Abstract
Glyphosate is a commonly used organophosphate herbicide that has an adverse impact on humans, mammals and soil microbial ecosystems. The redundant utilize of glyphosate to control weed growth cause the pollution of the soil environment by this chemical. The discharge of glyphosate in the agricultural drainage can also cause serious environmental damage and water pollution problems. Therefore, it is important to develop methods for enhancing glyphosate degradation in the soil through bioremediation. In this study, thirty bacterial isolates were selected from an agro-industrial zone located in Sadat City of Monufia Governorate, Egypt. The isolates were able to grow in LB medium supplemented with 7.2 mg/ml glyphosate. Ten isolates only had the ability to grow in a medium containing different concentrations of glyphosate (50, 100, 150, 200 and 250 mg/ml). The FACU3 bacterial isolate showed the highest CFU in the different concentrations of glyphosate. The FACU3 isolate was Gram-positive, spore-forming and rod-shape bacteria. Based on API 50 CHB/E medium kit, biochemical properties and 16S rRNA gene sequencing, the FACU3 isolate was identified as Bacillus aryabhattai. Different bioinformatics tools, including multiple sequence alignment (MSA), basic local alignment search tool (BLAST) and primer alignment, were used to design specific primers for goxB gene amplification and isolation. The goxB gene encodes FAD-dependent glyphosate oxidase enzyme that responsible for biodegradation process. The selected primers were successfully used to amplify the goxB gene from Bacillus aryabhattai FACU3. The results indicated that the Bacillus aryabhattai FACU3 can be utilized in glyphosate-contaminated environments for bioremediation. According to our knowledge, this is the first time to isolate of FAD-dependent glyphosate oxidase (goxB) gene from Bacillus aryabhattai.
Keywords: Glyphosate, Glyphosate-degrading bacteria, Bacillus aryabhattai, FAD-dependent glyphosate oxidase gene, BLAST, Multiple sequence alignment
1. Introduction
N-(phosphonomethyl) glycine, glyphosate (GP), is the most extensively applied herbicides in agriculture, post-emergent nonselective and broad-spectrum herbicide (Mbanaso et al., 2013). Glyphosate is used extensively for control of a great variety of plants including grasses and broad-leaved weeds. Its extensive use has been negatively linked to human health due to its toxicity (Goldstein et al., 2002). It has been reported that glyphosate is related to several health problems such as hypothyroidism, chronic kidney diseases, cancer and birth defects (Romano et al., 2012, Jayasumana et al., 2014). Because of the influence of glyphosate-containing herbicides and glyphosate on agriculture and microbial ecosystems in soil, it is very important to identify methods for enhancing glyphosate manipulation and bioremediation in soils (Romero et al., 2011).
Microorganisms have a significant role in pollutant removal in sediments, water and soil; especially for their use other than remediation methods (Demnerova et al., 2005). Leckie (2005) reported that bacteria are probably the most varied microorganisms due to their nutritional requirements. Bacteria are now widely used in the biological treatment process such as pesticides (Singh, 2008, Oliveira et al., 2015, Mohamed et al., 2018), polyaromatic hydrocarbon (Fulekar, 2017, Abdelhaleem et al., 2019) and heavy metals (Igiri et al., 2018, Mohamed et al., 2020). A lot of bacterial strains were isolated which able to degrade glyphosate and most of these strains have been isolated from sites already treated by the herbicide (Benslama and Boulahrouf, 2013).
Glyphosate is primarily decomposed in the soil by fungi and bacteria, which use glyphosate as a phosphorus source to produce a glycine or carbon source to produce an aminomethylphosphonic acid (AMPA) (Solomon et al., 2007). Cleansing soils contaminated with glyphosate herbicide using bacteria is considered an affordable and environmentally friendly bioremediation method (Kryuchkova et al., 2014). Many different strains of fungal and bacterial that can utilize glyphosate during metabolism as a source of phosphate, carbon or nitrogen have been isolated and characterized (Singh and Walker, 2006). Despite organophosphate-degrading microorganism diversity, very few reports have been published about bacteria-based technologies for the glyphosate-polluted soils cleanup.
The most active glyphosate-degrading strains were usually isolated from the soils contaminated with this herbicide. Many examples of bacteria have been isolated such as Bacillus cereus CB4 (Yang et al., 2012), Enterobacter cloacae, Enterobacter sp, Pseudomonas fluorescens (Andriani et al., 2017), Achromobacter sp. Kg 16 and Ochrobactrum anthropic GPK 3 (Ermakova et al., 2010), which possess a high degradative potential under environmental conditions and survive as well in the soil. Moneke et al. (2010) evaluated the abilities of five bacterial strains (Pseudomonas fluorescens, Azotobacter sp., Alcaligenes sp., Escherichia sp and Acetobacter sp.) to degrade glyphosate at different environmental conditions. Sviridov et al. (2011) have documented two different pathways for the glyphosate breakdown by Arthrobacter sp. GLP-1/Nit and Achromobacter sp. LW9. Therefore, the isolation and characterization of efficient glyphosate- degrading bacterial strains display great significance for application in bioremediation and wastewater treatment. So, the aim from this work was to isolate, characterize and identify a new bacterial strain capable of degrading high concentrations of glyphosate by using enrichment cultures. We also try to use bioinformatics tools to design specific primers for the isolation goxB gene that involved in glyphosate biodegradation.
2. Materials and methods
2.1. Chemicals and reagents
Glyphosate [N-(phosphonomethyl)glycine] 96% was purchased from Sigma-Aldrich (CAS Number: 1071-83-6, St. Louis, USA). Bacto-LB broth, Bacto-LB agar and mineral salts medium (MSM) were bought from Becton (Dickinson and Company, Sparks, MD, USA). The API 50 CHB/ medium for carbohydrate fermentation tests were purchased from (BioMérieux 50 430, Inc., Marcy l’Etoile, France).
