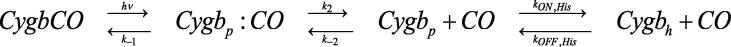Graphical abstract
1These authors contributed equally as First Author.
Abbreviations: CO, Carbon monoxide; Cygb, Cytoglobin; Cygb-1, Cytoglobin 1; Cygb-2, Cytoglobin 2; C.aceCygb-1, Cytoglobin-1 of C. aceratus; D.mawCygb-1, Cytoglobin-1 of D. mawsoni; DTT, Dithiothreitol; Hb, Hemoglobin; Cygbh, Hexa-coordinated bis-histidyl species; CYGB, Human Cygb; NGB, Human neuroglobin; MD, Molecular Dynamics; C.aceCygb-1*, Mutant of C.aceCygb-1; D.mawCygb-1*, Mutant of D.mawCygb-1; Mb, Myoglobin; NO, Nitric oxide; p50, O2 partial pressure required to achieve half saturation; Cygbp, Penta-coordinated Cygb; ROS, Reactive Oxygen Species; RNS, Reactive Nitrogen Species; rms, Root-mean square
Keywords: Cytoglobin, Cold-adaptation, Ligand properties, NO dioxygenase, X-ray structure
Abstract
While the functions of the recently discovered cytoglobin, ubiquitously expressed in vertebrate tissues, remain uncertain, Antarctic fish provide unparalleled models to study novel protein traits that may arise from cold adaptation. We report here the spectral, ligand-binding and enzymatic properties (peroxynitrite isomerization, nitrite-reductase activity) of cytoglobin-1 from two Antarctic fish, Chaenocephalus aceratus and Dissostichus mawsoni, and present the crystal structure of D. mawsoni cytoglobin-1. The Antarctic cytoglobins-1 display high O2 affinity, scarcely compatible with an O2-supply role, a slow rate constant for nitrite-reductase activity, and do not catalyze peroxynitrite isomerization. Compared with mesophilic orthologues, the cold-adapted cytoglobins favor binding of exogenous ligands to the hexa-coordinated bis-histidyl species, a trait related to their higher rate constant for distal-His/heme-Fe dissociation relative to human cytoglobin. At the light of a remarkable 3D-structure conservation, the observed differences in ligand-binding kinetics may reflect Antarctic fish cytoglobin-1 specific features in the dynamics of the heme distal region and of protein matrix cavities, suggesting adaptation to functional requirements posed by the cold environment. Taken together, the biochemical and biophysical data presented suggest that in Antarctic fish, as in humans, cytoglobin-1 unlikely plays a role in O2 transport, rather it may be involved in processes such as NO detoxification.
1. Introduction
Cytoglobin (Cygb) is a 21-kDa heme-protein belonging to the globin superfamily that is ubiquitously expressed in vertebrate tissues. It has been proposed to be involved in: (i) O2 supply to mitochondrial respiration [42], [50], (ii) nitric-oxide (NO) scavenging through its dioxygenase activity [40], (iii) nitrite reduction and NO generation under anaerobic conditions [53], (iv) modulation of NO levels and metabolism in the vascular walls [55], [56], (v) cell protection against nitrogen- and oxygen-reactive species [52], and (vi) fibrotic disorders [23], [62], [71]. Moreover, Cygb may function as tumor suppressor [63], [64], [77], and as potential tumor promoter in cells under stress [64].
In contrast to mammals, where Cygb occurs as a single-copy gene, in teleosts (e.g. Danio rerio, Oryzias latipes, Tetraodon nigroviridis, and Takifugu rubripes), two distinct paralogous Cygb genes (Cygb-1 and Cygb-2) were found, suggesting sub-functionalization of the two proteins after gene duplication [34]. Although both Cygb mRNAs find broad expression in many tissues, high levels of Cygb-2, also known as neuronal Cygb, were detected in brain and retina of D. rerio [34]. Cygb-2 is more closely related to mammalian Cygb than Cygb-1, with 67% sequence identity and 84% similarity, whereas Cygb-1 shares only 49% sequence identity and 78% similarity with human Cygb (CYGB) [34].
Similar to human neuroglobin (NGB) [66], the heme iron atom of CYGB - as well as of all mammalian orthologs - is hexa-coordinated [25], [26], being ligated to distal HisE7 and proximal HisF8 residues, both in the Fe(II) and Fe(III) states. In contrast, D. rerio Cygb-1 was found in the penta-coordinated state, more stable in the Fe(II) form, whereas D. rerio Cygb-2 exhibits hexa-coordination [22]. The slower autoxidation rate and a possible role as nitrite reductase of D. rerio Cygb-1 led to the conclusion that the two D. rerio paralogs might have acquired different biological functions after gene duplication, with Cygb-1 exhibiting properties consistent with an O2 carrier role [22].
Cygb-1 mRNA was recently also detected in some Antarctic notothenioid fish species: in the retina and liver of Dissostichus mawsoni, in the brain and liver of Chaenocephalus aceratus [24], in the muscle of Chionodraco hamatus [21] and in the retina, brain and gills of Trematomus bernacchii and C. hamatus (D. Giordano, personal communication). The Cygb-2 gene was annotated in the genome sequence of the red-blooded Antarctic fish Notothenia coriiceps (http://www.ncbi.nlm.nih.gov/nuccore/XM_010778246.1), and transcripts of Cygb-2 were also found in the brain, retina and gills of T. bernacchii and C. hamatus. In the latter species Cygb-1 and Cygb-2 display different expression levels in tissues exposed to hypoxia, thus supporting their involvement in specific and different functions (D. Giordano, personal communication).
One of the most important driving forces in the evolutionary adaptations of Antarctic marine organisms is the enhanced O2 solubility in the cold waters of the Southern Ocean. These environmental conditions promote the buildup of high reactive oxygen species (ROS) levels and related reactive nitrogen species (RNS), suggesting that cold-adapted organisms must develop effective defense mechanisms against oxidative and nitrosative stress. Evolutionary gain of new genes, changes in expression patterns, and loss of genetic information, especially in icefish, also reflect the specialization of Antarctic organisms to a narrow range of low temperatures. Remarkably, the red-blooded notothenioids can survive in these waters even though their blood hemoglobin (Hb) is poisoned by CO [27].
Modern Notothenioidei (icefish) belonging to the family Channichthyidae appear to be the end result of an extraordinary natural experiment, as they are the only vertebrates known to lack red blood cells and functional Hb genes, and hold the exceptional physiological features adopted by organisms permanently adapted to low temperatures [17]. Icefish may afford lack of myoglobin (Mb) and Hb through exploitation of high O2 solubility and low metabolic rates in the cold, where an enhanced fraction of O2 supply occurs through diffusive O2 flux. The homeostatic activity of NO probably facilitates the evolution of some compensatory characters. It was suggested that the evolution of the cardiovascular adaptations was ‘‘jump-started’’ by homeostatic NO response, leading to the hypothesis that in icefish other proteins (namely the tissue globins) may play the primary function of regulating NO degradation [74]. Six species, including C. aceratus, also lack Mb in skeletal muscle and heart [75]. Although the evolutionary pressure is against the maintenance of O2-binding proteins, they have retained Ngb [18] and Cygbs [24], thus suggesting that selective pressure kept alive the expression of proteins involved in NO detoxification in NO-active tissues.
Cygb-1 cDNAs of the red-blooded fish D. mawsoni (family Notothenidae) and of the related icefish C. aceratus (family Channichthyidae), which lacks both Mb and Hb, were cloned from retina and brain, respectively, over-expressed and characterized by Electron Paramagnetic Resonance, Resonance Raman and optical absorption spectroscopy [24]. The two Antarctic fish Cygbs-1 are composed of 179 amino acids, with mutual sequence identity of 98%. Although the Antarctic fish proteins display features in common with CYGB, e.g. hexa-coordination in the Fe(II) and Fe(III) forms, structural/dynamic properties of the heme pocket may account for the small differences observed in their redox potentials [24]. Furthermore, while in CYGB, similar to NGB, the integrity of an internal disulfide bond affects conformation, reactivity [41], [51], and dimerization [12], in cold-adapted fish Cygbs-1 the available Cys residues are too far apart for an intramolecular disulfide bond [24]. Nevertheless, covalent and non-covalent oligomers (up to pentamers) were found in Antarctic fish Cygbs-1, and association was shown to affect the heme-pocket environment [24].
Antarctic fish belonging to the suborder of Notothenioidei are excellent models, within vertebrates, to study the biological responses to low temperature [28], [37], [65]. Notothenioidei are teleosts largely endemic to the Southern Ocean where the temperatures on the shelf are constant, ranging from −1.9 to −1.7 °C throughout the water column [29]. Due to low temperature, the reduced thermal energy and the increased viscosity of water slow down all reaction rates, and may impact protein dynamics. Over the course of many million years, notothenioids developed many physiological and biochemical features in response to low temperature, proteins being the main target of adaptation mechanisms [28]. In this context, a thorough comparison of the functional and structural properties of cold-adapted Cygbs-1 vs CYGB may help us gain insight into the evolutionary adaptations of O2 transport processes and protein-mediated ligand reactivity. To this end, we investigated the spectral, ligand binding, and enzymatic properties of Cygb-1 from C. aceratus (C.aceCygb-1) and from D. mawsoni (D.mawCygb-1), together with the crystal structure of D.mawCygb-1. These data are presented here, for the first time, in a structure–function comparative analysis between cold-adapted Cygbs-1 and human CYGB.
2. Materials and Methods
2.1. Protein expression and purification
D.mawCygb-1 and C.aceCygb-1 were cloned from retina and brain tissues, respectively, and the recombinant proteins and their mutants (D.mawCygb-1* and C.aceCygb-1*), where Cys residues were replaced by Ser (Cys38Ser/Cys160Ser double mutants), were expressed and purified, as previously described [24].
2.2. O2 affinity
O2-binding curves of Fe(II) C.aceCygb-1 and D.mawCygb-1, as well as CYGB for comparison, were measured in 100 mM sodium phosphate, 1 mM EDTA, 5 mM DTT, 5 mM sodium ascorbate, 3000 U/ml catalase at pH 7.0, at 20 °C. Cygbs were at 30 μM concentration. A minimal amount of an antifoam agent (Antifoam 204, Sigma-Aldrich) was also added. O2 partial pressures in the range 0–7 Torr were generated with an Environics 4000 gas mixer by mixing pure helium with 0.187% and 1.19% O2/helium mixtures. The protocol was modified from previous applications [68] by bubbling the humidified gas mixture directly into the protein solution until equilibration was reached. Spectra were analyzed in the 380–480 nm range as a linear combination of reference spectra, i.e. the spectra of the pure deoxy and oxy forms [68]. The absorption spectra of the deoxyCygb forms were collected in the presence of 1 mM sodium dithionite, whereas those of the oxyCygb forms were collected after exposure to 7 Torr O2. In the presence of sodium ascorbate and catalase, which slowly reduces the oxidized heme, the Fe(II) form was stable enough to allow for the equilibration of each sample at 5 different O2 partial pressures with negligible formation of metCygb. Reversibility of the oxygenation was tested for each experiment.
2.3. Kinetics studies
Flash-photolysis and stopped-flow measurements were performed to gain insight into the kinetics of ligand rebinding and the equilibrium between bis-hystidyl hexa-coordinated and penta-coordinated states, respectively.
