Abstract
The development of CRISPR-Cas9 based genetic manipulation tools represents a huge breakthrough in life sciences and has been stimulating research on metabolic engineering, synthetic biology, and systems biology. The CRISPR-Cas9 and its derivative tools are one of the best choices for precise genome editing, multiplexed genome editing, and reversible gene expression control in microorganisms. However, challenges remain for applying CRISPR-Cas9 in novel microorganisms, especially those industrial microorganism hosts that are intractable using traditional genetic manipulation tools. How to further extend CRISPR-Cas9 to these microorganisms is being an urgent matter. In this review, we first introduce the mechanism and application of CRISPR-Cas9, then discuss how to optimize CRISPR-Cas9 as genome editing tools, including but not limited to how to reduce off-target effects and Cas9 related toxicity, and how to increase on-target efficiency by optimizing crRNA and sgRNA design.
Keywords: CRISPR-Cas9 system, Microorganisms, Off-target, Cas9 toxicity, sgRNA
1. Introduction
At present, three generations of nuclease-based genome editing technologies including ZFN, TALEN and the CRISPR-Cas systems have been developed [1]. All these technologies rely on double-strand break (DSB) induced DNA repair system at specific site for sequence modification [2]. However, ZFN technology generates cytotoxicity in cells and the production cost is high [3]. TALEN has advantage such as higher fidelity and less off-target, but it's module assembly is complicate [4]. Compared with the first two technologies, the CRISPR-Cas system has a larger target selection than the first two, avoid problems such as difficulty in assembly and off-target, and can effectively cleave any DNA site to achieve more accurate gene editing and modification [5].
CRISPR (clustered regularly interspaced short palindromic repeats) is an acquired immune system that protects against foreign virus or plasmid DNA in bacteria (>50%) and archaea (>90%) [6]. The system consists of clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated proteins (Cas). CRISPR sequences are composed of short, highly conserved repeats and different spacers. Repeat mostly has a palindrome structure. Spacer is homologous to foreign DNA (such as plasmid or virus), and can form crRNA, it can form a complex with Cas functional protein to specifically recognize and eliminate foreign plasmid or virus that invaded cells after being transcribed and processed with repeat to form crRNA. In 1987, when Ishino and colleagues studied the iap gene responsible for alkaline phosphatase isozyme conversion in E. coli, they found a 29 nt tandem repeat sequence downstream. Interestingly, the repeat units are separated by 32 nt non-repetitive sequences, which is the earliest report of CRISPR [7].
Cas and CRISPR sequences of the CRISPR-Cas system are highly diverse and dynamic. In 2015, Makarova et al. updated the CRISPR system classification and divided the CRISPR system into two categories (Class 1 and Class 2), 5 types and 16 subtypes [8]. The Class 1 system exists in bacteria and archaea including types Ⅰ, Ⅲ, and Ⅳ. And Class 1 includes effector complexes composed of four to seven Cas protein subunits. Most of the subunits of the Class 1 effector complexes contain RNA recognition motif domain [9,10]. Class2 including types II, V, Ⅵ system mainly in bacteria, requires only a single Cas protein forming effect module [11]. The characteristic effector molecule in Class 2 is Cas9, which has RNA-dependent endonuclease activity and contains two separate nuclease domains, HNH and RuvC, that function to cleave complementary and noncomplementary strands [12]. In the Cas9 protein, the HNH domain inserted between two similar RuvC domains in responsible to cleave the target strand, which is the base that matches the gRNA base.
Among the many types of CRISPR systems, the CRISPR-Cas9 system is relatively simple and more commonly used. In 2012, Doudna laboratories found that purified Cas9 from S. thermophilus can target and cleave gene by crRNA [13]. In addition, the Charpentier and Doudna labs also simplified the system. They combined tracrRNA (trans-activating CRISPR-associated RNA) and crRNA into a single guide RNA (gRNA). The combination of gRNA and Cas9 proteins can target specific sequences. In 2013, Zhang and Church groups applied this genome editing technology to eukaryotes almost simultaneously. Zhang's laboratory used S. thermophilus and S. pyogenes type II systems and found that Cas9 can cleave both mouse and human genomes under the guidance of gRNA [14]. When an exogenous donor is provided, Cas9 can also precisely edit target sites by homologous recombination. In addition, when multiple leader sequences are added to the CRISPR sequence, the system can simultaneously edit multiple sites in the mammalian genome. The Church laboratory also used the Type II system to edit the human genome and obtained similar results. These two articles created a new era of CRISPR genome editing technology, and then the world of life sciences blew up the CRISPR storm. Since then, the CRISPR-Cas9 system has been used by thousands of laboratories for genetic editing of various biological models.
Based on the CRISPR-Cas9 principle, many CRISPR-Cas9 technologies have been developed, for example, multiple genome editing technology CMGE and GTR-CRISPR for E. coli and Saccharomyces cerevisiae; convert Cas9 to DNA nicking enzymes for precise mediated editing; fuse dCas9 with EGFP and other fluorescent proteins to perform fluorescent localization at specific sites in the genome; and DNA-methylated or acetylated proteins through CRISPR-Cas technology introduced into the target genome for epigenetic regulation, etc. At present, the CRISPR-Cas9 system has been widely used in industrial microorganisms to increase the output of target products and other model or non-model microorganisms, animals and plants as gene editing tools. However, problems such as toxicity of Cas9 and off-target effect restricts its exploitation in new microorganisms.
In order to more efficiently apply the CRISPR-Cas9 system to bacteria based on the foregoing basic introduction, this article introduces the mechanism and application of the CRISPR-Cas9 system, this review details how to optimize CRISPR-Cas9 as genome editing tools, including but not limited to how to reduce off-target effects and Cas9 related toxicity, and how to increase on-target efficiency by optimizing crRNA and sgRNA design.
2. Mechanisms of CRISPR-Cas9 system
The three essential components of CRISPR-Cas9 system are the CRISPR locus composed of spacer and repeat sequences, tracrRNA, and Cas9 endonuclease. The Cas9 endonuclease including the RuvC domain located in N-terminal and the HNH nuclease active domain in the middle, is capable of inducing double-stranded cleavage of specific DNA sequences [12]. The length of the CRISPR site repeat sequence is usually 21 to 48 bp, and the spacer sequence which is responsible for recognizing foreign DNA is 26–72 bp [6]. The RNA duplex formed by tracrRNA and crRNA is responsible for scanning PAM (protospacer adjacent motif) on the target DNA with Cas9 to guide double-strand cleavage. PAM, the protospacer adjacent motif, was first proposed by Mojica et al., in 2009 [15]. They found that there is a 2-5 nt conserved sequence on the protospacer side of the spacer target sequence, either 5′upstream, or 3′downstream. And the sequence varies in different types of CRISPR systems. PAM is located 5′ upstream of protospacer in type I system, whereas, PAM is located 3′ downstream of protospacer in type II system. PAM has a special function on the identification and interference.