2.2. Isolation and purification of glyphosate manipulation bacteria
A Sampling of the glyphosate treated soil was conducted in June 2019 from an agro-industrial zone located in the industrial zone in Sadat City of Monufia Governorate, Egypt (30°23′01.7″N 30°31′38.0″E). Isolation of endogenous/associated glyphosate degrading bacteria form the soil samples were initiated by suspending the soil samples in sterile saline (1 g. 100 mL−1) under shaking at 150 rpm for 90 min at 30 °C. Isolation and further purification of the bacterial isolates were performed onto Luria Bertani (LB) agar medium supplemented with 7.2 mg/ml N-(phosphonomethyl) glycine in triplicate and incubated at 30 °C for 24 h. Up to 30 isolates (FACU1-FACU30) were isolated (Congeevaram et al., 2007). The thirty selected isolates were manipulated by spread-plated onto LB agar medium supplemented with 50 mg/ml glyphosate in triplicate and incubated at 30 °C for 24 h. The high concentration of glyphosate was added to ensure the capability and stability of the isolated bacteria to grow and degrading the glyphosate under a high concentration (Xu et al., 2011).
2.3. Enrichment and selection of glyphosate degrading strain
Up to 10 bacterial isolates were isolated under a high concentration of glyphosate (50 mg/ml glyphosate). The selected isolates were inoculated (5% v/v) in mineral salts medium (MSM) (g/L): K2HPO4, 5.8; NaMoO4, 0.002; (NH4)2SO4, 2; KH2PO4, 4.5; CaCl2, 20; MgCl2·2H2O, 0.16; FeSO4·7H2O, 0.001; MnCl2, 0.001. 50 mg/ml glyphosate was added in 250 mL Erlenmeyer flask as a sole carbon source and incubated under dark conditions with shaking at 150 rpm and 30 ± 2 °C for 7 days (Moneke et al., 2010). Biodegradability of the selected isolates was tested by inoculating (5% v/v) of the cultivated isolates into a fresh MSM containing various concentrations of glyphosate (50, 100, 150, 200 and 250 mg/ml) and incubated at the same conditions for 7 days. The CFU (Colony-Forming Unit) as a bacterial count of the ten selected isolates was measured after 7 days to detect the increasing/decreasing of the bacterial count under high concentrations of glyphosate. CFUs of the ten isolates were performed by the pure plate methods according to Sieuwerts et al. (2008).
2.4. Identification of the selected isolated glyphosate degradation bacteria
The most potential selected bacterial isolate for glyphosate degradation was initially identified to species level via morphologically, physiologically and biochemically confirmed by using API 50 CHB/E carbohydrate fermentation strips (Holding and Collee, 1971, Sankar et al., 2016). The kit was used according to the instructions of the manufacturer.
2.5. Molecular identification using 16S rDNA gene
PCR reaction was done to amplify the 16S rRNA gene of FACU3 isolate. The 16S rRNA region was amplified by using the universal primer set; 27F: 5′- AGAGTTTGATCMTGGCTCAG-3′ and 1492R: 5′-TACGGYTACCTTGTTACGACTT-3′. The primers were utilized to amplify a 1373 bp fragment using 10 ng genomic DNA.
2.6. DNA extraction
Qiagen Kit (Qiagen Sciences, USA; Cat. no 51304) was used for chromosomal DNA according to the manufacturer’s instruction manual. Both the alkaline lysis method (Sambrook et al., 1989) and QIAprep spin miniprep (Qiagen Sciences, USA; cat. no 27106) were used for plasmid DNA extraction.
2.7. Primer design for goxB gene
According to the DNA sequence of FAD-dependent glyphosate oxidase genes (GenBank accession no. GU479462, GU479463 and GU21 4711), GenBank Database (www.ncbi.nlm.nih.gov) was used for primers design of goxB gene (responsible for glyphosate manipulation) using the Primer3Plus (https://primer3plus.com/cgi-bin/dev/primer3plus.cgi). Multiple sequence alignment (MSA) was analyzed using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). The amino acid analysis was achieved using Expasy software (https://www.expasy.org/).
2.8. Polymerase chain reaction (PCR)
PCR reactions were done with a total volume of 50 μL, containing 5 μL bacterial DNA (50 ng/μL), 25 μL master mix (OnePCR™ genedirex, Cat. No. MB203-0100), 2.5 μL from each forward and reverse primers and 15 μL of nuclease-free water. PCR program, primary denaturation step at 94 °C for 5 min and 35 cycles including denaturation for 60 sec at 94 °C; annealing for 60 sec at 56 °C and extension for 120 sec at 72 °C, then final extension step for 5 min at 72 °C. Amplified products were visualized on a 1.2% agarose gel under ultraviolet (UV) light. The purification for the PCR product was done using ExoSAP-IT™ PCR Product Cleanup Reagent (applied biosystems, USA, cat. no 78201), after that the forward and reverse sequencing was done at HVD Company, Germany using PCR primers. The phylogenetic trees were constructed using Clustal Omega multiple sequence alignment tool (https://www.ebi.ac.uk/Tools/msa/clustalo/).
3. Results
The bioremediation process based on microbes mentioned as the potential methods to get rid of the environmental pollutions in an eco-environmental friendly way from the contaminated sites (Finley et al., 2010, Ibrahim et al., 2015). There are different strategies for microorganisms including co-metabolism, catabolism and metabolic enzymes in manipulation reaction towards pesticides in soils (Ortiz-Hernández and Sánchez-Salinas, 2010).
