3599. doi: 10.4049/jimmunol.1200860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Zemskov E.A., Lu Q., Ornatowski W., Klinger C.N., Desai A.A., Maltepe E., Yuan J.X., Wang T., Fineman J.R., Black S.M. Biomechanical forces and oxidative stress: implications for pulmonary vascular disease. Antioxidants Redox Signal. 2019;31(12):819–842. doi: 10.1089/ars.2018.7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Lee I.T., Yang C.M. Role of NADPH oxidase/ROS in pro-inflammatory mediators-induced airway and pulmonary diseases. Biochem. Pharmacol. 2012;84(5):581–590. doi: 10.1016/j.bcp.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 271.Ge X S.J., Fei A., Gao C., Pan S., Wu Z. Hydrogen sulfide treatment alleviated ventilator-induced lung injury through regulation of autophagy and endoplasmic reticulum stress. Int. J. Biol. Sci. 2019;15:2872–2884. doi: 10.7150/ijbs.38315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Bos E.M., Van Goor H., Joles J.A., Whiteman M., Leuvenink H.G.D. Hydrogen sulfide: physiological properties and therapeutic potential in ischaemia. Br. J. Pharmacol. 2015;172(6):1479–1493. doi: 10.1111/bph.12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Spassov S.G., Donus R., Ihle P.M., Engelstaedter H., Hoetzel A., Faller S. 2017. Hydrogen Sulfide prevents formation of reactive oxygen species through PI3K/Akt signaling and limits ventilator-induced lung injury. 2017, 1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Francis R.C., Vaporidi K., Bloch K.D., Ichinose F., Zapol W.M. Protective and detrimental effects of sodium sulfide and hydrogen sulfide in murine ventilator-induced lung injury. Anesthesiology. 2011;115(5):1012–1021. doi: 10.1097/ALN.0b013e31823306cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Esposito S., Tosco A., Villella V.R., Raia V., Kroemer G., Maiuri L. Manipulating proteostasis to repair the F508del-CFTR defect in cystic fibrosis. Mol.Cell.Pediatr. 2016;3(1) doi: 10.1186/s40348-016-0040-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Fraser-Pitt D., O’Neil D. Cystic fibrosis – a multiorgan protein misfolding disease. Future Science OA. 2015;1(2) doi: 10.4155/fso.15.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Maiuri L., Raia V., Kroemer G. Strategies for the etiological therapy of cystic fibrosis. Cell Death Differ. 2017;24(11):1825–1844. doi: 10.1038/cdd.2017.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Maiuri L., Luciani A., Giardino I., Raia V., Villella V.R., D'Apolito M., Pettoello-Mantovani M., Guido S., Ciacci C., Cimmino M., Cexus O.N., Londei M., Quaratino S. Tissue transglutaminase activation modulates inflammation in cystic fibrosis via. PPAR Down-Regulation. 2008;180(11):7697–7705. doi: 10.4049/jimmunol.180.11.7697. [DOI] [PubMed] [Google Scholar]
- 279.Pesce E., Sondo E., Ferrera L., Tomati V., Caci E., Scudieri P., Musante I., Renda M., Baatallah N., Servel N., Hinzpeter A., di Bernardo D., Pedemonte N., Galietta L.J.V. The autophagy inhibitor spautin-1 antagonizes rescue of mutant CFTR through an autophagy-independent and USP13-mediated mechanism. Front. Pharmacol. 2018;9:1464. doi: 10.3389/fphar.2018.01464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Yang A., Jiao Y., Yang S., Deng M., Yang X., Mao C., Sun Y., Ding N., Li N., Zhang M., Jin S., Zhang H., Jiang Y. Homocysteine activates autophagy by inhibition of CFTR expression via interaction between DNA methylation and H3K27me3 in mouse liver. Cell Death Dis. 2018;9(2) doi: 10.1038/s41419-017-0216-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Van Goor F., Hadida S., Grootenhuis P.D.J., Burton B., Stack J.H., Straley K.S., Decker C.J., Miller M., McCartney J., Olson E.R., Wine J.J., Frizzell R.A., Ashlock M., Negulescu P.A. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc. Natl. Acad. Sci. Unit. States Am. 2011;108(46):18843–18848. doi: 10.1073/pnas.1105787108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Gao L K.K., Yankaskas J.R., Forman H.J. Abnormal glutathione transport in cystic fibrosis airway epithelia. Am. J. Physiol. 1999;277:113–118. doi: 10.1152/ajplung.1999.277.1.L113. [DOI] [PubMed] [Google Scholar]
- 283.Leonard A.V.H., Velsor W., Day Brian J. Antioxidant imbalance in the lungs of cystic fibrosis transmembrane conductance regulator protein mutant mice. Am. J. Physiol. Lung Cell Mol. Physiol. 2001;281:31–38. doi: 10.1152/ajplung.2001.281.1.L31. [DOI] [PubMed] [Google Scholar]
- 284.Rahman I., Macnee W. Oxidative stress and regulation of glutathione in lung inflammation. Eur. Respir. J. 2000;16(3):534. doi: 10.1034/j.1399-3003.2000.016003534.x. [DOI] [PubMed] [Google Scholar]
- 285.Wree A., Bechmann L.P., Kumarasamy N., Sommerwerck U., Jochum C., Jakob H., Baba H.A., Gerken G., Kamler M., Canbay A. Elevated gamma-glutamyltransferase is associated with mortality in lung transplantation for cystic fibrosis. Transpl. Int. 2012;25(1):78–86. doi: 10.1111/j.1432-2277.2011.01376.x. [DOI] [PubMed] [Google Scholar]
- 286.Corti A., Griese M., Hector A., Pompella A. Increasing sputum levels of gamma-glutamyltransferase may identify cystic fibrosis patients who do not benefit from inhaled glutathione. J. Cyst. Fibros. 2017;16(3):342–345. doi: 10.1016/j.jcf.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 287.Zhang S., Stoll G., Pedro J.M.B.S., Sica V., Sauvat A., Obrist F., Kepp O., Li Y., Maiuri L., Zamzami N., Kroemer G. Evaluation of autophagy inducers in epithelial cells carrying the ΔF508 mutation of the cystic fibrosis transmembrane conductance regulator CFTR. Cell Death Dis. 2018;9(2) doi: 10.1038/s41419-017-0235-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Caution K., Pan A., Krause K., Badr A., Hamilton K., Vaidya A., Gosu H., Daily K., Estfanous S., Gavrilin M.A., Drew M.E., Cormet-Boyaka E., Chen X., Frankhouser D.E., Bundschuh R., Yan P., Dakhlallah D., Amer A.O. Methylomic correlates of autophagy activity in cystic fibrosis. J. Cyst. Fibros. 2019;18(4):491–500. doi: 10.1016/j.jcf.2019.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Reilly R., Mroz M.S., Dempsey E., Wynne K., Keely S.J., McKone E.F., Hiebel C., Behl C., Coppinger J.A. Targeting the PI3K/Akt/mTOR signalling pathway in Cystic Fibrosis. Sci. Rep. 2017;7(1) doi: 10.1038/s41598-017-06588-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Barnes P.J. Pulmonary diseases and ageing. Subcell. Biochem. 2019;91:45–74. doi: 10.1007/978-981-13-3681-2_3. [DOI] [PubMed] [Google Scholar]
- 291.Liao S.X., Sun P.P., Gu Y.H., Rao X.M., Zhang L.Y., Ou-Yang Y. Autophagy and pulmonary disease. Ther. Adv. Respir. Dis. 2019;13 doi: 10.1177/1753466619890538. 1753466619890538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Racanelli A.C., Kikkers S.A., Choi A.M.K., Cloonan S.M. Autophagy and inflammation in chronic respiratory disease. Autophagy. 2018;14(2):221–232. doi: 10.1080/15548627.2017.1389823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Richeldi L., Collard H.R., Jones M.G. Idiopathic pulmonary fibrosis. Lancet. 2017;389(10082):1941–1952. doi: 10.1016/S0140-6736(17)30866-8. [DOI] [PubMed] [Google Scholar]