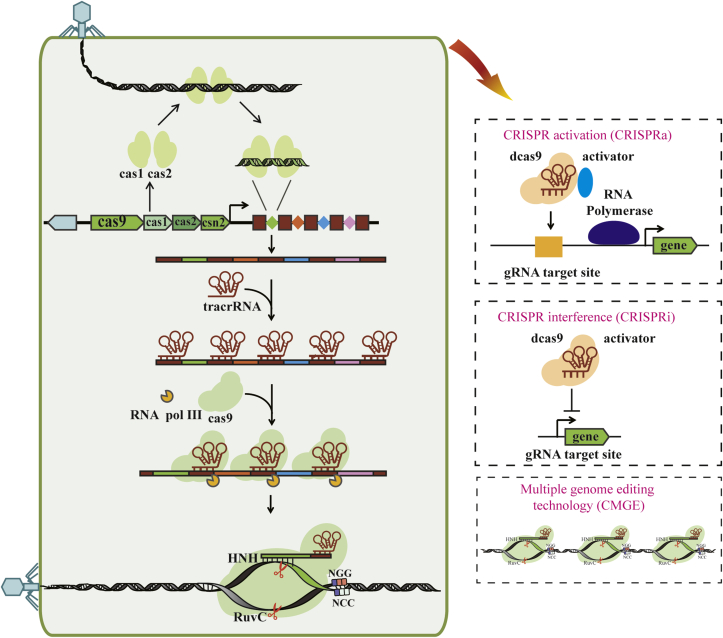
In 2007, Barrangou et al. first observed the CRISPR-Cas mediated adaptive immunity in S. thermophilus. They found a small number of surviving resistant clones in bacteriophage-infected industrial S. thermophilus strains, and found that their CRISPR structures acquired 1–4 new spacers that are partially matched the genomic sequences of the phage Φ858 and Φ2972 used in the infection experiment. The newly acquired spacers endowed these strains resistance to the corresponding phage [16,17]. The immune process of the CRISPR-Cas9 system is divided into three steps: adaptation, expression, and interference. As shown in Fig. 1.
Fig. 1.
Three stages of CRISPR-Cas9 system immunization and its applications. The CRISPR-cas9 immune process is divided into three phases: adaptation, expression (CRISPR RNA biogenesis), and interference. During the adaptation phase, virus or plasmid invasion, Cas1 and Cas2 protein captures the fragment and integrates into the CRISPR sequence to form a new spacer. In the expression phase, the CRISPR sequence is transcribed, the long-chain product is processed to obtain mature short-chain crRNA. In the interference phase, the crRNA forms an effector complex with the Cas9 protein, which directs the Cas9 protein to cleave the target sequence, thereby degrading the target.
Adaptation: exogenous DNA fragments are specifically selected and integrated into the CRISPR structure as new spacers to form the process of memory immunity [12]. When foreign DNA, such as viruses or plasmids, invade into prokaryotic cells, they are recognized by the Cas protein complexes (mainly Cas1 and Cas2) and specific fragments are cut out, This process is called “spacer selection”. Spacer region is usually obtained by specific recognition of PAM. Heler et al. found that Cas9 may be responsible for recognizing PAM and recruiting Cas1-Cas2 complexes in type II system [18]. Subsequently, the fragment was specifically integrated into the CRISPR DNA structure at a specific site (usually adjacent to the leader sequence) and the repeat at the integration site replicated precisely. Therefore, the periodicity of the CRISPR structure is maintained. This sub-stage can be called the “spacer integration process” [19].
Expression: The newly obtained spacers need to be transcribed and processed into mature small molecule crRNA to mediate specific immunity. This process is the second stage of CRISPR immunity, crRNA biosynthesis. Most type II systems encode a tracrRNA that is partially complementary to the repeat sequence. Generally, the leader sequence of a CRISPR structure contains promoter and other transcription elements. When the same virus or plasmid invades the bacteria again, the inserted new spacer sequence will be transcribed together with the repeat. Under the guidance of tracrRNA, the host-derived RNase III and Cas9 cut the repeat RNA, then a second cut will occur near the 5 'end inside the spacer sequence. The final mature crRNA molecule length is between 44 and 49 nt, of which the 20-22 nt 5′ end is derived from the spacer sequence, the 24-27 nt 3′end is derived from the repeat component [20].
Interference: a functional crRNA directs Cas9 nucleases to specifically recognize and cleave the viral DNA/RNA that is homologous to the spacer to achieve a specific immune process. After processing by RNase III, the mature crRNA and tracrRNA continue to maintain RNA duplex. RNA duplex and Cas9 rapidly scan the PAM sequence on the target DNA. The bases of the crRNA and protospacer DNA match and further extend the base match to the entire protospacer region, forming a stable R-loop structure [21]. The HNH domain and the Ruv-C domain cleave the two strands of the target DNA, resulting in a double-strand break.
3. Applications of CRISPR-Cas9 in microbes
The CRISPR-Cas9 system can transform strains and increase the output of industrial products through various editing methods, such as knockout, integration, CRISPRi, precise editing, etc., as shown in Table 1.
Table 1.
CRISPR-Cas technology transform bacteria to increase yield of target products The CRISPR/Cas9 system can modify the bacterial genome through a variety of editing methods, such as knockout, integration, multiplex gene editing, and random targeting, etc., to increase the output of industrial products or health products, as shown in Table 1.