3.1. Bacterial isolation
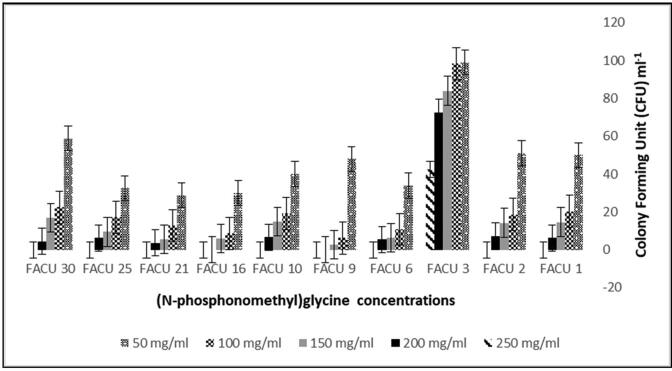
Bacterial isolates were collected from industrial areas contaminated with residual industries such as pesticides, detergents, disinfectants, chemicals, feed concentrates, veterinary medicines and agricultural fertilizers. Thirty bacterial isolates (FACU1 to FACU30) have been isolated and purified from the selected agro-industrial zone. The bacterial isolates were able to grow in LB medium supplemented with 7.2 mg/ml glyphosate. Ten bacterial isolates were capable to grow under different concentrations (50, 100, 150, 200 mg/ml) of glyphosate. Interestingly, the growth of selected isolates in the presence of herbicides was better than in natural conditions. The CFUs of the isolated bacteria referred to a significant variation that can be due to the variation of the capability of the isolated bacteria to grow and degrading the glyphosate under a high concentration of the glyphosate. The FACU3 isolate indicated to the highest CFU/ml at different concentrations of glyphosate (Fig. 1). These results suggested that all isolates were capable of glyphosate degradation at different concentrations. Especially, FACU3 isolate was the best isolate that showed a high ability to glyphosate degradation.
Fig. 1.
The CFU of the ten bacterial isolates at different concentrations of glyphosate.
3.2. Morphological and molecular characterization
The results of CFUs elucidated that the FACU3 isolate is the best glyphosate degradation. This isolate has been confirmed as a long rod bacteria of the spore-forming Gram-positive, motile, strictly aerobic, with optimum growth temperature at 30 °C, and non-pigmented on agar-containing media. The FACU3 isolate presented positive results for oxidase, catalase, amylase, gelatinase, urease, caseinase, tryptophane deaminase, lysine decarboxylase, ornithine decarboxylase, glutamate decarboxylase and acetic acid formation tests. Over and above, FACU3 isolate had the ability to utilize different carbon sources such as glycerol, D-arabinose, D- ribose, L-arabinose, D-fructose, D-glucose, D-mannitol, D-sucrose, D-galactose, D-melibiose and armygdalin (Table 1, Table 2). On the contrary, this isolate cannot utilize L-xylose and D-xylose, erythritol, D-adonitol, methyl-ßD-xylopyranoside, L-rhamnose, D-mannose, dulcitol, inositol, L-sorbose, D-sorbitol, methyl αD-mannopyranoside and methyl αD-glucopyranoside as a carbon source. Likewise, this isolate presented negative results for phosphatase, arginine dihydrolase, Voges–Proskauer, methyl red, indole production and nitrate reduction tests (Table 1, Table 2). According to the results of both the API identification profile and the Bergey's Manual of Systematic Bacteriology, FACU3 isolate was defined as a bacillus (Sneath et al., 1986).
Table 1.
Morphological and microscopic characters of FACU3 isolate.
| Result | Morphological test |
|---|---|
| + | Gram stain |
| Long rod-shaped | Cell shape |
| + | Spore forming |
| 5–8 mm | Colony size |
| + | Motility |
| White | Colony color |
Table 2.
Some Physiological, biochemical and chemotaxonomic characteristics using API 50 CHB Kit of FACU3 isolate.
| Test | Result* | Test | Result* |
|---|---|---|---|
| Oxidase | + | L-arabinose | + |
| Catalase | + | Erythritol | – |
| Amylase | + | D-arabinose. | + |
| Gelatinase | + | Glycerol | + |
| phosphatise | – | D-galactose | + |
| Tryptophane deaminase | + | L-xylose | – |
| Arginine dihydrolase | – | D-xylose | – |
| Lysine decarboxylase | + | D-Adonitol | – |
| Ornithine decarboxylase | + | Methyl-beta-D-xylopyranoside | – |
| Glutamate decarboxylase | + | D-ribose | + |
| Voges-Proskaure | – | D-glucose | + |
| Methyl red | – | D-mannitol | + |
| Nitrate reduction | – | D-mannose | – |
| Acetic acid | + | L-rhamnose | – |
| Hydrolysis of Casein | + | D-sorbitol | – |
| Hydrolysis of Gelatin | + | Dulcitol | – |
| Hydrolysis of Starch | + | Inositol | – |
| Hydrolysis of Urea | + | D-fructose | + |
| indole production | – | L-sorbose | – |
| Methyl-alpha-D-mannopyranoside | – | D-melibiose | + |
| Methyl-alpha-D-glucopyranoside | – | D-saccharose (sucrose) | + |
*+, positive test; −, Negative test.
The molecular identification at the genetic level was employed to confirm the morphological and biochemical identifications of the FACU3 isolate. 16S rRNA universal primers (27F and 1492R) were used in molecular identification. About 1373 bp of the 16S rRNA gene was amplified and sequenced (Fig. 2). A nucleotide sequence resulting from FACU3 16S rRNA gene sequencing was compared with the GenBank databases using BLAST. The isolated FACU3 displayed 99% of sequence homology with 16S rRNA gene from Bacillus aryabhattai. The obtained sequence was deposited to the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/WebSub/) under accession number MN749556 as Bacillus aryabhattai strain FACU. Then, the blast algorithm (http://www.ncbi.nlm.nih.gov /blast/Blast.cgi) was used for the alignment of the 16S rRNA sequence of Bacillus aryabhattai strain FACU and compared with the available sequences of 16S rRNA gene of several bacterial strains that deposited in NCBI databases. The phylogenetic tree was constructed (Fig. 3). Fig. 3 showed that the Bacillus aryabhattai strain FACU strain was grouped closely with Bacillus aryabhattai strain XT34, Bacillus sp. ATY1, Bacillus sp. ATY5 16S ribosomal RNA gene with 99.93% sequence similarity. The results of the 16S rRNA phenotypic characterization indicated that the Bacillus aryabhattai strain FACU is a species in the genus Bacillus.