For the study of CO-rebinding kinetics, flash-photolysis experiments were performed using a laser photolysis system (Edinburgh Instruments LP920) with a frequency-doubled Q-switched Nd:YAG laser (Spectra Physics Quanta-ray, 532 nm). Temperature-dependent experiments were performed at 10, 20 and 40 °C on solutions equilibrated with CO pressures of either 1 atm or 0.1 atm on a home-made laser flash photolysis setup that was previously described [1]. CO-bound samples were prepared in sealed 4x10 mm quartz cuvettes containing 1 ml of 100 mM potassium phosphate buffer, 1 mM EDTA at pH 7.0. The buffer was equilibrated with mixtures of CO and N2 at different ratios to obtain different CO concentrations (200, 400, 600 and 800 µM), using a High-Tech system (Bronkhorst). A saturated sodium dithionite solution (10 µl) was added to the buffer to scavenge residual O2. Protein was injected in a minimum amount to a final concentration of approximately 5 µM. The CO-ligated form was verified by UV/Vis spectroscopy. Recombination of photo-dissociated CO was monitored following the absorbance changes at 436 nm. At this wavelength, the absorbance change reflects only the spectral difference between the deoxy- and the carboxy- species, thus providing a measure of the concentration of deoxy- molecules as a function of time. At 436 nm, the spectral contribution of the bis-histidyl, hexa-coordinated species is negligible as demonstrated in other similar proteins [1]. Singular Value Decomposition of the spectral evolution after photolysis of other hexa-coordinated Hbs demonstrated that at 436 nm only the deoxy-carboxy spectral change contributes to the observed signal. However, it should be emphasized that the kinetics will any way reflect all reaction steps modulating rebinding of CO to the heme, including formation and decay of the bis-histidyl, hexa-coordinated intermediate. Data preprocessing was conducted as described [1]. Analysis of the kinetics with a sum of three exponential decay functions was performed using Origin (OriginLab Corporation, Northampton, MA, USA). From the lifetimes τi, apparent rates ki = 1/τi were calculated, and kON,CO was determined from the CO concentration dependence of the observed rate constant for the bimolecular phase. In order to improve confidence on retrieved parameters, kON, CO values obtained at each temperature from laser flash photolysis were used in the data fitting.
Stopped-flow studies were performed to determine the association and dissociation rate constants of distal His, kON,His and kOFF,His, respectively. Stopped-flow measurements were performed at 10, 20, and 40 °C in 100 mM degassed potassium phosphate buffer and 1 mM EDTA at pH 7.0, by using a thermostated stopped-flow apparatus (Applied Photophysics, Salisbury, UK). Sodium dithionite was added to both the protein and the CO solutions to a final concentration of 5 mM. Measurements were carried out at 418 nm. 5 µM protein solution was mixed with different CO concentrations (50, 100, 150, 200, 250 and 400 µM final concentrations). Analysis was performed using Origin (OriginLab Corporation, Northampton, MA, USA). Progress curves were fitted using an exponential decay. The observed rate constants kobs were then plotted vs CO concentration to retrieve kON,His and kOFF,His from the expression of the observed rate kobs for CO binding to hexa-coordinated Cygbs-1 [20].
2.4. Enzymatic studies
The enzymatic properties of Cygbs-1 were examined considering two chemical processes: the isomerization of peroxynitrite, and the nitrite-reductase activity.
Kinetics of peroxynitrite isomerization, in the absence and presence of C.aceCygb-1-Fe(III) and D.mawCygb-1-Fe(III), were recorded at 302 nm (ε = 1.705 × 103 M−1 cm−1) by rapidly mixing the buffer solution (1.0 × 10−1 M bis-Tris-propane buffer, pH 7.4) or the Antarctic Cygbs-1-Fe(III) (C.aceCygb-1 final concentration, 5.0 × 10−6 M to 2.5 × 10−5 M and D.mawCygb-1 final concentration 4.0 × 10−6 M to 2.0 × 10−5 M; 1.0 × 10−1 M bis-Tris-propane buffer, pH 7.4) with the peroxynitrite solution (final concentration, 2.5 × 10−4 M) [8], [38], [45], [46].
Values of the pseudo-first-order rate constant for peroxynitrite isomerization, in the absence and presence of C.aceCygb-1-Fe(III) and D.mawCygb-1-Fe(III), (i.e. k) were determined from the analysis of the time-dependent absorbance decrease at 302 nm, according to Eq (1):
| (1) |
With regard to nitrite reductase activity, kinetic data were obtained by rapid mixing the C.aceCygb-1-Fe(II) or D.mawCygb-1-Fe(II) solutions (final concentration 3.8 × 10−6 M and 3.6 × 10−6 M, respectively) with the NO2– solution (final concentration 2.0 × 10−3 M to 2.0 × 10−2 M for C.aceCygb-1, and 1.5 × 10−3 M to 2.0 × 10−2 M for D.mawCygb-1), in the presence of sodium dithionite (final concentration 2.0 × 10–3 M) at pH 7.4 (5.0 × 10−2 M bis-Tris-propane buffer) and 20 °C. Na2S2O4 concentration lower than 5.0 × 10–3 M does not effectively reduce NO2– to NO [39]. Kinetics were monitored between 370 and 460 nm. No gaseous phase was present.
Kinetics of NO2–-mediated conversion of C.aceCygb-1-Fe(II) and D.mawCygb-1-Fe(II) to Fe(II)–NO was analyzed in the framework of the minimum reaction Scheme 1 [9], [11], [39], [47], [48], [67], [73], [76], [78], [79].
Scheme 1.
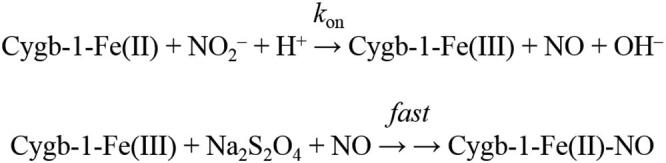
Values of the pseudo-first-order rate constant for the NO2–-mediated conversion of C.aceCygb-1-Fe(II) and D.mawCygb-1-Fe(II) to Fe(II)–NO (i.e., k) were determined from data analysis, according to Eq (2):
| (2) |
Values of the second-order rate constant for the NO2–-mediated conversion of Antarctic Cygbs-1-Fe(II) to Fe(II)–NO (i.e., kon) were obtained from the dependence of k on the NO2– concentration (i.e., [NO2–]), according to Eq (3):
| (3) |
The value of kon refers to the interaction of NO2– with C.aceCygb-1-Fe(II) and D.mawCygb-1-Fe(II), under the general assumption that this process is the rate-limiting step of the overall reaction (Scheme 1).
2.5. Crystallographic and molecular dynamics structural analyses
D.mawCygb-1* crystals grew at 293 K after several weeks using the hanging-drop vapor-diffusion setup under the following crystallization conditions: 2 M ammonium sulfate, 0.1 M Tris, pH 8.5. D.mawCygb-1* crystals were transferred to a 2.5 M ammonium sulfate stabilizing solution, supplemented with 20% (v/v) glycerol for cryoprotection, before data collection. X-ray diffraction data were collected at the ESRF (Grenoble, France – Beam line ID23-1) up to 3.0 Å resolution, reduced and scaled using XDS [49] and Scala [30].
D.mawCygb-1* 3D structure was solved by Molecular Replacement using Phaser [60], taking the CYGB 3D structure as search model (PDB code: 1UMO). The protein structure was refined with Refmac [61] to a final R-factor and R-free values of 18.9% and 24.5%, respectively. The D.mawCygb-1* atomic coordinates have been deposited in the Protein Data Bank (www.rscb.org) with access code 6Q6P.
Molecular Dynamics (MD) simulations of D.mawCygb-1 in the hexacoordinated bis-histidyl and ligand-bound states were performed with the AMBER14 package [16]. Simulations were performed at 283 and 298 K, keeping the pressure at 1-bar, using Berendsen thermostat and barostat. The proteins were simulated in the ligand bound form for 1.5 µs at each temperature. The ff99SBildn [54] force field was used for all protein residues, whereas previously developed and thoroughly tested parameters were used for the heme [59]. Every complex was immersed in an octahedral box of TIP3P water molecules and counter ions were added to neutralize the system. A minimum distance of 12 Å between any atom of the protein and the box limits was used to define the size of the simulated system. The system was energy minimized following a three-step protocol that involved first the hydrogen atoms of the protein, then the water molecules, and finally the whole system. At this point, the system was thermalized to 283.15, 298.15 and 313.15 K in 4 steps of 50 ps each from an initial temperature of 100 K. The last snapshot from the thermalization procedure was used as starting point for MD simulations using the NPT ensemble, a 2 fs time-step, periodic boundary conditions and keeping frozen all the bonds implying hydrogen atoms using SHAKE. Frames were collected at 1-ps intervals, taking the last 250 ns of every MD, which were subsequently used to analyse the trajectories.
The topology of inner cavities in the X-ray structure was determined with CAVER [19], using the default minimum radius of the probe, which is set to 0.9 Å. This allows to identify small pockets in the interior of the protein, which may be apt to bind single atoms (i.e., Xe atoms, with an atomic volume close to 40 Å3). In contrast, the occurrence of inner cavities and tunnels originated from protein fluctuations along the MD simulation was examined with MDpocket [72]. In this case, computational analyses were performed for the snapshots sampled in the last 250 ns of the trajectory using the default parameters that set the minimum radius of the probe to 3.0 Å, thus avoiding searching for too small pockets, and facilitating the identification of putative tunnels connecting inner cavities, or even access from the protein surface.
Finally, following previous studies [82], [83], [84], a series of 20 independent MD simulations were run to explore the migration of an unrestrained diatomic ligand through the interior of the protein matrix. Simulations were started from the last snapshot of the trajectory run for the ligand-bound hexa-coordinated state of C.aceCygb-1. The ligand was described using a three-site model [85] and was placed at random positions around the inner distal edge of the heme. MD simulations were run up to 50 ns at 283.15 K using the same conditions described above.
3. Results
3.1. Spectral properties and O2 affinity
The Fe(III) forms of C.aceCygb-1 and D.mawCygb-1 show the Soret band at 415 nm and the β and α Q-bands at 531 and 562 nm, respectively [24], typical of hexa-coordinated bis-histidyl species, as previously observed in Fe(III) CYGB [69]. The Fe(II) oxygenated forms of C.aceCygb-1 and D.mawCygb-1 have the Soret band at 420 nm, the β Q-band at 536 nm and a shoulder corresponding to the α Q-band at 563 nm (Supporting Information Figure S1). Upon exposure to helium for 3 h in the presence of 5 mM ascorbate and 3000 U/ml catalase, the spectrum of a typical internally hexa-coordinated heme is observed, with absorption maxima at 425 nm for the Soret band and 531 and 559 nm for the Q bands, as reported for the Fe(II) deoxy Cygb forms [24]. The spectra were identical to those obtained in the presence of 1 mM sodium dithionite (data not shown).
The binding isotherms fitted to the O2-binding curves of C.aceCygb-1, D.mawCygb-1 and CYGB were determined by equilibrating the protein solution with O2\helium mixtures at different O2 partial pressures at 20 °C and pH 7.0. The analysis of the binding curves (Fig. 1) yielded p50 (O2 partial pressure required to achieve half saturation) values of 0.08 ± 0.01 and 0.13 ± 0.02 Torr, for C.aceCygb-1 and D.mawCygb-1 respectively, which are 1.8- and 2.9-fold larger than the p50 value for CYGB measured under the same experimental conditions (0.045 ± 0.006 Torr). This finding is consistent with the general observation that cold-adapted globins exhibit lower O2 affinity relative to their mesophilic counterparts (28 and references within, [32].
Fig. 1.