| Industrial strains | Applied CRISPR–Cas9 gene editing technology | product | Product output | references |
|---|---|---|---|---|
| Synechococcus elongatu | Knock-out of glgc, knock-in of gltA-ppc | succinic acid | 0.58–0.63 mg/L | [22] |
| Saccharomyces cerevisiae | Knock-out of FAA1、FAA4、POX1、are1、ARE2、PAH1、LPP1and DPP1 | free fatty acid | 30-fold | [23] |
| E.coli | Modification of gltA 5′-UTR | n-butanol | 0.82 g/L | [24] |
| Corynebacterium glutamicum | inactivated pyk&ldhA | glutamic acid | 3-fold | [25] |
| Saccharomyces cerevisiae | Knock-down of ERG9, knock-out of ROX1, CRISPRa of HMG1 | β-carotene | 3-fold | [26] |
| Bacillus subtilis | Repression of Bpr and Vpr | BLA | 260-fold | [27] |
| Clostridium tyrobutyricum | Integration of adhE1 or adhE2 | n-butanol | 26.2 g/L | [28] |
| Corynebacterium glutamicum | Knock-out of Ncgl1221, gabT, gabP | γ-amino-butyric acid | 28.7 g/L | [29] |
| E.coli | Knock-in of fadR, delta9 and acc | fatty acids | 13% | [30] |
| E.coli | Knock-out of sucCD, hemB | 5-amino-levulinicacid | 2.81 g/L | [31] |
| E.coli | Knock-in and RBS substitution of thl, atoDA, adc, adh | isopropanol | 17.5 g/L | [32] |
| E.coli | Knock-in of almgs | β-carotene | 19.6 mg/L | [33] |
| Bacillus subtilis | Combined regulation of RbB, RIBA and RbH | riboflavin | 1.39 μg/L | [34] |
| Myceliophthora thermophile | Δcre1Δalp1Δres1 | cellulase | 5.1–13.3-fold | [35] |
| Filamentous fungus | Multiplex gene editing | mucic acid | 12.05 g/L | [36] |
| Aspergillus niger | Knock-out of aaA | galactaric acid | 4 g/L | [37] |
3.1. Genome editing
With the continuous development of genome sequencing technology and the progress of the human genome sequencing project, scientists have obtained a large amount of genome information, which brings opportunities for the development of basic science and industrial biology. The laborious traditional genome editing technologies via homologous recombination (HR)-restrict their use in numerous organisms. New evidence that DNA double-strand breaks (DSBs) can stimulate error-prone non-homologous end-joining (NHEJ) or homology-directed repair (HDR) at specific gene position, lays the foundation for the emergence of genome editing technology. CRISPR-Cas9 gene editing technology has significant advantages such as high efficiency, specificity, and simple design, which brings a huge breakthrough to gene editing technology.
Targeting gene sequences with double RNA (tracrRNA: crRNA) and introducing donor templates makes CRISPR-mediated insertions and deletions possible. Subsequently, replacing the double RNA (tracrRNA: crRNA) with sgRNA further simplified the system [12]. Song et al. developed the CRISPR-Cas9 D10A system for genomic engineering of Lactobacillus casei, which has been shown to mediate fast and efficient in-frame chromosome deletions and site-specific insertions/replacements [38]. As we all know, cyanobacteria are limited in their application as cell factories to produce biofuels and various biochemical products due to their oligoploidy nature and long-term instability of the introduced gene. Li et al. used the CRISPR-Cas9 system to effectively trigger a programmed DSB on the chromosome of the Synechococcus elongatus PCC7942, and achieved precise gene integration by co-transformation with a template plasmid harboring the gene cassette and flanking homology arms. Subsequently, Li et al. further used CRISPR-Cas9-assisted simultaneous glgc knock-out and gltA/ppc knock-in to modify the cyanobacteria to increase the succinic acid titer to 435.0 ± 35.0 μg/L, which is 11-fold increase compared with wild-type cells [22]. In order to develop microbial cell factories, multiple genome targets often need to be modified. Feng et al. developed a CRISPR-Cas9-assisted E. coli multiple genome editing technology (CMGE) based on a modular assembly strategy. The modification efficiencies of 2, 3 and 4 sites are 100%, 88.3% and 30% [39]. Zhang et al. developed a system for multiple genome editing of Saccharomyces cerevisiae, GTR-CRISPR, which uses reported effective gRNA to interrupt 8 genes simultaneously with efficiency up to 87%. They further developed an accelerated Lightning GTR-CRISPR system which can knock out 6 genes in 3 days. Using Lightning GTR-CRISPR to simplify the yeast lipid network, they can delete the previously identified 8 genes in only two rounds, and increase the production of free fatty acids by 30-fold in 10 days, which greatly promoted the development of synthetic biology [23].
The high specificity and efficiency of the CRISPR-Cas9 system has led to rapid development of precise gene editing technology. Cas9-induced DSB can be repaired by NHEJ or HDR. In the error-prone NHEJ pathway, the ends of DSBs are processed and recombined through endogenous DNA repair mechanisms, resulting in random mutations in the junction site. HDR is another major DNA repair pathway, which can produce precise modifications at the target location in the presence of exogenously introduced repair template [14]. While its efficiency may vary greatly depending on cell type and status, as well as genomic sites and repair template [40]. The repair template in the form of plasmid or ssODN can be high fidelity precise editing using HDR method. Single-strand nicks in DNA can induce HDR [41]. When Cas9 is converted to a DNA nickase (catalytic residue mutation, D10A in RuvC and H840A in HNH) [12,14,42], the single-strand gap is first repaired by a high-fidelity HDR pathway. In order to improve the CRISPR-Cas9 system, according to Ran et al., two Cas9 nickases guided by a pair of sgRNAs targeting opposite strands of a target locus mediate DSB. DSB can stimulate homology-directed repair (HDR) to achieve high-precision editing of target sites in genome [43]. Heo et al. used the CRISPR-Cas9 system to precisely edit the 5′-untranslated region sequence of gltA encoding citrate synthase to reduce its expression level, thereby carbon flux from acetyl-CoA to citric acid cycle was redirected to acetoacetyl-CoA. Finally, the butanol output reached 0.82 g/L [24].
3.2. Transcription control
CRISPR-Cas9 technology can also be applied to regulate gene expression at the level of transcription and translation, besides genome editing. Dcas9 (Cas9 mutated to inactivate nuclease activity) can bind to specific DNA sequences and regulate the expression of specific genes under the guidance of sgRNA. The method that dCas9 is used to inhibit gene expression under the guidance of sgRNA, called CRISPR interference technology (CRISPRi) [44]. According to the results of Bikard et al. [45] and Qi et al. [46], dCas9 mainly inhibits gene transcription in two ways: (1) restricting RNA polymerases binding to promoters to inhibit transcription initiation; (2) preventing RNA polymerases sliding on the DNA duplex to inhibit transcriptional extension. In order to increase the production of glutamic acid and fully transfer the carbon source to the final product, Wang et al. used the CRISPR-nCas9(D10A)-AID system to inactivative genes pyk, ldhA, odhA involved in competitive passway in Corynebacterium glutamicum. Finally, it was found that pyk&ldhA double-inactivated strain can increase glutamic acid production by 3 times [25]. In addition to transcriptional repression, dCas9 protein fused with a transcriptional activation domain can also activate transcription of specific genes. namely CRISPR activation (CRISPRa) [47]. The position of the target sequence affects the activation efficiency of CRISPRa. When the distance between the target sequence and the promoter is appropriate, the activation efficiency is higher; when the target sequence is further from the upstream of the promoter, the activation efficiency will be reduced to a certain extent. Inhibition occurs when the target sequence and the promoter are close or the target sequence is in an open reading frame. However, due to the lack of effective gene activators, methods for regulating bacterial cell gene expression programs are limited. To cope with this challenge, Chen et al. identified a variety of synthetic transcription activators compatible with CRISPRa in E. coli, including Soxs, TetD and ASIA. In particular, SOX interacts with the surface on the α subunit of RNA polymerase which is highly conserved among gammaproteobacteria, alphaproteobacteria, bacteroides, gram-positive bacteria. This conservative interface may allow the CRISPRa system developed in E. coli to be ported to non-model bacteria with a wide range of useful biological functions. It provides a basis for designing synthetic bacterial cell devices with applications in diagnostics, therapeutics and industrial biosynthesis [48].