Fig. 2.

16 s rDNA gene amplification using universal primers. M, DNA marker (1 Kb ladder); Lane 1, the isolate FACU3.
Fig. 3.
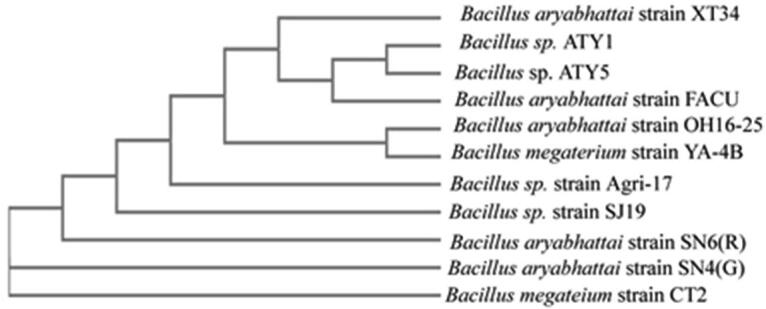
The phylogenetic tree of Bacillus aryabhattai strain FACU 16 s rRNA gene goxB gene localization.
Before gene amplification, the plasmid was isolated from Bacillus aryabhattai strain FACU to study the localization of goxB gene on the chromosome or in the plasmid, both manual preparation and Qiagen plasmid mini prep were used for plasmid extraction. The result exhibited that no plasmid has been found in the Bacillus aryabhattai strain FACU. This result showed the presence of goxB gene on the Bacillus aryabhattai strain FACU chromosome.
3.3. Primer designing and amplification of goxB gene
DNA nucleotide sequences, encoding FAD-dependent glyphosate oxidase enzyme, of three different bacterial species were used as published in the NCBI databases. All the DNA nucleotides sequences were complete open reading frames. BLAST was used with the selected nucleotide sequences recovered related DNA sequences of several bacterial species, including DNA sequence of the Ochrobactrum sp. To design goxB gene particular primers, initially, MSA analysis was performed between all recovered DNA sequences using Clustal Omega. Wide gaps and low levels of similarity were excluded using DNA sequences inspecting and filtering the MSA results. Then MSAs were used on the most similar DNA nucleotides sequences until the aligned sequences exhibited large areas of sequence similarity. The most appropriate primers of goxB gene amplification were selected and subjected to further analysis after alignment PCR analysis. To confirm the results of alignment PCR, the selected primers were BLAST with NCBI GenBank nucleotide databases. The primers BLAST displayed that the similarity percentage between targeted goxB gene DNA sequences and the designed primers was very high. The selected forward and reverse primers were used to amplify the expected segment of the goxB gene (Table 3).
Table 3.
the primers sequence used for goxB gene.
| Primer name | Primer sequence | Annealing temp. | Expected size |
|---|---|---|---|
| goxB-F | 5‘- ATGGCCGAGAACCATAAGAAGG-3‘ | 60 | 1296 bp |
| goxB-R | 5‘- TTAGGAGGCGGGGCCTGTTT-3‘ |
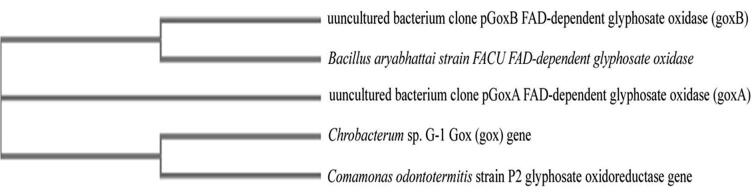
In this experiment, 1296 bp DNA fragment corresponding to the ORF of goxB gene was amplified from Bacillus aryabhattai strain FACU chromosomal DNA using PCR (Fig. 4). goxB gene was sequenced and then the sequence analysis was performed by blasting with available sequences in the NCBI. The results showed that the goxB gene was about 94.68% similarly with other closely related bacteria. The phylogenetic tree was constructed (Fig. 5). Fig. 5 showed that the goxB gene isolated from the Bacillus aryabhattai strain FACU strain was grouped closely with uncultured bacterium clone pGOXB FAD-dependent glyphosate oxidase (goxB).
Fig. 4.
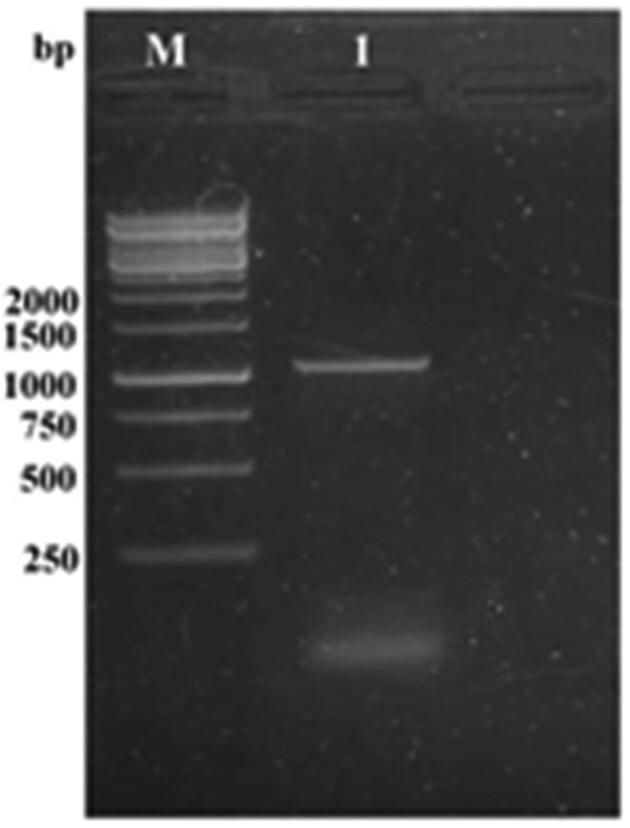
The goxB gene amplification (M); 1 Kb DNA ladder marker. Line 1: Bacillus aryabhattai strain FACU goxB gene.