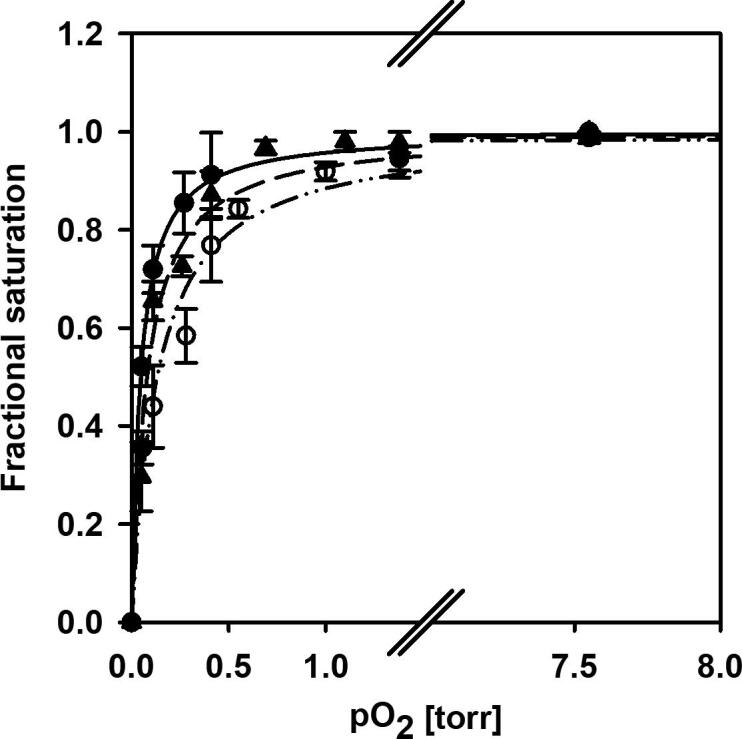
O2-binding curves of CYGB (closed circles), C.aceCygb-1 (closed triangles) and D.mawCygb-1 (open circles) at 20 °C in 100 mM sodium phosphate, 1 mM EDTA, 5 mM DTT, 5 mM sodium ascorbate, 3000 U/ml catalase at pH 7.0. The experimental points are the fractional saturations, determined by analysis of the spectra at each O2 partial pressure as a linear combination of the deoxy and oxy spectra in the range 380–480 nm. The lines present fittings of the experimental points to binding isotherms. p50 values of 0.08 ± 0.01, 0.13 ± 0.02 and 0.045 ± 0.006 Torr were determined for C.aceCygb-1, D.mawCygb-1 and CYGB, respectively.
3.2. CO-binding kinetics
Nanosecond laser-flash photolysis experiments using the CO model ligand were performed to determine the rate constants for binding of exogenous ligands to the heme, estimate the rate of escape to the photodissociated ligand to the solvent, and assess the competition with the internal distal His ligand.
The CO-rebinding kinetics of Cygb-1 solutions were recorded as a function of CO concentration and temperature at pH 7.0 (representative kinetics at 20 °C are shown in Fig. 2). The observed kinetics for D.mawCygb-1, C.aceCygb-1 and CYGB show the same general properties, with three kinetic phases clearly distinguishable. Experiments on D.mawCygb-1 and C.aceCygb-1 treated with dithiothreitol (DTT) to reduce the disulfide bridges yielded results indistinguishable from those obtained with untreated samples, and from those obtained with the mutants D.mawCygb-1* and C.aceCygb-1* (data not shown), suggesting that the intermolecular disulfide bridges, responsible only in part for their oligomerization [24], do not affect the kinetic properties of Antarctic Cygbs-1.
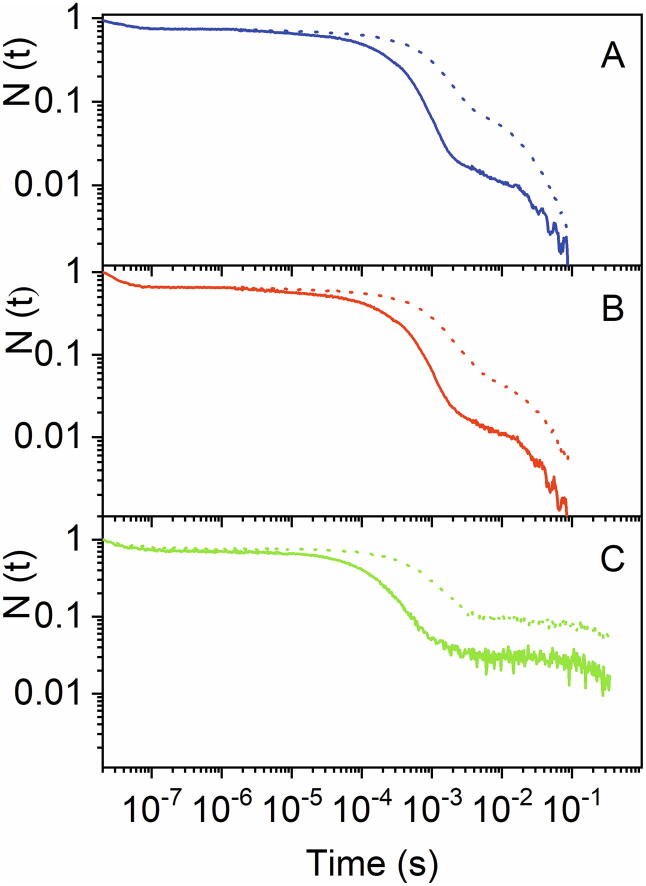
Fig. 2.
CO-rebinding kinetics of D.mawCygb-1 (panel A, blue) and C.aceCygb-1 (panel B, red) solutions equilibrated in 100 mM potassium phosphate buffer, 1 mM EDTA at pH 7.0, at 800 µM CO (solid lines) and 200 µM CO (dotted lines), and CYGB (panel C, green) equilibrated at 1 mM CO (solid line) and 100 µM CO (dotted lines), at 20 °C [35]. Data are reported as the progress curve representing the fraction of deoxy molecules, N(t), as a function of time after photolysis. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
From the CO-concentration dependence and steady-state absorption of the CO-bound and ligand-free Cygb samples, the nature of the three exponential decays can be identified as follows (Scheme 2). The geminate rebinding, with a CO-independent rate and amplitude, is observed on the nanosecond time scale. The bimolecular rebinding to the penta-coordinated Cygb (Cygbp) can be identified with a CO-dependent rate and amplitude on the long microsecond-to-millisecond time scale. However, the CO rebinding is competitive with the formation of the hexa-coordinated bis-histidyl species (Cygbh). Therefore, the last phase, occurring on the long milliseconds, corresponds to the distal His dissociation from the heme, concomitant with the full rebinding of CO to the heme. There is no indication of rebinding on faster time scales, as the overall absorbance change at the end of the laser pulse is roughly equal to the expected difference between carboxy- and deoxy- species. To completely rule out the existence of rebinding phases in the picosecond time scale, pump–probe experiments with sub-nanosecond excitation would be necessary and are beyond the scope of this general kinetic characterization.
Scheme 2.
Kinetic model for the analysis of the CO-rebinding kinetics to Cygb after nanosecond laser photolysis. Cygbp and Cygbh stand for penta- and bis-histidyl hexa-coordinated species, respectively. Cygbp: CO is the photodissociated Cygb in the penta-coordinated state, with CO still within the protein matrix.
The response of the rebinding curves to temperature shows the typical behavior observed for similar proteins. The amplitude of geminate rebinding decreases at increasing temperatures with a concomitant increase in the apparent rate. The bimolecular rebinding phase undergoes an increase in rate with temperature, but the increasing level of hexa-coordination suggests that the rate of His binding to the heme has a larger activation energy. Finally, His dissociation from the heme speeds up remarkably at increasing temperature.
CO binding to Cygbh was also investigated by stopped flow experiments as a function of CO concentration and temperature (Supporting Information Figure S2), this process being well described by a single exponential relaxation. The observed rate kobs for CO binding to the hexa-coordinated Cygbs is given by Eq (4) [20]:
| (4) |
where the overall rebinding rate to Cygbp (kON,CO) is related to microscopic rates k-1, k2, and k-2 (Scheme 2; values given in Supporting Information Table S1) as noted in Eq (5):
| (5) |
The integrated analysis of the kinetics yielded the kON,CO, kON,His, and kOFF,His (see Materials and Methods). The bimolecular rate constants of CO binding at 20 °C to penta-coordinated species (kON,CO) are similar in D.mawCygb-1 and C.aceCygb-1 (Table 1), with values only slightly smaller than in CYGB [35], [51], [80]. Data in Table 1 also show that the rate constants for His binding (kON,His) are similar in cold-adapted globins and CYGB. However, the rate constants of His dissociation (kOFF,His) are remarkably larger in D.mawCygb-1 and C.aceCygb-1 than in CYGB over the whole temperature range investigated (Supporting Information Figure S3). As a consequence, the resulting equilibrium binding constant (KHis) is much lower in D.mawCygb-1 and C.aceCygb-1 than in CYGB. The values of kobs, which afford an estimate of the apparent ligand-binding rate to hexa-coordinated proteins, at 20 °C (assuming [CO] = 1 mM; Table 1) are 28 s−1 in C.aceCygb-1 and 34 s−1 in D.mawCygb-1, much higher than in CYGB (1.8 s−1) [35]. Due to the very different temperature dependencies (Supporting Information Figure S3), the linear regression extrapolation to the physiological temperatures experienced by the Antarctic fish (around 0 °C) indicates that the distal His dissociation rates for D.mawCygb-1 and C.aceCygb-1 become more similar to that of CYGB, thus resulting in closer, although not identical, equilibrium binding constants.
Table 1.
Kinetic parameters of Antarctic fish Cygbs-1 and CYGB at 20 °C.
| kON,CO (µM−1 s−1)A | kgem(107 s−1) A | Fgem (%)A | kON,His(s−1) B | kOFF,His(s−1) BC | KHisB | kobs(s−1)D | |
|---|---|---|---|---|---|---|---|
| D.mawCygb-1 | 3.4 ± 0.3 | 5.0 ± 0.5 | 38 ± 3 | 149 ± 16 | 25 ± 1 | 6.4 ± 0.9 | 28 |
| C.aceCygb-1 | 2.7 ± 0.3 | 6.8 ± 0.5 | 47 ± 3 | 137 ± 16 | 38 ± 1 | 4.7 ± 0.8 | 34 |
| CYGBE | 6.5 ± 0.5 | 5.0 ± 0.5 | 27 ± 5 | 149 | 1.8 | 83 | 1.8 |
AData from laser flash photolysis
BData from stopped flow
cThe rate kOFF,His is identical to that obtained from fitting of the slowest kinetic phase in flash photolysis.
DCalculated for CO = 1 atm.
EData from [35].
The overall geminate rebinding rate kgem is defined as kgem = k2 + k-1, and the amplitude Fgem is related to rate constants in Scheme 1 through Eq (6):
| (6) |
The amplitudes of geminate rebinding in C.aceCygb-1 and in D.mawCygb-1 are only slightly larger than in CYGB (Table 1). The estimated exit rate k2 appears a little lower in C.aceCygb-1 and D.mawCygb-1 than in CYGB, a fact that favors geminate rebinding to solvent escape. Since the rate for the innermost binding step (k-1) is similar, the lower probability of escape to the solvent (k2) in the cold-adapted proteins could arise from topologically or dynamically hindered escape route through the articulated tunnels, connecting the distal pocket with the solvent (see below).
The activation free energies (Table 2, calculated at 20 °C) were estimated from the temperature dependence of the rate constants reported in Table 1 using the Eyring expression (ln(hki/kBT) = ΔSǂ/R − ΔHǂ/RT, where ΔSǂ is the activation entropy, ΔHǂ is the activation enthalpy, h is the Planck’s constant, kB is the Boltzmann constant, and R is the gas constant. The activation free energy barriers for geminate rebinding are quite similar (about 7 kcal/mol) and comparable to the one determined for CYGB [35]. However, the free energy barrier for kON,CO in CYGB is substantially smaller than those in D.mawCygb-1 and C.aceCygb-1, leading to faster CO rebinding to the penta-coordinated species in the human protein (see Table 1 and Fig. 2).
Table 2.