- 294.Bueno M., Lai Y.C., Romero Y., Brands J., St Croix C.M., Kamga C., Corey C., Herazo-Maya J.D., Sembrat J., Lee J.S., Duncan S.R., Rojas M., Shiva S., Chu C.T., Mora A.L. PINK1 deficiency impairs mitochondrial homeostasis and promotes lung fibrosis. J. Clin. Invest. 2015;125(2):521–538. doi: 10.1172/JCI74942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Sosulski M.L., Gongora R., Danchuk S., Dong C., Luo F., Sanchez C.G. Deregulation of selective autophagy during aging and pulmonary fibrosis: the role of TGFbeta1. Aging Cell. 2015;14(5):774–783. doi: 10.1111/acel.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Nho R.S., Hergert P. IPF fibroblasts are desensitized to type I collagen matrix-induced cell death by suppressing low autophagy via aberrant Akt/mTOR kinases. PloS One. 2014;9(4) doi: 10.1371/journal.pone.0094616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Ricci A., Cherubini E., Scozzi D., Pietrangeli V., Tabbi L., Raffa S., Leone L., Visco V., Torrisi M.R., Bruno P., Mancini R., Ciliberto G., Terzano C., Mariotta S. Decreased expression of autophagic beclin 1 protein in idiopathic pulmonary fibrosis fibroblasts. J. Cell. Physiol. 2013;228(7):1516–1524. doi: 10.1002/jcp.24307. [DOI] [PubMed] [Google Scholar]
- 298.Kato K., Hecker L. NADPH oxidases: pathophysiology and therapeutic potential in age-associated pulmonary fibrosis. Redox Biol. 2020:101541. doi: 10.1016/j.redox.2020.101541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Veith C., Boots A.W., Idris M., van Schooten F.J., van der Vliet A. Redox imbalance in idiopathic pulmonary fibrosis: a role for oxidant cross-talk between NADPH oxidase enzymes and mitochondria. Antioxidants Redox Signal. 2019;31(14):1092–1115. doi: 10.1089/ars.2019.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Tsubouchi K., Araya J., Minagawa S., Hara H., Ichikawa A., Saito N., Kadota T., Sato N., Yoshida M., Kurita Y., Kobayashi K., Ito S., Fujita Y., Utsumi H., Yanagisawa H., Hashimoto M., Wakui H., Yoshii Y., Ishikawa T., Numata T., Kaneko Y., Asano H., Yamashita M., Odaka M., Morikawa T., Nakayama K., Nakanishi Y., Kuwano K. Azithromycin attenuates myofibroblast differentiation and lung fibrosis development through proteasomal degradation of NOX4. Autophagy. 2017;13(8):1420–1434. doi: 10.1080/15548627.2017.1328348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Sato N., Takasaka N., Yoshida M., Tsubouchi K., Minagawa S., Araya J., Saito N., Fujita Y., Kurita Y., Kobayashi K., Ito S., Hara H., Kadota T., Yanagisawa H., Hashimoto M., Utsumi H., Wakui H., Kojima J., Numata T., Kaneko Y., Odaka M., Morikawa T., Nakayama K., Kohrogi H., Kuwano K. Metformin attenuates lung fibrosis development via NOX4 suppression. Respir. Res. 2016;17(1):107. doi: 10.1186/s12931-016-0420-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Fierro-Fernandez M., Busnadiego O., Sandoval P., Espinosa-Diez C., Blanco-Ruiz E., Rodriguez M., Pian H., Ramos R., Lopez-Cabrera M., Garcia-Bermejo M.L., Lamas S. miR-9-5p suppresses pro-fibrogenic transformation of fibroblasts and prevents organ fibrosis by targeting NOX4 and TGFBR2. EMBO Rep. 2015;16(10):1358–1377. doi: 10.15252/embr.201540750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Tian Y., Li H., Qiu T., Dai J., Zhang Y., Chen J., Cai H. Loss of PTEN induces lung fibrosis via alveolar epithelial cell senescence depending on NF-kappaB activation. Aging Cell. 2019;18(1) doi: 10.1111/acel.12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Schuliga M., Pechkovsky D.V., Read J., Waters D.W., Blokland K.E.C., Reid A.T., Hogaboam C.M., Khalil N., Burgess J.K., Prele C.M., Mutsaers S.E., Jaffar J., Westall G., Grainge