As more and more microbial cell factories are used to produce fuels, chemicals, etc., it is necessary to carry out metabolic engineering of microorganisms to maximize yield and productivity. In this case, researchers often need to modify multiple metabolic engineering targets with different regulatory modes, such as increasing expression of genes encoding rate-limiting enzymes, decreasing expression of essential genes, and removing expression of competing pathways. However, due to our limited understanding of cell metabolism regulation, developing a combinatorial metabolic engineering strategy to modify the host genome in a modular, parallel and high-throughput manner will be the key to optimize microbial cell factories. Lian et al. designed a combinatorial metabolic engineering strategy (CRISPR-AID) based on an orthogonal three-function CRISPR system, which combines transcription activation, transcription interference and gene deletion. Through CRISPR-AID technology, the production of β-carotene was increased by 3-fold in one step. And by combining and optimizing multiple metabolic engineering targets, the display of endoglucanase on the yeast surface was increased by 2.5-fold [26].
The fine-tuning of gene expression is essential for protein expression and pathway construction, but it still faces huge challenges due to the hierarchical gene regulation at multiple levels in a context-dependent manner. Lu et al. co-expressed dCas9 with transcriptional regulators α and ω, and designed position-specific gRNA to activate or represse the expression of different genes. Finally, by controlling the time of dCas9-expression, the expression of target genes can be efficiently regulated in multiple dimensions. By combining OAPS and dCas9-α, Lu et al. systematically evaluated the effects of promoter-based transcription, chaperone-assisted protein folding and protease-mediated degradation on the expression of amylase BLA in Bacillus subtilis. Finally, the production of BLA was increased 260-fold [27]. In order to explore the optimal intermediate level of gene expression, Matthew et al. established a method in Saccharomyces cerevisiae for rapid fine-tuning and hierarchical expression of enzymes through the regulation of dCas9. In this method, changing the position of the sgRNA target is regarded as the dominant parameter. By using the repressor and activator fused with dCas9, the medium-strength glycolytic promoter was modified to cover a nearly 40-fold expression range from near deletion to over-amplification. They finally achieved a 5.7-fold increase in titer applying this method to the glycerol biosynthetic pathway. They then identified and alleviated pathway bottlenecks resulting in a 7.8-fold increase in 3-dehydroshikimate titer and the identification of 3 unique targets for xylose catabolism by applying it to the pentose phosphate pathway in two distinct strain backgrounds. In addition, with the rapid decrease in the cost of DNA synthesis and the development of massively parallel sequencing technology, this technology may be used to complement genome-scale metabolic models in the future [49].
3.3. Other applications
DNA imaging: studying the interaction of specific genes with chromatin states requires a reliable method to visualize DNA in living cells. Traditional labeled DNA techniques, such as fluorescence in situ hybridization (FISH), require fixed samples and cannot capture live processes. Recently developed fluorescent marker Cas9 for specific DNA loci by Chen et al., is a viable alternative to DNA-FISH for live cell imaging [50]. When dCas9 is fused to EGFP and other fluorescent proteins, fluorescent localization can be realized at specific sites in the genome.
Instantaneous control: decomposing the two domains of Cas9 into two separate proteins and using chemical or light-induced dimerization methods can achieve instantaneous control of various genomic or epigenomic operations. Small molecule induction will facilitate the systematic control of Cas9, while optical regulation will allow more precise spatial perturbations. Konermann et al. have successfully used the light-induced dimerization domains CIB1 and CRY2 or the chemically-induced analogues ABI and PYL to construct inducible TALEs [51].
4. Optimization of CRISPR-Cas9 toolkit to improve editing efficiency
4.1. Improve editing efficiency by reduce off-target effects
Off-target effects occur when sgRNA is selected at a position with low specificity or the temporal, locus-specific and spatial control of Cas9 protein expression is not mastered, especially when Cas9 and gRNA are co-expressed on the same plasmid. At present, scientists have developed multiple CRISPR-Cas9 optimization schemes to increase the specificity of CRISPR-Cas9.
4.1.1. Reducing off-target effects by sgRNA design
The specificity of the CRISPR system can be improved by the modification of gRNA. Studies have shown that introducing deoxynucleotides to create RNA-DNA hybrids in the CRISPR system can obtain higher specificity [52]. Chemical modifications, such as 2′-O-methyl-3′-phosphonoacetate introduced at specific sites on the DNA recognition sequence of the gRNA ribose phosphate backbone also increases specificity [53]. Addition of nucleotides [54,55] or truncation of nucleotides [56,57] can reduce CRISPR system tolerance to base mismatches to reduce off-target rates. In addition, several algorithm-based tools, such as ChopChop and CRISPR Design, based on a range of factors including sequence similarity, number and location of mismatches, have been developed to avoid off-target effect [58,59]. The sgRNA that reduces the off-target efficiency depends on the selection of a target site with none or few similar genes. Furthermore, Ran et al. plotted the relationship between gRNA and Cas9 ratios and the number of off-target effects [56]. All of the above have provided a powerful effect for reducing off-target rate by modifying sgRNA.
4.1.2. Reducing off-target effects by Cas9 modifications
In addition, Cas9 can be transformed to reduce off-target effects. As mentioned earlier, using two sgRNAs for directing two copies of Cas9n (either HNH or Ruv-C active site was mutated) to two adjacent target sites, the simultaneous of double-strand breaks can greatly improve the specificity of gene targeting [43]. Although a single Cas9n can create gaps in DNA and introduce mutations much less efficiently than wild-type Cas9 theoretically. There are reports that high frequent insertions/deletions may still be caused at certain genomic loci, so the introduction of two copies of Cas9n: sgRNA may inhibit off-target effect. In order to overcome this problem, some research teams modified the above scheme. They replaced Cas9n with Cas9d (both HNH and Ruv-C active sites were mutated), which fused with FokI nuclease, to form RNA-guided FokI-Cas9d nuclease (RFN) under the guidance of gRNA. Because FokI needs to form a dimer to exert nuclease activity, two gRNAs can be used to direct two copies of RFN to adjacent sites to activate FokI dimerization and nuclease activity in the above scheme, thereby improving specificity and efficient cutting [60,61].