Overview of the activation free energies ΔGǂ (kcal/mol) calculated at 20 °C.
| D.mawCygb-1 | C.aceCygb-1 | CYGB | |
|---|---|---|---|
| kgem | 7 ± 1 | 7 ± 1 | 6.9 ± 0.2 |
| kON, CO | 13 ± 1 | 13 ± 1 | 7.9 ± 0.2 |
| KOFF, His | 15.3 ± 0.3 | 15.1 ± 0.2 | 16.3 ± 0.1 |
| KON, His | 14.2 ± 0.2 | 14.2 ± 0.1 | 14.1 ± 0.2 |
The free-energy barriers for binding and dissociation of the distal His are quite similar in D.mawCygb-1 and C.aceCygb-1. The energy barrier for His dissociation (~15.2 kcal/mol) is larger by about 1.0 kcal/mol than that for His binding (~14.2 kcal/mol). The free-energy barrier for His binding in CYGB is almost identical (14.1 kcal/mol), whereas the barrier for His dissociation is larger (16.3 kcal/mol), which leads to slower decay of the bis-histidyl hexa-coordinated species in Fig. 2C.
3.3. Enzymatic functionality
Peroxynitrite, the product of the reaction between NO and superoxide is a reactive and short-lived species that promotes oxidative molecular and tissue damage. The harmful effects of peroxynitrite and related reactive species are decreased by hemoproteins that are pivotal for scavenging peroxynitrite in vivo to protect cells from its detrimental effect [7] and references within). To evaluate whether the Antarctic proteins are involved in scavenging of RNS, we measured their ability to catalyze the isomerization of peroxynitrite, as well as their nitrite-reductase activity.
As reported for Fe(III) hexa-coordinated NGB [44]) and horse-heart cytochrome c [4], [5],D.mawCygb-1-Fe(III) and C.aceCygb-1-Fe(III) do not catalyze peroxynitrite isomerization (Fig. 3), which in turn appears to be a common feature of penta-coordinated Fe(III) heme proteins [2], [3], [4], [5], [6], [44]. On the other hand, D.mawCygb-1-Fe(II) and C.aceCygb-1-Fe(II) catalyze the conversion of NO2– to NO with concomitant nitrosylation of the metal center (Fig. 4), as in both Fe(II) penta- and hexa-coordinated hemoproteins [22], [43], [48], [53], [67], [76], [78], [79] (Table 3).
Fig. 3.
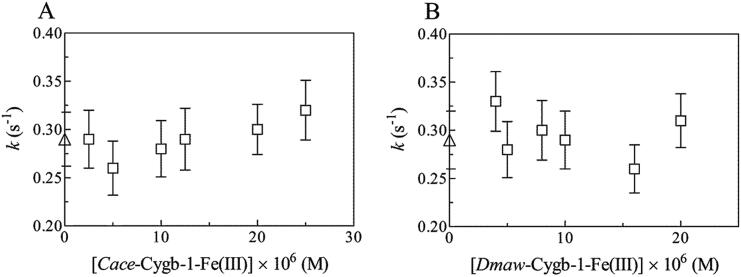
Dependence of k for peroxynitrite isomerization on the concentration of C.aceCygb-1-Fe(III) (A) and D.maw-Cygb-1-Fe(III) (B), at pH 7.4 and 20 °C. The triangle on the ordinate indicates the value of k in the absence of Antarctic Cygbs-1-Fe(III). The peroxynitrite concentration was 2.5 × 10−5 M.
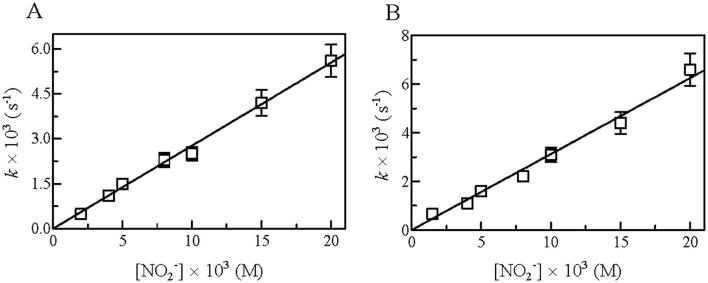
Fig. 4.
Dependence of k on the NO2– concentration for the NO2–-mediated nitrosylation of C.aceCygb-1-Fe(II) (A) and D.mawCygb-1-Fe(II) (B), at pH 7.4 and 20 °C. The analysis of data according to Eq(3) allowed the determination of kon values, (2.8 ± 0.3) × 10–1 M−1 s−1 and (3.1 ± 0.4) × 10–1 M−1 s−1) for the NO2–-mediated nitrosylation of C.aceCygb-1-Fe(II) (A) and D.mawCygb-1-Fe(II) (B), respectively.
Table 3.
Values of the second-order rate constant for the nitrite-reductase activity of hemoproteins.
| Heme-protein | kon (M−1 s−1) |
|---|---|
| C.aceCygb-1 | 2.8 × 10–1 |
| D.mawCygb-1 | 3.1 × 10–1 |
| D. rerio Cygb-1A | 14.2 |
| D. rerio Cygb-2A | 3.1 × 10–1 |
| Synechocystis HbB | 6.8 × 101 |
| Rice nonsymbiotic Hb class 1B | 3.3 × 101 |
| Arabidopsis thaliana Hb class 1C | 2.0 × 101 |
| Arabidopsis thaliana Hb class 2C | 4.9 |
| Carp Mb-1D | 5.3 |
| Carp Mb-2D | 1.8 |
| Horse-heart MbE | 2.9 |
| Sperm-whale MbF | 6.0 |
| Mouse NgbG | 5.1 |
| CYGBH | 1.4 × 10–1 |
| NGB CysCD4-CysD5I | 1.2 × 10–1 |
| NGB CysCD4/CysD5L | 1.2 × 10–2 |
| Human Hb - T stateF | 1.2 × 10–1 |
| Human Hb - R stateF | 6.0 |
pH 7.4, 25 °C [22]
pH 7.0; unknown temperature [76]
pH 7.4 and 25 °C [79]
pH 7.6 and 25 °C [43]
pH 7.4 and 25 °C [78]
pH 7.4 and 25 °C [47]
pH 7.4 and 25 °C [67]
pH 7.0 and 25 °C [53]
pH 7.4 and 25 °C [78]. “Human Ngb CysCD4-CysD5”, the CysCD4 and CysD5 residues form an intramolecular disulfide bond.
pH 7.4 and 25 °C. “Human Ngb CysCD4/CysD5”, the CysCD4 and CysD5 residues do not form the intramolecular disulfide bond.
The NO2–-mediated conversion of D.mawCygb-1-Fe(II) and C.aceCygb-1-Fe(II) to Fe(II)–NO corresponds to a monophasic process, and values of the pseudo-first-order rate constant (i.e. k) increase linearly with the NO2– concentration (Fig. 4). The analysis of data shown in Fig. 4 (Eq (3); see Materials and Methods) allowed the determination of kon values (corresponding to the slope of the linear plots) for the NO2–-mediated nitrosylation of D.mawCygb-1-Fe(II) and C.aceCygb-1-Fe(II), leading to values of (3.1 ± 0.3) × 10–1 M−1 s−1 and (2.8 ± 0.2) × 10–1 M−1 s−1, respectively. The formation of the transient D.mawCygb-1-Fe(III) and C.aceCygb-1-Fe(III) species (Scheme 1), which are quickly converted to Cygb-1-Fe(II) by dithionite, is the rate-limiting step of NO2–-mediated conversion of Cygb-1-Fe(II) to Fe(II)–NO.
Values of the apparent second-order rate constant for the nitrite reductase activity of ferrous heme-proteins and heme model compounds (i.e. kon) range between 10−2 M−1 s−1 and 102 M−1 s−1, reflecting the different structural features of the heme pocket and/or the structural arrangement of the energetic barriers along the ligand pathway toward the heme [10].
3.4. Structural analysis of Cygb-1
The crystal structure of D.mawCygb-1* Fe(III) form displays two molecules in the asymmetric unit (Table 4). However, their contact interface is minimal, largely solvent exposed, and based on a pair of salt bridges [Asp21(A2) - Lys111(F6), and Asp97(EF hinge) - Lys107(F2)], suggesting that the dimeric assembly is merely the result of crystal packing.
Table 4.
Data collection and refinement statistics for D.mawCygb-1.
| Data collection statistics | |
|---|---|
| Resolution limit (Å) | 81.3–3.0 |
| Space group | P212121 |
| Unit-cell parameter (Å) | a = 43.6, b = 105.7, c = 127.3 |
| Total reflections | 152,686 |
| Unique reflections | 12,427 |
| Completeness (%) | 99.9 (99.8)* |
| Multiplicity | 12.3 (12.1) |
| Rmerge† (%) | 24.2 (78.4) |
| Average I/σ(I) | 8.7 (4.2) |
| Refinement statistics and model quality | |
| Resolution range (Å) | 81.3 – 3.0 |
| Protein atoms | 1238 in chain A, 1243 in chain B |
| Heme | 1 per chain |
| Water molecules | 15 |
| R-factor/R-free (%) | 18.9/24.5 |
| R.m.s.d. deviation from ideal geometry: | |
| bond lengths (Å) | 0.010 |
| bond angles (°) | 1.46 |
| Ramachandran plot (%): | |
| most favored region | 95 |
| additional allowed region | 5 |
*Outer shell statistics (3.16 – 3.00 Å) are shown within parentheses.
†Rmerge = ΣhΣi |Ihi –< Ih >|/ΣhΣiIhi.
D.mawCygb-1 shares 59% overall sequence identity with CYGB (Fig. 5), which increases to 64.3% for 155 residues (Ser17 through Ser172) that build the 3-on-3 globin fold (81.9% if conservative mutations are accounted for in this region). Accordingly, overlay of the 3D structures yields a root-mean square (rms) deviation of 0.85 Å over 153 Cα atoms, the largest local deviations occurring in the DE and GH hinges (2 – 2.5 Å). If structural overlay of D.mawCygb-1* is run vs. the CYGB mutant bearing CysB2 [38] → Ser and CysE9 [83] → Ser substitutions (CYGB*; PDB ID 1URY; 25) in its Xe adduct, the rms deviation drops slightly (to 0.76 Å) for 146 Cα pairs, due to larger deviations (3.1 – 3.5 Å) in the GH hinge region that is excluded from the overlay. Comparison of the averaged B-factor profiles for human CYGB structures and D.mawCygb-1* does not show differences suggesting varied structural flexibility at both global and local levels for D.mawCygb-1* as far as the crystallographic data can support (Supporting Information Figure S4).
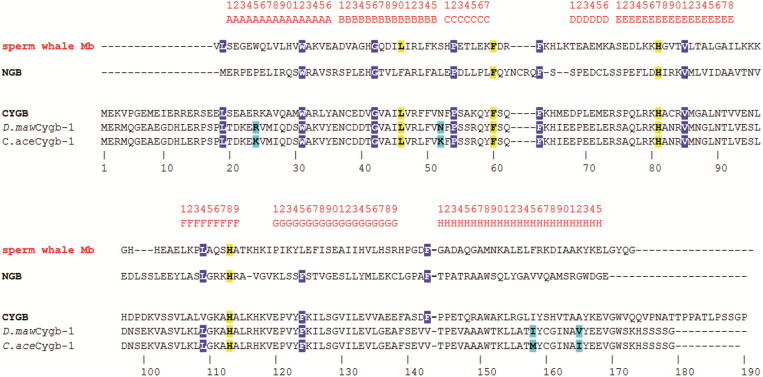
Fig. 5.
Structure-based sequence alignment of D.mawCygb-1, C.aceCygb-1 and CYGB versus sperm-whale Mb and NGB, taken as reference sequences for penta-coordinated and hexa-coordinated globins, respectively. Secondary structure topological sites (as defined in the classical globin fold) are shown in red in the top line. Conserved residues relevant for the distal and proximal sites are highlighted in yellow; residues relevant for structural conservation of the 3-over-3 globin fold are in blue; residue differences between the two Antarctic fish Cygbs-1 are in cyan. . (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Adapted from [27]
In agreement with the strong sequence and overall structural similarity between CYGB and D.mawCygb-1, non-conservative residue substitutions occur essentially on the protein surface. One exception is the CYGB Arg155(H12) mutation to Leu in D.mawCygb-1, which results in side chain re-orientation toward the protein core. Among substitutions that affect the globin core, the replacement of Met30(A11) in CYGB with Ser in D.mawCygb-1 is worth noting; residue 30 falls next to Trp151(H8) indole ring in D.mawCygb-1*, and the Ser30(A11) hydroxyl O atom forms a H-bond with the Trp151 indole NH group.