Fig. 5.
The phylogenetic tree of goxB gene Sequence analysis.
Expasy software was used to define the goxB gene amino acid sequences and the genetic distance then the amino acid sequence was blasted with available sequences in the NCBI (Fig. 6). Fig. 6 showed the goxB gene amino acid sequence. The amino acid sequence was aligned with the database using Clustal Omega multiple sequence alignment tool (Fig. 7). The goxB protein was 431 amino acids with 46.14 KDa molecular weight. The high amino acid composition was alanine (11.1%). The isoelectric point for goxB protein was 9.95.
Fig. 6.
The amino acid sequences of goxB gene.
Fig. 7.
The amino acid sequence alignment.
4. Discussion
More than sixty percent of the applied pesticides in agriculture are herbicides (Zimdahl, 2002). The herbicides affect the plant growth at the physiological and biochemical level. Those herbicides control plant growth by inhibiting different metabolism processes at the physiological level including blocking amino acid synthesis, photosynthesis, mimicking plant growth regulators, preventing cell division and cell elongation, etc. Understanding herbicide's mode of action is a remarkable tool in research to handle weed control problems, examine toxicological characteristics and develop application methods in different agricultural practices. Biochemical processes such as various enzymes and functionally proteins are important analysis for understanding the herbicide’s mode of action. Also, the understanding of the herbicide's mode of action is an important step for the management, organization and classification of those herbicides. It also provides an insight into herbicide resistance, which continues to be a problem in sustainable agricultural management. Bacteria treatment has been mightily utilized in the treatment process of wastewater. Although it is well documented that glyphosate is readily metabolized by soil bacteria (Rueppel et al., 1977), surprisingly few glyphosate-degrading bacteria have been isolated.
In this study, thirty bacterial isolates were able to grow in LB medium including 7.2 mg/ml glyphosate. From those isolates, ten isolated bacteria displayed the ability to grow at various glyphosate concentrations. Only one isolate named FACU3 showed the high CFU at various glyphosate concentrations. According to the biological method and the sequence analysis of the 16S rRNA gene, the isolate was identified as Bacillus aryabhattai FACU strain. The data showed that the Bacillus aryabhattai FACU strain was able to use glyphosate under different concentrations. Likewise, a lot of research has shown that glyphosate can be manipulated by microorganisms including the rhizosphere bacteria, the endophytic bacteria and plant growth promoting-bacteria. From the rhizoplane of various plants, Kryuchkova et al. (2014) isolated ten bacterial strains (including Pseudomonas sp. K3, Alcaligenes sp. K1, Azomonas sp. K5, Enterobacter cloacae K7 and Comamonas sp. K4) survive at a 10 mM concentration of glyphosate. Also, three bacterial strains, Pseudomonas putida, Pseudomonas aeruginosa and Acetobacter faecalis, were obtained from agricultural soil heavily polluted with glyphosate which is capable of degrading 1000 ppm glyphosate herbicide (Olawale and Akintobi, 2011). Furthermore, five bacterial species were purified by Moneke et al. (2010) which degraded glyphosate up to 250 mg/ml of glyphosate. Ezaka et al. (2018) purified some plant growth-promoting bacteria such as Bacillus cereus and Pseudomonas aeruginosa. At the same time, these strains exhibited significant ability to manipulate glyphosate of polluted soil. Their result reported that the percentage of degradation at 3.1 mg/ml of glyphosate were 85.8, 76.11, 75.8 and 49% for Bacillus cereus, Pseudomonas aeruginosa, their mixed culture and control respectively, while at 7.2 mg/ml concentration, the degradation percentage was 72.7, 84.9, 66.4% and 39.2% by Bacillus cereus, P. aeruginosa, mixed culture and control respectively. A Pseudomonas strain was isolated by Jacob et al. (1988) which showed a high ability to manipulate glyphosate about 2 g glyphosate/g dry biomass. Two bacterial isolates were reported to be efficient degraders of glyphosate, these strains are Bacillus cereus CB4 (Yang et al., 2012) and Ochrobacterium anthropic (Sviridov et al., 2011). Also, two plant growth-promoting bacteria, Pseudomonas fluoresces and Enterobacter sp were reported as glyphosate manipulating bacteria by Andriani et al. (2017). The rate of degradation depends on phosphate condition in bacteria culture growth phase, bacteria cell and the bacteria adaptation to herbicides (Kryuchkova et al., 2014). Travaglia et al. (2015) reported that Azospirilum and Pseudomonas able to degrade glyphosate due to the undergoing delayed phase of death and longer stationary phase.