Studies have found that the non-catalytic REC2 domain of Cas9 nuclease plays a crucial role in off-target recognition. Keewon et al. [62] used a single-molecule fluorescence method to study the conformational kinetics of the interaction between non-target DNA strands (NTS) and Cas9, and found that REC2 regulates NTS rearrangement through positively charged residues on its surface to perform cleavage reactions. This study promoted rationally designed highly specific Cas9 variants for genome editing.
4.2. Improve editing efficiency by reduce Cas9 toxicity effects
Due to the unique nature of the prokaryotic genetic profiles, the CRISPR-Cas9 system shows toxicity in a large number of microorganisms, which can easily lead to fatal chromosome breaks, resulting in low transformation efficiency and failure of gene editing. In the process of genome editing of Clostridium, Wang et al. [63] combined inducible expression of Cas9 and plasmid-borne editing templates. They observed severe vector integration (VIE) event, which has never been reported by other researchers in bacterial genetic engineering based on plasmid-edited templates for homologous recombination. This study offers two methods to reduce the toxicity of Cas9. On the one hand, the toxicity of this system is reduced by regulating the expression of cas9 gene; on the other hand, since most prokaryotes contain a natural CRISPR-Cas system, genome engineering can be achieved by using these endogenous immune systems to relieve Cas9 issues related to toxicity and low transformation efficiency.
4.2.1. Reducing Cas9 toxicity by regulates Cas9 protein expression
Due to absence of endogenous non-homologous end-joining (NHEJ) system, or deficiency of NHEJ, Cas9-induced double-strand breaks (DSB) is fatal to lots of bacteria. Therefore, regulating Cas9 expression is a crucial step for gene editing. Liu et al. developed a CRISPR-Cas9 genome editing toolbox for Streptomyces glutamine, in which Cas9 and gRNA expression cassettes were reconstituted to combat Cas9 toxicity and facilitate effective termination of gRNA transcription. Co-transformation of Cas9 and gRNA expression plasmids was exploited to overcome highly frequent mutation of Cas9, allowing not only highly efficient gene deletion and insertion with plasmid-borne editing templates (efficiencies up to 60.0 and 62.5%) but also simple and time-saving operation [64].
4.2.2. Reducing Cas9 toxicity by exploiting endogenous CRISPR-Cas
The CRISPR-Cas system has been extensively used for multifunctional genome editing and transcription regulation in various species since 2013. Most of these applications are based on the type II CRISPR-Cas9 system derived from Streptococcus pyogenes. However, the expression of heterologous Cas9 is highly toxic to a multitude of microorganisms [65,66], resulting in low transformation efficiency and failure of genome editing. Therefore, researchers proposed to use endogenous CRISPR-Cas system to reduce the toxicity of the CRISPR-Cas9 system [67]. At present, the endogenous CRISPR-Cas system has been used for genome editing and transcription regulation in a few bacteria and archaea [67], its transformation efficiency proved to be considerable. After several attempts to use CRISPR-Cas9 systems for genome editing, Zhang et al. successfully reused the IB-type CRISPR-Cas system in butyric-acid bacteria for genome editing and realized 100% efficiency for multiple genome editing [28]. Specifically, they replaced the leader sequence with a lactose-inducible promoter to drive the expression of the endogenous CRISPR-Cas system, resulting in a total transformation efficiency of 1.7 cfu/mL donor. In addition, any plasmid containing these heterologous nuclease (or nickelase) proteins such as Cas9 or nCas9 driven by the same inducible promoter could not be successfully transformed. This indicates that the endogenous CRISPR-Cas system can be applied to avoid toxicity caused by heterologous CRISPR-Cas9/nCas9 system. Finally, they replaced cat1 by the alcohol dehydrogenase gene adhE1 or adhE2 using the endogenous CRISPR-Cas system, resulting in a butanol production mutant with a butanol titer of 26.2 g/L, which is the highest level been reported.
4.3. Improve editing efficiency by optimizing crRNA
4.3.1. SOMACA
Wu et al. established a synthetic oligonucleotide-mediated assembly method (SOMACA) [68] for the construction of crRNA arrays, and achieved 100% efficiency of double genes in-frame knocking out, multiple point mutations (up to six), or single gene insertion using this tool. Finally, the authors used this method to perform nonsense mutations in the four genes (NAGA, NAGB, NAGP, GAMA) and the byproducts of the lactic acid (LDH) and acetic acid (PTA) synthesis pathways in the decomposition of N-acetylglucosamine, increasing N-acetylglucosamine titer by 50.9%.
4.3.2. Optimize crRNA length to improve editing efficiency
Liu et al. applied the bifunctional cluster regular spaced short palindromic repeat system (RE-CRISPR) to simultaneously edit and regulate genes in Corynebacterium glutamicum [69]. They used gfp, chromosome-integrated rfp, and lacZ as reporter genes to study the effect of crRNA length on the efficiency of transcriptional repression. It was found that crRNAs of 15 and 16 nt showed much higher repression efficiency than crRNAs of other lengths. Finally, the application of the RE-CRISPR system in the high-cysteine and serine metabolism engineering of Corynebacterium glutamicum was demonstrated. The combined use of the RE-CRISPR system simultaneously caused the deletion of AECD and the inhibition of mcbR, thereby cysteine titer further increased to 3.7-fold (42.8 mg/L). Simultaneous deletion of SDAA and inhibition of GlyA increased serine production by 2.5-fold.
4.4. Improve editing efficiency by optimizing sgRNA
The CRISPR-Cas9 system is a revolutionary genome editing tool in which the endonuclease Cas9 is guided to the genome target site by complementary base pairing of sgRNA. The sgRNA includes a 20 base targeting sequence at the 5'end. Cas9 mediates double-strand breaks after targeting the receptor genome with the sgRNA. Therefore, in this system, the sgRNA expression cassette needs to be carefully designed to ensure the formation of a functional Cas9−sgRNA complex. The promoter and structure of sgRNA are two important constraints during the construction of the sgRNA expression cassette.
4.4.1. Optimizing sgRNA promoter
Gene editing efficiency is connected with sgRNA promoters. In some cases, such as gene knockout, knockdown and etc., sgRNA does not need continuous expression. So when the system is first established in an organism, it is necessary to choose a proper sgRNA promoter. Gene editing efficiency of some sgRNA promoters varies with species. For example, the RNA Pol III promoter of the spliceosome U6 snRNA is widely used in the fungi CRISPR-Cas9 system, but useable U6 promoter has not been found in Aspergillus niger. Therefore, Zheng et al. established a new CRISPR-Cas9 system expressing sgRNA based on one endogenous U6 promoter and two heterologous U6 promoters in Aspergillus niger. All three U6 promoters can interrupt the Aspergillus niger polyketide synthase alba gene and efficiently insert the gene at the target genomic locus with 40 bp donor DNA [70]. Although the U6 small nuclear RNA (snRNA) promoter is one of the most commonly used promoters in eukaryotes, the low sequence conservation of the U6 promoter restricts its identification in many species [71,72]. Moreover, the U6 promoter requires guanosine nucleotides to initiate transcription, thereby reducing the available CRISPR-Cas target sites [73,74]. Because the 5S rRNA gene is highly conserved and expressed in eukaryotes, Zheng et al.used it as a promoter of sgRNA, and the editing efficiency reached 100% in dozens of gene editing system established in Aspergillus niger [75], and the system can also be used for simultaneous mutation of multiple genes in Aspergillus niger. Therefore, the 5S rRNA gene can be widely used as a guide RNA promoter in the eukaryotic CRISPR-Cas9 system.