As for CYGB, the heme Fe atom is hexa-coordinated in D.mawCygb-1*, the proximal and distal His being the axial ligands with coordination bonds of 2.1 Å. Both His ligands are staggered relative to the heme pyrrole N atoms, i.e. roughly oriented along the lines connecting opposing heme methinic bridges. A comparison of all hexa-coordinated CYGBs with D.mawCygb-1* shows that residues building the distal cavity are highly conserved, which in turn translates into conservation of the side chain conformations (for residues on the distal and proximal sides and within 10 Å from the heme Fe atom; Supporting Information Figure S5). The only notable residue change - CYGB Val109(F4) → D.mawCygb-1 Leu109(F4) - may provide a contact to a heme methyl group on pyrrole ring B, promoting a slight porphyrin ring rotation (roughly pivoting on the heme Fe atom) in D.mawCygb-1*. At the light of the different experimental resolutions associated with the crystal structures (3.0 Å and 1.68 Å, respectively), further fine conformational differences are difficult to establish.
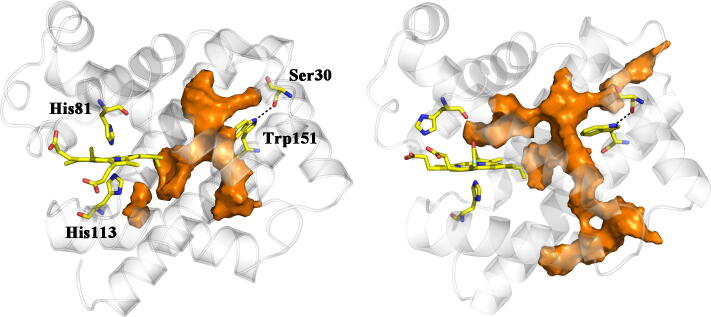
Inspection of D.mawCygb-1* protein core shows that a main cavity (124 Å3) is present in the deeper part of the heme pocket (Fig. 6), next to the heme vinyl group (on pyrrole ring B) and to Val85(E11), Leu89(E15), Leu109(F4), Phe124(G5), Leu127(G8), and Ile131(G12). Although not exactly matching, it resembles a slightly larger cavity (146 Å3) observed in CYGB*, which is the site for two bound Xe atoms (Xe2 and Xe3; 26). As for CYGB*, where smaller cavities are present (in fact the protein binds 4Xe atoms), three minor cavities are observed in D.mawCygb-1*, next to the inner distal heme face (30 Å3; this cavity has a match in CYGB*), and around Trp151(H8) (48 and 21 Å3, respectively, both likely related to the spatially close Met30(A11) → Ser mutation mentioned above).
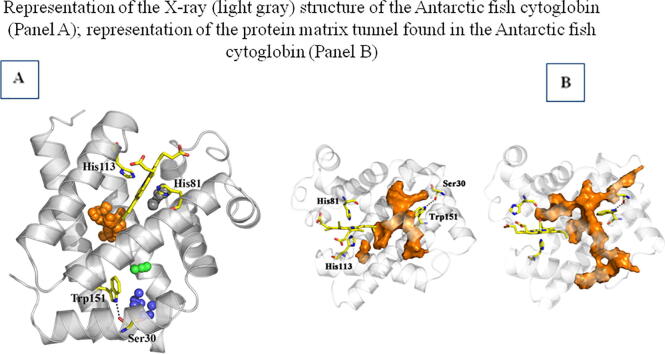
Fig. 6.
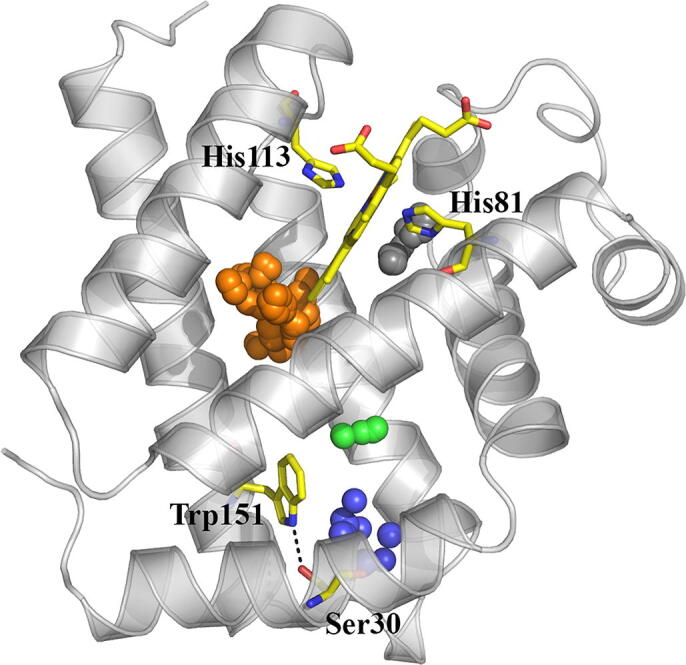
Representation of the X-ray (light gray) structure of D.mawCygb-1* (backbone skeleton shown as cartoon), together with the cavities observed in the X-ray structure (shown as clusters of spheres in orange, 124 Å3; blue, 48 Å3; black, 30 Å3; green, 21 Å3) mapped with CAVER. Selected residues are shown as yellow sticks, including the heme with distal and proximal His, and the H-bonded residues Ser30 --- Trp151 (dashed line). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In order to examine the effect of thermal fluctuations on the nature of inner cavities, MD simulations (1.5 μs) were performed for D.mawCygb-1 in both hexa-coordinated bis-histidyl and ligand (CO)-bound states. Inspection of the trajectories supports the stability of the overall protein fold, as noted in rms deviations of the backbone atoms generally lower than 2 Å compared to the X-ray structure (data not shown). The analysis of the snapshots sampled in the last 250 ns of the simulation performed for the hexa-coordinated bis-histidyl species supports the existence of a branched tunnel, which should facilitate the transit of small gaseous ligand through the protein matrix. This passage connects the innermost edge of the heme with two putative exits, which are located i) between helix A and the hinge formed by helices G and H, close to the Trp151-Ser30 hydrogen-bonded pair, and ii) between helix H and the hinge defined by helices E and F (Fig. 7).
Fig. 7.
Representation of the protein matrix tunnel found in D.mawCygb-1 (corresponding to a frequency level of 50% as determined from MDpocket calculations; shown in orange) at 283 K for the hexa-coordinated bis-histidyl (left) and ligand (CO)-bound (right) states. The protein backbone is shown as cartoon, and the heme together with selected residues (His151 and Ser30) are shown as yellow sticks. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Detachment of the distal HisE7 [81] from the heme, with the concomitant shift in helix E, reshapes the inner tunnel, leading to the occlusion of the small pockets in the proximal side of the heme and opening of a cavity on the distal side, as well as to a better definition of the passage to the bulk solvent. At this point, it is worth noting the reshaping of the branched tunnel, leading to wider passages above the hydrogen-bonded Trp151-Ser30 pair, close to the GH hinge, but especially regarding the passage located close to the EF hinge (Fig. 7). A similar tunnel system was found for simulations performed for the ligand-bound hexa-coordinated species of C.aceCygb-1 (Supporting Information Fig. 6). The gross features of the inner tunnel, nevertheless, are already present in the hexa-coordinated bis-histidyl species, which could thus facilitate the functional adaptation to the cold environment.
To examine the feasibility of ligand migration through the branched tunnel, a series of independent (50 ns) MD simulations providing a cumulative total of 1 μs simulation time were analyzed to explore the diffusion of unrestrained O2 in the ligand-bound form of C.aceCygb-1. MD simulations (18outofthe20) led to ligand egression from the protein interior within the simulation time. Egression primarily occurred through the long branch of the tunnel, which involves the migration through the passage leading to the EF hinge, though different passages to the bulk solvent were observed at the end of this tunnel branch. Only two trajectories led to ligand egression through the passage located close to the GH hinge, and two other trajectories led to the ligand egression via the open side of the heme cavity. Therefore, although caution is required for a quantitative analysis, these results provide qualitative support to the functional implication of the branched tunnel for ligand migration in the ligand-bound hexa-coordinated state.
The experimental kON,CO values for the fish Cygbs are consistent with the system of open cavities present in the CO-bound state (Fig. 7, right), which provides multiple access points for the entry of the ligand. This scenario is similar to that described for CYGB [35], where a comparable system of tunnels was detected from the analysis of classical MD simulations, which supported the existence of a branched tunnel consisting of passages through the terminal side of helix A and the GH hinge, and between helix A and loop EF. In the current kinetic analysis, the rate kON,CO (and the microscopic rates k2 and k-2) should be considered an average measure of the separate rates for the multiple entry/exit pathways through the articulate tunnels connecting the distal pocket with the solvent. The resulting overall rates for D.mawCygb-1 and C.aceCygb-1 appear quite similar to that for CYGB (within a factor 2), and this difference only contributes to a minor extent to the observed large change in kobs.
4. Discussion
The function of Cygb is still unknown, and many roles in O2 transport, redox signaling and NO regulation have been ascribed to this recently discovered protein [64]. While in skeletal and cardiac muscle, Mb is strongly involved in NO scavenging [57], the preferential expression of CYGB over Mb in the vascular smooth muscle, and the evidence that it is a potent NO dioxygenase, efficiently and rapidly reduced by cellular systems, suggest an important and exclusive role of CYGB in the O2-dependent NO metabolism [55].
In contrast to the penta-coordinated zebrafish Cygb-1, supposed to act as O2 carrier and nitrite reductase [22], Antarctic fish Cygbs-1 exhibit a fully hexa-coordinated heme group, similar to Antarctic Cygb-2 (D. Giordano, personal communication), with: i) a high-O2 affinity, unlikely to support an O2-delivery role, and ii) a slow rate constant for nitrite-reductase activity. In fact, the O2 affinities reported in this study (p50 0.08 ± 0.01 and 0.13 ± 0.02 Torr, for C.aceCygb-1 and D.mawCygb-1, respectively, at 20 °C) are much higher than that of human Hb (p50 2.61 ± 0.05 Torr at 15 °C, pH 7.0) [14], but also higher than that of Mb (p50 0.78 ± 0.02 Torr at 25 °C, 0.17 ± 0.01 at 10 °C) [70], suggesting that O2 transport is functionally unlikely in Antarctic fish Cygbs-1. Under our experimental conditions, the measured p50 for CYGB (0.045 ± 0.006 Torr) was lower than a previously reported value close to 1 Torr at 20 °C [31]. In this respect, it should be considered that the formation of a disulfide bond is known to affect affinity [31], but CYGB used for these experiments was an engineered species devoid of Cys.
Our results also suggest that D.mawCygb-1-Fe(III) and C.aceCygb-1-Fe(III) do not catalyze peroxynitrite isomerization. The values of the second-order rate constant for NO2–-mediated conversion of D.mawCygb-1-Fe(II) and C.aceCygb-1-Fe(II) to Fe(II)–NO are reminiscent of those of the hexa-coordinated slow-reacting forms of CYGB, mouse Ngb, NGB, and D. rerio Cygb-2 [22], [53], [67], [78]. In contrast, the rate constant of penta-coordinated D. rerio Cygb-1, possibly playing a nitrite-reductase role [22], is more similar to those of Fe(II) Mb and Hb [43], [47], [76], [78], [79].