C., Knight D.A. Mitochondrial dysfunction contributes to the senescent phenotype of IPF lung fibroblasts. J. Cell Mol. Med. 2018;22(12):5847–5861. doi: 10.1111/jcmm.13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Patel A.S., Lin L., Geyer A., Haspel J.A., An C.H., Cao J., Rosas I.O., Morse D. Autophagy in idiopathic pulmonary fibrosis. PloS One. 2012;7(7) doi: 10.1371/journal.pone.0041394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Kobayashi K., Araya J., Minagawa S., Hara H., Saito N., Kadota T., Sato N., Yoshida M., Tsubouchi K., Kurita Y., Ito S., Fujita Y., Takasaka N., Utsumi H., Yanagisawa H., Hashimoto M., Wakui H., Kojima J., Shimizu K., Numata T., Kawaishi M., Kaneko Y., Asano H., Yamashita M., Odaka M., Morikawa T., Nakayama K., Kuwano K. Involvement of PARK2-mediated mitophagy in idiopathic pulmonary fibrosis pathogenesis. J. Immunol. 2016;197(2):504–516. doi: 10.4049/jimmunol.1600265. [DOI] [PubMed] [Google Scholar]
- 307.Bueno M., Brands J., Voltz L., Fiedler K., Mays B., St Croix C., Sembrat J., Mallampalli R.K., Rojas M., Mora A.L. ATF3 represses PINK1 gene transcription in lung epithelial cells to control mitochondrial homeostasis. Aging Cell. 2018;17(2) doi: 10.1111/acel.12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Kurita Y., Araya J., Minagawa S., Hara H., Ichikawa A., Saito N., Kadota T., Tsubouchi K., Sato N., Yoshida M., Kobayashi K., Ito S., Fujita Y., Utsumi H., Yanagisawa H., Hashimoto M., Wakui H., Yoshii Y., Ishikawa T., Numata T., Kaneko Y., Asano H., Yamashita M., Odaka M., Morikawa T., Nakayama K., Kuwano K. Pirfenidone inhibits myofibroblast differentiation and lung fibrosis development during insufficient mitophagy. Respir. Res. 2017;18(1):114. doi: 10.1186/s12931-017-0600-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Bueno M., Zank D., Buendia-Roldan I., Fiedler K., Mays B.G., Alvarez D., Sembrat J., Kimball B., Bullock J.K., Martin J.L., Nouraie M., Kaufman B.A., Rojas M., Pardo A., Selman M., Mora A.L. PINK1 attenuates mtDNA release in alveolar epithelial cells and TLR9 mediated profibrotic responses. PloS One. 2019;14(6) doi: 10.1371/journal.pone.0218003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Ryu C., Sun H., Gulati M., Herazo-Maya J.D., Chen Y., Osafo-Addo A., Brandsdorfer C., Winkler J., Blaul C., Faunce J., Pan H., Woolard T., Tzouvelekis A., Antin-Ozerkis D.E., Puchalski J.T., Slade M., Gonzalez A.L., Bogenhagen D.F., Kirillov V., Feghali-Bostwick C., Gibson K., Lindell K., Herzog R.I., Dela Cruz C.S., Mehal W., Kaminski N., Herzog E.L., Trujillo G. Extracellular mitochondrial DNA is generated by fibroblasts and predicts death in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2017;196(12):1571–1581. doi: 10.1164/rccm.201612-2480OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Mora A.L., Bueno M., Rojas M. Mitochondria in the spotlight of aging and idiopathic pulmonary fibrosis. J. Clin. Invest. 2017;127(2):405–414. doi: 10.1172/JCI87440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Hancock L.A., Hennessy C.E., Solomon G.M., Dobrinskikh E., Estrella A., Hara N., Hill D.B., Kissner W.J., Markovetz M.R., Grove Villalon D.E., Voss M.E., Tearney G.J., Carroll K.S., Shi Y., Schwarz M.I., Thelin W.R., Rowe S.M., Yang I.V., Evans C.M., Schwartz D.A. Muc5b overexpression causes mucociliary dysfunction and enhances lung fibrosis in mice. Nat. Commun. 