During its degradation, different microorganisms can use glyphosate as a sole source of phosphorus, carbon, and nitrogen. Glyphosate degradation pathways and its metabolites have been frequently investigated, but the related enzymes and genes have been rarely studied. Two different pathways have been documented for glyphosate metabolism (Pipke et al., 1987, Rueppel et al., 1977). The first one including the C–N bond cleavage using the glyphosate oxidoreductase enzyme, the AMPA Pathway (Kishore and Barry, 1992). According to Pollegioni et al. (2011), the glyphosate oxidoreductase (GOX) enzyme breaks the C-N bond on the carboxyl side, producing the AMPA and glyoxylate. Glyphosate manipulation through the AMPA pathway by glyphosate oxidoreductase was reported in different bacteria, including Arthrobacter atrocyaneus ATCC 13752 (Pipke and Amrhein, 1988), Flavobacterium sp. strain GD1 (Balthazor and Hallas, 1986), Agrobacterium radiobacter (McAuliffe et al. 1990), etc. The other pathway is through the carbon phosphorus (C–P) bond cleavage by a C–P lyase activity which gives sarcosine, glycine and formaldehyde e.g. Pseudomonas sp (Sviridov et al., 2011). The C–P lyase activity could also degrade glyphosate to sarcosine, which eventually formed formaldehyde and glycine in a reaction catalyzed by sarcosine oxidase. Arthrobacter sp. strain GLP-1 (Pipke et al., 1987), Rhizobium meliloti 1021 (Liu et al., 1991), Pseudomonas pseudomallei 22 (Penaloza-Vazquez et al., 1995), Pseudomonas sp. GS (Albrecht et al., 1991), Streptomycete sp. StC (Obojska et al., 1999) are some examples of microbes that identified to have C-P lyases. This study revealed the glyphosate manipulation pathways by Bacillus aryabhattai FACU through FAD-glyphosate oxidoreductase enzyme. In this study, the goxB gene encoded for the FAD-glyphosate oxidoreductase enzyme was isolated. Glyphosate oxidoreductase (GOX) is the key enzyme of glyphosate degradation to AMPA via C-N bond cleavage. GOX-encoding genes have been identified in Ochrobactrum anthropi GPK 3, Comamonas odontotermitis P2 (KX980206.1) and Ochrobactrum sp. G1 (GU214711.1) with 99% similarity (Duke, 2010, Hadi et al., 2012). However, GOX purified from Ochrobactrum anthropi GPK 3, containing flavin adenine dinucleotide (FAD), belongs to bacterial flavin monooxygenase superfamily (Sviridov, 2012).
5. Conclusion
The bacterial biodegradation researches of herbicides have been extremely developed and a lot of bacterial strains that can manipulate herbicides have also been identified. However, the effective applications of bacterial biological treatment were limited due to the low efficiency of bacterial strains and sometimes environmental conditions. Bacillus aryabhattai FACU strain was isolated and identified. FACU strain showed the highest glyphosate manipulation activity. The bacteria used FAD-dependent glyphosate oxidase enzyme in catabolism of glyphosate. Two specific primers were designed and used for isolation FAD-dependent glyphosate oxidase (goxB) gene. So, in this study a new bacterial strain was added to the list of the glyphosate manipulating bacteria.
Authors’ contributions
NIE and AAA conceived and designed the study. NIE, AAA, IS, DSA, IA, GO and RHA performed experiments, drafted and edited the manuscript. All authors read and approved the final manuscript.
Funding
The author(s) received no specific funding for this work.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Peer review under responsibility of King Saud University.
References
- Abdelhaleem H.A.R., Zeinb H.S., Azeiza Abdel, Sharaf A.N., Abdelhadi A.A. Identification and characterization of novel bacterial polyaromatic hydrocarbon-degrading enzymes as potential tools for cleaning up hydrocarbon pollutants from different environmental sources. Environ. Toxicol. Pharmacol. 2019;67:108–116. doi: 10.1016/j.etap.2019.02.009. [DOI] [PubMed] [Google Scholar]
- Albrecht B., Weidhase R., Stock M., Weidhase R.A. In vivo utilization of N-(phosphonomethyl)-anilines and related substances by Pseudomonas spec. GS. J. Basic Microbial. 1991;31:403–411. doi: 10.1002/jobm.3620310602. [DOI] [PubMed] [Google Scholar]
- Andriani L.T., Aini L.Q., Hadiastono T. Glyphosate biodegradation by plant growth promoting bacteria and their effect to paddy germination in glyphosate contaminated soil. J. Degraded Mining Land Manage. 2017;5(1):995–1000. [Google Scholar]
- Balthazor T.M., Hallas L.E. Glyphosate-degrading microorganisms for industrial activated sludge. Appl. Environ. Microbiol. 1986;51:432–434. doi: 10.1128/aem.51.2.432-434.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benslama O., Boulahrouf A. Isolation and characterization of glyphosate-degrading bacteria from different soils of Algeria. African J. Microbiol. Res. 2013;7(49):5587–5595. [Google Scholar]
- Congeevaram S., Dhanarani S., Park J., Dexilin M., Thamaraiselvi K. Biosorption of chromium and nickel by heavy metal resistant fungal and bacterial isolates. J. Hazard. Mater. 2007;146:270–277. doi: 10.1016/j.jhazmat.2006.12.017. [DOI] [PubMed] [Google Scholar]
- de Sieuwerts S., Bok F.A.M., de Mols E., van Vos W.M., Hylckama J.E.T. A simple and fast method for determining colony forming units. Lett. Appl. Microbiol. 2008;47:275–278. doi: 10.1111/j.1472-765X.2008.02417.x. [DOI] [PubMed] [Google Scholar]