Ligand-binding kinetic studies highlight some remarkable differences between the rate constants of D.mawCygb-1 and C.aceCygb-1, on one hand, and CYGB on the other. First, it is worth noting the lack of effects of disulfide bridge reduction on ligand-binding kinetics of D.mawCygb-1 and C.aceCygb-1 (both globins can only form intermolecular disulfide bridges; this study and 24). In contrast, analysis of the effect of the internal disulfide bridge reduction in CYGB suggests major changes in kOFF,His. These distinctive features suggest that the functional role of CYGB can be regulated through the formation/disruption of the internal disulfide bridges depending on the cellular response to redox stress [12], while a similar regulation mechanism would be absent in the Antarctic Cygbs.
The amplitude of geminate rebinding in D.mawCygb-1 is intermediate between those observed in CYGB and C.aceCygb-1. The slightly larger rebinding observed in C.aceCygb-1 is due to a higher rebinding rate (k-1), which is only partially compensated by the escape rate (k2, Supporting Information Table S1). The changes in the rates k-1 and k2 between D.mawCygb-1 and C.aceCygb-1 may reflect subtle changes promoted by the topology of inner cavities mapped by MD, particularly related to the replacement of Ile158(H15) in D.mawCygb-1 by Met in C.aceCygb-1. Let us note that among the four mutations found between the sequences of the two proteins (Fig. 5), the Ile158 → Met mutation is the only one that faces the interior of the protein and contributes to shape the inner tunnel (Supporting Information Figure S7). Furthermore, Ile158 is located close to Leu155(H12), replacing Arg in CYGB, which is rearranged toward the hydrophobic interior of the protein, and is located close to the Trp151(H8) - Ser30(A11) hydrogen-bonded pair, an interaction absent in CYGB due to the Ser30(A11) → Met mutation. Overall, these differences could provide a favorable environment to host the ligand for longer times in fish Cygb-1, which could justify the slightly larger amplitudes of geminate rebinding relative to CYGB.
The most relevant difference between cold-adapted fish Cygb-1 and CYGB is the value of kOFF,His, which is 14- and 21-fold higher in D.mawCygb-1 and C.aceCygb-1 respectively, resulting from lower energy barriers for distal His dissociation in Cygb-1. This finding suggests that the structural and dynamic properties of the cold-adapted globins facilitate His(E7) dissociation, thus favoring the binding of exogenous ligands. Higher kOFF,His in D.mawCygb-1 and C.aceCygb-1, combined with the slightly lower kON,CO, yield kobs values (28 and 34 s−1 for D.mawCygb-1 and C.aceCygb-1, respectively) higher than in CYGB (1.8 s−1).
The values of kobs, for CO binding may suggest relevant consequences for multi-substrate reactions, e.g. NO dioxygenase, where sequential binding of O2 and NO is necessary to catalyze NO conversion to nitrate. Indeed, the binding of the first diatomic ligand (CO in our model system) in the hypothetic multi-substrate reaction may be much more efficient in cold-adapted fish than in Cygbs from mesophilic organisms at the same temperature. Similar features were also described in the mutants (Cys → Ser) of C.aceNgb* and D.mawNgb* when compared with NGB [36]). The values of kON were quite similar in C.aceNgb* (2.2 × 108 M−1 s−1) and D.mawNgb* (1.7 × 108 M−1 s−1), and about 2-fold higher than in human (7.7 × 107 M−1 s−1, [36], 5.0 × 107 M−1 s−1, [41] and zebrafish Ngbs (kON = 7 × 107 M−1 s−1, [33] . Furthermore, the combination of kON,His and kOFF,His values resulted in lower values of the equilibrium His-binding constant in cold-adapted Ngbs (KH = 476 in C.aceNgb*, KH = 255 in D.mawNgb*) than in Ngb from mesophilic organisms (KH = 1300 in NGB). It is therefore estimated that at 1 mM CO, kobs are 6 s−1 in C.aceNgb*, 3 s−1 in D.mawNgb*, and 0.2 s−1 in NGB [36]. We emphasize that these characteristics are similar to those reported in this work for Cygb-1 and CYGB, thus pointing to a general trend for cold-adapted hexa-coordinated globins.
Although the higher kobs values for the cold-adapted proteins are mainly a consequence of the high His dissociation rates, we suggest that the pre-formed tunnels in the hexa-coordinated state of Cygb-1 may also contribute to kobs by providing facile access of ligands to the distal pocket when the binding site becomes available (Fig. 7, left). Moreover, the structured cavities in the hexa-coordinated species may also assist multi-substrate reactions such as the NO dioxygenase, by providing a close-by reservoir of secondary reactants and sustaining catalytic turnover. Unfortunately, the very high auto-oxidation rates of most Antarctic O2-binding proteins [81], as well as that of D. rerio Cygb-2 [22], prevented measurements of NO di-oxygenation.
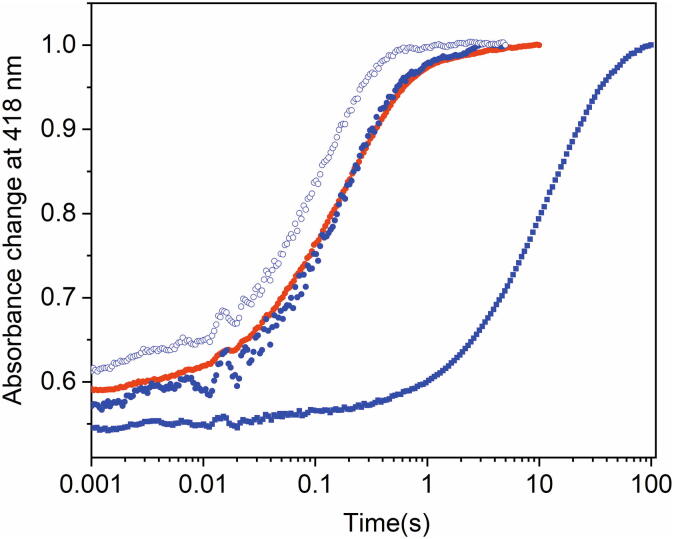
Overall, the general ligand binding mechanism in psychrophilic and mesophilic proteins discussed above appears to be conserved; however, our results show that in cold-adapted globins the combination of rate constants results in faster binding rates (kobs), suggesting that evolutionary pressure pushed these Cygbs towards an enhanced capability of exogenous ligand binding to the hexa-coordinated bis-histidyl state. Although the overall 3D structures of D.mawCygb-1* and CYGB appear very similar, cold adaptation of Antarctic fish Cygbs-1 may have encoded minor variations in the heme-pocket structure/dynamics, in line with the small differences observed in the redox potential, as well as minor changes in the (dynamic) pattern of inner cavities. Such considerations would be in keeping with the extreme Antarctic habitat conditions, and with the need to compensate for the low kinetic energy and motions dictated by the low temperature, as also shown by comparison of the stopped-flow traces of C.aceCygb-1 at 10 °C and CYGB at 40 °C (Fig. 8). The results obtained from these assays demonstrate that the cold-adapted proteins bind diatomic ligands at 10 °C with the same apparent rate as the human protein at 40 °C, thus suggesting that each protein shows similar, possibly optimal, performance at its physiological temperature.
Fig. 8.
Absorption changes at 418 nm after mixing Fe(II), deoxy C.aceCygb-1 (T = 10 °C, blue open circles), deoxy D.mawCygb-1 (T = 10 °C, blue filled circles) or CYGB (T = 40 °C, red filled circles; T = 10 °C, blue filled squares) solutions with CO to the same final concentration (200 μM). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
From a structural viewpoint, it has been shown that when CO is bound to the heme distal site (in CYGB) the distal HisE7 [81] residue swings by about 4 Å, affecting the conformation of residue PheCD4 [63] that moves away from the distal cavity by about 2 Å [58]. Binding of CO is at the same time reflected by a small displacement of the N-terminal region of the E-helix (maximum displacement 3.4 Å at residue ProE2 [76] to make room for the swung HisE7, as previously anticipated for a minor population of the ligand-free (6C) CYGB [25].
The structural analysis here presented does not point to specific details or marked residue differences that might promote a differential binding behavior for CO in human CYGB and in D.mawCygb-1*, e.g. explaining the different HisE7 off rates (kOFF,His) observed. Such difference may thus depend on distributed subtle details, as often suggested by the comparison of thermostable protein structures with their mesophilic homologues. Additionally, a key contributing factor may reside in the dynamics of selected structural elements, or of the protein as a whole; notably, the relevance of the CD-D region in regulating the transition between hexa- and penta-coordinated states in globins has been previously reported [13], [15]. In the case of D. mawsoni (and C. aceratus) Cygbs-1, the CD-loop and D-helix host five Glu residues, whereas in human CYGB two of these residues are mutated (to Asp and Ile, respectively). Additionally, two Met residues (62 and 66 in CYGB) are mutated into Ile and Leu, respectively, and Pro76 (at the N-terminus of E-helix) is mutated to Ala in D.mawCygb-1. All such structural features, although sequence-wise preceding HisE7 and the ligand binding site, can nevertheless affect the overall dynamics of the distal cavity, supporting different responses to the in-/out-coming diatomic ligand, such as differential rupture of the Fe distal coordination bond, or altered dynamics of CO diffusion. Indeed, the main differences between D. maw (and C. ace) Cygbs-1 functional properties are most apparent in the distal His dissociation rate (kOFF,His) that impacts on the observed rate constant (kobs) for CO binding to the hexacoordinated species (Table 1. As mentioned, it should also be recalled that human CYGB hosts a Cys38(B2)-Cys83(E9) disulfide bridge; notably, such disulfide, which can restrict dynamics of the E-helix and of the distal site [12], is absent in both D.mawCygb-1 and D.mawCygb-1*. Lastly, we remark that global control of Antarctic fish Cygb-1 function may also occur at the cellular level, involving changes in gene expression or in overall protein-synthesis.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was financially supported by the Italian National Programme for Antarctic Research (PNRA) (2016/AZ1.06-Project PNRA16_00043 and 2016/AZ1.20-Project PNRA16_00128). It was carried out in the framework of the SCAR Programmes “Antarctic Thresholds–Ecosystem Resilience and Adaptation” (AnT-ERA). SA and C Viappiani acknowledge support from Fondazione di Piacenza e Vigevano and Azienda USL di Piacenza. CS and FJL are grateful to the Ministerio de Economía y Competitividad (SAF2017-88107-R), Spanish Structures of Excellence María de Maeztu (MDM-2017-0767) and the Generalitat de Catalunya (2017SGR1746) for financial support, and the Barcelona Supercomputing Center (BCV-2016-3-0015) for computational resources. We wish to thank Luc Moens (University of Antwerp) for critically reading the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.08.007.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Abbruzzetti S., Bruno S., Faggiano S., Grandi E., Mozzarelli A., Viappiani C. Time-resolved methods in biophysics. 2. Monitoring haem proteins at work with nanosecond laser flash photolysis. Photochem Photobiol Sci. 2006;5(12):1109–1120. doi: 10.1039/b610236k. [DOI] [PubMed] [Google Scholar]