2018;9(1):5363. doi: 10.1038/s41467-018-07768-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Zhang Q., Wang Y., Qu D., Yu J., Yang J. The possible pathogenesis of idiopathic pulmonary fibrosis considering MUC5B. BioMed Res. Int. 2019;2019:9712464. doi: 10.1155/2019/9712464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Aggarwal S., Gross C.M., Sharma S., Fineman J.R., Black S.M. Reactive oxygen species in pulmonary vascular remodeling. Comp. Physiol. 2013;3(3):1011–1034. doi: 10.1002/cphy.c120024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Wedgwood S., Black S.M. Role of reactive oxygen species in vascular remodeling associated with pulmonary hypertension. Antioxidants Redox Signal. 2003;5(6):759–769. doi: 10.1089/152308603770380061. [DOI] [PubMed] [Google Scholar]
- 316.Grobe A.C., Wells S.M., Benavidez E., Oishi P., Azakie A., Fineman J.R., Black S.M. Increased oxidative stress in lambs with increased pulmonary blood flow and pulmonary hypertension: role of NADPH oxidase and endothelial NO synthase. Am. J. Physiol. Lung Cell Mol. Physiol. 2006;290(6):L1069–L1077. doi: 10.1152/ajplung.00408.2005. [DOI] [PubMed] [Google Scholar]
- 317.Rafikova O., Rafikov R., Kumar S., Sharma S., Aggarwal S., Schneider F., Jonigk D., Black S.M., Tofovic S.P. Bosentan inhibits oxidative and nitrosative stress and rescues occlusive pulmonary hypertension. Free Radic. Biol. Med. 2013;56:28–43. doi: 10.1016/j.freeradbiomed.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Sun X., Sharma S., Fratz S., Kumar S., Rafikov R., Aggarwal S., Rafikova O., Lu Q., Burns T., Dasarathy S., Wright J., Schreiber C., Radman M., Fineman J.R., Black S.M. Disruption of endothelial cell mitochondrial bioenergetics in lambs with increased pulmonary blood flow. Antioxidants Redox Signal. 2013;18(14):1739–1752. doi: 10.1089/ars.2012.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Rawat D.K., Alzoubi A., Gupte R., Chettimada S., Watanabe M., Kahn A.G., Okada T., McMurtry I.F., Gupte S.A. Increased reactive oxygen species, metabolic maladaptation, and autophagy contribute to pulmonary arterial hypertension-induced ventricular hypertrophy and diastolic heart failure. Hypertension. 2014;64(6):1266–1274. doi: 10.1161/HYPERTENSIONAHA.114.03261. [DOI] [PubMed] [Google Scholar]
- 320.Long L., Yang X., Southwood M., Lu J., Marciniak S.J., Dunmore B.J., Morrell N.W. Chloroquine prevents progression of experimental pulmonary hypertension via inhibition of autophagy and lysosomal bone morphogenetic protein type II receptor degradation. Circ. Res. 2013;112(8):1159–1170. doi: 10.1161/CIRCRESAHA.111.300483. [DOI] [PubMed] [Google Scholar]
- 321.Sakao S., Tatsumi K. The effects of antiangiogenic compound SU5416 in a rat model of pulmonary arterial hypertension. Respiration. 2011;81(3):253–261. doi: 10.1159/000322011. [DOI] [PubMed] [Google Scholar]
- 322.He Y., Cao X., Guo P., Li X., Shang H., Liu J., Xie M., Xu Y., Liu X. Quercetin induces autophagy via FOXO1-dependent pathways and autophagy suppression enhances quercetin-induced apoptosis in PASMCs in hypoxia. Free Radic. Biol. Med. 2017;103:165–176. doi: 10.1016/j.freeradbiomed.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 323.Zhang H., Gong Y., Wang Z., Jiang L., Chen R., Fan X., Zhu H., Han L., Li X., Xiao J., Kong X. Apelin inhibits the proliferation and migration of rat PASMCs via the activation of PI3K/Akt/mTOR signal and the inhibition of autophagy under hypoxia. J. Cell Mol. Med. 2014;18(3):542–553. doi: 10.1111/jcmm.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Wu K., Zhang Q., Wu X., Lu W., Tang H., Liang Z., Gu Y., Song S., Ayon R.J., Wang Z., McDermott K.M., Balistrieri A., Wang C., Black S.M., Garcia J.G.N., Makino A., Yuan J.X., Wang J. Chloroquine is a potent pulmonary vasodilator that attenuates hypoxia-induced pulmonary hypertension. Br. J. Pharmacol. 