- Demnerova K., Mackova M., Spevakova V., Beranova K., Kochankova L., Lovecká P., Ryslavá E., Macek T. Two approaches to biological decontamination of groundwater and soil polluted by aromatics characterization of microbial populations. Int. Microbiol. 2005;8:205–211. [PubMed] [Google Scholar]
- Duke S.O. Glyphosate degradation in glyphosate-resistant and-susceptible crops and weeds. J. Agric. Food Chem. 2010;59(11):5835–5841. doi: 10.1021/jf102704x. [DOI] [PubMed] [Google Scholar]
- Ermakova I.T., Kiseleva N.I., Shushkova T., Zharikov M., Zharikov G.A., Leontievsky A.A. Bioremediation of glyphosate-contaminated soils. Appl. Microbiol. Biotechnol. 2010;88(2):585–594. doi: 10.1007/s00253-010-2775-0. [DOI] [PubMed] [Google Scholar]
- Ezaka E., Akintokun A.K., Akintokun P.O., Taiwo L.B., Uthman A.C.O., Oyedele O.A., Aluko O.I. Glyphosate degradation by two plant growth promoting bacteria (PGPB) isolated from rhizosphere of maize. Microbiol. Res. J. Int. 2018;26(6):1–11. [Google Scholar]
- Finley S.D., Broadbelt L.J., Hatzimanikatis V. In silico feasibility of novel biodegradation pathways for 1, 2, 4-trichlorobenzene. BMC Syst. Biol. 2010;4(7):1–14. doi: 10.1186/1752-0509-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulekar M.H. Microbial degradation of petrochemical waste-polycyclic aromatic hydrocarbons. Bioresour. Bioprocess. 2017;4:28–36. doi: 10.1186/s40643-017-0158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein D.A., Acquavella J.F., Mannion R.M., Farmer D.R. An analysis of glyphosate data from the California environmental protection agency pesticide illness surveillance program. J. Toxicol. Clin. Toxicol. 2002;40:885–892. doi: 10.1081/clt-120016960. [DOI] [PubMed] [Google Scholar]
- Hadi F., Mousavi A., Salmanian A.H., Akbari N.K. Glyphosate tolerance in transgenic canola by a modified glyphosate oxidoreductase (gox) gene. Prog. Biol. Sci. 2012;2(1):50–58. [Google Scholar]
- Holding A.J., Collee J.G. Routine biochemical tests. Methods Microbiol. 1971;6A:2–32. [Google Scholar]
- Ibrahim G.A.G., Amin M.K., Hassan A.A., El-Sheikh E.A. Identification of pesticides degrading bacteria isolated from Egyptian soil. Zagazig J. Agric. Res. 2015;42(5):1129–1143. [Google Scholar]
- Igiri B.E., Okoduwa S.I.R., Idoko G.O., Akabuogu E.P., Adeyi A.O., Ejiogu I.K. Toxicity and bioremediation of heavy metals contaminated ecosystem from Tannery wastewater: A review. J. Toxicol. 2018;16 doi: 10.1155/2018/2568038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob G.S., Garbow J.R., Hallas L.E., Kimack N.M., Kishore G.M., Schaefer J. Metabolism of glyphosate in Pseudomonas sp. strain LBr. Appl. Environ. Microbiol. 1988;54:2953–2958. doi: 10.1128/aem.54.12.2953-2958.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasumana C., Gunatilake S., Senanayake P. Glyphosate, hard water and nephrotoxic metals: are they the culprits behind the epidemic of chronic kidney disease of unknown etiology in Sri Lanka? Int. J. Environ. Res. Public Health. 2014;11:2125–2147. doi: 10.3390/ijerph110202125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore, G.M., Barry, G.F. 1992. Glyphosate tolerant plants. International patent WO92/00377.
- Kryuchkova Y.V., Burygin G.L., Gogoleva N.E., Gogolev Y.V., Chernyshova M.P., Makarov O.E., Fedorov E.E., Turkovskaya O.V. Isolation and characterization of a glyphosate-degrading rhizosphere strain, Enterobacter cloacae K7. Microbiol. Res. 2014;169:99–105. doi: 10.1016/j.micres.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Leckie S.E. Methods of microbial community profiling and their application to forest soils. Forest Ecol. Manag. 2005;220:88–106. [Google Scholar]
- Liu, C.M., Mclean, P.A., Sookdeo, C.C., Cannon, F.C., 1991. Degradation of the herbicide glyphosate by members of the family Rhizobiaceae. Appl. Environ. Microbiol 57: 1799–1804. [DOI] [PMC free article] [PubMed]
- Mbanaso F.U., Coupe S.J., Charlesworth S.M., Nnadi E.O. Laboratory-based experiments to investigate the impact of glyphosate-containing herbicide on pollution attenuation and biodegradation in a model pervious paving system. Chemosphere. 2013;90:737–746. doi: 10.1016/j.chemosphere.2012.09.058. [DOI] [PubMed] [Google Scholar]
- McAuliffe K.S., Hallas L.E., Kulpa C.F. Glyphosate degradation by Agrobacterium radiobacter isolated from activated sludge. J. Ind. Microbiol. 1990;6:219–221. [Google Scholar]
- Mohamed M.S.M., Youssef A.F.A., Mohamed Y.A. The potentiality of Lysinibacillus sphaericus DM-3 and Bacillus cereus DM-5 in degrading dimethoate. Egypt. J. Bot. 2018;58(2):217–232. [Google Scholar]
- Mohamed M.S.M., El-Arabi N.I., El-Hussein A., Abu El-Maaty S., Abdelhadi A.A. Reduction of chromium-VI by chromium-resistant Escherichia coli FACU: a prospective bacterium for bioremediation. Folia Microbiologica (accepted) 2020 doi: 10.1007/s12223-020-00771-y. [DOI] [PubMed] [Google Scholar]
- Moneke A.N., Okpala G.N., Anyanwu C.U. Biodegradation of glyphosate herbicide in vitro using bacterial isolates from four rice fields. Afr. J. Biotechnol. 2010;9(26):4067–4074. [Google Scholar]