- 2.Ascenzi P., Bolli A., di Masi A., Tundo G.R., Fanali G., Coletta M. Isoniazid and rifampicin inhibit allosterically heme binding to albumin and peroxynitrite isomerization by heme albumin. J Biol Inorg Chem. 2011;16(1):97–108. doi: 10.1007/s00775-010-0706-2. [DOI] [PubMed] [Google Scholar]
- 3.Ascenzi P., Bolli A., Gullotta F., Fanali G., Fasano M. Drug binding to Sudlow’s site I impairs allosterically human serum heme-albumin-catalyzed peroxynitrite detoxification. IUBMB Life. 2010;62(10):776–780. doi: 10.1002/iub.381. [DOI] [PubMed] [Google Scholar]
- 4.Ascenzi P., Ciaccio C., Sinibaldi F., Santucci R., Coletta M. Cardiolipin modulates allosterically peroxynitrite detoxification by horse heart cytochrome C. Biochem Biophys Res Commun. 2011;404(1):190–194. doi: 10.1016/j.bbrc.2010.11.091. [DOI] [PubMed] [Google Scholar]
- 5.Ascenzi P., Ciaccio C., Sinibaldi F., Santucci R., Coletta M. Peroxynitrite detoxification by horse heart carboxymethylated cytochrome c is allosterically modulated by cardiolipin. Biochem Biophys Res Commun. 2011;415(3):463–467. doi: 10.1016/j.bbrc.2011.10.094. [DOI] [PubMed] [Google Scholar]
- 6.Ascenzi P., di Masi A., Coletta M., Ciaccio C., Fanali G., Nicoletti F.P. Ibuprofen impairs allosterically peroxynitrite isomerization by ferric human serum heme-albumin. J Biol Chem. 2009;284(45):31006–31017. doi: 10.1074/jbc.M109.010736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ascenzi P., di Masi A., Sciorati C., Clementi E. Peroxynitrite-An ugly biofactor? BioFactors. 2010;36(4):264–273. doi: 10.1002/biof.103. [DOI] [PubMed] [Google Scholar]
- 8.Ascenzi P., Fasano M. Abacavir modulates peroxynitrite-mediated oxidation of ferrous nitrosylated human serum heme–albumin. Biochem. Biophys. Res. Commun. 2007;353(2):469–474. doi: 10.1016/j.bbrc.2006.12.041. [DOI] [PubMed] [Google Scholar]
- 9.Ascenzi P., Marino M., Polticelli F., Santucci R., Coletta M. Cardiolipin modulates allosterically the nitrite reductase activity of horse heart cytochrome c. J Biol Inor Chem. 2014;19(7):1195–1201. doi: 10.1007/s00775-014-1175-9. [DOI] [PubMed] [Google Scholar]
- 10.Ascenzi P., Tundo G.R., Coletta M. The nitrite reductase activity of ferrous human hemoglobin: haptoglobin 1–1 and 2–2 complexes. J. Inorg. Biochem. 2018;187:116–122. doi: 10.1016/j.jinorgbio.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Ascenzi P., Tundo G.R., Fanali G., Coletta M., Fasano M. Warfarin modulates the nitrite reductase activity of ferrous human serum heme–albumin. J Biol Inor Chem. 2013;18(8):939–946. doi: 10.1007/s00775-013-1040-2. [DOI] [PubMed] [Google Scholar]
- 12.Beckerson P., Reeder B.J., Wilson M.T. Coupling of disulfide bond and distal histidine dissociation in human ferrous cytoglobin regulates ligand binding. FEBS Lett. 2015;589(4):507–512. doi: 10.1016/j.febslet.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 13.Boron I., Capece L., Pennacchietti F., Wetzler D.E., Bruno S., Abbruzzetti S. Engineered chimeras reveal the structural basis of hexacoordination in globins: a case study of neuroglobin and myoglobin. BBA. 2015;1850(1):169–177. doi: 10.1016/j.bbagen.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Caccia D., Ronda L., Frassi R., Perrella M., Del Favero E., Bruno S. PEGylation promotes hemoglobin tetramer dissociation. Bioconjug. Chem. 2009;20(7):1356–1366. doi: 10.1021/bc900130f. [DOI] [PubMed] [Google Scholar]
- 15.Capece L., Marti M.A., Bidon-Chanal A., Nadra A., Luque F.J., Estrin D.A. High pressure reveals structural determinants for globin hexacoordination: neuroglobin and myoglobin cases. Proteins. 2009;75(4):885–894. doi: 10.1002/prot.22297. [DOI] [PubMed] [Google Scholar]
- 16.Case DA, Babin V, Berryman JT, Betz RM, Cai Q, Cerutti DS, Cheatham TE, Darden TA, Duke RE, Gohlke H, Goetz AW, Gusarov S, Homeyer N, Janowski P, Kaus J, Kolossváry I, Kovalenko A, Lee TS, LeGrand S, Luchko T, Luo R, Madej BD, Merz KM, Paesani F, Roe DR, Roitberg A, Sagui C, Salomon-Ferrer R, Seabra G, Simmerling C, Smith W, Swails J, Walker RC, Wang J, Wolf RM, Wu X, Kollman PA. AMBER. Vol. 14. San Francisco: University of California, 2014.
- 17.Cheng C.-H.-C., Detrich H.W., III Molecular ecophysiology of Antarctic notothenioid fishes. Philos Trans R Soc Lond B Biol Sci. 2007;362(1488):2215–2232. doi: 10.1098/rstb.2006.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng C.-H.-C., di Prisco G., Verde C. The, “icefish paradox.” Which is the task of neuroglobin in Antarctic hemoglobin-less icefish? IUBMB Life. 2009;61(2):184–188. doi: 10.1002/iub.138. [DOI] [PubMed] [Google Scholar]
- 19.Chovancova E., Pavelka A., Benes P., Strnad O., Brezovsky J., Kozlikova B. 3.0: A tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput Biol. 2012;8 doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coletta M., Angeletti M., Sanctis G., Cerroni L., Giardina B., Amiconi G. Kinetic evidence for the existence of a rate-limiting step in the reaction of ferric hemoproteins with anionic ligands. Eur J Biochem. 1996;235:49–53. doi: 10.1111/j.1432-1033.1996.00049.x. [DOI] [PubMed] [Google Scholar]
- 21.Coppe A., Agostini C., Marino I.A.M., Zane L., Bargelloni L., Bortoluzzi S. Genome evolution in the cold: Antarctic icefish muscle transcriptome reveals selective duplications increasing mitochondrial function. Genome Biol Evol. 2013;5(1):45–60. doi: 10.1093/gbe/evs108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corti P., Ieraci M., Tejero J. Characterization of zebrafish neuroglobin and cytoglobins 1 and 2: zebrafish cytoglobins provide insights into the transition from six-coordinate to five-coordinate globins. Nitric Oxide. 2016;53:22–34. doi: 10.1016/j.niox.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 23.Cui W., Wang M., Maegawa H., Teranishi Y., Kawada N. Inhibition of the activation of hepatic stellate cells by arundic acid via the induction of cytoglobin. Biochem Biophys Res Commun. 2012;425(3):642–648. doi: 10.1016/j.bbrc.2012.07.126. [DOI] [PubMed] [Google Scholar]
- 24.Cuypers B., Vermeylen S., Hammerschmid D., Trashin S., Rahemi V., Konijnenberg A. Antarctic fish versus human cytoglobins – the same but yet so different. J Inorg Biochem. 2017;173:66–78. doi: 10.1016/j.jinorgbio.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 25.de Sanctis D., Dewilde S., Pesce A., Moens L., Ascenzi P., Hankeln T. Crystal structure of cytoglobin: the fourth globin type discovered in man displays heme hexa-coordination. J Mol Biol. 2004;336(4):917–927. doi: 10.1016/j.jmb.2003.12.063. [DOI] [PubMed] [Google Scholar]
- 26.de Sanctis D., Dewilde S., Pesce A., Moens L., Ascenzi P., Hankeln T. Mapping protein matrix cavities in human cytoglobin through Xe atom binding. Biochem Biophys Res Commun. 2004;316:1217–1221. doi: 10.1016/j.bbrc.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 27.di Prisco G., Macdonald J.A., Brunori M. Antarctic fishes survive exposure to carbon-monoxide. Experientia. 1992;48:473–475. doi: 10.1007/BF01928166. [DOI] [PubMed] [Google Scholar]
- 28.di Prisco G., Verde C. The Ross Sea and its rich life: research on molecular adaptive evolution of stenothermal and eurythermal Antarctic organisms and the Italian contribution Hydrobiologia. Biol Ross Sea. 2015;761:335–361. [Google Scholar]
- 29.Eastman J.T. Academic Press; San Diego: 1993. Antarctic Fish Biology: Evolution in a Unique Environment. [Google Scholar]
- 30.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 31.Fago A., Hundahl C., Dewilde S., Gilany K., Moens L., Weber R.E. Allosteric regulation and temperature dependence of oxygen binding in human neuroglobin and cytoglobin. Molecular mechanisms and physiological significance. J Biol Chem. 2004;279(43):44417–44426. doi: 10.1074/jbc.M407126200. [DOI] [PubMed] [Google Scholar]
- 32.Fago A., Wells R.M.G., Weber R.E. Temperature-dependent enthalpy of oxygenation in Antarctic fish hemoglobins. Comp Biochem Physiol. 1997;118B:319–326. [Google Scholar]
- 33.Fuchs C., Heib V., Kiger L., Haberkamp M., Roesner A., Schmidt M. Zebrafish reveals different and conserved features of vertebrate neuroglobin gene structure, expression pattern, and ligand binding. J Biol Chem. 2004;279(23):24116–24122. doi: 10.1074/jbc.M402011200. [DOI] [PubMed] [Google Scholar]
- 34.Fuchs C., Luckhardt A., Gerlach F., Burmester T., Hankeln T. Duplicated cytoglobin genes in teleost fishes. Biochem Biophys Res Commun. 2005;337(1):216–223. doi: 10.1016/j.bbrc.2005.08.271. [DOI] [PubMed] [Google Scholar]
- 35.Gabba M., Abbruzzetti S., Spyrakis F., Forti F., Bruno S., Mozzarelli A. CO rebinding kinetics and molecular dynamics simulations highlight dynamic regulation of internal cavities in human cytoglobin. PLoS ONE. 2013;8(1) doi: 10.1371/journal.pone.0049770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giordano D., Boron I., Abbruzzetti S., Van Leuven W., Nicoletti F.P., Forti F. Biophysical characterisation of neuroglobin of the icefish, a natural knockout for hemoglobin and myoglobin Comparison with human neuroglobin. PLoS ONE. 2012;7(12) doi: 10.1371/journal.pone.0044508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giordano D., Russo R., Coppola D., Altomonte G., di Prisco G., Bruno S. “Cool” adaptations to cold environments: globins in Notothenioidei Hydrobiologia. Biol. Ross Sea. 2015;761:293–312. [Google Scholar]
- 38.Goldstein S., Merényi G. The chemistry of peroxynitrite: implications for biological activity. Methods Enzymol. 2008;436:49–61. doi: 10.1016/S0076-6879(08)36004-2. [DOI] [PubMed] [Google Scholar]
- 39.Grubina R., Basu S., Tiso M., Kim-Shapiro D.B., Gladwin M.T. Nitrite reductase activity of hemoglobin s (sickle) provides insight into contributions of heme redox potential versus ligand affinity. J Biol Chem. 2008;283(6):3628–3638. doi: 10.1074/jbc.M705222200. [DOI] [PubMed] [Google Scholar]
- 40.Halligan KE, Jourd’heuil FL, Jourd’heuil D. Cytoglobin is expressed in the vasculature and regulates cell respiration and proliferation via nitric oxide dioxygenation. J Biol Chem 284 (13): 8539-8547, 2009. [DOI] [PMC free article] [PubMed]
- 41.Hamdane D., Kiger L., Dewilde S., Green B.N., Pesce A., Uzan J. The redox state of the cell regulates the ligand binding affinity of human neuroglobin and cytoglobin. J Biol Chem. 2003;278(51):51713–51721. doi: 10.1074/jbc.M309396200. [DOI] [PubMed] [Google Scholar]
- 42.Hankeln T., Ebner B., Fuchs C., Gerlach F., Haberkamp M., Laufs T.L. Neuroglobin and cytoglobin in search of their role in the vertebrate globin family. J Inor Biochem. 2005;99(1):110–119. doi: 10.1016/j.jinorgbio.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 43.Helbo S., Dewilde S., Williams D.R., Berghmans H., Berenbrink M., Cossins A.R. Functional differentiation of myoglobin isoforms in hypoxia-tolerant carp indicates tissue-specific protective roles. Am J Physiol Regul Integr Comp Physiol. 2012;302(6):R693–701. doi: 10.1152/ajpregu.00501.2011. [DOI] [PubMed] [Google Scholar]