2017;174(22):4155–4172. doi: 10.1111/bph.13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Ibe J.C., Zhou Q., Chen T., Tang H., Yuan J.X., Raj J.U., Zhou G. Adenosine monophosphate-activated protein kinase is required for pulmonary artery smooth muscle cell survival and the development of hypoxic pulmonary hypertension. Am. J. Respir. Cell Mol. Biol. 2013;49(4):609–618. doi: 10.1165/rcmb.2012-0446OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.He Y., Cao X., Liu X., Li X., Xu Y., Liu J., Shi J. Quercetin reverses experimental pulmonary arterial hypertension by modulating the TrkA pathway. Exp. Cell Res. 2015;339(1):122–134. doi: 10.1016/j.yexcr.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 327.Papi A., Brightling C., Pedersen S.E., Reddel H.K. Asthma, Lancet. 2018;391(10122):783–800. doi: 10.1016/S0140-6736(17)33311-1. [DOI] [PubMed] [Google Scholar]
- 328.Bullone M., Lavoie J.P. The contribution of oxidative stress and inflamm-aging in human and equine asthma. Int. J. Mol. Sci. 2017;18(12) doi: 10.3390/ijms18122612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Kleniewska P., Pawliczak R. The participation of oxidative stress in the pathogenesis of bronchial asthma. Biomed. Pharmacother. 2017;94:100–108. doi: 10.1016/j.biopha.2017.07.066. [DOI] [PubMed] [Google Scholar]
- 330.Hoshino T., Okamoto M., Takei S., Sakazaki Y., Iwanaga T., Aizawa H. Redox-regulated mechanisms in asthma. Antioxidants Redox Signal. 2008;10(4):769–783. doi: 10.1089/ars.2007.1936. [DOI] [PubMed] [Google Scholar]
- 331.Jarjour N.N., Calhoun W.J. Enhanced production of oxygen radicals in asthma. J. Lab. Clin. Med. 1994;123(1):131–136. [PubMed] [Google Scholar]
- 332.Lee J., Kim H.S. The role of autophagy in eosinophilic airway inflammation. Immune Netw. 2019;19(1):e5. doi: 10.4110/in.2019.19.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Liu J.N., Suh D.H., Trinh H.K., Chwae Y.J., Park H.S., Shin Y.S. The role of autophagy in allergic inflammation: a new target for severe asthma. Exp. Mol. Med. 2016;48(7):e243. doi: 10.1038/emm.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.McAlinden K.D., Deshpande D.A., Ghavami S., Xenaki D., Sohal S.S., Oliver B.G., Haghi M., Sharma P. Autophagy activation in asthma airways remodeling. Am. J. Respir. Cell Mol. Biol. 2019;60(5):541–553. doi: 10.1165/rcmb.2018-0169OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Poon A.H., Chouiali F., Tse S.M., Litonjua A.A., Hussain S.N., Baglole C.J., Eidelman D.H., Olivenstein R., Martin J.G., Weiss S.T., Hamid Q., Laprise C. Genetic and histologic evidence for autophagy in asthma pathogenesis. J. Allergy Clin. Immunol. 2012;129(2):569–571. doi: 10.1016/j.jaci.2011.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Ban G.Y., Pham D.L., Trinh T.H., Lee S.I., Suh D.H., Yang E.M., Ye Y.M., Shin Y.S., Chwae Y.J., Park H.S. Autophagy mechanisms in sputum and peripheral blood cells of patients with severe asthma: a new therapeutic target. Clin. Exp. Allergy. 2016;46(1):48–59. doi: 10.1111/cea.12585. [DOI] [PubMed] [Google Scholar]