4.4.2. Optimizing sgRNA structure
The secondary structure formed by partial sgRNA is necessary for Cas9 activity. There are two interdependent variable regions in the sgRNA gene. One region contains a 20 bp protospacer and the other is a 6 bp inverted repeating region that repeats the 5'end of the protospacer. The first 20 nucleotide sequence of the sgRNA is used to guide targeted DNA cleavage. Additional bases or other modifications at the 5'end of sgRNA may cancel the ability of gRNA to guide Cas9 to cut DNA [76], because eukaryote RNA polymerase II process and modify at both ends of RNA during transcription, so the transcribed RNA cannot be used directly as sgRNA. However, most of the well-characterized promoters are currently transcribed by RNA polymerase II, so post-transcriptional modification of sgRNA is important. In order to develop CRISPR-Cas9 systems which can perform directional mutations with spatial and temporal control in a wide range of organisms, methods for producing gRNA need to be improved. For example, when tending to sequentially interfere with different genes, it is necessary to use various promoters, such as hormone-responsive promoters and environmental signal-regulated promoters to control the time of gRNA production. Guo et al. proposed a general method for efficient production of gRNA in vitro and in vivo. They designed an artificial gene that produces an RGR with a ribozyme sequence at both ends of the gRNA after transcription. Through the action of two ribozymes (the 5′ end hammerhead and the 3′ end hepatitis D virus) on both sides of the gRNA, the mature gRNA self-catalyzed by primary transcript of RGR driven by any promoter can effectively guide the specific cleavage of the target gene in vitro and yeast [77]. Nodvig et al. also used this system to improve the efficiency of gene editing in filamentous fungi [78]. Therefore, if a suitable promoter is selected, cell and tissue specific gene expression can be achieved.
4.5. Improve editing efficiency by increasing recombination rates
Due to low efficiency of endogenous recombination system in many microorganisms, introducing a heterologous recombination system is a crucial step to apply CRISPR-Cas system for genome editing. Bassalo et al. provided a recombination strategy based on the coupling of the CRISPR and λ-red systems. They co-introduced a plasmid that containing Cas9 and a plasmid that containing λ-red recombinase into E. coli, and found that Cas9 can work effectively in different genomic environments with λ-red recombinase. Through this method, a 10 kb gene encoding the complete isobutanol production pathway can efficiently be labeled (less than 50%) in one day [79]. This powerful recombination ability makes rapid metabolic transformation of microorganisms a reality. Using this strategy, Cho et al. used CRISPR coupled with λ-red quickly and efficiently knock out multiple genes in Streptomyces glutamine [29]. Therefore, this genomic engineering system strategy is generally applicable to the development of genomic engineering systems for other microorganisms. This genomic engineering system accelerates the pace of metabolic engineering of industrial strains.
In another work, Jiang et al. co-transformed donor DNA provided in fragments and sgRNAs contained in pTargetF into E. coli cells, using the CRISPR-Cas9 system to achieve various precision genome modification, including gene deletions and insertions, with a maximum efficiency of 100%. Multiple genes can be simultaneously edited, using this method so as to improve the efficiency of recombination while reducing the time and labor required for construction. Using this system they also successfully targeted Tatumella Citrea chromosome deletion with highest efficiency up to 100% [80]. However, Jiang et al. also found that when increasing the number of target targets or reducing the length of homologous sequences (from 300 to 400 bp to 40 bp), the recombination efficiency was significantly reduced. We suspect that dsDNA has a low transformation efficiency in E. coli, and λ-red recombinase can promote the recombination of smaller single-stranded DNA fragments, so we can further improve the recombination efficiency by using single-stranded DNA as a donor, or increasing the length of the homologous sequence.
5. Concluding remarks and future perspectives
Just as recombinant DNA technology has benefited from basic research on restriction enzymes from microorganisms, the latest generation of Cas9-based genome engineering tools is also based on components of antiphage defense system from microorganisms. In recent years, the CRISPR-Cas9 system and various technologies derived from it, such as precise editing, multi-gene editing, and precise regulation, have catapulted innovation to provide integrated solutions, and have provided us with increased flexibility in bacterial genetic engineering. It plays an important role in the fields of single-stranded RNA editing and high-throughput gene screening. Nevertheless, the problems such as off-target rate and cytotoxicity should not be ignored. Since the DSB caused by the Cas9 protein is lethal to host cells, DNA base editor that does not introduce DSB has been developed. It is reported that an evolved Cas9 variant containing 7 mutations has relaxed the PAM requirement to NG or NNG [81]. The application of these variants is expected to significantly increase the number of targeted nucleotides in the genome. The existing base editor can only realize the base conversion between pyrimidine and between purine. Zhao et al. designed and constructed the cytosine deaminase-nCas9-Ung protein complex, creating a new glycosylase base editor (GBE). They developed a single-base gene editing system that can realize the transversion between pyrimidine and purine. Based on this system, it is the first time in the world to achieve arbitrary base editing in microorganisms and specific transversion of C-G bases in mammalian cells [82]. This is important in the construction of synthetic biological systems and modification of biological traits. As microbial cell factories are increasingly used to produce biochemical products, it will be a trend to use the CRISPR-Cas9 system to optimize the genome of microbial cells in a modular, parallel, and high-throughput manner to increase the yield of target products. Considering the striking progress of CRISPR-Cas9 system in eukaryotes, we anticipate that discoveries in the near future will expand its application in more microorganisms.
Funding
This work was supported by the National Key R&D Program of China (2018YFA0901600), the National Natural Science Foundation of China (31900052), Youth Innovation Promotion Association, Chinese Academy of Sciences (2020182).
Credit author statement
I would like to declare on behalf of my co-authors that the work described was original research that has not been published previously, and not under consideration for publication elsewhere, in whole or in part. All the authors listed have approved the manuscript that is enclosed.
Declaration of competing interest
The authors indicate that they have no conflict of interest.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Contributor Information
Jing Zhao, Email: zhaoj@tib.cas.cn.