- 44.Herold S., Fago A., Weber R.E., Dewilde S., Moens L. Reactivity studies of the Fe(III) and Fe(II)NO forms of human neuroglobin reveal a potential role against oxidative stress. J Biol Chem. 2004;279(22):22841–22847. doi: 10.1074/jbc.M313732200. [DOI] [PubMed] [Google Scholar]
- 45.Herold S., Shivashankar K., Matsui T., Watanabe Y. Mechanistic studies of the isomerization of peroxynitrite to nitrate catalyzed by distal histidine metmyoglobin mutants. J Am Chem Soc. 2004;126(22):6945–6955. doi: 10.1021/ja0493300. [DOI] [PubMed] [Google Scholar]
- 46.Herold S., Shivashankar K. Metmyoglobin and methemoglobin catalyze the isomerization of peroxynitrite to nitrate. Biochemistry. 2003;42(47):14036–14046. doi: 10.1021/bi0350349. [DOI] [PubMed] [Google Scholar]
- 47.Huang K.T., Keszler A., Patel N., Patel R.P., Gladwin M.T., Kim-Shapiro D.B. The reaction between nitrite and deoxyhemoglobin. Reassessment of reaction kinetics and stoichiometry. J Biol Chem. 2005;280(35):31126–31131. doi: 10.1074/jbc.M501496200. [DOI] [PubMed] [Google Scholar]
- 48.Huang Z., Shiva S., Kim-Shapiro D.B., Patel R.P., Ringwood L.A., Irby C.E. Enzymatic function of hemoglobin as a nitrite reductase that produces NO under allosteric control. J Clin Invest. 2005;115(8):2099–2107. doi: 10.1172/JCI24650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kabsch W. XDS. Acta Crystallogr.D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawada N., Kristensen D.B., Asahina K., Nakatani K., Minamiyama Y., Seki S. Characterization of a stellate cell activation-associated protein (STAP) with peroxidase activity found in rat hepatic stellate cells. J Biol Chem. 2001;276(27):25318–25323. doi: 10.1074/jbc.M102630200. [DOI] [PubMed] [Google Scholar]
- 51.Lechauve C., Chauvierre C., Dewilde S., Moens L., Green B.N., Marden M.C. Cytoglobin conformations and disulfide bond formation. FEBS J. 2010;277:2696–2704. doi: 10.1111/j.1742-464X.2010.07686.x. [DOI] [PubMed] [Google Scholar]
- 52.Li D., Chen X.Q., Li W.J., Yang Y.H., Wang J.Z., Yu A.C. Cytoglobin up-regulated by hydrogen peroxide plays a protective role in oxidative stress. Neurochem Res. 2007;32(8):1375–1380. doi: 10.1007/s11064-007-9317-x. [DOI] [PubMed] [Google Scholar]
- 53.Li H., Hemann C., Abdelghany T.M., El-Mahdy M.A., Zweier J.L. Characterization of the mechanism and magnitude of cytoglobin-mediated nitrite reduction and Nitric Oxide generation under anaerobic conditions. J Biol Chem. 2012;287:36623–36633. doi: 10.1074/jbc.M112.342378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins Struct Funct Bioinf. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu X., El-Mahdy M.A., Boslett J., Varadharaj S., Hemann C., Abdelghany T.M. Cytoglobin regulates blood pressure and vascular tone through nitric oxide metabolism in the vascular wall. Nat Commun. 2017;8:14807. doi: 10.1038/ncomms14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu X., Follmer D., Zweier J.R., Huang X., Hemann C., Liu K. Characterization of the function of cytoglobin as an oxygen-dependent regulator of nitric oxide concentration. Biochemistry. 2012;51(25):5072–5082. doi: 10.1021/bi300291h. [DOI] [PubMed] [Google Scholar]
- 57.Liu X., Tong J., Zweier J.R., Follmer D., Hemann C., Ismail R.S. Differences in oxygen-dependent nitric oxide metabolism by cytoglobin and myoglobin account for their differing functional roles. FEBS J. 2013;280(15):3621–3631. doi: 10.1111/febs.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Makino M., Sawai H., Shiro Y., Sugimoto H. Crystal structure of the carbon monoxide complex of human cytoglobin. Proteins. 2011;79(4):1143–1153. doi: 10.1002/prot.22950. [DOI] [PubMed] [Google Scholar]
- 59.Marti M.A., Crespo A., Capece L., Boechi L., Bikiel D.E.D.E., Scherlis D.A.D.A. Dioxygen affinity in heme proteins investigated by computer simulation. J. Inorg. Biochem. 2006;100:761–770. doi: 10.1016/j.jinorgbio.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 60.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the Maximum-Likelihood method. Acta Cryst D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 62.Nishi H., Inagi R., Kawada N., Yoshizato K., Mimura I., Fujita T. Cytoglobin, a novel member of the globin family, protects kidney fibroblasts against oxidative stress under ischemic conditions. Am. J. Pathol. 2011;178(1):128–139. doi: 10.1016/j.ajpath.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oleksiewicz U., Liloglou T., Field J.K., Xinarianos G. Cytoglobin: biochemical, functional and clinical perspective of the newest member of the globin family. Cell. Mol. Life Sci. 2011;68:3869–3883. doi: 10.1007/s00018-011-0764-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oleksiewicz U., Liloglou T., Tasopoulou K.M., Daskoulidou N., Bryan J., Gosney J.R. Cytoglobin has bimodal: tumour suppressor and oncogene functions in lung cancer cell lines. Hum. Mol. Genet. 2013;22:3207–3217. doi: 10.1093/hmg/ddt174. [DOI] [PubMed] [Google Scholar]
- 65.Patarnello T., Verde C., di Prisco G., Bargelloni L., Zane L. How will fish that evolved at constant sub-zero temperatures cope with global warming? Notothenioids as a case study. BioEssays. 2011;33:260–268. doi: 10.1002/bies.201000124. [DOI] [PubMed] [Google Scholar]
- 66.Pesce A., Dewilde S., Nardini M., Moens L., Ascenzi P., Hankeln T. “Human brain neuroglobin structure reveals a distinct mode of controlling oxygen affinity. Structure. 2003;11:1087–1095. doi: 10.1016/s0969-2126(03)00166-7. [DOI] [PubMed] [Google Scholar]
- 67.Petersen M.G., Dewilde S., Fago A. Reactions of ferrous neuroglobin and cytoglobin with nitrite under anaerobic conditions. J. Inorg. Biochem. 2008;102:1777–1782. doi: 10.1016/j.jinorgbio.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 68.Ronda L., Bruno S., Faggiano S., Bettati S., Mozzarelli A. Oxygen binding to heme proteins in solution, encapsulated in silica gels, and in the crystalline state. Meth Enzymol. 2008;437:311–328. doi: 10.1016/S0076-6879(07)37016-X. [DOI] [PubMed] [Google Scholar]
- 69.Sawai H., Kawada N., Yoshizato K., Nakajima H., Aono S., Shiro Y. Characterization of the heme environmental structure of cytoglobin, a fourth globin in humans. Biochemistry. 2003;42(17):5133–5142. doi: 10.1021/bi027067e. [DOI] [PubMed] [Google Scholar]
- 70.Schenkman KA, Marble DR, Burns DH, Feigl EO. Myoglobin oxygen dissociation by multiwavelength spectroscopy. J Appl Physiol (1985) 82 (1): 86-92, 1997. [DOI] [PubMed]
- 71.Schmidt M., Gerlach F., Avivi A., Laufs T., Wystub S., Simpson J.C. Cytoglobin is a respiratory protein in connective tissue and neurons, which is up-regulated by hypoxia. J. Biol. Chem. 2004;279:8063–8069. doi: 10.1074/jbc.M310540200. [DOI] [PubMed] [Google Scholar]
- 72.Schmidtke P., Bidon-Chanal A., Luque F.J., Barril X. MDpocket: Open-source cavity detection and characterization on molecular dynamics trajectories. Bioinformatics. 2011;27:3275–3285. doi: 10.1093/bioinformatics/btr550. [DOI] [PubMed] [Google Scholar]
- 73.Shiva S., Huang Z., Grubina R., Sun J., Ringwood L.A., MacArthur P.H. Deoxymyoglobin is a nitrite reductase that generates Nitric Oxide and regulates mitochondrial respiration. Circ. Res. 2007;100:654–661. doi: 10.1161/01.RES.0000260171.52224.6b. [DOI] [PubMed] [Google Scholar]
- 74.Sidell B.D., O'Brien K.M. When bad things happen to good fish: the loss of hemoglobin and myoglobin expression in Antarctic icefishes. J. Exp. Biol. 2006;209:1791–1802. doi: 10.1242/jeb.02091. [DOI] [PubMed] [Google Scholar]
- 75.Sidell B.D., Vayda M.E., Small D.J., Moylan T.J., Londraville R.L., Yuan M.-L. Variable expression of myoglobin among the hemoglobinless Antarctic icefishes. PNAS. 1997;94:3420–3424. doi: 10.1073/pnas.94.7.3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sturms R., DiSpirito A.A., Hargrove M.S. Plant and cyanobacterial hemoglobins reduce nitrite to nitric oxide under anoxic conditions. Biochemistry. 2011;50:3873–3878. doi: 10.1021/bi2004312. [DOI] [PubMed] [Google Scholar]
- 77.Thuy L.T.T., Morita T., Yoshida K., Wakasa K., Iizuka M., Ogawa T. Promotion of liver and lung tumorigenesis in DEN-treated cytoglobin-deficient mice. Am. J. Pathol. 2011;179:1050–1060. doi: 10.1016/j.ajpath.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tiso M., Tejero J., Basu S., Azarov I., Wang X., Simplaceanu V. Human neuroglobin functions as a redox-regulated nitrite reductase. J Biol Chem. 2011;286:18277–18289. doi: 10.1074/jbc.M110.159541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tiso M., Tejero J., Kenney C., Frizzell S., Gladwin M.T. Nitrite reductase activity of nonsymbiotic hemoglobins from Arabidopsis thaliana. Biochemistry. 2012;51:5285–5292. doi: 10.1021/bi300570v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Trent J.T., III, Hargrove M.S. A ubiquitously expressed human hexa-coordinate hemoglobin. J Biol Chem. 2002;277:19538–19545. doi: 10.1074/jbc.M201934200. [DOI] [PubMed] [Google Scholar]
- 81.Vitagliano L., Vergara A., Bonomi G., Merlino A., Verde C., di Prisco G. Spectroscopic and crystallographic characterization of a tetrameric haemoglobin oxidation reveals structural features of the functional intermediate R/T state. J Am Chem Soc. 2008;130:10527–10535. doi: 10.1021/ja803363p. [DOI] [PubMed] [Google Scholar]
- 82.Oliveira A., Singh S., Bidon-Chanal A., Forti F., Marti M.A., Boechi L. Role of PheE15 gate in ligand entry and nitric oxide detoxification function of Mycobacterium tuberculosis truncated hemoglobin N. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0049291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Oliveira A, Allegri A, Bidon-Chanal A, Knipp M, Roitberg AE, Abbruzzetti S, Viappiani C, Luque FJ. Kinetics and computational studies of ligand migration in nitrophorin 7 and its Δ1-3 mutant. Biochim. Biophys. Acta - Proteins Proteom. 1834: 1711-1721, 2013. [DOI] [PubMed]
- 84.Ruscio JZ, Kumar D, Shukla M, Prisant MG, murali TM, Onufriev AV. Atomic level computational identification of ligand migration pathways between solvent and binding site in myoglobin. Proc. Natl. Acad. Sci. USA 105: 9204-0209, 2008. [DOI] [PMC free article] [PubMed]
- 85.Cazade P.A., Meuwly M. Oxygen migration pathways in NI-bound truncated hemoglobin. ChemPhysChem. 2012;13:4276–4286. doi: 10.1002/cphc.201200608. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.