- 337.Martin L.J., Gupta J., Jyothula S.S., Butsch Kovacic M., Biagini Myers J.M., Patterson T.L., Ericksen M.B., He H., Gibson A.M., Baye T.M., Amirisetty S., Tsoras A.M., Sha Y., Eissa N.T., Hershey G.K. Functional variant in the autophagy-related 5 gene promotor is associated with childhood asthma. PloS One. 2012;7(4) doi: 10.1371/journal.pone.0033454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Suzuki Y., Maazi H., Sankaranarayanan I., Lam J., Khoo B., Soroosh P., Barbers R.G., James Ou J.H., Jung J.U., Akbari O. Lack of autophagy induces steroid-resistant airway inflammation. J. Allergy Clin. Immunol. 2016;137(5):1382–1389 e9. doi: 10.1016/j.jaci.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Pham D.L., Ban G.Y., Kim S.H., Shin Y.S., Ye Y.M., Chwae Y.J., Park H.S. Neutrophil autophagy and extracellular DNA traps contribute to airway inflammation in severe asthma. Clin. Exp. Allergy. 2017;47(1):57–70. doi: 10.1111/cea.12859. [DOI] [PubMed] [Google Scholar]
- 340.Pham D.L., Kim S.H., Losol P., Yang E.M., Shin Y.S., Ye Y.M., Park H.S. Association of autophagy related gene polymorphisms with neutrophilic airway inflammation in adult asthma. Korean J Intern Med. 2016;31(2):375–385. doi: 10.3904/kjim.2014.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Murai H., Okazaki S., Hayashi H., Kawakita A., Hosoki K., Yasutomi M., Sur S., Ohshima Y. Alternaria extract activates autophagy that induces IL-18 release from airway epithelial cells. Biochem. Biophys. Res. Commun. 2015;464(4):969–974. doi: 10.1016/j.bbrc.2015.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Weng C.M., Wang C.H., Lee M.J., He J.R., Huang H.Y., Chao M.W., Chung K.F., Kuo H.P. Aryl hydrocarbon receptor activation by diesel exhaust particles mediates epithelium-derived cytokines expression in severe allergic asthma. Allergy. 2018;73(11):2192–2204. doi: 10.1111/all.13462. [DOI] [PubMed] [Google Scholar]
- 343.Dickinson J.D., Alevy Y., Malvin N.P., Patel K.K., Gunsten S.P., Holtzman M.J., Stappenbeck T.S., Brody S.L. IL13 activates autophagy to regulate secretion in airway epithelial cells. Autophagy. 2016;12(2):397–409. doi: 10.1080/15548627.2015.1056967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Ghavami S., Cunnington R.H., Gupta S., Yeganeh B., Filomeno K.L., Freed D.H., Chen S., Klonisch T., Halayko A.J., Ambrose E., Singal R., Dixon I.M. Autophagy is a regulator of TGF-beta1-induced fibrogenesis in primary human atrial myofibroblasts. Cell Death Dis. 2015;6 doi: 10.1038/cddis.2015.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Aguilera-Aguirre L., Bacsi A., Saavedra-Molina A., Kurosky A., Sur S., Boldogh I. Mitochondrial dysfunction increases allergic airway inflammation. J. Immunol. 2009;183(8):5379–5387. doi: 10.4049/jimmunol.0900228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Reddy P.H. Mitochondrial dysfunction and oxidative stress in asthma: implications for mitochondria-targeted antioxidant therapeutics. Pharmaceuticals (Basel) 2011;4(3):429–456. doi: 10.3390/ph4030429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Iyer D., Mishra N., Agrawal A. Mitochondrial function in allergic disease. Curr. Allergy Asthma Rep. 2017;17(5):29. doi: 10.1007/s11882-017-0695-0. [DOI] [PubMed] [Google Scholar]
- 348.Qiu Y.N., Wang G.H., Zhou F., Hao J.J., Tian L., Guan L.F., Geng X.K., Ding Y.C., Wu H.W., Zhang K.Z. PM2.5 induces liver fibrosis via triggering ROS-mediated mitophagy. Ecotoxicol. Environ. Saf. 2019;167:178–187. doi: 10.1016/j.ecoenv.2018.08.050. [DOI] [PubMed] [Google Scholar]
- 349.Verbaanderd C., Maes H., Schaaf M.B., Sukhatme V.P., Pantziarka P., Sukhatme V., Agostinis P., Bouche G. Repurposing drugs in oncology (ReDO)-chloroquine and hydroxychloroquine as anti-cancer agents. Ecancermedicalscience. 2017;11:781. doi: 10.3332/ecancer.2017.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Kimura T., Takabatake Y., Takahashi A., Isaka Y. Chloroquine in cancer therapy: a double-edged sword of autophagy. Can. Res. 2013;73(1):3–7. doi: 10.1158/0008-5472.CAN-12-2464. [DOI] [PubMed] [Google Scholar]