Huan Fang, Email: fang_h@tib.cas.cn.
Dawei Zhang, Email: zhang_dw@tib.cas.cn.
References
- 1.Gaj T., Gersbach C., Barbas C. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013:31. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kok F., Lawson N., Wolfe S. Construction and application of site-specific artificial nucleases for targeted gene editing. Methods Mol Biol. 2014;1101:267–303. doi: 10.1007/978-1-62703-721-1_13. [DOI] [PubMed] [Google Scholar]
- 3.Reyon D. ZFNGenome: a comprehensive resource for locating zinc finger nuclease target sites in model organisms. BMC Genom. 2011;12:83. doi: 10.1186/1471-2164-12-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mussolino C. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burgess D. Technology: a CRISPR genome-editing tool. Nat Rev Genet. 2013;14 doi: 10.1038/nrg3409. [DOI] [PubMed] [Google Scholar]
- 6.Mojica F. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol Microbiol. 2000;36:244–246. doi: 10.1046/j.1365-2958.2000.01838.x. [DOI] [PubMed] [Google Scholar]
- 7.Ishino Y. Nucleotide sequence of the IAP gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1988;169:5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohanraju P. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016;353 doi: 10.1126/science.aad5147. aad5147-aad5147. [DOI] [PubMed] [Google Scholar]
- 9.Makarova K. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makarova K., Wolf Y., Koonin E. The basic building blocks and evolution of CRISPR-CAS systems. Biochem Soc Trans. 2013;41:1392–1400. doi: 10.1042/BST20130038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makarova K. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jinek M. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (New York, N.Y.) 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiedenheft B., Sternberg S., Doudna J. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 14.Cong L., Zhang F. Genome engineering using CRISPR-Cas9 system. Methods Mol Biol. 2015;1239:197–217. doi: 10.1007/978-1-4939-1862-1_10. [DOI] [PubMed] [Google Scholar]
- 15.Mojica F., Mojica F.J.M., Diez-Villasenor C., Garcia-Martinez J., Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. Microbiology (Reading, England), 733-40. [DOI] [PubMed] [Google Scholar]
- 16.Barrangou R. CRISPR provides acquired resistance against viruses in prokaryotes. Science (New York, N.Y.) 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 17.Datsenko K. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun. 2012;3:945. doi: 10.1038/ncomms1937. Nature communications. 3: p. 945. [DOI] [PubMed] [Google Scholar]
- 18.Heler R. Cas9 specifies functional viral targets during CRISPR–Cas adaptation. Nature. 2015;519(7542):199–202. doi: 10.1038/nature14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bolotin A. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology (Reading, Engl) 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 20.Deltcheva E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ivančić-Baće I., Al Howard J., Bolt E.L. Tuning in to interference: R-loops and cascade complexes in CRISPR immunity. J Mol Biol. 2012;422(5):607–616. doi: 10.1016/j.jmb.2012.06.024. [DOI] [PubMed] [Google Scholar]
- 22.Li H. CRISPR-Cas9 for the genome engineering of cyanobacteria and succinate production. Metab Eng. 2016;38 doi: 10.1016/j.ymben.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y. A gRNA-tRNA array for CRISPR-Cas9 based rapid multiplexed genome editing in Saccharomyces cerevisiae. Nat Commun. 2019;10(1):1053. doi: 10.1038/s41467-019-09005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heo M.-J. Controlling citrate synthase expression by CRISPR/Cas9 genome editing for n-butanol production in Escherichia coli. ACS Synth Biol. 2017;6(2):182–189. doi: 10.1021/acssynbio.6b00134. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y. MACBETH: multiplex automated Corynebacterium glutamicum base editing method. Metab Eng. 2018;47 doi: 10.1016/j.ymben.2018.02.016. [DOI] [PubMed] [Google Scholar]
- 26.Lian J. Combinatorial metabolic engineering using an orthogonal tri-functional CRISPR system. Nat Commun. 2017;8 doi: 10.1038/s41467-017-01695-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Z. CRISPR-assisted multi-dimensional regulation for fine-tuning gene expression in Bacillus subtilis. Nucleic Acids Res. 2019;47:e40. doi: 10.1093/nar/gkz072. -e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J. Exploiting endogenous CRISPR-Cas system for multiplex genome editing in Clostridium tyrobutyricum and engineer the strain for high-level butanol production. Metab Eng. 2018;47 doi: 10.1016/j.ymben.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 29.Cho J.S. CRISPR/Cas9-coupled recombineering for metabolic engineering of Corynebacterium glutamicum. Metab Eng. 2017:42. doi: 10.1016/j.ymben.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 30.Xia J. Expression of Shewanella frigidimarina fatty acid metabolic genes in E. coli by CRISPR/cas9-coupled lambda Red recombineering. Biotechnol Lett. 2016;38(1):117–122. doi: 10.1007/s10529-015-1956-4. [DOI] [PubMed] [Google Scholar]
- 31.Ding W. 5-Aminolevulinic acid production from inexpensive glucose by engineering the C4 pathway in Escherichia coli. J Ind Microbiol Biotechnol. 2017;44(8):1127–1135. doi: 10.1007/s10295-017-1940-1. [DOI] [PubMed] [Google Scholar]
- 32.Liang L. CRISPR EnAbled Trackable genome Engineering for isopropanol production in Escherichia coli. Metab Eng. 2017:41. doi: 10.1016/j.ymben.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 33.Tao W. Membrane engineering - a novel strategy to enhance the production and accumulation of β-carotene in Escherichia coli. Metab Eng. 2017;43 doi: 10.1016/j.ymben.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 34.Liu D. Development and characterization of a CRISPR/Cas9n-based multiplex genome editing system for Bacillus subtilis. Biotechnol Biofuels. 2019;12(1):197. doi: 10.1186/s13068-019-1537-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Q. Development of a genome-editing CRISPR/Cas9 system in thermophilic fungal Myceliophthora species and its application to hyper-cellulase production strain engineering. Biotechnol Biofuels. 2017;10 doi: 10.1186/s13068-016-0693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tong Z. Systems metabolic engineering for citric acid production by Aspergillus Niger in the post-genomic era. Microb Cell Factories. 2019;18(1):28. doi: 10.1186/s12934-019-1064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuivanen J., Wang Y.M., Richard P. Engineering Aspergillus Niger for galactaric acid production: elimination of galactaric acid catabolism by using RNA sequencing and CRISPR/Cas9. Microb Cell Factories. 2016;15 doi: 10.1186/s12934-016-0613-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song X. CRISPR-Cas9D10A nickase-assisted genome editing in Lactobacillus casei. Appl Environ Microbiol. 2017;83 doi: 10.1128/AEM.01259-17. AEM.01259-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng X. CRISPR/Cas9 assisted multiplex genome editing technique in Escherichia coli. Biotechnol J. 2017;13 doi: 10.1002/biot.201700604. [DOI] [PubMed] [Google Scholar]