- Obojska A., Lejczak B., Kubrak M. Degradation of phosphonates by streptomycete isolates. Appl. Microbiol. Biotechnol. 1999;51:872–876. doi: 10.1007/s002530051476. [DOI] [PubMed] [Google Scholar]
- Olawale A.K., Akintobi O.A. Biodegradation of glyphosate pesticide by bacteria isolated from agricultural soil. Report and Opinion. 2011;3(1):124–128. [Google Scholar]
- Oliveira B.R., Penetra A., Cardoso V.V., Benoliel M.J., Crespo M.B., Samson R.A., Pereira V.J. Biodegradation of pesticides using fungi species found in the aquatic environment. Environ. Sci. Pollut. Res. Int. 2015;22:11781–11791. doi: 10.1007/s11356-015-4472-0. [DOI] [PubMed] [Google Scholar]
- Ortiz-Hernández M.L., Sánchez-Salinas E. Biodegradation of The organophosphate pesticide tetrachlorvinphos by bacteria isolated from agricultural soils in México. Rev. Int. Contam. Ambient. 2010;26(1):27–38. [Google Scholar]
- Penaloza-Vazquez A., Mena G.L., Herrera-Estrella L., Bailey A.M. Cloning and sequencing of the genes involved in glyphosate utilization by Pseudomonas pseudomallei. Appl. Environ. Microbiol. 1995;61:538–543. doi: 10.1128/aem.61.2.538-543.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipke R., Amrhein N. Degradation of the phosphonate herbicide glyphosate by Arthrobacter atrocyaneus ATCC 13752. Appl. Environ. Microbiol. 1988;54:1293–1296. doi: 10.1128/aem.54.5.1293-1296.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipke R., Amrhein N., Jacob G.S., Schaefer J., Kishore G.M. Metabolism of glyphosate in an Arthrobacter sp. GLP-1. Eur. J. Biochem. 1987;165:267–273. doi: 10.1111/j.1432-1033.1987.tb11437.x. [DOI] [PubMed] [Google Scholar]
- Pollegioni L., Schonburn E., Siehl D. Molecular basis of glyphosate resistance: different approaches through protein engineering. FEBS J. 2011;278(16):2753–2766. doi: 10.1111/j.1742-4658.2011.08214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano R., Souza P., Nunes M., Romano M. Perinatal exposure to a commercial formulation of glyphosate reduces the mRNA expression and increases the protein content of beta TSH in the pituitary of male offspring. Endocr. Abstr. 2012;29:753. [Google Scholar]
- Romero D.M., Ríos de Molina M.C., Juárez Á.B. Oxidative stress induced by a commercial glyphosate formulation in a tolerant strain of Chlorella kessleri. Ecotoxicol. Environ. Saf. 2011;74:741–747. doi: 10.1016/j.ecoenv.2010.10.034. [DOI] [PubMed] [Google Scholar]
- Rueppel M.L., Brightwel B.B., Schaefer J., Marve J.T. Metabolism and degradation of glyphosate in soil and water. J. Agric. Food Chem. 1977;25:517–528. doi: 10.1021/jf60211a018. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritch E.F., Maniatis T. Molecular cloning laboratory manual, Vol. l. Cold spring harbor laboratory press, USA. Science. 1989;327:1139–1142. [Google Scholar]
- Sankar B.S., Kishore S., Saurav D., Madhumita B. Structural and functional analysis of glutamate decarboxylase system in Bacillus aryabhattai. Res. J. Biotechnol. 2016;11(11):1–11. [Google Scholar]
- Singh D.K. Biodegradation and bioremediation of pesticide in soil: concept, method and recent developments. Indian. J. Microbial. 2008;48:35–40. doi: 10.1007/s12088-008-0004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B.K., Walker A. Microbial degradation of organophosphorus compounds. FEMS Microbiol. Rev. 2006;33:428–471. doi: 10.1111/j.1574-6976.2006.00018.x. [DOI] [PubMed] [Google Scholar]
- Sneath P.H.A., Mair N.S., Sharpe M.E. vol 2. Williams & Wilkins Co; Baltimore, London: 1986. (Bergey’s manual of systematic bacteriology). [Google Scholar]
- Solomon K.R., Anadón A., Carrasquilla G., Cerdeira A.L. Coca and poppy eradication in Colombia: environmental and human health assessment of aerially applied glyphosate. Rev. Environ. Contam. Toxicol. 2007;190:43–125. doi: 10.1007/978-0-387-36903-7_2. [DOI] [PubMed] [Google Scholar]
- Sviridov A.V., Shushkova T.V., Zelenkova N.F., Vinokurova N.G., Morgunov I.G., Ermakova I.T., Leontievsky A.A. Distribution of glyphosate and methylphosphonate catabolism systems in soil bacteria Ochrobactrum an thropic and Achromobacter sp. Appl. Microbiol. Biotechnol. 2011;93:787–796. doi: 10.1007/s00253-011-3485-y. [DOI] [PubMed] [Google Scholar]
- Sviridov, A., 2012. Enzyme systems of organophosphonate catabolism of soil bacteria Achromobacter sp. and Ochrobactrum anthropi GPK3. PhD thesis (in Russian). Pushchinoa 152:120–132.
- Travaglia C., Masciarelli O., Fortuna J., Marchetti G., Cardozo P., Lucero M., Zorza E., Yu X.M., Yu T., Yin G.H., Dong Q.L., An M., Wang H.R., Ai C.X. Glyphosate biodegradation and potential soil bioremediation by Bacillus subtilis strain Bs-1 5. Genetic Mol. Resour. 2015;14:14713–14730. doi: 10.4238/2015.November.18.37. [DOI] [PubMed] [Google Scholar]
- Xu X., Ji F., Fan Z., He L. Degradation of glyphosate in soil photocatalyzed by Fe3O4/SiO2/TiO2 under solar light. Int. J. Environ. Res. Public Health. 2011;8(4):1258–1270. doi: 10.3390/ijerph8041258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.F.G., Zhao H., Shi G., Geng Y., Hou T., Tao K. Isolation, identification and characterization of a glyphosate-degrading bacterium, Bacillus cereus CB4, from soil. J. Gen. Appl. Microbiol. 2012;58:263–271. doi: 10.2323/jgam.58.263. [DOI] [PubMed] [Google Scholar]
- Zimdahl R.L. My view. Weed Sci. 2002;50:687. [Google Scholar]