- 351.Shoji-Kawata S., Sumpter R., Leveno M., Campbell G.R., Zou Z., Kinch L., Wilkins A.D., Sun Q., Pallauf K., MacDuff D., Huerta C., Virgin H.W., Helms J.B., Eerland R., Tooze S.A., Xavier R., Lenschow D.J., Yamamoto A., King D., Lichtarge O., Grishin N.V., Spector S.A., Kaloyanova D.V., Levine B. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013;494(7436):201–206. doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Rubinsztein D.C., Codogno P., Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 2012;11(9):709–730. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Choi A.M., Ryter S.W., Levine B. Autophagy in human health and disease. N. Engl. J. Med. 2013;368(19):1845–1846. doi: 10.1056/NEJMc1303158. [DOI] [PubMed] [Google Scholar]
- 354.Giampieri F., Afrin S., Forbes-Hernandez T.Y., Gasparrini M., Cianciosi D., Reboredo-Rodriguez P., Varela-Lopez A., Quiles J.L., Battino M. Autophagy in human health and disease: novel therapeutic opportunities. Antioxidants Redox Signal. 2019;30(4):577–634. doi: 10.1089/ars.2017.7234. [DOI] [PubMed] [Google Scholar]
- 355.Azad M.B., Chen Y., Gibson S.B. Regulation of autophagy by reactive oxygen species (ROS): implications for cancer progression and treatment. Antioxidants Redox Signal. 2009;11(4):777–790. doi: 10.1089/ars.2008.2270. [DOI] [PubMed] [Google Scholar]
- 356.Kondo Y., Kondo S. Autophagy and cancer therapy. Autophagy. 2006;2(2):85–90. doi: 10.4161/auto.2.2.2463. [DOI] [PubMed] [Google Scholar]
- 357.Kanzawa T., Kondo Y., Ito H., Kondo S., Germano I. Induction of autophagic cell death in malignant glioma cells by arsenic trioxide. Can. Res. 2003;63(9):2103–2108. [PubMed] [Google Scholar]
- 358.Chen Y., Gibson S.B. Is mitochondrial generation of reactive oxygen species a trigger for autophagy? Autophagy. 2008;4(2):246–248. doi: 10.4161/auto.5432. [DOI] [PubMed] [Google Scholar]
- 359.Jin S., White E. Role of autophagy in cancer: management of metabolic stress. Autophagy. 2007;3(1):28–31. doi: 10.4161/auto.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Hardie D.G. AMPK: a target for drugs and natural products with effects on both diabetes and cancer. Diabetes. 2013;62(7):2164–2172. doi: 10.2337/db13-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Palikaras K., Lionaki E., Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat. Cell Biol. 2018;20(9):1013–1022. doi: 10.1038/s41556-018-0176-2. [DOI] [PubMed] [Google Scholar]
- 362.Masuda G.O., Yashiro M., Kitayama K., Miki Y., Kasashima H., Kinoshita H., Morisaki T., Fukuoka T., Hasegawa T., Sakurai K., Toyokawa T., Kubo N., Tanaka H., Muguruma K., Masaichi O., Hirakawa K. Clinicopathological correlations of autophagy-related proteins LC3, beclin 1 and p62 in gastric cancer. Anticancer Res. 2016;36(1):129–136. [PubMed] [Google Scholar]
- 363.Yang M., Zhao H., Guo L., Zhang Q., Zhao L., Bai S., Zhang M., Xu S., Wang F., Wang X., Zhao B. Autophagy-based survival prognosis in human colorectal carcinoma. Oncotarget. 2015;6(9):7084–7103. doi: 10.18632/oncotarget.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Winardi D., Tsai H.P., Chai C.Y., Chung C.L., Loh J.K., Chen Y.H., Hsieh C.L. Correlation of altered expression of the autophagy marker LC3B with poor prognosis in astrocytoma. BioMed Res. Int. 2014;2014:723176. doi: 10.1155/2014/723176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Subramani S., Malhotra V. Non-autophagic roles of autophagy-related proteins. EMBO Rep. 2013;14(2):143–151. doi: 10.1038/embor.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]