- 40.Helleday T. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 2004;32:3683–3688. doi: 10.1093/nar/gkh703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen F. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods. 2011;8:753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gasiunas, G., et al., Gasiunas, G, Barrangou, R, Horvath, P and Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA 109: E2579-E2586. Proceedings of the national Academy of sciences of the United States of America, 2012. : p. E2579-86. [DOI] [PMC free article] [PubMed]
- 43.Ran F. Double nicking by RNA-Guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013:154. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larson M. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bikard D. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013:41. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qi L. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng A. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23 doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong C. Synthetic CRISPR-Cas gene activators for transcriptional reprogramming in bacteria. Nat Commun. 2018;9(1):2489. doi: 10.1038/s41467-018-04901-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deaner M., Alper H. Systematic testing of enzyme perturbation sensitivities via graded dCas9 modulation in Saccharomyces cerevisiae. Metab Eng. 2017:40. doi: 10.1016/j.ymben.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 50.Chen B. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Konermann S. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500(7463):472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yin H. Partial DNA-guided Cas9 enables genome editing with reduced off-target activity. Nat Chem Biol. 2018;14 doi: 10.1038/nchembio.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ryan D.E. Improving CRISPR-Cas specificity with chemical modifications in single-guide RNAs. Nucleic Acids Res. 2018;46(2):792–803. doi: 10.1093/nar/gkx1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ryu S.-m. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat Biotechnol. 2018;36 doi: 10.1038/nbt.4148. [DOI] [PubMed] [Google Scholar]
- 55.Cho S.W. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2013;24 doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fu Y. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32 doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lei C. The CCTL (Cpf1-assisted Cutting and Taq DNA ligase-assisted Ligation) method for efficient editing of large DNA constructs in vitro. Nucleic Acids Res. 2017;45 doi: 10.1093/nar/gkx018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hsu P. DNA targeting specificity of RNA-guided CAS9 nucleases. Nat Biotechnol. 2013:31. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jennifer, et al., Approaches to reduce CRISPR off-target effects for safer genome editing.Applied biosafety: Journal of the American Biological Safety Association, 2017,7-13.
- 60.Guilinger J., Thompson D., Liu D., Guilinger J.P., Thompson D.B., Liu D.R. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. Nature biotechnology, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bolukbasi M.F. DNA-binding-domain fusions enhance the targeting range and precision of Cas9. Nat Methods. 2015;12 doi: 10.1038/nmeth.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sung K. Target specificity of Cas9 nuclease via DNA rearrangement regulated by the REC2 domain. J Am Chem Soc. 2018;140 doi: 10.1021/jacs.8b03102. [DOI] [PubMed] [Google Scholar]
- 63.Wang Y. Bacterial genome editing with CRISPR-cas9: deletion, integration, single nucleotide modification, and desirable “clean” mutant selection in Clostridium beijerinckii as an example. ACS Synth Biol. 2016;5(7):721–732. doi: 10.1021/acssynbio.6b00060. [DOI] [PubMed] [Google Scholar]
- 64.Liu J. Development of a CRISPR/Cas9 genome editing toolbox for Corynebacterium glutamicum. Microb Cell Factories. 2017;16 doi: 10.1186/s12934-017-0815-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Charpentier E., Doudna J. Biotechnology: rewriting a genome. Nature. 2013;495:50–51. doi: 10.1038/495050a. [DOI] [PubMed] [Google Scholar]
- 66.Wang H. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-Mediated genome engineering. Cell. 2013;153 doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pyne M. Harnessing heterologous and endogenous CRISPR-Cas machineries for efficient markerless genome editing in Clostridium. Sci Rep. 2016;6:25666. doi: 10.1038/srep25666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu Y. CAMERS-B: CRISPR/Cpf1 assisted multiple-genes editing and regulation system for Bacillus subtilis. Biotechnol Bioeng. 2020:1–9. doi: 10.1002/bit.27322. [DOI] [PubMed] [Google Scholar]
- 69.Liu W. Combined genome editing and transcriptional repression for metabolic pathway engineering in Corynebacterium glutamicum using a catalytically active Cas12a. Appl Microbiol Biotechnol. 2019;103 doi: 10.1007/s00253-019-10118-4. [DOI] [PubMed] [Google Scholar]
- 70.Zheng X. Heterologous and endogenous U6 snRNA promoters enable CRISPR/Cas9 mediated genome editing in Aspergillus Niger. Fungal Biology and Biotechnology. 2018;5 doi: 10.1186/s40694-018-0047-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu R. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov. 2015;1:1–11. doi: 10.1038/celldisc.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng Y.-M. Development of a versatile and conventional technique for gene disruption in filamentous fungi based on CRISPR-Cas9 technology. Sci Rep. 2017;7 doi: 10.1038/s41598-017-10052-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ranganathan V. Expansion of the CRISPR–Cas9 genome targeting space through the use of H1 promoter-expressed guide RNAs. Nat Commun. 2014;5(1):4516. doi: 10.1038/ncomms5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mali P. RNA-guided human genome engineering via Cas9. Science. 2013:339. doi: 10.1126/science.1232033. New York, N.Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zheng X. 5S rRNA promoter for guide RNA expression enabled highly efficient CRISPR/Cas9 genome editing in Aspergillus Niger. ACS Synth Biol. 2018;8 doi: 10.1021/acssynbio.7b00456. [DOI] [PubMed] [Google Scholar]
- 76.Haurwitz R. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gao Y., Zhao Y. Self- processing of ribozyme- flanked RNAs into guide RNAs in vitro and in vivo for CRISPR- mediated genome editing. J Integr Plant Biol. 2013;56 doi: 10.1111/jipb.12152. [DOI] [PubMed] [Google Scholar]
- 78.Nødvig C. A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PloS One. 2015;10 doi: 10.1371/journal.pone.0133085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bassalo M.C. Rapid and efficient one-step metabolic pathway integration in E. coli. ACS Synth Biol. 2016;5(7):561–568. doi: 10.1021/acssynbio.5b00187. [DOI] [PubMed] [Google Scholar]
- 80.Jiang Y. Erratum for Jiang et al., Multigene Editing in the Escherichia coli Genome via the CRISPR-Cas9 System. Appl Environ Microbiol. 2016;82 doi: 10.1128/AEM.01181-16. 3693-3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hu J. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018:556. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao D. New base editors change C to A in bacteria and C to G in mammalian cells. Nat Biotechnol. 2020:1546–1696. doi: 10.1038/s41587-020-0592-2. [DOI] [Google Scholar]